 Open Access
Open Access
ARTICLE
De Novo Transcriptome Analysis in Hevea brasiliensis to Unveil Genes Involved in Low Temperature Stress Response
1 College of Agriculture, Ludong University, Yantai, 264025, China
2 The Engineering Research Institute of Agriculture and Forestry, Ludong University, Yantai, 264025, China
* Corresponding Author: Meixia Liang. Email:
# These authors contribute equally to this work
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(2), 559-575. https://doi.org/10.32604/phyton.2022.024266
Received 27 May 2022; Accepted 20 July 2022; Issue published 12 October 2022
Abstract
Low temperature is one of the adversities threatening the growth and development and reduces the yield of rubber trees. However, molecular mechanisms toward rubber trees in response to low temperature are largely unclear. In this study, 7,159 and 7,600 differentially expressed genes (DEGs) were identified in ‘Reyan 73397’ rubber trees. Through GO analysis, the catalytic activity was the representative of the GO term in the only DEGs at the two studied temperatures (room temperature and 4°C, respectively), while KEGG analysis showed that carbon metabolism was the most important grouping under the comparison of these two temperatures. In addition, expression of 9 members of transcription factor MYB family genes were further verified by qRT-PCR, and MYB family genes may play important roles in the regulation of rubber trees under low temperature stress. This study provided a theoretical foundation for (1) revealing the molecular mechanisms of rubber trees in response to low temperature and (2) breeding of tolerant varieties of rubber trees.Keywords
Rubber trees (Hevea brasiliensis) are important tropical economic crops, which were originated from Amazon tropical rain forest, and their latex can be used as industrial raw materials and military strategic materials [1]. The rubber tree planting area in China is located in the northern edge of the tropics, which is not regarded as a traditional rubber planting area. It often encounters cold wave and suffers from freezing damage, resulting in a decline in yield and quality. Under low temperature conditions, rubber trees will change a series of physiological and biochemical indexes, including free water content and peroxidase activity. This will help them to protect from injury [2]. Meanwhile, low temperature is also a widely existing limiting factor in tropical and subtropical crop industry areas, and tropical economic crops in China are universal in having to face low temperatures. Therefore, it is worth exploring the resistance mechanisms of rubber trees systematically under low temperature stress, which also lays a foundation for further revealing the low temperature response mechanism of tropical crops.
Low temperature is one of the major environmental factors that limiting plant growth, development, yield and geographical distribution [3]. Plants will carry out a series of physiological mechanisms, including biofilm and osmotic adjustment systems, protective enzyme systems and fatty acids, to resist low temperature adversities [4]. Notably, transcription factor families of MYBs, WRKYs and ZATs also play important roles in plant response to cold or low temperature stress. In particular, MYB family is a larger family among plant transcription factor families [5], which have been confirmed to be involved in plant tolerance to biotic stresses [6] and abiotic stresses [7,8]. In rice, OsMYB2 is involved in the response to three kinds of abiotic stress, including cold, high salt and drought [9]. The expression of OsMYB4 was induced by cold stress, and the overexpression of OsMYB4 in Arabidopsis enhanced plant tolerance to low temperature or freezing [10]. OsMYB4 can regulate the target genes of transcription factors such as MYB, bZIP, NAC, ARF and CCAAT-HAP [11]. Overexpression of GmMYB76 and GmMYB177 in soybean enhanced plant tolerance to low temperatures [12]. Interestingly, AtMYB15 was a negative regulator in cold resistance and could inhibit the expression of CBF1/DREB1 [13].
In plant tissues or organs, the accumulation of flavonoids is an important characteristic of plants in response to low temperature. Flavonoids in plants have the functions in protecting radiation damage, combining plant toxins, enhancing plant cold resistance and insect resistance. Plants have great differences in tolerance under low temperature stress, which is closely related to the content, type and structure of flavonoids in their bodies, and MYB transcription factors have been widely involved in the regulation of flavonoid metabolic pathways [14,15].
With the development of high-throughput sequencing technology, RNA-seq is a new research approach to quantify the genes related to stress response, including low temperature, and to explore stress response mechanisms and structural networks [16,17]. In recent years, transcriptome analyses have been observed in eggplant [18], wheat [19], citrus [20], pepper [21], rapeseed [22] under low temperature stress. However, transcriptome analysis of tropical woody plants under low temperature stress is essentially rare.
Notably, genes related to motallothionein, calcium-dependent protein kinase, zinc finger protein transcription factors, protein synthesis and degradation related ubiquitin, 26S proteasome, mitochondrial substrate carrier family protein, galactose-1-phosphate uridyl transferase were screened in rubber trees using cDNA array or rapid amplification of cDNA ends (RACE) technique [23,24]. In this study, transcriptome analysis was carried out in rubber trees under low temperature stress, with the intent to reveal differential expressed genes (DEGs), especially of MYB family genes, that may be involved in plant resistance to low temperature. This study provides a theoretical basis for revealing the molecular mechanisms of rubber tolerance to low temperature stress, which contributes to uncover the biological function of MYB transcription factors in tropical crops.
2.1 Plant Materials and Stress Treatment
One-year-old tissue culture seedings of ‘Reyan 73397’ rubber trees were used throughout the whole study. Seedlings were transplanted into the pot, and placed in a growth chamber with 12 h light at 25°C followed by 12 h dark at 20°C (with 60% relative humidity). For low temperature treatment, the temperature was set up at 4°C (with 60% relative humidity) for 48 h before transcriptome analysis. Leaf samples from the top layer of rubber tree seedlings were collected for transcriptome analysis and quantitative real-time PCR experiments. Same experiments were carried out with three repeats, each with thirty seedlings of equal growth status.
2.2 RNA Extraction and Transcriptome Sequencing
The total RNA (TIANGEN, Beijing) of the leaf sample was extracted by Polysaccharide Polyphenol RNA Extraction kit (Tiangen, China). RNA samples were undergone transcriptome sequencing analysis in Biomaker Biotechnology Co., Ltd. (Beijing, China). The total RNA was enriched by magnetic beads with Oligo (dT). Fragmentation buffer was added to break mRNA into short fragments. The first strand of cDNA was synthesized by Random Hexamers (six-base random primers). Then the second cDNA strand was synthesized by adding buffer, dNTPs, RNase H and DNA polymerase I, purified by QiaQuick PCR kit (Qiagen, China), eluted by EB buffer, and then repaired at the end. Poly (A) was added and sequencing linker was connected. Then the fragment was selected by gel electrophoresis, and finally PCR amplification was carried out. The library was sequenced with Illumina HiSeq 4000 [25].
2.3 Gene Function Annotation and Expression Analysis
After gene splicing, sequence comparison was carried out with protein sequences from 8 public databases (COG, GO, KEGG, KOG, Pfam, Swissport, eggNOG and Nr), and the threshold value was set as e ≤ e−10. Functional annotation was carried out by sequence similarity, and BLAST algorithm was mainly used for sequence similarity alignment. RPKM (Reads Per Kilobase of exon model per Million mapped reads) was used to count the expression of known genes. The larger RPKM, the higher the expression level [26].
2.4 Comparative Analysis of DEGs
DEGs were screened by setting threshold via FDR (false discovery rate) [27]. In the comparison of the two groups, log2 fold change ≥ 2 and FDR < 0.5 were considered as differential expression.
2.5 Isolation of MYB Family Genes
According to the annotations of 8 public databases (COG, GO, KEGG, KOG, Pfam, Swissport, eggNOG and Nr), the members containing MYB domain were obtained with different expression levels [28], and the top 9 family genes were screened out according to the expression levels from high to low. With the help of HMMER software, 9 MYB family members that containing the conserved domain of MYB family (PF00249, http://pfam.xfam.org/) were verified [29].
2.6 Bioinformatics Analysis of MYB Family Members
The online software ExPASY ProtParam (https://web.expasy.org/protparam/) was used to predict the isoelectric point and molecular weight of MYB proteins of rubber trees (Table 1) [30]. Subcellular localization was carried out using the online website (http://www.csbio.sjtu.edu.cn/) [31]. Protein tertiary structure was predicted throughout Phyre2 software (http://www.sbg.bio.ic.ac.uk/phyre2/html/). Sequence alignment used software DANMAN. Phylogenetic tree of MYB homologs from rubber tree, Arabidopsis, apple and poplar was constructed by MEGA11 software [32].
2.7 Quantitative Real-Time PCR Verification
Specific primers for MYB family genes and control gene Actin of rubber tree were designed using NCBI/Primer-BLAST on-line server. Quantitative Real-Time PCR (qRT-PCR) was carried out on 7500 Real-Time PCR System (Applied Biosystems, New York, USA), using SYBR Green fluorescence labelling kit (TaKaRa, Kyoto, Japan), to verify the accuracy of RNA-seq data [33].
3.1 Transcriptome Sequencing and Assembly of DEGs in Rubber Tree Seedlings under Low Temperature
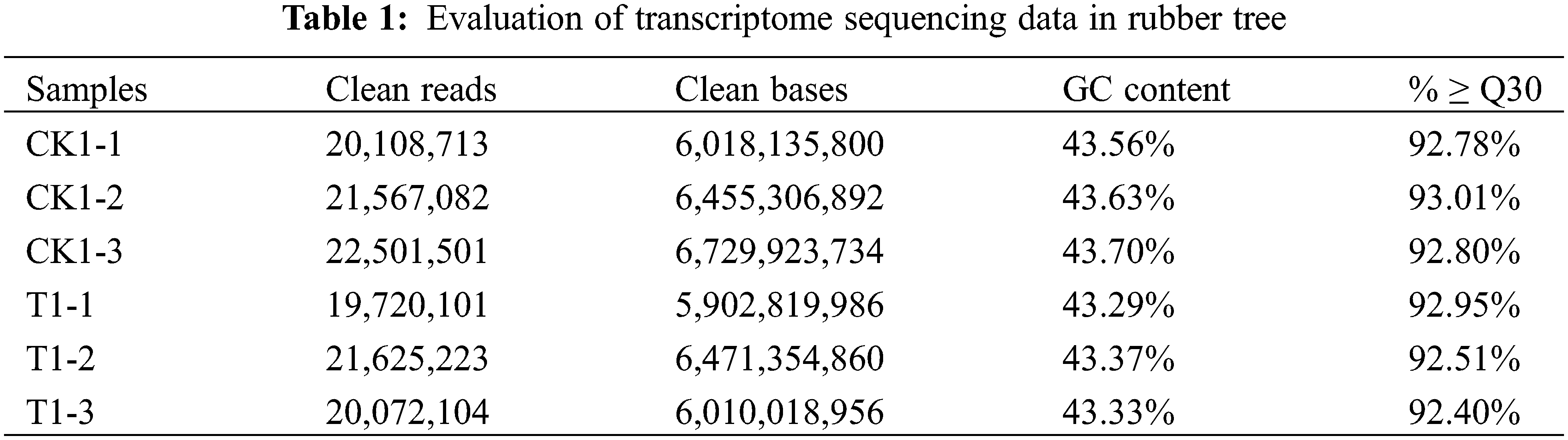
After 48 h of low temperature treatment, there was no significant growth difference between after and before low temperature treatment (Fig. 1). In order to comprehensively study the transcriptome and gene expression profiles of rubber under control condition and low temperature stress, a total of 37.59 Gb Clean Data was obtained after removing low-quality readings by sequencing quality control using Trinity program [25], and the percentage of Q30 bases in each sample was not less than 92.40% (Table 1).
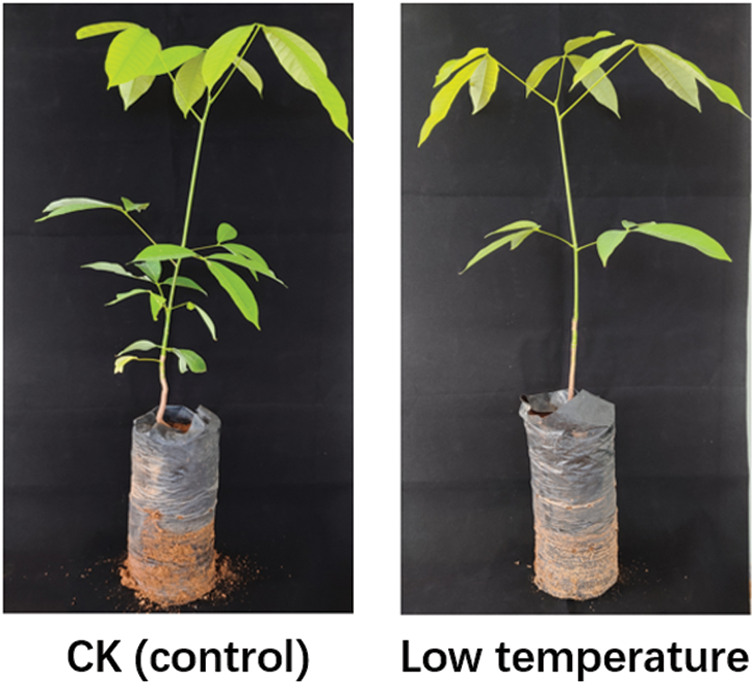
Figure 1: Phenotype of rubber trees under low temperature. 1-year-old tissue culture seedings of ‘Reyan 73397’ rubber trees were exposed to 4°C for 48 h before transcriptome sequencing
Taking the expression threshold of 1 as the screening criterion, Group 1 (control conditions) and Group 2 (low temperature stress) were used as data to draw Wayne diagram (Fig. 2), and 24369 DEGs with expressive intersection were obtained. By carrying out DESeq2 analysis, with Fold Change ≥ 2 and FDR < 0.01, a total of 14,759 DEGs were obtained, of which 7,159 were up-regulated and 7,600 were down-regulated.
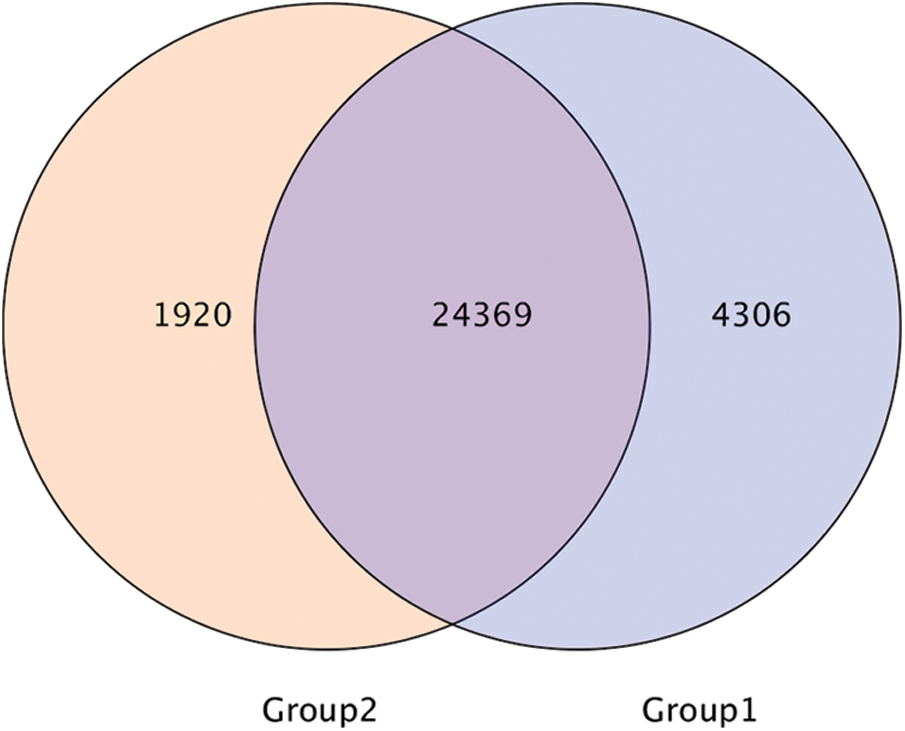
Figure 2: Wayne diagram of gene expression set in rubber trees under low temperature. Group 1, control conditions, Group 2, low temperature stress
3.2 GO Classification and KOG Classification of DEGs in Rubber Tree Seedlings under Low Temperature
In order to better tap the potential functions of DEGs, GO analysis of DEGs in rubber trees under low temperature was carried out. The possible functions of GO distribution system on rubber genes can be classified into three main categories, including cell component (CC), molecular function (MF) and biological process (BP). In total, 10,650 genes were annotated (Fig. 3). Among the three main categories, 47 processes were involved. The top three places of cell component category were cell (4,555), cell part (4,522) and membrane (4,008); the top three places of molecular function category were catalytic activity (5,870), binding (5,160) and transport activity (679), and the top three places of biological process category were metabolic process (6,214), cellular process (5,861) and single-organization process (4,289). In addition, the functions of membrane part (3,112) and organelle (2,943) were also highly representative.
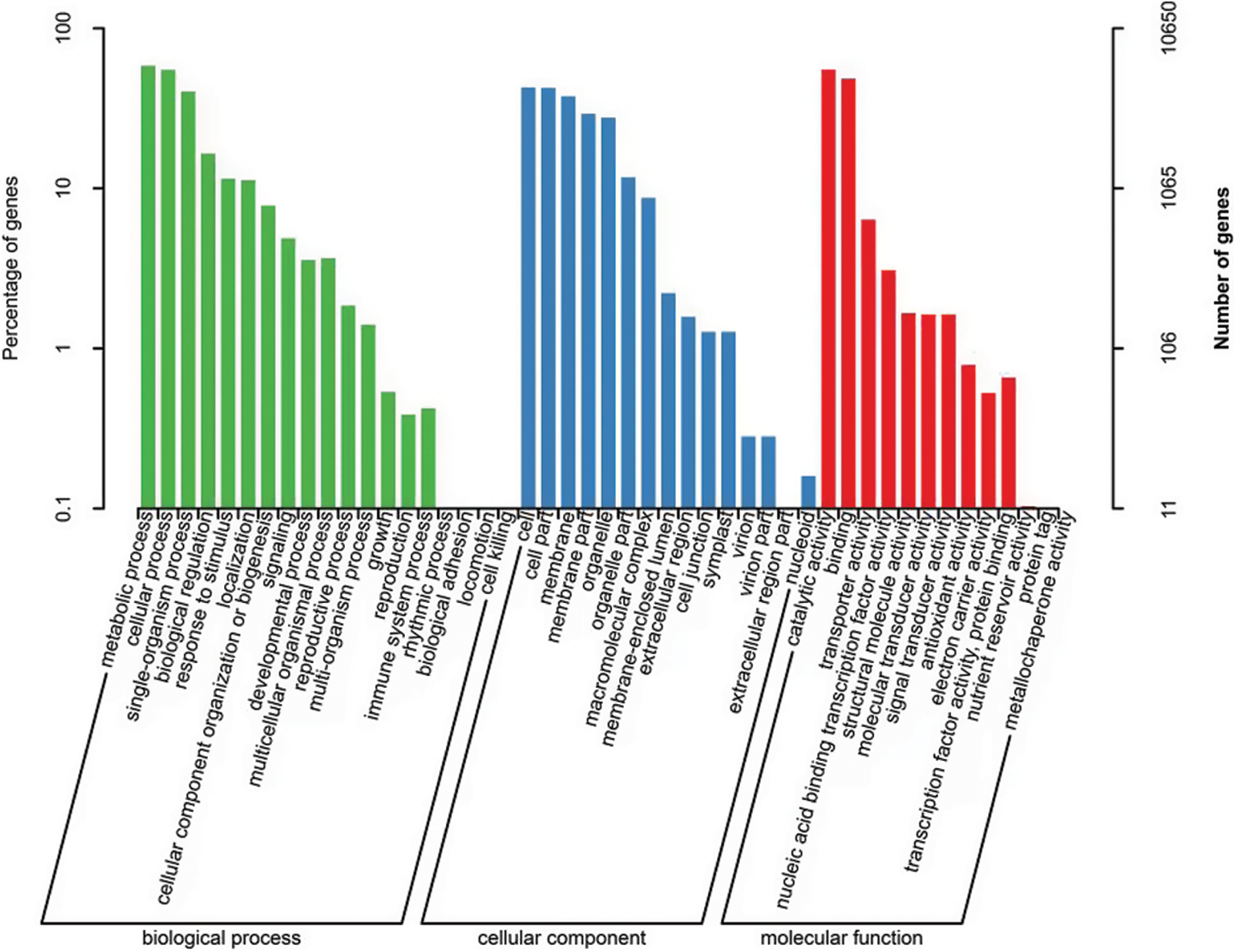
Figure 3: GO analysis of DEGs in rubber trees under low temperature
In addition, DEGs were also annotated in KOG database. A total of 9,527 Unigenes are divided into 25 functional classifications (Fig. 4). Among these classifications, the largest category is general function prediction only (1,863), followed by signal translation mechanisms (1,109), post-translational modification, protein turnover, chaperones (964), carbohydrate transport and metabolism (677).
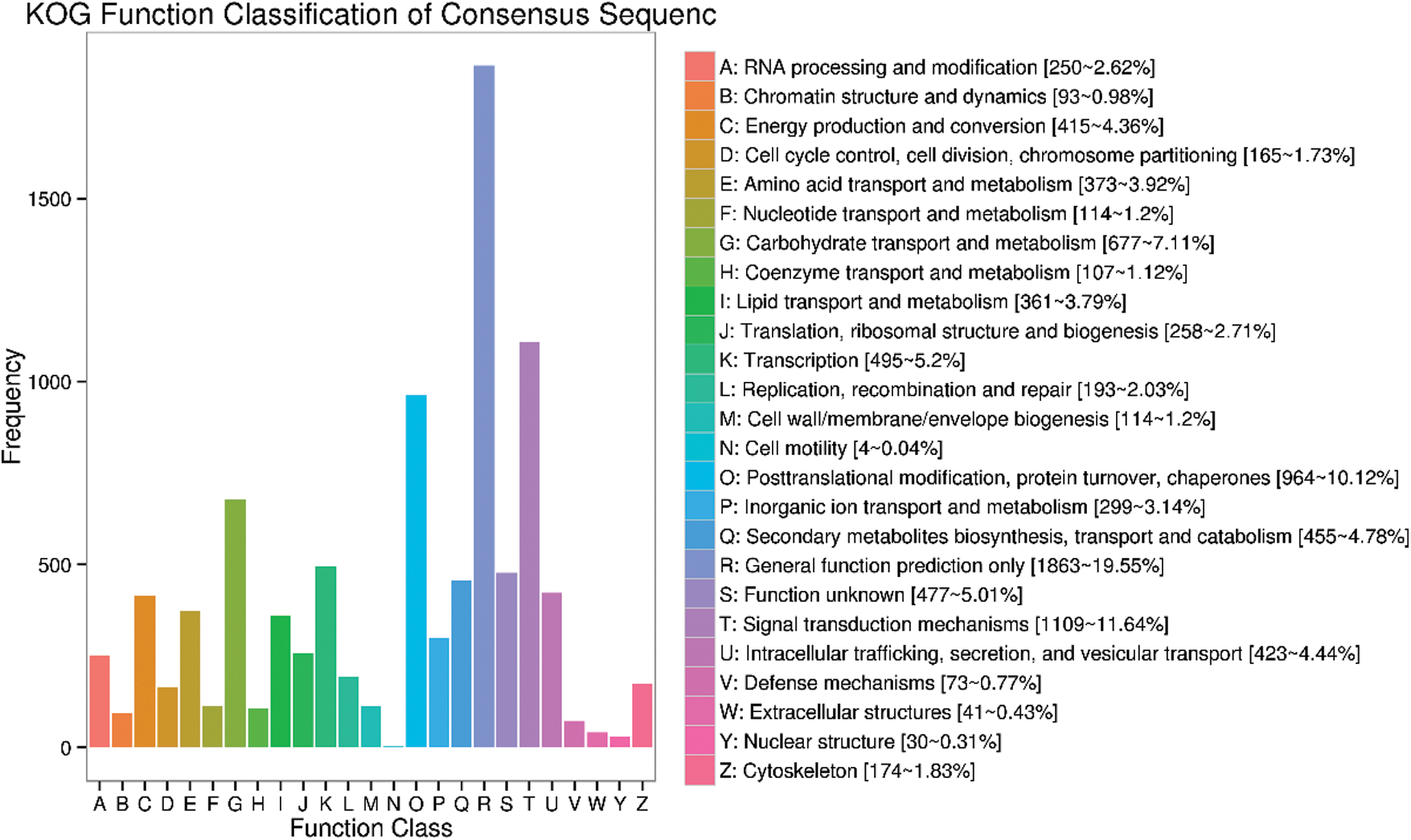
Figure 4: KOG classification of DEGs in rubber trees under low temperature
3.3 KEGG Pathway Analysis of DEGs in Rubber Tree Seedlings under Low Temperature
In order to determine the main signaling pathways and biological pathways as well as potential target genes of DEGs, we carried out KEGG pathway analysis in this study. After comparing DEGs with KEGG database, a KEGG classification map were obtained (Fig. 5). Notably, 122 metabolic pathways were enriched, in which human diseases were excluded. These pathways were divided into 5 categories, including metabolism, genetic information processing, environmental information processing, cellular processes and organic systems, among which more DEGs were donated as carbon metabolism, starch and sucrose metabolism, biosynthesis of amino acids and plant hormone signal transformation (Fig. 5).
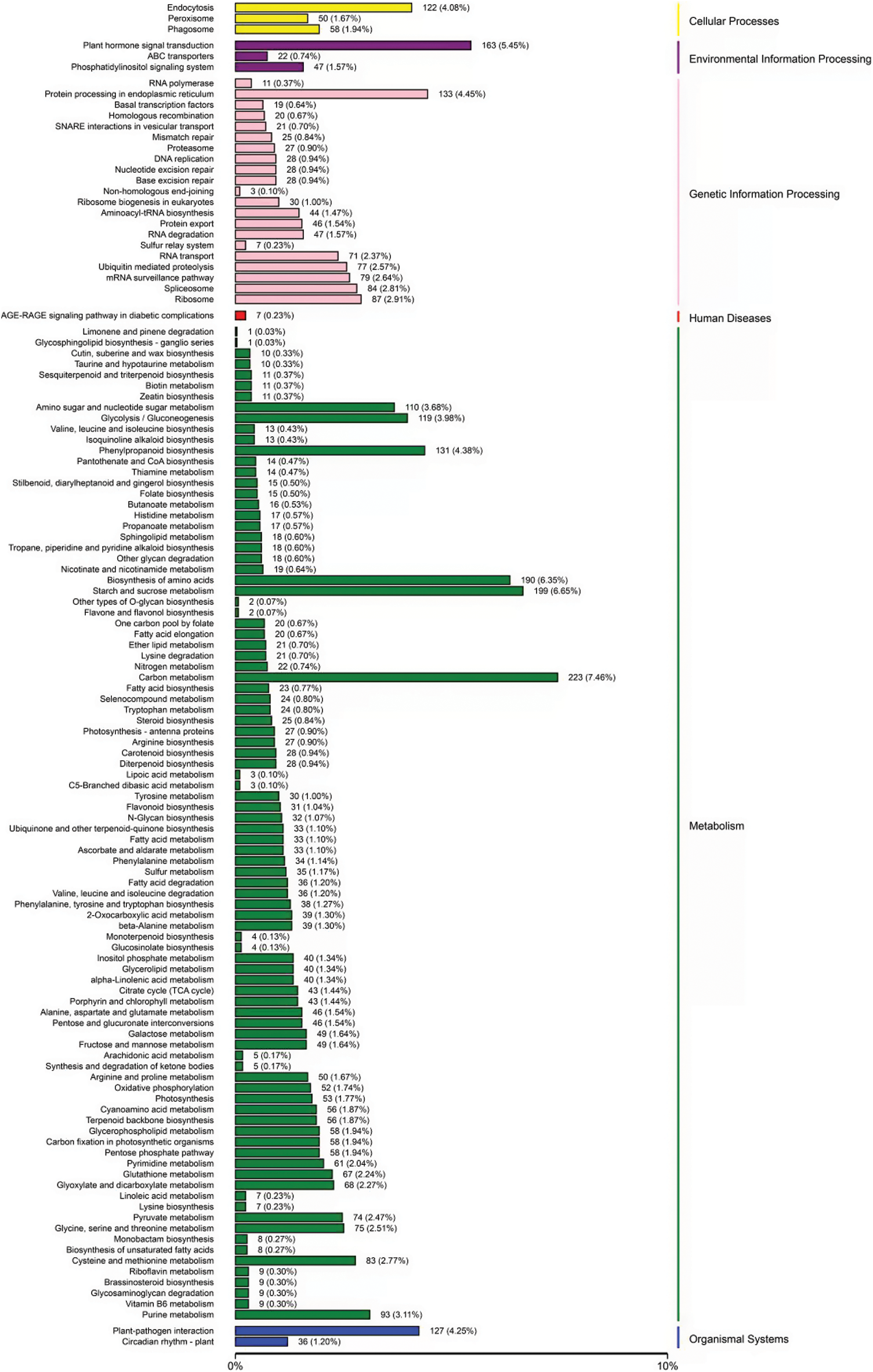
Figure 5: KEGG classification analysis of DEGs in rubber trees under low temperature
3.4 Transcription Factors of DEGs in Rubber Tree Seedlings under Low Temperature
Transcription factors play vital roles in responding to abiotic stresses. DEGs were compared with PlantTFDB database, and results showed that AP2/ERF family (76), WRKY family (73), NAC family (60), MYB family (53) and GRAS related family (46) were the major transcription factors in rubber tree under low temperature (Fig. 6).
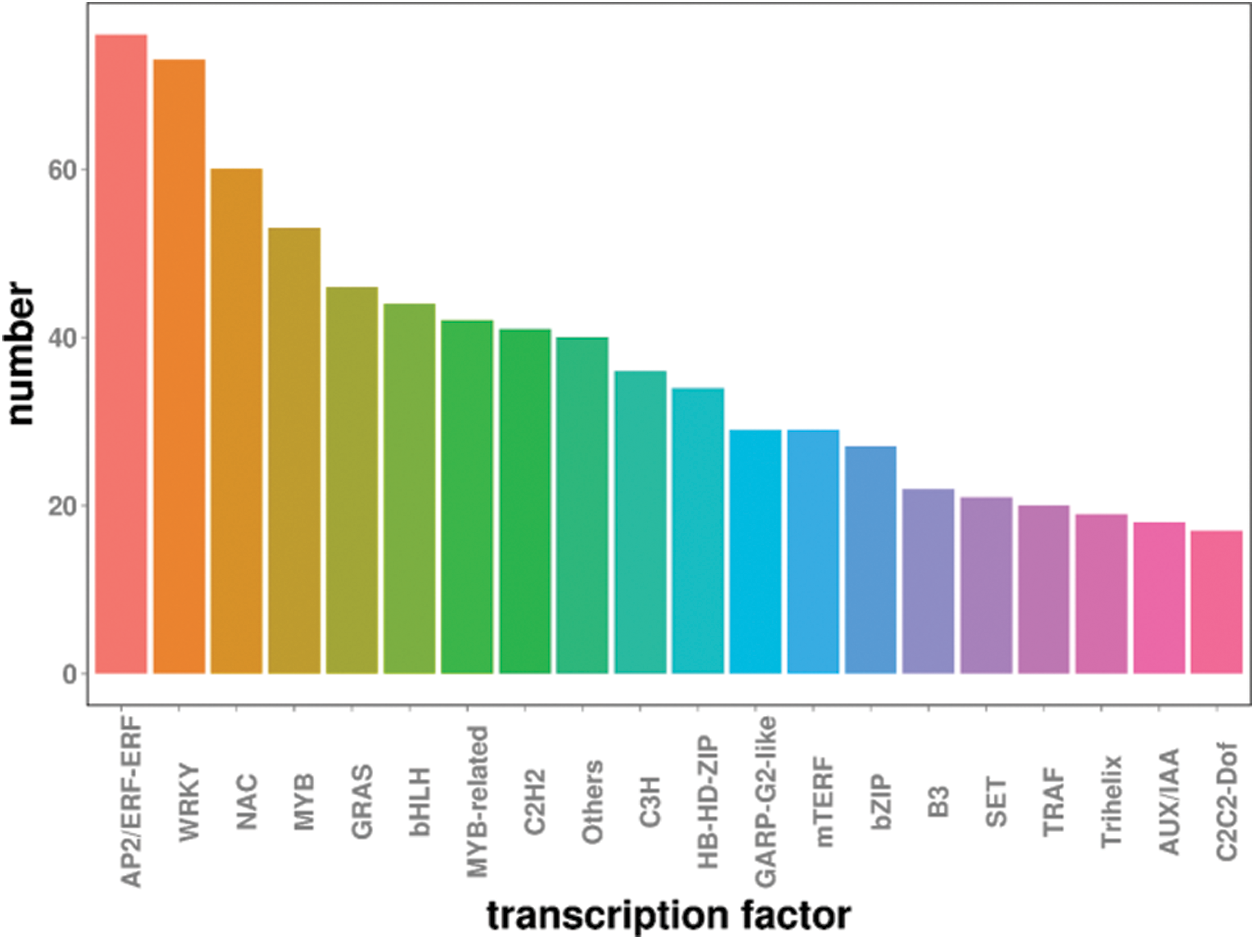
Figure 6: Transcription factors prediction of DEGs in rubber trees under low temperature
3.5 Identification of Differentially Expressed MYB Genes in Rubber Tree Seedlings under Low Temperature
3.5.1 Basic Physicochemical Properties of Highly Expressed MYB Members in Rubber Tree Seedlings under Low Temperature
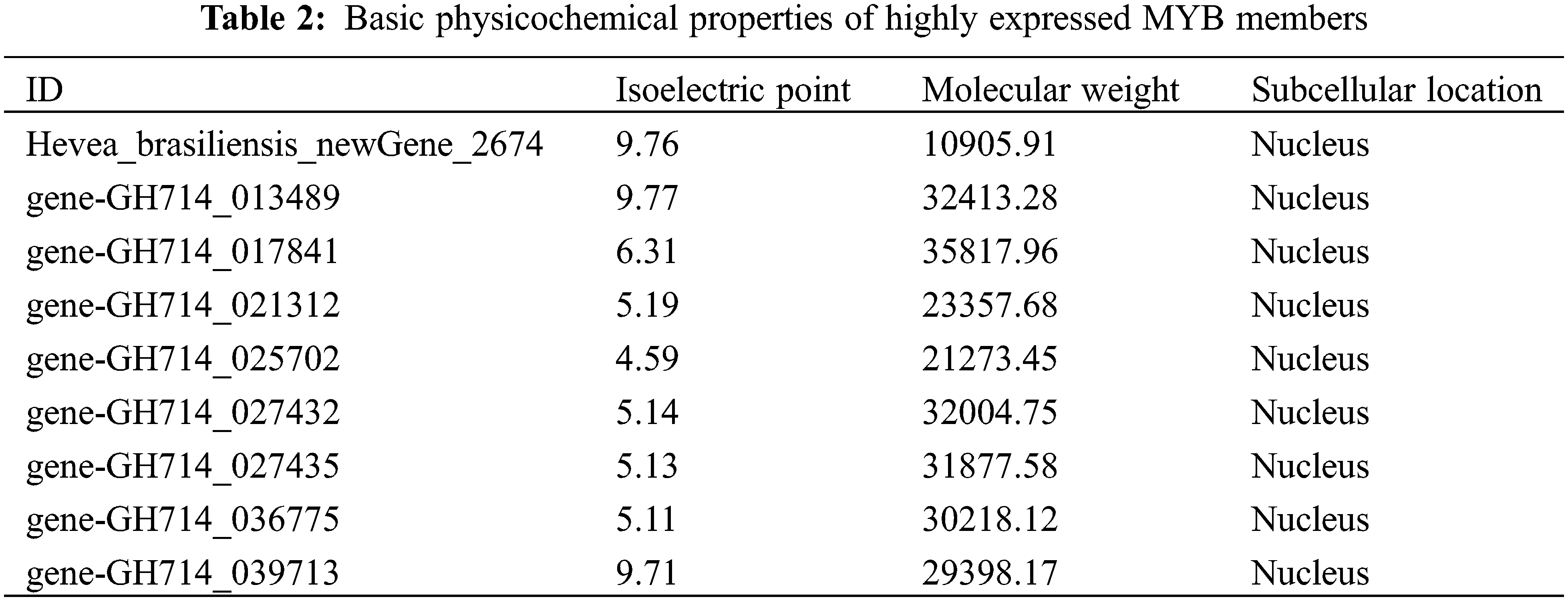
In total, there are 53 differentially expressed MYB family genes in rubber tree seedlings under low temperature, and the top 9 differential expressed MYB genes with the largest expression changes under low temperature were screened for further studies. The isoelectric points of the nine MYB family proteins were between 5.11 and 9.77, and molecular weights ranged from 10.905 to 35.817 KD (Table 2). Subcellular localization prediction indicated that the 9 MYB family genes mainly existed in nucleus, which was consistent with the characteristics of previously reported MYB family genes.
In plants, MYB family proteins can be simply divided into 3 subclasses (R1-MYB, R2R3-MYB and R1R2R3-MYB). In this study, all 9 MYB family proteins obvious structural characteristics, like target DNA recognition by R2R3-type MYB2, and there were 1~3 tandem and incomplete repetitive MYB domains (R1, R2 and R3) in MYB proteins (Fig. 7).
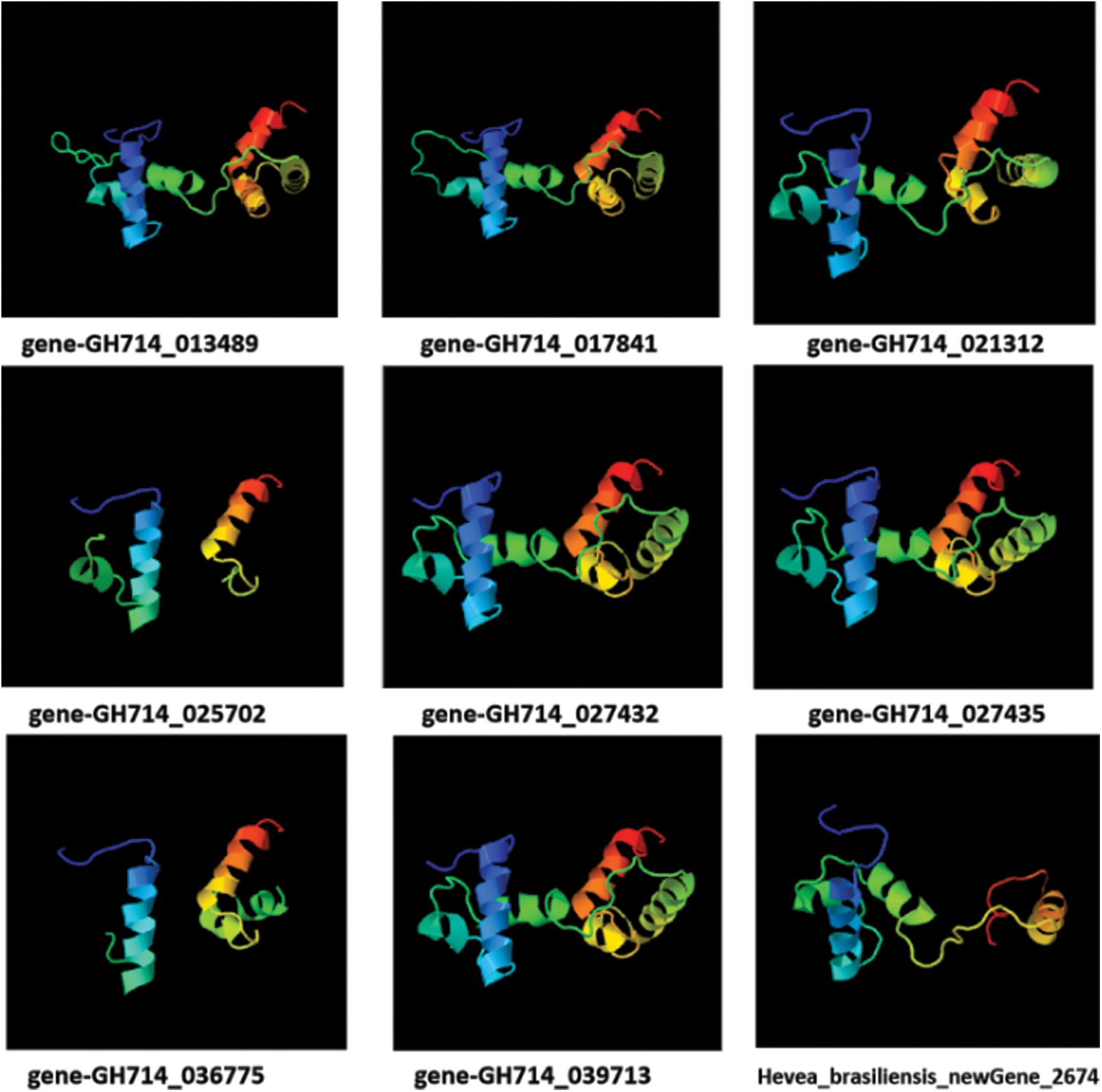
Figure 7: Tertiary structure prediction of 9 MYB family proteins
3.5.2 Phylogenetic Tree Analysis of Known Plant MYB Homologs
Multisequence alignment showed that rubber MYB proteins had high similarity with the known plant MYB homologs (Fig. 8). The phylogenetic tree of MYB homologs from rubber tree, Arabidopsis, apple and poplar was constructed (Fig. 9). As a woody plant, rubber tree MYB family proteins were closely clustered with poplar and apple homologs, but far from Arabidopsis homologs in the evolutionary relationship. In addition, MYB members within rubber trees were prone to be clustered together with each other. All MYB homologs were divided into 4 Groups, I–IV, and all these 9 rubber tree MYB proteins were concentrated in Group IV (Fig. 10).
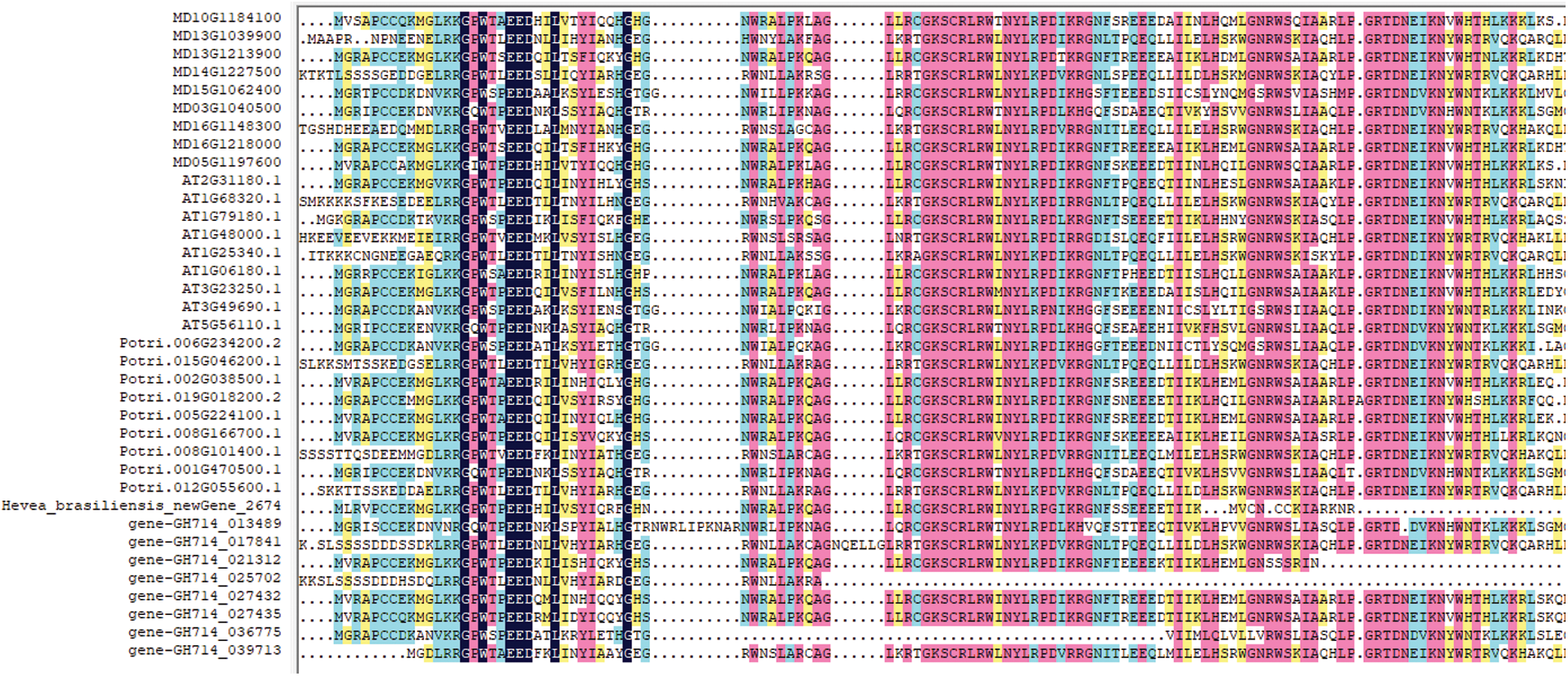
Figure 8: Multiple sequence alignment of known plant MYB homologsgure
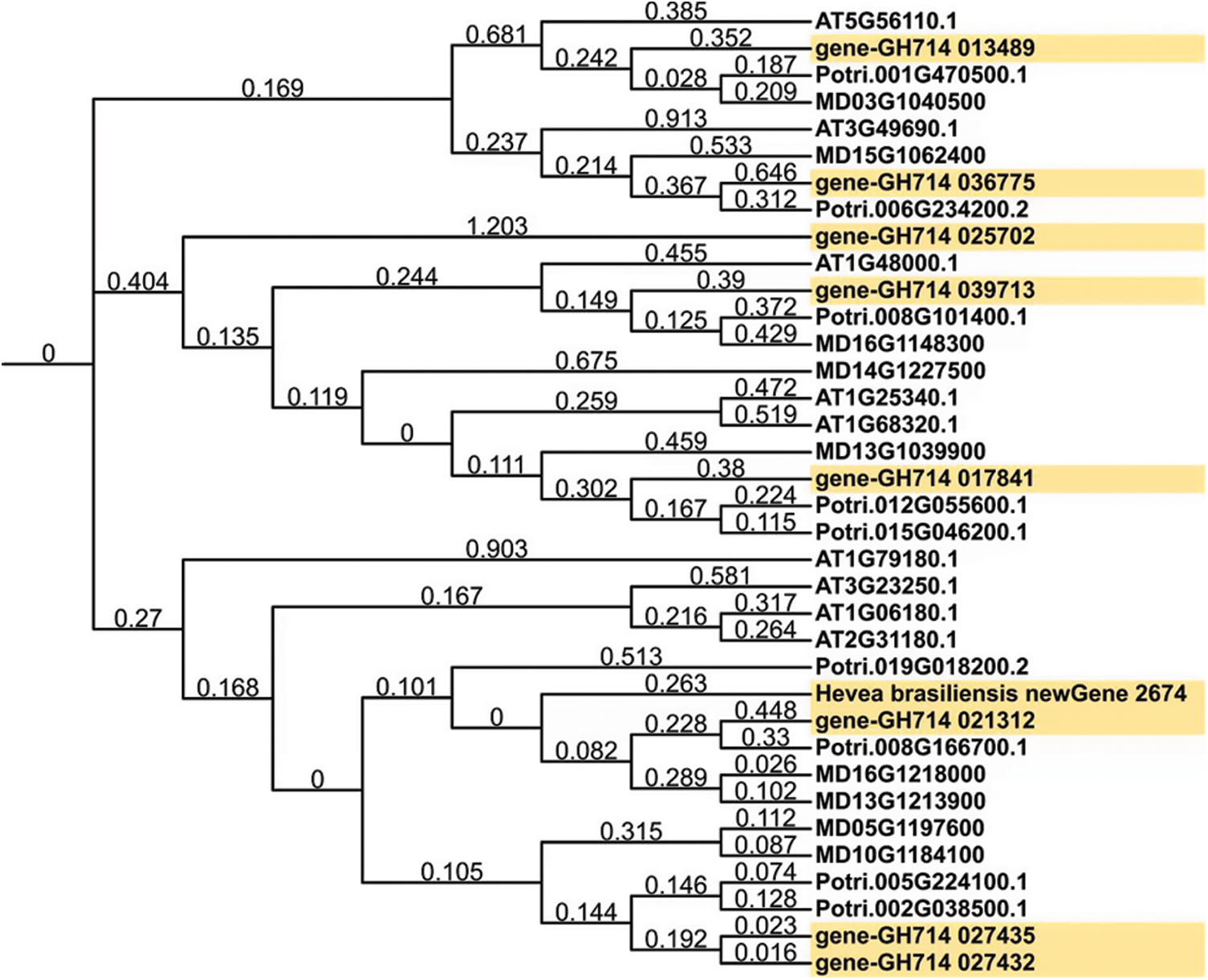
Figure 9: Phylogenetic tree of MYB family proteins of rubber tree
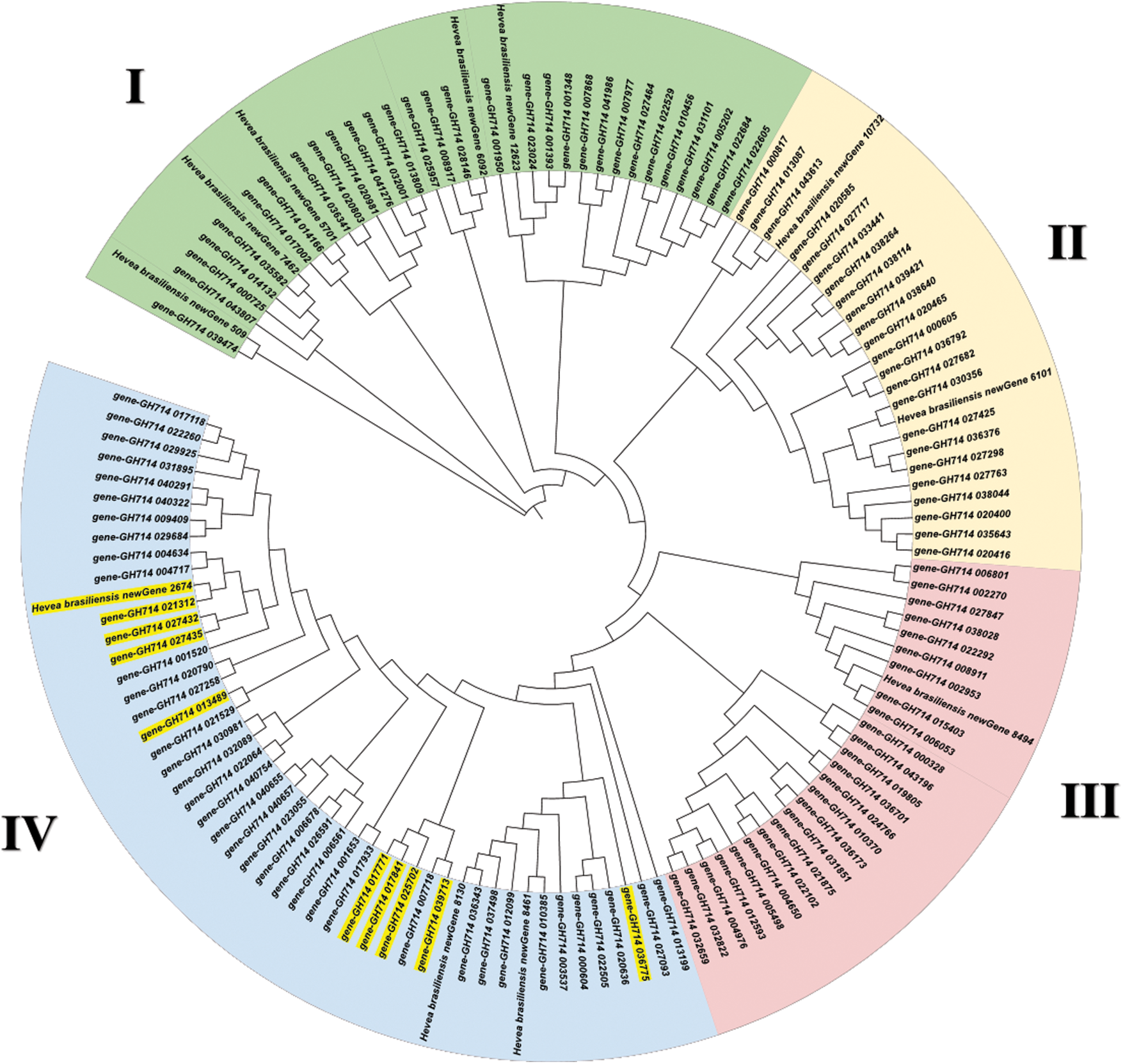
Figure 10: Phylogenetic tree analysis of known plant MYB homologs
3.5.3 qRT-PCR Verification of Differential Expressed MYB Family Genes in Rubber Tree Seedlings under Low Temperature
In order to validate the reliability of the transcriptome data, expression levels of 9 MYB family genes were analyzed by qRT-PCR. Results showed that 9 MYB genes were up-regulated under low temperature (Fig. 11), which was basically consistent with that of transcriptome data.
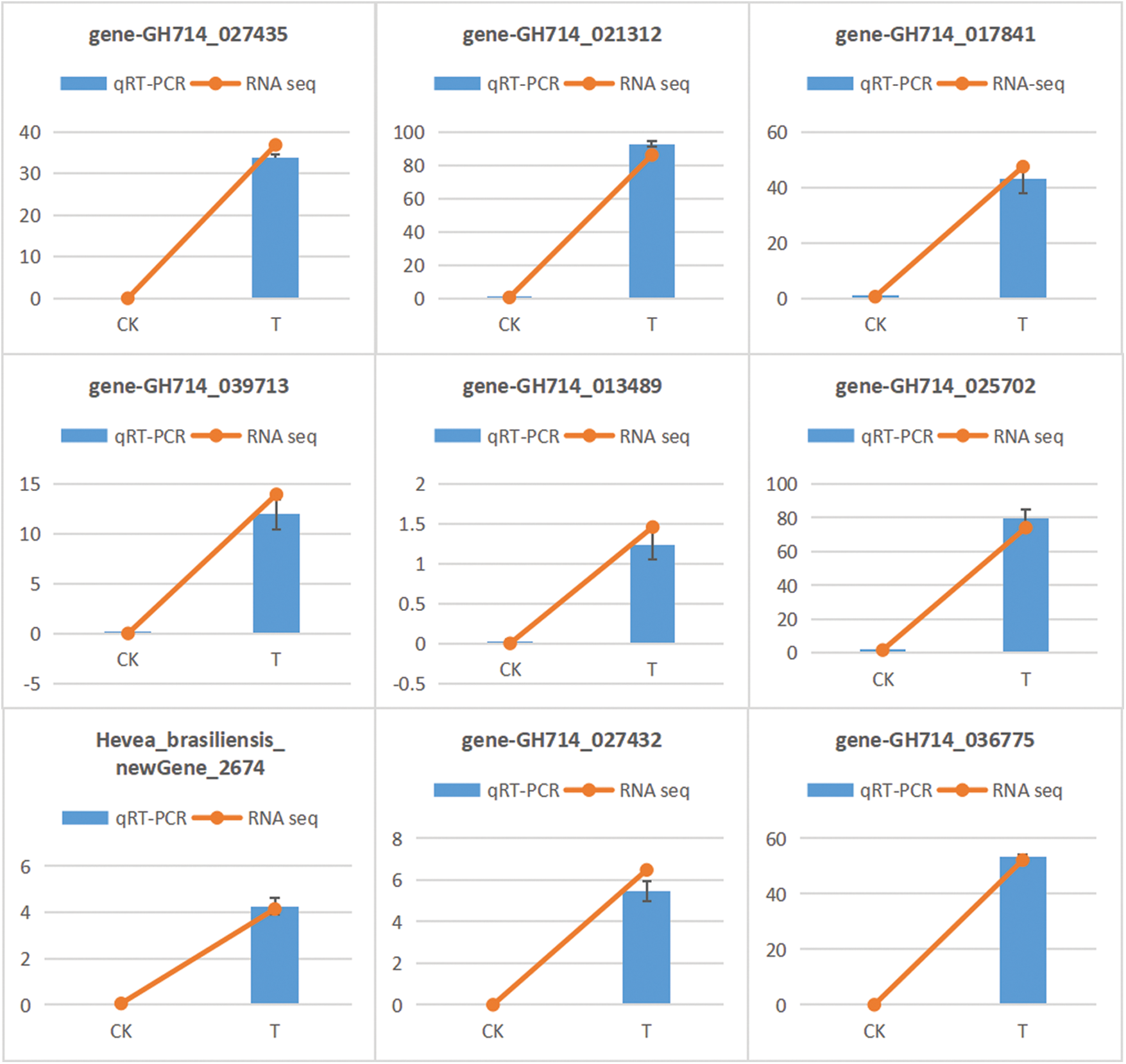
Figure 11: qRT-PCR verification of differential expressed MYB family genes in rubber trees under low temperature stress. CK, control conditions, T, low temperature treatment. 1-year-old tissue culture seedings of ‘Reyan 73397’ rubber trees were exposed to 4°C for 48 h before qRT-PCR analysis
As tropical plants, rubber trees were vulnerable to low temperature stress or cold conditions, which adversely inhibited rubber yield and even premature death. The response of plants under low temperature stress is quite complex, which requires multiple gene families and regulatory mechanisms to participate in many biological pathways, including metabolism, signal transduction, energy production and transportation, ion permeation and transportation [34]. With the development of high-throughput sequencing technology, molecular mechanisms towards rubber response under low temperature stress are being able to fulfill. Comprehensive transcriptome analysis of rubber trees contributes to reveal the molecular mechanisms of tropical plants under cold stress.
The unique DEGs are annotated with GO analysis. Under low temperature, the most abundant terms of rubber tree species are catalytic activity, metabolic process. These terms belong to biological processes and molecular functions, and the annotations of these terms are reported to be related to the low temperature response of plants. These GO conditions always affect the biochemical content, and then affect the tolerance of rubber trees. Overexpressed of CsWRKY46 in cucumber exhibited more proline accumulation and lower malondialdehyde level, which confers transgenic plants with stronger cold tolerance [35]. Notably, membrane is also a rich term. When the temperature drops, the phase change of biofilm first occurs, which may lead to the increase of membrane permeability and electrolyte leakage. The damage of cell membrane system induced by low temperature stress may be related to membrane lipid peroxidation induced by free radicals and reactive oxygen species (ROS). Malondialdehyde (MDA) is a product of membrane lipid peroxidation, and the combination of MDA with protein may cause denaturation of membrane proteins [36]. Increasing the proportion of unsaturated fatty acids in membrane is beneficial to the fluidity of membrane, thus reducing the phase transition temperature of membrane [37]. Therefore, the content of membrane fatty acids and membrane fluidity are closely related to the cold resistance of plants.
In this present study, KEGG analysis showed that carbon metabolism, starch and sucrose metabolism, amino acid biosynthesis and plant hormone signal transduction may favorably contribute to plant tolerance of rubber trees under low temperature, which need further verification. As an osmotic regulator and anti-dehydration agent, soluble sugar can reduce cell water potential and improve water retention capacity [38]. Sugar can provide carbon source and substrate to induce other related cold resistance and physiological and biochemical processes, which is helpful to improve cold resistance. Sugar can also avoid protein coagulation caused by low temperature and further improve the cold resistance of plants [39,40]. The expression of genes that involved in plant hormone formation, including ABA, ET, SA and JA, can be affected by the DEGs that involved in signal cascade mechanism. Then, these hormones may amplify cascade reactions or start some new signaling pathways [41]. In this study, KEGG analysis showed that ABA and auxin are the two largest response hormone signal transduction pathways in rubber trees, which needs a large number of DEGs. In particular, ABA can regulate stomatal closure, reduce water loss and maintain cell growth, and PLY and PP2C genes were mainly regulated by ABA signaling pathway [42]. In this current study, the expression of PLY and PP2C genes were up-regulated under cold stress (Fig. 12), which may further promote stomatal closure, reduce transpiration, and thus reduce water loss to adapt to low temperature stress. These KEGG analysis results were in line with previous studies in Arabidopsis [43] and rice [44]. Moreover, Auxin is a plant growth-promoting hormone [45]. Down-regulation of AUX1 gene may reduce the response of plants to low temperature stress. In this study, all Auxin synthesis related genes were down-regulated (Fig. 12). However, how these plant hormone-mediated signal transducers participate in the chilling injury of rubber trees still needs further studies.
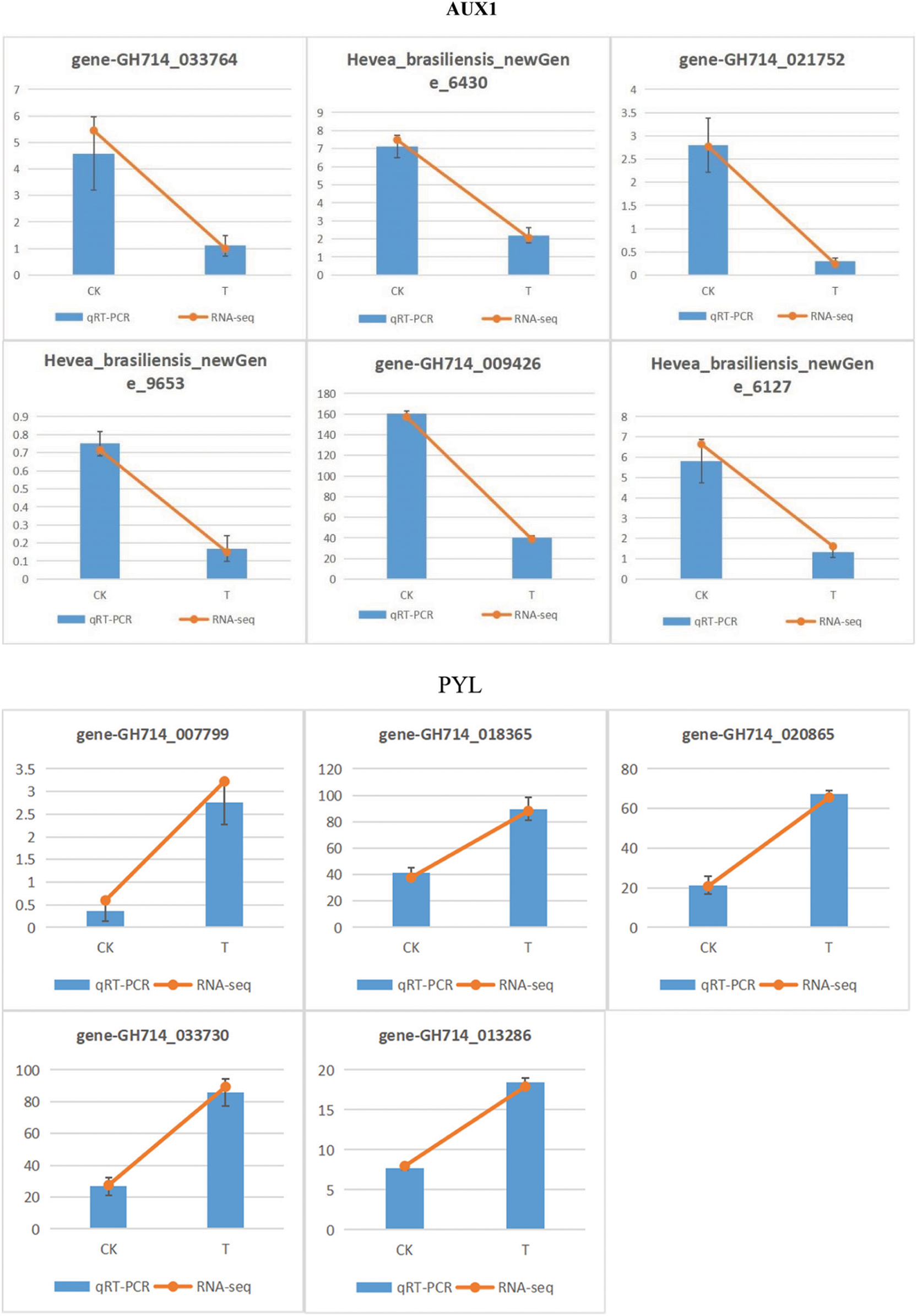
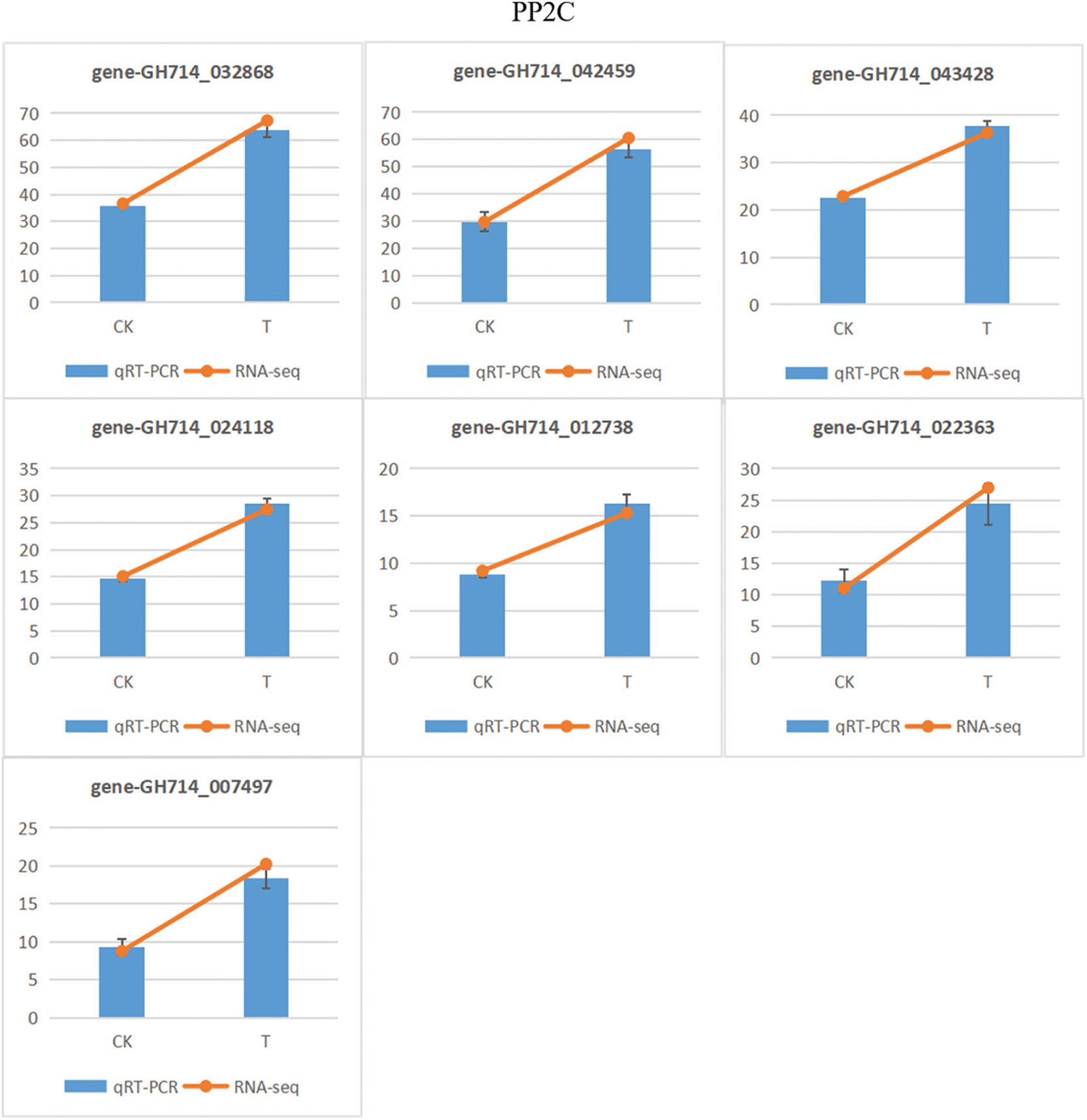
Figure 12: qRT-PCR verification of selected DEGs in rubber trees under low temperature stress. CK, control conditions, T, low temperature treatment. 1-year-old tissue culture seedings of ‘Reyan 73397’ rubber trees were exposed to 4°C for 48 h before qRT-PCR analysis
Under low temperature, transcriptome analysis indicated that transcription factors were mainly identified as AP2/ERF family, WRKY family, NAC family, MYB family and GRAS family. In this study, we focused on the highly differentially expressed MYB family genes, which were mainly up-regulated and even some non-expressed genes responded to low temperature stress. These findings were in accordance with previous studies in Arabidopsis [9], rice [10,11], and soybean [46], but contrast to that of AtMYB15 [12]. Nonetheless, this study helps to unveil resistance mechanisms of tropical crops under low temperature stress.
Transcriptome analysis revealed that 7,159 DEGs were up-regulated and 7,600 DEGs were down-regulated under low temperature treatment. GO enrichment analysis showed that DEGs were mainly enriched in metabolic process, catalytic activity and cellular process. In addition, 9 MYB family genes were up-regulated under low temperature, which was basically consistent with that of transcriptome data.
Acknowledgement: We are grateful to Prof. Zhizhong Song and Prof. Hongxia Zhang, Ludong University, for technique support and valuable suggestions.
Data Availability: The data used to support the findings of this study are included within the article.
Authorship:The authors confirm contribution to the paper as follows: study conception and design: Meixia Liang, Xiaomeng Shi; data collection: Bingsong Yang, Tingting Guo; analysis and interpretation of results: Bingsong Yang, Tingting Guo; draft manuscript preparation: Meixia Liang, Xiaomeng Shi. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This work was jointly supported by the following grants: the National Key R&D Program of China (2019YFD1000500), the National Natural Science Foundations of Shandong Province (ZR2020MC138), and the Agricultural Variety Improvement Project of Shandong Province (2020LZGC007).
Conflicts of Interest: We declare that we do not have any commercial or associative interest that represents a conflict of interest with the work submitted.
References
1. Beilen, J., Poirier, Y. (2007). Establishment of new crops for the production of natural rubber. Trends in Biotechnology, 25(11), 522–529. [Google Scholar]
2. Mai, J., Stéphane, H., Vandame, M., Kositsup, B., Kasemsap, P. et al. (2009). Effect of chilling on photosynthesis and antioxidant enzymes in Hevea brasiliensis MuellArg. Trees, 23, 863–874. [Google Scholar]
3. Pearce, R. S. (2001). Plant freezing and damage. Annal of Botany, 87, 417–424. [Google Scholar]
4. Theocharis, A., Clément, C., Barka, A. (2012). Physiological and molecular changes in plants grown at low temperatures. Planta, 235(6), 1091–1105. [Google Scholar]
5. Riechmann, J., Heard, J., Martin, G., Reuber, L., Jiang, C. et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science, 290(5499), 2105–2110. [Google Scholar]
6. Raffaele, S., Rivas, S., Roby, D. (2006). An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Letters, 580(14), 3498–3504. [Google Scholar]
7. Hoeren, F. U., Dolferus, R., Wu, Y., Peacock, W. J., Dennis, E. S. (1998). Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics, 149(2), 479–490. [Google Scholar]
8. Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T. et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biology, 15(13), 1196–2000. [Google Scholar]
9. Yang, A., Dai, X. Y., Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold and dehydration tolerance in rice. Journal of Experimental Botany, 63(7), 2541–2556. [Google Scholar]
10. Pasquali, G., Biricolti, S. (2008). Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Reports, 27(10), 1677–1686. [Google Scholar]
11. Agarwal, M., Hao, Y., Kapoor, A., Dong, C., Fujii, H. et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry, 281(49), 37636–37645. [Google Scholar]
12. Liao, Y., Zou, H. F., Wang, H. W., Zhang, W. K., Ma, B. et al. (2008). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Research, 18(10), 1047–1060. [Google Scholar]
13. Chen, Y. H., Zhang, X. B., Wu, W., Chen, Z. L., Gu, H. Y. et al. (2006). Overexpression of the wounding-responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. Journal of Integrative Plant Biology, 48(9), 1084–1095. [Google Scholar]
14. Boddu, J., Jiang, C., Sangar, V., Olson, T., Peterson, T. et al. (2006). Comparative structural and functional characterization of sorghum and maize duplications containing orthologous MYB transcription regulators of 3-deoxyflavonoid biosynthesis. Plant Molecular Biology, 60(2), 185–199. [Google Scholar]
15. Ithal, N., Reddy, A. R. (2004). Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Science, 166(6), 1505–1513. [Google Scholar]
16. Sawicki, M., Jeanson, E., Celiz, V., Clement, C., Jacquard, C. et al. (2012). Adaptation of grapevine flowers to cold involves different mechanisms depending on stress intensity. PLoS One, 7(10), e46976. [Google Scholar]
17. Taylor A. O., Rowley J. A. (1971). Plants under climatic stress I. low temperature, high light effects on photosynthesis. Plant Physiology, 47(5), 713–718. DOI 10.1104/pp.47.5.713. [Google Scholar] [CrossRef]
18. Yang Y., Liu J., Zhou X., Liu S., Zhuang Y. (2020). Transcriptomics analysis unravels the response to low temperature in sensitive and tolerant eggplants. Scientia Horticulturae, 271(20), 109468. DOI 10.1016/j.scienta.2020.109468. [Google Scholar] [CrossRef]
19. Zhao, Y., Zhou, M., Xu, K., Li, J. H., Li, S. S. et al. (2019). Integrated transcriptomics and metabolomics analyses provide insights into cold stress response in wheat. The Crop Journal, 7(6), 857–866. [Google Scholar]
20. Yun, Z., Jin, S., Ding, Y., Wang, Z., Gao, H. J. et al. (2012). Comparative transcriptomics and proteomics analysis of citrus fruit, to improve understanding of the effect of low temperature on maintaining fruit quality during lengthy post-harvest storage. Journal of Experimental Botany, 63(8), 2873–2893. [Google Scholar]
21. Wang, J., Lv, J., Liu, Z., Liu, Y., Song, S. et al. (2019). Integration of Transcriptomics and Metabolomics for pepper Capsicum annuum L. in response to heat stress. International Journal of Molecular Sciences, 20(20), 5042. [Google Scholar]
22. Raza, A., Su, W., Hussain, M. A., Mehmood, S. S., Zhang, X. et al. (2021). Integrated analysis of metabolome and transcriptome reveals insights for cold tolerance in rapeseed Brassica napus L. Frontiers in Plant Science, 12, 721681. [Google Scholar]
23. Cheng, H., Zhang, Y. Y., Cai, H. B., An, Z. W., Hu, Y. S. et al. (2016). A preliminary identification of cold resistant genes using cDNA arrays in Hevea Brasiliensis. Chinses Journal of Tropical Crops, 37(6), 1092–1097 (in Chinese). [Google Scholar]
24. Li, Y. (2011). Cloning and expressing patterns of genes related to drought-and cold-resistance from rubber tree (Hevea brasiliensis Muell. Arg.) (Master Thesis). Hainan University, China (in Chinese). [Google Scholar]
25. Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A. et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29(7), 644–652. [Google Scholar]
26. Trapnell, C., Pachter, L., Salzberg, S. L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics, 25(9), 1105–1111. [Google Scholar]
27. Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biology, 11(10), R106. DOI 10.1186/gb-2010-11-10-r106. [Google Scholar] [CrossRef]
28. Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J. H., Zhang, Z. et al. (1997). Gapped BLAST and PSI BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402. [Google Scholar]
29. Bateman, A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. et al. (2000). The Pfam protein families database. Nucleic Acids Research, 28(1), 263–266. [Google Scholar]
30. Panu, A., Manohar, J., Konstantin, A., Delphine, B., Gabor, C. et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research, 40, W597–W603. [Google Scholar]
31. Stothard, P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques, 28(6), 1102–1104. DOI 10.2144/00286ir01. [Google Scholar] [CrossRef]
32. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. et al. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology & Evolution, 28(10), 2731–2739. [Google Scholar]
33. Livak, K. J., Schmittgen, T. D. (2011). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods, 25(4), 402–408. [Google Scholar]
34. Miao, M. J., Tan, H. Q., Liang, L., Huang, H. T., Chang, W. et al. (2022). Comparative transcriptome analysis of cold-tolerant and-sensitive asparagus bean under chilling stress and recovery. PeerJ, 10, e13167. [Google Scholar]
35. Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J. S. et al. (2016). WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 59(2), 86–101. [Google Scholar]
36. Peng, X. (2007). Progress in the study of cold resistance in plant. Biotechnology Bulletin (in Chinese). DOI 10.3969/j.issn.1002-5464.2007.04.004. [Google Scholar] [CrossRef]
37. Zhou, Z., Wang, M. J., Zhao, S. T., Hu, J. J., Lu, M. Z. (2010). Changes in freezing tolerance in hybrid poplar caused by up- and down-regulation of PtFAD2 gene expression. Transgenic Research, 19, 647–654. [Google Scholar]
38. Ma, Y. Y., Xiao, X., Zhang, W. N. (2012). Research progress of plants under cold stress. Journal of Anhui Agricultural Science, 40, 7007–7008 (in Chinese). DOI 10.3969/j.issn.0517-6611.2012.12.007. [Google Scholar] [CrossRef]
39. Liu, W. Y., Liu, J. X., Han, B., Zhang, R. (2013). Determination of reduced sugar content in potatoes. Agriculture & Technology, 10(26), 33 (in Chinese). DOI 10.3969/j.issn.1671-962X.2013.10.003. [Google Scholar] [CrossRef]
40. Xu, C. X. (2012). Research progress on the mechanism of improving plant cold hardiness. Acta Ecologica Sinica, 32(24), 7966–7980. [Google Scholar]
41. Yadav, S. K. (2010). Cold stress tolerance mechanisms in plants. A review. Agronomy for Sustainable Development, 30, 515–527. [Google Scholar]
42. Peleg, Z., Blumwald, E. (2011). Hormone balance and abiotic stress tolerance in crop plants. Current Opinion in Plant Biology, 14(3), 290–295. [Google Scholar]
43. Tähtiharju, P., Tapio, S. (2010). Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant Journal, 26(4), 461–470. [Google Scholar]
44. Guan, S. G., Xu, Q., Ma, D. R., Zhang, W. Z., Xu, Z. J. et al. (2019). Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene, 685(15), 96–105. [Google Scholar]
45. Nemhauser, J. L., Hong, F., Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell, 126(3), 467–475. [Google Scholar]
46. Guan, Q., Wu, J., Zhang, Y., Jiang, C., Liu, R. et al. (2013). A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell, 25(1), 342–356. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools