 Open Access
Open Access
ARTICLE
Identification and Evaluation of Insect and Disease Resistance in Transgenic Cry1Ab13-1 and NPR1 Maize
Joint Laboratory of International Cooperation in Modern Agriculture Technology of Ministry of Education, Plant Biotechnology Center, Jilin Agricultural University, Changchun, China
* Corresponding Author: Piwu Wang. Email:
(This article belongs to the Special Issue: Abiotic and Biotic Stress Tolerance in Crop)
Phyton-International Journal of Experimental Botany 2023, 92(4), 1257-1274. https://doi.org/10.32604/phyton.2023.025918
Received 05 August 2022; Accepted 21 October 2022; Issue published 06 January 2023
Abstract
PCR detection, quantitative real-time PCR (q-RTPCR), outdoor insect resistance, and disease resistance identification were carried out for the detection of genetic stability and disease resistance through generations (T2, T3, and T4) in transgenic maize germplasms (S3002 and 349) containing the bivalent genes (insect resistance gene Cry1Ab13-1 and disease resistance gene NPR1) and their corresponding wild type. Results indicated that the target genes Cry1Ab13-1 and NPR1 were successfully transferred into both germplasms through tested generations; q-PCR confirmed the expression of Cry1Ab13-1 and NPR1 genes in roots, stems, and leaves of tested maize plants. In addition, S3002 and 349 bivalent gene-transformed lines exhibited resistance to large leaf spots and corn borer in the field evaluation compared to the wild type. Our study confirmed that Cry1Ab13-1 and NPR1 bivalent genes enhanced the resistance against maize borer and large leaf spot disease and can stably inherit. These findings could be exploited for improving other cultivated maize varieties.Keywords
As a food crop, maize (Zea mays) is crucial for the food security of the world, since it is a source of feed and energy [1,2]. Moreover, it has significantly increased the access of humankind to food. During the period from jointing to heading, if the rainfall is concentrated, the field humidity is high, and the temperature is suitable, it can cause an epidemic of diseases and insect pests, which will severely affect the yield and quality of corn. This will restrict the healthy development of the corn industry.
Corn borers belong to the order Lepidoptera, family Moth. There are mainly two kinds of corn border of Asian corn borer Ostrinia furnacalis (Guenée) and European corn borer Ostrinia nubilalis (Hubner) in the world. The Asian corn borer is widely distributed in the world, mainly in Southeast Asia, East Asia, India, Northeast Asia, Australia, and the Western Pacific Ocean; while the European corn borer is mainly distributed in Europe, North America and Northern Africa, and so on [3]. Spring maize in Northern China is mainly affected by the Asian corn borer. The surface of the earth has warmed by 0.74°C on average during the past 100 years as a result of the greenhouse effect [4]. High temperature prolongs the growth period of insects. With the continuous increase of temperature, the temperature is positively correlated with the growth and development of insects. However, after the temperature exceeds the economic threshold, the temperature gets negatively correlated with the growth and development of insects [5,6]. The risk of harm to the Asian corn borer has significantly increased as a result of the rise in effective accumulated temperature brought on by global warming. The Asian corn borer found in the central region of Jilin Province has changed from having an incomplete second generation to having a full second generation every year. As a result, the problem of insect pests will become more and more serious, which will have a greater impact on the production of corn. Corn borer is a destructive pest of crops such as corn, sorghum, millet, and cotton. It is found throughout China, ranging from the southern tropical region to the northern freezing zone, and it may reproduce year-round at a rate of seven or more generations per year. Since the corn borer damages the plant by burrowing it as it begins to erode the corn plant, it will display a straight line of insect feeding holes and so on, posing a serious threat to the growth and development of the corn. Every year, the crop will experience significant harvest losses, with at least 10% yield loss in spring corn and 20%–30% loss in summer corn. In some instances, it may even exceed 30% [7]. The breeding of insect-resistant corn is a significant means of resolving the issue, because the corn borer poses a serious threat to the growth state of corn while affecting food security in China [8]. This results in an annual reduction of millions of tons of corn production, causing enormous economic losses to the corn production and farming industry of China, and severely restricting the economic development of the country [9,10].
Grey leaf spot (GLS) disease is one of the most prevalent ailments that affect maize during its growth and development, and it may be found everywhere where maize is cultivated. The GLS produces a productivity loss of approximately 30%, which poses a serious threat to the corn harvest [11]. Numerous variables such as disease resistance, crop rotation scheme, meteorological circumstances, and management practices of the inbred line have an impact on the prevalence of maize big patches. The large spot is most affected by them when it comes to temperature and humidity in the weather. Due to the cold temperatures and high air humidity in northern China, grey leaf Spot disease is more common. Generally, the relative humidity is above 80%, the daily temperature is 14°C~25°C, sunlight is available for less than 6 h, and continuous cloudy days are 5D~7D. These conditions will greatly increase the probability of grey leaf Spot disease [12]. After the maize tasseling, lesions initially appeared on the lower leaves. Later, the virus slowly spread to the upper leaves of the maize plant. When a large leaf spot occurs in maize, the symptoms are blue-gray water-soaked spots on the leaves. Later, the spots are longitudinally split, and several small spots are connected into larger and irregular mold spots.
Traditional chemical pesticide spraying and control not only pollutes the environment but is also time-consuming and labor-intensive. Moreover, they cannot solve the problem from the root cause. Discovering resistant resources and improving the understanding of corresponding defense mechanisms are particularly important for breeding resistant varieties, but traditional breeding techniques are time-consuming, time-consuming, and labor-intensive [13]. To open up a new and effective way to prevent and control these pests and diseases, and improve yield and quality, my country is also actively carrying out the development and application of genetic engineering in crops [14,15].
The utilization of genetically modified disease-resistant and insect-resistant varieties has become one of the most economical, effective, and environmentally friendly control methods [16]. The use of transgenic breeding technology to breed corn varieties with strong and durable resistance to corn borer and great leaf spot disease provides a powerful reference for molecular breeding of corn [17].
NPR1 gene was discovered from Arabidopsis mutants. NPR1 gene is a key gene in plant systemic acquired disease resistance (SAR) and plays an important role in regulating the broad-spectrum disease resistance of plants [18–20]. NPR1 homologous gene exists widely in plants and has been studied as a disease resistance-related gene. It plays an important role in regulating salicylic acid signaling in plant disease resistance and pathogenesis in Arabidopsis [21], and in wheat, (Triticum aestivum) [22], Rice (Oryza Sativa) [23], Barley (Hordeum vulgare) [24] have been reported in other species.
The Cry1Ab13-1 gene is a member of the Bacillus thuringiensis (Bt) gene family. After spore formation, Cry1Ab13-1 can produce proteins that are toxic to pests. These include Lepidoptera, Hymenoptera, Homoptera, Diptera, Coleoptera, etc. Foreign research on Bt gene insect-resistant maize started early [25], and GM Insect-Resistant Corn Begins Commercially in 1996 [26]. Many recent studies have found that Bt corn can reduce 99% of borer tunnels and 84% of leaf damage.
In the initial stage of this experiment, Cry1Ab13-1 and NPR1 pCAMBIA3301-Cry1Ab13-1-NPR1 genes were transformed into maize varieties S3002 and 349, which improved the disease resistance and insect resistance of maize. Therefore, this experiment was used to identify and evaluate the disease resistance and insect resistance of T2, T3, and T4 of the transgenic Cry1Ab13-1 and NPR1 bivalent maize varieties S3002 and 349.
The tested varieties are S3002 (Cry1Ab13-1+NPR1), 349 (Cry1Ab13-1+NPR1), three generations of maize lines T2, T3, and T4, and their corresponding wild types provided by the Plant Biotechnology Center of Jilin Agricultural University (China). The insect taken into consideration was the Asian corn borer, provided by the Institute of Plant Protection, Jilin Academy of Agricultural Sciences (China). The test agent was 10% Glufosinate Ammonium (10% Solution).
A split-plot experimental design was adopted, with inbred varieties as the main plot, generations as the subplots, and three-row plots with a row length of 5.5 m, repeated three times. The transgenic T2, T3, and T4 generation seeds and wild-type recipient seeds of lines S3002 and 349 were planted in the transgenic experimental field of Jilin Agricultural University, with three rows per generation, and three rows of wild type were planted beside the corresponding lines. 20 rows of corn were sown. The transgenic plants were examined at the seedling stage, and 10 plants that tested positive were identified by rubbing herbicide (glufosinate-ammonium) on their leaves.
2.3 PCR Detection of Transformed Plants
The DNA of the positive plant leaves was extracted using a plant genomic DNA extraction kit (CWBio, Jiangsu, China). The Cry1Ab13-1 and NPR1 plasmids were used as positive controls, wild-type genome was used as a negative control, and water was used as blank control. Perform PCR testing. PCR amplification conditions and primer sequences are demonstrated in the table given below (Table S1).
2.4 Fluorescence Quantitative Real-Time PCR Detection of Transgenic Positive Plants
In reference to the method suggested by Xie et al. [10], to extract the total RNA of the roots, stems, and leaves of the plants that tested positive in 1.2.1, a fluorescent quantitative reverse transcription kit (TakaraBio, RR036A) was used to reverse transcribe into cDNA and use the online design primer website (Primer Quest Tool) (https://sg.idtdna.com/pages/tools). The Mx3000P fluorescence quantitative PCR instrument (Agilent Technologies, United States) was used to detect the relative expression levels of the target genes NPR1 and Cry1AB13-1 in the roots, stems, and leaves of the three-leaf stage of transformed plants, and the expression was calculated as
2.5 Outdoor Insect Resistance Test Method of Transgenic Positive Plants
When corn grew to the six to eight leaf stage, 20 positive plants were randomly selected from each line, and 20 to 30 2-instar corn borer larvae were placed in the corn core leaves. Later, they were inoculated again after 3D to resist insects. The sex identification standard adopted the nine-level grading standard formulated by the International Corn Borer Collaborative Group [29] (Table S2).
2.6 Outdoor Disease Resistance Test Method of Transgenic Positive Plants
The large leaf blight bacteria (Exserohilum turcicum (Pass.) Leonay et Suggs) was cultivated on the Potato Dextrose Agar (PDA) medium and then inoculated on the sorghum grain medium with the cultured strains. Later, the sorghum grains were covered with bacteria spare with silk. The Grey spot inoculation test on the genetically modified land was conducted on June 25th. In reference to the method suggested by Xia et al. [30], at the 9–11 leaf stage of the corn plant, sorghum grains covered with mycelium were sprinkled into the corn core leaves, and 10–15 grains were sprinkled into each corn plant. 10 corn plants were randomly investigated in each row, and the survey objects were three leaves above the ear and three leaves below the ear. According to the nine-level evaluation system (Table S3), each maize plant was investigated for GLS at the maize milk maturity stage [30].
All the experiments were carried out with replicates of three and the same was repeated three times. The statistical evaluation was performed to determine the significant difference by one-way ANOVA Duncan test and two-way ANOVA Duncan test. Values are means ± SE of independent biological replicates of three. The mean values under each treatment were compared at 0.01 < p < 0.05. All the analyses were performed by GraphPad Prism 6.0 software and SPSS 23.0 software.
3.1 PCR Detection of Transgenic Plants
3.1.1 PCR Detection of T0, T2, T3, and T4 Generation Transgenic Plants
By employing seamless cloning technology and double enzyme digestion, the original GUS region of 3301 was replaced with NPR1, and Cry1Ab13-1 was inserted by exploiting the various cloning sites on the vector, as demonstrated in Fig. S1.
The size of the amplified target bands was 500, 552, 205, 525, and 1657 bp, which was consistent with the size of the target gene 35S, Bar, Nos, NPR1, and Cry1Ab13-1 fragments, indicating that the functional genes and marker gene fragments have been integrated into T0, T2, T3, and T4 generation lines (Figs. S2–S5). However, due to the small number of seeds obtained from T0 generation plants, subsequent analysis of these plants was not carried out.
3.2 Fluorescence Quantitative Real-Time PCR Detection of Transgenic Maize
3.2.1 Fluorescence Quantitative Real-Time PCR Detection of Cry1Ab13-1 Gene in Transgenic Maize
By detecting the relative expression levels of Cry1Ab13-1 gene in roots, stems and leaves of S3002 and 349 T2, T3 and T4 generations at V3 stage, the contents of T2 generation stems of S3002 and 349 were used as controls, respectively, the quantitative Real-time PCR (qRT-PCR) was performed, and the results were shown in Figs. 1A and 1B. By calculating the mean value of Cry1Ab13-1 gene in roots, stems and leaves of T2, T3 and T4 generations of S3002 and 349, it could be seen that there was no significant difference in the expression of Cry1Ab13-1 gene in the same tissue of T2, T3 and T4 generations of S3002 and 349. There were significant differences in the expression levels of Cry1Ab13-1 gene in different tissues of T2, T3 and T4 generations of S3002 and 349. The results showed that the highest relative expression of Cry1Ab13-1 gene in S3002 was in leaves, followed by roots, with the lowest content in stems, the highest relative expression of Cry1Ab13-1 gene in 349 was in leaves, followed by stems, with the lowest content in roots.
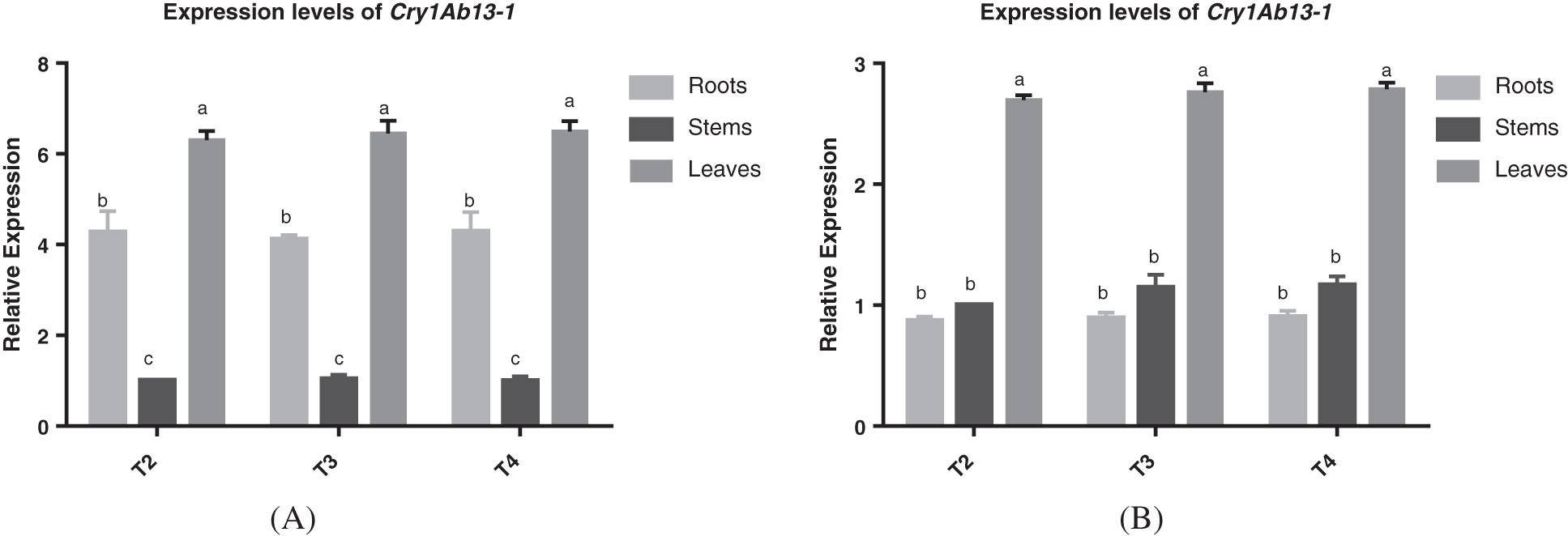
Figure 1: (A) Relative expression of Cry1Ab13-1 gene in roots stems and leaves of V3 transgenic maize of S3002 generations T2, T3, and T4; (B) Relative expression of Cry1Ab13-1 gene in roots stems and leaves of V3 transgenic maize of 349 generations T2, T3, and T4. All values were the mean of three replicates (mean ± SE). Different letters represent significant differences between each treatment at 0.01 < p < 0.05, calculated using two-way ANOVA Duncan test. The small letter (i.e., a and b)indicated significant differences between different organizations
3.2.2 Fluorescence Quantitative Real-Time PCR Detection of NPR1 Gene in Transgenic Maize
The relative expression levels of NPR1 gene in roots, stems and leaves of S3002 and 349 T2, T3 and T4 generations at V3 stage were detected. The content of T2 stem of S3002 and 349 was used as the control for qRT-PCR detection, and the results were shown in Figs. 2A and 2B. By calculating the mean value of NPR1 gene in roots, stems and leaves of T2, T3 and T4 generations of S3002 and 349, it could be seen that there was no significant difference in the expression of NPR1 gene in the same tissue of T2, T3 and T4 generations of S3002 and 349. The expression levels of NPR1 gene in different tissues of T2, T3 and T4 generations of S3002 were significantly different. The expression of NPR1 gene in roots and leaves of 349 T2, T3 and T4 generations was significantly different from that in stems. The results showed that the highest relative expression of NPR1 gene in S3002 and 349 was in leaves, followed by roots, with the lowest in stems.
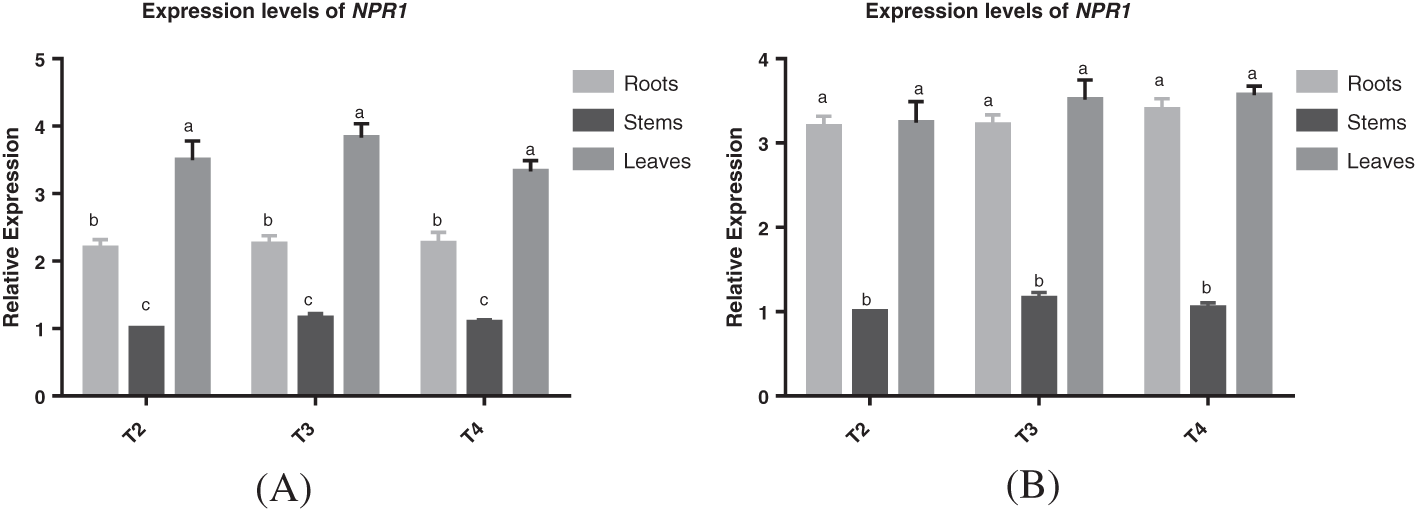
Figure 2: (A) Relative expression of NPR1 gene in roots, stems, and leaves of V3 stage of transgenic maize of S3002 T2, T3, and T4 generations; (B) Relative expression of NPR1 gene in roots, stems, and leaves of V3 stage of transgenic maize of 349 T2, T3, and T4 generations. All values were the mean of three replicates (mean ± SE). Different letters represent significant differences between each treatment at 0.01 < p < 0.05, calculated using two-way ANOVA Duncan test. The small letter (i.e., a and b) indicated significant differences between different organizations
The results indicated that Cry1Ab13-1 and NPR1 genes were expressed in the roots, stems, and leaves of the T2, T3, and T4 progeny of transgenic S3002 and 349. The expression levels of the Cry1Ab13-1 gene and NPR1 gene in the same tissues of the same cultivar and different generations have no significant difference indicating that Cry1Ab13-1 and NPR1 genes can be expressed in various parts of the transgenic maize plant and can be stably inherited.
3.3 Identification of Outdoor Insect Resistance of Transgenic Maize
After the 7D, 14D, and 21D field investigations (Fig. 3), the plants were processed for data processing (Figs. 4 and 5). At the 7D after inoculation, the T2, T3, and T4 generations of S3002 all reached the resistance (R) level, while the wild insect resistance of the type was the susceptible (S) level. T2, T3, and T4 generations of the 349 line had no significant differences in leaf-eating grades, and their insect resistance reached the resistance (R) level, while the insect resistance of the 349 wild-types was at the insect-susceptible (S) level.

Figure 3: Picture of external transplanting of transformed plants. Note: (A) Untransformed inbred line 349; (B) Transformed inbred line 349; (C) Untransformed inbred line S3002; (D) Transformed inbred line S3002
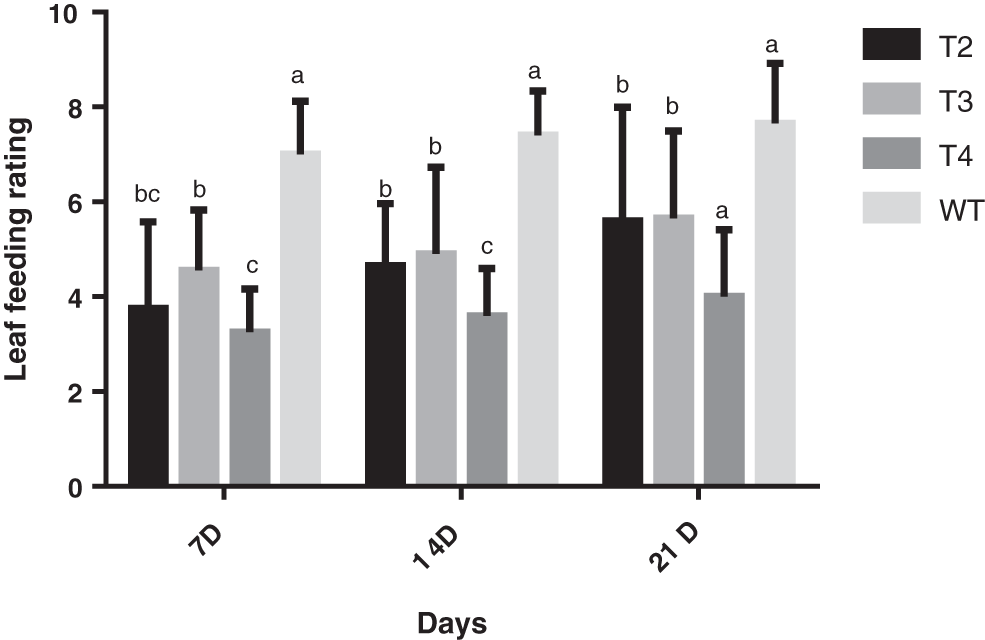
Figure 4: Leaf feeding levels of T2, T3, and T4 of transgenic positive plants S3002. All values were the mean of three replicates (mean ± SE). Different letters represent significant differences between each treatment at 0.01 < p < 0.05, calculated using two-way ANOVA Duncan test. The small letter (i.e., a and b) indicate significant differences among different generations
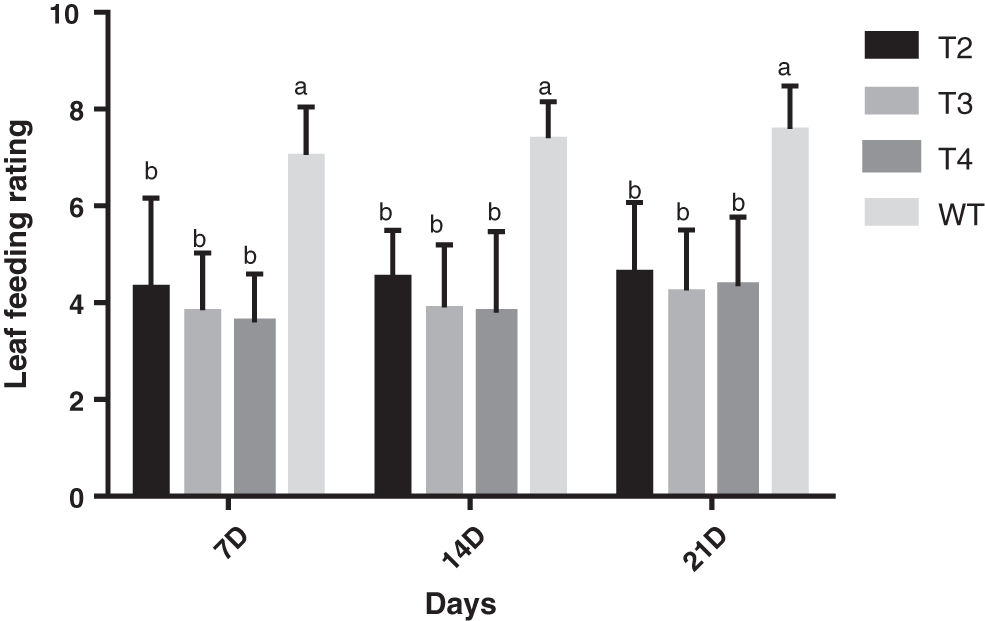
Figure 5: Leaf feeding levels of T2, T3, and T4 of transgenic positive plants 349. All values were the mean of three replicates (mean ± SE). Different letters represent significant differences between each treatment at 0.01 < p < 0.05, calculated using two-way ANOVA Duncan test. The small letter (i.e., a and b) indicated significant differences among different generations
On the 14D after inoculation, the insect resistance of the three generations T2, T3, T4, and the wild type of S3002 was the insect resistance (R) level, while the wild type was all susceptible (S) level, respectively. Among them, the differences in the leaf-eating grades of the T2 and T3 generations were not significant, the differences in the leaf-eating grades between the T2 and T4, with the T3 and T4 generations were significant, and the three generations of the S3002 line and the wild type had significant leaf-eating grades. The insect resistance of the three generations of 349 continued at the level of insect resistance (R), while the insect resistance of the wild type was at the susceptible (S) level, and there were significant differences in the leaf-eating levels of the three generations of T2, T3, and T4 from wild type.
On the 21D after inoculation, the insect resistance of each line of S3002 and 349 and the wild type was consistent with the insect resistance after inoculation 14D. The transgenic lines were all at the resistance (R) level, while the wild type was all at the susceptible (S) level, and there is no change in leaf-feeding level with the wild type, all of which are significantly different.
3.4 Identification of Outdoor Disease Resistance of Transgenic Positive Plants
As demonstrated in Fig. 6, it is the survey pictures of S3002, 349 transgenic maize and wild-type maize large leaf spot, and Figs. 7 and 8 demonstrated the diseased resistance grade of outdoor transformed plant grey leaf Spot disease. In the late stage of maize milk maturity, 10 transgenic maize plants were selected for S3002 and 349 for investigation, and there were wild-type plants. The disease resistance grades are depicted in Fig. 7. The average disease resistance grades of S3002 and 349 transgenic lines were 4.00 and 3.20, respectively, and the disease resistances were both resistant (R). By variance analysis, S3002, 349 transgenic maize significantly improved disease resistance compared with wild-type maize.

Figure 6: Pictures of transformed plants exposed to disease outside. Note: (A) Untransformed inbred line 349; (B) Transformed inbred line 349; (C) Untransformed inbred line S3002; (D) Transformed inbred line S3002
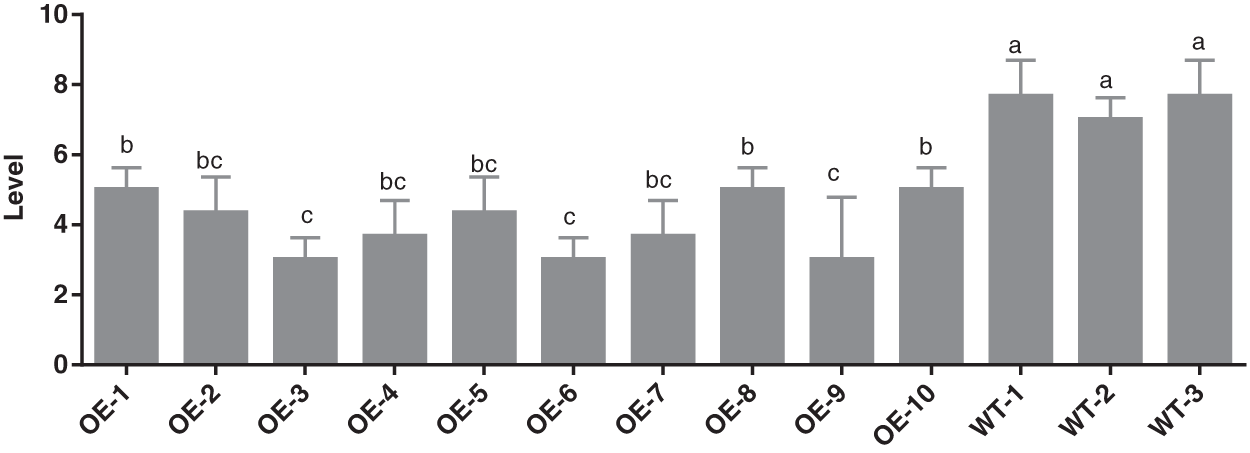
Figure 7: Identification of resistance grade of untransformed inbred line S3002 and transformed inbred line S3002. All values were the mean of three replicates (mean ± SE). Different letters represent significant differences between each treatment at 0.01 < p < 0.05, calculated using one-way ANOVA Duncan test. The small letter (i.e., a and b) indicated significant differences among different lines
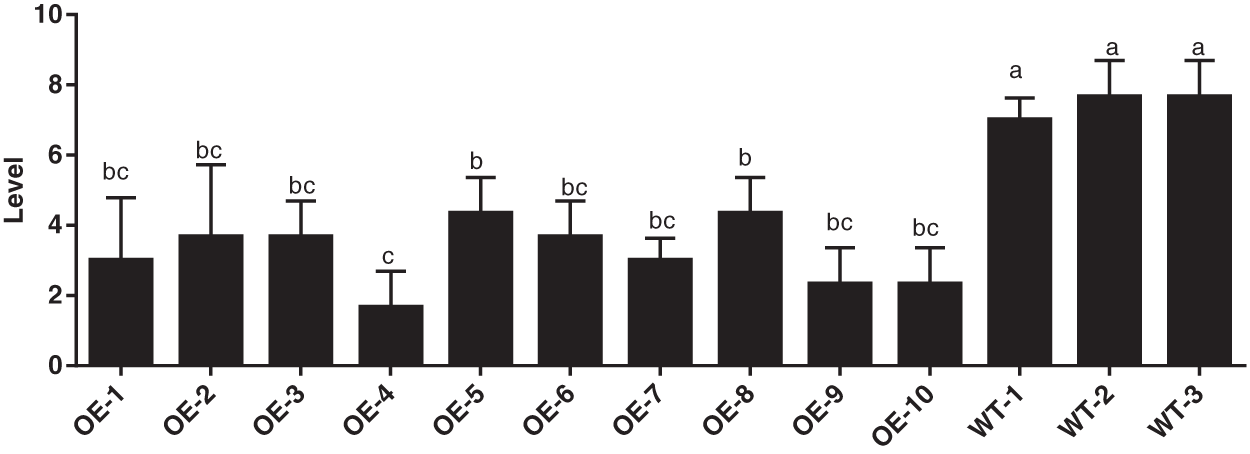
Figure 8: Identification of resistance grade of untransformed inbred line 349 and transformed inbred line 349. All values were the mean of three replicates (mean ± SE). Different letters represent significant differences between each treatment at 0.01 < p < 0.05, calculated using one-way ANOVA Duncan test. The small letter (i.e., a and b) indicated significant differences among different lines
This study confirmed that the T2, T3, and T4 generation of maize varieties S3002 and 349 containing NPR1 and Cry1Ab13-1 bivalent gene maize materials can better enhance insect resistance and disease resistance. The progeny of T2, T3, and T4 transgenic lines of maize varieties S3002 and 349 were tested by conventional PCR, qRT-PCR, outdoor insect resistance, and disease resistance. Through the identification of outdoor insect resistance and disease resistance of the two receptors and wild-type plants and the fact that transgenic maize has insect resistance and disease resistance, it is confirmed that the bivalent gene polymerized maize line is more resistant to insect and disease than the wild type. The disease resistance ability has been improved, and it can be stably inherited for multiple generations. The results obtained from this research provided a good foundation for breeding new maize germplasm with insect resistance and disease resistance.
There are few research reports on bivalent plant expression vectors in maize. In this study, we examined the genetic stability and resistance in the T2, T3, and T4 generation maize materials containing bivalent genes and their corresponding wild types transformed with the insect-resistance gene Cry1Ab13-1 and the disease-resistance gene NPR1 double-resistance gene sick ability. The results showed that the bivalent gene polymerized maize line had improved insect and disease resistance compared with the wild type.
Sprays with the insecticidal protein Bt have been used for decades to control pests in agriculture, forestry, and public health. Due to its precise and very effective target features, transgenic Bt maize is well-liked by growers [31,32]. Since 1996, Bt crops have been recognized for their ability to control target pests and reduce impacts on non-target organisms [33]. Many researchers have transferred the Bt gene into the maize backbone line and obtained insect-resistant transformants and Bt-transgenic insect-resistant maize inbred lines.
In China, transgenic Cry1Ab maize has good field insect resistance to Asian corn borer [34]. In 1995, the study by Armstrong et al. proved that the structure of Cry1Ab toxin protein endowed it with strong insecticidal activity against the diamondback moth Plutella xylostella and the tobacco hawkmoth Manduca sexta. Tang et al. [35] demonstrated that the structure of Cry1Ab toxin protein endowed it with strong insecticidal activity against rice. Subsequently, many researchers from China and other countries introduced the Cry1Ab gene into maize and proved that the transgenic maize expressing the Cry1Ab protein was effective against the Asian corn borer Ostrinia furnacalis and European corn borer Ostrinia nubilalis has good field resistance and can effectively control the damage of corn borer at the heart leaf stage and ear stage. The results of this experiment are consistent with the previous results, the transgenic plants obtained after the Cry1Ab13-1 gene was transformed into the recipient improved the insect resistance from the susceptible to the resistant.
Numerous crops exhibit the broad-spectrum of disease resistance of the NPR1 gene. In grapes (Vitis vinifera L.), rice (Oryza sativa) [23], apples (Malus domestica) [36], bananas (Musa nana Lour.) [37], and other plants, several NPR1 homologous genes have been cloned consecutively. The results from the research conducted by Quilis et al. [38] demonstrated that NPR1-mediated rice had a positive regulatory effect on the defense response of biological and abiotic stress in rice growth and development, and the plants with transformed NPR1 gene developed resistance to fungal pathogens that caused rice blast and rice seedling disease, respectively, and resistance to bacterial pathogens that caused foot rot, filling the gap in maize research. There are few research reports on bivalent plant expression vectors in maize. This study verifies that NPR1 and Cry1Ab13-1 bivalent gene maize materials can better enhance insect resistance and disease resistance than non-transgenic positive plants and can be more stable in inheritance from generation to generation.
The experimental findings of this work support the use of bivalent gene transformation vectors in maize. Currently, bivalent gene transformation vectors are employed in soybeans, sweet potatoes, alfalfa, and kale rape. However, there remain few research reports on corn. New germplasm resources are provided for disease-resistant and insect-resistant varieties, and the bivalent expression vector has a good breeding prospect in disease-resistant and insect-resistant breeding.
Authors Contribution: Yongjing Xi drafted the original manuscript. Zhou Yang reviewed and edited the manuscript. Yukun Jin, Jing Qu, and Shuyan Guan conducted the experiments and performed the analysis. Piwu Wang conducted a formal analysis. Siyan Liu supervised the experiments. All authors contributed to the article and approved the submitted version.
Funding Statement: This research was supported by the National Key Research and Development Program of China (2019YFD1002603-1) and we also thank Jilin Agricultural University, Biotechnology Center for facilitating the working environment to conduct the experiment.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Shiferaw, B., Prasanna, B. M., Hellin, J., Bänziger, M. (2011). Crops that feed the world 6 past successes and future challenges to the role played by maize in global foodsecurity. Food Security, 3(3), 307–327. [Google Scholar]
2. Amon, T., Amon, B., Kryvoruchko, V., Machmüller, A., Hopfner-Sixt, K. et al. (2007). Methane production through anaerobic digestion of various energy crops grown in sustainable crop rotations. Bioresource Technology, 98(17), 3204–3212. [Google Scholar]
3. Mutuura, A., Munroe, E. (1970). Taxonomy and distribution of the European corn borer and allied species: Genus ostrinia (lepidoptera: pyralidae). The Memoirs of the Entomological Society of Canada, 71(S71), 1–112. [Google Scholar]
4. St Louis, M. E., Hess, J. J. (2008). Climate change: Impacts on and implications for global health. American Journal of Preventive Medicine, 35(5), 527–538. [Google Scholar]
5. Vucic-Pestic, O., Ehnes, R. B., Rall, B. C., Brose, U. (2011). Warming up the system: Higher predator feeding rates but lower energetic efficiencies. Global Change Biology, 17(3), 1301–1310. [Google Scholar]
6. Clusella-Trullas, S., Blackburn, T. M., Chown, S. L. (2011). Climatic predictors of temperatu-re performance curve parameters in ectotherms imply complex responses to climate change. The American Naturalist, 177(6), 738–751. [Google Scholar]
7. Du, D., Geng, C., Zhang, X., Zhang, Z., Zheng, Y. et al. (2014). Transgenic maize linesexp-ressing acry1c*gene are resistant to insect pests. Plant Molecular Biology Reporter, 32(2), 549–557. [Google Scholar]
8. Dowd, P. F., Lagrimini, L. M., Herms, D. A. (1998). Differential leaf resistance to insects of transgenic sweetgum (Liquidambar styraciflua) expressing tobacco anionic peroxidase. Cellular and Molecular Life Sciences, 54(7), 712–720. [Google Scholar]
9. Archer, T. L., Schuster, G., Patrick, C., Cronholm, G., Bynum, E. D. et al. (2000). Whorland stalk damage by European and Southwestern corn borers to four events of bacillus thuringiensis transgenic maize. Crop Protection, 19(3), 181–190. [Google Scholar]
10. Xie, W., Ali, T., Cui, Q., Huang, J. K. (2017). Economic impacts of commercializing insect-resistant gm maize in China. China Agricultural Economic Review, 9(3), 340–354. [Google Scholar]
11. Weems, J. D., Bradley, C. A. (2018). Exserohilum turcicum race population distribution in the North Central United States. Plant Disease, 102(2), 292–299. DOI 10.1094/PDIS-01-17-0128-RE. [Google Scholar] [CrossRef]
12. Levy, Y., Pataky, J. K. (1992). Epidemiology of northern leaf blight on sweet corn. Phytoparasitica, 20(1), 53–66. DOI 10.1007/BF02995636. [Google Scholar] [CrossRef]
13. Zhao, W., Wang, T., Qi, R. D. (2015). Ypt1 gene-based detection of phytophthora sojae in a loop-mediated isothermal amplification assay. Journal of Plant Diseases and Protection, 122(2), 66–73. DOI 10.1007/BF03356533. [Google Scholar] [CrossRef]
14. Liu, Q., Hallerman, E., Peng, Y., Li, Y. (2016). Development of Bt rice and Bt maize in China and their efficacy in target pest control. International Journal of Molecular Sciences, 17(10), 1561. DOI 10.3390/ijms17101561. [Google Scholar] [CrossRef]
15. Dale, P. J., Clarke, B., Fontes, E. M. G. (2002). Erratum: Potential for the environmental impact of transgenic crops. Nature Biotechnology, 20(8), 843. DOI 10.1038/nbt0802-843b. [Google Scholar] [CrossRef]
16. Munkvold, G. P., Martinson, C. A., Shriver, J. M., Dixon, P. M. (2001). Probabilities for profitable fungicide use against gray leaf spot in hybrid maize. Phytopathology, 91(5), 477–484. DOI 10.1094/PHYTO.2001.91.5.477. [Google Scholar] [CrossRef]
17. Haudenshield, J. S., Song, J. Y., Hartman, G. L. (2017). A novel, multiplexed, probe-based quantitative PCR assay for the soybean root- and stem-rot pathogen, Phytophthora sojae, utilizes its transposable element. PLoS One, 12(4), e0176567. DOI 10.1371/journal.pone.0176567. [Google Scholar] [CrossRef]
18. Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell, 88(1), 57–63. DOI 10.1016/S0092-8674(00)81858-9. [Google Scholar] [CrossRef]
19. Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y. et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell, 173(6), 1454–1467.e15. DOI 10.1016/j.cell.2018.03.044. [Google Scholar] [CrossRef]
20. Kinkema, M., Fan, W., Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. The Plant Cell, 12(12), 2339–2350. DOI 10.1105/tpc.12.12.2339. [Google Scholar] [CrossRef]
21. Castelló, M. J., Medina-Puche, L., Lamilla, J., Tornero, P. (2018). NPR1 paralogs of Arabid-opsis and their role in salicylic acid perception. PLoS One, 13(12), e0209835. DOI 10.1371/journal.pone.0209835. [Google Scholar] [CrossRef]
22. Wang, X. D., Bi, W. S., Gao, J., Yu, X. M., Wang, H. Y. et al. (2018). Systemic acqui-red resistance, NPR1, and pathogenesis-related genes in wheat and barley. Journal of Integrative Agriculture, 17(11), 2468–2477. DOI 10.1016/S2095-3119(17)61852-5. [Google Scholar] [CrossRef]
23. Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y. et al. (2007). Functional analysis of rice NPR1 like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnology Journal, 5(2), 313–324. DOI 10.1111/j.1467-7652.2007.00243.x. [Google Scholar] [CrossRef]
24. Gao, J., Bi, W., Li, H., Wu, J., Yu, X. et al. (2018). WRKY transcription factors associated with NPR1-mediated acquired resistance in barley are potential resources to improve wheat resistance to Puccinia triticina. Frontiers in Plant Science, 9, 1486. DOI 10.3389/fpls.2018.01486. [Google Scholar] [CrossRef]
25. Gordon-Kamm, W. J., Spencer, T. M., Mangano, M. L., Adams, T. R., Daines, R. J. et al. (1990). Transformation of maize cells and regeneration of fertile transgenic plants. The Plant Cell, 2(7), 603–618. DOI 10.2307/3869124. [Google Scholar] [CrossRef]
26. Denolf, P., Hendrickx, K., van Damme, J., Jansens, S., Peferoen, M. et al. (1997). Cloning and characterization of Manduca sexta and Plutella xylostella midgut aminopeptidase Nenzymes related to Bacillus thuringiensis toxin-binding proteins. European Journal of Biochemistry, 248(3), 748–761. [Google Scholar]
27. Javdi, M., Haghnazari, A., Tohidfar, M., Negari, S., Ghareyazi, B. (2010). Zygosity identifica-tion in transgenic cotton (Gossypium hirsutum) by real-time quantitative PCR. Euphytica, 173(2), 185–191. [Google Scholar]
28. Lu, S., Zhang, M., Zhang, Z., Wang, Z., Wu, N. et al. (2018). Screening and verification of genes associated with leaf angle and leaf orientation value in inbred maize lines. PLoS One, 13(12), e0208386. [Google Scholar]
29. Abel, C. A., Berhow, M. A., Wilson, R. L., Binder, B. F., Hibbard, B. E. (2000). Evaluation of conventional resistance to European corn borer (Lepidoptera: Crambidae) and western corn rootworm (Coleoptera: Chrysomelidae) in experimental maize lines developed from abackcross breeding program. Journal of Economic Entomology, 93(6), 1814–1821. [Google Scholar]
30. Xia, H., Gao, W., Qu, J., Dai, L., Gao, Y. et al. (2020). Genetic mapping of northern corn leaf blight-resistant quantitative trait loci in maize. Medicine, 99(31), e21326. [Google Scholar]
31. Milne, A. E., Bell, J. R., Hutchison, W. D., van den Bosch, F., Mitchell, P. D. et al. (2015). The effect of farmers’ decisions on pest control with Bt crops: A billion dollar game of strategy. PLoS Computational Biology, 11(12), e1004483. [Google Scholar]
32. Hutchison, W. D., Burkness, E. C., Mitchell, P. D., Moon, R. D., Leslie, T. W. et al. (2010). Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science, 330(6001), 222–225. [Google Scholar]
33. Klümper, W., Qaim, M. (2014). A meta-analysis of the impacts of genetically modified crops. PLoS One, 9(11), e111629. [Google Scholar]
34. He, K., Wang, Z., Zhou, D., Wen, L., Song, Y. et al. (2003). Evaluation of transgenic Bt corn for resistance to the Asian corn borer (Lepidoptera: Pyralidae). Journal of Economic Entomology, 96(3), 935–940. [Google Scholar]
35. Tang, W., Lin, Y. J. (2007). Field experiment of transgenic cry1Ab insect resistant rice. Yi Chuan = Hereditas, 29(8), 1008–1012. [Google Scholar]
36. Zhao, J. T., Huang, X., Chen, Y. P., Chen, Y. F., Huang, X. L. (2009). Molecular cloningand characterization of an ortholog of NPR1 gene from dongguan dajiao (Musa spp. ABB). Plant Molecular Biology Reporter, 27(3), 243–249. DOI 10.1007/s11105-008-0074-z. [Google Scholar] [CrossRef]
37. Chupeau, M. C., Pautot, V., Chupeau, Y. (1994). Recovery of transgenic trees after electr-oporation of poplar protoplasts. Transgenic Research, 3(1), 13–19. DOI 10.1007/BF01976022. [Google Scholar] [CrossRef]
38. Quilis, J., Peñas, G., Messeguer, J., Brugidou, C., San Segundo, B. (2008). The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viralpathoge-ns while conferring hypersensitivity to abiotic stresses in transgenic rice. Molecular Plant-Microbe Interactions, 21(9), 1215–1231. DOI 10.1094/MPMI-21-9-1215. [Google Scholar] [CrossRef]
Appendix
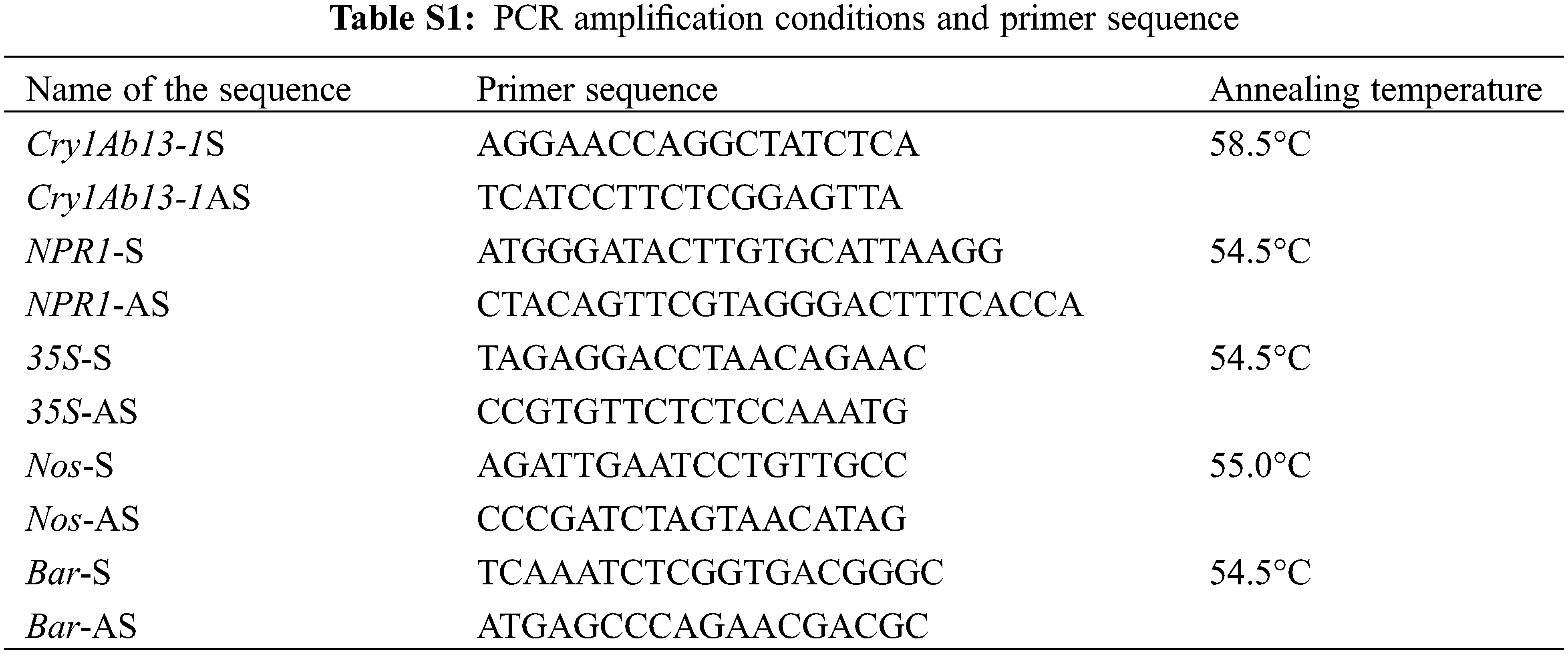
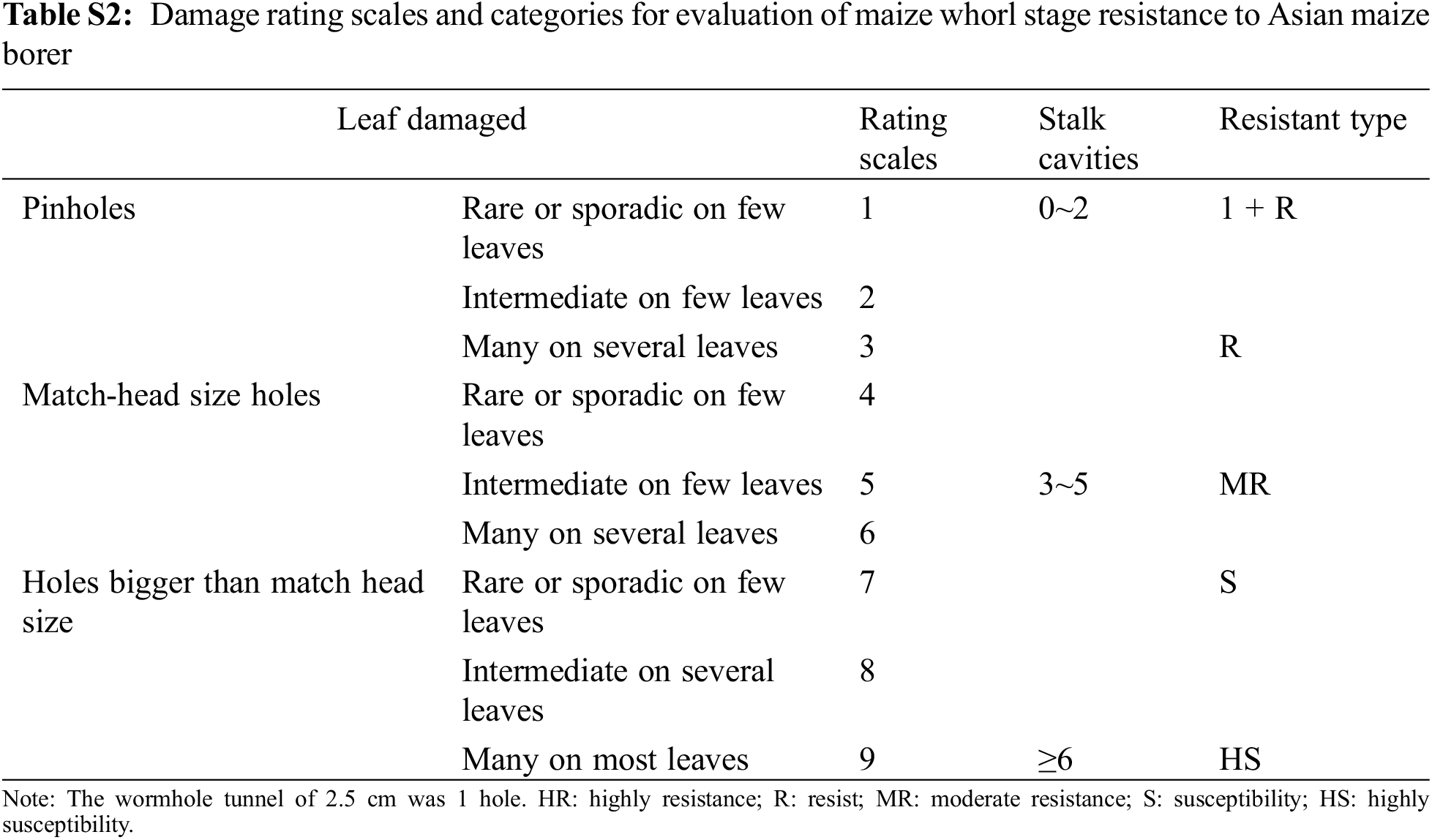
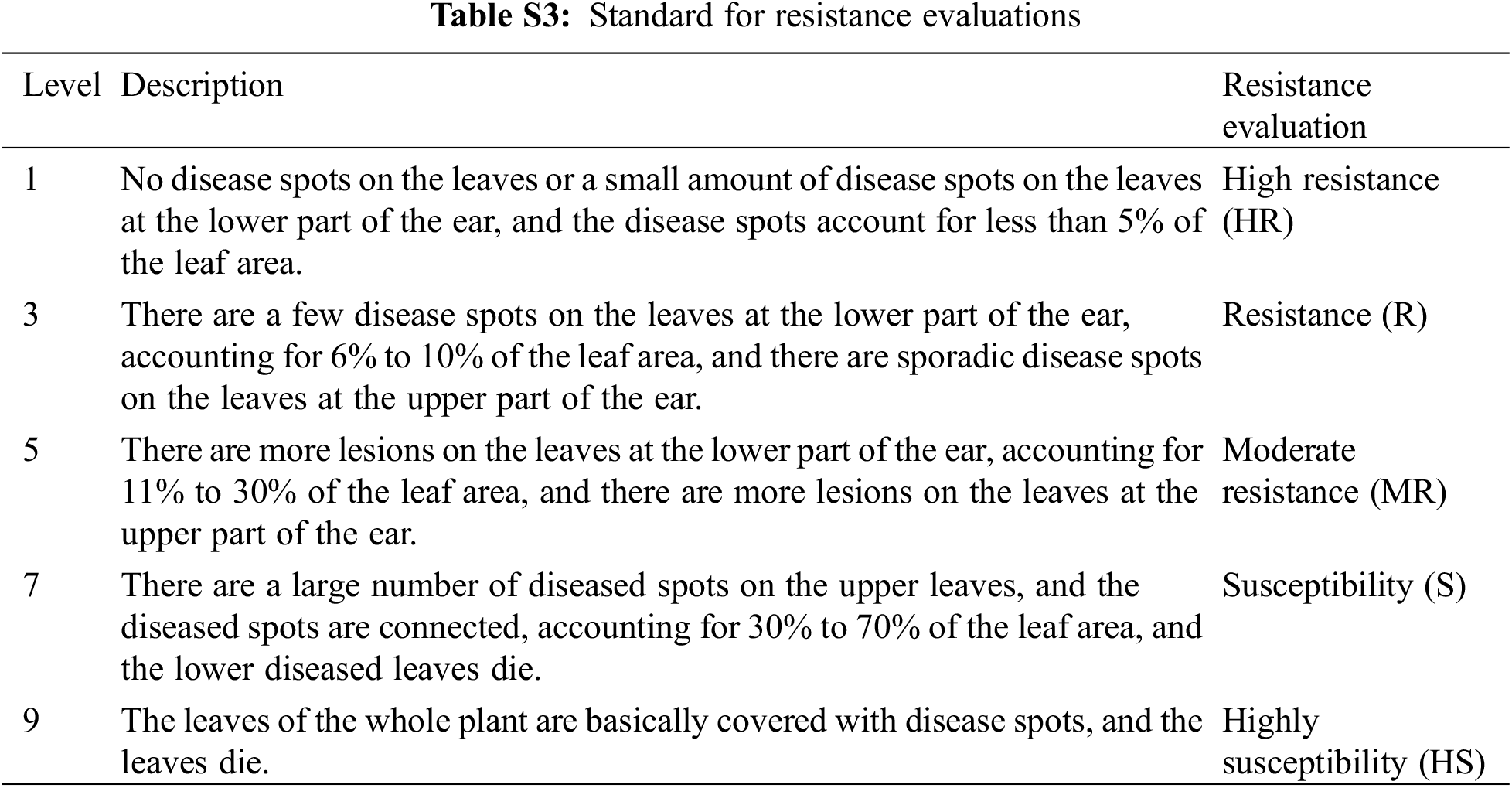
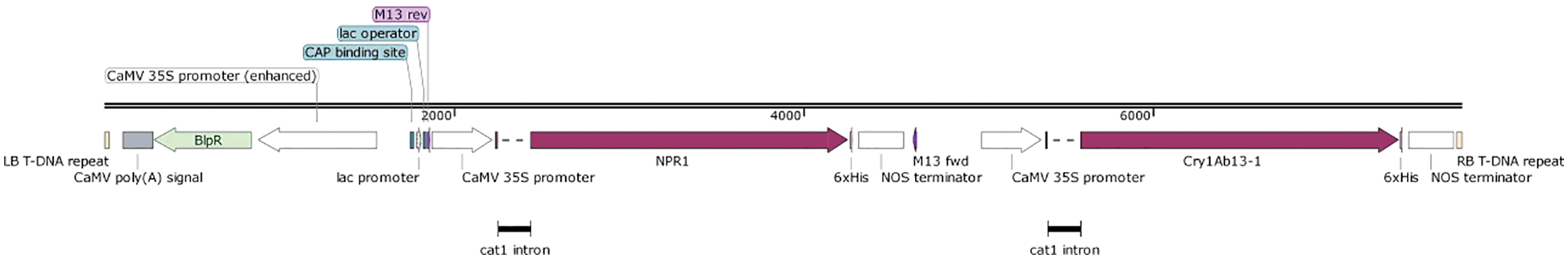
Figure S1: Structure of recombinant plant expression vector PCAMBIA3301-Cry1Ab13-1-NPR1
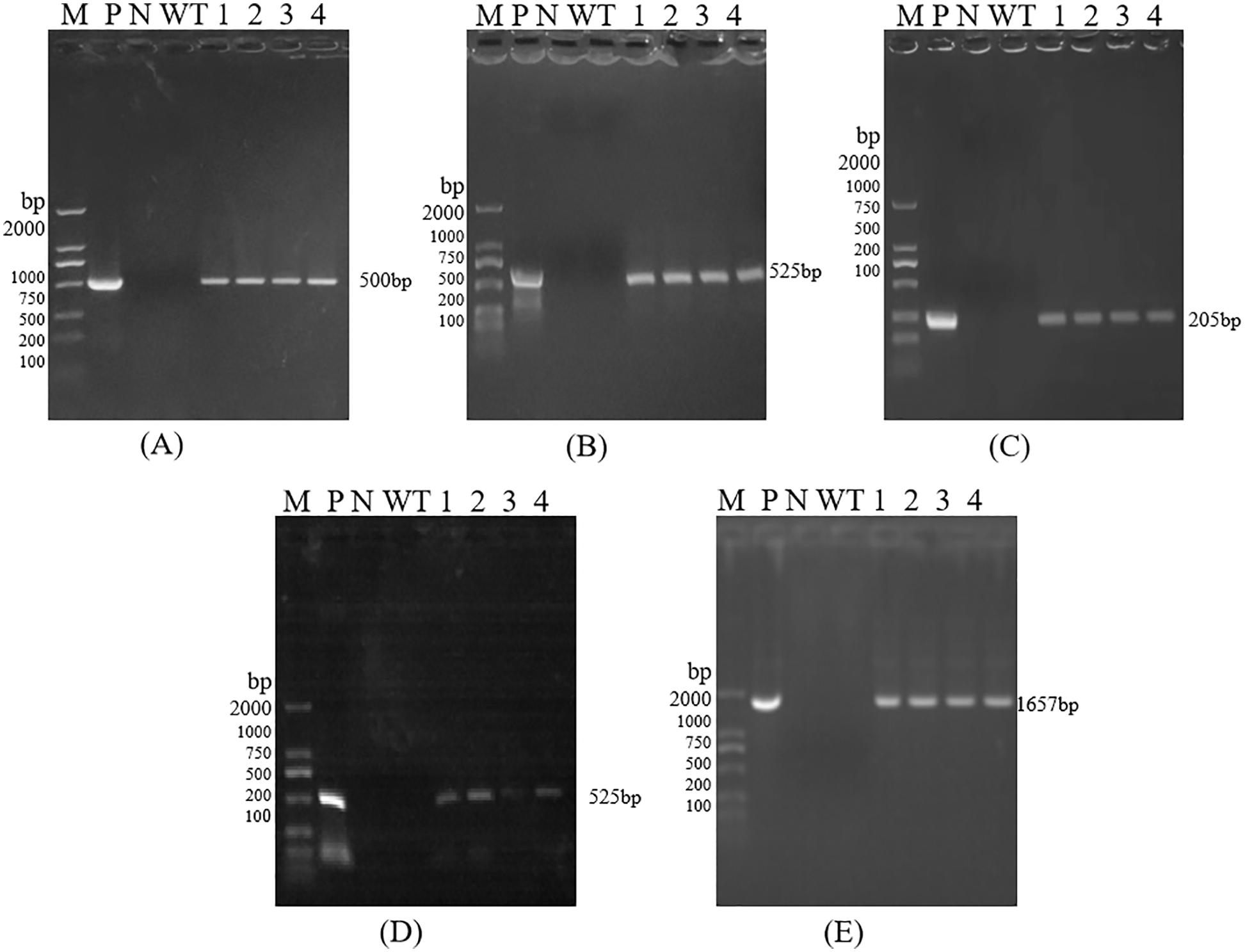
Figure S2: PCR detection of T0 Transgenic Maize; Each part of electrophoretogram is 35S (3A), Bar (3B), Nos (3C), NPR1 (3D), Cry1Ab13-1 (3E). Note: A–E are PCR detection of 35S, Bar, Nos, NPR1, Cry1Ab13-1; M: DL 2000 DNAmarker; N: H2O; P: Positive control; WT: Untransformed plants; 1–4: Samples
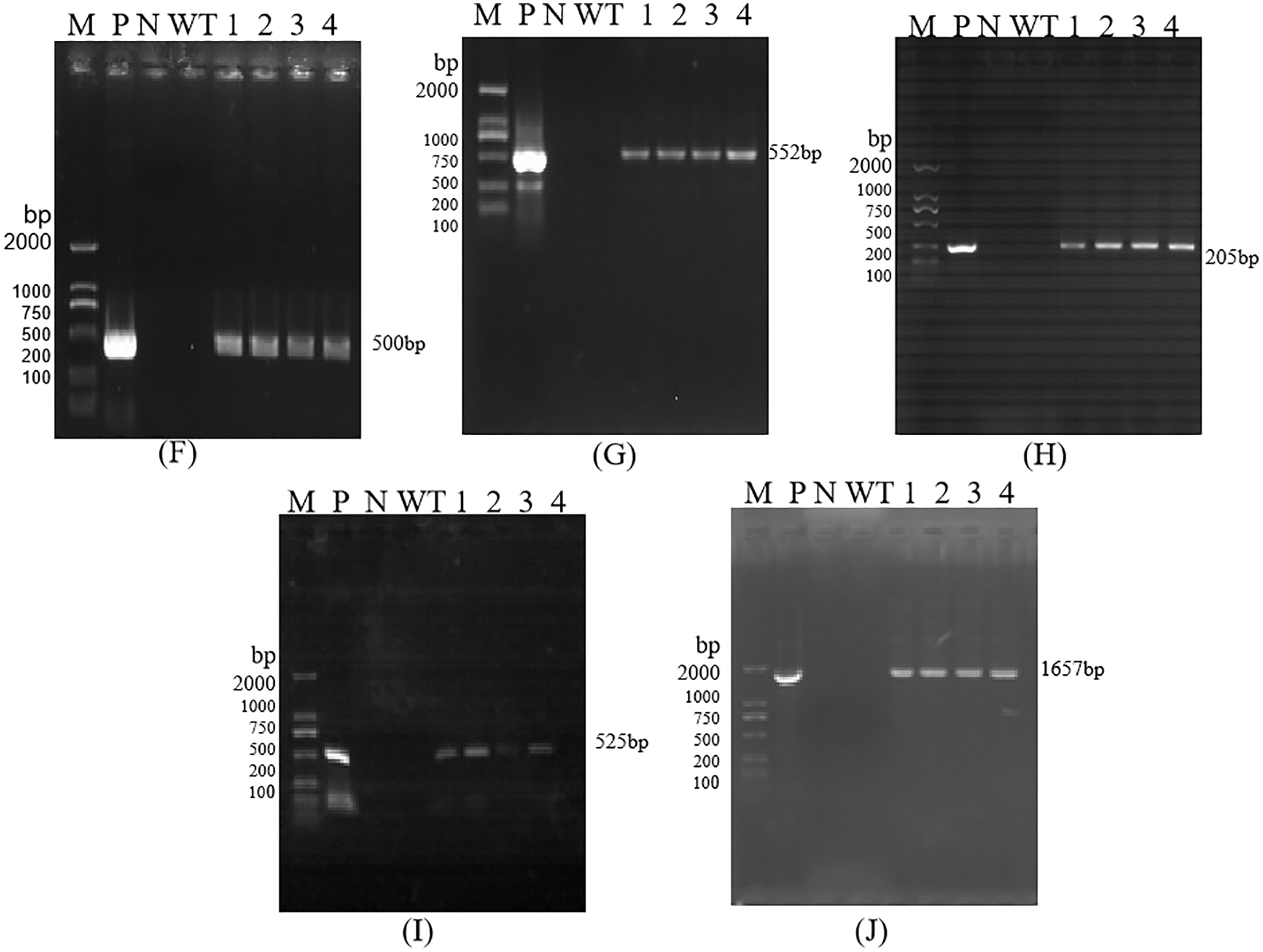
Figure S3: PCR detection of T2 Transgenic Maize; Each part of electrophoretogram is 35S (3F), Bar (3G), Nos (3H), NPR1 (3I), Cry1Ab13-1 (3J). Note: F–J are PCR detection of 35S, Bar, Nos, NPR1, Cry1Ab13-1; M: DL 2000 DNA marker; N: H2O; P: Positive control; WT: Untransformed plants; 1–4: Samples
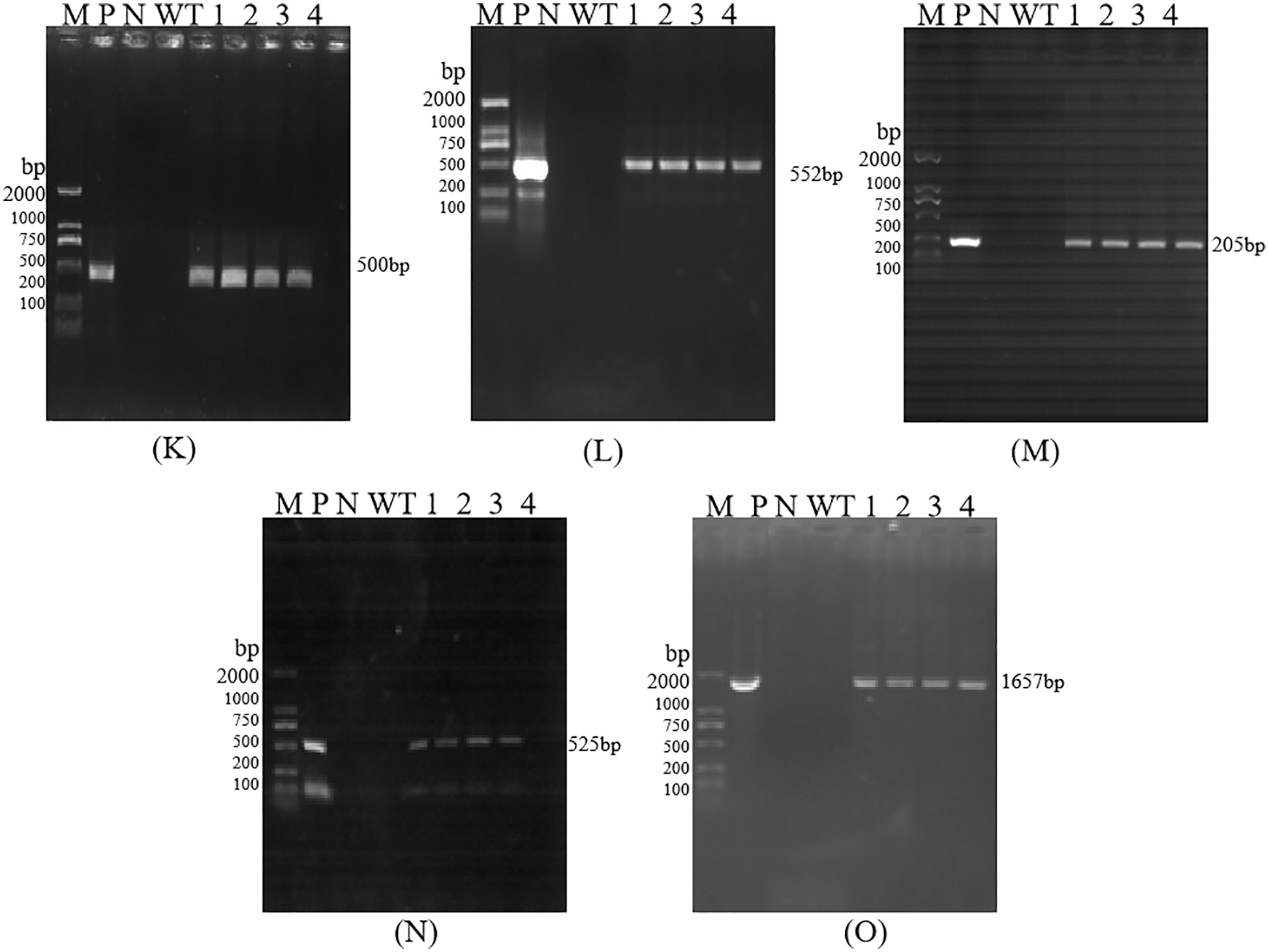
Figure S4: PCR detection of T3 Transgenic Maize. Each part of electrophoretogram is 35S (3K), Bar (3L), Nos (3M), NPR1 (3N), Cry1Ab13-1 (3O). Note: K–O are PCR detection of 35S, Bar, Nos, NPR1, Cry1Ab13-1; M: DL 2000 DNA marker; N: H2O; P: Positive control; WT: Untransformed plants; 1–4: Samples
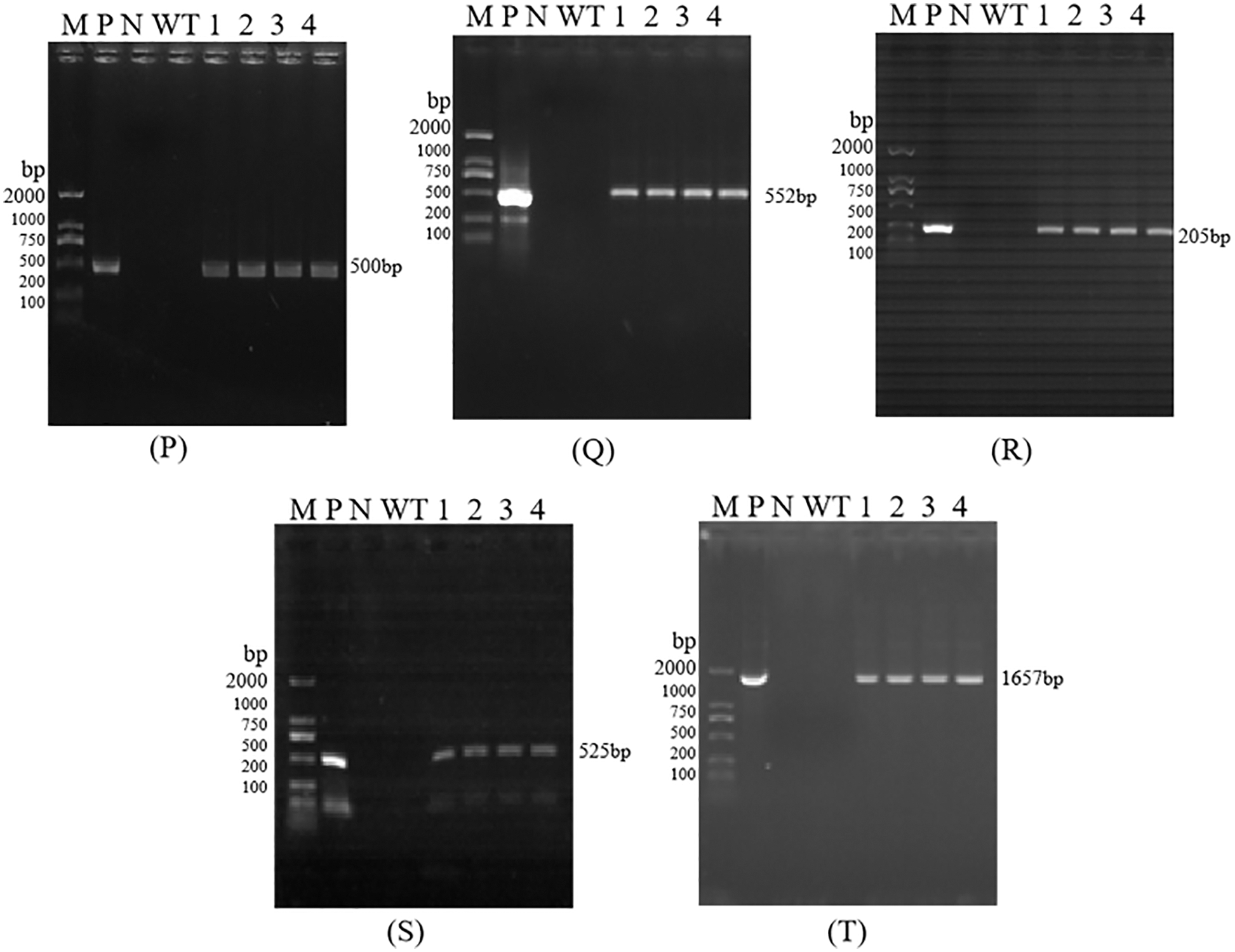
Figure S5: PCR detection of T4 Transgenic Maize. Each part of electrophoretogram is 35S (3P), Bar (3Q), Nos (3R), NPR1 (3S), Cry1Ab13-1 (3T). Note: P–T are PCR detection of 35S, Bar, Nos, NPR1, Cry1Ab13-1; M: DL 2000 DNA marker; N: H2O; P: Positive control; WT: Untransformed plants; 1–4: Samples
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools