 Open Access
Open Access
ARTICLE
Light Promotes Protein Stability of Auxin Response Factor 7
Laboratory of Plant Molecular Genetics & Crop Gene Editing, School of Life Sciences, Linyi University, Linyi, 276000, China
* Corresponding Author: Shucai Wang. Email:
Phyton-International Journal of Experimental Botany 2023, 92(4), 1153-1160. https://doi.org/10.32604/phyton.2023.026355
Received 25 August 2022; Accepted 24 October 2022; Issue published 06 January 2023
Abstract
Light is an environmental signaling, whereas Aux/IAA proteins and Auxin Response Factors (ARFs) are regulators of auxin signalling. Aux/IAA proteins are unstable, and their degradation dependents on 26S ubiquitin-proteasome and is promoted by Auxin. Auxin binds directly to a SCF-type ubiquitin-protein ligase, TIR1, facilitates the interaction between Aux/IAA proteins and TIR1, and then the degradation of Aux/IAA proteins. A few studies have reported that some ARFs are also unstable proteins, and their degradation is also mediated by 26S proteasome. In this study, by using of antibodies recognizing endogenous ARF7 proteins, we found that protein stability of ARF7 was affected by light. By expressing MYC tagged ARF activators in protoplasts, we found that degradation of ARF7 was inhibited by 26 proteasome inhibitors. In addition, at least ARF5 and ARF19 were also unstable proteins, and degradation of ARF5 via 26S proteasome was further confirmed by using stable transformed plants overexpressing ARF5 with a GUS tag.Keywords
Aux/IAA proteins and Auxin Response Factors (ARFs), two different families of transcription factors, regulate auxin signal transduction in plants [1–3]. ARF activators are able to activate the expression of auxin responsive genes including Aux/IAA genes via binding to the TGTCTC auxin response elements (AuxREs) in their promoters. In turn, Aux/IAA proteins are able to dimerize with ARF activators via their conserved C-terminal dimerization domain (CTD) to repress the functions of ARF activators [1,2,4,5]. Yet the interaction of ARF activators and Aux/IAA proteins may not be strictly required for auxin response gene expression [6].
Aux/IAA proteins are well-known short-living proteins and their degradation is regulated by auxin. Binding of auxin to the receptor Transport Inhibitor Response 1 (TIR1), who is also serving as part of the SCF ubiquitin ligase complex, promotes their binding to the degron of Aux/IAA proteins. Binding of TIR1 facilitates the entry of Aux/IAA proteins into the ubiquitin-proteasome pathway for degradation, therefore releases the repression of ARF activators [7–10].
On the other hand, ARF1 and ARF2, two ARF repressors have also been reported to be unstable proteins whose degradation is mediated by 26S proteasome [11,12]. Whereas the stability of the ARF activators ARF5, ARF6 and ARF19 and the ARF repressor ARF10 was affected by several factors including low and high temperature, salt treatment as well as plant hormone ABA but not auxin [13]. Very recently, it has been shown that E3 ubiquitin ligase AUXIN RESPONSE FACTOR F-BOX1 (AFF1) is able to mediate the degradation of both ARF7 and ARF19 via 26S proteasome pathway [14]. Eventhough ARF7 functions as key regulator of auxin signaling, the arf7 mutant was initially identified as a mutant with phototropic response defect, therefore was named nonphototropic hypocotyl 4 (nph4) [15,16], suggesting that ARF7 may mediate cross-talk between auxin and light signals.
The objectives of this study were to examine protein stability of ARF activators and if light may affect protein stability of ARF activators especially ARF7. We found independently that the ARF activators ARF5, ARF7 and ARF19 are unstable proteins and their degradation is 26S proteasome dependent. Most importantly, we found that light promotes protein stability of ARF7.
2.1 Plant Materials and Growth Conditions
The Columbia-0 (Col) ecotype Arabidopsis was used as a wild type. The nph4-1, arf19-3, arf19-4, nph4-1 arf19-3 and nph4-1 arf19-4 mutants in the Col background have been described previously [17,18]. For plant transformation and protoplasts isolation, seeds of the Col wild type were germinated and grown in 3 × 3 inch pots containing moistened Pro-Mix (Premier Horticulture Inc., Red Hill, PA 18076). The plants were grown at 20°C under continuous light as described previously [17]. Leaves from plants that were 3–5-week-old were used for protoplasts isolation. Plants ~6-week-old plants with several mature flowers in the main inflorescence were used for plant transformation.
For endogenous ARF7 proteins detection, seeds of the Col wild type and the mutants were surface sterilized and grown on ½ Murashige & Skoog (MS) media [19] containing 1× Gamborg vitamins (Sigma, St. Louis, MO, USA), 1% sucrose, 0.7% agar (Type A, Sigma, St. Louis, MO, USA) in sterile Petri dishes at 20°C under continuous light or dark for 3–5 days as indicated in the figures.
Rabbit polyclonal anti-ARF7 antibodies were generated by using the middle region (MR) of ARF7 proteins as described previously [17]. Anti-MYC monoclonal antibodies were obtained from Roche Diagnostics (Mannhein, Germany).
MYC epitope tagged ARFs full length constructs used have been described previously [17]. The GUS-ARF5 construct used for plant transformation was generated by fusing ARF5 in frame with a N-terminal GUS and cloned into pUC19 vector under the control of the 35S promoter [4,17]. The pUC19-GUS-ARF5 construct was then digested with EcoR1 to obtain the whole cassette including 35S promoter, GUS-ARF insert and nos, and cloned into the binary vector pPZP211 [20].
2.4 Protoplasts Isolation and Transfection
Protoplasts isolation and transfection assays were performed as described previously [4,17]. Briefly, protoplasts were isolated from leaves of 3–5-week-old Col wild type plants. Plasmids of the MYC-ARFs were transfected into the protoplasts, and incubated in darkness for 20–22 h, or incubated for 16 h, then treated with MG132 + CHX for 6 h as described in 2.6. The experiments were repeated at least two times with similar results.
2.5 Plant Transformation and Selection of Transgenic Plants
About 6-week-old plants were transformed with the GUS-ARF5 construct in Agrobacterium tumefaciens GV3101 by using the floral dip method [21]. To select transgenic plants, T1 seeds were surface sterilized and grown on ½ MS media containing 1× Gamborg vitamins, 1% sucrose, 0.7% agar, 50 μg/mL kanamycin (Amersham Life Science, Piscataway, NJ, USA) and 200 μg/mL timentin (Smithkline Beecham Pharmaceuticals, Philadelphia, PA, USA) in sterile Petri dishes at 20°C under continuous light for 7–10 days.
Transgenic plants were then transferred to pots containing moistened Pro-Mix (Premier Horticulture Inc., Red Hill, PA, USA) and grown at 20°C under long-day conditions (16 h light/8 h dark). Homozygous lines were identified in T3 generations and used for proteasome inhibitors treatment and GUS staining.
2.6 Proteasome Inhibitors Treatment and GUS Staining
To examine protein stability in transfected protoplasts, the transfected protoplasts were incubated in darkness for 16 h, then mock treated or treated with 50 µM MG132 at the present of 100 µM CHX for 6 h by adding corresponding chemicals to the WI solution. Then protoplasts were collected and lysesed with SDS buffer and boiled for 3 min as described previously [17].
To examine protein stability in seedlings, 3- or 5-day-old light- or dark-grown seedlings were collected and treated 50 µM MG132 for 4 h, and then seedlings were frozen in liquid nitrogen and ground in SDS buffer and boiled for 3 min [17]. Or, MG132 was removed after 4 h and then 200 µg CHX was added and samples were collected at indicated time and ground in SDS buffer for western blot.
5-day-old light-grown seedlings of transgenic plant overexpressing GUS-ARF5 were used for histochemical staining. The seedlings were first mock treated or treated with 50 µM MG132 or 10 µM β-lactone for 4 h, then GUS activity was monitored using X-gluc as described previously [22].
Western blot was performed as described previously [17]. Briefly, proteins were separated by SDS gel electrophoresis, transferred to an immobilon-P membrane (Millipore Intertech, Bedford, MA, USA) using a semi-dry transfer cell (BioRad, Richmond, CA, USA). After blocked with 5% nonfat dry milk in phosphate buffered saline plus 0.1% Tween 20 (PBST), the membrane was probed with rabbit polyclonal anti-ARF7 or mouse monoclonal anti-MYC antibodies in PBST. The primary antibodies were then detected by using a horseradish peroxidase-labeled donkey anti-rabbit (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) or sheep anti-mouse second antibody (Jackson ImmunoResearch Laboratories) and western lightningTM Chemiluminescence reagent (PerkinElmer Life Sciences, Inc., Boston, MA, USA) as described by the manufacturer.
3.1 Light Grown Arabidopsis Seedlings Accumulated More ARF7 Proteins
Previously we have generated anti-ARF7 antibodies by using the MR of ARF7 proteins, a non-conserved domain between the conserved DNA binging domain (DBD) and CTD of ARF7 proteins [17]. To test the specificity of those antibodies, we expressed the MYC tagged ARF activator constructs in Arabidopsis protoplasts and then tested the proteins with anti-ARF7 and anti-MYC antibodies respectively. As shown in Fig. 1, proteins of all the five MYC tagged ARF activators were detectable by anti-MYC antibodies, similar as reported previously [17]. On the other hand, the anti-ARF7 antibodies were only able to detect the MYC tagged ARF7 proteins. These results suggested that anti-ARF7 antibodies were able to detect ARF7 proteins specifically, indicating that specific antibodies can be generated by using non-conserved domains of ARF proteins.
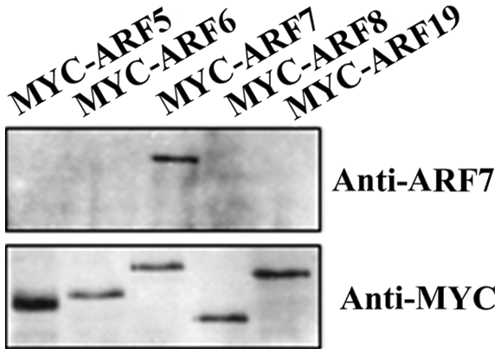
Figure 1: Specificity of anti-ARF7 antibodies. The anti-ARF7 antibodies were raised in rabbit using the MR of the ARF7 proteins peptides expressed in Escherichia coil. Plasmids of the 35S:MYC-ARF effector genes were transfected into Arabidopsis mesophyll protoplasts isolated from leaves of the Col wild type plants. The transfected protoplasts were incubated in darkness for 20–22 h, and then whole cell extracts were tested with anti-ARF7 and anti-MYC antibodies, respectively
We then examined endogenous ARF7 proteins using the anti-ARF7 antibodies. As shown in Fig. 2, ARF7 proteins were not detectable in nph4/arf7 mutant seedlings, consistent with our previously results [17], but were detectable in the Col wild type and the arf19 mutant seedlings. ARF7 proteins were also not detectable in the nph4 arf19 double mutant seedlings (Fig. 2). Consistent with the nature that nph4 and arf19 are loss-of-function mutants caused by T-DNA insertions for ARF7 and ARF19, respectively [17,18].
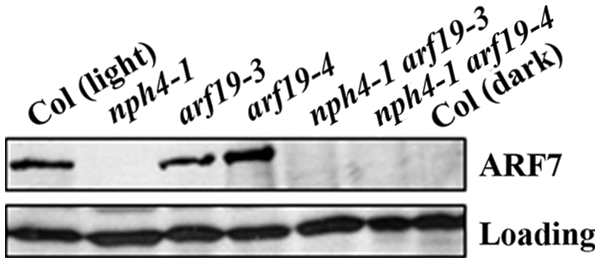
Figure 2: Detection of endogenous ARF7 proeins in light-grown seedlings of the Col wild type and arf mutants and dark grown seedlings of the Col wild type. Whole cell extracts from five-day-old light-or dark-grown Col wild type and light-grown mutant seedlings were tested by western blotting with anti-ARF7 antibodies. Loading control was a nonspecific band detected in the seedlings by the antibodies
ARF7 has been shown to be required for phototropic response [15,16], and previously research has shown that accumulation of ARF2, an ARF repressor, is affected by ethylene. Whereas light is able to affect the stability of ethylene-regulated HOOKLESS1 (HLS1), thereby affecting accumulation of ARF2 [11]. We therefore examined if light may affect the accumulation of ARF7 proteins by comparing endogenous protein levels of ARF7 in light- and dark-grown seedlings. We found that ARF7 proteins were accumulated in the light-grown Col wild type seedlings, whereas that in the dark-grown seedlings was almost undetectable (Fig. 2).
3.2 Decreased Level of ARF7 Proteins in Dark-Grown Seedling Was Due to Protein Degradation
The observation that light-grown seedlings accumulated more ARF7 proteins indicates that either the transcription level of ARF7 was reduced in dark-grown seedlings, or ARF7 proteins were unstable in dark-grown seedlings.
We therefore compared the endogenous protein level of ARF7 in dark-grown seedlings that were mock treated and treated with a 26S proteasome inhibitor MG132. As shown in Fig. 3, it is clear that MG132 treatment dramatically increased the endogenous protein level of ARF7. On the other hand, once MG132 was removed and CHX, a protein synthesis inhibitor was added, a decreased level of ARF7 proteins was observed (Fig. 3). These results suggest that ARF7 proteins are unstable proteins, and their degradation is mediated by 26S proteasome, similar to the results reported recently [14]. Most importantly, our results indicate that light is able to stabilize the ARF7 proteins, whereas dark promoted their degradation. These results provided new lights into the cross-talk between auxin and light signals.
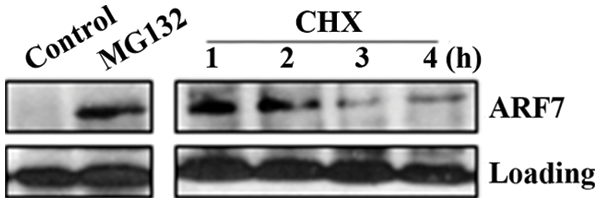
Figure 3: Accumulation of ARF7 proteins in response to 26 proteasome inhibitors. Three-day-old dark-grown seedlings were mock treated or treated with 50 μM MG132 for 4 h, then MG132 was removed and seedlings were treated with 200 μg/mL CHX for indicated time. Whole cell extracts were prepared and tested with anti-ARF7 antibodies. Loading control is a nonspecific band detected in the seedlings by the antibodies
3.3 Some of the Other ARF Activator Proteins Accumulated in Response to 26 Proteasome Inhibitors
Having shown that ARF7 is an unstable protein and darkness promoted its degradation, we further examined if other ARF activator proteins may also be unstable proteins by using transfected protoplasts incubated in darkness. Plasmids of MYC tagged ARF activator constructs were transfected into Arabidopsis protoplasts, and incubated in darkness for 16 h to allow gene expression and protein accumulation. Then the protoplasts were mock treated or treated with MG132 in the present of CHX for 6 h before proteins were detected by using anti-MYC antibodies.
As shown in Fig. 4, proteins for all the 5 ARF activators were detectable in the mock treated protoplasts, and a clearly increase in protein levels of ARF5, ARF7 and ARF19 was observed in the MG132 and CHX treated protoplasts. It should be note that in the recent publication, ARF19 proteins have also been reported to be unstable proteins and their degradation is mediated by 26S proteasome [14].
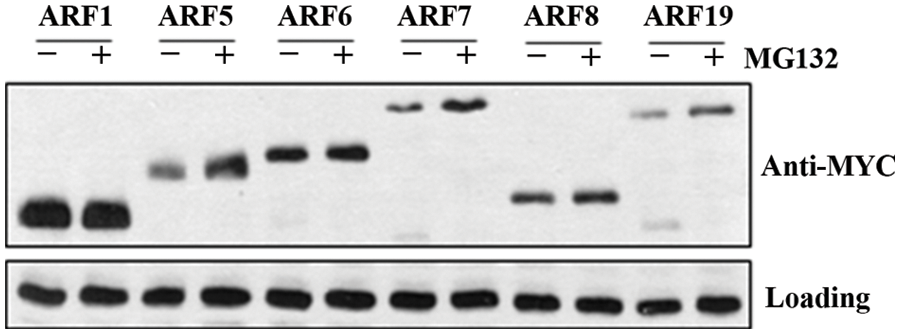
Figure 4: Accumulation of ARF activator proteins in response to a 26 proteasome inhibitor. 35S:MYC-ARF effector genes encoding full-length ARF activators were transfected into Arabidopsis mesophyll protoplasts isolated from leaves of the Col wild type plants. The transfected protoplasts were incubated in darkness for 16 h, then protoplasts were mock treated or treated with 50 µM MG132 for 6 h in the present of 100 μM CHX. Whole cell extracts were tested with anti-MYC antibodies. Loading control is a band detected in protoplasts with the antibodies
On the other hand, little if any increase in protein levels was observed for ARF6 and ARF8 in the MG132 and CHX treated protoplasts (Fig. 4). In addition, we also included ARF1, a repressor ARF in our experiments. High protein level of ARF1 was detected in transfected protoplasts, but no difference was observed in mock treated and MG132 and CHX treated protoplasts. Even though ARF1 has been reported to be an unstable protein and its degradation is mediated by 26S proteasome [12]. It is possibly that ARF1 proteins accumulated at a high level in the transfected protoplasts, thus it is difficult to observe their degradation. On the other hand, we could not rule out the possibility that the tag fused to the ARF proteins may have some effects on protein stability. Therefore it will be of great interest to generate specific antibodies to examine the proteins stability of the different ARF proteins as for ARF7, thus to reveal the true situation in plants.
To further test the stability of ARF5 proteins in plant, we generated stable transformed plants overexpressing GUS-ARF5 which will enable to directly see the protein level of ARF5 by GUS staining. As shown in Fig. 5, the level of GUS staining was relative lower in mock treated seedlings, especially in cotyledons. However, treatment with proteasome inhibitor MG132 or β-lactone increased the level of the staining, both in cotyledons and roots. These results further confirmed that ARF5 is an unstable protein and its degradation was mediated by 26 proteasome. On another hand, these results also suggested that even though light was able to stabilize ARF activator proteins, it may not able to full block their degradation.
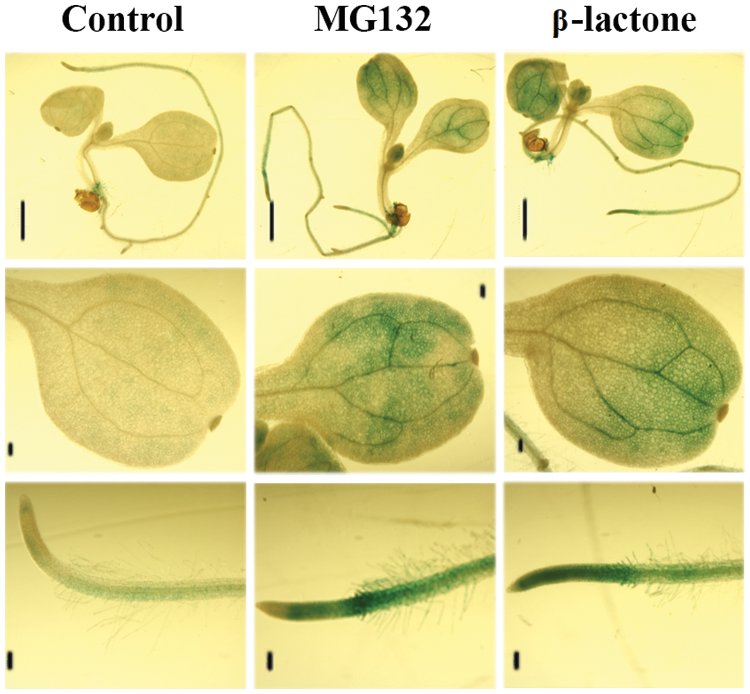
Figure 5: Accumulation of ARF5 proteins in stable transformed plants. Five-day-old light grown seedlings overexpressing GUS-ARF5 were mock treated or treated with 50 μM MG132 or 10 μM β-lactone for 4 h, then GUS activity were histochemical stained with X-gluc. The scale line segment represents 1 mm
The results in this study show that light is able to stabilize ARF7 proteins, and several ARF activators including ARF5, ARF7 and ARF19 are unstable proteins and their degradation are mediated by 26 proteasome. These results may shed light into the mechanisms of the cross talking between light and auxin signals in plants.
Authorship: The author confirms contribution to the paper as follows: study conception and design: S.W.; data collection: S.W.; analysis and interpretation of results: S.W.; draft manuscript preparation: S.W. The author reviewed the results and approved the final version of the manuscript.
Acknowledgement: The experiments were performed in Prof. Tom Guilfoyle’s Lab at University of Missouri-Columbia (MU). As a great mentor, Prof. Guilfoyle showed me not only how to doing science, but also the importance of independence, creativity and critical thinking. He led me to the area of transcription factor regulated gene expression, which is still the main focus in my own laboratory. My great appreciate to his help, and he will always be remembered. I would also thank Prof. Gretchen Hagen, Prof. Guilfoyle’s wife and research partner, for her kindness and helps during my stay at MU. May she continue to live a good life when Prof. Guilfoyle is no longer there with her.
Funding Statement: The author received no specific funding for this study.
Conflicts of Interest: The author declares that he has no conflicts of interest to report regarding the present study.
References
1. Guilfoyle, T. J., Hagen, G. (2007). Auxin response factors. Current Opinion in Plant Biology, 10(5), 453–460. DOI 10.1016/j.pbi.2007.08.014. [Google Scholar] [CrossRef]
2. Guilfoyle, T. J., Hagen, G. (2012). Getting a grasp on domain III/IV responsible for auxin response factor-IAA protein interactions. Plant Science, 190, 82–88. DOI 10.1016/j.plantsci.2012.04.003. [Google Scholar] [CrossRef]
3. Cancé, C., Martin-Arevalillo, R., Boubekeur, K., Dumas, R. (2022). Auxin response factors are keys to the many auxin doors. New Phytologist, 235(2), 402–419. DOI 10.1111/nph.18159. [Google Scholar] [CrossRef]
4. Tiwari, S. B., Hagen, G., Guilfoyle, T. J. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell, 15(2), 533–543. DOI 10.1105/tpc.008417. [Google Scholar] [CrossRef]
5. Ulmasov, T., Hagen, G., Guilfoyle, T. J. (1999). Dimerization and DNA binding of auxin response factors. Plant Journal, 19(3), 309–319. DOI 10.1046/j.1365-313X.1999.00538.x. [Google Scholar] [CrossRef]
6. Wang, S., Hagen, G., Guilfoyle, T. J. (2013). ARF-Aux/IAA interactions through domain III/IV are not strictly required for auxin-responsive gene expression. Plant Signaling & Behavior, 8(6), e24526. [Google Scholar]
7. Gray, W., Kepinski, S., Rouse, D., Leyser, O., Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature, 414(6861), 271–276. [Google Scholar]
8. Ramos, J., Zenser, N., Leyser, O., Callis, J. (2001). Rapid degradation of auixn/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell, 13(10), 2349–2360. [Google Scholar]
9. Tiwari, S. B., Wang, X. J., Hagen, G., Guilfoyle, T. J. (2001). Aux/IAA proteins are active repressors and their stability and activity are modulated by auxin. Plant Cell, 13(12), 2809–2822. [Google Scholar]
10. Zenser, N., Ellsmore, A., Leasure, C., Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. PNAS, 98(20), 111795–111800. [Google Scholar]
11. Li, H., Johnson, P., Stepanova, A., Alonso, J. M., Ecker, J. R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Development Cell, 7(2), 193–204. DOI 10.1016/j.devcel.2004.07.002. [Google Scholar] [CrossRef]
12. Salmon, J., Ramos, J., Callis, J. (2008). Degradation of the auxin response factor ARF1. Plant Journal, 54(1), 118–128. DOI 10.1111/j.1365-313X.2007.03396.x. [Google Scholar] [CrossRef]
13. Li, K., Wang, S., Wu, H., Wang, H. (2020). Protein levels of several Arabidopsis auxin response factors are regulated by multiple factors and ABA promotes ARF6 protein ubiquitination. Interinal Journal of Molecular Sciences, 21(24), 9437. DOI 10.3390/ijms21249437. [Google Scholar] [CrossRef]
14. Jing, H., Korasick, D. A., Emenecker, R. J., Morffy, N., Wilkinson, E. G. et al. (2022). Regulation of AUXIN RESPONSE FACTOR condensation and nucleo-cytoplasmic partitioning. Nature Communications, 13(1), 4015. DOI 10.1038/s41467-022-31628-2. [Google Scholar] [CrossRef]
15. Stowe-Evans, E. L., Harper, R. M., Motchoulski, A. V., Liscum, E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiology, 118(4), 1265–1275. DOI 10.1104/pp.118.4.1265. [Google Scholar] [CrossRef]
16. Harper, R. M., Stowe-Evans, E. L., Luesse, D. R., Muto, H., Tatematsu, K. et al. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell, 12(5), 757–770. [Google Scholar]
17. Wang, S., Tiwari, S. B., Hagen, G., Guilfoyle, T. J. (2005). AUXIN RESPONSE FACTOR7 rostores the expression of auxin-responsive genes in mutant Arabidopsis leaf medophyll protoplansts. Plant Cell, 17(7), 1979–1993. [Google Scholar]
18. Wilmoth, J. C., Wang, S., Tiwari, S. B., Joshi, A. D., Hagen, G. et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant Journal, 43(1), 118–130. [Google Scholar]
19. Murashige, T., Skoog, F. A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum, 15(3), 473–497. [Google Scholar]
20. Hajdukiewicz, P., Svab, Z., Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molocular Biology, 25(6), 989–994. [Google Scholar]
21. Clough, S., Ben, A. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal, 16(6), 735–743. [Google Scholar]
22. Ulmasov, T., Murfett, J., Hagen, G., Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell, 9(11), 1963–1971. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools