 Open Access
Open Access
ARTICLE
Genome-Wide Identification and Expression Analysis of the Phytocyanin Gene Family in Nicotiana tabacum
1 School of Agronomy and Biotechnology, Engineering Research Center of South Upland Agriculture, Ministry of Education, Southwest University, Chongqing, 400715, China
2 Tobacco Science Research Institute of Chongqing Tobacco Company, Chongqing, 400715, China
3 Chongqing Qianjiang Tobacco Branch Company Chongqing Institute of Tobacco Science, Chongqing, 400715, China
* Corresponding Author: Liquan Zhu. Email:
# Co-first authors
(This article belongs to the Special Issue: Integrating Agronomy and Plant Physiology for Improving Crop Production)
Phyton-International Journal of Experimental Botany 2023, 92(5), 1469-1492. https://doi.org/10.32604/phyton.2023.027133
Received 19 October 2022; Accepted 26 December 2022; Issue published 09 March 2023
Abstract
Phytocyanin (PC) is a class of plant-specific blue copper proteins involved in electron transport, plant growth, development, and stress resistance. However, PC proteins have not been systematically evaluated in tobacco plants. We determined the whole-genome sequences of the PC family in the tobacco cultivar ‘K326.’ The transcriptome data were used to analyze the expression of the NtPC family at different development stages and tissue-specific genes. Real-time fluorescence quantitative analysis was used to analyze the expression of the NtPC gene family under low temperature and methyl jasmonate stress. The tobacco NtPC family contained 110 members and was divided into four subfamilies: early nodulin-like protein (NtENODL), uclacyanin-like protein, stellacyanin1-like protein, and plantacyanin-like protein. According to phylogenetic and structural analyses, the NtPC family could be divided into eight structural types. Fifty-three NtPCs were randomly distributed on 22 of 24 tobacco chromosomes. Collinearity analysis revealed 33 pairs of genes belonging to the NtPC family. Gene ontology analysis showed that the PC genes are components of the plasma membrane and may participate in plasma membrane-related functions. The NtPC family contained numerous elements related to hormonal and abiotic stress responses and was specifically expressed in the tobacco prosperous, maturation, and budding periods. Tissue-specific expression analysis showed that some genes were tissue specific. The expression of NtENODL58 and other genes was significantly induced by low-temperature and methyl jasmonate stress. Thus, the NtPC gene family plays an important role in plant stress response.Keywords
Plastocyanin (PC) is a plant-specific blue copper protein containing a single type-I mononuclear copper site that can bind only to a single copper atom. The PC protein family contains a plastocyanin-like domain (PCLD). Based on their copper-binding sites, ligand compositions, glycosylation states, domain structures, and spectral characteristics, these proteins can be divided into four subfamilies: uclacyanin-like proteins (UCL), stellacyanin-like proteins (SCL), plantacyanin-like proteins (PLCL), and early nodulin-like proteins (ENODL) [1,2]. PC proteins function as electron transporters in various biological systems [3] and participate in the growth, development, and anti-stress responses of plants.
During plant growth and development, the blue protein, a member of the PC family isolated from the lily stigma, can induce chemotaxis in pollen tubes [4]. OsUCL8 is an uclacyanin gene of the PC family highly expressed in the pistils, young panicles, developing seeds, and inflorescence meristems of rice, and the downregulation of OsUCL8 by OsmiR408 regulates grain yield [5]. A recent study showed that phytochrome interacting factor 3/4/5 (PIF3/4/5) and miR408 promote leaf senescence by regulating the plantacyanin-senescence-associated gene 14 (PCY-SAG14) plant cyanoprotein module, demonstrating that intracellular copper homeostasis mediated by the PCY-SAG14 module plays an important role in dark-induced leaf senescence [6]. Members of the ENODL subfamily are expressed in the root nodules of leguminous plants and may be involved in cell differentiation, cell wall reorganization, and intercellular signal transduction during the nodulation of leguminous plants [7]. In addition, members of the ENODL subfamily affect other organs in some plants. In Arabidopsis, AtENODL14 specifically interacts with the extracellular domain of the receptor-like kinase FERONIA and precisely controls pollen tube reception [8]. Overexpression of AtENODL15 interferes with pollen tube guidance and reduces fertility [9].
Abiotic sources of stress, such as exposure to aluminum and oxidation, can induce the expression of the blue copper-binding protein (AtBCB/AtSC3) in Arabidopsis [10]. The AtBCB product can inhibit aluminum absorption and protect the cell wall and membrane from aluminum toxicity [11]. The expression of AtENODL2/18 is induced by osmotic and salt stress [12]. BcBCP1, an early nodulin-like protein gene from Boea crassifolia, can enhance the osmotic tolerance of transgenic tobacco and is highly expressed under drought, salt, and abscisic acid stress [13]. Overexpression of GhENODL6 in cotton was recently shown to significantly enhance the expression of salicylic acid (SA)-related transcription factors and genes related to fighting pathogens, as well as the content of hydrogen peroxide (H2O2) and salicylic acid, which has been shown to improve the resistance of transgenic Arabidopsis to Verticillium wilt in cotton [14].
The PC gene family has been analyzed in Arabidopsis [12], rice [2], Chinese cabbage [15], maize [16], Phalaenopsis [17], Medicago tribulus [18], Populus tomentosa [19], Dendrobium candidum [20], and rape [21]. In the past, most studies focused on the biochemical characteristics of the PC gene family, namely spectroscopy and redox properties [1]. In recent years, some studies also found that the PC gene family plays an important role in stress resistance and plant growth and development [10–14]. Nicotiana tabacum (tobacco) is an important cash crop in China. Biological and abiotic stressors greatly impact the quality and yield of tobacco. However, the PC gene family in tobacco has not been examined. We studied the transcriptomes of tobacco plants at different developmental stages. Based on genome-wide identification, chromosome distribution, and evolutionary analyses of members of the tobacco PC family, we performed Gene Ontology (GO) analysis to clarify the biological functions of PC family members expressed during tobacco development and under stress conditions. This work provides a foundation for further research on PCs.
2.1 Identification and Bioinformatic Analysis of NtPC Family
All protein files for the general tobacco cultivar ‘K326’ were downloaded from the Solanaceae database (https://solgenomics.net/organism/Nicotiana_tabacum/genome). The hidden Markov model (HMM) with PCLD domain (PF02365, e-value ≤ 1.1e-10) was searched in the Pfam database (http://pfam.sanger.uk/) [22]. The candidate sequences were submitted in the Pfam database and NCBI Conserved Domains Database for verification (https://www.ncbi.nlm.nih.gov/cdd). All proteins were examined to determine whether signal peptides (SPs) were present using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP) [23]. We predicted the glycosylphosphatidylinositol anchor signal (GAS) using the big-PI plant predictor [24]. N-glycosylation sites were predicted using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/). The arabinogalactan protein-like domain (ALR) was manually predicted based on previously described criteria [25,26].
2.2 Multiple Sequence Alignment and Phylogenetic Analysis
The amino acid sequences were aligned using MEGA X (http://www.megasoftware.net), and the resulting sequence alignment files were aligned and manually edited using the GENEDOC software. Then, MEGA X was used to create a neighbor-joining phylogenetic tree. The bootstrap value was set at 1,000.
2.3 Chromosomal Localization and Collinearity Analysis
Data for the corresponding chromosomal location of each PC gene in N. tabacum (NtPC) were downloaded from the Solanaceae database. A map of the NtPC gene on the 24 chromosomes of tobacco was drawn using the MG2C website (http://mg2c.iask.in/mg2c_v2.1/). Collinearity relationships between NtPC genes were analyzed using One-Step MCScanX in TBtools software and visualized using the Amazing Super-Circos and Dual System Plotter in TBtools software, a visualization tool for gene structure prediction [27].
2.4 Analysis of Exon/Intron Structures and Conserved Motifs
The genomic and coding sequences of the NtPC genes were downloaded from the Solanaceae database, and the NtPC gene structure was mapped using TBtools [27]. The conserved motifs (motifs) of NtPC proteins were analyzed using the online website MEME Suite5.1.0 (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). The number of motifs was set to 10, and other parameters were set to default values [28].
2.5 Promoter Cis-Element Analysis
A sequence of 2,000 base pairs upstream of the NtPC gene start codon was downloaded from the Solanaceae database. Cis elements in this region were analyzed using the online website PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [29] and visualized using the TBtools software [27]. Cis-acting elements related to abiotic stress and hormones were selected for statistical analysis.
GO annotation of the functions of genes in the PC proteins family was performed using eggNOG-mapper to obtain the GO-annotated ID. The TBtools software was used for GO enrichment analysis of over-representation analysis (ORA) patterns [27].
2.7 Transcriptome Data and Gene Expression Pattern Analysis
Based on field transcriptome data obtained in our laboratory for different developmental stages of tobacco cultivar ‘K326’, the expression of some NtPC genes was analyzed during the prosperous, maturation, and budding periods. We downloaded TobEA data of 19 different tissues of tobacco K326, including the seed, closed flower bud, open flower bud, floral apex, flower, young shoot, vegetative shoot apex, upper stem, lower stem, young leaf, cauline leaf, cotyledon, early senescent leaf, mid-early senescent leaf, mid-late senescent leaf, late senescent leaf, mature leaf, young root, and mature root from the EMBL-EBI website (http://www.ebi.ac.uk/arrayexpress/experiments). For this analysis, we considered the whole developmental process of tobacco, from seed germination to plant senescence. We drew an expression heat map of different genes at each developmental stage using an illustrator in the TBtools software [27].
2.8 Low Temperature and Methyl Jasmonate Stress Treatments
Tobacco cultivar ‘K326’ was seeded in soil and cultured at room temperature (25°C) until the plants reached the 6–7 leaf stage. Then, the plants were divided into two groups. The plants in the first group were placed in a light incubator (GDN-560D-2, Southeast Ningbo, China) for low-temperature treatment (4°C), under light/dark conditions for 16 and 8 h, respectively, with a relative humidity of 70% and illuminance of 16 klx. The plants in the second group were sprayed with methyl jasmonate (MeJA) (diluted at 1:10 in 95% ethanol and further diluted with distilled water containing 0.1% Triton X-100 to a final concentration of 100 µmol L−1) and cultured at room temperature (25°C). At 0, 1, 6, 12, and 24 h, we collected leaf samples from the same parts of the low-temperature- and MeJA-treated plants, including two treatments and three biological replicates per period for 24 samples. Samples were immediately frozen in liquid nitrogen and stored at −80°C until further use.
2.9 Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
The frozen samples were powdered in liquid nitrogen, and total RNA was extracted from each sample using a Spin Column Plant Total RNA Purification Kit (Sangon Biotech, China). The RNA quality was detected using 1% agarose gel electrophoresis. Then, we performed reverse transcription using All-In-One 5X RT Master Mix (abm, China). The resulting cDNA was stored at −20°C until further use.
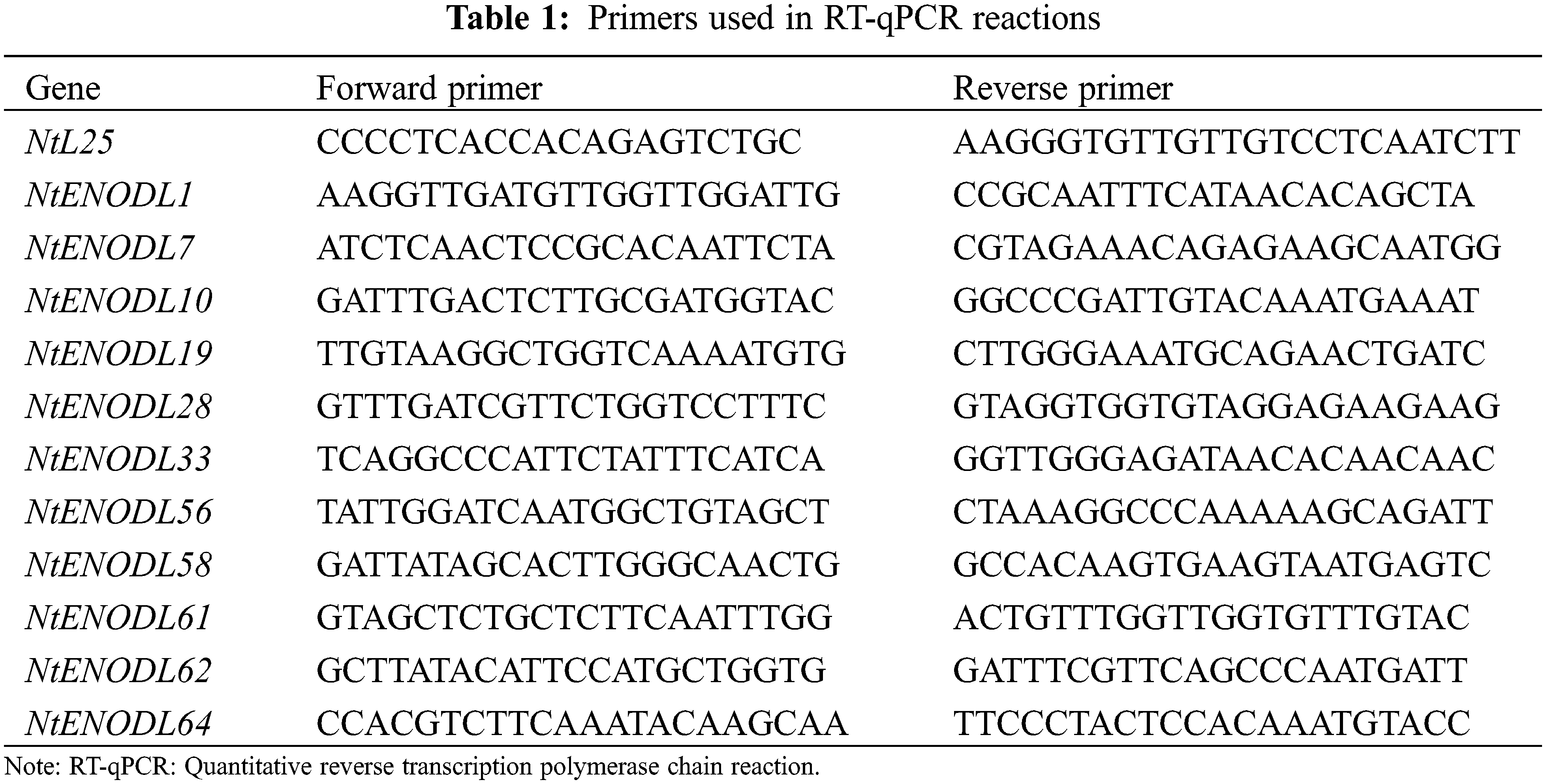
The primers used in the RT-qPCR were designed using the qPrimerDB qPCR Primer Database-online website, followed by primer-specific detection using the Primer-BLAST tool in The National Center for Biotechnology Information (NCBI) (Table 1). The tobacco ribosomal protein gene, L25 (GenBank No. l18908) was used as an internal control. The RT-qPCR with SYBR Premix Ex Taq II (TaKaRa, China) was repeated at least three times on a CFX-1000 Real Time System (BioRad). Briefly, the RT-qPCR was performed as follows: 95°C for 30 s, 95°C for 5 s, 57°C for 30 s, and 40 cycles above, 95°C for 10 s, 65°C for 5 s, 95°C for 5 min, repeat three times. The results were analyzed using the 2−ΔΔCt method [30]. We used the SPSS 25.0 data processing software to conduct variance analysis and difference significance experiments on experimental data and the Prism software to conduct mapping analysis on experimental data.
3.1 Identification and Classification of NtPC Family Members
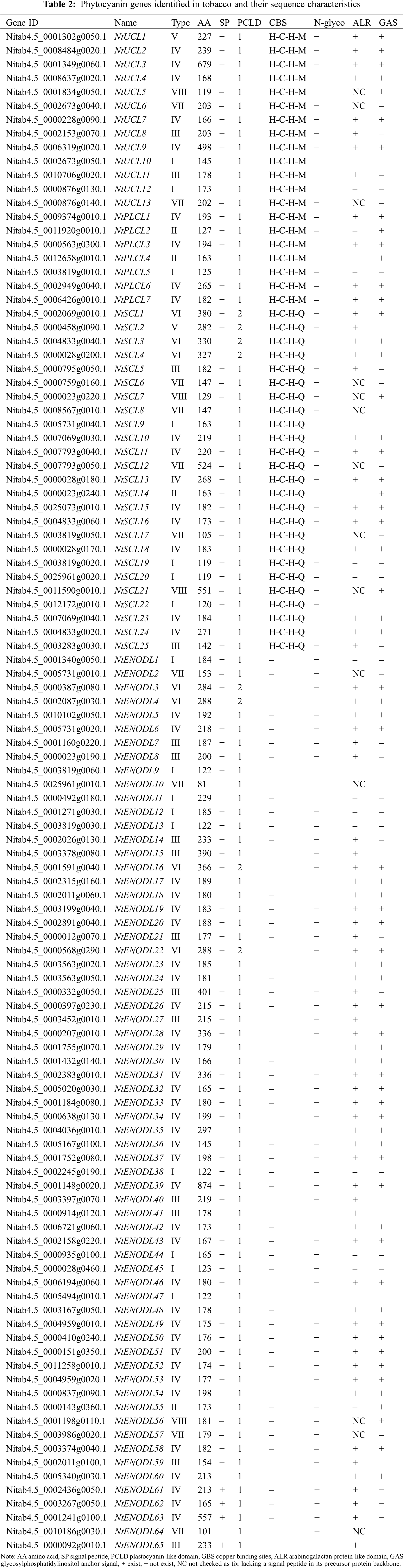
We identified 110 NtPC genes in the tobacco K326 genome (Fig. 1, Table 2). Multiple sequence alignment of the PCLD domains of the NtPC family members revealed that all of them contained both Cys residues highly conserved in their PCLD domain. Four of the 45 NtPC proteins had intact copper-binding ligands (His, Cys, His, and Met/Gln). Twenty NtPC proteins contained the H-C-H-M copper-binding motif, whereas twenty-five contained the H-C-H-Q copper-binding motif. The NtPC proteins were divided into four subfamilies based on whether they contained copper-binding and glycosylation sites. Of these, 13 members belonged to the early nodulin-like protein (NtUCL) class, 7 belonged to the NtPLCL class (although the four conserved residues were identical between NtPLCL and NtUCL, there were no predicted glycosylation sites on the NtPLCL backbone), 25 belonged to the NtSCL class, and the remaining 65 NtPCs had no copper-binding sites and belonged to the NtENODL class. Alignment also revealed several more conserved motifs in the NtENODL subfamily, similar to the copper-binding sites in other subfamilies. However, other amino acid residues were substituted for His, Cys, His, and Met/Gln, which are also presumably involved in copper binding.
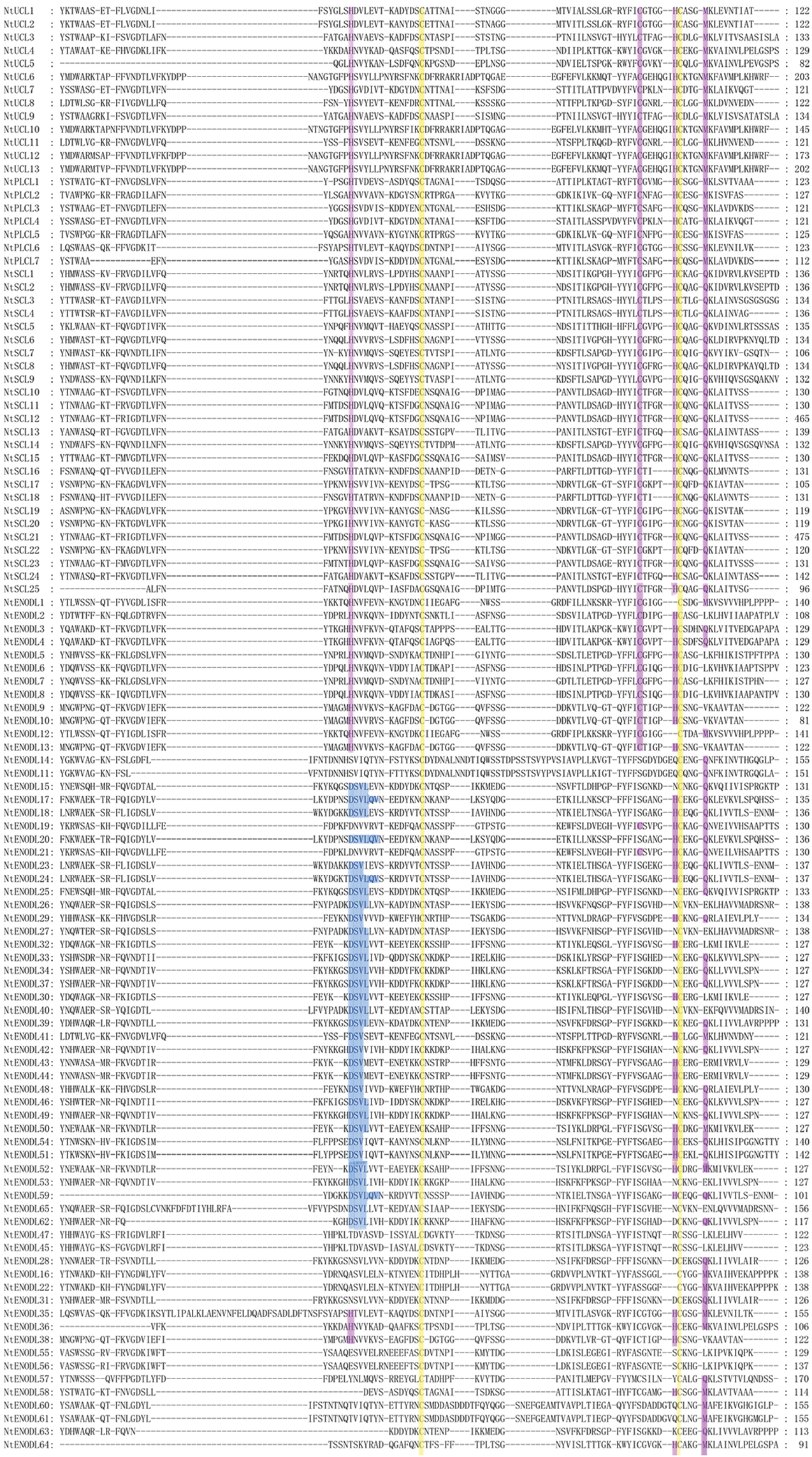
Figure 1: Multiple sequence alignment of plastocyanin-like domains in NtPCs. The purple background represents conserved amino acid residues (His, Cys, His, and Gln/Met) that may be involved in the binding of copper ions. The yellow background represents the cysteine residues involved in disulfide bond formation. The blue background represents conserved amino acid residues in the ENODL subfamily. NtPCs, PC gene in Nicotiana tabacum
3.2 Phylogenetic and Structural Analysis of the NtPC Family
To further understand their structural features, the N-terminal SP, C-terminal GAS, arabinogalactan glycoprotein (AG) glycomodule, and N-glycosylation sites were predicted by using a bioinformatics website (Table 2). SignalP 4.0 predicted that 96 NtPCs had N-terminal SPs. The big-PI Plant Predictor predicted that 68 NtPCs had GAS. In addition, 88 NtPCs were predicted to contain N-glycosylation sites in their PCLD-rich regions.
Based on the previous results, the presence or absence of the [Ala/Ser/Thr/Gly]-Pro-X(0,10)-[Ala/Ser/Thr/Gly]-Pr and [Ala/Ser/Thr/Gly]-Pro3-4 motifs were used as the predictive criteria for determining whether putative AG glycomodules were present [25,26]. Arabinogalactan proteins (AGPs) were classified based on the presence or absence of an N-terminal signal peptide. An analysis of 96 NtPCs containing an N-terminal signal peptide suggesting the presence of at least one AG module revealed that 76 NtPCs contained AG modules. Of these, 8, 4, 14, and 50 had AG modules in NtUCL, NtPLCL, NtSCL, and NtENODL, respectively, indicating that these proteins are members of the AGP superfamily.
The NtPC proteins were classified into eight types (I–VIII) based on whether several components, including an SP, PCLD, ALR, and GAS, were present in their backbones (Fig. 2). Of the 96 NtPC proteins with N-terminal signal peptides, only types VII and VIII did not possess an N-terminal SP. Type I had one fewer GAS domain compared to type II. Finally, types III, IV, V, and VI were classified as AGPs because they contained both SP and ALR domains, and types V and VI were composed of eight NtPC proteins with two PCLD domains.
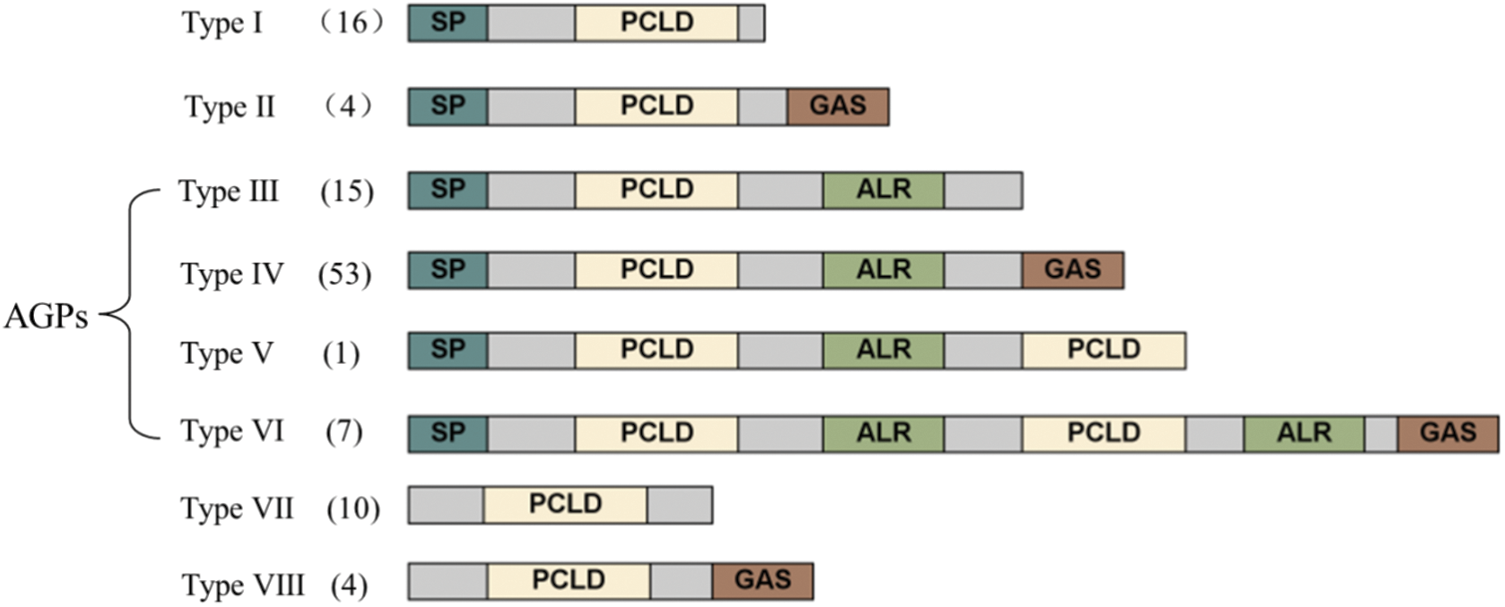
Figure 2: Schematic representations of eight groups of NtPCs. The diagram showing the features of NtPC domains was generated with IBS (http://ibs.biocuckoo.org). AGPs, arabinogalactan proteins; ALR, arabinogalactan protein-like domain; GAS, glycosylphosphatidylinositol anchor signal; PCLD, plastocyanin-like domain; SP, signal peptide
A phylogenetic tree was constructed using the multiple sequence alignment results (Fig. 3), which showed that the 110 NtPCs were divided into seven clades, with 34, 15, 3, 10, 15, 21, and 12 NtPCs in each clade, respectively. All the NtPCs in Clades A and C belonged to the NtENODL subfamily. There were 5, 4, 5, 4, and 8 NtENODLs in the remaining five clades, respectively, forming at least two subfamilies. Except for NtENODL44, the genes in Clade A were AGPs, whereas none of the NtENODLs in Clade C were AGPs. The NtSCL subfamily had 8, 4, and 15 members in Clades B, D, and F, respectively. In clade F, NtSCLs other than NtSCL21 were AGPs. There were 5, 4, and 4 members of the NtUCL subfamily in clades E, F, and G, respectively. All of them belonged to the AGPs except for the NtUCL5 in Clades E and F. Two members of the NtPLCL subfamily were grouped into Clade D. In contrast, five members of this family were grouped into clade E. All of them were AGPs except for NtPLCL4 in Clade E.
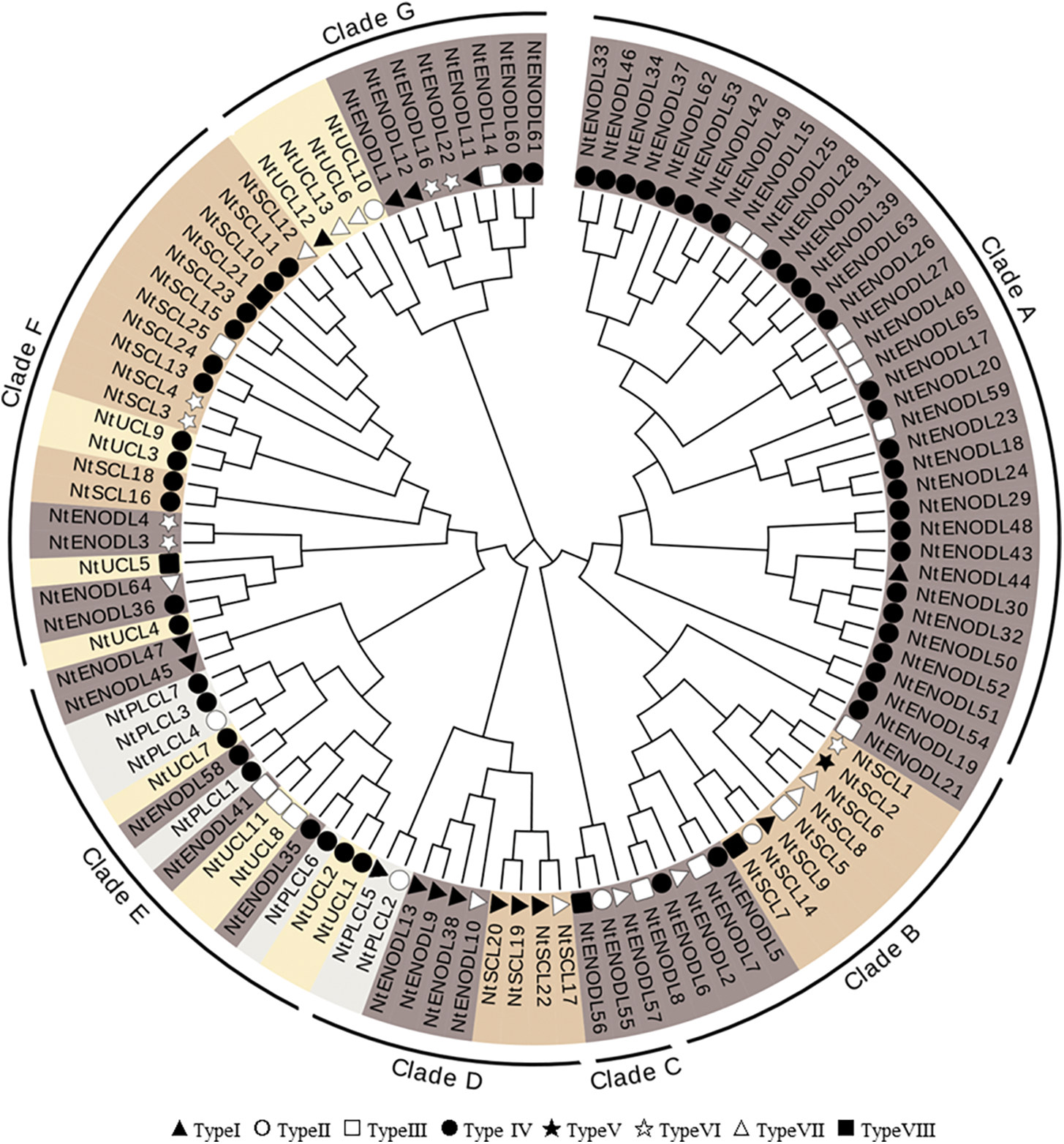
Figure 3: Phylogenetic analysis of PC family in Nicotiana tabacum. The tree amplified 1,000 Bootstrap replicates using the Neighbor-Joining (NJ) method using MEGA X. The tree divided these NtPC proteins into 7 groups, named Clade A to Clade G. Different subfamilies are indicated by different colors. Different symbols represent different types
3.3 Chromosomal Localization and Collinearity Analysis in NtPC Families
Chromosomal localization analysis revealed that 53 NtPCs were randomly distributed on 22 of the 24 tobacco chromosomes. Chromosomes 16 and 18 contained no mapped NtPC genes, whereas chromosome 19 contained the largest number of NtPC genes, with a total of 7. The remaining NtPCs were mapped on chromosomal scaffolds (Fig. 4). Thirty-three pairs of genes were collinear within the tobacco genome (Fig. 5). Further analysis of collinearity between the tobacco and Arabidopsis genome PC gene families showed a total of 18 pairs of genes (Fig. 6); three genes in the tobacco PC gene family showed collinearity and two genes in the Arabidopsis PC gene family presented collinearity between NtENODL16 and NtENODL22, respectively. Furthermore, NtENODL16 and AtENODL17 in Arabidopsis showed collinearity. In Arabidopsis, NtENODL39 exhibited a co-linear relationship with both AtENODL1 and AtENODL2.
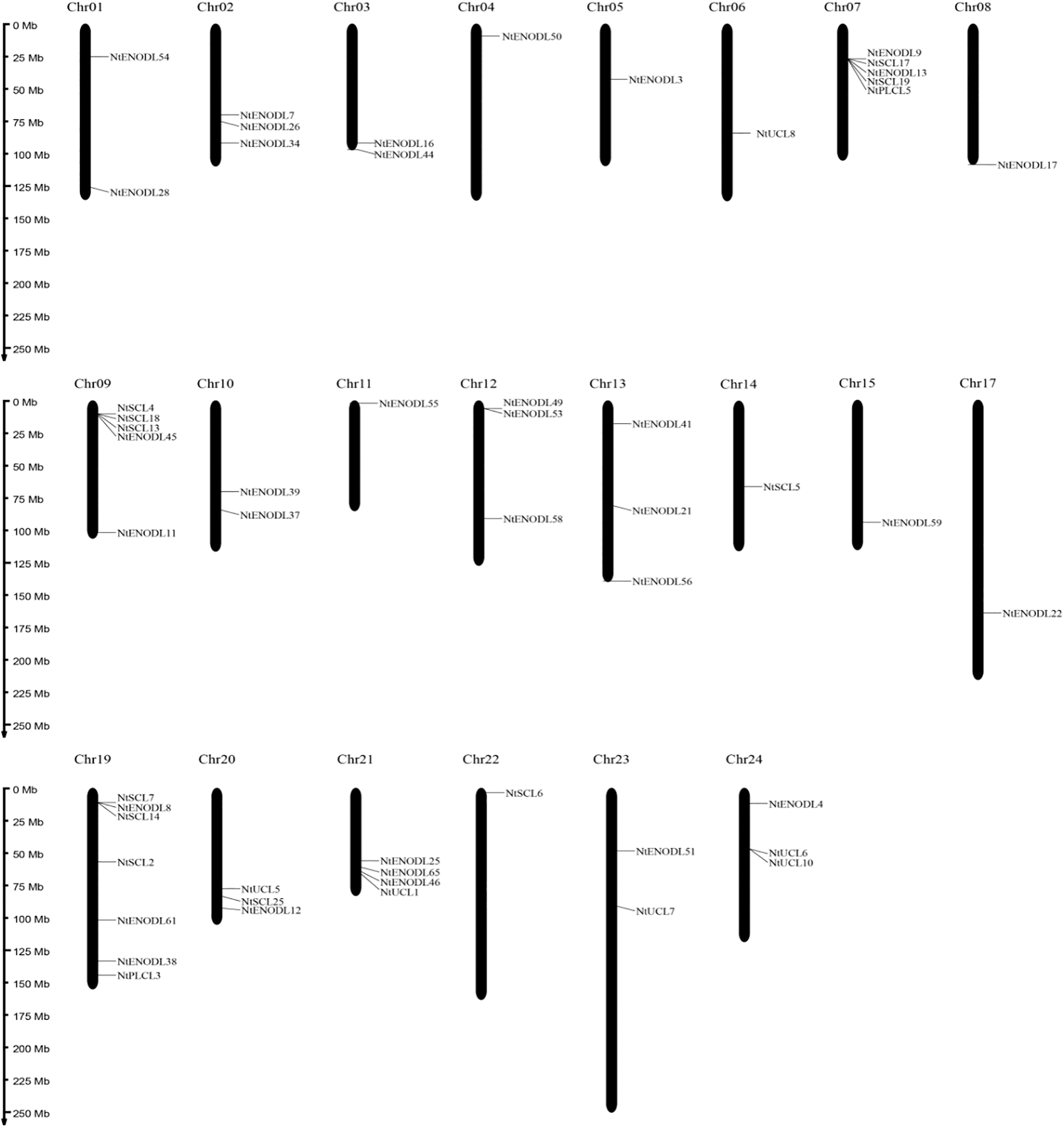
Figure 4: Distribution of PC genes on the chromosomes of Nicotiana tabacum. Vertical bars represent the chromosomes within the tobacco genome. The chromosome number is indicated at the top of each chromosome. The scale on the left is in millions of bases (Mb) and indicates the physical length of each linkage group. The positions of each NtPC gene are represented by black lines. Chr, chromosome
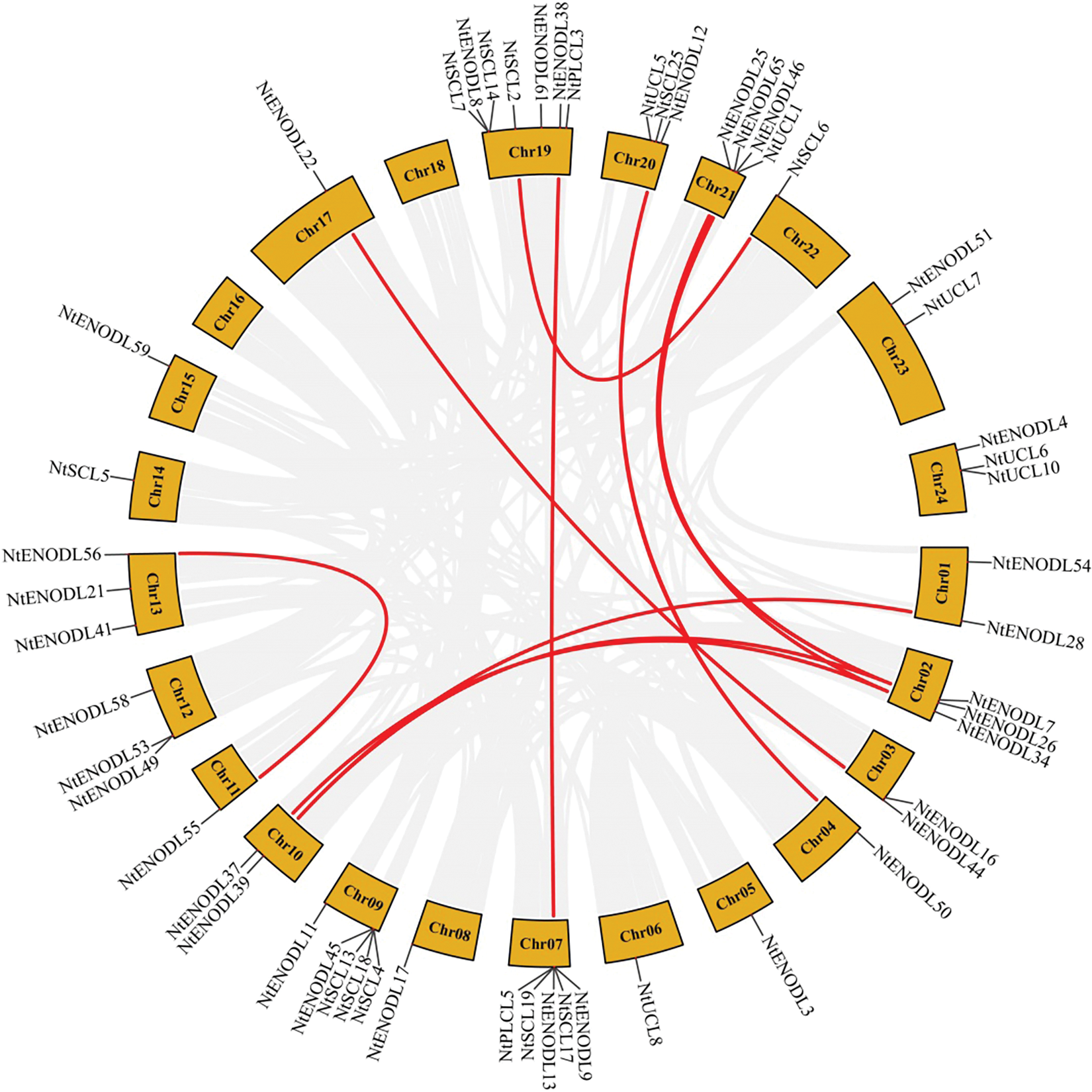
Figure 5: Syntenic relationship of PC genes in Nicotiana tabacum. The red lines indicate segmentally duplicated gene pairs
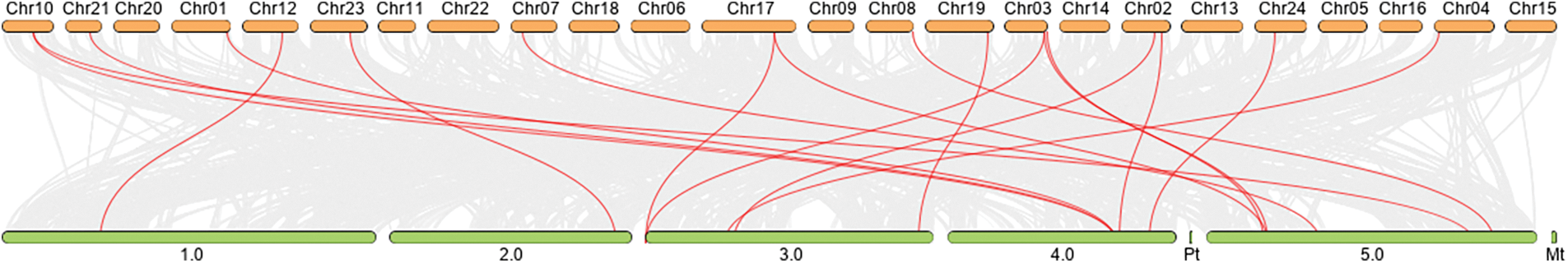
Figure 6: Collinearity analysis of Nicotiana tabacum and Arabidopsis PC gene families. The red lines indicate segmentally duplicated gene pairs
3.4 Analysis of Gene Structure and Conserved Motifs in the NtPC Family
Analysis of the composition of NtPC proteins using MEME identified 10 conserved motifs (Fig. 7) that were 10–50 amino acids in length. The structure of the NtPC members was similar within the same clade and contained the 1/2/3/4/5/7/10 motifs, which are all parts of the PCLD domain present in all NtPC members.

Figure 7: Motif analysis and gene structures of NtPC family members. A: Motif analysis of NtPC family members; B: Gene structures of NtPC family members
Structural analysis of the coding region sequences of the 110 NtPC family gene members (Fig. 7) showed that the number of exons ranged from 1 to 5, with 88 NtPC genes containing 2 exons, 12 NtPC genes containing 3 exons, and 8 NtPC genes containing 4 exons. Only NtENODL16 contained 5 exons, and NtUCL5 contained 1 exon and no introns.
3.5 Analysis of Cis-Acting Elements of NtPC Family Genes
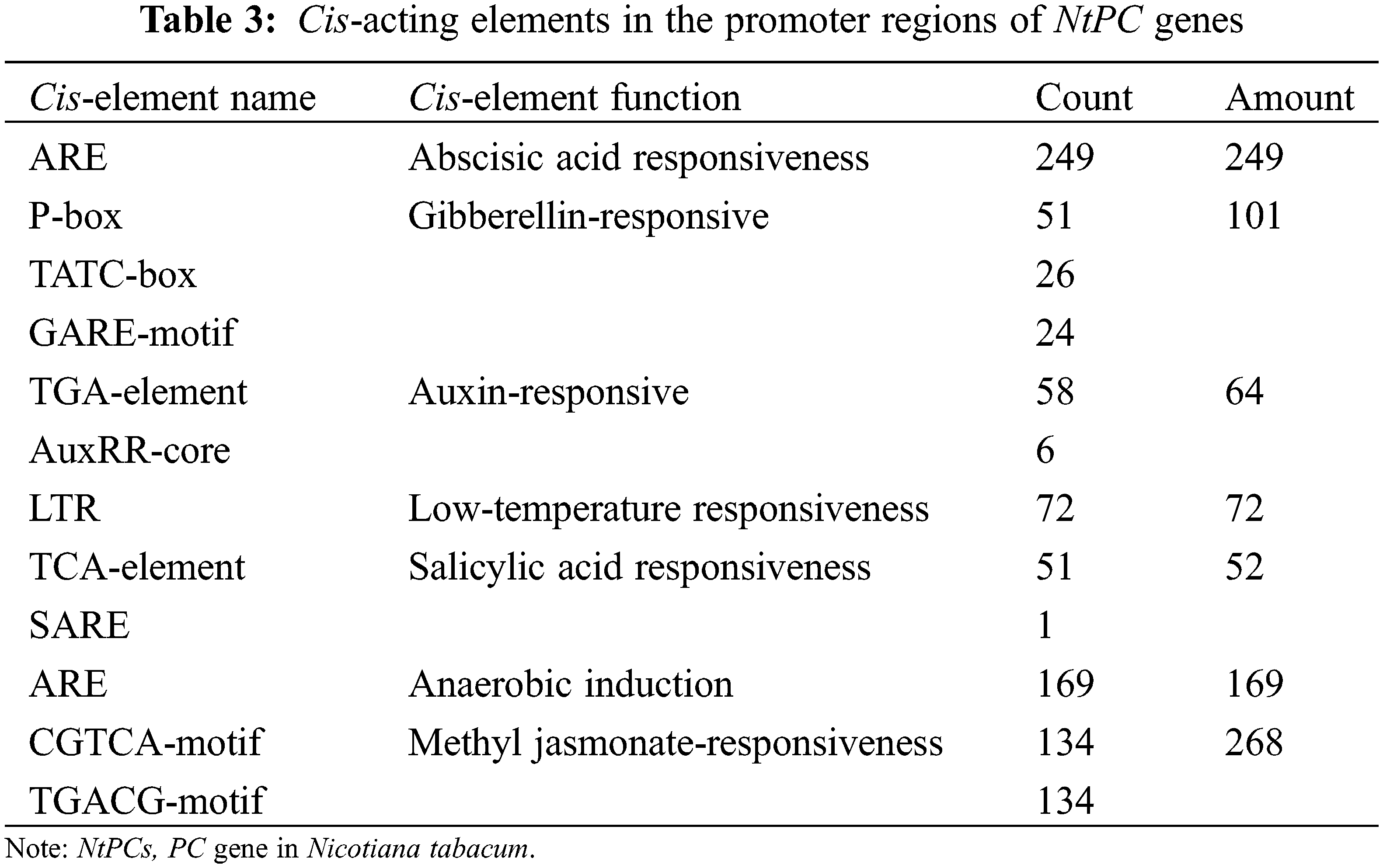
We used the PlantCARE software to profile cis-elements in the promoter regions of the 110 NtPC genes. Various cis-acting elements (Fig. 8, Table 3) were identified, including those associated with plant growth and development, such as numerous light-responsive elements (TCT, Box4, and G-box); elements associated with abiotic stress responses, such as low-temperature responses and anaerobic regulatory elements (ARE), necessary for anaerobic induction; and elements associated with hormone responses, such as those associated with gibberellin (P-box, TATC-box, and GARE-motif), auxin (TGA-element and AuxRR-core), abscisic acid, MeJA (CGTCA-motif and TGACG-motif), and salicylic acid (TCA-element, SARE) response. Among them, the elements associated with the response to MeJA were the largest in number, followed by those associated with the abscisic acid response and basic promoter elements in eukaryotes, such as CAAT boxes and TATA boxes. These results suggest that NtPCs regulate plant growth and development, responses to abiotic stresses, and hormonal responses.

Figure 8: Cis-acting elements in the promoter regions of NtPC genes
3.6 GO Analysis of NtPC Family Genes
NtPC family genes were classified as belonging to one major group of cellular components based on the similarity of their amino acid sequences. Most PC family members belonged to the components of the cytoplasmic membrane and might be involved in plasma membrane-related functions (Fig. 9).
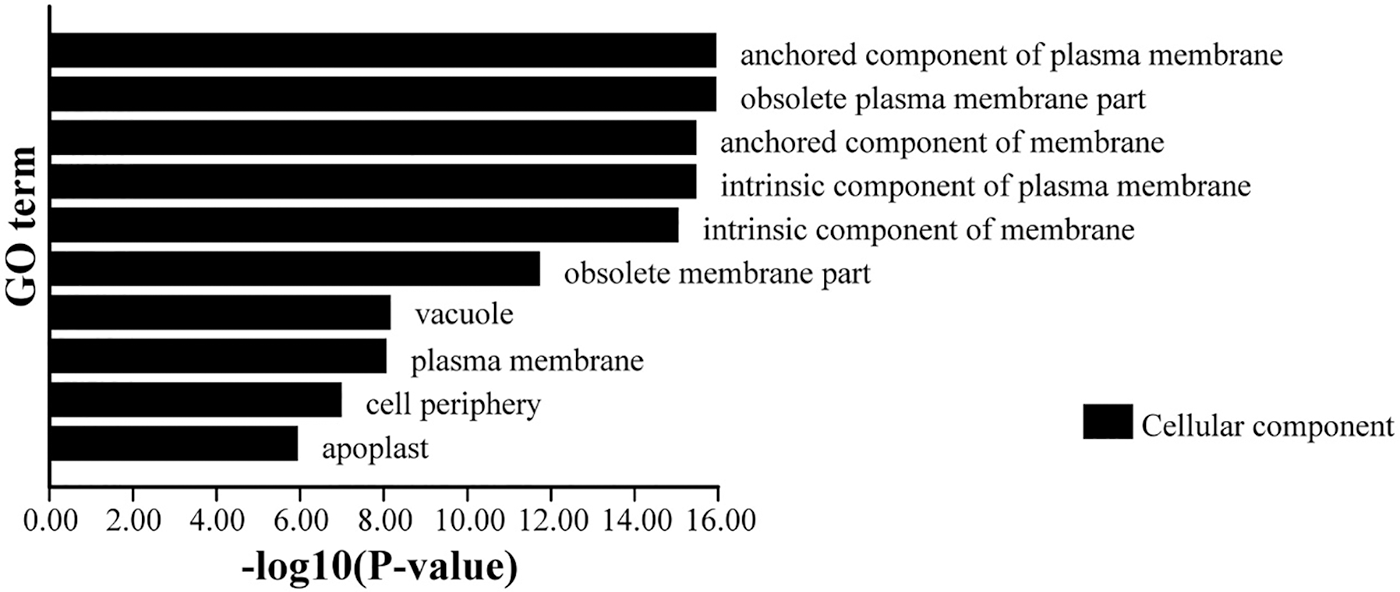
Figure 9: Gene Ontology (GO) analysis of NtPC genes. NtPCs, PC gene in Nicotiana tabacum
3.7 Expression Patterns and Spatiotemporal Expression Patterns of Ntpc Family Genes in Different Developmental Stages of Tobacco
To explore the expression levels of NtPC genes during the prosperous, maturation, and budding periods of tobacco, we analyzed gene expression based on transcriptome data from the experimental group (Fig. 10). All NtPC genes (NtENODL56, NtENODL54, NtSCL3, NtSCL15, and NtENODL51) were expressed at higher levels in the budding period than in the maturation and prosperous periods, with NtENODL51, NtSCL23, NtENODL14, NtENODL11, NtENODL22, NtSCL11, and NtSCL4 showing minimal expression in the maturation period. In addition, NtENODL25, NtENODL15, NtENODL61, NtSCL24, and NtSCL13 showed higher expression in the maturation and budding periods than in the prosperous period. The expression of NtENODL31 was higher in the maturation period than in the budding period, with minimal expression observed in the prosperous period. The remaining genes showed higher expression during the prosperous period than the maturation and budding periods, with NtSCL5 showing minimal expression in the budding period and NtENODL28 showing minimal expression in the maturation period.
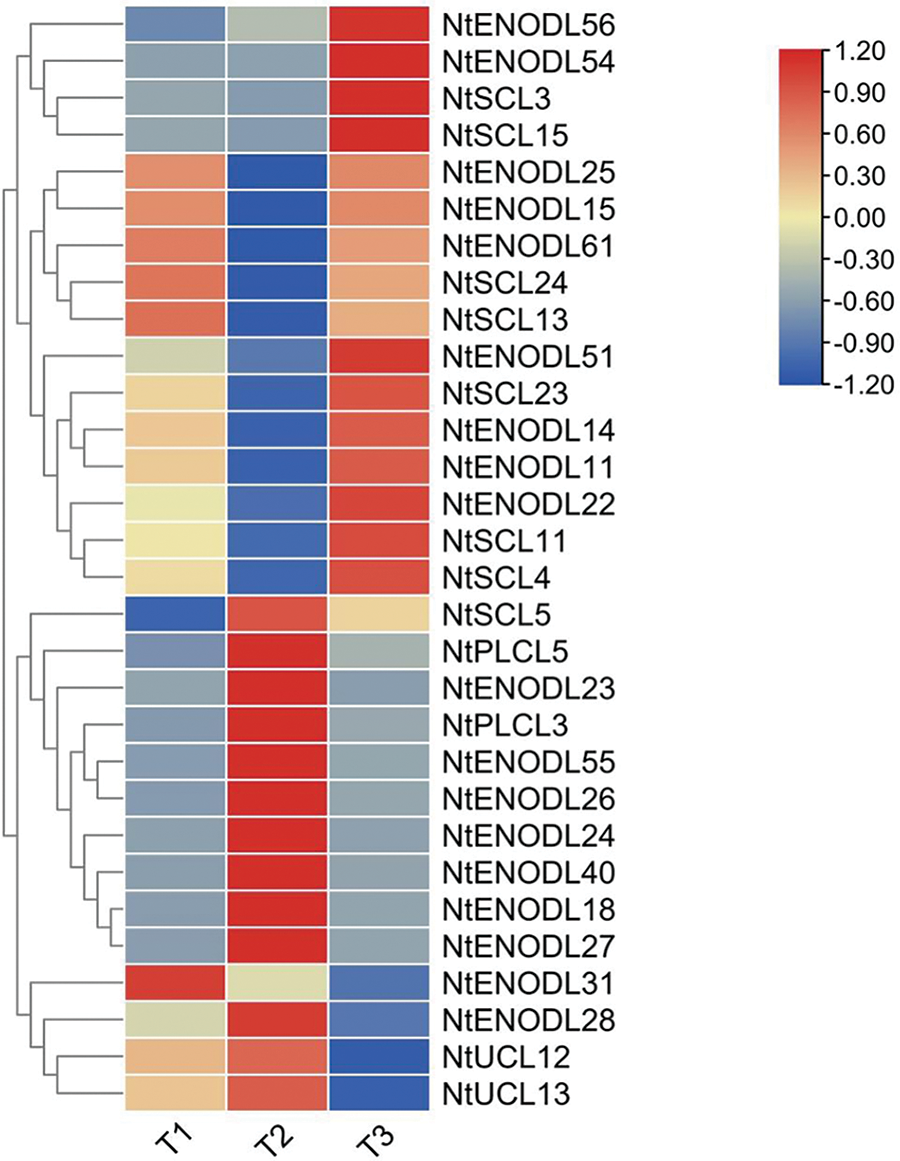
Figure 10: Gene expression of NtPCs. T1 indicates the maturation period, T2 the prosperous period, and T3 the budding period
According to the tissue-specific expression characteristics (Fig. 11), NtPC genes can be divided into four types. The first type has low or no expression in all tissues and organs. The second type is expressed in almost all tissues and organs with no significant difference in the expression level of each tissue and organ, such as NtENODL15, NtENODL25, and NtENODL56 genes. The third type is expressed in most tissues but is highly expressed in specific tissues, such as NtPLCL4 and NtUCL7, which are more expressed in roots. The expression of the fourth type of gene showed strong tissue-organ specificity. For example, NtPLCL3, NtPLCL7, and NtENODL45 genes were specifically expressed in roots, while NtENODL44, NtENODL30, and NtENODL32 were specifically expressed in flower buds.
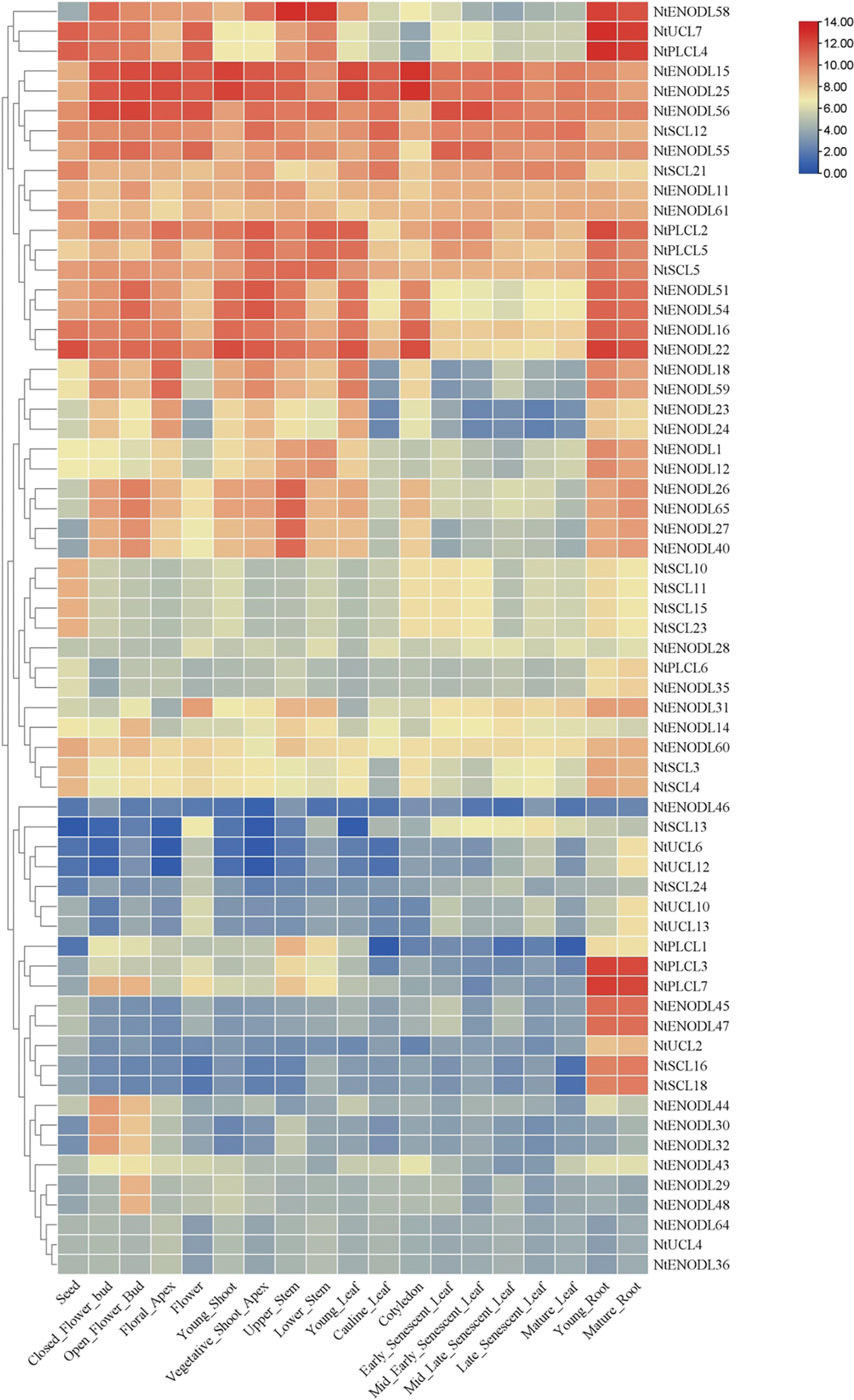
Figure 11: Expression pattern of NtPC genes in different tissues or organs
3.8 Expression Pattern Analysis of NtPC Family Genes under Stress Treatments
Early studies found that ENODL genes are involved in plant responses to abiotic stress. However, the role of ENODL genes in responding to low-temperature stress remains unclear [12,14]. Promoter element prediction revealed numerous response elements for MeJA. To further explore whether NtENODL is involved in response to low-temperature stress and stress induced by MeJA exposure, 11 genes of the NtENODL subfamily with cold cis-acting elements were selected, of which 6 genes had both cold and MeJA cis-acting elements, and were analyzed under two types of stress. The expression levels of these genes differed at 0, 1, 6, 12, and 24 h.
During low-temperature treatment (4°C) (Fig. 12), the expression levels of NtENODL7, NtENODL10, NtENOD L19, NtENODL62, and NtENODL64 tended to zero after 24 h. The total expression level of NtENOD L28 and NtENOD L33 was still lower than that at 0 h. The expression level of NtENODL1 first decreased, increased, and then decreased again after 24 h. The expression levels of NtENODL58 and NtENODL61 increased first and then decreased, reaching the highest value at 6 h, when the expression level was 46 times higher than that at 0 h. The maximum expression of NtENODL61 was observed at 12 h, which was about 6 times that at 0 h. The maximum expression level of NtENODL56 was observed at 6 h, about 4 times that at 0 h.
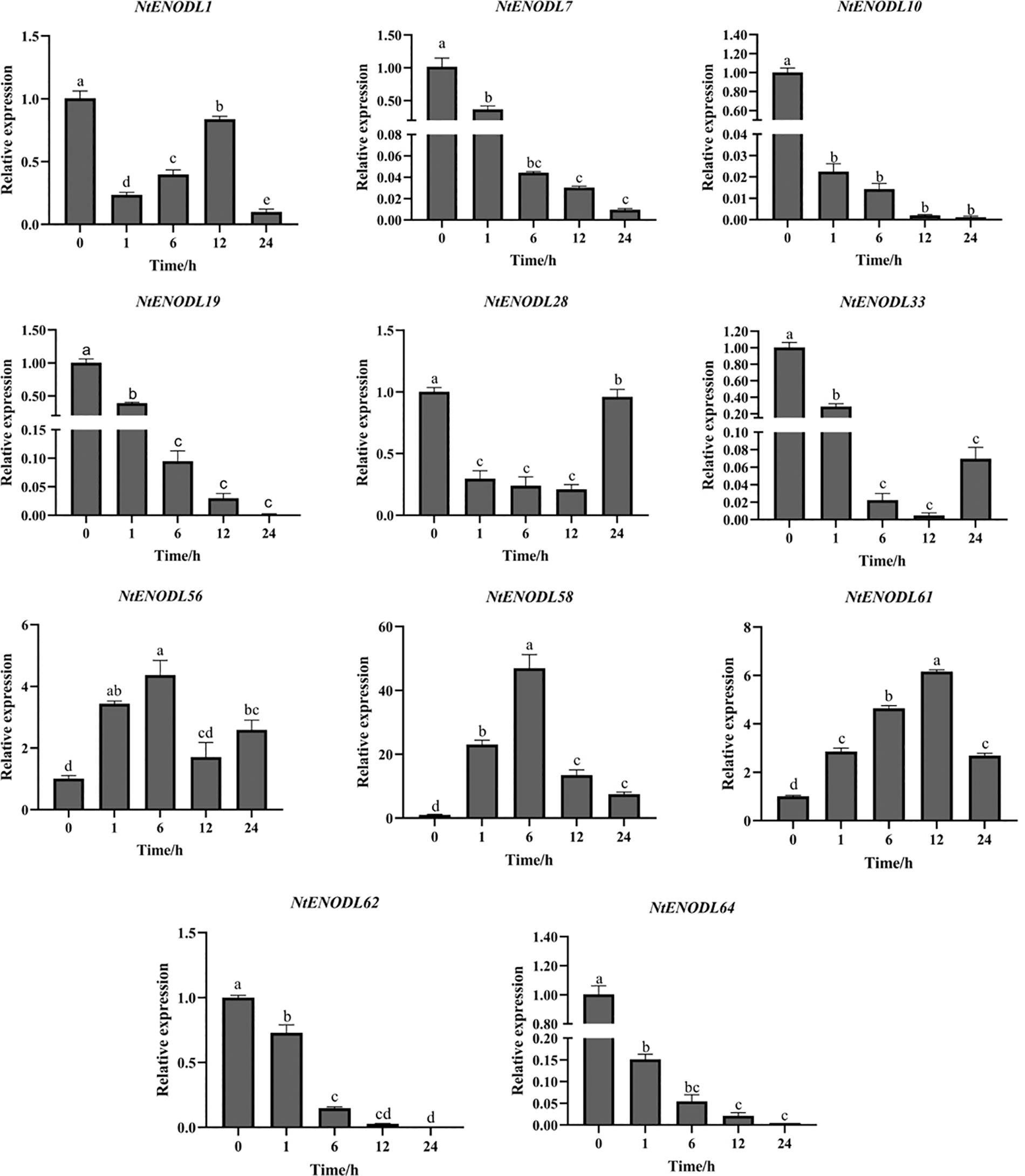
Figure 12: Relative expression levels of NtPC family members under low-temperature stress. Error bars represent means ± SE (n = 3). Three independent experiments were performed for each sample. Letters indicate significant differences (p < 0.05)
Under MeJA stress (Fig. 13), the overall expression of NtENODL61 was higher than that at 0 h, reaching its maximum at 1 h, which is about 5 times higher than that at 0 h. The expression levels of NtENODL1 and NtENODL56 reached the maximum at 1 h. The NtENODL58 expression reached its highest value at 12 h, when it was approximately 35-fold higher than it was at 0 h. The expression levels of NtENODL7, NtENODL10, NtENODL19, NtENODL28, NtENODL33, and NtENODL62 first decreased, then increased, and finally decreased again at 12 h. The expression of NtENODL28 reached its highest level at 12 h, when it was approximately 1.8-fold higher than it was at 0 h. The expression levels of the remaining genes were lower than those at 0 h. The expression of NtENODL64 first decreased and then increased, but its overall expression was also lower than it was at 0 h.
The above results show that the NtENODL58 expression levels had greater variation than those of other genes in the control group under the same conditions. Thus, NtENODL58 might play an important role in low-temperature and MeJA stress resistance and should be further evaluated.
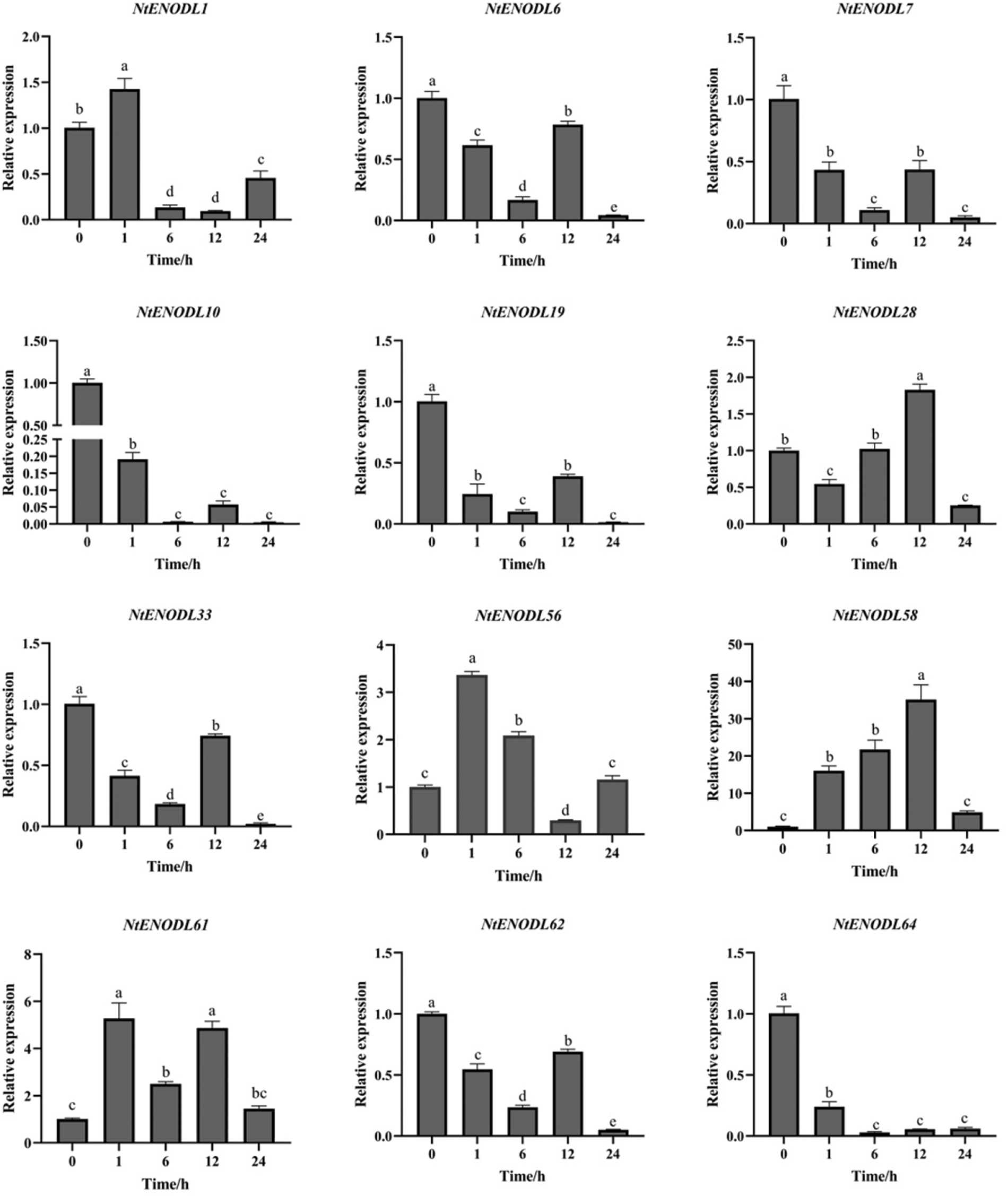
Figure 13: Relative expression levels of NtPC family members under stress induced by methyl jasmonate treatment. Error bars represent means ± SE (n = 3). Three independent experiments were performed for each sample. Letters indicate significant differences (p < 0.05). NtPCs, PC gene in Nicotiana tabacum
Previous whole-genome studies in plants have suggested that PC genes are involved in plant growth, development, and response to abiotic stress [2,12,15,17–21]. However, the role of PC genes in tobacco has not been evaluated. In this study, 110 NtPC genes were identified in the tobacco K326 genome. Because ordinary tobacco is allotetraploid and gene replication may occur in the tobacco genome [31], the number of NtPCs in tobacco is higher than that in Arabidopsis, rice, Chinese cabbage, and other plants.
Based on the differences in their copper-binding and glycosylation sites, NtPC proteins could be divided into four subfamilies: NtUCL, NtSCL, NtPLCL, and NtENODL. Among these, the NtENODL and NtPLCL subfamilies contained the largest and smallest number of genes, respectively. The NtENODL subfamily did not contain four complete copper-binding ligands (His, Cys, His, and Met/Gln). However, other subfamilies had some conserved motifs in positions corresponding to the copper-binding sites, which might be related to binding copper. In addition, NtPCs could be divided into seven evolutionary branches, similar to the results previously observed for Chinese cabbage, Arabidopsis, and P. tomentosa [12,15,19], indicating that the PC family in dicotyledons is conserved and their subfamily members cluster together in phylogenetic trees. The functions of the 4 subfamilies are thought to differ.
The NtPC proteins were divided into eight types based on the presence of the SP, PCLD, ALR, and GAS domains in the NtPC protein skeleton. Most PCs contain AGPs, such as ALR and SP. Therefore, the PC gene family is typically classified as a subfamily of the AGP superfamily [2]. In the NtPC gene family, 76 NtPCs have AG modules and thus may be AGPs. The largest number of promoter regions were light-responsive elements, supporting the idea that NtPC genes are induced by light [15], participate in photosynthesis, and play key roles in plant growth and development. In addition, we detected numerous hormone and abiotic stress response elements, showing that the NtPC family is involved in the tobacco stress response. Most PC family members are components of the cell plasma membrane and may participate in plasma membrane-related functions. However, pathway analysis could not be performed because the Kyoto Encyclopedia of Genes and Genomes-related notes are incomplete.
NtPC genes showed specific expression patterns during the prosperous, maturation, and budding periods of tobacco, indicating that these genes regulate and participate in the growth and development of tobacco from the mature to the budding period. Through the analysis of tissue-specific expression mode, we found that some genes have tissue-specific expression. For example, NtPLCL3, NtPLCL7, and NtENODL45 genes are specifically expressed in the root, and most NtENODL subfamily genes are expressed in the root. Previous studies showed that NtENODL genes are highly expressed in the root and participate in the nodulation of leguminous plants [18]. We found that most NtENODL genes are also highly expressed in roots, indicating that the function of NtENODL genes in roots may be critical. The ENODL gene plays a role in responding to abiotic stress [13]. However, its function in response to low-temperature stress has not been evaluated.
We identified numerous MeJA response elements and analyzed the expression levels of NtENODL subfamily genes under low-temperature and MeJA stresses. The expression levels of 11 genes of the tobacco plants under low-temperature and MeJA stresses were significantly different from those in the control group. The expression level of NtENODL58 under low-temperature and MeJA stress first increased and then decreased, and at a particular time, its expression level showed greater differences than other genes in the control group, and the highest expression level was 46 times and 35 times that of the control group. Secondly, the expression level of NtENODL56 and NtENODL61 under low temperature and MeJA stress differed significantly from that of the control group. These genes may play an important role in low-temperature and MeJA stress resistance and should be further evaluated. In addition, a previous study showed that miR408 was upregulated in Taxus seeds under cold stress and that this microRNA could regulate the genes encoding plant PC family proteins [32]. In Arabidopsis, AtPLCs are also believed to be target genes of miR408 [33]. Therefore, genes belonging to the NtPC family might be regulated by miR408 under low-temperature stress. Thus, the regulation of the NtPC family by miR408 should be investigated in the future.
We identified 110 PC family genes in tobacco cultivar ‘K326’ and phylogenetically classified these genes into 4 subfamilies: NtENODL, NtUCL, NtSCL, and NtPLCL. The NtPC genes were classified into 8 types based on family structure analysis. In addition, we found that 53 NtPCs were randomly distributed on 22 of the 24 tobacco chromosomes, with 33 pairs of collinear genes belonging to the NtPC family. All PC genes were components of the cytoplasmic membrane and might be involved in functions related to the cytoplasmic membrane. The NtPC family contained many hormone response-related and abiotic stress-responsive elements. The NtPC family was specifically expressed during the prosperous, vigorous growth, maturation, and budding periods of tobacco development, and some genes have tissue-specific expression. Most of these genes were downregulated under low-temperature and MeJA induction, and both conditions significantly induced NtENODL58 expression. In conclusion, the NtPC gene family might play an important role in the plant stress response. Thus, our results provide insights into the functions of NtPC family genes and a foundation for further study.
Acknowledgement: We thank the developers of the Tbtools software. We also thank all the team members who helped us with the experimental process.
Funding Statement: This study was supported by the Tobacco Science Research Institute of the Chongqing Tobacco Company (A20201NY01-1305).
Author Contributions: Study conception and design: Pei-Ling Wang, Xiao-Hong Xu; data collection: Pei-Ling Wang, Xiao-Hong Xu, Yong Li, He-Cui Zhang, Xue-Jie Zhang; analysis and interpretation of results: Pei-Ling Wang, Si-Ru Zhou, Yi-Mei Liu, Yun-Yan Feng, Tong-Hong Zuo; draft manuscript preparation: Pei-Ling Wang, Xiao-Hong Xu; revised the manuscript: Li-Quan Zhu, He-Cui Zhang, Xue-Jie Zhang. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Nersissian, A. M., Immoos, C., Hill, M. G., Hart, P. J., Williams, G. et al. (1998). Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: Plant-specific mononuclear blue copper proteins. Protein Science, 7, 1915–1929. [Google Scholar] [PubMed]
2. Ma, H. L., Zhao, H. M., Liu, Z., Zhao, J. (2011). The phytocyanin gene family in rice (Oryza sativaL.Genome-wide identification, classification and transcriptional analysis. PLoS One, 6, e25184. [Google Scholar]
3. Giri, A. V., Anishetty, S., Gautam, P. (2004). Functionally specified protein signatures distinctive for each of the different blue copper proteins. BMC Bioinformatics, 5, 127. [Google Scholar] [PubMed]
4. Kim, S., Mollet, J. C., Dong, J., Zhang, K., Park, S. Y. et al. (2003). Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. PNAS, 100, 16125–16130. [Google Scholar] [PubMed]
5. Zhang, F., Zhang, Y. C., Zhang, J. P., Yu, Y., Zhou, Y. F. et al. (2018). Rice UCL8, a plantacyanin gene targeted by miR408, regulates fertility by controlling pollen tube germination and growth. Rice, 11, 1–6. [Google Scholar]
6. Hao, C., Yang, Y. Z., Du, J. M., Deng, X. W., Li, L. (2022). The PCY-SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. PNAS, 119, e2116623119. [Google Scholar] [PubMed]
7. Kato, T., Kawashima, K., Miwa, M., Mimura, Y., Tamaoki, M. et al. (2002). Expression of genes encoding late nodulins characterized by a putative signal peptide and conserved cysteine residues is reduced in ineffective pea nodules. Molecular Plant-Microbe Interactions, 15, 129–137. [Google Scholar] [PubMed]
8. Escobar-Restrepo, J. M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, G. et al. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science, 317, 656–660. [Google Scholar] [PubMed]
9. Hou, Y., Guo, X., Cyprys, P., Zhang, Y., Bleckmann, A. et al. (2016). Maternal ENODLs are required for pollen tube reception in Arabidopsis. Current Biology, 26, 2343–2350. [Google Scholar] [PubMed]
10. Richards, K. D., Schott, E. J., Sharma, Y. K., Davis, K. R., Gardner, R. C. (1998). Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiology, 116, 409–418. [Google Scholar] [PubMed]
11. Ezaki, B., Sasaki, K., Matsumoto, H., Nakashima, S. (2005). Functions of two genes in aluminium (Al) stress resistance: Repression of oxidative damage by the AtBCB gene and promotion of efflux of Al ions by the NtGDI1 gene. Journal of Experimental Botany, 56, 2661–2671. [Google Scholar] [PubMed]
12. Mashiguchi, K., Asami, T., Suzuki, Y. (2009). Genome-wide identification, structure and expression studies, and mutant collection of 22 early nodulin-like protein genes in Arabidopsis. Bioscience, Biotechnology, and Biochemistry, 73, 2452–2459. [Google Scholar] [PubMed]
13. Wu, H., Shen, Y., Hu, Y., Tan, S., Lin, Z. (2011). A phytocyanin-related early nodulin-like gene, BcBCP1, cloned from Boeacrassifolia enhances osmotic tolerance in transgenic tobacco. Journal of Plant Physiology, 168, 935–943. [Google Scholar] [PubMed]
14. Zhang, M., Wang, X., Yang, J., Wang, Z., Chen, B. et al. (2022). GhENODL6 isoforms from the phytocyanin gene family regulated verticillium wilt resistance in cotton. International Journal of Molecular Science, 23, 2913. [Google Scholar]
15. Li, J., Gao, G., Zhang, T., Wu, X. (2013). The putative phytocyanin genes in Chinese cabbage (Brassica rapaL.Genome-wide identification, classification and expression analysis. Molecular Genetics and Genomics, 288, 1–20. [Google Scholar] [PubMed]
16. Cao, J., Li, X., Lv, Y., Ding, L. (2015). Comparative analysis of the phytocyanin gene family in 10 plant species: A focus on Zea mays. Frontiers in Plant Science, 6, 515. [Google Scholar] [PubMed]
17. Xu, L., Wang, X. J., Wang, T., Li, L. B. (2017). Genome-wide identification, classification, and expression analysis of the phytocyanin gene family in Phalaenopsis equestris. Biologia Plantarum, 61, 445–452. [Google Scholar]
18. Sun, Y., Wu, Z., Wang, Y., Yang, J., Wei, G. et al. (2019). Identification of phytocyanin gene family in legume plants and their involvement in nodulation of Medicago truncatula. Plant Cell Physiology, 60, 900–915. [Google Scholar] [PubMed]
19. Luo, S., Hu, W., Wang, Y., Liu, B., Yan, H. et al. (2018). Genome-wide identification, classification, and expression of phytocyanins in Populu strichocarpa. Planta, 247, 1133–1148. [Google Scholar] [PubMed]
20. Song, Z., Li, L. B., Liang, L. X., Tao, W. (2018). Genome-wide analysis of phytocyanin gene family in Dendrobium officinale. Forest Research, 31, 98–106. [Google Scholar]
21. Zhang, S., Lin, K. Y., Hou, C. P., Fang, Y. J., Wang, Y. P. (2022). Genome-wide identification and expression analysis of phytocyanin family in Brassica napus. Chinese Journal of Oil Crop Sciences, 44, 116–129. [Google Scholar]
22. Finn, R. D., Mistry, J., Schuster-Bōckler, B., Griffiths-Jones, S., Hollich, V. et al. (2006). Pfam: Clans, web tools and services. Nucleic Acids Research, 34, D247–D251. [Google Scholar] [PubMed]
23. Petersen, T. N., Brunak, S., von Heijne, G., Nielsen, H. (2011). SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods, 8, 785–786. [Google Scholar] [PubMed]
24. Eisenhaber, B., Wildpaner, M., Schultz, C. J., Borner, G. H. H., Dupree, P. et al. (2003). Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiology, 133, 1691–1701. [Google Scholar] [PubMed]
25. Schultz, C. J., Rumsewicz, M. P., Johnson, K. L., Jones, B. J., Gasper, Y. M. et al. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiology, 129, 1448–1463. [Google Scholar] [PubMed]
26. Ma, H. L., Zhao, J. (2010). Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativaL.). Journal of Experimental Botany, 61, 2647–2668. [Google Scholar] [PubMed]
27. Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020(13), 1194–1202. [Google Scholar]
28. Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E. et al. (2009). MEMESUITE: Tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. [Google Scholar] [PubMed]
29. Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y. et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30, 325–327. [Google Scholar] [PubMed]
30. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [Google Scholar] [PubMed]
31. Sierro, N., Battey, J. N. D., Ouadi, S., Bakaher, N., Bovet, L. et al. (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nature Communications, 5, 3833. [Google Scholar] [PubMed]
32. Fu, J., Wan, L., Song, L., He, L., Jiang, N. et al. (2021). Identification of microRNAs in Taxillus chinensis (DC.) Danser seeds under cold stress. Biomedical Research International, 2021, 5585884. [Google Scholar]
33. Sunkar, R., Zhu, J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell, 16, 2001–2019. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools