 Open Access
Open Access
ARTICLE
Introgression of Drought Tolerance into Elite Basmati Rice Variety through Marker-Assisted Backcrossing
1
National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, 38000, Pakistan
2
Rice Research Institute, Kala Shah Kaku, 39020, Pakistan
3
National Institute for Genomics and Advanced Biotechnology (NIGAB), Islamabad, 44000, Pakistan
4
Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, 100081, China
5
Department of Biotechnology, COMSATS University Islamabad, Abbottabad, 22020, Pakistan
6
Department of Biotechnology, University of Sialkot, Sialkot, 51310, Pakistan
7
Pakistan Institute of Engineering and Applied Sciences, Islamabad, 44000, Pakistan
* Corresponding Authors: Shahzad Amir Naveed. Email: ; Muhammad Arif. Emial:
# Both authors contributed equally
Phyton-International Journal of Experimental Botany 2023, 92(5), 1421-1438. https://doi.org/10.32604/phyton.2023.025801
Received 31 July 2022; Accepted 28 October 2022; Issue published 09 March 2023
Abstract
Drought is one of the major abiotic threat to rice production in the context of climate change. Super Basmati is an elite, fine grain basmati rice variety grown in Punjab, Pakistan. Due to drought sensitive in nature, its yield has been facing an alarming situation in production because of gradual decrease in irrigated water for a couple of years. Three reported novel QTLs for drought tolerance were selected for incorporation into Super Basmati by employing marker assisted selection strategy. IR55419-04 with novel QTLs was used as a donor parent. Foreground selection was performed by applying PCR based QTL linked SSR markers followed by recombinant selection by using 2–4 flanking markers. Background selection was exercised by using polymorphic SSR markers for maximum genome recovery of the Super Basmati. The individuals homozygous at the target QTLs and with maximum background of Super Basmati at the rest of the non-target genome was selected for evaluation of drought tolerance. Under drought stress conditions, the yields of all introgressed lines (ILs) were 44.2%–125.7% higher than recurrent parent. Six superior ILs that are drought tolerant and very similar to Super Basmati in terms of agronomic and grain quality traits are marked for release as drought-tolerant varieties in arid regions or for use in breeding programs of high grain quality and drought-tolerant parents.Keywords
Irrigation water value as a function of field crops and freshwater scarcity is a challenge to world agricultural production. This water deficit situation is mainly caused by shrinking water resources, global warming, and fluctuating rainfall [1]. Sustainable rice production is endangered by increasing irrigation water scarcity too. It is of high importance for rice because of its range of growing ecosystems. Water scarcity at specific growth stages in rice can cause a considerable damage to its productivity. A vegetative stage drought stress of a certain level can result in a moderate loss of yield while incidence of such stress at the time of fertilization or pollen meiosis may cause a clean sweep in terms of production. Therefore, pyramiding of drought tolerance genes in mega varieties of rice is obligatory. Marker assisted breeding (MAB) effectiveness has earlier been successfully practiced in rice for the introgression of bacterial blight resistance genes salt resistance, submergence tolerance Sub1 gene [2] and green revolution sd1 gene [3]. Drought stress at the seedling and flowering stages of rice leads to severe reduction in rice yield and affects grain quality, Marker-assisted selection and backcross breeding approaches can combine drought tolerance with grain quality traits in elite rice varieties [4,5].
In Pakistan, Rice (Oryza sativa L.) is grown on an area of 2.3 million hectares which contributes in total world rice trade about 11%, while by quantity is 8.9%. Amongst the rice grown in Pakistan, Basmati rice fetches a high price in the international market due to its premium quality [6]. It is grown in the famous “Kallar” tract of the Punjab, Pakistan. The influence of climate change on Basmati rice cultivation cannot be ignored, because all Basmati rice varieties are highly sensitive to drought stress [7]. It is being anticipated that climate change may explain around 20% increase in water deficit this century globally [8]. The future of food security and water availability is endangered in this scenario worldwide. Therefore, introgression of drought tolerant QTLs into Basmati genetic background is in high demand for sustainable Basmati rice production suitable to withstand under water limited environments [9]. Dixit et al. [10] Applied marker-assisted backcross breeding approach to introgression drought QTLs (qDTY 3.2 and qDTY 12.1) in the genetic background of the popular rice variety Sabitri grown in rainfed regions of Nepal. They combined the phenotypic and marker assisted selection approaches and developed the drought tolerance NILs for Sabitri. A similar study was conducted by Janaki et al. [11] a susceptible elite rice variety was used as recurrent parent to three major genes (Pi9, Xa21 and Gm8), and three major QTLs qDTY1.1, qDTY2.2 and qDTY4.1) and Naveen conferred increased yield under drought conditions. Dhawan et al. [12] used the drought-susceptible elite cultivar Pusa Basmati 1 (recurrent parent) to cross with drought-tolerant N22 (donor parent) and a major drought-tolerant QTL (qDTY1.1) was introgressed into the genetic background of Pusa Basmati 1. A panel of 113 SSRs polymorphic primers was studied to find genomic recovery of recurrent parents and eighteen drought tolerant near isogenic lines (NILs) were developed.
Over the past two decades, scientists around the world have identified hundreds of drought QTLs, but few of them have been introgressed into elite varieties to improve grain quality and quantity [13]. The objectives of the present study are introgression of putative drought tolerant QTLs/genes in Super Basmati rice using DNA markers. In turn, the drought tolerant introgression lines (DTILs) developed through this research effort will be utilized in the future rice breeding programs in the country. Ultimately, sustainable rice production will bring more opportunities for the prosperity of the country through food security and poverty alleviation.
IR55419-04 line outcome of a cross of IR12979-24-1 (BROWN) /UPL RI 5 at the International Rice Research Institute (IRRI), Philippines developed for drought prone ecosystem was used as the donor parent [14]. Super basmati an elite rice variety release by Rice Research Institute, Kala Shah Kaku, Pakistan, possessing drought sensitivity was used as recipient parent.
2.2 QTL Selection and Marker Assisted Back Crossing Scheme
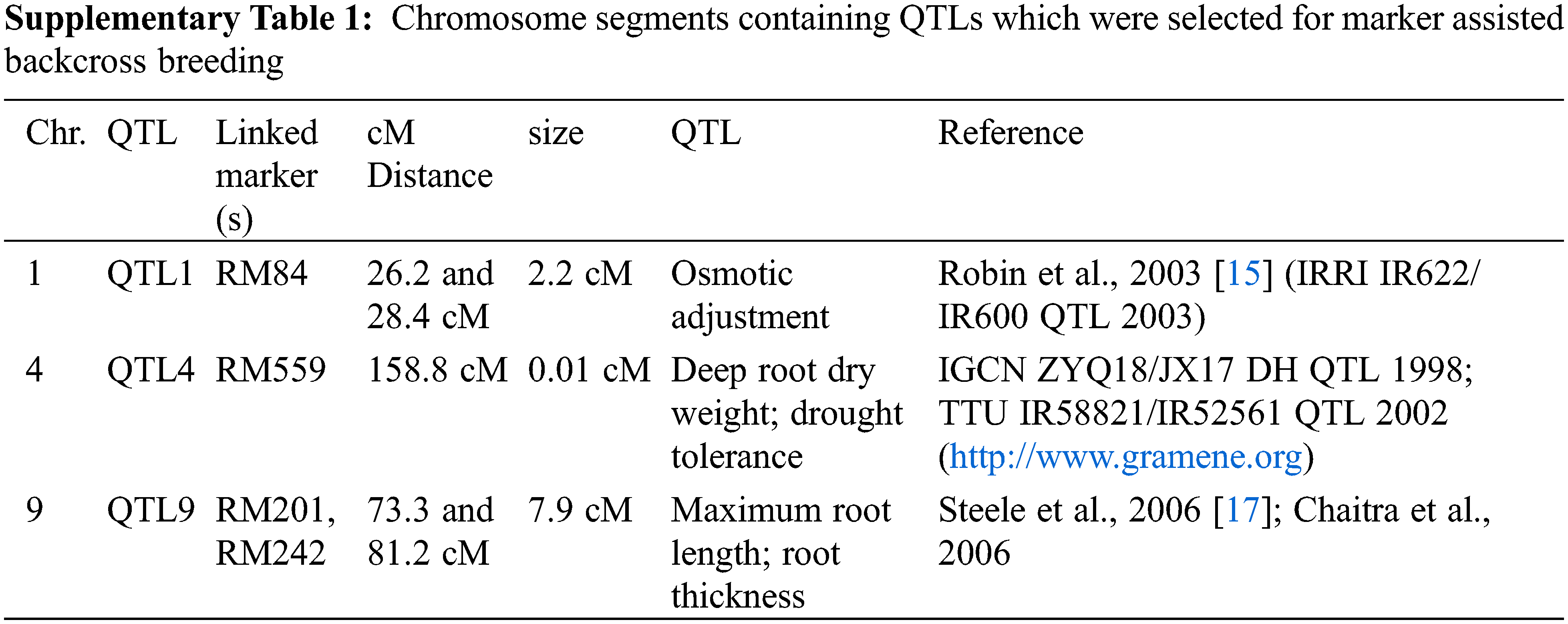
From the review of previous findings, three segments located on chromosome (Chr.) 1 [15], 4 (www.gramene.com) and 9 [16,17] were selected as target QTLs, named as QTL1 (Chr. 1) QTL4 (Chr. 4) and QTL9 (Chr. 9), respectively for introgression (SupplementaryTable 1). A crossing was performed between IR55419-04 and Super Basmati rice genotypes (Fig. 1A). The F1 plants were screened by applying QTL linked markers and true F1 were backcross with recurrent parent. The BC1F1 and BC2F1 were screened for foreground selection by using PCR based SSR markers and BC2F1 plants were advanced by self-crossing to raise BC2F2. The BC2F2 plants harboring homozygous alleles of the target QTLs and possessing phenotypic resemblance with the recurrent parent were selected for recurrent parent genome (RPG) by applying ninety-nine polymorphic SSR markers (Fig. 1B) and the value for the recurrent parent genome recovery (RPGR) was given by using the following equation. Selected BC2F2 plants were advanced by self-crossing to raise BC2F3 and this population was used for field drought assessment (Fig. 1C).
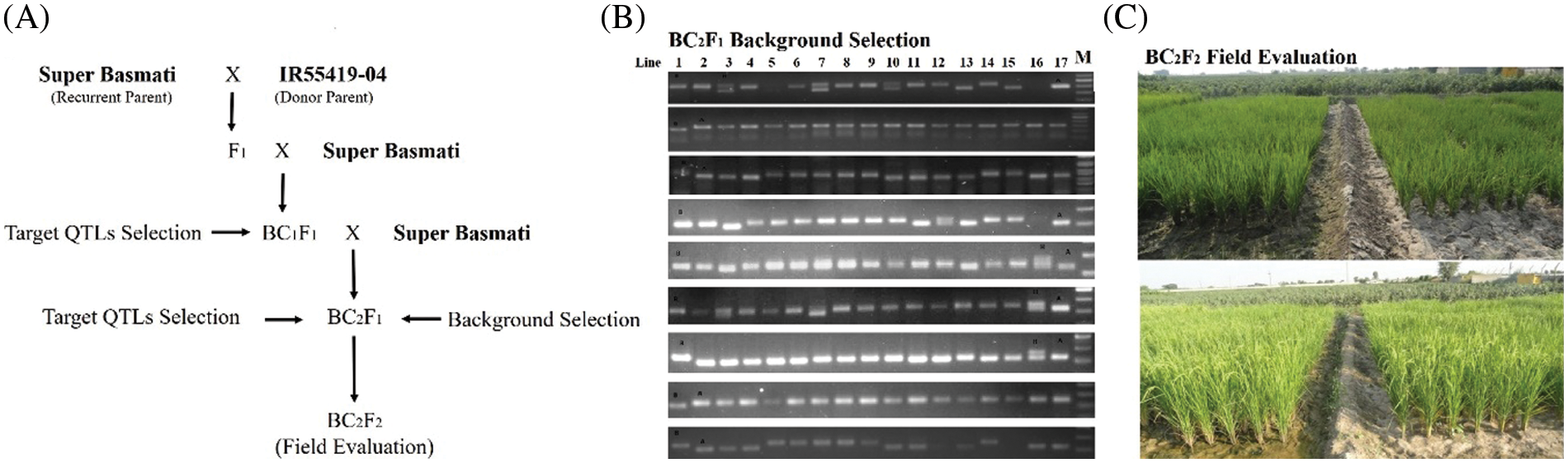
Figure 1: (A) Schematic summary of procedures for introgression of drought tolerant QTLs in super basmati background, (B) Agarose gel image showing DTIL polymorphism compared to parental lines, (C) Performance comparison of DTILs under irrigation and drought stress conditions
where, p = number of recurrent parent alleles; h = number of heterozygotes; and t = total number of alleles.
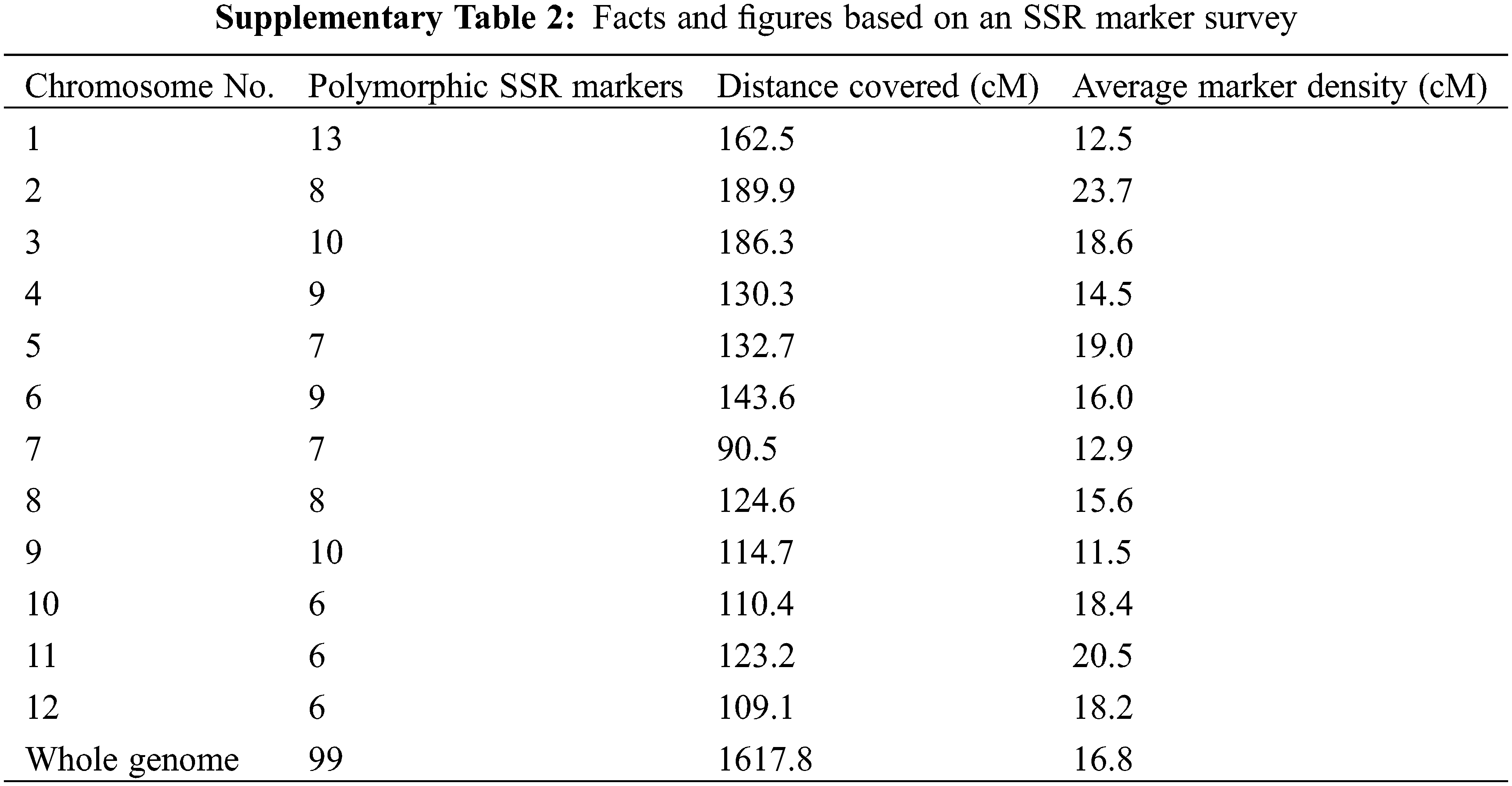
Genomic DNA of the drought tolerance introgression rice genotypes, donor and recurrent parents was extracted. PCR was performed for detecting selected QTLs by utilizing QTL linked SSR markers as presented in SupplementaryTable 1 by following PCR protocol as described by Shah et al. 2013. Two gene specific markers, i.e., Wx and STS aroma genes were used to monitor the inheritance of amylose and aroma, respectively [18,19]. For background selection a set of 207 microsatellite markers straddling on the 12 chromosomes were surveyed between the two parents and identified polymorphic markers were used to find a maximum RPG from developed genotypes for further selection. Graphical Genotype (GGT) Version 2.0 software was used to develop a schematic map, by using the polymorphic marker data, depicting the genomic contribution of donor and recurrent parents to identify DTILs having maximum recovery of RPG.
2.4 Evaluation of DTILs for Drought
Selected DTILs along with donor and recipient parent were evaluated under both irrigated and stress conditions in the research area of the National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan, by following a split plot design. Each entry was replicated thrice at a spacing of 22.5 cm × 22.5 cm between plants and rows for water treatment levels. Recommended crop management practices were adopted. In the drought stress treatment, the field was drained out at the tillering stage 35 days after transplanting. Fifteen days water stress was imposed for the genotypic responses. In the irrigated control, irrigation was applied every 3–4 days until the lines reached maturity. The irrigation was stopped 2 weeks before harvesting. At the maturity, five representative plants in each plot were measured for plant height, number of productive tillers per plant, number of filled-grain per panicle, thousand-grain weight, and spikelet fertility.
2.5 Evaluation of Grain Quality
Grain samples of DTILs, along with its recurrent and donor parent, were analyzed for physical and chemical attributes. Number of filled-grain per panicle, thousand-grain weight, spikelet fertility, grain length, width, thickness, length to width ratio, the shape of the polished grain, aroma and amylose content were measured applying standard procedures [20].
The analysis of variances (two-way ANOVA) and t-tests were performed to compare the differences for the measured traits among rice genotypes within each population and genotype (parents/DT-ILs) treatments (stress and non-stress) interactions using the Statistix v8.1 (USA).
3.1 Polymorphism between Parents and Drought Tolerance QTLs
Out of two hundred and seven microsatellite markers, ninety-nine markers were found polymorphic (SupplementaryTable 2). The linkage map proposed by Temnykh et al. [19] was used as reference map.
3.2 Marker Assisted Introgression of Drought
In each backcross generation, target QTLs were monitored using foreground markers linked to the specific loci. In BC1F1, 151 individuals were genotyped using foreground markers and found 69 plants heterozygous for target QTLs in different combinations. One plant from each QTLs combination was selected based on target and recombinant selections for further Back crossing. A total of 678 BC2F1 seeds from the three selected plants was sown in the field. A foreground selection of these BC2F1 individuals was based on that they were heterozygous at the foreground marker loci. Foreground selection resulted in selection of 376 BC2F1 individuals based on the presence of QTLs either individually or in 2–3 QTLs combination. As a result of recombinant selection, 19 plants were selected to raise BC2F2.
Out of these 19 plants, target QTLs in individual QTL1, QTL4 and QTL9 were present in 3, 4 and 2 plants, respectively. The number of plants with two QTLs combinations, i.e., QTL1 + QTL4, QTL1 + QTL9, and QTL4 + QTL9 were 1, 2 and 2, respectively, whereas, 5 plants possessed selected three QTLs (Table 1 and Fig. 2). All these 19 plants were allowed to self-pollinate to obtain BC2F3 seeds and were evaluated in the field as drought tolerant introgression lines (DTILs) to assess the response through screening under water stress condition.
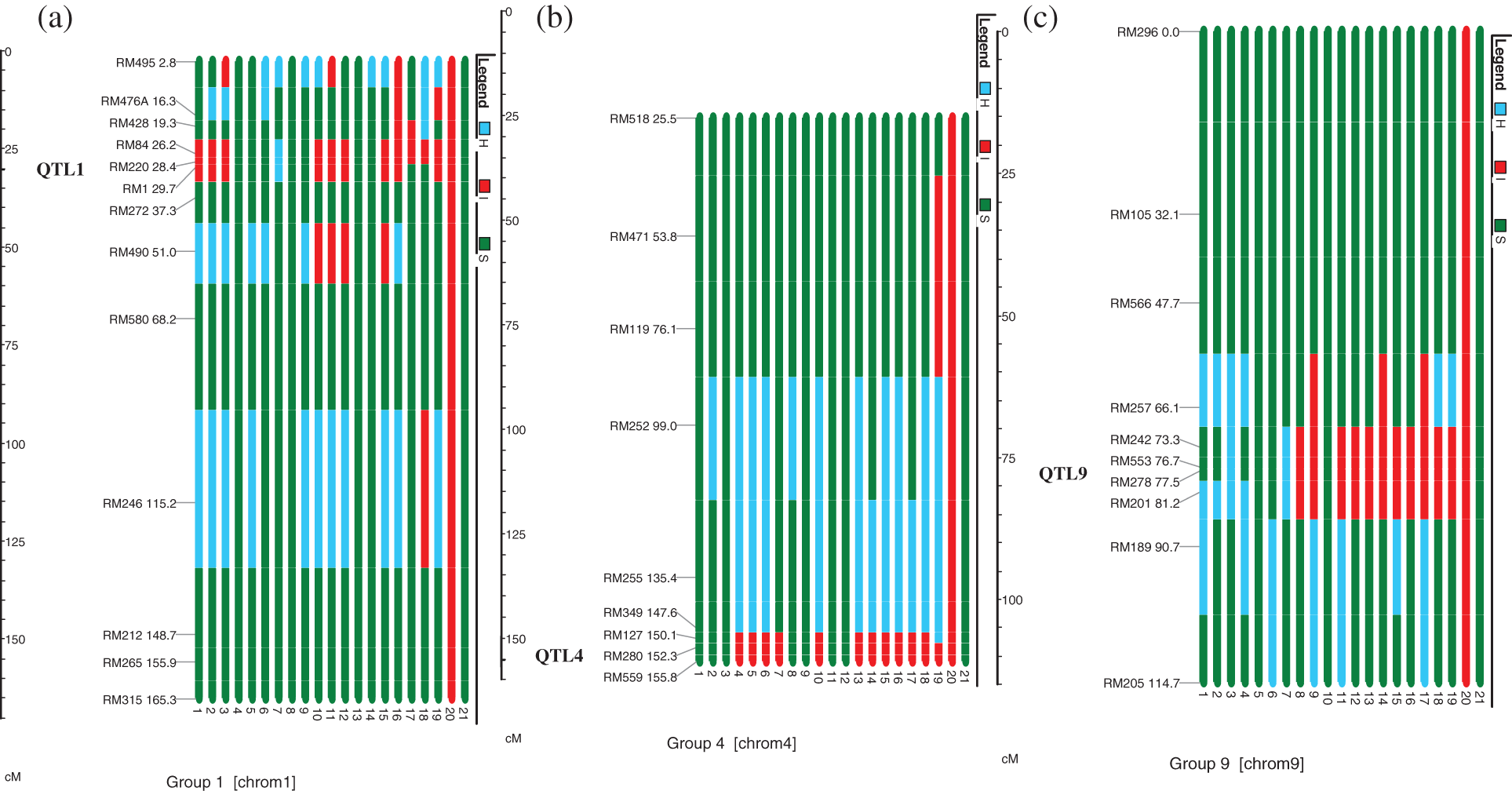
Figure 2: Graphical representation of (a) Chromosome 1, (b) Chromosome 4 and (c) Chromosome 9 carrying target QTLs, i.e., QTL1, QTL4 and QTL9. Each bar is the respective introgression line or variety. Bar Nos. 1–19 (given at the bottom of each bar) are the introgression lines. Bars No. 20 and 21 show the IR55419-04 (donor parent) and Super Basmati (recipient parent), respectively. Red and green colors depict the homozygous alleles of IR55419-04 and Super Basmati, respectively while light blue color indicates the heterozygous
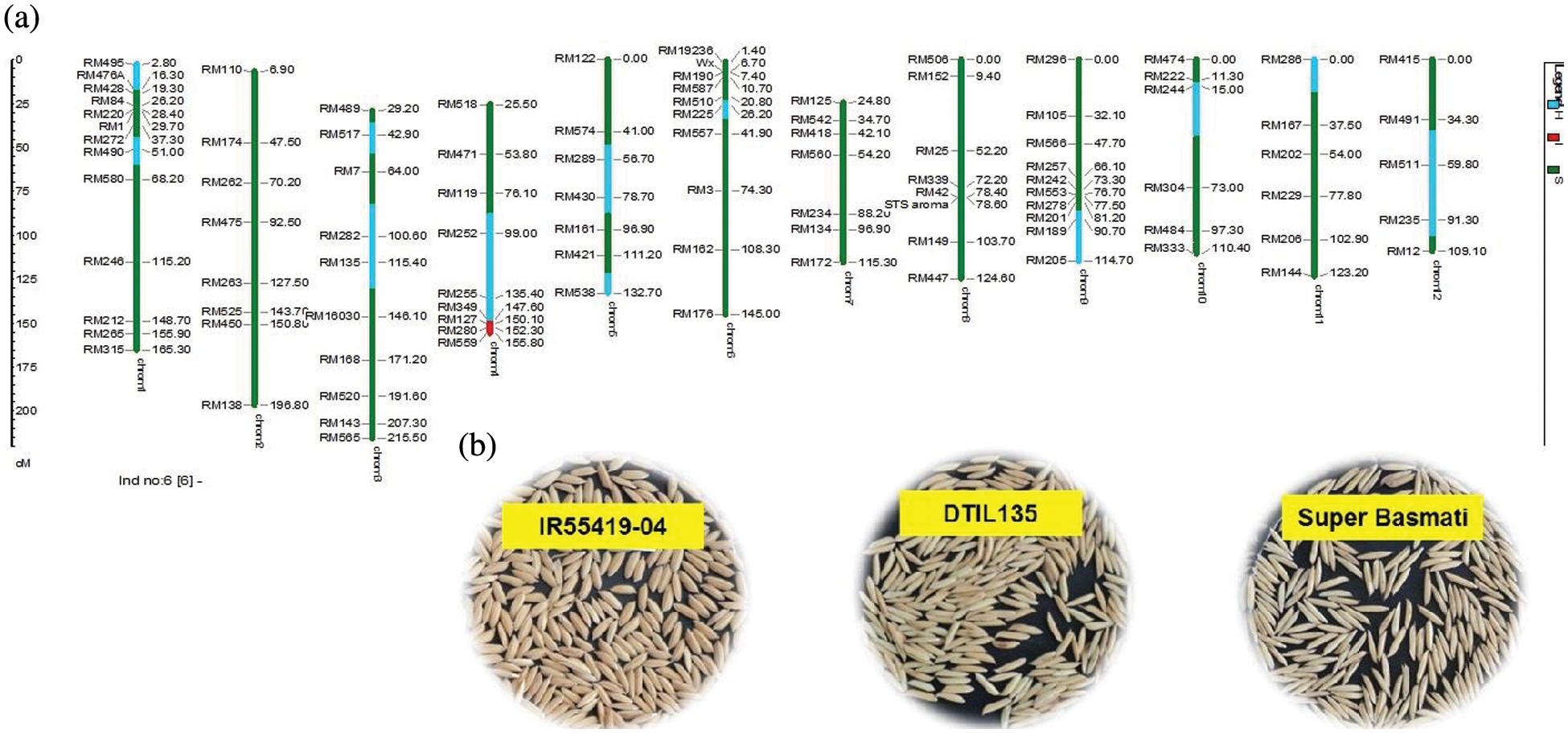
As a whole, 99 polymorphic microsatellite markers unlinked to target QTLs covering all the rice chromosomes were used for background selection to recover the recipient genome. The graphical genotypes of DTIL60 is illustrated in Fig. 3 While the graphical genotypes of DTIL 60, DTIL 135, DTIL 56, DTIL 269 and DTIL 146 is given in Supplementary Figs. 1–5.
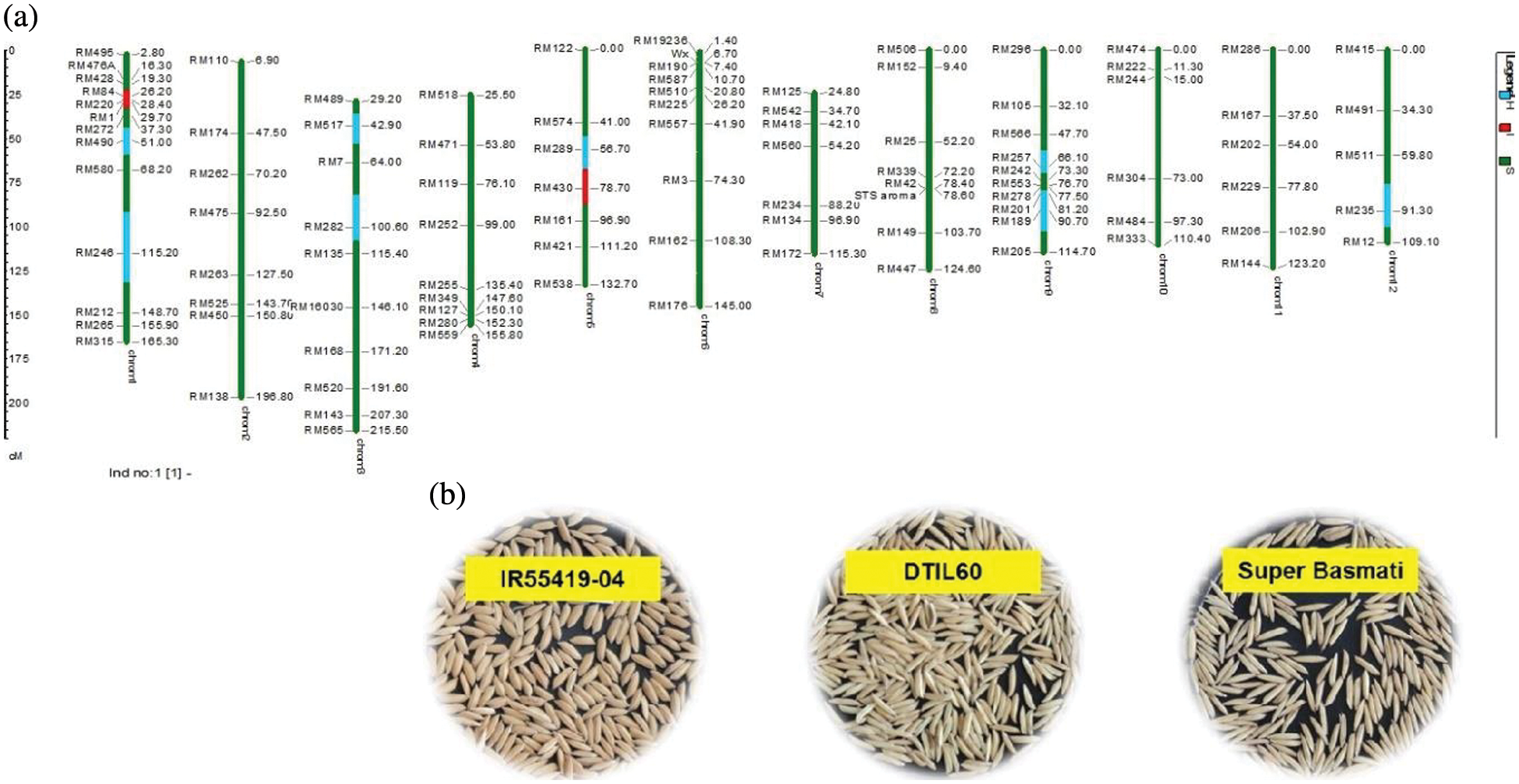
Figure 3: (a) Graphical genotype of DTIL60 (Q1). Green and red colors depict the homozygous alleles of Super Basmati (recipient) and IR55419-04 (donor), respectively while light blue color indicates the heterozygous status of specific marker loci. Both the size scale indicating chromosome lengths in centiMorgans (cM) and marker names are on the left. (b) Paddy rice of IR55419-04 (left), DTIL60 (center) and Super Basmati (right)
3.4 Genome Recovery by Recurrent Parent in DTILs
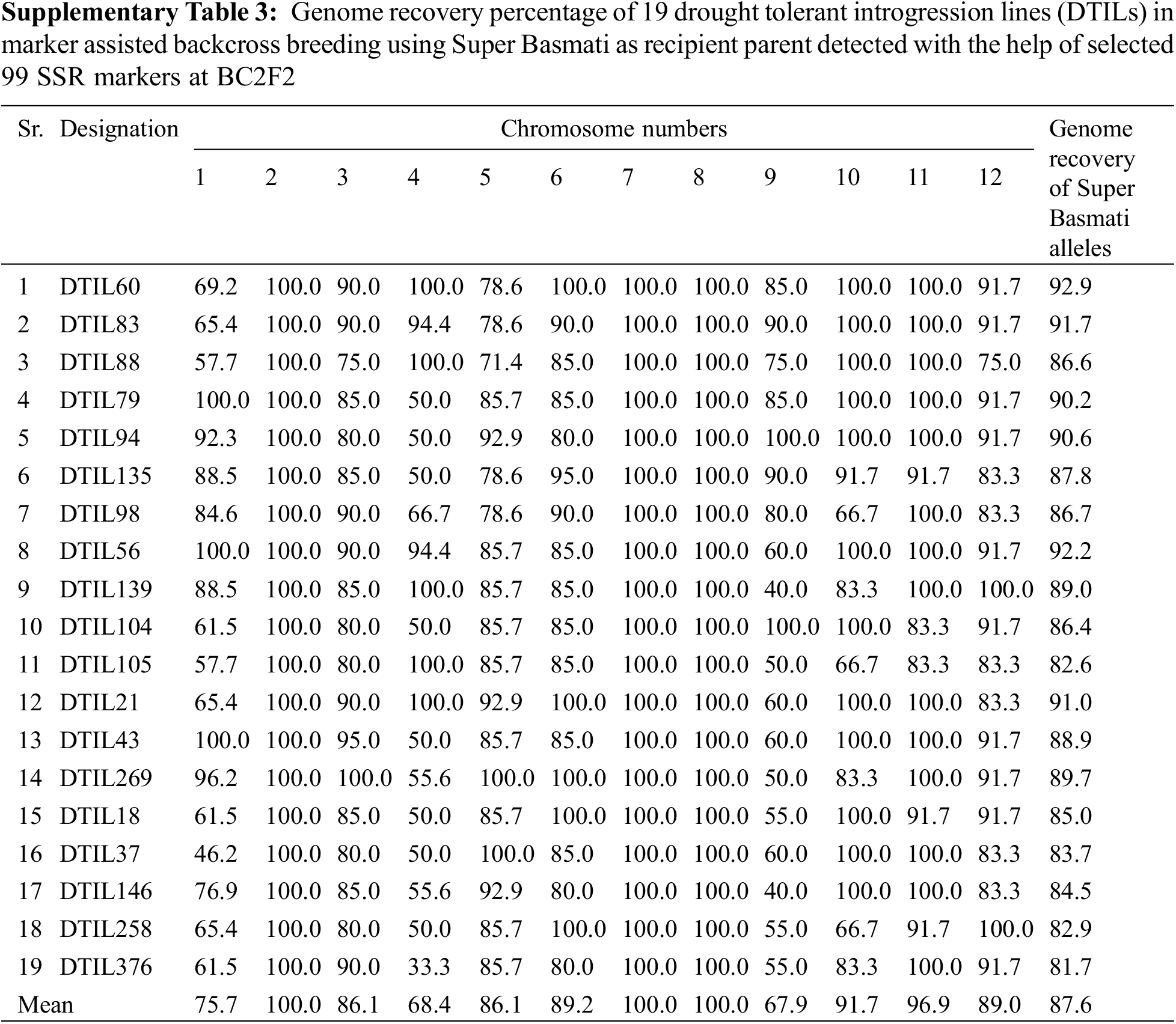
The genome recovery percentage of the DTILs was assessed by using 99 polymorphic SSR markers between parents was presented in SupplementaryTable 3. Minimum mean genome recovery, i.e., 67.9% was recorded for chromosome 9 whereas chromosomes 2, 7 and 8 were 100.0% similar to Super Basmati across selected DTILs.
3.5 Performance of DTILs under Water Stress
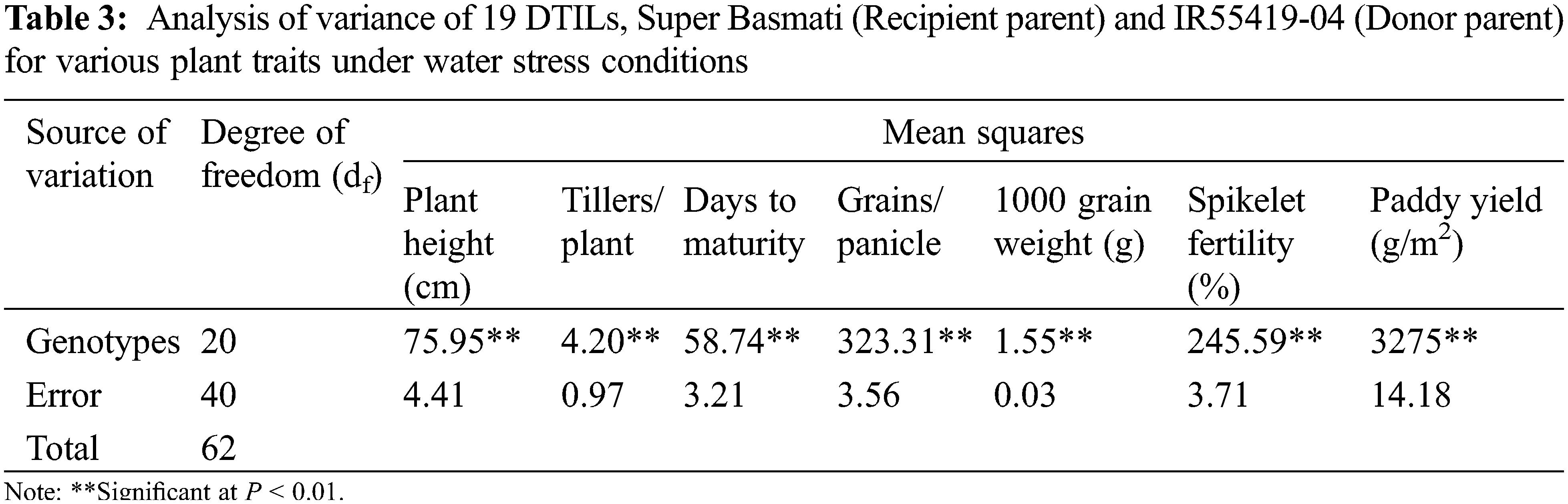
New DTILs possessed mono, di and tri QTLs integration with different combinations (Table 1). The analysis of variance for various traits was performed for two water regimes, i.e., well-watered control and water stress (15 days) separately and are presented in Tables 2 and 3. The mean squares due to genotypes showed significant differences at P < 0.05 for all the traits studied. Based on the performance, a considerable range of variation was exhibited by all the plant traits studied at both control and water stress levels (Table 4).
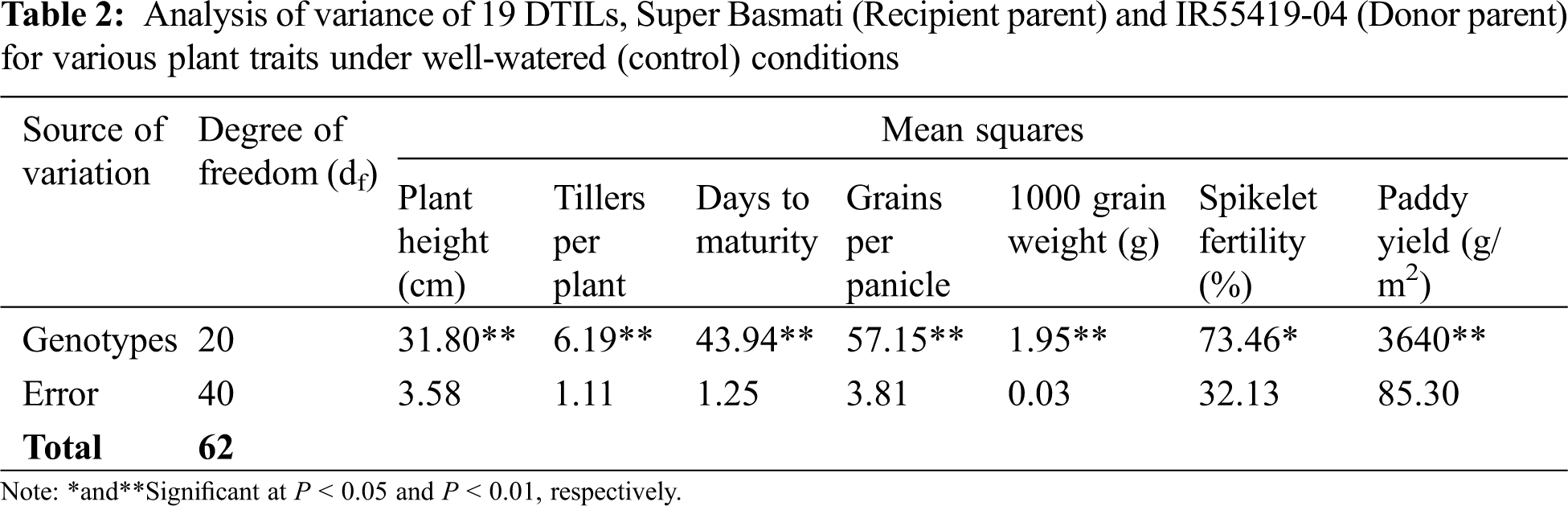
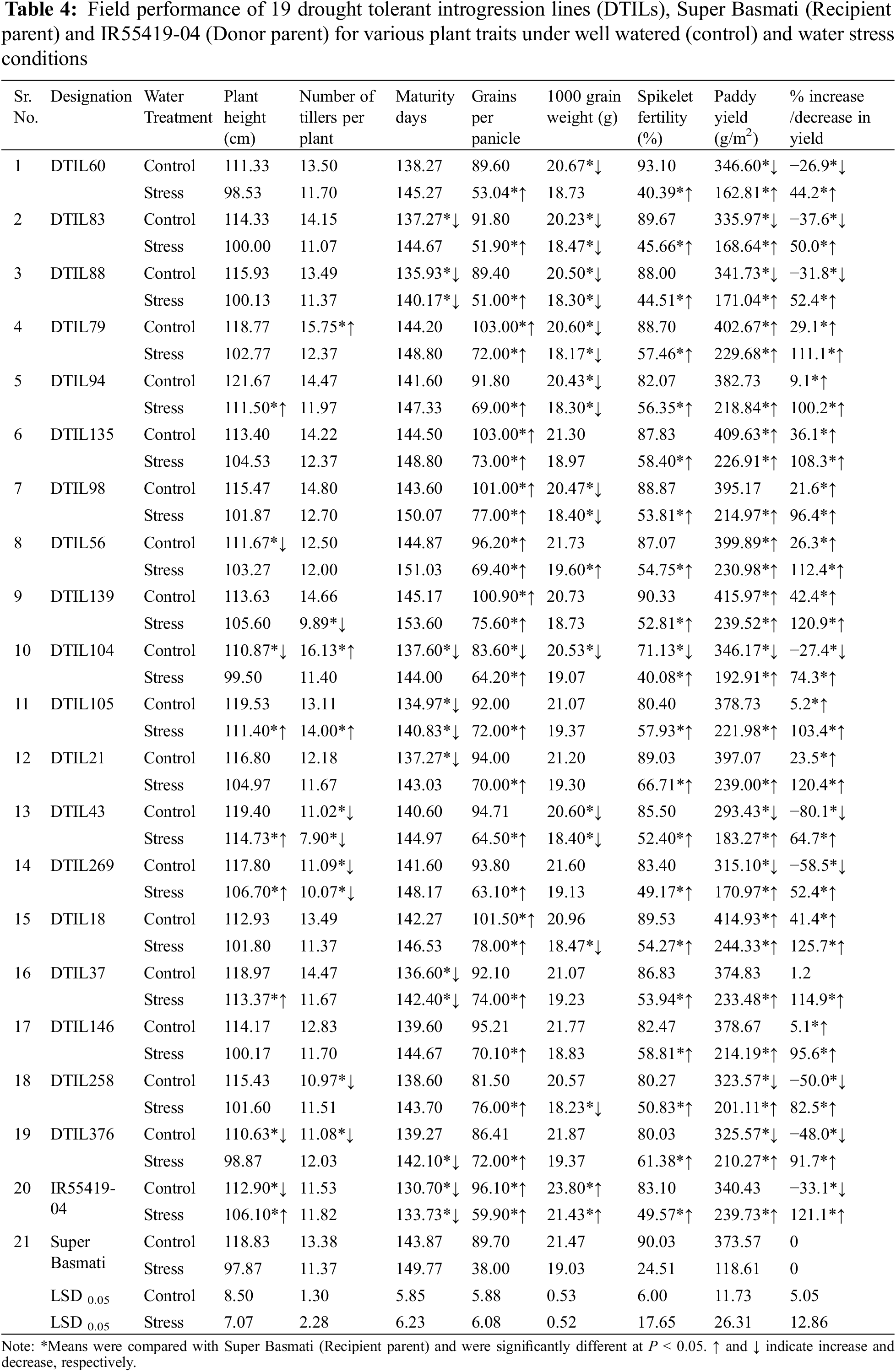
Under well-watered conditions, significant differences were observed between the two parents for plant height, maturity days, number of grains per panicle and 1000 grain weight, whereas there were non-significant differences in other traits. Three DTILs viz. DTIL376, DTIL104 and DTIL56 gained significantly decreased plant height of 110.63, 110.87, and 111.67, respectively, while the rest of the DTILs produced plant height statistically similar to Super Basmati.
Under water stress treatment, parental lines, i.e., Super Basmati and IR55419-04 exhibited significant differences in all the plant traits studied except number of tillers per plant (Table 4). Plant height (cm) attained by five introgression lines viz. DTIL94 (111.50), DTIL105 (111.40), DTIL43 (114.73), DTIL269 (106.70) and DTIL37 (113.37) was significantly more than Super Basmati (97.87). The introgression lines exhibited less, or similar number of tillers as compared to Super Basmati (11.37) whereas only one DTIL105 (14.00) showed a significantly more number of tillers per plant.
Based on introgressed QTLs, nineteen DTILs were grouped into 7 categories, i.e., G1 (3 DTILs carrying QTL1), G2 (4 DTILs carrying QTL4), G3 (2 DTILs carrying QTL9), G4 (1 DTIL carrying QTL1 and QTL4), G5 (2 DTILs carrying QTL1 and QTL4), G6 (2 DTILs carrying QTL4 and QTL9) and G7 (5 DTILs carrying QTL1, QTL4 and QTL9) as shown in Table 4. Under well-watered control, average paddy yield (gram/meter2) of G2 (397.6) and G3 (404.8) were significantly higher than Super Basmati whereas G4 (346.2) and G6 (304.3) produced less with significant impact. When exposed to water stress conditions, average paddy yield (range 167.5–239.7) of all the groups was significantly higher than Super Basmati (118.6). Data revealed that the relative paddy yield reduction was considerably less than Super Basmati that was affected up to a yield reduction of 68.2% (Table 4).
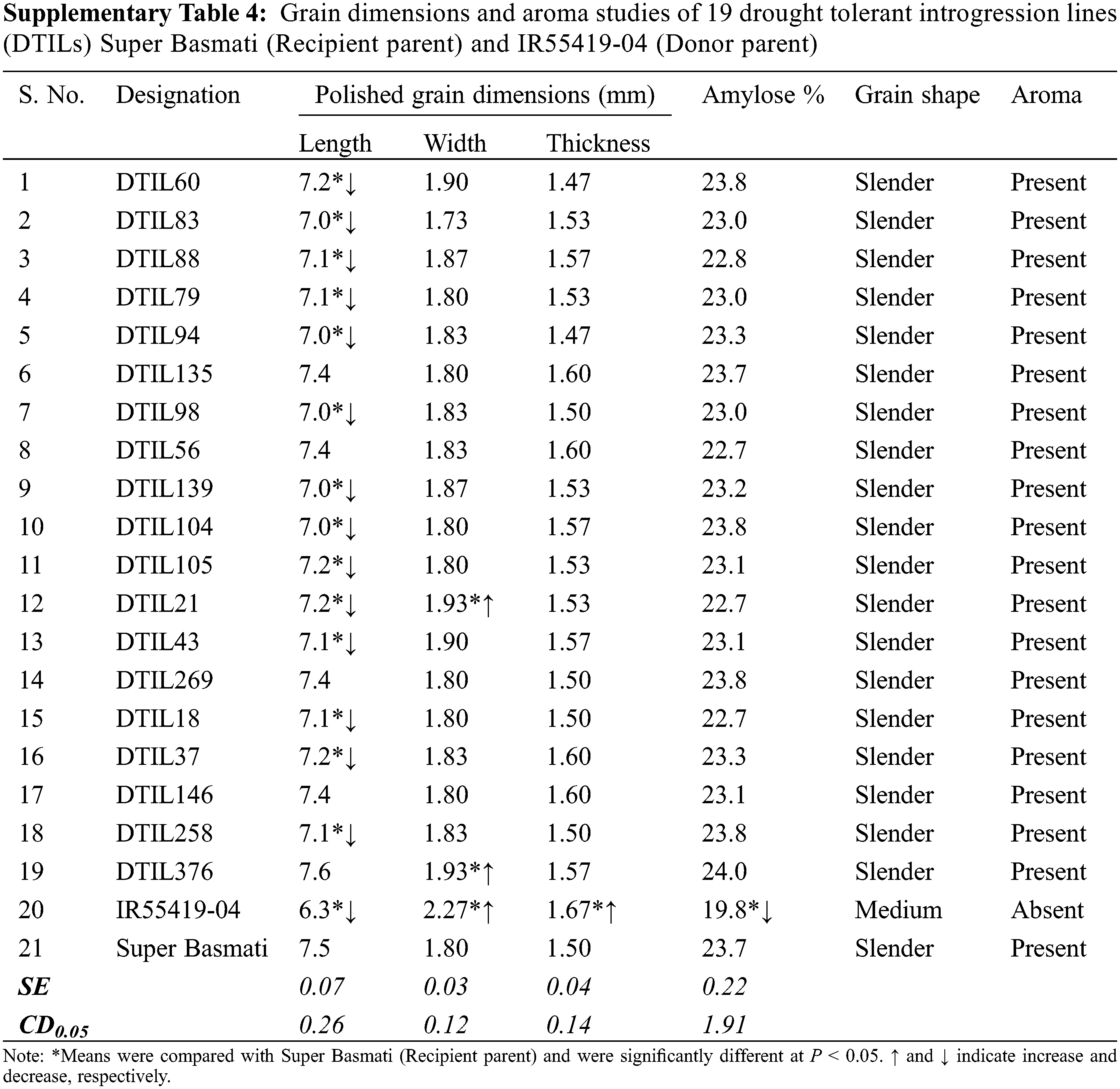
Significant differences were recorded for polished grain dimensions, i.e., grain length, width and thickness between the recipient and donor parents. Polished grain length, grain width and grain thickness were compared with Super Basmati to assess the grain dimensions of the DTILs. Significant differences were observed for grain length and grain width of DTILs (SupplementaryTable 4). Selected DTILs exhibited a significant decrease (range 7.0–7.2) in grain length (mm) as compared to Super Basmati (7.5) except DTIL135 (7.4), DTIL56 (7.4), DTIL146 (7.4) and DTIL376 (7.6). Polished grain width (mm) of DTIL21 (1.93) and DTIL376 (1.93) were statistically higher than Super Basmati (1.80) while the rest of the DTILs exhibited grain width similar to Super Basmati. Polished grain thickness (mm) of selected lines (range 1.47–1.60) was similar to Super Basmati (1.50). Grains of the DTILs were slender shape and were similar to Super Basmati. The presence of aroma was recorded for the selected DTILs as Super Basmati.
4.1 Parental Survey to Check Polymorphism
Backcross breeding is a recognized approach for introgression a specific chromosomal region (gene/QTL) into a recipient genotype (mostly a well-adapted variety) from a donor genotype. For the genome recovery of the recipient parent, repeated Back crossing remained as the main objective of this approach [21]. That is achieved by whole genomic survey based on polymorphic markers between the parents. Therefore, for the current study an SSR marker survey was carried out that resulted in 99 polymorphic microsatellites out of 207 that counted 47.8% polymorphism between the two parents. The level of polymorphism found was good enough indicating the genetic distance between the two parents for the marker loci used for the survey.
The tendency of genes or loci that are situated adjacently on a chromosome to be inherited in a group together during meiosis is referred as genetic linkage and in marker assisted breeding is known as linkage drag. Such loci that exhibit genetic linkage are less likely to be separated onto different chromatids during chromosomal crossover [21,22]. In this study, this might be due to this strong genetic linkage between the loci RM220 and RM1. The frequency of recombination between two locations is directly proportional to the distance between the loci. The level of recombination between RM220 and RM1 was comparatively much lower than the recombination between RM84 and RM428 indicating that the genetic distance between RM220 and RM1 (1.3 cM) is less than the distance between RM84 and RM428 (6.9 cM) on chromosome 1 (Fig. 2). Young et al. [23] pointed out that the donor allele/linkage drag on the carrier chromosome were the most difficult to eliminate [24]. Non targeted donor segments at RM495 and RM246 on carrier chromosome 1 whereas at RM471 and RM119 on chromosome 9 were also observed (Fig. 2).
4.3 Linkage Drag and Genome Recovery
In the present study, the linkage drag in proximity to the target QTLs was minimized with the help of more polymorphic markers around the target loci. Even then, the linkage drag was observed in the target loci. This indicated the limitation of introgression for more than one segment in one genetic background while selecting an individual in two cycles Back crossing. DTILs were developed within two generations of Back crossing and one generation of self-pollination as outlined in MABB scheme (Fig. 1).
Due to selection for the donor segment at the target loci in each generation, the rate of recovery for recipient genotype on carrier chromosome is observed to be slower than on non-carrier chromosomes [25]. Genome recovery percentage of recipient parent alleles on carrier chromosomes 1, 4 and 9 carrying 3 targets QTLs were low as compared to other chromosomes. Similar findings have been reported by earlier MAB cases in rice [26–28].
4.4 Background Selection and Phenotypic Selection
Accumulating the number of background selection markers in early generations on non-carrier chromosomes was not much effective due to probability of crossover in later generations. An increase in the number of markers should be considered as a way of optimizing selection [29]. In our study, the whole genome background selection was done at BC2F2 and the average marker density for background selection was 16.8 cM with 6 to 9 markers per chromosome. Previous studies suggested that, one marker every 5 Mb (20 cM) or a total of 4 to 9 well distributed markers per chromosome is enough to monitor the donor introgressions [30]. This was also reported in other MABB simulation studies [31,32].
The selection scheme was adopted in a modified way in which before background selection, phenotypic selection for traits like plant height, leaf morphology, plant type, maturity days and especially the grain dimensions was done that reduced the population size for background selection. Somehow, in two backcrosses, we able to restore up to 92.9% alleles of Super Basmati that reflects the successful application of phenotypic selection cycle prior to background selection. Moreover, this approach not only saved the time, but also the resources in term of reduced cost of genotyping.
About 90% dry matter contents of a milled rice kernel is made up of starch and amylose content (range 15% to 35%) of starches determines the rice cooking quality in the sense of volume expansion. As Basmati rice is famous in the global market for its typical aroma and unique grain quality and our objective was to develop introgression lines in basmati background, therefore, the inheritance of these two traits was ensured by using the linked markers in MABB.
4.5 Yield Improvement under Water Stress
Grain yield under drought has been proposed as selection criteria for water stress breeding programs [33]. Following this approach, improved genotypes has been evolved in various rice growing countries like the Philippines, India, Cambodia [31,34,35]. We also adopted the same strategy for evaluation of DTILs. Under water stress treatment, a significant improvement in yield under drought was observed in all the DTILs. DTILs groups showed significantly less yield reduction percentage (range 40.6–51.9) as compared to Super Basmati which was mainly contributed by increased number of grains per panicle and gain in spikelet fertility percentage. Marker-aided breeding can also significantly improve genetic gain for the traits that are difficult to phenotype because of its dependence on specific environmental conditions or its expense [36,37]. Under well watered control, field evaluation results showed that most of the DTILs performed similar to the Super Basmati for all the observed plant traits. Significant variation in trait expression may be due to pleotropic or epistatic effect of introgressed loci from the donor parent [38,39]. Reduction in plant height is a desired trait in all breeding programs [40] and hence we can say that this trait has been improved. Similarly, yield potential of some DTILs was improved that as well might be the result of some heterozygous loci transgression present in the genome. Although, a decrease in grain size was observed in most of the DTILs but there were some DTILs that gave grain dimensions like Super Basmati. This means that grain length linked markers should be used in future. Grain quality parameters like amylose contents, grain shape and aroma were similar to Super Basmati.
The present study results showed the efficiency of MABB that resulted in the development of introgression lines with two backcross cycles. Six superior (DTIL56, DTIL60, DTIL135, DTIL146, DTIL269 and DTIL376) ILs that are drought tolerant and very similar to Super Basmati in terms of agronomic and grain quality traits can be released as drought-tolerant varieties in arid regions. This approach has been successfully practiced in earlier rice breeding programs for the introgression of bacterial blight resistance gene Xa38 [41], QTLs controlling root traits [17], submergence tolerance Sub1 gene [28], and the green revolution sd1 gene [3].
In all, six drought tolerant introgression lines viz., DTIL56, DTIL60, DTIL135, DTIL146, DTIL269 and DTIL376 are found to be superior and are very similar to Super Basmati for agronomic and grain quality traits. These well characterized drought tolerant introgression lines developed in the present study can be released as variety or used as donors to develop new drought tolerant basmati rice varieties in the future breeding programs.
Acknowledgement: We acknowledge Michael J. Thomson and Reza Mohammadi for provision of drought tolerant lines. We pay our special thanks to Maria Ymber Reveche, Socorro Carandang, and Pauline Capistrano for their guidance and help during research.
Funding Statement: We thank Higher Education Commission of Pakistan and Green Super Rice Project a Component of Productivity Enhancement of Rice (PSDP No. 0754), Pakistan for providing financial support.
Author Contributions: M.S. and M.A. designed the experiments and obtained funding for the research. M.S., S.A.N., S.M.S. and A.R., contributed to compiling and analyzing the data and wrote the manuscript. M.M.S., T.A., and M.R.K. conducted statistical analysis. S.A.N., J.X., and M.A. participated in the data analysis and supervised the writing of the manuscript. All authors read and approved the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Khoso, S., Wagan, F. H., Tunio, A. H., Ansari, A. A. (2015). An overview on emerging water scarcity in Pakistan, its causes, impacts and remedial measures. Journal of Applied Engineering Science, 13(1). https://doi.org/10.5937/jaes13-6445 [Google Scholar] [CrossRef]
2. Rahman, H., Dakshinamurthi, V., Ramasamy, S., Manickam, S., Kaliyaperumal, A. K. et al. (2018). Introgression of submergence tolerance into CO 43, a popular rice variety of India, through marker-assisted backcross breeding. Czech Journal of Genetics and Plant Breeding, 54(3), 101–108. https://doi.org/10.17221/149/2017-CJGPB [Google Scholar] [CrossRef]
3. Tomita, M. (2009). Introgression of green revolution sd1 gene into isogenic genome of rice super cultivar Koshihikari to create novel semidwarf cultivar ‘Hikarishinseiki’ (Koshihikari-sd1). Field Crops Research, 114(2), 173–181. https://doi.org/10.1016/j.fcr.2009.05.004 [Google Scholar] [CrossRef]
4. Singh, U. M., Dixit, S., Alam, S., Yadav, S., Prasanth, V. V. et al. (2022). Marker-assisted forward breeding to develop a drought-, bacterial-leaf-blight-, and blast-resistant rice cultivar. The Plant Genome, 15(1), e20170. https://doi.org/10.1002/tpg2.20170 [Google Scholar] [PubMed] [CrossRef]
5. Shamsudin, N. A., Swamy, B. P., Ratnam, W. et al. (2016). Marker assisted pyramiding of drought yield QTLs into a popular Malaysian rice cultivar, MR219. BMC Genet, 17, 30. https://doi.org/10.1186/s12863-016-0334-0 [Google Scholar] [PubMed] [CrossRef]
6. Rehman, A., Jingdong, L., Chandio, A. A., Shabbir, M., Hussain, I. (2017). Economic outlook of rice crops in Pakistan: A time seried analysis (1970–2015). Financial Innovation, 3(13). https://doi.org/10.1186/s40854-017–0063-z [Google Scholar] [CrossRef]
7. Yang, X., Wang, B., Chen, L., Li, P., Cao, C. (2019). The different influences of drought stress at the flowering stage on rice physiological traits, grain yield, and quality. Scientific Reports, 9(1), 3742. https://doi.org/10.1038/s41598-019-40161-0 [Google Scholar] [PubMed] [CrossRef]
8. Anser, M. K., Hina, T., Hameed, S., Nasir, M. H., Ahmad, I. et al. (2020). Modeling adaptation strategies against climate change impacts in integrated rice-wheat agricultural production system of Pakistan. International Journal of Environmental Research and Public Health, 17(7), 2522. https://doi.org/10.3390/ijerph17072522 [Google Scholar] [PubMed] [CrossRef]
9. Misra, A. K. (2014). Climate change and challenges of water and food security. International Journal of Sustainable Built Environment, 3(1), 153–165. https://doi.org/10.1016/j.ijsbe.2014.04.006 [Google Scholar] [CrossRef]
10. Dixit, S., Yadaw, R. B., Mishra, K. K., Kumar, A. (2017). Marker-assisted breeding to develop the drought-tolerant version of Sabitri, a popular variety from Nepal. Euphytica, 213(8), 184. https://doi.org/10.1007/s10681-017-1976-3 [Google Scholar] [PubMed] [CrossRef]
11. Janaki, R. P., Vinukonda, V. P., Singh, U. M., Alam, S., Venkateshwarlu, C. et al. (2021). Marker-assisted forward and backcross breeding for improvement of elite Indian rice variety Naveen for multiple biotic and abiotic stress tolerance. PLoS One, 16(9), e0256721. https://doi.org/10.1371/journal.pone.0256721 [Google Scholar] [PubMed] [CrossRef]
12. Dhawan, G., Kumar, A., Dwivedi, P., Gopala Krishnan, S., Pal, M. et al. (2021). Introgression of qDTY1. 1 governing reproductive stage drought tolerance into an elite basmati rice variety “Pusa Basmati 1” through marker assisted backcross breeding. Agronomy, 11(2), 202. https://doi.org/10.3390/agronomy11020202 [Google Scholar] [CrossRef]
13. Panda, D., Mishra, S. S., Behera, P. K. (2021). Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Science, 28(2), 119–132. https://doi.org/10.1016/j.rsci.2021.01.002 [Google Scholar] [CrossRef]
14. Kumar, A., Dixit, S., Ram, T., Yadaw, R., Mishra, K. et al. (2014). Breeding high-yielding drought-tolerant rice: Genetic variations and conventional and molecular approaches. Journal of Experimental Botany, 65(21), 6265–6278. https://doi.org/10.1093/jxb/eru363 [Google Scholar] [PubMed] [CrossRef]
15. Robin, S., Pathan, M., Courtois, B., Lafitte, R., Carandang, S. et al. (2003). Mapping osmotic adjustment in an advanced back-cross inbred population of rice. Theoretical and Applied Genetics, 107(7), 1288–1296. https://doi.org/10.1007/s00122-003-1360-7 [Google Scholar] [PubMed] [CrossRef]
16. Barik, S. R., Pandit, E., Monhanty, S. P., Nayak, D. K., Pradhan, S. K. (2020). Genetic mapping of physiological traits associated with terminal stage drought tolerance in rice. BMC Genet, 21(76). https://doi.org/10.1186/s12863-020-00883-x [Google Scholar] [PubMed] [CrossRef]
17. Steele, K., Price, A. H., Shashidhar, H., Witcombe, J. (2006). Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theoretical and Applied Genetics, 112, 208–221. [Google Scholar] [PubMed]
18. Amarawathi, Y., Singh, R., Singh, A. K., Singh, V. P., Mohapatra, T. et al. (2008). Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Molecular Breeding, 21, 49–65. [Google Scholar]
19. Temnykh, S., DeClerck, G., Lukashova, A., Lipovich, L., Cartinhour, S. et al. (2001). Computational and experimental analysis of microsatellites in rice (Oryza sativa L.Frequency, length variation, transposon associations, and genetic marker potential. Genome Research, 11, 1441–1452. [Google Scholar] [PubMed]
20. Cruz, N. D., Khush, G. (2000). Rice grain quality evaluation procedures. Aromatic Rices, 3, 15–28. [Google Scholar]
21. Bouchez, A., Hospital, F., Causse, M., Gallais, A., Charcosset, A. (2002). Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics, 162, 1945–1959. [Google Scholar] [PubMed]
22. Kurata, N., Nonomura, K., Harushima, Y. (2002). Rice genome organisation: The centromere and genome interactions. Annals of Botany, 90(4). https://doi.org/10.1093/aob/mcf218 [Google Scholar] [PubMed] [CrossRef]
23. Young, N., Tanksley, S. (1989). RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theoretical and Applied Genetics, 77(3), 353–359. https://doi.org/10.1007/BF00305828 [Google Scholar] [PubMed] [CrossRef]
24. Randhawa, H. S., Mutti, J. S., Kidwell, K., Morris, C. F., Chen, X. et al. (2009). Rapid and targeted introgression of genes into popular wheat cultivars using marker-assisted background selection. PLoS One, 4(6), e5752. https://doi.org/10.1371/journal.pone.0005752 [Google Scholar] [PubMed] [CrossRef]
25. Frisch, M., Bohn, M., Melchinger, A. A. (1999). Minimum sample size and optimal positioning of flanking markers in marker-assisted backcrossing for transfer of a target gene. Crop Science, 39(4), 967–975. https://doi.org/10.2135/cropsci1999.0011183X003900040003x [Google Scholar] [CrossRef]
26. Hasan, M. M., Rafii, M. Y., Ismail, M. R., Mahmood, M., Rahim, H. A. et al. (2015). Marker-assisted backcrossing: A useful method for rice improvement. Biotechnology & Biotechnological Equipment, 29(2), 237–254. https://doi.org/10.1080/13102818.2014.995920 [Google Scholar] [PubMed] [CrossRef]
27. Joseph, M., Gopalakrishnan, S., Sharma, R., Singh, V., Singh, A. et al. (2004). Combining bacterial blight resistance and Basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Molecular Breeding, 13(4), 377–387. https://doi.org/10.1023/B:MOLB.0000034093.63593.4c [Google Scholar] [CrossRef]
28. Neeraja, C. N., Maghirang-Rodriguez, R., Pamplona, A., Heuer, S., Collard, B. C. et al. (2007). A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theoretical and Applied Genetics, 115(6), 767–776. https://doi.org/10.1007/s00122-007-0607-0 [Google Scholar] [PubMed] [CrossRef]
29. Maazou, M. A., Tu, J., Qiu, J. M., Liu, Z. (2016). Breeding for drought tolerance in Maize (Zea mays L.). American Journal of Plant Science, 7(1858–1870). https://doi.org/10.4236/ajps.2016.714172 [Google Scholar] [CrossRef]
30. Septiningsih, E. M., Pamplona, A. M., Sanchez, D. L., Neeraja, C. N., Vergara, G. V. et al. (2009). Development of submergence-tolerant rice cultivars: The Sub1 locus and beyond. Annals of Botany, 103(2), 151–160. https://doi.org/10.1093/aob/mcn206 [Google Scholar] [PubMed] [CrossRef]
31. Khan, G. H., Shikari, A. B., Vaishnavi, R., Najeeb, S., Padder, B. A. et al. (2018). Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Scientific Reports, 8(1), 1–13. https://doi.org/10.1038/s41598-018-22246-4 [Google Scholar] [PubMed] [CrossRef]
32. Servin, B., Martin, O. C., Mézard, M., Hospital, F. (2004). Toward a theory of marker-assisted gene pyramiding. Genetics, 168(1), 513–523. https://doi.org/10.1534/genetics.103.023358 [Google Scholar] [PubMed] [CrossRef]
33. Bernier, J., Atlin, G. N., Serraj, R., Kumar, A., Spaner, D. (2008). Breeding upland rice for drought resistance. Journal of the Science of Food and Agriculture, 88, 927–939. https://doi.org/10.1002/(ISSN)1097-0010 [Google Scholar] [CrossRef]
34. Bernier, J., Kumar, A., Venuprasad, R., Spaner, D., Verulkar, S. et al. (2009). Characterization of the effect of a QTL for drought resistance in rice, qtl12.1, over a range of environments in the Philippines and eastern India. Euphytica, 166(2), 207–217. https://doi.org/10.1007/s10681-008-9826-y [Google Scholar] [CrossRef]
35. Ouk, M., Basnayake, J., Tsubo, M., Fukai, S., Fischer, K. et al. (2006). Use of drought response index for identification of drought tolerant genotypes in rainfed lowland rice. Field Crops Research, 99(1), 48–58. https://doi.org/10.1016/j.fcr.2006.03.003 [Google Scholar] [CrossRef]
36. Moose, S. P., Mumm, R. H. (2008). Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiology, 147(3), 969–977. https://doi.org/10.1104/pp.108.118232 [Google Scholar] [PubMed] [CrossRef]
37. Hasan, N., Choudhary, S., Naaz, N., Sharma, N., Laskar, R. A. (2021). Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. Journal of Genetic Engineering and Biotechnology, 19(1), 1–26. https://doi.org/10.1186/s43141-021-00231-1 [Google Scholar] [PubMed] [CrossRef]
38. Liu, X., Jin, J., Wang, G., Herbert, S. (2008). Soybean yield physiology and development of high-yielding practices in Northeast China. Field Crops Research, 105, 157–171. [Google Scholar]
39. Beerelli, K., Balakrishnan, D., Addanki, K. R., Surapaneni, M., Neelamraju, S. (2022). Mapping of QTLs for yield traits using F 2:3:4 populations derived from two alien introgression lines reveals qTGW8. 1 as a consistent QTL for grain weight from Oryza nivara. Frontiers in Plant Science, 13, 790221. [Google Scholar] [PubMed]
40. Vikram, P., Swamy, B., Dixit, S., Singh, R., Singh, B. P. et al. (2015). Drought susceptibility of modern rice varieties: An effect of linkage of drought tolerance with undesirable traits. Scientific Reports, 5, 1–18. [Google Scholar]
41. Yugander, A., Sundaram, R. M., Singh, K., Ladhalakshmi, D., Subba Rao, L. V. et al. (2018). Incorporation of the novel bacterial blight resistance gene Xa38 into the genetic background of elite rice variety Improved Samba Mahsuri. PLoS One, 13, e0198260. [Google Scholar] [PubMed]
Supplementary Materials
Supplementary Figure 1: (a) Graphical genotype of DTIL135 (Q4). Green and red colors depict the homozygous alleles of Super Basmati (recipient) and IR55419-04 (donor), respectively while light blue color indicates the heterozygous status of specific marker loci. Both the size scale indicating chromosome lengths in centiMorgans (cM) and marker names are on the left. (b) Paddy rice of IR55419-04 (left), DTIL135 (center) and Super Basmati (right)
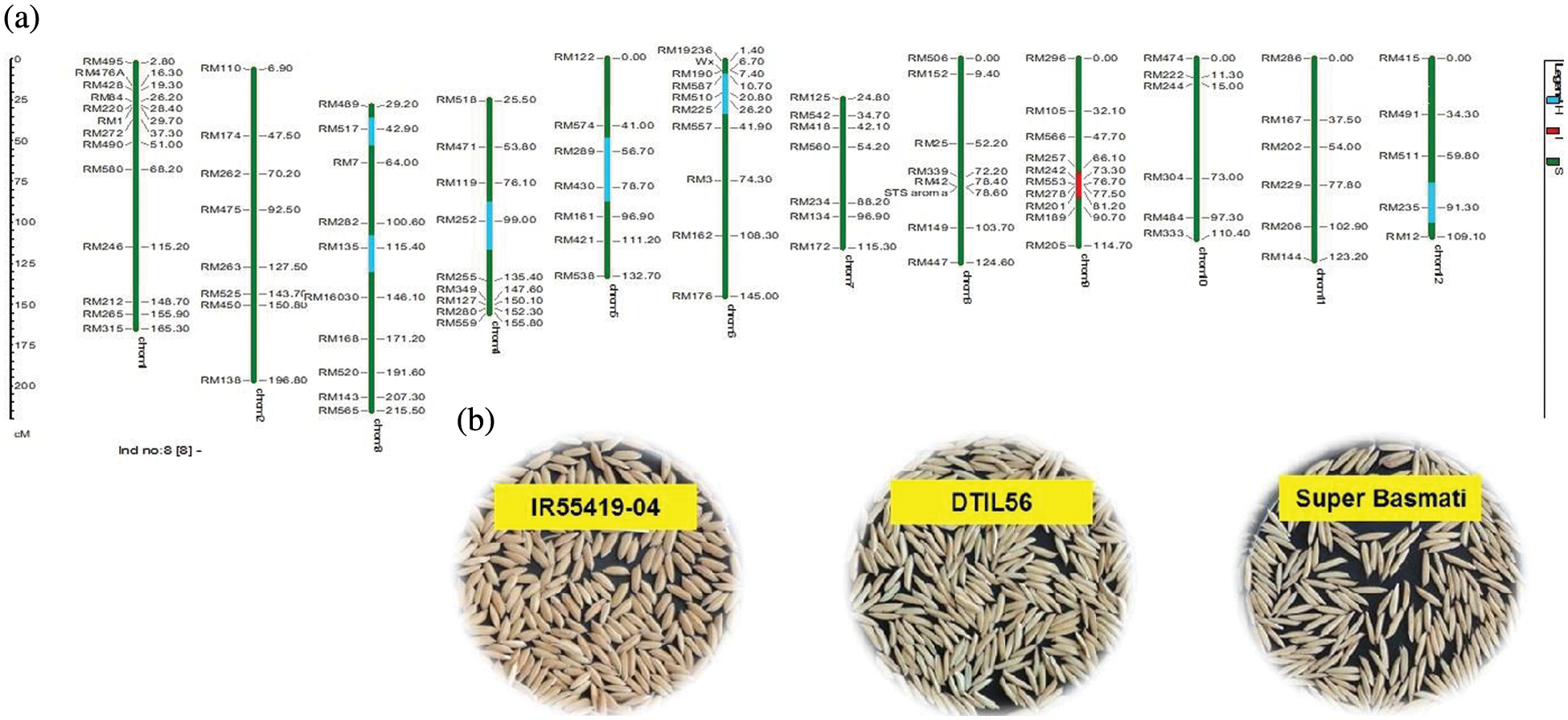
Supplementary Figure 2: (a) Graphical genotype of DTIL56 (Q9). Green and red colors depict the homozygous alleles of Super Basmati (recipient) and IR55419-04 (donor), respectively while light blue color indicates the heterozygous status of specific marker loci. Both the size scale indicating chromosome lengths in centiMorgans (cM) and marker names are on the left. (b) Paddy rice of IR55419-04 (left), DTIL56 (center) and Super Basmati (right)
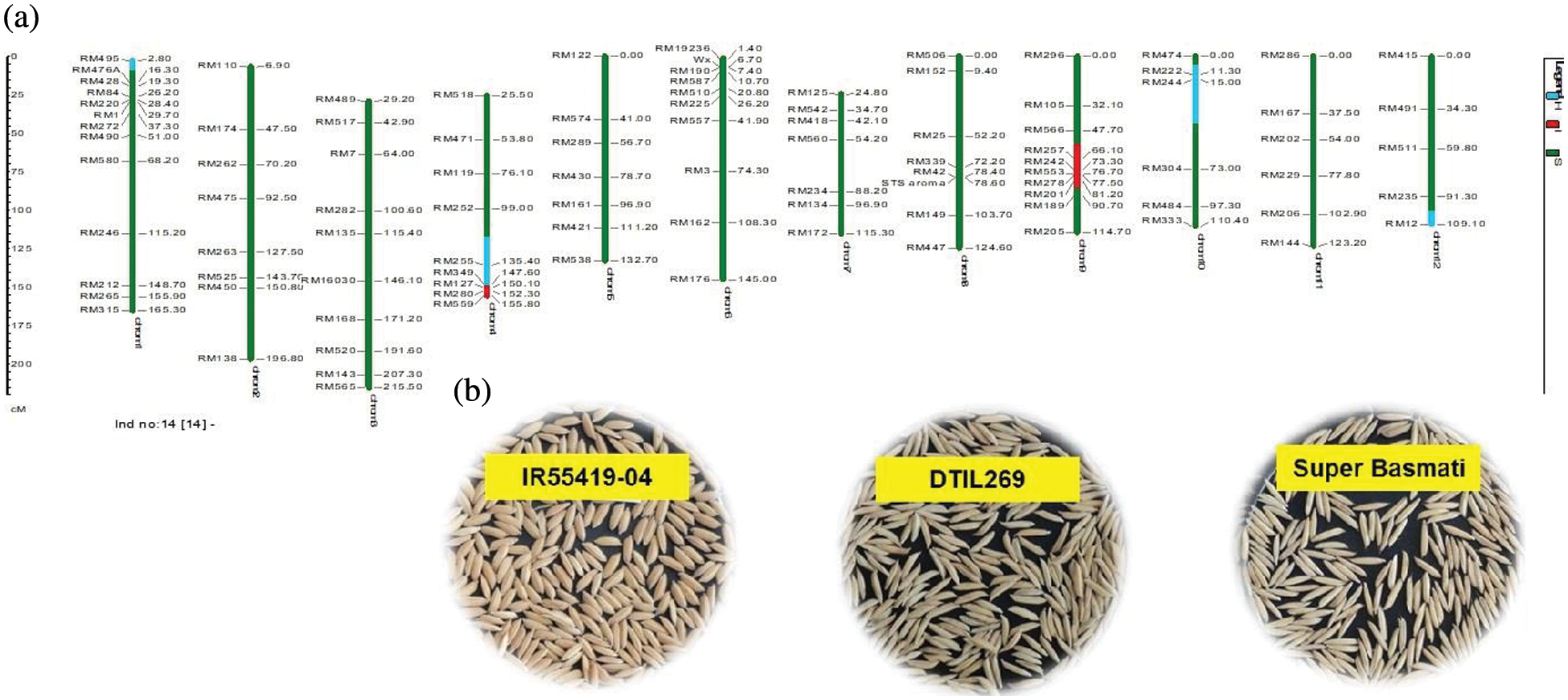
Supplementary Figure 3: (a) Graphical genotype of DTIL269 (Q4+Q9). Green and red colors depict the homozygous alleles of Super Basmati (recipient) and IR55419-04 (donor), respectively while light blue color indicates the heterozygous status of specific marker loci. Both the size scale indicating chromosome lengths in centiMorgans (cM) and marker names are on the left. (b) Paddy rice of IR55419-04 (left), DTIL269 (center) and Super Basmati (right)
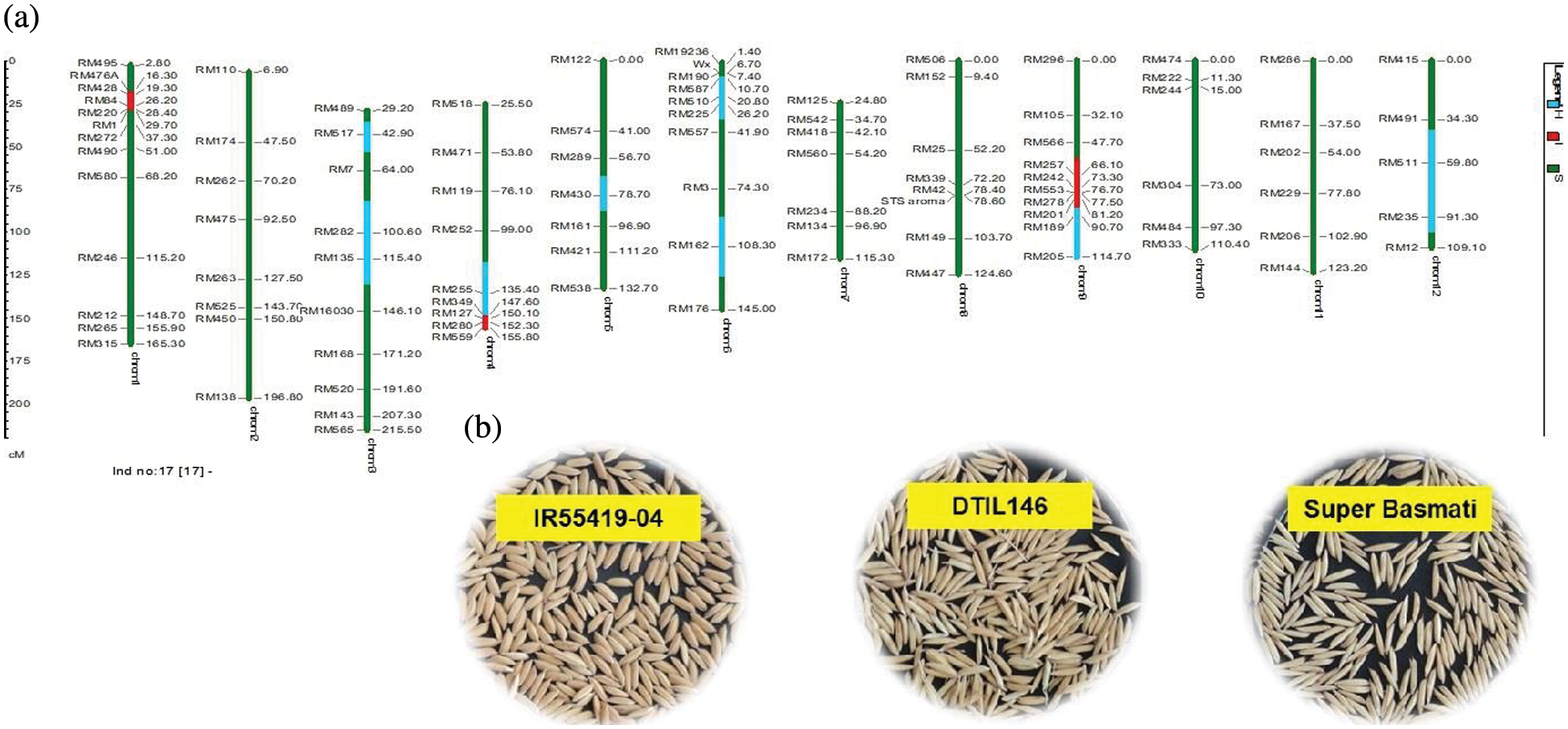
Supplementary Figure 4: (a) Graphical genotype of DTIL146 (Q1+Q4+Q9). Green and red colors depict the homozygous alleles of Super Basmati (recipient) and IR55419-04 (donor), respectively while light blue color indicates the heterozygous status of specific marker loci. Both the size scale indicating chromosome lengths in centiMorgans (cM) and marker names are on the left. (b) Paddy rice of IR55419-04 (left), DTIL146 (center) and Super Basmati (right)
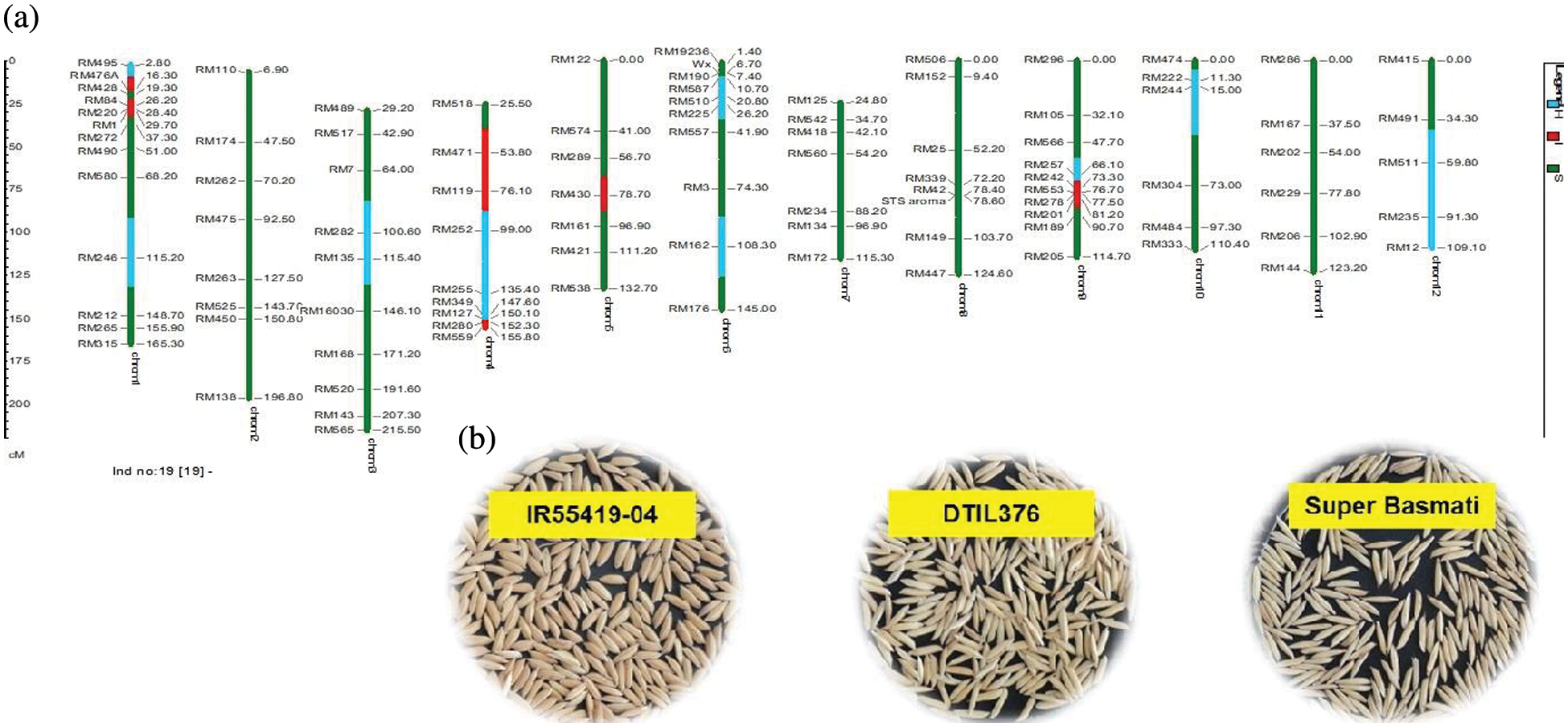
Supplementary Figure 5: (a) Graphical genotype of DTIL376 (Q1+Q4+Q9). Green and red colors depict the homozygous alleles of Super Basmati (recipient) and IR55419-04 (donor), respectively while light blue color indicates the heterozygous status of specific marker loci. Both the size scale indicating chromosome lengths in centiMorgans (cM) and marker names are on the left. (b) Paddy rice of IR55419-04 (left), DTIL376 (center) and Super Basmati (right)
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools