 Open Access
Open Access
ARTICLE
Pre-Breeding Genetic Diversity Assessment of Tomato (Solanum lycopersicum L.) Cultivars Based on Molecular, Morphological and Physicochemical Parameters
1
Department of Agricultural Biotechnology, College of Agriculture and Food Sciences, King Faisal University, Al-Ahsa, 31982,
Saudi Arabia
2
Department of Biology, College of Science, University of Jeddah, Jeddah, 21959, Saudi Arabia
3
Department of Arid Land Agriculture, College of Agriculture and Food Sciences, King Faisal University, Al-Ahsa, 31982,
Saudi Arabia, Plant Pests, and Diseases Unit, College of Agriculture and Food Sciences, King Faisal University, Al-Ahsa, 31982,
Saudi Arabia
4
Department of Horticulture, Faculty of Agriculture, Kafrelsheikh University, Kafr El-Sheikh, 33516, Egypt
5
Department of Biochemistry, Faculty of Agriculture, Zagazig University, Zagazig, 44511, Egypt
6
Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671,
Saudi Arabia
7
Central Laboratories, King Faisal University, Al-Ahsa, 31982, Saudi Arabia
8
Department of Botany and Microbiology, Faculty of Science, South Valley University, Qena, 83523, Egypt
9
Department of Genetics, Faculty of Agriculture, Zagazig University, Zagazig, 44511, Egypt
* Corresponding Authors: Jameel M. Al-Khayri. Email: ; Abdallah A. Hassanin. Email:
,
Phyton-International Journal of Experimental Botany 2023, 92(5), 1493-1512. https://doi.org/10.32604/phyton.2023.027375
Received 27 October 2022; Accepted 05 January 2023; Issue published 09 March 2023
Abstract
Appropriate knowledge of the parental cultivars is a pre-requisite for a successful breeding program. This study characterized fruit yield, quality attributes, and molecular variations of ten tomato cultivars during three consecutive generations under greenhouse conditions. Peto 86, Castle Rock, and Red Star cultivars showed the highest fruit yield (kg/plant), total phenolic compounds (TPC), and sap acidity. Principal component analysis categorized the evaluated fruit yield into three groups based on their quality attributes. A robust positive correlation appeared among traits inside each group. A positive correlation was likewise noticed between the first and the second groups. However, a negative correlation was detected between the first, the second and the third group. Molecular profiling, using seven inter-simple sequence repeat (ISSR) primers, produced 60 loci, including 49 polymorphic loci. The molecular analysis also pinpointed the highest genetic similarity (0.92) between P73 and Moneymaker, while the lowest genetic similarity (0.46) was observed between Castle Rock and Moneymaker. The cultivars P73 and Moneymaker showed the lowest genetic distance (2.24), while the highest genetic distance (5.92) was observed between Super Marmand and Peto86, on the one hand, and between Castle Rock and Moneymaker, on the other hand. The chemical analysis of fruit sap indicated the highest levels of TPC, total flavonoids, anthocyanin, ascorbic acid and total soluble solids in Peto 86 and Castle Rock cultivars. Phylogeny analysis of tomato cultivars based on morphological and molecular attributes indicated four distinct clades. Peto 86, Castle Rock, and Red star cultivars can be recommended for the tomato hybridization breeding programs in the future, with other tomato cultivars as potentially high-yielding parents.Keywords
Tomato (Solanum lycopersicum L.) is ranked as the 2nd most-produced and consumed vegetable crop after potato and a rich source of vital minerals and antioxidants [1]. Besides its global nutritional and economic value in agriculture as a vegetable crop, tomato is a suitable model for studying the genetics of flowering plants [2,3]. In the research studies, the morphological attributes of tomatoes should be combined with the relevant physicochemical and molecular characteristics, particularly in studies aimed to minimize the effects of environmental conditions under which the plant genotype is grown [4]. For example, different plant characteristics such as fruit weight, inflorescence length, number of flowers and fruits per inflorescence can influence the total tomato yield [5]. In addition, developmental processes, including inflorescence formation, floral development, and fruit ripening also considerably impact tomato yield [6].
Recently, breeding for fruit quality, including flavor and morphological features, has attracted the attention of seed producers due to its significant influence on consumers’ demand and choice [7–9]. In the past decades, tomato breeding has passed through three main goals: breeding for yield, prolonged shelf-life, and fruit taste. Recently, tomato breeders started to put more efforts to develop new tomato cultivars with high nutritional value [7,10]. Conventional plant breeding based on morphological selection and progeny testing effectively improved crop quality and quantity during the past several decades [6,11]. However, conventional breeding approaches are tedious and extravagant, therefore alternate approaches, such as use of molecular markers, have been successfully employed to speed-up conventional breeding approaches. The use of molecular markers is an effective approach for rapid and accurate estimation of genetic diversity and genetic similarity among different crop cultivars for specific traits. Inter-varietal genetic diversity of different tomato varieties has been successfully studied using many PCR-based DNA markers such as random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), simple sequence repeats (SSRs), inter-simple sequence repeat polymorphism (ISSR) and others [12]. Comparatively, ISSR-based molecular markers provide a robust analysis of genetic variations and has been efficiently used to study the genetic variability at very low levels [1]. The ISSR is an attractive method to generate molecular genetic markers and, thus, molecular profiling by combining most of the advantages of simple sequence repeat (SSR) and amplified fragment length polymorphism (AFLP) to random amplified polymorphic DNA (RAPD) [13]. Furthermore, using long primers enables the ISSR technique to employ higher annealing temperatures, achieving high annealing specificity [14]. Practically, knowing yield and quality attributes and using molecular markers and marker-assisted selection can facilitate the breeding of tomatoes, leading to new distinct hybrids with improved features such as quality and yield [15].
This study aimed to compare ten featured tomato cultivars for their yield-related molecular, morphological, and physicochemical profiles. The potentially generated information is supposed to support the subsequent yield and quality-targeted breeding programs and hybrid seed production.
All cultivars were selected based on their advantages relevant to hybrid seed production, e.g., earliness, resistance to diseases, good yield, and sweet flavor. The seeds of ten international tomato cultivars, i.e., UC 97-3 (USA), Super Marmande (France), Super Queen (USA), Castel Rock (USA), Super Strain B (USA), Red Star (USA), Peto 86 (USA), and Strain B (USA) were obtained from the Egyptian National Gene Bank, while the seeds of Moneymaker and P73 cultivars were obtained from Spain. The plants were cultivated for three consecutive generations (September 2018, March 2019, and September 2019) at the greenhouse.
Forty seeds of each cultivar were sown in peat moss. Twelve days old seedlings were transplanted into 5.0 L pots filled with peat moss and silt soil mixture at the rate of 50:50 (V:V). Three replicates were used per treatment, and the completely randomized design (CRD) was followed. The plants were irrigated as needed and fertilized every week with 0.82 g/l Ca(NO3)2, 0.24 g/L MgSO4, 0.2 g/L KNO3, 0.35 g/L K2SO4, 0.15 g/L H3PO4.
Morphological data of tomato genotypes, i.e., plant height (cm), number of leaves/plant, and number of inflorescences/plant, were calculated in 10 randomly selected plants per cultivar. The fruit weight (g) was recorded as the average of 100 fruits collected from 10 plants. The plant yield (kg/plant) was calculated by multiplying the number of fruits by the average fruit weight of 10 plants per cultivar. Physicochemical data, i.e., total phenolic compounds (TPC) (mg/100 g), total flavonoids (TF) (mg/100 g), anthocyanin (mg/g), lycopene (mg/100 g), β-carotene (mg/100 g), pH, acidity (mg/100 g), ascorbic acid (mg/100 g), total soluble solids (TSS) (%) and total sugars (TS) (g/100 g) were calculated. All data were recorded at the complete maturity stage (Fig. 1).
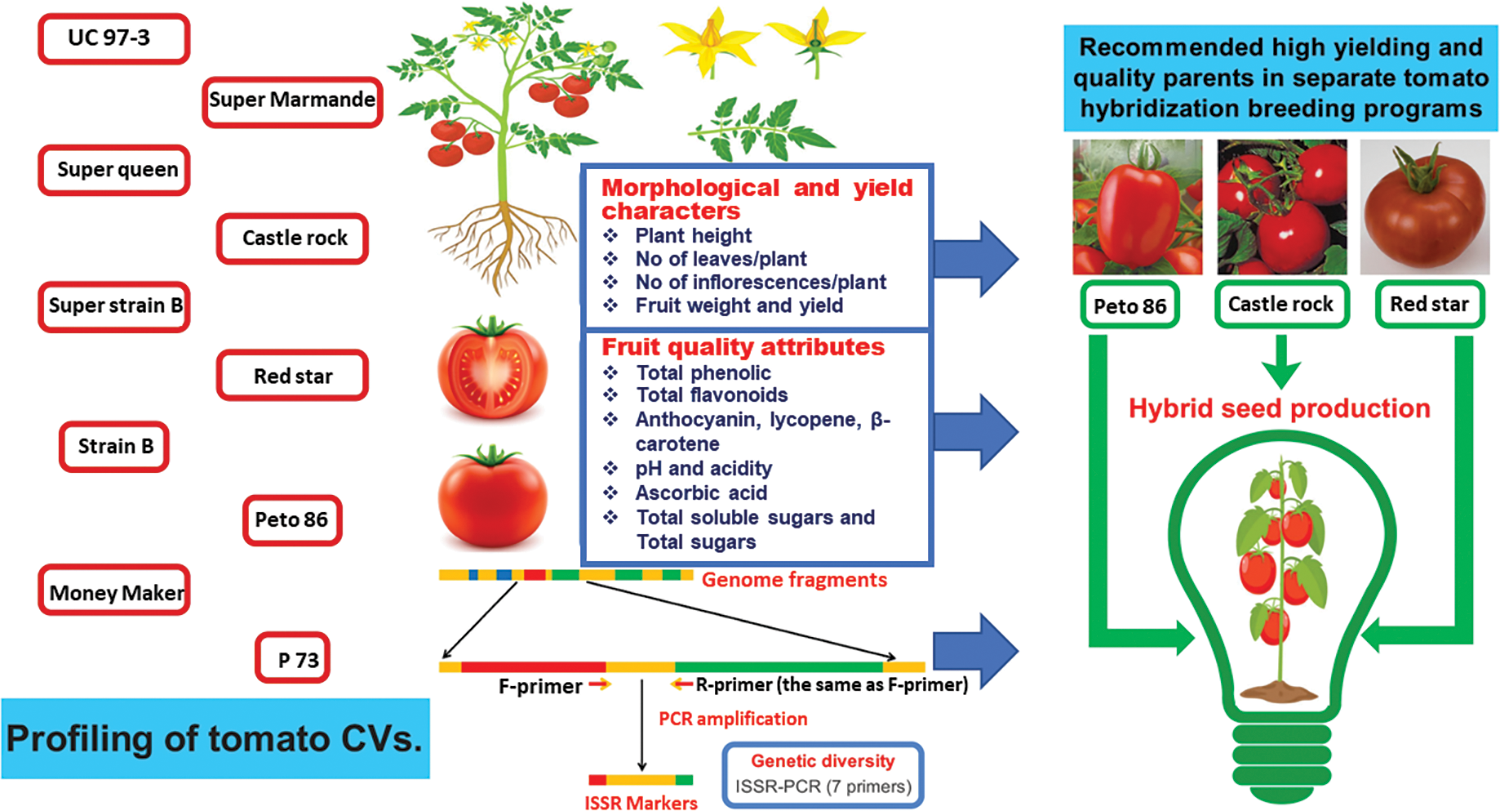
Figure 1: Graphical illustration of data collection and experimental objectives
Young leaves (~0.5 g) from ten tomato cultivars were collected and used to extract genomic DNA using Plant DNeasy Mini Kit (Qiagen, Santa Clarita, CA) according to the provided protocol from the manufacturer. DNA extracts were subjected to RNase-A (100 mg/ml, Sigma, USA) treatment for 30 min at 37°C [16]. The extracted DNA concentration was quantified by a Nano Drop 2000 (Thermo Scientific™, Waltham, MA, USA) and adjusted to 50 ng/μl using TrisEDTA (TE) buffer and subsequently employed for PCR amplifications.
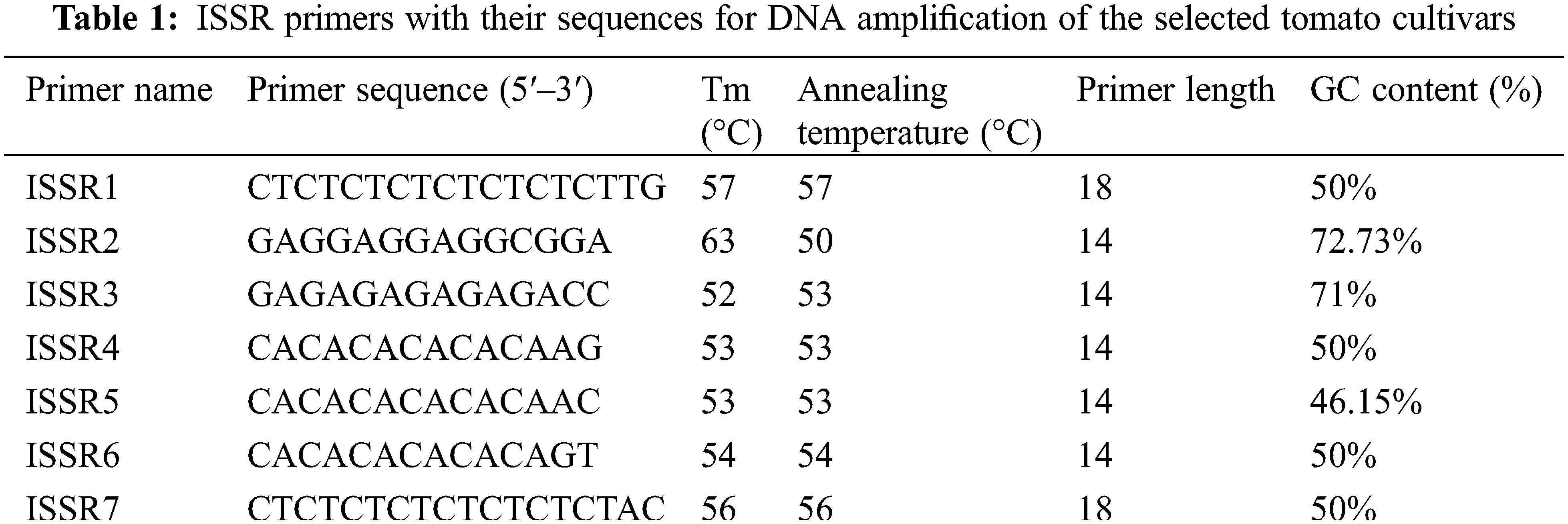
PCR amplification was performed on the extracted tomato genomic DNA using seven ISSR primers (Table 1). The reactions were performed in a thermal cycler (Veriti™ 96-Well Fast) with 25 μl reaction mixture containing 3 μl genomic DNA (50 ng/μl), 1.5 μl of each primer (5 μM), 2.5 μl of 10x reaction buffer, 2 units of Taq DNA polymerase (Promega) (5 U/μl), 0.5 mM dNTPs, and 5 mM MgCl2. The following amplification program was followed: incubation for 5 min at 95°C for initial denaturation, then 30 cycles at 95°C for 30 s for denaturation, 30 s at annealing temperature (Table 1), 2 min at 72°C for extension, followed by five minutes final extension at the same temperature [17].
The amplified PCR products were separated at 90 volts for 30 min using electrophoresis on a 1.5% agarose gel containing ethidium bromide (MP Biomedicals, Goddard Irvine, CA, USA) for staining. A ladder of 100 bp (GeneRuler 100 bp Plus DNA Ladder, Thermo Fischer Scientific, USA) was used as a standard to identify the molecular sizes of the amplicons. The resultant amplicons were visualized using Gel doc apparatus (BIO-RAD Gel DOCTM EZ Imager).
2.7.1 Preparation of Tomato Extracts
Approximately, 200 g of each tomato fruit sample were dried at 50°C and finely ground to make powder. The tomato powder (10 g) was mixed with 100 mL of 50% ethanol (1:10) and the mixture was agitated for 3 h at room temperature before filtering through Whatman No. 1 paper [18]. The solvent in the filtrate was recovered using a rotary evaporator, and for later analysis, the concentration of the free solvent extracts was set at 100 mg/mL. The samples were stored at 4°C for the subsequent use.
2.7.2 Physicochemical Characterization
A pH meter was used to determine the pH of tomato extracts. The Abbe Refractometer was utilized to determine the TSS concentration (Brix%) of the extracts. Citric acid (%) was used to determine the titratable acidity [19]. Following AOAC, ascorbic acid was determined by redox titration with iodine. Following Saad et al. [18], the TS, TF and TPC were determined in the samples. For beta-carotene measurement, One gram of tomato samples was carefully weighed in a glass test tube and the tube was then filled with 5 mL of cold acetone and maintained at 4°C for 15 min with periodic shaking before being vortexed at high speed for 10 min and centrifuged at 1300 xg for 10 min. The supernatant was transferred to a separate test tube, and the desired component was re-extracted using 5 mL of acetone followed by another centrifugation step. All supernatants were combined and filtered using Whatman filter paper No. 42. The absorbance was measured at 449 nm using a UV-Vis spectrophotometer [20]. For lycopene quantification, juice extract from the tomato samples was mixed with a solvent combination of hexane: ethanol: acetone (1:8, v:v) and incubated in the dark for 10 min as described by Popescu et al. [21]. After adding water, the mixture was settled for 10 min at room temperature before taking the top layers to measure the absorbance at 503 nm.
Morphological and yield measurements were recorded. Data were subjected to analysis of variance (Two way ANOVA) to locate the significant differences among means squares. The significant differences among mean values at p < 0.05 were obtained by L.S.D0.05 according to Snedecor et al. [22] using Statistix 9 software. Principal component analysis (PCA) was performed using the statistical program R version 3.6on the means of the studied traits to identify their relationship.
For each treatment, the ISSR-based PCR locus detection was scored as present (1) or absent (0), comparing the band patterns across all genotypes allowed for the determination of genetic diversity. The levels of polymorphism were calculated by dividing the total number of scored loci by the number of polymorphic loci according to Dice coefficient measurement [23]. The genetic similarities among cultivars were calculated using IBM SPSS statistics software [24]. The phylogeny dendrogram was generated by the clustering analysis [25] in STATISTICA 8 software [26].
3.1 Analysis of Morphological and Yield Characters
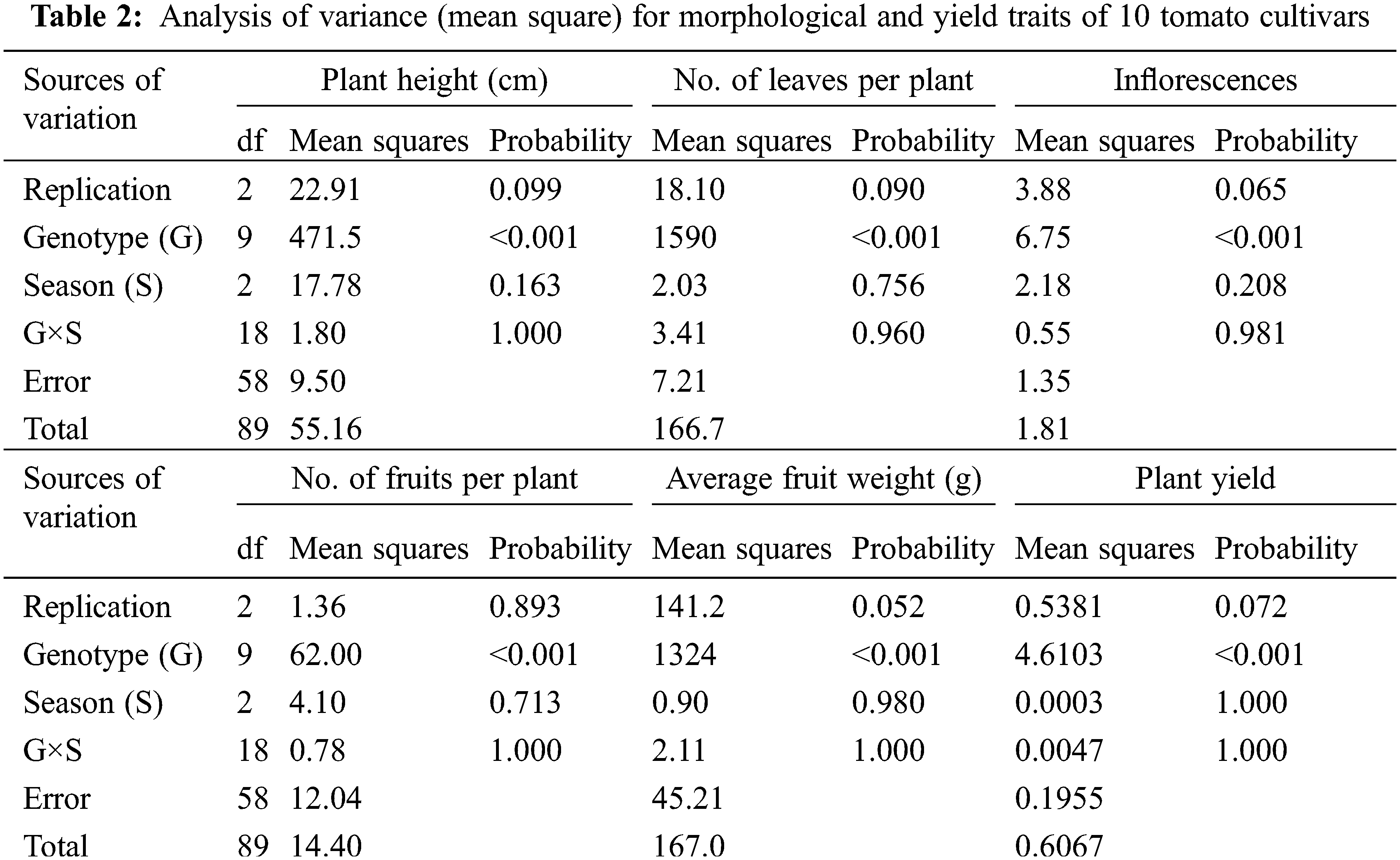
Ten tomato cultivars were cultivated in the greenhouse and evaluated for morphological and yield characteristics as shown in Fig. 2. The significant differences were found for morphological and yield characters between the ten tomato cultivars based on the LSD values. The analysis of variance of morphological and yield characters revealed that the differences among the ten cultivars were highly significant (p < 0.001) for all studied traits, the differences among the three cultivation generations were insignificant (p > 0.05) for all traits. The analysis also revealed that the interaction between genotypes and seasons was not significant for all traits (p > 0.05) in Table 2. According to L.S.D0.05 analysis, the tested cultivars showed highly significant differences for plant height except for three cases, i.e., Super Marmande vs. UC 97-3, Moneymaker vs. Super Queen vs. Red Star, and Peto 86 vs. Strain B vs. Super Strain B cultivars, which showed non-significant differences. Similarly, all cultivars showed statistically significant differences for the number of leaves/plants, with three exceptions recorded between Super Marmande vs. Super Queen, P73 vs. Peto 86 vs. Strain B, and Super Strain B vs. Peto 86 vs. Strain B. The differences for the number of inflorescences/plant among all cultivars were not highly significant except Super Strain B and Money Maker (10 inflorescences/plant) (Fig. 2C). Nevertheless, the number of fruits showed significant differences among all cultivars except between Super Marmande, Castle Rock, and Moneymaker; Super Marmande and Super Queen; Red Star and both of Strain B and P73; Super Queen and both of Strain B and P73; Super Strain B and both of Castle Rock and Moneymaker; Super Queen, Strain B, and P73 (Fig. 2D). For average fruit weight, the cultivar Red Star produced significant results with the highest fruit weight as compared to all other cultivars, for which the average fruit weights were significantly or non-significantly different from each other (Fig. 2E).The cultivar Pet 86 produced the highest average plant yield, which was significant from all other cultivars (Fig. 2F).
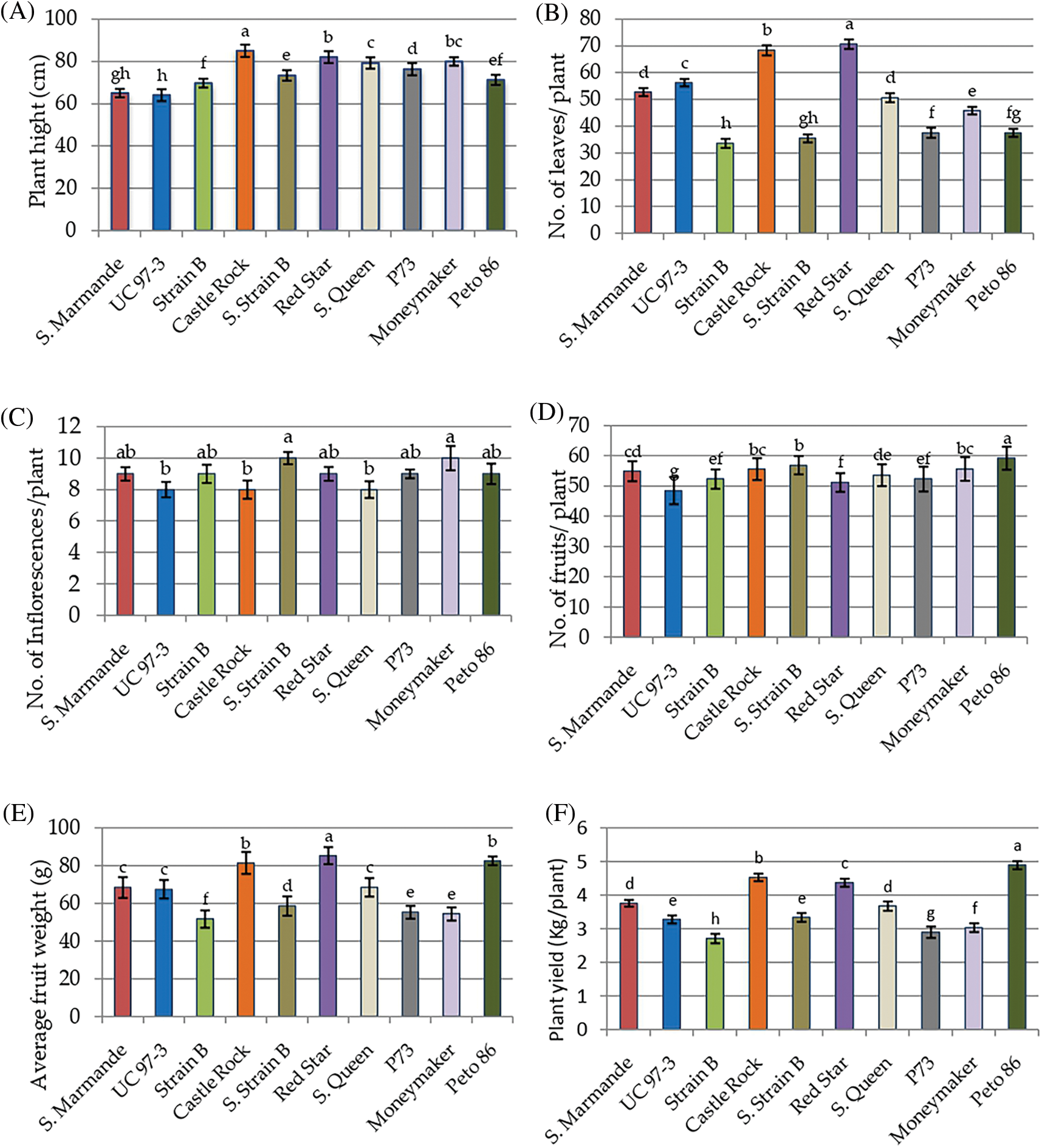
Figure 2: Mean values of morphological and yield characters of ten tomato cultivars measured under greenhouse conditions during three consecutive generations. (A) Plant height. (B) Numbers of leaves/plant. (C) Numbers of inflorescences/plant. (D) Numbers of fruits/plant. (E) Fruit weight. (F) Plant yield
Notes: Values with the same letter in the same character are not significantly different at p < 0.05.
3.2 Diversity Analysis of Physicochemical Characters
Physicochemical characters TPC, TF, Anthocyanin, lycopene, β-carotene, pH, acidity, ascorbic acid, TSS and TS were recorded to measure the fruit quality of the ten studied tomato cultivars. The analysis of variance of physicochemical characters revealed that the differences among the ten cultivars were highly significant (p < 0.001) for all studied traits, except for pH and acidity, the differences among the three cultivation generations were insignificant (p > 0.05) for all traits. The analysis also revealed that the interaction between genotypes and seasons was not significant for all traits (p > 0.05) in Table 3. According to L.S.D0.05 analysis, the accumulation of total phenol contents (TPC, TF, and anthocyanin) were highly significant in Peto 86 and Castle Rock with relative increases of 43% and 42%, respectively (Figs. 3A–3C). The lycopene content was also significantly higher in Peto 86 (7.2 mg/100 g), Red Star (6.7 mg/100 g) and Castle Rock (6.5 mg/100 g). Nevertheless, β carotene content was significantly higher in P73 (1.3 mg/100 mg) and Super Queen (1.1 mg/100 mg). Differences in pH values among all cultivars were presented in Fig. 3F. It was observed that the pH value of P73 was significantly higher among all cultivars while significantly lowest pH value was found in Castle Rock. However, Super Marmande and super strain B cultivars were more acidic with Super Marmande showing significantly highest values for acidity among all tomato cultivars (Fig. 3G). The accumulation of ascorbic acid content was found significantly higher in Castle Rock (66 mg/100 g), Peto 86 (62 mg/100 g), Red Star (55 mg/100 g), and Super Marmande (52 mg/100 g) (Fig. 3H). TSS value determined the firmness and freshness of the fruits. Fruits that contain a low TSS value can be used in juices production, while hard fruits can be used to manufacture ketchup. The cultivar Castle Rock, Peto 86, Super Strain B, and Strain Bshowed the significantly highest TSS value, while Super Strain B, Strain B, and Super Marmande had the significantly lowest TS content among all the cultivars (Figs. 3I and 3J).
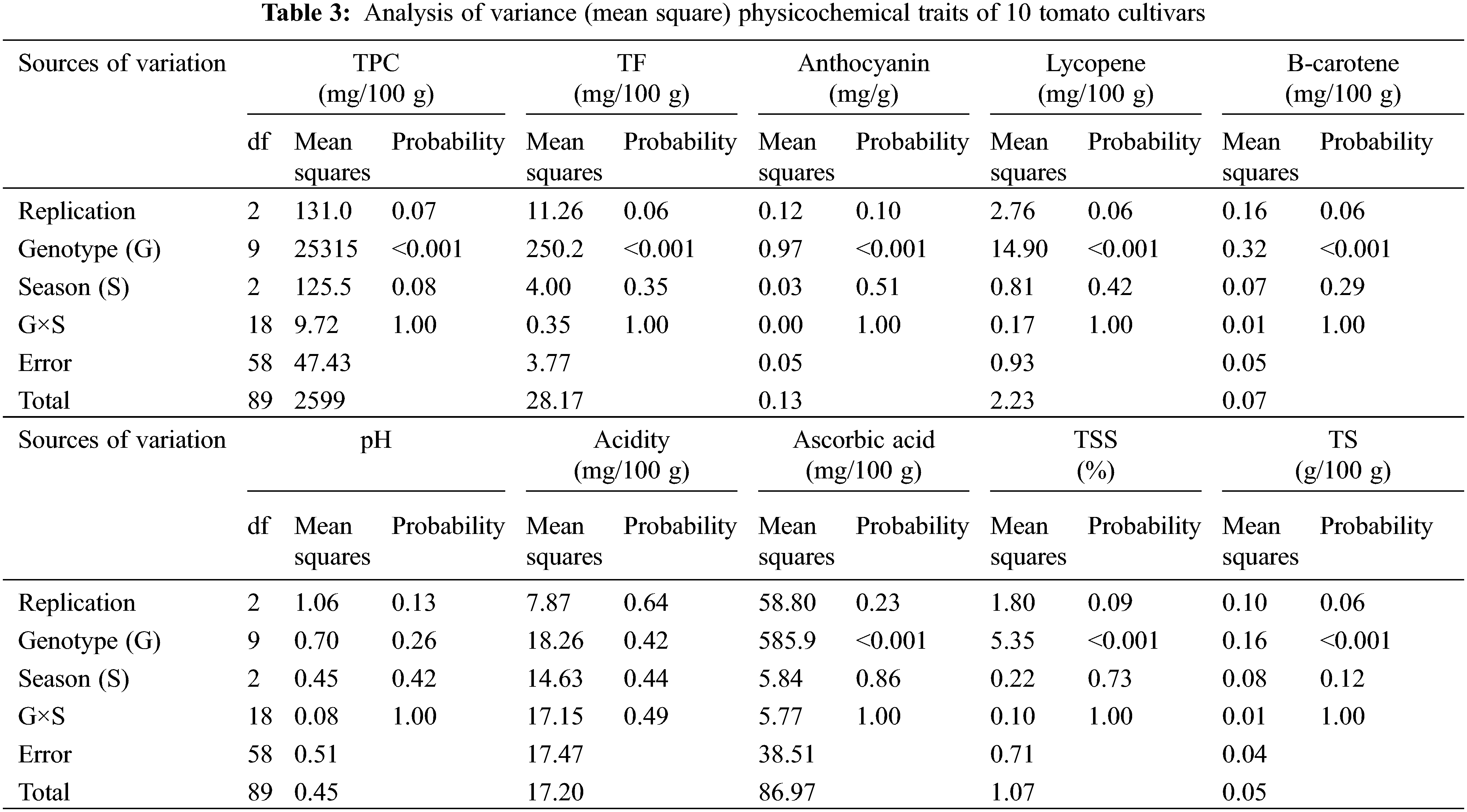
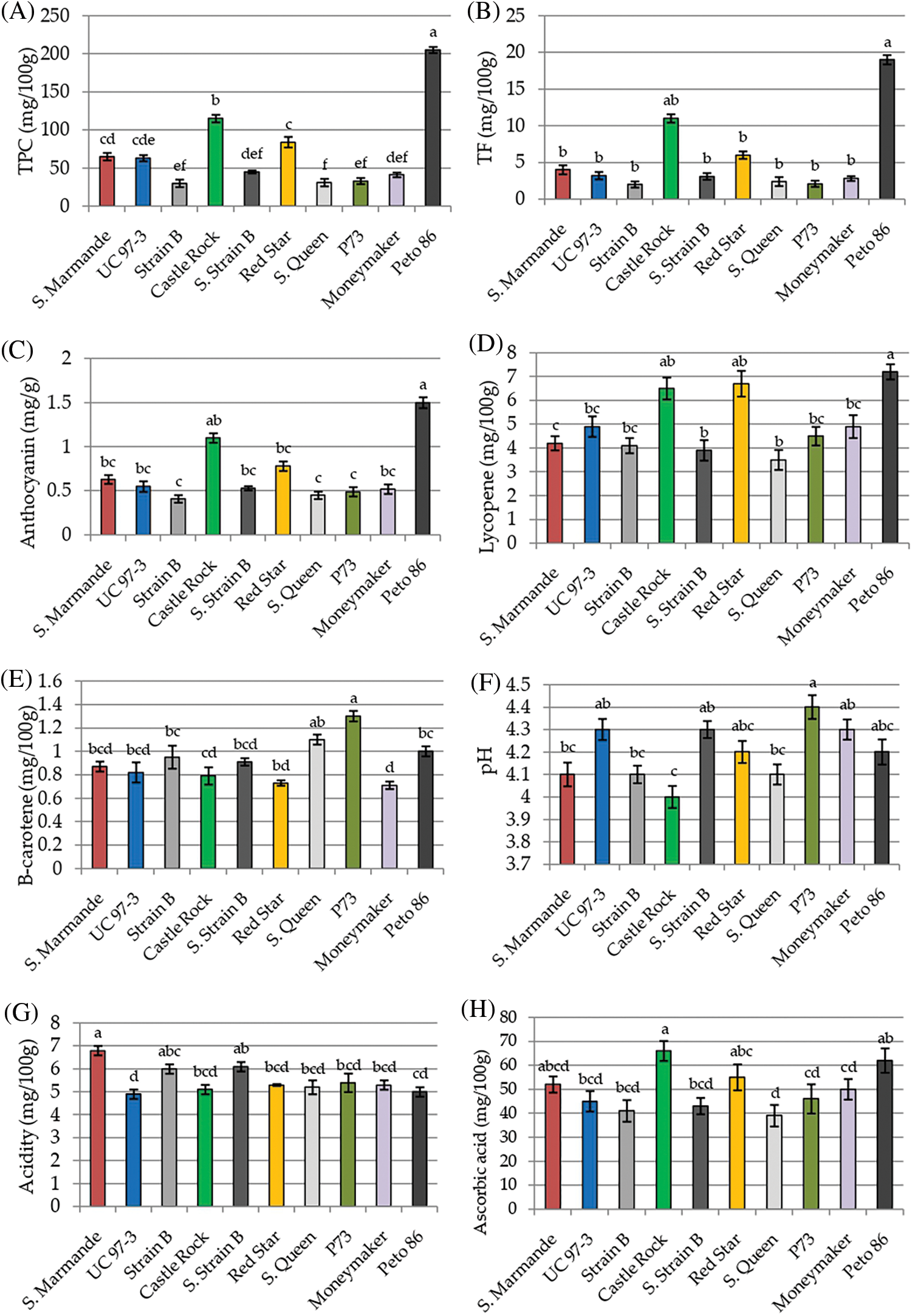
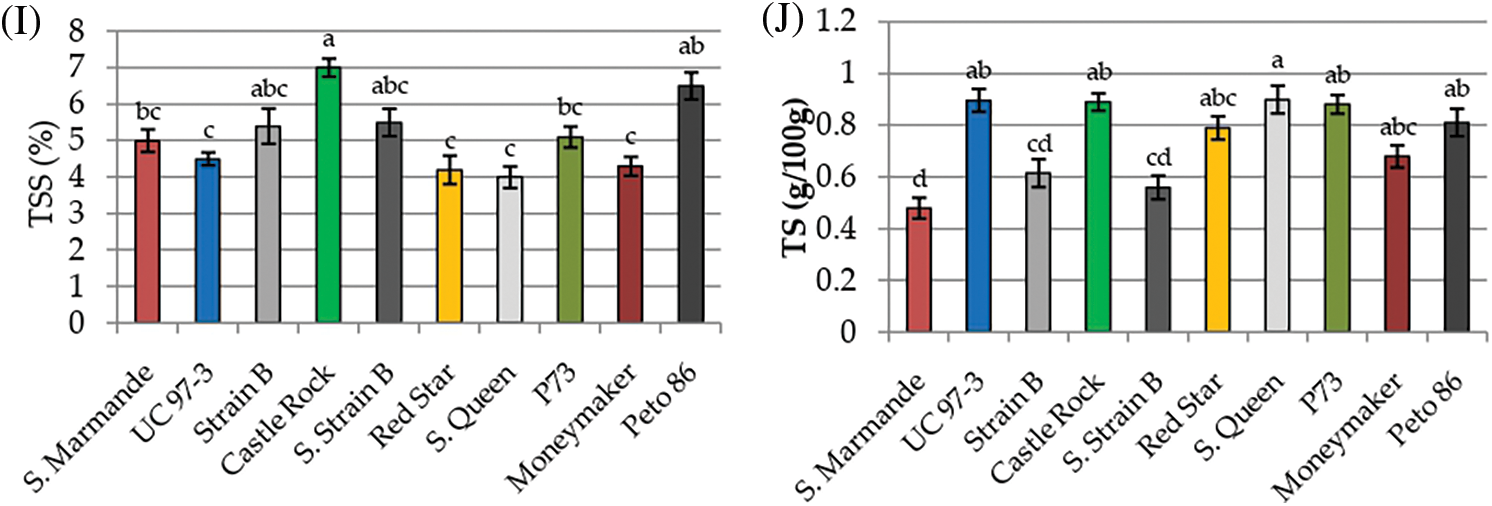
Figure 3: Mean values of physicochemical characters of fresh juice of ten tomato cultivars grown under greenhouse conditions during three consecutive generations. (A) TPC (mg/100 g). (B) TF (mg/100 g). (C) Anthocyanin (mg/g). (D) Lycopene (mg/100 g). (E) β-carotene (mg/100 g). (F) pH. (G) Acidity (mg/100 g). (H) Ascorbic acid (mg/100 g). (I) TSS (%). (J) TS (g/100 g)
Notes: Values with the same letter in the same character are not significantly different at p > 0.05.
3.3 Interrelationship among Evaluated Characters
Principal component analysis (PCA) was performed to illustrate the association among different traits of the tomato cultivars. The PC1 and PC2 displayed the most variance, registering around 77.41% (65.37% and 12.04% by PC1 and PC2, respectively), and were used to construct the PC-biplot (Fig. 4). The character vectors form acute angles, implying positive correlations. The measured characters could be classified into three groups. Group 1 included No. of fruits/ plant, TSS, TF, TPC, anthocyanin, ascorbic acid, plant yield, lycopene, and average fruit weight. Group 2 consisted of plant height, No. of leaves/plant, and TS, while Group 3 comprised no. of inflorescences/plant, acidity, β-carotene, and pH. There was a clear positive correlation between the traits in each group. The first and second groups showed a positive correlation, while the third group showed a negative correlation with the first and the second groups (Fig. 4B).
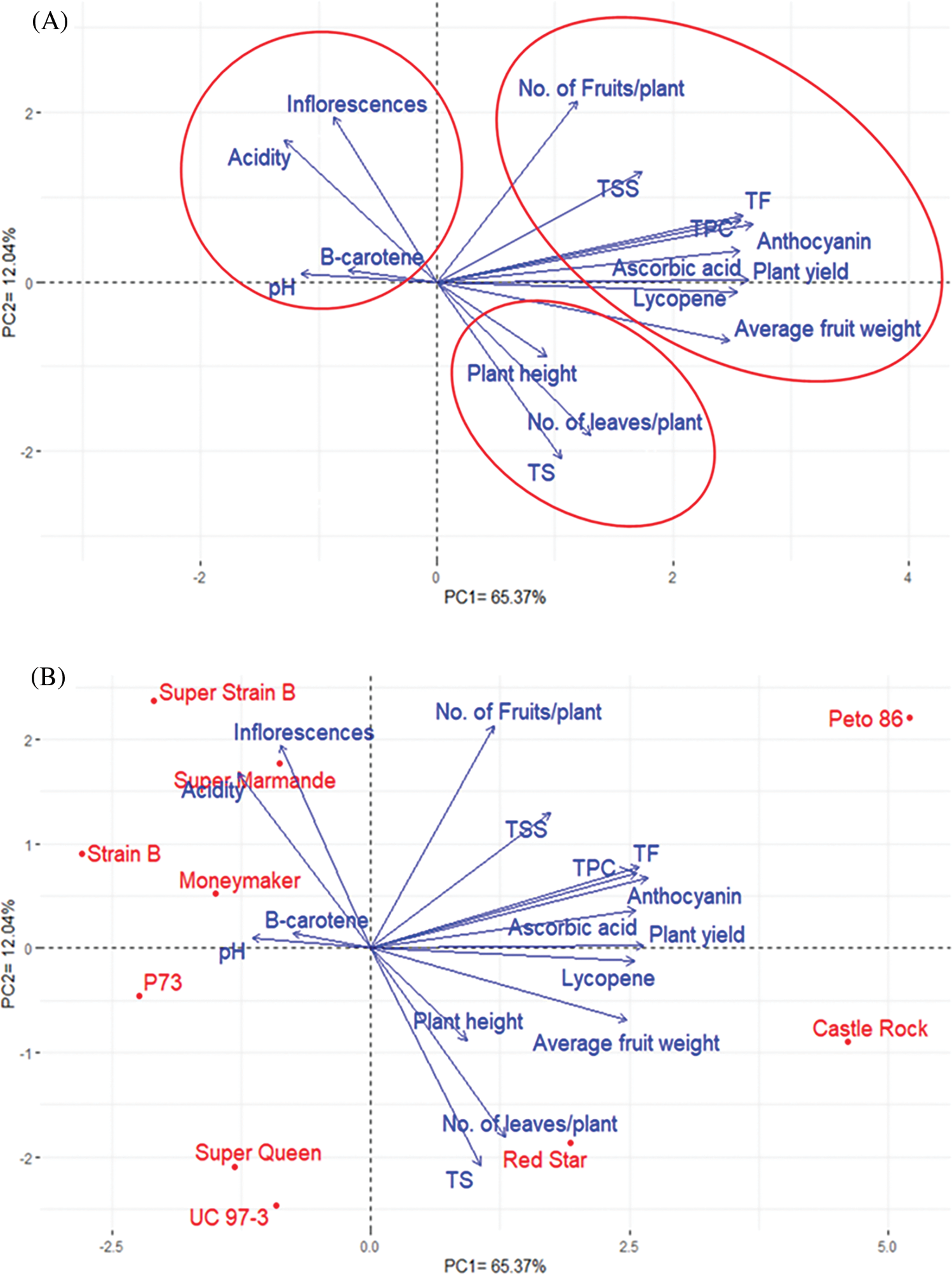
Figure 4: Biplot of the principal components (PC1 and PC2) of ten tomato cultivars. (A) PC1 and PC2for yield and quality traits without cultivars. (B) PC1 and PC2, including cultivars
Interestingly, there was a strong positive correlation between plant yield and its components (no. of fruits/plant and average fruit weight). These results are essential for plant breeders when building their selection criteria on strong, positively correlated characters. Notably, PC1 (65.73%) analysis divided the ten tomato cultivars according to their yield and quality parameters into two groups; the first group included Castel Rock, Red Star, UC97-3, Super Queen, and P73 cultivars, while the second group included Peta 86, Super strain B, Super Marmand, Strain B, and Moneymaker cultivars. On the other hand, PC2 (12.04%) divided the cultivars as Peto 86, Castel Rock, and Red Star in one group and the rest of the cultivars in another group.
Genetic divergence analysis of ten tomato genotypes using seven ISSR primers showed a total of 60 loci with 49 polymorphic, while only 11 were monomorphic loci. The number of amplified loci per single primer varied from 5 to 12, with an average of 8.57 loci per primer (Table 4). All the tested primers successfully generated reliable polymorphic bands within all studied genotypes (Fig. 5A). The polymorphism ranged from 50% (with ISSR3 primer) to 100% (with ISSR4, ISSR6, and ISSR7 primers) (Table 4). The ISSR markers used in this study, especially ISSR4, ISSR6, and ISSR7, can be considered highly effective in determining genetic diversity and distinguishing genotypes according to the levels of their polymorphic bands. These primers also provide important clues in understanding the origin and relationships between species. The phylogeny analysis (Fig. 5B) shows four clusters according to the data scored from the seven ISSR primers, and clustering analysis grouped the ten tomato cultivars into four groups (Fig. 5B). Super Marmande formed a separate cluster (cluster I), while both Super Queen and UC 97-3 cultivars were grouped into cluster II while Cluster III included Castle Rock and Red Star cultivars. Cluster IV included Super Strain B, Strain B, P73, and Moneymaker cultivars. The cluster analysis showed that genotypes from the exact origin were grouped in one cluster (cluster II, cluster III), while in some cases, they were placed in different clusters (Super Strain B). In this context, the Super Marmande genotype is clustered independently due to its different origins (France).
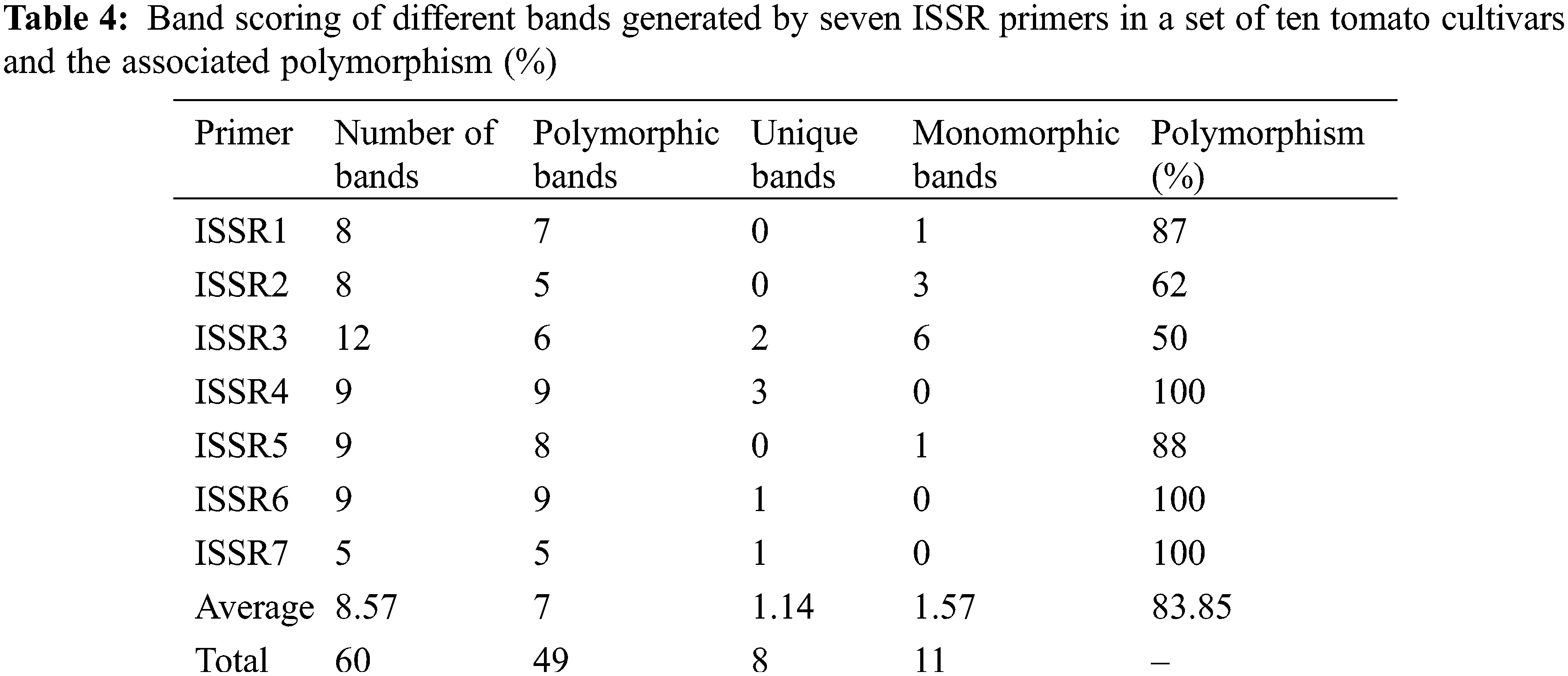
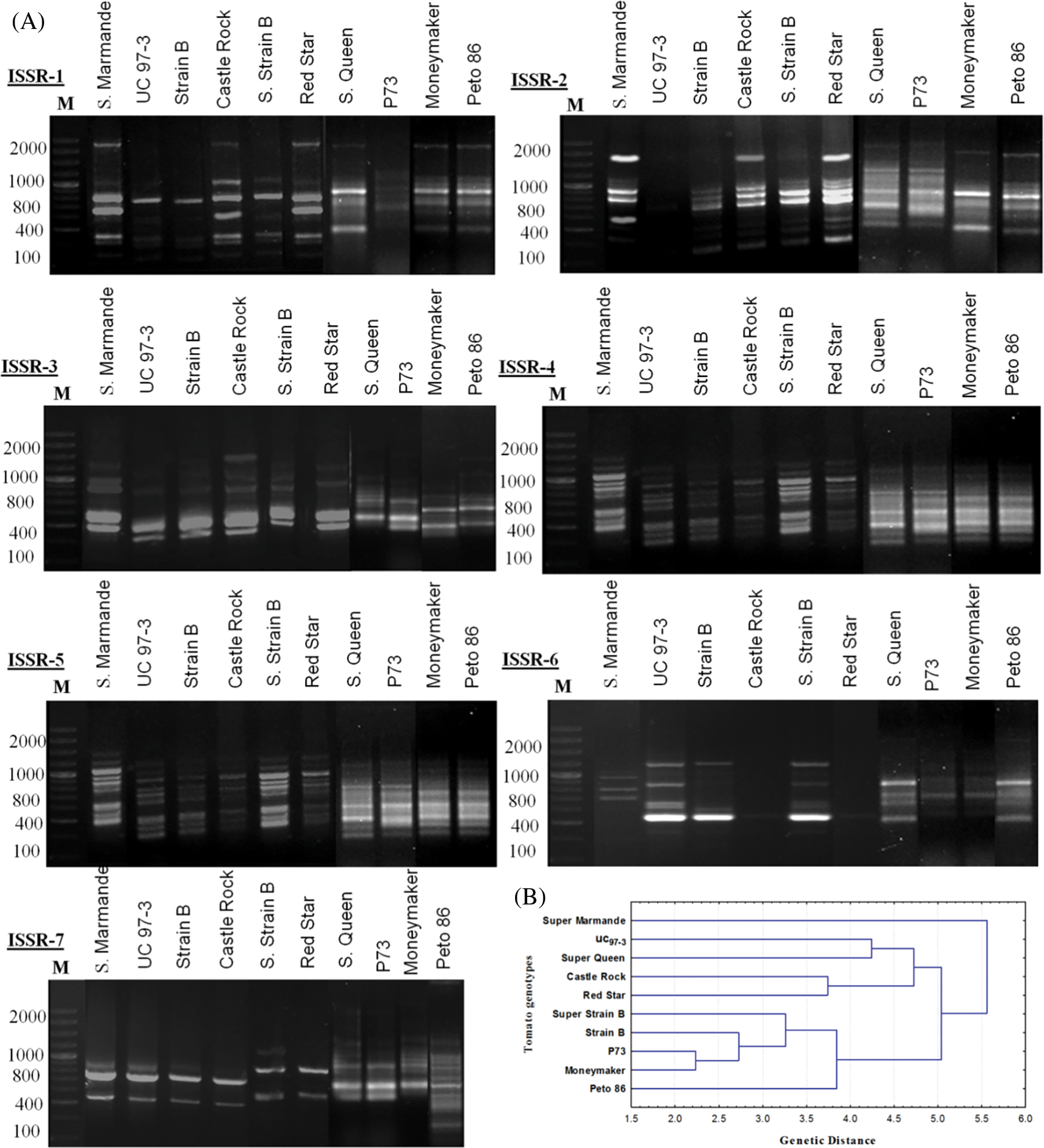
Figure 5: (A) ISSR-PCR banding patterns of ten S. lycopersicon cultivars. Lane M is showing a 100 bp ladder. (B) Phylogenetic tree (Linkage dendrogram) of the ten tomato cultivars based on differences in ISSR-PCR profiles
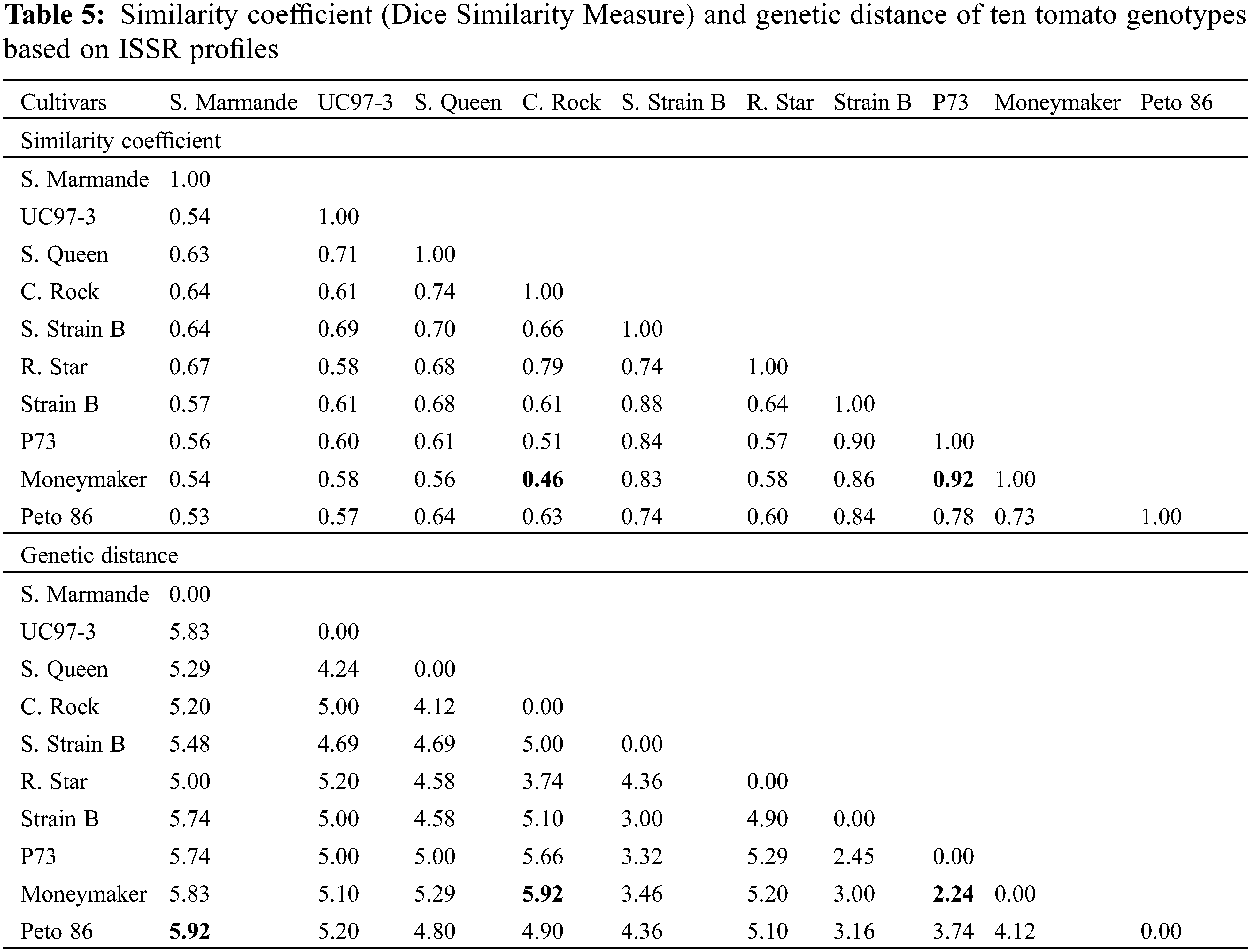
3.5 Genetic Distances and Similarities
Studying ISSR-PCR-based variation among tomato genotypes indicated the highest genetic similarity (0.92) between P73 and Moneymaker, while the lowest (0.46) was noted between Castle Rock and Moneymaker cultivars (Table 5). Ultimately, the least genetic distance (2.24) was observed between P73 and Moneymaker cultivars. Maximum genetic distance (5.92) was observed between Super Marmande and Peto 86 cultivars, on the one hand and between Castle Rock and Moneymaker cultivars on the other hand (Table 5).
The data recorded from morphological and physicochemical characters were combined and used to construct a dendrogram (Fig. 6), qualifying the relatedness distance among the tested tomato cultivars. The clustering analysis of morphological and physicochemical data grouped the tomato cultivars into four clusters (I, II, III, and IV). Cluster I included Super Marmande, UC 97-3, and Super Queen, while four cultivars Super Strain B, P73, Strain B, and Moneymaker were included into cluster II, respectively. Interestingly, cluster III comprised only Peto 86, while cluster IV included Castle Rock and Red Star, which showed less similarity with the other cultivars tested. On the other hand, generally, the conducted analysis did not show the exact clustering pattern according to the origin of the cultivars. The cluster analysis makes it evident that cultivars from the same origin could be clustered, while in some cases, they were placed in different clusters.
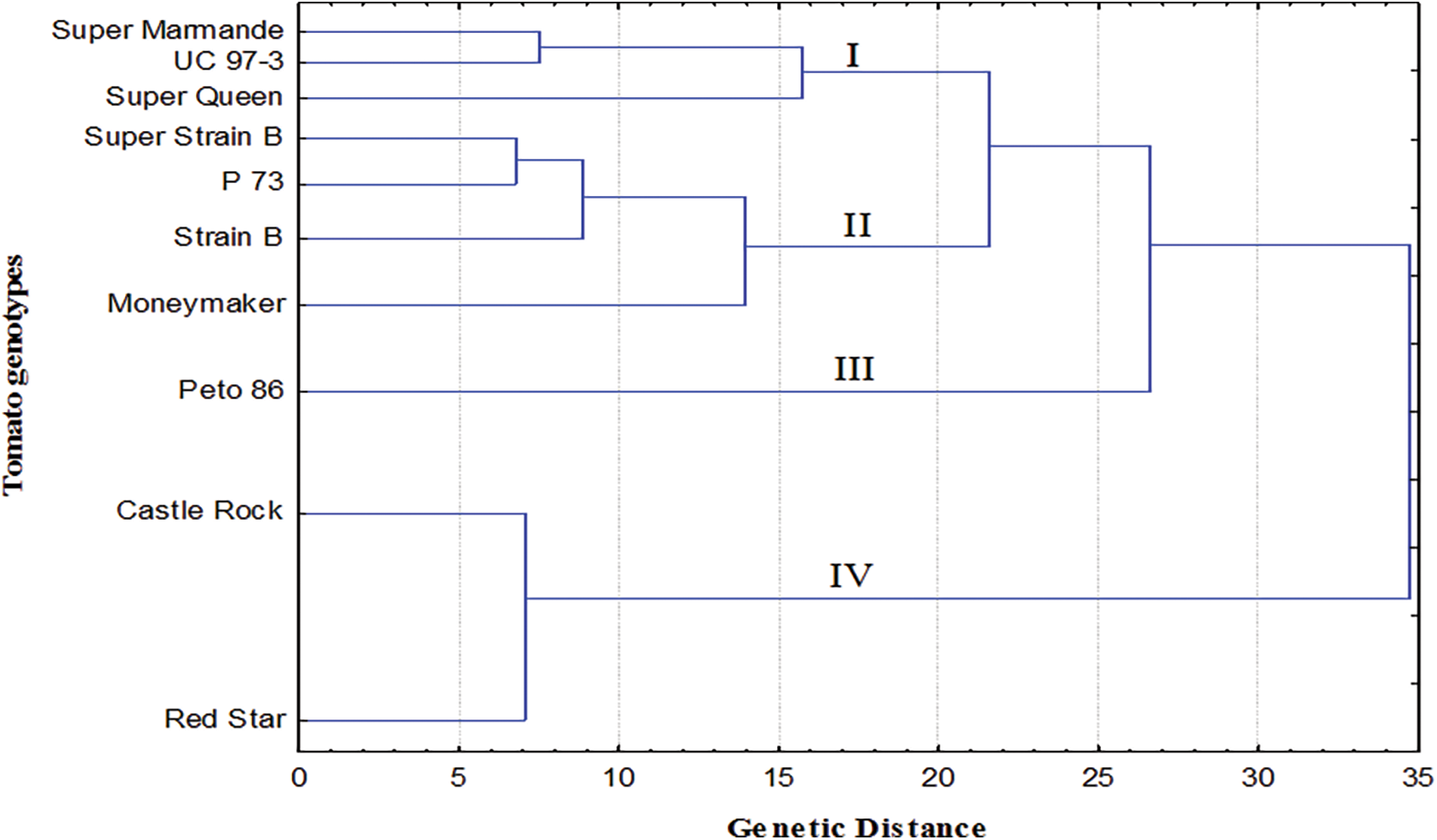
Figure 6: Phylogenetic tree of ten tomato genotypes based on their morphological and physicochemical attributes
The assessment of the genetic diversity based on the measuring of morphological characteristics [5]. To design a breeding program for a specific character in tomato especially plant yield, the variation in plant yield is crucial among the available cultivars. Analysis of variance revealed significant differences (Table 2) among the studied cultivars for all the traits. Development of high yield tomato cultivars is one of the most important breeding targets in Egypt to match the huge usage of tomato in daily life. Among the studied cultivars, Peto 86, Castle Rock and Red Star showed highest fruit yield. Therefore, these cultivars can be used in cultivars development programs. Similar results were reported by Islam et al. [5]; they found that the genotypes, GPB0107 and GPB0120 showed early flowering and early fruiting characteristics that qualify them to be used as parents in cultivars development programs.
The phenotypic and physicochemical characterization may require additional approaches, such as molecular studies, for confirmation and precision. Molecular profiling can be reliably used in plant improvement and breeding programs and in elucidating the genetic diversity among organisms [27]. The significant differences among cultivars may support the breeding and crop improvement programs [28,29].
In this study, the morphological, physiochemical and molecular attributes of the ten tomato cultivars revealed considerable phenotypic variability among them. The cultivars Peto 86, Castle Rock, and Red Star were distinctly featured in their morphological, yield and physicochemical attributes. Total phenolic compounds are natural antibiotics due to antioxidant activity and their potential role in specific defense mechanisms against phytopathogens [30]. Tomato fruits are considered one of the essential sources of phenolic compounds along with other vital nutrients in the human diet [31]. Lycopene and β-carotene are the major constituents of antioxidants produced by tomatoes. Agricultural practices, biotic and abiotic stresses, genetic variations, environmental conditions, cultivation technologies, and postharvest storage significantly affect the chemical composition of tomato fruits [32]. The commercial potential of tomato fruits is of significant interest in tomato breeding, although for fresh consumption, the primary factors attracting customers are those with visual impacts, such as size, shape, color and flavor. In the current study, highly significant levels of phenolic compounds were detected in the cultivars Peto 86 and Castle Rock. In an analysis performed by George et al. [33], the values of phenolic compounds varied as a genotype-related function from 188 to 465 mg/100 g. Since tomato fruits of all cultivars were harvested at the ripening stage in the current investigation, the pH should surpass the 4.5 value, according to El-Attar et al. [34]. Contrarily, the pH of tomato fruits ranged from 4 to 4.4 in our study (Fig. 2), where Castle Rock showed the lowest pH value among all other cultivars. A pH lower than 4.5 is a good feature because it increases the antimicrobial activity of the fruits and presents well-quality sustenance [35,36]. Moreover, the best flavor requires a high acidity value and Super Marmande surpass all the cultivars for high acidity value. High fruit acidity is advantageous as it protects the fruits against fungal infections [37]. Moreover, tomatoes with high sugar and low acid content produced best flavor [38] and vice versa [39]. Ascorbic acid (or vitamin C) is present in fruits and vegetables and is important for human health. Its primary activities include scurvy prevention and skin and blood vessel maintenance [40]. Ascorbic acid (mg/100 g) and citric acid contents were cultivar dependent. Peto 86, Castle Rock and Red Star performed significantly better for acidic properties among all cultivars and thus can be suitable parents in the breeding program to improve these traits in tomato.
Some quality parameters, such as fruit form and size, lycopene concentration, vitamin C, and soluble solids, are strongly linked to genotypes. These parameters have different heritability and are highly dependent on genotype expression in conjunction with the environment (crop peculiarities). The genetic makeup of a tomato cultivar is cumulatively influenced by its genetic background and the surrounding environment [41]. The most significant quality parameters for tomatoes are red color, firmness, juicy texture, and good flavor [39]. Additionally, TSS, juice content, acidity, vitamin C content, and other factors contribute to these qualities. The TSS range of fresh tomatoes is 4.5% to 8.5% of their fresh weight. The main components of soluble solids are glucose and fructose. When the goal is dehydration, pulp concentration, or both, TSS is critical for industrialization since product yield is directly connected to Brix. Lycopene, ascorbic acid (vitamin C), and potassium are some of the essential nutrients in tomatoes that positively impact human health [38].
The data on genetic distance and similarity between cultivars is beneficial in breeding programs as it helps to select suitable parental cultivars for hybridization and development of plant populations [42,43]. In this study, the clustering analysis revealed clear relationships between the studied tomato cultivars (Table 3 and Figs. 4 and 5). The results of clustering analysis and phylogeny have great importance in determining the genetic association between the crop cultivars [6,44–47] and selecting the most appropriate parents in hybrid production programs based on a low similarity coefficient as in the case of Red star and Strain B (0.89) or a vast genetic distance as in the case of Red Star and Strain B (51.5). Tomato hybrids are not exclusively produced to benefit from heterosis per se but also to support the investment of breeders via combining valuable features such as disease resistance and cultivar production with ripening attenuating genes in the heterozygous state [48]. However, heterosis in tomatoes has been reported for many vital traits [48].
Increasing tomato fruit quality and yield is a global aim of most current tomato improvement experiments. The best way to accomplish this is to choose additional advantageous features, such as abiotic tolerance, disease resistance and earliness. For example, in the breeding for fresh market tomato production, increasing fruit yield has been combined with heat tolerance breeding through cultivation under humid and hot conditions [49]. However, breeders frequently designate particular factors that may uncertainly contribute to production and emphasis the selection for such traits. Therefore, selection for yield is rarely very effective. The correlation analysis may play an essential role in predicting the performance of some positively correlated traits to another trait in the indirect selection and vice versa. Hence, one or more traits in the same group would be a reliable parameter for the selection of plants having desirable content of the other positively correlated traits [50]. In the same context, a trait that negatively correlates with other traits would also be a reliable indicator as a de-selection criterion. On the other hand, identifying molecular profiles potentially and considerably associated with fruit yield and quality may enable reliable early selection of inbred lines or hybrids with more reliably predictable high fruit yield and quality based on the presence or the absence of a specific molecular profile.
The association between molecular profiles and morphological and physicochemical traits was previously investigated in several studies [51,52], showing a significant association between molecular markers (quantitative trait loci) and morphological and physicochemical traits [53].
The current study supports the genetic improvement of tomato cultivars through traditional breeding programs. However, more recent approaches such genome editing approaches [54,55], molecular markers, bioinformatics and phylogeny analysis [56–59], and using of mutagens [60] could effectively be utilized to breed and develop tomato for many desirable traits.
This study comprehensively characterized ten tomato cultivars based on morphological, physicochemical, and molecular profiles. Peto 86, Castle Rock, and Red Star cultivars showed the highest fruit yield (kg/plant), TPC, and sap acidity. The results also showed that the cultivars had several specific features relevant to their use in local markets, processing, and genetic improvement and breeding programs. Peto 86, Castle Rock and Red star cultivars may get considerable attention because of their richness of total phenolic compounds, high acidity, low pH, and increased plant yield (kg/plant). Significant differences were also observed in pH values between P73, Super Queen, Castle Rock, Strain B, Super Marmande, Castle Rock and UC 97-3, Super Strain B, and Moneymaker cultivars. Molecular profiling showed that the lowest genetic distance (2.24) was between P73 and Moneymaker cultivars, while the highest (5.92) was observed between Super Marmand and Peto 86, on the one hand, and between Castle Rock and Moneymaker on the other hand. Finally, it is concluded that Peto 86, Castle Rock, and Red star can be recommended as featured parents in future tomato breeding programs.
Acknowledgement: The authors extend their appreciation for the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia [Project No. GRANT805], and Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2023R318), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. Also, the authors extend their appreciation for Zagazig University and South Valley University, Egypt for providing facilities to conduct this research.
Funding Statement: This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia [Project No. GRANT805], and the Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2023R318), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Al-Shaal, A., Murshed, R. A., Albiski, F. A. (2021). Molecular characterization of some tomato genotypes using inter simple sequence repeats (ISSR) technique. Journal of Genetic and Environmental Resources Conservation, 9(1), 134–142. [Google Scholar]
2. Chandel, R., Sadashiva, A., Ravishankar, K. V., Das, A., Rout, B. M. et al. (2022). Genetic combining, heterosis analysis for horticultural traits in tomato (Solanum lycopersicum L.) using ToLCV-resistant lines and molecular validation of Ty genes. Plant Genetic Resources, 19(6), 512–521. https://doi.org/10.1017/S1479262121000630 [Google Scholar] [CrossRef]
3. Salim, M. M. R., Rashid, M. H., Hossain, M. M., Zakaria, M. (2020). Morphological characterization of tomato (Solanum lycopersicum L.) genotypes. Journal of the Saudi Society of Agricultural Sciences, 19, 233–240. [Google Scholar]
4. Panthee, D. R., Labate, J. A., McGrath, M. T., Breksa, A. P., Robertson, L. D. (2013). Genotype and environmental interaction for fruit quality traits in vintage tomato varieties. Euphytica, 193(2), 169–182. https://doi.org/10.1007/s10681-013-0895-1 [Google Scholar] [CrossRef]
5. Islam, S., Hassan, L., Hossain, M. A. (2022). Breeding potential of some exotic tomato lines: A combined study of morphological variability, genetic divergence, and association of traits. Phyton-International Journal of Experimental Botany, 91(1), 97–114. https://doi.org/10.32604/phyton.2022.017251 [Google Scholar] [CrossRef]
6. Hassanin, A. A., Saad, A. M., Bardisi, E. A., Salama, A., Sitohy, M. Z. (2020). Transfer of anthocyanin accumulating delila and rosea1 genes from the transgenic tomato micro-tom cultivar to moneymaker cultivar by conventional breeding. Journal of Agricultural and Food Chemistry, 68(39), 10741–10749. https://doi.org/10.1021/acs.jafc.0c03307 [Google Scholar] [PubMed] [CrossRef]
7. Duangjit, J., Causse, M., Sauvage, C. (2016). Efficiency of genomic selection for tomato fruit quality. Molecular Breeding, 36(3), 29. https://doi.org/10.1007/s11032-016-0453-3 [Google Scholar] [CrossRef]
8. Hassanin, A. A., Eldomiaty, A. S., Ujjan, J. A., Al-Mushhin, A. A. M., Alrashidi, A. A. et al. (2022). Assessment of the R2R3 MYB gene expression profile during tomato fruit development using in silico analysis, quantitative and semi-quantitative RT-PCR. Saudi Journal of Biological Sciences, 9, 71. https://doi.org/10.1016/j.sjbs.2022.02.021 [Google Scholar] [CrossRef]
9. Hassanin, A. A., Soliman, S. S. A., Ismail, T. A., Amin, M. K. A. (2017). The role of slmyb gene in tomato fruit development. Zagazig Journal of Agricultural Research, 44(3), 969–988. https://doi.org/10.21608/zjar.2017.52295 [Google Scholar] [CrossRef]
10. Bai, Y., Lindhout, P. (2007). Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Annals of Botany, 100(5), 1085–1094. https://doi.org/10.1093/aob/mcm150 [Google Scholar] [PubMed] [CrossRef]
11. El-Saadony, M. T., Abuljadayel, D. A., Shafi, M. E., Albaqami, N. M., Desoky, E. S. M. et al. (2021). Control of foliar phytoparasitic nematodes through sustainable natural materials: Current progress and challenges. Saudi Journal of Biological Sciences, 28(12), 7314–7326. https://doi.org/10.1016/j.sjbs.2021.08.035 [Google Scholar] [PubMed] [CrossRef]
12. Tiwari, J. K., Yerasu, S. R., Rai, N., Singh, D. P., Singh, A. K. et al. (2022). Progress in marker-assisted selection to genomics-assisted breeding in tomato. Critical Reviews in Plant Sciences, 41(5), 321–350. https://doi.org/10.1080/07352689.2022.2130361 [Google Scholar] [CrossRef]
13. Al-Khayri, J. M., Mahdy, E. M. B., Taha, H. S. A., Eldomiaty, A. S., Abd-Elfattah, M. A. et al. (2022). Genetic and morphological diversity assessment of five kalanchoe genotypes by SCoT, ISSR and RAPD-PCR markers. Plants, 11(13), 1722. https://doi.org/10.3390/plants11131722 [Google Scholar] [PubMed] [CrossRef]
14. Poczai, P., Varga, I., Laos, M., Cseh, A., Bell, N. et al. (2013). Advances in plant gene-targeted and functional markers: A review. Plant Methods, 9(1), 1–32. https://doi.org/10.1186/1746-4811-9-6 [Google Scholar] [PubMed] [CrossRef]
15. Ghani, M. A., Abbas, M. M., Amjad, M., Ziaf, K., Ali, B. et al. (2020). Production and characterisation of tomato derived from interspecific hybridisation between cultivated tomato and its wild relatives. The Journal of Horticultural Science and Biotechnology, 95(4), 506–520. https://doi.org/10.1080/14620316.2019.1689182 [Google Scholar] [CrossRef]
16. El-Sayed, A. S., Shindia, A. A., AbouZaid, A. A., Yassin, A. M., Ali, G. S. et al. (2019). Biochemical characterization of peptidylarginine deiminase-like orthologs from thermotolerant Emericella dentata and Aspergillus nidulans. Enzyme and Microbial Technology, 124, 41–53. https://doi.org/10.1016/j.enzmictec.2019.02.004 [Google Scholar] [PubMed] [CrossRef]
17. El-Sayed, A. S., Yassin, M. A., Ali, G. S. (2015). Transcriptional and proteomic profiling of Aspergillus flavipes in response to sulfur starvation. PLoS One, 10(12), e0144304. https://doi.org/10.1371/journal.pone.0144304 [Google Scholar] [PubMed] [CrossRef]
18. Saad, A. M., Mohamed, A. S., El-Saadony, M. T., Sitohy, M. Z. (2021). Palatable functional cucumber juices supplemented with polyphenols-rich herbal extracts. LWT, 148(7), 111668. https://doi.org/10.1016/j.lwt.2021.111668 [Google Scholar] [CrossRef]
19. AOAC (2012). Official method of analysis: Association of analytical chemists, 19th editionpp. 121–130. Washington DC. [Google Scholar]
20. Safdarian, M., Hashemi, P., Ghiasvand, A. (2021). A fast and simple method for determination of β-carotene in commercial fruit juice by cloud point extraction-cold column trapping combined with UV-Vis spectrophotometry. Food Chemistry, 343(5), 128481. https://doi.org/10.1016/j.foodchem.2020.128481 [Google Scholar] [PubMed] [CrossRef]
21. Popescu, M., Iancu, P., Pleşu, V., Bîldea, C. S., Todasca, C. M. (2022). Different spectrophotometric methods for simultaneous quantification of lycopene and β-carotene from a binary mixture. LWT, 160(2), 113238. https://doi.org/10.1016/j.lwt.2022.113238 [Google Scholar] [CrossRef]
22. Snedecor, G. W., Cochran, W. G. (1989). Statistical methods, 8th editionpp. 1191. Ames, Iowa: Iowa state University Press. [Google Scholar]
23. Dice, L. R. (1945). Measures of the amount of ecologic association between species. Ecology, 26(3), 297–302. https://doi.org/10.2307/1932409 [Google Scholar] [CrossRef]
24. Norusis, M. J. (1993). SPSS for windows: Professional statistics, release 6. Spss Inc. [Google Scholar]
25. Rokach, L., Maimon, O. (2005). Clustering methods. In: Data mining and knowledge discovery handbook, pp. 321–352. Springer. [Google Scholar]
26. Weiß, C. H. (2007). StatSoft, version 8. In: AStA advances in statistical analysis, vol. 91, pp. 339–341. Tulsa, OK: StatSoft, Inc. [Google Scholar]
27. Zhani, K., Hamdi, W., Sedraoui, S., Fendri, R., Lajimi, O. et al. (2015). A comparative study of morphological characterization of Tunisian accessions of chili pepper (Capsicum frutescens L.). International Research Journal of Engineering and Technology, 2(4), 87–94. [Google Scholar]
28. Moustafa, E. S. A., Ali, M. M. A., Kamara, M. M., Awad, M. F., Hassanin, A. A. et al. (2021). Field screening of wheat advanced lines for salinity tolerance. Agronomy, 11(2), 281. https://doi.org/10.3390/agronomy11020281 [Google Scholar] [CrossRef]
29. Dagnoko, S., Yaro-Diarisso, N., Sanogo, P. N., Adetula, O., Dolo-Nantoumé, A. et al. (2013). Overview of pepper (Capsicum spp.) breeding in West Africa. African Journal of Agricultural Research, 8(13), 1108–1114. https://doi.org/10.5897/AJAR2012.1758 [Google Scholar] [CrossRef]
30. Sharma, T., Khandelwal, V., Gupta, S., Singh, S. (2021). Secondary metabolites, boon for plants; Their role in defence mechanism and antioxidant activity of anthocephalus cadamba. In: Antioxidants in plant-microbe interaction, pp. 413–424. Singapore: Springer. [Google Scholar]
31. Taveira, M., Ferreres, F., Gil-Izquierdo, A., Oliveira, L., Valentão, P. et al. (2012). Fast determination of bioactive compounds from Lycopersicon esculentum Mill. leaves. Food Chemistry, 135(2), 748–755. https://doi.org/10.1016/j.foodchem.2012.05.016 [Google Scholar] [PubMed] [CrossRef]
32. Hernandez, M. F., Antonio-Ordonez, E., Preciado-Rangel, P., Gallegos-Robles, M. A., Vazquez-Vazquez, C. et al. (2021). Effect of substrates formulated with organic materials on yielding, commercial and phytochemical quality, and benefit-cost ratio of tomato (Solanum lycopersicum L.) produced under greenhouse conditions. Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 49(1), 11999. https://doi.org/10.15835/nbha49111999 [Google Scholar] [CrossRef]
33. George, B., Kaur, C., Khurdiya, D., Kapoor, H. (2004). Antioxidants in tomato (Lycopersium esculentum) as a function of genotype. Food Chemistry, 84(1), 45–51. https://doi.org/10.1016/S0308-8146(03)00165-1 [Google Scholar] [CrossRef]
34. El-Attar, E., El-Gazaar, N., El-Magoli, S. (2018). Quality characteristics of black olive paste with natural flavors dried basil and tomatoes. Bulletin of the National Nutrition, 51(1), 92–111. https://doi.org/10.21608/bnni.2018.14165 [Google Scholar] [CrossRef]
35. Sibanda, S., Workneh, T. S. (2019). Effects of indirect air cooling combined with direct evaporative cooling on the quality of stored tomato fruit. CyTA-Journal of Food, 17(1), 603–612. https://doi.org/10.1080/19476337.2019.1622595 [Google Scholar] [CrossRef]
36. Tigist, M., Workneh, T. S., Woldetsadik, K. (2013). Effects of variety on the quality of tomato stored under ambient conditions. Journal of Food Science and Technology, 50(3), 477–486. https://doi.org/10.1007/s13197-011-0378-0 [Google Scholar] [PubMed] [CrossRef]
37. Martinez, J. A. (2012). Natural fungicides obtained from plants. In: Fungicides for plant and animal diseases. Spain: IntechOpen. [Google Scholar]
38. Calafiore, R., Aliberti, A., Ruggieri, V., Olivieri, F., Rigano, M. M. et al. (2019). Phenotypic and molecular selection of a superior Solanum pennellii introgression sub-line suitable for improving quality traits of cultivated tomatoes. Frontiers in Plant Science, 10, 190. https://doi.org/10.3389/fpls.2019.00190 [Google Scholar] [PubMed] [CrossRef]
39. Felföldi, Z., Ranga, F., Roman, I. A., Sestras, A. F., Vodnar, D. C. et al. (2022). Analysis of physico-chemical and organoleptic fruit parameters relevant for tomato quality. Agronomy, 12(5), 1232. https://doi.org/10.3390/agronomy12051232 [Google Scholar] [CrossRef]
40. Lee, S. K., Kader, A. A. (2000). Preharvest and postharvest factors influencing vitamin C content of horticultural crops. Postharvest Biology and Technology, 20(3), 207–220. https://doi.org/10.1016/S0925-5214(00)00133-2 [Google Scholar] [CrossRef]
41. Toor, R., Savage, G., Lister, C. (2006). Seasonal variations in the antioxidant composition of greenhouse grown tomatoes. Journal of Food Composition and Analysis, 19(1), 1–10. https://doi.org/10.1016/j.jfca.2004.11.008 [Google Scholar] [CrossRef]
42. Jordan, C. Y., Lohse, K., Turner, F., Thomson, M., Gharbi, K. et al. (2018). Maintaining their genetic distance: little evidence for introgression between widely hybridizing species of Geum with contrasting mating systems. Molecular Ecology, 27(5), 1214–1228. https://doi.org/10.1111/mec.14426 [Google Scholar] [PubMed] [CrossRef]
43. Goulet, B. E., Roda, F., Hopkins, R. (2017). Hybridization in plants: Old ideas, new techniques. Plant Physiology, 173(1), 65–78. https://doi.org/10.1104/pp.16.01340 [Google Scholar] [PubMed] [CrossRef]
44. Zhang, L., Hong, Y., Liao, Y., Tian, K., Sun, H. et al. (2022). Dietary Lasia spinosa Thw. improves growth performance in broilers. Frontiers in Nutrition, 13(8), 1188. [Google Scholar]
45. Raza, S. H. A., Hassanin, A. A., Dhshan, A. I. M., Abdelnour, S. A., Khan, R. et al. (2021). In silico genomic and proteomic analyses of three heat shock proteins (HSP70, HSP90-α, and HSP90-β) in even-toed ungulates. Electronic Journal of Biotechnology, 53(1), 61–70. https://doi.org/10.1016/j.ejbt.2021.07.002 [Google Scholar] [CrossRef]
46. Hassanin, A. A., Haidar Abbas Raza, S., Ahmed Ujjan, J., Aysh Alrashidi, A., Sitohy, B. M. et al. (2021). Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: Benefits of getting vaccinated and common questions. Saudi Journal of Biological Sciences. [Google Scholar]
47. Hassanin, A. A., Osman, A., Atallah, O. O., El-Saadony, M. T., Abdelnour, S. A. et al. (2022). Phylogenetic comparative analysis: Chemical and biological features of caseins (alpha-S-1, alpha-S-2, beta- and kappa-) in domestic dairy animals. Frontiers in Veterinary Science, 9, 952319. https://doi.org/10.3389/fvets.2022.952319 [Google Scholar] [PubMed] [CrossRef]
48. Wang, B., Smith, S. M., Li, J. (2018). Genetic regulation of shoot architecture. Annual Review of Plant Biology, 69(1), 437–468. https://doi.org/10.1146/annurev-arplant-042817-040422 [Google Scholar] [PubMed] [CrossRef]
49. Ruggieri, V., Calafiore, R., Schettini, C., Rigano, M. M., Olivieri, F. et al. (2019). Exploiting genetic and genomic resources to enhance heat-tolerance in tomatoes. Agronomy, 9(1), 22. https://doi.org/10.3390/agronomy9010022 [Google Scholar] [CrossRef]
50. Bernard, A., Lheureux, F., Dirlewanger, E. (2018). Walnut: Past and future of genetic improvement. Tree Genetics & Genomes, 14(1), 1–28. https://doi.org/10.1007/s11295-017-1214-0 [Google Scholar] [CrossRef]
51. Cebolla-Cornejo, J., Roselló, S., Nuez, F. (2013). Phenotypic and genetic diversity of Spanish tomato landraces. Scientia Horticulturae, 162, 150–164. https://doi.org/10.1016/j.scienta.2013.07.044 [Google Scholar] [CrossRef]
52. Raza, S. H. A., Abdelnour, S. A., Dhshan, A. I. M., Hassanin, A. A., Noreldin, A. E. et al. (2021). Potential role of specific microRNAs in the regulation of thermal stress response in livestock. Journal of Thermal Biology, 96(11), 102859. https://doi.org/10.1016/j.jtherbio.2021.102859 [Google Scholar] [PubMed] [CrossRef]
53. Frary, A., Doganlar, S., Daunay, M. C., Tanksley, S. D. (2003). QTL analysis of morphological traits in eggplant and implications for conservation of gene function during evolution of solanaceous species. Theoretical and Applied Genetics, 107(2), 359–370. https://doi.org/10.1007/s00122-003-1257-5 [Google Scholar] [PubMed] [CrossRef]
54. Abdelnour, S. A., Xie, L., Hassanin, A. A., Zuo, E., Lu, Y. (2021). The potential of CRISPR/Cas9 gene editing as a treatment strategy for inherited diseases. Frontiers in Cell and Developmental Biology, 9, 699597. https://doi.org/10.3389/fcell.2021.699597 [Google Scholar] [PubMed] [CrossRef]
55. Raza, S. H. A., Hassanin, A. A., Pant, S. D., Bing, S., Sitohy, M. Z. et al. (2022). Potentials, prospects and applications of genome editing technologies in livestock production. Saudi Journal of Biological Sciences, 29(4), 1928–1935. https://doi.org/10.1016/j.sjbs.2021.11.037 [Google Scholar] [PubMed] [CrossRef]
56. Fang, E. F., Hassanien, A. A. E., Wong, J. H., Bah, C. S. F., Soliman, S. S. et al. (2010). Purification and modes of antifungal action by Vicia faba cv. Egypt trypsin inhibitor. Journal of Agricultural and Food Chemistry, 58(19), 10729–10735. https://doi.org/10.1021/jf102277k [Google Scholar] [PubMed] [CrossRef]
57. Fathy, M., Eldomiaty, A. S., Abd El-Fattah, H., Mahgoub, E. I., A Hassanin, A. (2021). Morphological, biochemical and molecular characterization of rhizobia of faba bean plants grown in North Nile Delta Egypt. Pakistan Journal of Biological Sciences, 24(6), 672–679. https://doi.org/10.3923/pjbs.2021.672.679 [Google Scholar] [CrossRef]
58. AL-Khayri, J., Latef, A. A., Taha, H., Eldomiaty, A., Abd-elfattah, M. et al. (2022). In silico profiling of proline biosynthesis and degradation related genes during fruit development of tomato. SABRAO Journal of Breeding and Genetics, 54(3), 549–564. https://doi.org/10.54910/sabrao2022.54.3.8 [Google Scholar] [CrossRef]
59. Fang, E. F., Hassanien, A. A., Wong, J. H., Bah, C. S., Soliman, S. S. et al. (2011). Isolation of a new trypsin inhibitor from the faba bean (Vicia faba cv. Giza 843) with potential medicinal applications. Protein & Peptide Letters, 18(1), 64–72. https://doi.org/10.2174/092986611794328726 [Google Scholar] [PubMed] [CrossRef]
60. Ghareeb, Y. E., Soliman, S. S., Ismail, T. A., Hassan, M. A., Abdelkader, M. A. et al. (2022). Improvement of German chamomile (Matricaria recutita L.) for mechanical harvesting, high flower yield and essential oil content using physical and chemical mutagenesis. Plants, 11(21), 2940. https://doi.org/10.3390/plants11212940 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools