 Open Access
Open Access
ARTICLE
Proteomic Study of Differentially Expressed Proteins in Seeds between Parents and Offspring of Castor Bean (Ricinus communis L.)
1 College of Life Science and Food Engineering, Inner Mongolia Minzu University, Tongliao, 028000, China
2 Key Laboratory of Castor Breeding of State Ethnic Affairs Commission, Tongliao, 028000, China
3 Research Center for Engineering and Technology of Castor Industry by Universities in Inner Mongolia Autonomous Region, Tongliao, 028000, China
4 Key Laboratory of Castor Breeding of Inner Mongolia Autonomous Region, Tongliao, 028000, China
5 Collaborative Innovation Center of Castor Industry of Inner Mongolia Autonomous Region, Tongliao, 028000, China
6 Engineering Research Center of Castor Industry Technology Innovation, Inner Mongolia Autonomous Region, Tongliao, 028000, China
* Corresponding Author: Fenglan Huang. Email:
# These authors contributed equally to the work
(This article belongs to the Special Issue: Integrating Agronomy and Plant Physiology for Improving Crop Production)
Phyton-International Journal of Experimental Botany 2023, 92(6), 1765-1792. https://doi.org/10.32604/phyton.2023.026935
Received 10 October 2022; Accepted 27 December 2022; Issue published 11 April 2023
Abstract
Castor bean (Ricinus communis L.), is one of the top 10 oilseed crops in the world and, therefore, of high economic value. Hybridization is one of the most effective ways to breed new varieties with high yield, high oil content, and better stress resistance. Therefore, prediction of desired traits in castor hybrid offspring is particularly important. In this study, proteomic analysis was performed to identify differentially expressed proteins (DEPs) in seeds between castor hybrid offspring and their female (Lm female line aLmAB2) and male parents (CSR·181). Among the DEPs upregulated in the seeds of hybrid offspring, the majority were related to seed yield and stress tolerance, while some were related to oil synthesis and fatty acid synthesis and metabolism in seeds. In other words, the hybrid offspring showed heterosis for seed yield, stress tolerance, oil synthesis, and fatty acid synthesis and metabolism when compared with their parents. Further, real-time quantitative polymerase chain reaction assays were performed on 12 genes encoding DEPs involved in oil synthesis, pollen abortion, yield, and stress tolerance of seeds. The results showed that the expression levels of the 12 genes were consistent with those of the DEPs.Keywords
Supplementary Material
Supplementary Material FileCastor bean (Ricinus communis L., Euphorbiaceae). Due to its strong environmental adaptability, castor bean can grow in harsh conditions such as saline-alkali soil and heavy metal-contaminated soil and is considered a valuable plant resource for phytoremediation. Therefore, it has been widely planted in tropical, subtropical, and warm-temperature countries, especially in India, China, and Brazil [1,2]. China has more than 2,300 germplasm resources of castor collected both at home and abroad, and in-depth studies have been conducted to further explore the application of castor bean. In Inner Mongolia, castor bean is a characteristic oilseed crop pivotal to local economic and social development [3]. It is a special industrial oil crop, with prominent intraspecific heterosis.
Seed storage substances gradually accumulate during seed development and maturation. Protein, one of the seed storage substances, can help a seed to form cellular structures and provide a nitrogen source for normal metabolism and development of seeds, thereby playing a vital role in cellular structure and function [4]. In 1994, Wilkins and Williams first proposed the concept of the proteome, which refers to all proteins expressed in a cell, tissue, or organism [5]. The methods used for the proteomic analysis of seeds mainly include two categories: gel and longer analysis. Gel analysis is the most commonly used, including two-dimensional gel electrophoresis (2-DE) and two-dimensional difference gel electrophoresis (2D-DIGE) [6]. After the differentially expressed spots are determined by using 2-DE and 2D-DIGE, a complete protein atlas can be obtained by mass spectrometry (MS), and this can better reflect the changes in protein expression and posttranslational modification [7,8]. Xia et al. [9] conducted a proteomic analysis to investigate the impact of temperature on seed metabolism in dormant and germinating seeds and deciphered the regulation of central metabolism activity during seed germination. Gu et al. [10] studied seed germination vigor in Brassica napus seeds differing in oil content by an integrated proteomic and genomic approach, and their research laid a foundation for studying the regulatory mechanism of seed germination vigor in Brassica napus.
In this study, the parent material (aLmAB2) is a Chinese castor standard female plant material with independent intellectual property rights in our laboratory. This material has good yield quality and plant stress resistance, and because it is a standard female line, it can eliminate castor hermaphrodite inflorescences at an early stage of the plant to avoid producing self-pollinated offspring seeds. The parent material (CSR·181) used in this experiment is from France, which has long fruit spikes, high yield and excellent drought resistance. The use of these two materials can take advantage of distant crosses. In this study, seeds from the female castor parent (Lm female line aLmAB2), the male parent (CSR·181) and their hybrid progeny were used as study material and the hybrid traits of the progeny and the parent were identified by proteomic analysis. In addition, genes associated with these traits were determined and tested by reverse transcription-quantitative PCR (RT-qPCR). The results of this study can provide a basis for guiding castor production practices and breeding new castor varieties with high yield, high oil and resistance to adversity.
The plant materials used in this study were the castor seeds from the female parent (aLmAB2), male parent (CSR·181), and their hybrid offspring F0 and F1. Castor plants at the flowering stage had their inflorescences manually pollinated and bagged to avoid any further risk of additional natural cross pollination. Subsequently, the seeds that had a developmental time of 20 days (Fig. 1A), 40 days (Fig. 1B), and 60 days post-pollination (DPP) (Fig. 1C) were collected. After removing their testa, the seeds were wrapped in tinfoil paper and stored in an ultralow-temperature freezer at −80°C for subsequent use. The experimental materials were provided by Tongliao Research Institute of Agriculture and Animal Husbandry (China).
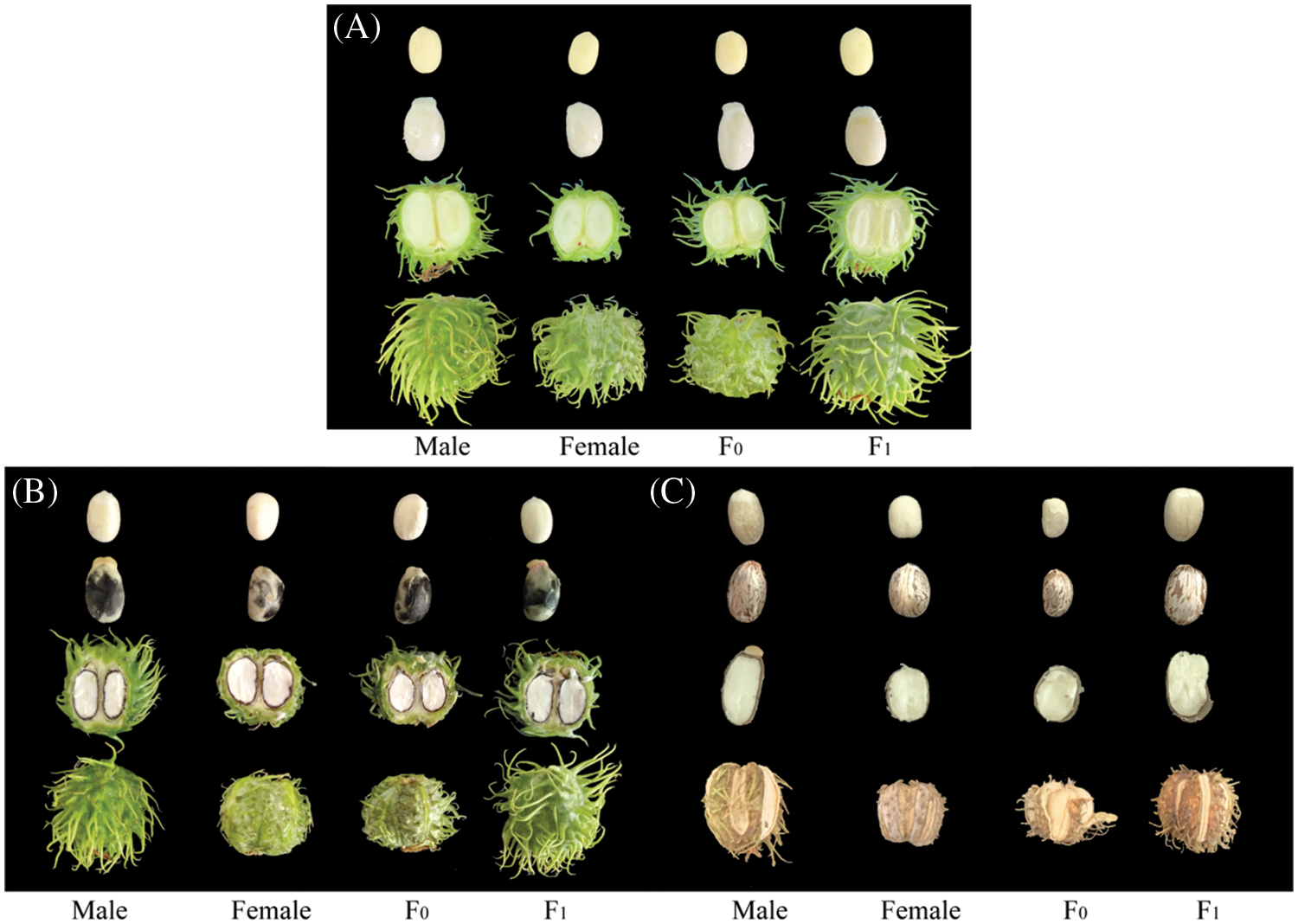
Figure 1: Castor seeds at different developmental stages
The reagents used in this experiment included Tris-HCI solution, ethylene diamine tetraacetic acid stock solution, 0.4% β-mercaptoethanol solution, 0.9 M sucrose, balanced phenol solution, extraction solution, ammonium acetate/methanol solution, acetone solution, lysis solution, NaOH solution, HCl solution, lysine solution, Amersham CyDye DIGE Fluors (minimal dyes) for the Ettan DIGE kit (GE), the 2-D Clean-up kit (GE), the 2-D Quant kit (GE), hydration solution, 87% glycerol, covering oil, monomer storage solution, 4×separating gel buffer, 10% sodium dodecyl sulfate (SDS) solution, N,N,N′,N′-tetramethylethylenediamine (TEMED), 10% ammonium persulfate solution, 75% alcohol, gel-sealing liquid, equilibrium solution, bromophenol blue dye, 1,4-dithiothreitol (DTT), iodoacetamide (IAA), premixed protein marker, SDS electrophoresis buffer, stationary liquid, amplification solution, Coomassie brilliant blue dye, decolorizing solution, RNA Rapid Extraction Kit for plants (ZOMANBIO), GoTaq® qPCR and RT-qPCR Systems (Promega), and reverse transcription system (Promega). All reagent formulas are shown in Table S1. The instruments used in this study included high-speed and low-speed refrigerated centrifuges (Thermo), an Infinite M200 Pro multifunctional microplate reader (TECAN), a gel imaging system and electrophoresis apparatus (Bio-Rad), the Ettan IPGphor 3 for electrophoresis by isoelectric focusing (IEF) in the first dimension, a MultiTemp IV water bath, a FLA9500 multifunctional biomolecular imager (GE), and a real-time fluorescence-based quantitative PCR system (ABI 7500, ABI Company, USA).
An optimized phenol extraction method was used to extract protein from castor seeds. The obtained dry protein powder was aliquoted into 1.5-mL centrifuge tubes. Each tube was added with a 20-fold volume of lysis solution (each 1 mL of lysis solution was added to 0.0062 g DTT, 20 μL of immobilized pH gradient (IPG) buffer, and 40 μL of protease inhibitor before use). Then, the tube was vortexed at a low temperature for 30 min, followed by an ultrasound ice-bath for 20 min. The process was repeated until dry protein powder was fully dissolved. After centrifugation at 13,000 g for 1 h at 4°C, the supernatant was transferred to a 1.5-mL centrifuge tube. The 2-D Clean-up kit was used to remove impurities from the sample. The 2-D Quant kit was used for sample quantitation, which derived a standard curve: y = −0.0041x + 0.3751 (R2 = 0.999), where ‘y’ was the absorbance value of the post-quantitation sample at 480 nm and ‘x’ was the protein concentration (μg/μL).
For the first dimension, 450 μL of the sample solution containing 800 μg of protein was loaded for electrophoresis by IEF. First, 800 μg of the lysed sample was transferred into a centrifuge tube, which was then added to hydration solution to achieve a total volume of 450 μL (each 1 mL of hydration solution was added with 5 μL of NL3-10 IPG buffer and 0.0028 g of DTT). Then, the sample solution was vacuum-centrifuged at 13,000 rpm for 1 hour at 4°C. After the centrifuged sample solution was transferred to the strip groove, a 24-cm-long dry gel strip (pH 3–10 NL) was placed into the groove; after ensuring that the gel surface was fully in contact with the sample, the covering oil was added, and then a lid was placed to cover the gel strip groove. Finally, after placing the gel strips of all samples into the PROTEAN IEF instrument at a constant temperature of 20°C, run the IEF protocol in Table S2.
For the second-dimension SDS-PAGE, 12.5%T polyacrylamide gel was used. First, the gel strips after the first-dimension IEF were equilibrated for 15 min in each of 2% (w/v) DTT and 2.5% (w/v) IAA equilibration buffer. Then the well-equilibrated strips were sealed with sealing liquid in a gel plate, which was then placed in the Ettan DALT 12 unit (12 gels per run) (GE-061-TY7119, GE Healthcare). Electrophoresis was performed at 2 W per gel for 1 h and then 16 W per gel until the bromophenol blue band was 0.5–1 cm away from the bottom of the gel.
After electrophoresis, the gel was taken out and sequentially submerged in stationary liquid for 30 min, amplification solution for 20 min, Coomassie brilliant blue dye for 10 min, and finally, the decolorizing solution until only protein spots appeared on the gel, without other, interfering dye residues. The Image Scanner III image scanning system (28-9076-07 GE Healthcare, USA) was used for gel imaging, and the Image Master 2-D Platinum 7.0 software was used for gel image analysis.
The sample was lysed using the lysis solution without DTT or IPG buffer. After lysis, the sample solution was adjusted to pH 8.0–9.0 using 100 mM NaOH and 1 M HCl, with a target pH of 8.5. The subsequent procedures were the same as those described in 2.2.1.
2.4.2 Preparation of CyDye DIGE Fluor Minimal Dyes
Preparation of 1 nmol/μL fluorescent dye stock solution: After the dye-storing tube was taken out of the −80°C refrigerator, each tube received 5 μL of N,N-dimethylformamide solution and then vortexed for 30 s to fully dissolve the dye. Next, it was centrifuged at 12,000 g for 30 s to push the dye stock solution to the bottom of the tube. Preparation of 400 pmol/μL fluorescent dye working solution: Five microliters of fluorescent dye stock solution was diluted with 7.5 μL of N,N-dimethylformamide solution to achieve a final concentration of 400 pmol/μL, followed by centrifugation at 12,000 g for 30 s to push the solution to the bottom of the tube. All the above procedures were performed at 4°C in the dark.
A certain volume of the sample solution (containing 50 μg of protein) was taken out and mixed with 1 μL of the dye-working solution through vortexing, followed by brief centrifugation. Then, as a quench-labeling reaction, the sample was kept on ice in the dark for 30 min and then mixed with 1 μL of 10 mM lysine solution through vortexing. After a brief centrifugation, the sample was placed on ice in the dark for 10 min.
According to the experimental design, the internal standard sample and the target samples labeled with Cy2, Cy3, or Cy5 were mixed in a centrifuge tube and then added to the hydration solution (every 1 mL of hydration solution had been added with 5 μL of NL3-10 IPG buffer and 0.0028 g of DTT) up to a total volume of 450 μL. After being placed on ice in the dark for 10 min, the sample solution was treated as in 2.2.2 for the first-dimension IEF.
For the second-dimension SDS-PAGE, 12.5% polyacrylamide gel was prepared using low-fluorescence glass plates, followed by the same procedures as in 1.2.2. Light exposure was avoided during the entire process. The Typhoon FLA9500 multifunctional biomolecular imager was used for gel imaging, and the DeCyder software for 2D-DIGE analysis was used to identify and annotate the differentially expressed spots (ratio ≥ 1.5 and Significance of Anova ≤ 0.05).
2.5 MS Identification and Analysis of the DEP Spots
The DEP spots in the gel were recovered and sent to Shanghai Bioclouds Biological Tech Co., Ltd. (China), for MS identification. The identification steps of MS are as follows.
In-gel digestion: (according to Katayama et al. [11], Wash the gel spot twice; Remove the water and de-stained the gel spot with CBB de-staining solution at room temperature for 30 min; Remove the de-stained solution and add dehydration solution 1 for 30 min; Remove the dehydration solution 1 and add dehydration solution 2 for 30 min; Remove the dehydration solution 2 and add 10 μL working solution for 30 min; Covered with cover solution for digestion overnight (about 16 h) at 37°C; The supernatants were transferred into another tube; Add extracting solution to the gel at 37°C for 30 min, then centrifuge 5 min; The peptide extracts and the supernatant of the gel spot were combined and then completely dried.
2.5.3 Mass Spectrometry and Database Search
Samples were re-suspended with 5 µL 0.1% TFA followed by mixing in 1:1 ratio with a matrix consisting of a saturated solution of α-cyano-4-hydroxy-trans-cinn-amic acid in 50% ACN, 0.1% TFA; 1 μL mixture was spotted on a stainless-steel sample target plate. Peptide MS and MS/MS were performed on an ABI 5800 MALDI-TOF/TOF Plus mass spectrometer (Applied Biosystems, Foster City, USA); Data were acquired in a positive MS reflector using a CalMix5 standard to calibrate the instrument (ABI5800 Calibration Mixture); Both the MS and MS/MS data were integrated and processed by using the GPS Explorer V3.6 software (Applied Biosystems, USA) with default parameters.
2.5.4 Database Search and Functional Classification of Differentially Expressed Proteins (DEPs)
The information on the DEPs identified by MS was obtained by searching the database of castor bean proteins (https://www.ncbi.nlm.nih.gov/). The proteins were functionally classified into Gene Ontology (GO) categories according to UniProt (http://www.UniProt.org/) and QuickGO (https://www.ebi.ac.uk/QuickGO/), and their roles in metabolic pathways were categorized according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database (https://www.kegg.jp/kegg/pathway.html). The histograms of GO classification were generated on the Omics Cloud platform established by GENE DENOVO (http://www.omicshare.com/index.php). The heatmaps were drawn using GraphPad Prism7.04 software.
RNA was extracted from castor seeds using the Plant RNA Rapid Extraction Kit. extracted RNA was reverse transcribed into cDNA. primers were designed using SnapGene 2.8.0 software, and primer sequences are listed in Table S3 in the Supplementary materials.
The GoTaq® qPCR system was used for qPCR. The reaction system consisted of 10 μL of SYBR Premix Ex Taq, 0.4 μL of Rox reference dye II, 1.0 μL of each of the upstream and downstream primers, 2.0 μL of template DNA, and ddH2O up to 20 µL. The amplification program was by 45 cycles of 95°C for 30 s, 95°C 10 s, melting temperature for 45 s, and 72°C for 10 s. The results were calculated using the 2−ΔΔCt method.
One-way ANOVA was performed with SPSS version 19.0 software, and the relevant histograms were generated in Excel 2019 software.
3.1 Optimization of the 2-DE System
To determine whether it is necessary to remove impurities from protein samples in this experiment, the same protein samples were divided into two groups, one with impurity removal and the other without impurity removal, before 2-DE (24-cm dry gel strips, pH 3–10 NL, protein loading amount 800 μg). The results are presented in Fig. S1 To determine the optimal pH range for this experiment, the protein samples with the same treatment (with or without impurity removal) were subjected to IEF and SDS-PAGE using the 24-cm dry gel strips with pH 3–10 NL or pH 4–7. The results are shown in Fig. S2 To determine the optimal protein loading amount, different amounts of protein (800, 1,000, and 1,200 μg) were loaded for 2-DE (24-cm dry gel strips; pH 3–10 NL) under the same conditions. The comparison results are shown in Fig. S3 Based on the results of these preliminary experiments, the optimal experimental conditions were impurity removal, 24-cm dry gel strips with pH 3–10 NL, and a loading amount of 800 μg. These conditions were used in the following experiments.
The castor seeds at different developmental stages from parents and offspring were subjected to 2D-DIGE. The results are shown in Fig. 2. Note: A: Male, F0, 20 d. B: Female, F1, 20 d, A & B when they had Developed to 20 DPP. C: Male, F0, 40 d. D: Female, F1, 40 d, C & D when they had Developed to 40 DPP. E: Male, F0, 60 d. F: Female, F1, 60 d, E & F when they had Developed to 40 DPP.
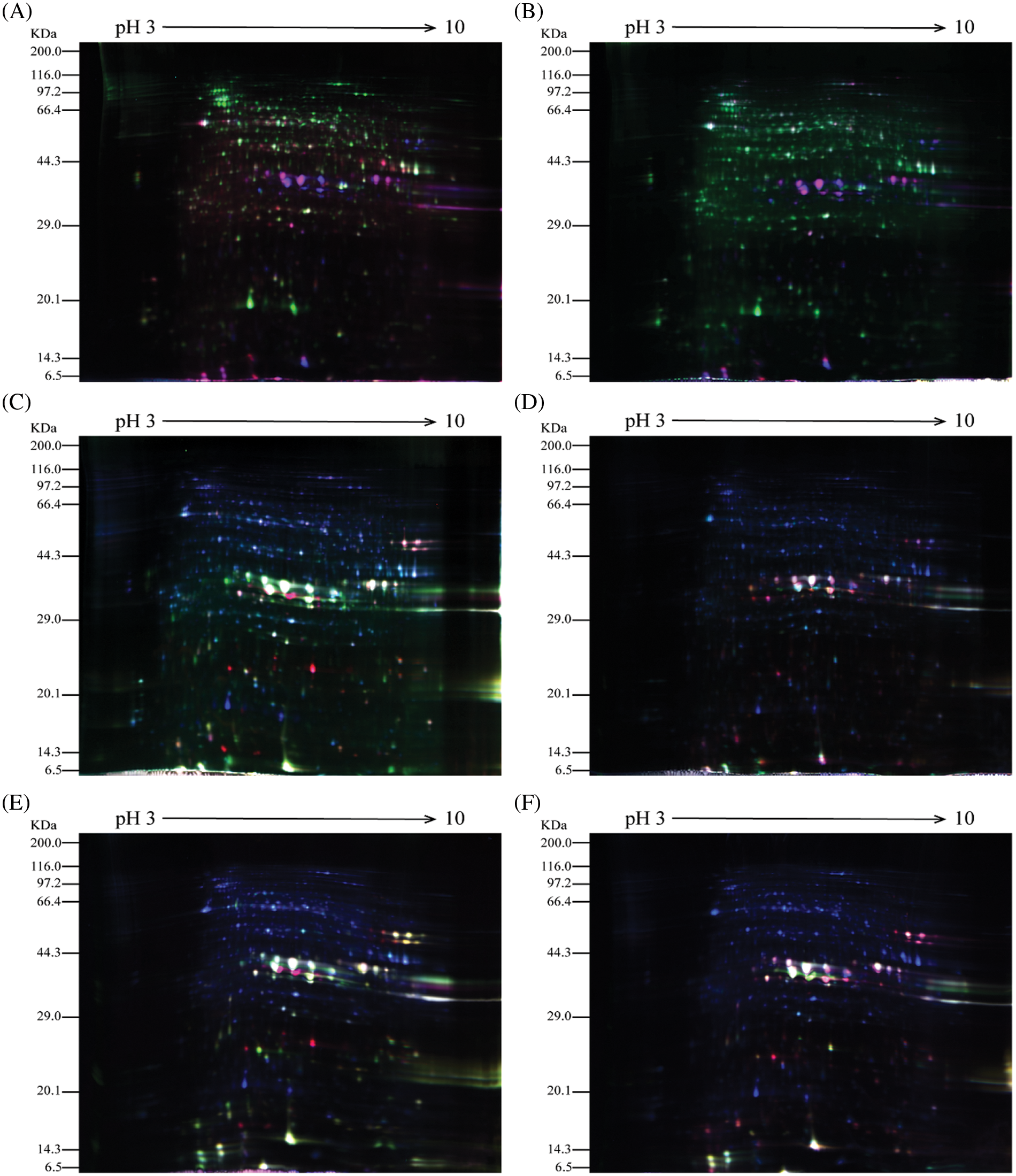
Figure 2: 2D-DIGE results of castor parents and their offspring
In the figures, the blue protein spots denote the internal standard samples labeled with Cy2; the green protein spots are the Cy3-labeled samples from male and female parents; and the red protein spots are the Cy5-labeled samples from castor offspring. In Fig. 2, the 2D-DIGE protein spots for the seeds at different development stages from both castor parents and offspring indicate that the dye labeling of all three samples was successful and met the requirements for experimental analysis.
3.3 MS Identification and Functional Classification Results of DEPs
3.3.1 MS Identification Results
The 2D-DIGE results of castor seeds at different development stages from parents and their offspring were analyzed for differences using DeCyder 2-D software. A total of 122 differentially expressed spots (ratio ≥ 1.5 and Significance of Anova ≤ 0.05) were determined and recovered, of which 121 spots containing 93 proteins were successfully identified by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF-MS). The results are shown in Fig. S4 and Table S4. The sources/names and classification of these DEPs are listed in Table S5. Among the DEPs between the castor parents and their offspring, 69 were present in seeds at 20 DPP, 46 in seeds at 40 DPP, and 20 in seeds at 60 DPP. Some proteins had a significantly different expression in the castor seeds at two or all three development stages between castor parents and their offspring, such as starch synthase, D-7 late embryogenesis abundant (LEA) protein, superoxide dismutase (SOD), and aspartic proteases (APs).
3.3.2 Functional Classification Results of the DEPs
The 93 DEPs identified were classified into five categories (Fig. 3) related to stress responses (61.29%), seed nutrient storage (17.20%), fatty acid synthesis and metabolism (12.90%), amino acid metabolism (6.45%), and others (2.15%). The DEPs were most often related to stress responses. These included SOD, APs, D-7 LEA protein, and glutathione peroxidase. The DEPs associated with nutrient storage in seeds included starch synthase (chloroplast/starch), 2S albumin precursor, and vicilin GC72-A. The DEPs associated with fatty acid synthesis and metabolism included stearoyl-ACP desaturase, oil body–associated protein 2C (OBAP2C), and acetyl-CoA carboxylase. The DEPs associated with amino acid metabolism included transaldolase, transketolase (chloroplast), S-adenosylmethionine synthase (SAMS), and others.
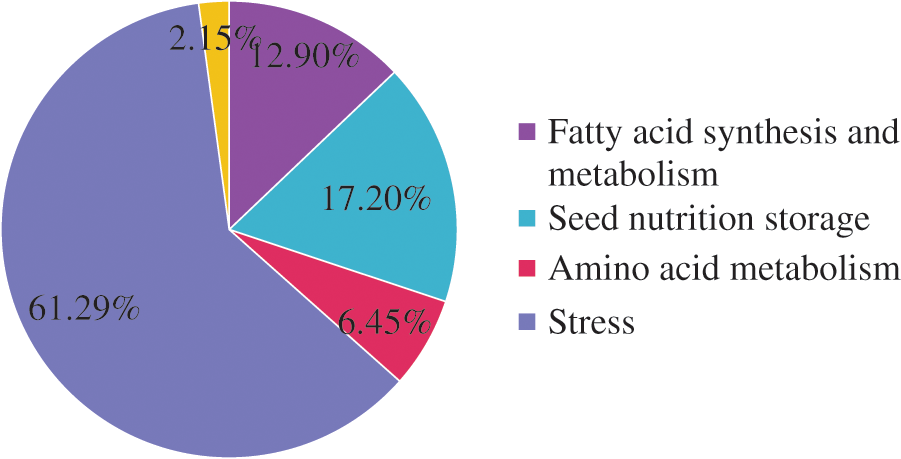
Figure 3: Functional classification of the differentially expressed proteins between castor parents and their offspring
3.3.3 Functional Classification Results of the DEPs in Seeds at 20 DPP between Castor Parents and Their Offspring
A total of 69 DEPs were identified in seeds at 20 DPP between castor parents and their offspring. The seeds from F1 offspring had 16 upregulated DEPs (e.g., OBAP2C, acetyltransferase NATA1, and 2S albumin precursor) when compared with their male parent and 13 upregulated DEPs (e.g., protein disulfide isomerase (PDI), luminal-binding protein 5, and triphosphate isomerase) when compared with their female parent. The seeds from F1 offspring had 23 downregulated DEPs (e.g., dihydrolipoyl dehydrogenase 1, MLP-like protein 34, and salicylic acid-binding protein 2) when compared with their male parent and nine downregulated DEPs (e.g., l-ascorbate peroxidase, adenosine kinase 2, and the 36-kDa porin in the mitochondrial outer membrane) when compared with their female parent. In the F0 seeds, 11 DEPs (e.g., legume protein A, SOD, and hydroxyl-ACP dehydrase) were upregulated when compared with their male parent, and five DEPs (i.e., transaldolase, stearoyl-ACP desaturase, 60S acidic ribosomal protein P2, SOD, and uncharacterized protein) were upregulated when compared with their female parent. The F0 seeds had 11 downregulated DEPs (e.g., the major latex allergen Hev b5, MLP-like protein 31, and GDP-mannose 3,5-epimerase 1) when compared with their male parent and four downregulated DEPs (i.e., adenosyl homocysteinase, xylem serine proteinase 1 precursor, cytochrome c oxidase subunit 6b-1, and lactoylglutathione lyase GLX1) when compared with their female parent. The relative expression levels of DEPs are shown in Fig. S5, and the results regarding their involvement in KEGG metabolic pathways are shown in Fig. S6.
Fig. 4 shows the GO functional annotations of the 69 DEPs in the three categories of biological process, cellular component, and molecular function. Note: A: Male, F1, 20 DPP; B: Female, F1, 20 DPP, C: Male, F0, 20 DPP; D: Female, F0, 20 DPP. The DEPs between F1 and parents and DEPs between F0 and parents were mostly associated with biological processes, including starch synthase, ribulose bisphosphate carboxylase small chain, and glutathione peroxidase. A smaller number of DEPs were associated with molecular functions, including glutathione peroxidase, APs, and acetyltransferase NATA1. The fewest DEPs were associated with cellular components, including 60S acidic ribosomal protein P2, stearoyl-ACP desaturase, and PDI. The analysis of KEGG metabolic pathways showed that the DEPs between F1 and parents and the DEPs between F0 and parents were mostly involved in rcu01100 (metabolic pathways) and rcu01110 (biosynthesis of secondary metabolites), such as alcohol dehydrogenase 1 (ADH1), starch synthase, and hydroxyl-ACP dehydrase. A number of DEPs were involved in rcu01230 (biosynthesis of amino acids), rcu00710 (carbon fixation in photosynthetic organisms), and rcu00061 (fatty acid biosynthesis).
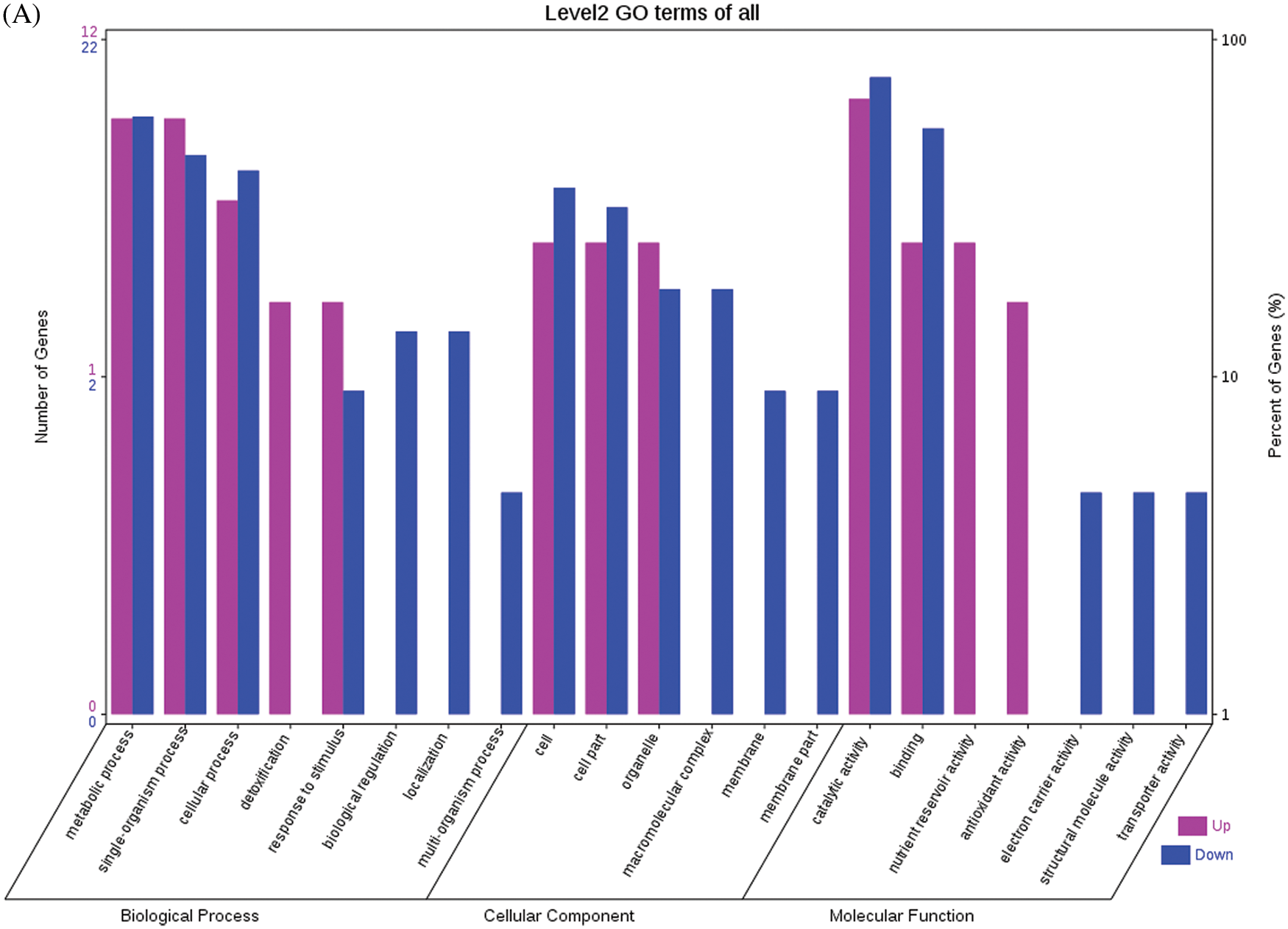
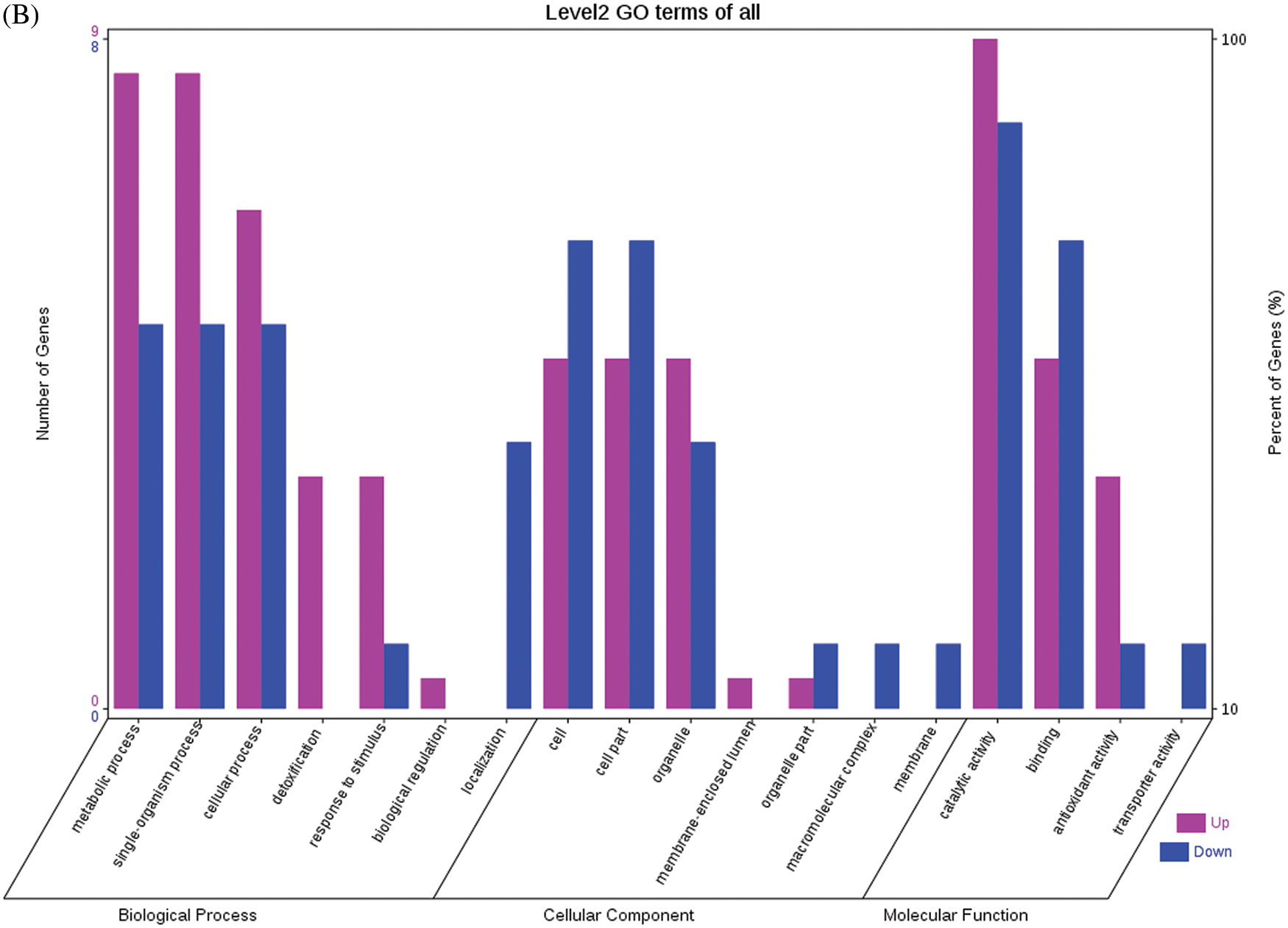
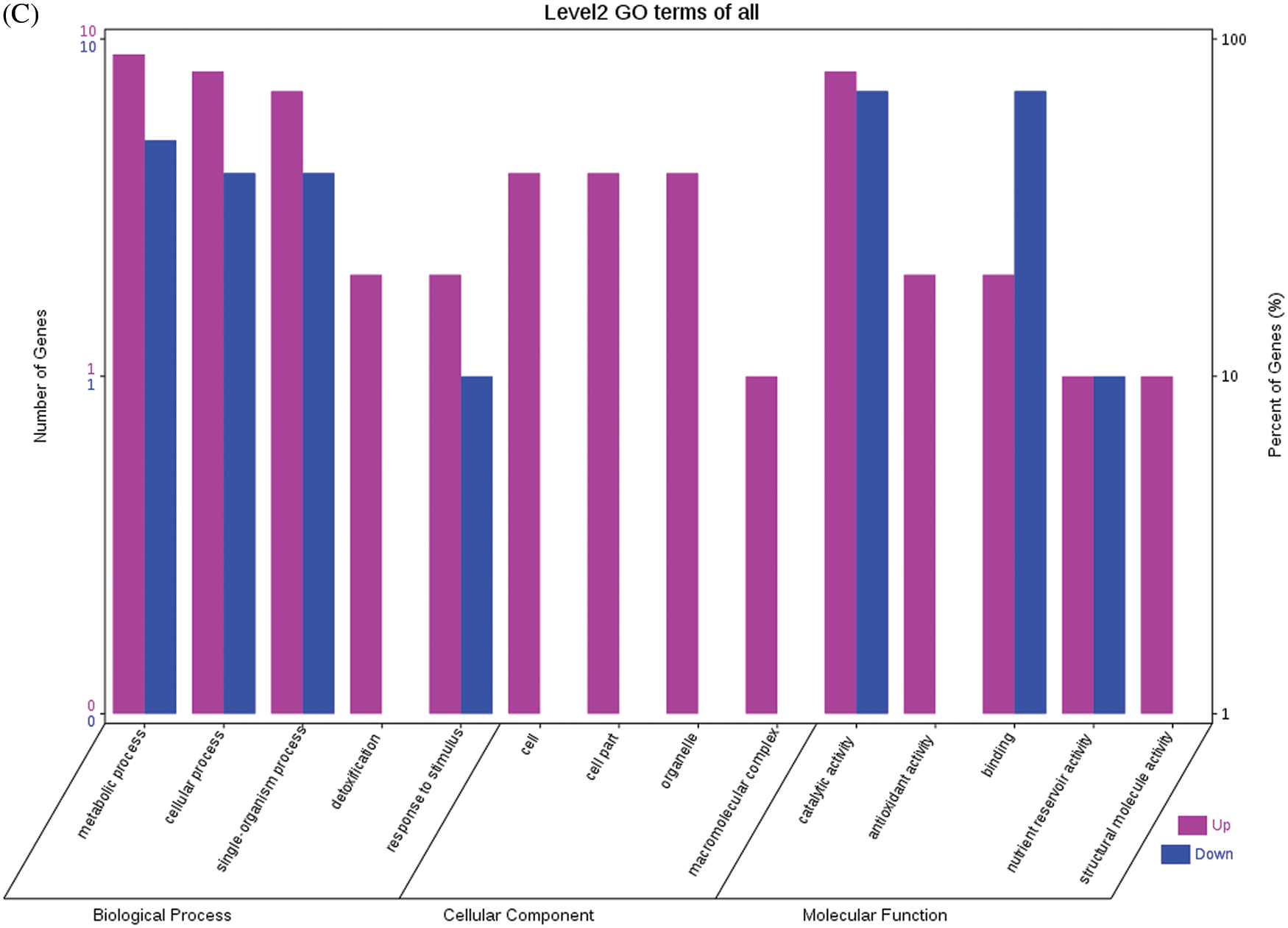
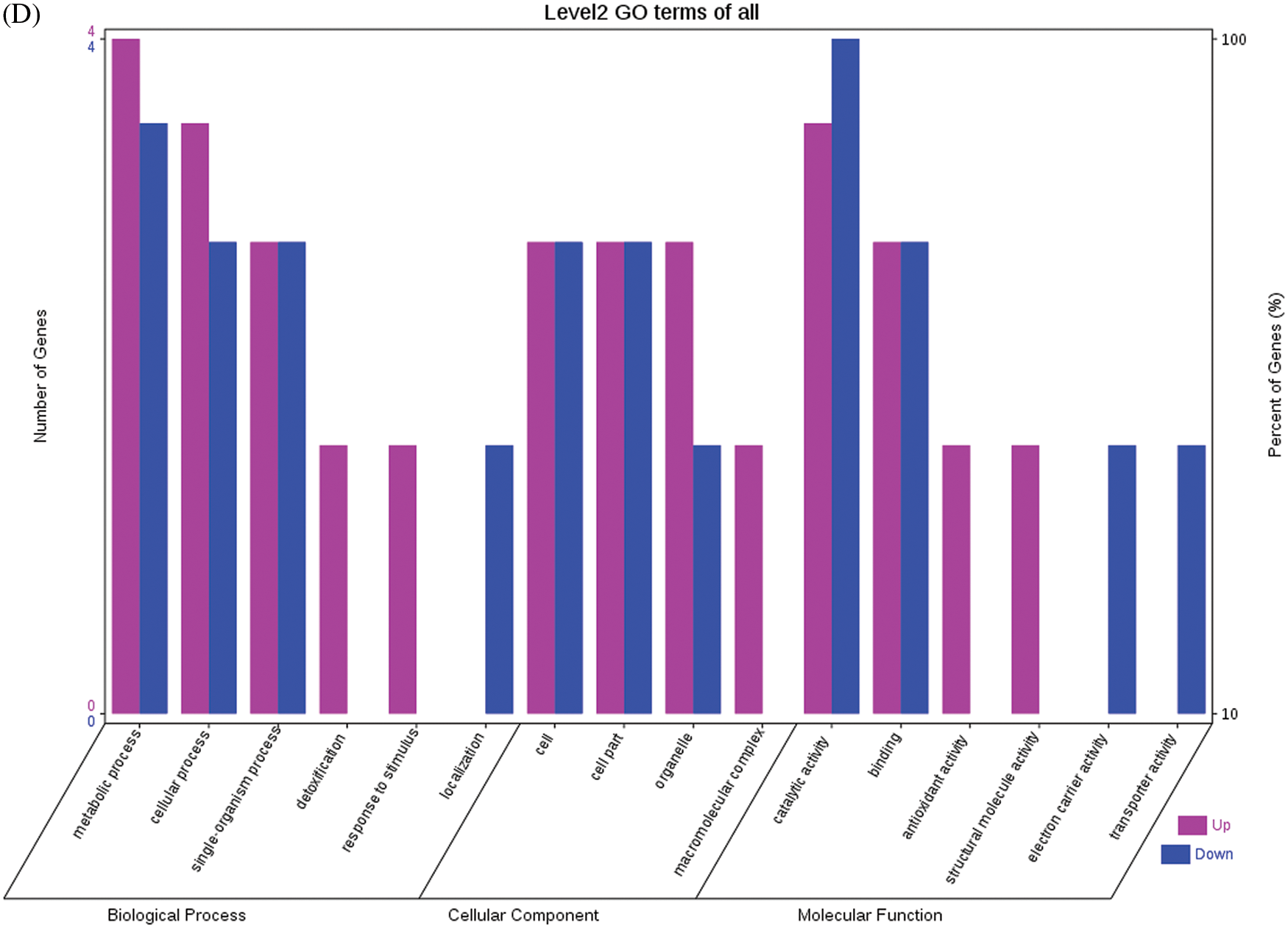
Figure 4: GO functional annotation of differentially expressed proteins in seeds at 20 DPP between castor parents and their offspring
3.3.4 Functional Classification Results of the DEPs in Seeds at 40 DPP between Castor Parents and Their Offspring
A total of 46 DEPs were identified in seeds at 40 DPP between castor parents and their offspring. The F1 seeds had three upregulated DEPs when compared with their male parent, including D-7 LEA protein, legume protein A, and heat shock protein (HSP); and they had 10 upregulated DEPs when compared with their female parent, including the large subunit of 2S sulfur-rich seed storage protein, nucleolar protein 56 (NOP56), and reduced nicotinamide adenine dinucleotide phosphate (NADPH)-dependent aldehyde reductase 1. Compared with their male parents, four DEPs were downregulated in the F1 seeds, including ATP synthase subunit β, HSP, ADH1, and plastid 3-keto-acyl-ACP synthase; when compared with female parents, six DEPs were downregulated, including starch synthase, fructose-bisphosphate aldolase, GroES chaperonin, etc. In the F0 seeds, seven DEPs (e.g., APs, large subunits of 2S sulfur-rich seed storage protein, and nucleoside diphosphate kinase (NDPK)) and nine DEPs (e.g., SOD, D-7 LEA protein, and glutelin type-A precursor) were upregulated when compared with the male and female parents, respectively. One DEP (enoyl-ACP reductase (EAR) precursor) and three DEPs (i.e., translation-controlled tumor protein homolog, pre-pro-ricin, and GroES chaperonin) were downregulated when compared with the male and female parents, respectively. Compared with the F0 generation, the seeds of the F1 generation had seven upregulated DEPs (e.g., legume protein A, HSP, and 14-3-3-like protein D isoform X1) and seven downregulated DEPs (e.g., fructose-bisphosphate aldolase 1, adenosine kinase 2, and adenosyl homocysteinase). The relative expression levels of the DEPs mentioned above are depicted in Fig. S7, and the results regarding their involvement in the KEGG metabolic pathways are presented in Fig. S8.
The GO functional annotations for the 46 DEPs are presented in Fig. 5. Note: A: Male, F1, 40 DPP; B: Female, F1, 40 DPP; C: Male, F0, 40 DPP; D: Female, F0, 40 DPP; E: F1 vs. F0, 40 DPP. The majority of DEPs in seeds between F1 and male parent was related to biological processes and molecular functions; compared with the male parent, only legume protein A was upregulated, while all other DEPs were downregulated in the F1 seeds. Most of the DEPs in seeds between F1 and the female parent (e.g., NADPH-dependent aldehyde reductase 1 and rRNA N-glycosidase) were associated with biological processes and molecular functions. Similarly, most of the DEPs between F0 and parents were related to biological processes and molecular functions, including cysteine protease inhibitor and glutelin type-A precursor. The DEPs between F1 and F0 were mostly related to biological processes, including SAMS and adenosyl homocysteinase; the DEP categorized under cellular component was enolase, and those categorized under molecular function included HSP and 14-3-3-like protein D isoform X1. According to the KEGG metabolic pathways, some HSP family members involved in rcu04141 (protein processing in endoplasmic reticulum), including HSP, were differentially expressed in seeds between F1 and parents, between F0 and parents, and between F1 and F0. In addition, among the upregulated DEPs in the F0 seeds vs. the male parent, NDPK was involved in rcu04016 (MAPK signaling pathway), and betaine aldehyde dehydrogenase 1 (BADH1) was involved in rcu00260 (glycine, serine, and threonine metabolism). SOD, an upregulated DEP in the F0 seeds compared with their female parent, was involved in rcu04146 (peroxisome); and chaperonin, an upregulated DEP in the F1 generation compared with F0, was involved in rcu03018 (RNA degradation).
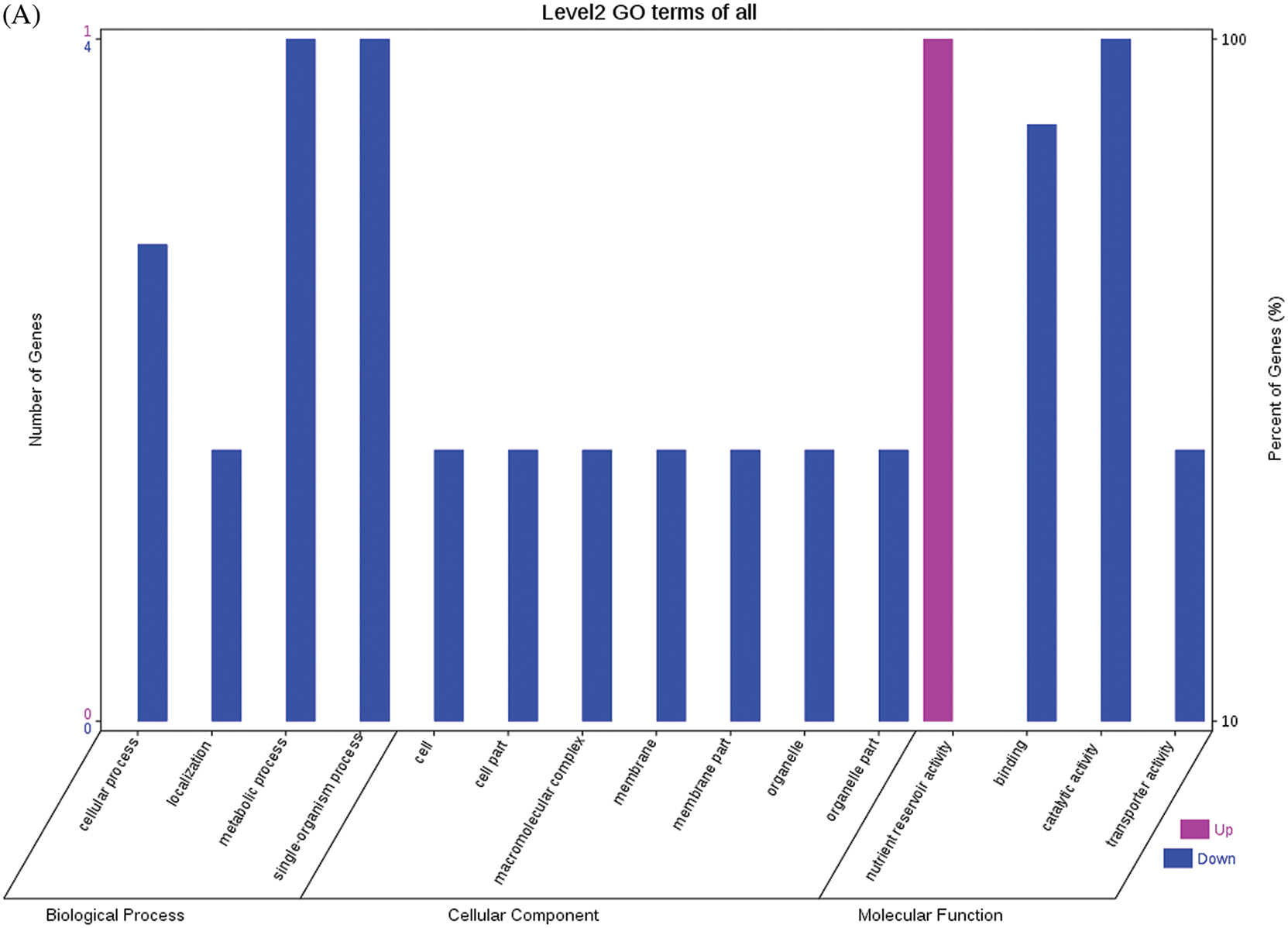
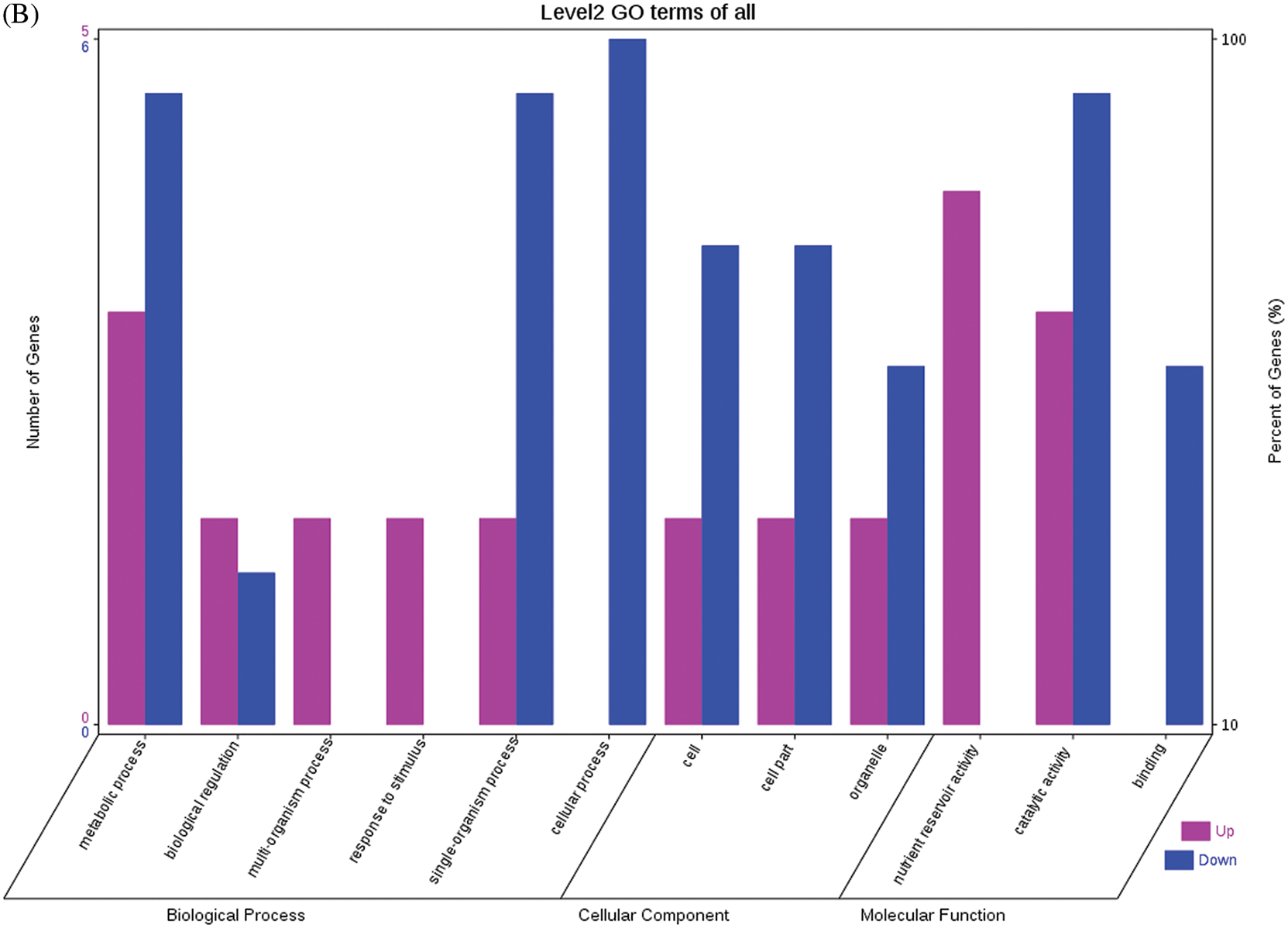
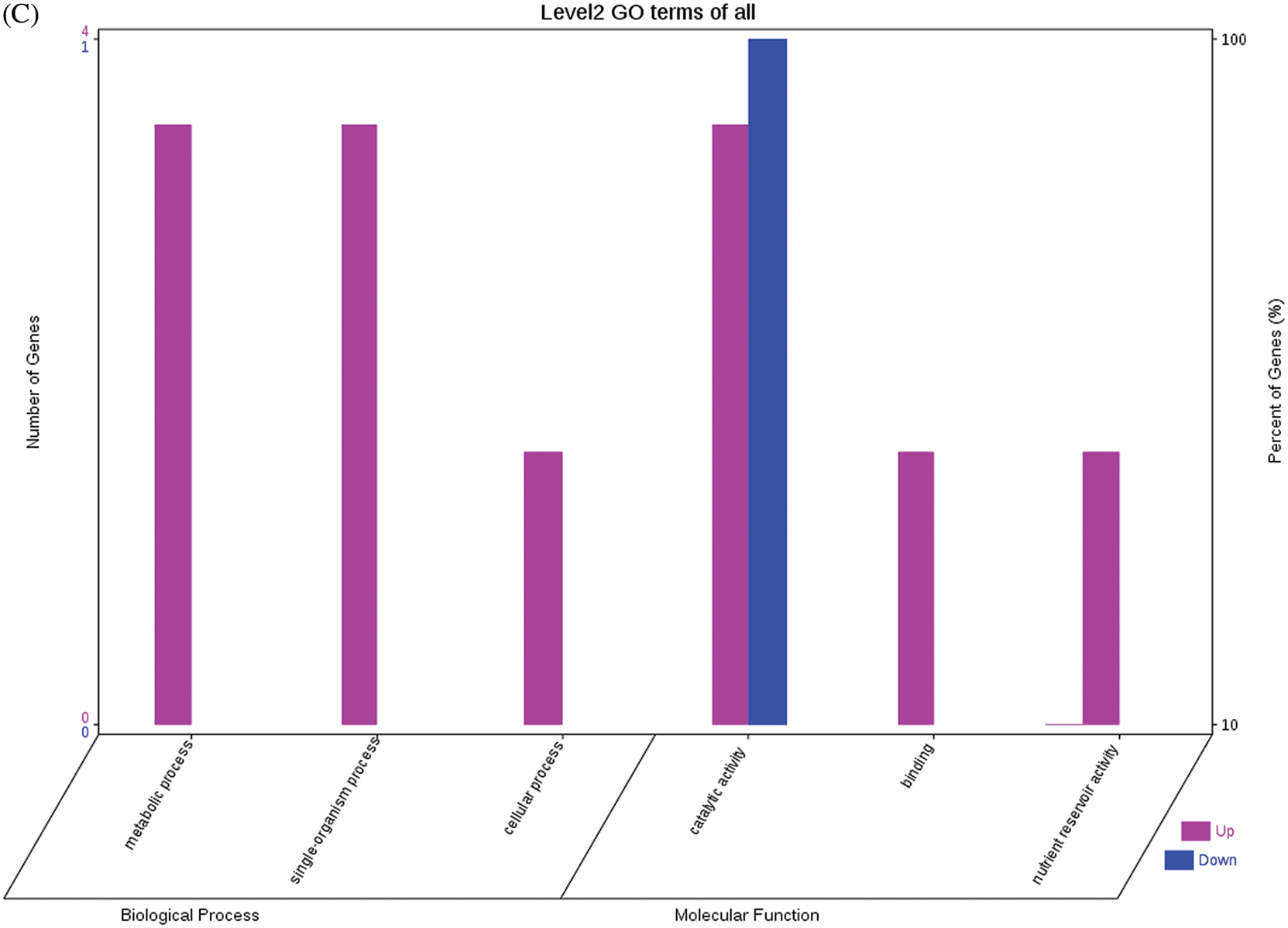
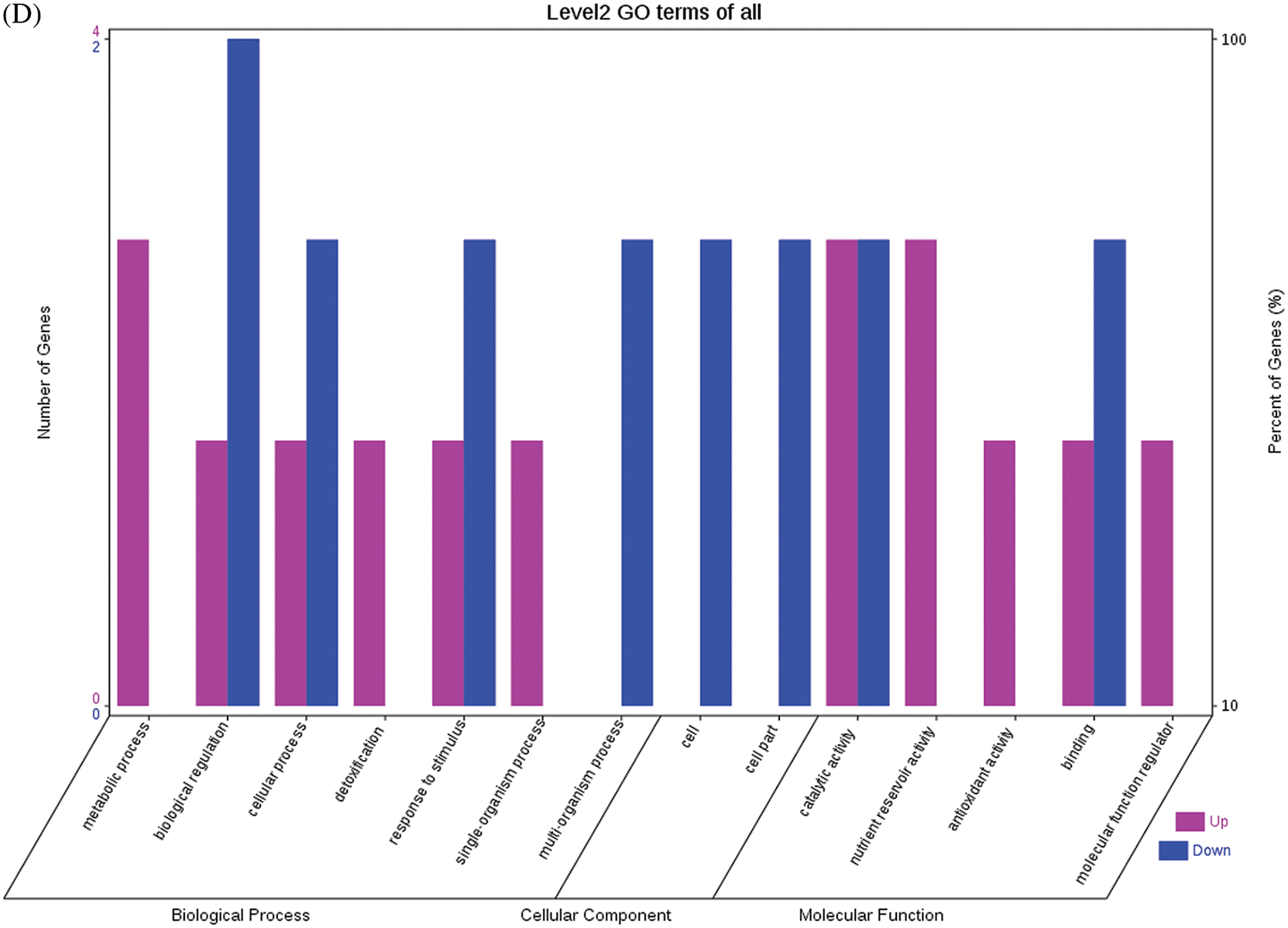
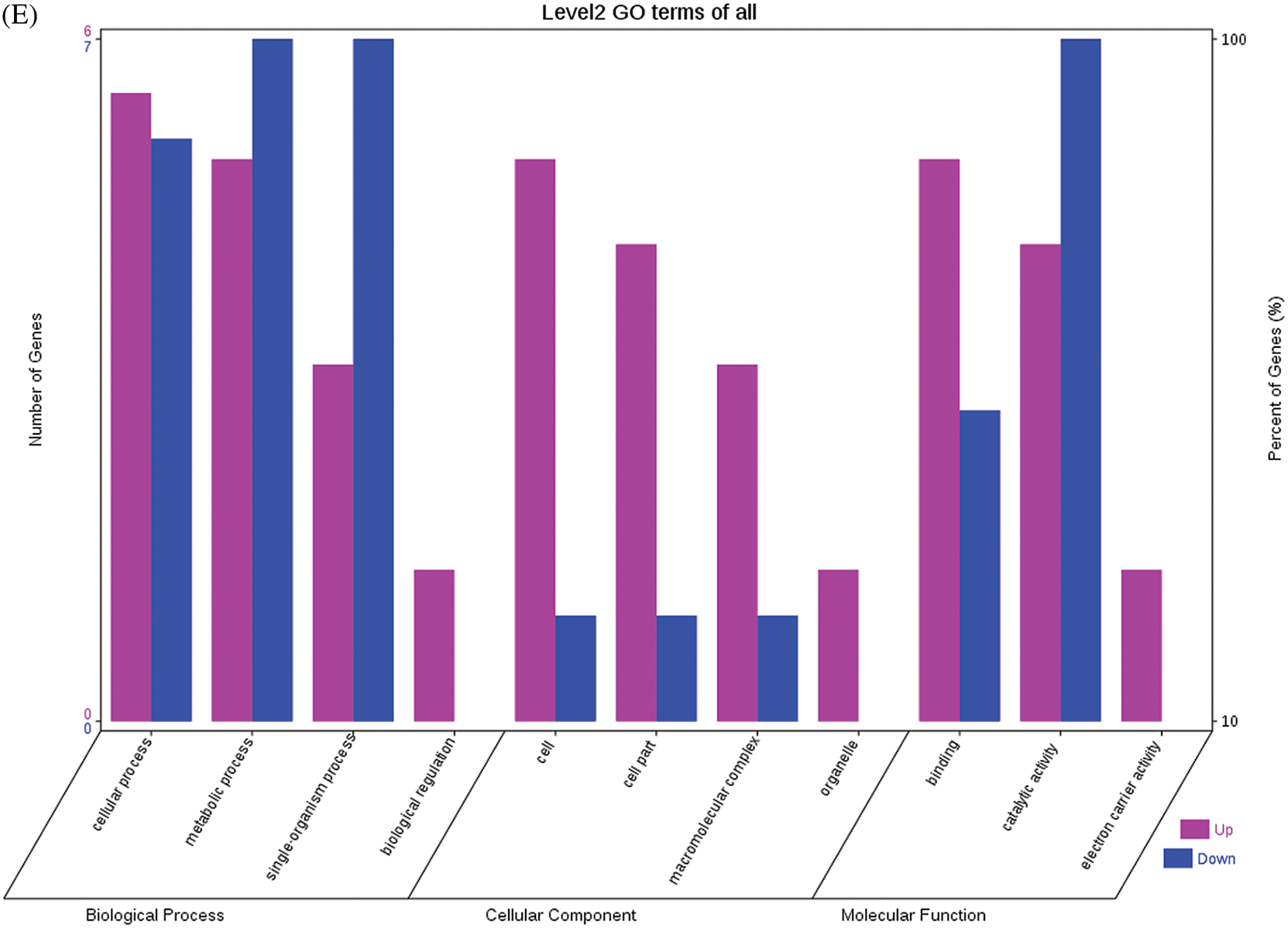
Figure 5: GO functional annotation of differentially expressed proteins in seeds at 40 DPP between castor parents and their offspring
3.3.5 Functional Classification Results of the DEPs in Seeds at 60 DPP between Castor Parents and Their Offspring
A total of 25 DEPs were identified in seeds at 60 DPP between castor parents and their offspring. The F1 seeds had two upregulated DEPs when compared with their male parent, NOP56 and 5-methyltetrahydroproylglutamate-homocysteine methyltransferase; and they had six upregulated DEPs when compared with their female parent, including D-7 LEA protein, NDPK, and large subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase. Five DEPs were downregulated in the F1 seeds compared with their male parent, including malate dehydrogenase, phospho-glucosidase, and enolase, and no DEP showed detectable downregulation in expression in the F1 seeds compared to their female parent. In the seeds of the F0 generation, three DEPs (i.e., vicilin GC72-A, legume protein A, and seed storage protein) and eight DEPs (e.g., 6-phosphogluconate dehydrogenase, large subunits of 2S sulfur-rich seed storage protein, and D-7 LEA protein) were upregulated compared to the male and female parents, respectively. No DEP showed detectable downregulation in the F0 seeds compared with either of their parents. Compared with F0, the F1 seeds had five upregulated DEPs (e.g., 2S albumin precursor, phospho-glucosidase, and malate dehydrogenase) but no downregulated DEP. The relative expression levels of the abovementioned DEPs are shown in Fig. S9, and the results on their involvement in KEGG metabolic pathways are shown in Fig. S10. The DEPs between F0 and the male parent were not involved in any metabolic pathways.
The GO functional annotations of the 25 DEPs are shown in Fig. 6. Note: A: Male, F1, 60 DPP; B: Female, F1, 60 DPP; C: Male, F0, 60 DPP; D: Female, F0, 60d; E: F1 vs. F0, 60 DPP. Most of the DEPs between F1 and parents and between F0 and parents (e.g., ADH1, NOP56, and NDPK) were related to biological processes and molecular functions. Most of the DEPs between F1 and F0 (e.g., phospho-glucosidase, malate dehydrogenase, and enolase) were related to biological processes. From the KEGG metabolic pathway analysis, the DEPs between F1 and parents, between F0 and parents, and between F1 and F0 were primarily involved in rcu01100 (metabolic pathways) and rcu01110 (biosynthesis of secondary metabolites), including 6-Phosphate gluconate dehydrogenase, ADH1, and 5-N-methyl-dl-glutamicf1 ACID-Homocysteine methyltransferase. In addition, the DEPs upregulated in F1 and F0 over their parents were also involved in the pathways of rcu00270 (cysteine and methionine metabolism), rcu00592 (alpha-Linolenic acid metabolism), rcu00230 (purine metabolism), etc. The DEPs upregulated in the generation compared with F0 also participated in the pathways related to rcu01200 (carbon metabolism) and rcu01230 (biosynthesis of amino acids).
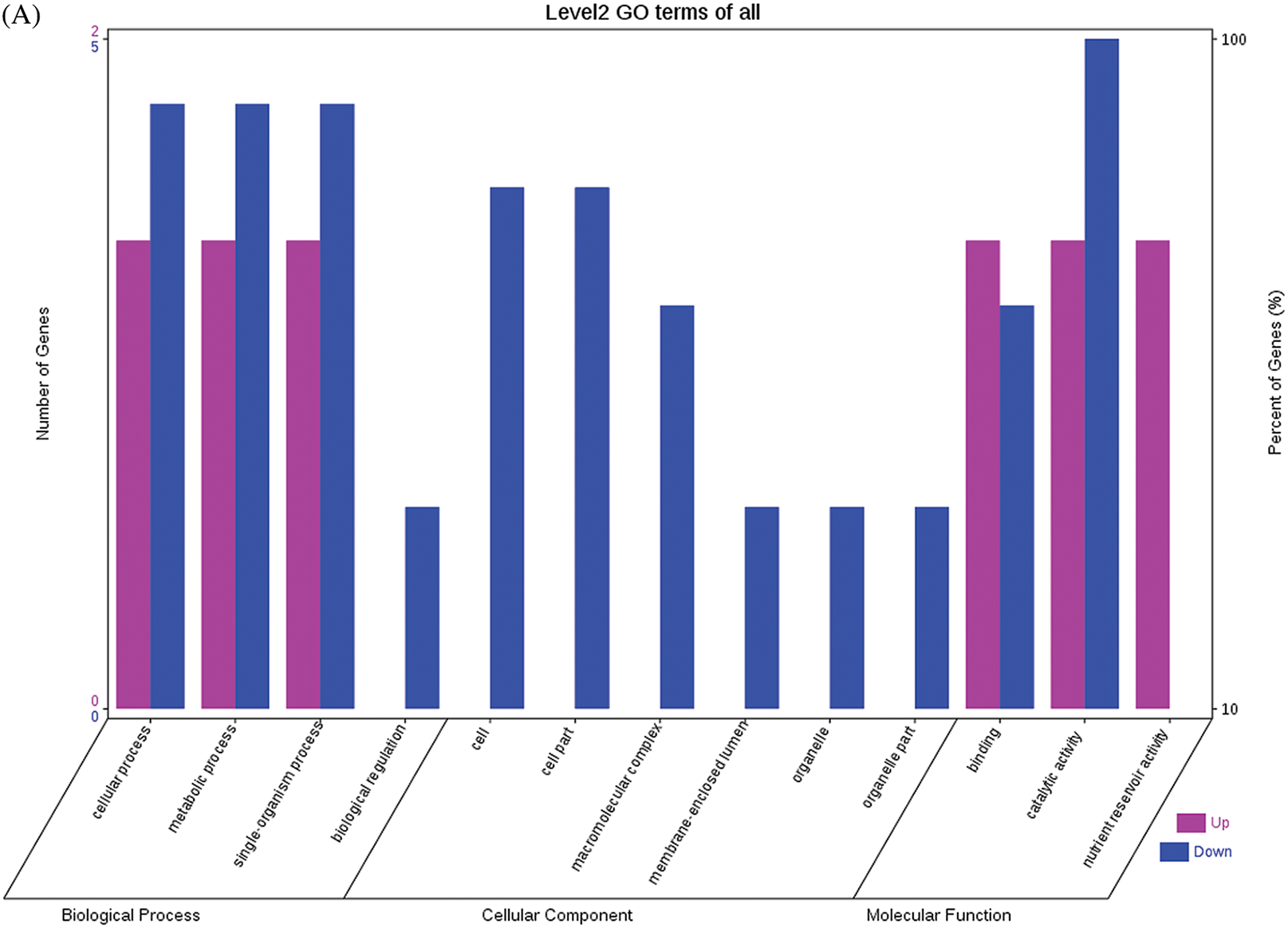
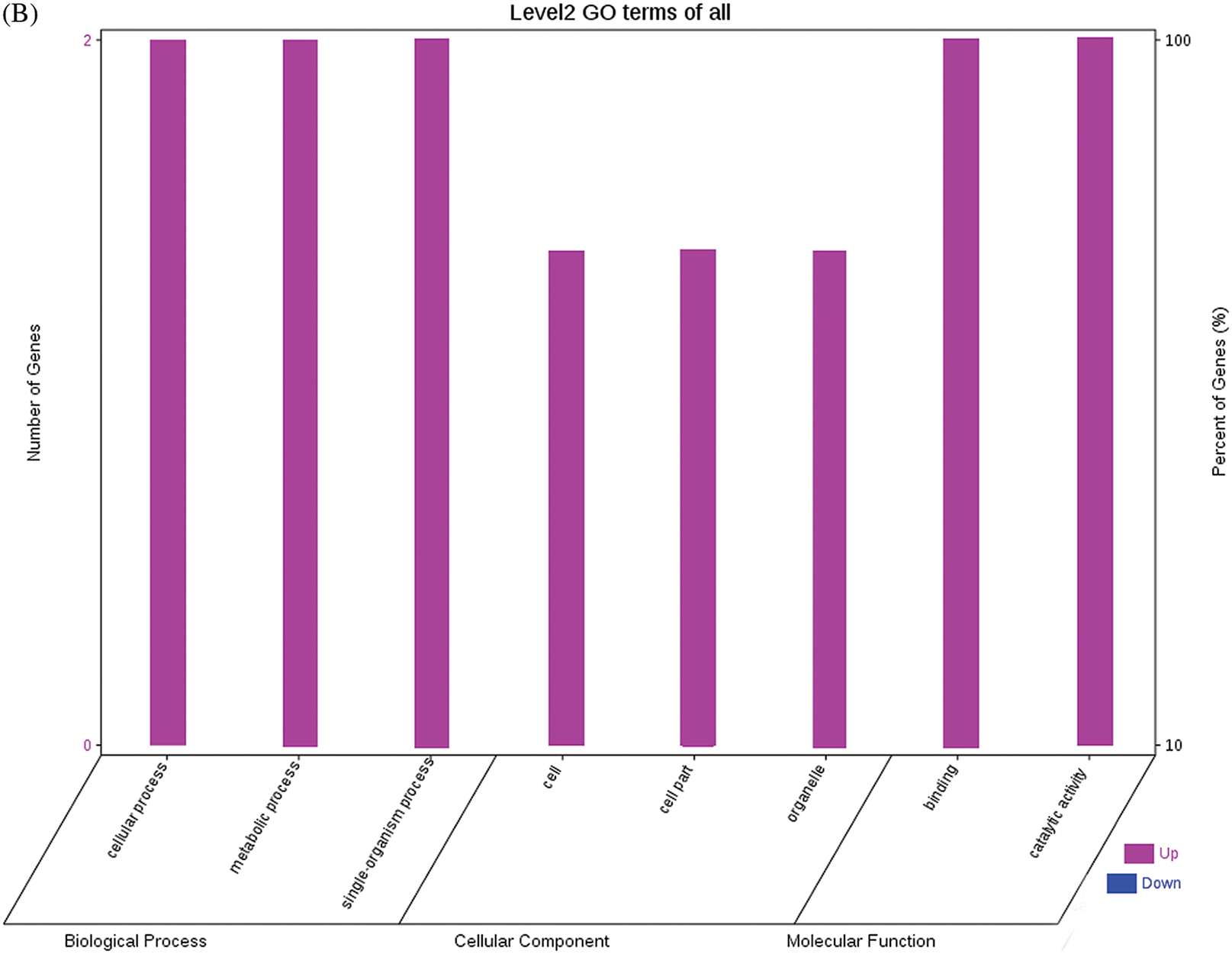
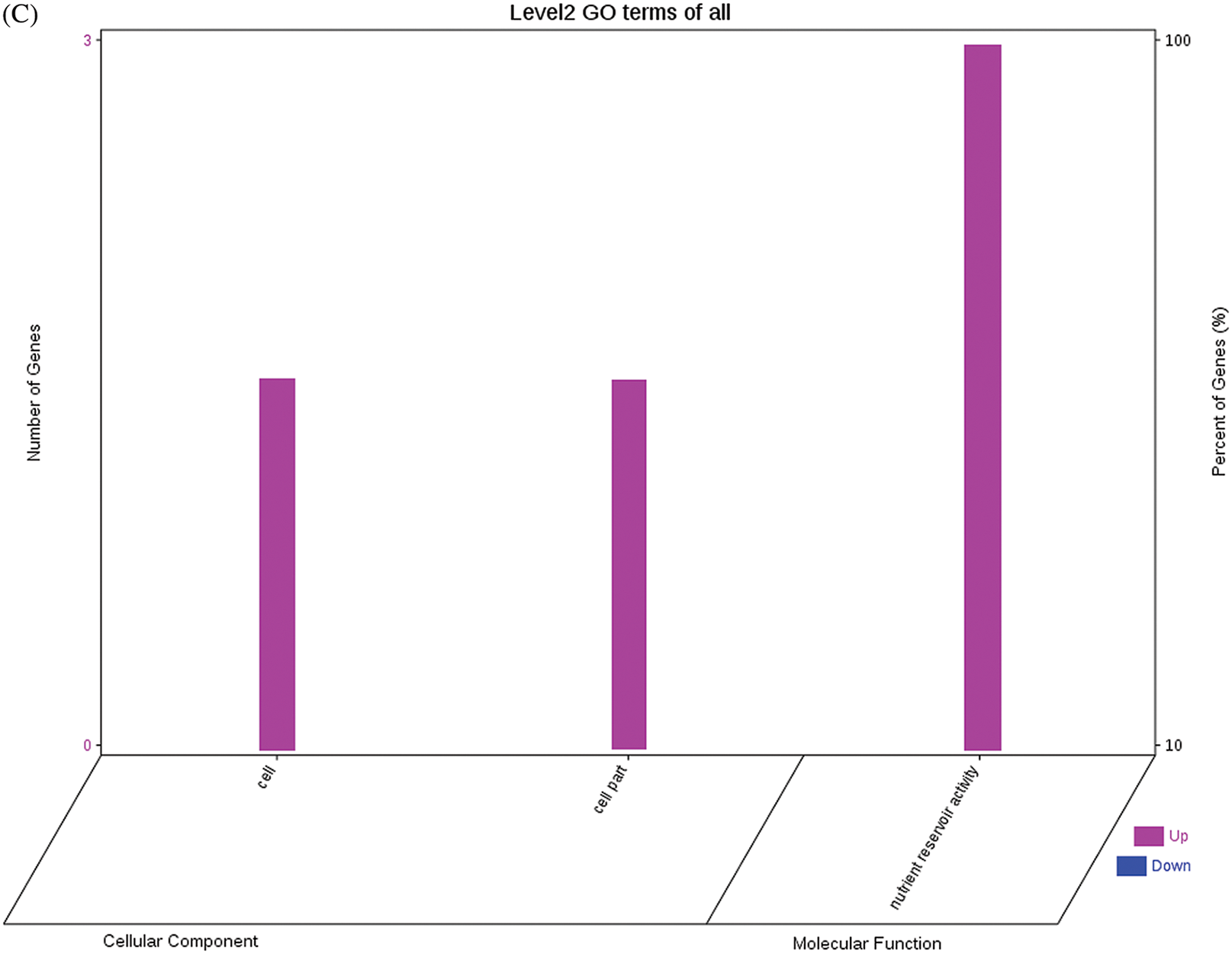
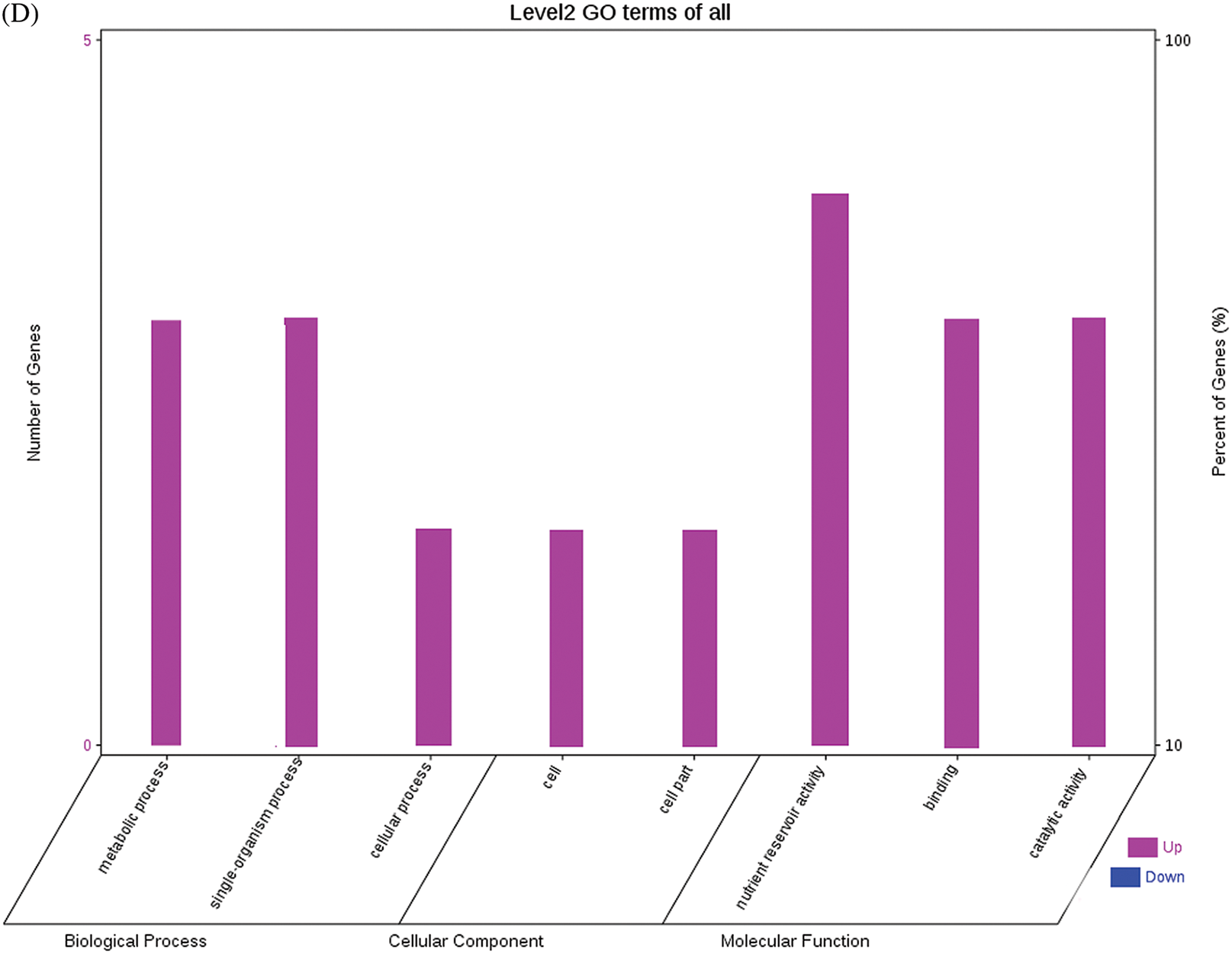
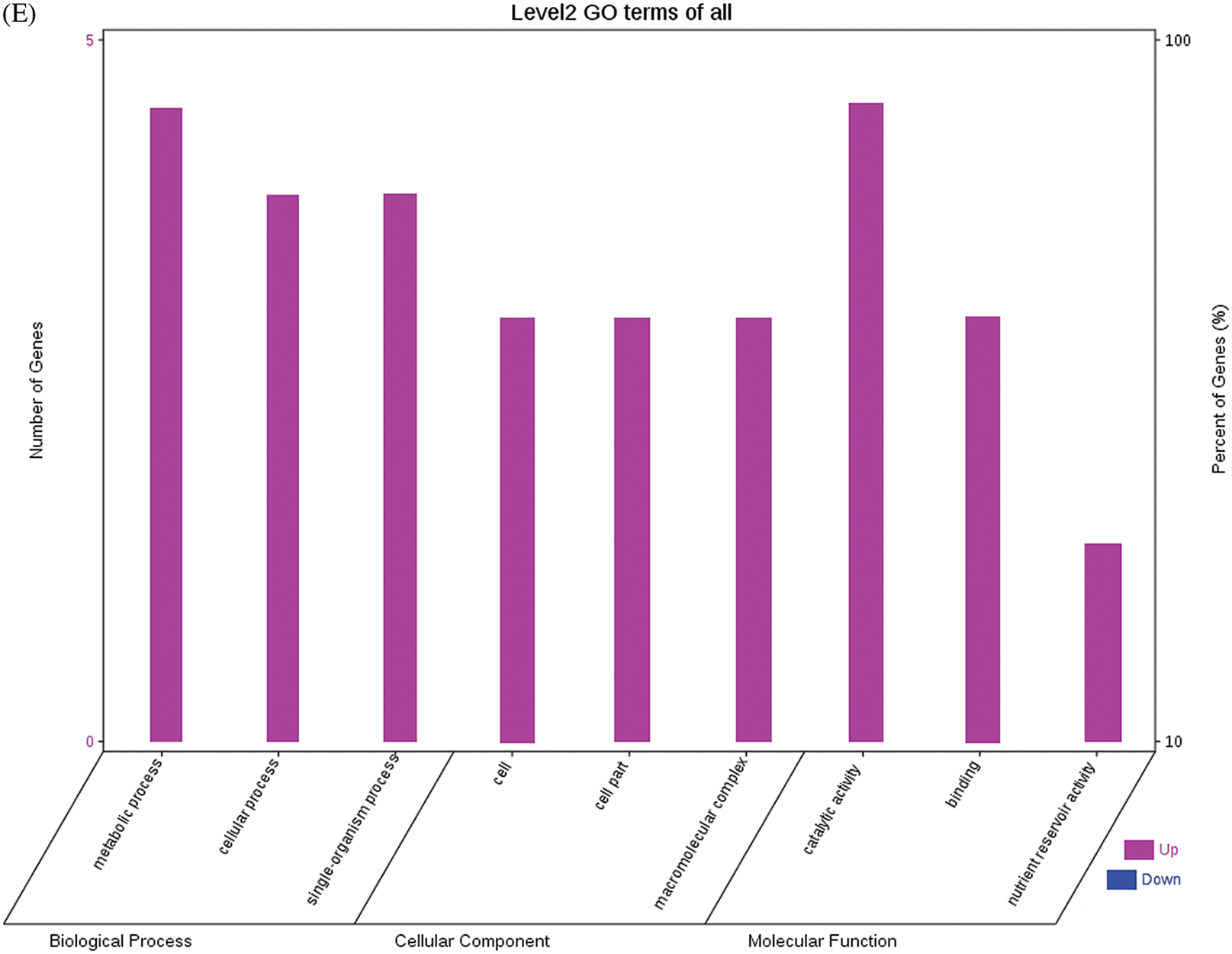
Figure 6: GO functional annotation of differentially expressed proteins in seeds at 60 DPP between castor parents and their offspring
3.4 RT-qPCR Results of the DEPs
Twelve DEPs in the seeds between castor parents and their offspring (Table 1) were selected for RT-qPCR analysis, and the results are shown in Fig. 7. The expression of the genes corresponding to the 12 DEPs all showed differences consistent with those of the DEPs.
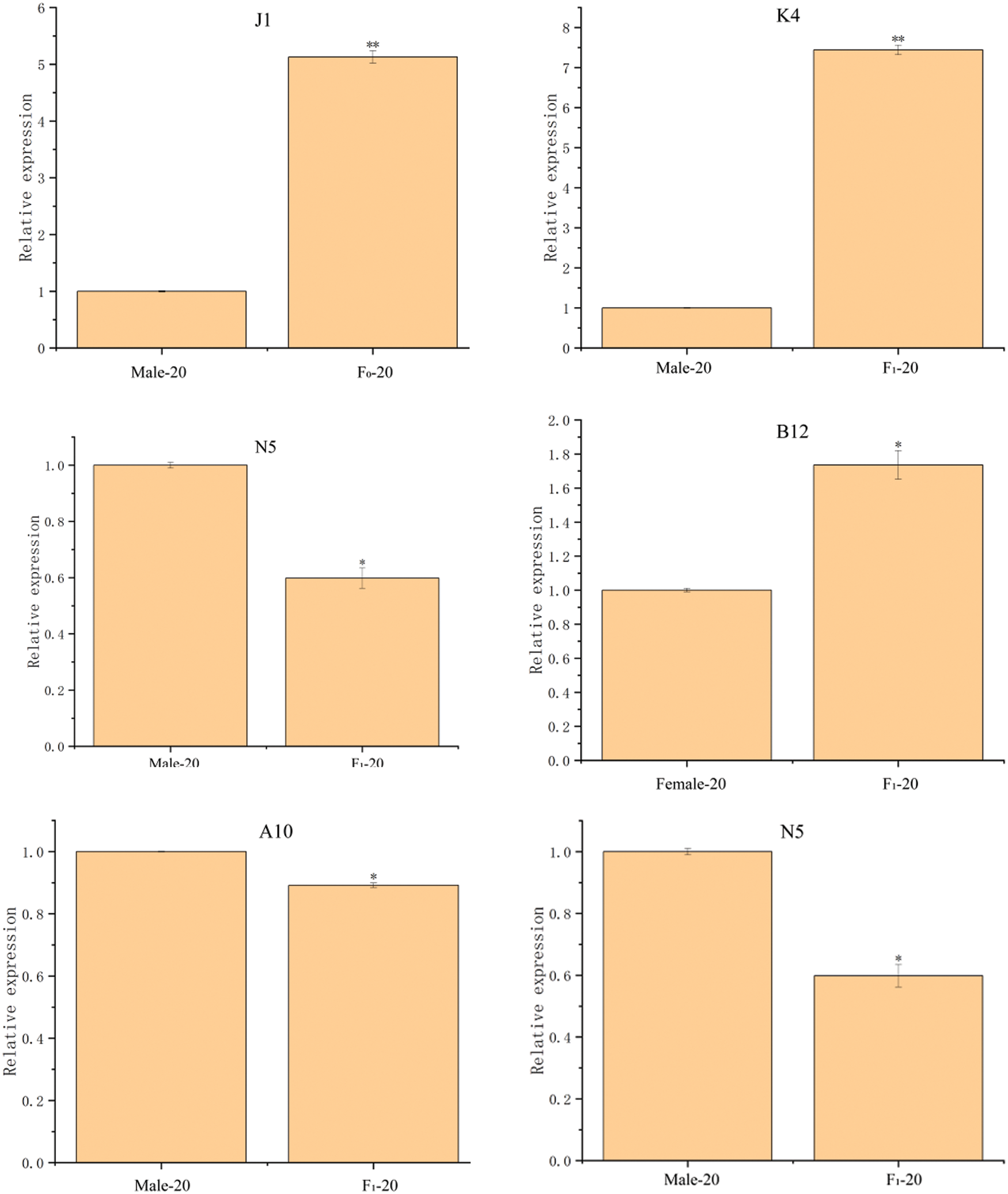
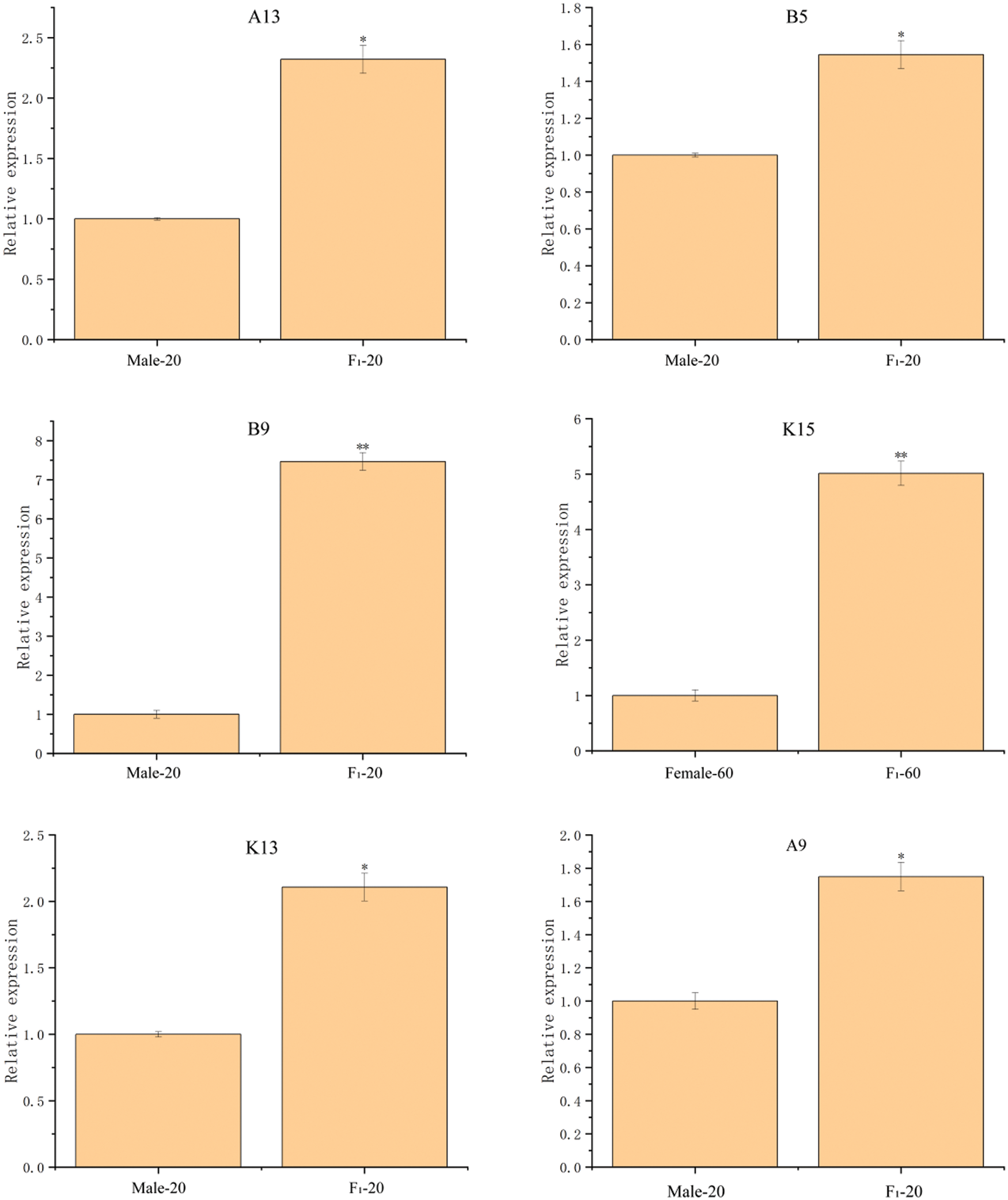
Figure 7: The RT-qPCR results of partially differentially expressed proteins between castor parents and their offspring
Note: *: 0.01 < p ≤ 0.05, **: 0 < p ≤ 0.01.
For more than a century, the goal of castor breeding research is to breed new varieties with high yield, high oil content and resistance to adversity. To achieve this goal, the most common method is hybridization, because most hybrid plants will show hybrid advantage, castor hybrid advantage is mainly expressed in plant height, above-ground dry matter accumulation, capsule shell rate, 100 grain weight and seed crude fat content, etc. With the increasing demand for castor oil in many countries, the selection and genetic improvement of castor varieties are receiving more and more attention from breeders.
The crude fat and fatty acid content in the seeds of oilseeds directly affects their germination, growth and storage, and is closely related to plant yield. As one of the top ten oil crops in the world, castor has an oil content of 37%–60% in seeds, which is significantly better than other oil crops, and the study of its crude fat and fatty acid content is crucial. The molecular formula of castor oil is C57H104O9, which is one of the few pure natural glycerides. Its fatty acid ricinoleic acid accounts for more than 70%, and also contains a small amount of linoleic acid, oleic acid, stearic acid, palmitic acid, linolenic acid, etc. The composition of fatty acids is directly related to the oil content of castor seeds.
In order to guide the practice of castor production, and to lay the foundation for cultivating new varieties of castor with high yield, high oil and resistance to adversity. In this experiment, the castor female Lm-type female line aLmAB2, the parent CSR·181 and their hybrid progeny were used as plant material for quality index determination and then proteomics technology was used to study which traits of castor progeny showed hybrid advantage compared with the parents; the genes related to these traits were screened and RT-qPCR assay was performed to use The genes associated with these traits were screened and RT-qPCR was performed to predict the heterozygous advantage of the five castor crosses that performed well in production.
Determination of quality indicators of castor parents and their progeny seeds and prediction of hybrid advantage in five castor hybrids using marker genes are added as supplementary chapter in the supplementary document.
The results of quality indexes of castor parents and their offspring seeds at different development times showed that with the extension of development time, the fresh seeds and fresh seed kernel weight showed a trend of increasing and then decreasing, which was due to the increasing free water content at the early stage of seed development, and the free water content at the later stage of development turned from increasing to decreasing. The fresh seed kernel rate, dried seed kernel rate and 100-kernel weight of dried seed kernels all gradually increased, which was related to the gradual accumulation of various substances during the development of seeds. The crude fat content of seeds increased and then decreased with the development time, which was related to the accumulation of substances and oil synthesis during the development of seeds. The content of ricinoleic acid, oleic acid, stearic acid, arachidonic acid, and arachidonic acid in seeds all increased and then decreased with developmental time, while the content of palmitic acid, palmitoylenic acid, linoleic acid, and linolenic acid all gradually decreased with developmental time, and the content of heptadecanoic acid gradually increased with developmental time, which is because fatty acids transform into each other during seed development, so the development trend of various fatty acid contents with developmental time The trend of fatty acid content with development time was not the same.
The predicted analysis of the hybrid advantage of the five castor hybrids that performed better in production showed that: in terms of seed oil synthesis ability, Tong Castor No.16 and Tong Castor No.13 showed greater hybrid advantage compared with Tong Castor No.18, Tong Castor No.11 and Tong Castor No.17; although the acetyl coenzyme A carboxylase content in the seeds of Tong Castor No.16 and Tong Castor No.13 was higher than that of the progeny of group A, the oil body-related protein 2C Although the content of acetyl coenzyme A carboxylase in the seeds of Tong Castor No.16 and Tong Castor No.13 was higher than that of the progeny of group A, the content of oil body-related protein 2C was lower than that of the progeny of group A. Therefore, whether Tong Castor No.16 and Tong Castor No.13 were better than the progeny of group A in oil synthesis needs further study. In terms of reducing pollen abortion, Tong Castor No.18 and Tong Castor No.11 showed more obvious hybrid advantage compared with Tong Castor No.16, Tong Castor No.13 and Tong Castor No.17; moreover, the contents of 26S protease regulatory subunit 6a and cysteine protease in the seeds of Tong Castor No.18 and Tong Castor No.11 were lower than those of the F1 generation of group A. Therefore, Tong Castor No.18 and Tong Castor No.11 were superior to the F1 generation of group A in the trait of reducing pollen abortion. Group A F1 generation. In terms of yield and stress resistance, there were no significant differences among Tong Castor No.18, Tong Castor No.16, Tong Castor No.13, Tong Castor No.11 and Tong Castor No.17 are superior to the progeny of group A in terms of yield and stress resistance.
Through comparative proteomic analysis of seeds at different developmental stages between castor parents and their offspring, 122 DEP spots were detected by MS, though they only contained 93 DEPs. This is because a number of proteins had significant differential expression at all developmental stages of seeds, such as starch synthase, D-7 LEA protein, SOD, and APs.
Most of the DEPs were associated with stress responses (61.29%), and they were predominantly upregulated in the seeds of offspring. SOD is an antioxidant enzyme in the reactive oxygen species scavenging system that effectively prevents peroxides (e.g., H2O2) from damaging the structure and function of plant cells. This enzyme participates in the rcu04146 pathway (peroxisome). The peroxisome, an essential organelle of plants, plays a key role in redox signal transduction and lipid homeostasis [12–16]. APs are mainly involved in such processes as metabolism, precursor protein processing, protein degradation, and programmed cell death, as well as pollen germination and pollen tube growth in plants [17–19]. D-7 LEA, as such, is named late embryonic abundant, a stress-related-responding protein that massively accumulates during the seed maturation drying stage, is also generated during plant growth and development under diverse adverse conditions. It regulates the metabolic balance of plants through mechanisms such as stabilizing the cell membrane and binding metal ions [20–22]. Glutathione peroxidase contains sulfhydryl groups and can effectively block the free radicals of reactive oxygen species from further damaging the body by scavenging H2O2 and lipid peroxides [23,24]. ADH1, a medium-chain zinc-containing enzyme widely found in various tissues in plants, binds zinc ions. Zinc plays an essential role in the synthesis of indole-3-acetic acid (IAA), an auxin in plants. Zinc deficiency can lead to decreased tryptophan and IAA content, hindering the growth of plant roots. Therefore, ADHs are closely related to plant growth and development [25–27]. In addition to those five proteins, several DEPs between offspring and parents were associated with stress responses, including 14-3-3-like protein, BADH1 (chloroplast), and l-ascorbate peroxidase.
Among the DEPs between castor offspring and parents, 17.20% were involved in seed nutrient storage, most of which were upregulated in the offspring seeds. Starch, the main storage carbohydrate of plant seeds, may account for 50% of the dry matter in storage organs (tubers or seeds), and it is an important energy source for plant growth and development, so its content directly determines the yield of crops [28–30]. Starch synthase is a key enzyme involved in starch synthesis and metabolism. 2S albumin precursor, a storage protein, can gradually be degraded to produce various amino acids and thereby provide essential nitrogen, carbon, and sulfur sources for plant growth during the seed germination and early growth stages, which is of great significance for plant growth and development [31]. Vicilin GC72-A belongs to the family of antimicrobial peptides, a class of small-molecule polypeptides synthesized by plants to defend against pathogen invasion; these antimicrobial peptides can inhibit or kill a variety of microorganisms and fungi [32,33]. Other DEPs associated with seed nutrient storage of plants included adenosyl homocysteinase, NOP56, and salicylic acid-binding protein 2.
Of the DEPs between castor offspring and parents, 12.90% were associated with the synthesis and metabolism of plant fatty acids, most of which were upregulated in the offspring seeds. Stearoyl-ACP desaturase is a key enzyme in the conversion between stearic and unsaturated fatty acids, so its upregulation may increase the content of unsaturated fatty acids in plants, thus improving their resistance to low temperatures [34]. OBAP2C plays a crucial role in the seed development of plants. Oil bodies are subcellular organelle particles in plant seeds that store lipids, provide nutrients to the plant, and are essential for plant growth and development [35–37]. Acetyl-CoA carboxylase is a biotinase that catalyzes the carboxylation of acetyl-CoA in the initial step of fatty acid synthesis to form malonyl-CoA. fatty acids are membrane lipid components of all cells and are important energy reserves for multicellular organisms. Thus, acetyl-CoA carboxylase is a catalyst for the first step of fatty acid biosynthesis and plays a key role in plant growth and development [38–40]. Other DEPs related to fatty acid synthesis and metabolism include EAR precursors, plastidic 3-ketoacyl-ACP synthase I and hydroxy-ACP dehydratase.
6.45% of the differential proteins between offspring and parents were related to plant amino acid metabolism, and most of them were up-regulated in the offspring seeds. Both transaldolase and transketolase (chloroplast) are enzymes involved in the non-oxidative phase of the pentose phosphate pathway (PPP). Transaldolase catalyzes the transfer of the dihydroxyacetone fraction of siderophore heptulose-7-phosphate to glyceraldehyde-3-phosphate, which regulates the balance between the oxidative and non-oxidative phases of PPP by regulating the production of ribose 5-phosphate and NADPH by the PPP and linking the PPP and glycolytic pathways. Trans-ketohydrolase is a “non-regulatory” enzyme that catalyzes the reversible transfer of the glycoalkalyl group from the activated ketose (donor) to the activated aldose (acceptor), and studies have shown that increased expression of trans-ketohydrolase can promote carbon fixation and growth and development in plants [41–45]. SAMS is the only enzyme synthesizing SAM in organisms. SAM, a metabolite from the interaction of methionine and adenosine triphosphate, has been widely found in animals, plants, and microorganisms. It is involved in up to 40 types of biochemical reactions and plays a significant role in improving plant drought tolerance [46–49]. All three enzymes mentioned above were involved in rcu01230 (biosynthesis of amino acids). In addition, NADP-dependent isocitrate dehydrogenase, l,l-diaminopimelate aminotransferase (chloroplast), and 5-methyltetrahydroproylglutamate-homocysteine methyltransferase in the amino acid biosynthesis pathway were also differentially expressed in seeds between castor offspring and parents.
The high expression of cysteine protease and 26S protease regulatory subunit 6a can lead to abnormal degradation of some regulatory proteins in anther tissue and may cause pollen abortion. In this study, these two proteins were downregulated in F1 compared with their male parent [50]. Importantly, we found that cysteine protease inhibitor was upregulated in the offspring seeds compared with their parents. This enzyme can bind to cysteine protease to inhibit its activity and reduce the risk of pollen abortion, and it is involved in the stress responses of plants under adverse conditions and participates in physiological processes such as senescence of plants [51].
The quality indexes of castor parents and their hybrid progeny seeds at different development times were determined, which showed that: in terms of seed 100 grain weight, seed kernel rate, crude fat content, and various fatty acid contents, the castor hybrid progeny seeds showed a hybrid advantage over the parents, and the F1 generation was better than the F0 generation.
To conduct a proteomic analysis to compare the differences in castor seeds at different developmental stages between parents and their offspring, we first optimized our two-dimensional electrophoresis system. The protein atlas obtained from SDS-PAGE following IEF was the best when using samples after impurity removal, 24-cm dry gel strips (pH 3–10 NL), and a protein loading amount of 800 μg.
By 2D-DIGE analysis under these optimized two-dimensional electrophoresis, 93 DEPs were identified in the seeds at different developmental stages between castor parents and offspring. They belonged to five categories: proteins related to stress response (61.29%), seed nutrient storage (17.20%), fatty acid synthesis and metabolism (12.90%), amino acid metabolism (6.45%), and others (2.15%).
RT-qPCR was performed on 12 DEPs, including (1) stearoyl-ACP desaturase, OBAP2C, and acetyl-CoA carboxylase, which were upregulated in the offspring seeds compared with their parents and are involved in oil synthesis and fatty acid synthesis and metabolism; (2) 26S protease regulatory subunit 6a and cysteine protease, which was downregulated in the offspring seeds compared with their parents and is involved in pollen abortion; and (3) SOD, starch synthase, D-7 LEA protein, glutathione peroxidase, ADH1, 2S albumin precursor, and APs, which were upregulated in the offspring seeds compared with their parents and are involved in plant resistance to adverse conditions. The RT-qPCR results showed that the expression levels of the genes encoding these 12 DEPs were consistent with the expression of the DEPs.
The predicted analysis of the hybrid advantage of five castor hybrids using marker genes showed that Tong Castor No.16 and Tong Castor No.13 showed greater hybrid advantage in oil synthesis; Tong Castor No.18 and Tong Castor No.11 showed greater hybrid advantage in reducing pollen defeat; there was no significant difference in yield and resistance between Tong Castor No.18, Tong Castor No.16, Tong Castor No.13, Tong Castor No.11 and Tong Castor No.17.
In summary, this study can help guide the production of castor beans and lays the foundation for cultivating new castor varieties with high yield, high oil content, and strong stress resistance.
Funding Statement: 1. National Natural Science Foundation of China (31860071); 2. Research and Reform Practice Project in New Agricultural Sciences of the Ministry of Education in 2020 (2020114); 3. Natural Science Foundation of Inner Mongolia Autonomous Region (2021MS03008); 4. Inner Mongolia Autonomous Region Grassland Talents Innovation Team-Castor Molecular Breeding Research Innovative Talent Team Rolling Support Project (2022); 5. Higher Education Teaching Reform Research Project of National Ethnic Affairs Commission in 2021 (21082); 6. Fundamental Research Funds in Higher Education Institutions of Inner Mongolia in 2022 (237); 7. Autonomous Region Basic Scientific Research Business Fee Project of Inner Mongolia Minzu University in 2023 (225,227,244); 8. Inner Mongolia Autonomous Region Castor Industry Collaborative Innovation Center Construction Project (MDK2021011, MDK2022014).
Author Contributions: All listed authors should have substantially contributed to the manuscript and have approved the final submitted version, which should include a description of each author’s specific work and contributorship. The authors confirm contribution to the paper as follows: study conception and design: Fenglan Huang; data collection: Rui Luo, Mingda Yin, Zhiyan Wang, and Yumiao Huo; analysis and interpretation of results: Yanxin Zhang, Yanpeng Wen, and Xuemei Hu; draft manuscript preparation: Xiaotian Liang, and Qi Wen. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Yu, A., Li, F., Liu, A. (2020). Comparative proteomic and transcriptomic analyses provide new insight into the formation of seed size in castor bean. BMC Plant Biology, 20, 48. [Google Scholar] [PubMed]
2. Olivares, A. R., Carrillo-González, R., González-Chávez, M. C. A., Soto Hernández, R. M. (2013). Potential of castor bean (Ricinus communis L.) for phytoremediation of mine tailings and oil production. Journal of Environmental Management, 114, 316–323. [Google Scholar]
3. Yasur, J., Rani, P. U. (2013). Environmental effects of nanosilver: Impact on castor seed germination, seedling growth, and plant physiology. Environmental Science and Pollution Research, 20(12), 8636–8648. [Google Scholar] [PubMed]
4. Zhu, M. M., Liu, E. Q., Bao, Y., Duan, S. L., She, J. et al. (2019). Low concentration of corn steep liquor promotes seed germination, plant growth, biomass production and flowering in soybean. Plant Growth Regulation, 87, 29–37. [Google Scholar]
5. Iimure, T., Sato, K. (2013). Beer proteomics analysis for beer quality control and malting barley breeding. Food Research International, 54(1), 1013–1020. [Google Scholar]
6. Kim, S. T., Kim, S. G., Agrawal, G. K., Kikuchi, S., Rakwal, R. (2014). Rice proteomics: A model system for crop improvement and food security. Proteomics, 14(4–5), 593–610. [Google Scholar] [PubMed]
7. Sghaier-Hammami, B., Castillejo, M. Á., Baazaoui, N., Jorrín-Novo, J. V., Escandón, M. (2021). GeLC-Orbitrap/MS and 2-DE-MALDI-TOF/TOF comparative proteomics analysis of seed cotyledons from the non-orthodox Quercus ilex tree species. Journal of Proteomics, 233, 104087. [Google Scholar] [PubMed]
8. Jian, M., Zhang, D., Wang, X., Wei, S., Zhao, Y. et al. (2020). Differential expression pattern of the proteome in response to cadmium stress based on proteomics analysis of wheat roots. BMC Genomics, 21(1), 1–13. [Google Scholar]
9. Xia, Q., Ponnaiah, M., Cueff, G., Rajjou, L., Prodhomme, D. et al. (2018). Integrating proteomics and enzymatic profiling to decipher seed metabolism affected by temperature in seed dormancy and germination. Plant Science., 269, 118–125. [Google Scholar] [PubMed]
10. Gu, J., Hou, D., Li, Y., Chao, H., Zhang, K. et al. (2019). Integration of proteomic and genomic approaches to dissect seed germination vigor in Brassica napus seeds differing in oil content. BMC Plant Biology, 19(1), 1–20. [Google Scholar]
11. Katayama, H., Nagasu, T., Oda, Y. (2001). Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry, 15(16), 1416–1421. [Google Scholar] [PubMed]
12. Azarabadi, S., Abdollahi, H., Torabi, M., Salehi, Z., Nasiri, J. (2017). ROS generation, oxidative burst and dynamic expression profiles of ROS-scavenging enzymes of superoxide dismutase (SODcatalase (CAT) and ascorbate peroxidase (APX) in response to Erwinia amylovora in pear (Pyrus communis L). European Journal of Plant Pathology, 147(2), 279–294. [Google Scholar]
13. Lu, B. W., An, F. X., Cao, L. J., Yang, Y. J., Liu, P. M. et al. (2020). Proteomic profiling uncovered the cytosolic superoxide dismutase BsSOD1 associated with plant defence in the herbal orchid Bletilla striata. Functional Plant Biology, 47(10), 937–944. [Google Scholar] [PubMed]
14. Jafari, S., Garmdareh, S. E. H., Azadegan, B. (2019). Effects of drought stress on morphological, physiological, and biochemical characteristics of stock plant (Matthiola incana L.). Scientia Horticulturae, 253, 128–133. [Google Scholar]
15. Pour-Aboughadareh, A., Etminan, A., Abdelrahman, M., Siddique, K. H. M., Tran, L. S. P. (2020). Assessment of biochemical and physiological parameters of durum wheat genotypes at the seedling stage during polyethylene glycol-induced water stress. Plant Growth Regulation, 92, 81–93. [Google Scholar]
16. Che, Y., Zhang, N., Zhu, X., Li, S., Wang, S. et al. (2020). Enhanced tolerance of the transgenic potato plants overexpressing Cu/Zn superoxide dismutase to low temperature. Scientia Horticulturae, 261, 108949. [Google Scholar]
17. Soares, A., Carlton, S. M. R., Simões, I. (2019). A typical and nucellin-like aspartic proteases: Emerging players in plant developmental processes and stress responses. Journal of Experimental Botany, 70, 2059–2076. [Google Scholar] [PubMed]
18. Yang, Y., Feng, D. (2020). Genome-wide identification of the aspartic protease gene family and their response under powdery mildew stress in wheat. Molecular Biology Reports, 47, 8949–8961. [Google Scholar] [PubMed]
19. Sebastián, D. I., Fernando, F. D., Raúl, D. G., Gabriela, G. M. (2020). Overexpression of Arabidopsis aspartic protease APA1 gene confers drought tolerance. Plant Science, 292, 110406. [Google Scholar] [PubMed]
20. Park, S. C., Kim, Y. H., Jeong, J. C., Kim, C. Y., Lee, H. S. et al. (2011). Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta, 233, 621–634. [Google Scholar] [PubMed]
21. Li, N., Zhang, S., Liang, Y., Qi, Y., Chen, J. et al. (2018). Label-free quantitative proteomic analysis of drought stress-responsive late embryogenesis abundant proteins in the seedling leaves of two wheat (Triticum aestivum L.) genotypes. Journal of Proteomics, 172, 122–142. [Google Scholar] [PubMed]
22. Zhao, W., Yao, F., Zhang, M., Jing, T., Zhang, S. et al. (2016). The potential roles of the G1LEA and G3LEA proteins in early embryo development and in response to low temperature and high salinity in Artemia sinica. PLoS One, 11, e0162272. [Google Scholar] [PubMed]
23. Eshdat, Y., Holland, D., Faltin, Z., Ben-Hayyim, G. (1997). Plant glutathione peroxidases. Physiologia Plantarum, 100, 234–240. [Google Scholar]
24. Kammoun, M., Essid, M. F., Ksouri, F., Rokka, V. M., Charfeddine, M. et al. (2020). Assessment of physiological age and antioxidant status of new somatic hybrid potato seeds during extended cold storage. Journal of Plant Physiology, 254, 153279. [Google Scholar] [PubMed]
25. Kim, Y. H., Huh, G. H. (2019). Overexpression of cinnamyl alcohol dehydrogenase gene from sweetpotato enhances oxidative stress tolerance in transgenic Arabidopsis. In Vitro Cellular & Developmental Biology-Plant, 55, 172–179. [Google Scholar]
26. Zhuang, M., Lam, S. K., Zhang, J., Li, H., Shan, N. et al. (2019). Effect of full substituting compound fertilizer with different organic manure on reactive nitrogen losses and crop productivity in intensive vegetable production system of China. Journal of Environmental Management, 243, 381–384. [Google Scholar] [PubMed]
27. Rattanapolsan, L., Nakbanpote, W., Sangdee, A. (2020). Zinc- and cadmium-tolerant endophytic bacteria from Murdannia spectabilis (Kurz) Faden. studied for plant growth-promoting properties, in vitro inoculation, and antagonism. Archives of Microbiology, 203, 1131–1148. [Google Scholar] [PubMed]
28. Wang, C. L., Zhang, Z. P., Miao, M. M. (2016). SNF1-related protein kinase (SnRK) 1 involved in the regulation of raffinose family oligosaccharide metabolism in cucumber (Cucumis sativus L.) Calli. Journal of Plant Growth Regulation., 35, 851–864. [Google Scholar]
29. Mehrpouyan, S., Menon, U., Tetlow, I. J., Emes, M. J. (2021). Protein phosphorylation regulates maize endosperm starch synthase IIa activity and protein−protein interactions. Plant Journal, 105, 1098–1112. [Google Scholar]
30. do Carmo, C. D., Sousa, M. B., Brito, A. C., de Oliveira, E. J. (2020). Genome-wide association studies for waxy starch in cassava. Euphytica, 216, 82. [Google Scholar]
31. Nguyen, T. P., Cueff, G., Hegedus, D. D., Rajjou, L., Bentsink, L. (2015). A role for seed storage proteins in Arabidopsis seed longevity. Journal of Experimental Botony, 66, 6399–6413. [Google Scholar] [PubMed]
32. Lopes, F. E. S., da Costa, H. P. S., Souza, P. F. N., Oliveira, J. P. B., Ramos, M. V. et al. (2019). Peptide from thaumatin plant protein exhibits selective anticandidal activity by inducing apoptosis via membrane receptor. Phytochemistry, 159, 46–55. [Google Scholar] [PubMed]
33. Wang, C., Yang, C., Gao, C., Wang, Y. (2009). Cloning and expression analysis of 14 lipid transfer protein genes from Tamarix hispida responding to different abiotic stresses. Tree Physiology, 29, 1607–1619. [Google Scholar] [PubMed]
34. Whittle, E. J., Cai, Y., Keereetaweep, J., Chai, J., Buist, P. H. et al. (2020). Castor staroyl-ACP desaturase can synthesize a vicinal diol by dioxygenase chemistry. Plant Physiology, 182, 730–738. [Google Scholar] [PubMed]
35. de Oliveira, T. S., Freitas-Silva, O., Kluczkovski, A. M., Campelo, P. H. (2020). Potential use of vegetable proteins to reduce Brazil nut oil oxidation in microparticle systems. Food Research International, 137, 109526. [Google Scholar]
36. Zhou, L. Z., Chen, F. S., Hao, L. H., Du, Y., Liu, C. (2019). Peanut oil body composition and stability. Journal of Food Science, 84, 2812–2819. [Google Scholar] [PubMed]
37. Jolivet, P., Acevedo, F., Boulard, C., d’Andréa, S., Faure, J. D. et al. (2013). Crop seed oil bodies: From challenges in protein identification to an emerging picture of the oil body proteome. Proteomics, 13, 1836–1849. [Google Scholar] [PubMed]
38. Parvy, J. P., Napal, L., Rubin, T., Poidevin, M., Perrin, L. et al. (2012). Drosophila melanogaster Acetyl-CoA-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genetics, 8, e1002925. [Google Scholar] [PubMed]
39. Vijayakumar, A., Olson, I., Wang, T., Hollenback, D., Toteva, M. et al. (2020). Efficacy and safety of an acetyl CoA carboxylase inhibitor are improved in combination with PPAR agonists in a dyslipidemic rat model. Journal of Hepatology, 73, S656. [Google Scholar]
40. Thelen, J. J., Ohlrogge, J. B. (2002). Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme A carboxylase in Arabidopsis thaliana. Plant Journal, 32, 419–431. [Google Scholar]
41. Oaks, Z., Jimah, J., Grossman, C. C., Beckford, M., Kelly, R. et al. (2020). Transaldolase haploinsufficiency in subjects with acetaminophen-induced liver failure. Journal of Inherited Metabolic Disease, 43, 496–506. [Google Scholar] [PubMed]
42. Zheng, M., Zhu, C., Yang, T., Qian, J., Hsu, Y. F. (2020). GSM2, a transaldolase, contributes to reactive oxygen species homeostasis in Arabidopsis. Plant Molecular Biology, 104, 39–53. [Google Scholar] [PubMed]
43. Tylki-Szymanska, A., Wamelink, M. M. C., Stradomska, T. J., Salomons, G. S., Taybert, J. et al. (2014). Clinical and molecular characteristics of two transaldolase-deficient patients. European Journal of Pediatrics, 173, 1679–1682. [Google Scholar] [PubMed]
44. Li, M., Zhang, X., Lu, Y., Meng, S., Quan, H. et al. (2020). The nuclear translocation of transketolase inhibits the farnesoid receptor expression by promoting the binding of HDAC3 to FXR promoter in hepatocellular carcinoma cell lines. Cell Death Discover, 11, 31. [Google Scholar]
45. Bi, H., Li, F., Wang, H., Ai, X. (2019). Overexpression of transketolase gene promotes chilling tolerance by increasing the activities of photosynthetic enzymes, alleviating oxidative damage and stabilizing cell structure in Cucumis sativus L. Physiologia Plantarum, 167, 502–515. [Google Scholar] [PubMed]
46. Seong, E. S., Jeon, M. R., Choi, J. H., Yoo, J. H., Lee, J. G. et al. (2020). Overexpression of S-Adenosylmethionine synthetase enhances tolerance to cold stress in Tobacco. Russian Journal of Plant Physiology, 67, 242–249. [Google Scholar]
47. Mo, H. J., Sun, Y. X., Zhu, X. L., Wang, X. F., Zhang, Y. et al. (2016). Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta, 243, 1023–1039. [Google Scholar] [PubMed]
48. Elango, R. (2020). Methionine nutrition and metabolism: Insights from animal studies to inform human nutrition. Journal of Nutrition, 150, 2518S–2523S. [Google Scholar] [PubMed]
49. Kovács, L., Mendel, Á., Szentgyörgyi, A., Fekete, S., Söre, F. et al. (2020). Comparative analysis of overexpressed Fragaria vesca S-adenosyl-l-methionine synthase (FvSAMS) and decarboxylase (FvSAMDC) during salt stress in transgenic Nicotiana benthamiana. Plant Growth Regulation, 91, 53–73. [Google Scholar]
50. Passarge, A., Demir, F., Green, K., Depotter, J. R. L., Scott, B. et al. (2021). Host apoplastic cysteine protease activity is suppressed during the mutualistic association of Lolium perenne and Epichloë festucae. Journal of Experimental Botany, 72(9), 3410–3426. [Google Scholar] [PubMed]
51. Kaur, H., Gill, R. S., Kaur, S. (2020). Rice bean (Vigna umbellata Thunb. Ohwi and Ohashi) protection against Callosobruchus maculatus F.” by presence of protein profile. Journal of Stored Products Research, 86, 101574. [Google Scholar]
Supplementary Materials
Table S1:Reagent formula
Table S2:IEF program
Table S3:RT-qPCR primer sequence
Table S4:Differential protein mass spectrometry identification result
Table S5:Different protein sources and classification results
Figure S1:The 2-DE results comparison of removing protein sample impurities or not
Figure S2:The 2-DE results comparison of using different pH dry strips
Figure S3:The 2-DE results comparison of different sample loadings
Figure S4:2-DE gel protein punctuation
Figure S5:The relative expression of differentially expressed protein in 20 d castor parents and their offspring
Figure S6:The KEGG pathway of differentially expressed protein in 20 d castor parents and their offspring
Figure S7:The relative expression of differentially expressed protein in 40 d castor parents and their offspring
Figure S8:The KEGG pathway of differentially expressed protein in 40 d castor parents and their offspring
Figure S9:The relative expression of differentially expressed protein in 60 d castor parents and their offspring
Figure S10:The KEGG pathway of differentially expressed protein in 60 d castor parents and their offspring
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools