 Open Access
Open Access
ARTICLE
VvAGAMOUS Affect Development of Four Different Grape Species Ovary
1 College of Horticulture, Shanxi Agricultural University, Taigu, 030801, China
2 Pomology Institute, Shanxi Agricultural University, Shanxi Academy of Agricultural Sciences, Taigu, 030801, China
* Corresponding Authors: Pengfei Wen. Email: ; Jinjun Liang. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(4), 1125-1138. https://doi.org/10.32604/phyton.2023.026227
Received 25 August 2022; Accepted 19 October 2022; Issue published 06 January 2023
Abstract
Grape pistil has an important influence on fruit size and quality. However, there were few studies on grape ovary, and the development process of the ovary is still unclear. Therefore, in this paper, four different grape varieties with different lengths of small inflorescences, namely ‘Musct Hambourg’ grape (Vitis vinifera), ‘Concord’ grape (Vitis labrusca), ‘ShanPuTao’ grape (Vitis amurensis) and ‘GongNiang2Hao’ grape (Vitis amurensis × Vitis vinifera) were used as test materials. Four varieties ovary were significant differences by means of stereomicroscope, paraffin section. The expression of ovary determining gene VvAGAMOUS (VvAG) and its development related genes VvCRABS CLAW (VvCRC) and VvAGAMOUS-LIKE 11 (VvAGL11) with similar functions during the development of different grape varieties were preliminarily explored using fluorescence quantitative test. The relationship between VvAG and VvCRC, VvAG and VvAGL11 were analyzed using Y1H assay. Our results showed that there were obvious abdominal sutures on the surface of expect for ‘Musct Hambourg’ grape, and existing poly carpels. The ovary development of ‘ShanPuTao’ and ‘GongNiang2Hao’ grape was completed when the inflorescence length was less than 1 cm, while the ‘Concord’ and ‘Musct Hambourg’ grape were fully developed when the length of inflorescence was 3–4 and 4–5 cm, respectively. VvAG and VvCRC began to express in large quantities after the formation of stamen primordia, while VvAGL11 during the forming of ovule primordia. Therefore, VvAG and VvCRC mainly regulated the development of stamens and carpels and also promote the development of ovules, while VvAGL11 major regulated the development of ovules. The promoters of VvCRC and VvAGL11 were bound by VvAG. This study provides an important theoretical basis for further research on the molecular mechanism of grape ovary development.Keywords
Vitis genus (Vitis L.) is a woody vine, that contains multiple grape species, which have various characteristics. V. vinifera grapes are the main cultivars in the world, mostly used for fresh food and vintage, but the cold and disease resistance are poor. V. labrusca grapes are mostly used for rootstocks due to their strong disease resistance. The cold resistance of V. amurensis grapes is the strongest among all the grape varieties, often cultivated in cold regions [1,2]. V. amurensis × V. vinifera grapes are mainly used for wine making, juice making and inherits the characteristics of strong cold resistance of V. amurensis grapes. Grape ovary, as a precursor to fruit, usually forms an internal space in the flower organ by folding and curling, in which the ovule was wrapped for development and finally forms the seed [3–6]. The ovary is divided into monocarpellary ovary and multicarpellary ovary, of which the multicarpellary is divided into li-berate ovary and coalescent ovary. Most grapes are fruits with 2 carpels and 4 ovules, and contain up to 4 seeds, but a few varieties form multiple carpels in the process [7]. Ovary characters affect fruit size and quality [8,9].
At present, the development of ovary had been extensively studied in several model plants, however there was little research in grapes. AGAMOUS (AG) is a class C gene in the ABCDE model of plant flower organs development, which is involved in the third and fourth rounds of flower organs development [10–12]. AG is a transcription regulator that regulates the development of stamens and carpels primordia, participates in the formation of reproductive flower organs and the decisive control of meristems, and is the earliest cloned flower development regulatory gene [13,14]. In tomato, when TAG1 gene was inhibited, stamens of tomato became petal-like organs, and carpels development abnormal [15,16]. During the formation and development of rose stamens, RhAG was significantly and massively expressed. Silencing RhAG could increase the number of rose petals, indicating that RhAG, like AG in Arabidopsis, was mainly involved in stamen development [17].
CRABS CLAW (CRC), a member of YABBY family gene, is one of the regulators, as downstream genes of AG, involved in the carpel development of Arabidopsis [18]. It was expressed in the abaxial side of the carpel and participates in the development of the carpel primordium abaxial tissue and pistil, key developmental processes for ensuring successful plant reproduction and crop production [19,20]. Studies had shown that CRC and SPATULA (SPT) genes have similar functions to class C genes and can promote the development of carpels and ovules [21]. For example, in the process of termination regulation of Arabidopsis flower meristem, AG gene could directly activate the expression of its downstream target gene CRC and participate in pistil formation [22].
AGAMOUS-LIKE 11 (AGL11), also known as SEEDSTICK (STK), is classified as class D according to its gene function. It was highly expressed in the central tissue of seeds and young fruits, and plays a crucial role in ovule development and seed formation [23–26]. AGL11 not only controlled Arabidopsis ovule morphogenesis, but also controlled the development of embryo stalk and seed shedding [27,28]. Silencing SlyAGL11 gene in tomato produced a seedless fruit, which indicated that SlyAGL11 plays a direct and important role in the seed development of fleshy fruits [29]. In the process of studying the molecular mechanism of controlling seedless grapes found VvAGL11 transcription was essential for seed morphogenesis in grapevine during berry development [30]. On the formation process of ovule primordia of ‘Xiangfei’ grape, it was found that VvAG2 and VvAGL11 were involved in the formation of grape ovules [31]. In ‘Muscadine’ grape, VroAGL11 was found that the gene was most expressed in the berries with the highest seed content (weight) and almost not in seedless berries, indicating that VroAGL11 gene played an important role in controlling grape seed morphogenesis [32].
In this study, the inflorescences of four different grape varieties with different lengths were used as test materials. The development process of four grape varieties ovaries were studied by means of stereoscope and paraffin section. At the same time, to clarify the expression of VvAG, VvCRC and VvAGL11, related to carpel and ovule development, during the development of grape ovary, quantitatively were detected by fluorescence. The interactions between VvAG and VvCRC, VvAG and VvAGL11 were further proved by Y1H assay.
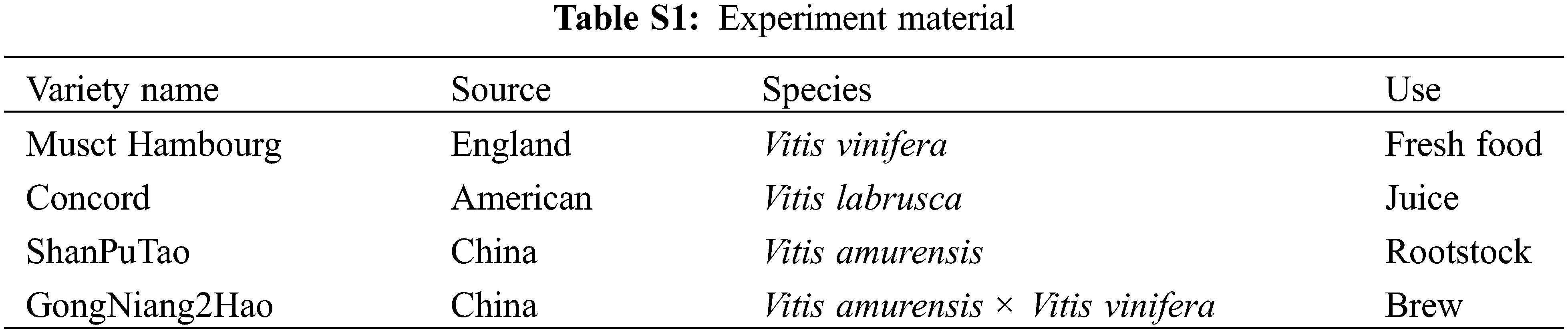
The experimental materials were collected from the national “Grape Germplasm Resource nursery” Pomology Institute, Shanxi Agricultural University/Shanxi Academy of Agricultural Sciences on the morning of May 01, 2021. The resource garden is located at (37 °N, 112 °E) with temperate continental climate and four distinct seasons. The average sunshine duration in this area is 2500~2600 h, and the annual average temperature and precipitation are about 5°C~10°C and 458 mm, respectively. The cultivation mode and growth environment of the four different grape varieties were the same, which would not affect the phenotype. Inflorescences of four different grape varieties, ‘Musct Hambourg’ grape (V. vinifera), ‘Concord’ grape (V. labrusca), ‘ShanPuTao’ grape (V. amurensis) and ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera), were collected (Table S1). Sampling was performed at different developmental stages, according to the length of inflorescences: <1, 1–2, 2–3, 3–4, 4–5 cm (Fig. 1) [33]. Basal position small inflorescences of samples were taken, one of which was immediately frozen in liquid nitrogen and taken back to the laboratory for storage in an ultra-low temperature refrigerator at –80°C; one copy was kept in FAA [50% (v/v) ethanol, 5% (v/v) glacial acetic acid and 5% (v/v) formaldehyde] fixative.
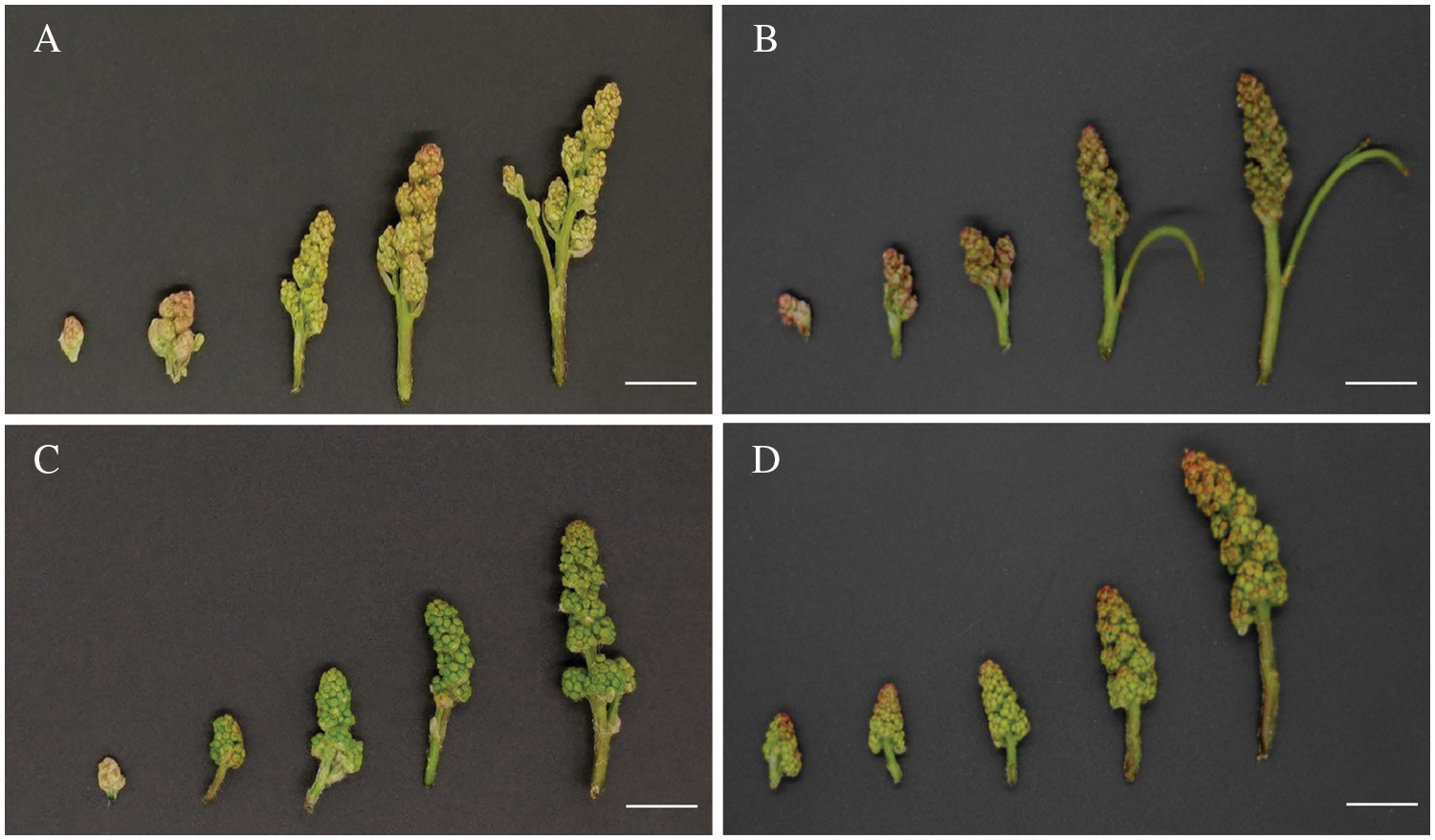
Figure 1: Inflorescences from four grape varieties with different lengths. A. The ‘Musct Hambourg’ grape inflorescences (V. vinifera). B. The ‘Concord’ grape inflorescences (V. labrusca). C. The ‘ShanPuTao’ grape inflorescences (V. amurensis). D. The ‘GongNiang2Hao’ grape inflorescences (V. amurensis × V. vinifera). White bars indicate the scale = 1 cm
At the full flowering stage, 30–50 small fruits of four different grape varieties were selected. The appearance characters were observed by stereomicroscope, and then the young fruits were crosscut to observe and determine the young fruits phenotypic characters, carpels and ovules number.
The florets of basal of inflorescences with four varieties grape with different lengths were selected for paraffin sectioning [34]. Materials were fixed in FAA, dehydrated with different ethanol concentration gradients, then place in a mixture of different proportions ethanol and n-butanol gradients. Afterwards dipped with pure paraffin and embedded in paraffin. The samples were cut into 8 μm sections using a microtome, after which they were stained with Safranin O-Fast Green. Finally, used a microscope to observe and take pictures.
Total RNA was extracted from the florets occupying the basal positions of the inflorescences of four grape varieties with different lengths. RNA was extracted by modified CTAB method [35], the concentration was detected, and the clarity and integrity of band was checked using 1.2% agarose gel electrophoresis. RNA was reverse transcribed into cDNA by using reverse transcription Kit (Takara Bio, Beijing, China). The primers for amplification of the VvAG gene full-length cDNA of the coding region were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). (AGF-TGGGAAGGGGGAAGATCGAG; AGR- TTACACTAATTGAAGAGCTGGTTGG). PCR reaction conditions were: heat denaturation at 94°C for 5 min; 40 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 1.5 min; extension at 72°C for 5 min. The obtained cDNA fragments were recovered with the gel recovery kit (Real-Times Biotechnology Co., Ltd., Beijing, China) and then connected to the pMD-19T vector (Takara Bio, Beijing, China), then transformed into Escherichia coli Trans5α (TransGen Biotech, Beijing, China), plated and cultured, strains were selected, positive clones were identified by PCR, and sent to Sangon Biotech Co., Ltd. (Shanghai, China) sequencing.
2.5 RNA Extraction and qRT-PCR
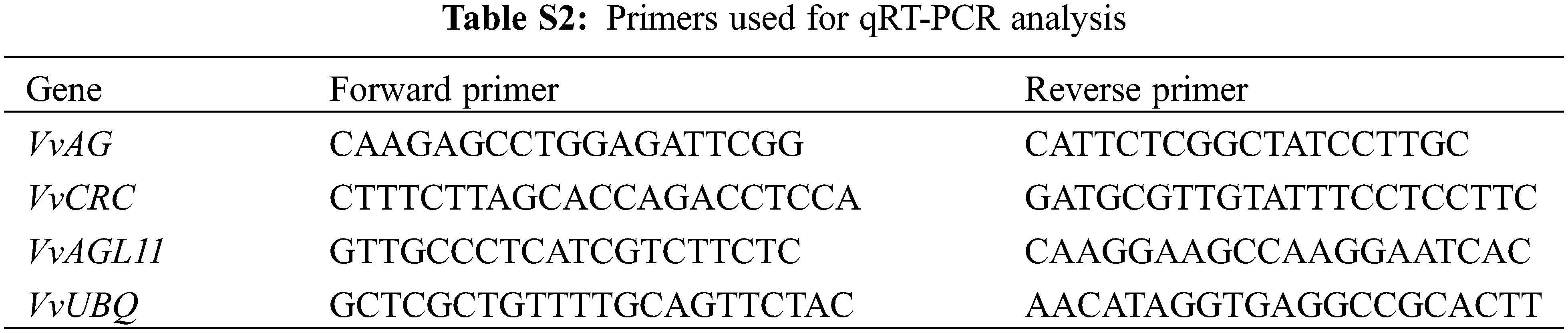
RNA from roots, tendrils, young leaves, mature leaves, flowers and young fruits of ‘Musct Hambourg’ grapes were extracted for tissue-specific analysis. Using cDNA as template, the expression levels of VvAG, VvCRC and VvAGL11 in different grape tissues and the expression levels of four different grape varieties with different inflorescence lengths were detected by qRT-PCR. Real time qRT-PCR reaction was carried out with SYBR Green PCR Master Mix Kit (Mei5 Biotechnology Co., Ltd., Beijing, China). Fluorescent quantitative primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Table S2). 20 μL reaction system: 2× realtime PCR super mix 10 μL + primers 1.0 μL + 1.0 μL sample cDNA + 8.0 μL ddH2O. qRT-PCR reaction conditions: predenaturation at 95°C for 60 s; denaturation at 95°C for 15 s, annealing at 58°C for 15 s, extension at 72°C for 60 s, 40 cycles; 72°C for 5 min. Using VvUBQ as the internal reference gene, the relative expression of the corresponding gene was calculated by 2−∆∆CT method. Each sample was repeated 3 times.
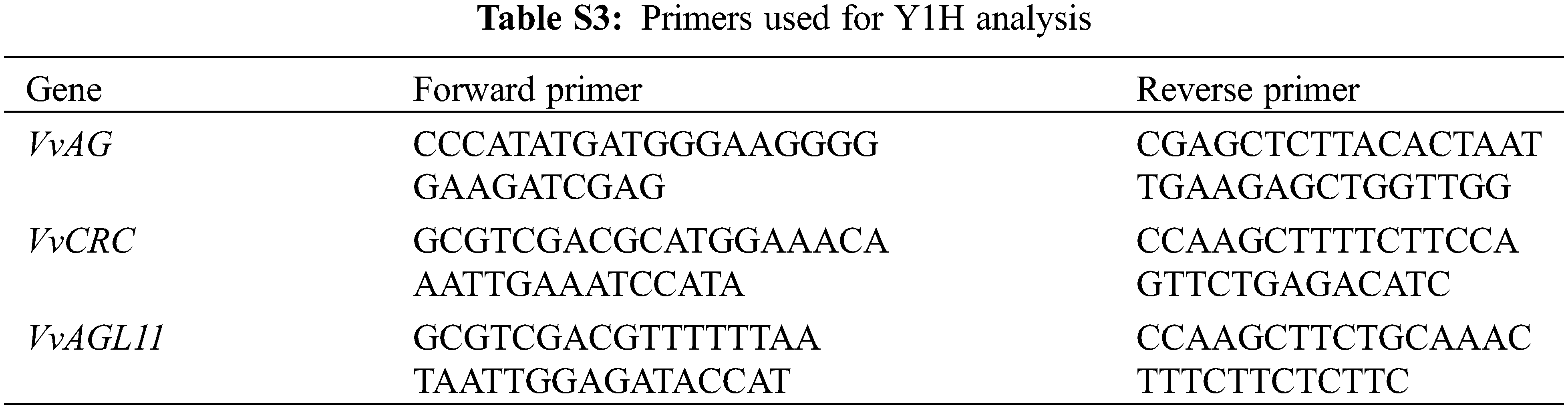
The VvAG-coding sequences were cloned into the pGADT7 vector. The cis-acting elements of promoter VvCRC and VvAGL11 were predicted according to PlantCARE online website. The fragments containing CArG-box elements were inserted into pAbAi vector to construct pAbAi-VvCRC and pAbAi-VvAGL11 vectors. The mutation-related vectors pAbAi-Vvcrc and pAbAi-Vvagl11 were constructed by Sangon Biotech Co., Ltd. (Shanghai, China). The resultant constructs were transformed into the yeast Y1H Gold strain and selected by optimal AbA (Aureobasidin A) concentration on the SD/-LEU (Synthetic Dropout Medium/-Leucine) medium. The pGADT7-VvAG + pAbAi-VvCRC, pGADT7-VvAG + pAbAi-VvAGL11 were experimental group and pGADT7-VvAG + pAbAi-Vvcrc, pGADT7-VvAG + pAbAi-Vvagl11 were negative control (Table S3) [36].
3.1 Differences of Ovary Morphology among Four Grape Varieties
The young fruits of four grape varieties were observed and photographed under the stereomicroscope. It was found that the ‘Musct Hambourg’ grape had different appearances compared with the ‘Concord’, ‘ShanPuTao’ and the ‘GongNiang2Hao’. The ‘Musct Hambourg’ grape fruits had an irregular appearance, with obvious abdominal sutures on the surface, and the more abdominal sutures, the more carpels and ovules. As shown in Fig. 2 A1, A3 and A5, there were two, three and four abdominal sutures, respectively. The observation of the corresponding young fruits crosscut were shown in Fig. 2 A2, A4 and A6, in which are two carpels four ovules, three carpels six ovules and four carpels eight ovules, respectively. The remaining three grape varieties are round or oval in appearance, with smooth pericarp surface and no abdominal sutures. Two ovaries were observed after cross cutting, and each ovary contains two ovules.
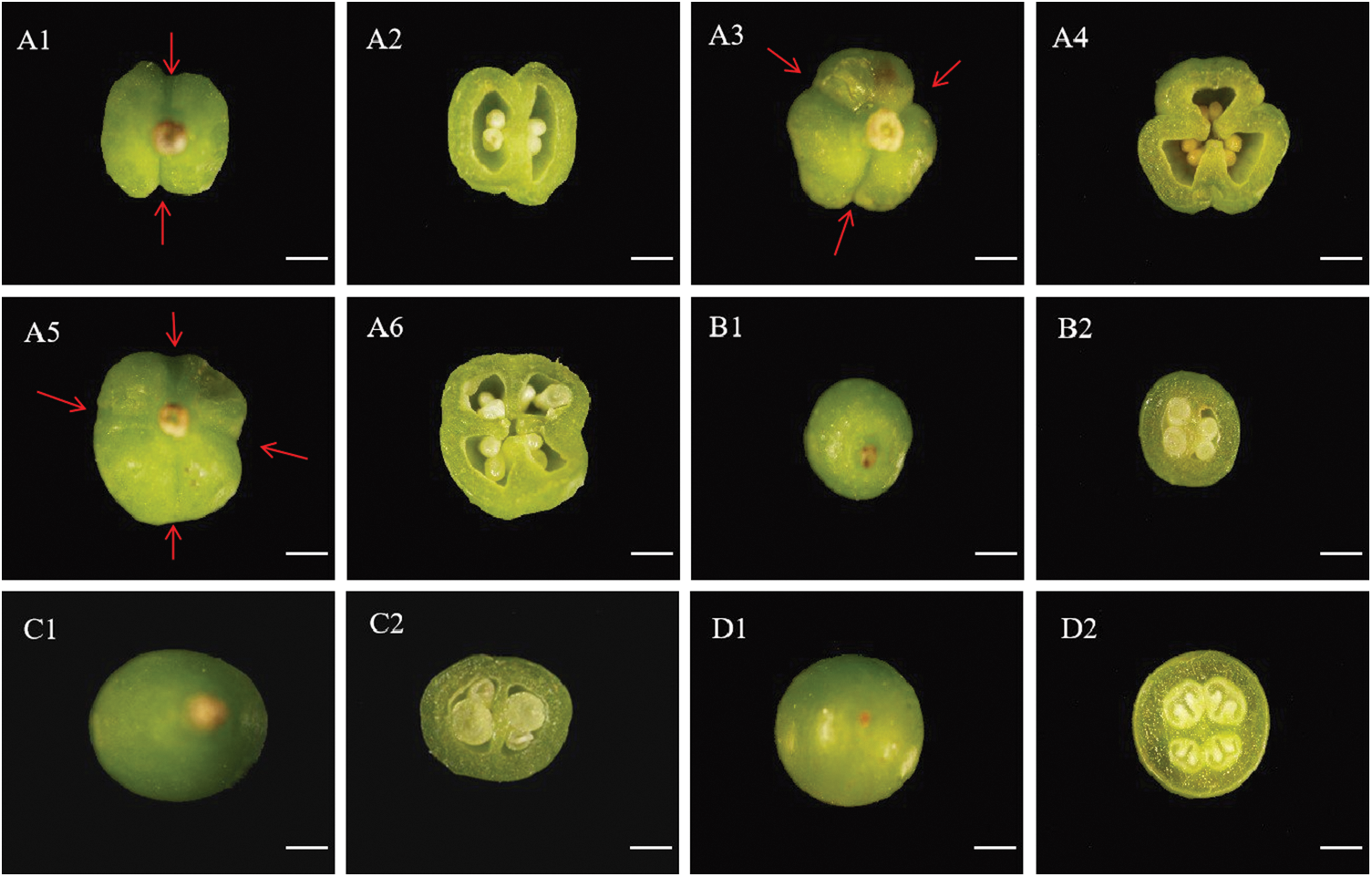
Figure 2: Observation on young fruits appearance and ovary morphology of four grape varieties. A1 and A2. 2 abdominal sutures and 2 carpels of ‘Musct Hambourg’ grape (V. vinifera). A3 and A4. 3 abdominal sutures and 3 carpels of ‘Musct Hambourg’ grape (V. vinifera). A5 and A6. 4 abdominal sutures and 4 carpels of ‘Musct Hambourg’ grape (V. vinifera). B1 and B2. 2 abdominal sutures and 2 carpels of ‘Concord’ grape (V. labrusca). C1 and C2. 2 abdominal sutures and 2 carpels of ‘ShanPuTao’ grape (V. amurensis). D1 and D2. 2 abdominal sutures and 2 carpels of ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera). Red arrow indicates abdominal suture. White bars indicate the scale = 100 μm
3.2 Anatomical Observation on Florets of Four Grape Varieties at Different Stages
To determine the key period of grape ovary formation, four grape varieties inflorescences with different lengths were observed through paraffin sections (Fig. 3). The flower development of ‘Musct Hambourg’ grape (V. vinifera) was the slowest. At the early stage of inflorescence development, when the inflorescence length was less than 1 cm, the sepals and flower caps had been fully developed and the stamen primordia began to form (Fig. 3 A1). When the inflorescence length was 1–2 cm, the stamens were completed development and the carpel primordia began to form (Fig. 3 A2). In inflorescences 2–3 cm in length, stamens had entered the stage of sporogenous tissue differentiation, and the two carpel primordia were crescent shaped and high protuberant (Fig. 3 A3). The carpel primordia continued to grow and the ovule primordia began to form in the 3–4 cm inflorescences (Fig. 3 A4). Ovule primordia further differentiated when the inflorescences were 4–5 cm long (Fig. 3 A5).
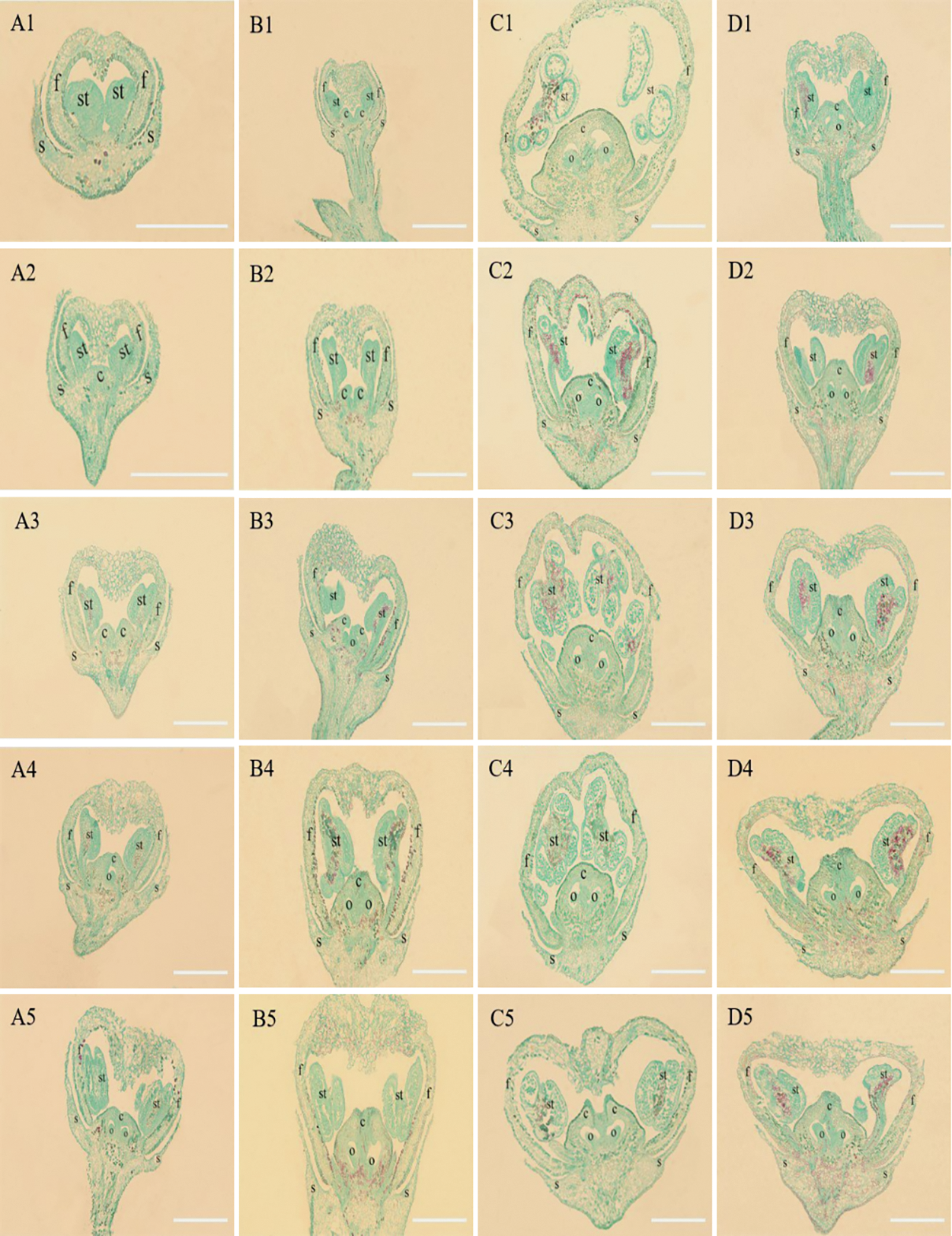
Figure 3: Anatomical structure of four grape varieties at different developmental stages. A1–A5. The ‘Musct Hambourg’ grape (V. vinifera). B1–B5. The ‘Concord’ grape (V. labrusca). C1–C5. The ‘ShanPuTao’ grape (V. amurensis). D1–D5. The ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera). 1. < 1 cm; 2. 1–2 cm; 3. 2–3 cm; 4. 3–4 cm; 5. 4–5 cm. s. sepals; f. flower caps; st. stamens; c. carpels; o. ovules. White bars indicate the scale = 100 μm
At the early stage of inflorescences development, the ‘Concord’ grape (V. labrusca) formed sepals, flower caps, stamens and the two carpel primordia began to form (Fig. 3 B1). Carpel primordia was further developed, extending around and widening at the bottom, when the inflorescences were 1–2 cm long. The two symmetrical segments form the carpel marginal primary meristem, and the stamens undergo protoplast differentiation (Fig. 3 B2). The primary meristems on both sides were close to the center, and the carpels gradually fused in the 2–3 cm inflorescences, and the ovule primordia began to form (Fig. 3 B3). In inflorescences 3–4 cm in length, the ovule primordia further differentiated, carpels fully fused, and stigma cells began to develop (Fig. 3 B4). The stigma form and the ovules entered the inversion stage, when the inflorescences were 4–5 cm long (Fig. 3 B5).
The flower development of ‘ShanPuTao’ grape (V. amurensis) was the fastest, the carpels and ovules both fully development in inflorescence lengths less than 1 cm (Fig. 3 C1).
The sepals, flower caps, stamens and carpels of ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera) were formed when the inflorescences length was 0–1 cm, in which the ovule primordia began to form (Fig. 3 D1). Ovules were completed development in the 1–2 cm inflorescences (Fig. 3 D2). In inflorescences 2–3 cm in length, the ovules entered the inversion stage and the stigma cells began to develop (Fig. 3 D3). The stigma had formed, in the small florets of the 3–4 cm inflorescences. (Fig. 3 D4).
3.3 Sequence Analysis of VvAG Gene
The results showed that bands with a molecular size of 680 bp were amplified, respectively, which were consistent with the sequence size published by NCBI. There was only one amino acid difference in the non-conserved domain of the VvAG genes of the four grape varieties, which will not affect the function of the gene. The encoded protein contains a MADS domain of 57 amino acids, a K domain of 67 amino acids, and a C domain of 75 oxyacids (AGI and AGII). The M region is highly conserved, K regions are relatively conserved, and the C-terminal specificity is strong. They had highly similar to the amino acid sequences of Arabidopsis, Nicotiana and Malus domestic (Fig. 4).
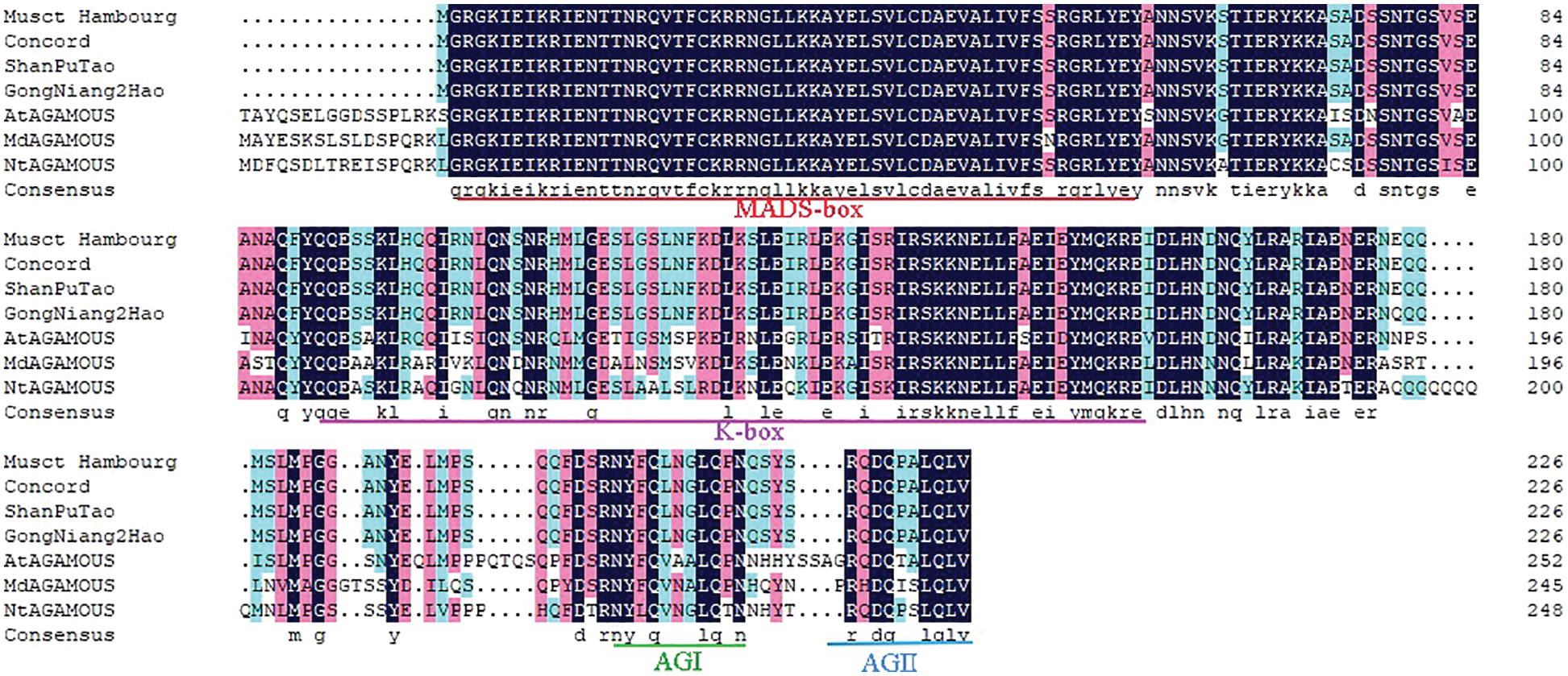
Figure 4: Analysis of VvAG gene amino acid sequences of four different grape varieties and other representative species
3.4 VvAG, VvCRC and VvAGL11 Tissue Specificity
The expression of VvAG, VvCRC and VvAGL11 in different tissues of grapes was detected with qRT-PCR (Fig. 5). It was found that the expression of VvAG and VvCRC was the highest in flowers, followed by young fruits, a small amount of expression in young leaves and mature leaves, and almost no expression in stems and tendrils. VvAGL11 was highly expressed in young fruits, followed by flowers, but not in stems, tendrils, young leaves and mature leaves.
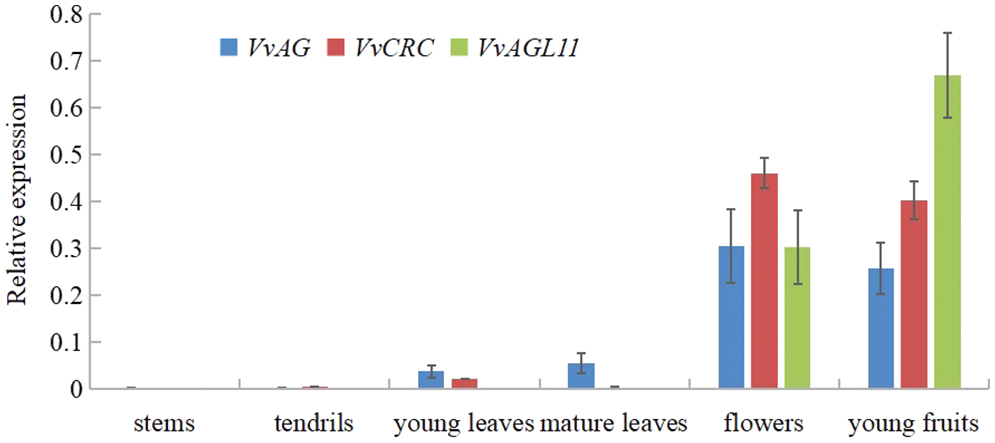
Figure 5: The relative expression levels of VvAG, VvCRC and VvAGL11 in different grape tissues
3.5 Expression Differences of VvAG, VvCRC and VvAGL11 in Florets at Different Developmental Stages
The expression levels of VvAG, VvCRC and VvAGL11 genes were measured at different inflorescences length of grapes in different varieties (Fig. 6). VvAG was expressed when the ‘Musct Hambourg’ grape inflorescence length was 0–1 cm, and the stamens forming. In inflorescences 1–2 and 2–3 cm in length, VvAG began to express in large quantities, and the carpel primordia began to form and rapidly formation, respectively. The expression of VvCRC gene increased followed the increase of VvAG gene expression. The expression of VvAGL11 gene was low between inflorescences 0–1 and 1–2 cm in length. However, the expression of VvAGL11 gene was increased quickly in inflorescences length 2–3 and 3–4 cm, in which ovule primordia began to form and gradually developed, respectively (Fig. 6A).
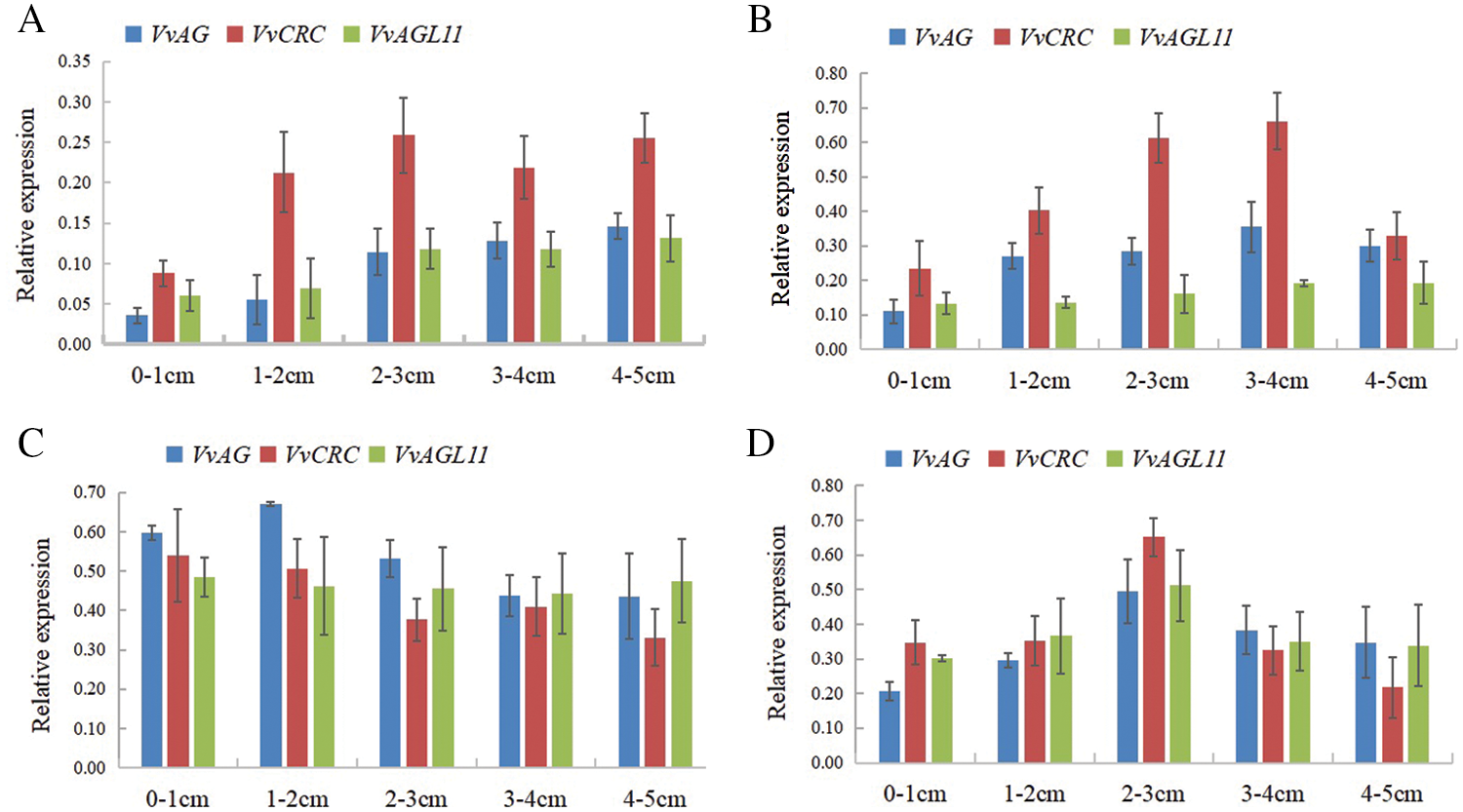
Figure 6: The relative expression levels of VvAG, VvCRC, VvAGL11 at different inflorescence developmental stages. A. The ‘Musct Hambourg’ grape (V. vinifera). B. The ‘Concord’ grape (V. labrusca). C. The ‘ShanPuTao’ grape (V. amurensis). D. The ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera)
The expression of VvAG and VvCRC genes were increasing in ‘Concord’ grape (V. labrusca) inflorescences 0–1 cm length, which the formation of carpel primordia. The carpel was completely fused, and the expression reached the highest level, when the inflorescences were 3–4 cm long. The expression of VvAGL11 was low in inflorescences 0–1 and 1–2 cm in length, slightly increased in inflorescences 2–3 cm long, in which the ovule primordia began to form. The expression increased significantly in inflorescences 3–4 cm in length, in which the ovule primordia began to develop (Fig. 6B).
The expression of VvAG gene was reached the highest when the ‘ShanPuTao’ grape (V. amurensis) inflorescence length was 1–2 cm. The expression of VvCRC and VvAGL11 reached the maximum in the 0–1 cm inflorescence, in which the carpels and ovules were fully developed (Fig. 6C).
The expression of VvAG, VvCRC and VvAGL11 genes in the inflorescence of ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera) reached the highest when the inflorescence were 2–3 cm long. During this period, the carpels and ovules were fully developed. After that, the expression began to decline, and the genes regulating the development of subsequent flower organs began to express (Fig. 6D).
3.6 Interaction of VvAG with VvCRC and VvAGL11 Promoters
Specific binding sites (CArG-boxes) of VvAG were found in promoters of VvCRC and VvAGL11. Protein-DNA interactions were examined with Y1H assays. Our data showed that both the empty vector and mutant control successfully inhibited the growth on SD/−leu+50 ng AbA plate, while the experimental group grew on SD/−leu+100 ng AbA plate, indicating that VvAG can directly bind to the CArG-boxes acting elements of VvCRC and VvAGL11 promoters (Fig. 7).
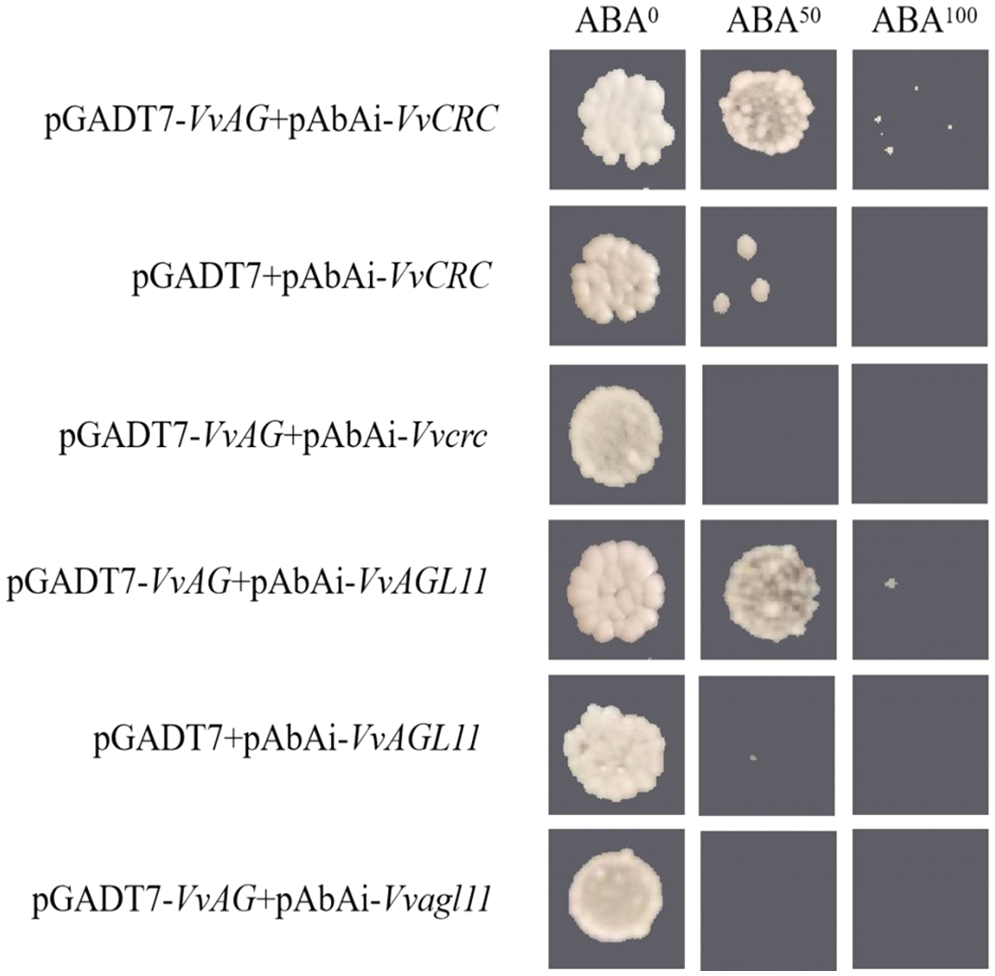
Figure 7: The interaction between VvAG and the promoters of VvCRC and VvAGL11 was analyzed by Y1H assay
4.1 Ovary Phenotypic Differences of Four Grape Varieties from Different Species
The formation and development of ovaries are very important to the number of grape seeds, fruit size and quality. However, there are few studies on grape ovaries, mainly focusing on V. vinifera species. In this study, four grapes varieties from different species were used to provide a theoretical basis for the follow-up studies. The experiment found that the ‘Musct Hambourg’ grape (V. vinifera) had multiple carpels, while the other three grape varieties only had two carpels (Fig. 2). V. vinifera species had abdominal sutures obvious, while that of other species is not. A researcher had investigated the carpel number of seven different grape cultivars for three consecutive years and found that six grape cultivars have two carpels or little three carpels. Only the ‘Xiangfei’ grape (V. vinifrea) has a high ratio of multi carpels, and its multi carpel ratio is stable at more than 50% annually [33]. Therefore, the polycarpels grape could be selected to cultivate large fruit grains grape varieties, but there is no in-depth study on this aspect.
4.2 Ovary Formation of Four Grape Varieties from Different Species
This study found that ‘ShanPuTao’ had the fastest ovary development, then ‘GongNiang2Hao’ also had a faster ovary development, next was ‘Concord’, and ‘Musct Hambourg’ was the slowest. Grapes need to be buried into the soil to spend the cold winter in northern of China. They will not be dug out again until the weather warms up around April of the next year. However, ‘ShanPuTao’ and ‘GongNiang2Hao’ grapes need not to be buried into the soil due to their strong cold resistance. Therefore, they have begun to carry out physiological activities to form flower primordial when other grapes are still in dormancy. That is why the rapid ovary development of V. amurensis and V. amurensis × V. vinifera grapes.
The carpels and ovules of the ‘Musct Hambourg’ were developed in inflorescences 3–4 and 4–5 cm in length, respectively (Fig. 3 A4 A5). Inflorescences of different lengths of ‘Xiangfei’ (V. vinifrea) were observed by paraffin slices and found that the carpels were fully fused when the inflorescences were 4–5 cm long, in which the ovule primordia were begun to form [33]. It showed that the ovary fully developed in the 4–5 cm long inflorescence of V. vinifrea grapes, and slightly differences may be existed in different varieties.
There were little studies on ovary development of V. labrusca grapes. When the inflorescences of the ‘Concord’ were 2–3 and 3–4 cm length, the carpels and ovules were fully developed (Fig. 3 B3 B4). Its ovaries developed faster than V. vinifrea.
4.3 VvAG, VvCRC and VvAGL11 Participate in Grape Ovary Development
Some studies had found that the expression of VvAG2 was the highest in flowers, instead low in roots, mature leaves and young leaves [31]. There were some studies indicated that VviAGL11 gene highly expressed in flower and fruit tissues of grapes, but inhibited in roots, branches, leaves, buds and tendrils [26]. These results consistent with the tissue-specific expression test of VvAG and VvAGL11 genes in this study. VvCRC had been rarely studied on grapes, but CRC was mainly expressed at the distal axis of the carpel and nectary in Arabidopsis [19]. This result was consistent with the expression of VvCRC in this study.
At the same time, the fluorescence quantitative results of ‘Musct Hambourg’ showed that VvAG gene began to express in large quantities during the inflorescences 2–3 cm length, in which carpel primordia began to develop (Fig. 5 A). The expression signal of VvAG gene had been found in 1–2 cm stamen primordium using in situ hybridization, the signal could still be observed when the inflorescences were 5–6 cm long [33]. Similarly, other study also found that VvAG2 was expressed on carpel and ovule primordia in 4–5 cm inflorescences, which had strong hybridization signal on the ovule primordia [31]. Therefore, VvAG not only regulates carpel development, but also ovule development.
In this study, we found that VvAGL11 began to express abundantly from the development of ovule primordia. A weak VvAGL11 gene signal could be observed in the ovule primordia in the 4–5 cm inflorescences at the stage of ovule primordia formation through in situ hybridization [31]. Reduced expression of VvAGL11 is responsible for stenospermocarpic seedlessness in bunch grapes [32]. The VroAGL11 gene controlled the seed morphogenesis after ovule formation in ‘Muscadine’ grapes by analyzed its divergence from other plants of molecular level [32]. VvAGL11 transcripts exhibited a high accumulation in seeds after 2 and 4 weeks of development in ‘Chardonnay’ using in situ hybridization [30]. Therefore, VvAGL11 might be a major gene for regulating ovule development.
In Arabidopsis, CRC gene is essential for carpel and nectary development [37]. PfCRC was found that it was mainly expressed in the carpel, and the loss of PfCRC function would change the determination of carpel meristem, carpel closure and ovule number [38]. The strong expression of EcCRC was detected at the flower development stage after the beginning of pistil using in situ hybridization. VIGS test proved that EcCRC gene participated in the initiation of ovule [39]. This is consistent with the results that VvCRC gene mainly regulated pistil development. In tomato, in situ hybridization analysis of flowers at different stages found that SICRCa began to accumulate uniformly when the carpel primordia was initiated, and persisted in carpel growth and primordium of placenta emerged [40].
4.4 The Interactions between VvAG and VvCRC, VvAG and VvAGL11
We observed that VvAG can directly activate VvCRC and VvAGL11 promoters by Y1H assay. There had less research on the interaction of VvAG and VvCRC in grapes. But Some studies found that the grape MADS-box family transcription factor complexes VvAG2/VvSEP3 and VvAGL11/VvSEP3 form tetramers that may be involved in the formation of ovules [31].
This study found that the multi carpel existed in ‘Musct Hambourg’ grape (V. vinifera), instead, other varieties not. The speed of ovary development was ‘ShanPuTao’ grape (V. amurensis) > ‘GongNiang2Hao’ grape (V. amurensis × V. vinifera) > ‘Concord’ grape (V. labrusca) > ‘Musct Hambourg’ grape (V. vinifera). VvAG and VvCRC genes mainly regulated the development of stamens and carpels, but also promoted the development of ovules, while VvAGL11 might be a major gene for regulating ovule development. The CArG-boxes acting elements of VvCRC and VvAGL11 promoters were bound by VvAG.
Authorship: Study conception and design: Jinjun Liang, Pengfei Wen and Tiequan Niu; data collection: Yuqin Zhang, Qifeng Zhao; analysis and interpretation of results: Pengfei Zhang and Yuqin Zhang; draft manuscript preparation: Yuqin Zhang and Pengfei Zhang. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This work was financially supported from the Shanxi Youth Science Research Project (Grant No. 20210302124067), the Shanxi Agricultural University Doctoral Research Startup Project (Grant No. 2021BQ32), the Shanxi Provincial Doctoral Graduates and Postdoctoral Researchers Working in Shanxi Reward Fund Research Project (Grant No. SXBYKY2021075), the National Youth Science Foundation Project (Grant No. 32202456), and the earmarked fund for Modern Agro-Industry Technology Research System.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Liu, L. Y., Hua, L. (2013). Review: Research progress in amur grape, Vitis amurensis Rupr. Canadian Journal of Plant Science, 93(4), 565–575. DOI 10.1023/A:1025001402838. [Google Scholar] [CrossRef]
2. Zhao, Q., Duan, C. Q., Wang, J. (2010). Anthocyanins profifile of grape berries of Vitis amurensis, its hybrids and their wines. International Journal of Molecular Sciences, 11(5), 2212–2228. DOI 10.3390/ijms11052212. [Google Scholar] [CrossRef]
3. Smyth, D. R., Bowman, J. L., Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell, 2(8), 755–767. DOI 10.1105/tpc.2.8.755. [Google Scholar] [CrossRef]
4. Endress, P. K., Doyle, J. A. (2009). Reconstructing the ancestral flower and its initial specializations. American Journal of Botany, 96(1), 22–66. DOI 10.3732/ajb.0800047. [Google Scholar] [CrossRef]
5. Doyle, J. A., Endress, P. K. (2000). Morphological phylogenetic analysis of basal angiosperms: Comparison and combination with molecular data. International Journal of Plant Sciences, 161(S6), S121–S153. DOI 10.1086/317578. [Google Scholar] [CrossRef]
6. Endress, P. K. (2015). Patterns of angiospermy development before carpel sealing across living angiosperms: Diversity, and morphological and systematic aspects. Botanical Journal of the Linnean Society, 178(4), 556–591. DOI 10.1111/boj.12294. [Google Scholar] [CrossRef]
7. Hu, J. F., He, H. Y., Leng, P. (2004). Carpel morphology in ‘Kyoho’ grapevine. Acta Horticulturae Sinica, 31(2), 155–159. [Google Scholar]
8. Ferrándiz, C., Fourquin, C., Prunet, N., Scutt, C. P., Vialette-Guiraud, A. C. M. (2010). Carpel development. Advances in Botanical Research, 55(402), 1–73. DOI 10.1016/B978-0-12-380868-4.00001-6. [Google Scholar] [CrossRef]
9. Xing, J. Y., Lu, L., Wen, T. J., Hu, J. F. (2016). The correlation between expression and the ontogeny of tricarpellate fruit in ‘Xiangfei’ grapevine. Journal of Horticultural Science & Biotechnology, 91(3), 308–315. DOI 10.1080/14620316.2016.1157006. [Google Scholar] [CrossRef]
10. Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature, 353(6339), 31–37. DOI 10.1038/353031a0. [Google Scholar] [CrossRef]
11. Günter, T., Heinz, S. (2001). Plant biology floral quartets. Nature, 409(6819), 469–471. DOI 10.1038/35054172. [Google Scholar] [CrossRef]
12. Smaczniak, C., Immink, R., Angenent, G. C., Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development, 139(17), 3081–3098. DOI 10.1242/dev.074674. [Google Scholar] [CrossRef]
13. Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A. et al. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature, 346(6279), 35–39. DOI 10.1038/346035a0. [Google Scholar] [CrossRef]
14. ÓMaoiléidigh, D. S., Wuest, S. E., Rae, L., Raganelli, A., Ryan, P. T. et al. (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell, 25(7), 2482–2503. DOI 10.1105/tpc.113.113209. [Google Scholar] [CrossRef]
15. Pnueli, L., Hareven, D., Rounsley, S. D., Lifschitz, Y. E. (1994). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. The Plant Cell Online, 6(2), 163–173. DOI 10.1105/tpc.6.2.163. [Google Scholar] [CrossRef]
16. Busi, M. V., Bustamante, C., D’Angelo, C., Hidalgo-Cuevas, M., Zabaleta, E. (2003). MADS-box genes expressed during tomato seed and fruit development. Plant Molecular Biology, 52(4), 801–815. DOI 10.1023/A:1025001402838. [Google Scholar] [CrossRef]
17. Ma, N., Chen, W., Fan, T. G., Tian, Y. R., Zhang, S. et al. (2015). Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biology, 15(1), 237. DOI 10.1186/s12870-015-0623-1. [Google Scholar] [CrossRef]
18. Gross, T., Broholm, S., Becker, A. (2018). CRABS CLAW acts as a bifunctional transcription factor in flower development. Frontiers Plant Science, 9, 835. DOI 10.3389/fpls.2018.00835. [Google Scholar] [CrossRef]
19. Gomez-Mena, C., Foher, S., Costa, M. M., Angenent, G. C., Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development, 132(3), 429–438. DOI 10.1242/dev.01600. [Google Scholar] [CrossRef]
20. Ng, K. H., Yu, H., Ito, T. (2009). AGAMOUS Controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biology, 7(11), 1000251. DOI 10.1371/journal.pbio.1000251. [Google Scholar] [CrossRef]
21. Breuil-Broyer, S., Trehin, C., Morel, P., Boltz, V., Sun, B. et al. (2016). Analysis of the arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male-female boundary, flower meristem termination and carpel compartmentalization. Annals of Botany, 117(5), 905–923. DOI 10.1093/aob/mcw023. [Google Scholar] [CrossRef]
22. Wei, M. M., Zeng, X., An, Z. W., Hu, Y. S., Huang, X. et al. (2020). Advances in the maintenance and termination of floral meristem regulated by c-type floral organ gene AGAMOUS (AG). Biotechnology Bulletin, 36(1), 9. [Google Scholar]
23. Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L. et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell, 15(11), 2603–2611. DOI 10.1105/tpc.015123. [Google Scholar] [CrossRef]
24. Battaglia, R., Brambilla, V., Colombo, L. (2008). Morphological analysis of female gametophyte development in the bel1 stk shp1 shp2 mutant. Giornale Botanico Italiano, 142(3), 643–649. DOI 10.1080/11263500802411098. [Google Scholar] [CrossRef]
25. Chen, Y. Y., Lee, P. F., Hsiao, Y. Y., Wu, W. L., Pan, Z. J. et al. (2012). C- and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Applied Surface Science, 53(6), 1053–1067. DOI 10.1093/pcp/pcs048. [Google Scholar] [CrossRef]
26. Díaz-Riquelme, J., Grimplet, J., Martínez-Zapater, J. M., Carmona, M. J. (2012). Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biology, 12(1), 181. DOI 10.1186/1471-2229-12-181. [Google Scholar] [CrossRef]
27. Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E. et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature, 424(6944), 85–88. DOI 10.1038/nature01741. [Google Scholar] [CrossRef]
28. Lu, H., Klocko, A. L., Brunner, A. M., Ma, C., Magnuson, A. C. et al. (2019). RNA interference suppression of AGAMOUS and SEEDSTICK alters floral organ identity and impairs floral organ determinacy, ovule differentiation, and seed-hair development in Populus. New Phytologist, 222(2), 923–937. DOI 10.1111/nph.15648. [Google Scholar] [CrossRef]
29. Ocarez, N., Mejía, N. (2016). Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits. Plant Cell Reports, 35(1), 239–254. DOI 10.1007/s00299-015-1882-x. [Google Scholar] [CrossRef]
30. Malabarba, J., Buffon, V., Mariath, J., Gaeta, M. L., Dornelas, M. C. et al. (2017). The MADS-box gene Agamous-like 11 is essential for seed morphogenesis in grapevine. Journal of Experimental Botany, 68(7), 1493–1506. DOI 10.1093/jxb/erx025. [Google Scholar] [CrossRef]
31. Wang, Y., Liu, Z. H., Wu, J., Hong, L., Liang, J. J. et al. (2021). MADS-box protein complex VvAG2, VvSEP3 and VvAGL11 regulates the formation of ovules in Vitis vinifera L. cv. ‘Xiangfei’. Genes, 12(5), 647. DOI 10.3390/genes12050647. [Google Scholar] [CrossRef]
32. Rahman, M. A., Balasubramani, S. P., Basha, S. M. (2021). Molecular characterization and phylogenetic analysis of MADS-box gene VroAGL11 associated with stenospermocarpic seedlessness in muscadine grapes. Genes, 12(2), 232. DOI 10.3390/genes12020232. [Google Scholar] [CrossRef]
33. Liang, J. J., Guan, P. Y., Liu, Z. H., Wang, Y., Xing, J. Y. et al. (2020). The VvSUPERMAN-like gene is differentially expressed between bicarpellate and tricarpellate florets of Vitis vinifera L. Cv. ‘Xiangfei’ and its heterologous expression reduces carpel number in tomato. Plant and Cell Physiology, 61(10), 1760–1774. DOI 10.1093/pcp/pcaa103. [Google Scholar] [CrossRef]
34. Lu, L. (2016). The study on the molecular mechanism of gibberellin-induced parthenocarpy and improved fruit set in grapevine (Ph.D. Thesis). China Agricultural University, China. [Google Scholar]
35. Yang, B., Liu, H. X., Niu, T. Q., Zhang, P. F., Liang, C. M. et al. (2021). Transient expression analysis of VvANR gene in grape leaves mediated by TRV. Journal of Nuclear Agricultural Science, 35(4), 826–836. [Google Scholar]
36. Niu, T. Q., Dong, Y. M., Liu, H. X., Zhang, X. J., Gao, Y. et al. (2017). The regulations of the MYBA1 in UFGT and DFR from the grape berries. Scientia Agricultura Sinica, 51(12), 2368–2377. [Google Scholar]
37. Lee, J. Y., Baum, S. F., Oh, S. H., Jiang, C. Z., Chen, J. C. et al. (2005). Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development, 132(22), 5021–5032. DOI 10.1242/dev.02067. [Google Scholar] [CrossRef]
38. Gong, P., Song, C., Liu, H., Li, P., Zhang, M. et al. (2021). Physalis floridana CRABS CLAW mediates neofunctionalization of GLOBOSA genes in carpel development. Journal of Experimental Botany, 72(20), 6882–6903. DOI 10.1093/jxb/erab309. [Google Scholar] [CrossRef]
39. Orashakova, S., Lange, M., Lange, S., Wege, S., Becker, A. (2009). The CRABS CLAW ortholog from California poppy (Eschscholzia californica, PapaveraceaeEcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant Journal, 58(4), 682–693. DOI 10.1111/j.1365-313X.2009.03807.x. [Google Scholar] [CrossRef]
40. Castañeda, L., Giménez, E., Pineda, B., García-Sogo, B., Ortiz-Atienza1, A. et al. (2021). Tomato CRABS CLAW paralogues interact with chromatin remodelling factors to mediate carpel development and floral determinacy. New Phytologist, 234(3), 1059–1074. DOI 10.1101/2021.08.19.456989. [Google Scholar] [CrossRef]
Appendix
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools