 Open Access
Open Access
ARTICLE
Genome Wide Characterization of CBL-CIPK Family Genes and Their Responsive Expression in Rosa chinensis
1 The Engineering Research Institute of Agriculture and Forestry, Ludong University, Yantai, 264025, China
2 College of Agriculture, Ludong University, Yantai, 264025, China
* Corresponding Authors: Chunyan Yu. Email: ; Xiaotong Guo. Email:
# These authors contribute equally to this work
Phyton-International Journal of Experimental Botany 2023, 92(2), 349-368. https://doi.org/10.32604/phyton.2022.025467
Received 14 July 2022; Accepted 29 August 2022; Issue published 12 October 2022
Abstract
Calcium (Ca2+) plays a pivotal role in various signal transduction pathways. Calcineurin B-like proteins (CBLs) are a unique group of Ca2+ sensors that decode Ca2+ signals by activating the plant specific protein kinase known as the CBL-interacting protein kinase (CIPK). In plants, the CBL-CIPK signaling network regulates multiple signals in response to different extracellular cues including abiotic stress. However, the genome wide annotation and expression patterns of CBLs and CIPKs in woody cutting flower plants are still unclear. In this study, a total number of 7 CBLs (RcCBLs) and 17 CIPKs (RcCIPKs) genes, divided into four and five subfamilies, respectively, were identified from the rose genome. All RcCBLs possess a classic elongation factor-hand (EF-hand) domain, while all RcCIPKs possess both the classic kinase and NAF domains. Most RcCBLs were predicted to be plasma membrane localized, whereas most RcCIPKs were predicted to be cytoplasmic localized. Synteny analysis showed that one RcCBL gene pair and five RcCIPK gene pairs have gone through whole genome duplication events. Promoter cis-element prediction assays indicated that RcCBLs and RcCIPKs could function in different abiotic stress responses in rose plants. Further quantitative real-time PCR analysis demonstrated that RcCBLs and RcCIPKs were expressed in different organs with overlapped but distinct patterns in response to various abiotic stresses. The findings in this work will provide fundamental information and gene resources for further functional research on RcCBLs and RcCIPKs.Keywords
Supplementary Material
Supplementary Material FileCalcium (Ca2+) functions as a second messenger to regulate physiological and developmental processes, and its level in plants is affected by biotic and abiotic stresses [1]. A diverse of environmental factors can evoke Ca2+ responses with specific temporal and spatial characteristics. These Ca2+ signals encoded as spikes, waves and oscillations, were interpreted by Ca2+ sensors and effectors, leading to specific responses [2]. Ca2+ signals are first perceived by Ca2+ sensors, such as calmodulin-like proteins (CMLs), calmodulins (CaMs), calcium-dependent protein kinases (CDPKs) and calcineurin B-like proteins (CBLs), and then relayed into the downstream responses by interacting with the downstream proteins and phosphorylation cascades [3,4].
CBLs are a class of proteins that sense and transmit Ca2+ signals. Typically, a CBL protein contains three or four EF-hands (elongation factor-hands) to capture Ca2+ signals and interact with other proteins [1,4–6]. Usually, CBL does not act alone but forms a complex with a protein called CIPK (CBL-interacting serine-threonine protein kinase), which possesses a serine/threonine protein kinase structural domain, a 24-amino acid NAF structural domain, and a PPI structural domain. The NAF domain mediates the interaction with CBL, while the kinase structural domain performs other functions [5,7]. In the CBL-CIPK pathway, CBL protein binds with Ca2+, thereby activates CIPK and forms a CBL-CIPK complex to phosphorylate and modify the downstream acting proteins, thus achieves the transduction of various signals [8–10]. CBLs contain N-myristoylation and palmitoylation sites and are generally localized on cell membranes, whereas CIPKs are found free in cytoplasm [6,7,11,12]. Both CBLs and CIPKs were first identified in Arabidopsis and subsequently studied in different plant species such as rice, honeysuckle, wheat, poplar, pepper, walnut, rapeseed and grape [6,13–16]. To date, a total number of 10 CBLs and 26 CIPKs in Arabidopsis, 10 CBLs and 33 CIPKs in rice, 10 CBLs and 27 CIPKs in poplar, 9 CBLs and 30 CIPKs in pecan, 6 CBLs and 17 CIPKs in honeysuckle, and 8 CBLs and 20 CIPKs in grape, have identified [6,13–18]. Previous studies have reported that CBL-CIPK complex plays a vital role in plant growth and response to numerous stresses. The response of CBLs and CIPKs in plants have been found to be different under different stress conditions such as salinity, osmotic, drought, cold, oxidative, pathogenic, and other abiotic and biotic stresses [11,15,19,20]. In Arabidopsis, expressions of both AtCBL4 and AtCBL10 were sensitive to salt stress. AtCIPK24 phosphorylated AtCBL10 at the Ser237 site, and the interaction of AtCBL4 and AtCIPK24 activated the activity of Na+/H+ antiporter located on cell membrane to alleviate salt stress [21–23]. Meanwhile, AtCIPK8, a homolog of AtCIPK24, bound to AtCBL10 and activated AtSOS1 to alleviate salt stress [24–26]. AtCBL1 was also induced by various stresses, and overexpression of AtCBL1 increased salt and drought tolerance but decreased cold resistance in transgenic plants [27–31]. In rice, silencing of OsCIPK23 improved the salt and drought resistance of transgenic plants [32]. In apple, MdCIPK13 and MdCIPK22 acted on the Ser254 and Ser381 phosphorylation sites of MdSUT2.2, and enhanced drought and salt resistance [33,34]. In Hordeum brevisubulatum, HbCIPK2 was induced by salt, drought and ABA treatments, and ectopic expression of HbCIPK2 in wild type Arabidopsis and sos2-1 mutant increased the salt tolerance of transgenic plants [35]. CIPK can bind with CBL even in the absence of Ca2+ ions. In Arabidopsis, Ca2+ bound with AtCBL1 and AtCBL9 to activate AtCIPK3 under low iron condition [36]. Overexpression of AtCBL10 influenced potassium (K+) the transport, seed germination, stomatal opening and closing, and lateral root development [37–44]. In cotton, GhTST2 interacted with GhCIPK6 to regulate plasma membrane sugar homeostasis [45].
Due to its commercial, economic and application values, Rose (Rosa chinensis) have been grown worldwide as one of the most popular ornamental flower crops. However, its shoot growth and flower production are severely affected by adverse environmental changes. Although the functions of CBLs and CIPKs in several plant species have been studied, the genome wide characterization and the regulatory roles of CBLs and CIPKs in rose still remain unknown. We identified 7 CBL and 17 CIPK proteins in the rose genome, and analyzed their gene structure, chromosome position, evolutionary relationships, promoter cis-acting elements and gene expression pattern in response to different abiotic stresses. The information presented here will provide a theoretical basis for the future researches on the accurate functions of CBL-CIPK proteins in response to adverse abiotic stress in woody cutting flower plants.
2.1 Identification and Classification of CBL-CIPK Genes in Rose
The information for genomic analysis and gene annotation of rose CBL and CIPK genes were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/data-hub/taxonomy/74649/) [46]. The gene sequences of CBLs and CIPKs were obtained from the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/) and the Rice Genome Annotation Project (RGAP) (http://rice.uga.edu/annotation_community_families.shtml), respectively [6,5,17]. The calcium binding EF-hand motif (PF00036) of CBL proteins was obtained based on the hidden Markov models (HMMs) in the Pfam database (http://pfam.xfam.org/) [47–49], and the kinase domain (PF00069) and NAF domain (PF03822) of CIPK proteins were determined based on the HMMs [50]. The candidate CBL-CIPK genes were identified using the HMM parameters of E-value < 0.05 and identity > 50% for the non-redundant sequences [51]. All the candidate genes were further validated using SMART (http://smart.embl-heidelberg.de) and NCBI protein BLAST tool in SWISS-PORT database (E-value < 0.05, Identity > 50%) (https://blast.ncbi.nlm.nih.gov/Blast.cgi ) [16,52,53]. The molecular weight (MW) and isoelectric point (pI) were calculated using the online tool ExPASy (https://web.expasy.org/compute_pi/) [54], and subcellular localization of proteins was predicted using the online tool Plant-PLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) [55].
2.2 Phylogenetic Analysis and Chromosomal Mapping of CBL-CIPK Members
The full length amino acid sequences of all the identified CBLs and CIPKs from Rosa chinensis (Rc), Arabidopsis thaliana (At), and Oryza sativa (Os) were analyzed with Clustal X [56–58]. The phylogenetic trees were constructed using the MEGA7.0 software following the neighbor-joining (NJ) statistical method, with 1000 bootstrap replicates and the Poisson model [56]. The online tool iTOL was used to display the phylogenetic trees [59]. The chromosomal localization of RcCBLs and RcCIPKs were obtained using the TBtools software, based on the rose genome annotation data [60].
2.3 Gene Structure and Conserved Motif Analysis
The online tool SMART (http://smart.embl-heidelberg.de) was used to analyze the gene structure based on the untranslated regions (UTR), full length coding sequences (CDS), and genomic sequences [52]. The online program MEME (Multiple Em for Motif Elicitation) (https://meme-suite.org/) was used to assess the conserved motifs with the number of repetition set to zero or one, the maximum number of motif set at 15 (RcCIPKs) or 7 (RcCBLs), and all other parameter set as default [61].
To explore the potential interaction of RcCBLs and RcCIPKs, the cis-acting elements in the 2.0 kb upstream sequences of their CDS were predicted using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [46,62].
2.5 Synteny Analysis of RcCBLs and RcCIPKs
The syntenic relationship between pairs of chromosomes from rose, rice and Arabidopsis were analyzed and visualized using the MCScanX program [63]. The tandem repeat events within species were displayed using the Advanced Circos, and the non-synonymous substitution rate (Ka), synonymous substitution rate (Ks) and Ka/Ks values were calculated using the TBtools software [60,64]. Whole genome duplication (WGD) events depending on the Ka/Ks values were judged with KaKs Calculator software [65–67].
2.6 Protein Interaction Prediction
Using Arabidopsis as the background, the STRING database (http://stringdb.org/) was used to predict the interaction between RcCBLs and RcCIPKs (minimum required interaction score: highest confidence 0.900). Cytoscape was used to visualize the interaction networks [68].
2.7 Plant Materials, Growth Conditions and Stress Treatments
In this study, rose (Rosa chinensis) cv. Red Leonardo da Vinci was used. One-year-old cutting plants at the same size and growth state were selected and cultured in greenhouse set at 28°C/26°C day/night temperature with a 16 h/8 h light/dark photoperiod cycle and 65% relative humidity. For gene expression analysis, root, stem, leaf, flower, petiole and calyx samples were collected. To analyze the gene expression in response to different abiotic stresses, plantlets were respectively subjected to 4°C, 37°C, 300 mM NaCl, 15% (w:v) PEG 6000 and 5 µM ABA treatments for 0, 3, 6, 9, 12, 24 and 48 h. Subsequently, leaf samples were collected at relative time points, frozen in liquid nitrogen, and stored at −80°C for further RNA extraction and quantitative real-time PCR (qRT-PCR) analysis.
2.8 RNA Extraction and qRT-PCR Assays
Total RNA was extracted using the Plant RNA Kit (OMEGA, USA) following the manufacturer’s instructions. First-strand cDNA was synthesized using HiScript® III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). qRT-PCR was performed in 96-well plates on a BIO-RAD CFX Connect Real-Time System (BIO-RAD, USA) using the SYBR qPCR Master Mix (Vazyme, China) according to the manufacturer’s instructions. Primers used for qRT-PCR were designed using the Primer3plus online tool (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi) and validated by NCBI-primer blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi). The rose RcActin gene (LOC112192124) was used as the reference gene to calculate the relative expression levels of RcCBLs and RcCIPKs [69]. Specific primers used for qRT-PCR are listed in Table S1.
All data were analyzed for variance using the IBM SPSS Statistics 21. Differences between mean values were analyzed using the Student’s t-test and considered statistically significant at a probability level of 5% (*P < 0.05) or 1% (**P < 0.01) [70].
3.1 Seven RcCBLs and Seventeen RcCIPKs Are Identified in the Rose Genome
To identify the CBL and CIPK genes in rose genome, we blast searched the NCBI database (https://www.ncbi.nlm.nih.gov/data-hub/taxonomy/74649/), using the amino acid sequences of 10 Arabidopsis CBLs and 23 CIPKs as references. A total number of 7 sequences containing the calcium-binding and EF-hand domain, and 17 sequences containing the kinase and NAF domains, were confirmed as rose CBL and CIPK genes, respectively. They were named as RcCBL1 to RcCBL7 and RcCIPK1 to RcCIPK17 (Table 1; Table S2). We investigated the chromosome distribution of these RcCBLs and RcCIPKs, and found that they were mapped to four and seven chromosomes, respectively. Among the seventeen RcCIPKs, six of them, RcCIPK10 to RcCIPK15, were located on chromosome 5. Physiological and biochemical property analyses indicated that the seven identified RcCBL proteins were comprised of 211 to 231 amino acids, with a predicted isoelectric points (pI) from 4.55 to 4.9 and molecular weight (MW) from 24.45 to 28.54 kDa, whereas the seventeen identified RcCIPK proteins were comprised of 336 to 494 amino acids, with a predicted pI from 5.67 to 9.15 and MW from 37.78 to 53.47 kDa. The pI points of all RcCBL proteins were less than 7, while the pI points of most RcCIPK proteins (88%) were greater than 7 (Table 1). Subcellular localization prediction with the online tool Plant-PLoc server revealed that six out of the seven RcCBLs (RcCBL1 to RcCBL6) were plasma membrane localized, except RcCBL7, which was localized to both plasma membrane and cytoplasm. Whereas most RcCIPKs (RcCIPK1, RcCIPK3 to RcCIPK8, RcCIPK10 and RcCIPK14 to RcCIPK17) were predicted to be cytoplasmic localized, except RcCIPK2, RcCIPK9, RcCIPK11 to RcCIPK13, which were predicted to be both cytoplasmic and nucleic localized (Table 1). In addition, alternative splicing was predicted in RcCBL1, RcCBL3, RcCBL5, RcCIPK1, RcCIPK4, RcCIPK8, RcCIPK9 and RcCIPK13 (Table 1).
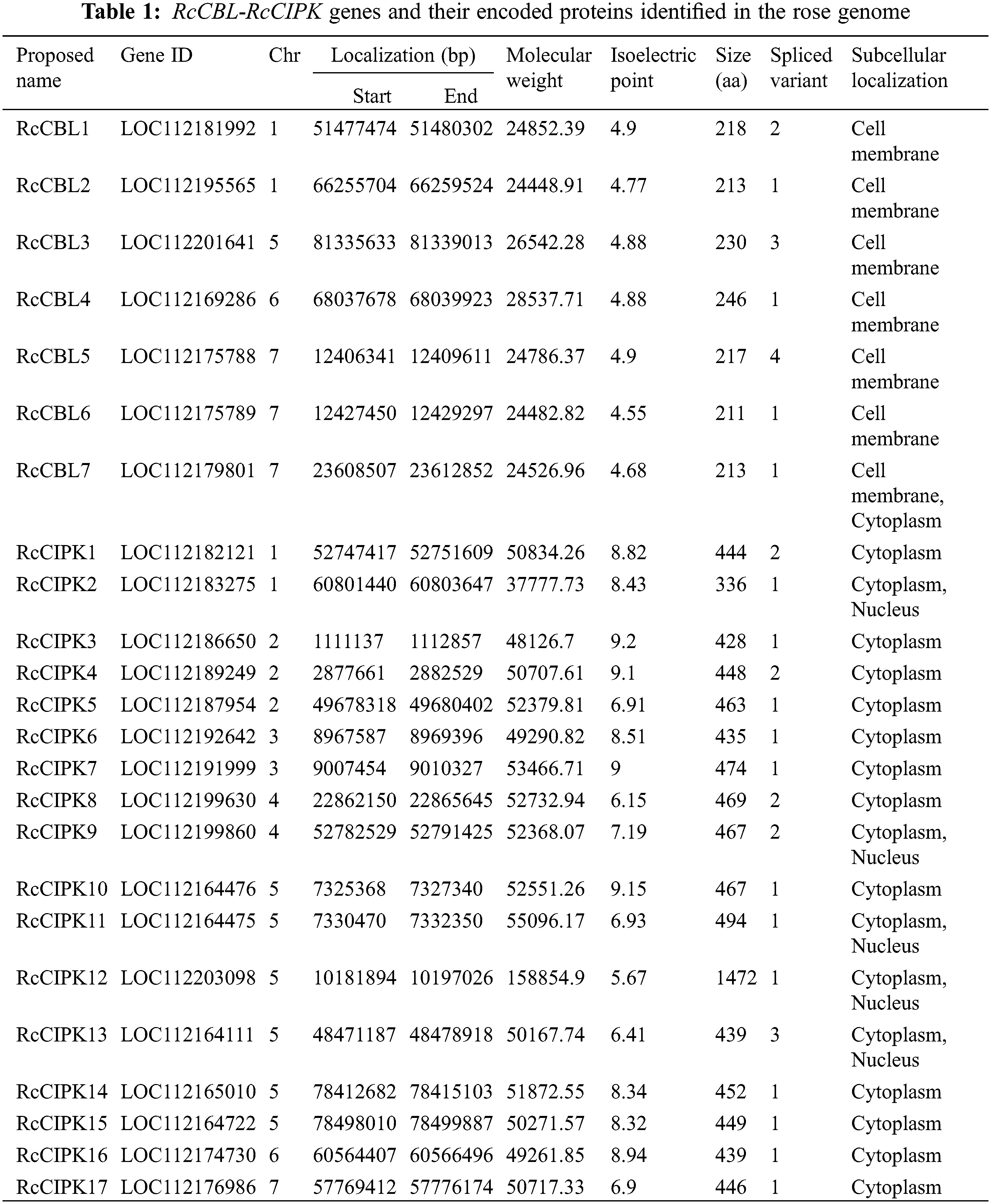
3.2 CIPK and CBL Proteins Are Respectively Classified into Four and Five Subgroups
To understand the possible functions and evolutionary relationships of the CBL and CIPK proteins in rose, we constructed a rootless phylogenetic tree consisting of 27 CBLs and 75 CIPKs from Arabidopsis thaliana, Oryza sativa and Rosa chinensis (Fig. 1). Based on their sequence similarity, the seven RcCBLs were classified into four subgroups, including 2 RcCBLs (RcCBL2, RcCBL6) in group I, 1 RcCBL (RcCBL3) in group II, 2 RcCBLs (RcCBL4, RcCBL5) in group III and 2 RcCBLs (RcCBL1, RcCBL7) in group IV (Fig. 1A). The seventeen RcCIPKs were divided into five subgroups, including 2 RcCIPKs (RcCIPK6, RcCIPK15) in group I, 3 RcCIPKs (RcCIPK5, RcCIPK11, RcCIPK16) in group II, 1 RcCIPK (RcCIPK3) in group III, 4 RcCIPKs (RcCIPK2, RcCIPK7, RcCIPK10, RcCIPK14) in group IV and 7 RcCIPKs (RcCIPK1, RcCIPK4, RcCIPK8, RcCIPK9, RcCIPK12, RcCIPK13, RcCIPK17) in group V (Fig. 1B).
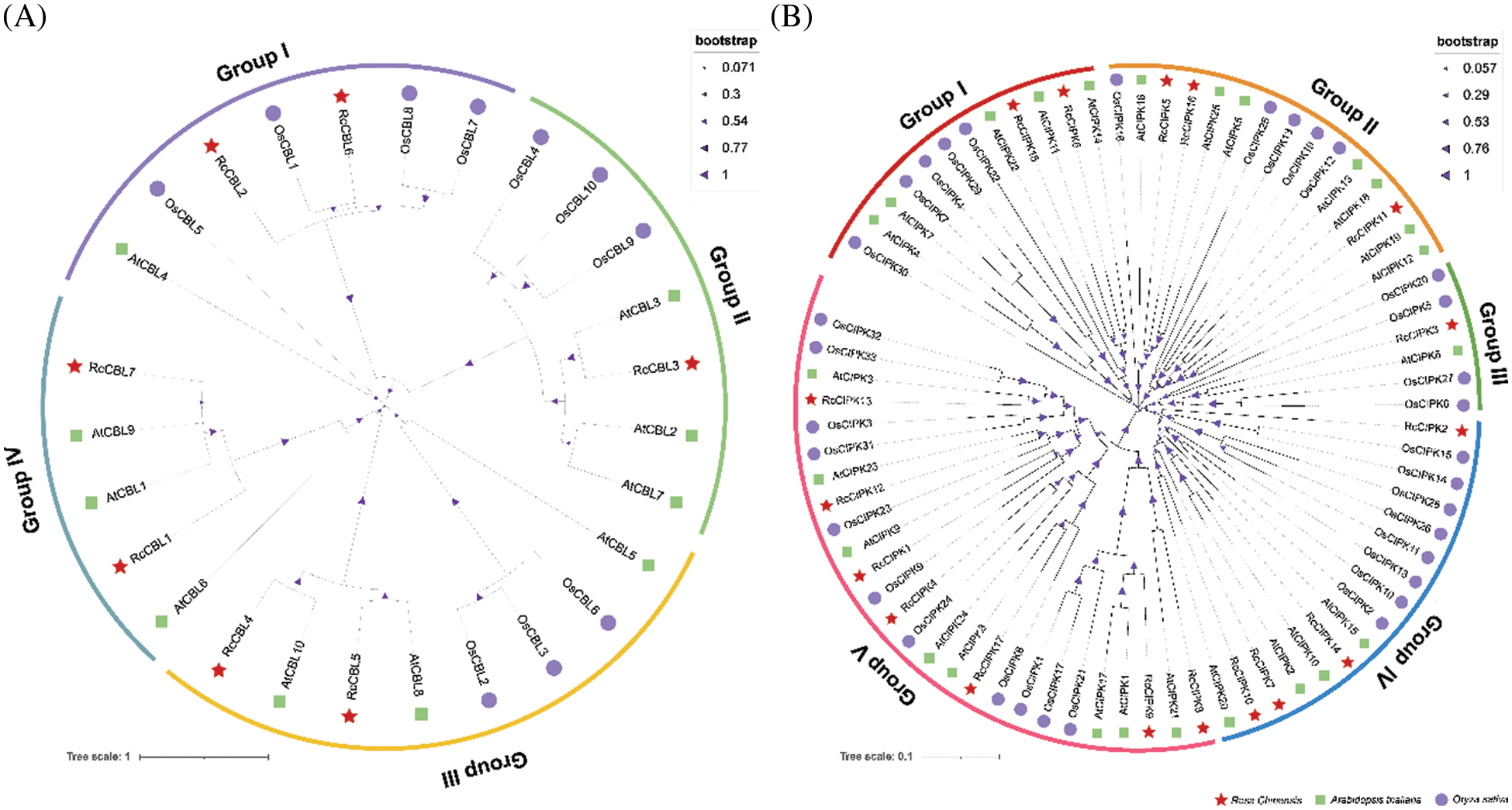
Figure 1: Phylogenetic relationship analysis of CBL and CIPK genes from Arabidopsis, rice and rose. Full length amino acid sequences of CBLs and CIPKs were used to construct the neighbor-joining (NJ) tree using MEGA7.0 with 1000 bootstrap replicates. Subfamilies are highlighted in different colors. (A) Phylogenetic tree of CBL proteins. (B) Phylogenetic tree of CIPK proteins. At, Arabidopsis thaliana; Os, Oryza sativa; Rc, Rosa chinensis
3.3 Gene Structures of RcCBLs and RcCIPKs Are Highly Conserved
We further analyzed the structures of RcCBL and RcCIPK genes and found that members in the same subfamily demonstrated similar motif distribution and gene structures (Fig. 2). A total number of seven and fifteen conserved motifs in RcCBL and RcCIPK proteins were respectively identified (Fig. 2A). All RcCBL proteins, except RcCBL4, which lacked motif 7, possessed all the seven motifs (motifs 1 to 7). In RcCBL proteins, motif 1, motif 3 and motif 4 contained the essential phosphorylation site for the calcium ion EF-hand structure, and motif 7 was identified as the myristoylation site (Fig. 2C). Almost all RcCIPK proteins, except RcCIPK2, which lacked motifs 5 and 6, possessed motifs 1 to 7. Six of the seven RcCIPKs (RcCIPK1, RcCIPK4, RcCIPK8, RcCIPK12, RcCIPK13, and RcCIPK17) in group V contained motif 11 (Fig. 1C). Motif 12 was not only observed in the C-terminus of RcCIPK7, RcCIPK10 and RcCIPK14, but also in the N-terminal of RcCIPK5 and RcCIPK15. In addition, motifs 8, 9 and 13 were detected in most RcCIPK proteins. However, RcCIPK2, RcCIPK5 and RcCIPK15 lacked motif 8, RcCIPK2, RcCIPK8, RcCIPK9 and RcCIPK15 lacked motif 9, and RcCIPK6, RcCIPK7, RcCIPK14 and RcCIPK15 lacked motif 13 (Fig. 2A). All the motifs in RcCIPK proteins were listed in Table S3, and the detailed sequences of the six representative motifs were exhibited in Fig. 2D. Motifs 1, 2 and 5 possessed the kinase catalytic activation loop, while motifs 7, 8 and 14 contained the basic NAF motif regulatory domain.
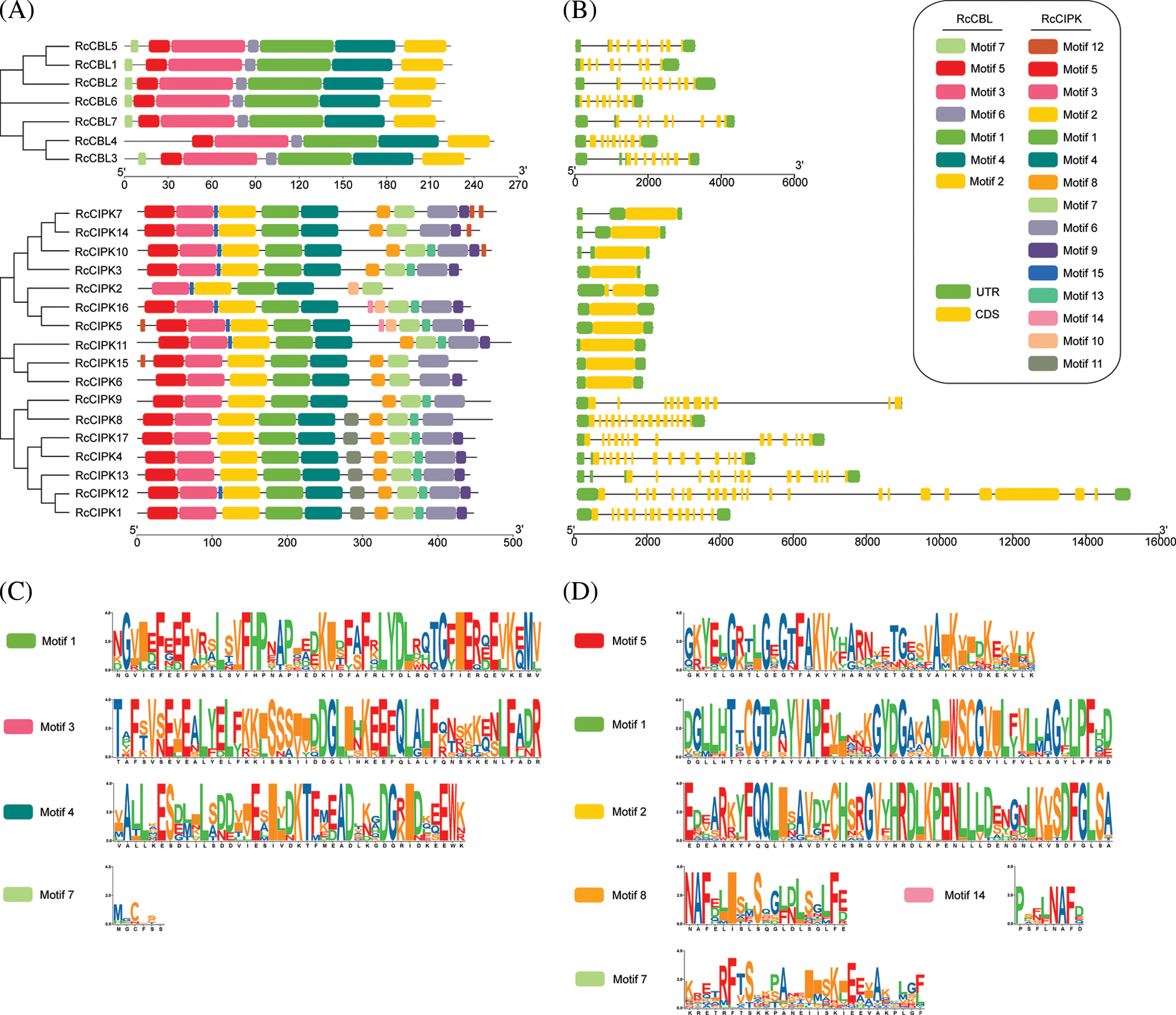
Figure 2: Phylogenetic relationship, conserved motifs and gene structure of RcCBLs and RcCIPKs. A. Phylogenetic trees of RcCBL and RcCIPK proteins generated with the MEGA 7.0. The conserved motifs of RcCBLs and RcCIPKs were analyzed with the MEME program. B. Exon-intron structures of RcCBLs and RcCIPKs. Untranslated regions (UTR), exons, and introns are represented with green boxes, yellow boxes and gray lines, respectively. C. Sequence logos of the four core motifs of RcCBLs. Motif 1, motif 3 and motif 4, the EF-hand motifs; motif 7, the transmembrane domain. D. Sequence logos of the six core motifs of the RcCIPKs. Motifs 1, 2 and 5, the serine/threonine protein kinases; motifs 8 and 14, the NAF motifs; motif 7, the PPI motif
Furthermore, the CDS and UTR of the RcCBL and RcCIPK genes were analyzed to determine the exon and intron distribution. We observed that all RcCBLs contained 8–9 introns, whereas most RcCIPKs contained 0–1 intron, except those in group V, which contained 10–22 introns (Fig. 2B). Consistent with the results of phylogenetic analysis, RcCBLs and RcCIPKs on the same developmental branch showed similar exon-intron organization, despite the differences of intron numbers and distances in each gene (Fig. 2B). In particular, RcCIPK3, RcCIPK5, RcCIPK6, RcCIPK7, RcCIPK10, RcCIPK11, RcCIPK14, RcCIPK15 and RcCIPK16 contained contiguous CDS sequences that converged on two evolutionary branches at the same time. The continuity or discontinuity of the CDS region of RcCIPKs might be correlated with the evolutionary changes.
3.4 Promoters of RcCBLs and RcCIPKs Contain the Key Cis-Elements for Plant Hormone and Stress Response
We further analyzed the promoter regions of RcCBL and RcCIPK genes. We found that they all contained the basic cis-acting elements related to stress response, light response, anaerobic regulation, plant hormone (ABA, SA, MeJA, GA), mechanical injury defense and developmental regulation (Fig. 3, Table S4). All cis-acting elements detected in the promoter regions of RcCIPKs, except HD-Zip 1, were also found in the promoter regions of RcCBLs. In addition, a number of light responsive elements, ABA responsive and MYB binding sites were found in the promoter region of RcCBLs and RcCIPKs, implying their multiple roles in flowering, morphogenesis and stress response in rose.
Figure 3: Phylogenic relationship and cis-acting elements in the promoter regions of RcCBLs and RcCIPKs. Phylogenetic tree of RcCBL and RcCIPK proteins was constructed with the MEGA 7.0 software. The cis-acting elements were predicted with PlantCARE and displayed with boxes in different colors
3.5 Gene Duplication Events in RcCBLs and RcCIPKs Are Observed
We investigated the chromosome distribution of these RcCBLs and RcCIPKs, and found that RcCBLs were mapped to chromosome 1, chromosome 5, chromosome 6 and chromosome 7, whereas RcCIPKs were distributed unevenly on all the seven chromosomes (Fig. 4). We also examined gene duplication events in RcCBLs and RcCIPKs using MCScanX. Collinear analyses based on the annotation information revealed that 2 gene pairs RcCBL5 and RcCBL6, and RcCIPK10 and RcCIPK11, have undergone tandem duplication events, and 6 gene pairs RcCBL2 and RcCBL5, RcCIPK2 and RcCIPK6, RcCIPK2 and RcCIPK14, RcCIPK3 and RcCIPK16, RcCIPK7 and RcCIPK13, and RcCIPK6 and RcCIPK14, have undergone whole genome duplication events (Fig. 4).
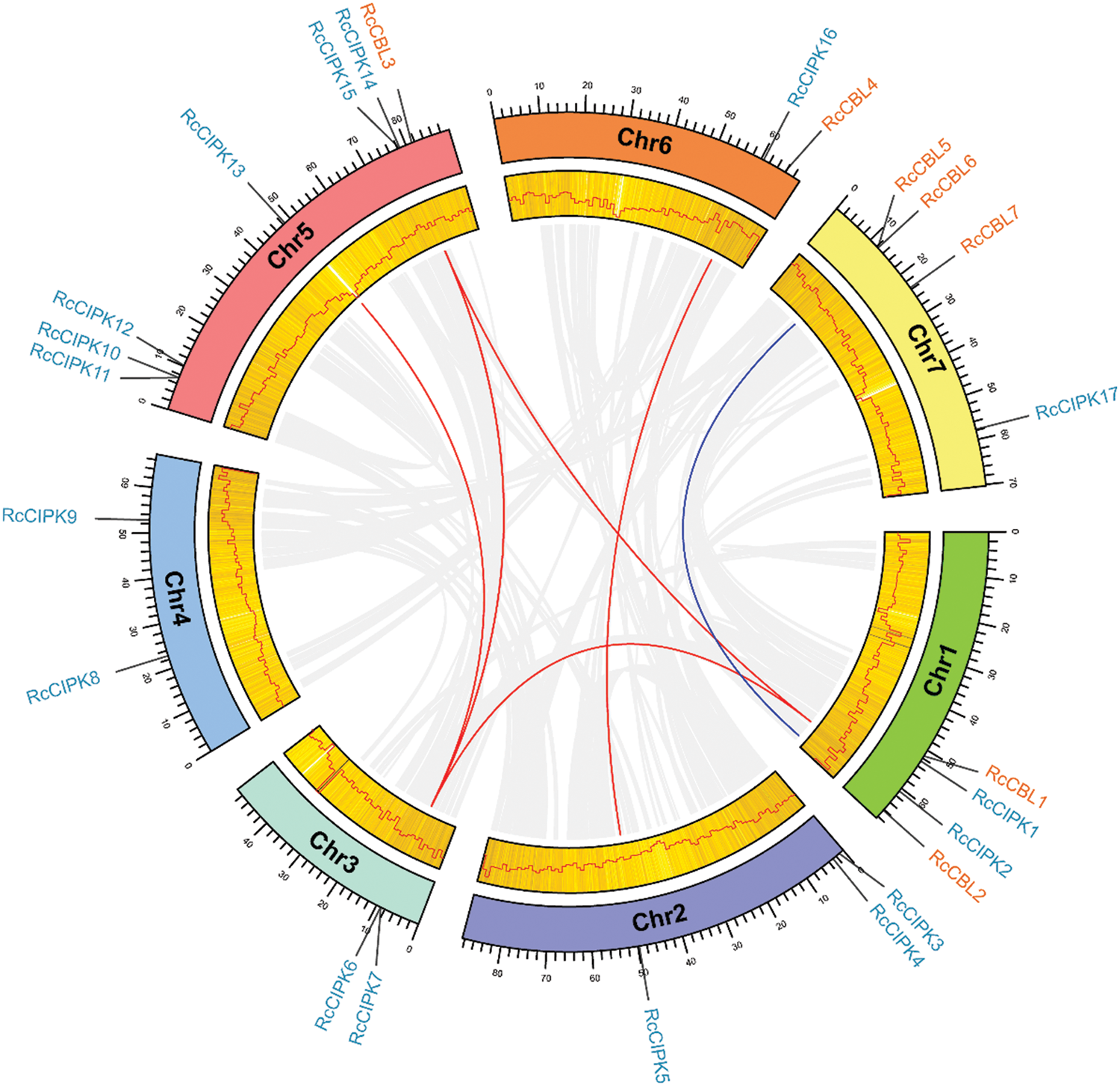
Figure 4: Chromosome distribution and syntenic analysis of CBLs and CIPKs. Syntenic relationships between RcCBLs and RcCIPKs were visualized using Circos. The rose chromosomes are represented with colored bars. The length of chromosomes is marked in scale, and the density of genes is demonstrated in a heatmap and a line chart. Gray lines indicate synteny blocks in the rose genome. Duplicated RcCBL and RcCIPK gene pairs were indicated with blue and orange lines, respectively
To further understand the evolution of RcCBL and RcCIPK genes in details, Ka/Ks ratios were calculated. The Ka/Ks values of all the seven gene pairs were less than 1, indicating the influence of purifying selection on the evolution of these genes with limited functional divergence after the duplication events (Table 2). Therefore, the replication events in these seven gene pairs have increased the number and functional diversity of RcCBL and RcCIPK family members. Then, we constructed a comparative syntenic map using the CBL and CIPK genes of Arabidopsis, rice and rose. We found that 19 RcCBL and RcCIPK genes had homologs in rice, while 24 RcCBL and RcCIPK genes had homologs in Arabidopsis, implying that these genes might have played significant roles in the evolution of CBL and CIPK gene families (Fig. 5).
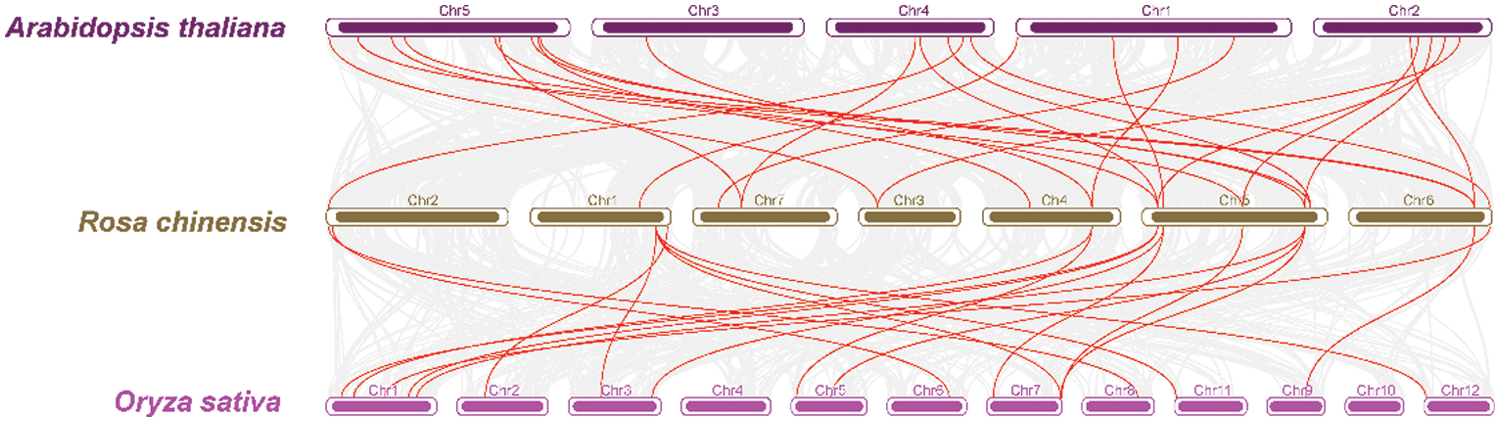
Figure 5: Collinear correlations of Arabidopsis, rice, and rose. The five chromosomes of Arabidopsis, seven chromosomes of rose and 12 chromosomes of rice were presented in different colors. The collinear blocks and syntenic RcCBLs and RcCIPKs gene pairs among Arabidopsis, rice, and rose are respectively shown in gray and red lines
3.6 Protein Interaction Are Predicted in RcCBLs and RcCIPKs
In the protein interaction network, RcCBL and RcCIPK proteins showed 34 nodes and 44 edges among themselves (Fig. 6). In particular, RcCBL3, RcCBL6, RcCIPK12 and RcCIPK15 demonstrated potential interactions with most other members. In addition, ten proteins were predicted to potentially interact with RcCBLs and RcCIPKs (Table S5).
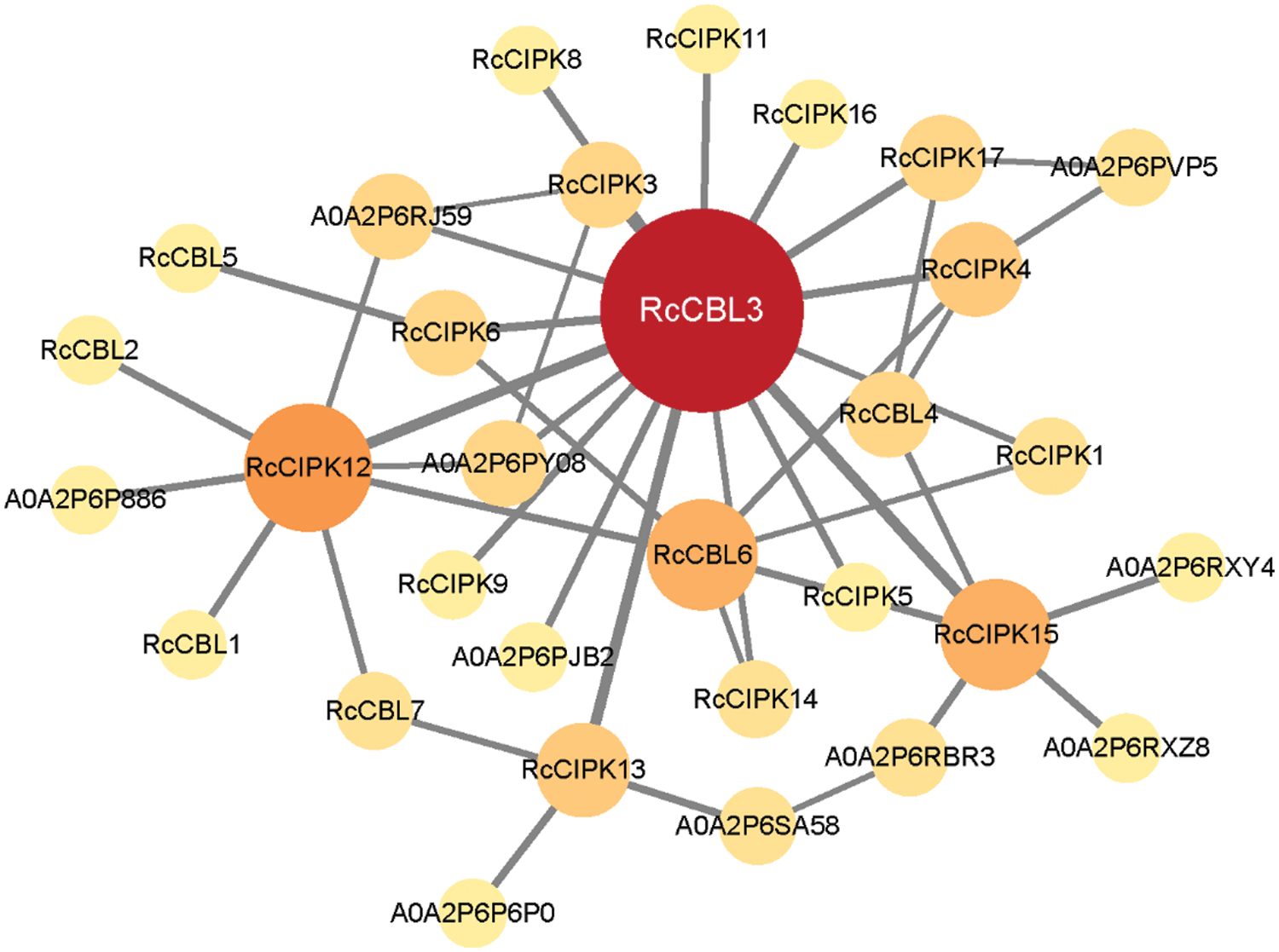
Figure 6: A predicted protein interaction network of RcCBLs and RcCIPKs. Knot sizes in different colors indicate the frequency of interacting proteins. The higher the protein-protein interaction frequency is, the deeper the color and the larger the size are. The line thickness shows the intension of data support
3.7 RcCBL and RcCIPK Genes Show Overlapped but Distinct Expression Patterns in Rose
To understand the possible functions of RcCBLs and RcCIPKs in the growth and development of rose, we examined their expression levels in various organs including roots, stems, leaves, petioles, flowers and calyxes. They showed overlapped but distinct expression patterns (Fig. 7, Table S6). RcCIPK3 and RcCIPK7 were highly expressed in all tested organs, while expressions of RcCIPK2, RcCIPK4 and RcCIPK10 were hardly detected. All RcCBLs displayed a higher expression in the vegetative organs such as roots, stems, leaves and petioles, and a lower expression in flowers. A higher expression of RcCBL2, RcCIPK4, RcCIPK5, RcCIPK12, RcCIPK13 and RcCIPK14 in leaves, RcCBL5 in roots, RcCBL4 and RcCIPK3 in stems, and RcCBL6, RcCIPK1, RcCIPK2, RcCIPK7, RcCIPK9 and RcCIPK10 in petioles, was detected than in the other organs. Meanwhile, RcCIPK15 and RcCIPK16 were predominantly expressed in flowers, indicating their possible role in flowering regulation.
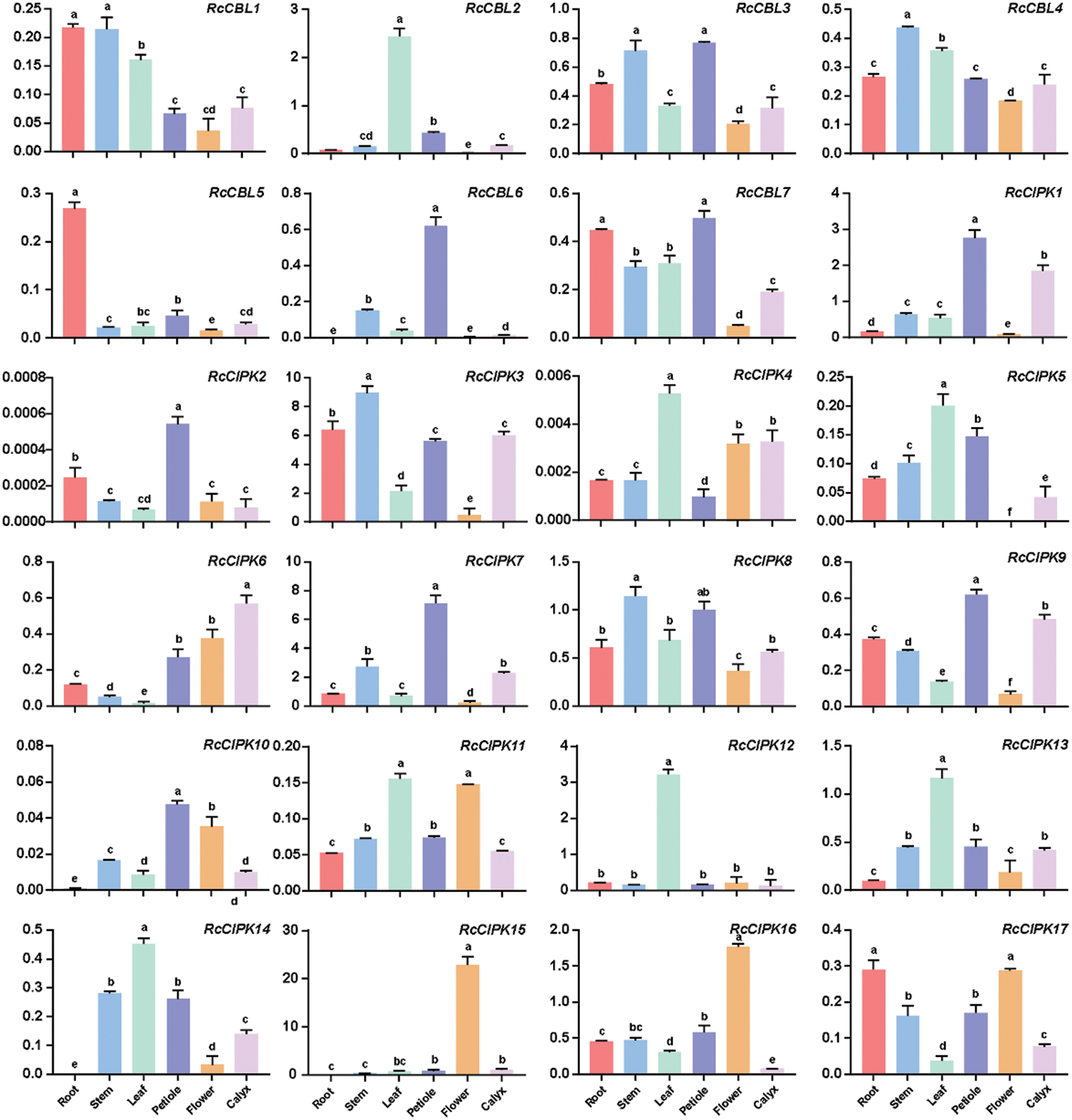
Figure 7: Expression patterns of RcCBLs and RcCIPKs in rose plants grown under normal condition. The relative expression levels of RcCBL and RcCIPK genes in roots, stems, leaves, flowers, petioles and calyxes were normalized with the reference gene RcActin. Data are represented as the mean ± SD of three biological replicates. Different letters (a, b, c, d, and e) indicate significant differences in the gene expression levels among roots, stems, leaves, flowers, petioles and calyxes (n = 9; P < 0.05, Student’s t-test)
3.8 Expressions of RcCBLs and RcCIPKs Are Regulated by Different Abiotic Stresses
To further understand the possible functions of RcCBLs and RcCIPKs in the response to environment stresses, we examined their expression levels in the leaves of rose plants in response to 4°C, 37°C, NaCl, PEG and ABA treatments (Fig. 8, Table S7). One-year-old rose plants grown in green house were subjected to 4°C, 37°C, 300 mM NaCl, 15% PEG and 5 µM ABA treatments for 0, 3, 6, 12, 24 and 48 h, respectively. All RcCBLs and RcCIPKs showed responsive expression to at least one treatment. They all showed up- or down-regulated expression in response to low temperature (4°C), high temperature (37°C), salt stress, PEG and ABA treatments. However, expression of RcCBL4 was not affected by high temperature, and RcCBL2 not by ABA and heat stress. Compared with that at 0 h, low temperature induced RcCBL5 expression at 6, 9, 12 h, RcCBL3 expression at 6 h, and RcCBL2 expression at 6 and 9 h, whereas suppressed RcCBL1 and RcCBL4 expression at 48 h, RcCBL2 and RcCBL7 expression at 3 h, and RcCBL6 expression at 9 h. Meanwhile, heat stress induced the expression of RcCBL1, RcCBL3, RcCBL5, RcCBL6 and RcCBL7 at 6, 3, 3, 3 and 9 h, and suppressed the expression of RcCBL6 at 12 h and 24 h, and RcCBL3 at 12 h, respectively. ABA treatment increased the expression levels of RcCBL1, RcCBL3, RcCBL6 and RcCBL7 at 3 h, and RcCBL1, RcCBL5 and RcCBL7 at 6 h, but suppressed the expression of RcCBL5 at 48 h. Salt stress increased the expression of RcCBL2, RcCBL3, RcCBL4 and RcCBL7 at 3 h, and RcCBL4, RcCBL5 and RcCBL6 at 9 h, but suppressed the expression of RcCBL4 at 24 and 48 h. PEG treatment increased the expression of all seven RcCBLs at one or more time points. Most RcCIPKs showed different expressions upon different stress treatments, and at least six of the seventeen RcCIPKs showed fluctuated expression levels under each stress treatment. Only RcCIPK9 and RcCIPK15 showed responsive expression to all the five treatments, while fifteen RcCIPKs responded to at least one stress treatment. RcCIPK11 responded to low temperature. RcCIPK7 was up-regulated by PEG and salt stress. Expression of RcCIPK2 was induced by high temperature, salt stress and PEG treatment. Expression of RcCIPK5 was up- or down-regulated by high temperature, salt stress, PEG and ABA treatments (Fig. 8, Table S7).
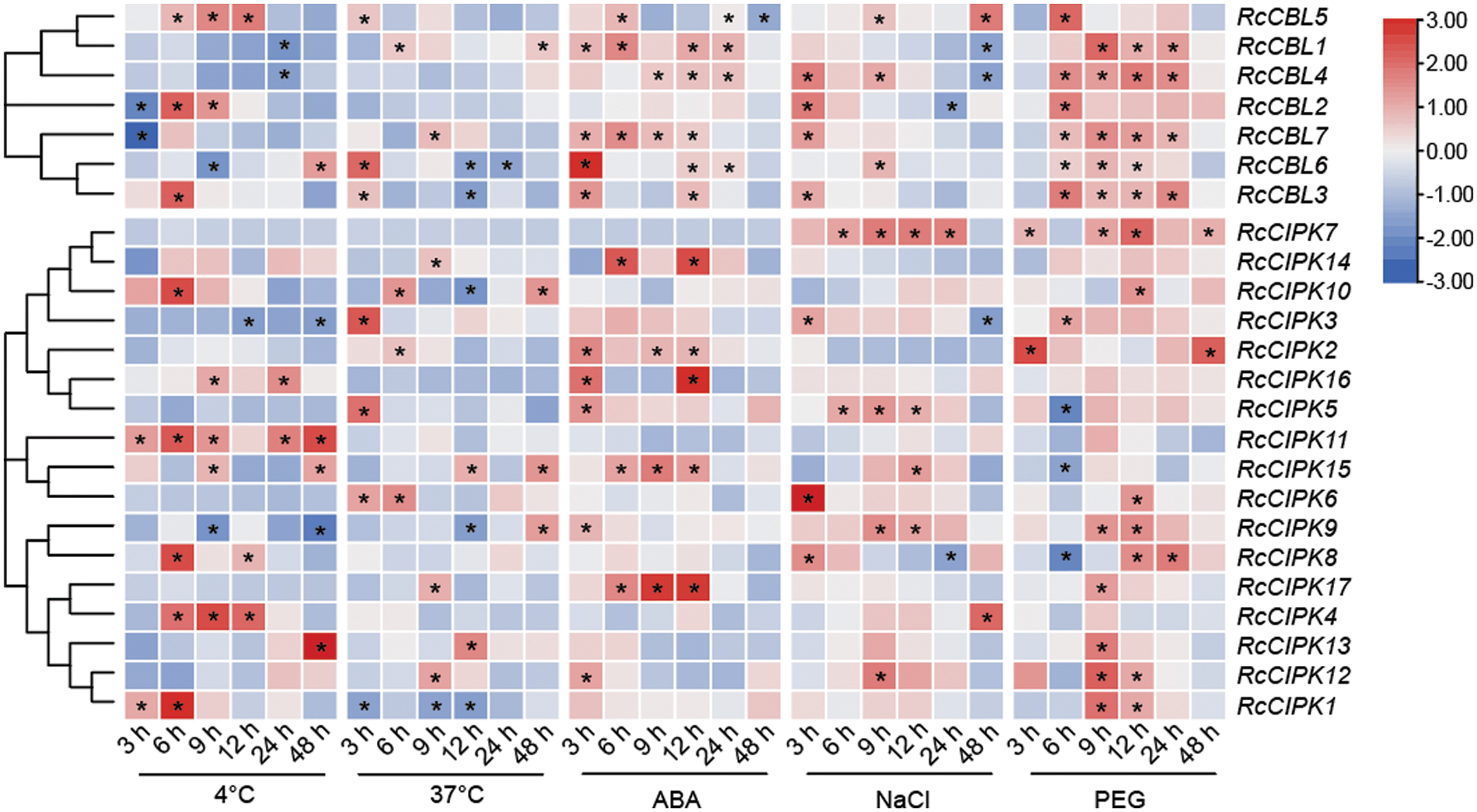
Figure 8: Hierarchical clustering of the relative expressions of RcCBLs and RcCIPKs at six time points under different stress (4°C, 37°C, ABA, NaCl and PEG) conditions. The transcript levels at 0 h were used as the internal control. The expression of each gene relative to RcActin was used as the internal standard. The average of three independent experiments was used to analyze the SD (n = 9; P < 0.05, Student’s t-test)
The CBL-CIPK family is a class of Ca2+ signaling complexes widely found in plants to perceive Ca2+ levels under various external environments [3,4]. Previous studies have identified the CBL-CIPK members in many species and reported their responses to low temperature, ABA treatment and salt stress [23,41,71,72]. However, research on their possible roles in cutting flower crops is still limited. In this study, 7 RcCBLs and 17 RcCIPKs in the rose genome were identified, which is consistent with the number of CBLs but less than the CIPKs in other species [3,6,14]. We also compared their protein characteristics, such molecular weight, isoelectric point, protein size, spliced variant and subcellular localization (Table 1). Consistent with the Arabidopsis CBLs, RcCBLs were also predicted to be localized on plasma membrane and could activate CIPK upon the receiving of calcium signals [9,73].
Phylogenetic analysis has been used to analyze species evolution history and kinship. We constructed the phylogenetic trees which sorted RcCBLs into four subgroups and RcCIPKs into five subgroups (Fig. 1). Consistent with the grouping in other species, RcCBL proteins were evenly distributed among the four groups, while RcCIPK were mainly distributed in the fifth subgroup (42% of the total) [15,18]. Further gene structure analysis revealed that, like the CBLs in A. thaliana and grape, the rose CBLs also contained four EF-hand motifs (Fig. 2A) [15]. Consistent with those in other species, all the RcCIPKs identified in this study have the basic kinase domain and NAF motif [1,6,17,18]. RcCIPK2, in which motif 6 and motif 9 in the C-terminal and motif 5 in the N-terminal were missing, was shorter than the other RcCIPKs (Fig. 2A, Table S3). The distribution of motifs varied among RcCIPKs, while the members on the same branch in the phylogenetic tree shared similar motif distribution. However, the length and distribution of the motifs in RcCIPK12 were extremely different from the length and distribution of the motifs in the other RcCIPKs due to the absence of a protein structural domain at the C-terminus (Table 1). The loss of this domain may probably be due to a stop codon error in genome sequencing [46]. Moreover, the phylogeny and gene structure of RcCBLs and RcCIPKs indicated a potential functional association among them, and the number and relative position of the introns showed a correlation with the phylogeny. For example, RcCBL3 and RcCBL4 were on the same branch, and they showed similar gene structures. While RcCIPK was intuitively divided into intron poor and intron rich groups according to the phylogenetic relationship (Fig. 2B). This property is highly conserved among various species, such as rice, Arabidopsis, poplar and soybean [18,74]. The presence of introns is often accompanied by variable splicing, especially in response to environmental changes [75]. We also observed variable splicing in RcCBLs and RcCIPKs, with a quantitative distribution similar to that detected in other species, indicating that CBLs and CIPKs are conserved across species in terms of gene structure (Table 1) [13,15].
The chromosome distribution of CBLs and CIPKs varies and the regulatory relationship of them changes depending on their biological functions [76,77]. We analyzed the phylogenetic relationships and their regulatory elements of RcCBLs and RcCIPKs (Fig. 3, Table S4). Unfortunately, we did not find a significant association between phylogeny and regulatory elements in the promoters of RcCBLs and RcCIPKs, not even for the tandem repeat gene pairs (e.g., RcCIPK10 and RcCIPK11) or the gene pairs with high homology but located on different chromosomes (e.g., RcCBL3 and RcCBL4). However, numerous stress responsive cis-acting elements were found in the promoter regions of RcCBLs and RcCIPKs, which have been also identified in the promoters of CBLs and CIPKs from other plant species, and some of them have been experimentally confirmed to respond to transcription factors [78–81].
To infer evolutionary divergences of genes, covariance analysis and Ka/Ks calculation have been used to gain detailed information on the approximate timing of the replication events [65]. RcCBLs were present only on chromosomes 1, 5, 6 and 7, whereas RcCIPKs were distributed on all chromosomes. This distribution is probably related to the evolutionary function and the complex gene exchange. Consistent with previous reports in other plant species, both tandem and genome wide duplication repeats were detected in RcCBLs and RcCIPKs (Fig. 4, Table 2) [78,79]. Based on the Ka/Ks values (≤1), we inferred that RcCBL and RcCIPK genes have gone through strong purifying selection during the evolution. Furthermore, interspecies covariance analysis also indicated that both CBL and CIPK genes were continuously and unequally distributed in Arabidopsis, rice and rose (Fig. 5) [3,6]. CBL and CIPK can form a complex to perceive Ca2+ signal [17]. We predicted the interaction between RcCBLs and RcCIPKs based on previous reports and found that RcCBL3 and RcCIPK12 were highly related to other members, indicating that they may function as the core proteins in the CBL-CIPK protein interaction network (Fig. 6).
During different developmental stages, gene expression patterns may vary among different organs. Some genes are expressed only at the early growth and development stages of plants, while some gene act during the formation of organs. Therefore, study on gene expression patterns can provide important information for the understanding of their biological functions [69,82,83]. We observed differential expression patterns of RcCBLs and RcCIPKs in rose plants (Fig. 7). The expressions of RcCBL2 and RcCIPK12 were significantly higher in leaves than in other parts, implying their possible roles in leaf development. RcCIPK15 and RcCIPK16 were predominantly expressed in flowers, indicating their possible role in flowering regulation. Meanwhile, some genes, such as RcCBL4 and RcCIPK8, were constitutively expressed in all tested organs, which indicated that they may function in the whole developmental process of rose plants [69].
Numerous studies have demonstrated that CBL, CIPK or CBL-CIPK complex play important roles in plant growth and response to abiotic stress. In Arabidopsis, AtCBL1 was induced by cold, mannitol and salt treatments, and were involved in ABA-mediated salt, cold and drought stress response [18,25,27,84–87]. AtCBL4-AtCIPK24 activated the membrane Na+/H+ ion channels to alleviate the toxic damage caused by salt stress in an ABA-dependent manner [87–89]. Meanwhile, AtCBL3-AtCIPK9 was reported to target PAT10 and participate in ABA-dependent stomata divergence under stress condition [43,44]. In addition, AtCBL10 and AtCIPK8 were associated with AtSOS1 in alleviating salt stress, while AtCBL5 and AtCIPK11 targeted by SLAC1 were associated with ABA signaling [12]. Overexpression of OsCIPK23 in rice and ZmCIPK8 in maize respectively increased plant tolerance to salt and drought [32,90]. We observed that most RcCBLs and RcCIPKs responded to cold, heat, salt and ABA treatment, although the response patterns varied among different abiotic stresses (Fig. 8). Most RcCBL and RcCIPK genes were up-regulated under osmotic stress condition, but down-regulated by high temperature treatment. These results indicate that RcCBL and RcCIPK genes may have different functions in response to different abiotic stresses at different growth stages in rose.
Taken together, a total number of 7 CBL and 17 CIPK genes in the rose genome were identified and comprehensively analyzed. The identified RcCBLs and RcCIPKs were respectively divided into four and five subfamilies. Expression pattern analyses revealed that most RcCBLs and RcCIPKs were constitutively expressed and all members were either up- and/or down-regulated by cold, heat, salt, osmotic stress and/or ABA treatment. Our findings provide valuable information for the future study on the biological functions of CBL and CIPK genes in the growth, development and response to abiotic stress in flowering crops.
Data Availability Statement: Data are contained within the article and supplementary material.
Authorship: The authors confirm contribution to the paper as follows: study conception and design: Lunzeng Huang, Hongsheng Gao and Ning Jiang; data collection: Yunhong Xu, Zijian Gong, Lele Chen, Shijie Xue; analysis and interpretation of results: Xiaoyan Li, Ruichao Liu and Bei Li; draft manuscript preparation: Hongxia Zhang, Chunyan Yu, and Xiaotong Guo. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This research was funded by the following grants: the Natural Science Foundation of Shandong Province of China (ZR2021MC169); the Cooperation Project of University and Local Enterprise in Yantai of Shandong Province (2021XDRHXMPT09).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ma, X., Li, Q., Yu, Y., Qiao, Y., Gong, Z. (2020). The CBL-CIPK pathway in plant response to stress signals. International Journal of Molecular Sciences, 21(16), 5668. [Google Scholar]
2. Tian, W., Wang, C., Gao, Q., Li, L., Luan, S. (2020). Calcium spikes, waves and oscillations in plant development and biotic interactions. Nature Plants, 6(7), 750–759. [Google Scholar]
3. McAinsh, M. R., Pittman, J. K. (2009). Shaping the calcium signature. New Phytologist, 181(2), 275–294. [Google Scholar]
4. Weinl, S., Kudla, J. (2009). The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytologist, 184(3), 517–528. [Google Scholar]
5. Luan, S., Kudla, J., Rodríguez-Concepción, M., Yalovsky, S., Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. The Plant Cell, 14, S389–S400. [Google Scholar]
6. Mohanta, T. K., Mohanta, N., Mohanta, Y. K., Parida, P., Bae, H. (2015). Genome-wide identification of Calcineurin B-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biology, 15(6), 189–204. [Google Scholar]
7. Kolukisaoglu, U., Weinl, S., Blazevic, D., Kudla, O. B. (2004). Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiology, 134(1), 43–58. [Google Scholar]
8. Sanchez-Barrena, M. J., Fujii, H., Angulo, I., Martinez-Ripoll, M., Zhu, J. et al. (2007). The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Molecular Cell, 26(3), 427–435. [Google Scholar]
9. Shi, J., Kim, K., Ritz, O., Albrecht, V., Gupta, R. et al. (1999). Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. The Plant Cell, 11(12), 2393–2405. [Google Scholar]
10. Tang, R., Wang, C., Li, K., Luan, S. (2020). The CBL-CIPK calcium signaling network: Unified paradigm from 20 years of discoveries. Trends in Plant Science, 25(6), 604–617. [Google Scholar]
11. Batistic, O., Waadt, R., Steinhorst, L., Held, K., Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. The Plant Journal, 61(2), 211–222. [Google Scholar]
12. Saito, S., Hamamoto, S., Moriya, K., Matsuura, A., Sato, Y. et al. (2018). N-myristoylation and S-acylation are common modifications of Ca2+-regulated Arabidopsis kinases and are required for activation of the SLAC1 anion channel. New Phytologist, 218(4), 1504–1521. [Google Scholar]
13. Ma, X., Gai, W. X., Qiao, Y. M., Ali, M., Wei, A. M. et al. (2019). Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genomics, 20(1), 775–793. [Google Scholar]
14. Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y. et al. (2015). Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biology, 15(269), 269. [Google Scholar]
15. Xi, Y., Liu, J., Dong, C., Cheng, Z. (2017). The CBL and CIPK gene family in grapevine (Vitis viniferaGenome-wide analysis and expression profiles in response to various abiotic stresses. Frontiers in Plant Science, 8, 978. [Google Scholar]
16. Zhang, H., Yang, B., Liu, W., Li, H. W., Wang, L. et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biology, 14(8), 1769–1781. [Google Scholar]
17. Kanwar, P., Sanyal, S. K., Tokas, I., Yadav, A. K., Pandey, A. et al. (2014). Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium, 56(2), 81–95. [Google Scholar]
18. Yu, Y., Xia, X., Yin, W., Zhang, H. (2007). Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regulation, 52(2), 101–110. [Google Scholar]
19. Hu, W., Xia, Z., Yan, Y., Ding, Z., Tie, W. et al. (2015). Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Frontiers in Plant Science, 6, 914. [Google Scholar]
20. Liu, H., Wang, Y., Li, H., Teng, R., Wang, Y. et al. (2019). Genome-wide identification and expression analysis of calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase family genes in tea plant. DNA and Cell Biology, 38(8), 824–839. [Google Scholar]
21. Halfter, U., Ishitani, M., Zhu, J. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proceedings of the National Academy of Sciences of the United States of America, 97(7), 3735–3740. [Google Scholar]
22. Liu, J., Zhu, J. (1998). A calcium sensor homolog required for plant salt tolerance. Science, 280(5371), 1943–1945. [Google Scholar]
23. Qiu, Q., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Sciences of the United States of America, 99(12), 8436–8441. [Google Scholar]
24. Kim, B., Waadt, R., Cheong, Y. H., Pandey, G. K., Dominguez-Solis, J. R. et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. The Plant Journal, 52(3), 473–484. [Google Scholar]
25. Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y. et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. The Plant Cell, 21(5), 1607–1619. [Google Scholar]
26. Yang, Y., Zhang, C., Tang, R., Xu, H., Lan, W. et al. (2019). Calcineurin B-like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. International Journal of Molecular Sciences, 20(10), 2421. [Google Scholar]
27. Cheong, Y. H., Kim, K., Pandey, G. K., Gupta, R., Grant, J. J. et al. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. The Plant Cell, 15(8), 1833–1845. [Google Scholar]
28. Li, Z., Xu, Z., Chen, Y., He, G., Yang, G. et al. (2013). A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development. PLoS One, 8(2), e56412. [Google Scholar]
29. Ligaba-Osena, A., Fei, Z., Liu, J., Xu, Y., Shaff, J. et al. (2017). Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis. New Phytologist, 214(2), 830–841. [Google Scholar]
30. Maehs, A., Steinhorst, L., Han, J., Shen, L., Wang, Y. et al. (2013). The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Molecular Plant, 6(4), 1149–1162. [Google Scholar]
31. Straub, T., Ludewig, U., Neuhaeuser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. The Plant Cell, 29(2), 409–422. [Google Scholar]
32. Yang, W., Kong, Z., Omo-Ikerodah, E., Xu, W., Li, Q. et al. (2008). Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). Journal of Genetics and Genomics, 35(9), 531–543. [Google Scholar]
33. Ma, Q., Sun, M., Kang, H., Lu, J., You, C. et al. (2019). A CIPK protein kinase targets sucrose transporter MdSUT2.2 at Ser254 for phosphorylation to enhance salt tolerance. Plant, Cell & Environment, 42(3), 918–930. [Google Scholar]
34. Ma, Q., Sun, M., Lu, J., Kang, H., You, C. et al. (2019). An apple sucrose transporter MdSUT2.2 is a phosphorylation target for protein kinase MdCIPK22 in response to drought. Plant Biotechnology Journal, 17(3), 625–637. [Google Scholar]
35. Ma, Q., Sun, M., Lu, J., Liu, Y., You, C. et al. (2017). An apple CIPK protein kinase targets a novel residue of AREB transcription factor for ABA-dependent phosphorylation. Plant Cell And Environment, 40(10), 2207–2219. [Google Scholar]
36. Tian, Q., Zhang, X., Yang, A., Wang, T., Zhang, W. (2016). CIPK23 is involved in iron acquisition of Arabidopsis by affecting ferric chelate reductase activity. Plant Science, 246, 70–79. [Google Scholar]
37. Ren, X., Qi, G., Feng, H., Zhao, S., Zhao, S. et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. The Plant Journal, 74(2), 258–266. [Google Scholar]
38. Kim, K., Cheong, Y. H., Grant, J. J., Pandey, G. K., Luan, S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. The Plant Cell, 15(2), 411–423. [Google Scholar]
39. Meena, M. K., Vishwakarma, N. K., Tripathi, V., Chattopadhyay, D. (2019). CBL-interacting protein kinase 25 contributes to root meristem development. Journal of Experimental Botany, 70(1), 133–147. [Google Scholar]
40. Pandey, G. K., Cheong, A. Y. H., Kim, K. N., Grant, B. J. J., Li, A. L. (2004). The calcium sensor calcineurin B-Like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. The Plant Cell, 16(7), 1912–1924. [Google Scholar]
41. Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L. G. et al. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Molecular Plant, 1(2), 238–248. [Google Scholar]
42. Sanyal, S. K., Kanwar, P., Yadav, A. K., Sharma, C., Kumar, A. et al. (2017). Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Science, 254, 48–59. [Google Scholar]
43. Song, S., Feng, Q., Li, C., Li, E., Liu, Q. et al. (2018). A tonoplast-associated calcium-signaling module dampens ABA signaling during stomatal movement. Plant Physiology, 177(4), 1666–1678. [Google Scholar]
44. Tang, R., Zhao, F., Garcia, V. J., Kleist, T. J., Yang, L. et al. (2015). Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proceedings of the National Academy of Sciences, 112(10), 3134–3139. [Google Scholar]
45. Deng, J., Yang, X., Sun, W., Miao, Y., He, L. et al. (2020). The calcium sensor CBL2 and its interacting kinase CIPK6 are involved in plant sugar homeostasis via interacting with tonoplast sugar transporter TST2. Plant Physiology, 183(1), 236–249. [Google Scholar]
46. Hibrand Saint-Oyant, L., Ruttink, T., Hamama, L., Kirov, I., Lakhwani, D. et al. (2018). A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nature Plants, 4(7), 473–484. [Google Scholar]
47. Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J. et al. (2008). The Pfam protein families database. Nucleic Acids Research, 32(1), D138. [Google Scholar]
48. Kretsinger, R. H., Nakayama, S. (1993). Evolution of EF-hand calcium-modulated proteins. IV. Exon shuffling did not determine the domain compositions of EF-hand proteins. Journal of Molecular Evolution, 36(5), 477–488. [Google Scholar]
49. Nakayama, S., Moncrief, N. D., Kretsinger, R. H. (1992). Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. Journal of Molecular Evolution, 34(5), 416–448. [Google Scholar]
50. Finn, B. E., Forsén, S. (1995). The evolving model of calmodulin structure, function and activation. Structure, 3(1), 7–11. [Google Scholar]
51. Kai, W., Mingyao, L., Hadley, D., Rui, L., Glessner, J. et al. (2007). PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research, 17(11), 1665–1674. [Google Scholar]
52. Letunic, I. (2006). SMART 5: Domains in the context of genomes and networks. Nucleic Acids Research, 34(90001), D257–D260. DOI 10.1093/nar/gkj079. [Google Scholar] [CrossRef]
53. Zhu, K., Chen, F., Liu, J., Chen, X., Hewezi, T. et al. (2016). Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Scientific Reports, 6(1), 28225. DOI 10.1038/srep28225. [Google Scholar] [CrossRef]
54. Wilkins, M. R., Gasteiger, E., Bairoch., A., Sanchez, J. C., Williams., K. L. et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods in Molecular Biology, 112, 531–552. [Google Scholar]
55. Chou, K., Shen, H. (2008). Cell-PLoc: A package of web servers for predicting subcellular localization of proteins in various organisms. Nature Protocols, 3(2), 153–162. DOI 10.1038/nprot.2007.494. [Google Scholar] [CrossRef]
56. Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J. et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research, 31(13), 3497–3500. DOI 10.1093/nar/gkg500. [Google Scholar] [CrossRef]
57. Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. DOI 10.1093/molbev/msw054. [Google Scholar] [CrossRef]
58. Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A. et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948. DOI 10.1093/bioinformatics/btm404. [Google Scholar] [CrossRef]
59. Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Research, 49(W1), 293–296. DOI 10.1093/nar/gkab301. [Google Scholar] [CrossRef]
60. Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. DOI 10.1016/j.molp.2020.06.009. [Google Scholar] [CrossRef]
61. Bailey, T. L., Mikael, B., Buske, F. A., Martin, F., Grant, C. E. et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Research, 37, 202–208. DOI 10.1093/nar/gkp335. [Google Scholar] [CrossRef]
62. Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y. et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30(1), 325–327. DOI 10.1093/nar/30.1.325. [Google Scholar] [CrossRef]
63. Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J. et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40(7), e49. DOI 10.1093/nar/gkr1293. [Google Scholar] [CrossRef]
64. Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R. et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Research, 19(9), 1639–1645. DOI 10.1101/gr.092759.109. [Google Scholar] [CrossRef]
65. Peterson, G. I., Masel, J. (2009). Quantitative prediction of molecular clock and Ka/Ks at short timescales. Molecular Biology and Evolution, 26(11), 2595–2603. DOI 10.1093/molbev/msp175. [Google Scholar] [CrossRef]
66. Zhang, C., Wang, J., Long, M., Fan, C. (2013). gKaKs: The pipeline for genome-level Ka/Ks calculation. Bioinformatics, 29(5), 645–646. [Google Scholar]
67. Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K. et al. (2006). KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics, 4(4), 259–263. [Google Scholar]
68. Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., Demchak, B. (2019). Cytoscape automation: Empowering workflow-based network analysis. Genome Biology, 20(1), 185. [Google Scholar]
69. Han, Y., Wan, H., Cheng, T., Wang, J., Yang, W. et al. (2017). Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis. Scientific Reports, 7(1), 43382. [Google Scholar]
70. Gao, H., Huang, H., Lu, K., Wang, C., Liu, X. et al. (2021). OsCYP714D1 improves plant growth and salt tolerance through regulating gibberellin and ion homeostasis in transgenic poplar. Plant Physiology and Biochemistry, 168, 447–456. [Google Scholar]
71. Hu, H., Wang, Y., Tsay, Y. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. The Plant Journal, 57(2), 264–278. [Google Scholar]
72. Pandey, G. K., Kanwar, P., Singh, A., Steinhorst, L., Pandey, A. et al. (2015). Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiology, 169(1), 780–792. [Google Scholar]
73. Guo, Y., Halfter, U., Ishitani, M., Zhu, J. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. The Plant Cell, 13(6), 1383–1399. [Google Scholar]
74. Xiang, Y., Huang, Y., Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiology, 144(3), 1416–1428. [Google Scholar]
75. Filichkin, S. A., Hamilton, M., Dharmawardhana, P. D., Singh, S. K., Sullivan, C. et al. (2018). Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Frontiers in Plant Science, 9, 5. [Google Scholar]
76. Tran, L. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y. et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell, 16(9), 2481–2498. [Google Scholar]
77. Urao, T., Yamaguchi Shinozaki, K., Urao, S., Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. The Plant Cell, 5(11), 1529–1539. [Google Scholar]
78. Du, W., Yang, J., Ma, L., Su, Q., Pang, Y. (2021). Identification and characterization of abiotic stress responsive CBL-CIPK family genes in medicago. International Journal of Molecular Sciences, 22(9), 4634. [Google Scholar]
79. Wang, Q., Zhao, K., Gong, Y., Yang, Y., Yue, Y. (2022). Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). Genes, 13(5), 795. [Google Scholar]
80. Cao, Y., Li, K., Li, Y., Zhao, X., Wang, L. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biology, 9(3), 61. [Google Scholar]
81. Lai, Y., Li, H., Yamagishi, M. (2013). A review of target gene specificity of flavonoid R2R3-MYB transcription factors and a discussion of factors contributing to the target gene selectivity. Frontiers in Biology, 8(6), 577–598. [Google Scholar]
82. Monihan, S. M., Magness, C. A., Yadegari, R., Smith, S. E., Schumaker, K. S. (2016). Arabidopsis CALCINEURIN B-LIKE10 functions independently of the SOS pathway during reproductive development in saline conditions. Plant Physiology, 171(1), 369–379. [Google Scholar]
83. Zhou, L., Lan, W., Chen, B., Fang, W., Luan, S. (2015). A calcium sensor-regulated protein kinase, CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19, is required for pollen tube growth and polarity. Plant Physiology, 167(4), 1351–1360. [Google Scholar]
84. Huang, C., Ding, S., Zhang, H., Du, H., An, L. (2011). CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Science, 181(1), 57–64. [Google Scholar]
85. Huang, Y., Xu, P., Hou, B., Shen, Y. (2019). Strawberry tonoplast transporter, FaVPT1, controls phosphate accumulation and fruit quality. Plant Cell and Environment, 42(9), 2715–2729. [Google Scholar]
86. Li, J., Long, Y., Qi, G., Li, J., Xu, Z. et al. (2014). The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. The Plant Cell, 26(8), 3387–3402. [Google Scholar]
87. Ishitani, M., Liu, J., Halfter, U., Kim, C. S., Shi, W. et al. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. The Plant Cell, 12(9), 1667–1677. [Google Scholar]
88. Quintero, F., Ohta, M., Shi, H., Zhu, J., Pardo, J. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proceedings of the National Academy of Sciences, 99(13), 9061–9066. [Google Scholar]
89. Quintero, F. J., Martinez Atienza, J., Villalta, I., Jiang, X., Kim, W. Y. et al. (2011). Activation of the plasma membrane Na+/H+ antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proceedings of the National Academy of Sciences, 108(6), 2611–2616. [Google Scholar]
90. Tai, F., Yuan, Z., Li, S., Wang, Q., Liu, F. et al. (2016). ZmCIPK8, a CBL-interacting protein kinase, regulates maize response to drought stress. Plant Cell Tissue and Organ Culture, 124(3), 459–469. [Google Scholar]
Supporting Information:
Table S1: Primer sequences used for qRT-PCR analysis
Table S2: CBL and CIPK genes from Arabidopsis and rice
Table S3: Motifs in RcCBLs and RcCIPKs
Table S4: Cis-acting elements in the promoter regions of RcCBLs and RcCIPKs
Table S5: Predication of RcCBL-RcCIPK protein-protein interaction network
Table S6: Expression of RcCBLs and RcCIPKs in different organs under normal condition
Table S7: Expression of RcCBLs and RcCIPKs under various abiotic stress conditions
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools