 Open Access
Open Access
ARTICLE
HD-Zip Transcription Factor is Responsible for No-Lobed Leaf in Watermelon (Citrullus lanatus L.)
1 College of Horticulture, Henan Agricultural University, Zhengzhou, 450002, China
2 Cotton Institute, Chinese Academy of Agricultural Sciences, Anyang, 455000, China
* Corresponding Authors: Junling Dou. Email: ; Huayu Zhu. Email:
# These authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2023, 92(5), 1311-1328. https://doi.org/10.32604/phyton.2023.026928
Received 03 October 2022; Accepted 14 November 2022; Issue published 09 March 2023
Abstract
Leaf is a vital organ of plants that plays an essential role in photosynthesis and respiration. As an important agronomic trait in leaf development, leaf shape is classified into lobed, entire (no-lobed), and serrated in most crops. In this study, two-lobed leaf watermelon inbred lines WT2 and WCZ, and a no-lobed leaf watermelon inbred line WT20 were used to create two F2 populations. Segregation analysis suggested that lobed leaves were dominant over the no-lobed leaves, and it was controlled by a signal gene. A locus on watermelon chromosome 4 controlling watermelon lobed/no-lobed leaves was identified through BSA-seq strategy combined with linkage analysis. The candidate gene was fine-mapped to a 61.5 kb region between 21,224,481 and 21,285,957 bp on watermelon chromosome 4 using two F2 populations. Four functional genes were annotated in the candidate region, while sequences blast showed that there was a single-base deletion (A/-) only in the exon of Cla018360, which resulted in premature termination of translation in the no-lobed leaf lines. Function prediction showed that Cla018360 encodes an HD-Zip protein that has been reported to regulate the development of leaf shape. The single-base deletion also occurred in the HD-Zip domain. We inferred that the Cla018360 gene is the candidate gene for regulating the development of lobed/no-lobed leaves in watermelon. Gene expression analysis showed that Cla018360 was highly expressed in young leaves. Phylogenetic analysis showed that Cla018360 had a close genetic relationship with AtHB51, which had been reported to regulate the formation of leaf shape in Arabidopsis. Furthermore, transcriptome analysis showed that a total of 333 differentially expressed genes were identified between WT2 and WT20, of which 115 and 218 genes were upregulated and downregulated in no-lobed leaved watermelon WT20. This study not only provides a good entry point for studying leaf development but also provides foundational insights into breeding for special plant architecture in watermelon.Keywords
Supplementary Material
Supplementary Material FileAs an essential organ in flowering plants, leaves play important roles in photosynthesis, respiration, gas exchange, nutrient supply, water transport, fruit quality, and yield. In addition, there is remarkable variety in leaf size and shape among different crops and different varieties, which are used for breeding and domestication [1–5]. The leaf margin is an important leaf shape trait that has been classified into lobed, entire (no-lobed), and serrated leaves [6]. A previous study reported that lobe-leafed plants are more suitable for mechanical production with high-density cultivation because of excellent ventilation and sufficient light [7]. The leaf margin is mainly affected by genetic regulators and can be easily distinguished at the seedling stage in most crops [8,9]. Therefore, it can also be used as an effective morphological marker for cultivar identification and improvement, thereby ensuring the purity of the seeds [9,10]. Therefore, the study of the genetic mechanism and regulatory pathway of leaf shape formation is beneficial to breeders in the marker-assisted selection, manipulation of leaf shape, improvement of plant architecture, and increase of yield during crop breeding.
Plant leaves are initiated from the peripheral region of the shoot apical meristem (SAM) and then develop into great variations in size and shape. In the early stage of leaf development in Arabidopsis, leaf shape is affected by the differentiation of SAM cells and the split of polar cells. After the formation of leaf margin cells from SAM, the cells undergo rapid lateral stretching, development, and differentiation. Finally, the regulation of near axis-distant axis and asymmetry increasing of cells determines the formation of the entire leaf [6]. In the leaf formation stage of tomato, the marginal blastozone has the potential to develop side apical meristems, which can maintain the activity of the apical meristem; after leaf maturation, the cell division and differentiation process is blocked and affected by other factors, resulting in differentiation of local cells, ultimately forming the phenotype of lobed leaf at the maturity stage [11].
At present, genes regulating leaf margin development have been cloned or identifiedin many crops, providing a theoretical and genetic basis for further research on the regulatory mechanism of leaf development. In Arabidopsis, the redundancy of CUC1, CUC2, and CUC3 genes resulted in the regularity of cotyledon margins and the formation of embryonic shoot meristems through the regulation of the CUP-SHAPED COTYLEDON transcription factor [12]; the CUC2 gene regulates the extent of leaf serration mainly through the balance between CUC2 and its target gene miR164A [13]. The LATE MERISTEM IDENTITY 1 (LMI1) gene, which encodes the homeodomain leucine-zipper transcription factor class I (HD-Zip), plays an important role in the formation of serrated leaves [14]. Other LMI1-like genes, such as the Tendril-less (TL) gene in pea and the REDUCED COMPLEXITY (RCO) gene in Cardamine hirsuta, have been reported to affect the formation of lateral organs and then regulate lateral organ genesis in compound leaves [15,16]. In tomato, the number of leaflets and the leaf shape could be regulated by the expression level of the KNOX gene (Class I KNOTTED1-like homeobox through CK (cytokinin hormone) signal regulation), and KNOX gene can be re-pressed by TRIPINNATE and CLAUSA genes. The interaction of these three genes can regulate the development of leaf shape [17–19]. In rapeseed (Brassica napus L.), two tandemly duplicated LMI1-like genes, BnA10.LMI1 and BnA10.RCO, on chromosome A10 have been identified to positively regulate the formation of lobed leaves [20–22]. In ornamental kale (B. oleracea var. acephala), the homolog of LIM1 has been reported as the possible candidate gene controlling the development of lobed leaves [23,24]. In addition, another semi-dominant gene BoFL, which encodes an alpha-1,2-glucosyltransferase, has been shown to regulate the phenotype of feathered leaves in ornamental kale [25]. In cotton, the study revealed that the formation of lobed, broad, and compound leaves was controlled by KNOX1 and LMI1-like genes [26]. In Medicago truncatula, the loss-of-function mutation of the Mt-AGO7/LOBED LEAFLET1 gene resulted in the phenotype of lobed leaves, which demonstrated that the gene could play an important role in the formation of leaf shape [27]. The mutation of a YUC homolog in Medicago sativa suppresses lateral leaflet development. The expression of YUCs in the leaves mediates the development of leaf margins and subsequently promotes blade outgrowth [28]. In lettuce (Lactuca sativa var. ramosa Hort.), the latest research showed that LsKN1 promotes leafyheads by downregulating LsAS1, and the upregulated LsKN1 gene transforms pinnately to palmately lobed leaves through auxin, gibberellin, and leaf dorsiventrality pathways [29,30].
Many studies have reported the genetic regulation of leaf shape in Cucurbitaceae. The leaf shape of pumpkin (Cucurbita maxima) is diverse among Cucurbitaceae. The initial report in pumpkin showed that the entire leaf was dominant to the lobed leaf, while the entire leaf in pumpkin was recessive to the deeply lobed leaf in Cucurbita ecuadorensis [31,32]. In melon (Cucumis melo L.), it was reported that the lobed leaf gene was controlled by a dominant gene [33], while the deeply lobed leaf was controlled by a recessive gene. A single recessive gene (pll) was fine-mapped and cloned in the following studies as regulating the phenotype of the deeply lobed leaf [2]. In zucchini (Cucurbita pepo L.), major QTL Li-10 was identified as controlling leaf incision [34]. Furthermore, the CpDll gene, which encodes transcription factor HD-Zip I, has been reported to positively regulate the formation of a deeply lobed leaf shape because of a mutation in the promoter region [7]. However, the genetic basis of leaf margin development and regulatory network are largely unknown in Cucurbitaceae.
Watermelon (Citrullus lanatus) is grown widely and very popular with consumers all over the world. It has 11 chromosomes (2n = 2X = 22) with an estimated genome size of 425 Mb. Watermelon reference genome ‘97103’ v1 was sequenced and released in 2013 [35]. Subsequently, the ‘Charleston Gray’ reference genome and ‘97103’ v2 were also released [36]. The availability of these reference genomes makes it possible to apply next-generation sequencing and fine-mapping to identify functional genes. To adapt to the current situation of insufficient labor, simplified cultivation will be the main direction of watermelon cultivation in the future. The breeding of watermelon varieties with ideal plant architecture plays an important role in the simplified cultivation and mechanical production of watermelon. To date, some genes controlling watermelon plant architecture have been reported: the plant dwarf genes Cldf [37,38] and Cldw-1 [39], the branchless gene ClTFL1 [40]. Although leaf shape is an important plant architecture trait, there are few studies on leaf shape, especially for leaf margin; only one report showed that lobed leaf gene ClLL1 mapped to a 127.6-kb region [41].
In this study, we found that watermelon lobed/no-lobed leaves were regulated by a single gene according to the inheritance analysis of two F2 populations (WCZ × WT20 F2 and WT2 × WT20 F2), which was named it Clnl (Citrullus lanatus no-lobed leaf). A BSA-seq strategy combined with linkage analysis was used for mapping the candidate gene. Finally, a candidate gene Cla018360, which belongs to the HD-Zip transcription factor, was identified to regulate the development of watermelon lobed/no-lobed leaf. Transcriptome analysis also revealed that many leaf development genes had differential expression between two parents. This study will help understand the regulatory pathway of leaf shape formation and organ development.
2.1 Plant Materials and Phenotypic Measurement
Watermelon accessions WT2 (P1, lobed leaf), WT20 (P2, no-lobed leaf), and WCZ (P3, lobed leaf) are homozygous inbred lines (Fig. 1) that have been manually self-pollinated for at least five generations. Three accessions were used to construct a segregating population of watermelon with lobed leaves. P1 and P2 were planted in a greenhouse to derive WT2 × WT20 F1 generation by crossing. Then, WT2 × WT20 F1 was self-pollinated to obtain the F2 population (WT2 × WT20 F2). Another F2 population (WCZ × WT20 F2) was obtained by crossing WT20 and WCZ in the same way. Backcross population of BC1P2 or BC1P3 was created by hybridizing between WCZ × WT20 F1 and parents (WT20 and WCZ).

Figure 1: Watermelon leaf phenotypes of inbred lines WT2, WT20, WCZ, and two F1. Bar = 2 cm
For segregation analysis, the WCZ × WT20 F2 population was grown and investigated in three experiments conducted in the spring and autumn of 2020 and spring of 2021, with 80,217, and 992 F2 individuals, respectively. The WT2 × WT20 F2 population was grown and investigated in two experiments conducted in spring and autumn of 2021 with 231 and 255 F2 individuals, respectively. Moreover, 113 BC1P2 and 69 BC1P3 individuals were planted and investigated in the spring of 2020. Parents with 20 individuals and F1 with 10 individuals were included in each experiment. The leaf phenotype of each individual was observed and recorded for three times independently during plant growth and development, and the segregation ratio was tested using the chi-squared test (Table S1). All watermelon materials used in this study were sown in nursery trays, and the seedlings were transplanted to greenhouses in the Science and Education Park of Henan Agricultural University, Zhengzhou, China.
2.2 Bulked-Segregation Analysis of DNA
Genomic DNA from fresh leaves of the WCZ × WT20 F2 population in the spring of 2020 was extracted using the CTAB method [42], which was used for sequencing and molecular marker analysis. We constructed and re-sequenced two DNA pools, the lobed leaf pool (L-pool) and the no-lobed leaf pool (N-pool), by mixing equal amounts of DNA from 20 lobed leaf individuals and 20 no-lobed leaf individuals in the WCZ × WT20 F2 population from the spring of 2020. Using an Illumina Genome Analyzer IIx, a paired-end sequencing library (read length of 100 bp) with approximately 500-bp insert sizes was prepared for sequencing. For the sequencing results in L- and N-pools, all short reads were aligned to reference genome ‘97103’ using BWA software [35,43]. Using SAM tools, we converted alignment files into SAM/BAM files [44]. To increase the accuracy of SNP calling, we applied the SNP-calling filter Coval, as previously developed [45]. All SNP positions were calculated using the SNP-index. The SNP-index is the proportion of the difference between the reads harboring SNPs and the reference sequence. SNP positions with a read depth <5 and an SNP-index <0.4 from two sequences were excluded because they may represent pseudo-SNPs due to sequencing/alignment errors or genomic repeat sequences.
According to the SNP-index, we calculated Δ(SNP-index) to identify candidate intervals for lobed leaf loci [45,46]. The Δ(SNP-index) was calculated by subtracting the SNP-index of the L-pool from that of the N-pool. Therefore, if the SNP-index was 0, all short reads were from the no-lobed leaf line; if the SNP-index was 1, all short read fragments were from the ‘97103’ reference genome. A sliding window analysis with a window size of 1 Mb and an increment of 10 kb was used to calculate the average SNP-index of SNPs located in a given genomic region. The SNP-index graphs and the corresponding Δ(SNP-index) graphs were drawn according to the above.
To generate confidence intervals of the SNP-index value under the null hypothesis of no QTL, we conducted a computer simulation. First, we obtained two bulks of offspring through random sampling according to a given number of individuals. A given number of alleles, according to the read depth, were sampled from each bulk. We then calculated the SNP-index of each bulk and obtained the Δ(SNP-index). For each reading depth, this process was repeated 10,000 times and generated a confidence interval. The interval was plotted for all genomic regions with variable read depth.
2.3 Marker Development and Fine Mapping
The whole genome sequence of watermelon ‘97103’ was released to the Cucurbit Genomics Database (http://www.icugi.org) in 2013 [35]; 32,869 SSR markers distributed across the watermelon genome were then developed according to the ‘97103’ genome [47]. To confirm the candidate regions from BSA-seq, SSR markers on chromosome 4 were screened to analyze the polymorphism between the parents WT2 and WT20. Moreover, we constructed two DNA pools in the WT2 × WT20 F2 population from the spring of 2021, the L1-pool and the N1-pool, by mixing equal amounts of DNA from 20 lobed leaf individuals and 20 no-lobed leaf individuals. The SSR markers, which were polymorphic between WT2 and WT20, were used to screen the polymorphism between two DNA pools. Finally, nine polymorphic markers were used for linkage analysis and primarily mapped through 157 WT2 × WT20 F2 individuals. Then, eight Indel, five dCAPS, and two SNP markers were developed during the interval of primary mapping according to the re-sequencing data for fine-mapping in the WT2 × WT20 F2 population, which contained a total of 486 individuals from the spring and autumn of 2021 (Table S2). The Indel and dCAPS markers were designed based on the Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/) and dCAPS Finder 2.0 [48], respectively. To further narrow the candidate interval, the WCZ × WT20 F2 population with a total of 1289 individuals was used for linkage analysis and fine-mapping with the same methods as above. The PCR reaction system, reaction procedure, restriction enzyme digestion reactions, and subsequent gel electrophoresis were conducted as described by Dou et al. [40]. Linkage analysis was performed using the Kosambi mapping function of JoinMap 4.0, with the threshold logarithm 10 of the odds (LOD) score of 5.0. The important markers used for linkage analysis in this study are listed in Table S2.
2.4 Prediction and Verification of Candidate Genes
The predicted genes in the candidate region were retrieved from the ‘97103’ watermelon reference genome (http://cucurbitgenomics.org/). Gene-specific primers were designed according to the DNA and CDS sequence of the candidate gene. The DNA and CDS sequence of candidate genes were amplified from young leaves of three parents, WCZ, WT20, and WT2; then, PCR products were sequenced and aligned between the ‘97103’ reference genome and the parents (WCZ, WT20, and WT2) for detecting sequence differences. The function of the candidate gene was predicted based on NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Cucurbitaceae Genome Database (http://cucurbitgenomics.org/). To verify the candidate gene, the co-segregation marker was tested on 100 F2 individuals (randomly selected in different years) and 94 lobed leaf watermelon germplasm resources (Table S3), which were derived from Guo et al. [49].
2.5 Quantitative Real-Time PCR
To check the expression of candidate gene Clnl, the different tissues (root, stem, male flower, female flower, young leaf, mutant leaf) of watermelon WT2 and WT20 were used to extract RNA for investigating the expression pattern of the candidate gene. Total RNA was extracted using the plant RNA purification kit (Omega, Norcross, USA) according to the manufacturer’s protocol. The concentration and quality of the RNA samples were determined and assayed using NanoDrop 2000 (Thermo) and 1.5% agarose gel, respectively. cDNA was synthesized using reverse transcriptase M-MLV (RNase H-) based on the instructions of the manufacturer (Takara, Kyoto, Japan). The primers of genes for real-time PCR were designed for amplifying products of 100–250 bp using Primer 5.0 online software without any interference with the conserved region. The Actin primer [50] and gene primers for real-time PCR are listed in Table S4. The reaction of qRT-PCR was carried out in accordance with a previous report [40]. The experiments were performed with three biological and three technical replicates, and the relative expression pattern was calculated using the 2−
The amino acid sequence of Clnl was retrieved from the Cucurbitaceae Genome Database. A total of 102 homologs of Clnl were identified from A. thaliana, tomato (Solanum lycopersicum), rice (Oryza sativa. L.), and Cucurbitaceae crops (cucumber ‘Chinese Long’ v2, melo ‘DHL92’, watermelon ‘97103’ v1). All information on the 102 homologs of Clnl used for phylogenetic analysis is listed in Table S5. According to the sequence alignments, the phylogenetic tree was constructed based on MEGA7.0 software [52]. Neighbor-joining (NJ) was generated with the model of Poisson correction, 1,000 bootstrap replicates, and pairwise alignment [38].
To study the transcriptional difference of watermelon in leaf development, young leaves of WT2 and WT20 at the three-leaf stage were used for RNA-seq by Biomarker Technologies Co., Ltd. (Beijing, China). Total RNA was extracted from fresh samples according to the CTAB method [53]. The sample contained 1.0 g, and three biological replicates were performed for each sample. RNA concentrations and quality were determined and assayed using NanoDrop 2000 (Thermo) and 1.5% agarose gel, respectively. The RNA-seq library was used to prepare the sample libraries according to the constructed flow path, and then the RNA was sequenced on an Illumina HiSeq2000 platform. Clean reads, obtained by filtering the contaminant sequences and low-quality reads, were aligned to watermelon reference genome ‘97103’ according to HISAT2 [54]. The transcript data were assembled using the software Stringtie [55]. The heatmap was generated using TBtools. The RNA-seq data are available with accession number PRJNA483539 on NCBI, and the detailed statistics and different expression genes of RNA-seq are listed in Tables S6 and S7.
3.1 Inheritance Features of Lobed Leaves in Watermelon
Watermelon accessions WT2 and WCZ were homozygous inbred lines with the normal lobed phenotype in all mature leaves, while watermelon accession WT20 had a no-lobed phenotype in all mature leaves (Fig. 1). The phenotype of the leaf could be easily distinguished from the seedling period to the maturation period. All three accessions were manually self-pollinated for at least five generations and were genetically stable. To study the genetic regulation of lobed leaves in watermelon, WT2, WT20, and WCZ were used as parents to construct genetic populations. No-lobed leaf watermelon WT20 (P2) was crossed with WT2 (P1) and WCZ (P3) for constructing segregation populations. All F1 individuals had lobed leaves, suggesting that watermelon lobed leaves are dominant over no-lobed leaves (Fig. 1). Segregation populations were constructed through a cross between WCZ and WT20. WT2 was used for constructing the F2 population by crossing with WT20. The segregation ratio of lobed/no-lobed leaves among different populations in two years is presented in Table S1. The χ2 test showed that the segregation ratio in the F2 and BC1 populations were in line with 3:1 and 1:1, respectively, indicating that the no-lobed leaf phenotype in watermelon is a qualitative trait and regulated by a single recessive gene, Clnl (Citrullus lanatus no-lobed leaf).
3.2 BSA-Seq Revealed that Clnl Gene Was Located on Chromosome 4
Two pools (L-pool and N-pool) were constructed from the F2 population in the spring of 2020 for BSA-seq analysis using 20 lobed leaf individuals and 20 no-lobed leaf individuals, respectively. Two pools were sequenced using the Illumina HiSeqTM PE150 platform. A total of 36.5 GB data were generated from both pools, with approximately 20× depth and more than 99% coverage for each. All data from L- and N-pools were mapped to the watermelon reference genome ‘97103’ v1 (http://cucurbitgenomics.org/) and 5634 SNPs were detected between the two pools. Each identified SNP was computed using an SNP-index. We calculated the average SNP-index in the interval of 1 Mb using a sliding window of 1 kb. Graphs of the SNP-index with the L- and N-pools were generated by plotting the average SNP-index relative to the position of each sliding window in the watermelon genome reference ‘97103’ v1 assembly. The Δ(SNP-index) graph was drawn and the genome locations were calculated by combining the information on the SNP-index in L- and N-pools (Fig. 2a). Only the interval on chromosome 4 between 19.8 and 23.8 Mb had an average Δ(SNP-index) that was higher than 0.4 with a distinct peak. This suggests that candidate gene Clnl was located at an interval of 19.8–23.8 Mb on chromosome 4 in watermelon (Fig. 2a).
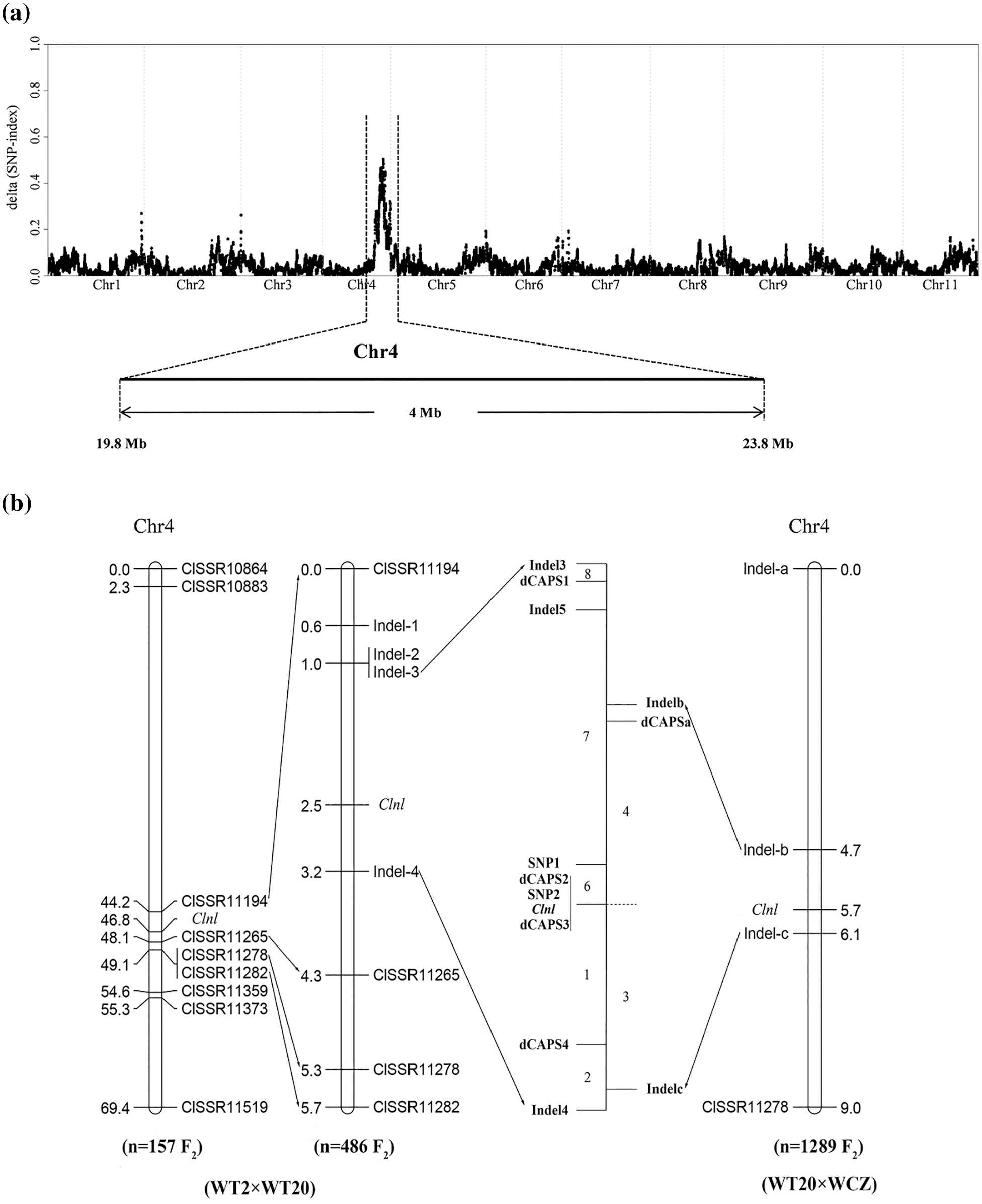
Figure 2: Gene mapping of watermelon no-lobed leaf trait. (a) Watermelon no-lobed leaf was located at an interval of 19.8–23.8 Mb on chromosome 4 through BSA-seq. (b) Linkage analysis based on different individuals of two F2 populations (WT2 × WT20 F2, WCZ × WT20 F2). The candidate gene was mapped between the SNP1 and dCAPS4 markers on chromosome 4
3.3 Fine Mapping of the Clnl Gene
To check the BSA-seq results and further fine-map of the candidate gene of watermelon lobed leaves, two F2 populations (WT2 × WT20 F2 and WCZ × WT20 F2) were used for linkage analysis and fine-mapping. The polymorphism SSR markers were screened on chromosome 4 among the parents according to genome-wide SSR markers identified in the watermelon reference genome [47]. A total of nine SSR markers (Table S2) were clearly polymorphic among the three parents (WT2, WT20, and WCZ) and were used for polymorphic analysis in 157 individuals of the WT2 × WT20 F2 population. The Clnl gene was mapped to 3.9 cM genetic distance between ClSSR11194 and ClSSR11265 (Fig. 2b), which was consistent with the BSA-seq result.
To further narrow the candidate region of the Clnl gene, four Indel markers (Table S2) with significant polymorphisms were developed between ClSSR11194 and ClSSR11265 makers according to the re-sequencing data of the parents. A total of 486 individuals of the WT2 × WT20 F2 population were used for fine-mapping. The Clnl gene was mapped between Indel3 and Indel4. Furthermore, another four Indel, five dCAPS, and two SNP markers (Table S2) were developed and used for polymorphic analysis in 486 individuals of the WT2 × WT20 F2 population. To further narrow the predicted region using more F2 individuals, 1289 individuals from the WCZ × WT20 F2 population of three cultivated seasons (spring and autumn of 2020 and spring of 2021) were also used for linkage analysis and fine-mapping based on the above markers. The linkage analysis from two F2 populations (WT2 × WT20 F2 and WCZ × WT20 F2) showed that the Clnl gene was located between SNP1 and dCAPS4 markers, with a genetic distance of 1.4 cM. According to the physical position of the markers, the Clnl gene was fine-mapped to the 61.5 kb region between 21,224,481 and 21,285,957 bp on watermelon chromosome 4 (Fig. 2b).
3.4 Identification of the Clnl Gene
According to the annotation database of the watermelon reference genome ‘97103’ v1 (http://cucurbitgenomics.org/), four functional genes (Cla018360, Cla018361, Cla018362, Cla018363) were annotated in the candidate region. The sequences of these four genes were amplified and aligned among the three parents. The sequences of the three genes were consistent among three parents, while Cla018360 with three exons and total 2387 bp DNA sequence had sequence differences among the three parents. A single-base deletion (A/-) at the 391st base of Cla018360 CDS only existed in the no-lobed leaf watermelon WT20 (Fig. 3). The deletion resulted in premature translation termination in WT20 (Fig. 3b). The amino acid sequence of Cla018360 was 233 aa in WT2, WCZ, and reference genome ‘97103’, while it was only 158 aa in WT20 (Fig. 3c). The function annotation from the watermelon reference genome and BLAST-P search in the Arabidopsis database (https://www.arabidopsis.org/) showed that Cla018360 belongs to the HD-Zip gene family, which has been reported to regulate leaf development [41,56]. Therefore, we further suggested that Cla018360 was the candidate gene controlling the lobed/no-lobed leaf phenotype in watermelon.
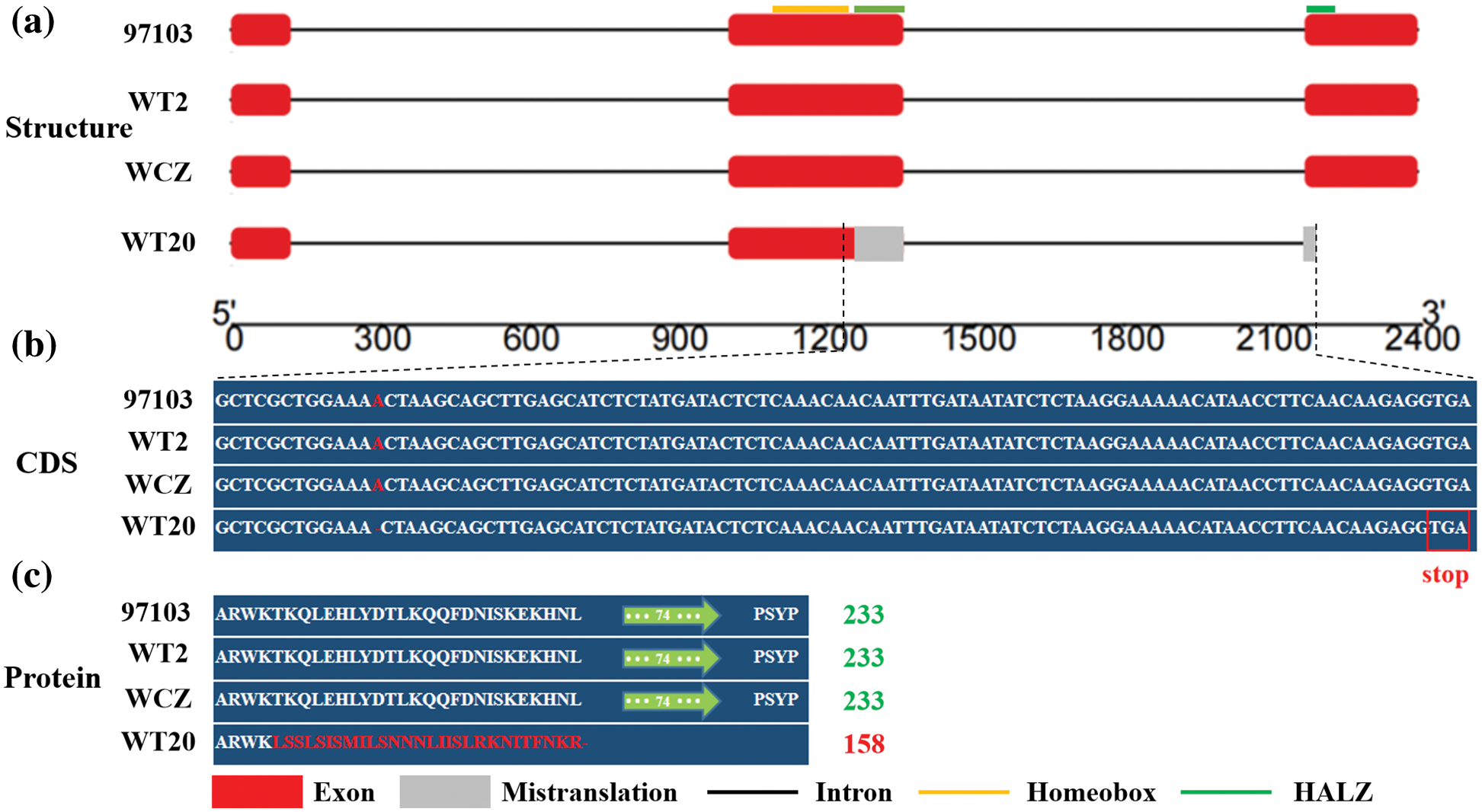
Figure 3: Identification of the candidate gene Cla018360. (a) Structure analysis of the Cla018360 gene. Red boxes represent exons, while black lines denote introns; gray box indicates that the single-base deletion (A/-) results in premature translation terminate in WT20. (b) Sequence information of the Cla018360 gene among three parents and the reference genome. (c) The amino acid sequence difference of Cla018360 among the three parents and the reference genome
To validate the candidate gene, we selected 100 F2 individuals (randomly selected in different years) for testing for polymorphisms according to the co-segregation marker of Cla018360. The results showed that all 77 lobed leaf individuals were dominant (24 individuals for homogeneous and 53 individuals for heterozygous dominance), and 23 no-lobed leaf individuals were homozygous recessive, which was consistent with the phenotypes. Furthermore, 94 lobed leaf watermelon germplasm resources [44] were used for analysis of the sequence difference of Cla018360. The results showed that there was no mutation of Cla018360 in all 94 accessions (Table S3), which indicated that the normal Cla018360 may regulate watermelon lobed leaves. These results further indicate that the Cla018360 gene is the candidate gene Clnl controlling watermelon lobed/no-lobed leaves, and mutation of Cla018360 CDS may result in a no-lobed leaf phenotype in watermelon.
3.5 Cla018360 Was Highly Expressed in Young Leaves
The expression patterns of Cla018360 were investigated via qRT-PCR in different tissues (root, stem, male flower, female flower, young leaves, and mature leaves) between WT2 and WT20. The specific primers of Cla018360 and the reference gene Actin are listed in Table S4. The results showed that the expression level of Cla018360 in lobed leaf watermelon WT2 was higher than in no-lobed leaf watermelon WT20. The young leaves had the highest expression among all tested tissues, with a significant difference between WT2 and WT20 (Fig. 4). The lobed/no-lobed leaves can be distinguished at the watermelon seedling stage. This corresponded to the expression pattern of young leaves, suggesting that the Cla018360 gene plays an important role in the leaf formation stage.
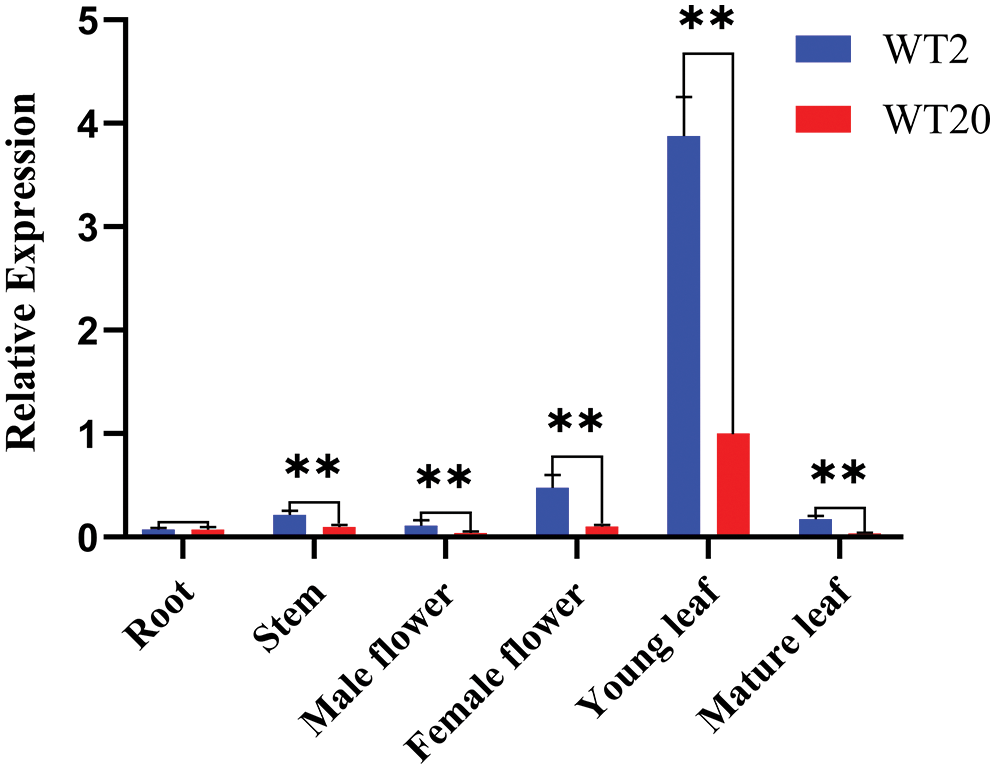
Figure 4: Expression analysis of the Cla018360 gene in watermelon tissues
3.6 Phylogenetic Analysis of Cla018360
To investigate the classification and function and to gain insights into the potential function of the HD-Zip gene family, 102 HD-Zip proteins from Arabidopsis thaliana, S.lycopersicum, O. sativa, Cucumis sativus, Cucumis melo, and Citrullus lanatus were identified for phylogenetic analysis (Table S5). The phylogenetic tree was constructed according to amino acid sequences of all 102 HD-Zip proteins, which were divided into four groups (I, II, III, and IV) (Fig. 5). There were similar numbers of genes in the four groups. The gene Cla018360 was clustered into group I according to phylogenetic analysis and had a close genetic relationship with AtHB51, which has been reported to regulate the formation of leaf shape in Arabidopsis [14,56]. These results further suggested that the Cla018360 gene may play an important role in regulating lobed/no-lobed leaf formation in watermelon.
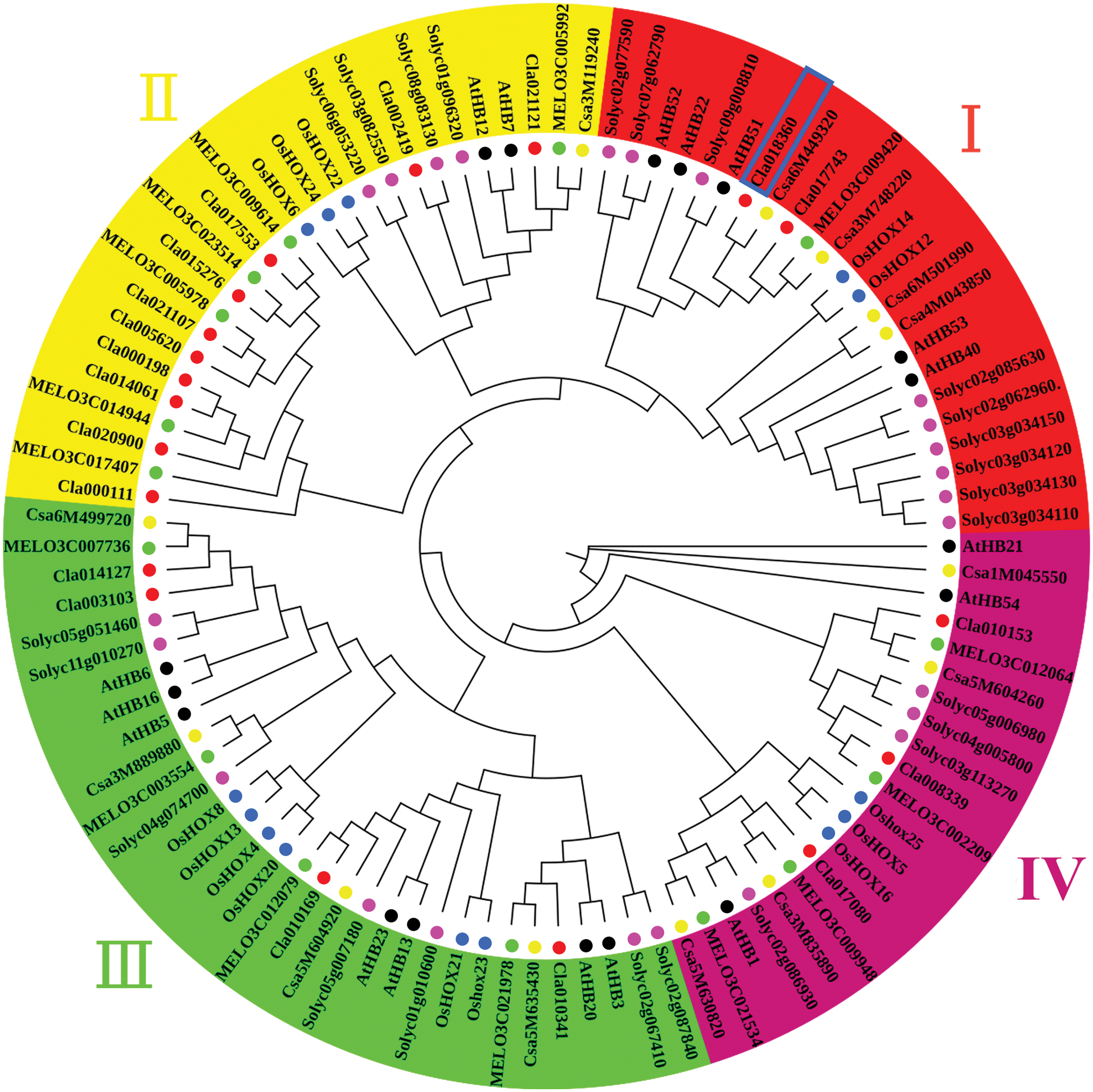
Figure 5: Phylogenetic tree analysis of Cla018360 among Citrullus lanatus, Cucumis melo, Cucumis sativus, O. sativa L, Lycopersicon esculentum, and A. thaliana. I, II, III, and IV represent the four sub-groups
3.7 RNA-Seq Analysis and qRT-PCR Validation
To explore the regulatory pathway by which the Clnl gene regulates the formation of leaf shape in watermelon, according to the expression pattern of Clnl in different tissues, the young leaves from WT2 and WT20 at the three-leaf period were used for RNA-seq. The expression profiles of 333 genes were more than two-fold between WT2 and WT20, of which 115 genes were upregulated, and 218 genes were downregulated in no-lobed leaf watermelon WT20 (Table S7). We selected 23 transcription factors from 333 differential expressed genes to create the heatmap (Fig. 6), of which nine transcription factors were upregulated, and 14 transcription factors were downregulated in no-lobed leaf watermelon WT20. Furthermore, eight genes, which were reported to regulate leaf development, were selected to validate the expression level between lobed leaf watermelon WT2 and no-lobed leaf watermelon WT20 using the qRT-PCR (Fig. 7). The results showed that ClERF5, ClADP, ClCUC2, ClKNAT, and TCP-like genes had higher expression level in WT20 compared with WT2, in conformity with the RNA-seq results. In contrast, the transcript levels of two HP-zip genes and the ClCUC3 gene were more highly expressed in WT2 compared with WT20, similar to the RNA-seq data. These results indicate that the Clnl gene and other leaf development genes together regulate the formation of watermelon leaf shape.
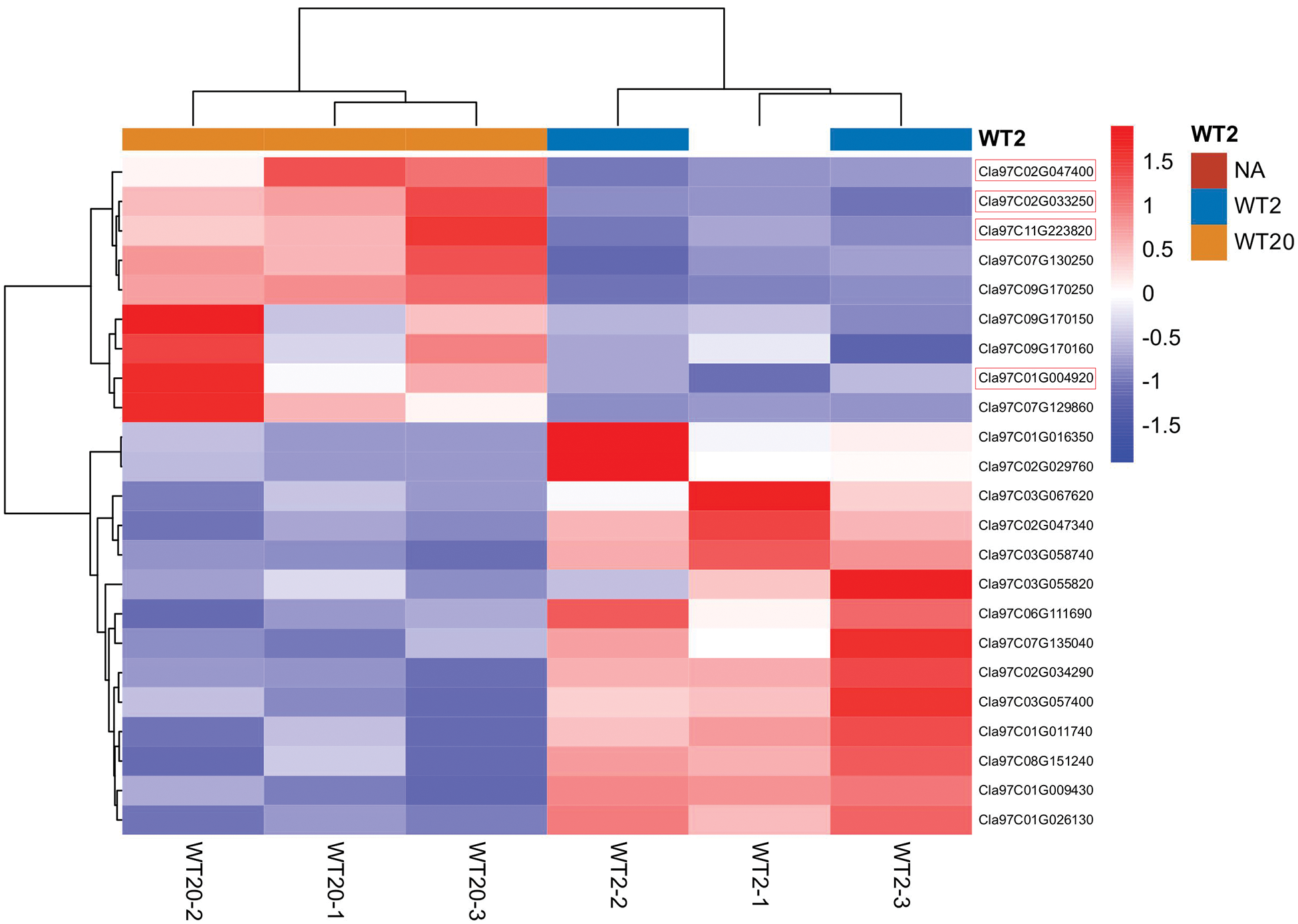
Figure 6: Heatmap analysis of transcription factors between WT2 and WT20 according to RNA-seq. Three replicates of each sample were analyzed
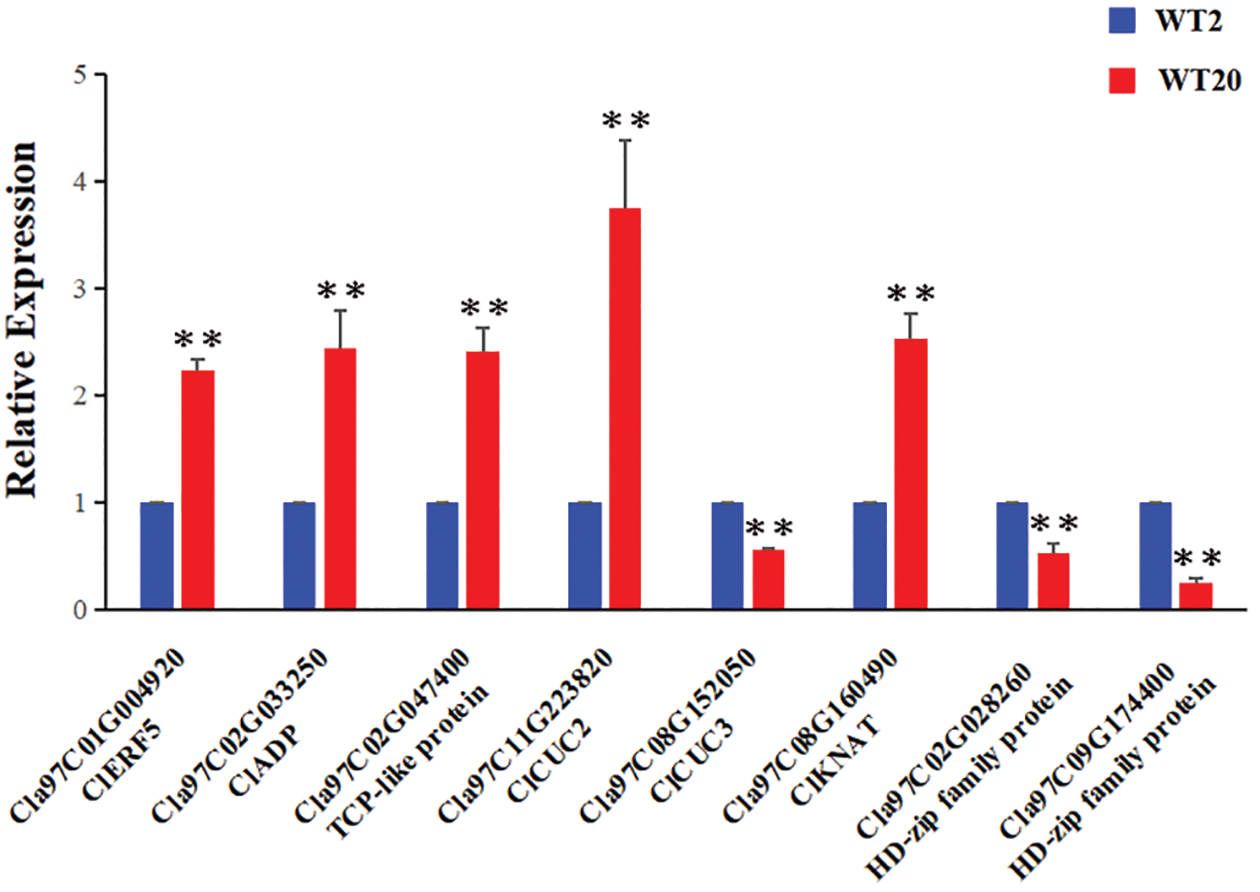
Figure 7: Expression analysis of genes related to leaf development between WT2 and WT20
Unlike other Cucurbitaceae with diverse leaf shapes, most watermelon varieties are lobed leaf shapes in the current market. There are few accessions with an entire (no-lobed) leaf shape among existing watermelon germplasm resources. In the present study, the no-lobed leaf accession WT20 was collected and preserved from our lab, which had been manually self-pollinated for at least five generations (Fig. 1). WT20 was the ideal material for researching watermelon leaf shape based on its stable inheritance without trait segregation during cultivation. Two F2 populations from WT20 were constructed by crossing with WT2 and WCZ, respectively. Segregation in two F2 populations exhibited a ratio of three lobed leaf individuals to one no-lobed leaf individual (Table S1), which indicated that watermelon lobed leaf was a qualitative trait and regulated by a single dominant gene. Inheritance in this study was also the same as in previous studies in cotton [26,57,58], B. napus [20], B. juncea [59], and watermelon [41]. In turn, there are reports showing that lobed leaves are controlled by a single recessive gene in melon [2] and Cucurbita maxima [31]. These studies support that the regulatory pathway and molecular mechanisms of leaf shape are probably diverse in different crops.
With the release of the watermelon reference genome, gene mapping and cloning was accelerated using the next-generation sequencing (NGS) technology, and the efficiency of genotyping was also improved. NGS-assisted BSA provides an effective and simple method to identify molecular markers linked to target genes/QTLs. Based on BSA-seq technology, several genes in watermelon have been mapped and cloned, such as the ClFS1 gene controlling fruit shape [60], the ClCGMenG gene controlling rind color [61], the ClERF4 gene controlling rind hardness [62], and the Cldf gene controlling plant dwarfism [38]. In addition, according to the watermelon reference genome ‘97103’ v1, 32,869 SSR markers were developed for polymorphism and linkage analysis [47]. In this study, BSA-seq technology combined with linkage analysis were used for mapping the Clnl gene, and the candidate gene was delimited in the 61.5 kb region between 21,224,481 and 21,285,957 bp on watermelon chromosome 4 (Fig. 2). Sequence alignment among three parents (WT2, WT20, WCZ) showed that the sequence of Cla018360 CDS represented a single-base deletion (A/-) at the second exon, which resulted in premature translation termination in WT20 (no-lobed leaf) with only 158 amino acids (Fig. 3). Function annotation showed that the Cla018360 gene was a member of the HD-Zip transcription factor family, and the single-base deletion (A/-) occurred in the HD-Zip domain. Furthermore, 100 F2 individuals and 94 lobed leaf watermelon germplasm resources from Guo et al. [49] were demonstrated to co-segregate with the Cla018360 gene (Table S3), which showed that the Cla018360 gene is the candidate gene Clnl for lobed leaves in watermelon.
The homeodomain leucine zipper (HD-Zip) is a large family of transcription factors present in plants that is composed of 60–61 conserved amino acids. HD-Zip proteins exhibit a leucine zipper structure through folding into a three α-helix structure with the singular combination of a homeodomain [63,64]. The important roles of HD-Zip transcription factors in abiotic stress, sugar signaling, de-etiogenesis, and plant embryogenesis have been intensively studied [64]. In most researches, HD-Zip has also been shown to regulate the development of leaves. In Arabidopsis, the HD-Zip protein plays an important role in regulating leaf polarity. The AtHB16 gene, which encodes HD-Zip protein, participates in leaf cell expansion [65]. As target genes of the HD-Zip transcription factor, the mutation of miR165 and miR166 could result in the gain-of-function of PHB and PHV genes, which indicate their important roles in meristem maintenance and leaf polarity determination [66–68]. In cotton, HD-Zip transcription factor GhOKRA has been found to control the formation of deeply lobed leaves, while the GhOKRA mutation results in the formation of broad leaves [1,26,47]. In addition, there are many reports about the regulation of HD-Zip transcription factors in leaf shape [7,15,16]. Phylogenetic analysis showed that Clnl has a close genetic relationship with AtHB51 (Fig. 5), which has been reported to regulate the formation of leaf shape in Arabidopsis [14,56]. Expression pattern analysis showed that the expression level of Clnl in lobed leaf watermelon was higher than that of in no-lobed leaf watermelon, with the highest expression level in young leaves (Fig. 4). The leaf shape of watermelon can be distinguished at the seedling stage; the expression pattern implied that the regulation of the Clnl gene occurred in the seedling period.
Genetic variants started from gene mutations with extremely low frequency, which appear in an individual from a specific population. Then, the variants or mutations were transformed into common ones during the evolutionary process through migration, selection, and genetic drift [40]. In watermelon, genome speciation events occurred between 15 and 23 million years ago [35], and wild watermelon originated in the desert area, which exhibited the phenotype of a deeply lobed leaf [49]. We speculated that the Clnl gene may occur naturally in the long-term process of evolution and domestication, and the variation was preserved through artificial selection. After many generations of selection and purification, the locus of the no-lobed leaf was preserved and inherited stably. In this study, it was identified that the Clnl gene plays an important role in the development of watermelon leaf shape. RNA-seq showed that HD-Zip genes and ClCUC3 gene, which are related to leaf margin development, were more expressed in lobed leaf watermelon, while ClERF5, ClADP, ClCUC2, ClKNAT, and TCP-like genes were more expressed in no-lobed leaf watermelon (Figs. 6 and 7). We speculated that the formation of watermelon leaf shape is a complex process and regulated by multiple genes. However, the molecular mechanisms and regulatory network of the Clnl gene in the formation of lobed/no-lobed leaves need further investigation. The phenotype of a no-lobed leaf can be used as the morphological marker for cultivar identification and improved breeding efficiency. This study provides a good entry point for studying the development of leaves, as well as providing foundational insights into breeding for special plant architecture in watermelon.
Funding Statement: This research was supported by the National Natural Science Foundation of China (32102389, 32172602), the Zhongyuan Youth Talent Support Program (ZYQR201912161), the Program for Science & Technology Innovation Talents in Universities of Henan Province (21HASTIT038), the Funding of Joint Research on Agricultural Varietie Improvement of Henan Province (2022010503), the Major Science and Technology Project of Henan Province (221100110400), and the Science and Technology Innovation Fund of Henan Agricultural University (KJCX2021A14).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Zhu HY, Dou JL; data collection: Duan SX, Guo YM, Yang S, Umer MJ; analysis and interpretation of results: Liu DM, Niu HH, Sun SR; draft manuscript preparation: Duan SX, Wang YP, Yang LM. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Andres, R. J., Bowman, D. T., Kaur, B., Kuraparthy, V. (2014). Mapping and genomic targeting of the major leaf shape gene (L) in Upland cotton (Gossypium hirsutum L.). Theoretical and Applied Genetics, 127, 167–177. [Google Scholar] [PubMed]
2. Gao, X., Ning, X., Wang, Y., Wang, X., Yan, W. et al. (2014). Fine mapping of a gene that confers palmately lobed leaf (pll) in melon (Cucumis melo L.). Euphytica, 200, 337–347. [Google Scholar]
3. Jiao, K., Li, X., Guo, W., Yuan, X., Cui, X. et al. (2016). Genome re-sequencing of two accessions and fine mapping the locus of lobed leaflet margins in mungbean. Molecular Breeding, 36, 128. [Google Scholar]
4. Nicotra, A. B., Leigh, A., Boyce, C. K., Jones, C. S., Niklas, K. J. et al. (2011). The evolution and functional significance of leaf shape in the angiosperms. Functional Plant Biology, 38, 535–552. [Google Scholar] [PubMed]
5. Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annuual Review of Plant Biology, 57, 477–496. [Google Scholar] [PubMed]
6. Kessler, S., Sinha, N. (2004). Shaping up: The genetic control of leaf shape. Current Opinion in Plant Biology, 7(1), 65–72. [Google Scholar] [PubMed]
7. Bo, K., Duan, Y., Qiu, X., Zhang, M., Shu, Q. et al. (2022). Promoter variation in a homeobox gene, CpDll, is associated with deeply lobed leaf in Cucurbita pepo L. Theoretical and Applied Genetics, 135(4), 1223–1234. [Google Scholar] [PubMed]
8. Tsukaya, H. (2005). Leaf shape: Genetic controls and environmental factors. International Journal of Developmental Biology, 49, 547–555. [Google Scholar] [PubMed]
9. Ni, X., Huang, J., Ali, B., Zhou, W., Zhao, J. (2015). Genetic analysis and fine mapping of the LOBED-LEAF 1 (BnLL1) gene in rapeseed (Brassica napus L.). Euphytica, 204, 29–38. [Google Scholar]
10. Rodriguez, R. E., Debernardi, J. M., Palatnik, J. F. (2014). Morphogenesis of simple leaves: Regulation of leafsize and shape. Wiley Interdisciplinary Reviews: Developmental Biology, 3(1), 41–57. [Google Scholar] [PubMed]
11. Dengler, N. G., Tsukaya, H. (2001). Leaf morphogenesis in dicotyledons: Current issues. International Journal of Plant Sciences, 162(3), 459–464. [Google Scholar]
12. Hibara, K. I., Karim, M. R., Takada, S., Taoka, K. I., Furutani, M. et al. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristemand organ boundary formation. The Plant Cell, 18(11), 2946–2957. [Google Scholar] [PubMed]
13. Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H. et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. The Plant Cell, 18, 2929–2945. [Google Scholar] [PubMed]
14. Saddic, L. A., Huvermann, B., Bezhani, S., Su, Y., Winter, C. M. et al. (2006). The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of Cauliflower. Development, 133(9), 1673–1682. [Google Scholar] [PubMed]
15. Hofer, J., Turner, L., Moreau, C., Ambrose, M., Isaac, P. et al. (2009). Tendril-less regulates tendril formation in pea leaves. The Plant Cell, 21, 420–428. [Google Scholar] [PubMed]
16. Vlad, D., Kierzkowski, D., Rast, M. I., Vuolo, F., Ioio, R. D. et al. (2014). Leaf shape evolution through uplication, regulatory diversification, and loss of a homeobox gene. Science, 343, 780–783. [Google Scholar] [PubMed]
17. Jasinski, S., Kaur, H., Tattersall, A., Tsiantis, M. (2007). Negative regulation of KNOX expression in tomato leaves. Planta, 226, 1255–1263. [Google Scholar] [PubMed]
18. Hay, A., Tsiantis, M. (2009). A KNOX family TALE. Current Opinion in Plant Biology, 12, 593–598. [Google Scholar] [PubMed]
19. Shani, E., Ben-Gera, H., Shleizer-Burko, S., Burko, Y., Weiss, D. et al. (2010). Cytokinin regulates compound leaf development in tomato. The Plant Cell, 22, 3206–3217. [Google Scholar] [PubMed]
20. Ni, X., Liu, H., Huang, J., Zhao, J. (2017). LMI1-like genes involved in leaf margin development of Brassica napus. Genetica, 145, 269–274. [Google Scholar] [PubMed]
21. Hu, L., Zhang, H., Sun, Y., Shen, X., Amoo, O. et al. (2020). BnA10.RCO, a homeobox gene, positively regulates leaf lobed formation in Brassica napus L. Theoretical and Applied Genetics, 133, 3333–3343. [Google Scholar] [PubMed]
22. Hu, L., Zhang, H., Yang, Q., Meng, Q., Han, S. et al. (2018). Promoter variations in a homeobox gene, BnA10.LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theoretical and Applied Genetics, 131, 2699–2708. [Google Scholar] [PubMed]
23. Ren, J., Liu, Z., Du, J., Fu, W., Hou, A. et al. (2019). Fine mapping of a gene for the lobed leaf, BoLl, in ornamental kale (Brassica oleracea L. var. acephala). Molecular Breeding, 39, 40. [Google Scholar]
24. Zhang, B., Chen, W., Li, X., Ren, W., Chen, L. et al. (2021). Map-based cloning and promoter variation analysis of the lobed leaf gene BoL-MI1a in ornamental kale (Brassica oleracea L. var. acephala). BMC Plant Biology, 21, 456. [Google Scholar] [PubMed]
25. Feng, X., Li, X., Yang, X., Zhu, P. (2020). Fine mapping and identification of the leaf shape gene BoFL in ornamental kale. Theoretical and Applied Genetics, 133, 1303–1312. [Google Scholar] [PubMed]
26. Chang, L., Mei, G., Hu, Y., Deng, J., Zhang, T. (2019). LMI1-like and KNOX1 genes coordinately regulate plant leaf development in dicotyledons. Plant Molecular Biology, 99, 449–460. [Google Scholar] [PubMed]
27. Zhou, C., Han, L., Fu, C., Wen, J., Cheng, X. et al. (2013). The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. The Plant Cell, 25, 4845–4862. [Google Scholar] [PubMed]
28. Zhao, B., He, L., Jiang, C., Liu, Y., He, H. et al. (2020). Lateral leaflet Suppression 1 (LLS1encoding the MtYUCCA1 protein, regulates lateral leaflet development in Medicago truncatula. New Phytologists, 227, 613–628. [Google Scholar] [PubMed]
29. Yu, C., Yan, C., Liu, Y., Liu, Y., Jia, Y. et al. (2020). Upregulation of a KN1 homolog by transposon insertion promotes leafyhead development in lettuce. Proceedings of the National Academy of Sciences of the United States of America, 117(52), 33668–33678. [Google Scholar] [PubMed]
30. Wang, M., Lavelle, D., Yu, C., Zhang, W., Cheng, J. et al. (2022). The upregulated LsKN1 gene transforms pinnately to palmately lobed leaves through auxin, gibberellin, and leaf dorsiventrality pathway. Plant Biotechnology Journal, 20, 1756–1769. [Google Scholar] [PubMed]
31. Dyutin, K. E. (1980). Spontaneous mutant of Cucurbita maxima Duch. squash with lobed leaves. Genetika, 16, 176–178. [Google Scholar]
32. Herrington, M. E., Brown, P. J. (1988). Inheritance of leaf and fruit characteristics in Cucurbita maxima Duch. Cv Queensland Blue × C. ecuadorensis Cutler and Whitaker. Queensl Journal of Agricultural & Animic Science, 45, 45–48. [Google Scholar]
33. Ganesan, J., Sambandam, C. N. (1985). Inheritance of leaf shape in muskmelon (Cucumis melo L.) I. A qualitative approach. Annamalai University Agricultural Research Annual, 12, 53–58. [Google Scholar]
34. Montero-Pau, J., Blanca, J., Esteras, C., Martínez-Pérez, E. M., Gómez, P. et al. (2017). An SNP-based saturated genetic map and QTL analysis of fruit-related traits in Zucchini using genotyping-by-sequencing. BMC Genomics, 18, 94. [Google Scholar] [PubMed]
35. Guo, S., Zhang, J., Sun, H., Salse, J., Lucas, W. J. et al. (2013). The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nature Genetics, 45, 51–58. [Google Scholar] [PubMed]
36. Wu, S., Wang, X., Reddy, U., Sun, H., Bao, K. et al. (2019). Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the US National Plant Germplasm System watermelon collection. Plant Biotechnology Journal, 17, 2246–2258. [Google Scholar] [PubMed]
37. Gebremeskel, H., Dou, J., Li, B., Zhao, S., Muhammad, U. et al. (2020). Molecular mapping and candidate gene analysis for GA3 responsive short internode in watermelon (Citrullus lanatus). International Journal of Molecular Sciences, 21, 290. [Google Scholar]
38. Wei, C., Zhu, C., Yang, L., Zhao, W., Ma, R. et al. (2019). A point mutation resulting in a 13 bp deletion in the coding sequence of Cldf leads to a GA-deficient dwarf phenotype in watermelon. Horticulture Research, 6, 132. [Google Scholar] [PubMed]
39. Zhu, H., Zhang, M., Sun, S., Yang, S., Li, J. et al. (2019). A single nucleotide deletion in an ABC transporter gene leads to a dwarf phenotype in watermelon. Frontiers in Plant Science, 10, 1399. [Google Scholar] [PubMed]
40. Dou, J., Yang, H., Sun, D., Yang, S., Sun, S. et al. (2022). The branchless gene Clbl in watermelon encoding a TERMINAL FLOWER1 protein regulates the number of lateral branches. Theoretical and Applied Genetics, 135, 65–79. [Google Scholar] [PubMed]
41. Wei, C., Chen, X., Wang, Z., Liu, Q., Li, H. et al. (2017). Genetic mapping of the LOBED LEAF 1 (ClLL1) gene to a 127.6-kb region in watermelon (Citrullus lanatus L.). PLoS One, 12(7), e0180741. [Google Scholar] [PubMed]
42. Porebski, S., Bailey, L., Baum, B. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter, 15, 8–15. [Google Scholar]
43. Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14), 1754–1760. [Google Scholar] [PubMed]
44. Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J. et al. (2009). The sequence alignment/map format and SAM tools. Bioinformatics, 25(16), 2078–2079. [Google Scholar] [PubMed]
45. Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H. et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology, 30, 174–178. [Google Scholar] [PubMed]
46. Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S. et al. (2013). QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. The Plant Journal, 74, 174–183. [Google Scholar] [PubMed]
47. Zhu, H., Song, P., Koo, D. H., Guo, L., Li, Y. et al. (2016). Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genomics, 17, 557. [Google Scholar] [PubMed]
48. Neff, M. M., Neff, J. D., Chory, J., Pepper, A. E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. The Plant Journal, 14(3), 387–392. [Google Scholar] [PubMed]
49. Guo, S., Zhao, S., Sun, H., Wang, X., Wu, S. et al. (2019). Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nature Genetics, 51, 1616–1623. [Google Scholar] [PubMed]
50. Kong, Q., Yuan, J., Gao, L., Zhao, L., Cheng, F. et al. (2015). Evaluation of appropriate reference genes for gene expression normalization during watermelon fruit development. PLoS One, 10, e0130865. [Google Scholar] [PubMed]
51. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [Google Scholar] [PubMed]
52. Shi, P., Guy, K. M., Wu, W., Fang, B., Yang, J. et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biology, 16, 85. [Google Scholar] [PubMed]
53. Liu, D., Yang, H., Yuan, Y., Zhu, H., Zhang, M. et al. (2020). Comparative transcriptome analysis provides insights into yellow rind formation and preliminary mapping of the Clyr (yellow rind) gene in watermelon. Frontiers in Plant Science, 11, 192. [Google Scholar] [PubMed]
54. Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. [Google Scholar] [PubMed]
55. Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcriptlevel expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature Protocols, 11, 1650–1667. [Google Scholar] [PubMed]
56. Sluis, A., Hake, S. (2015). Organogenesis in plants: Initiation and elaboration of leaves. Trends in Genetics, 31(6), 300–306. [Google Scholar] [PubMed]
57. Andres, R. J., Coneva, V., Frank, M. H., Tuttle, J. R., Samayoa, L. F. et al. (2017). Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of upland cotton (Gossypium hirsutum L.). Proceedings of the National Academy of Sciences of the United States of America, 114(1), 57–66. [Google Scholar]
58. He, D., Zhao, X., Liang, C., Zhu, T., Abid, M. A. et al. (2018). Genetic variation in LBL1 contributes to depth of leaf blades lobes between cotton subspecies, Gossypium barbadense and Gossypium hirsutum. Journal of Integrative Agriculture, 17(11), 2394–2404. [Google Scholar]
59. Heng, S., Huang, H., Cui, M., Liu, M., Lv, Q. et al. (2020). Rapid identification of the BjRCO gene associated with lobed leaves in Brassica juncea via bulked segregant RNA-seq. Molecular Breeding, 40, 42. [Google Scholar]
60. Dou, J., Zhao, S., Lu, X., He, N., Zhang, L. et al. (2018). Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theoretical and Applied Genetics, 131, 947–958. [Google Scholar] [PubMed]
61. Li, B., Zhao, S., Dou, J., Ali, A., Gebremeskel, H. et al. (2019). Genetic mapping and development of molecular markers for a candidate gene locus controlling rind color in watermelon. Theoretical and Applied Genetics, 132, 2741–2753. [Google Scholar] [PubMed]
62. Liao, N., Hu, Z., Li, Y., Hao, J., Chen, S. et al. (2020). Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biotechnology Journal, 18, 1066–1077. [Google Scholar] [PubMed]
63. Ariel, F. D., Manavella, P. A., Dezar, C. A., Chan, R. L. (2007). The true story of the HD-Zip family. Trends in Plant Science, 12(9), 419–426. [Google Scholar] [PubMed]
64. Liu, W., Fu, R., Li, Q., Li, J., Wang, L. et al. (2013). Genome-wide identification and expression profile of homeodomain-leucine zipper Class I gene family in Cucumis sativus. Gene, 531(2), 279–287. [Google Scholar] [PubMed]
65. Wang, Y., Henriksson, E., Söderman, E., Henriksson, K. N., Sundberg, E. et al. (2003). The Arabidopsis homeobox gene, ATHB16, regulates leaf development and the sensitivity to photoperiod in Arabidopsis. Developmental Biology, 264, 228–239. [Google Scholar]
66. Emery, J. F., Floyd, J. S., Alvarez, K., Eshed, Y., Hawker, N. P. et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-Zip and KANADI genes. Current Biology, 13, 1768–1774. [Google Scholar] [PubMed]
67. McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J. et al. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature, 411, 709–713. [Google Scholar] [PubMed]
68. Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Clark, D. S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. The Plant Cell, 17, 61–76. [Google Scholar] [PubMed]
Appendix
Table S1: The segregation ratio of lobed leaf and no-lobed leaf individuals among different populations (WT20 × WT2) and (WT20 × WCZ)
Table S2: Information on the remaining important markers on chromosome 4 used to analyze the polymorphic
Table S3: Watermelon accessions from Guo et al. [49] for validating the candidate gene
Table S4: Information on qRT-PCR markers
Table S5: Detailed information on phylogenetic analysis in different varieties
Table S6: Detailed statistics of RNA-seq data for two parental lines, WT2 and WT20
Table S7: Genes with differential expression in RNA-seq
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools