 Open Access
Open Access
REVIEW
Research Progress and Application of Plant Branching
1 Shanghai Key Laboratory of Bio-Energy Crops, School of Life Sciences, Shanghai University, Shanghai, 200444, China
2 Department of Applied Sciences, University of the West of England, Bristol, BS16 1QY, UK
* Corresponding Author: Xiangyang Hu. Email:
Phyton-International Journal of Experimental Botany 2023, 92(3), 679-689. https://doi.org/10.32604/phyton.2023.024904
Received 13 June 2022; Accepted 16 September 2022; Issue published 29 November 2022
Abstract
Plant branching development plays an important role in plant morphogenesis (aboveground plant type), the number and angle of branches are important agronomic characters that determine crop plant type. Effective branches determine the number of panicles or pods of crops and then control the yield of crops. With the rapid development of plant genomics and molecular genetics, great progress has been made in the study of branching development. In recent years, a series of important branching-related genes have been validated from Arabidopsis thaliana, rice, pea, tomato and maize mutants. It is reviewed that plant branching development is controlled by genetic elements and plant hormones, such as auxin, cytokinin and lactones (or lactone derivatives), as well as by environment and genetic elements. Meanwhile, shoot architecture in crop breeding was discussed in order to provide theoretical basis for the study of crop branching regulation.Keywords
Phenotypic plasticity in plants is dramatic. Their plasticity of growth enables plants to thrive in a myriad of environments and respond to biotic and abiotic stresses through changes in growth and development [1]. Shoot architecture is an excellent example of this and remains one of the most variable determinants of yield in many crops. Plant type refers to the morphological characteristics and spatial arrangement of plant objects, which is mainly determined by the morphological characteristics and spatial arrangement of roots, stems, leaves, branches, flowers and so on. For crops, plant type is an important factor that determines crop yield, and an ideal plant type can effectively improve plant yield [2].
In the 1950s, some research experts put forward the concept of ideal breeding but did not make a specific boundary. Chinese scholars believe that the ideal plant type mainly refers to the morphological characteristics and arrangement of plants in space. At the same time, it also believes that the physiological and ecological role of plants in the process of growth. Plant type mainly refers to the growth form of plant leaves or the posture of photosynthesis. The ideal plant type is mainly to analyze the shape of the whole plant growth environment to ensure that plants can use good growth posture to carry out photosynthesis in the process of growth and increase economic yield [3].
Plant branching is such an important factor that it determines its morphogenesis. It not only affects the distribution of leaf curtain, the number and yield of flowers and fruits, but also it affects the competition between plants and weeds or insects [4]. It is a common growth phenomenon of plants, axillary buds after formation, further grows to form lateral branches, and axillary buds on lateral branches can also develop into lateral branches, and then form strains.
It not only affects crop yield, but also affects crop cultivation density and nutrient distribution. More branches will lead to poor ventilation, which can easily lead to crop diseases and reduce yield. In addition, branching is also an important plant type character of ornamental plants, and the growth state of branches can affect the landscaping effect of ornamental plants. In recent years, the aboveground character gradually preferred by breeders is branching.
2 Branching Mode and Influencing Factors
Because of the nature of plants which are differences in quality and species, by observing the branching morphology of plants, all branches can be divided into five categories: monopodial branching (such as Arabidopsis and Cucumis sativus, etc.), Sympodial branching, false dichotomous branching (Solanum melongena), dichotomous branching (Capsicum spp.) and tiller (Fig. 1).
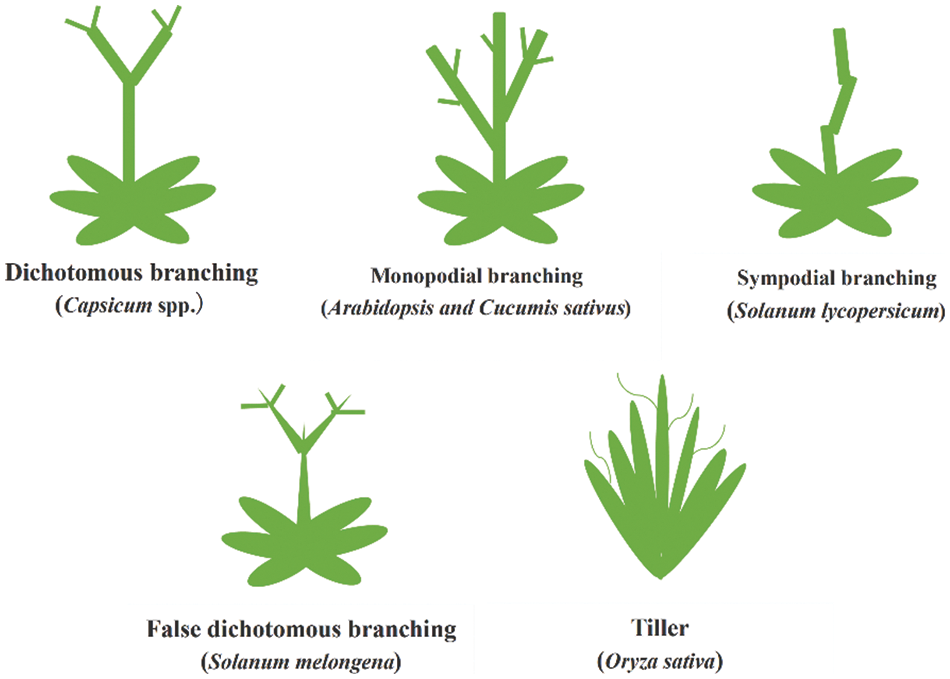
Figure 1: Plant branching type
2.2 Plant Branches and Plant Endogenous Hormones
Plant hormone is an essential factor to break the dormancy of lateral buds and the continuous growth of branches [5]. The change of external environment can make the plant hormone change accordingly, promote the dynamic balance of plant hormone, and then stimulate the expression of plant branching genes.
2.2.1 Plant Branches and Auxin
It was first found that the hormone acting on plant branches is indole-3-acetic acid (IAA). After increasing the level of IAA in bean plants, the number of lateral branches decreased significantly [6,7]. The number of lateral branches decreased significantly after increasing the level of IAA in plants. The results showed that IAA in cucumber shoot tips could not inhibit the growth of axillary buds independently, needed to act together with other substances that inhibited the growth of axillary buds [8]. After spraying IAA on rice with spikelets removed, the tiller elongation of rice was inhibited [9].
It is found that the lowest IAA content is the key to the initiation of axillary meristem (AM) [10]. The mutants of two important genes YUCCA (YUC) and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) in the IAA synthesis pathway do not have a strong ability to synthesize IAA, which results in the inhibition of apical dominance and the promotion of Arabidopsis branching [11]. YUCCA (YUC) gene in Arabidopsis thaliana is related to auxin biosynthesis and plays an important role in all aspects of development. In maize, SPI1 is a specific member of YUC gene family in monocotyledons and is also important for maize inflorescence development. The functional loss of SPI1, a homologous gene of YUC in maize, will lead to defects in the initiation and lateral organ development of AM, thus reducing branching [12]. Mutations or deletions of key genes IAA3, IAA5 and IAA15 in IAA signal transduction promote tomato lateral branch growth by affecting IAA signal transduction [13].
2.2.2 Plant Branches and Cytokinin
Cytokinin (CTK) can activate lateral buds and promote plant branches by promoting cell division. Spraying CTK directly on plant lateral buds could inhibit apical dominance and make lateral buds grow. It was found that the content of CTK in A. thaliana mutants as sho and hoc was high, indicating that CTK was regulating plant branching [14]. Applying CTK to the base of A. thaliana could weaken the apical dominance and promote the growth of lateral buds [15]. CTK can also promote the initiation and lateral bud formation of AM. The increase of CTK level in Arabidopsis multibranched mutant supershoot (sps) leads to the formation of multiple AM in the leaf axils of rosette and cauline leaves, which increases the number of branches and promotes the elongation of lateral branches [16]. The high expression of ISOPRENYLTRANSFERASE (IPT), the key gene of CTK synthase, can promote the activation of CTK on lateral buds, and then promote the branching of A. thaliana [17].
2.2.3 Plant Branches and Strigolactone
Strigolactones (SLs) is a new type of plant hormone ubiquitous in plants, which can stimulate seed germination of parasitic plants and inhibit lateral bud germination directly or indirectly. SLs can be used as a long-distance signal substance to inhibit the growth and development of lateral buds, and as a graft transfer substance to regulate the branch inhibitory transcription factors BRC1, teosinte of TCP family. TEOSINTE BRANCHED 1 (TB1) and FINE CULM 1 (FC1), thereby inhibiting lateral bud elongation [18,19]. SLs can inhibit lateral bud growth by down regulating the level of CTK in axillary buds of pea [20].
There are three inhibitors in the SLs signal pathway, namely SMXL6, SMXL7 and SMXL8. It has been found that without SLs, SMXL6 and TPL protein can directly bind to the promoters of SMXL6, SMXL7 and SMXL8 and inhibit their expression to increase the content of SLs, thus inhibiting Arabidopsis branching.
At the same time, smxl6 can also form complexes with unknown transcription factors, which are expected to recognize and bind the promoters of BRC1, TCP1 or PAP1, thereby inhibiting their transcription and promoting Arabidopsis branching [21].
2.2.4 Plant Branches and Other Hormones
It has been found that gibberellin (GA), abscisic acid (ABA) and brassinolide (BR) can regulate plant branching. Rice TILLER ENHANCER (TE) gene can inhibit tillering, and high levels of GA can activate APC/CTE (E3 ubiquitin ligase complex) and promote MONOCULM 1 (MOC1) degradation of AM, thus inhibiting rice tillering [22]. High concentration of ABA could inhibit the germination of Arabidopsis lateral buds and thus inhibit the branching [23]. BRASSINAZOLE RESISTANT (BZR1) ncodes a positive regulator of the brassinosteroid (BR) signaling pathway that mediates both downstream BR responses and negative feedback regulation of BR biosynthesis. BZR1 inhibits the expression of CUP-SHAPED COTYLEDON (CUC), the key gene of AM initiation in A. thaliana, thus inhibiting AM initiation and branching [24].
2.3 Plant Branches and Environmental Factors
As the growth position of the plant cannot be moved, the change of it surrounding environment will have a great impact on the plant [25]. Therefore, plants can adapt to the changing environment only by constantly changing their developmental state according to various signals. There is growing evidence that the environment can affect the branching of plants. Understanding the molecular mechanism of plant branching response to environmental changes will help us to cultivate excellent crop varieties with high and stable yield under special environmental conditions. The following mainly discusses the effects of light, temperature, water and nutrition on crop branching.
In general, high intensity light promotes the growth of lateral buds. Studies have shown that high intensity light can promote the growth of lateral buds of vaccinium and thus promote branching [26]. Low light quantum flux density reduces A. thaliana branching [27]. When the ratio of red light to far red light decreased, the expression of branch inhibitor BRANCHED 1 (BRC1) increased, which led to the decrease of branching in A. thaliana [28,29]. The decrease of photosynthetic area could promote the expression of branch suppressor gene TB1 and reduce the tillering of maize [30]. Beyond a certain range of light density, the content of monopodium lactone (SLs) and the expression of SLs synthesis gene CAROTENOID CLEAVAGE DIOXYGENASE 7 (CCD7) in tomato increased, thus inhibiting tomato branching [31]. In addition, the increase of light/dark (7 h/7 h) transition frequency could inhibit the development of axillary buds in A. thaliana and reduce the branching [32].
Temperature is also one of the important factors affecting branching. Low temperature will induce the expression of SLs signal gene MORE AXILLARY GROWTH 1 (RwMAX1) in rose, promote it to form a gradient in stem nodes, and then affect the germination rate of lateral branches [33]. Low temperature also promoted the transcription of SLs receptor gene DWARF 14 (D14), which inhibited tillering in rice [34].
Water is also an important factor affecting the formation of branches. When rice is under drought in the early stage of development, the number of tillers can be reduced by SL and OsTB1 pathway to adapt to drought conditions [35].
Nutrition is also essential for plant branching. When the plant is undernourished, SL will be synthesized in the roots of the plant, and part of the SL will be transported upward to inhibit the production of lateral buds and branches, thus reducing the consumption of nutrients [36]. Nitrogen is the most important nutrient, and the main sources of nitrogen are nitrate and ammonium fertilizer. Studies have shown that nitrate signals can promote carbon and nitrogen metabolism and plant endogenous hormone synthesis [37], and then promote the growth of axillary buds. Nitrate transporter (nitrate and peptide transporter family, NPF) promotes nitrogen uptake and regulates plant growth and development. OsNPFs gene can promote nitrate absorption and increase the number of tillers in rice. Phosphate also helps to tiller rice [38]. When phosphorus was deficient in the plant, the content of SL and the growth of tiller buds were inhibited. When there was enough phosphorus in the plant, the content of SL decreased and promoted the growth of tiller buds.
These studies show that many genes can combine stress response with branching regulation to make plants better adapt to the dynamic environment. Generally, the branching of environmental impact is ultimately involved in the branching of plants by affecting the balance of endogenous hormones.
Besides environmental factors and plant hormones, there are also genetic factors that affect plant branching. Genetic factors fundamentally determine the branching characteristics of plants. The branching of the plant is differentiated from the axillary meristem, and the formation of lateral buds is first the expression of genes for special traits in the axils of leaves; then, the axillary meristem grows; then it grows into lateral buds. In this process of life activities, the genes that control branching and plant hormones coordinate to control the branching traits of plants. Many genes that affect plant branching have been reported one after another, and their mechanisms are different. Therefore, the internal regulatory genes are divided into three types: one is the lateral bud initiation development regulation gene; the second is the lateral bud elongation development regulation gene; the third is the lateral bud initiation and elongation development co-regulation gene.
2.4.1 Genes Regulating the Initiation of Lateral Bud Development
The initiation of plant lateral buds is mainly regulated by genes. EXCES SIVE BRANCHES1 (EXB1) belongs to the WRKY transcription factor family, which can promote the initiation of AM and produce more lateral buds to promote the branching of A. thaliana [39]. REGULATORS OF AXILLARY MERISTEMS1-3 (RAX1-3) belongs to the family of MYB transcription factors [40] and regulates A. thaliana branching. EXB1 upstream in RAXs, is regulating the transcription of RAXs to promote AM initiation, thus promoting Arabidopsis branching. Blind gene in tomato is a homologous gene of RAX and participates in AM initiation.
NAC transcription factor CUC is activated with Arabidopsis AM. CUC3 plays an important role in Arabidopsis AM formation. It was found that CUC2 and CUC3 directly bind to the promoter of DA1 (its peptidase substrate UBP15 can inhibit the initiation of AM) and activate its expression to form a CUC2/CUC3-DA1-UBP15 regulatory module to control the initiation of AM in A. thaliana [41]. LATERAL SUPPRESOR (LAS) belongs to the GRAS transcription factor family, which promotes AM formation and regulates Arabidopsis branching [42].
Studies have shown that during the initiation and establishment of AM, LAS and MOC1 play a role in the upstream of Arabidopsis SHOOTMERISTEMLESS (STM). REVOLUTA (REV) belongs to the HDZIP transcription factor family and acts on Arabidopsis AM initiation [43]. REV can up-regulate the expression of SAM gene to start Arabidopsis AM [44]. STM belongs to the KNOX transcription factor family, which promotes AM formation in A. thaliana by regulating the differentiation of AM cells [45].
WUSCHEL (WUS) is a homeobox gene that controls the stem cell pool. Expressed in the center of the stem cell tissue of the meristem. Stem cells need to be kept in an undifferentiated state. The regulation of WUS transcription is the core checkpoint in stem cell control, which promotes axillary bud primordium formation by maintaining stem cell activity and regulating downstream genes. And reactive oxygen species (ROS) affects WUS gene, which in turn affects cell maintenance and differentiation. It is worth noting that the activities of stem tip meristem (SAM) and root tip meristem (RAM) have been proved to be affected by the interaction between ROS, redox components and plant hormones [46]. BARREN STALK1 (Ba1) gene in maize and REGULATOR OF AXILLARY MERISTEM FORMATION (ROX) gene in Arabidopsis are homologous genes of LAX1, which regulate tillering or branching by coding proteins [47,48].
2.4.2 Genes Regulating the Development of Lateral Bud Elongation
BRC1, TB1 and FC1 are homologous genes in Arabidopsis, maize and rice, respectively, and they are all members of the TCP transcription factor family, which inhibit the growth of branches and tillers. Through QTL mapping, it was found that the TB1 gene of Yumi caused the structural difference between maize and multi-tiller Teosinte maize. TB1 is mainly expressed in AM to coordinate maize bud growth [49]. FC1 is expressed at the base of AM and SAM, which inhibits rice tillering by promoting bud dormancy [50]. The BRC1 gene of A. thaliana is expressed in axillary buds and inhibits axillary bud elongation and development. It is the downstream gene of multiple regulatory pathways. Cucumber branching inhibitor CsBRC1 cooperates with auxin polar transport gene CsPIN3 to regulate cucumber branching. CYP831 gene belongs to the cytochrome P450 family, which promotes Arabidopsis branching by reducing IAA content and inhibiting apical dominance. In A. thaliana, AXR1 gene is an IAA responsive gene that can inhibit the growth of axillary buds. PINOID (PID) gene negatively regulates IAA signal transduction, which can promote Arabidopsis branching. AtMYB2 gene negatively regulates CTK content, thus inhibiting Arabidopsis branching [51].
BUD1 gene belongs to member of plant mitogen-activated protein kinase group D. Bud1 mutants have been reported, in which MAP kinase 7 (MKK7) is structurally activated, resulting in multiple phenotypic changes. It is known that mitogen-activated protein kinase (MAPK/MPK) plays an important role in a series of signals such as biotic and abiotic stress and hormone-mediated development in higher plants. Genetic analysis showed that MKK7-MPK6 cascade is specifically responsible for the regulation of shoot branching, hypocotyl gravitropism, filament elongation, and lateral root formation [52]. D27 gene participates in SLs synthesis in rice to inhibit branching [53]. MAX1 gene encodes cytochrome P450, which acts on the downstream of MAX3 and MAX4. It is the key gene of SLs signal transduction and inhibits Arabidopsis branching. D3 gene in rice is homologous to MAX2 gene in A. thaliana, which is an important motif in SLs regulatory genes. It plays a role in the downstream of MAX1, MAX3 and MAX4 and can inhibit branching [54]. HTD1 gene in rice is homologous to MAX3 gene in Arabidopsis and allele to D17, which is the key gene encoding SLs synthetase and inhibits branching, while D10 gene in rice is homologous to MAX4 gene and participates in SLs biosynthesis and inhibits branch [55].
In rice, three alleles D14, D88 and HTD2 play a role in SLs signal transduction and are negative regulators of tillering [56]. D53 is the suppressor of SLs signal transduction and is positively regulating rice branching [57].
2.4.3 Co-Regulatory Genes of Lateral Bud Initiation and Elongation Development
The genes that play a role in both AM initiation and lateral bud development are SPS and BUSHY (BUS). They are homologous genes. SPS gene is mainly strongly expressed in leaf axils and controls the synthesis of IAA to regulate Arabidopsis branching [58]. The sps mutant of A. thaliana is characterized by an increase in the number of meristems formed in leaf axils and the stagnant release of lateral buds, resulting in many lateral buds, thus forming repeatable branches. Some studies have also shown that SPS gene reduces the concentration of CTK in leaf axils by accelerating the oxidative degradation of CTK, which inhibits the initiation of AM and the development of axillary buds [59], resulting in reduced branching of A. thaliana (Fig. 2).
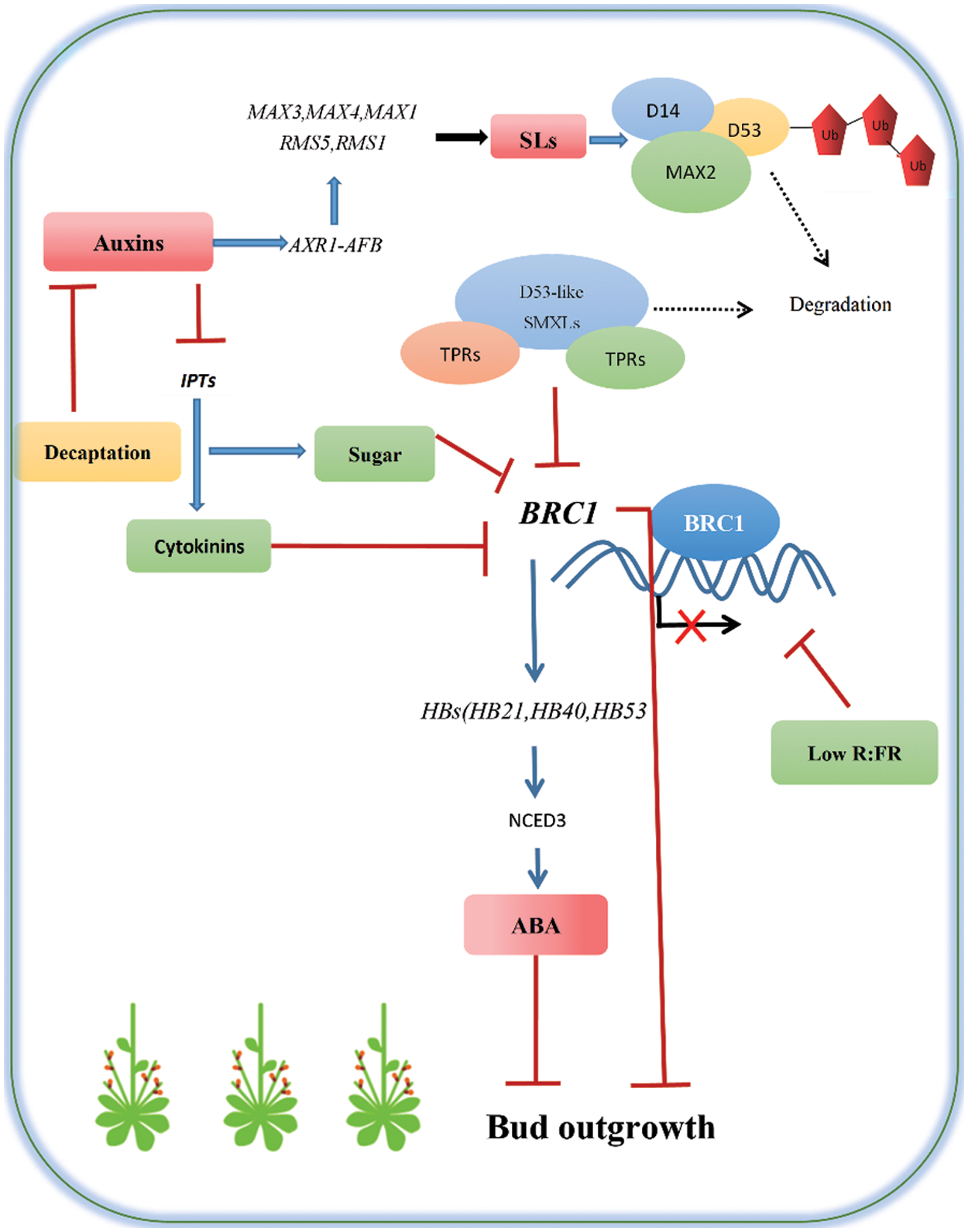
Figure 2: Plant hormones, environmental conditions and key regulatory genes control bud outgrowth
Plant type is a comprehensive and important agronomic character. Through the optimization of individual plant type, we can construct the optimal population under specific environmental conditions, including adversity, and make effective use of light, heat, water, fertilizer and other resources to obtain the highest yield and best quality. It is the direction of agricultural breeding and cultivation in the future. Compared with conventional breeding methods, molecular breeding methods represented by molecular marker-assisted selection, transgenic and gene editing cannot only shorten the breeding cycle, but also achieve accurate improvement of crop specific traits with higher efficiency (Oryza sativa L., Triticum aestivum, Zea mays, etc.). In the aspect of crop plant type, we should further excavate the new key genes controlling crop plant type and the explanation of their regulatory mechanism, develop molecular markers closely linked to plant type, transfer several key genes controlling crop plant type to good parents by means of molecular breeding and molecular design, and cultivate new crop varieties with ideal plant type, which will be an important direction of crop genetic improvement in the future, which has important practical significance.
3.1 Make Great Efforts to Improve Plant Type
Plant type is a comprehensive and important agronomic character, which is an important character affecting the population structure and yield formation of plant. Plant type plays a common role at both individual and population levels. In a specific natural environment, plant ontogeny interacts with population structure to obtain reasonable individual plant type and population structure, which can not only increase biological yield, but also improve light transmission and ventilation conditions, reduce the occurrence of germs and insect pests, reduce the use of pesticides and so on. Plant type determines plant yield potential, harvest index, disease resistance and its adaptability to growth environment.
Through the optimization of individual plant type, we can construct the optimal population under specific environmental conditions, including adversity, and make effective use of light, heat, water, fertilizer and other resources to obtain the highest yield and best quality. it is the direction of agricultural breeding and cultivation in the future.
Identify the key genes controlling crop plant type, develop molecular markers closely linked to plant type, transfer several key genes controlling crop plant type to good parents by means of molecular breeding and molecular design, and cultivate new crop varieties with ideal plant type will be an important direction of crop genetic improvement in the future.
The key genes controlling plant type through map-based cloning or homologous cloning and the development of molecular markers linked to plant type are the premise and basis for the molecular improvement of plant type of these crops.
3.2 Continue to Increase Crop Yields
The output value of vegetables in China is about 2.2 trillion yuan, which has surpassed that of food crops to become the largest industry in the planting industry. Solanum lycopersicumL. Cucumis sativus and Solanum melongenaL. are the main vegetable crops under protected cultivation in China. Because of more branches, pruning, branching, hanging vines and other labor-consuming operations need to be carried out frequently in the production of eggplant fruits and melons, which increases the labor cost. Secondly, more branches are easy to cause uneven distribution of plant nutrients and diseases, thus reducing the yield and quality of vegetable crops [60]. With the gradual increase of labor cost, the cultivation of eggplant fruit and melon vegetable varieties with ideal plant type (few or weak lateral branches) and suitable for light and simplified cultivation is one of the urgent problems to be solved in China’s vegetable industry. In the production process, in addition to selecting the ideal plant type varieties, the branching of eggplant fruits and melons can also be regulated according to the above research results.
First, plant branching can be regulated by changing the growth environment. For example, branching can be reduced by appropriately increasing far red light, lowering temperature or applying more nitrogen fertilizer in the cultivation environment, or by reducing potash and sugars.
Second, plant hormones can be used to regulate branching. The branching was reduced by spraying IAA, SLs, GA, ABA, BRs and so on, and the branching was also reduced by CTK inhibitor treatment.
Third, using the identified plant branching regulatory genes to develop molecular markers, through molecular marker-assisted breeding to cultivate new varieties of ideal plant type.
Fourth, using the cloned plant branching regulatory genes, new germplasm with different branching types can be obtained by gene editing technique. In cucumber, Professor Zhang Xiaolan of China Agricultural University cloned CsBRC1, a key gene regulating branching development of cucumber [61]. Because cucumber cultivars in China tend to select varieties with small number of lateral branches, short length and late germination, we can use CsBRC1 gene information to develop molecular markers or identify and create cucumber germplasm resources with different branching types by gene editing techniques.
With the deepening of people’s understanding of plant branching regulation, it will effectively solve the problem of more branches in the production of eggplant fruits and melons, innovate germplasm resources, and cultivate new varieties of high-quality and high-yield vegetables that meet the goal of breeding. Effectively solve the problems of difficult and expensive labor in the process of vegetable production, reduce the cost of vegetable production, improve vegetable production and simplify cultivation.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Adriani, D. E., Dingkuhn, M., Dardou, A., Adam, H., Luquet, D. et al. (2016). Rice panicle plasticity in Near Isogenic Lines carrying a QTL for larger panicle is genotype and environment dependent. Rice, 9(1), 1–15. [Google Scholar]
2. Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G. et al. (2003). Control of tillering in rice. Nature, 422(6932), 618–621. [Google Scholar]
3. Donald, C. T. (1968). The breeding of crop ideotypes. Euphytica, 17(3), 385–403. [Google Scholar]
4. Wang, Y., Li, J. (2005). The plant architecture of rice (Oryza sativa). Plant Molecular Biology, 59(1), 75–84. [Google Scholar]
5. Assuero, S. G., Tognetti, J. A. (2010). Tillering regulation by endogenous and environmental factors and its agricultural management. American Journal of Plant Sciences. Biotechnol, 4(1), 35–48. [Google Scholar]
6. Romano, C. P., Cooper, M. L., Klee, H. J. (1993). Uncoupling auxin and ethylene effects in transgenic tobacco and Arabidopsis plants. The Plant Cell, 5(2), 181–189. [Google Scholar]
7. Domagalska, M. A., Leyser, O. (2011). Signal integration in the control of shoot branching. Nature Reviews Molecular Cell Biology, 12(4), 211–221. [Google Scholar]
8. Shimizu-Sato, S., Tanaka, M., Mori, H. (2009). Auxin-cytokinin interactions in the control of shoot branching. Plant Molecular Biology, 69(4), 429–435. [Google Scholar]
9. Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M. et al. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal, 51(6), 1019–1029. [Google Scholar]
10. Gallavotti, A. (2013). The role of auxin in shaping shoot architecture. Journal of Experimental Botany, 64(9), 2593–2608. [Google Scholar]
11. Cheng, Y., Dai, X., Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & Development, 20(13), 1790–1799. [Google Scholar]
12. Phillips, K. A., Skirpan, A. L., Liu, X., Christensen, A., Slewinski, T. L. et al. (2011). Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. The Plant Cell, 23(2), 550–566. [Google Scholar]
13. Deng, W., Yan, F., Liu, M., Wang, X., Li, Z. (2012). Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signaling & Behavior, 7(8), 911–913. [Google Scholar]
14. Catterou, M., Dubois, F., Smets, R., Vaniet, S., Kichey, T. et al. (2002). hoc: An Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity. The Plant Journal, 30(3), 273–287. [Google Scholar]
15. Chatfield, S. P., Stirnberg, P., Forde, B. G., Leyser, O. (2000). The hormonal regulation of axillary bud growth in Arabidopsis. The Plant Journal, 24(2), 159–169. [Google Scholar]
16. Tantikanjana, T., Yong, J. W., Letham, D. S., Griffith, M., Hussain, M. et al. (2001). Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes & Development, 15(12), 1577–1588. [Google Scholar]
17. Guo, Y., Gan, S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiology, 156(3), 1612–1619. [Google Scholar]
18. Aguilar-Martínez, J. A., Poza-Carrión, C., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant Cell, 19(2), 458–472. [Google Scholar]
19. Shinohara, N., Taylor, C., Leyser, O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biology, 11(1), e1001474. [Google Scholar]
20. Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A. et al. (2008). Strigolactone inhibition of shoot branching. Nature, 455(7210), 189–194. [Google Scholar]
21. Wang, L., Wang, B., Yu, H., Guo, H., Lin, T. et al. (2020). Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature, 583(7815), 277–281. [Google Scholar]
22. Lin, Q., Zhang, Z., Wu, F., Feng, M., Sun, Y. et al. (2020). The APC/CTE E3 ubiquitin ligase complex mediates the antagonistic regulation of root growth and tillering by ABA and GA. The Plant Cell, 32(6), 1973–1987. [Google Scholar]
23. Pei, Z. M., Ghassemian, M., Kwak, C. M., McCourt, P., Schroeder, J. I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science, 282(5387), 287–290. [Google Scholar]
24. Gendron, J. M., Liu, J. S., Fan, M., Bai, M. Y., Wenkel, S. et al. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. PNAS, 109(51), 21152–21157. [Google Scholar]
25. Snowden, K. C., Napoli, C. A. (2003). A quantitative study of lateral branching in petunia. Functional Plant Biology, 30(9), 987–994. [Google Scholar]
26. Kawamura, K., Takeda, H. (2002). Light environment and crown architecture of two temperate Vaccinium species: Inherent growth rules versus degree of plasticity in light response. Canadian Journal of Botany, 80(10), 1063–1077. [Google Scholar]
27. Su, H., Abernathy, S. D., White, R. H., Finlayson, S. A. (2011). Photosynthetic photon flux density and phytochrome B interact to regulate branching in Arabidopsis. Plant, Cell & Environment, 34(11), 1986–1998. [Google Scholar]
28. Franklin, K. A., Whitelam, G. C. (2005). Phytochromes and shade-avoidance responses in plants. Annals of Botany, 96(2), 169–175. [Google Scholar]
29. González-Grandío, E., Poza-Carrión, C., Sorzano, C. O. S., Cubas, P. (2013). BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. The Plant Cell, 25(3), 834–850. [Google Scholar]
30. Kebrom, T. H., Mullet, J. E. (2015). Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant, Cell & Environment, 38(8), 1471–1478. [Google Scholar]
31. Koltai, H., Cohen, M., Chesin, O., Mayzlish-Gati, E., Bécard, G. et al. (2011). Light is a positive regulator of strigolactone levels in tomato roots. Journal of Plant Physiology, 168(16), 1993–1996. [Google Scholar]
32. Jouve, L., Greppin, H., Degli Agosti, R. (1998). Arabidopsis thaliana floral stem elongation: Evidence for an endogenous circadian rhythm. Plant Physiology and Biochemistry, 36(6), 469–472. [Google Scholar]
33. Djennane, S., Hibrand-Saint Oyant, L., Kawamura, K., Lalanne, D., Laffaire, M. et al. (2014). Impacts of light and temperature on shoot branching gradient and expression of strigolactone synthesis and signalling genes in rose. Plant, Cell & Environment, 37(3), 742–757. [Google Scholar]
34. Chen, L., Zhao, Y., Xu, S., Zhang, Z., Xu, Y. et al. (2018). Os MADS 57 together with Os TB 1 coordinates transcription of its target Os WRKY 94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytologist, 218(1), 219–231. [Google Scholar]
35. Du, H., Huang, F., Wu, N., Li, X., Hu, H. et al. (2018). Integrative regulation of drought escape through ABA-dependent and-independent pathways in rice. Molecular Plant, 11(4), 584–597. [Google Scholar]
36. Kohlen, W., Charnikhova, T., Liu, Q., Bours, R., Domagalska, M. A. et al. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiology, 155(2), 974–987. [Google Scholar]
37. Sakakibara, H., Takei, K., Hirose, N. (2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends in Plant Science, 11(9), 440–448. [Google Scholar]
38. Fang, Z., Xia, K., Yang, X., Grotemeyer, M. S., Meier, S. et al. (2013). Altered expression of the PTR/NRT 1 homologue Os PTR 9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnology Journal, 11(4), 446–458. [Google Scholar]
39. Guo, D., Zhang, J., Wang, X., Han, X., Wei, B. et al. (2015). The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. The Plant Cell, 27(11), 3112–3127. [Google Scholar]
40. Müller, D. R., Schmitz, G., Theres, K. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. The Plant Cell, 18(3), 586–597. [Google Scholar]
41. Li, Y., Xia, T., Gao, F., Li, Y. (2020). Control of plant branching by the CUC2/CUC3-DA1-UBP15 regulatory module. The Plant Cell, 32(6), 1919–1932. [Google Scholar]
42. Greb, T., Clarenz, O., Schäfer, E., Müller, D., Herrero, R. et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes & Development, 17(9), 1175–1187. [Google Scholar]
43. Long, J. A., Moan, E. I., Medford, J. I., Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature, 379(6560), 66–69. [Google Scholar]
44. Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N., Clark, S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal, 25(2), 223–236. [Google Scholar]
45. Williams, L., Fletcher, J. C. (2005). Stem cell regulation in the Arabidopsis shoot apical meristem. Current Opinion in Plant Biology, 8(6), 582–586. [Google Scholar]
46. Schippers, J. H. M., Foyer, C. H., van Dongen, J. T., (2016). Redox regulation in shoot growth, SAM maintenance and flowering. Current Opinion in Plant Biology, 29, 121–128. [Google Scholar]
47. Yang, F., Wang, Q., Schmitz, G., Müller, D., Theres, K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. The Plant Journal, 71(1), 61–70. [Google Scholar]
48. Gallavotti, A., Zhao, Q., Kyozuka, J., Meeley, R. B., Ritter, M. K. et al. (2004). The role of barren stalk1 in the architecture of maize. Nature, 432(7017), 630–635. [Google Scholar]
49. Finlayson, S. A. (2007). Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant and Cell Physiology, 48(5), 667–677. [Google Scholar]
50. Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M. et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. The Plant Journal, 33(3), 513–520. [Google Scholar]
51. Michniewicz, M., Zago, M. K., Abas, L., Weijers, D., Schweighofer, A. et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell, 130(6), 1044–1056. [Google Scholar]
52. Jia, W. Y., Li, B. H., Li, S. J., Liang, Y., Wu, X. W. et al. (2016). Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biology, 14(9), 24. [Google Scholar]
53. Lin, H., Wang, R., Qian, Q., Yan, M., Meng, X. et al. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. The Plant Cell, 21(5), 1512–1525. [Google Scholar]
54. Zou, J., Zhang, S., Zhang, W., Li, G., Chen, Z. et al. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal, 48(5), 687–698. [Google Scholar]
55. Wang, Y., Li, J. (2011). Branching in rice. Current Opinion in Plant Biology, 14(1), 94–99. [Google Scholar]
56. Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M. et al. (2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology, 50(8), 1416–1424. [Google Scholar]
57. Jiang, L., Liu, X., Xiong, G., Liu, H., Chen, F. et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature, 504(7480), 401–405. [Google Scholar]
58. Reintanz, B., Lehnen, M., Reichelt, M., Gershenzon, J., Kowalczyk, M. et al. (2001). bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. The Plant Cell, 13(2), 351–367. [Google Scholar]
59. Grbić, V., Bleecker, A. B. (2000). Axillary meristem development in Arabidopsis thaliana. The Plant Journal, 21(2), 215–223. [Google Scholar]
60. Saqib, M., Anjum, M. A. (2021). Applications of decision support system: A case study of solanaceous vegetables. Phyton-International Journal of Experimental Botany, 90(2), 331–352. DOI 10.32604/phyton.2021.011685. [Google Scholar] [CrossRef]
61. Shen, J., Zhang, Y., Ge, D., Wang, Z., Song, W. et al. (2019). CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. PNAS, 116(34), 17105–17114. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools