 Open Access
Open Access
ARTICLE
Chickpea C2H2-Type Zinc Finger Protein ZF2 is a Positive Regulator in Drought Response in Arabidopsis
1 Department of Life Sciences and Health, Huzhou College, Huzhou, 313000, China
2 State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, 210095, China
3 Department of Crop and Soil Sciences, North Carolina State University, Raleigh, 27695-7620, USA
4 Institute of Desert in the Arid Areas, College of Grassland and Environment Sciences, Xinjiang Agricultural University, Urumqi, 830052, China
* Corresponding Author: Hao Ma. Email:
# Sushuang Liu and Yanmin Liu contributed equally to this work
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(2), 577-590. https://doi.org/10.32604/phyton.2022.023738
Received 12 May 2022; Accepted 26 July 2022; Issue published 12 October 2022
Abstract
Drought is a major abiotic stress limiting agricultural crops production worldwide. In our study, we isolated a novel C2H2-type zinc finger protein gene ZF2 from chickpea. ZF2 consisted of 232 amino acids with two QALGGH motifs in Cys2/His2 zinc finger domain. Transient expression analysis of ZF2:GFP fusion protein showed that ZF2 was a nuclear localized protein. In the yeast assay system, the full-length of ZF2 did not show transcriptional activation. Expression of ZF2 gene was enhanced by treatments of several abiotic stresses and phytohormones. The promoter region of ZF2 contained multiple stress- and hormone-related cis-elements. Overexpression of ZF2 in Arabidopsis significantly improved the root length and fresh weight at seedling stage and enhanced the survival rates and proline content under drought treatment. These results indicated that ZF2 functioned as a positive regulator in drought response.Keywords
Abbreviation
| ABA | abscisic acid |
| cDNA | DNA complementary to RNA |
| DRE | dehydration responsive element |
| GFP | green fluorescent protein |
| LTRE | low-temperature-responsive element |
| MeJA | jasmonic acid |
| PCR | polymerase chain reaction |
| qRT-PCR | quantitative reverse transcriptase PCR |
| X-α-Gal | 5-bromo-4-chloro-3-indolyl β-α-D-galactopyranoside |
Abiotic stresses, including drought, high salinity and extreme temperature, influence the growth and development of plants. The plants have evolved efficient mechanisms in response to these unfavorable conditions to achieve an optimal adaptation to adverse environment. In these acclimation mechanisms, transcriptional modulation is considered to play an important role [1]. Transcription factors could control the expression of downstream genes as positive or negative regulators [2]. A great deal of transcription factors have been proved to be involved in abiotic stresses, such as NAC, MYB, WRKY zinc-finger proteins [3–8].
Zinc finger represents a sequence of protein domains in which cysteines and/or histidines coordinate a zinc atom to form a compact finger structure. Zinc finger protein has been divided into C2H2, C3H, C3HC4 and other types on the grounds of the order and number of Cys and His residues [9,10]. Thereinto, C2H2 zinc finger protein is a common and important class of transcription factors. The motif sequence of C2H2 zinc finger proteins is CX2-4CX3FX5LX2HX3-5H (X: any amino acid; number: amino acid amounts), which two pairs of specific cysteines and histidines combine with a Zn ion, folding to form a finger shaped structure composed of an α-helix and a β-sheet. Most C2H2 zinc finger proteins have a constant domain QALGGH, which is unique to plant zinc finger proteins [11]. So far, a lot of genes encoding C2H2 zinc finger proteins from different plants have been demonstrated to be associated with stress-responsive [12,13].
SCOF-1 and GmZF1, C2H2 zinc finger protein genes from soybean, had been proved to improve the tolerance to low temperature in Arabidopsis [14,15]. The Arabidopsis C2H2 zinc finger protein gene ZFP3 had been demonstrated to increase salt and osmotic stresses tolerance [16]. A C2H2 zinc finger protein gene from soybean, GmWRKY16, improves resistance to salt and drought tolerance through an ABA-mediated pathway. The wild soybean C2H2 zinc finger protein GsZFP1 played the role of positive regulation in plant tolerance to drought and cold stress [17]. A potato C2H2 zinc finger protein gene, StZFP1, was related with plant response to dehydration and salt stresses [11]. Overexpression of a tomato C2H2-type zinc finger protein gene SlCZFP1 in rice and Arabidopsis increased plant resistance to cold [18]. In addition, C2H2-type zinc finger proteins had been found to negatively regulate plants response to stress. Soybean C2H2-type zinc finger protein GmZFP3 acted as a passive role in plant tolerance to drought stress [16].
Chickpea (Cicer arietinum L.) is an important legume crop in the arid and semi-arid regions. It is rich in environmental stress resistance genes because of long-term growth in extreme environments. To date, a few genes of chickpea, such as CarNAC2, CarNAC4, CarNAC5 and CarNAC6 have been confirmed to be related with tolerance to abiotic stress [6,15,19]. However, studies have been rarely done about C2H2-type zinc finger protein genes of chickpea. In the present study, we isolated a putative C2H2-type zinc finger gene, named ZF2, from the complementary DNA (cDNA) library analysis of chickpea leaves under drought stress treatment [20]. Drought stress could induce the expression of ZF2 gene, and ZF2 transgenic Arabidopsis plants obtained significantly increased tolerance to drought treatment.
2.1 Plant Materials and Stress Treatments
Chickpea (Cicer arietinum L.) cultivar Xj-209 was used for isolation and expression analysis of ZF2 gene in this study. Seeds were grown under greenhouse conditions using the following conditions: 25°C, 60% relative humidity, 12 h daylength. Arabidopsis wild type (WT) was grown in a greenhouse with 70% relative humidity, 22°C, 16 h daylength.
The 14-day-old chickpea plants were subjected to stress and chemical treatments [6,7]. The chickpea roots were immersed in aqueous solution of 200 mM NaCl, 20% (w/v) PEG 6000, 100 μM MeJA, 100 μM ABA, 20 μM IAA and 50 μM H2O2, respectively, with H2O as control. Chickpea seedlings were transferred to 4°C or 37°C in growth chamber for cold and heat treatment, respectively. The leaves in the same position were sampled at designated time intervals after treatments.
2.2 Isolation and Sequence Analysis of ZF2 Gene
Fresh leaves of chickpea were used to extract total RNA with the TRIZOL Kit (Invitrogen, China), and the total RNA was reverse-transcribed according to the instructions of PrimeScriptTM RT reagent Kit with gDNA Eraser (TAKARA, Japan). The ZF2 gene specific primers were applied to clone the ZF2 full-length sequences from chickpea leaves tissue cDNA. ZF2 sequence was analyzed by blast at NCBI (http://www.ncbi.nlm.nih.gov/blast/). Alignment of ZF2 with homologous proteins from other species was obtained with DNAMAN 8.0. MEGA 5.0 was used to build a phylogenetic tree with the Neighbor-Joining (NJ) method (bootstrap 1000). The name and accession number of 19 proteins used in this section are listed in Supplementary Table 1.
2.3 Isolation and in Silico Sequence Analysis of ZF2 Promoter
The cetyltrimethylammonium bromide (CTAB) method was used to extract the genomic DNA from fresh leaves of chickpea. The number and location of cis-elements in the promoter sequence were analyzed by the online search tool PLACE (http://www.dna.affrc.go.jp/PLACE/) [21].
2.4 Gene Expression Profile Analysis
The quantitative Real-time Polymerase Chain Reaction (qRT-PCR) was performed on each cDNA template using SYBR ®Premix Ex TaqTM II (TAKARA, Japan). The specific primers, ZF2-Q-F and ZF2-Q-R, were used to examine the expression of ZF2 gene. AtTublin and CarACTIN1 were used as endogenous control genes in Arabidopsis thaliana and chickpea, respectively. The primer sequences used in this part were listed in Supplementary Table 1. The qRT-PCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 56°C for 20 s, and 72°C for 20 s. A standard curve of each primer pair was performed to make sure the primer pair conformed the requirement and melting curve analysis was generated to check the specificity of amplification. All data were obtained with three independent biological replicates, each including three technical replicates. Relative quantification values calculated using 2−∆∆Ct method were used to represent the relative expression levels of candidate genes.
2.5 Subcellular Location of ZF2 Protein
To research the localization of ZF2 protein, we inserted the full-length coding sequence of ZF2 into PA7 vector [5], which contains a sequence of coding GFP proteins. Particle bombardment was executed to transfer the recombinant construct and negative control vector into tobacco epidermal cells. After incubation on MS medium for 1 d, the tobacco epidermal cells were observed using a confocal microscope (ZEISS, LSM780).
The coding region of ZF2 full-length (1-693 bp), the N-terminus without DLN coding sequence (1-543 bp), and the C-terminus (544-693 bp) were inserted into the yeast expression vector pGBKT7 (pBD) to obtain pBD-ZF2-FL (1-232 aa), pBD-ZF2-N (1-181 aa) and pBD-ZF2-C (182-232 aa), respectively. According to the lithium acetate method, expression vectors and pBD empty vector (the negative control) were introduced into the yeast strain AH109. The transformed strains were streaked onto SD/-Trp or SD/-His plates and their growth status and the activity of α-galactosidase were recorded to estimate the trans-activation activity of each protein.
2.7 Transformation of Arabidopsis
PCR was used to amplify the coding region of ZF2 gene, which was inserted into PBI121-GFP vector to obtain the overexpression vector PBI121-ZF2 [15]. To study the function of ZF2 promoter, the promoter sequence of ZF2 was used to replace the CaMV 35S of PBI121-GUS vector to create the recombinant vector PBI121-ZF2-P, in which the GUS gene was driven by ZF2 promoter. The PBI121-ZF2, PBI121-ZF2-P vectors and PBI121-GUS vector (as a control) were transferred into the Argobacterium tumefaciens strain EHA105. Using the floral dip method, the Argobacterium tumefaciens strains with PBI121-ZF2, PBI121-ZF2-P and PBI121-GUS were transformed into Arabidopsis WT plants, respectively. Positive transgenic plants were screened on selective plates containing kanamycin (50 mg/mL), detected through PCR, RT-PCR.
2.8 Assay for Drought Tolerance
1/2 MS plates containing 200 mM mannitol were used to grow WT and transgenic seeds, with blank plates as control. After 14 d, root length and fresh weight of each plant was measured. Three-week-old soil-planted WT and transgenic plants were subjected to drought stress by withholding water until most of the WT plants presented fatal effects of dehydration. The survival rates of WT and transgenic lines were calculated after 3 d re-watering. Approximately 20 plants of each line were applied to the experiments. Fresh leaves from drought-treated and well-watered plants were collected to measure the content of free proline. All experiments were repeated independently at least three times.
Student’ t test was applied to compare mean values. Statistically significant differences were determined at a value of P < 0.05 or P < 0.01.
3.1 ZF2 is a C2H2-Type Zinc Finger Protein
Phylogenetic analysis of ZF2 and other zinc finger proteins indicated that ZF2 was orthologous to C2H2-type ZFPs of soybean SCOF-1 and GmZAT10-like (Fig. 1A). Multiple sequence alignment of ZF2 with the other typical C2H2-type zinc finger proteins showed that ZF2 protein included two canonical Cys2/His2 zinc finger domains and two QALGGH motifs in domains, which were conserved plant-specific sequences (Fig. 1B). In addition, a putative nuclear localization signal sequence and DLN box/EAR-motif were founded in amino acid sequence (Fig. 1B).
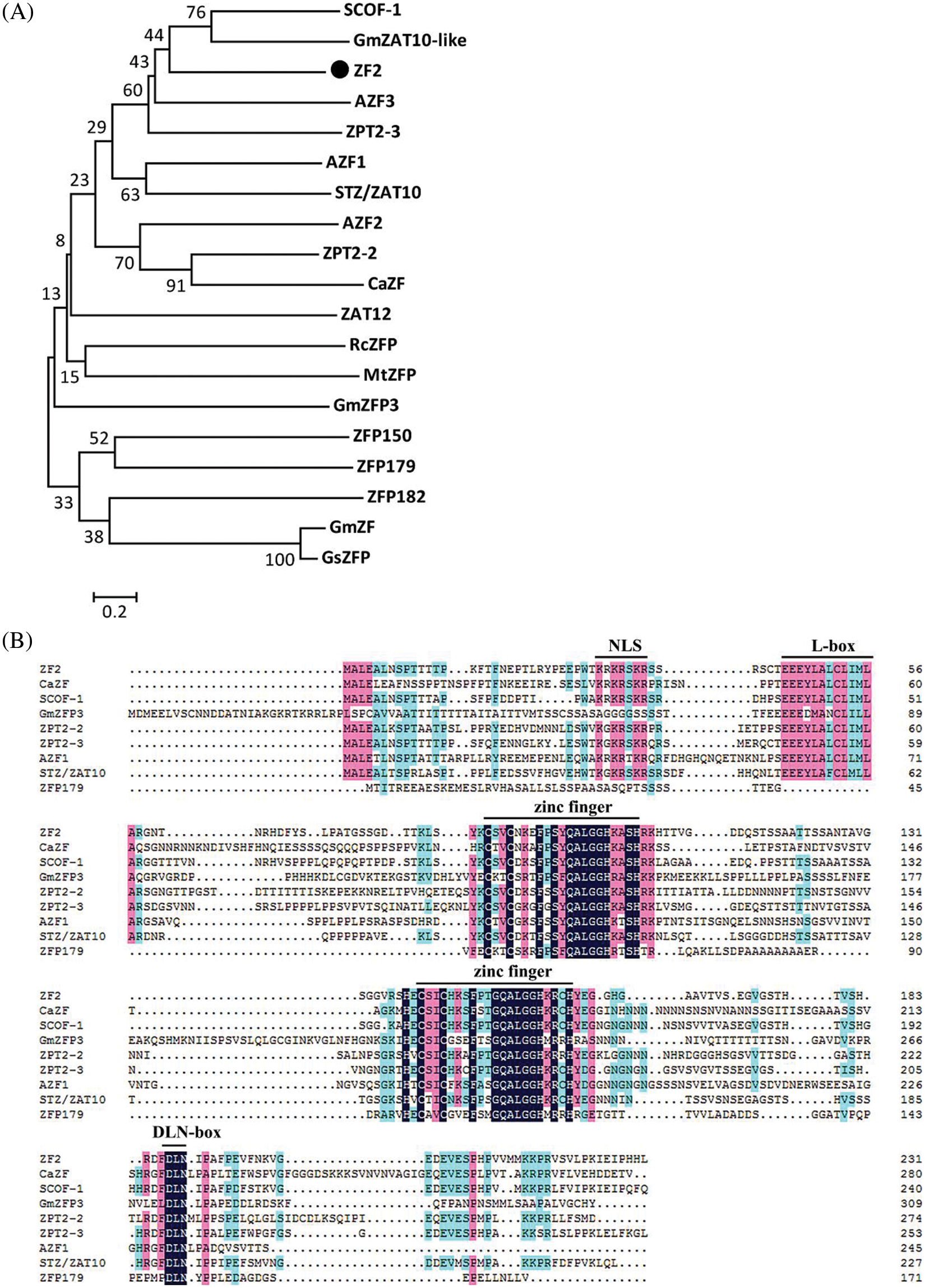
Figure 1: Sequence analysis of ZF2 and related C2H2-type zinc finger proteins. (A) Phylogenic relationship between ZF2 and other zinc finger proteins from Glycine max (SCOF-1; GmZF; GmZAT10-like; GmZFP3), Petunia hybrida (ZPT2-2; ZPT2-3), Arabidopsis thaliana (AZF1; AZF2; AZF3; STZ/ZAT10; ZAT12), Oryza sativa Japonica group (ZFP150; ZFP179; ZFP182), Glycine soja (GsZFP), Ricinus communis (RcZFP), Medicago truncatula (MtZFP), Cicer arietinum (CaZF). (B) Multiple sequence alignment of ZF2 with typical zinc finger proteins. Identical and conserved amino acids are displayed in black and light backgrounds, respectively. The conserved domain is underlined
3.2 Localisation of ZF2 Protein is in the Nucleus of Plant Cells
To validate the location of ZF2 protein, we fused its open reading frame to a gene encoding GFP protein and introduced this construct into tobacco epidermal cells to express ZF2::GFP transiently. The ZF2::GFP fluorescence signal was found to be distributed in the nucleus, while the distribution of free GFP signal was ubiquitous (Fig. 2). This result indicated that ZF2 was located in the nucleus of plant cells.
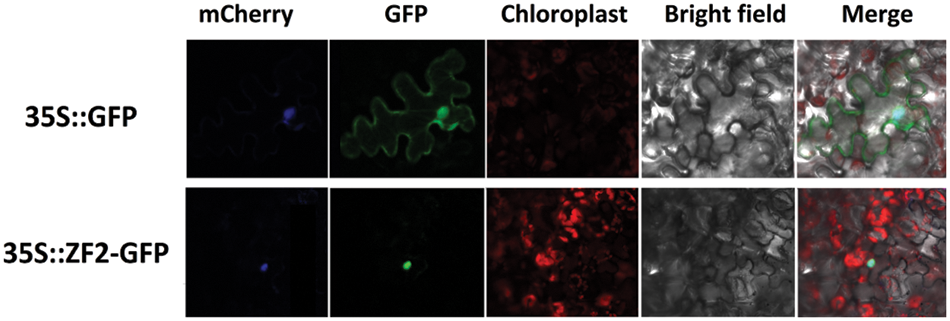
Figure 2: Subcellular localization of ZF2. The 35::GFP and 35S::ZF2-GFP fusion proteins were transiently expressed in tobacco epidermal cells and observed by a laser scanning confocal microscope. mCherry was used to indicate the position of nucleus
3.3 ZF2 Lacks Transcriptional Activation Activity in Yeast Cells
We fused the ZF2-FL (full-length), ZF2-N (N-terminal without DLN domain), ZF2-C (DLN domain) to the GAL4 DNA binding domain to assay the transcriptional activation ability of ZF2 protein. The fusion plasmids, ZF2-FL-BD, ZF2-N-BD, ZF2-C-BD and empty vector BD, were transformed into yeast strain AH109, respectively. As shown in Fig. 3, only the cells containing ZF2-C-BD could grow well on SD/-His plate and possessed α-galactosidase activity, whereas the yeast cells containing ZF2-FL-BD, ZF2-N-BD and empty vector BD could not grow. It indicated that ZF2 full-length protein could not activate expression of HIS reporter gene in yeast cells, whereas the C-terminal with DLN domain has transcriptional activation activity in yeast cells.
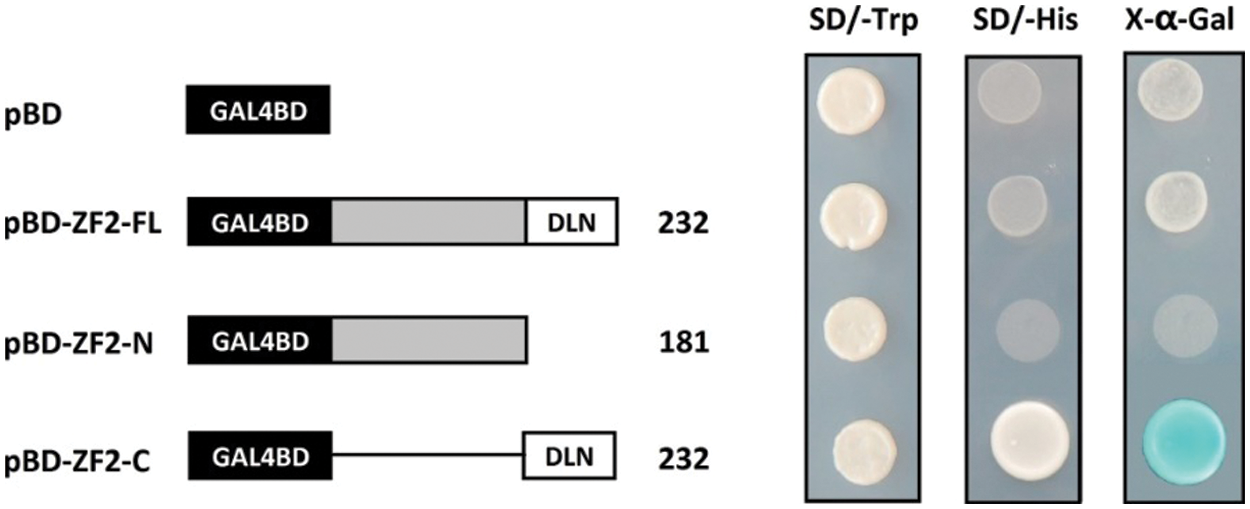
Figure 3: Transactivation assay of ZF2 in yeast cells. The full-length (ZF2-FL), N-terminal (ZF2-N), and C-terminal (ZF2-C) with GAL4-DB fusion effector were introduced into the yeast strain AH109. Transformants were streaked on SD/-Trp or SD/-His plates and pGBKT7 were used as a negative control. X-α-Gal was used to detect the activity of α-galactosidase
3.4 Various Stress Treatments Could Induce the Expression of ZF2
The expression pattern of ZF2 gene under diverse stress was investigated by qRT-PCR. There were no significant changes of ZF2 expression levels under control conditions (H2O treatment) at sampling points (data not shown). The expression of ZF2 gene was significantly induced under dehydration stress, and peaked to 11.4-folds at 48 h. The relative expression levels of ZF2 gene were found to up-regulate under the salt, heat and cold stress treatments (Fig. 4A). In addition, we discovered that the transcription levels of ZF2 were also induced by various phytohormones and signaling moleculars, such as ABA, IAA, MeJA and H2O2 (Fig. 4B).
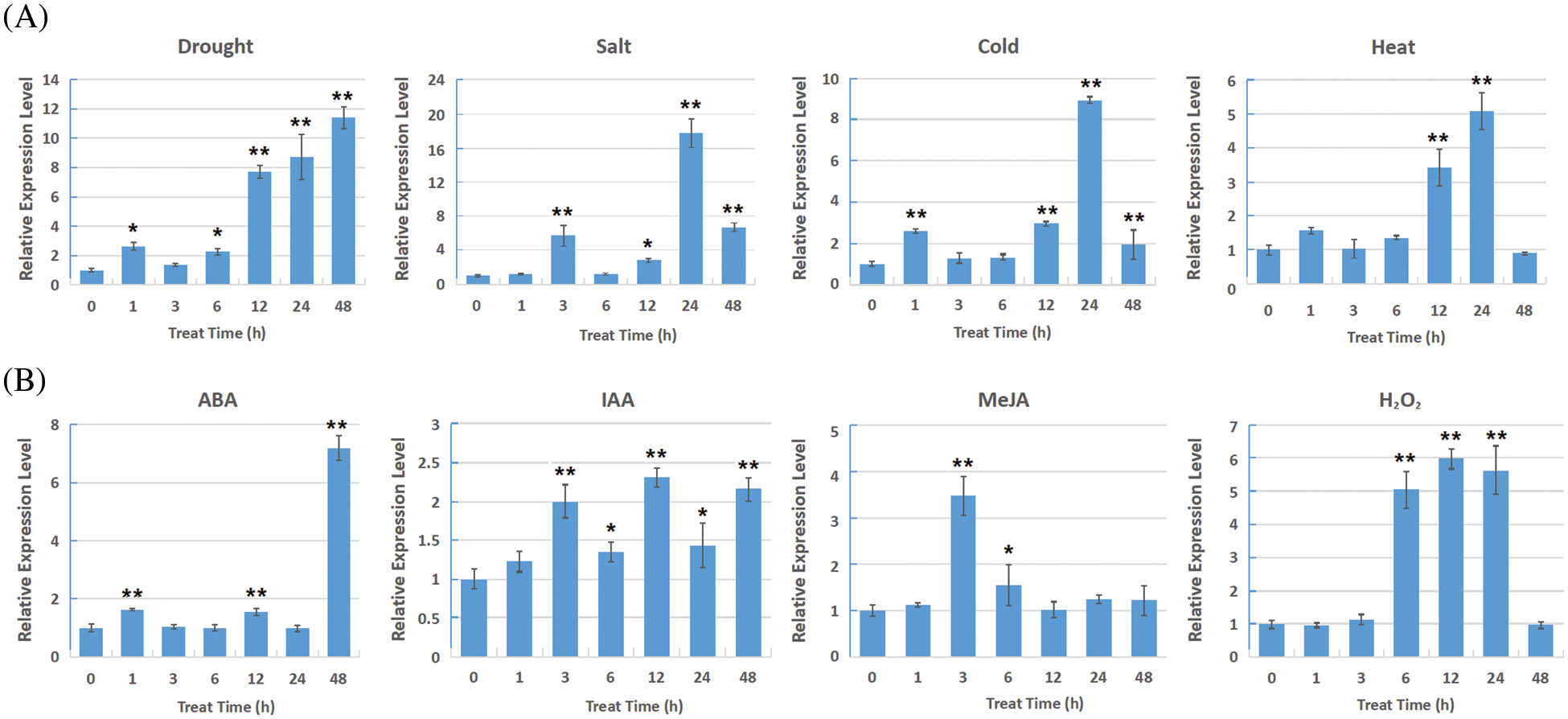
Figure 4: The expression pattern of ZF2 under various stress treatments. (A) Expression patterns under drought, salt, heat and cold, respectively. (B) Expression patterns under ABA, IAA, MeJA and H2O2, respectively. Error bars indicate SD, and asterisks indicate a significant difference (*P < 0.05; **P < 0.01) compared to the non-treatment controls
3.5 Promoter Analysis of ZF2 Gene
A 2461 bp upstream fragment (A in initiation codon ATG as +1 and the first base before ATG as −1) of ZF2 gene was isolated from the genomic of chickpea leaf. Using PLACE database, we analyzed the key cis-acting elements and their position on ZF2 promoter. Multiple stress- and hormone-related cis-acting elements were found (Table 1). Three MYB recognition sites, eleven MYC recognition sites and one DRE/CRT motif associated with drought stress response were discovered. Six sequences of salt- and pathogenesis-related cis-acting elements GT-1 were detected. Two PRE cis-acting elements, which mainly participate in proline synthesis and response to osmotic stress were found. Four ABA responsive elements (ABRE) and one GA responsive element (GARE) were discovered. Moreover, there were one low temperature responsive element (LTRE) and one light-responsive element (I-box) in ZF2 promoter (Fig. 5A).
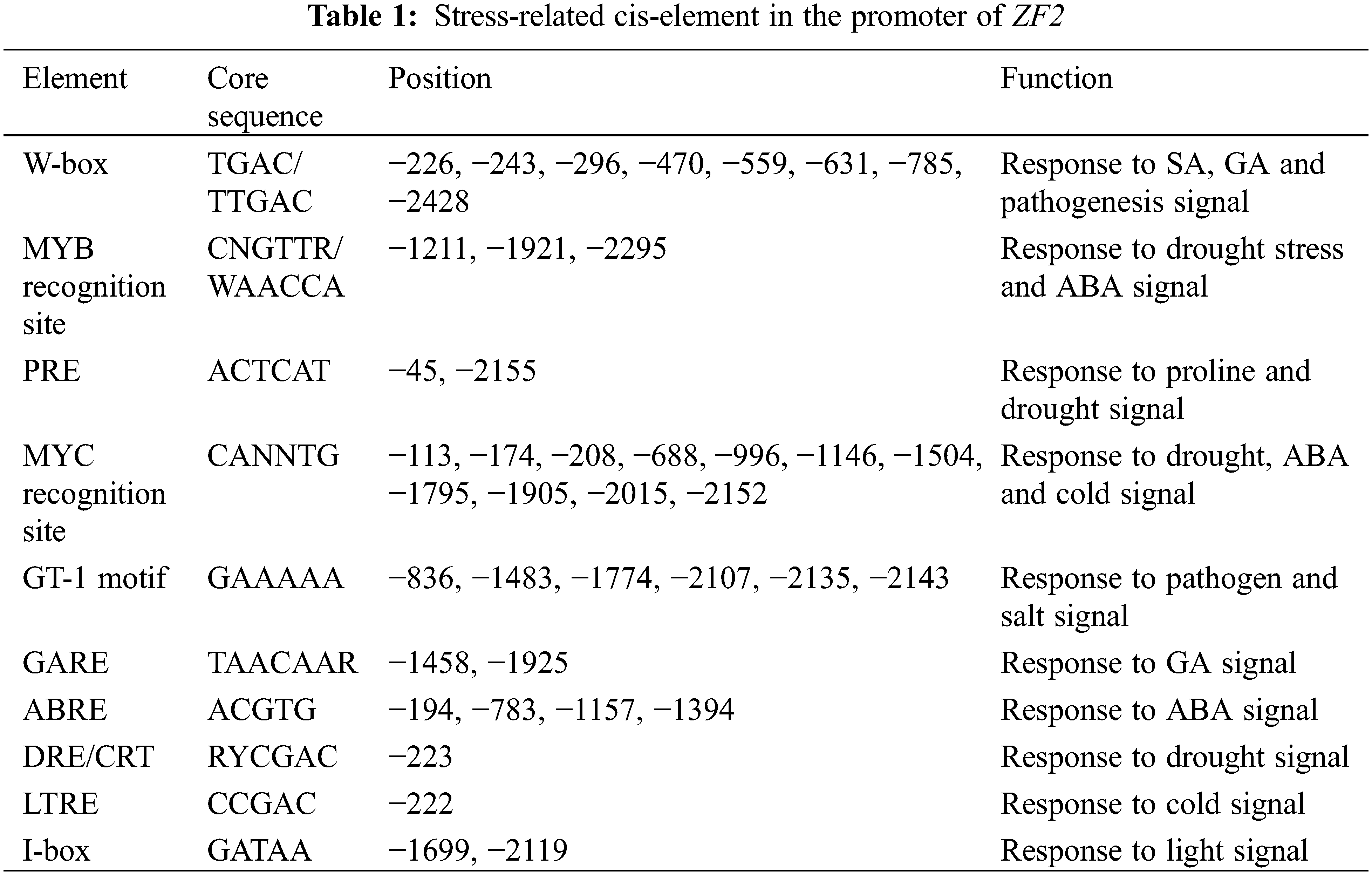
Figure 5: Isolation and analysis of ZF2 promoter. (A) Schematic map of cis-elements on the ZF2 promoter sequence. The arrow indicates the direction of transcription. The putative cis-elements are marked in the boxes and their positions are directed by the lateral lines. (B) Histochemical expression of GUS gene driven by ZF2 promoter in transgenic Arabidopsis. (a) 7-day old seedling. (b) Leaf. (c) Flower. (d) pod
Promoter plays a crucial part in regulating the expression level and controlling the spatial and temporal expression pattern of a gene [22]. Histochemical β-glucuronidase (GUS) staining of transgenic Arabidopsis plants containing a ZF2 promoter::GUS reporter construct was used to determine the expression pattern of ZF2. In 7-day old seedlings, expression of GUS was observed mainly in leaves and hypocotyl as shown in Fig. 5B. In mature plants, GUS primarily expressed in leaves, petals and puerile pods, whereas only little expression could be observed in stems and mature pods.
3.6 ZF2 Transgenic Arabidopsis Obtain Increased Drought Tolerance
To further study the function of ZF2 gene, we obtained transgenic Arabidopsis plants, in which ZF2 gene was overexpressed under the control of CaMV35S promoter. Three independent homozygous transgenic lines (ZF2-1, ZF2-2, and ZF2-3) were chosen to be used in subsequent experiments. Under normal conditions, there was no obvious morphological difference between wild-type plants and ZF2 transgenic plants. At the seedling stage, the root length and fresh weight of transgenic and wild type under normal and drought treatments were measured. As shown in Fig. 6A, there was no difference on the growth of primary roots of wild-type and transgenic plants under normal condition. Whereas, the root length and fresh weight of transgenic plants were significantly higher than those of wild-type plants under the drought treatment.
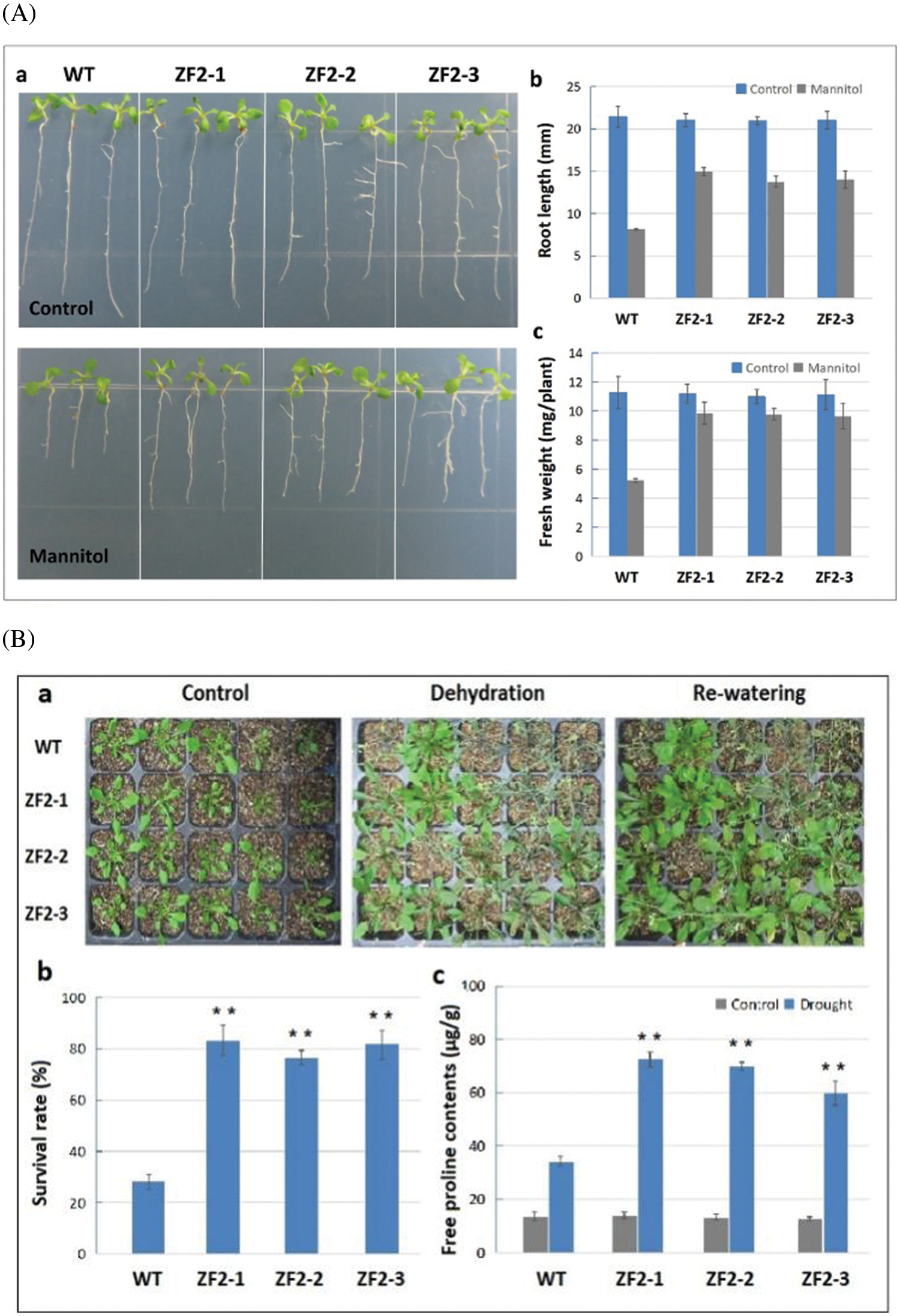
Figure 6: Drought stress tolerance analysis of ZF2 transgenic Arabidopsis. (A) Responses of WT and transgenic Arabidopsis seedlings cultured on 1/2 MS medium supplemented with mannititol. (a) Phenotypes of WT and transgenic seedlings grown on MS medium containing 0 or 200 mM mannitol. (b, c) Root length and fresh weight of 2-week-old seedlings. (B) Overexpression of ZF2 enhanced drought tolerance in Arabidopsis. (a) Phenotypes of WT and transgenic plants under drought stress. 3-week-old WT and transgenic plants were subjected to 14 days drought stress followed by 3 days re-watering. (b) Survival rate of WT and transgenic plants in drought tolerance assays. (c) Free proline content of WT and three transgenic lines. Error bars indicate SD, and asterisks indicate a significant difference (*P < 0.05; **P < 0.01) between the transgenic and WT plants
The survival rates of soil-planted wild-type and ZF2 transgenic lines under drought treatment were also investigated. After 14 days of dehydration (the soil moisture content dropped to approximately 20%), most of the Arabidopsis plants displayed wilting. However, the performance of ZF2 transgenic plants was superior to that of the wild-type plants (Fig. 6B). After 3 days re-watering, only 28% of wild-type lines could continue to grow, and the survival rates of three transgenic lines were 83%, 76%, and 82%, respectively.
Proline is an important osmotic adjustment substance in plants, which could be used as an indicator in the stress evaluation of plants [23]. Under normal condition, the free proline content did not show significant difference between the wild-type and transgenic plants. Nevertheless, the content of proline in ZF2 transgenic plants was significantly (P < 0.01) higher than that of wild-type plants under drought conditions (Fig. 6B). Together, our data demonstrated that ZF2 overexpressing Arabidopsis plants obtained enhanced tolerance to drought stress, and ZF2 might be a positive regulator in response to such stress.
Abiotic stresses usually lead to the action of large number of genes, including both functional genes (encoding structural and enzymatic proteins) and regulatory genes (encoding signal transduction proteins and transcription factors) [24–26]. These genes improve tolerance to stress in plants through diversified pathways. It has been confirmed that C2H2-type zinc finger transcription factors were concerned with plant response to various environment stresses [27–32]. It has reported that SCOF-1 encoded a C2H2-type zinc finger transcription factor with two zinc finger domains, and cold stress could induce the expression of SCOF-1 [14,33]. However, the member of zinc finger proteins was rarely reported in chickpea, and the function of zinc finger proteins related to chickpea response to stress is still not clearly defined.
In the present study, a novel C2H2-type zinc finger protein gene ZF2 from the cDNA library constructed from chickpea leaves under drought treatment was isolated. ZF2 gene encodes a polypeptide of 232 amino acids, which has a spacer composed of 34 amino acids between two Cys2/His2 zinc finger domains. The length of spacers is different in various zinc finger proteins and the diversity may be connected with the specificity of target DNA sequences [9,10]. ZF2 protein contained conserved QALGGH motifs, which has been found in many of C2H2-type zinc finger proteins. GmZFP3 contained this motif, which has been shown to reduce tolerance to drought in plant [16]. The overexpression of GmZF1 gene, which could encode a C2H2-type zinc finger protein with QALGGH motif, enhanced the tolerance to cold stress in Arabidopsis [15]. Whereas, a lot of C2H2-type zinc finger proteins without QALGGH motif were identified from different plants in recent years. GsZFP1 protein from wild soybean (Glycine soja) lacking the QALGGH motif was proved to play an important part in enhanced tolerance to drought and cold stresses [17]. These results indicated the QALGGH motif might play different roles in different plants.
ABA acts as a signal regulator in plants response to abiotic stresses [2]. In our study, exogenous ABA treatment induced the expression of ZF2 gene, which suggested that ZF2 might respond to abiotic stresses through ABA-dependent signal transduction pathway. ABRE (ABA responsive element) is a typical cis-acting element in mediating ABA-dependent signaling [1]. There were four ABRE in the promoter region of ZF2. Furthermore, ABA-independent signal transduction pathway also plays a crucial part in plant stress responses [34]. LTRE and DRE cis-acting elements, which have been identified to participate in abiotic stresses response via ABA-independent pathway, also existed in ZF2 promoter [35,36]. Therefore, we hypothesized that ZF2 gene might be involved in plant response to abiotic stresses through both ABA-dependent and -independent signal transduction pathways.
The full-length and N-terminal of ZF2 protein did not show transcriptional activation. It is interesting that the deletion mutants, ZF2-C (the DLN domain separately), had transcriptional activation activity in yeast cells. The DLN-box had been reported to play the function of transcriptional suppressor, such as STZ/ZAT 10 from Arabidopsis [37] and ZPT2-3 from petunia [29]. However, a part of zinc finger proteins with DLN-box, such as CaZF from chickpea and ZFP179 from rice [38,39], were related to transcriptional activation. Based on our results, we propose the following hypotheses. First, the ZF2 protein may serve as a transcriptional repressor and activate the expression of stress responsive genes by inhibiting the inhibitors of these genes [37]. Second, the ZF2 protein might play different roles in yeast and plant. It was considered that the activation function of yeast systems was different to those on plants [40]. Third, ZF2 may need post-translational modification or environmental stimuli to function as a transcriptional activator.
Although the expression of ZF2 gene was induced by various stress treatments, our research focused on revealing the molecular mechanism of ZF2 in response to drought stress. We obtained ZF2 overexpressed Arabidopsis thaliana lines to analyze biological functions of the ZF2 gene. Under drought stress simulated by mannitol, the root length and fresh weight of transgenic plants at the seedling stage were significantly higher than those of wild-type plants. The survival rates of wild-type plants were lower to those of ZF2 transgenic lines after the dehydration treatment. These results demonstrated that the negative impact of drought stress on the growth and development of wild-type plants was higher than that on ZF2 transgenic plants. Environment stresses often cause direct and indirect dehydration stress on plants, and osmotic adjustment substance are accumulated to improve the stress tolerance [6,7]. As the most widely distributed multifunctional osmolyte in many organisms, the accumulation of proline can be used as an indicator of plant stress tolerance [41,42]. In our study, the content of free proline in transgenic Arabidopsis lines was higher than that on the wild-type under drought condition, indicating that the ZF2 gene might be involved in proline metabolism. Based on the above results, we speculate that the ZF2 gene improved the tolerance to drought in plants by proline accumulation.
In summary, we isolated a novel C2H2-type zinc finger protein gene ZF2 from chickpea. Expression of ZF2 was induced by various abiotic stresses. ZF2 functioned as a positive regulator in plant response to drought. Although chickpea is widely cultivated in the world, there is little research on the function of genes in chickpea. This study broadens our understanding on the function of zinc finger protein in chickpea and offers a potential candidate for molecular breeding in improving the tolerance to abiotic stresses in staple crops. With the development of follow-up work, we will have a clearer understanding on the biological and molecular functions of ZF2.
Authors Contributions: The authors confirm contribution to the paper as follows: study conception and design: H.M., S.L., Y.L.; data collection: X.Y.; analysis and interpretation of results: S.L., Y.L., C.L.; draft manuscript preparation: H.M. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement:This work was sponsored by the National Natural Science Foundation of China (31160306 and 30860152), Natural Science Foundation of Zhejiang Province (LQ20C130003) and Scientific Research Fund of Zhejiang Provincial Education Department (Y202145972 and Y202248468), Zhejiang Students’ Technology and Innovation Program (Xinmiao Program) (2022R485A003).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Fujita, Y., Fujita, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. Journal of Plant Research, 124(4), 509–525. DOI 10.1007/s10265-011-0412-3. [Google Scholar] [CrossRef]
2. Yang, X. W., Wang, X. Y., Ji, L., Yi, Z. L., Fu, C. X. et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Reporters, 34(6), 943–958. DOI 10.1007/s00299-015-1756-2. [Google Scholar] [CrossRef]
3. Liu, K., Wang, L., Xu, Y., Chen, N., Ma, Q. et al. (2007). Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta, 226(4), 1007–1016. DOI 10.1007/s00425-007-0548-5. [Google Scholar] [CrossRef]
4. Cong, L., Chai, T. Y., Zhang, Y. X. (2008). Characterization of the novel gene BjDREB1B encoding a DRE-binding transcription factor from Brassica juncea L. Biochemical Biophysical Researh Communications, 371(4), 702–706. DOI 10.1016/j.bbrc.2008.04.126. [Google Scholar] [CrossRef]
5. Zheng, M., Wang, Y., Wang, Y., Wang, C., Ren, Y. et al. (2015). DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytologist, 206(4), 1476–1490. DOI 10.1111/nph.13318. [Google Scholar] [CrossRef]
6. Yu, X. W., Liu, Y. M., Wang, S., Tao, Y., Wang, Z. K. et al. (2016). A chickpea stress-responsive NAC transcription factor, CarNAC5, confers enhanced tolerance to drought stress in transgenic Arabidopsis. Plant Growth Regulation, 79(2), 187–197. DOI 10.1007/s10725-015-0124-0. [Google Scholar] [CrossRef]
7. Yu, X. W., Liu, Y. M., Wang, S., Tao, Y., Wang, Z. K. et al. (2016). CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Reports, 35(3), 613–627. DOI 10.1007/s00299-015-1907-5. [Google Scholar] [CrossRef]
8. Xu, H. F., Wang, N., Wang, Y. C., Jiang, S. H., Fang, H. C. et al. (2018). Overexpression of the transcription factor MdbHLH33 increases cold tolerance of transgenic apple callus. Plant Cell Tissue and Organ Culture, 134(1), 131–140. DOI 10.1007/s11240-018-1406-9. [Google Scholar] [CrossRef]
9. Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N. et al. (2020). C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Frontiers in Plant Science, 11, 115. [Google Scholar]
10. Han, G., Qiao, Z., Li, Y., Wang, C., Wang, B. (2021). The roles of CCCH zinc-finger proteins in plant abiotic stress tolerance. International Journal of Molecular Sciences, 22(15), 8327. [Google Scholar]
11. Tian, Z. D., Zhang, Y., Liu, J., Xie, C. H. (2010). Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biology, 12(5), 689–697. DOI 10.1111/j.1438-8677.2009.00276.x. [Google Scholar] [CrossRef]
12. Zhang, H., Liu, Y. P., Wen, F., Yao, D. M., Wang, L. et al. (2014). A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. Journal Experimental Botany, 65(20), 5795–5809. DOI 10.1093/jxb/eru313. [Google Scholar] [CrossRef]
13. Teng, K., Tan, P. H., Guo, W. E., Yue, Y. S., Fan, X. F. et al. (2018). Heterologous expression of a novel Zoysia japonica C2H2 zinc finger gene, ZjZFN1, improved salt tolerance in Arabidopsis. Frontiers in Plant Science, 9, 1159. DOI 10.3389/fpls.2018.01159. [Google Scholar] [CrossRef]
14. Kim, J. C., Jeong, J. C., Park, H. C., Yoo, J. H., Koo, Y. D. et al. (2001). Cold Accumulation of SCOF-1 transcripts is associated with transcriptional activation and mRNA stability. Molecular Cells, 12(2), 204–208. [Google Scholar]
15. Yu, G. H., Jiang, L. L., Ma, X. F., Xu, Z. S., Liu, M. M. et al. (2014). A Soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic Arabidopsis. PLoS One, 9(10), e109399. DOI 10.1371/journal.pone.0109399. [Google Scholar] [CrossRef]
16. Zhang, A. D., Liu, D. D., Hua, C. M., Yan, A., Liu, B. H. et al. (2016). The Arabidopsis gene zinc finger protein 3(ZFP3) is involved in salt stress and osmotic stress response. PLoS One, 11(12), e0168367. DOI 10.1371/journal.pone.0168367. [Google Scholar] [CrossRef]
17. Luo, X., Bai, X., Zhu, D., Li, Y., Ji, W. et al. (2012). GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta, 235(6), 1141–1155. DOI 10.1007/s00425-011-1563-0. [Google Scholar] [CrossRef]
18. Zhang, X., Guo, X. P., Lei, C. L., Cheng, Z. J., Lin, Q. B. et al. (2011). Overexpression of SlCZFP1, a novel TFIIIA-type zinc finger protein from tomato, confers enhanced cold tolerance in transgenic Arabidopsis and rice. Plant Molecular Biology, 29(1), 185–196. DOI 10.1007/s11105-010-0223-z. [Google Scholar] [CrossRef]
19. Liu, Y. M., Yu, X. W., Liu, S. S., Peng, H., Mijiti, A. et al. (2017). A chickpea NAC-type transcription factor, CarNAC6, confers enhanced dehydration tolerance in Arabidopsis. Plant Molecular Biology Reporter, 35(1), 83–96. DOI 10.1007/s11105-016-1004-0. [Google Scholar] [CrossRef]
20. Gao, W. R., Wang, X. S., Liu, Q. Y., Peng, H., Chen, C. et al. (2008). Comparative analysis of ESTs in response to drought stress in chickpea (C. arietinum L.). Biochemical Biophysical Researh Communications, 376(3), 578–583. DOI 10.1016/j.bbrc.2008.09.030. [Google Scholar] [CrossRef]
21. Higo, K., Ugawa, Y., Iwamoto, M., Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research, 27(1), 297–300. DOI 10.1093/nar/27.1.297. [Google Scholar] [CrossRef]
22. Guo, L. L., Yu, Y. H., Xia, X. L., Yin, W. L. (2010). Identification and functional characterisation of the promoter of the calcium sensor gene CBL1 from the xerophyte Ammopiptanthus mongolicus. BMC Plant Biology, 10(1), 18. DOI 10.1186/1471-2229-10-18. [Google Scholar] [CrossRef]
23. Movahedi, A., Zhang, J. X., Yin, T. M., Zhuge, Q. (2015). Functional analysis of two orthologous NAC genes, CarNAC3, and CarNAC6 from Cicer arietinum, involved in abiotic stresses in poplar. Plant Molecular Biology Reporter, 33(5), 1539–1551. DOI 10.1007/s11105-015-0855-0. [Google Scholar] [CrossRef]
24. Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E. et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant and Cell Physiology, 49(8), 1135–1149. DOI 10.1093/pcp/pcn101. [Google Scholar] [CrossRef]
25. Abogadallah, G. M., Nada, R. M., Malinowski, R., Quick, P. (2011). Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifolium alexandrinum L. Planta, 233(6), 1265–1276. DOI 10.1007/s00425-011-1382-3. [Google Scholar] [CrossRef]
26. Finkelstein, R., Lynch, T., Reeves, W., Petitfils, M., Mostachetti, M. (2011). Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. Journal of Experimental Botany, 62(11), 3971–3979. DOI 10.1093/jxb/err093. [Google Scholar] [CrossRef]
27. Iida, A., Kazuoka, T., Torikai, S., Kikuchi, H., Oeda, K. (2000). A zinc finger protein RHl41 mediates the light acclimatization response in Arabidopsis. Plant Journal, 24(2), 191–203. DOI 10.1046/j.1365-313x.2000.00864.x. [Google Scholar] [CrossRef]
28. Rizhsky, L., Davletova, S., Liang, H., Mittler, R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. Journal of Biological Chemistry, 279(12), 11736–11743. DOI 10.1074/jbc.M313350200. [Google Scholar] [CrossRef]
29. Sakamoto, H., Maruyama, K., Sakuma, Y., Meshi, T., Iwabuchi, M. et al. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiology, 136(1), 2734–2746. DOI 10.1104/pp.104.046599. [Google Scholar] [CrossRef]
30. Davletova, S., Schlauch, K., Coutu, J., Mittler, R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology, 139(2), 847–856. DOI 10.1104/pp.105.068254. [Google Scholar] [CrossRef]
31. Mittler, R., Kim, Y., Song, L., Coutu, J., Coutu, A. et al. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Letters, 580(28–29), 6537–6542. DOI 10.1016/j.febslet.2006.11.002. [Google Scholar] [CrossRef]
32. Huang, J., Yang, X., Wang, M. M., Tang, H. J., Ding, L. Y. et al. (2007). A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochimica ET Biophysica Acta-Biomemebranes, 1769(4), 220–227. DOI 10.1016/j.bbaexp.2007.02.006. [Google Scholar] [CrossRef]
33. Kim, Y. H., Kim, M. D., Park, S. C., Yang, K. S., Jeong, J. C. et al. (2011). SCOF-1-expressing transgenic sweet potato plants show enhanced tolerance to low-temperature stress. Plant Physiology and Biochemistry, 49(12), 1436–1441. DOI 10.1016/j.plaphy.2011.09.002. [Google Scholar] [CrossRef]
34. Yoshida, T., Mogami, J., Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Current Opinion Plant Biology, 21, 133–139. DOI 10.1016/j.pbi.2014.07.009. [Google Scholar] [CrossRef]
35. Shinozaki, K., Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Current Opinion Plant Biology, 3(3), 217–223. [Google Scholar]
36. Yamaguchi-Shinozaki, K., Shinozaki, K. (2005). Organization of cisacting regulatory elements in osmotic- and cold-stress responsive promoters. Trends in Plant Science, 10(2), 88–94. DOI 10.1016/j.tplants.2004.12.012. [Google Scholar] [CrossRef]
37. Sugano, S., Kaminaka, H., Rybka, Z., Catala, R., Salinas, J. et al. (2003). Stress responsive zinc finger gene ZPT2-3 plays a role in drought tolerance in petunia. Plant Journal, 36(6), 830–841. DOI 10.1046/j.1365-313x.2003.01924.x. [Google Scholar] [CrossRef]
38. Jain, D., Roy, N., Chattopadhyay, D. (2009). CaZF, a plant transcription factor functions through and parallel to HOG and calcineurin pathways in Saccharomyces cerevisiae to provide osmotolerance. PLoS One, 4(4), e5154. DOI 10.1371/journal.pone.0005154. [Google Scholar] [CrossRef]
39. Sun, S. J., Guo, S. Q., Yang, X., Bao, Y. M., Tang, H. J. et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. Journal of Experimental Botany, 61(10), 2807–2818. DOI 10.1093/jxb/erq120. [Google Scholar] [CrossRef]
40. Heinekamp, T., Kuhlmann, M., Lenk, A., Strathmann, A., Droge-Laser, W. (2002). The tobacco bZIP transcription factor BZI-1 binds to G-box elements in the promoters of phenylpropanoid pathway genes in vitro, but it is not involved in their regulation in vivo. Molecular Genetics and Genomics, 267(1), 16–26. DOI 10.1007/s00438-001-0636-3. [Google Scholar] [CrossRef]
41. Hasegawa, P. M., Bressan, R. A., Zhu, J. K., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology, 51(1), 463–499. DOI 10.1146/annurev.arplant.51.1.463. [Google Scholar] [CrossRef]
42. Ashraf, M., Foolad, M. R. (2007). Roles of glycinebetaine and proline in improving plant abiotic stress resistance. Environmental and Experimental Botany, 59(2), 206–216. DOI 10.1016/j.envexpbot.2005.12.006. [Google Scholar] [CrossRef]
Supplementary 1. The name of C2H2-type zinc finger proteins used for constructing phylogenetic tree
SCOF-1 (AAB39638.1); ZPT2-2 (BAA05077.1); ZPT2-3 (BAA05079.1); AZF1 (BAA85108.1); AZF2 (BAB02542.1); AZF3 (NP_199131.1); STZ/ZAT10 (NP_174094.1); ZAT12 (AAM65582.1); ZFP150 (AAP42460.1); ZFP179 (AAL76091.1); ZFP182 (AAP42461.1); GmZF (AAZ03389.1); GmZAT10-like (NP_001254622.1); GsZFP (ACJ48970.1); RcZFP (XP_002515449.1); MtZFP (XP_003604205.2); CaZF (ACD13216.1); GmZFP3 (AHN52234.1); ZF2 (ACO54859.10).

Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools