 Open Access
Open Access
ARTICLE
Development of Molecular Marker Linked with Cercospora Leaf Spot (CLS) Disease Resistance in Vigna radiata, Cloning, and Expression for Evaluating Antifungal Activity against Cercospora canescens
1 Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture Faisalabad, Faisalabad, 38040, Pakistan
2 Department of Plant Pathology, University of Agriculture Faisalabad, Faisalabad, 38040, Pakistan
* Corresponding Author: Siddra Ijaz. Email:
(This article belongs to the Special Issue: Agricultural Intensification, Climate Change, and Food Security)
Phyton-International Journal of Experimental Botany 2023, 92(4), 1289-1300. https://doi.org/10.32604/phyton.2023.026469
Received 07 September 2022; Accepted 11 November 2022; Issue published 06 January 2023
Abstract
We developed a molecular marker for MAS of mungbean resistant varieties against CLS from the consensus sequence (MB-CLsRG) of identified RGAs (MB-ClsRCaG1 and MB-ClsRCaG2). The MB-CLsRG sequence-specific primer pair was used to screen Cercospora leaf spot (CLS) resistant varieties of mungbean in genomic analysis that showed congruency with phenotypic screening. Validation of molecular marker linkage with CLS resistance was performed using rtPCR in transcriptomic analysis. The sequenced PCR products showed 100% homology with MB-CLsRG sequence and putative disease resistance proteins that confirmed the linkage of molecular marker with CLS resistance in mungbean. The antifungal potential of MB-CLsRG gene encoding protein was assessed. The MB-CLsRG gene sequence was cloned in the E. coli expression vector for recombinant protein production. The recombinant protein was then investigated for its in vitro antifungal potential against Cercospora canescens. The in vitro investigation showed strong antifungal activity of recombinant protein as it restricted the growth of fungal mycelial mass. The results validated the linkage of developed marker with CLS-resistant mungbean varieties; therefore, it can be used to screen resistant varieties from a large population in MAS. Moreover, the recombinant protein of the MB-CLsRG gene sequence revealed antifungal potential, which proved the gene sequence could be suitable to use in transgenic plants technology to develop fungal-resistant transgenic crops.Keywords
Supplementary Material
Supplementary Material FileVarious phytopathogens greatly affect agriculture crops, resulting in massive annual losses of 13 to 22 percent in various food crops [1,2]. They pose a danger to the long-term availability of high-quality food, raising worries about food security. Moreover, global climate change has contributed to creating such atmospheric conditions, which lead to the increased spread of various diseases. Phytopathogens, especially fungi, are responsible for many plant diseases, ultimately contributing to high yield losses [3]. Over the years, phytopathogens have evolved to subdue plant defense mechanisms [4], which caused devastating effects on crop production, resulting in heavy economic losses [5].
In order to overcome crop losses, various approaches are in practice for sustainable disease management. Resistant plant sources and agrochemicals are the common possible ways of controlling plant diseases [6]. Fungicides are used on a large scale for controlling fungal pathogens. The agrochemicals negatively impact the ecosystem and develop resistance in fungal pathogens against fungicides [7]. Hence, environment-friendly alternative agents should be used instead of chemical control strategies. Natural plant proteins, exhibiting antifungal activity, are known to be an eco-friendly and sustainable approach to controlling plant fungal disease [8]. Naturally, plants have evolved diverse arrays of defense mechanisms against pathogens. Among them are the innate plant proteins possessing antifungal potential. These plant antifungal proteins (AFPs) are strong agents for sustainable disease management as they are nontoxic to humans, act antagonistically, are biodegradable, and have undergone millions of evolutionary processes against fungal pathogens [9]. Moreover, antifungal plant proteins act against rapidly evolving fungal pathogens through their innate mechanisms [10].
DNA markers linked with disease resistance genes can be used in marker-assisted selection (MAS) programs to speed up the screening of large populations for developing resistant varieties [11] against biotic and abiotic stresses. MAS is relatively more efficient and reliable than phenotypic screening as it involves DNA markers that eliminate environmental errors. Various quantitative trait loci (QTL) markers linked to Cercospora leaf spot disease and Mungbean yellow mosaic virus (MYMV) disease resistance in V. species have been documented [12,13]. Using molecular markers linked to the resistance genes is a promising approach in MAS. Resistant Gene Analog Polymorphism (RGAP) technique is an efficient tool for identifying molecular markers for disease resistance. RGAP markers are developed from the conserved domains of resistance genes, including nucleotide-binding site (NBS), leucine-rich repeat (LRR), and protein kinase domain of R genes. Various resistance gene analogs (RGAs) have been used successfully to develop molecular markers linked to resistance genes in wheat, sorghum, and rice [14], but in mungbean, no RGA marker against Cercospora leaf spot (CLS) disease has yet been reported. Hence, this study attempted to fill the gap that will support and strengthen the MAS in mungbean.
The availability of mungbean genome sequence has facilitated the development of molecular markers and the identification of resistance candidate genes against various biotic stresses [15]. Our study investigated molecular marker identification against CLS disease in mungbean (V. radiata L. Wilczek). The causal agent of CLS is the fungal pathogen C. canescens. The Cercospora toxin (a photo activator) causes the Cercospora leaf spot in mungbean [16], a significant threat to mungbean production and per annum yield. It can cause approximately 60%–100% loss in yield [17]. The disease spread rate is relatively high in susceptible varieties, resulting in grain and pod size decrement and plant defoliation [18]. The present study focused on identifying molecular marker for MAS in mungbean breeding programs. The cloning and expression of the developed molecular marker gene were performed to investigate its antifungal potential against C. canescens.
2.1 Development of Molecular Marker and Screening of Mungbean Germplasm
Primer Designing
We identified three RGAs (MB-ClsRCaG1, MT886216; MB-ClsRCaG2, MT919519, and MB-ClsRCaG3, MT919520) in V. radiata under Cercospora leaf spot (CLS) disease stress. The transcriptome analysis under CLS stress showed the amplification of these RGAs in a resistant mungbean variety, while no amplification was in a susceptible variety [19]. The translated sequences of identified RGAs, MB-ClsRCaG1, and MB-ClsRCaG2, possess NB-ARC and RPW8 domains that are the conserved domains of disease resistance genes. Considering the expression of these gene sequences in mungbean during CLS disease, we investigated their contribution to CLS resistance to develop molecular markers for MAS.
The DNA sequences of MB-ClsRCaG1 and MB-ClsRCaG2 were aligned through Clustal W, a consensus sequence was generated using Lasergene v 7.1.0 SeqMan pro (SeqManTM II), and a primer pair was designed through PrimerQuest Tool (Integrated DNA Technologies, Inc., Iowa, US). The contig deposited in GenBank was designated as MB-CLsRG.
2.2 Germplasm Collection and Maintenance
Thirty-six mungbean varieties, were collected from different institutes in Pakistan (Table S1). Seeds were grown in earthen pots, filled with compost, and maintained in the Fungal Molecular Biology laboratory (FMB Lab) greenhouse, Department of Plant Pathology, University of Agriculture, Faisalabad, Pakistan.
2.3 Mungbean Population Screening
One-month-old plants of 36 mungbean varieties were inoculated using C. canescens conidial suspension (5 × 105 concentration per mL) through the foliar spray method [20]. Relative humidity was maintained by covering all inoculated plants with polythene bags. The experiment was laid out as three biological repeats and three technical repeats of each variety for phenotypic screening under a completely randomized design. Disease data was recorded following the disease rating scale described by Abbas et al. [16].
DNA extraction: 100 mg leave sample of each mungbean variety was taken. Each leave sample was ground into a fine powder using liquid nitrogen. The ground material was treated as per the protocol provided by the manufacturer in the GeneJET Plant Genomic DNA Purification Mini Kit (K0791, Thermo Scientific Massachusetts, USA). The extracted DNA sample of each variety was quantified (NanoDrop 8000 Thermo SCIENTIFIC, Massachusetts, USA) and diluted in nuclease-free water (50 ng/μL).
Polymerase Chain Reaction analysis: PCR analysis was carried out in a thermal cycler (peqSTAR, Erlangen, Germany) using a designed specific primer pair (Forward primer: 5’GATGCTCTTGTTGGACACGTGCTTGA3’; Reverse primer: 5’GATGCTGACTCTGGCAAAACACCGTC3’) for genotypic screening of mungbean germplasm. The reaction mixture contained template DNA, Phusion High-Fidelity PCR Master Mix, HF Buffer (Thermo Scientific, Massachusetts, USA), forward and reverse primers, and deionized water, in a total volume of 25 µL. The optimized thermal profile was comprised of initial denaturation (95°C for 3 minutes), a loop of 40 cycles including denaturation (95°C, 1 min), annealing (57°C, 45 s), extension (72°C, 1 min) and final extension (72°C for 10 min). PCR products were resolved on 1% UltrapureTM agarose gel (INVITROGEN) through gel electrophoresis (WEALTEC, ELITE 300 PLUS, Nevada, USA) and visualized on a Gel Documentation system (GDS, Bio-Rad, California, USA) to get DNA profiling.
2.4 Validation of Molecular Marker Linkage with CLS Resistance in V. radiata
The mungbean varieties that gave PCR amplification in the genomic analysis were inoculated with C. canescens. After one week of post-inoculation and six weeks of post-inoculation (the days at which symptoms started to appear on susceptible variety), the total RNA of each genotype was extracted (GeneJET Plant RNA Purification kit) and treated to remove genomic DNA with RapidOut DNA Removal kit. Extracted purified RNA samples were quantified through UV visible NANODROP (8000 Spectrophotometer, Thermo Scientific, Massachusetts, USA). The cDNA was synthesized (RevertAid First Strand cDNA Synthesis Kit) and diluted ten times. Reverse transcription PCR (rtPCR) analysis was performed using primer pair (mentioned above). The PCR products were purified (FavorPrep PCR Clean-Up Kit) and sequenced (Eurofins Genomics DNA sequencing services, Pennsylvania, USA). The generated sequences were trimmed through BioEdit version 7.2.6.1. For comparison, these DNA sequences were searched against non-redundant sequences in the GenBank database by the BLASTX program. After validating the linkage of identified molecular marker with CLS resistance in mungbean, the DNA sequence of the molecular marker linked to CLS resistance was submitted to National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) database for the acquisition of GenBank accession number.
2.5 Development of E. coli Expression Vector
We synthesized the MB-CLsRG gene sequence flanked by two restriction sites, SacI and BamHI, in the pUC57 backbone. The MB-CLsRG/ pUC57 construct was delivered to the TOP10 E. coli strain cells by heat shock to multiply the plasmid DNA. The treated cells were cultured on Luria-Bertani (LB) broth containing ampicillin (100 μg/mL) at 37°C on moderate shaking. The plasmid DNA extraction was made using GeneJET plasmid Kit (Thermo Scientific, Massachusetts, USA) and digested with SacI and BamHI restriction enzymes. The restricted product was run on 0.6% agarose gel, and the DNA fragment was eluted using a Gel Purification kit (FavorPrep, Taiwan). The purified fragment was ligated into SacI- and BamHI- digested pET28a, E. coli expression vector [20,21]. The ligated product was delivered into BL21 E. coli strain through heat shock, cultured on LB medium containing kanamycin (50–100 μg/mL). The bacterial colony was picked and cultured on LB broth (having kanamycin) for plasmid DNA isolation. The constructed plasmid (MB-CLsRG/pET28a) was confirmed through double digestion with SacI and BamHI restriction enzymes. A sequence encoding a His-tag was present 33 bp upstream from the SacI restriction site on the pET28a vector; therefore, it produced recombinant protein with a fused C-terminal His-tag.
2.6 Expression and Purification of Recombinant Protein
The pETMBRg clone (MB-CLsRG/ pET28a) was cultured on LB broth supplemented with isopropyl-β-D-thiogalactopyranoside (IPTG) for inducing recombinant protein expression, and 100 μg/mL kanamycin by following the protocol described by Boshtam et al. [22]. The purification of expressed recombinant protein (MBRg protein) was performed by adopting the protocols described by Zandvakili et al. [23] and Ryan [24]. The purified expressed protein was run on Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie Brilliant Blue R 250 (ROTH) protein stain. The DNA degradation assay was performed according to Xie et al. [25] to evaluate the functionally active purified protein. The DNase activity was visualized on a gel documentation system after running the treated samples on 1% agarose through gel electrophoresis.
The effect of purified recombinant protein against C. canescens was investigated using the radial diffusion method following Brogue et al. [26] with few modifications. C. canescens was cultured on sterilized petri plates containing potato dextrose agar (PDA) medium and let at 28°C ± 2°C until mycelial growth. Holes (5 mm) were made in the PDA medium containing petri plates with the help of a sterilized cork borer. Different concentrations (25, 35, 45, and 50 µg) of purified protein were mixed with conidial suspension (5 × 105 concentration/m) of C. canescens and added to holes of each plate, along with negative control (C. canescens+buffer), and incubated at 28°C ± 2°C. The experiment was repeated thrice.
3.1 Screening of the Mungbean Population
The screening of mungbean germplasm was done on both phenotypic and genotypic basis. The consensus sequence of 815 bp was generated through Lasergene v. 7.1.0 SeqMan pro (SeqManTM II) by aligning MB-ClsRCaG1 (MT886216; 708 bp) and MB-ClsRCaG2 (MT919519; 615 bp) sequences in Clustal W. The contig was named MB-CLsRG. Primer pair was developed from the sequence and evaluated its association with CLS resistance in mungbean. For this purpose, PCR analysis was carried out on the genomic DNA of the collected mungbean population. The amplification of DNA fragments in PCR analysis by this primer pair revealed its presence in CLS-resistant and tolerant mungbean germplasm.
The one-month-old mungbean population was inoculated with C. canescens conidial suspension. The symptoms started to appear after one week of post-inoculation. The information on some mungbean varieties regarding CLS resistance and susceptibility was found in the literature. In the greenhouse screening experiment, disease recording was done after one-week post-inoculation, two-week of post-inoculation, four-week of post-inoculation, and six-week of post-inoculation (Table S1; Fig. S1). Phenotypic screening data showed congruency with the literature.
PCR analysis was performed using a DNA template of each of the 36 mungbean varieties with a designed primer to screen the mungbean population at the genotypic level. The designed primer amplified the samples that had come out as CLS resistant or tolerant mungbean germplasm and displayed congruency to greenhouse screening and literature. The PCR results showed amplification in 24 mungbean genotypes (Fig. 1).
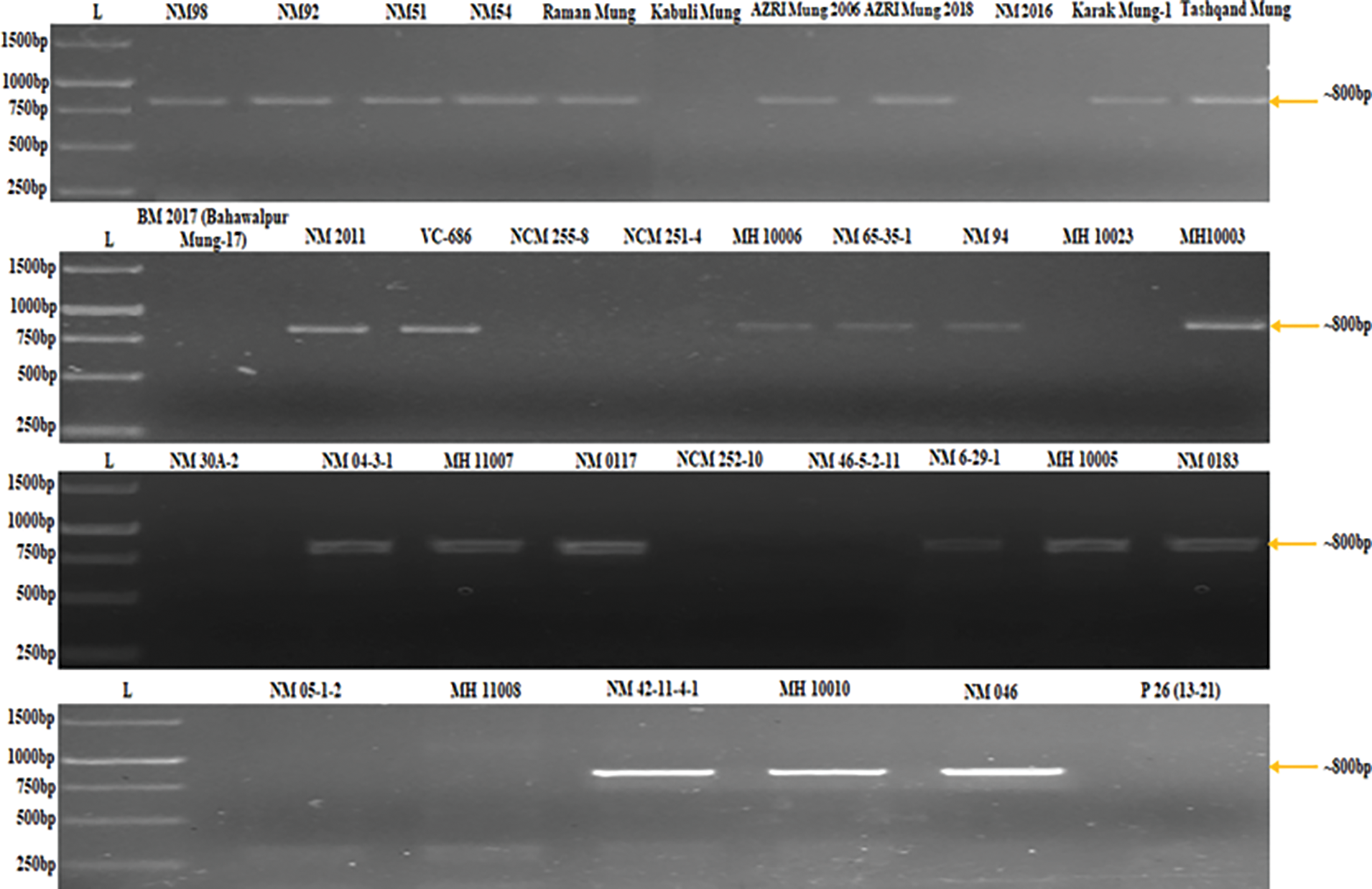
Figure 1: Genotypic screening of mungbean population using primer pair (MB-CLsRG-F1/ MB-CLsRG-R1) to determine the linkage of identified RGA (MB-CLsRG) to Cercospora leaf spot resistance in mungbean
3.2 Validation of Molecular Marker Linkage with CLS Resistance
The linkage of the molecular marker was validated by reverse transcription PCR (rtPCR) analysis. The rtPCR analysis amplified all genotypes using primer pair (above-mentioned). The sequence homology search through BLASTn and BLASTX displayed 100% homology with the sequence of MB-CLsRG and putative disease resistance proteins. The molecular marker linked to CLS resistance got the GenBank accession number OP369046.
3.3 Antifungal Activity of rMB-CLsRG
The recombinant protein (rMB-CLsRG) was expressed by delivering MB-CLsRG/ pET28a expression plasmid in BL21 E. coli cells. The pETMBRg clone (6,196 bp) was confirmed with double digestion through SacI and BamHI restriction enzymes. The length of the MB-CLsRG sequence cloned into the pET28a vector was 815 bp, and after digestion of the pETMBRg (MB-CLsRG/ pET28a) expression plasmid, ~821 bp, and 5,375 bp fragments (DNA bands) were resolved on agarose gel in the electrophoresis of a restriction-digestion reaction product. The ~15.4 kDa protein band was shown on SDS-PAGE. The DNase activity of purified recombinant protein against the mungbean genomic DNA revealed the complete degradation of mungbean DNA compared to the negative control (buffer without protein).
The antifungal activity of the purified protein was assessed through the modified radial diffusion method. The purified protein (~15.4 kDa) exhibited a strong inhibitory effect on the mycelial growth of C. canescens. In incubated plates, significant growth inhibition was observed for all purified protein concentrations (25, 35, 45, and 50 µg). No visible mycelial growth was seen after 24 h due to its slow growth rate [24]. On culture plates, the mycelial mass ring continued to increase proportionally with time in control well; however, a clear inhibitory effect of recombinant protein with concentrations of 25, 35, 45, and 50 µg was observed after 48, 72, 96, 120 and 144 h of C. canescens culturing (Fig. 2). The visual presentation of plates depicted that even the lowest concentration (25 µg) of purified protein possessed high antifungal potential and thereby restricted the growth of fungal mycelial mass.
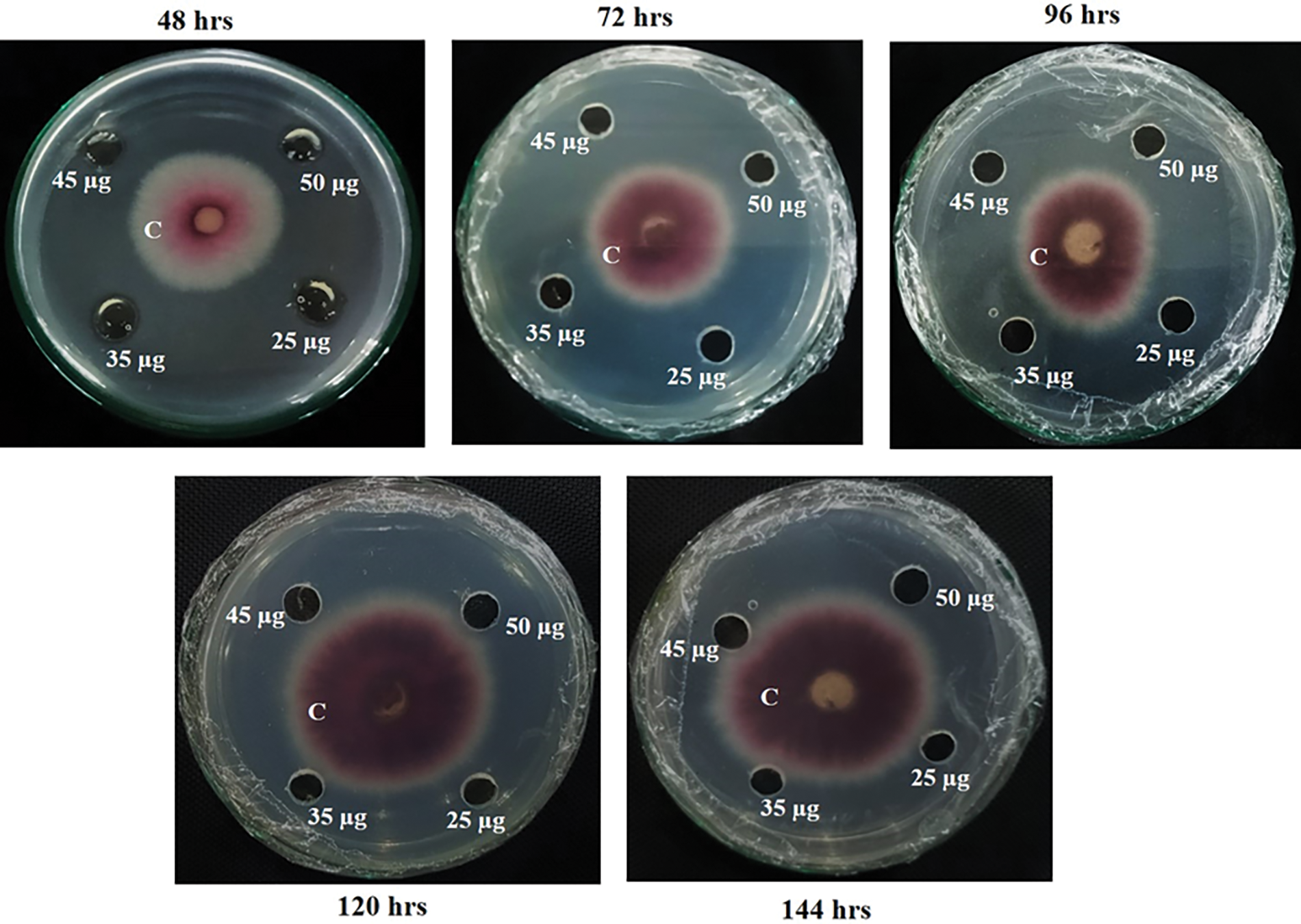
Figure 2: In vitro assessment of the antifungal potential of recombinant purified protein (rMB-CLsRG) at concentrations of 25, 35, 45, and 50 µg. The fungal growth inhibition data recorded after 48, 72, 96, 120, and 144 h of culturing showed the increasing mycelial mass in control well (conidial suspension of C. canescens with concentration 5 × 105 per mL+ protein-free buffer) However, no fungal growth was observed in treatment wells (recombinant protein +conidial suspension of C. canescens with concentration 5 × 105 per mL). * C = negative control
Phytopathogens are responsible for multiple plant diseases, affecting their productivity, physiology, and development. Nearly 5,000 diseases are discovered every year that cause a drastic impact on economically important crops [27]. Therefore, the chief goal of all crop improvement projects is mainly sustainable disease management. Three main approaches are considered for plant disease management, i.e., chemical control, disease prevention, and genetic resistance. Chemical control against pathogens is the most important and prevailing control. However, its misuse has triggered an alarming situation in the form of environmental contamination. A resistant source is the most reliable, safest, and practical approach for sustainable disease management. Using resistant varieties can cut down expenses of chemicals (insecticides, pesticides, fungicides) for many farmers who rely largely on chemicals for crop protection. In crop plants, resistance is a key step in all disease protection programs [28].
The novelty of DNA molecular markers has opened new avenues for scientists in evolutionary studies [29], plant systematics studies, and tagging and tracking genes encode for different traits, i.e., agronomic and stress-related traits [30].
Molecular marker technology has greatly contributed to agronomically important traits like disease resistance in mungbean due to its high selection efficiency [31]. MAS seemed to be not efficient for different agronomically crucial genes as markers are indirectly linked with target genes that might get separated during recombination. However, recent studies showed that it is possible to develop disease-linked markers using resistance (R) genes. Therefore, such molecular markers are developed using R genes or sequences containing conserved domains that can confer resistance in plants against pathogens [32].
Molecular marker technology assists in screening the resistance genotypes in a shorter period, even without the disease [33]. Crop improvement programs for mungbean through MAS require molecular markers, and tightly linked markers are suitable enough to make the selection procedure quite easy and more reliable [34]. Lately, MAS has seemed to be a powerful tool for CLS resistance in legumes [11]. In mungbean, the molecular marker identification linked with major genes mainly targeted fungal diseases, i.e., CLS disease and powdery mildew disease [34]. Therefore, this study aimed to develop a molecular marker linked to the CLS disease of the mungbean. RGA markers designed from the conserved motifs and domains of various known resistance genes [14–35] are also used in MAS. Basak et al. [36] used 24 R genes-based RGA markers to screen V. mungo germplasm, out of which one marker distinguished the resistant genotype from other susceptible genotypes. In our study, a molecular marker was developed from the consensus sequence of resistance gene analogs identified by Babar et al. [19]. The consensus DNA sequence was evaluated for its linkage to CLS resistance in mungbean through PCR analysis of the mungbean population at the genomic level. The marker exhibited amplification in those varieties that showed resistance against CLS disease during phenotypic screening, thus confirming its linkage with the CLS disease in mungbean. Papan et al. [37] experimented with validating a molecular marker linked to CLS disease in mungbean. They reported three new simple sequence repeat (SSR) markers and a InDel marker, along with six previously reported markers (linked to qCLSC72V18-1 QTL, controlling CLS resistance). They carried out the experimentation to further identify markers located nearer to qCLSC72V18-1. The previously reported marker (I16274) and VrTAF5_indel markers, flanking QTL of interest 41.56%–60.38% of the phenotypic variation, with genetic distances of 4.0 and 5.0 cM from the resistance gene, respectively. Similarly, Yundaeng et al. [12] conducted experiments in which they investigated the molecular basis of qCLS for CLS resistance in mungbean through fine mapping. They developed the InDel marker (VrTAF5_indel) and SSR marker (Vr6gCLS133) from an intergenic region of VrTAF5 candidate gene controlling the CLS resistance and shown to be co-segregated with the resistance.
Maiti et al. [32] identified a molecular marker linked to MYMV resistance genes through ISSR methodology, and its validation was performed on black gram germplasm. A study by Naik et al. [38] validated two markers (CEDG097 and CEDG172) that flanked qYMV1 QTL in 25 resistant and susceptible genotypes of black gram. The phenotypic screening revealed that, out of 25 genotypes, 15 genotypes showed resistance against YMV, while the remaining ten genotypes showed susceptibility to YMV disease. In genotypic screening, CEDG097 amplified a resistant allele in 10 black gram genotypes and a susceptible allele in one genotype, thus showing 91% linkage to known resistant and susceptible genotypes. The marker CEDG172 exhibited 74% linkage in known varieties against YMV disease.
Similarly, Binyamin et al. [39] validated two SCAR markers, one of which was reported to be linked with the MYMV resistance gene in V. radiata, while the other was reported to be linked with the MYMV disease in V. mungo. Both markers showed amplification only in resistant and moderately resistant genotypes, and no amplification was observed in susceptible genotypes, confirming their linkage with the MYMV disease resistance gene. In another study, SCAR markers, developed from RAPD markers, were employed to tag the MYMV resistance gene in mungbean [40]. Similarly, in a study by Sai et al. [41], two SCAR markers (CM9 and CM815) were validated to assure their linkage with MYMV disease in V. radiata. Two RGA markers (RGASF1 and RGASF2) were linked with MYMV resistance genes in black gram and mungbean [32].
In addition to the marker development, our study focused on recombinant protein expression of MB-CLsRG, its purification, and investigating its antifungal activity against C. canescens, the causal agent of CLS disease. The pET expression system was employed for cloning and expressing the recombinant protein in E. coli. It is the most prevailing system used due to high polymerase activity, high rate of translation mediated by the T7 gene, and ten translation initiation signals [42]. Moreover, we used IPTG as an inducing agent to provide better induction [43]. The expressed protein was purified according to the method described by Zanvakili et al. [23]. The recombinant proteins expressed in the bacterial system ultimately form inclusion bodies containing a majority of the expressed protein [44] due to the absence of post-translational modification in the bacterial system [45]. The protein present in inclusion bodies cannot be used directly due to the absence of biological activity [46]. The activity of purified MBRg protein (~15.4 kDa) was checked by mungbean genomic DNA degradation, which revealed that our purified protein was functionally active, as reported by Zanvakili et al. [23] and Xie et al. [25].
Plant AFPs are powerful tools for crop protection and sustainable disease management. The recombinant protein was further investigated for its antifungal activity against C. canescens. The in vitro comparison of mycelial growth in control (C. canescens + protein-free buffer) and various concentrations of recombinant protein clearly showed inhibition of fungal mycelial growth, thus indicating that our expressed protein possesses antifungal potential (Fig. 2). A 15 kDa protein from Lippia sidoides (rosemary pepper) flowers, with sequence homology of NBS-LRR, showed antifungal potential against Botrytis cinerea and inhibited its growth [47]. The mechanism of action of protein to inhibit mycelial growth includes damage to cellular ribosomes, inhibition of DNA synthesis and cell cycle, and fungal cell wall degradation. Enzymes like catalase and superoxide dismutase were involved in resistance in mungbean against C. canescens. These enzymes also play a role in detoxifying cercosporin, which might help mungbean’s resistance mechanism against pathogen attack [48]. The genes that encode proteins possessing antifungal potential are widely used in agribusiness to enhance the production of plants that can endure fungal attacks in fields and cut down economic losses [49].
Authorship:: The authors confirm their contribution to the paper as follows: Study conception and design: Siddra Ijaz and Imran Ul Haq; Data collection: Maria Babar, Siddra Ijaz and Imran Ul Haq; Analysis and interpretation of results: Siddra Ijaz, Imran Ul Haq and Maria Babar; Draft manuscript preparation: Maria Babar, Siddra Ijaz, Imran Ul Haq and Muhammad Sarwar Khan.
Acknowledgement: We highly acknowledge Higher Education Commission (HEC) for funding to conduct this study under the Project “Transcriptomics Based Understanding of Cercospora Leaf Spot Resistance in Mungbean and Disease Management through Nanotechnology,” Project No. 7425; Special thanks to Center for Advances Studies (CAS), University of Agriculture Faisalabad, Pakistan for providing the facility of SDS-PAGE.
Funding Statement: The Higher Education Commission (HEC) funded this work under “Transcriptomics Based Understanding of Cercospora Leaf Spot Resistance in Mungbean and Disease Management through Nanotechnology,” Project No. 7425.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Oerke, E. C. (2006). Crop losses to pests. Journal of Agricultural Science, 144(1), 31–43. DOI 10.1017/S0021859605005708. [Google Scholar] [CrossRef]
2. Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N. et al. (2019). The global burden of pathogens and pests on major food crops. Nature Ecology and Evolution, 3(3), 430–439. DOI 10.1038/s41559-018-0793-y. [Google Scholar] [CrossRef]
3. Wang, Y., Kausch, A. P., Chandlee, J. M., Luo, H., Ruemmele, B. A. et al. (2003). Co-transfer and expression of chitinase, glucanase, and bar genes in creeping bentgrass for conferring fungal disease resistance. Trends in Plant Science, 165(3), 497–506. DOI 10.1016/S0168-9452(03)00198-5. [Google Scholar] [CrossRef]
4. Zvereva, A. S., Pooggin, M. M. (2012). Silencing and innate immunity in plant defense against viral and non-viral pathogens. Virus, 4(11), 2578–2597. DOI 10.3390/v4112578. [Google Scholar] [CrossRef]
5. Agrios, G. N. (2005). Plant pathology. Fifth edition. Amsterdam, Netherlands: Elsevier Academic Press. [Google Scholar]
6. Burdon, J. J., Zhan, J. (2020). Climate change and disease in plant communities. PLoS One, 18(11), 3000949. DOI 10.1371/journal.pbio.3000949. [Google Scholar] [CrossRef]
7. Pandey, A. K., Burlakoti, R. R., Kenyon, L., Nair, R. M. (2018). Perspectives and challenges for sustainable management of fungal diseases of mung bean [Vigna radiata (L.) R. Wilczek var. radiata]: A review. Frontiers in Environmental Science, 6(3), 53. DOI 10.3389/fenvs.2018.00053. [Google Scholar] [CrossRef]
8. Marcos, J. F., Gandía, M., Garrigues, S., Manzanares, P., Coca, M. (2019). Antifungal peptides and proteins with activity against fungi causing postharvest decay. In: Postharvest pathology of fresh horticultural produce, pp. 757–792. USA: CRC Press. [Google Scholar]
9. Zaker, M. (2016). Natural plant products as eco-friendly fungicides for plant diseases control–A review. The Agriculturist, 14(1), 134–141. DOI 10.3329/agric.v14i1.29111. [Google Scholar] [CrossRef]
10. Chiu, T., Poucet, T., Li, Y. (2022). The potential of plant proteins as antifungal agents for agricultural applications. Synthetic and Systems Biotechnology, 7(4), 1075–1083. DOI 10.1016/j.synbio.2022.06.009. [Google Scholar] [CrossRef]
11. Kasettranan, W., Somta, P., Srinives, P. (2010). Mapping of quantitative trait loci controlling powdery mildew resistance in mungbean (Vigna radiata (L.) Wilczek). Journal of Crop Science and Biotechnology, 13(3), 155–161. DOI 10.1007/s12892-010-0052-z. [Google Scholar] [CrossRef]
12. Yundaeng, C., Somta, P., Chen, J., Yuan, X., Chankaew, S. et al. (2021). Fine mapping of QTL conferring Cercospora leaf spot disease resistance in mungbean revealed TAF5 as candidate gene for the resistance. Theoretical and Applied Genetics, 134(2), 701–714. DOI 10.1007/s00122-020-03724-8. [Google Scholar] [CrossRef]
13. Dharajiya, D. T., Ravindrababu, Y. (2019). Identification of molecular marker associated with mungbean yellow mosaic virus resistance in mungbean [Vigna radiata (L.) Wilczek]. Vegetos, 32(4), 532–553. DOI 10.1007/s42535-019-00063-. [Google Scholar] [CrossRef]
14. Chen, X. M., Line, R. F., Leung, H. (1998). Genome scanning for resistance-gene analogs in rice, barley, and wheat by high-resolution electrophoresis. Theoretical and Applied Genetics, 97(3), 345–355. DOI 10.1007/s001220050905. [Google Scholar] [CrossRef]
15. Kaewwongwal, A., Liu, C., Somta, P., Chen, J., Tian, J. et al. (2020). A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41. Molecular Genetics and Genomics, 295(2), 275–286. DOI 10.1007/s00438-019-01619-y. [Google Scholar] [CrossRef]
16. Abbas, H., Iqbal, M. A., Kamran, M., Shahbaz, M. U., Kamber, H. U. et al. (2020). Evaluation of advanced mung bean germplasm against Cercospora leaf spot and its in-vitro management by different fungicides. Pakistan Journal of Agricultural Research, 33(4), 692–956. DOI 10.17582/journal.pjar/2020/33.4.872.877. [Google Scholar] [CrossRef]
17. Iqbal, S. M., Ghafoor, A., Bashir, M., Malik, B. A. (1995). Estimation of losses in yield components of mungbean due to Cercospora leaf spot. Pakistan Journal of Phytopathology, 7(1), 80–81. [Google Scholar]
18. Grewal, J. S. (1978). Diseases of mungbean in India. The 1st International Mungbean Symposium, Asian Vegetable Research and Development Center (AVRDC) Shanhua, Tainan, Taiwan. [Google Scholar]
19. Babar, M., Ijaz, S., Khan, M. S., Ul Haq, I. (2021). Computational genomics based probing of resistance gene analogs (RGAs) in mungbean under Cercospora leaf spot disease challenge. Pakistan Journal of Agricultural Sciences, 58(5), 1523–1536. DOI 10.21162/PAKJAS/21.439. [Google Scholar] [CrossRef]
20. Babar, M., Ijaz, S., Ul Haq, I., Khan, I. A. (2022). Morpho-functional characterization, in vitro study, and tripartite interaction assay evaluate the potential of biosynthesized silver nanoparticles to manage Cercospora leaf spot disease in Vigna radiata. Journal of Plant Pathology, 104, 1371–1381. DOI 10.1007/s42161-022-01168-1. [Google Scholar] [CrossRef]
21. Walia, R., Deb, J. K., Mukherjee, K. J. (2007). Development of expression vectors for Escherichia coli based on the pCR2 replicon. Microbial Cell Factories, 6(2), 1–9. DOI 10.1186/1475-2859-6-14. [Google Scholar] [CrossRef]
22. Boshtam, M., Shahreza, H. K., Feizollahzadeh, S., Rahimmanesh, I., Asgary, S. (2018). Expression and purification of biologically active recombinant rabbit monocyte chemoattractant protein1 in Escherichia coli. FEMS Microbiology Letters, 365(9), fny070. DOI 10.1093/femsle/fny070. [Google Scholar] [CrossRef]
23. Zandvakili, N., Zamani, M., Motallebi, M., Jahromi, Z. M. (2017). Cloning, overexpression and in vitro antifungal activity of Zea mays PR10 protein. Iranian Journal of Biotechnology, 15(1), 42–49. DOI 10.15171/ijb.1357. [Google Scholar] [CrossRef]
24. Ryan, B. J. (2011). Differential precipitation and solubilization of proteins. In: Protein chromatography, pp. 203–213. Totowa, New Jersey, USA: Humana Press. [Google Scholar]
25. Xie, Y. R., Chen, Z. Y., Brown, R. L., Bhatnagar, D. (2010). Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. Journal of Plant Physiology, 167(2), 121–130. DOI 10.1016/j.jplph.2009.07.004. [Google Scholar] [CrossRef]
26. Brogue, K., Chet, I., Holliday, M., Cressman, R., Biddle, P. et al. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science, 254(5035), 1194–1197. DOI 10.1126/science.254.5035.1194. [Google Scholar] [CrossRef]
27. Lucas, P. M. (1992). Health and disease in the late prehistoric Southeast. In: Verano, J. W., Ubelaker, D. H. (Eds.Disease and demography in the Americas. Washington: Smithsonian Institute Press. [Google Scholar]
28. Khan, A. H., Hassan, M., Khan, M. N. (2020). Conventional plant breeding program for disease resistance. In: Plant disease management strategies for sustainable agriculture through traditional and modern approaches, pp. 27–51. Cham, Switzerland: Springer. [Google Scholar]
29. Zargar, S. A., Fayaz, H., Wani, A. A., Saggoo, M. I. S., Mir, R. A. et al. (2021). Genetic and genomic resources in rice bean (Vigna umbellata Thunb.Availability, advancements, and applications. In: Neglected and underutilized crops-towards nutritional security and sustainability, pp. 191–202. Singapore: Springer. [Google Scholar]
30. Negi, M. S., Devic, M., Delseny, M., Lakshmikumaran, M. (2000). Identifcation of AFLP fragments linked to seed coat colour in Brassica juncea and conversion to a SCAR marker for rapid selection. Theoretical and Applied Genetics, 101(2), 146–152. [Google Scholar]
31. Sahoo, J. P., Samal, K. C., Tripathy, S. K., Lenka, D., Mishra, P. et al. (2022). Understanding the genetics of Cercospora leaf spot (CLS) resistance in mung bean (Vigna radiata L. Wilczek). Tropical Plant, 47(6), 1–5. [Google Scholar]
32. Maiti, S., Basak, J., Kundagrami, S., Kundu, A., Pal, A. (2011). Molecular marker-assisted genotyping of mungbean yellow mosaic India virus resistant germplasms of mungbean and urdbean. Molecular Biotechnology, 47(2), 95–104. DOI 10.1007/s12033-010-9314-1. [Google Scholar] [CrossRef]
33. Souframanien, J., Gopalakrishna, T. (2006). ISSR and SCAR markers linked to the mungbean yellow mosaic virus (MYMV) resistance gene in blackgram [Vigna mungo (L.) Hepper]. Plant Breeding, 125(6), 619–622. DOI 10.1111/j.1439-0523.2006.01260.x. [Google Scholar] [CrossRef]
34. Kasettranan, W., Somta, P., Srinives, P. (2010). Mapping of quantitative trait loci controlling powdery mildew resistance in mungbean (Vigna radiata (L.) Wilczek). Journal of Crop Science and Biotechnology, 13(3), 155–161. DOI 10.1007/s12892-010-0052-z. [Google Scholar] [CrossRef]
35. Yan, G., Chen, X., Line, R., Wellings, C. (2003). Resistance gene-analog polymorphism markers co-segregating with the Yr5 gene for resistance to wheat stripe rust. Theoretical and Applied Genetics, 106(4), 636–643. DOI 10.1007/s00122-002-1109-8. [Google Scholar] [CrossRef]
36. Basak, J., Kundagrami, S., Ghose, T. K., Pal, A. (2005). Development of yellow mosaic virus (YMV) resistance linked DNA marker in Vigna mungo from populations segregating for YMV-reaction. Molecular Breeding, 14(4), 375–383. DOI 10.1007/s11032-005-0238-6. [Google Scholar] [CrossRef]
37. Papan, P., Chueakhunthod, W., Poolsawat, O., Arsakit, K., Tharapreuksapong, A. et al. (2021). Validation of molecular markers linked to Cercospora leaf spot disease resistance in mungbean (Vigna radiata [L.] WILCZEK). SABRAO Journal of Breeding & Genetics, 53(4), 749–757. DOI 10.54910/sabrao2021.53.4.16. [Google Scholar] [CrossRef]
38. Naik, D. B. V., Bhaskara, R., Srividhya, A., Jayalakshmi, D., Reddypogu, P. et al. (2022). Quantitative trait loci (QTL) detection and validation of linked markers associated with yellow mosaic virus resistance in Urdbean [Vigna mungo (L.) Hepper] in Andhra Pradesh. Indian Journal of Agricultural Research, 10(2). DOI 10.18805/IJARe.A-5895. [Google Scholar] [CrossRef]
39. Binyamin, R., Khan, M. A., Khan, N. A., Khan, A. I. (2015). Application of SCAR markers linked with mungbean yellow mosaic virus disease-resistance gene in Pakistan mungbean germplasm. The Genetics and Molecular Research Journal, 14(1), 2825–2830. DOI 10.4238. [Google Scholar]
40. Dhole, V. J., Reddy, K. S. (2013). Development of a SCAR marker linked with a MYMV resistance gene in mungbean (Vigna radiata L. Plant Breeding, 132(1), 127–132. DOI 10.1111/pbr.12006. [Google Scholar] [CrossRef]
41. Sai, C. B., Nagarajan, P., Raveendran, M., Rabindran, R., KannanBapu, J. R. et al. (2017). Understanding the inheritance of mungbean yellow mosaic virus (MYMV) resistance in mungbean (Vigna radiata L. Wilczek). Molecular Breeding, 37(5), 1–5. DOI 10.1007/s11032-017-0650-8. [Google Scholar] [CrossRef]
42. Liu, H., Naismith, J. H. (2009). A simple and efficient expression and purification system using two newly constructed vectors. Protein Expression and Purification, 63(2), 102–111. DOI 10.1016/j.pep.2008.09.008. [Google Scholar] [CrossRef]
43. Ramos, C. R. R., Abreu, P. A. E., Nascimento, A. L. T. O., Ho, P. L. (2004). A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Brazilian Journal of Medical and Biological Research, 37(8), 1103–1109. DOI 10.1590/s0100-879x2004000800001. [Google Scholar] [CrossRef]
44. Kirubakaran, S. I., Sakthivel, N. (2007). Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Expression and Purification, 52(1), 159–166. DOI 10.1016/j.pep.2006.08.012. [Google Scholar] [CrossRef]
45. Baneyx, F., Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnology, 22(11), 1399–1408. DOI 10.1038/nbt1029. [Google Scholar] [CrossRef]
46. Saeidi, M., Zareie, R. (2020). Prediction, isolation, overexpression and antifungal activity analysis of Medicago truncatula var. truncatula putative thaumatin like proteins (TLP-1, -2, -3, -4 and -5). Turkish Journal of Biology, 44(4), 176–187. DOI 10.3906/by-1912-18. [Google Scholar] [CrossRef]
47. Moreira, J. S., Almeida, R. G., Tavares, L. S., Santos, M. O., Viccini, L. F. et al. (2011). Identification of botryticidal proteins with similarity to NBS-LRR proteins in rosemary pepper (Lippia sidoides Cham) flowers. Protein Journal, 30(1), 32–38. DOI 10.1007/s10930-010-9299-4. [Google Scholar] [CrossRef]
48. Maurya, A. K., Navathe, S., Mohapatra, C., Chand, R. (2018). Antioxidants elevates the resistance to Cercospora canescens in interspecific cross of Vigna radiata (Kopergaon) Vigna mungo (Pant Urd 31). Indian Phytopathology, 71(4), 519–528. DOI 10.1007/s42360-018-0101-4. [Google Scholar] [CrossRef]
49. Selitrennikoff, C. P. (2001). Antifungal proteins. Applied and Environmental Microbiology, 67(7), 2883–2894. DOI 10.1128/AEM.67.7.2883-2894.2001. [Google Scholar] [CrossRef]
Appendix
Figure S1: The phenotypic screening of mungbean population against CLS disease under greenhouse condition
Table S1: The phenotypic screening data of mungbean population
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools