 Open Access
Open Access
ARTICLE
Cloning and Function Identification of a Phytoene Desaturase Gene from Eucommia ulmoides
1 The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), College of Tea Sciences, College of Life Sciences, Guizhou University, Guiyang, 550000, China
2 Plant Conservation Technology Center, Guizhou Key Laboratory of Agricultural Biotechnology, Guizhou Academy of Agricultural Sciences, Guiyang, 550000, China
* Corresponding Authors: Yichen Zhao. Email: ; Degang Zhao. Email:
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(5), 1377-1389. https://doi.org/10.32604/phyton.2023.026830
Received 27 September 2022; Accepted 28 November 2022; Issue published 09 March 2023
Abstract
The phytoene desaturase (PDS) encodes a crucial enzyme in the carotenoid biosynthesis pathway. Silencing or inhibiting PDS expression leads to the appearance of mottled, chlorosis, or albino leaves. In this study, the CDS sequence of EuPDS (Eucommia ulmoides Phytoene Desaturase) was first cloned and then PDS was silenced in Nicotiana benthamiana. Result showed the expression level of EuPDS in leaves was higher than that in the roots and stems. In N. benthamiana leaves, which were treated by Agrobacterium for 24 h, photo-bleaching was shown on the fresh leaves one week after injection and the transcript level of PDS was down-regulated during the period of emersion. This suggested that EuPDS could silence PDS of N. benthamiana, so as to cause the phenotype of leaf whitening. PDS is the main reporter gene involved in virus-induced gene silencing (VIGS). This study offered molecular evidence for identifying PDS gene involved in Carotenoid’s biosynthesis pathway and the regulation networks in E. ulmides. It also laid a useful foundation for study on leaf discoloration mechanism of other woody plants.Keywords
Eucommia ulmoides Oliver, a medicinal edible tonic herb used in traditional chinese medicine (TCM) [1], is a single Eucommiagenus species of the Eucommiaceae family [2,3]. The results demonstrated that secondary metabolites such as lignin, naphthenes, phenylpropane, polysaccharides, flavonoids, amino acids and gutta-percha were of great importance in the pharmacological effects [4–6]. Recent pharmacological researches indicated that E. ulmoides has anti-aging, anti-cancer, bone cell proliferation, cardiovascular and immune control functions [7–9]. In recent years, Agrobacterium-mediated regenerative transformation has been successfully applied in E. ulmoides [10]. However, because of taking root and endophytic fungal contamination problems, it is dificult to obtain transgenic plants and to verify the gene function of E. ulmoides, which requires the development of genetic transformation strategies that bypass their inherent barriers.
Virus-induced gene silencing (VIGS), associated with post-transcriptional gene silencing, is a useful tool to protect plants and other organisms from infections that are naturally present [11]. Double-stranded RNA (dsRNA), the viral vectors with unique exogenous genes, is involved in VIGS. In plants, RNA interference (RNAi) treated by dsRNA are processed by Dicer-like nuclease, the dsRNA degraded into small interfering RNA (siRNAs) ranging in length from 21 to 25 nt. These plant siRNAs reduced the corresponding targeted mRNAs genes, resulting in its degradation [11]. VIGS vectors originated from tobacco rattle virus (TRV) are widely used because of their distinct symptoms of infection, the ability to silently clone fragments and infect meristems [12].
Carotenoids are significant pigments that contribute to the variety of colors found in plant organs. The different colors of pigment match known pigments, which make up the red, green, and yellow colors of petals. Carotenoids are lipid-soluble pigments produced by photosynthetic organs, which vary in solubility depending on the species and environment [13]. Currently, the major carotenoids involved in photosynthesis are the lutein and carotene [14]. Phytoene desaturase (PDS) is the first discovered enzyme in the plant carotenoid biosynthesis pathway. PDS encodes a phytoene desaturase, which catalyzes a critical step in the carotenoid biosynthesis pathway, thus, recognized as an indicator for VIGS [15]. The silent phenotype of VIGS-PDS was utilized as a visual indicator of a VIGS efficacy in numerous plants, such as tomato [16], Forsythia [17] and Nicotiana benthamiana [18]. In the color of flowers or fruits, it has been catalyzed the synthesis colorless of colored carotenoids and, at the transcriptional level regulates the accumulation of carotenoids [19]. At the moment, PDS has been cloned and reported in a variety of plants, such as SrPDS (Streptocarpus rexii) [20], SmPDS (Solanum melongena) [21] and PhPDS (Petunia hybrida) [22].
Nowadays, the complete sequence of E. ulmoides PDS (EuPDS) is available in the E. ulmoides database [1]. However, the EuPDS gene has not been used till now. Therefore, the purpose of this study was to cause phenotypic function loss in order to better understand gene function and to facilitate a comparative approach. In order to obtain albinized plants and identify the biological function of EuPDS in tobacco plants, the research first cloned the EuPDS’s cDNA sequence and developed the plant viral vector pTRV2-EuPDS, which was injected into tobacco leaves by Agrobacterium-mediated infection. The findings not only establish a platform for further investigation of this gene’s mechanism of color creation, but also give a reporter gene for the construction of an E. ulmoides VIGS system.
2.1 Plant Materials and Reagents
All E. ulmoides seeds were cultivated at the Agricultural Biotechnology Base of Guizhou University (Guizhou, China). The pTRV2-EuPDS constructed in our laboratory is also the repository of A. tumefaciens GV3101 (Guizhou, China). RNA pure Kit and RNA extraction Kit from Kangwei Century (Beijing, China), the SYBR® Select Master Mix from Novoprotein (Suzhou, China), Taq Green PCR Master Mix (2×) and T4 DNA Ligase from Thermo Fisher Scientific (Massachusetts, USA), FastDigest BamHI and FastDigest XbaI (Massachusetts, USA), Acetone from China National Pharmaceutical Group Corporation (Beijing, China), MES from BIOBOMEI (Hefei, China), and Migic Neo Hih-Fidelity DNA from Mei5bio, and the BeyoRT™ III First Strand cDNA Synthesis Master Mix (5×) from Beyotime (Dalian, China) were obtained from the cited locations and countries.
N. benthamiana and E. ulmoides seeds were sown on regular trays with medium: vermiculite (weight/weight, w/w; 4:1) to keep the soil wet. After two weeks, fully cotyledons of tobacco seedlings were transplanted into new pots of potting soil of the same formula.
All work containing virus-infected material was conducted in artificial climate room, at 25°C (±2.0°C). To sustain a 16-h day/8-h night cycle, additional lighting of 3000 LX intensity from metal halide lamps was recruited. The humidity level was 65%.
This study identified the conserved domain of PDS gene from the Pfam (http://pfam.xfam.org/) database, compared the homologous sequence with the amino acid sequence in E. ulmoides database, and successfully found EuPDS gene (ID: GWHTAAAL009905). Total RNA was obtained from E. ulmoides leaves with RNA extraction kit from Kangwei Century and then reversed transcribed to cDNA from Beyotime. Primers were designed based on the PDS sequence of EuPDS from E. ulmoides database. High-fidelity thermostable DNA polymerase was employed to produce one complete fragment. The PDS gene was employed in PCR using E. ulmoides cDNA templates to produce two overlapping fragments that were sequenced. We successfully cloned the CDS sequence of EuPDS. Positive clones of each fragment were identified via DNA sequencing (BGI, Beijing, China).
2.4 VIGS Vector Construction and Agro-Infiltration
The pTRV1 and pTRV2-VIGS vectors were obtained from Bio-Transduction Lab Co., Ltd. (China). This paper made use of one constructed, including pTRV2-EuPDS. EuPDS fragments up to 398 bp in length were amplified from the Eucommia cDNA library using M5 Magic Neo high-quality DNA polymerase. 100 ng of purified PCR product was inactivated by T4 DNA polymerase in 1× buffer containing 5 mM/L dATP at 12°C for 15 min, then at 75°C inactivate for 20 min. Meanwhile, 500 ng of linearized TRV2 (YY13) vector digested with XbaI was treated by dTTP for T4 DNA polymerase (Fig. S1). Transfer 2 μL of the treated TRV2 vector into 3 μL of the treated PCR product and incubate for 10 min at 70°C and 10 min at 22°C. The entire reaction solution was transformed into competent E. coli DH5α cells. Positive clones were determined via PCR amplification and sequencing (Fig. S2).
By the freeze-thaw procedure, the plasmid pTRV2-EuPDS was transformed into Agrobacterium, and positive colonies were found using colony PCR (Fig. S3). Amplification of the PCR product was performed using the signed verification primers (Table 1). The Agrobacterium tumefaciens-mediated infiltration method used was that of Burch-Smith et al. [23]. The Agrobacterium pTRV1 and pTRV2, as well as pTRV1 and TRV-EuPDS, were then mixed 1:1 and cultured at room temperature for 3 h. Leaf infiltration was used to infect the first or second pair of newly emerging leaves.
2.5 Virus-Induced Gene Silencing
The PDS participated in chlorophyll biosynthesis was silenced in N. benthamiana. After a seedling grew six true leaves, the undersides of the leaves were gently rubbed with a 10 µL tip and then inoculated with A. tumefaciens using a 2.5 mL needless syringe. The plant that was not silenced served as controls (infiltrated with A. tumefaciens of empty vectors pTRV1 and pTRV2).
2.6 Virus Detection and Quantitative Real-Time PCR (qPCR)
Plant leaves samples were collected and immediately frozen in liquid nitrogen, and total RNA was obtained from fresh leaf tissue of silenced with the RNA plant extracting Kit. First-strand cDNA was synthesized via the BeyoRT™ III First Strand cDNA Synthesis Master Mix (5×). For RT-PCR, 1 µL of first-strand cDNA was diluted. As a template in a reaction using the KAPA SYBR Quantitative PCR Kit in a Step One Plus Real-Time PCR System. The cDNA was diluted to 5 ng/µL. To determine the presence of TRV, two sets of particular primers were used to amplify pTRV1 and pTRV2 (Table 1). The qPCR was carried out in three biological replicates using the NovoProtein SYBR® Select Master Mix in 10 µL reactions. The following parameters were utilized for amplification: 95°C pre-denaturation for 3 min; 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s and extension at 72°C for 30 s.
2.7 Quantification of Pigments
Place 0.5 g of fresh uninjected tobacco leaves, no-load injection leaves, and leukized fresh leaves in a sterile treated 15 mL centrifuge tube, add 5 mL extract (80% acetone: 95% ethanol = 1:1) and soak in the dark. After two days, the leaves were almost entirely decorated, and the supernatant was collected in a cuvette. Photometric measurements were performed at 662, 645 and 470 nm [24,25].
SPSS21.0 software was used to analyze the qRT-PCR data in all of these experiments. p < 0.05 is considered significant difference. Each group was repeated independently at least three times. Data analysis and sequence analysis were carried out with Excel 2007, and DNAMAN 5.0 and a public website (https://www.ncbi.nlm.nih.gov/) database, respectively.
3.1 Cloning of EuPDS Gene in E. ulmoides
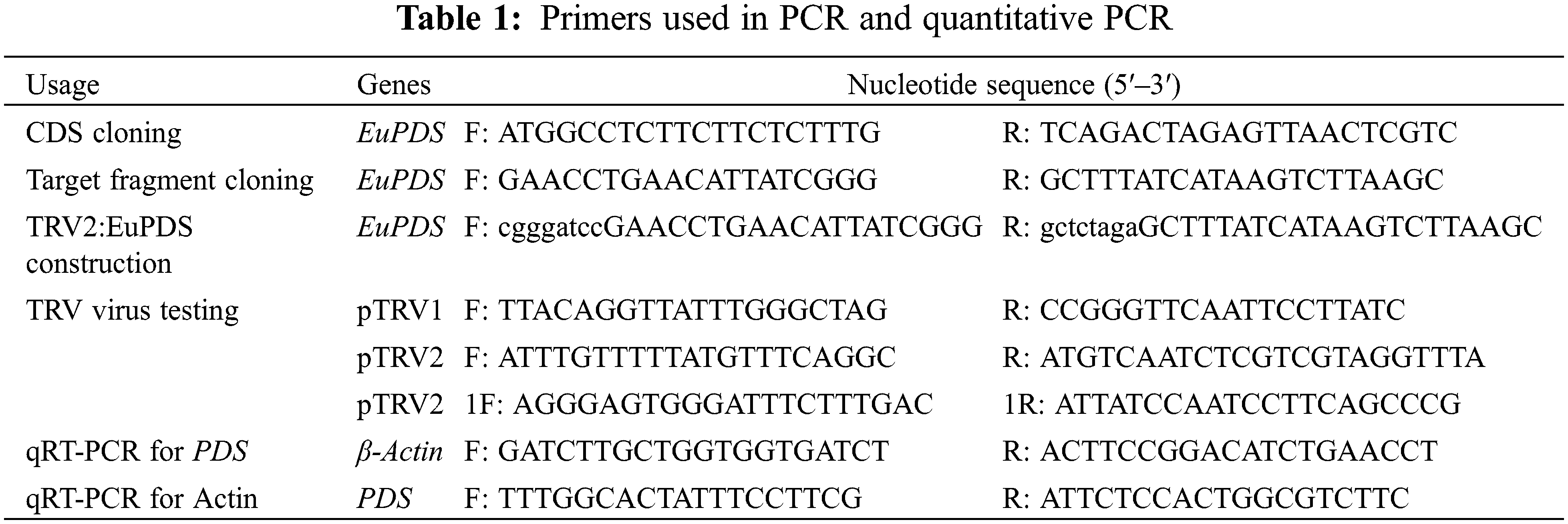
When PDS is deleted, silencing of PDS induce a characteristic photobleaching. The whole sequence of this gene was cloned successfully using PCR technology and re-confirmed the sequence by Sanger sequencing (Fig. 1).
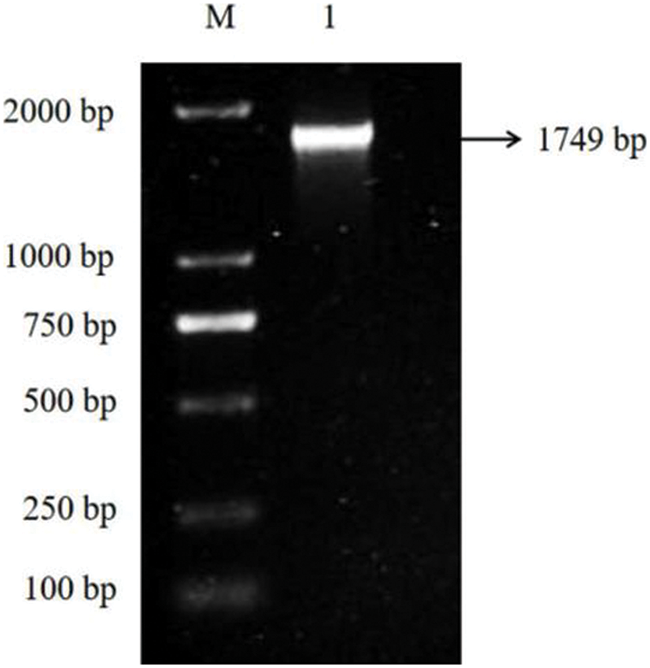
Figure 1: A full-length cDNA of PDS gene from E. ulmoides was obtained through PCR. M: DL 2000 Marker 1: PCR amplification product of EuPDS gene
3.2 Cloning and Bioinformatics Analysis of EuPDS Gene
According to the sequence information in the genome database of E. ulmoides constructed earlier in our laboratory, the cDNA fragment (1,749 bp) of E. ulmoides phytoene desaturase (PDS) gene encoding 583 amino acids was obtained by RT-PCR. The predicted molecular weight of EuPDS protein is 64,455.10 Da, and its theoretical isoelectric point is 8.79. The total number of negatively charged residues (Asp and Glu) is 64; the total number of positively charged residues (Arginine and Lysine) is 72. The instability index of EuPDS protein is 40.86, suggesting that EuPDS is an unstable protein. The secondary structure analysis with SOPMA indicated that putative EuPDS protein contained an α-helix (40.21%), an extended chain (14.78%), a random coil (40.38%), and a β-turn (4.64%). The average protein hydrophilicity value of EuPDS is −0.141, which is moderately hydrophilic. The protein is predicted to contain an amino acid oxidase structure without transmembrane domain and signal peptide. The encoded protein was found to be 69.70% homologous to the PDS protein of N. benthamiana, A. thaliana, Capsicum annuum, Bitter Gourd, and Populus trichocarpa (Fig. 2). The effectiveness of silencing is based on length of a gene fragment, with the best suitable fragment lengths between 150 and 500 bp, and the minimum sequence length for gene silencing is between 21 and 25 bp. The homology of tobacco and E. ulmoides PDS genes was compared, the size of these two fragments is consistent with the size of the smallest interference fragment, so the EuPDS fragment with two fragments consistent with tobacco PDS was selected, and the fragment size was 398 bp (201 to 599 bp).

Figure 2: Homology alignment of EuPDS amino acid sequence. EuPDS: E. ulmoides Oliver, GWHTAAAL009905; NtPDS: N. benthamiana, ABE99707.1; AtPDS: A. thaliana, AAA20109.1; CaPDS: C. annuum, CAA48195.1; McPDS: Momordica charantia, AAR86105.1; PtPDS: P. trichocarpa, XP_002321104.3
In order to analyze the expression pattern of EuPDS in different tissues of E. ulmoides, the RNA from roots, stems and leaves was analyzed by quantitative real-time PCR technology. The result has shown that the expression level of EuPDS in leaves was higher than that in the roots and stems (Fig. 3).
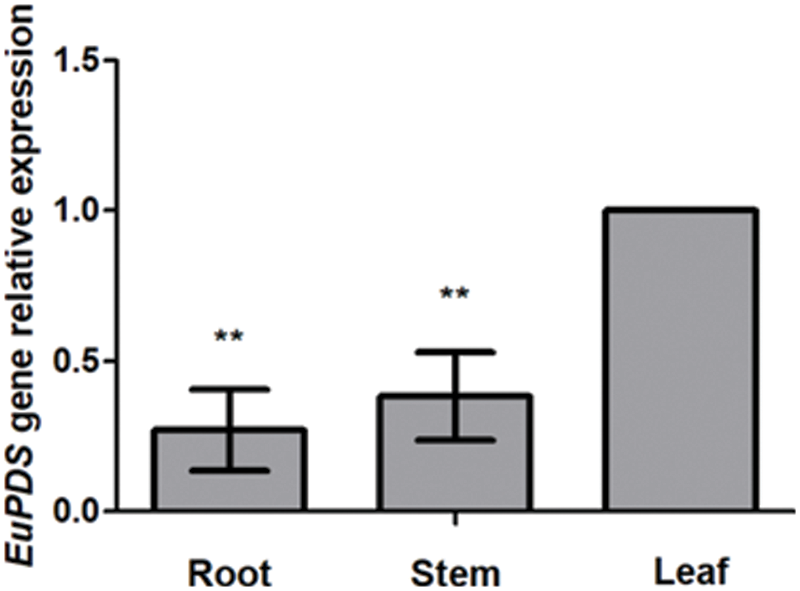
Figure 3: The expression pattern of EuPDS in different tissues of E. ulomides
3.3 Silencing of the PDS Gene in N. benthamiana
To explore whether EuPDS could cause NtPDS gene silencing in N. benthamiana plants, the study generated pTRV2-EuPDS recombinant to silence the endogenous N. benthamiana PDS, followed the operating approach of Martin-Hernandez et al. [14], and evaluated the gene silencing phenotypes 1 week after infiltration.
All treated plants were pruned below the infected site. Ten days later, sprouting young infected leaves harboring with the mix of pTRV1 and pTRV2-EuPDS exhibited total photobleaching, whereas control plants treated with the mix of pTRV2 and pTRV1 preserved their green color. New shoots and leaves of untreated plants (WT, wild-type plants), TRV2 plants (infiltrated with mixture of pTRV1 and pTRV2), silent plants with albino leaves (infiltrated with mixture of pTRV1 and pTRV2-EuPDS), and silent plants with new green leaves (mixed infiltration of pTRV1 and pTRV2-EuPDS) were collected after pruning (Fig. S4). Two strands of the virus TRV indicated the virus’s presence in newly grown leaves of infected N. benthamiana plants (Fig. 4).
Figure 4: The detection of the two strands of viral pTRV2 and pTRV1 in N. benthamiana leaves. (A–C) Wild-type plants; (D–F) empty plants; (G–I) albino tobacco plants; (J) RT-PCR detection of RNA1 of TRV in tobacco, note: 1–3: wild-type plants; 4–6: empty plants; 7–9: albino tobacco plants; (K) RT-PCR detection of RNA2 of TRV in tobacco, note: 1–3: wild-type plants; 4–6: empty plants; 7–9: albino tobacco plants
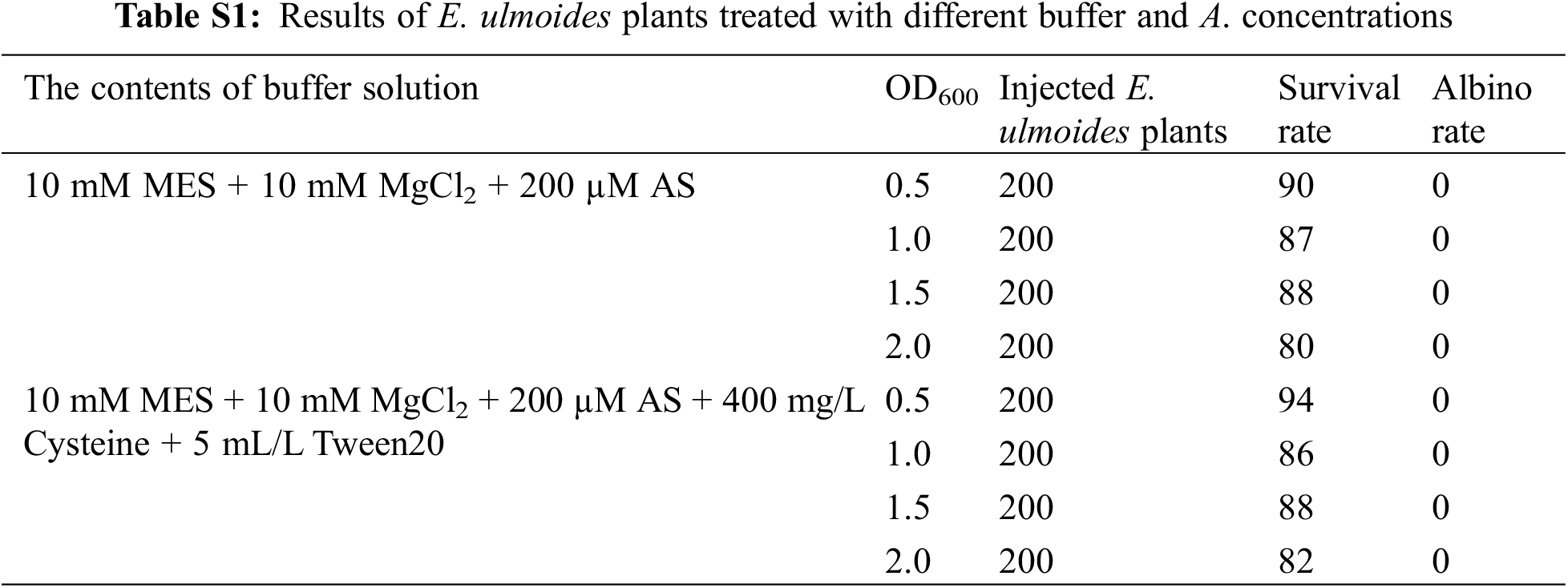
1,600 E. ulmoides plants were injected to investigate the effects of different Agrobacterium concentration and different buffers on the survival rate and albino rate of E. ulmoides seedlings. The experimental results demonstrated that different Agrobacterium concentrations had different effects on the survival rate of E. ulmoides seedlings, and there were significant differences (Table S1). However, albino symptoms did not appear, and the RNA1 and RNA2 chains of the virus were not detected, indicating that the TRV virus could not replicate in E. ulmoides, so the VIGS system could not be established in E. ulmoides (Fig. 5). The reason may be that the in vitro transcription is unstable and the recombinant vector is not effectively infiltrated into the plant; it may also be related to the genetic characteristics and temperature selection of E. ulmoides itself. In addition, it contains a lot of E. ulmoides gum, the leaves are rough, and the growth is slow, which also hinders the virus from spreading in diffusion on E. ulmoides leaves.
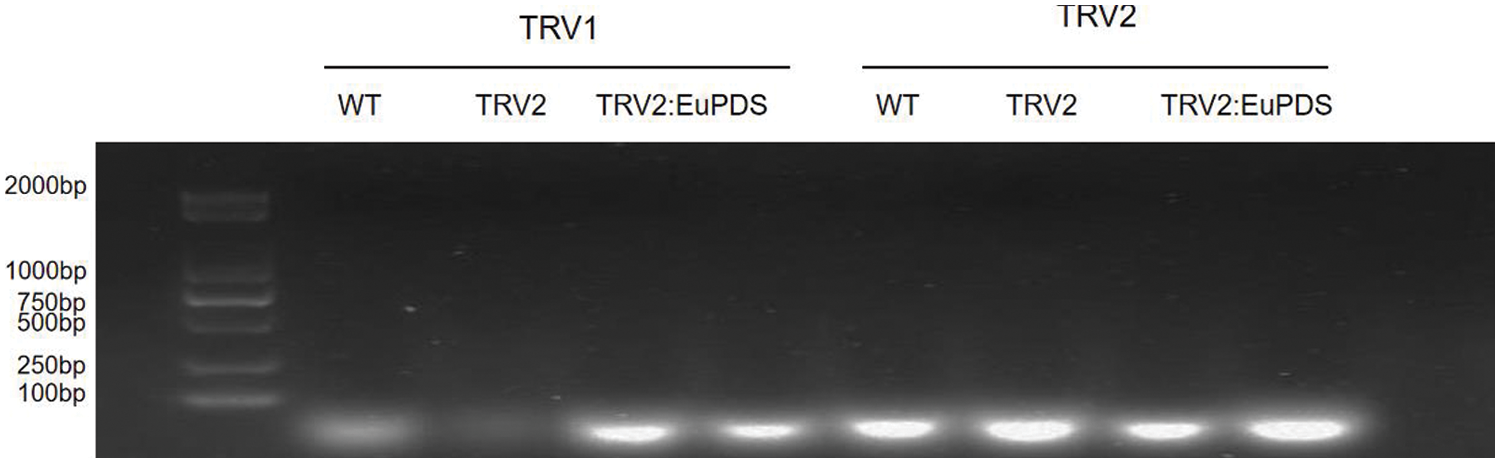
Figure 5: RT-PCR detection of RNA1 and RNA2 of TRV in E. ulmoides. WT: wild-type plant; TRV2: no-load control plant; TRV2: EuPDS: E. ulmoides plant injected by TRV2: EuPDS
3.4 Validation of the Knockdown of PDS with RT-qPCR
To verify the suppression of PDS in N. Benthamiana at the molecular level, quantitative real-time polymerase chain reaction (RT-qPCR) analysis was used. PDS mRNA was decreased by more than 97.6% in plants infected with pTRV2-EuPDS compared with controls infected with TRV alone (Fig. 6). β-actin functioned as an internal control for RNA quality for the RT-qPCR.
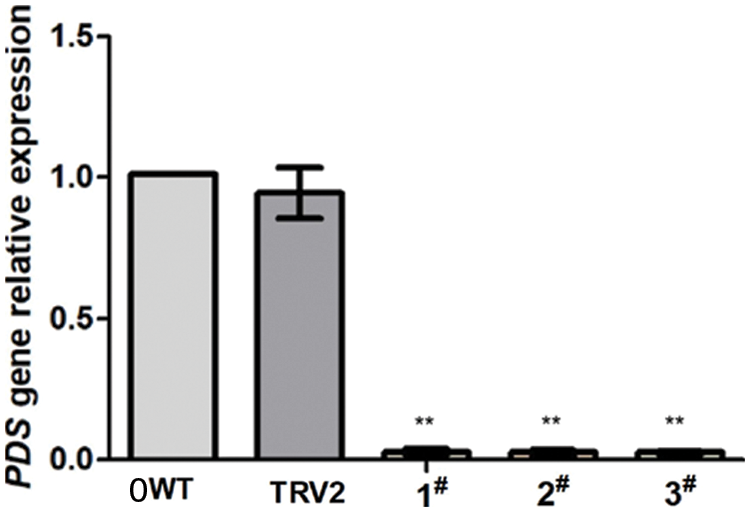
Figure 6: In the silenced plant was quantified using RT-qPCR. Error bars show standard error. WT: wild-type plant; TRV2: transfer vector plant with pTRV1 and pTRV2; 1#, 2# and 3#: transfer vector plant with pTRV1 and pTRV2: EuPDS “**” indicates extremely significant level
3.5 Chlorophyll Content Measurement
Chlorophyll was obtained from green control leaves and complete photobleached leaves to verify the alterations in chlorophyll following silencing. Chlorophyll in tobacco was extracted with 95% ethanol to determine its absorbance at 470 and 665 nm, respectively (Fig. 7). Excel2010, spass22 and GraphPad Prism were used to analyze chlorophyll a, chlorophyll b, total chlorophyll content; chlorophyll a, chlorophyll b, chlorophyll b and carotenoid content were lower than that in wild-type tobacco plants. Compared with the blank control plants (pTRV2), the contents of chlorophyll a (Fig. 7B), total chlorophyll b (Fig. 7C), chlorophyll (Fig. 7D) and carotenoids (Fig. 7E) were all obviously lower than those of the blank control plants. This shows that the change in leaf color in albino tobacco plants may be due to a change in chlorophyll concentration in response to the strength of gene expression.
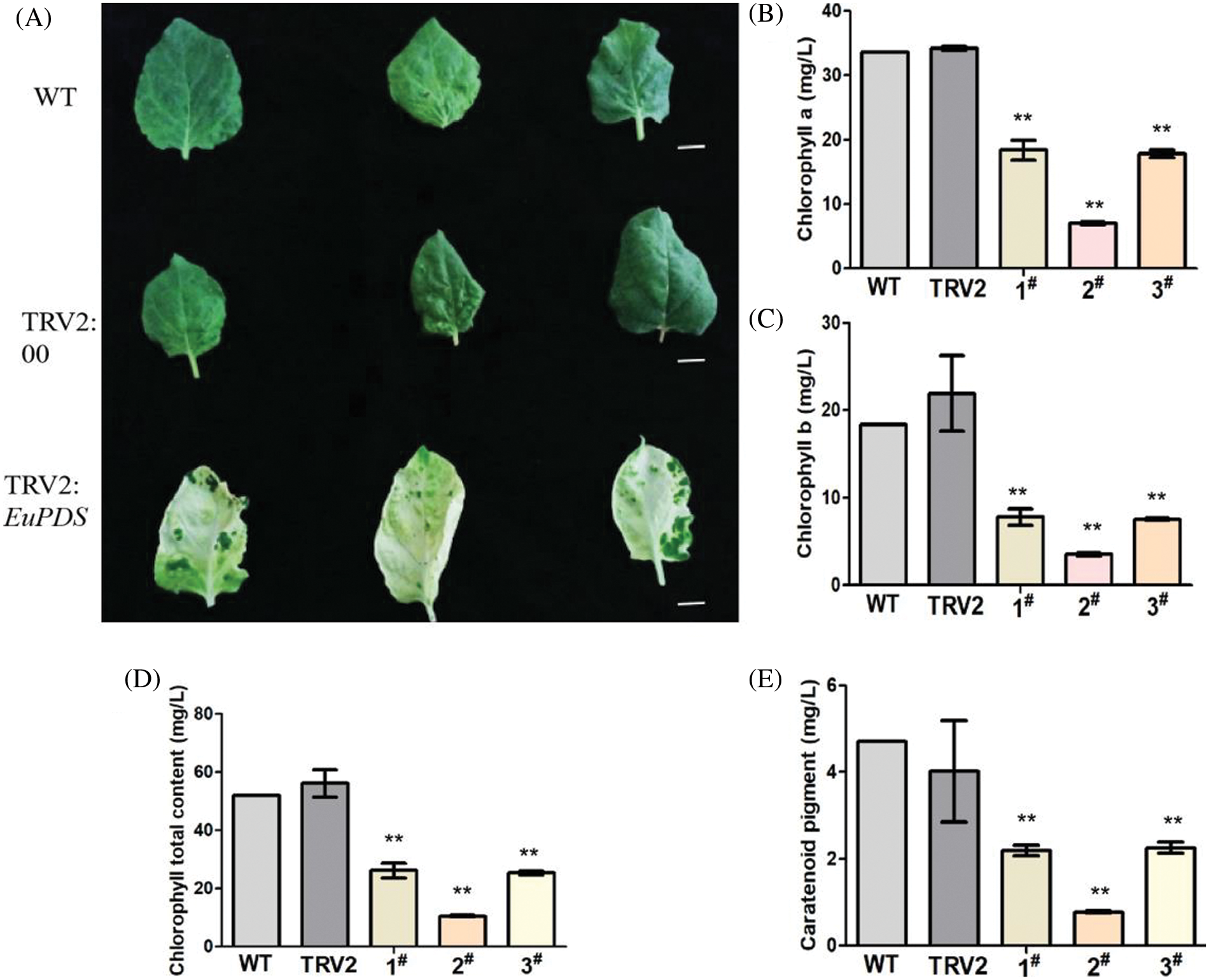
Figure 7: ChlorophyII content in leaves of albino tobacco lines. (A) Tobacco phenotypes observed. (B) The results of Chlorophyll a. (C) The results of Chlorophyll b. (D) The results of Chlorophyll total content. (E) The results of Caratenoid pigment. Notes: WT: Wild-type plants; TRV2: empty vector plants, 1#, 2# and 3#: EuPDS infected three albino tobacco plants; “**” indicates extremely significant level
The Cha/b ratio of each chromoprotein complex from wild-type Arabidopsis thaliana remained unchanged according to Hirashima et al. [26]. However, the research made by Hu demonstrated that the content of carotenoids and chlorophyll in albino leaves would be reduced by silencing PDS gene in P. hybrida, but the ratio of Cha to Chb altered [27]. In this study, similar results were obtained in Albino tobacco plants. White tobacco plants showed a greater chlorophyll a/b ratio than CK control plants and blank control plants, as shown in Fig. 8.
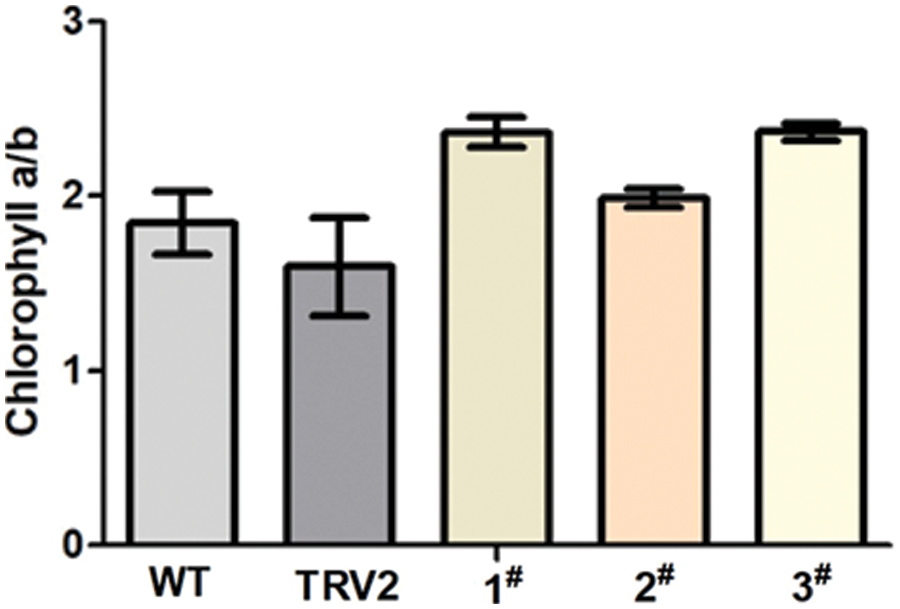
Figure 8: The ratio of chlorophyll a/b. WT: Wild-type plants; TRV2: empty vector plants, 1#, 2# and 3#: EuPDS infected three albino tobacco plants
E. ulmoides Oliver is a commercially significant medicinal tree species that warrants further exploration considering its diverse genetic functions. In comparison to woody plants such as Camellia oleifera and Populus, the identification of the function of its genes is sluggish, and it is critical to develop a rapid, efficient, and suitable approach system for high-throughput gene function identification.
In recent years, studies on A. thaliana and Vernicia fordii have shown that PDS is the rate-limiting enzyme involved in carotenoid biosynthesis, which converted into colored carotenoids by introducing two double bonds into the symmetric structure of colorless phytoene substrates [28]. Plants are often able to produce phenotypes that can be visually identified after silencing the endogenous PDS gene or upregulating its expression. In this study, the same appearance was observed in N. benthamiana. Therefore, PDS is often used as a positive reference for gene silencing or gene editing systems, and is widely used for the regulation and construction of carotenoids biosynthesis [29].
The study cloned the whole sequence of EuPDS gene using PCR techonology and charaterized its function. The pTRV2-EuPDS was constructed based on TRV and injected in the leaves of N. benthamiana. It was shown that PDS is a detectable marker for VIGS in N. benthamiana leaves. The successful application of the VIGS of N. benthamiana enables us to conduct functional investigations within E. ulmoides rather than depending on tedious, time-consuming, and small-scale transformation. Meanwhile, E. ulmoides is a significant Chinese medicine with a high pharmacological value that is important in disease resistance [30]. Thus, this research provides a reporter gene for the developing VIGS technology of E. ulmoides, which will provide help for further study on E. ulmoides gene function. The successful identification of these genes will provide the basis for the yield improvement of E. ulmoides rubber.
This research first cloned EuPDS gene and identified its function. The expression pattern of EuPDS in different tissues of E. ulmoides has shown that the expression level of EuPDS in leaves was higher than that in roots and stems. This study constructed an efficient TRV vector using this gene. This method successfully induced a photobleaching phenotype in newly grown leaves of N. benthamiana. The sprouting fresh tobacco leaves showed strong photobleaching. Photometric measurements indicated that the contents of chlorophyll a, chlorophyll b, total chlorophyll and carotenoids were all obviously lower than those of the blank control plants, but the ratio of Cha to Chb altered. This result not only provides a platform for further study of the gene’s color-forming mechanism, but also provides a reporter gene for the construction of E. ulmoides VIGS systems.
Funding Statement: This study was funded by the National Natural Science Foundation of China (Nos. 31870285, 31660076 & 32160384), the Open Fund for Key Laboratory of Ministry of Education and Science (No. KY [2022]366), and Guizhou Province High-Level Innovative Talent Training Program Project (No. [2016]4003).
Author Contributions: ZDG, ZYC and HXZ planned and conducted the experiments. WJL and CXM implemented the experiments. WJL wrote the manuscript. All authors reviewed and approved the manuscript.
Conflicts of Interest: The authors declare that there are no conflicts of interest in this study.
References
1. He, X. R., Wang, J. H., Li, M. X., Hao, D. J., Yang, Y. et al. (2014). Eucommia ulmoides Oliv.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. Journal of Ethnopharmacology, 151(1), 78–92. [Google Scholar] [PubMed]
2. Wu, J. M., Chen, H. X., Li, H., Tang, Y., Yang, L. et al. (2016). Antidepressant potential of chlorogenic acid-enriched extract from Eucommia ulmoides Oliver bark with neuron protection and promotion of serotonin release through enhancing synapsin I expression. Molecules, 21(3), 260. [Google Scholar] [PubMed]
3. Zhu, M. Q., Sun, R. C. (2018). Eucommia ulmoides Oliver: A potential feedstock for bioactive products. Journal of Agricultural and Food Chemistry, 66(22), 5433–5438. [Google Scholar] [PubMed]
4. Hao, S., Xiao, Y., Lin, Y., Mo, Z. T., Chen, Y. et al. (2016). Chlorogenic acid-enriched extract from Eucommia ulmoides leaves inhibits hepatic lipid accumulation through regulation of cholesterol metabolism in HepG2 cells. Pharmaceutical Biology, 54(2), 251–259. [Google Scholar] [PubMed]
5. Takamura, C., Hirata, T., Ueda, T., Ono, M., Migashita, H. et al. (2006). Iridoids from the green leaves of Eucommia ulmoides. Journal of Natural Products, 70(8), 1312–1316. [Google Scholar]
6. Wang, W. C., Yang, G. Q., Deng, X., Shao, F. Q., Li, Y. Q. et al. (2020). Molecular sex identification in the hardy rubber tree (Eucommia ulmoides Oliver) via ddRAD markers. International Journal of Genomics, 10, 2420976. [Google Scholar]
7. Hirata, T., Kobayashi, T., Wada, A., Ueda, A., Fujikawa, T. et al. (2011). Anti-obesity compounds in green leaves of Eucommia ulmoides. Bloorganic & Medicinal Chemistry Letters, 21(6), 1786–1791. [Google Scholar]
8. Ho, J. N., Lee, Y. H., Park, J. S., Jun, W. J., Kim, H. K. et al. (2005). Protective effects of aucubin isolated from Eucommia ulmoides against UVB-induced oxidative stress in human skin fibroblasts. Biological & Pharmaceutical Bulletin, 28(7), 1244–1248. [Google Scholar]
9. Zhang, N. D., Han, T., Huang, B. K., Rahman, K., Jiang, Y. P. et al. (2016). Traditional Chinese medicine formulas for the treatment of osteoporosis: Implication for antiosteoporotic drug discovery. Journal of Ethnopharmacology, 189, 61–80. [Google Scholar] [PubMed]
10. Feng, Y. Z., Zhang, L., Cao, H. P., Tan, X. F., Li, F. D. (2015). Transcriptome sequencing reveals fatty acid synthesis pathway in Eucommia ulmoides Oliv. Hortscience, 50(9), 371–372. [Google Scholar]
11. Bone, R. A., Landnum, J. T., Femandez, L., Taesis, S. L. (1988). Analysis of the macular pigment by HPLC: Retinal distribution and age study. Investigative Ophthalmology Visual Science, 29(6), 843–849. [Google Scholar] [PubMed]
12. Fleischmann, P., Watanabe, N., Peter, W. (2003). Enzymatic carotenoid cleavage in star fruit (Averrhoa carambola). Phytochemistry, 63(2), 131–137. [Google Scholar] [PubMed]
13. Robertson, D. (2004). VIGS vectors for gene silencing: Many targets, many tools. Annual Review of Plant Biology, 55, 495–519. [Google Scholar] [PubMed]
14. Martin-Hernandez, A. M., Baulcombe, D. C. (2008). Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. Journal of Virology, 82(8), 4064–4071. [Google Scholar] [PubMed]
15. Cunningham, F. X., Gantt, E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Plant Physiology, 49, 557–583. [Google Scholar]
16. Liu, Y., Schiff, M., Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant, 31(6), 777–786. [Google Scholar]
17. Shen, J. H., Si, W. J., Wu, Y. T., Xu, Y., Wang, J. et al. (2021). Establishment and verification of an efficient virus-induced gene silencing system in Forsythia. Horizontal Plant Journal, 7(1), 81–88. [Google Scholar]
18. Ratcliff, F., Martin-Hernandez, A. M., Baulcombe, D. C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant Journal, 25, 237–245. [Google Scholar]
19. Chamovitz, D., Sandmann, G., Hirschberg, J. (1993). Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate- limiting step in carotenoid biosynthesis. Journal of Biological Chemistry, 268(23), 17348–17353. [Google Scholar] [PubMed]
20. Nishii, K., Fei, Y., Hudson, A., Moller, M., Molnar, A. (2020). Virus-induced gene silencing in Streptocarpus rexii (Gesneriaceae). Molecular Biotechnology, 62(6–7), 317–325. [Google Scholar] [PubMed]
21. Liu, P. F., Wang, A. L., Du, Q. X., Du, H. Y. (2020). Chemotype classification and biomarker screening of male Eucommia ulmoides Oliv. flower core collections using UPLC-QTOF/MS-based non-targeted metabolomics. PeerJ, 8, e9786. [Google Scholar] [PubMed]
22. Zhou, Y. J., Deng, Y. T., Liu, D., Wang, H. Z., Zhang, X. et al. (2021). Promoting virus-induced gene silencing of pepper genes by a heterologous viral silencing suppressor. Plant Biotechnology Journal, 19(12), 2398–2400. [Google Scholar] [PubMed]
23. Burch-Smith, T. M., Anderson, J. C., Martin, G. B., Dinesh-Kumar, S. P. (2014). Applications and advantages of virus-induced gene silencing for gene function studies in plants. The Plant Journal: For Cell and Molecular Biology, 39(5), 734–746. [Google Scholar]
24. Yan, J., Xiang, F., Yang, P., Li, X., Zhao, X. (2021). Overexpression of BnGA2ox2, a rapeseed gibberellin 2-oxidase, cause dwarfism and increased chlorophyll and anthocyanin accumulation in Arabidopsis and rapeseed. Plant Growth Regulation, 93(1), 1–13. [Google Scholar]
25. Yang, P., Li, Y. X., He, C. S., Yang, J. D., Zhang, W. et al. (2020). Phenotype and TMT-based quantitative proteomics analysis of Brassica napus reveals new insight into chlorophyll synthesis and chloroplast structure. Journal of Proteomics, 214(C), 103621. [Google Scholar] [PubMed]
26. Hirashima, M., Satoh, S., Tanaka, R., Tanaka, A. (2006). Pigment shuffling in antenna systems achieved by expressing prokaryotic chlorophyllide a oxygenase in Arabidopsis. The Journal of Biological Chemistry, 281(22), 15385–15893. [Google Scholar] [PubMed]
27. Hu, L. (2017). Effects of silent PhPDS gene on growth and development of Petunia hybrida. Huazhong Agricultural University, China. [Google Scholar]
28. Bramley, P. M. (2002). Regulation of carotenoid formation during romato fruit ripening and development. Journal of Experimental Botany, 53(377), 2107–2113. [Google Scholar] [PubMed]
29. Gemmecker, S., Schaub, P., Koschmieder, J., Brauseman, A., Drepper, F. et al. (2015). Phytoene desaturase from Oryza sativa: Oligomeric assembly, membrane association and preliminary 3D-analysis. PLoS One, 10(7), e0131717. [Google Scholar] [PubMed]
30. Fang, C., Chen, L. Y., He, M. Z., Luo, Y. Y., Zhou, M. J. et al. (2019). Molecular mechanistic insight into the anti-hyperuricemic effect of Eucommia ulmoides in mice and rats. Pharmaceutical Biology, 57(1), 112–119. [Google Scholar] [PubMed]
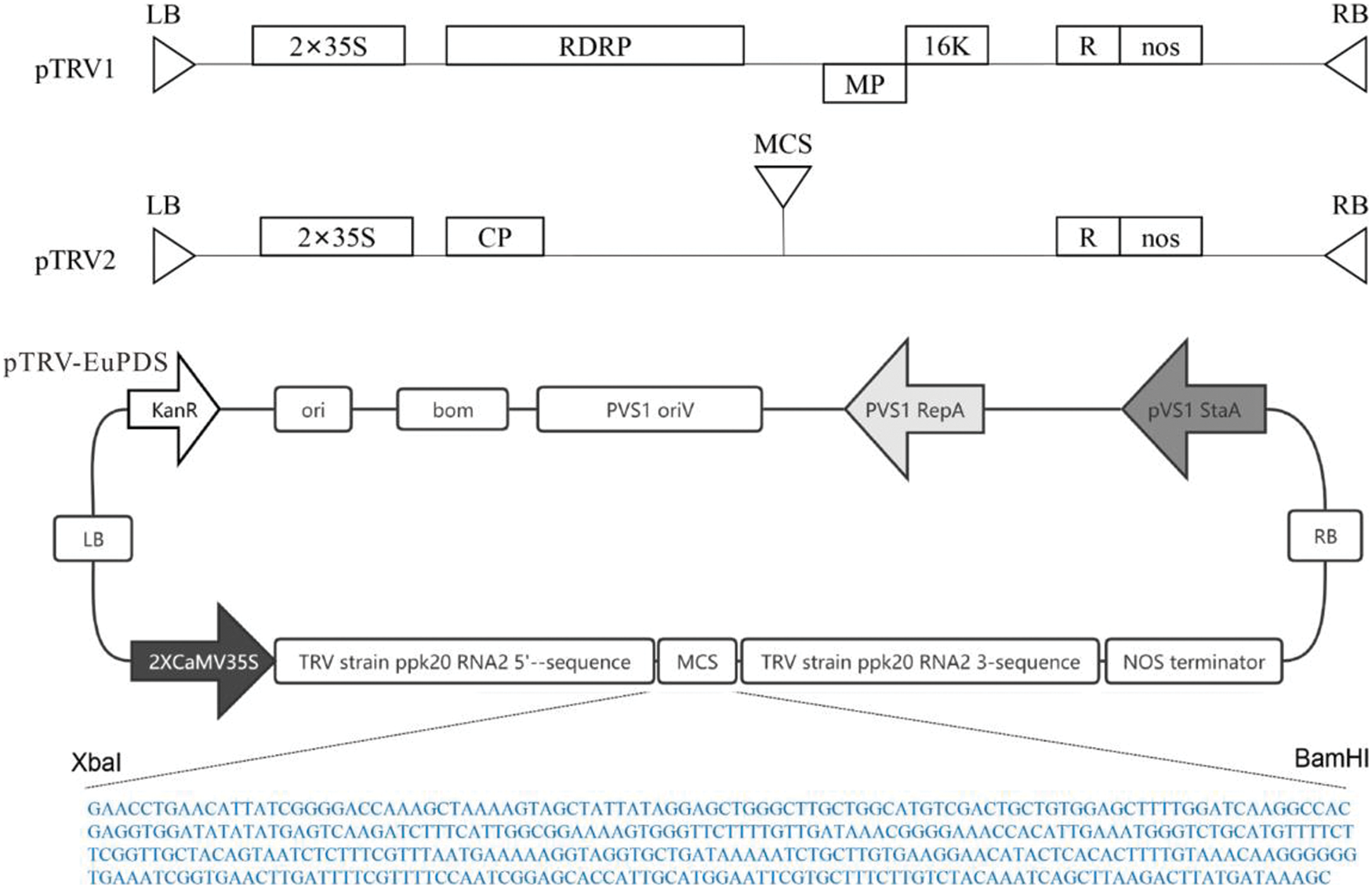
Figure S1: Schematic representation of the pTRV2-EuPDS vector
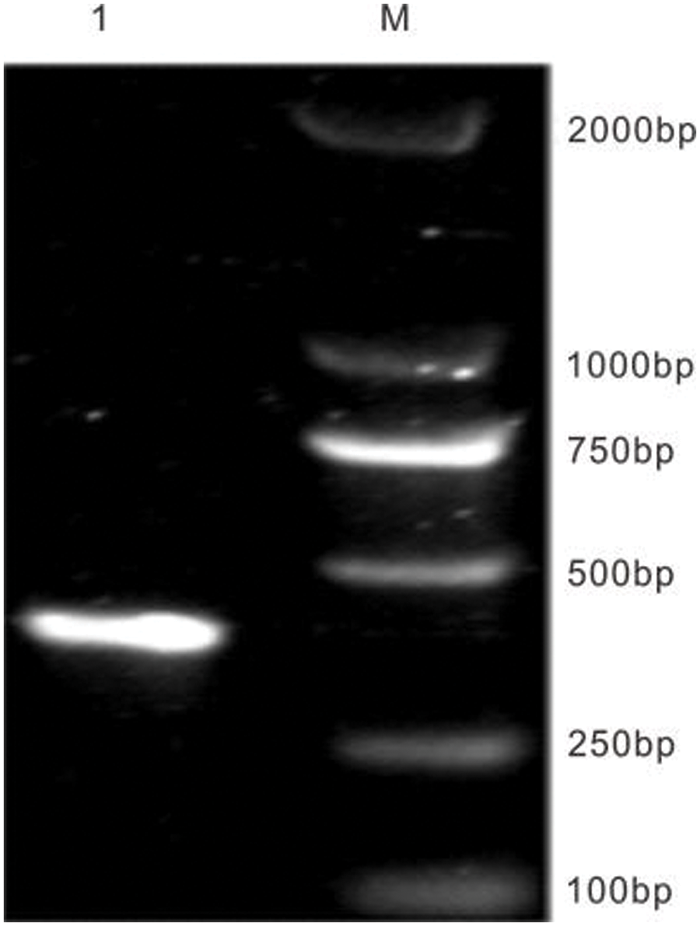
Figure S2: The amplification of EuPDS gene special segment. M: DNA marker DL 2000; 1: EuPDS special segment
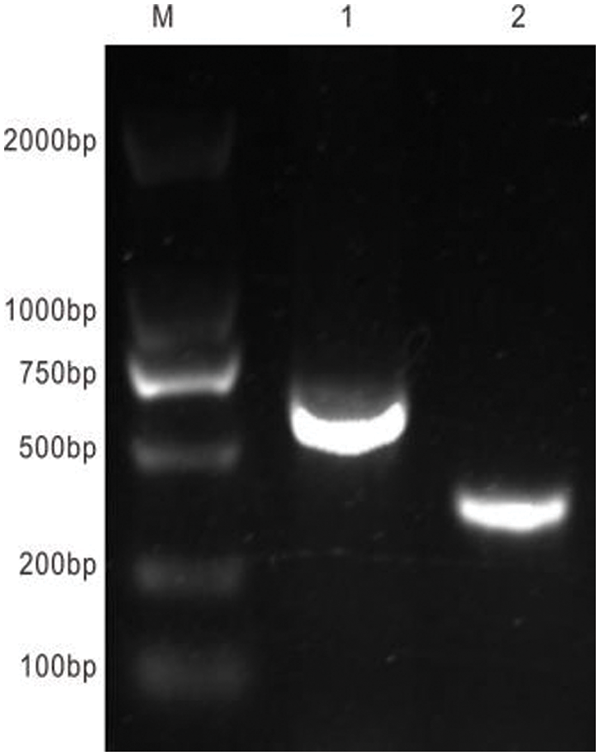
Figure S3: Results of construction of plant interference vector. M: DNA marker DL 2000; 1: Amplification segment of RNA2 premier; 2: EuPDS special segment
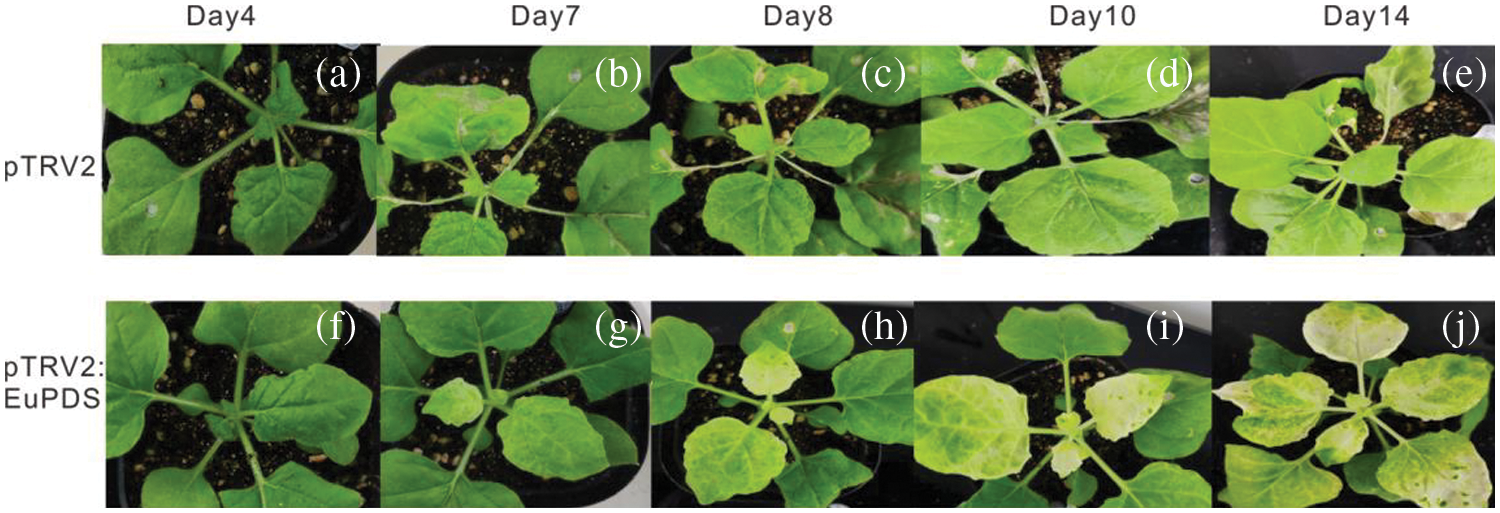
Figure S4: VIGS of N. benthamiana phytoene desaturase ortholog PDS results in varying degrees of leaf photobleaching. Note: a/f: The sprouted young leaves of 4 days after injection; b/g: The sprouted young leaves of 7 days after injection; c/h: The sprouted young leaves of 8 days after injection; d/i: The sprouted young leaves of 10 days after injection; e/j: The sprouted young leaves of 14 days after injection
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools