 Open Access
Open Access
ARTICLE
Conventional Breeding and Molecular Markers for Blast Disease Resistance in Rice (Oryza sativa L.)
1
Agricultural Biotechnology Department, College of Agricultural and Food Science, King Faisal University, Al-Ahsa, 31982, Saudi Arabia
2
Plant Pathology Research Institute, Agriculture Research Center, Giza, 12619, Egypt
3
Rice Research and Training Center, Sakha, Field Crops Research Institute, Agricultural Research Center, Kafr El-Sheikh, 33717, Egypt
4
Biochemistry Department, Faculty of Agriculture, Cairo University, Giza, 12613, Egypt
5
Center of Excellence in Biotechnology Research, King Saud University, Riyadh, 11451, Saudi Arabia
6
Rice Biotechnology Laboratory, Rice Research Department, Field Crops Research Institute, ARC, Kafr EL-Sheikh, 33717, Egypt
* Corresponding Author: Hossam S. El-Beltagi. Email:
(This article belongs to the Special Issue: Plant–Environment Interactions)
Phyton-International Journal of Experimental Botany 2023, 92(3), 725-746. https://doi.org/10.32604/phyton.2022.024645
Received 03 June 2022; Accepted 10 August 2022; Issue published 29 November 2022
Abstract
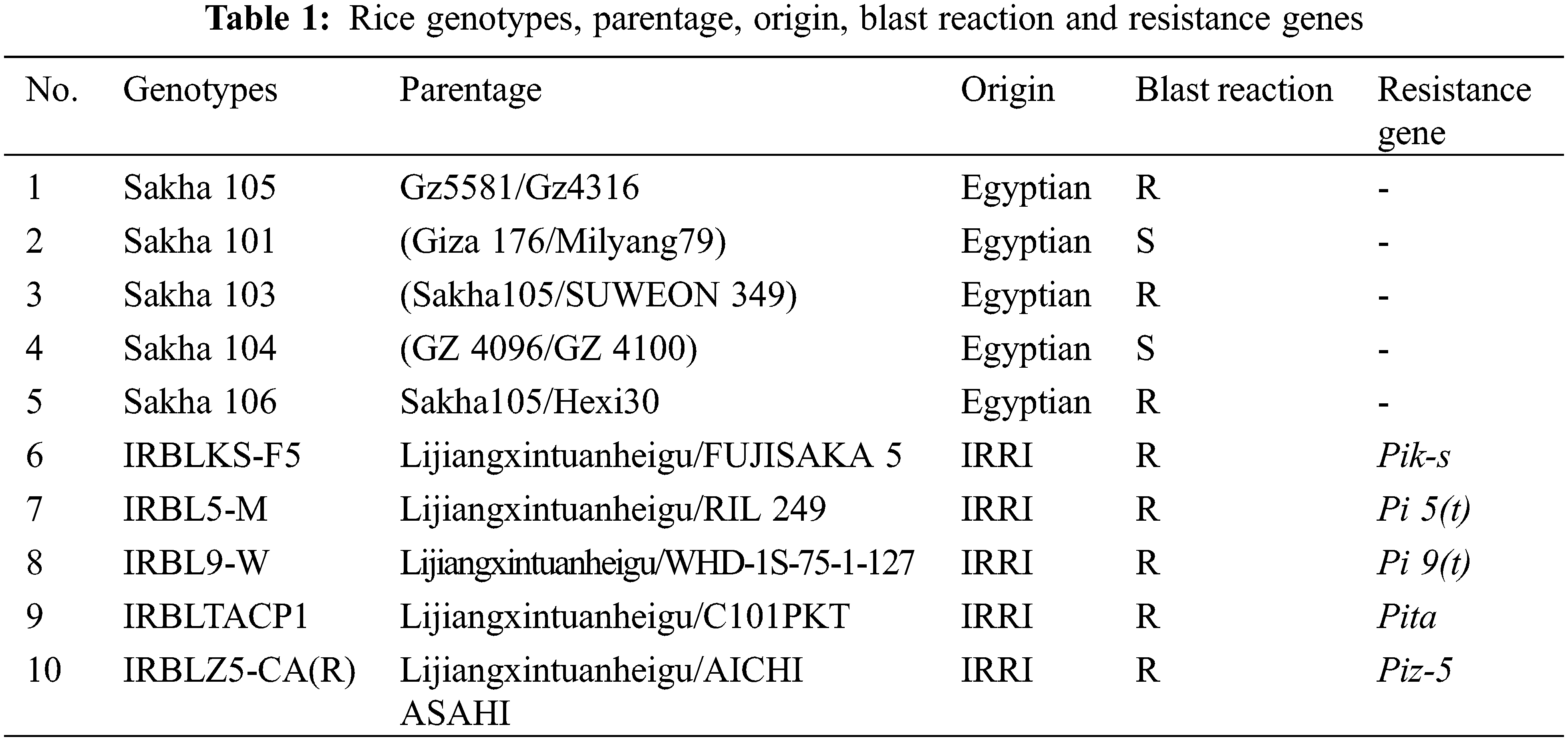
Monogenic lines, which carried 23 genes for blast resistance were tested and used donors to transfer resistance genes by crossing method. The results under blast nursery revealed that 9 genes from 23 genes were susceptible to highly susceptible under the three locations (Sakha, Gemmeza, and Zarzoura in Egypt); Pia, Pik, Pik-p, Piz-t, Pita, Pi b, Pi, Pi 19 and Pi 20. While, the genes Pii, Pik-s, Pik-h, Pi z, Piz-5, Pi sh, Pi 3, Pi 1, Pi 5, Pi 7, Pi 9, Pi 12, Pikm and Pita-2 were highly resistant at the same locations. Clustering analysis confirmed the results, which divided into two groups; the first one included all the susceptible genes, while the second one included the resistance genes. In the greenhouse test, the reaction pattern of five races produced 100% resistance under artificial inoculation with eight genes showing complete resistance to all isolates. The completely resistant genes: Pii, Pik-s, Piz, Piz-5 (=bi2) (t), Pita (=Pi4) (t), Pita, Pi b and Pi1 as well as clustering analysis confirmed the results. In the F1 crosses, the results showed all the 25 crosses were resistant for leaf blast disease under field conditions. While, the results in F2 population showed seven crosses with segregation ratio of 15 (R):1 (S), two cross gave segregated ratio of 3 R:1 S and one gave 13:3. For the identi- fication of blast resistance genes in the parental lines, the marker K3959, linked to Pik-s gene and the variety IRBLKS-F5 carry this gene, which was from the monogenic line. The results showed that four genotypes; Sakha 105, Sakha 103, Sakha 106 and IRBLKS-F5 were carrying Pik-s gene, while was absent in the Sakha 101, Sakha 104, IRBL5-M, IRBL9-W, IRBLTACP1 and IRBL9-W(R) genotypes. As for Pi 5 gene, the results showed that it was present in Sakha 103 and Sakha 104 varieties and absent in the rest of the genotypes. In addition, Pita-Pita- 2 gene was found in the three Egyptian genotypes (Sakha 105, Sakha 101 and Sakha 104) plus IRBLTACP1 monogenetic. In F2 generation, six populations were used to study the inheritance of blast resistance and specific primers to confirm the ratio and identify the resistance genes. However, the ratios in molecular markers were the same of the ratio under field evaluation in the most population studies. These findings would facilitate in breeding programs for gene pyramiding and gene accumulation to produce durable resistance for blast using those genotypes.Keywords
Many pathogens attack the rice crop, causing diseases with different symptoms that vary from species to species [1,2]. However, the most devastating one is blast disease, caused by Magnaporthe oryzae, which infects the leaves and panicles results in a huge loss in yield [3]. The estimation of the yield losses was recorded from 5% to 70%, according to the severity of infection [4–6]. Crop loss through blast disease has been estimated worldwide as sufficient as food for approximately 60 million people, and this crop loss was also estimated to be approximately $70 billion.
The blast disease infection is affected by different factors such as the number of resistance genes in the host cultivar, cultural practices, and climate change [7]. Resistance genes are used as a genetic tool to control the inheritance of blast disease characteristics and are considered as a cost-effective and environmentally beneficial mean for minimizing crop losses caused by disease. So far, more than 100 major blast resistance genes and 300 quantitative trait loci (QTL) have been identified and documented [8,9]. Many rice varieties have complete resistance to the blast, however, after a few cultivated years; the resistance has been broken down due to blast race changes and the appearance of newer, more virulent isolates of rice blast fungus [10]. Conventional breeding, thus, is still the mainstay for managing blast resistant cultivars to develop highly resistant cultivars to manage the disease. In addition it is a good method to understand the relationships between races and the resistance genes studied [11,12]. As a result, one of the goals of the breeding programs is to use resistance varieties as donors to transfer resistance genes to offspring, which can also be used to obtain durable resistance through gene pyramiding. The developed Japanese differential varieties (JDV) were the first varieties used as donors for blast resistance genes and have also been used to identify the individual genes responsible for blast resistance [13]. Development of monogenic lines with a single blast resistance genes were produced through backcross breeding method. Varieties and lines with known gene(s) for resistance were used as donors. Lijianxintuanheigu (LTH) was used as a recurrent parent in the backcrossing, through this method 23 resistance genes were transfer as individual Lijianxintuanheigu variety [14]. Therefore, the identification of blast resistance genes in the cross-parents is very important to transfer the resistance trait and accumulating many different resistance genes in a new variety, which can produce durable resistance. The molecular marker technology was used to identify and confirm the blast resistance genes and detect the chromosomal regions associated with such characters [15,16]. Many different molecular markers have been used, such as microsatellite or simple sequence repeat (SSR), inter-simple sequence repeat (ISSR), intron-exon splice junction (ISJ), Single Nucleotide Polymorphism (SNP), Sequence Tagged Microsatellites (STMS) and Sequence Tagged Sites (STS) to identify blast resistance genes [17–19]. The objectives of this study were to: 1) Use hybridization to transfer the blast resistance genes from monogenic lines to some Egyptian rice varieties; 2) Investigate the transmission of blast traits through the second generation; and 3) Use SNP and SSR markers to identify blast resistance genes in the parents under study and confirm the ration in the F2 generation.
This study was conducted at the experimental farm of the Rice Research and Training Center (RRTC), Sakha, Kafr El-Sheikh, Egypt and Agricultural Biotechnology Department, King Faisal University during the years 2018 to 2021. Ten parents were used as the genetic materials in this investigation (Table 1), including five Egyptian commercial varieties as parental lines, and five monogenic lines (carry different resistance genes) as “testers.”
The blast resistance evaluations for the thirty monogenic lines were achieved during the 2018 season with both natural infections at blast nursery test in three different locations (Sakha, Gemmeza, and Zarzoura) and artificial inoculation in the greenhouse and five of them were selected as testers (Table 1).
2.2 Greenhouse Evaluation under Artificial Inoculation
Seeds of monogenic lines varieties were seeded in plastic trays (30 cm × 20 cm × 15 cm). Each tray included 20 rows representing fifteen monogenic lines and five local varieties. The rest of the monogenic lines were sown in other trays. The trays were kept in the greenhouse and fertilized with Urea 46.5% N (5 g/tray). Seedlings were inoculated after 29 days from the sowing date. The seedlings were artificially inoculated with five rice blast isolates, i.e., Eg-5, IG-1, IB-45, 367, and 374. All tested lines were sprayed with spore suspension (100 mL) adjusted to 5 × 104 spores/mL. Each isolate was sprayed using an electrical spray gun. The inoculated seedlings were held in a moist chamber with more than 90% R.H. and 25°C–28°C for 24 h, and then moved to the greenhouse. Seven days after inoculation, blast reactions were recorded according to the standard evaluation system using a 0–9 scale [20].
Ten parental varieties (5 Egyptian as lines and selected 5 monogenics as testers) in this study were sown at three sowing dates at fifteen-day intervals to overcome the difference in heading dates among the parental varieties. At 30 days of age, the seedlings of the parents were transplanted to the experimental field in 3 rows, each five meters long and 20 cm apart between plants in each row. A line × tester cross was conducted among the 10 parents to produce 25 crosses. The hybridization technique was done according to Jodon [21] and Butany [22]. All the materials were arranged in a randomized complete block design (RCBD) with three replications; each replication contained 25 individual plants. The 25 populations of F2 materials, each consisting of more than 240 individual plants, were planted and evaluated for blast nursery.
2.4 Blast Scoring at Blast Nursery Test
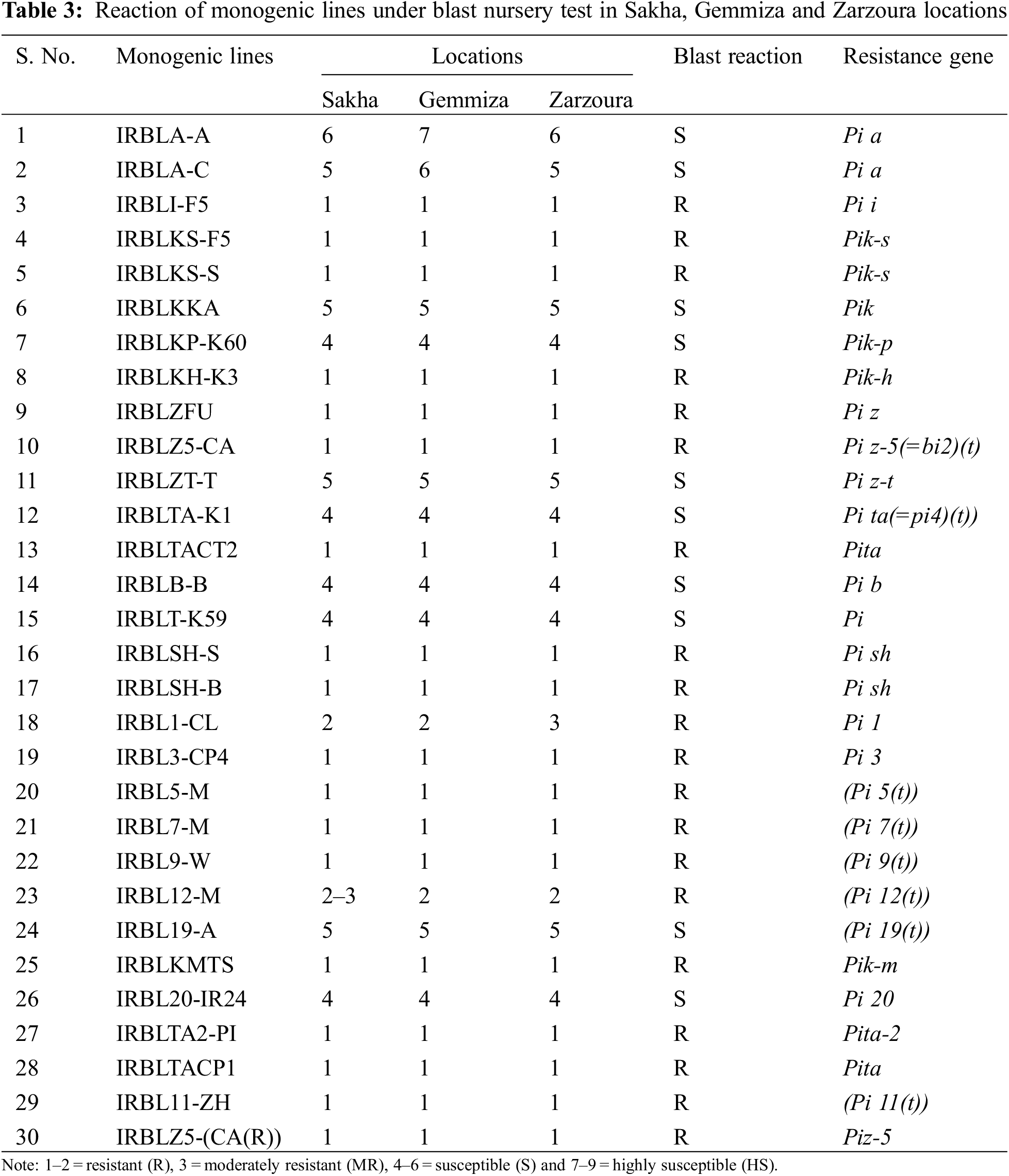
In the blast nursery, all the breeding materials (parents, F1 and F2) were tested against the natural infestation of blast races under the optimum conditions for blast infection, like the delay of sowing dates, increasing the amount of nitrogen fertilization, growing susceptible check (blast spreader), growing and applying in different locations. The plants’ materials were evaluated for their reaction against M. grisea at the blast nursery in three locations: Sakha, Gemmiza, and Zarzora during 2019, 2020, and 2021 seasons for blast resistance at the seedling stage. About forty days from the sowing date, the typical blast lesions were scored according to the Standard Evaluation System using a 0–9 scale [20] as follows: 1–2 = resistant (R), 3 = moderately resistant (MR), 4–6 = susceptible (S), and 7–9 = highly susceptible (HS).
The quantities and qualitative characters; leaf blast reaction at seedling stage (LBR), heading date (DH-day), plant height (PH-cm), number of panicles/plant (NPP), Spikelet fertility% (SF), 1000-grain weight (1000GW-g), grain yield/plant (GY-g), panicle weight (PW-g) and number of primary branches were evaluated for F1 according to the Standard Evaluation System IRRI [20]. As well as 25 F2 populations at an adult stage were evaluated to study the inheritance of blast resistance traits in the segregating populations. The Chi-square x2 test was computed as follows [23]. The t-test was used to examine the existence of genetic variance between parental means.
Correlation of traits: Correlation among the values of the traits estimated was calculated using Pearson correlation coefficients and plotted via the packages corrplot and Performance Analytics [24].
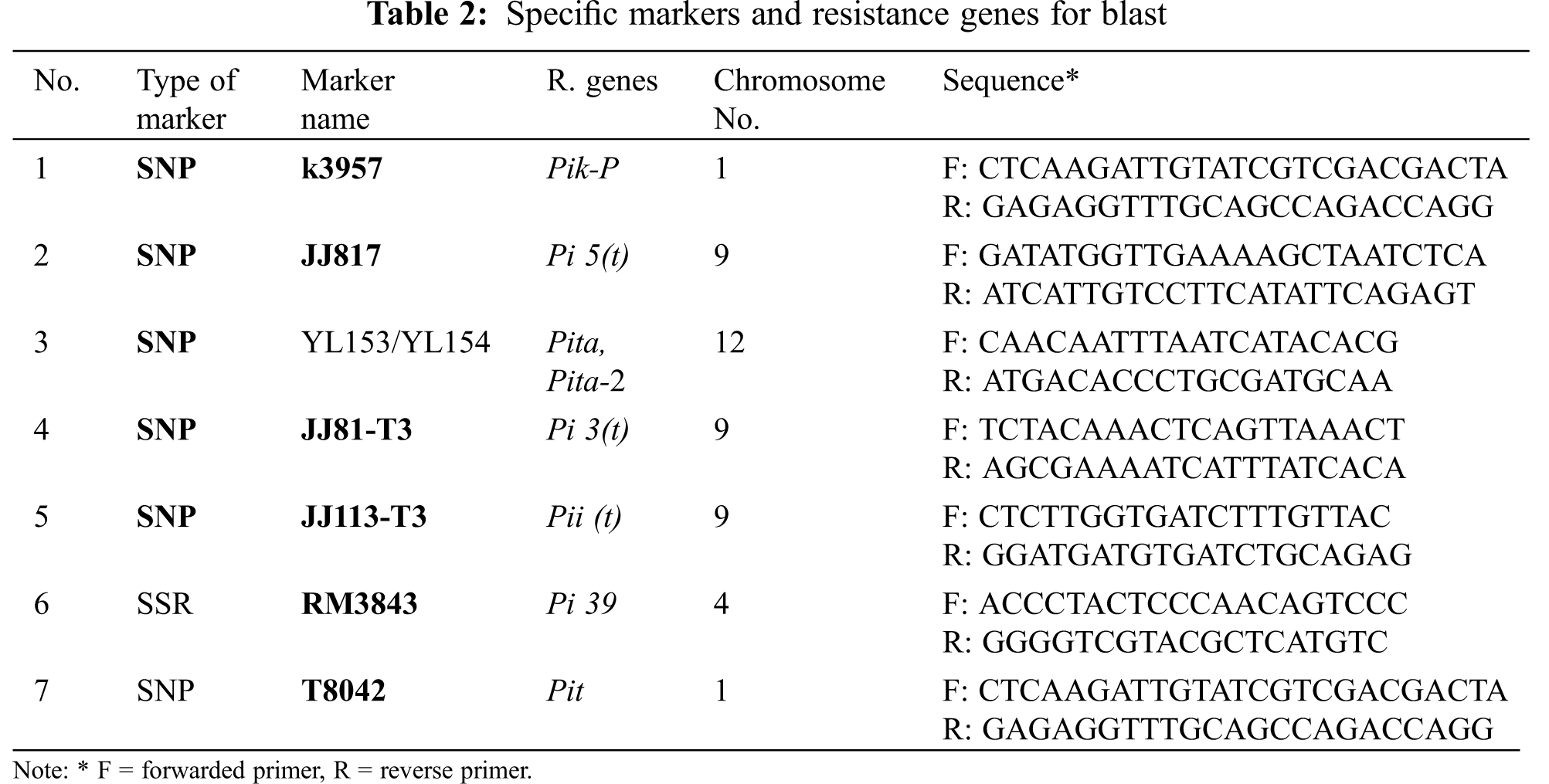
Ten parental genotypes and six F2 populations were employed in the study to identify the blast resistance genes using seven specific primers purchased from Sangon Company, China. DNA Genomic extraction of the parental genotypes and F2 were carried out [25]. Seven markers (Table 2) were used for this investigation. Six of them were Single Nucleotide Polymorphism (SNP) and one was an SSR marker.
2.6 Polymerase Chain Reaction (PCR) Assay
The reaction mixture (25 μL) consisted of: 12.5 μL of 2× master mix ready to use {0.1 U/μL Taq Polymerase, 20 mM Tris-HCl (pH8.3), 100 mM KCl, 3 mM MgCl2, and stabilizer and enhancer)}, 10 Pmol of each primer (1.0 μL), 1.0 uL of DNA (50 ng) and 9.5 μL of PCR grade water. Amplifications were performed in a thermocycler (Bio-Rad, C-1000, California, USA) as follows: (1) initial denaturation at 94°C for 5 min; (2) 30 s of denaturation at 94°C; (3) primers’ annealing temperatures differ according to their Tm, for 1 min; (4) one-minute extension at 72°C; (5) Steps 2, 3, and 4 are repeated 40 times; (6) a final extension of 10 min at 72°C was given. The amplified products were analyzed on a 2% agarose gel electrophoresis containing ethidium bromide and then photographed and analyzed using BioDoc Analysis software (Biometra, Germany).
3.1 Scoring of Monogenic Lines under Natural Infection
The blast reactions of monogenic lines were tested at three locations in the blast nursery and the results are shown in Table 3. The tested monogenic lines exhibited different reactions to the dominant races in the blast nurseries and natural fields according to the response of their genes (Table 3). However, the reaction revealed that 10 out of 23 genes were susceptible to highly susceptible conditions at three locations, while the rest of the genes were highly resistant at the same locations (Table 3).
Some alleles of the Pik gene were resistant, while others are susceptible at the same location. The results demonstrated that the most effective genes under both natural and artificial inoculation were Piz and Pii. These two genes are resistant under Egyptian conditions in the three locations. Therefore, they can be employed to solve the broken-down resistance varieties. Clustering analysis divided all the genotypes into two groups (Fig. 1). The first group included all the susceptible genes, while the second group included the resistance genes. Generally, resistance genes for blast commonly called Pi were helping and providing a broad-spectrum resistance against the most prevalent races and can be extremely valuable in rice breeding efforts [6,26].
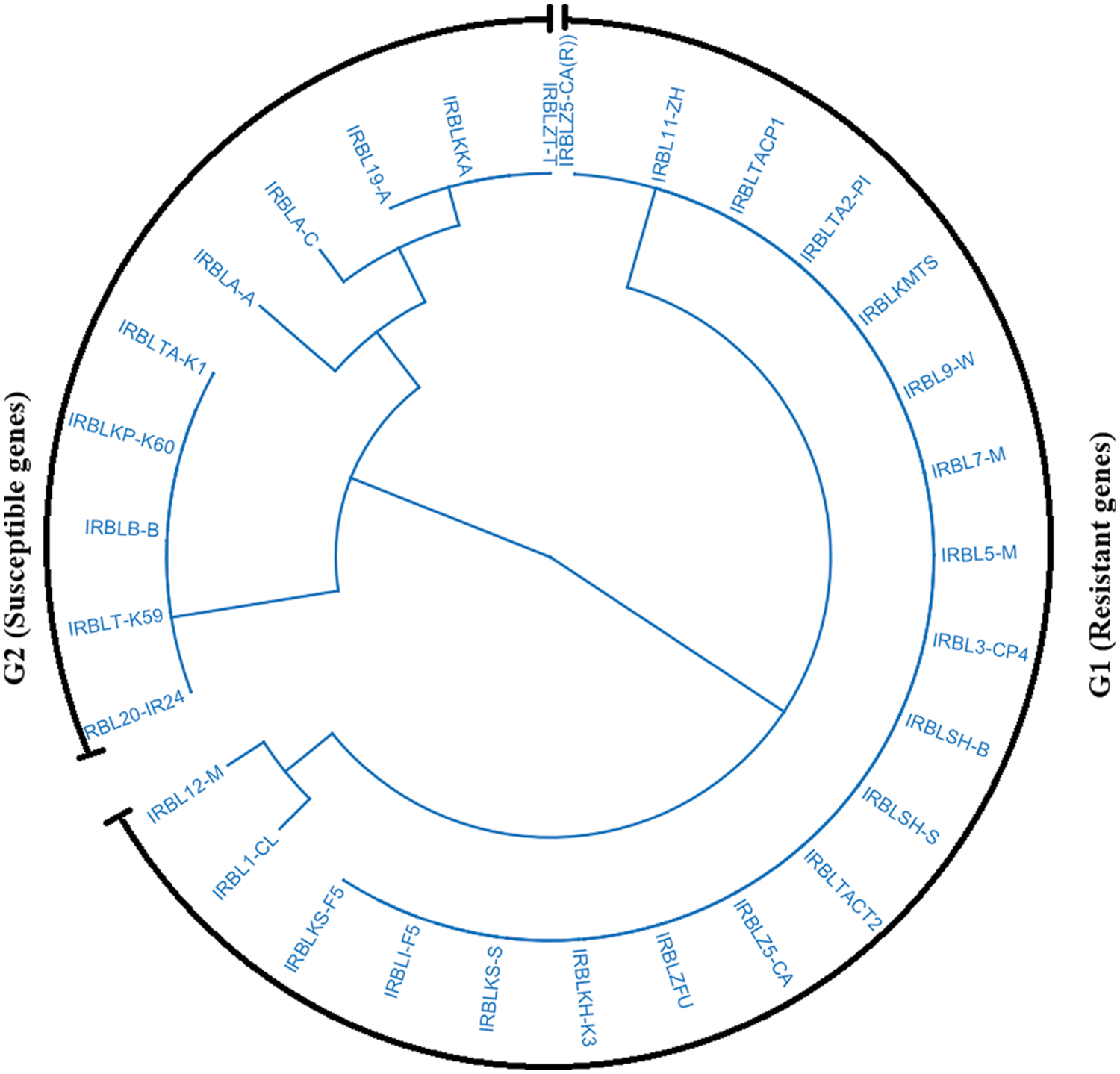
Figure 1: Dendrogram for monogenic lines according to the reactions in different locations
In addition, it has become necessary to know and identify these genes, to be used in hybridization to transfer the resistance trait into new varieties [3,11]. In another context, the modern or new varieties are better to carry multiple resistance genes to display broad-spectrum and durable resistance in the field or gene accumulation and gene pyramiding. In any case, the traditional method was used for producing monogenetic lines to make crosses between them and the commercial varieties whose resistance is required to be improved, and with molecular markers to identify resistance genes.
3.2 Evaluation of Monogenic Lines under Artificial Infection
Five specific blast races were used to evaluate monogenic lines under greenhouse conditions. The selected five isolates were classified as pathogenic and were used as differential blast isolates to estimate resistance genes by reaction pattern. The isolate Eg-5 was used as a highly virulent and most aggressive isolate for different local varieties (Table 4).
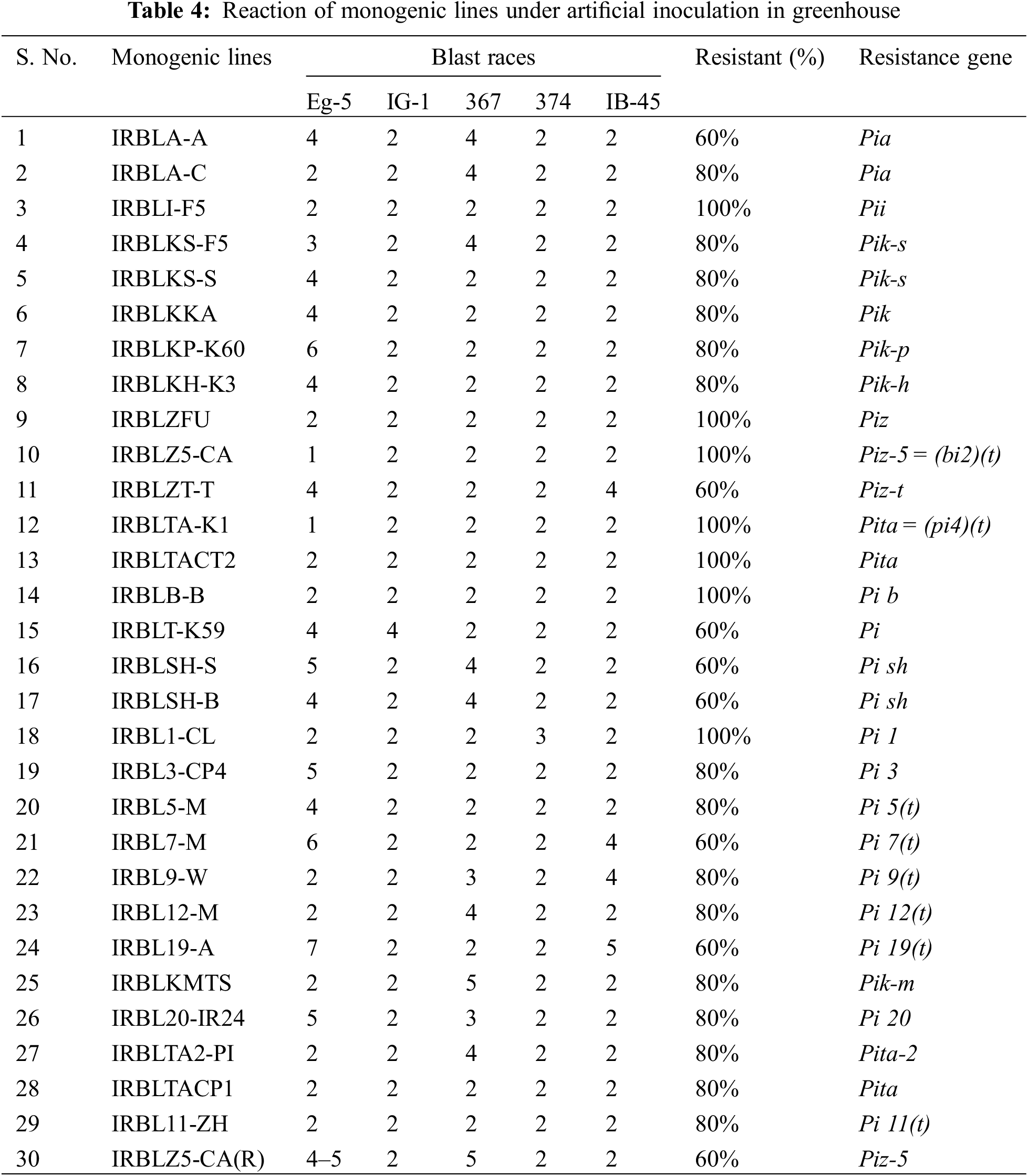
The results showed that, according to the reaction patterns of five races under artificial inoculation, seven genes (Pii, Piz, Piz-5 = (bi2)(t), Pita = (Pi4)(t), Pita, Pi b, and Pi 1) showed complete resistance to all isolates. Only five of the seven completely resistant genes (Pii, Pik-s, Piz, Piz-5 = (bi2)(t), and Pita = (Pi4)(t)) were resistant and effective under both natural and artificial blast nursery conditions. These results indicated that the monogenic lines that carry these genes could be recommended to be used as donors in breeding programs to improve resistance to blast disease. On the other hand, the genes Pia, Pik-s, Pik, Pik-p, Pik-h, Pi 3, Pi 5(t), Pi 9(t), Pi 12(t), Pik-m, Pita-2, Pita and Pi 11(t) gave 80% resistance, while the genes Pia, Piz-t, Pi, Pi sh, Pi 7(t), Pi 19(t) and Piz-5 gave 60% resistance (Table 4). These results were found with [27–30].
Clustering analysis (Fig. 2) confirmed the results, which were divided into two major groups. The first one included IRBL7-M and IRBL19-A, which carry the Pi 7 and Pi 19 genes, and the resistance percentage was 60% under the five races in the greenhouse test, while, the second group was divided into two sub-groups. The first sub-group included Pia, Pik-s, Pik-p, and Pi 5 genes, and the second sub-group included Pia, Pita-2, Pik-m, Pii, Piz, Pi b, Piz-5, and Pi (t).
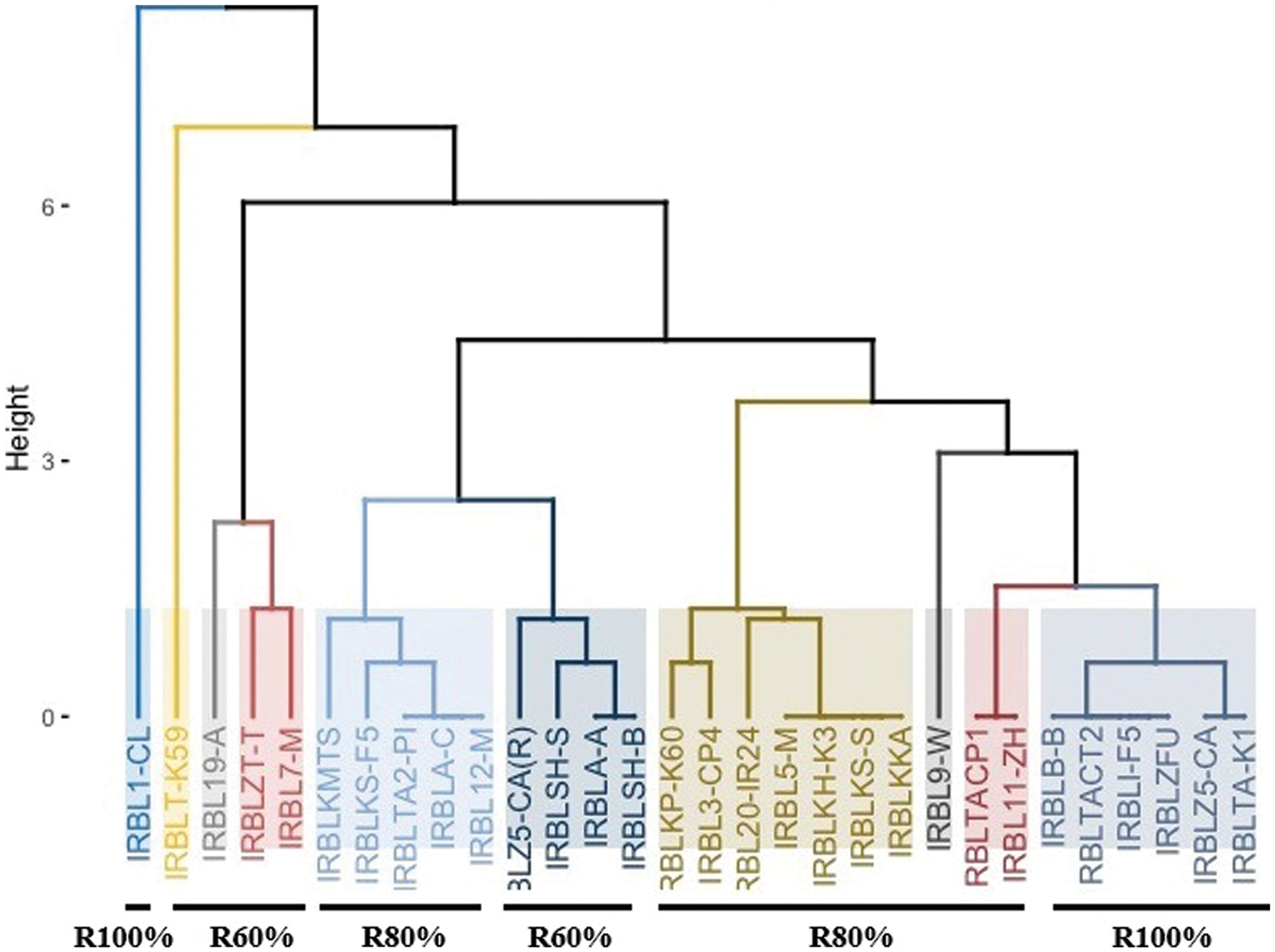
Figure 2: Cluster analysis for monogenic lines according to the artificial inoculation in greenhouse
The rice breeder can use the parents between clusters as parents for future hybridization rather than within clusters for a successful breeding program and selection of genetically diverse parents as an important pre-requisite to obtain better and desirable recombinants. These results agreed well with earlier researchers [31–33].
The result of ANOVA analysis (Table 5) was performed to test the difference between the parents and hybrids for all the studied traits. Results revealed that mean squares due to genotypes were significant for all the traits. The mean squares due to genotypes were further partitioned into parents, crosses and parents vs. crosses. The differences among parents were highly significant for all traits, indicating the presence of wide genetic variability among parents for almost all the traits. The mean square values due to crosses for all traits were found to be significant at 0.01 levels. Parents vs. crosses mean square values further revealed highly significant differences in all crosses. In addition, male testers exhibited highly significant differences for all traits. The highly significant mean squares of lines × testers for all traits indicated that they interacted and produced markedly different combining ability effects, and this might be due to the wide genetic diversity of lines and testers. In addition, the mean squares due to lines vs. testers were significant for all traits, which indicated that female and male parents differed significantly for these traits. These results are similar to those obtained by [34,35].

The results in Table 6 showed the mean performance of parents, lines, and their hybrids for the studied blast resistance traits. All testers showed high resistance to leaf blast and carried different resistance genes (Pik-s, Pi 5 (t), Pi 9 (t), Pita, and Piz-5, respectively). Furthermore, all varieties were resistant, except the Sakha 101 and Sakha 104 varieties, which were highly susceptible to the leaf blast trait, while F1 crosses recorded resistance to leaf blast disease under field conditions. Moreover, the results revealed that all crosses that were driven by parents’ resistance × resistance parents and resistance × susceptible parents were all resistant in F1. These results indicated that the parents carried dominant genes for resistance and that resistance was completely dominant over susceptibility to blast. For agronomic traits, the mean performance of parents, lines, and their hybrids for studied traits are presented in Table 6.

Testers showed a considerable number of early values, and lines showed a lower number of values than testers. The genotypes IRBLKS-F5 and IRBLTACP1 had the shortest duration among the tester lines, while the Sakha 103 and Sakha 105 varieties were the earliest maturing varieties among the lines. The best combinations were Sakha 103 × IRBLKS-F5 (116 days), Sakha 105 × IRBL9-W (R) (117), Sakha 103 × IRBL9-W (117 days) and Sakha 103 × IRBL9-W (117 days). Regarding the yield traits for testers, lines, and hybrids (Table 6), in general, lines demonstrated higher values for panicle/plant, spikelet fertility%, 1000-grain weight, and grain yield/plant than testers. However, some hybrids indicated high values for yield traits, such as Sakha 105 × IRBL9-W and Sakha 101 × IRBL5-M, which gave the highest values (62.1 and 61.5, respectively). Heatmap analysis data (Fig. 3) observed the significant differences among genotypes in the studied traits (Fig. 3). Moreover, the heatmap analysis classified the genotypes based on their mean performance and their response to blast reaction. The figure showed that only two susceptible varieties were grouped and other genotypes were resistant.
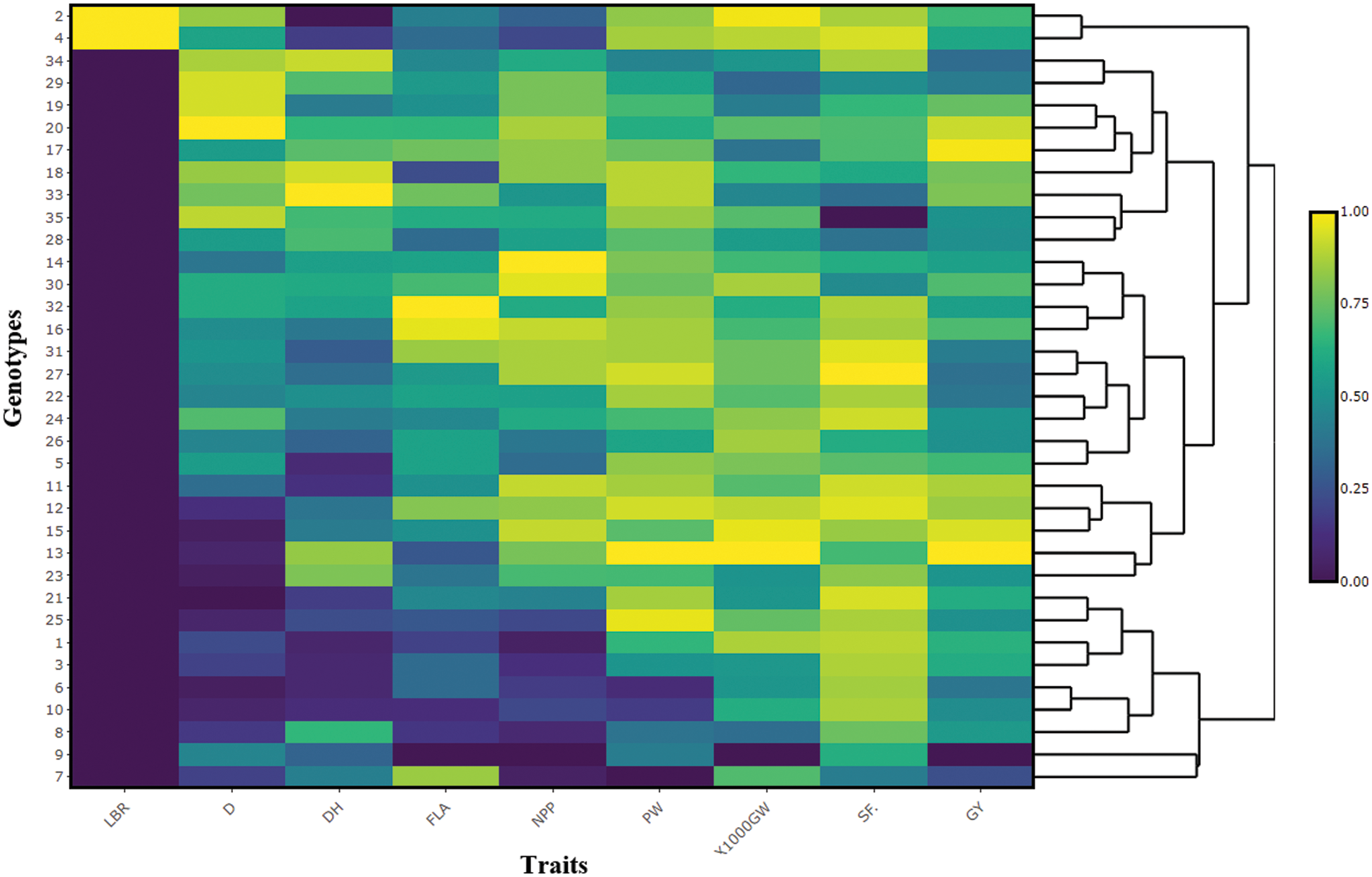
Figure 3: Heat map analysis of nine traits across 35 genotypes
Correlation analysis of the nine studied traits revealed that there was negative correlation between blast reaction (LBR) and D, PW, and 1000 GW, respectively (Fig. 4).
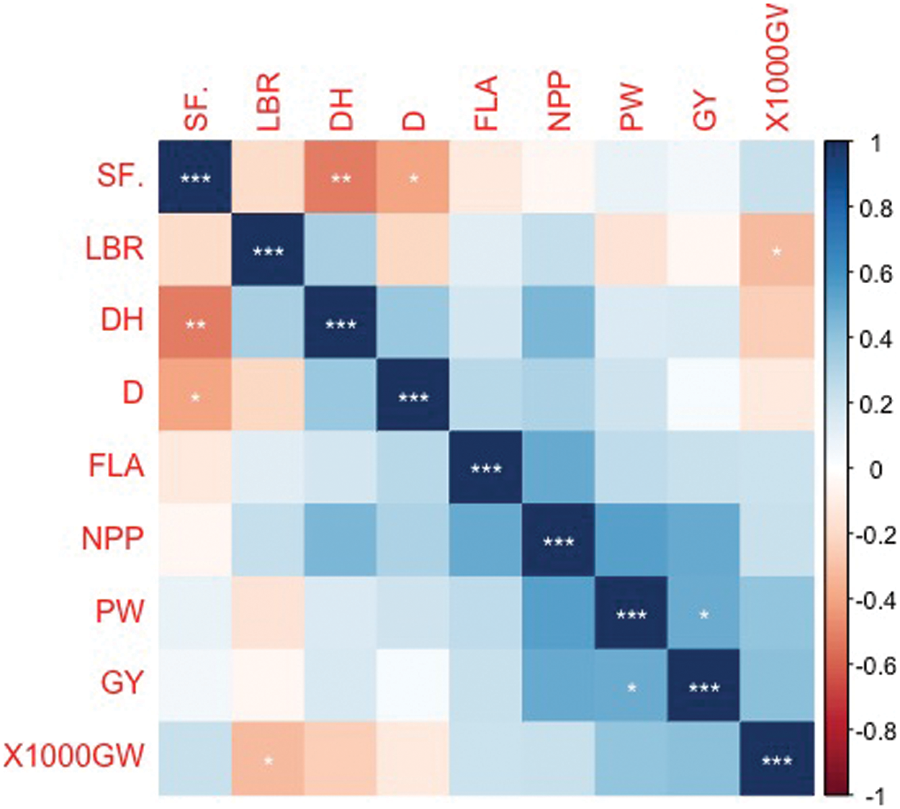
Figure 4: Corrplot depicting Pearson’s correlation among nine traits across 35 genotypes. Red squares indicate a negative correlation; blue squares indicate a positive correlation; and white squares indicate no correlation. The asterisks indicate significant correlations using a two-tailed t-test (* and ** = P < 0.05; and *** P = < 0.01)
The present finding was also supported [36,37]. On the other hand, clustering analysis confirmed the results and divided the genotypes into two groups. The first one included all the susceptible genes, while the second one included the resistance genes. The ordinary analysis of variance for lines, tester and line × tester were shown to be highly significant mean squares for all studied traits, which indicated overall differences among the lines, tester and line × tester (Table 5). These results are in agreement with [38,39].
3.5 Phenotypic Traits and Their Segregation in the F2 Population
Twenty-five segregation F2 populations are presented in Table 7, and the results showed that eight crosses showed a segregation ratio of 15 (R) to 1 (S).

This indicates the presence of two resistance genes to leaf blast segregating in these crosses, and each gene can express resistance in the genetic background. In addition, each parent in these crosses contained one of these genes, and the allelic relationship was complete dominance. On the other hand, two cross gave a segregated ratio of 3 (R) to 1 (S), which indicated the presence of one dominant major resistance gene transferred from these resistant parents to their offspring that controlled the resistance against blast (Table 7). Concerning the second cross, the expected ratio of 13:3 and the percentage of type reaction 139 (R) % and 21% (S) %, respectively, with P-value (0.10–0.50), indicated the presence of two complementary dominance genes in these crosses. On the other hand, the last one contained fourteen crosses that were found to be resistant in F1 and all the F2 plants were resistant without segregation, which indicates that the resistance genes in those parents could be the same or allelic. Finally, phenotypic segregation in F2 was used to study the inheritance of resistance in offspring, which will help plant breeders in the process of resistance. However, the segregation ratios were depending for the number of genes controlling these traits such as [15 (R) to 1 (S)] means that there are two resistance genes and each gene can express resistance. While, 3 (R) to 1 (S) indicated the presence of one dominant major resistance gene transferred from these resistant parents to their offspring that controlled the blast resistance [40,41]. Also, two complementary dominance genes were found in some crosses and clarified the ratio of 13:3 [42,43].
3.6.1 Identification of Blast Resistance Genes in the Parental Lines
Three specific markers (K3959, JJ817 and YL153/YL154) were used to identify blast resistance genes in the ten parents. Those markers are linked to Pik-s, Pi 5 and Pita-Pita-2 genes, respectively. The results as in Fig. 5 showed that the marker K3957 was linked to the Pik-s gene that is harbored in the IRBLKS-F5 monogenic line. This monogenic line was used as a control to check the presence of Pik-s gene in the Egyptian varieties. In any case, the results showed that the three Egyptian varieties (Sakha 105, Sakha 103, Sakha 106) and IRBLKS-F5 were positive to Pik-s gene (Fig. 5A) and two alleles (size 450 and 600 bp) were detected. While, was these were absent in Sakha 101, Sakha 104, IRBL5-M, IRBL9-W, IRBLTACP1 and IRBL9-W(R) genotypes. As for JJ817 marker, which linked with Pi 5 gene, the monogenic line IRBL5-M was carrying this gene and was used as a donor for Pi 5 gene in this study. The results as in Fig. 5B showed that Pi 5 gene was present in Sakha 103 and Sakha 104 parents and absent in the rest of the genotypes.
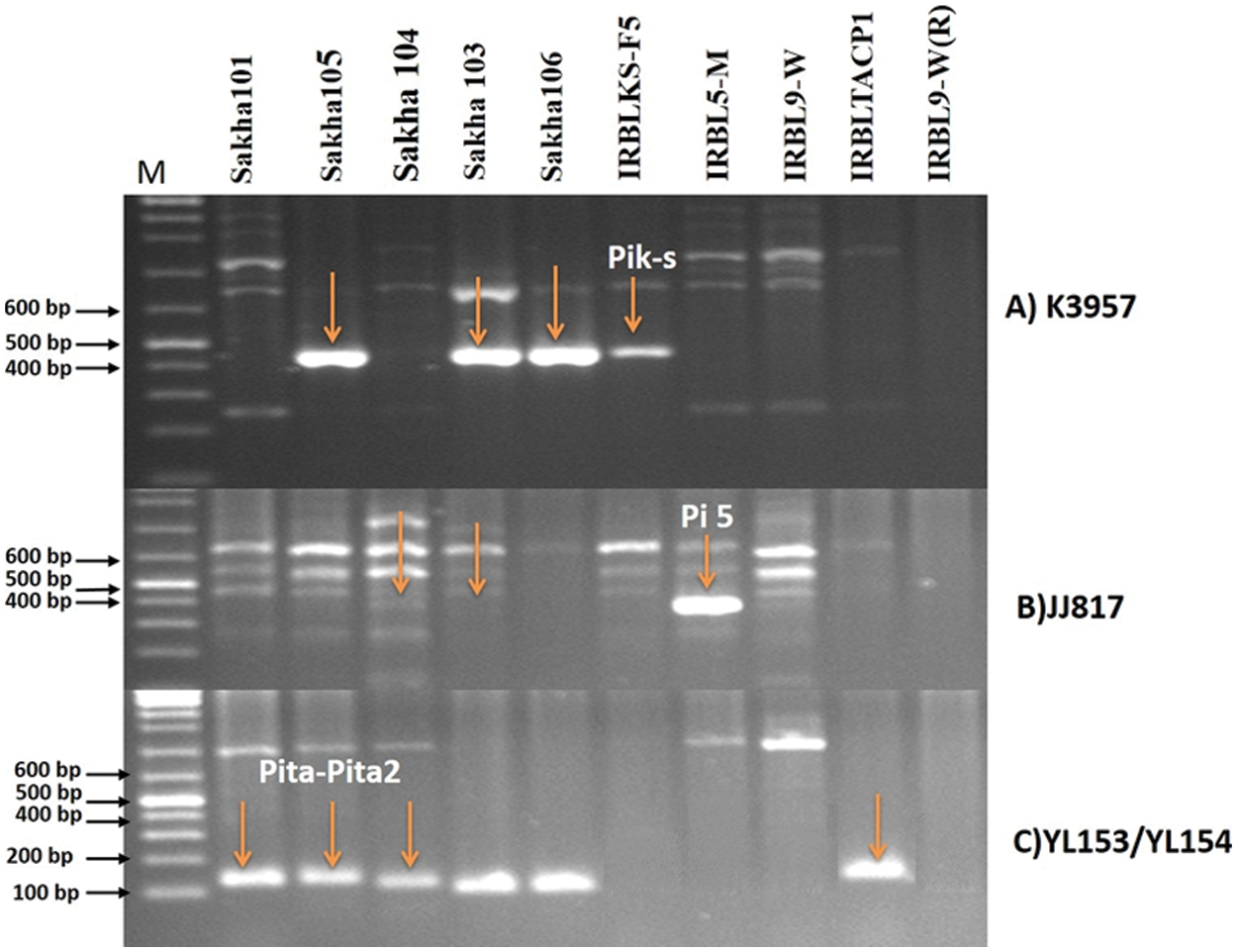
Figure 5: Amplification pattern of markers. A) K3957, B) JJ817and C) YL153/YL154, M: 100 bp DNA ladder, 1–10: denotes 10 rice genotypes included in the study, arrows indicate the specific amplified alleles
Although Pi 5 resistant gene is detected in Sakha 104 while it was susceptible to rice blast infection in the field evaluation. This means that this gene may have no effect under the Egyptian conditions or there are any other gene(s) is affecting the expiration of this gene. Similar results were obtained with previous studies [44–47]. Positive amplified fragments for YL153/YL154 marker that linked to Pita-Pita-2 gene were detected in three Egyptian genotypes (Sakha 105, Sakha 101, and Sakha 104) and IRBLTA-CP1 monogenic line. This monogenic line was used as a donor for Pita gene (Fig. 5C). On the other hand, this gene was absent in other four monogenic lines {IRBLks-F5, IRBL5-M, IRBL9-W Pi 5(t) and IRBLz5-CA (R)}. There are many studies that showed the importance of identifying the blast resistance genes, which helped in rice breeding programs for resistance [48–50].
In any case, this study showed the presence of Pita gene in cultivar Sakha 101, which was susceptible to blast disease in the field. This means that this gene is inactive under Egyptian conditions or maybe working alone to show resistance in Sakha 101. Whereas, Sakha 105 and Sakha 103 were resistant and the results showed the presence of Pik-s gene in those two varieties, explaining that the action of this gene is through its presence in genetic groups or that it works in complementary genes. Some of these genes were also found to be highly effective against rice blast either under Egyptian conditions [15,16] or under Chinese conditions [18].
3.6.2 Genetic Analysis in F2 Population
Six populations and four markers were used to confirm the expected ratio and study the inheritance of blast resistance trait. The populations namely; Sakha 101 × IRBLKS-F5, Sakha 101 × IRBL5-M, Sakha 101 × IRBLTACP1, Sakha 104 × IRBL9-W(R), Sakha 104 × IRBL9-W and Sakha 104 × IRBLTACP1. The result in Table 8 showed that all the population produced 15 positives: 1 negative band with the markers JJ81-T3, JJ113-T3, RM3843 and T8042 (Fig. 6). Except for the population Sakha 104 × IRBL9-W the ratio was 3 positive: 1 negative with all the primers [51–56]. However, the data for F2 generations were categorized into two groups according to segregation ratios. The first group, was segregated in a ratio of 15 (R) to 1 (S). The ratio 15 (R) to 1 (S) suggested that the two genes of leaf blast resistance were segregating in this cross. In addition, the same data was found in molecular markers and it confirmed that two genes were controlled in blast resistance in this cross. On the other hand, the second group of the F2 population showed segregation of 3 (R) to 1 (S). This indicated that the resistance was transferred from resistant varieties that carried one major gene for resistance to blast disease. These results were in agreement with those of [11,16].


Figure 6: PCR products of gene markers JJ81-T3, JJ113-T3, RM3843 and T8042 for F2 segregations produced from some crosses R = resistance line and S = susceptible line
In conclusion results of segregation ratio in group one (15 (R) to 1 (S)) in the F2 generations suggested that resistant varieties carried two dominant resistance genes for leaf blast (e.g., A and B), while the susceptible variety carried their recessive alleles. While, in the second group, which showed a segregation ratio 3 to 1 in F2 the data suggested that the genetic constitution of resistance monogenic lines could be carried by one dominant gene (AAbb). The current study clarified the importance of using the monogenic lines that are resistant to blast and use to produce and improve new varieties through the traditional breeding methods. Identification of blast resistance genes are very important in breeding program to improvement new resistance varieties [57,58]. Monogenic lines were produced to help the breeders for identifying and transfer resistance genes by conventional breeding method. In any case, there are two types of resistance: vertical resistance, which is controlled by a large number of genes with a minor effect, and horizontal resistance, which is controlled by a small number of genes with major effect. The fluctuation in effective resistance genes from one season to another and between this study and other studies may be due to the prevalence of common physiological races in every season and every location, this is agreeable by many investigators. Also, the described this phenomenon in different countries [59–61] and observed that virulent strains existed for all the identified genes of vertical resistance and most of the strains possessed virulent genes which were not necessary for their survival. In any case, Tables 3 and 4 explained the same differences between the locations under study and also the different degrees of infection between the races or strains under artificial inoculation, which confirms the theory of gene for gene.
Thirteen genes for blast were resistant under this study and the most effective genes under both natural and artificial inoculation were Pi-Z and Pi-I genes under field conditions. While, under artificial inoculation, seven genes showed complete resistance to all isolates and gave 100% resistance. Pii, Piz, and Piz-5 = (bi2) (t), Pita = (Pi4) (t), Pita, Pib, and Pi1 are the completely resistant genes. The genotypes IRBLKS-F5 and IRBLTACP1, Sakha 103 and Sakha 105 varieties were the earliest varieties among the lines. The best combinations were Sakha 103 × IRBLKS-F5, Sakha 105 × IRBL9-W (R), Sakha 103 × IRBL9-W and Sakha 103 × IRBL9-W. Different variation was also noted among the F2 population and identification of different resistance genes against blast is an effective way of improving the resistance of rice varieties. Four rice varieties namely; Sakha 105, Sakha 103, Sakha 106 and IRBLKS-F5 carry Pik-s gene. While, Pita-Pita2 gene the results showed positive amplification in the five Egyptian genotypes; Sakha 105, Sakha 101, Sakha 103, Sakha 104 and Sakha 106. The genotypes could be used for gene pyramiding and gene accumulation to produce durable resistance to blast.
Authorship: The authors confirm contribution to the paper as follows: study conception and design: Adel A. Rezk, Mohamed M. El-Malky, analysis and interpretation of results Mohamed M. El-Malky Mohamed Al-daej and Kotb A. Attia, draft manuscript preparation Adel A. Rezk, Kotb A. Attia and Hossam S. El-Beltagi. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgement: Authors extend their appreciation to the Deanship of Scientific Research, King Faisal University, Saudi Arabia, for supporting this research for work through Project No. NA000112.
Funding Statement: Authors extend their appreciation to Deanship of Scientific Research, King Faisal University, Saudi Arabia, for supporting this research (NA000112).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Zhu, D., Kang, H., Li, Z., Liu, M., Zhu, X. et al. (2016). A genome–wide association study of field resistance to Magnaporthe oryzae in rice. Rice, 9(44), 1–9. DOI 10.1186/s12284-016-0116-3. [Google Scholar] [CrossRef]
2. Xiao, N., Pan, C., Li, Y., Wu, Y., Cai, Y. et al. (2021). Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biology, 22(232), 1–22. DOI 10.1186/s13059-021-02488-8. [Google Scholar] [CrossRef]
3. Nizolli, V. O., Pegoraro, C., Deoliveira, A. C. (2021). Rice blast: Strategies and challenges for improving genetic resistance. Crop Breeding and Applied Biotechnology, 21(9), 1–15. DOI 10.1590/1984-70332021v21Sa22. [Google Scholar] [CrossRef]
4. Shehab, G. G., Ahmed, O. K., El-Beltagi, H. S. (2010). Effects of various chemical agents for alleviation of drought stress in rice plants (Oryza sativa L.). Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 38, 139–148. [Google Scholar]
5. Nalley, L., Tsiboe, F., Durand-Morat, A., Shew, A., Thoma, G. (2016). Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLoS One, 11(12), 1–15. e0167295. DOI 10.1371/journal.pone.0167295. [Google Scholar] [CrossRef]
6. Agbowuro, G. O., Afolabi, M. S., Olamiriki, E. F., Awoyemi, S. O. (2020). Rice blast disease (Magnaporthe oryzaeA menace to rice production and humanity. International Journal of Pathogen Research, 4(3), 32–39. DOI 10.9734/ijpr/2020/v4i330114. [Google Scholar] [CrossRef]
7. Elbasiouny, H. Y., Elbehiry, F. (2020). Rice production in Egypt: The challenges of climate change and water deficiency. In: Climate change impacts on agriculture and food security in Egypt, pp. 295–319. Switzerland AG: Springer International Publishing. DOI 10.1007/978-3-030-41628-7. [Google Scholar] [CrossRef]
8. Angeles-Shim, R. B., Reyes, V. P., del Valle, M. M., Lapis, R. S., Shim, J. et al. (2020). Marker-assisted introgression of quantitative resistance gene pi21 confers broad spectrum resistance to rice blast. Rice Science, 27(2), 113–123. [Google Scholar]
9. Mutiga, S. K., Rotich, F., Were, V. M., Kimani, J. M., Mwongera, D. T. et al. (2021). Integrated strategies for durable rice blast resistance in Sub-Saharan Africa. Plant Disease, 10(105), 2749–2770. DOI 10.1094/PDIS-03-21-0593-FE. [Google Scholar] [CrossRef]
10. Simkhada, K., Thapa, R. (2022). Rice blast, A major threat to the rice production and its various management techniques. Turkish Journal of Agriculture-Food Science and Technology, 10(2), 147–157. DOI 10.24925/turjaf.v10i2.147-157.4548. [Google Scholar] [CrossRef]
11. El-Malky, M. M., Hassan, H. M., Metwali, E. M. R., Hadifa, A. A. (2016). Identifying the genes of blast resistance in rice (Oryza sativa L.) using line × tester analysis. Journal of Plant Production, Mansoura University, 7(12), 1269–1280. DOI 10.21608/J2016.47016. [Google Scholar] [CrossRef]
12. Ijaz, S., Ul Haq, I. (2020). Genome editing technologies for resistance against phytopathogens: Principles, applications and future prospects. In: Ijaz, S., Ul Haq, I. (Eds.Plant disease management strategies for sustainable agriculture through traditional and modern approaches, pp. 237–245. AG Switzerland: Springer. DOI 10.1007/978-3-030-35955-3_11. [Google Scholar] [CrossRef]
13. Mary Jeanie, T. Y., Koide, Y., Fukuta, Y., Imbe, T., Kato, H. et al. (2010). Development of near-isogenic lines of japonica-type rice variety Lijiangxintuanheigu as differentials for blast resistance. Breeding Science, 60(5), 629–638. DOI 10.1270/jsbbs.60.629. [Google Scholar] [CrossRef]
14. Tsunematsu, H., Mary Jeanie, T. Y., Ebron, L. A., Nagao Hayashi, I. A., Hiroshi, K. et al. (2000). Development of monogenic lines of rice for blast resistance. Breeding Science, 50(3), 229–234. DOI 10.1270/jsbbs.50.229. [Google Scholar] [CrossRef]
15. Hasana, M. M., Rafiib, M. Y., Ismail, M. R., Mahmood, M., Rahime, H. A. et al. (2018). Genetic analysis of the resistance to rice blast in the BC2F1 population derived from MR263 × Pongsu Seribu 1. Biotechnology & Biotechnological Equipment, 32(5), 1134–1140. DOI 10.1080/13102818.2018.1506266. [Google Scholar] [CrossRef]
16. Al-Daej, M. I., Ismail, M., Rezk, A. A., El-Malky, M. M. (2019). Molecular identification of blast resistance genes in rice genotypes using gene-specific markers. Journal of Biotechnology, Computational Biology and Bionanotechnology, 100(3), 311–322. DOI 10.5114/bta.2019.87590. [Google Scholar] [CrossRef]
17. Vieira, M. L. C., Santini, L., Diniz, A. L., Munhoz, C. D. F. (2016). Microsatellite markers: What they mean and why they are so useful. Genetics and Molecular Biology, 39(3), 312–328. DOI 10.1590/1678-4685-GMB-2016-0027. [Google Scholar] [CrossRef]
18. Xiao, N., Wu, Y. Y., Li, A. H. (2020). Strategy for use of rice blast resistance genes in rice molecular breeding. Rice Science, 27(4), 263–277. DOI 10.1016/j.rsci.2020.05.003. [Google Scholar] [CrossRef]
19. Alzahrani, O., Abouseadaa, H., Abdelmoneim, T. K., Alshehri, M. A., Elmogy, M. M. et al. (2021). Agronomical, physiological and molecular evaluation reveals superior salt-tolerance in bread wheat through salt-induced priming approach. Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 49, 12310. DOI 10.15835/nbha49212310. [Google Scholar] [CrossRef]
20. International Rice Research Institute (2014). Standard evaluation system for rice. LosBanos, Laguna, Philippines: IRRI, International Rice Research Institute, Manila, Philippines. 522014. [Google Scholar]
21. Jodon, N. E. (1938). Experiments on artificial hybridization of rice. Agronomy Journal, 30(4), 294–305. DOI 10.2134/agronj1938.00021962003000040004x. [Google Scholar] [CrossRef]
22. Butany, W. T. (1961). Mass emasculation in rice. Rice News Teller, 9, 9–13. [Google Scholar]
23. Gomes, K. A., Gomes, A. A. (1983). Statistical procedures for agricultural research, 2nd edition. USA, New York: Wiley-liss Inc., NY. [Google Scholar]
24. Wei, T., Simko, V., Levy, M., Xie, Y., Jin, Y. et al. (2018). Corrplot: Visualization of a correlation matrix. http://cran.r-project.org/package=corrplot. [Google Scholar]
25. Maixner, M., Ahrens, U., Seemüller, E. (1995). Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. European Journal of Plant Pathology, 101(5), 241–250. DOI 10.1007/BF01874780. [Google Scholar] [CrossRef]
26. Breseghello, F., Coelho, A. S. G. (2013). Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). Journal of Agricultural and Food Chemistry, 61(35), 8277–8286. DOI 10.1021/jf305531. [Google Scholar] [CrossRef]
27. Jia, Y., Moldenhauer, K. (2010). Development of monogenic and digenic rice lines for blast resistance genes Pi-ta, Pi-kh/Pi-ks. Journal of Plant Registrations, 4(2), 163–166. DOI 10.3198/jpr2009.04.0223crmp. [Google Scholar] [CrossRef]
28. Shallan, M. A., El-Beltagi, H. S., Mona, A. M., Amera, T. M., Sohir, N. A. (2010). Effect of amylose content and pre-germinated brown rice on serum blood glucose and lipids in experimental animal. Australian Journal of Basic Applied Science, 4(2), 114–121. [Google Scholar]
29. Shabana, Y. M., El-Wahsh, S. M., Abdelkhalik, A. F., Fayzalla, S. A., Hassan, A. A. (2013). Physiological races of rice blast pathogen and host resistant genes under Egyptian conditions. Journal of Plant Protection and Pathology, Mansoura University, 4(8), 709–720. DOI 10.21608/jp2013.87469. [Google Scholar] [CrossRef]
30. Thu Nguyen, T. T., Truong, H. T. H., Nguyen, T. L., Nguyen, L. H. K. (2015). Identification of rice blast resistance genes in south central coast of Vietnam using monogenic lines under field condition and pathogenicity assays. Journal of Agricultural Science and Technology, A5(12), 491–500. DOI 10.17265/2161-6256/2015.10.007. [Google Scholar] [CrossRef]
31. Tejaswini, K. L. Y., Manukonda, S., Ramana, P. V., Kumar, B. R., Mohammad, L. A. et al. (2016). Cluster analysis studies in rice (Oryza sativa L.using wards minimum variance method. Journal of Agricultural and Crop Research, 4(9), 129–139. [Google Scholar]
32. Abdollahi Elyasi, S. S., Mollasadeghi, V. (2016). Cluster analysis of 24 genotypes of modified rice according to qualitative and quantitative traits. Bulletin of Environment, Pharmacology and Life Sciences, 3(8), 109–111. [Google Scholar]
33. Gaballah, M. M., Attia, K. A., Ghoneim, A. M., Khan, N., EL-Ezz, A. F. et al. (2022). Assessment of genetic parameters and gene action associated with heterosis for enhancing yield characters in novel hybrid rice parental lines. Plants, 11(3), 266. DOI 10.3390/plants11030266. [Google Scholar] [CrossRef]
34. Namrata, R., Singh, P., Verma, R., Bisen, P., Singh, P. et al. (2019). Inheritance of blast disease resistance in the cross HUR 3022 × TETEP of rice (Oryza sativa L.). Journal of Experimental Biology and Agricultural Sciences, 7(6), 529–535. DOI 10.18006/2019.7(6). 529.535. [Google Scholar] [CrossRef]
35. Al-Daej, M. I., El-Malky, M. M., Sattar, M. N., Rezk, A. A., Naqqash, M. N. et al. (2022). Rice (Oryza sativa L.) breeding among hassawi landrace and Egyptian genotypes for stem borer (Chilo agamemnon Bles.) resistance and related quantitative traits. Phyton-International Journal of Experimental Botany, 91(9), 1905–1922. DOI 10.32604/phyton.2022.020629. [Google Scholar] [CrossRef]
36. Ponnaiah, G., Shalini, T., Manonmani, S., Robin, S., Vishnu, V. N. et al. (2017). Genetic variability for yield and yield components characters in bacterial blight and blast resistance genes introgressed backcross populations in rice. International Journal of Current Microbiology and Applied Sciences, 6(2), 100–103. DOI 10.20546/ijcmas.2017.602.014. [Google Scholar] [CrossRef]
37. Sabri, R. S., Rafii, M. Y., Ismail, M. R., Yusuff, O., Chukwu, S. C. et al. (2020). Assessment of agro-morphologic performance, genetic parameters and clustering patternof newly developed blast resistant rice lines tested in four environments. Agronomy, 10(8), 1098. DOI 10.3390/agronomy10081098. [Google Scholar] [CrossRef]
38. Shallan, M. A., El-Beltagi, H. S., Mona, A. M., Amera, T. M. (2010b). Chemical evaluation of pre-germinated brown rice and whole grain rice bread. Electronic Journal of Environmental, Agricultural and Food Chemistry, 9, 958–971. [Google Scholar]
39. El-Malky, M. M., Al-Daej, M. I. (2018). Studies of genetic parameters and cluster analysis of some quantitative characters through diallel analysis of rice (Oryza sativa L.). Vegetos–An International Journal of Plant Research, 31(1), 1–10. DOI 10.4172/2229-4473.1000377. [Google Scholar] [CrossRef]
40. Yu, Z. H., Mackill, D. J., Bonman, J. M. (1987). Inheritance of resistance to blast in some traditional and improved rice cultivars. American Phytopathological Society, 77, 323–326. DOI 10.1094/Phyto-77-323. [Google Scholar] [CrossRef]
41. El-Beltagi, H. S., Basit, A., Ullah, I., Mohamed, H. I., Alqurashi, R. M. (2021). Assessment of biochemical diversity, estimation of mineral composition and nutritional status in different wheat (Triticum aestivum L.) genotypes. Fresenius Environmental Bulletin, 30(7A), 9490–9507. [Google Scholar]
42. Miah, G., Rafii, M. Y., Ismail, M. R., Puteh, A. B., Rahim, H. A. et al. (2012). Blast resistance in rice: A review of conventional breeding to molecular approaches. Molecular Biology Reports, 40(3), 2369–2388. DOI 10.1007/s11033-012-2318-0. [Google Scholar] [CrossRef]
43. Ghazzawy, H. S., Mahdy, E. M., Dinar, H. M. A., El-Beltagi, H. S. (2021). Impact of geographical distribution on genetic variation of two date palm cultivars in arid regions. Fresenius Environmental Bulletin, 30(10), 11513–11523. [Google Scholar]
44. Thippeswamy, S., Chandramohan, Y., Samreen, Z., Srinivas, B., Padmaja, D. et al. (2016). Identification of blast resistant rice (Oryza sativa L.) genotypes in indigenous and exotic germplasm and validation of pi gene linked molecular markers. Electronic Journal of Plant Breeding, 7(1), 21–29. DOI 10.5958/0975-928X.2016.00003.X. [Google Scholar] [CrossRef]
45. Thulasinathan, T., Kambale, R., Ayyenar, B., Manonmani, S., Muthurajan, R. (2020). Evaluation of blast resistance genes Pi9 and Pi54 in rice against local isolates of Tamil Nadu. Electronic Journal of Plant Breeding, 11(4), 1153–1158. DOI 10.37992/2020.1104.186. [Google Scholar] [CrossRef]
46. de la Concepcion, J. C., Maidmentm, J. H., Longyam, A., Xiaom, G., Franceschettim, M. et al. (2021). The allelic rice immune receptor pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface. PLoS Pathogoms, 17(3), e1009368. DOI 10.1371/journal.ppat.1009368. [Google Scholar] [CrossRef]
47. Frontini, M., Boisnard, A., Frouin, J., Ouikene, M., Morel, J. B. et al. (2021). Genome-wide association of rice response to blast fungus identifies loci for robust resistance under high nitrogen. BMC Plant Biology, 21(99), 1–12. DOI 10.1186/s12870-021-02864-3. [Google Scholar] [CrossRef]
48. Koide, Y., Kobayashi, N., Xu, D., Fukuta, Y. (2009). Resistance genes and selection DNA markers for blast disease in rice (Oryza sativa L.). Japan Agricultural Research Quarterly, 43(4), 255–280. DOI 10.1016/j.crvi.2014.11.003. [Google Scholar] [CrossRef]
49. Ashkani, S., Rafii, M. Y., Shabanimofrad, M., Miah, G., Sahebi, M. et al. (2015). Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Frontiers in Plant Science, 6, 886. DOI 10.3389/fpls.2015.00886. [Google Scholar] [CrossRef]
50. Fang, X., Snell, P., Barbetti, M. J., Lanoiselet, V. (2017). Races of Magnaporthe oryzae in Australia and genes with resistance to these races revealed through host resistance screening in monogenic lines of Oryza sativa. European Journal of Plant Pathology, 148(3), 647–656. DOI 10.1007/s10658-016-1122-4. [Google Scholar] [CrossRef]
51. Gouda, P., Surap, A. A., Varmav, M. K. C., Kancharla, N., Sankathip, P. et al. (2013). Marker-assisted breeding of Pi-1 and Piz-5 genes imparting resistance to rice blast in PRR78, restorer line of pusa RH-10 basmati rice hybrid. Plant Breeding, 132, 61–69. DOI 10.1111/pbr.12017. [Google Scholar] [CrossRef]
52. Singh, A. K., Singh, P. K., Arya, M., Singh, N. K., Singh, U. S. (2015). Molecular screening of blast resistance genes in rice using SSR markers. Plant Pathology Journal, 31(1), 12–24. DOI 10.5423/PPJ.OA.06.2014.0054. [Google Scholar] [CrossRef]
53. Xing, J., Jia, M. H., Correll, J. C., Yuan, L., Deng, H. G. et al. (2015). Confirming and identifying new loci for rice blast disease resistance using Magnaporthe oryzae field isolates in the US. Crop Science, 55(6), 2620–2627. DOI 10.2135/cropsci2015.02.0077. [Google Scholar] [CrossRef]
54. Su, J., Wang, W. J., Han, J. L., Chen, S., Wang, C. Y. et al. (2015). Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theoretical and Applied Genetics, 128(11), 2213–2225. DOI 10.1007/s00122-015-2579-9. [Google Scholar] [CrossRef]
55. Vasudevan, K., Gruissem, W., Bhullar, N. K. (2015). Identification of novel alleles of the rice blast resistance gene Pi54. Scientific Reports, 5(15678), 1–11. DOI 10.1038/srep15678. [Google Scholar] [CrossRef]
56. Zheng, W., Wang, Y., Wang, L., Ma, Z., Zhao, J. et al. (2016). Genetic mapping and molecular marker development for Pi65(t), a novel broad–spectrum resistance gene to rice blast using next–generation sequencing. Theoretical and Applied Genetics, 129(5), 1035–1044. DOI 10.1007/s00122-016-2681-7. [Google Scholar] [CrossRef]
57. Yin, J., Zou, L., Zhu, X., Cao, Y., He, M. et al. (2021). Fighting the enemy: How rice survives the blast pathogen’s attack. Crop Journal, 9(3), 543–552. DOI 10.1016/j.cj.2021.03.009. [Google Scholar] [CrossRef]
58. Wang, L., Ma, Z., Kang, H., Gu, S., Mukhina, Z. et al. (2022). Cloning and functional analysis of the novel rice blast resistance gene Pi65 in japonica rice. Theoretical and Applied Genetics, 135(1), 1–11. DOI 10.1007/s00122-021-03957-1. [Google Scholar] [CrossRef]
59. Nutsugah, S. K., Twumasi, G. K., Chipili, G., Sere, Y., Sreenivasaprasad, S. (2008). Diversity of the rice blast pathogen population in Ghana and strategies for resistance management. Journal of Plant Pathology, 7(1), 109–113. DOI 10.3923/ppj.2008.109.113. [Google Scholar] [CrossRef]
60. El-Beltagi, H. S., Mohamed, H. I., Aldaej, M. I., Al-Khayri, J. M., Rezk, A. A. et al. (2022). Production and antioxidant activity of secondary metabolites in Hassawi rice (Oryza sativa L.) cell suspension under salicylic acid, yeast extract, and pectin elicitation. In Vitro Cellular & Developmental Biology-Plant, 58, 615–629. DOI 10.1007/s11627-022-10264-x. [Google Scholar] [CrossRef]
61. Rajashekara, H., Jeevan, B., Krishna, K. M., Subbanna, A. R., Kant, L. (2021). Blast disease: Historical importance, distribution, and host infectivity across cereal crops. In: Blast disease of cereal crops. Springer Nature Switzerland AG. DOI 10.1007/978-3-030-60585-8_1. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools