 Open Access
Open Access
ARTICLE
Construction of EMS-Induced Peanut Mutant Libraries and Identification of Pod-Related Traits Mutant Lines
1 Institute of Crop Sciences, Fujian Academy of Agricultural Sciences, Fujian Research Station of Crop Gene Resource & Germplasm Enhancement, Ministry of Agriculture and Rural Affairs of People’s Republic of China, Fujian Engineering Research Center for Characteristic Upland Crops Breeding, Fujian Engineering Laboratory of Crop Molecular Breeding, Fuzhou, 350013, China
2 Cash Crops Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007, China
* Corresponding Authors: Faqian Xiong. Email: ; Guoqiang Lin. Email:
Phyton-International Journal of Experimental Botany 2023, 92(2), 537-557. https://doi.org/10.32604/phyton.2022.023912
Received 15 May 2022; Accepted 22 June 2022; Issue published 12 October 2022
Abstract
Peanut (Arachis hypogaea L.) is an oil and economic crop of vital importance, and peanut pod is the key organ influencing the yield and processing quality. Hence, the Pod-related traits (PRTs) are considered as important agronomic traits in peanut breeding. To broaden the variability of PRTs in current peanut germplasms, three elite peanut cultivars were used to construct Ethyl methane sulfonate (EMS)-induced mutant libraries in this study. The optimal EMS treatment conditions for the three peanut varieties were determined. It was found that the median lethal dose (LD50) of EMS treatment varied greatly among different genotypes. Finally, the EMS-induced peanut mutant libraries were constructed and a total of 124 mutant lines for PRTs were identified and evaluated. Furthermore, “M-8070”, one of the mutant lines for pod constriction, was re-sequenced via high-throughput sequencing technology. The genome-wide variations between “M-8070” and its wild parent “Fuhua 8” (FH 8) were detected. 2994 EMS-induced single nucleotide polymorphisms (SNPs) and 1188 insertion-deletions (InDels) between “M-8070” and its wild parent were identified. The predominant SNP mutation type was C/G to T/A transitions, while the predominant InDel mutation type was “1-bp”. We analyzed the distribution of identified mutations and annotated their functions. Most of the mutations (91.68% of the SNPs and 77.69% of the InDels) were located in the intergenic region. 72 SNPs were identified in the exonic region, leading to 27 synonymous, 43 non-synonymous and 2 stop-gain variation for gene structure. 13 Indels were identified in the exonic region, leading to 4 frame-shift, 8 non-frame-shift and 1 stop-gain variations of genes. These mutations may lead to the phenotypic variation of “M-8070”. Our study provided valuable resources for peanut improvement and functional genomic research.Keywords
Peanut (Arachis hypogaea L.), which is rich in vegetable fat, proteins, vitamins, mineral elements and other nutrients, supplies excellent raw material for oil processing industry and food productions [1]. It is one of the most significant oil and economic crops worldwide, with a global production of approximately 47 million metric tons during the year 2020 [2]. Breeding new peanut varieties with high yield production and good quality will be meaningful for consumers, peanut producers, manufacturers and all allied industries.
Pod is the most important organ in peanut from an agronomic perspective. Pod-related traits (PRTs) are also considered as important agronomic traits in peanut breeding. PRTs, such as pod weight, pod length, and pod width have been reported to directly influence peanut yield production [3]. Furthermore, some of the PRTs, such as pod length-width ratio and constriction trait co-determine pod size, shape and appearance of peanut, which are also important in in-shell consumption and processing industry [4,5]. Therefore, it is important for us to broaden the variability of PRTs in the current peanut breeding resource in order to meet various requirements.
Induced mutagenesis, which provides the possibility of generating desired phenotypes that are not present in nature, is a useful tool in plant breeding by broadening the genetic diversity in the existing varieties. It is also widely applied in plant functional genome research [6]. Ethyl methane sulfonate (EMS) is one of the most commonly used mutagens. It induces point base substitutions of guanine-cytosine (G/C) to adenine-thymine (A/T) with high efficiency [7,8]. Compared with other mutant strategies which give rises to chromosomal deletions or aberrations, EMS-induced random point mutations are less detrimental to the organism, and consequently the treated organism has a higher frequency to survive. As a result, it is easier to obtain a saturated mutant population [9]. In peanut, efforts have been made to construct the EMS-induced mutant library, and a lot of mutants with desirable mutant traits, such as high-oleate content [10–12], allergen reduction [13], oil content [14], different testa color [15], salt tolerance [16] and yield-related traits [17,18], have been identified. Some of the new peanut varieties have been developed based on the EMS mutants [19]. However, more mutants with different genetic backgrounds are still urgent needed. It is important for both peanut breeding programs and functional genomic research.
In this study, three elite peanut varieties were treated by EMS to produce new chemical mutants. The optimal EMS concentrations for mutagenesis for different peanut varieties have been investigated. A series of PRTs mutant lines were selected and identified from the mutant libraries. Furthermore, one of the mutants “M-8070”, which showed a deeper constriction phenotype than its wild type, was re-sequenced and analyzed.
Three peanut cultivars, “Fuhua 6” (FH 6), “Fuhua 8” (FH 8) and “Kanghuang 1” (KH 1), were used in this study. These three cultivars were widely cultivated in Fujian Province, South of China. “FH 6” is a peanut cultivar with high-oil; “FH 8” is a high-protein content peanut cultivar, and “KH 1” is one of the earliest released Aspergillus flavus-resistant peanut cultivars in China [20]. All the seeds used for EMS treatment were propagated by selfing for over five generations to guarantee the purity and homozygous.
2.2 EMS Treatment and Measurement of the Germination Rate under EMS Treatment
The EMS treatments consisted of 0.3%, 0.6% and 0.9% (v/v) EMS solutions (Sigma-Aldrich (Shanghai) Trading Co., Ltd., Shanghai, China) (0.1 M phosphate buffer (pH 7.0) was used as dissolvent of EMS solutions). In general, peanut seeds were soaked and shaken at low speed in the EMS solution for 8 h. The control seeds were treated by 0.1 M phosphate buffer (pH 7.0) without EMS. Then, all the treated seeds were washed via running water for 4 h. After basically drain surface moisture of seeds, the treated seeds were sowed in the field of Fuzhou (E: 119.28; N: 26.08). In order to calculate the germination rate under the EMS treatments, each treatment had three parallel replications and each replication included 300 seeds. The experiment was carried out in a complete randomized-block design. The germination rates of the treated seeds and their control were evaluated at 17 days after sowing (EMS treatment delayed the emergence of some treated seeds). Germination rate was calculated by dividing the number of germinated seeds by the total number of tested seeds, and then multiplying the number by 100 to get a percentage. The relative germination rate of different treatments in this study was calculated by dividing the germination rate of any treated group by the germination rate of the control group, and then multiplying the number by 100 to get the percentage. The LD50 values for peanut mutagenesis with different EMS concentrations in this study were estimated by linear regression analysis described in a previous study [17], based on the relative germination rate.
The mutants for PRTs were firstly detected by visual observation during the harvest. The pods from mutants were harvested and dried. The 100-pod weight (HPW), pod length, and pod width were analyzed via a Wanshen seed testing instrument, using at least 10 typical pods. The pod length/width ratio was calculated based on the data of pod length and width. The pod constriction extent was also firstly visual evaluated in the field, and then calculated by a pod constriction index (PCI) described in a previous study [5] (The width of the constricted area was measured by a Vernier caliper, and at least 5 typical pods were measured for a line).
2.4 Identification and Annotation of EMS-Induced Mutations
The young leaves from the mutant line “M-8070” (in the M4 generation) were collected for DNA extraction. The DNA library was constructed following the protocol of NEB Next Ultra II DNA Library Prep Kit for Illumina. A high-throughput sequencing of “M-8070” DNA library was carried out by Illumina NovaSeq platform with NovaSeq 6000 S4 Reagent Kit. In addition, we got the high-throughput sequencing data of “FH 8” from a previous study [21]. Then, the clean data of “M-8070” and “FH 8” were mapped to a reference genome of cultivated peanut (Tifrunner_V20190521, https://www.peanutbase.org/data/public/Arachis_hypogaea/Tifrunner.gnm2.J5K5/) using the tool of BWA [22], and the comparison results were obtained in a SAM format. “SAMtools” [23] was applied to transform the comparison results from sam to bam format. Picard was used to sort duplicates, add read group information and index the bam file (http://broadinstitute.github.io/picard/). Any possible EMS-induced mutation was detected by the “HaplotypeCaller” module of “GATK” tools software [24]. The annotation and effects of mutations on gene function were predicted by “ANNOVAR” [25]. The raw data were deposited on https://www.ncbi.nlm.nih.gov/sra/PRJNA839216.
The data obtained from different types of mutants were statistically analyzed by one-way ANOVA analysis, using SPSS 16.0 software (SPSS, Chicago, IL, USA). The analyzed data were presented as means (±SD) for at least three replications.
3.1 Determination of the Optimal EMS Concentrations for Mutagenesis in Three Peanut Varieties
To determine the optimal EMS concentration of mutagenesis for peanut in this study, three EMS solution concentrations (v/v) of 0.3%, 0.6% and 0.9% were applied to treat seeds from three peanut cultivars, “FH 6”, “FH 8” and “KH 1” for 8 h, respectively. Field germination rates of the treated seeds and their control were evaluated. It was found that the germination rate was negatively correlated with the EMS concentration. However, different genotypes showed different sensitivity to the EMS treatment. Under the treatment of 0.3% EMS concentration for 8 h, about 80% “FH 6” seeds, over 70% “FH 8” seeds and over 40% “KH 1” seeds were germinated. Under the treatment of 0.6% EMS concentration for 8 h, over 60% “FH 6” seeds, about 50% “FH 8” seeds and about 2% “KH 1” seeds were germinated. When the seeds were treated with 0.9% EMS concentration for 8 h; the germination rate was about 58.33%, 38.77% and 0% for “FH 6”, “FH 8” and “KH 1”, respectively (Fig. 1). Hence, the median lethal dose (LD50) of EMS concentration for “FH 6” was around 0.9% (0.96%), for “FH 8” was around 0.6% (0.68%), and for “KH 1” was around 0.3% (0.34%) under the 8-h treatment.
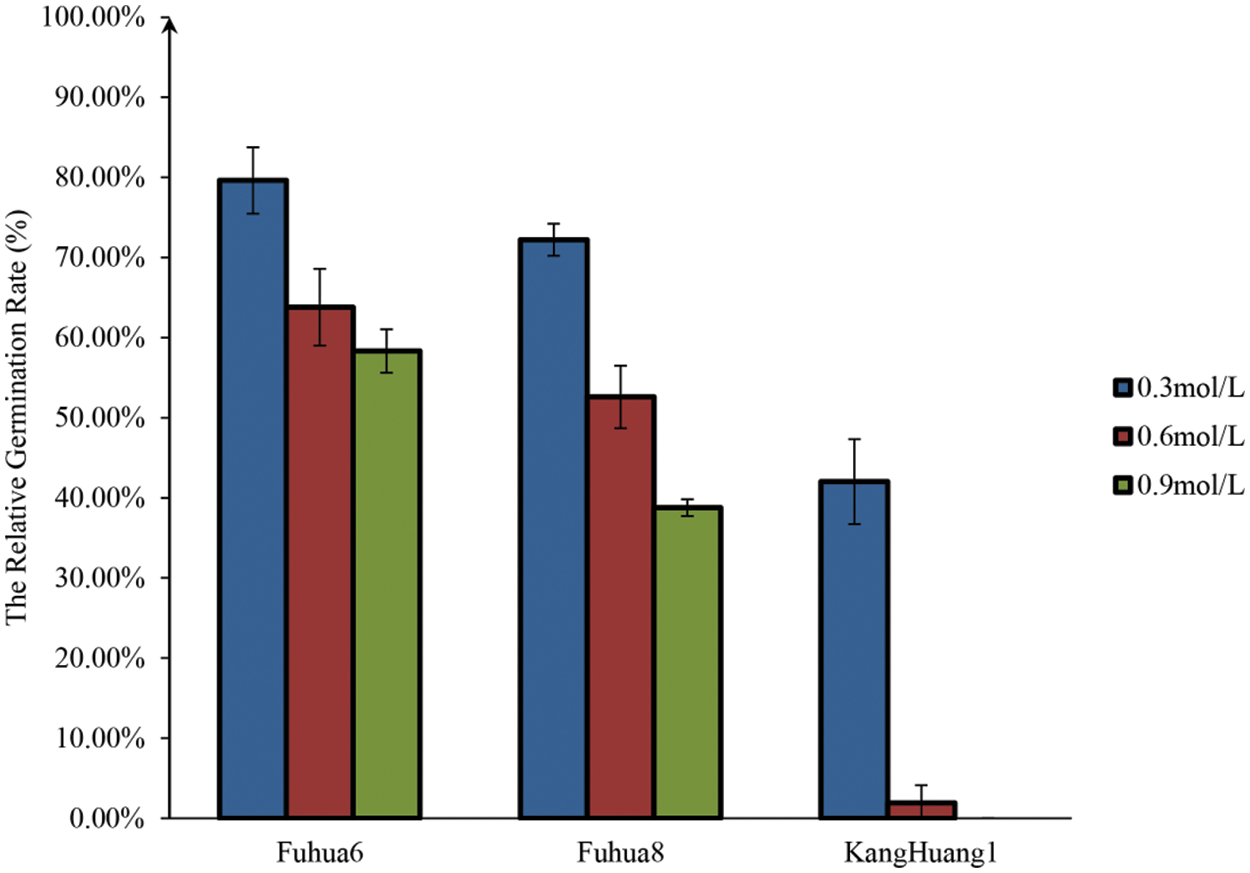
Figure 1: The relative germination rate of M0 seeds treated by different EMS concentrations. Each histogram represented the means of relative germination rate for different EMS concentration treatments for various peanut genotypes, n = 3 replications, each replication included 300 seeds. Vertical bars represented means ± standard deviations of the results of three independent replications
3.2 Development of the Mutant Peanut Libraries and Identification of Mutants for PRTs
Approximately 5000 treated seeds were germinated as M0 Plants. 3502 of these plants (1562 plants for “FH 6”, 1711 plants for “FH 8” and for 229 plants for “KH 1”, respectively) were harvested with seeds. The remaining plants did not produce seeds for various reasons, such as sterility, severe dwarfing and all kinds of diseases in the field. The seeds harvested from the M0 plants which survived were planted as M1 lines. Preliminary field observations for PRTs variation (including pod length, width, the length-width ratio and pod constriction) were conducted during the harvest period of M1 lines. The seeds of the M1 plants which showed variation in PRTs were harvested and planted as M2 lines. The pods from each of the M2 lines were evaluated to confirm the results of the M1 population. As a result, 124 mutant lines (51 lines for “FH 6”, 70 lines for “FH 8” and 3 lines for “KH 1”, respectively) with stable inheritance variation in PRTs were obtained (Fig. 2).
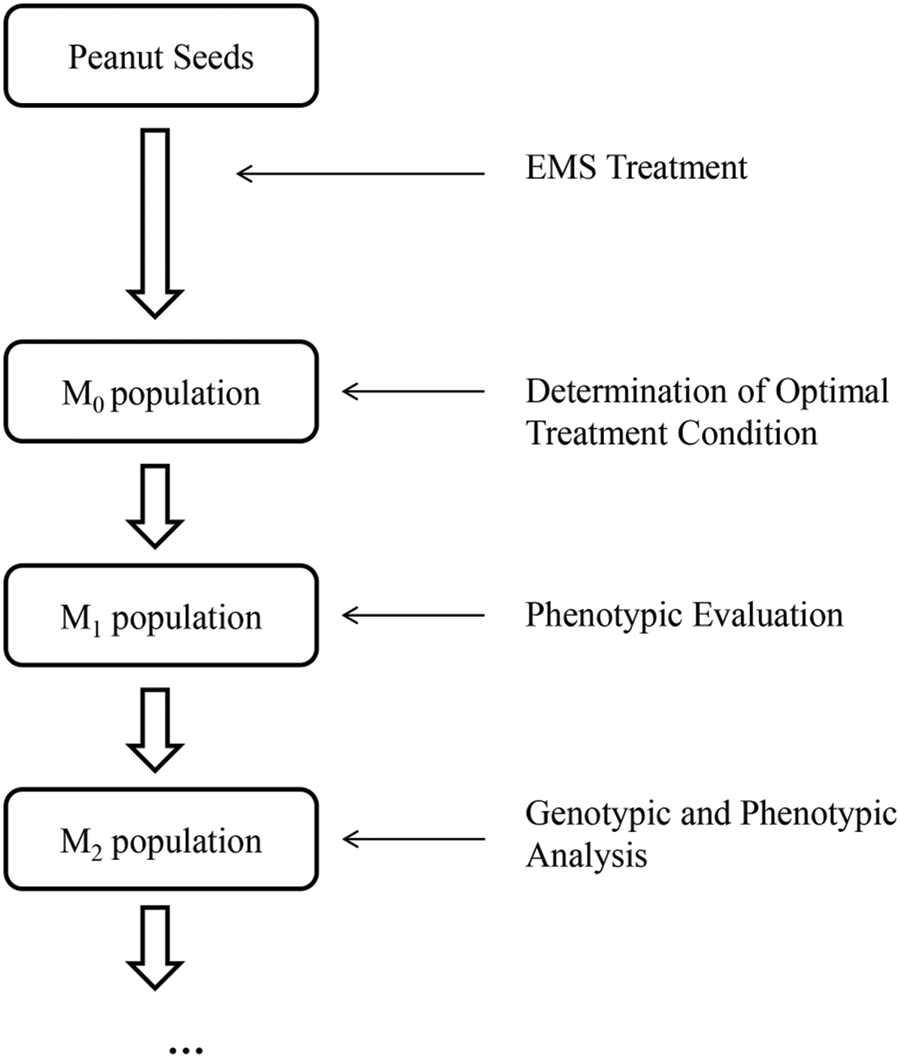
Figure 2: Development of the mutagenized peanut libraries
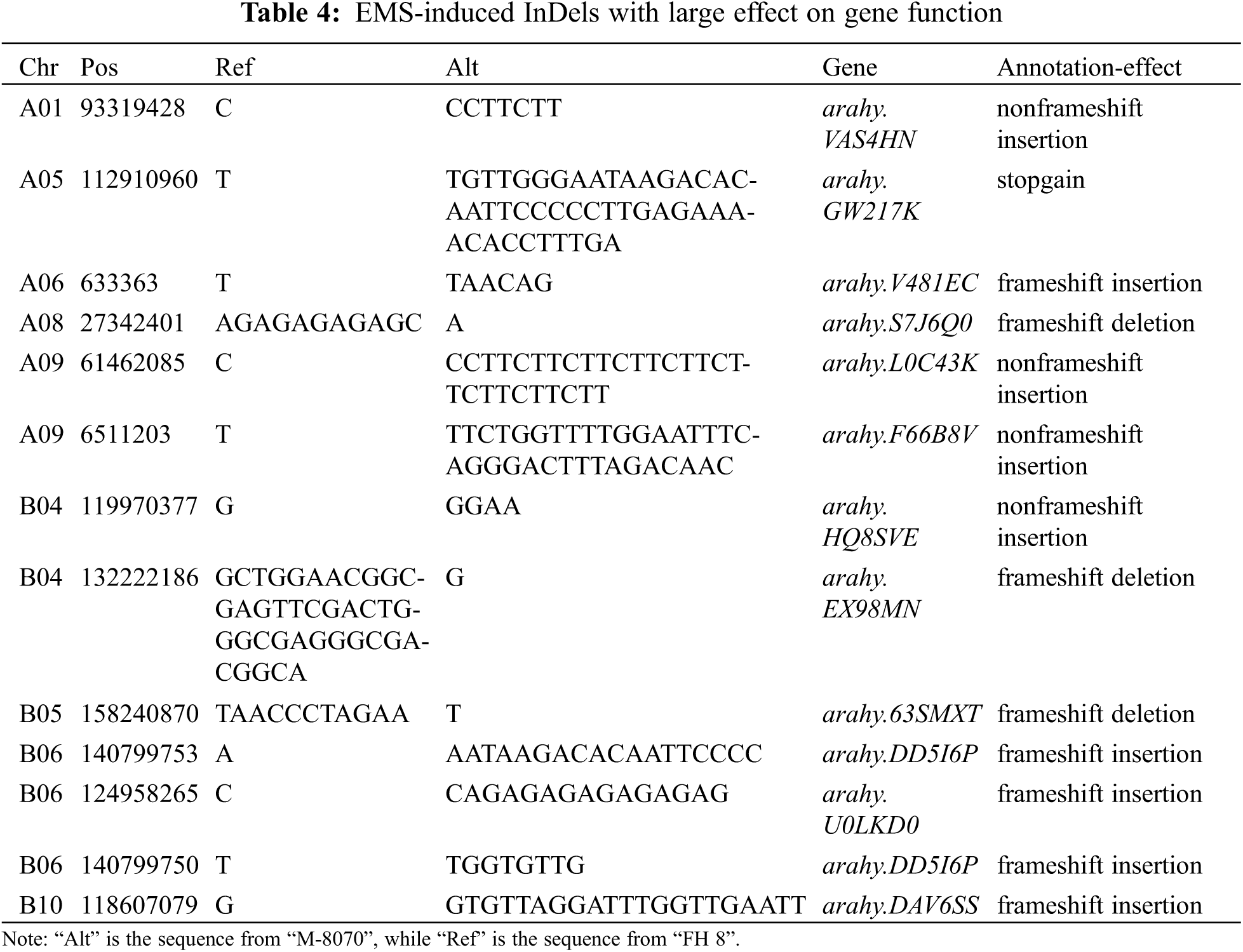
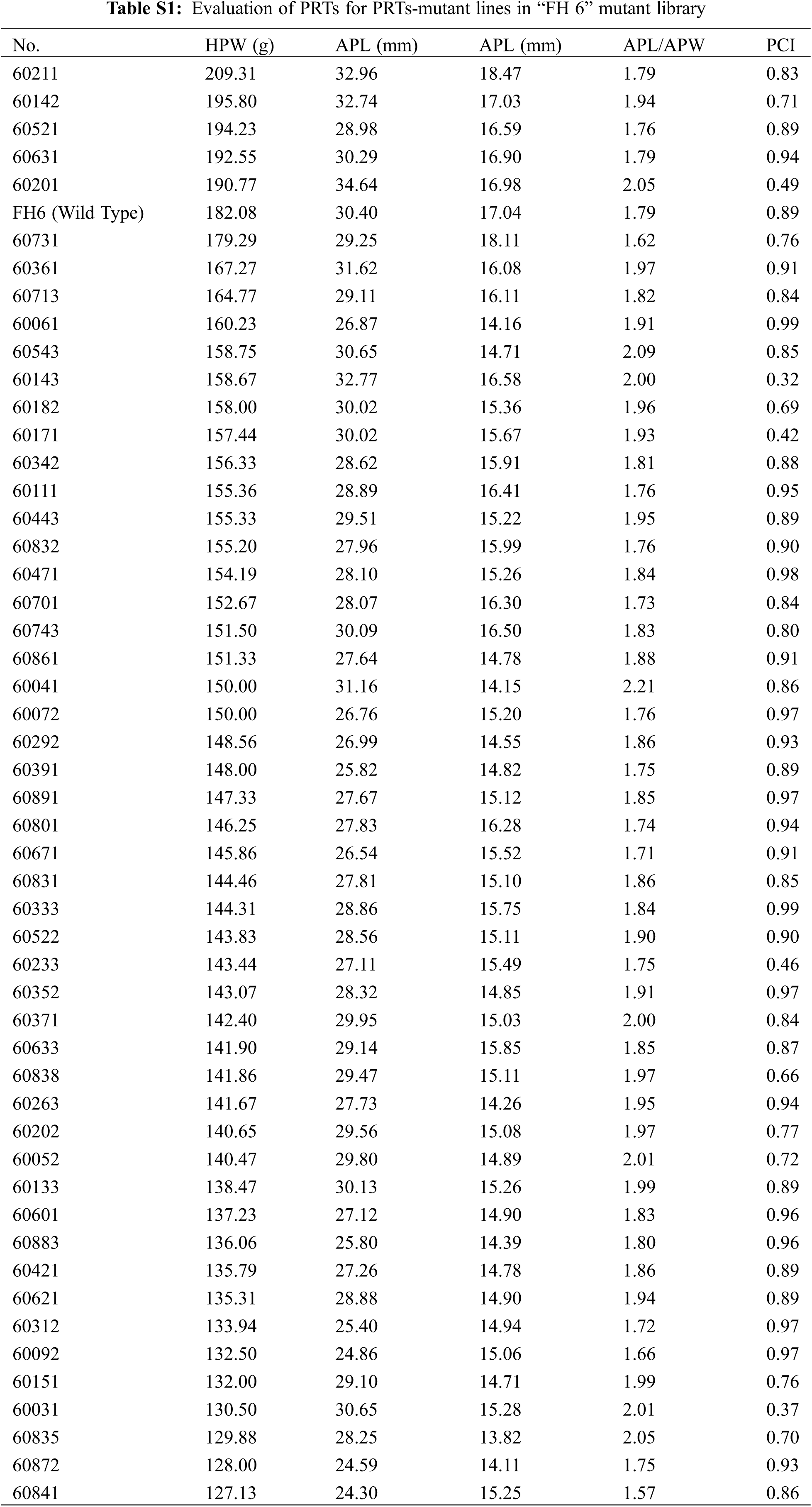
We analyzed the PRTs (including pod weight, pod length, pod width, the length-width ratio and pod constriction) variation in all identified mutants of “FH 6” and “FH 8”. Among accessions of the identified “FH 6” PRTs mutants, the one hundred pod weight ranged from 127.13 to 209.31 g, 5 lines showed heavier pods, and 46 lines showed less pod weights than their wild type. The pod length ranged from 24.30 to 34.64 mm, and 8 lines showed an increase, while the remaining 38 lines showed a decrease in pod length than “FH 6”. The pod width of the “FH 6”-based mutants ranged from 13.82 to 18.47 mm; 2 lines increased in pod width, and the remaining lines decreased in pod length than “FH 6” (Figs. 3A and 3C; Table S1). 14 lines exhibited a visual change in length-width ratio, and 5 lines varied in pod constriction extent (Table S1).
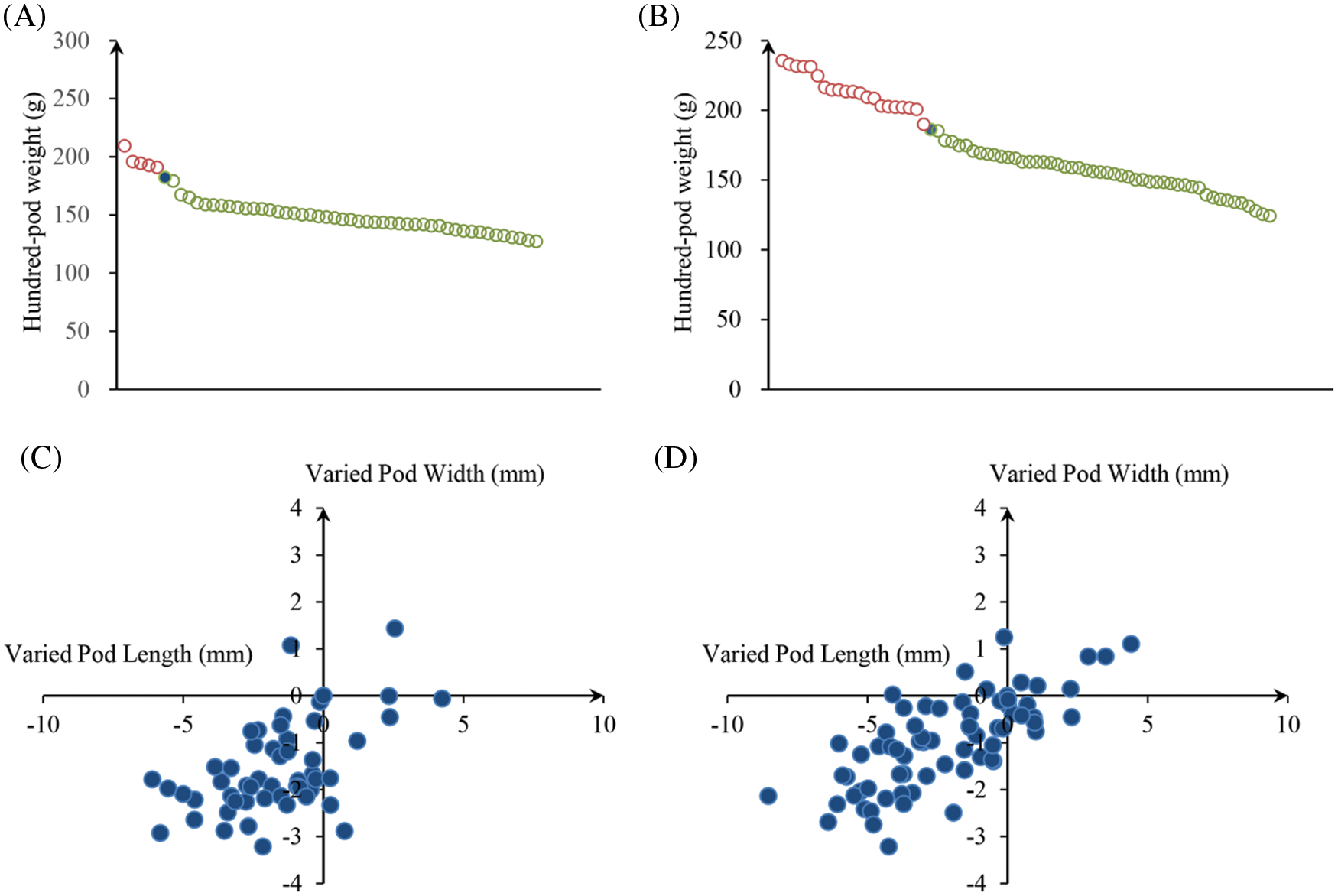
Figure 3: Phenotypic evaluation of PRTs mutants for the “FH 6” and “FH 8” mutagenesis libraries. (A) One hundred-pod weight of PRTs mutant lines from the “FH 6” mutant library. The blue spot represents the wild type parent “FH 6”; (B) One hundred-pod weight of PRTs mutant lines from the “FH 8” mutant library. The blue spot represents “FH 8” (C) Variation in pod length and pod width of PRTs mutant lines from the “FH 6” mutant library. (D) Variation in pod length and pod width of PRTs mutant lines from the “FH 8” mutant library
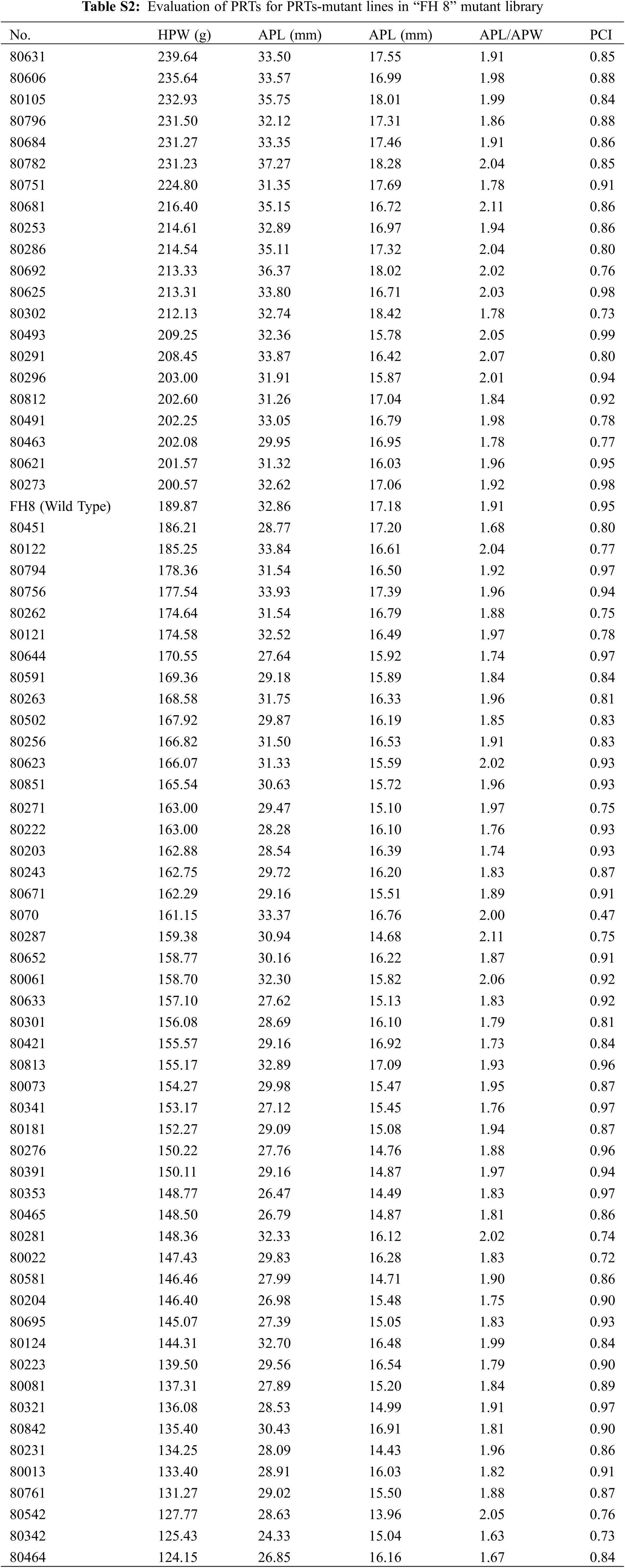
In the “FH 8”-based mutant, the one hundred pod weight ranged from 124.15 to 239.64 g, and 21 lines increased in pod weight, while 49 lines decreased in it. The pod length of “FH 8”-based mutant lines ranged from 26.47 to 36.37 mm, 16 lines increased in pod length, and 54 lines decreased in pod length. The pod width of the “FH 8”-based mutant lines rangeed from 13.96 to 18.42 mm. Among the identified lines, 12 showed and increased pod width, while 58 showed a decrease on it (Figs. 3B and 3D; Table S2). In addition, we also identified 5 lines with a varied length-width ratio mutant, and 1 line with a different pod constriction extent in this “FH 8”-based mutant library (Table S2). The information of PRTs mutants in the “KH 1” background was listed in Table S3.
3.3 Identification of a Pod-Constriction Mutant and Its Whole-Genome Sequencing
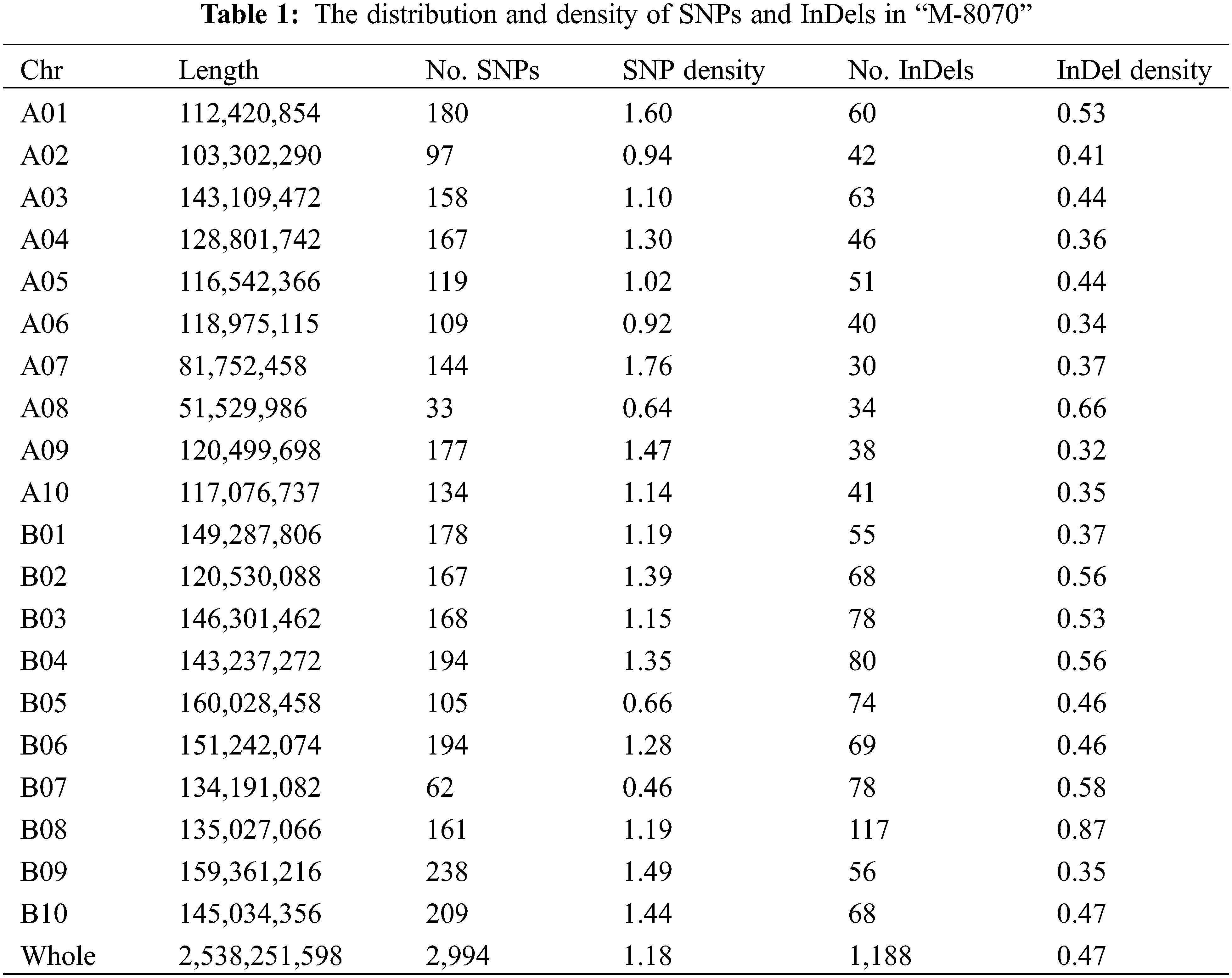
Among the identified mutant, “M-8070”, one “FH 8”-based mutant showed deeply constricted pods, while its wild type “FH 8” exhibited slightly or no constriction (Fig. 4). This mutant was re-sequenced, and a total of 33.25 Gb paired-end reads were obtained with 11.69 × average sequencing depth. The obtained clean short-read sequences were mapped to the Tifrunner reference genome, and the average genome coverage was 95.64%. Meanwhile, the “FH 8” re-sequencing data were obtained from our previous study (also used Tifrunner as a reference genome). After filtering out the common base change shared by “M-8070” and “FH 8”, a total of 4,182 loci were considered as EMS-induced sites. Among them, 2,994 were single nucleotide polymorphisms (SNPs), and 1,188 were InDels. As expected, 2,258 SNPs were G/C to A/T transitions, and they were the predominant SNP base changes (75.41%) in our mutant. This is also the typical point base substitutions induced via EMS. The frequency was followed by C/G to A/T transversions (265, 8.85%), A/T to G/C transitions (201, 6.71%), A/T to T/A transversions (165, 5.51%), C/G to G/C transversions (58, 1.94%), and A/T to C/G transversions (57, 1.90%) (Fig. 5A). Among the 1,188 identified InDels, the most frequent InDel length was 1-bp (593), it accounted for 49.92% of the total InDels (Fig. 5B). We further analyzed the distribution and density of EMS-induced sites. It demonstrated that the mutant frequency was 1.65 mutant site/Mb, and those sites were evenly distributed in the whole genome (Figs. 6 and 7, Table 1).
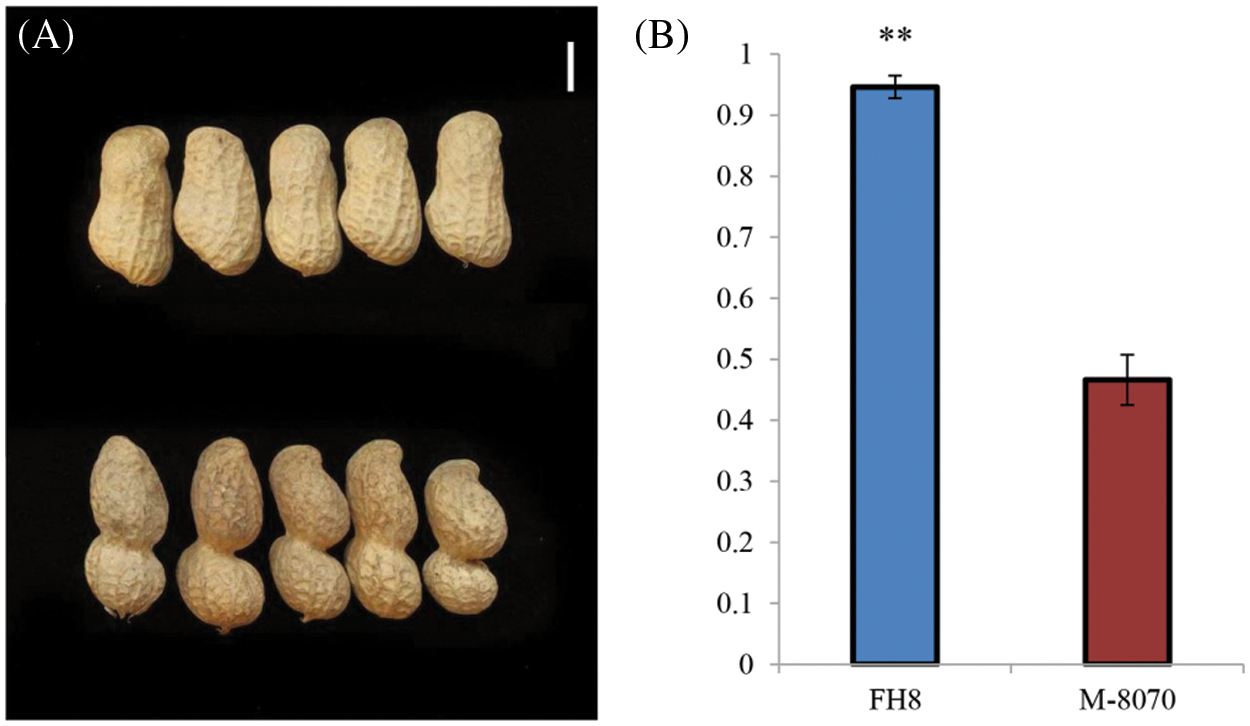
Figure 4: The phenotype of “M-8070” and its wild type parent “FH 8”. (A) Pods of “M-8070” and its wild type parent “FH 8”. The pod of “FH 8” is present in the upper line, while the pod of “M-8070” is present in the lower line. The white bar represents the length of 1 cm. (B) The PCI index of “M-8070” and “FH 8”. The error bars represent the mean ± SD of five biological replicates. The Student t-test was applied to assess the difference of means between the two varieties. “**” indicates a significant difference between “M-8070” and “FH 8” (P =< 0.01)
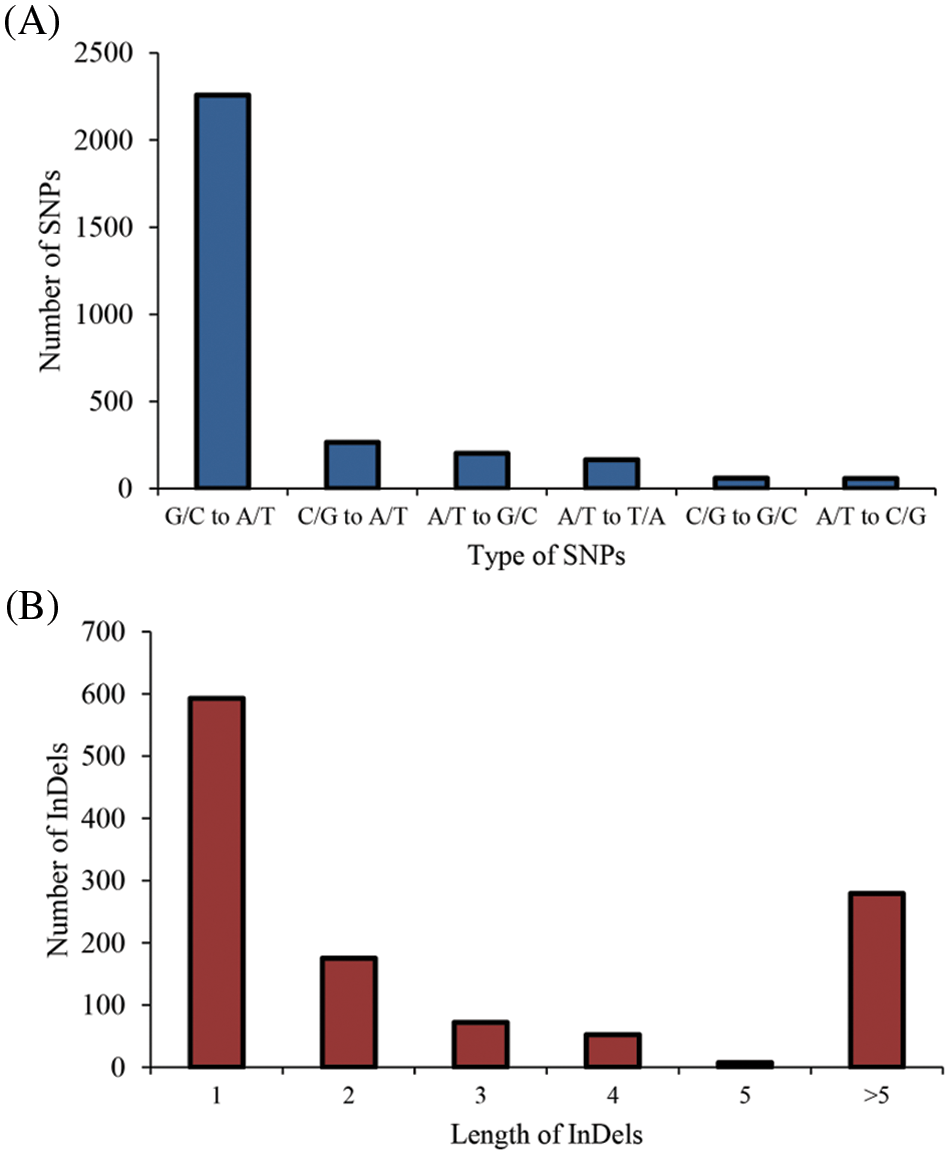
Figure 5: Type of mutations identified in “M-8070”. (A) The type of SNPs identified in “M-8070”. (B) The type of InDels identified in “M-8070”

Figure 6: The identified mutation of “M-8070” in chromosomes view

Figure 7: The distribution of identified mutation in the genome of “M-8070”
3.4 Determination of the Functional Effects of EMS-Induced Mutation
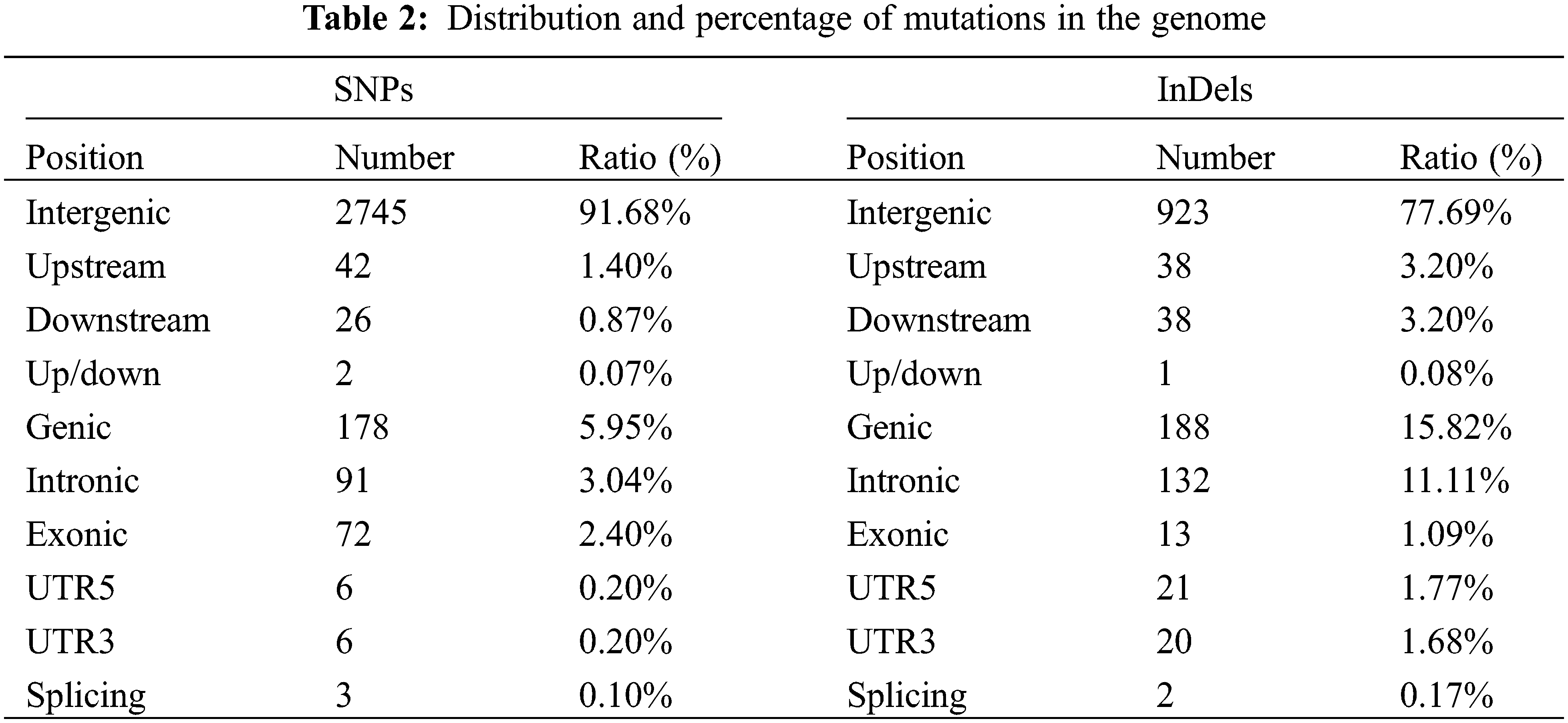
We used the software “ANNOVAR” to annotate these mutations, and predicted the effect of mutations on gene function. Most SNPs (2,745 of 2,994, 91.68%) mutations were located in the intergenic region. 70 of 2,994 SNPs (2.37%) were identified in the upstream/downstream region. There were 178 of 2,994 SNPs (5.95%) identified in the genic region. 91 of them were in the intronic region, 72 were in the exonic region, and the rest were identified in the UTR region or splicing region (Table 2). The 72 SNPs identified in the exonic region led to 27 synonymous, 43 non-synonymous and 2 stop-gain variations of genes. For InDels, most of them (923 of 1,188, 77.69%) also occurred in the intergenic region. Nevertheless, 77 InDels (6.48%) occurred in the upstream/downstream region, and 188 InDels (15.82%) were identified in the genic region (Table 2). 13 Indels occurred in the exonic region led to 4 frame-shift, 8 non-frame-shift and 1 stop gain variations of genes.
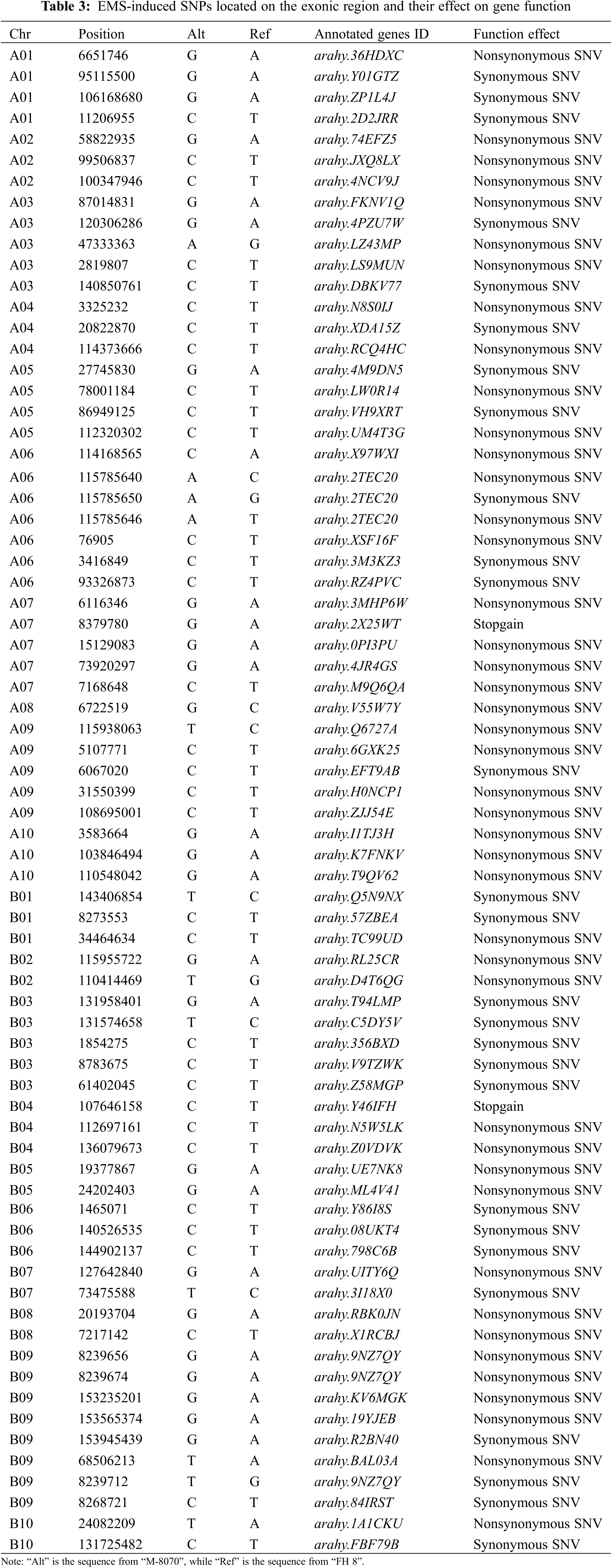
Some of the EMS-induced SNPs were located on the exonic region of genes; we collected the information of these genes and annotated them (Table 3). In addition, we further annotated genes which EMS-induced Indels mutations led to the “stop-gain”, “nonframeshift insertion”, “frameshift insertion”, or “frameshift-deletion” effect on gene structure (Table 4). Among these genes, two “stop-gain” type genes induced by SNPs, which encode “zinc finger MYM-type protein 1-like” and “probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1, 3-glucosyltransferase-like”, respectively, showed the typical point base substitutions of guanine-cytosine (G/C) to adenine-thymine (A/T) induced via EMS treatment. We speculated that they were the possible candidate genes for the deep constriction phenotype in our mutant.
In this study, we investigated the optimal EMS treatment condition for different peanut genotypes, and constructed an EMS-induced mutant library. The mutants underlying PRTs were identified. One of the PRTs mutants which exhibited deeply constricted pods was selected for re-sequencing.
According to our study, there were great differences in EMS sensitivity among peanut varieties. Although previous studies have reported that the sensitivity to EMS mutagens varied with genotype in peanut [17], such great variation in EMS sensitivity was not addressed. It is reported that a higher frequency of nucleotide variation would be induced by a higher EMS concentration treatment [26,27]. Since the multiple mutations may mask the possible varied phenotype induced by EMS treatment, and the extraordinary high frequency mutations may lead to a higher possibility for lethal phenotypes in the mutant library [28], an optimal EMS treatment concentration should be determined [29]. The LD50 value is regarded as the evaluating indicator for the EMS treatment. Hence, it is worthwhile to evaluate the LD50 value for different peanut genotypes before large scale application of EMS inducement for the new genotypes.
Pod-related traits (PRTs) are important agronomic traits not only influencing peanut yield, but also affecting pod appearance and processing quality. Although many QTLs underlying PRTs were identified through natural populations [30–35], more information was still needed for elucidating the genetic mechanism underlying these traits. EMS-induced mutagenesis is a suitable choice to supplement the study in the natural population. On the one hand, mutants may create haplotypes not existing in the natural environment. On the other hand, fewer variations in the genome and typical point mutations in the EMS-induced mutant make it easier to find the candidate gene than using the natural materials. In this study, we also produced a variety of peanut PRTs mutants via this method. They exhibited a varying degree of variations in PRTs, and they will be good materials for further peanut breeding programs and functional genomics research.
Whole-genome sequencing is a powerful tool for surveying the mutations induced by EMS mutagens [36]. It has been successfully applied in several crop species. However, few reports were listed about the whole-genome sequencing for EMS induced mutants in peanut. In our study, whole-genome sequencing was applied to survey the mutations in one of our identified mutants. A total of 2,994 SNPs and 1,188 InDels have been identified and characterized. The mutation frequency in our study was 1.65 mutant site/Mb, and this was similar to that of in other crop species, such as Arabidopsis (11.24 mutant site/Mb) [37], foxtail millet (4.69 mutant site/Mb) [28], rice (3.77 mutant site/Mb) [38], maize (2.06 mutant site/Mb) [39], sorghum (1.90 mutant site/Mb) [40], and tomato (1.36 mutant site/Mb) [41]. It indicates that our EMS mutagens may induce proper density of mutation in the genome. On one hand, it can induce enough mutations to create variation in phenotypes. On the other hand, a moderate frequency of mutations can minimize the possible masked phenotypes resulted by multiple mutations. Based on our result, the majority of mutations were C/G to T/A transitions in our mutant. It is consistent with the previous studies, and fit the working mechanism of EMS alkylating agent. However, a series of non-C/G to T/A transitions have also been detected in our mutant. This condition has also been reported in other studies [27,36,42]. What is certain is that some of the non-C/G to T/A transitions may be the false positive result during re-sequencing and data analysis. However, the mechanism on how other non-C/G to T/A transitions are induced is still unclear to us. Nevertheless, in the traditional reverse-genetic study, non-C/G to T/A type mutations were filtered out, and only C/G to T/A type mutations were considered to be the reason for variation [36,43]. In this study, after filtering out the non-C/G to T/A type mutations, 2,248 C/G to T/A-type mutations remained. 2,057 of the 2,248 SNPs were identified in the intergenic region, 54 were in the upstream/downstream of gene, and 137 were identified in the genic region. Among the 137 SNPs identified in the genic region, 68 are identified in the intronic region, 11 were identified in the UTR region or splicing region and the remaining 58 were identified in the exonic region, leading to 27 synonymous, 29 non-synonymous and 2 stop-gain variations of genes, respectively.
The degree of pod constriction influences seed development in peanut. It is also an important agronomic trait affecting peanut processing, especially for peanut in-shell processing industry. The genetic mechanism for peanut constriction is known to be very complex [5,44,45]. Although some QTLs underlying the peanut pod constriction degree have been identified via linkage analysis [5], more genes controlling constriction should be addressed to dissect its genetic mechanism. In this study, we identified the deep constriction mutant lines, and re-sequenced one of them. We propose that those mutations leading to the stop-gain variations are the most likely cause for the phenotype variation. In our study, two genes exhibited the “stop-gain” variation in gene structure resulting from the typical G/C to A/T transitions point base substitutions induced via EMS treatment. We speculated that they were the possible candidate genes for deep constriction phenotypes in our mutant. These two genes encode “zinc finger MYM-type protein 1-like” and “probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1, 3-glucosyltransferase-like”, respectively. Their specific roles in peanut pod development are still not known based on the current knowledge. According to the study in other species, the zinc finger MYM-type proteins are involved in a protein-protein interaction module, a function in regulating the transcriptional ability [46], telomeric fusions [47], and DNA repair [48]. The dolichyl pyrophosphate Man9GlcNAc2 alpha-1, 3-glucosyltransferase catalyzes the addition of the second glucose residue to the lipid-linked oligosaccharide precursor for N-linked glycosylation of proteins [49]. Of course, the candidate gene for phenotypic variation should be confirmed via further study, such as mut-map, and so on.
Authorship: The authors confirm contribution to the paper as follows: study conception and design: H.C, G.L, Z.T; data collection: H.C, F.X, R.X, X.C, H.Z, Y.Z, X.L, Y.C, R.H; analysis and interpretation of results: H.C, F.X; draft manuscript preparation: H.C, F.X. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This research was funded by the Basic Scientific Research Special Project for Fujian Provincial Public Research Institutes (2020R10310011; 2019R1031-12); the National Natural Science Foundation of China (32001577); the Foreign Cooperation Projects for FAAS (DWHZ2021-20); the Opening Foundation of Fujian Key Laboratory for Vegetable Breeding (FJVRC2020-02); the Free Explore Program for FAAS (ZYTS2019007); the Guangxi Natural Science Foundation Program (2018GXNSFDA281027), and the Science and Technology Innovative Team in Fujian Academy of Agricultural Sciences (CXTD2021008-3).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ojiewo, C. O., Janila, P., Bhatnagar-Mathur, P., Pandey, M. K., Desmae, H. et al. (2020). Advances in crop improvement and delivery research for nutritional quality and health benefits of groundnut (Arachis hypogaea L.). Frontiers in Plant Science, 11, 29. [Google Scholar]
2. FAO (2021). FAOSTAT statistical database. Rome, Italy: FAO. [Google Scholar]
3. Gomes, R. L. F., de Almeida Lopes, Â. C. (2005). Correlations and path analysis in peanut. Crop Breeding and Applied Biotechnology, 5, 105–112. [Google Scholar]
4. Zhang, S., Hu, X., Miao, H., Chu, Y., Cui, F. et al. (2019). QTL identification for seed weight and size based on a high-density SLAF-seq genetic map in peanut (Arachis hypogaea L.). BMC Plant Biology, 19(1), 1–15. [Google Scholar]
5. Patil, A. S., Popovsky, S., Levy, Y., Chu, Y., Clevenger, J. et al. (2018). Genetic insight and mapping of the pod constriction trait in Virginia-type peanut. BMC Genetics, 19(1), 1–9. [Google Scholar]
6. Oladosu, Y., Rafii, M. Y., Abdullah, N., Hussin, G., Ramli, A. et al. (2016). Principle and application of plant mutagenesis in crop improvement: A review. Biotechnology Biotechnological Equipment, 30(1), 1–16. [Google Scholar]
7. Krieg, D. R. (1963). Ethyl methanesulfonate-induced reversion of bacteriophage T4rII mutants. Genetics, 48(4), 561–580. [Google Scholar]
8. Taheri, S., Abdullah, T. L., Jain, S. M., Sahebi, M., Azizi, P. (2017). TILLING, high-resolution melting (HRMand next-generation sequencing (NGS) techniques in plant mutation breeding. Molecular Breeding, 37(3), 1–23. [Google Scholar]
9. Gilchrist, E., Haughn, G. (2010). Reverse genetics techniques: Engineering loss and gain of gene function in plants. Briefings in Functional Genomics, 9(2), 103–110. [Google Scholar]
10. Karaman, K., Kizil, S., Başak, M., Uzun, B., Yol, E. (2021). Development of EMS-induced mutagenized groundnut population and discovery of point mutations in the ahFAD2 and Ara h 1 genes by TILLING. Journal of Oleo Science, 70(11), 1631–1640. [Google Scholar]
11. Fang, C. Q., Wang, C. T., Wang, P. W., Tang, Y. Y., Wang, X. Z. et al. (2012). Identification of a novel mutation in FAD2B from a peanut EMS mutant with elevated oleate content. Journal of Oleo Science, 61(3), 143–148. [Google Scholar]
12. Mondal, S., Badigannavar, A. (2013). A narrow leaf groundnut mutant, TMV2-NLM has a G to A mutation in AhFAD2A gene for high oleate trait. Indian Journal of Genetics and Plant Breeding, 73(1), 105–109. [Google Scholar]
13. Knoll, J. E., Ramos, M. L., Zeng, Y., Holbrook, C. C., Chow, M. et al. (2011). TILLING for allergen reduction and improvement of quality traits in peanut (Arachis hypogaea L.). BMC Plant Biology, 11(1), 1–13. [Google Scholar]
14. Tang, Y., Wang, X., Wu, Q., Fang, C., Guan, S. et al. (2013). Identification of differentially expressed genes from developing seeds of a normal oil peanut cultivar and its high oil EMS mutant. Research Crops, 14(2), 511–516. [Google Scholar]
15. Wan, L., Li, B., Pandey, M. K., Wu, Y., Lei, Y. et al. (2016). Transcriptome analysis of a new peanut seed coat mutant for the physiological regulatory mechanism involved in seed coat cracking and pigmentation. Frontiers in Plant Science, 7, 1491. [Google Scholar]
16. Zhao, M., Sun, H., Ji, R., Hu, X., Sui, J. et al. (2013). In vitro mutagenesis and directed screening for salt-tolerant mutants in peanut. Euphytica, 193(1), 89–99. [Google Scholar]
17. Chen, T., Huang, L., Wang, M., Huang, Y., Zeng, R. et al. (2020). Ethyl methyl sulfonate-induced mutagenesis and its effects on peanut agronomic, yield and quality traits. Agronomy, 10(5), 655. [Google Scholar]
18. Wang, C., Wang, X., Tang, Y., Chen, D., Zhang, J. et al. (2010). High yielding mutants achieved by injecting EMS into peanut flower organs. Journal of Nuclear Agricultural Sciences, 24(2), 239–242. [Google Scholar]
19. Wu, L., Li, Z., Qiu, Q., Miao, H. (2002). Application of mutation techniques in peanut breeding in China. Journal of Nuclear Agricultural Sciences, 16(5), 334–336. [Google Scholar]
20. Liao, B., Zhuang, W., Tang, R., Zhang, X., Shan, S. et al. (2009). Peanut aflatoxin and genomics research in China: Progress and perspectives. Peanut Science, 36(1), 21–28. [Google Scholar]
21. Chen, H., Chen, X., Xu, R., Liu, W., Liu, N. et al. (2021). Fine-mapping and gene candidate analysis for AhRt1, a major dominant locus responsible for testa color in cultivated peanut. Theoretical and Applied Genetics, 134(11), 3721–3730. [Google Scholar]
22. Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. [Google Scholar]
23. Etherington, G. J., Ramirez-Gonzalez, R. H., MacLean, D. (2015). bio-samtools 2: A package for analysis and visualization of sequence and alignment data with SAMtools in Ruby. Bioinformatics, 31(15), 2565–2567. [Google Scholar]
24. McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K. et al. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9), 1297–1303. [Google Scholar]
25. Wang, K., Li, M., Hakonarson, H. (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research, 38(16), e164. [Google Scholar]
26. Wang, N., Wang, Y., Tian, F., King, G. J., Zhang, C. et al. (2008). A functional genomics resource for Brassica napus: Development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytologist, 180(4), 751–765. [Google Scholar]
27. Tang, S., Liu, D., Lu, S., Yu, L., Li, Y. et al. (2020). Development and screening of EMS mutants with altered seed oil content or fatty acid composition in Brassica napus. Plant Journal, 104(5), 1410–1422. [Google Scholar]
28. Sun, J., Luu, N. S., Chen, Z., Chen, B., Cui, X. et al. (2019). Generation and characterization of a foxtail millet (Setaria italica) mutant library. Frontiers in Plant Science, 10, 369. [Google Scholar]
29. Yadav, P., Meena, H., Meena, P., Kumar, A., Gupta, R. et al. (2016). Determination of LD50 of ethyl methanesulfonate (EMS) for induction of mutations in rapeseed-mustard. Journal of Oilseed Brassica, 1(1), 77–82. [Google Scholar]
30. Chen, W., Jiao, Y., Cheng, L., Huang, L., Liao, B. et al. (2016). Quantitative trait locus analysis for pod-and kernel-related traits in the cultivated peanut (Arachis hypogaea L.). BMC Genetics, 17(1), 1–9. [Google Scholar]
31. Chavarro, C., Chu, Y., Holbrook, C., Isleib, T., Bertioli, D. et al. (2020). Pod and seed trait QTL identification to assist breeding for peanut market preferences. G3: Genes, Genomes, Genetics, 10(7), 2297–2315. [Google Scholar]
32. Gangurde, S. S., Wang, H., Yaduru, S., Pandey, M. K., Fountain, J. C. et al. (2020). Nested-association mapping (NAM)-based genetic dissection uncovers candidate genes for seed and pod weights in peanut (Arachis hypogaea). Plant Biotechnology Journal, 18(6), 1457–1471. [Google Scholar]
33. Luo, H., Ren, X., Li, Z., Xu, Z., Li, X. et al. (2017). Co-localization of major quantitative trait loci for pod size and weight to a 3.7 cM interval on chromosome A05 in cultivated peanut (Arachis hypogaea L.). BMC Genomics, 18(1), 1–12. [Google Scholar]
34. Luo, H., Guo, J., Ren, X., Chen, W., Huang, L. et al. (2018). Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.). Theoretical and Applied Genetics, 131(2), 267–282. [Google Scholar]
35. Gomez Selvaraj, M., Narayana, M., Schubert, A. M., Ayers, J. L., Baring, M. R. et al. (2009). Identification of QTLs for pod and kernel traits in cultivated peanut by bulked segregant analysis. Electronic Journal of Biotechnology, 12(2), 3–4. [Google Scholar]
36. Shirasawa, K., Hirakawa, H., Nunome, T., Tabata, S., Isobe, S. (2016). Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnology Journal, 14(1), 51–60. [Google Scholar]
37. Martín, B., Ramiro, M., Martínez-Zapater, J. M., Alonso-Blanco, C. (2009). A high-density collection of EMS-induced mutations for TILLING in Landsberg erecta genetic background of Arabidopsis. BMC Plant Biology, 9(1), 1–9. [Google Scholar]
38. Suzuki, T., Eiguchi, M., Kumamaru, T., Satoh, H., Matsusaka, H. et al. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Molecular Genetics Genomics, 279(3), 213–223. DOI 10.1007/s00438-007-0293-2. [Google Scholar] [CrossRef]
39. Till, B. J., Reynolds, S. H., Weil, C., Springer, N., Burtner, C. et al. (2004). Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biology, 4(1), 1–8. DOI 10.1186/1471-2229-4-12. [Google Scholar] [CrossRef]
40. Xin, Z., Li, W., Barkley, N. A., Burow, G., Franks, C. et al. (2008). Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biology, 8(1), 1–14. DOI 10.1186/1471-2229-8-103. [Google Scholar] [CrossRef]
41. Okabe, Y., Asamizu, E., Saito, T., Matsukura, C., Ariizumi, T. et al. (2011). Tomato TILLING technology: Development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiology, 52(11), 1994–2005. DOI 10.1093/pcp/pcr134. [Google Scholar] [CrossRef]
42. Lu, X., Liu, J., Ren, W., Yang, Q., Chai, Z. et al. (2018). Gene-indexed mutations in maize. Molecular Plant, 11(3), 496–504. DOI 10.1016/j.molp.2017.11.013. [Google Scholar] [CrossRef]
43. Uchida, N., Sakamoto, T., Kurata, T., Tasaka, M. (2011). Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiology, 52(4), 716–722. [Google Scholar]
44. Cofflet, T. A., Hammons, R. O. (1974). Inheritance of pod constriction in peanuts. Journal of Heredity, 65(2), 94–96. [Google Scholar]
45. Hake, A. A., Shirasawa, K., Yadawad, A., Sukruth, M., Patil, M. et al. (2017). Mapping of important taxonomic and productivity traits using genic and non-genic transposable element markers in peanut (Arachis hypogaea L.). PLoS One, 12(10), e0186113. [Google Scholar]
46. Guzzo, C. M., Ringel, A., Cox, E., Uzoma, I., Zhu, H. et al. (2014). Characterization of the SUMO-binding activity of the myeloproliferative and mental retardation (MYM)-type zinc fingers in ZNF261 and ZNF198. PLoS One, 9(8), e105271. [Google Scholar]
47. Xhemalce, B., Riising, E. M., Baumann, P., Dejean, A., Arcangioli, B. et al. (2007). Role of SUMO in the dynamics of telomere maintenance in fission yeast. PNAS, 104(3), 893–898. [Google Scholar]
48. Kunapuli, P., Somerville, R., Still, I. H., Cowell, J. K. (2003). ZNF198 protein, involved in rearrangement in myeloproliferative disease, forms complexes with the DNA repair-associated HHR6A/6B and RAD18 proteins. Oncogene, 22, 3417–3423. DOI 10.1038/sj.onc.1206408. [Google Scholar] [CrossRef]
49. Burda, P., Aebi, M. (1999). The dolichol pathway of N-linked glycosylation. Biochimica et Biophysica Acta (BBA)-General Subjects, 1426(2), 239–257. DOI 10.1016/S0304-4165(98)00127-5. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools