 Open Access
Open Access
ARTICLE
Transcriptome Analysis of Auxin Drives Cone Size Regulation in Fokienia hodginsii
1
Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, National Engineering Laboratory for Tree Breeding,
Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological
Sciences and Technology, Beijing Forestry University, Beijing, 100083, China
2
Key Laboratory of Timber Forest Breeding and Cultivation for Mountainous Areas in Southern China Key Laboratory of Forest
Culture and Forest Product Processing Utilization of Fujian Province, Fuzhou, 350012, China
3
Basic Forestry and Proteomics Research Center, College of Forestry, Fujian Provincial Key Laboratory of Haixia Applied Plant
Systems Biology, Fujian Agriculture and Forestry University, Fuzhou, 350002, China
* Corresponding Authors: Xiangyang Kang. Email: ; Renhua Zheng. Email:
# These authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2023, 92(6), 1713-1723. https://doi.org/10.32604/phyton.2023.027586
Received 04 November 2022; Accepted 27 February 2023; Issue published 11 April 2023
Abstract
As the reproductive organ of the endangered species Fokienia hodginsii, the size of the cones is a constraint on the reproductive renewal of the population. In this study, the molecular basis of the influence of cone size on F. hodginsii was elucidated by comparing the phenotype, biochemistry, and transcriptome of two cultivars of F. hodginsii (‘FJ431’ and ‘FJ415’). The two cultivars differed significantly in cone size, with FJ431 having a significantly larger cone size and weight than FJ415, 1.32 and 1.90 times that of FJ415, respectively. RNA-Seq analysis of both cultivars retrieved 75,940 genes whose approximate functions were classified as the pathway of response to endogenous stimulus and response to hormone and showed significant differences in the auxin-activated signaling pathway, particularly the MAPK signaling pathway-plant. Furthermore, the endogenous IAA content was significantly higher in FJ431 than in FJ415, and 1.58 and 1.29 times more IAA was present in immature and mature cones, respectively. Moreover, exogenous IAA treatment significantly induced the expression of the MAPK pathway-related gene TRINITY_DN10564_c0_g1 and significantly inhibited the expression of the MAPK pathwayrelated gene TRINITY _DN17056_c0_g1. Our work suggests that IAA can affect the cone size of F. hodginsii, most probably through the MAPK pathway. This has high theoretical and practical significance for the improvement of genetic breeding and the further cultivation of quality germplasm resources of F. hodginsii.Keywords
Fokienia hodginsii (Dunn) A. Henry & H. H. Thomas (Cupressaceae) is an endemic tree species in China that plays an important role in maintaining soil moisture and improving the micro-ecological environment and is considered valuable fast-growing timber with high economic development value [1,2]. Due to its outstanding advantages in timber, landscaping, and ecological restoration, many subpopulations have been destroyed by illegal and logical logging [2]. Currently, F. hodginsii is listed as a Grade II protected plant in China and a vulnerable species on the International Union for Conservation of Nature (IUCN) Red List of Threatened Species [3]. To protect and utilize F. hodginsii, Chinese researchers have established seed orchards, but the small number of seeds and low yields have led to the increasingly prominent problem of low and unstable seed orchard yields, and its germplasm resources have become a constraint to the reproduction and renewal of F. hodginsii populations [4].
The size of the cones, as reproductive structures of F. hodginsii, can influence seed dispersal and germination by affecting seed size and morphology, which is important for the species’ phylogeny, community succession, population renewal, and ecological adaptation [5–7]. In terms of exogenous environmental signals, cone size is closely linked to its geographical variation, habitat climate, and population characteristics [8–10]. For example, plant adaptation to external factors such as light, temperature, moisture, and geographical differences can influence cone yield. In addition, cone size varies considerably within individuals and even within and between populations [11]. In terms of endogenous regulatory mechanisms, some pathways that can regulate cone size have been demonstrated, mainly the phytohormone pathway, MAPK pathway, G protein signaling pathway, IKU pathway, ubiquitin-proteasome pathway, and transcriptional regulation [12,13].
Plant hormones, especially growth hormones, are important factors that influence seed quality and cone size. Hormones play an important role in plant growth, development, and reproduction processes, and major plant hormones, such as Indole-3-acetic acid (IAA), Gibberellins (GA), Abscisic Acid (ABA), Cytokinin (CTK), and Brassinosteroids (BR), are involved in regulating seed development [14,15]. High levels of phytohormones such as GA, CTK, and BR are effective in increasing cone production in mature trees, while ABA is antagonistic to GA and IAA, and low levels of ABA contribute to cone genesis and growth [16,17]. These phytohormone signaling pathways interact with each other and regulate seed size in plants.
Auxin plays an important physiological role in initiating cell division, promoting cell elongation, and delivering nutrients to the fruit and seeds, and combined with other hormones regulates the ultimate form and size of the embryo [16,18,19]. According to current works on those model plants, auxin initiates central cell division during endosperm development and influences endosperm cell proliferation and development by regulating the expression of auxin synthetic genes, which influence seed size and grain weight [20,21]. The involvement of auxin in endosperm formation differs between angiosperms and gymnosperms in woody plants. It has also been shown that auxin levels increase synchronously with embryo development in angiosperms such as Malus pumila and Rosa roxburghii Tratt [22,23]. In contrast, in gymnosperms, such as Ginkgo biloba, auxin plays an important role in promoting the meiosis of female gametes and the formation of cell walls in endosperm cells, but auxin levels do not show a similar pattern of synchronous increase in the latter stages of seed development [24]. The significant association between cone enlargement in response to exogenous auxin in Cupressaceae species indicates that auxin is involved in the development of seeds and plays a vital function in the formation and growth of cones gymnosperms [24]. However, how auxin affects the endosperm development though the size of the cones in gymnosperms (including F. hodginsii) remains largely unknown.
Both positive and negative MAPK pathways are involved in auxin signaling [25]. For example, auxin induction increases MAPK activity in plant roots and regulates the phosphorylation of downstream ERF13 by activating MPK14, which in turn affects the synthesis of very long-chain fatty acids and regulates lateral root development [26,27]. NPK1 (a specific plant MAPK kinase, MAP-KKK) activates the MAPK signaling cascade and represses early auxin response gene transcription, and NPK1 overexpression can cause defects in embryo and endosperm development [28]. However, the role of the MAPK pathway in the auxin response that affects seed size is unclear.
In this study, the transcriptome and phytohormone content of two cultivars of F. hodginsii with significant differences in cone size were compared to uncover the underlying molecular basis of the size of the cones of F. hodginsii, thereby improving its breeding and further cultivation of high-quality germplasm resources.
2.1 Plant Material and Growth Conditions
The two cultivars of F. hodginsii involved in this study (‘FJ431’ and ‘FJ415’) were collected from the Fengtian State Plantation in Anxi, Fujian, China (118°0′5.24″–118°2′14.38″E, 25°16′25.45″–25°20′53.41″N). FJ431 and FJ415 were grafted in 2001 from seed trees originating from Longyan and Anxi, Fujian, China, respectively. Four mature cones of a single individual of each cultivar were randomly sampled (four individuals for each cultivar), and their radius and weights were measured immediately after collection. After counting the size and weight of the cones, the cones were peeled to count the number of seeds, the seed weight and these seeds were used for germination tests to calculate germination rates. Combined with the fact that FJ431 cones have a larger radius and weight than FJ415 cones, we inferred that cone size was substantially connected with the quantity of seeds or reproductive capability.
The seedlings of F. hodginsii were cultured in nutrient solution (10 g/L Huaduoduo 2# Peters®10-30-20+TE ICL Specialty Fertilizers) for a month before further treatment. Plant materials were grown at 22°C, with a light: dark cycle of 16:8 h, photon flux density of 60 µmol·m2·s−1, and relative humidity of 85%.
2.2 RNA-Seq Analysis and Bioinformatics
The samples for RNA-Seq comprised the mature cones of FJ431 and FJ415 (n = 3). RNA extractions and RNA-Seq analysis of the leaves from F. hodginsii followed previous methods [29]. The trimming of raw data resulted in only the uniquely aligned reads, and a fold-change in expression of >1.3 and a false-discovery rate (FDR) <0.01 were set as the thresholds for designating differentially expressed genes [30].
For the raw reads obtained from transcriptome sequencing, cutadapt and Perl scripts were used to remove splice sequences, low-quality sequences with a length <100 bp and <5% unknown bases, and bases with a quality value Q ≤ 10 accounting for more than 20% of the whole sequence in the raw data, followed by verification of data quality using FASTQ. Analysis without a reference transcriptome was used, as no reference genome was available or the genome annotation information was incomplete.
The unigenes were annotated and classified according to Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). Based on this, differentially expressed genes were identified and subjected to GO term and KEGG enrichment analyses. All samples were filtered using the transcriptome assembly software Trinity (Broad Institute, USA), and the resulting clean reads were assembled from scratch to obtain long fragments (contigs) that were assembled into sequences that could not be further extended at both ends, which were then finally normalized to unigenes. The unigenes were then evaluated for assembly quality, including length, GC content (GC%), and N50 [31].
The obtained unigenes were annotated against GO and KEGG for functional annotation and classification, and differential expression analysis was performed on unigenes from different groups to identify differentially expressed genes among the samples from the different localities of F. hodginsii.
2.3 IAA Treatments and Quantification of Endogenous Auxin (IAA)
One-month-old seedlings of FJ431 and FJ415 were used for exogenous IAA treatment experiments. Thirty seedlings for each treatment were cultured in Hoagland solution containing different concentrations (0, 0.5, 1 or 2 μM) of IAA (I2886, Sigma), and DMSO (D4540, Sigma) was used as the negative control [32].
Fifteen mature cones and fifteen immature cones were collected from FJ431 and FJ415, respectively. After being placed in liquid nitrogen, all mature and immature cones from each cultivar were combined. The IAA content was measured with the Plant Auxin (IAA; Auxin) ELISA Kit (JLC11517-96T, Gelatins), with three repeats of each sample.
The reasons why seedling was chosen for the exogenous treatment of growth hormone are as follows: Firstly, performing follow-up verification (auxin response) experiments in isolated cones after ripening is difficult [33–35]. Secondly, more than 90% of genes are expressed in seedling which led to a more sensitive system for testing our hypothesis [33,36]. Also, plant responses to auxin are similar at different development stages of woody plant [33,36]. Furthermore, this strategy was widely used in different research and produced reliable outputs [12,37].
A polysaccharide polyphenol FastPure Plant Total RNA Isolation Kit (Vazyme) was used to extract the total RNA of different samples, and the concentration and mass of RNA (A260/230, A260/280) were determined by DeNovix (DS-11 FX+). For qRT-PCR, 1 μg total RNA was used for cDNA synthesis using the cDNA Synthesis SuperMix for qPCR Kit (YEASEM, CAT:1123ES60). The primers used in this study are listed in Table S1. qPCR was performed with SYBR-Green PCR Mastermix (CAT:11202ES08). The expression levels of the tested genes were estimated using the relative
3.1 Morphological Differences of F. hodginsii Cultivars FJ431 and FJ415
Although FJ431 and FJ415 were planted in the same seed garden, their cone phenotypes were significantly different (Fig. 1A). Based on phenotypic observations and data analysis, significant differences were observed in cone weight and cone size. The average radius of the cones of FJ431 and FJ415 were 0.48 ± 0.026 cm and 0.36 ± 0.039 cm (p < 0.05), respectively, and the weight of cones was 0.16 ± 0.006 g and 0.08 ± 0.004 g (p < 0.001), respectively (Fig. 1B). FJ431 had a significantly higher radius and cone weight than FJ415. In addition, this study correlated the number, weight and germination rate of seeds collected from each cone and found that there were significant differences in the number of seeds between FJ431 and FJ415, but not in seed weight and germination rate. The average number of seeds in cones of FJ431 and FJ415 was 16.00 ± 0.57 and 16.00 ± 0.57 (p < 0.001), the mean seed weight in cones was 0.0094 ± 0.0003 g and 0.0102 ± 0.0006 g (p > 0.05), and the mean seed germination rate was 35.73 ± 1.20% and 33.73 ± 5.16% (p > 0.05), respectively (Fig. S1).
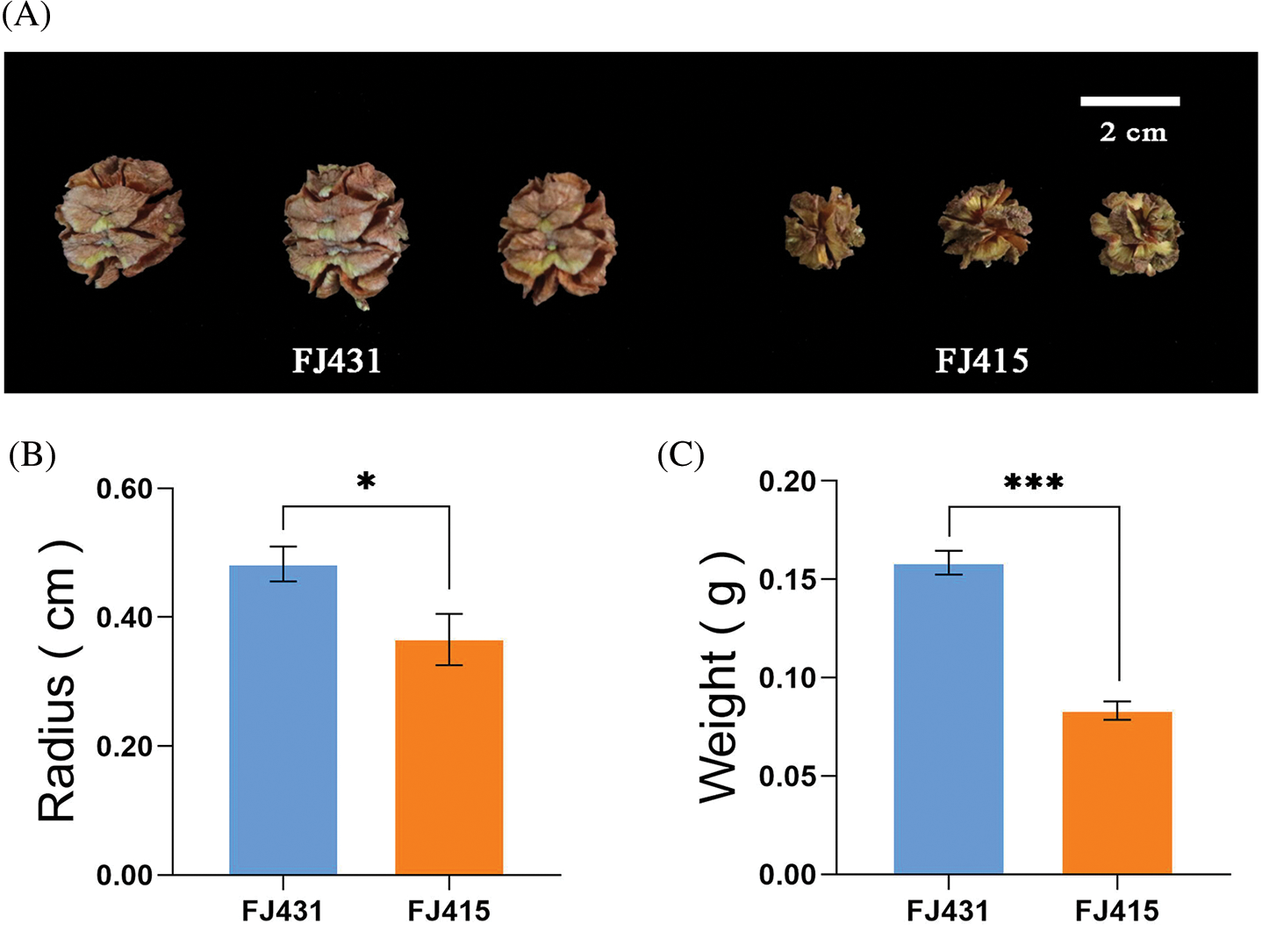
Figure 1: Cone size of two Fokienia hodginsii cultivars. (A) Illumination of cones of two F. hodginsii cultivars, FJ431 and FJ415. (B) and (C) The radius (cm) and weight (g) of the cones were measured for FJ431 and FJ415. Data are shown as the mean ± SD (n = 16). ‘*’ and ‘***’ indicate significant differences (p < 0.05 and p < 0.0001, respectively) between samples based on one-way ANOVA with multiple comparisons using Tukey’s test
3.2 Transcriptional Differences of F. hodginsii Cultivars FJ431 and FJ415
Comparing the transcriptome of FJ431 with that of FJ415, 75,940 differentially expressed genes were screened (FPKM ≥ 1) by the Illumina RNA-Seq results of FJ431 and FJ415. Of these, FJ431 and FJ415 had a total of 22,800 duplicated genes, 22,226 genes unique to FJ431, and 30,914 genes unique to FJ415 (Fig. 2A). Gene ontology analysis and KEGG enrichment analysis were performed on the differential genes obtained from the analysis.
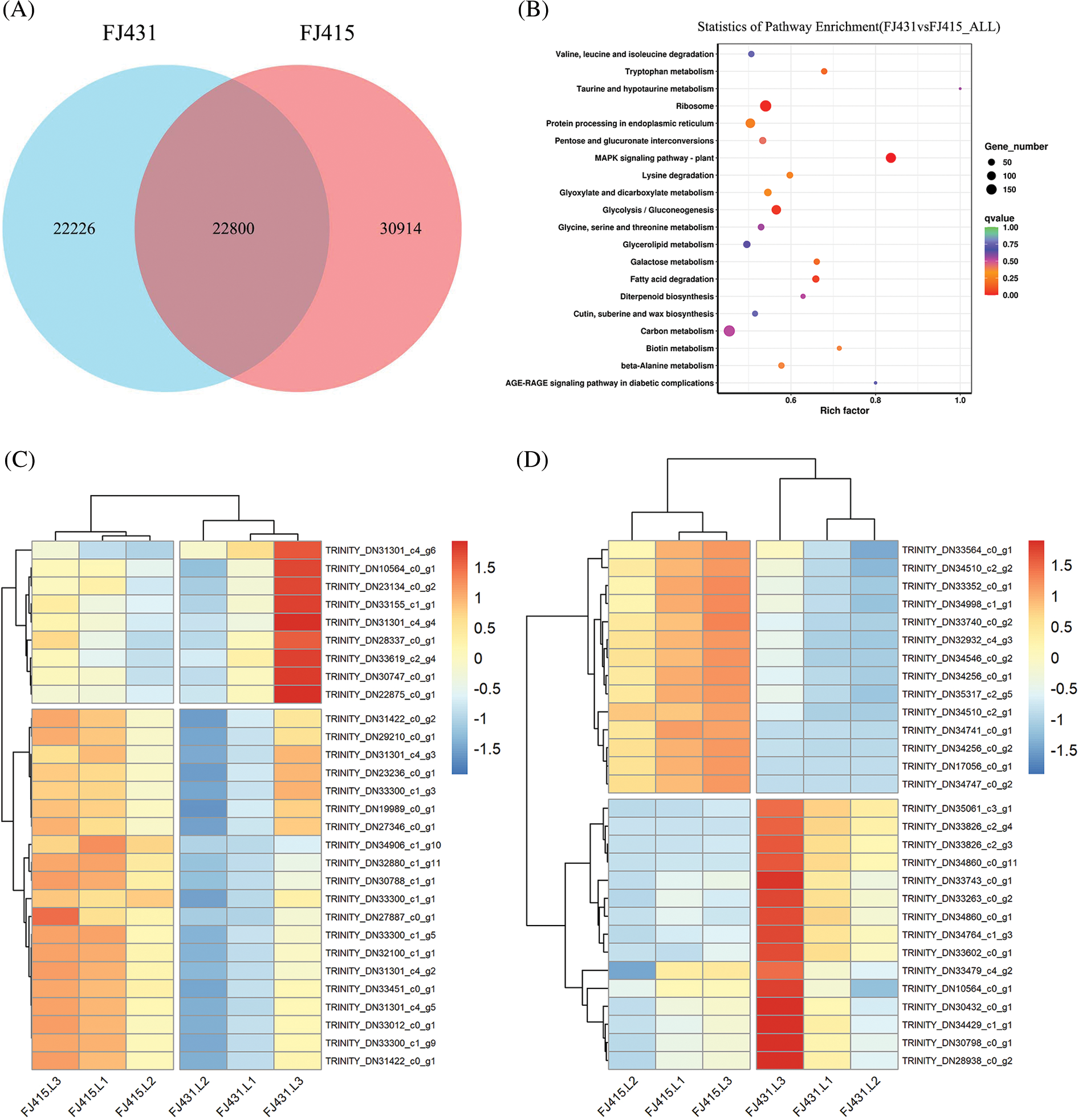
Figure 2: Transcriptome analysis of FJ431 and FJ415. (A) Venn diagram of expression genes in FJ431 and FJ415. Numbers represent the number of transcripts. KEGG enrichment analysis and analysis of IAA and MAPK pathway-related genes. (B) The functions of differentially expressed genes between FJ431 and FJ415 were predicted by KEGG enrichment analysis. The vertical axis indicates the pathway name; the horizontal axis indicates the Rich factor; the size of the dots indicates the number of differentially expressed genes in the pathway; and the color of the dots corresponds to the different Q-value ranges. (C) and (D) The expression levels of IAA and MAPK pathway-related genes were plotted in a heatmap. Each row represents one gene, each column represents one sample, and the data results are homogenized by row. The color ranges from blue to red, indicating log10(FPKM + 1) from small to large
In the GO term analysis of FJ431 and FJ415, the pathways of plant responses to hormones were significantly different (p < 0.01). Twenty signaling pathways were identified as auxin-related pathways, with a total of 33 genes enriched, mainly for the cellular response to auxin stimulus, response to auxin stimulation, auxin-activated signaling pathway, response to auxin, auxin biosynthetic process, and other signaling pathways. Among them, the auxin-activated signaling pathway enriched the most genes. In the KEGG enrichment analysis of FJ431 and FJ415, the enrichment of auxin-related MAPK signaling pathway-plant was the highest, with a Rich factor of 0.8 (p < 0.001) (Fig. 2B). A total of 143 genes were enriched in the MAPK signaling pathway plant.
3.3 Contents of Endogenous IAA in F. hodginsii Cultivars FJ431 and FJ415
To verify the endogenous IAA content in two cultivars of F. hodginsii, which have obvious differences in cone phenotype, changes in IAA in the mature and immature cones of FJ431 and FJ415 F. hodginsii were detected. The mean IAA content of the immature cones of FJ431 and FJ415 was 167.02 ± 24.9 μg/g and 105.68 ± 15.7 μg/g, respectively. The mean IAA content of the mature cones of FJ431 and FJ415 was 105.96 ± 37.7 μg/g and 81.96 ± 3.8 μg/g, respectively. The IAA content in mature cones was significantly lower than that in immature cones (p < 0.05), and the IAA content in FJ431 cones was significantly higher than in FJ415 (p < 0.05) (Fig. 3A).
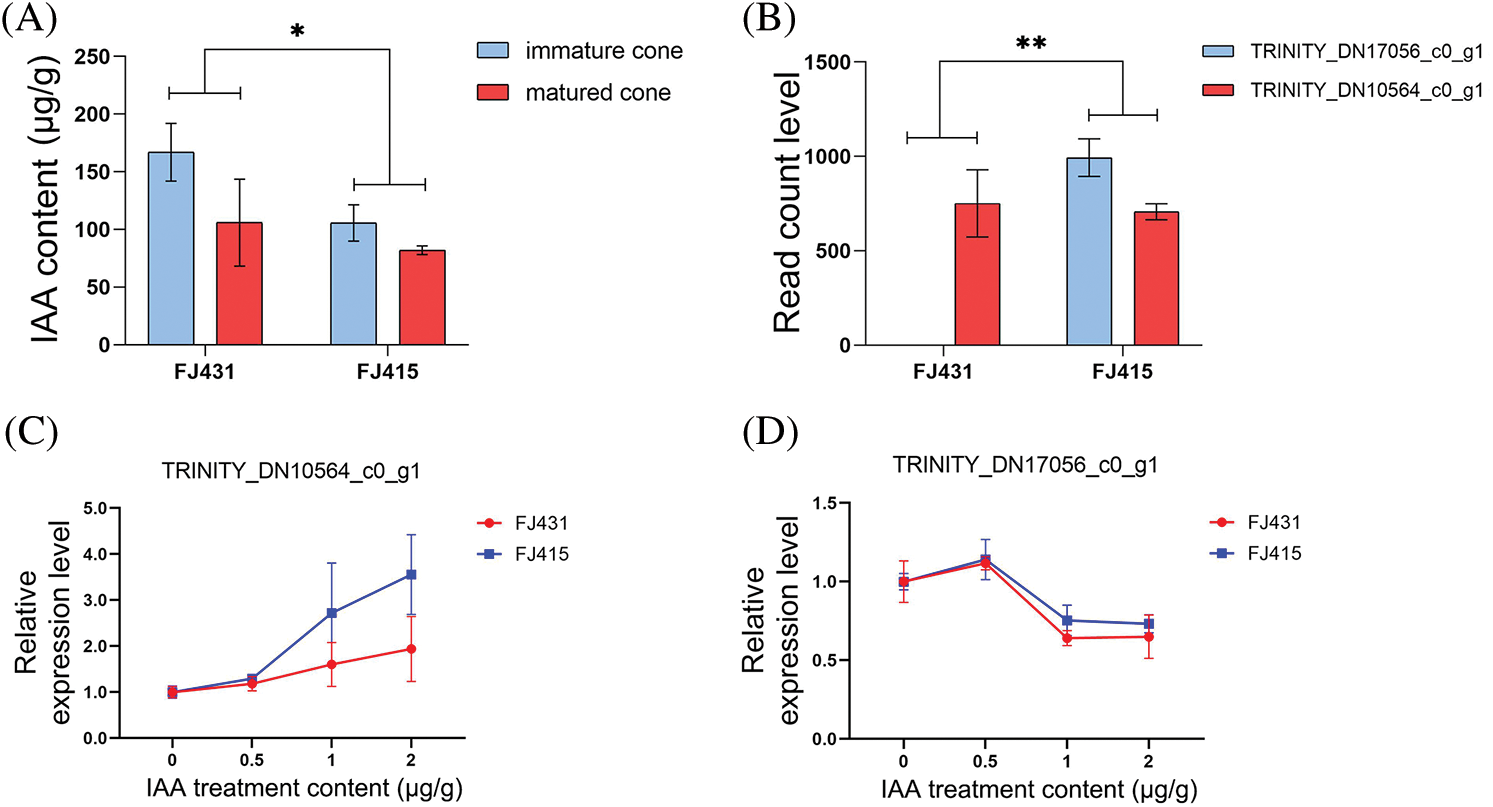
Figure 3: IAA accumulation enlarged the cone size of Fokienia hodginsii. (A) The level of IAA in both mature and immature cones of FJ431 and FJ415 (n = 15) was measured. (B) Read count level of IAA- (TRINITY_DN17056_c0_g1) and MAPK- (TRINITY_DN10564_c0_g1) related genes in FJ431 and FJ415 were represented by the mean ± SD. (C) and (D) Relative expression dynamics of IAA- (TRINITY_DN17056_c0_g1) and MAPK- (TRINITY_DN10564_c0_g1) related genes in FJ431 and FJ415 under different concentrations of IAA. Data are shown as the mean ± SD (n = 30). Different lowercase letters indicate the significance of different treatments (p < 0.05) between samples based on one-way ANOVA with multiple comparisons using Tukey’s test
3.4 Different IAA-Related Response Genes
According to the results of RNA-Seq and IAA content, all differential genes in the auxin-activated signaling pathway were visualized using a heatmap (Fig. 2C). Fifteen upregulated genes and fourteen downregulated genes in the MAPK signaling pathway plant were visualized using a heatmap (Fig. 2D). By screening the overlapping genes of auxin-activated signaling pathway and MAPK signaling pathway plant, a related upregulated gene was identified as TRINITY_DN10564_c0_g1 (p < 0.001). In addition, the gene with the greatest difference was screened in the MAPK signaling pathway plant, which was the downregulated gene TRINITY_DN17056_c0_g1 (p < 0.001). The mean read count levels of TRINITY_DN10564_c0_g1 in FJ431 and FJ415 were 751 and 707, respectively, and the read count levels of TRINITY_DN17056_c0_g1 in FJ431 and FJ415 were 0 and 994, respectively (Fig. 3B). To determine whether the expression of TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 was affected by IAA, we treated 1-month-old seedlings with various concentrations of hormones for 2 h. qRT-PCR results showed that the relative expression level of TRINITY_DN10564_c0_g1 in both FJ431 and FJ415 increased after IAA treatment at different concentrations, and the upward trend of FJ415 was significantly higher than that of FJ431 (Fig. 3C). By contrast, the relative expression level of TRINITY_DN17056_c0_g1 decreased, and the trend was similar in FJ415 and FJ431 (Fig. 3D).
As the reproductive structure of gymnosperms, the size and quality of cones are crucial to gymnosperms. Numerous studies have reported on the internal and external mechanisms regulating plant cone size. RNA-Seq is an effective method for assessing transcriptomic changes in plants with different cone sizes and can elucidate the intrinsic mechanisms regulating cone size.
In this study, significant differences in cone radius and weight were found between the two clones of F. hodginsii grown in the same seed orchard, and the cone size and weight of FJ431 were significantly higher than those of FJ415. Based on these data, we speculated that there were biochemical differences between FJ431 and FJ415 after excluding geographic differences. The GO analysis in RNA-Seq showed that FJ431 and FJ415 have significant differences in the pathway that responds to plant hormones, and 20 pathways were identified as auxin-related pathways. Among them, the cellular response to auxin stimulus, auxin-activated signaling pathway, and other pathways was significantly different. The KEGG analysis of RNA-Seq showed that the MAPK signaling pathway plant associated with the auxin pathway also showed significant differences. In addition, we screened the upregulated gene TRINITY_DN10564_c0_g1, which is a repetitive gene of the auxin-activated signaling pathway and MAPK signaling pathway plant, and the downregulated gene TRINITY_DN17056_c0_g1, which is the most differential gene in the MAPK signaling pathway-plant. Based on these results, we speculated that TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 were the key counterpart genes linking their auxin pathway and MAPK pathway, and auxin may participate in the molecular mechanism of plant regulation of cone size through the MAPK pathway.
Therefore, we measured the IAA content of two clones of F. hodginsii and found that the IAA content of FJ431 with larger cones was significantly higher than that of FJ415 with smaller cones in both mature and immature cones. Furthermore, we performed exogenous IAA treatment on two clones of F. hodginsii seedlings and found that exogenous auxin treatment also led to differences in RNA levels with similar cone sizes; that is, TRINITY_DN10564_c0_g1, which is upregulated in the auxin and MAPK pathway in FJ415, showed a significantly higher upward trend than that in FJ431 after IAA treatment, and by contrast, TRINITY_DN17056_c0_g1, which is a related downregulated gene, showed a downward trend in FJ431 and FJ415. This is consistent with previous RNA-Seq results and speculation that auxin may be involved in the molecular mechanism of plant regulation of cone size through the MAPK pathway. Auxin, one of the most important hormones in plants, can regulate the auxin-responsive elements of transcription factors and their downstream genes to regulate seed size [39,40]. In addition, auxin can interact with the MAPK pathway and other plant hormones to affect downstream genes or proteins that regulate plant growth and development [25,41,42]. TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 were highly correlated with the cone size phenotype and IAA, which further confirms the previous speculation that TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 genes are regulated by auxin and MAPK pathways, presumably downstream of the growth hormone pathway, and that some proteins that may affect the MAPK pathway are jointly involved in the regulation of cone size. However, this experiment did not continue to study the effects of auxin on plant metabolism and other hormones. Its relevance can be explored in future experimental plans by measuring the levels of other hormones, such as GA and ABA, after the application of exogenous auxin and by quantifying the key genes in these gene-related pathways.
Here, two signaling pathways were identified using RNA-Seq and included the auxin-activated signaling pathway and MAPK signaling pathway-plant, both of which are closely related to auxin. By screening the overlapping upregulated genes in these two signaling pathways and the most differentially downregulated genes in the related pathways, we obtained two related genes. Based on qRT-PCR validation after exogenous application of auxin, we elucidated that TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 were the key counterpart genes linking the auxin pathway and the MAPK pathway. For the first time, we demonstrated that auxin can interact with the MAPK signaling pathway to regulate the downstream genes TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 to regulate the size of cypress cones, which may also be the basis for the adaptive evolution of F. hodginsii.
In this study, the molecular basis of cone size on F. hodginsii was elucidated by comparing the phenotype, biochemistry, and transcriptome of two cultivars (FJ431 and FJ415). There were significant differences in cone size between the two clones, with FJ431 having a significantly larger cone size and weight than FJ415. After excluding extrinsic geographical differences, it was concluded that cone size differences were due to biochemical differences seen in the different clones. RNA-Seq analysis of both cultivars showed significant differences in the auxin-activated signaling pathway and MAPK signaling pathway-plant and screened out an upregulated gene TRINITY_DN10564_c0_g1 and a downregulated gene TRINITY_DN17056_c0_g1. Furthermore, the endogenous IAA content was significantly higher in FJ431 than in FJ415. TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 expressions were highly correlated with IAA treatment and cone-sized fruit size through exogenous IAA treatment. This suggests that TRINITY_DN10564_c0_g1 and TRINITY_DN17056_c0_g1 are regulated by the IAA and MAPK pathways to influence cone size in F. hodginsii and are presumably downstream genes of the growth hormone pathway, most likely by affecting some of the MAPK pathways to regulate cone size.
Funding Statement: 1. The “Eagle Program” of Fujian Province, funded by the Department of Human Resources and Social Security of Fujian Province; 2. The “Fujian Cypress 1st Generation Core Breeding Population Construction Research” (No. 2021R1010004), funded by the Department of Science and Technology of Fujian Province.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Gao, Z. W. (1994). A precious timber species—Fokienia hodginsii. Journal of Fujian Forestry Science and Technology, 21(2), 62–66. [Google Scholar]
2. Yuan, J. G., Ding, B. Y., Yu, M. J. (2005). A preliminary study of phytocoenosis characteristic of Fokienia hodginsii in Fengyangshan Nature Reserve. Journal of Zhejiang Forestry College, 22(2), 133–138. [Google Scholar]
3. Ding, M. Y., Meng, K. K., Fan, Q., Tan, W. Z., Liao, W. B. et al. (2017). Development and validation of EST-SSR markers for Fokienia hodginsii (Cupressaceae). Applications in Plant Sciences, 5(3), 1600152. [Google Scholar]
4. Zhou, C. C., Xu, W. D., Chen, L. Y., He, T. Y., Rong, J. D. et al. (2019). Research progress on protection and utilization of Fokienia hodginsii germplasm resources. Subtropical Agriculture Research, 15(4), 271–278. [Google Scholar]
5. Leslie, A. B., Beaulieu, J. M., Mathews, S. (2017). Variation in seed size is structured by dispersal syndrome and cone morphology in conifers and other nonflowering seed plants. New Phytologist, 216(2), 429–437. [Google Scholar] [PubMed]
6. Xu, Y. M. (2015). The impact of the fertilization and plant growth regulators on seed orchard production in Fokienia (Master Thesis).Fujian Agriculture and Forestry University, China. [Google Scholar]
7. Zheng, R. H., Huang, D. L., Li, J. L., Yang, Z. W., Shi, J. L. et al. (2004). Studies on the selection and phenotypic variations on seed and cone traits of plus-trees of Fokienia hodginsii. Journal of Fujian Forestry Science and Technology, 21(S1), 1–6+10. [Google Scholar]
8. Gil, L., Climent, J., Nanos, N., Mutke, S., Ortiz, I. et al. (2002). Cone morphology variation in Pinus canariensis Sm. Plant Systematics and Evolution, 235(1–4), 35–51. [Google Scholar]
9. Leslie, A. B., Beaulieu, J. M., Crane, P. R., Donoghue, M. J. (2014). Cone size is related to branching architecture in conifers. New Phytologist, 203(4), 1119–1127. [Google Scholar] [PubMed]
10. Xu, L., Bao, W. K., He, Y. H. (2004). Morphological characters and geographical variation of cones and seeds of four Cupressus chengiana S.Y. Hu populations. Chinese Journal of Applied and Environmental Biology, 10(6), 707–711. [Google Scholar]
11. Haymes, K. L., Fox, G. A. (2012). Variation among individuals in cone production in Pinus palustris (Pinaceae). American Journal of Botany, 99(4), 640–645. [Google Scholar] [PubMed]
12. Li, N., Li, Y. H. (2016). Signaling pathways of seed size control in plants. Current Opinion in Plant Biology, 33, 23–32. https://doi.org/10.1016/j.pbi.2016.05.008 [Google Scholar] [PubMed] [CrossRef]
13. Wang, H. Q., Wang, K., Du, Q. G., Wang, Y. F., Fu, Z. Y. et al. (2018). Maize Urb2 protein is required for kernel development and vegetative growth by affecting pre-ribosomal RNA processing. New Phytologist, 218(3), 1233–1246. https://doi.org/10.1111/nph.15057 [Google Scholar] [PubMed] [CrossRef]
14. Bartrina, I., Otto, E., Strnad, M., Werner, T., Schmulling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell, 23(1), 69–80. https://doi.org/10.1105/tpc.110.079079 [Google Scholar] [PubMed] [CrossRef]
15. Ohto, M. A., Floyd, S. K., Fischer, R. L., Goldberg, R. B., Harada, J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sexual Plant Reproduction, 22(4), 277–289. https://doi.org/10.1007/s00497-009-0116-1 [Google Scholar] [PubMed] [CrossRef]
16. Basunia, M. A., Nonhebel, H. M. (2019). Hormonal regulation of cereal endosperm development with a focus on rice (Oryza sativa). Functional Plant Biology, 46(6), 493–506. https://doi.org/10.1071/FP18323 [Google Scholar] [PubMed] [CrossRef]
17. Mao, J. W., Yang, Y. H., Chen, K. P., Tan, X. L. (2021). Research progress in molecular mechanisms of plant hormone and microRNA regulating seed size and development. Plant Physiology, 57(2), 274–292. [Google Scholar]
18. Klosinska, M., Picard, C. L., Gehring, M. (2016). Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nature Plants, 2(10), 1–8. https://doi.org/10.1038/nplants.2016.145 [Google Scholar] [PubMed] [CrossRef]
19. Gomes, G. L. B., Scortecci, K. C. (2021). Auxin and its role in plant development: structure, signalling, regulation and response mechanisms. Plant Biology, 23(6), 894–904. https://doi.org/10.1111/plb.13303 [Google Scholar] [PubMed] [CrossRef]
20. Figueiredo, D. D., Batista, R. A., Roszak, P. J., Kohler, C. (2015). Auxin production couples endosperm development to fertilization. Nature Plants, 1(12), 15184. https://doi.org/10.1038/nplants.2015.184 [Google Scholar] [PubMed] [CrossRef]
21. Zhang, J., Niu, B. X., E, Z. G., Chen, C. (2021). Towards understanding the genetic regulations of endosperm development in rice. Chinese Journal of Rice Science, 35(4), 326–341. [Google Scholar]
22. Guo, W. F., Ming, H. A., Qin, G. L., Tao, S. H., Ping, J. L. (2004). Changes in endogenous hormone contents during seed germination of Anemone rivularis var. flore-minore. Scientia Agricultura Sinica, 24(5), 728–733. [Google Scholar]
23. Jun, Y. Z., Xi, Y. W. (2002). Research progress on the regulation of endogenous hormones on the fruit growth and development of Malus pumila. Shaanxi Journal of Agricultural Sciences, 48(10), 13–15. [Google Scholar]
24. Boboto, A. L. (2006). Studies on changes of plant hormone content and tissue structure during growth and development of seed in Ginkgo Biloba L. (Master Thesis). Yangzhou University, China. [Google Scholar]
25. Mishra, N. S., Tuteja, R., Tuteja, N. (2006). Signaling through MAP kinase networks in plants. Archives of Biochemistry and Biophysics, 452(1), 55–68. [Google Scholar] [PubMed]
26. Mockaitis, K., Howell, S. H. (2008). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. The Plant Journal, 24(6), 785–796. [Google Scholar]
27. Huang, R. F., Xu, T. D. (2021). Auxin regulates the lateral root development through MAPK-mediated VLCFAs biosynthesis. Chinese Bulletin of Botany, 56(1), 6–9. [Google Scholar]
28. Kovtun, Y., Chiu, W. L., Zeng, W., Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature, 395(6703), 716–720. https://doi.org/10.1038/27240 [Google Scholar] [PubMed] [CrossRef]
29. Wang, Z., Gerstein, M., Snyder, M. (2009). RNA-Seq: A revolutionary tool for transcriptomics. Nature Reviews Genetics, 10(1), 57–63. https://doi.org/10.1038/nrg2484 [Google Scholar] [PubMed] [CrossRef]
30. Zhao, H., Lin, K., Ma, L., Chen, Q., Gan, S. et al. (2020). Arabidopsis NUCLEAR FACTOR Y A8 inhibits the juvenile-to-adult transition by activating transcription of MIR156s. Journal of Experimental Botany, 71(16), 4890–4902. https://doi.org/10.1093/jxb/eraa197 [Google Scholar] [PubMed] [CrossRef]
31. Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5(7), 621–628. https://doi.org/10.1038/nmeth.1226 [Google Scholar] [PubMed] [CrossRef]
32. Li, Y., Li, J. L., Chen, Z. H., Wei, Y., Qi, Y. H. et al. (2020). OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnology Journal, 18(10), 2015–2026. https://doi.org/10.1111/pbi.13360 [Google Scholar] [PubMed] [CrossRef]
33. Shun, S., Mei, D. H., Hong, W. M., Shui, J. W., Xi, Z. Z. et al. (2018). Auxin response factor (ARF) and plant genetic breeding. Molecular Plant Breeding, 16(8), 2638–2645. [Google Scholar]
34. Hai, W. S., Yong, J. M., Xiao, F. L., Feng, C., Ying, Q. et al. (2016). The roles of auxin in seed dormancy and germination. Hereditas, 38(4), 314–322. [Google Scholar]
35. Liu, X. D., Zhang, H., Zhao, Y., Feng, Z. Y., Li, Q. et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 110(38), 15485–15490. https://doi.org/10.1073/pnas.1304651110 [Google Scholar] [PubMed] [CrossRef]
36. Su, Y. H., Liu, Y. B., Bai, B., Zhang, X. S. (2015). Establishment of embryonic shoot-root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Frontiers in Plant Science, 5, 792. [Google Scholar] [PubMed]
37. Long, Z. H., Zhen, Z., Ming, Y. X., Rui, W., Nan, X. W. et al. (2021). Cloning of circadian clock gene LHY and its effect on photosynthesis under exogenous hormone treatment in Camellia oleifera. Non-wood Forest Research, 39(4), 1–9. [Google Scholar]
38. Chang, S. J., Chen, W. C., Yang, J. C. (2009). Another formula for calculating the gene change rate in real-time RT-PCR. Molecular Biology Reports, 36(8), 2165–2168. https://doi.org/10.1007/s11033-008-9430-1 [Google Scholar] [PubMed] [CrossRef]
39. Wyllie-Echeverria, S., Cox, P. A., Churchill, A. C., Brotherson, J. D., Wyllie-Echeverria, T. (2003). Seed size variation within Zostera marina L. (Zosteraceae). Botanical Journal of the Linnean Society, 142(3), 281–288. https://doi.org/10.1046/j.1095-8339.2003.00180.x [Google Scholar] [CrossRef]
40. Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S. et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell, 17(3), 776–790. https://doi.org/10.1105/tpc.104.024950 [Google Scholar] [PubMed] [CrossRef]
41. He, Y. Q., Zhao, J., Yang, B., Sun, S., Peng, L. L. et al. (2020). Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnology Journal, 18(9), 1933–1945. https://doi.org/10.1111/pbi.13353 [Google Scholar] [PubMed] [CrossRef]
42. Jia, F. L., Wang, C. P., Liu, S., Jiao, Z. Y., Yin, W. L. et al. (2017). Effects of exogenous BR and lAA on drought tolerance of Populus deltoides× P. nigra. Journal of Beijing Forestry University, 39(7), 31–39. [Google Scholar]
Appendix

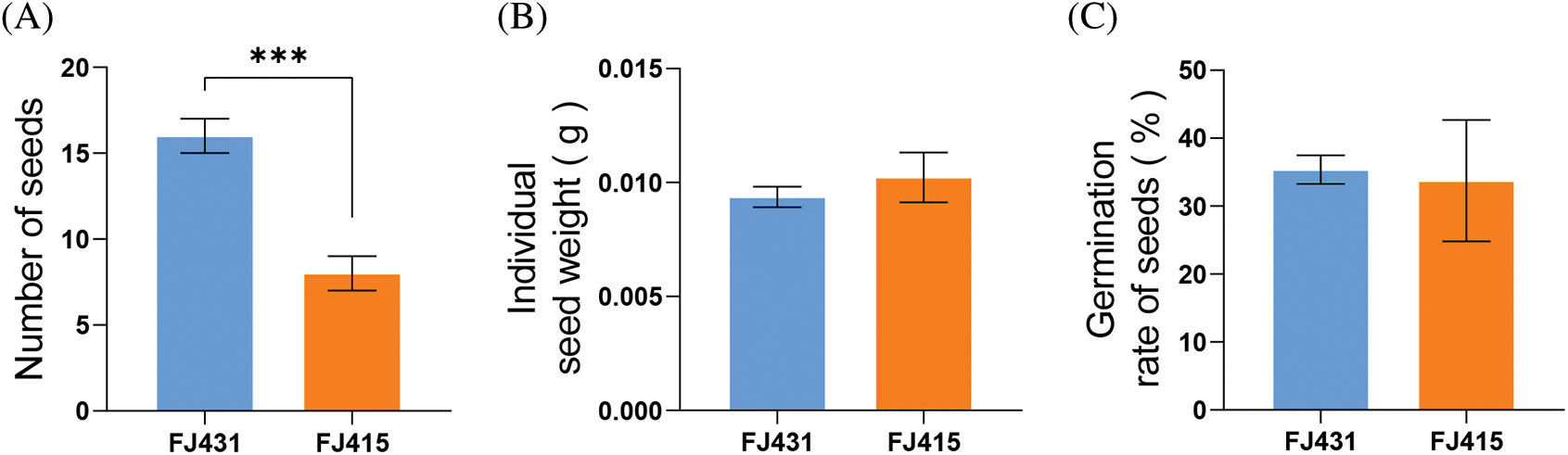
Figure S1: Seeds number and weight and germination of two Fokienia hodginsii cultivars cones. The seeds were peeled from cones of two F. hodginsii cultivars and the numbers, weight, and germination rate were determined. (A) The number of seeds of two F. hodginsii cultivars cones, FJ431 and FJ415. (B) and (C) The weight (g) and germination (%) of the seeds were measured for FJ431 and FJ415. Data are shown as the mean ± SD (n = 16). ‘***’ indicate significant differences (p < 0.0001) between samples based on one-way ANOVA with multiple comparisons using Tukey’s test
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools