 Open Access
Open Access
REVIEW
The Genetic and Biochemical Mechanisms Underlying Cereal Seed Dormancy
1 Shanghai Key Laboratory of Bio-Energy Crops, Research Center for Natural Products, Plant Science Center, School of Life Sciences, Shanghai University, Shanghai, 200444, China
2 State Key Laboratory for Managing Biotic and Chemical Treats to the Quality and Safety of Agro-products, Research Center for Crop Molecular Breeding at Zhejiang Province, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Science, Hangzhou, 310021, China
3 Department of Applied Sciences, University of the West of England, Bristol, BS16 1QY, UK
* Corresponding Authors: Ying Zhu. Email: ; Ping Li. Email:
Phyton-International Journal of Experimental Botany 2023, 92(4), 1203-1214. https://doi.org/10.32604/phyton.2023.026305
Received 29 August 2022; Accepted 11 November 2022; Issue published 06 January 2023
Abstract
The crop seeds have been a staple food for humans, and seed yield is important for sustaining agriculture development and enhancing human adaptability to food risks. The phenomenon of pre-harvest sprouting (PHS), caused by seed dormancy deficiency, and the phenomenon of low seedling emergence caused by seed deep dormancy, will lead to a reduction in agricultural production. Therefore, it is particularly important to understand the regulation mechanisms of seed dormancy. There are many studies on the regulation of seed dormancy in rice, but there are few studies on the regulation of seed dormancy in other crops, and the research on its mechanism is not thorough enough. In this paper, we comprehensively summarize the regulation mechanisms of cereal seed dormancy, including rice, barley and wheat, discussing the integral mechanism of seed dormancy. This information should provide new insights for developing versatile cultivated lines to improve crop yield and economic benefits.Keywords
Seed dormancy is an innate seed characteristic, which helps determines the environmental conditions for seed germination [1]. It is genetically determined and influenced by the environment, which is mediated at least in part by the plant hormones abscisic acid and gibberellin. The dormancy state of seed is not only affected by the environment of seed maturation, but also changes with time after seed shedding, and the change way depends on the surrounding environment. Since seed dormancy occurs in all major climate zones, it comes as a result of adaptation to different environments. Factors that inhibit germination are present in the embryo and surrounding structures, such as the seed coat [2] and endosperm [3]. In summary, dormancy is a quantitative trait whose depth and duration are regulated by genetic and environmental factors.
Seed germination is achieved through three processes: tissue differentiation, cell enlargement and maturation dehydration. The changes of any one of these links may affect seed germination and thus show seed dormancy. First, the unicellular zygote divides and differentiates into young embryos, and eventually forms the endosperm, completing the whole process of tissue differentiation. Then cell division stops, storage accumulates, cells expand, and finally seed development stops due to dehydration, metabolism is reduced, and finally the embryo enters a static or inactive state. To some extent, seed germination is noticable and completed by radicle emergence, which is derived by the balance of the embryo and the covering tissues, such as the endosperm and the testa [4]. Primary dormancy can be assigned either by the embryo or by surrounding tissues. During seed germination, surrounding tissues such as endosperm, radicle or inner seed coat can produce enzymes that promote seed coat rupture, thus promoting seed germination [5]. It has been supposed that the properties of the primary cell wall might affect the ABA-GA balance through a-xylosidase (XYL1). There may be an influence of the cutin layer covering the entire endosperm. Interestingly, cutin biosynthesis or deposition affects the expression of nine-cis-epoxycarotenoid dioxygenase 6 (NCED6) and nine-cis-epoxycarotenoid dioxygenase 9 (NCED9), which are necessary for dormancy [6].
Seed dormancy can be classified as primary dormancy and secondary dormancy, based on when the induction of dormancy occurs. Primary dormancy is established during seed maturation on the mother plant. When nondormant seeds are exposed to certain unfavorable environmental conditions, it may also reenter into dormancy, namely secondary dormancy [7]. Primary dormancy contains five types by Baskin’s formula system, including physiological, morphological, morphological physiological, physical and compound dormancy [8]. Secondary dormancy induction occurs due to the environmental conditions and genetic background of the seeds, which is poorly understood. Nevertheless, an overlap between primary and secondary dormancy pathways likely takes place [9].
China is a big agricultural country, and seed dormancy is very important for agricultural production. On the one hand, if the seeds are in a dormant state, the growth of seed will be uneven. On the other hand, in rainy weather during harvest season, if seed dormancy is lost, earlier germination is likely to happen [10]. Activity of α-amylase in wheat with early germination increases, and starch is partly hydrolyzed. As a result, the physical and chemical properties of wheat change [11], leading to the decline in grain yield and quality, resulting in huge economic losses. Therefore, it is very important to control the dormancy time of crop seed. The dormancy mechanism of crop seed is briefly summarized in this paper.
2 Mechanisms Regulating Seed Dormancy
Seed dormancy is regulated by many factors, among which abscisic acid (ABA) is one of the main regulators of seed dormancy [12]. In Arabidopsis thaliana, the seed germination rate of ABA-related mutants is faster than that of the wild-type [13], and the overexpression of ABA biosynthesis rate-limiting gene NCED6 leads to deep seed dormancy [14]. Exogenous nitrate can regulate the expression of the ABA catabolism gene CYP707A2. The influence of different nitrate nitrogen concentrations on CYP707A2 mRNA levels during the growth and development of female plants was significantly positive. CYP707A2 gene plays a significant role in nitrate-mediated seed development and germination [15]. ATPER 1 is a seed specific peroxiredoxin that enhances seed dormancy and reduces seed germination by inhibiting ABA catabolism and GA biosynthesis in Arabidopsis seeds. atper1-1 and atper1-2 inhibited seed initial dormancy and increased germination rates compared with wild type. Further studies showed that atper1-1 and atper1-2 decreased ABA content and increased GA content in seeds. In addition, atper1 seeds were insensitive to abiotic stress during seed germination. The expression of several ABA catabolic genes (CYP707A1, CYP707A2, CYP707A3) and GA biosynthesis genes (GA20ox1, GA20ox3 and KAO3) in atper1 seed was higher than that in wild type seed [16]. In addition, miRNA is also involved in regulating seed dormancy. MiR156 mutants enhance seed dormancy by inhibiting the gibberelin (GA) pathway by inhibiting the miR156 target gene IPA1 (Ideal Plant Architecture 1). IPA1 directly regulates multiple genes in the GA pathway [17].
During seed germination, almost all other plant hormones, including ethylene (ET), brassinosteroids (BRS), jasmonic acid (JA), salicylic acid (SA), cytokinins (CTKs) and phytolactones (SLS), maintain the ABA/GA balance. Transcription factors including ARF, MYB96, ABI3, ABI4, and ABI5 regulate ABA biosynthesis by interacting with CYP707A1 and CYP707A2, while GA negative regulation is ensured by DELLA genes. This balance is maintained until the stage of seed emergence [18]. The classic ABA signaling pathway is that PP2C inhibits SnRK2 when ABA, PYR/PYL/RCAR and PP2C are integrated into a complex. Subsequently, activated SnRK2 phosphorylates downstream targets such as ABA-insensitive factor 3 (ABI3), ABI4, ABI5, and ABA response element (ABRE), binding factor (ABF). These ABA metabolic enzymes and core ABA signaling pathway components both play vital roles in ABA-mediated dormancy and germination regulation [19]. In addition, ABI4 enhances ABA content by inhibiting CYP707A1/2 expression [20], and MYB96 directly binds promoters of NCED2 and NCED6 to promote ABA biosynthesis [21]. RELATIVE OF EARLY FLOWERING6 (REF6) directly binds to CYP707A1 and CYP707A3 and reduces their H3K27me3 levels, thereby increasing ABA concentration [22].
Environmental factors such as moisture, temperature and light also affect seed dormancy. Sorghum seed developed under drought conditions showed earlier germination ability at maturity than control seeds, and thus they were less resistant to spike germination when exposed to high humidity conditions. Matured seed on the mother plant under water stress have higher ABA content in the early stage of development [23]. Cold induces seed dormancy through a variety of mechanisms, including up-regulation of DOG1 transcription and the role of DOG1 in promoting GA catabolism, while C-repeat BINDING FACTORS (CBFs) are required to regulate dormancy induced by low temperature and GA2OX6 and DOG1 expression [24]. In both Bur and Cvi ecotypes of Arabidopsis thaliana, high temperature reduced seed dormancy and seed yield. Bur decreased by more than 90% and Cvi decreased by more than 50% [25]. Studies have shown that DNA (demethylation) may be an important regulatory process of seed germination under heat stress [26]. Increasing nitrate supply to mother plants reduces ABA content and dormancy level of seed offspring [13].
Reactive oxygen species (ROS) also participate in the regulation of seed dormancy, which regulates catalase activity and thus the mutual transformation of GSH and GSSG, resulting in changes in GA pathway [27]. ROS are associated with dormancy release through direct oxidation of biomolecules in dry-stored seed. ROS are involved in the perception and transduction of environmental conditions during seed imbibition. When these conditions allow germination, ROS levels are maintained at a level that triggers cellular events related to germination, such as hormonal signaling. The spatio-temporal regulation of ROS production synergistically interacts with hormonal signals to regulate the cellular events involved in germination related cell expansion [28]. Light also regulates seed germination, and red light promotes seed germination. There are two conformation photochromes in plants: Pr (inactive form) and Pfr (active form). Pr is converted into Pfr by absorbing red light, while Pfr is converted into Pr by absorbing far-red light [29]. De-etiolated1 (DET1) acts upstream of hypocotyl in far-red 1 (HFR1) and phytochrome interacting factors1 (PIF1), which is a key positive and negative transcriptional regulator in seed germination. DET1, COP10 (Constitutive Photomorphogenic 10) and Cullin4 E3 ligase form complex to degrade HFR1 [30]. PHOTOPERIODIC CONTROL OF HYPOCOTYL 1 (PCH1) and pCH1-like (PCHL) regulates seed dormancy by stabilizing the active form of PhyB and regulating different light responses through direct interactions with PIF1 and COP1 [31]. In barley, red and far-red light had no effect on dormancy, while blue light promoted dormancy [32]. In wheat, blue light and far-red light promote kernel dormancy, while red light reduces it [33].
Active nitrogen molecules also contribute to the regulation of seed hypnosis, and NO can regulate ABA by regulating ABI5 and thus participate in the regulation of seed dormancy. ABI5 is a type of leucine zipper, and NO can nitrosylate the CYS153 of ABI5 and promote seed germination [34]. Several miRNA regulatory pathways involved in seed dormancy and germination have been identified in Arabidopsis thaliana. For example, auxin response factor (ARF) is a transcription factor involved in auxin signal transduction at multiple stages of plant growth and development. ARF10, ARF16, and ARF17 are targeted by Arabidopsis microRNA160 (MiR160) [35]. In germinating Arabidopsis seeds, ABA induces the accumulation of microRNA159 (MiR159) in an ABI3-dependent manner, a dynamic balancing mechanism that guides the degradation of MYB33 and MYB101 transcription, thereby reducing sensitivity to hormone signaling in seedling stress response [36]. DOG1 gene can regulate seed dormancy and flowering time of lettuce and Arabidopsis at miR156 and miR172 levels [37].
Hydrogen sulfide promotes seed germination and alleviates these stresses under various abiotic stresses. These stress responses are also associated with ionic imbalance, osmotic balance, lipid peroxidation, protein alterations, and oxidative damage to macromolecular biomolecules (DNA and proteins) [38]. Exogenous HRW treatment of rice seeds could alleviate the inhibition of salt stress on seed germination and seedling growth. Under normal and stress conditions, any H2S donor treatment could significantly increase seed germination rate, and NaHS treatment had the best effect [39]. In addition, hydrogen sulfide also plays a catalytic role in embryonic development and lateral root formation [40].
3 Mechanisms of Seed Dormancy and Germination in Crop
3.1 Regulation of Dormancy of Rice Seed
The dormancy of rice is also controlled by hormones and environmental factors. For example, the expression of OsDOG1L-3 is positively correlated with seed dormancy induced by ABA. OsbZIP 75 and OsbZIP 78 directly bind to the OsDOG1L-3 promoter and induces its expression. Overexpression of OsbZIP 75 increased the abundance of OsDOG1L-3 protein and enhanced seed dormancy. OsDOG1l-3 up-regulated the expression of ABA-related genes and increased ABA content [41]. WRKY transcription factor also regulates an ABA signaling pathway. OsWRKY29 showed enhanced seed dormancy, while overexpression showed reduced seed dormancy. OsWRKY 29 directly down-regulated OsABF1 and OsVP1 in seed to inhibit seed dormancy [42]. Rice gene Germin-like protein 2-1 (OsGLP-2-1) promotes seed dormancy. Also, overexpression of OsGLP-2-1 leads to increased seed dormancy [43]. Squamosa-promoter BINDING protein-like (SPL) SPL12 and IPA1 enhance seed dormancy by directly regulating related genes in GA pathway. In addition, seed-specific overexpression of IPA1 also increases seed grain size, thus improving grain productivity [44].
Rice quantitative trait locus SDR4 plays an important role in seed dormancy differences among rice varieties. The expression of SDR4 is positively regulated by OsVP1, which in turn positively regulates the potential regulator of seed dormancy and inhibits the expression of genes after germination. Only the Nippon allele SDR4-N in japonica rice varieties endows less dormancy, while SDR4-N and SDR4-K are widely distributed in indica rice populations [45]. In Arabidopsis thaliana, the transcription level of ODR1, a homolog of rice SDR4, is directly inhibited by ABI3, and bHLH57 promotes the expression of NCED6 and NCED9, furthering increasing ABA levels. ODR1 interacts with bHLH57 to inhibit the binding of bHLH57 to NCED6 and NCED9, thereby negatively regulating seed dormancy [46]. Seed dormancy1-2 (QSD1-2) is a gene involved in endosperm induced dormancy and plant height. QSD1-2 is located in a 20 KB region containing OsGA20ox2, and QSD1-2 controls primary dormancy through a GA-regulated dehydration mechanism [47]. OsAP2-39 also regulates ABA-GA balance. Osap2-39 up-regulated the transcription of the ABA biosynthesis gene OsNCED1, leading to an increase in endogenous ABA levels. OsAP2-39 also enhances expression of GA inactivated genes OsEUI (EUI Elongation of Upper most Internode (EUI) protein), resulting in a decline in endogenous GA content [48]. Protein phosphatase 2C cladaryA (PP2CA) is a major signaling component of the ABA-dependent signaling cascade that regulates seed germination. OsPP2C51 is extremely specific in embryos. Studies have shown that OsPP2C51 plays a positive regulatory role in seed germination by directly inhibiting phosphorylated OsbZIP10 (ABI5) [49]. In addition, OsLOL1 (a C2C2 zinc finger protein) interacts with OsbZIP58 to activate OsKO2. OsLOL1 also reduces the expression of SOD1 gene in rice aleurone layer, accelerates programmed cell death (PCD) in aleurone layer, and ultimately promotes rice seed germination [50]. PHS9 encodes a higher plant-specific TYPE CC glugoredoxin that interacts with OsGAP, an interaction partner of ABA receptor OsRCAR1. PHS9 or OsGAP overexpressed plants showed decreased ABA sensitivity during seed germination, while PHS9 or OsGAP knockout mutant plants showed increased ABA sensitivity during seed germination, suggesting that PHS9 and OsGAP negatively regulated ABA signal transduction during seed germination [51].
Serine carboxypeptidase (SCP) is a member of the largest enzyme group that catalyzes protein functional maturation. SCP46 is mainly expressed in developing seeds, especially in the embryo, endosperm and aleurone layer, and can be induced by ABA. After SCP46 gene was knocked out, grain size became smaller and seed germination was enhanced. Numerous grain filling and seed dormancy related genes, such as suc synthase protein kinase (SPK), viviparous 1 (VP1) and ADP-glucose pyrophosphorylase (AGPs) were down-regulated in scp46. SCP 46 may be a major regulator of grain filling and seed germination by participating in ABA signal transduction [52].
3.2 Regulation of Seed Dormancy in Wheat
The regulation of seed dormancy in wheat is also achieved by regulating the balance between ABA and GA. Other hormones are also regulated by these two hormones. For example, jasmonate regulates wheat seed dormancy by regulating the balance between GA and ABA [53]. Wheat protein phosphatase PP2C-A10 interacts with TaDOG1L1 and TaDOG1L4 to promote the germination of transgenic Arabidopsis seeds and decrease their drought tolerance [54].
In Arabidopsis, AFPs are negative regulators of ABA signal transduction, promoting ABI5 protein degradation and attenuating ABA signal regulation by targeting the ABI5 gene [55]. TaAFP in wheat and AFPs in Arabidopsis have a conserved domain, and two alleles of TaAFP were identified on chromosome 2BS of common wheat, named TaAFP-B1A and TaAFP-B1B, respectively. There is a 4-base insertion in the 5′ non-coding region of TaAFP-B gene, which leads to a higher average germination rate of TaAFP-B1A than TaAFP-B1B [56]. Seed dormancy allele TaSDR-A1A mainly exists in the local wheat varieties in China and is associated with spike germination. Researchers have cloned TaSDR-A1 on chromosome 2A. Sequence analysis of TaSDR-A1 showed that there was a SNP at site 643, and the G allele was present in the genotype with low germinating index (GI) value, while the A allele was present in the genotype with high GI [57]. Temperature during wheat seed development had a strong influence on seed dormancy. The lower the temperature, the higher the seed dormancy level. The expression of homologous genes of MOTHER OF FT and TFL1 (MFT) was up-regulated during seed development at low temperature. MFT (MFT-3A) on chromosome 3A co-locates with qPHS.OCS-3a1, indicating that MFT plays an essential regulatory role in wheat germination [58].
TaSDR4, a homolog of rice OsSDR4, is located on the second set of homologous chromosomes and is named as TaSDR-A1, TasDR-B1 and TaSDR-D1, respectively. Sequence analysis of TaSDR-B1 showed that there was A SNP in the −11 position upstream of the initiation codon, which was A base in the varieties with low germination rate and G base in the varieties with the high germination rate. Therefore, wheat TaSDR4 controlled seed germination [59].
3.3 Regulation of Seed Dormancy in Barley
ROS release seed dormancy and strengthen seed germination in several food crops. Studies have shown that ABA up-regulates catalase activity through transcriptional activation of HvCAT2. Hydrogen peroxide (H2O2) treatment did not affect ABA sensitivity, but upregulated the expression of the GA-induced gene HvExpA 11, inhibited the expression of HvGA2ox3 related to GA catabolism, and increased the expression of HvGA20ox1 related to GA synthesis. In barley, H2O2 may be involved in alleviating dormancy through activation of GA signals and synthesis rather than through inhibition of ABA signals [27]. In contrast, non-dormant seed produced higher levels of H2O2 than dormant seed after imbibition of barley seed embryos. H2O2 regulates ABA content in embryos by ABA-80 hydroxylase, an ABA catabolic enzyme. The changes of ABA and ROS balance in barley seed embryos after imbibition are dynamic, and play a regulatory role in dormancy and germination of barley seeds [60]. Studies have isolated the main seed dormancy gene QSD1 from wild barley, which encodes alanine aminotransferase (AlaAT), and alanine aminotransferase controls seed dormancy of barley [61]. The homologue of Arabidopsis DOG1 gene is labeled on chromosome 3H of barley, but HvDOG1 plays little role in barley seed dormancy. The gibberellic acid oxidase gene HvGA20ox1 promoted the release of dormancy [62]. ANT28 encodes Hvmyb10, which is a critical factor in barley dormancy, and Ant 28 reduces dormancy [63].
The dormancy of barley was also determined by environmental factors. In barley, seed of primary dormancy did not germinate at air temperature of 30°C, and germinated at air temperature of 15°C. Although high temperature has an indirect effect on the availability of O2, data show that high temperature has no effect on the expression of proline-4-hydroxylase. Compared with high temperature induction, GA has a stronger induction effect and ABA has a smaller regulatory effect under hypoxia induction at 15°C [64]. Light also regulated seed dormancy, and white light promoted the expression of ABA biosynthesis gene HvNCED1 in embryos. Afterripening had no effect on the expression of ABA biosynthetic genes, but promoted the expression of ABA catabolism genes (HvABA 8 ′OH1), GA biosynthetic genes (HvGA3ox2) and GA decomposition genes (HvGA2ox3) [32]. By inducing the expression of ABA biosynthesis gene 9-cis-epoxyoid dioxygenase and inhibiting the expression of ABA 8′-hydroxylase, blue light inhibits the germination of barley and thus increases ABA content in grains. CRY 1 is a key receptor for sensing and transmitting blue light signals in dormant grains [65]. In addition, studies have shown that HvNCED 1 and HvNCED 2 have certain regulatory effects on ABA synthesis during secondary seed dormancy [66].
3.4 Regulation of Seed Dormancy in Other Crops
Soybean is a significant oil crop and primary dietary protein resource. The limited understanding of soybean oil biosynthesis has become a vital obstacle to increase soybean oil yield. The transcription factor abscisic acid insensitivity 3 (ABI3) plays an important role in growth and development and seed dormancy in many crops. Content of GmABI3 in soybean seed and leaves changed with time at 4°C, 42°C, ABA treatment and JA treatment, respectively. GmABI3 has the effect of regulating stress resistance of soybean and is suitable for various crops to deal with biological stress [67]. ABA biosynthesis inhibitor fluuron reversed the auxin induced delayed germination phenotype, while the GA biosynthesis inhibitor paclobutrazol inhibited soybean seed germination. Exogenous auxin inhibits soybean seed germination by mediating ABA and GA biosynthesis [68].
In Arabidopsis, DOG1 plays a key role in seed dormancy and germination. Soybean GmDOG1L family consists of 40 members, which have different expression changes in different tissues. GmDOG1-l1, GmDOG1-L2, GmDOG1-L3 and GmDOG1-L39 have the highest expression in pod, accompanied by dry seed. The expression levels of GmDOG1-L11, GmDOG1-L27, GmDOG1-L30 and GMDOG1-L37 in dry seed were the highest. Gibberellin (GA) significantly inhibited the expression of most GmDOG1Ls genes. Abscisic acid (ABA) inhibited the expression of some GmDOG1Ls (GmDOG1-L11, GmDOG1-L26, GmDOG1-L39) and promoted the expression of other GmDOG1Ls (GmDOG1-L10, GmDOG1-L27, GmDOG1-L30). Therefore, it is speculated that GmDOG1Ls are directly or indirectly related to ABA and GA pathways, and regulate dormancy and germination of soybean seeds through complex interactions [69].
In maize, the Arabidopsis ABI4 homolog ZmABI4 was also isolated and the ZmABI4 protein was shown to bind to CE1 elements in various ABA-related genes. ZmABI4 also binds to the promoter of the sugar-responsive ADH1 gene, indicating that the protein has the ability of ABA and sugar regulation [70]. Raffinose is an oligosaccharide present in many seeds and plays an important role in seed vigor. ZmABI5 is a direct physical interaction with ZmVP1, and directly binds to ZmGOLS 2 (GALACTINOL SYNTHASE 2) promoter to regulate its expression and promote raffinose accumulation, thus regulating seed dormancy [71].
Dormancy of crops is regulated by multiple factors, including internal factors such as hormones, ROS and nutrients, and external factors such as temperature, light and moisture. At present, it is known that seed dormancy is mainly regulated by ABA and GA signaling pathways in rice, wheat, barley, soybean and maize, but the specific mechanism is not completely clear, especially in corn and soybean. It has been found that the functions of some Arabidopsis homologous genes, such as DOG1 and ABI3, are relatively conserved in crops, and work can continue to explore regulatory mechanisms in other crops based on this. Based on the above literature, a signal pathway diagram of seed dormancy regulation in crops can be drawn, as shown in Fig. 1.
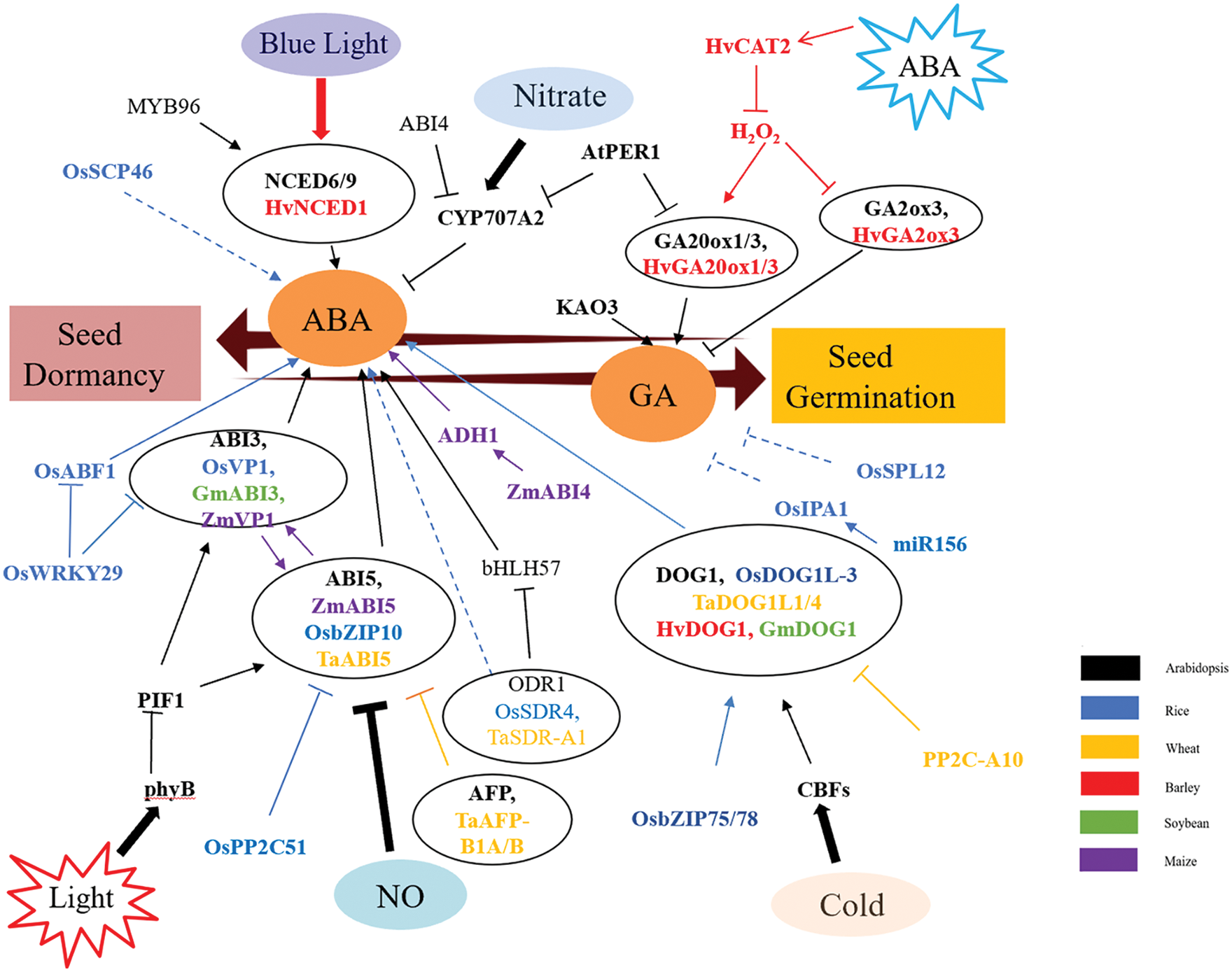
Figure 1: Preliminary network of phytohormone functions in seed dormancy
ABA and GA play an important role in seed dormancy. High ABA levels lead to seed dormancy, and high GA levels lead to seed germination. NCED6/9 is a rate-limiting enzyme in ABA synthesis. CYP707A2 degrades ABA. Various factors promote the formation of NCED6/9 and inhibit the synthesis of CYP707A2, thus increasing ABA content and promoting seed dormancy. Many transcription factors regulate seed dormancy, such as ABI3 and ABI5, and environmental factors such as light and NO regulate seed dormancy by regulating the levels of ABI3 and ABI5. ABI3 and ABI5 homologous genes were found in rice, wheat and maize, which also regulated seed dormancy. In addition, many other genes also regulate seed dormancy and germination, such as OsSCP46, OsPP2C51, OsSPL12, etc. Arrows indicate positive regulation and bars indicate negative regulation. Solid lines indicate direct effects, dotted lines indicate indirect effects.
In rice, some other genes regulate seed dormancy through other pathways. For example, rice OsBT1 regulates seed dormancy through the glucose metabolism pathway, and the deletion of 1 base of OsBT1 leads to the decreased seed dormancy of H470. Most of the sugar content of H470 seed is higher than that of the wild type. h470 mutation affects glucose metabolism [72]. In the presence of exogenous glucose, OsABA8ox gene expression was inhibited, especially OsABA8ox2 and OsABA8ox3. In contrast, the expression level of OsNCED gene, which controls the limiting step of ABA biosynthesis, do not change significantly at low glucose level. Glucose-induced delay in seed germination is caused by inhibition of ABA catabolism rather than promotion of ABA biosynthesis during rice seed germination [73]. MKKK62 negatively regulated dormancy of rice seeds, ABA sensitivity and OsMFT transcription were negatively regulated by MKKK62 during seed germination and later maturation. This reflects the role of phosphorylation pathway in seed dormancy regulation [74].
In addition, the rapid development of modern molecular biology technology has provided more channels for the study of seed dormancy regulation. For example, in barley, cDNA-AFLP was used to study the molecular regulation of dormancy in dormant and non-dormant embryos to identify transcripts that were differentially expressed in dormant and non-dormant embryos. Seven transcription source fragments (TDF) were found to be differentially expressed in dormant and non-dormant seeds. In particular, HV12D, HV42B and HV32B are potential signal elements. HV12D is homologous to the Ariadne gene, which may be linked to the ABA signaling pathway [75]. In maize, GA, ABA or double distilled water treated maize embryos served as controls to construct small RNA libraries. Bioinformatics tools were used to predict the target genes of differentially expressed miRNAs and miRNA-mediated gene expression was found to affect GA and ABA signaling pathways during seed germination [76]. In conclusion, the study of seed dormancy in future crops has broad prospects and huge research potential, and can further explore the specific mechanism of seed dormancy with the help of a variety of modern molecular biology techniques.
Acknowledgement: The authors confirm contribution to the paper as follows: Sasa Jing wrote the text, Yuan Tian and Heng Zhang prepared for the figures, Yin Zhu and Ping Li revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This work was funded by the National Natural Science Foundation of China (Grant No. 32170562).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Finch-Savage, W. E., Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytologist, 171(3), 501–523. DOI 10.1111/j.1469-8137.2006.01787.x. [Google Scholar] [CrossRef]
2. Debeaujon, I., Leon-Kloosterziel, K. M., Koornneef, M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology, 122(2), 403–414. DOI 10.1104/pp.122.2.403. [Google Scholar] [CrossRef]
3. Hilhorst, H. W. (2007). Development, dormancy and germination. In: Bradford, K. J., Nonogaki, H. (Eds.Definitions and hypotheses of seed dormancy, vol. 27, 50–71. [Google Scholar]
4. Bewley, J. D., Bradford, K. J., Hilhorst, H. W. M., Nonogaki, H. (2013). Dormancy and the control of germination. In: Seeds: Physiology of development. 3rd editionpp. 247–297. New York: Springer. [Google Scholar]
5. William, E. F., Gerhard, L. (2010). Seed dormancy and the control of germination. New Phytologist, 173(3), 501–523. [Google Scholar]
6. Nonogaki, H. (2019). Seed germination and dormancy: The classic story, new puzzles, and evolution. Journal of Integrative Plant Biology, 61(5), 541–563. DOI 10.1111/jipb.12762. [Google Scholar] [CrossRef]
7. Buijs, G. (2020). A perspective on secondary seed dormancy in Arabidopsis thaliana. Plants, 9(6), 749. DOI 10.3390/plants9060749. [Google Scholar] [CrossRef]
8. Baskin, J. M., Baskin, C. C. (2008). Some considerations for adoption of Nikolaeva’s formula system into seed dormancy classification. Seed Science Research, 18(3), 131–137. [Google Scholar]
9. Tuan, P. A., Sun, M., Nguyen, T. N., Park, S., Ayele, B. T. (2019). Molecular mechanisms of seed germination. In: Sprouted grains, pp. 1–24. Elsevier. [Google Scholar]
10. Shu, K., Liu, X. D., Xie, Q., He, Z. H. (2016). Two faces of one seed: Hormonal regulation of dormancy and germination. Molecular Plant, 9(1), 34–45. DOI 10.1016/j.molp.2015.08.010. [Google Scholar] [CrossRef]
11. Simsek, S., Ohm, J. B., Lu, H., Rugg, M., Berzonsky, W. et al. (2014). Effect of pre-harvest sprouting on physicochemical properties of starch in wheat. Foods, 3(2), 194. DOI 10.3390/foods3020194. [Google Scholar] [CrossRef]
12. Vaistij, F. E., Gan, Y., Penfield, S., Gilday, A. D., Dave, A. et al. (2013). Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proceedings of the National Academy of Sciences, 110(26), 10866–10871. DOI 10.1073/pnas.1301647110. [Google Scholar] [CrossRef]
13. Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F. et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant Journal, 70(3), 501–512. DOI 10.1111/j.1365-313X.2011.04887.x. [Google Scholar] [CrossRef]
14. Martinez-Andujar, C., Ordiz, M. I., Huang, Z., Nonogaki, M., Beachy, R. N. et al. (2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. PNAS, 108(41), 17225–17229. DOI 10.1073/pnas.1112151108. [Google Scholar] [CrossRef]
15. Matakiadis, T., Alboresi, A., Jikumaru, Y., Tatematsu, K., Pichon, O. et al. (2009). The arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiology, 149(2), 949–960. DOI 10.1104/pp.108.126938. [Google Scholar] [CrossRef]
16. Chen, H., Tong, J., Fu, W., Liang, Z., Ruan, J. et al. (2020). The H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 suppresses seed dormancy by inducing abscisic acid catabolism. Plant Physiology, 184(4), 1969–1978. DOI 10.1104/pp.20.01255. [Google Scholar] [CrossRef]
17. Chen, H., Ruan, J., Chu, P., Fu, W., Liang, Z. et al. (2020). AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant Journal, 101(2), 310–323. DOI 10.1111/tpj.14542. [Google Scholar] [CrossRef]
18. Miao, C., Wang, Z., Zhang, L., Yao, J., Hua, K. et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nature Communication, 10(1), 3814–3822. DOI 10.1038/s41467-019-11830-5. [Google Scholar] [CrossRef]
19. Sohn, S. I., Pandian, S., Kumar, T. S., Zoclanclounon, Y. A. B., Muthuramalingam, P. et al. (2021). Seed dormancy and pre-harvest sprouting in rice-an updated overview. International Journal of Molecular Sciences, 22(21), 11804–11825. DOI 10.3390/ijms222111804. [Google Scholar] [CrossRef]
20. Tai, L., Wang, H. J., Xu, X. J., Sun, W. H., Ju, L. et al. (2021). Pre-harvest sprouting in cereals: Genetic and biochemical mechanisms. Journal of Experimental Botany, 72(8), 2857–2876. DOI 10.1093/jxb/erab024. [Google Scholar] [CrossRef]
21. Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y. et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genetics, 9(6), e1003577–e1003590. DOI 10.1371/journal.pgen.1003577. [Google Scholar] [CrossRef]
22. Lee, H. G., Lee, K., Seo, P. J. (2015). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Molecular Biology, 87(4–5), 371–381. DOI 10.1007/s11103-015-0283-4. [Google Scholar] [CrossRef]
23. Benech Arnold, R. L., Fenner, M., Edwards, P. J. (1991). Changes in germinability, ABA content and ABA embryonic sensitivity in developing seeds of Sorghum bicolor (L.) Moench. Induced by water stress during grain filling. New Phytologist, 118(2), 339–347. DOI 10.1111/j.1469-8137.1991.tb00986.x. [Google Scholar] [CrossRef]
24. Kendall, S. L., Hellwege, A., Marriot, P., Whalley, C., Graham, I. A. et al. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell, 23(7), 2568–2580. DOI 10.1105/tpc.111.087643. [Google Scholar] [CrossRef]
25. Huang, Z., Footitt, S., Finch-Savage, W. E. (2014). The effect of temperature on reproduction in the summer and winter annual Arabidopsis thaliana ecotypes Bur and Cvi. Annals of Botany, 113(6), 921–929. DOI 10.1093/aob/mcu014. [Google Scholar] [CrossRef]
26. Malabarba, J., Windels, D., Xu, W., Verdier, J. (2021). Regulation of DNA (de)methylation positively impacts seed germination during seed development under heat stress. Genes, 12(3), 457. DOI 10.3390/genes12030457. [Google Scholar] [CrossRef]
27. Bahin, E., Bailly, C., Sotta, B., Kranner, I., Corbineau, F. et al. (2011). Crosstalk between reactive oxygen species and hormonal signalling pathways regulates grain dormancy in barley. Plant, Cell & Environment, 34(6), 980–993. [Google Scholar]
28. Bailly, C. (2019). The signalling role of ROS in the regulation of seed germination and dormancy. Biochemical Journal, 476(20), 3019–3032. DOI 10.1042/BCJ20190159. [Google Scholar] [CrossRef]
29. Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M. et al. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. PNAS, 93(15), 8129–8133. DOI 10.1073/pnas.93.15.8129. [Google Scholar] [CrossRef]
30. Shi, H., Wang, X., Mo, X., Tang, C., Zhong, S. et al. (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. PNAS, 112(12), 3817–3822. DOI 10.1073/pnas.1502405112. [Google Scholar] [CrossRef]
31. Cheng, M. C., Enderle, B., Kathare, P. K., Islam, R., Hiltbrunner, A. et al. (2020). PCH1 and PCHL directly interact with PIF1, promote its degradation, and inhibit its transcriptional function during photomorphogenesis. Molecular Plant, 13(3), 499–514. DOI 10.1016/j.molp.2020.02.003. [Google Scholar] [CrossRef]
32. Gubler, F., Hughes, T., Waterhouse, P., Jacobsen, J. (2008). Regulation of dormancy in barley by blue light and after-ripening: Effects on abscisic acid and gibberellin metabolism. Plant Physiology, 147(2), 886–896. DOI 10.1104/pp.107.115469. [Google Scholar] [CrossRef]
33. Jacobsen, J. V., Barrero, J. M., Hughes, T., Julkowska, M., Taylor, J. M. et al. (2013). Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta, 238(1), 121–138. DOI 10.1007/s00425-013-1878-0. [Google Scholar] [CrossRef]
34. Albertos, P., Romero-Puertas, M. C., Tatematsu, K., Mateos, I., Sanchez-Vicente, I. et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nature Communication, 6(1), 8669. DOI 10.1038/ncomms9669. [Google Scholar] [CrossRef]
35. Liu, P. P., Montgomery, T. A., Fahlgren, N., Kasschau, K. D., Nonogaki, H. et al. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant Journal, 52(1), 133–146. DOI 10.1111/j.1365-313X.2007.03218.x. [Google Scholar] [CrossRef]
36. Reyes, J. L., Chua, N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant Journal, 49(4), 592–606. DOI 10.1111/j.1365-313X.2006.02980.x. [Google Scholar] [CrossRef]
37. Huo, H., Wei, S., Bradford, K. J. (2016). DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. PNAS, 113(15), E2199–2206. DOI 10.1073/pnas.1600558113. [Google Scholar] [CrossRef]
38. Sharma, P., Meyyazhagan, A., Easwaran, M., Sharma, M. M. M., Mehta, S. et al. (2022). Hydrogen sulfide: A new warrior in assisting seed germination during adverse environmental conditions. Plant Growth Regulation, 98(3), 401–420. DOI 10.1007/s10725-022-00887-w. [Google Scholar] [CrossRef]
39. Xu, S., Zhu, S., Jiang, Y., Wang, N., Wang, R. et al. (2013). Hydrogen-rich water alleviates salt stress in rice during seed germination. Plant and Soil, 370(1–2), 47–57. DOI 10.1007/s11104-013-1614-3. [Google Scholar] [CrossRef]
40. Mei, Y., Chen, H., Shen, W., Shen, W., Huang, L. (2017). Hydrogen peroxide is involved in hydrogen sulfide-induced lateral root formation in tomato seedlings. BMC Plant Biology, 17(1), 162–173. DOI 10.1186/s12870-017-1110-7. [Google Scholar] [CrossRef]
41. Wang, Q., Lin, Q., Wu, T., Duan, E., Huang, Y. et al. (2020). OsDOG1L-3 regulates seed dormancy through the abscisic acid pathway in rice. Plant Science, 298, 110570–110580. DOI 10.1016/j.plantsci.2020.110570. [Google Scholar] [CrossRef]
42. Zhou, C., Lin, Q., Lan, J., Zhang, T., Liu, X. et al. (2020). WRKY transcription factor OsWRKY29 represses seed dormancy in rice by weakening abscisic acid response. Frontiers in Plant Science, 11, 691–705. DOI 10.3389/fpls.2020.00691. [Google Scholar] [CrossRef]
43. Wang, H., Zhang, Y., Xiao, N., Zhang, G., Wang, F. et al. (2020). Rice GERMIN-LIKE PROTEIN 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiology, 183(3), 1157–1170. DOI 10.1104/pp.20.00253. [Google Scholar] [CrossRef]
44. Qin, M., Zhang, Y., Yang, Y., Miao, C., Liu, S. (2020). Seed-specific overexpression of SPL12 and IPA1 improves seed dormancy and grain size in rice. Frontiers in Plant Science, 11, 532771–532777. DOI 10.3389/fpls.2020.532771. [Google Scholar] [CrossRef]
45. Sugimoto, K., Takeuchi, Y., Ebana, K., Miyao, A., Hirochika, H. et al. (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. PNAS, 107(13), 5792–5797. DOI 10.1073/pnas.0911965107. [Google Scholar] [CrossRef]
46. Liu, F., Zhang, H., Ding, L., Soppe, W. J. J., Xiang, Y. (2020). REVERSAL OF RDO5 1, a homolog of rice seed Dormancy4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis. Plant Cell, 32(6), 1933–1948. DOI 10.1105/tpc.20.00026. [Google Scholar] [CrossRef]
47. Ye, H., Feng, J., Zhang, L., Zhang, J., Mispan, M. S. et al. (2015). Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiology, 169(3), 2152–2165. [Google Scholar]
48. Yaish, M. W., El-Kereamy, A., Zhu, T., Beatty, P. H., Good, A. G. et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genetics, 6(9), e1001098–e1001109. DOI 10.1371/journal.pgen.1001098. [Google Scholar] [CrossRef]
49. Bhatnagar, N., Min, M. K., Choi, E. H., Kim, N., Moon, S. J. et al. (2017). The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Molecular Biology, 93(4–5), 389–401. DOI 10.1007/s11103-016-0568-2. [Google Scholar] [CrossRef]
50. Wu, J., Zhu, C., Pang, J., Zhang, X., Yang, C. et al. (2014). OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. Plant Journal, 80(6), 1118–1130. DOI 10.1111/tpj.12714. [Google Scholar] [CrossRef]
51. Xu, F., Tang, J., Gao, S., Cheng, X., Du, L. et al. (2019). Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant Journal, 100(5), 1036–1051. DOI 10.1111/tpj.14501. [Google Scholar] [CrossRef]
52. Li, Z., Tang, L., Qiu, J., Zhang, W., Wang, Y. et al. (2016). Serine carboxypeptidase 46 regulates grain filling and seed germination in rice (Oryza sativa L.). PLoS One, 11(7), e0159737. DOI 10.1371/journal.pone.0159737. [Google Scholar] [CrossRef]
53. Nguyen, T. N., Tuan, P. A., Ayele, B. T. (2022). Jasmonate regulates seed dormancy in wheat via modulating the balance between gibberellin and abscisic acid. Journal of Experimental Botany, 73(8), 2434–2453. DOI 10.1093/jxb/erac041. [Google Scholar] [CrossRef]
54. Yu, X., Han, J., Li, L., Zhang, Q., Yang, G. et al. (2020). Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis. Plant Cell Reporter, 39(5), 635–651. DOI 10.1007/s00299-020-02520-4. [Google Scholar] [CrossRef]
55. Lopez-Molina, L., Mongrand, S., Kinoshita, N., Chua, N. H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Development, 17(3), 410–418. DOI 10.1101/gad.1055803. [Google Scholar] [CrossRef]
56. Feng, Y., Liu, M., Wang, Z., Zhao, X., Han, B. et al. (2019). A 4-bp deletion in the 5′UTR of TaAFP-B is associated with seed dormancy in common wheat (Triticum aestivum L.). BMC Plant Biology, 19(1), 349. DOI 10.1186/s12870-019-1950-4. [Google Scholar] [CrossRef]
57. Zhang, Y., Xia, X., He, Z. (2017). The seed dormancy allele TaSdr-A1a associated with pre-harvest sprouting tolerance is mainly present in Chinese wheat landraces. Theoretical and Applied Genetics, 130(1), 81–89. DOI 10.1007/s00122-016-2793-0. [Google Scholar] [CrossRef]
58. Nakamura, S., Abe, F., Kawahigashi, H., Nakazono, K., Tagiri, A. et al. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell, 23(9), 3215–3229. DOI 10.1105/tpc.111.088492. [Google Scholar] [CrossRef]
59. Zhang, Y., Miao, X., Xia, X., He, Z. (2014). Cloning of seed dormancy genes (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theoretical and Applied Genetics, 127(4), 855–866. DOI 10.1007/s00122-014-2262-6. [Google Scholar] [CrossRef]
60. Ishibashi, Y., Aoki, N., Kasa, S., Sakamoto, M., Kai, K. et al. (2017). The interrelationship between abscisic acid and reactive oxygen species plays a key role in barley seed dormancy and germination. Frontiers in Plant Science, 8(77), 275–284. DOI 10.3389/fpls.2017.00275. [Google Scholar] [CrossRef]
61. Sato, K., Yamane, M., Yamaji, N., Kanamori, H., Tagiri, A. et al. (2016). Alanine aminotransferase controls seed dormancy in barley. Nature Communication, 7(1), 11625–11633. DOI 10.1038/ncomms11625. [Google Scholar] [CrossRef]
62. Nagel, M., Alqudah, A. M., Bailly, M., Rajjou, L., Pistrick, S. et al. (2019). Novel loci and a role for nitric oxide for seed dormancy and preharvest sprouting in barley. Plant Cell Environment, 42(4), 1318–1327. DOI 10.1111/pce.13483. [Google Scholar] [CrossRef]
63. Himi, E., Yamashita, Y., Haruyama, N., Yanagisawa, T., Maekawa, M. et al. (2011). Ant28 gene for proanthocyanidin synthesis encoding the R2R3 MYB domain protein (Hvmyb10) highly affects grain dormancy in barley. Euphytica, 188(1), 141–151. DOI 10.1007/s10681-011-0552-5. [Google Scholar] [CrossRef]
64. Hoang, H. H., Bailly, C., Corbineau, F., Leymarie, J. (2013). Induction of secondary dormancy by hypoxia in barley grains and its hormonal regulation. Journal of Experimental Botany, 64(7), 2017–2025. DOI 10.1093/jxb/ert062. [Google Scholar] [CrossRef]
65. Barrero, J. M., Downie, A. B., Xu, Q., Gubler, F. (2014). A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell, 26(3), 1094–1104. DOI 10.1105/tpc.113.121830. [Google Scholar] [CrossRef]
66. Leymarie, J., Robayo-Romero, M. E., Gendreau, E., Benech-Arnold, R. L., Corbineau, F. (2008). Involvement of ABA in induction of secondary dormancy in barley (Hordeum vulgare L.) seeds. Plant Cell Physiology, 49(12), 1830–1838. DOI 10.1093/pcp/pcn164. [Google Scholar] [CrossRef]
67. Manan, S., Zhao, J. (2021). Role of Glycine max ABSCISIC ACID INSENSITIVE 3 (GmABI3) in lipid biosynthesis and stress tolerance in soybean. Functional Plant Biology, 48(2), 171–179. DOI 10.1071/FP19260. [Google Scholar] [CrossRef]
68. Shuai, H., Meng, Y., Luo, X., Chen, F., Zhou, W. et al. (2017). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Science Reporter, 7(1), 12620–12630. DOI 10.1038/s41598-017-13093-w. [Google Scholar] [CrossRef]
69. Yang, Y., Zheng, C., Chandrasekaran, U., Yu, L., Liu, C. et al. (2020). Identification and bioinformatic analysis of the GmDOG1-like family in soybean and investigation of their expression in response to gibberellic acid and abscisic acid. Plants, 9(8), 937. DOI 10.3390/plants9080937. [Google Scholar] [CrossRef]
70. Niu, X., Helentjaris, T., Bate, N. J. (2002). Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell, 14(10), 2565–2575. DOI 10.1105/tpc.003400. [Google Scholar] [CrossRef]
71. Zhang, Y., Sun, Q., Zhang, C., Hao, G., Wang, C. et al. (2019). Maize VIVIPAROUS1 interacts with ABA INSENSITIVE5 to regulate GALACTINOL SYNTHASE2 expression controlling seed raffinose accumulation. Journal of Agriculture and Food Chemistry, 67(15), 4214–4223. DOI 10.1021/acs.jafc.9b00322. [Google Scholar] [CrossRef]
72. Song, W., Hao, Q., Cai, M., Wang, Y., Zhu, X. et al. (2020). Rice OsBT1 regulates seed dormancy through the glycometabolism pathway. Plant Physiology and Biochemistry, 151, 469–476. DOI 10.1016/j.plaphy.2020.03.055. [Google Scholar] [CrossRef]
73. Zhu, G., Ye, N., Zhang, J. (2009). Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiology, 50(3), 644–651. DOI 10.1093/pcp/pcp022. [Google Scholar] [CrossRef]
74. Mao, X., Zhang, J., Liu, W., Yan, S., Liu, Q. et al. (2019). The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice. Rice, 12(1), 2–15. DOI 10.1186/s12284-018-0260-z. [Google Scholar] [CrossRef]
75. Leymarie, J., Bruneaux, E., Gibot-Leclerc, S., Corbineau, F. (2006). Identification of transcripts potentially involved in barley seed germination and dormancy using cDNA-AFLP. Journal of Experimental Botany, 58(3), 425–437. DOI 10.1093/jxb/erl211. [Google Scholar] [CrossRef]
76. Liu, J., Guo, X., Zhai, T., Shu, A., Zhao, L. et al. (2020). Genome-wide identification and characterization of microRNAs responding to ABA and GA in maize embryos during seed germination. Plant Biology, 22(5), 949–957. DOI 10.1111/plb.13142. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools