 Open Access
Open Access
ARTICLE
Lack of Tocopherol Inhibits Rice Growth by Triggering an Ectopic Stress Response and the Accumulation of DELLA Protein
1 Huaiyin Institute of Agricultural Sciences in Xuhuai Region of Jiangsu, Huai’an, 223001, China
2 School of Life Sciences, Huaiyin Normal University, Huai’an, 223300, China
3 Key Laboratory of Eco-Agricultural Biotechnology around Hongze Lake, Regional Cooperative Innovation Center for Modern Agriculture and Environmental Protection, Huaiyin Normal University, Huai’an, 223300, China
* Corresponding Authors: Di Wang. Email: ; Xi Liu. Email:
# Equivalent contributing authors
(This article belongs to the Special Issue: Advances in Molecular Genetics and Physiology towards a Better Understanding of Agricultural Crop Plants)
Phyton-International Journal of Experimental Botany 2023, 92(4), 1173-1183. https://doi.org/10.32604/phyton.2023.026526
Received 09 September 2022; Accepted 02 November 2022; Issue published 06 January 2023
Abstract
Although tocopherols are essential for rice development, the molecular details by which their absence affects development remain to be determined. To study how tocopherols function during rice development, we performed a transcriptome deep sequencing (RNA-seq) analysis of the rice cultivar Nipponbare (Nip) and the tocopherol-deficient mutant small grain and dwarf 1-2 (sgd1-2). We identified 563 differentially expressed genes that were enriched in Gene Ontology categories associated with metabolism, stress, cellular responses, and transcriptional regulation. We determined that the total fatty acid composition of Nip and sgd1-2 was comparable, although cell membrane penetrability in sgd1-2 was significantly higher than in Nip under optimal growth conditions, indicating that tocopherol deficiency induces cell membrane damage. The expression levels of dehydration-responsive element binding 1 (DREB1) genes and free proline content in sgd1-2 were also higher than those in Nip. We also showed that the DELLA protein SLENDER RICE1 (SLR1) accumulated in sgd1-2, resulting in significant changes in the global transcriptome. Our study confirms that the lack of tocopherol accumulation in rice induced ectopic stress responses and limited growth by enhancing SLR1 abundance through increasing SLR1 transcript levels. These results provide new insights into tocopherol during rice development.Keywords
Supplementary Material
Supplementary Material FileOne of the vital functions of vitamin E, which is an integral part of the human diet, is as an antioxidant that protects membranes and helps prevent the onset of cardiovascular diseases [1,2]. Tocopherols (α, β, γ, and δ) and tocotrienols (α, β, γ, and δ) are two of the eight forms of vitamin E. Tocopherols are found in cell membranes and are linked to highly polyunsaturated fatty acids (PUFAs), through which they influence membrane characteristics such as permeability and stability [3–6]. Only certain cyanobacteria and plants can naturally produce tocopherols. The single distinguishing feature between tocopherols and tocotrienols is the presence of isoprenoid side chains in tocotrienols, which can be produced from either phytyl-diphosphate (PDP) or geranylgeranyl-diphosphate (GGDP) [7]. The second step in tocopherol biosynthesis is the condensation of homogentisate (HGA) and PDP by homogentisate phytyltransferase (HPT/VTE2) [8]. The subsequent methylation and cyclization reactions that lead to the biosynthesis of γ- and δ-tocopherols, respectively, are catalyzed by VITAMIN E DEFECTIVE 3 (VTE3) and VTE1 [9,10]. VTE4 then catalyzes the conversion of both γ- and δ-tocopherols into α- and β-tocopherols, respectively [11].
Numerous plant tocopherol biosynthesis mutants have been identified and characterized [12–14]. The tocopherol-deficient vte2 mutants in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) are susceptible to cold stress, despite the different phenotypes displayed by these mutants when grown under ideal growth conditions [15,16]. Under non-freezing low-temperature conditions, Arabidopsis vte2 mutants exhibit substantial growth retardation [17]. Linoleic acid desaturation in the vte2 mutant is lower, primarily in membrane lipids produced in the endoplasmic reticulum (ER) rather than in chloroplasts before low-temperature treatment. Exposure to low temperature worsens this ER membrane lipid phenotype, which can be fully rescued by the introduction of mutant alleles of the ER-resident oleate desaturase genes FATTY ACID DESATURASE2 (FAD2) and the ER-to-plastid lipid transporter genes TRIGALACTOSYLDIACYLGLYCEROL1-4 (TGD1-4) [18,19]. Tocopherols may directly interact with ER-resident enzymes, as demonstrated by trans-organellar complementation, and may therefore play a role in determining membrane composition [20]. After exposure to cold stress, the survival rate of the rice mutant small grain and dwarf1 (sgd1, also named rice tocopherol deficiency 1 [rtd1]) lacking the function of a VTE2-like enzyme, was much lower than that of the wild type [15,16]. These findings suggested that tocopherols may influence cold stress tolerance by changing membrane composition. However, the chemical mechanisms underlying the link between low-temperature sensitivity and tocopherol deficiency need to be better understood.
Plant adaptation to cold temperatures is mostly regulated by the C-REPEAT BINDING FACTOR (CBF)/DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN1 (DREB1) cold-responsive pathway. The transcription of many stress-inducible genes is under the control of the transcription factors CBF/DREB1, which bind to the DRE/CRT cis-acting element [21]. The Arabidopsis genome encodes three CBF proteins: CBF1/DREB1B, CBF2/DREB1C, and CBF3/DREB1A; the rice genome harbors 10 potential DREB1 homologs (OsDREB1A to OsDREB1J). Cold stress was previously shown to induce the expression of 6 of these 10 genes (OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1E, OsDREB1F, and OsDREB1G). Moreover, transgenic rice plants overexpressing OsDREB1 genes showed improved resistance to drought, high-salt, and low-temperature stressors [22,23]. The above examples underscore how OsDREB1 genes are essential for the rice cold response pathway.
Gibberellins (GAs) are a class of tetracyclic diterpenoid plant hormones that have a variety of biological roles, including the promotion of flowering and seed germination, stem and leaf growth, and seed germination [24]. Aspartic Acid–Glutamic Acid–Leucine–Leucine–Alanine Protein (DELLA) proteins are crucial regulators of GA responses and essential components of the GA signaling cascade. DELLAs negatively regulate the GA response by preventing transcription factors from directly activating the expression of their downstream genes, while also indirectly regulating GA biosynthesis genes to increase GA responsiveness by promoting the transcription of the GA receptor gene GIBBERELLIN INSENSITIVE DWARF1 (GID1). GA communicates extensively and establishes crosstalk with other plant hormones to regulate plant growth and development [25]. The Arabidopsis genome encodes five DELLAs: GA-INSENSITIVE (GAI), REPRESSOR of GA1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3 [26]. The GA response phenotype of the rice mutant slender rice 1-1 (slr1-1) demonstrated that the single DELLA protein SLR1 of rice also regulates GA signaling [27]. The discovery that growth constraints caused by exposure to various types of abiotic stresses is at least partially mediated by DELLAs represents a significant advance in our understanding of the role of these proteins in regulating plant development. Moreover, Arabidopsis seedlings exposed to salt or cold stress displayed lower levels of endogenous bioactive GAs and a concomitant accumulation of DELLAs [28,29]. CBF1 was also demonstrated to control plant growth via a DELLA-dependent signaling cascade [29].
We previously showed that tocopherols are crucial for rice growth, based on the characterization of the sgd1-2 mutant. Indeed, the sgd1-2 mutant displayed a pronounced dwarf phenotype and produced small grains relative to its wild-type Nipponbare (Nip). The sgd1-2 mutant also showed hypersensitivity to cold stress [15]. However, how rice development is affected by the loss of tocopherol accumulation is unknown. To begin to answer this question, we explored the effects of the loss of tocopherol on global gene expression using transcriptome deep sequencing (RNA-seq) of the wild-type Nip and the sgd1-2 mutant. We identified 563 differentially expressed genes (DEGs) that were enriched in Gene Ontology (GO) categories related to transcriptional control, stress, metabolism, and other cellular processes. We observed no differences in the total fatty acid contents of Nip and sgd1-2. However, cell membrane penetrability was much higher in sgd1-2 than in Nip when grown under optimal growth conditions, indicating that the absence of tocopherol damaged the cell membrane. The expression levels of CBF/DREB1 genes and free proline content were also higher in sgd1-2 compared to Nip, indicating that an ectopic stress response was induced in sgd1-2. Finally, the accumulation of SLR1 in the sgd1-2 mutant suggested that tocopherol deficiency affects rice growth via the GA signaling pathway.
The previously characterized sgd1-2 mutant in the rice (O. sativa L.) Nip background was used in this study [15]. The wild-type cultivar Nip and the sgd1-2 mutant were grown in a growth chamber under a 16-h-light/8-h-dark photoperiod and 30/22°C day/night temperature cycle.
2.2 RNA-Seq and RT-qPCR Analysis
Samples were collected from 2-week-old Nip and sgd1-2 seedlings for RNA-seq. Total RNA extraction, library construction and sequencing, and subsequent analysis were carried out by Nanjing Genepioneer Institute (Nanjing, China). Three biological replicates were analyzed for each genotype. One-week-old Nip and sgd1-2 seedlings were also collected for RT-qPCR analysis. The internal reference transcript was OsACTIN1. Primers used for qPCR of DREB1 genes were reported in an earlier work [22]. Table S1 provides the list of other primers used in this investigation.
2.3 Fatty Acid Composition Analysis
One-week-old Nip and sgd1-2 seedlings were harvested and their fatty acid methyl esters determined in accordance with a previously published method [30]. Briefly, Nip and sgd1-2 seedlings were frozen in liquid nitrogen, crushed coarsely, and extracted in acidic methanol. The extracts were combined with 1 mL of hexane and 1 mL of 0.9% (w/v) NaCl after incubation at 80°C for 2 h. The top organic layer was collected and dried under nitrogen gas flow. Hexane (25 mL) was used to reconstitute the samples. The fatty acid methyl esters were evaluated by gas chromatography and quantified by flame ionization detection largely as previously reported, using internal heptadecanoic acid (C17:0) as standard [31,32].
2.4 Cell Membrane Penetrability Assay
Electrolyte leakage (EL) was determined as EL1/EL2 of the cell membrane as previously published with a few changes and was used to assess the permeability of the cell membrane [33].
Proline contents were measured using a proline assay kit according to the manufacturer’s recommendations (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) from three biological replicates.
The α-amylase test was carried out essentially as previously described [34]. An assay based on second leaf sheath elongation was conducted to test the sensitivity to GA, largely according to the published method [35]. Surface-sterilized rice seeds were incubated for 2 days at 30°C in complete darkness. Germinated seedlings were then transferred to half-strength Murashige and Skoog medium contained various concentrations of GA3 (0.01, 0.1, 1, 10, and 100 μM), an grew under a 16-h-light/8-h-dark photoperiod and a 28/22°C day/night temperature cycle, with 15 seeds per plate. After 7 days of growth, the lengths of the second leaf sheaths were measured.
The leaves of 10-day-old seedlings (0.5 g) were frozen in liquid nitrogen, crushed finely, and extracted overnight at 4°C in 80% (v/v) methanol. Levels of endogenous GA1 were quantified as described previously [36].
A polyclonal anti-SLR1 antibody was produced by ABclonal (www.abclonal.com). Ten-day-old seedlings of Nip and sgd1-2 were treated with 10 mM GA3 or 10 mM PAC (paclobutrazol) for 6 h or with double distilled water as control. Samples were then collected and processed for immunoblot analysis with the anti-SLR1 antibody, using an anti-EF-1 (Agrisera) antibody as loading control.
3.1 Expression Analysis of Nip and the sgd1-2 Mutant
We conducted an RNA-seq analysis of 2-week-old Nip and sgd1-2 seedlings grown under optimal growth conditions to identify DEGs between the two genotypes. We selected this time point based on the clear dwarf phenotype exhibited by sgd1-2 relative to Nip. Table S2 provides a summary of the RNA-seq analysis. We defined a gene as being differentially expressed when its expression level differed by at least two-fold between sgd1-2 and Nip. We obtained 563 DEGs between Nip and sgd1-2, of which 383 genes were upregulated in sgd1-2 and the remaining 180 genes were downregulated (with a false discovery rate [FDR] P < 0.05). 9 DEGs were selected for RT-qPCR validation of the RNA-seq data. The RT-qPCR results were consistent with those of the RNA-seq data (Table S3). We then performed a GO enrichment analysis with these DEGs, which revealed an enrichment for genes related to metabolism, stress, cellular processes, and transcriptional control in the mutant (Fig. 1).
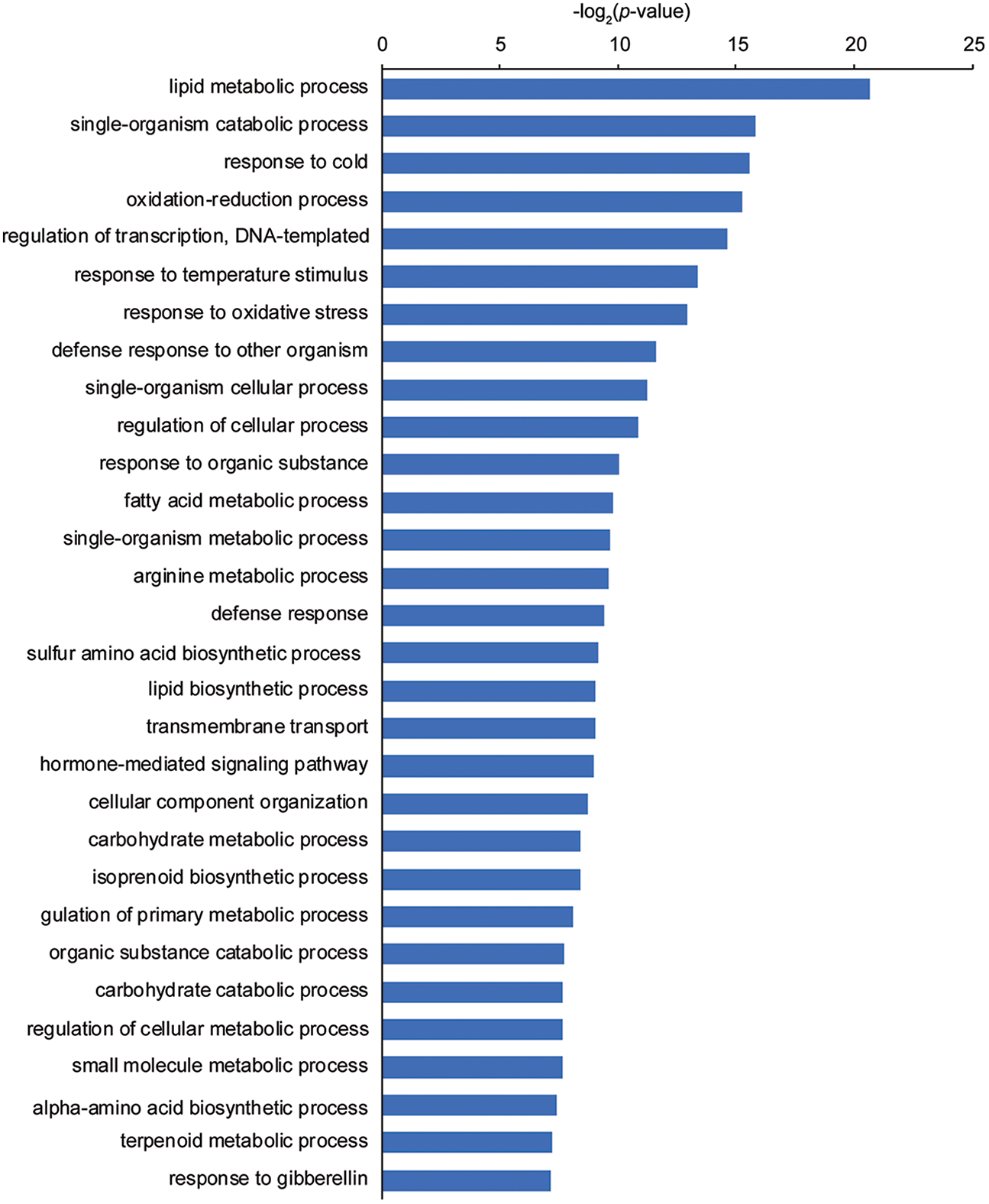
Figure 1: GO analysis of DEGs between Nip and the sgd1-2 mutant. GO categories enriched among the DEGs between 2-week-old Nip and sgd1-2 seedlings grown under optimal growth conditions are shown (P < 0.05)
3.2 Tocopherol Deficiency Induces Cell Membrane Damage
We collected 1-week-old Nip and sgd1-2 seedlings to quantify their membrane lipid profiles and membrane integrity. We chose this earlier time point because of the many DEGs that were enriched in GO categories known to be related to lipid and fatty acid metabolism. We did not observe differences between Nip and sgd1-2 in terms of their total fatty acid contents (Fig. 2A). Tocopherols were thought to solely affect membrane lipids produced in the ER, but not those produced in plastids, as shown in Arabidopsis. We examined the cell membrane penetrability of Nip and sgd1-2 seedlings grown under ideal conditions. We determined that the cell membrane of the sgd1-2 mutant was more permeable than that of Nip even under optimal growth conditions, indicative of damage to the membrane caused by the lack of tocopherols (Fig. 2B).
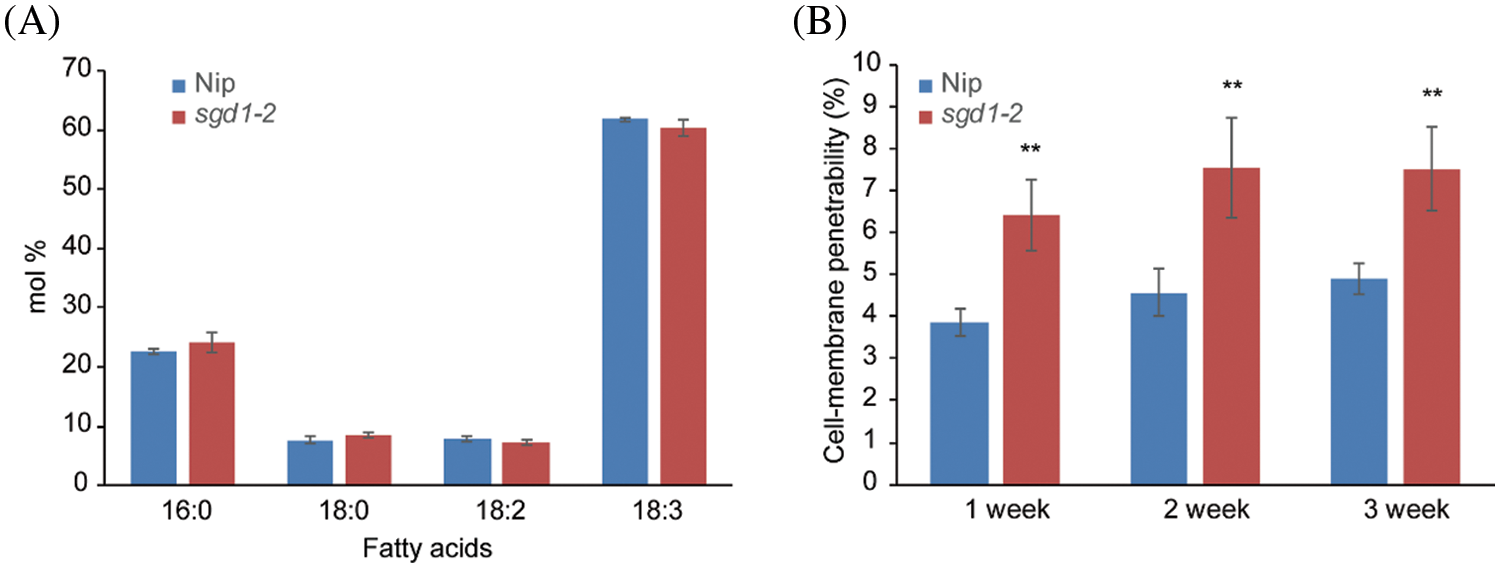
Figure 2: Tocopherol deficiency caused cell membrane damage in sgd1-2. (A) Total fatty acid composition in the leaves of 1-week-old Nip and sgd1-2 seedlings. Values for each fatty acid (number of carbons: number of unsaturated bonds) are means ± standard deviation (SD, n = 3 biological replicates) and are expressed as mol %. (B) Cell membrane penetrability in leaves of Nip and sgd1-2 seedlings under optimal growth conditions. Values are means ± SD, n = 5 biological replicates, **P < 0.01
3.3 Tocopherol Deficiency Induces Ectopic Stress Response
Relative conductivity measures ion leakage and thus reflects damage to cell membranes caused by cold stress. We asked if sgd1-2 displayed symptoms typically associated with a cold stress response when grown under normal growth conditions. We measured CBF/DREB1 expression levels in 1-week-old Nip and sgd1-2 seedlings. Under normal growth conditions, the six CBF/DREB1 genes (OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1E, OsDREB1F, and OsDREB1G) were expressed to higher levels in sgd1-2 relative to Nip (Fig. 3A). Moreover, the free proline content of the sgd1-2 mutant was almost two times that of Nip (Fig. 3B). These findings suggest that sgd1-2 evoked an ectopic stress response due to the lack of tocopherols.
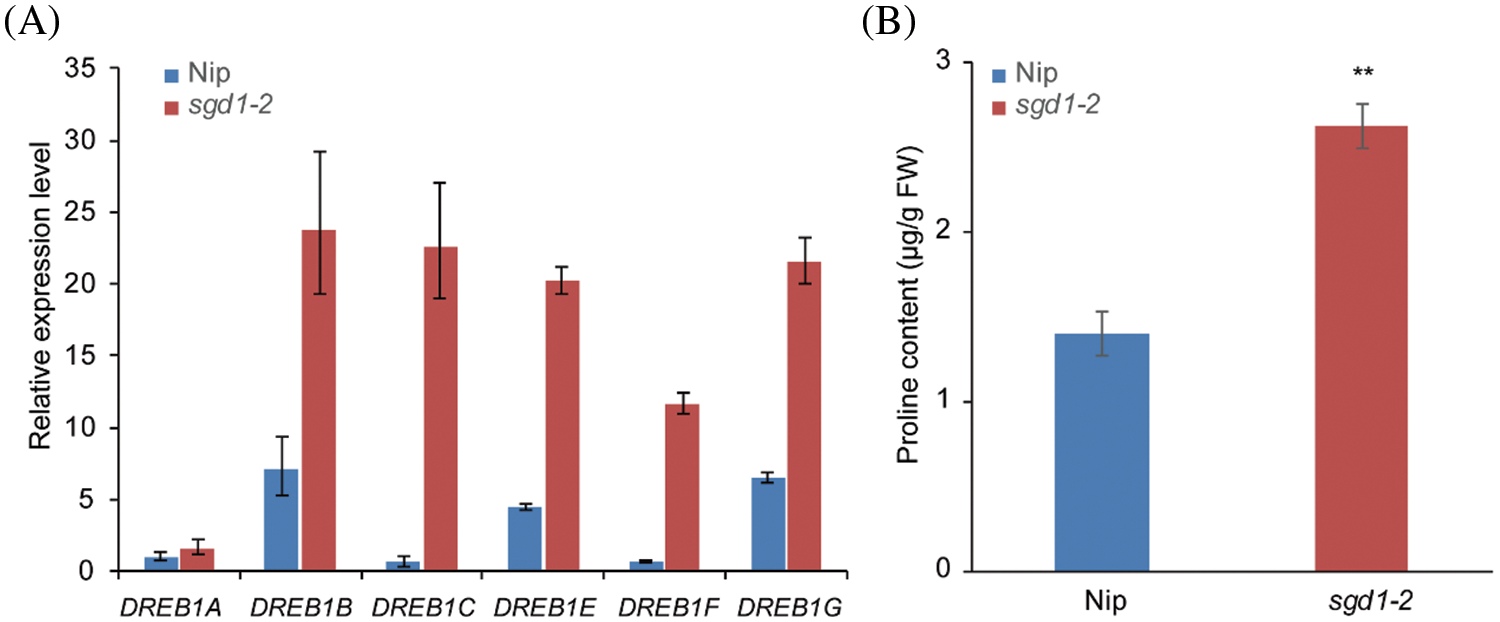
Figure 3: The sgd1-2 mutant showed an ectopic stress response. (A) Expression levels of CBF/DREB1 genes in 1-week-old Nip and sgd1-2 seedlings grown under natural growth conditions. Values are means ± SD, n = 3 biological replicates. (B) Free proline contents in one-week-old Nip and sgd1-2 seedlings grown under natural growth conditions. Values are means ± SD, n = 3 biological replicates, **P < 0.01
3.4 Tocopherol Deficiency Induces the Accumulation of SLR1
Previous research demonstrated that CBF/DREB1 proteins prevent Arabidopsis development by modulating the abundance of DELLA proteins [29]. We thus applied exogenous GA3 to Nip and sgd1-2 seedings and tested them for α-amylase activity using half-seeds without the embryo. We determined that both Nip and sgd1-2 seeds formed plaques, reflecting α-amylase activity, but the sgd1-2 mutant formed plaques of smaller diameter than Nip (Fig. 4A). We also treated Nip and sgd1-2 seedlings with exogenous GA3, finding that the mutant was less responsive to the treatment than Nip, as evidenced by the shorter stature of the mutant and the modest sheath elongation seen over time (Fig. 4B).
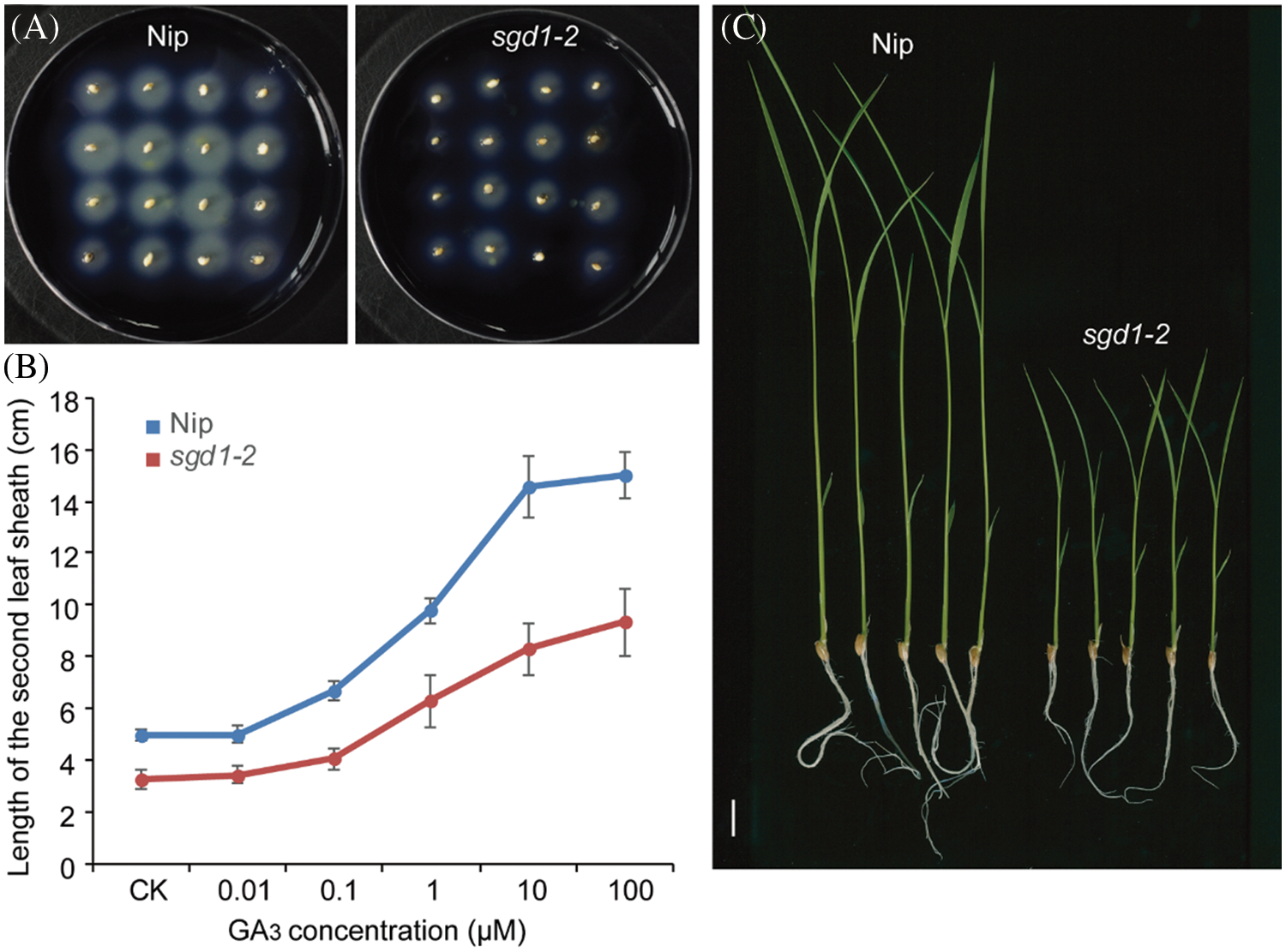
Figure 4: The sgd1-2 mutant had reduced sensitivity to GA. (A) α-Amylase production from half-seeds of Nip and sgd1-2 without the embryo. (B) Length of the second leaf sheath in response to GA3 treatment in 7 days old Nip and sgd1-2 seedlings. Values are means ± SD, n = 15. (C) Image of Nip and sgd1-2 seedlings treated with 100 μM GA3. Scale bar, 2 cm
We carried out an immunoblot analysis to ascertain SLR1 abundance in Nip and slr1-2 and discovered that sgd1-2 accumulated more SLR1 than Nip, in agreement with the lower sensitivity of this mutant to GA (Fig. 5A). Indeed, lower bioactive GA levels resulted in an accumulation of DELLAs, leading to growth inhibition [37,38]. We also measured the levels of endogenous GA1 in 10-day-old Nip and sgd1-2 seedlings to investigate whether the higher SLR1 abundance in the sgd1-2 mutant was caused by altered GA metabolism. However, we observed no difference for the GA1 levels of Nip and sgd1-2 (Fig. 5B, Table S4). We also measured SLR1 expression in the two genotypes and determined that SLR1 was expressed to much higher levels in the sgd1-2 mutant than in Nip (Fig. 5C). We conclude that the lack of tocopherols inhibits rice development via the GA signaling pathway.
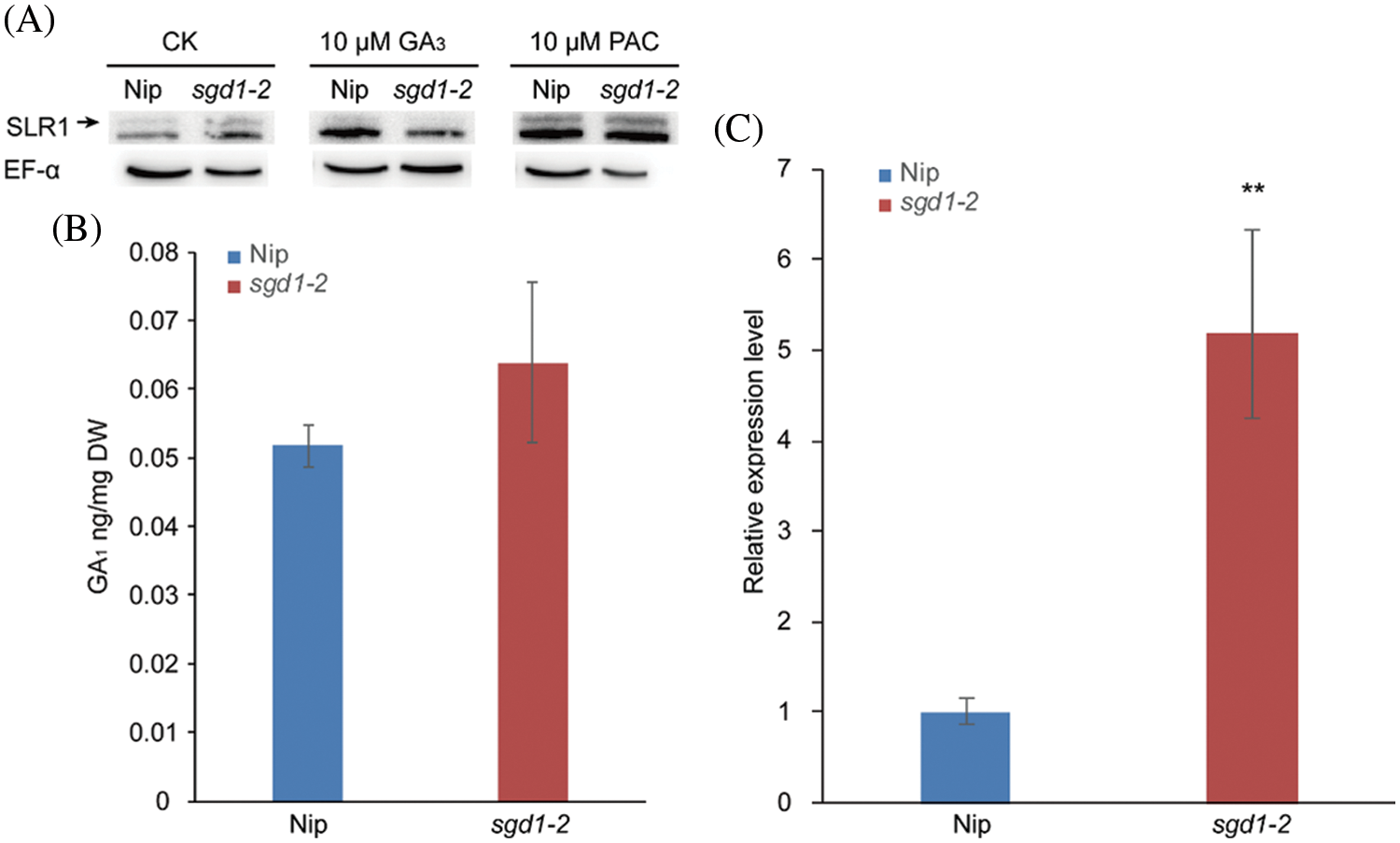
Figure 5: SLR1 abundance and SLR1 expression levels in sgd1-2. (A) Immunoblot analysis of SLR1 in 10-day-old Nip and sgd1-2 seedlings. The anti-SLR1 antibody detected two bands, with the upper band being degraded upon GA3 treatment but increasing in intensity following PAC treatment, indicating that the upper band (black arrow) was SLR1. (B) Endogenous GA1 contents in 10-day-old Nip and sgd1-2 seedlings. Values are means ± SD, n = 3 biological repeats. DW, dry weight. (C) Relative SLR1 expression levels in 10-day-old Nip and sgd1-2 seedlings. Values are means ± SD, n = 3 biological repeats. **P < 0.01
Tocopherols have been shown to be crucial for rice growth and development. The sgd1-2 mutant showed substantial growth retardation throughout its life cycle. Although tocopherols are considered to have antioxidant properties [39], we did not detect evidence for oxidative damage in the sgd1-2 mutant, indicating that the growth retardation phenotype of this mutant was not a result of oxidative damage. The sgd1-2 mutant of rice therefore underscores the fundamental differences in the functions of tocopherols in rice and Arabidopsis. The maize (Zea mays) sucrose export defective1 (sed1) mutant exhibits growth retardation reminiscent of the rice sgd1-2 mutant under ideal growth conditions [13]. Tocopherols may therefore be crucial for the growth of monocots but not dicots.
Many of the DEGs were enriched in GO categories known to be involved in lipid and fatty acid metabolisms, suggesting that tocopherol deficiency may influence lipid composition in rice. However, the total fatty acid composition was identical in Nip and sgd1-2. Previous studies have indicated that tocopherols can change membrane lipid composition by modulating lipid biosynthesis in Arabidopsis. Lipidomic analyses using electrospray ionization triple quadrupole mass spectrometry demonstrated that tocopherols specifically affect ER fatty acid desaturation, but not that taking place in plastids [18,19]. Tocopherols may directly interact with ER-resident enzymes, influencing how the composition of membranes is determined [20]. Further investigation is needed to uncover whether and how tocopherols affect ER fatty acid desaturation in rice. Although membrane lipid composition did not show clear changes in sgd1-2, cell membrane penetrability was significantly higher in sgd1-2 than in Nip, suggesting that tocopherol deficiency induced cell membrane damage in rice. Previous studies have reported how changes in tocopherol levels can affect the stability and fluidity of membranes [5,6]. Due to the comparable geometries of tocopherol and asymmetric phospholipids, research in model phospholipid membranes has suggested that tocopherol stabilizes membranes [40,41]. In rice, tocopherol is also necessary for the regular operation of cell membranes. More research is needed to determine the molecular mechanism underlying the association between the lack of tocopherol and cell membrane penetrability in rice.
The CBF/DREB1 cold response pathway is crucial for plant adaptation to cold [42]. The expression of CBF/DREB1 genes is quickly induced in response to cold stress and causes the transcription of genes (the CBF regulon) that encode a variety of proteins to shield plants from cold stress [43]. We observed that the expression levels of CBF/DREB1 genes were markedly higher in sgd1-2 relative to Nip when grown under normal growth conditions. Free proline also accumulated in sgd1-2. In addition, many of the DEGs in sgd1-2 were enriched in GO categories associated with stress responses, suggesting that sgd1-2 seedlings underwent an ectopic stress response caused by tocopherol deficiency. The cell membrane is the primary site of perception for some stresses in plants [44]. Notably, we detected evidence of cell membrane damage in sgd1-2; together with the notion that tocopherols can change membrane lipid composition in Arabidopsis, we propose that the lack of tocopherols in the mutant may modulate the function of the cell membrane and result in the induction of a stress response even in the absence of a stimulus. Both rice and Arabidopsis plants overexpressing CBF/DREB1 genes display substantial growth retardation even under ideal growth conditions, much like the sgd1-2 mutant, in which CBF/DREB1 genes were highly expressed, offering one possible explanation for the dwarf phenotype of the mutant.
CBF1 overexpression inhibits growth in Arabidopsis via the GA signaling pathway. Based on the results of the α-amylase activity assay and leaf sheath elongation following GA3 treatment, we concluded that sgd1-2 responded less favorably to GA3, indicating that tocopherol deficit may also inhibit development in rice via the GA signaling pathway. In Arabidopsis, the accumulation of CBF/DREB1s in response to cold induces the expression of RGL3, stabilizes DELLAs, and decreases bioactive GA levels, thus leading to growth inhibition [29]. The contents of bioactive GA1 were similar in Nip and sgd1-2, but we detected higher expression levels for the DELLA-encoding gene SLR1, as well as greater SLR1 abundance. That bioactive GA levels were not affected may be another difference between rice and Arabidopsis. We propose that rice growth is restricted by tocopherol deficiency via the transcriptional regulation of SLR1 expression levels, directly leading to the accumulation of SLR1 without invoking a posttranslational mechanism. These results provide fresh perspectives on how tocopherols operate during rice growth and development.
Authorship: Di Wang, Hao Gao and Xi Liu designed and conceived the research. Di Wang, Hao Gao, Jian Wang, Baoshan Cheng, Gang Li and Weijun Xu performed the experiments. Di Wang and Xi Liu wrote and revised the manuscript.
Acknowledgement: We are grateful to Prof. Xiangning Jiang (Beijing Forestry University) for the GA quantification.
Funding Statement: This work was supported by funding from the Natural Science Foundation of Jiangsu Province (BK20190239), Huai’an Academy of Agricultural Sciences Initiation and Development of Scientific Research Fund for High-Level Introduced Talents (0062019016B), the Scientific Research Fund Development Project of Huai’an Academy of Agricultural Sciences (HNY202102).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Collakova, E., DellaPenna, D. (2001). Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiology, 127(3), 1113–1124. DOI 10.1104/pp.010421. [Google Scholar] [CrossRef]
2. Schledz, M., Seidler, A., Beyer, P., Neuhaus, G. (2001). A novel phytyltransferase from Synechocystis sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Letters, 499(1–2), 15–20. DOI 10.1016/s0014-5793(01)02508-x. [Google Scholar] [CrossRef]
3. Wang, X., Quinn, P. J. (2000). The location and function of vitamin E in membranes (review). Molecular Membrane Biology, 17(3), 143–156. DOI 10.1080/09687680010000311. [Google Scholar] [CrossRef]
4. Kagan, V. E. (1989). Tocopherol stabilizes membrane against phospholipase A, free fatty acids, and lysophospholipids. Annals of the New York Academy of Sciences, 570, 121–135. DOI 10.1111/j.1749-6632.1989.tb14913.x. [Google Scholar] [CrossRef]
5. Mokrosnop, V. M. (2014). Functions of tocopherols in the cells of plants and other photosynthetic organisms. Ukrainskii Biokhimicheskii Zhurnal, 86(5), 26–36. DOI 10.15407/ubj86.05.026. [Google Scholar] [CrossRef]
6. Munné-Bosch, S., Alegre, L. (2002). The function of tocopherols and tocotrienols in plants. Critical Reviews in Plant Sciences, 21(1), 31–57. DOI 10.1016/S0735-2689(02)80037-5. [Google Scholar] [CrossRef]
7. DellaPenna, D., Pogson, B. J. (2006). Vitamin synthesis in plants: Tocopherols and carotenoids. Annual Review of Plant Biology, 57(1), 711–738. DOI 10.1146/annurev.arplant.56.032604.144301. [Google Scholar] [CrossRef]
8. Collakova, E., DellaPenna, D. (2003). Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiology, 131(2), 632–642. DOI 10.1104/pp.015222. [Google Scholar] [CrossRef]
9. Porfirova, S., Bergmuller, E., Tropf, S., Lemke, R., Dormann, P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proceedings of the National Academy of Sciences, 99(19), 12495–12500. DOI 10.1073/pnas.182330899. [Google Scholar] [CrossRef]
10. Cheng, Z., Sattler, S., Maeda, H., Sakuragi, Y., Bryant, D. A. et al. (2003). Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell, 15(10), 2343–2356. DOI 10.1105/tpc.013656. [Google Scholar] [CrossRef]
11. Cela, J., Chang, C., Munné-Bosch, S. (2011). Accumulation of γ-rather than α-tocopherol alters ethylene signaling gene expression in the vte4 mutant of Arabidopsis thaliana. Plant & Cell Physiology, 52(8), 1389–1400. DOI 10.1093/pcp/pcr085. [Google Scholar] [CrossRef]
12. Savidge, B., Weiss, J. D., Wong, Y. H., Lassner, M. W., Mitsky, T. A. et al. (2002). Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiology, 129(1), 321–332. DOI 10.1104/pp.010747. [Google Scholar] [CrossRef]
13. Russin, W. A., Evert, R. F., Vanderveer, P. J., Sharkey, T. D., Briggs, S. P. (1996). Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. Plant Cell, 8(4), 645–658. DOI 10.1105/tpc.8.4.645. [Google Scholar] [CrossRef]
14. Hofius, D., Hajirezaei, M. R., Geiger, M., Tschiersch, H., Melzer, M. et al. (2004). RNAi-mediated tocopherol deficiency impairs photoassimilate export in transgenic potato plants. Plant Physiology, 135(3), 1256–1268. DOI 10.1104/pp.104.043927. [Google Scholar] [CrossRef]
15. Wang, D., Wang, Y., Long, W., Niu, M., Zhao, Z. et al. (2017). SGD1, a key enzyme in tocopherol biosynthesis, is essential for plant development and cold tolerance in rice. Plant Science, 260, 90–100. DOI 10.1016/j.plantsci.2017.04.008. [Google Scholar] [CrossRef]
16. Zhang, Y., Liu, K., Zhu, X., Wu, Y., Zhang, S. et al. (2018). Rice tocopherol deficiency 1 encodes a homogentisate phytyltransferase essential for tocopherol biosynthesis and plant development in rice. Plant Cell Reports, 37(5), 775–787. DOI 10.1007/s00299-018-2266-9. [Google Scholar] [CrossRef]
17. Maeda, H., Song, W., Sage, T. L., DellaPenna, D. (2006). Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell, 18(10), 2710–2732. DOI 10.1105/tpc.105.039404. [Google Scholar] [CrossRef]
18. Maeda, H., Sage, T. L., Isaac, G., Welti, R., Dellapenna, D. (2008). Tocopherols modulate extraplastidic polyunsaturated fatty acid metabolism in Arabidopsis at low temperature. Plant Cell, 20(2), 452–470. DOI 10.1105/tpc.107.054718. [Google Scholar] [CrossRef]
19. Song, W., Maeda, H., DellaPenna, D. (2010). Mutations of the ER to plastid lipid transporters TGD1, 2, 3 and 4 and the ER oleate desaturase FAD2 suppress the low temperature-induced phenotype of Arabidopsis tocopherol-deficient mutant vte2. Plant Journal, 62(6), 1004–1018. DOI 10.1111/j.1365-313X.2010.04212.x. [Google Scholar] [CrossRef]
20. Mehrshahi, P., Stefano, G., Andaloro, J. M., Brandizzi, F., Froehlich, J. E. et al. (2013). Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proceedings of the National Academy of Sciences, 110(29), 12126–12131. DOI 10.1073/pnas.1306331110. [Google Scholar] [CrossRef]
21. Hirayama, T., Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant Journal, 61(6), 1041–1052. DOI 10.1111/j.1365-313X.2010.04124.x. [Google Scholar] [CrossRef]
22. Mao, D., Chen, C. (2012). Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS One, 7(10), e47275. DOI 10.1371/journal.pone.0047275. [Google Scholar] [CrossRef]
23. Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M. et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant & Cell Physiology, 47(1), 141–153. DOI 10.1093/pcp/pci230. [Google Scholar] [CrossRef]
24. Fleet, C. M., Sun, T. P. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Current Opinion in Plant Biology, 8(1), 77–85. DOI 10.1016/j.pbi.2004.11.015. [Google Scholar] [CrossRef]
25. Xue, H., Gao, J., He, P., Xiao, G. (2022). Origin, evolution, and molecular function of DELLA proteins in plants. The Crop Journal, 10(2), 287–299. DOI 10.1016/j.cj.2021.06.005. [Google Scholar] [CrossRef]
26. Sun, T. P., Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annual Review of Plant Biology, 55(1), 197–223. DOI 10.1146/annurev.arplant.55.031903.141753. [Google Scholar] [CrossRef]
27. Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M. et al. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell, 13(5), 999–1010. DOI 10.1105/tpc.13.5.999. [Google Scholar] [CrossRef]
28. Achard, P., Cheng, H., de Grauwe, L., Decat, J., Schoutteten, H. et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science, 311(5757), 91–94. DOI 10.1126/science.1118642. [Google Scholar] [CrossRef]
29. Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P. et al. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell, 20(8), 2117–2129. DOI 10.1105/tpc.108.058941. [Google Scholar] [CrossRef]
30. Focks, N., Benning, C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiology, 118(1), 91–101. DOI 10.1104/pp.118.1.91. [Google Scholar] [CrossRef]
31. Chapman, K. D., Trelease, R. N. (1991). Acquisition of membrane lipids by differentiating glyoxysomes: Role of lipid bodies. Journal of Cell Biology, 115(4), 995–1007. DOI 10.1083/jcb.115.4.995. [Google Scholar] [CrossRef]
32. Zhang, D., Pirtle, I. L., Park, S. J., Nampaisansuk, M., Neogi, P. et al. (2009). Identification and expression of a new delta-12 fatty acid desaturase (FAD2-4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiology and Biochemistry, 47(6), 462–471. DOI 10.1016/j.plaphy.2008.12.024. [Google Scholar] [CrossRef]
33. Peixoto, M. D. M., Sage, R. F. (2016). Improved experimental protocols to evaluate cold tolerance thresholds in Miscanthus and switchgrass rhizomes. GCB Bioenergy, 8(2), 257–268. DOI 10.1111/gcbb.12301. [Google Scholar] [CrossRef]
34. Li, W., Wu, J., Weng, S., Zhang, Y., Zhang, D. et al. (2010). Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta, 232(6), 1383–1396. DOI 10.1073/pnas.97.21.11638. [Google Scholar] [CrossRef]
35. Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y. et al. (2000). Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proceedings of the National Academy, 97(21), 11638–11643. DOI 10.1073/pnas.97.21.11638. [Google Scholar] [CrossRef]
36. Liu, S., Chen, W., Qu, L., Gai, Y., Jiang, X. (2013). Simultaneous determination of 24 or more acidic and alkaline phytohormones in femtomole quantities of plant tissues by high-performance liquid chromatography-electrospray ionization-ion trap mass spectrometry. Analytical and Bioanalytical Chemistry, 405(4), 1257–1266. DOI 10.1007/s00216-012-6509-2. [Google Scholar] [CrossRef]
37. Fu, X., Richards, D. E., Fleck, B., Xie, D., Burton, N. et al. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell, 16(6), 1406–1418. DOI 10.1105/tpc.021386. [Google Scholar] [CrossRef]
38. Silverstone, A. L., Jung, H. S., Dill, A., Kawaide, H., Kamiya, Y. et al. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell, 13(7), 1555–1566. DOI 10.1007/s00425-003-1126-0. [Google Scholar] [CrossRef]
39. Munn Bosch, S., Falk, J. (2004). New insights into the function of tocopherols in plants. Planta, 218(3), 323–326. DOI 10.1007/s00425-003-1126-0. [Google Scholar] [CrossRef]
40. Salgado, J., Villalain, J., Gómez-Fernández, J. C. (1993). Magic angle spinning 13C-NMR spin-lattice relaxation study of the location and effects of α-tocopherol, ubiquinone-10 and ubiquinol-10 in unsonicated model membranes. European Biophysics Journal, 22(2), 151–155. DOI 10.1007/BF00196919. [Google Scholar] [CrossRef]
41. Salgado, J., Villalain, J., Gomez-Fernandez, J. C. (1993). Alpha-tocopherol interacts with natural micelle-forming single-chain phospholipids stabilizing the bilayer phase. Archives of Biochemistry and Biophysics, 306(2), 368–376. DOI 10.1006/abbi.1993.1525. [Google Scholar] [CrossRef]
42. Cook, D., Fowler, S., Fiehn, O., Thomashow, M. F. (2004). A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proceedings of the National Academy of Sciences, 101(42), 15243–15248. DOI 10.1073/pnas.0406069101. [Google Scholar] [CrossRef]
43. Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H. et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry, 281(49), 37636–37645. DOI 10.1074/jbc.M605895200. [Google Scholar] [CrossRef]
44. Vaultier, M. N., Cantrel, C., Vergnolle, C., Justin, A. M., Demandre, C. et al. (2006). Desaturase mutants reveal that membrane rigidification acts as a cold perception mechanism upstream of the diacylglycerol kinase pathway in Arabidopsis cells. FEBS Letters, 580(17), 4218–4223. DOI 10.1016/j.febslet.2006.06.083. [Google Scholar] [CrossRef]
Appendix:
Supplementary Table S1: Primers used in this study
Supplementary Table S2: DEGs in sgd1-2 relative to Nip seedlings grown under optimal growth conditions
Supplementary Table S3: Validation of RNA-seq data by RT-qPCR analysis
Supplementary Table S4: Summary of endogenous GA contents
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools