 Open Access
Open Access
ARTICLE
Transcriptome Analysis of a Wild Eggplant Germplasm M239 in Response to Verticillium dahliae Infection
1 Institute of Horticultural Crops, Yunnan Academy of Agricultural Sciences, Kunming, 650205, China
2 Institute of Plant Resources, Yunnan University, Kunming, 650500, China
* Corresponding Authors: Min Gui. Email: ; Guanghui Du. Email:
Phyton-International Journal of Experimental Botany 2023, 92(2), 591-609. https://doi.org/10.32604/phyton.2022.023481
Received 28 April 2022; Accepted 04 July 2022; Issue published 12 October 2022
Abstract
In this study, wild eggplant germplasm No. M239, which is highly susceptible to Verticillium wilt, was used as the experimental material. The physiological and biochemical indices (SOD, PAL, MDA and soluble protein) of M239 roots were measured at different times (0, 12, 24, 36, 48, 60 and 72 h) post inoculation with Verticillium dahliae, and the key time points for the M239 response to Verticillium wilt infection were screened. Then, RNA-Seq technology was used to screen the differentially expressed genes (DEGs) in M239 roots at 0, 12 and 48 h post-inoculation (hpi). The transcriptional results of M239 were also compared with those resistance genes from some reported wild relative Solanum species (S. sisymbriifolium and S. aculeatissimum). Then some DEGs were chosen for validation by qRT–PCR. The results showed that 12 and 48 hpi were the turning points in the changes in all physiological and biochemical indices. A total of 6,783 DEGs were identified by RNA-Seq, including 6,141 DEGs (3,046 upregulated and 3,095 downregulated) at the M_12 h vs. M_0 h, 1,903 DEGs (792 upregulated and 1,111 downregulated) at M_48 h vs. M_12 h, and 1,261 DEGs that appeared simultaneously in both stages. KEGG enrichment analysis showed that there were 5 metabolic pathways enriched from DEGs, which were mostly related to primary metabolism, such as glycolysis, amino acid and ribosome biogenesis. Compared with the NCBI non-redundant protein (NR) database, one Ve2 homologous gene and 8 PR protein-related genes were screened. Transcription factor analysis showed that there were a large number of DEGs, such as MYB, AP2-EREBP, bHLH, NAC and Orphans in the two stages. Compared with the reported Verticillium wilt-resistant wild eggplant species, it was found that there were fewer genes and enriched metabolic pathways in the M239 response to Verticillium wilt infection, and it also lacked the response of some known key resistance genes. These results proved that the above resistance genes and metabolic pathways played a key role in the wild eggplant response to V. dahliae infection.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| DEG | Differentially expressed gene |
| EVW | Eggplant Verticillium wilt |
| FPKM | Fragments per kilobase per million fragments |
| hpi | Hours post inoculation |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MDA | Malondialdehyde |
| NCBI | National center for biotechnology information |
| NR | NCBI non-redundant protein |
| PAL | Phenylalanine ammonia-lyase |
| PR | Pathogenesis-related protein |
| qRT-PCR | Real-time quantitative PCR |
| SRA | Sequence Read Archive |
| SOD | Superoxide dismutase |
| TFs | Transcription factors |
Eggplant (Solanum melongena L.), a common vegetable crop, is native to Southeast Asia and India. Eggplant was introduced into China as early as the 4th~5th century Anno Domini (A.D.). It is generally believed that China is the second origin of eggplant. At present, China has the largest planting area of eggplant and the largest number of eggplant germplasm resources [1]. Eggplant Verticillium wilt (EVW) is a soil-borne vascular disease caused by Verticillium spp. Once it occurs, eggplant can be seriously reduced or can even fall out of production; thus, EVW has become one of the main diseases threatening eggplant production. According to some domestic reports, EVW in China is mainly caused by Verticillium dahliae [2].
In recent years, researchers have been trying to reveal the interaction mechanism between V. dahliae and different plants, such as Arabidopsis [3], tomato [4–6], cotton [7–10] and potato [11]. Meanwhile, some resistance genes to Verticillium wilt have been screened and identified in eggplant and its wild relatives, such as SlVe [12], StVe [13] and StDAHP [14]. Especially with the application of RNA-Seq technology [15–18], relevant data at the transcriptional level of plant Verticillium wilt have been obtained and utilized.
Many studies have shown that EVW-resistant materials mainly exist in the wild relatives Solanum species and semicultivated eggplant types [1,19]. There are rich wild eggplant resources in Yunnan Province and its surrounding areas of China [20]. However, research on Verticillium wilt resistance is mainly focused on several wild eggplant species, such as S. torvum [16], S. aculeatissimum [17], and S. sisymbriifolium [18], and there is little research on Verticillium wilt-susceptible materials in wild eggplants. The above wild eggplant resistance materials are not only distantly related to cultivated eggplant (i.e., widely incompatible with distant hybridization with cultivated eggplant), but also have great differences in their growth environments. These factors make it difficult to solve the disease problem in the actual production of eggplant. The study of wild eggplant materials close to cultivated eggplant can eliminate the huge interference caused by the differences in genetic background and growth environments between wild and cultivated eggplant to a certain extent and speed up the research and utilization of wild eggplant resources. Among them, a wild eggplant germplasm (No. V06B1239 of national vegetable medium-term data, M239) was collected from Dali City, Yunnan Province by the Institute of Horticultural Crops, Yunnan Academy of Agricultural Sciences in the 1990s. In our preliminary study [2], it was found that M239 was more susceptible to the disease than the cultivated eggplant after inoculation by V. dahliae, and its genetic background and morphology were close to those of the cultivated eggplants [21]. Therefore, it is speculated that M239 had a history of cultivation, but it was eliminated because of its poor disease resistance and gradually became wild.
Based on the previously screened Verticillium wilt highly susceptible material M239 [2], this study analysed its roots at different time points (0, 12 and 48 h) after M239 was inoculated with V. dahliae at the transcriptional level by the Illumina high-throughput sequencing method and established a transcriptome database of M239 in response to V. dahliae infection. The key pathways and resistance genes of plant resistance to Verticillium wilt were screened by comparative analysis with the reported resistant materials (S. sisymbriifolium and S. aculeatissimum, etc.) of wild eggplant species. These results are expected to fully reveal the transcriptome information characteristics of highly susceptible wild eggplant materials in response to Verticillium wilt, clarify the expression differences of transcriptional levels between highly resistant and highly susceptible materials of wild eggplants, provide a reference for studying eggplant Verticillium wilt resistance, screen resistant eggplant germplasm resources, and also lay a foundation for the screening and identification of Verticillium wilt resistance genes.
The highly pathogenic strain QZ-S of V. dahliae and the highly susceptible material M239 (Fig. 1) screened in a preliminary experiment by our research group were used as the test materials [2]. The above test materials were all collected and preserved by the Institute of Horticultural Crops, Yunnan Academy of Agricultural Sciences.

Figure 1: Wild eggplant M239, a highly susceptible material to Verticillium wilt. (A) the whole plant; (B) normal seedlings; (C) infected seedlings
2.2 Determination of Physiological Indices in M239 after Inoculation with V. dahliae
Key measures for the sowing and growing of M239 seedlings were used as described by the method of Wu et al. [2]. At the plant’s four true leaves stage, the Verticillium wilt pathogen was artificially inoculated according to the method of Wu et al. [18]. Plants at 0, 12, 24, 36, 48, 60 and 72 h post-inoculation (hpi) were selected, and the roots of 5~10 plants with consistent growth were taken. After liquid nitrogen treatment, they were immediately stored at −80°C for the next physiological and biochemical index determination and transcriptome sequencing. The plants at 0 hpi were used as the control (CK), and each treatment was repeated three times.
The physiological indices were measured with a kit from the Nanjing Jiancheng Bioengineering Institute. Total SOD enzyme activity was determined by an A001-1 kit (product number, the same below), PAL enzyme activity was determined by an A137-1 kit, MDA content was determined by an A003-1 kit, and soluble protein content was determined by an A145-1 kit. A UV1600 ultraviolet spectrophotometer (Shanghai Yoke Instrument Co., Ltd., China) was used to measure the absorbance value. The specific enzyme activity determination and calculations were performed according to the instructions of the corresponding kit.
2.3 Sequencing and Transcriptome Analysis of M239 after Inoculation with V. dahliae
According to the results of physiological and biochemical indices, the roots of M239 at the key time points (0, 12, and 48 hpi) after inoculation with V. dahliae were selected for transcriptome sequencing and analysis. The experiment was divided into three groups with three biological repetitions in each group.
2.3.1 Extraction and Quality Detection of Total RNA
Total RNA from each sample was extracted using an RNAout 1.0 kit (Beijing Tianenze Gene Technology Co., Ltd., China). A 1% agarose gel was used to detect the quality of RNA, and its purity was detected by NanoDrop (Implen, USA). The RNA concentration was measured by a Qubit RNA analysis kit and a Qubit 2.0 fluorometer (Life Technologies, USA). RNA integrity was evaluated using an RNA Nano 6000 analysis kit and an Agilent 2100 biological analyser (Agilent Technologies, USA). The RNA was then used for the next step of database construction.
2.3.2 Transcriptome Sequencing, Data Assembly and Functional Annotation
The construction and sequencing of the cDNA library were completed by Beijing Novogene Bioinformatics Technology Co., Ltd., China. A cDNA library was successfully constructed, and high-throughput sequencing was carried out using the Illumina sequencing platform (Illumina HiSeq 2000). The raw data (or raw reads) generated by sequencing were obtained after removing the connector, poly-N and low-quality reads (Q < 20). Subsequent analysis was based on the above high-quality clean reads.
Compared with the eggplant reference genome [22], unigenes were obtained by applying HISAT software (v2.0.4), and the unmatched sequences were assembled with Cufflinks software (v2.2.1). Then new transcripts were predicted by comparing the output results of Cufflinks with known gene models. The assembly, sequencing and splicing were all completed by Beijing Novogene Bioinformatics Technology Co., Ltd., China.
2.3.3 Identification of Differentially Expressed Genes (DEGs)
The reads per kilobase of exon model per million mapped reads (FPKM) value was used to quantify the gene expression level. DEGs were screened by the DESeq R software package (v1.10.1). For the experiments with biological duplication, DESeq eliminated the biological variation. Therefore, the threshold standard of DEGs set by transcriptome data was padj < 0.05 in this study. KEGG enrichment analysis of DEGs was carried out by KOBAS software [23]. A corrected p value (q value) <0.05 was used as the standard for enrichment and screening. The BLAST procedure in NCBI was used for sequence alignment and searching homologous genes.
RNA from M239 roots was reverse transcribed into cDNA by a kit (Easyscript®First Strand cDNA Synthesis SuperMax, Trans, Beijing, China), and 18S RNA from eggplant was used as a reference gene. Twenty DEGs were randomly selected, and the transcriptome data were verified by qRT–PCR. The specific primer sequences are shown in Table 1. The qRT–PCR procedure was performed according to the kit instructions (Transstart® Tip Green qPCR SuperMax, Trans, Beijing, China) and run on an Applied Biosystems 7500 real-time fluorescence quantitative PCR (USA). Relative gene expression levels were calculated using the
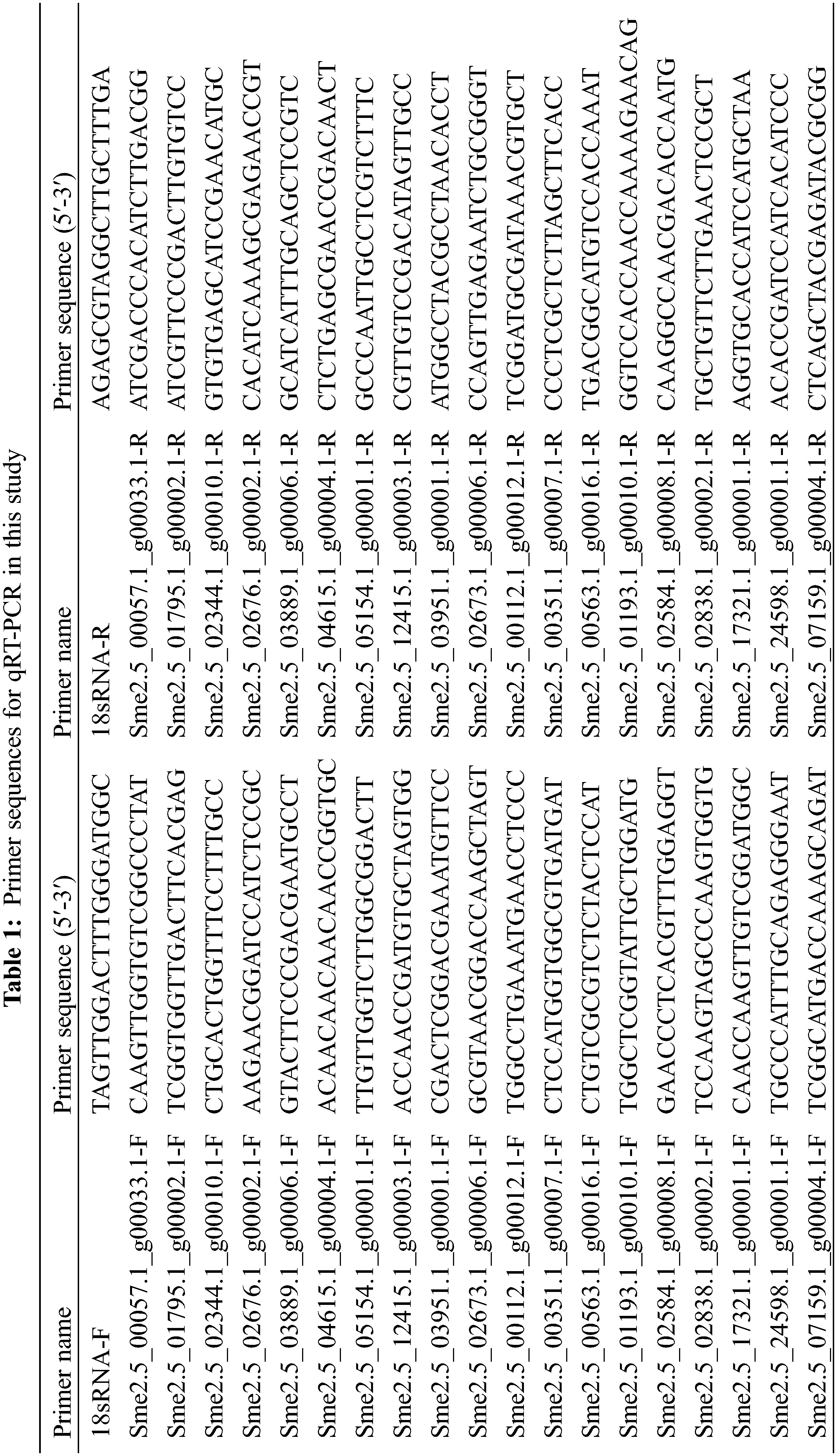
2.5 Data Statistics and Analysis
The software programs Excel 2010 and SPSS 19.0 were used for data processing and analysis, and the figures were drawn in Sigmaplot 10.0 software.
3.1 Physiological Response of M239 within 72 h after Inoculation with V. dahliae
Four physiological and biochemical indices (SOD, PAL, MDA, and soluble protein) were measured within 72 h after M239 was inoculated with V. dahliae to study the physiological response mechanism of M239 inoculated with V. dahliae and screen the accurate sampling and analysis time for the next transcriptome sequencing.
As shown in Fig. 2, it can be seen that within 72 h after M239 was inoculated with V. dahliae, all physiological and biochemical indices had changed significantly compared with CK (0 hpi). Among them, SOD activity increased rapidly after inoculation and began to decrease after reaching a peak at 12 hpi. PAL activity increased rapidly after inoculation, began to change slowly after 6 hpi, reached the highest value at 12 hpi, then began to decrease rapidly, and reached a low point at 48 hpi. The change in MDA content was not obvious, and there were only two small peaks and valleys at 12 and 48 hpi, respectively. The content of soluble protein first increased, then decreased and finally increased after inoculation, there were two peaks at 6 and 48 hpi and an inflection point at 24 hpi.
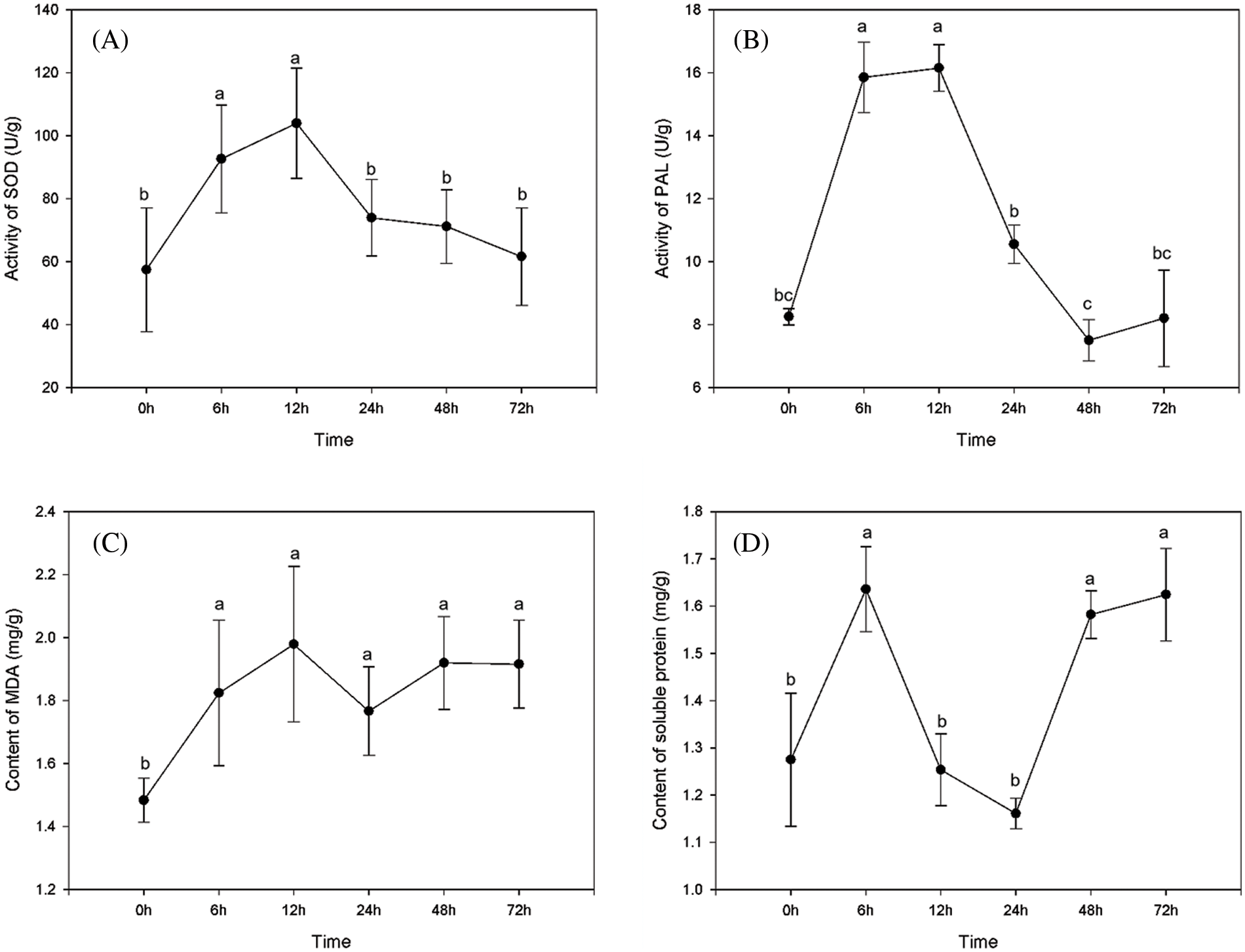
Figure 2: Changes in physiological and biochemical indices in M239 within 72 h post-inoculation with V. dahliae. (A) SOD; (B) PAL; (C) MDA; (D) Soluble protein. Each point on the graph represents the average of 3 replicates, and the vertical lines on each point are standard deviations. Different lowercase letters show significant level at 0.05
The above results showed that the physiological and biochemical characteristics of M239 after inoculation with V. dahliae changed to varying degrees. In addition, 12 and 48 hpi appeared as inflection points in many indices, suggesting that they were two important plant disease interaction time points. Therefore, 12 and 48 hpi were used as the key time points for subsequent transcriptome sequencing.
3.2 Transcriptional Response of M239 Inoculated with V. dahliae
3.2.1 Transcriptome Sequencing and Splicing
The output of the sequencing data is shown in Table 2. An average of 51, 343, 125 clean reads from 9 databases were obtained by transcriptome sequencing of M239 roots at 0, 12 and 48 h post-inoculation with V. dahliae, and its average comparison rate with the eggplant reference genome was 89.61% (Table 3). The raw sequencing data have been stored in the SRA (Sequence Read Archive) database (https://www.ncbi.nlm.nih.gov/sra/PRJNA495025), and the accession number is PRJNA495025.
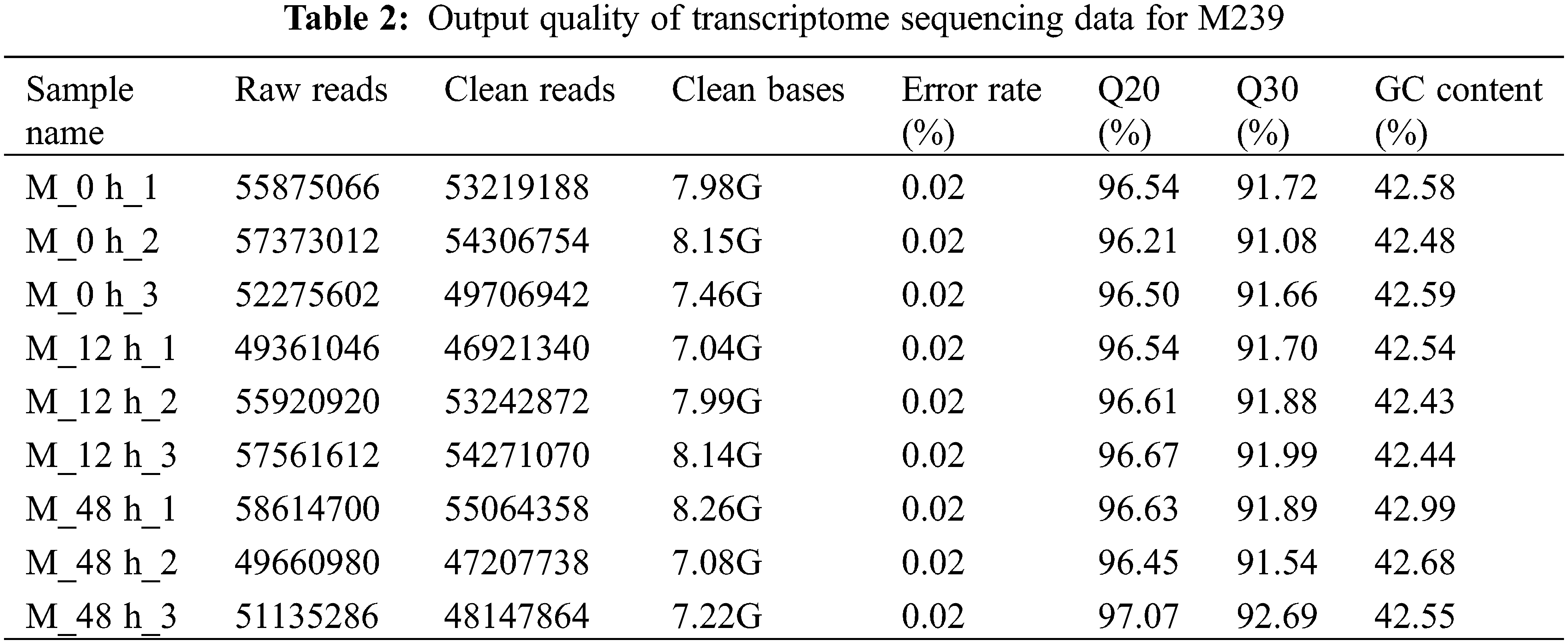
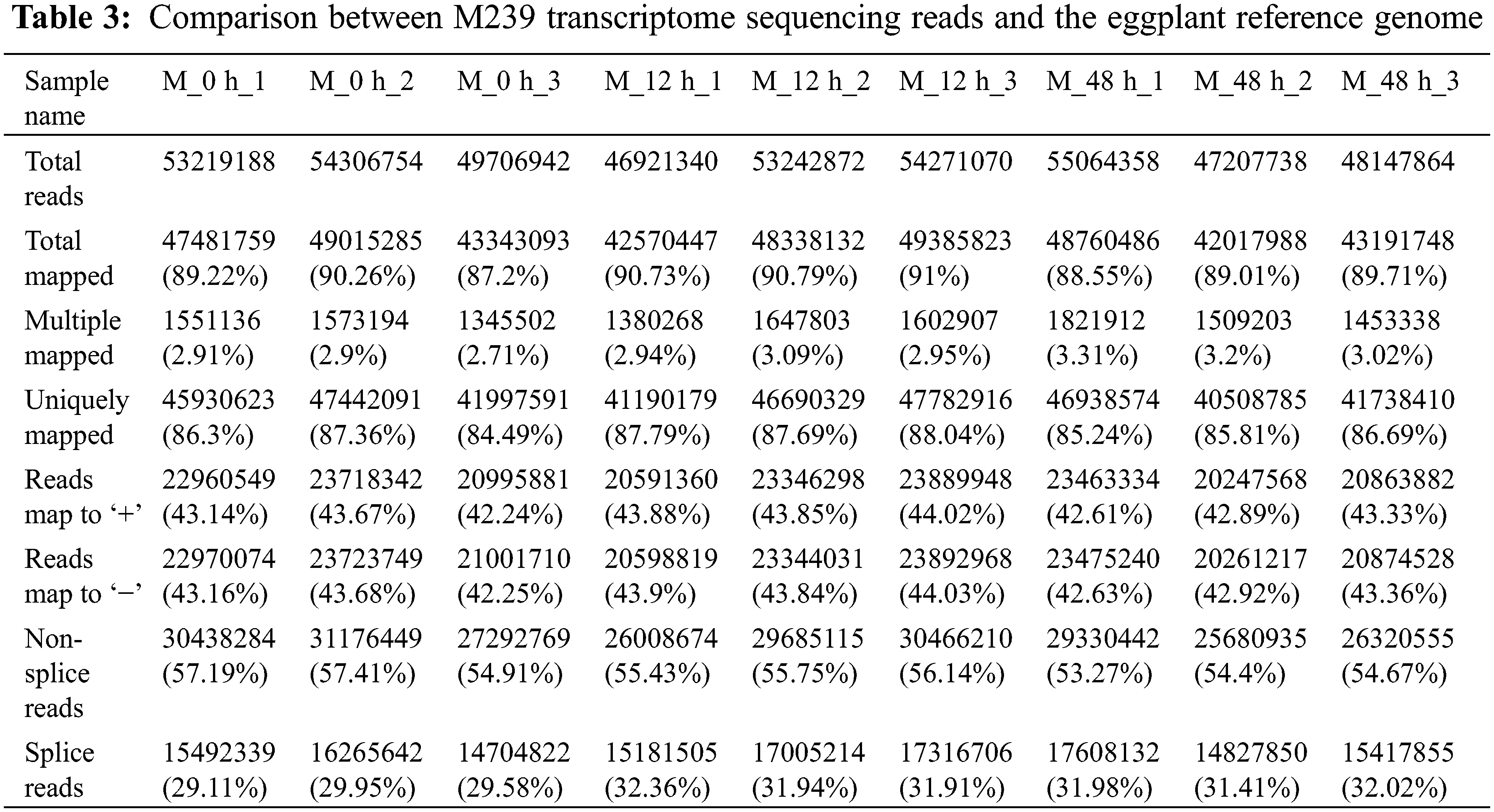
Fig. 3 shows that there were 6,141 DEGs in M239 at 12 hpi compared with 0 hpi (the first stage, M_12 h vs. M_0 h, the same below), of which 3,046 were upregulated and 3,095 were downregulated. Compared with 12 hpi (the second stage, M_48 h vs. M_12 h, the same below), there were 1,903 DEGs at 48 hpi, of which 792 were upregulated and 1,111 were downregulated. A total of 1,261 DEGs appeared at the same time in both stages. Compared with 0 hpi (M_48 h vs. M_0 h, the same below), there were 3,329 DEGs, of which 1,272 were upregulated and 2,057 were downregulated.
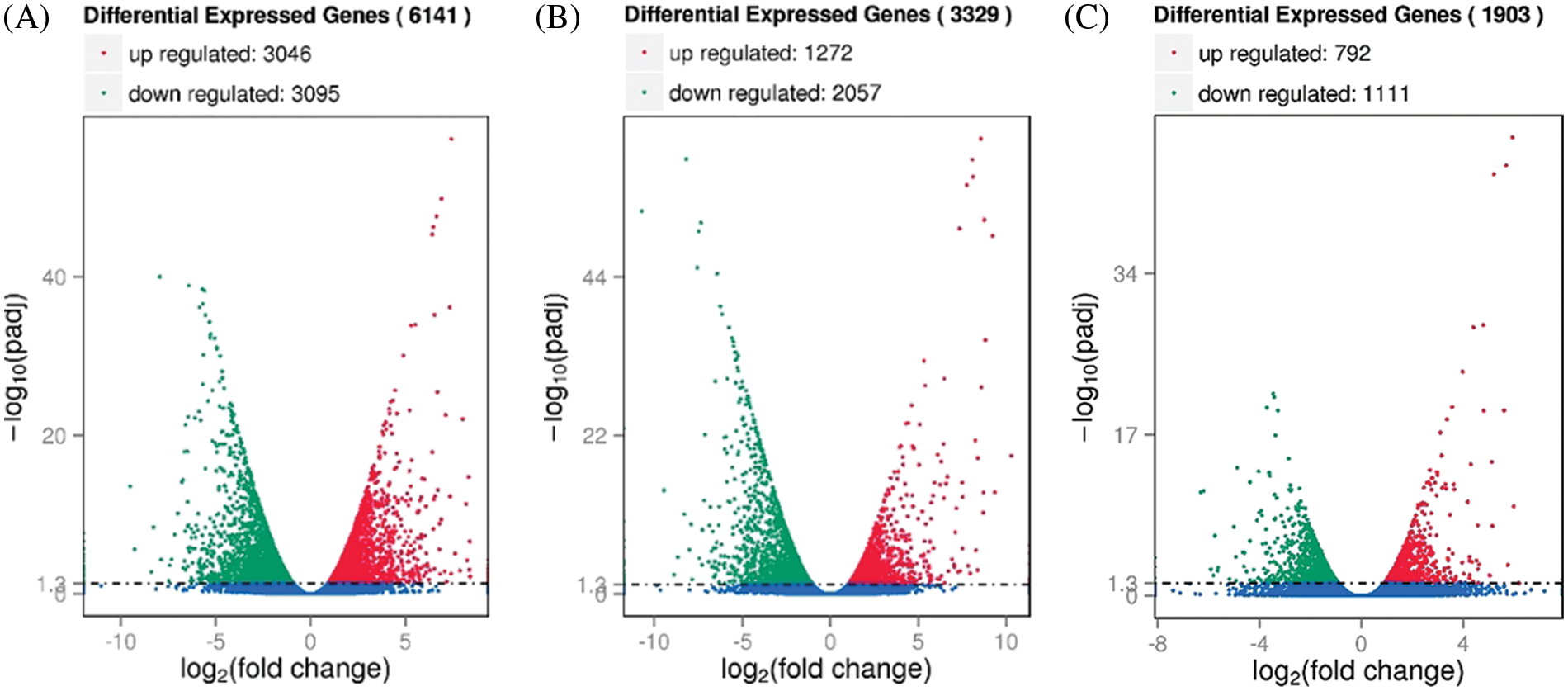
Figure 3: Volcano map of differentially expressed genes (DEGs) in M239. (A) M_12 h vs. M_0 h; (B) M_48 h vs. M_0 h; (C) M_48 h vs. M_12 h
3.2.3 KEGG Enrichment Pathway Analysis of DEGs
After M239 was inoculated with V. dahliae, the KEGG enrichment pathways (q value < 0.05) of DEGs were less and more related to the primary metabolism, such as glycolysis, amino acid and ribosome production. Compared with 0 hpi (CK), only one pathway, “ribosome”, was enriched at 12 hpi (Fig. 4A), and only one pathway, “glycolysis/gluconeogenesis”, was enriched at 48 hpi (Fig. 4B). Compared with 12 hpi, three pathways, namely, “circadian rhythm-plant”, “brassinosteroid biosynthesis”, and “ribosome biogenesis in eukaryotes”, were enriched at 48 hpi (Fig. 4C).
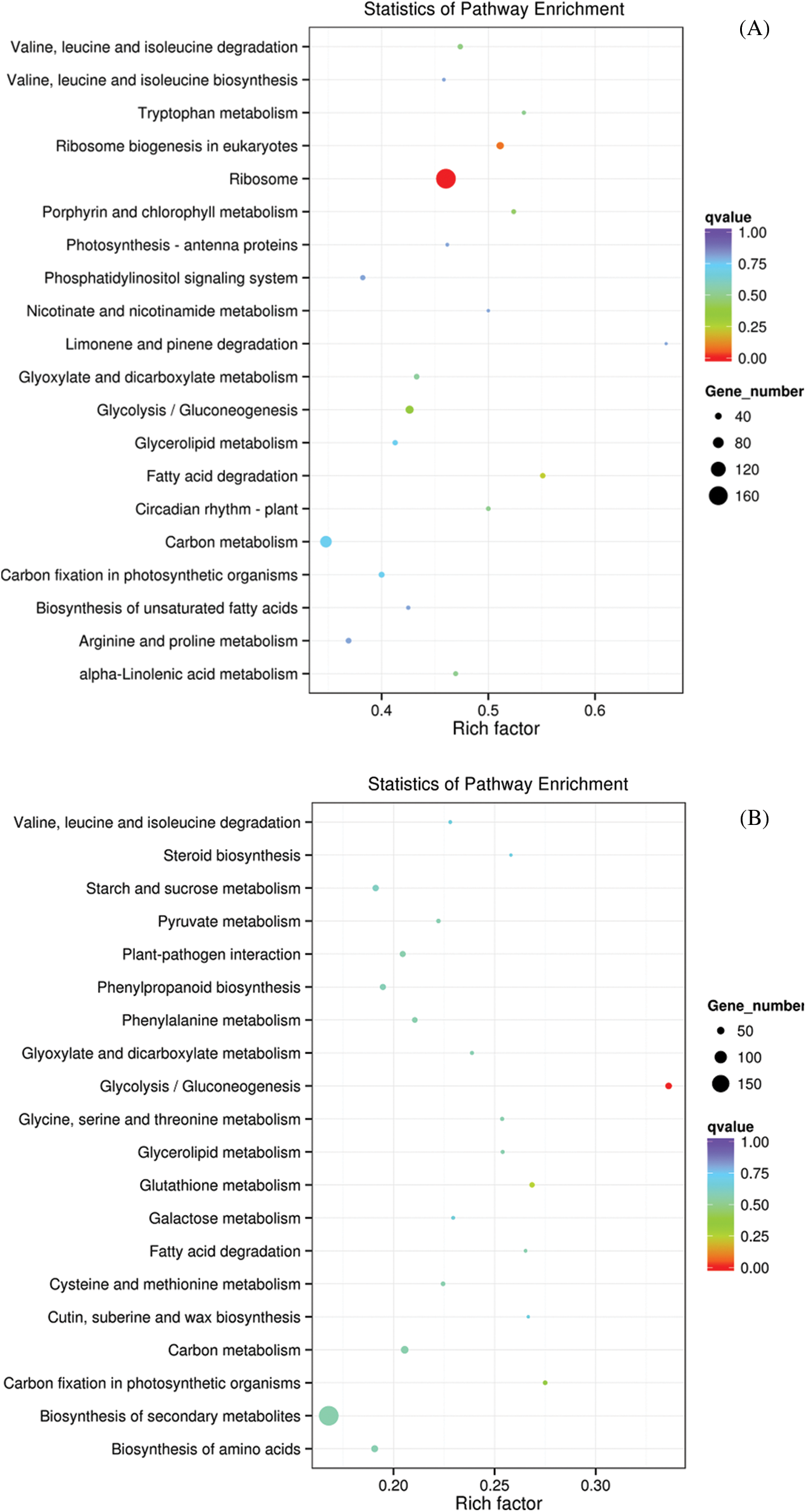
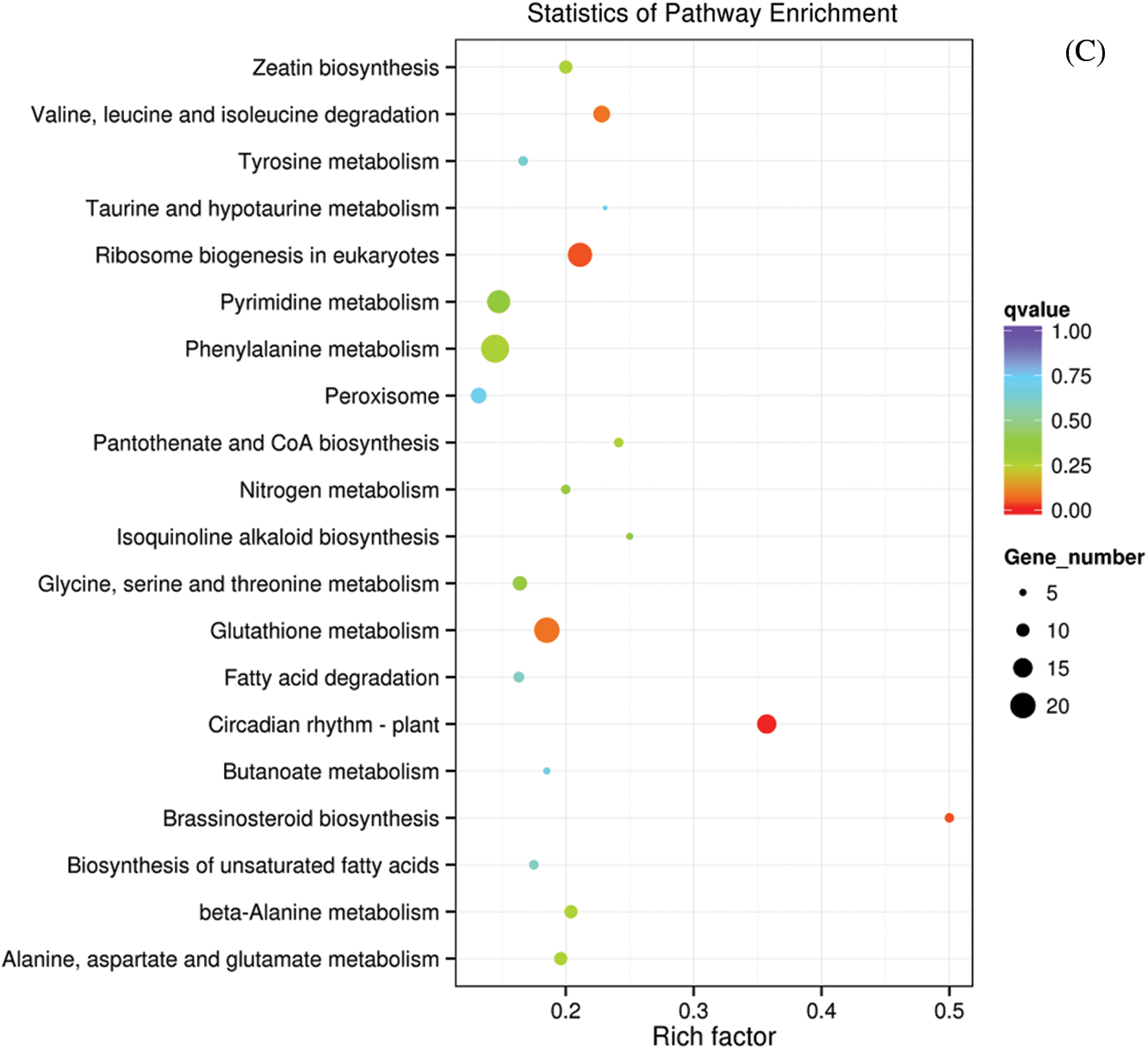
Figure 4: Scatter diagram of KEGG enrichment pathways in M239. (A) M_12 h vs. M_0 h; (B) M_48 h vs. M_0 h; (C) M_48 h vs. M_12 h
3.2.4 Screening of Resistance Genes to Verticillium Wilt
Verticillium Wilt Resistance Gene (Ve) and Pathogenesis-Related Protein (PR) Genes
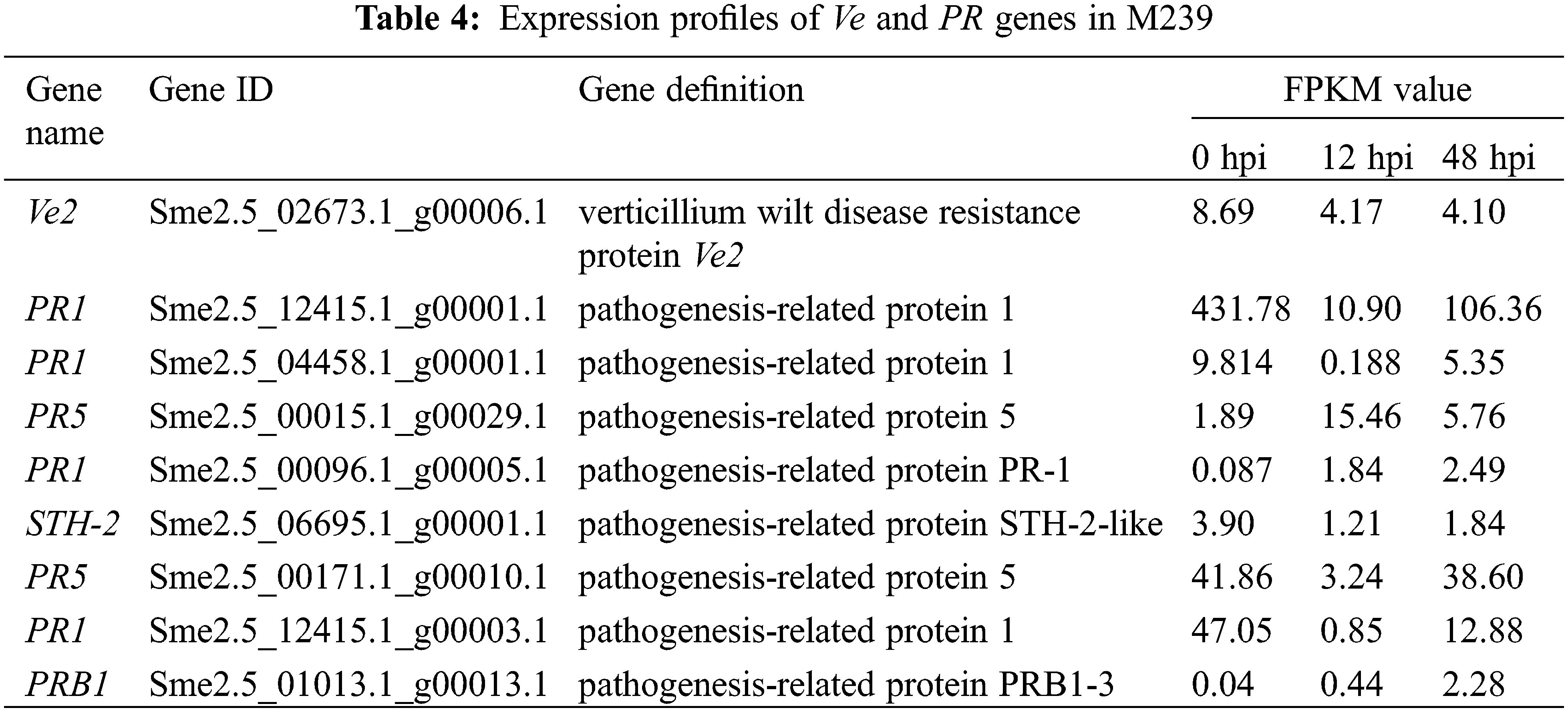
In M239 (Table 4), one gene was compared as Ve2 (sme2.5_02673.1_g00006.1) by the NR (non-redundant protein sequence) database. After BLAST through NCBI, it was found that the identified gene was the most similar (similarity 98.2%) to the sequence of the Verticillium wilt resistance gene SaVe2 in S. aethiopicum (accession number: AY598746.1). At the same time, eight PR genes were screened from the DEGs of M239. Among them, four genes encoded PR1 proteins, two genes encoded PR5 proteins, and two genes encoded STH-2 and PRB1 proteins, respectively.
Transcription Factors (TFs)
Through the comparison of DEGs, we found that M239 had 537 and 277 TFs in the first stage (Fig. 5A) and the second stage (Fig. 5B), which were classified into 58 and 50 gene families, respectively. After inoculation, MYB, AP2-EREBP, WRKY, bHLH, NAC, and Orphans were the most enriched gene families (Tables S1 and S2). The main TFs in the two stages were relatively similar, and MYB, AP2-EREBP, bHLH, NAC, and Orphans appeared in large numbers in the two stages.
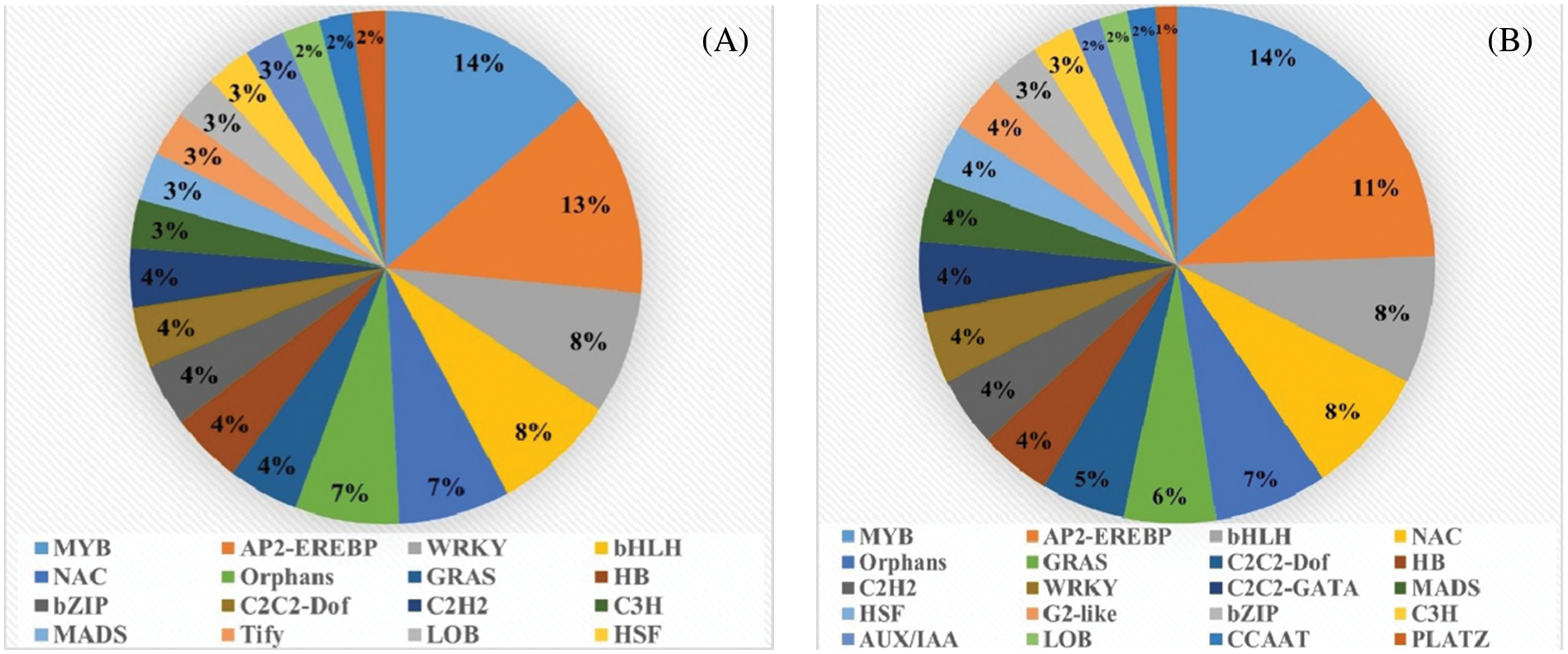
Figure 5: Classification of transcription factors in the differentially expressed genes (DEGs). (A) M_12 h vs. M_0 h, (B) M_48 h vs. M_12 h
The validation results showed that some gene expression trends of M239 identified by qRT–PCR were consistent with those in the transcriptome data (Fig. 6), indicating that the results of this transcriptome sequencing are accurate and reliable and can be used for further analysis. Importantly, these validated genes belonged to three KEGG pathways (Table 5), namely, “plant hormone signal transduction”, “plant-pathogen interaction”, and “phenylpropanoid biosynthesis”, which are important pathways in response to biotic stress. Meanwhile, some candidate genes (PR1, Ve2, HSP90) were validated (Table 5).
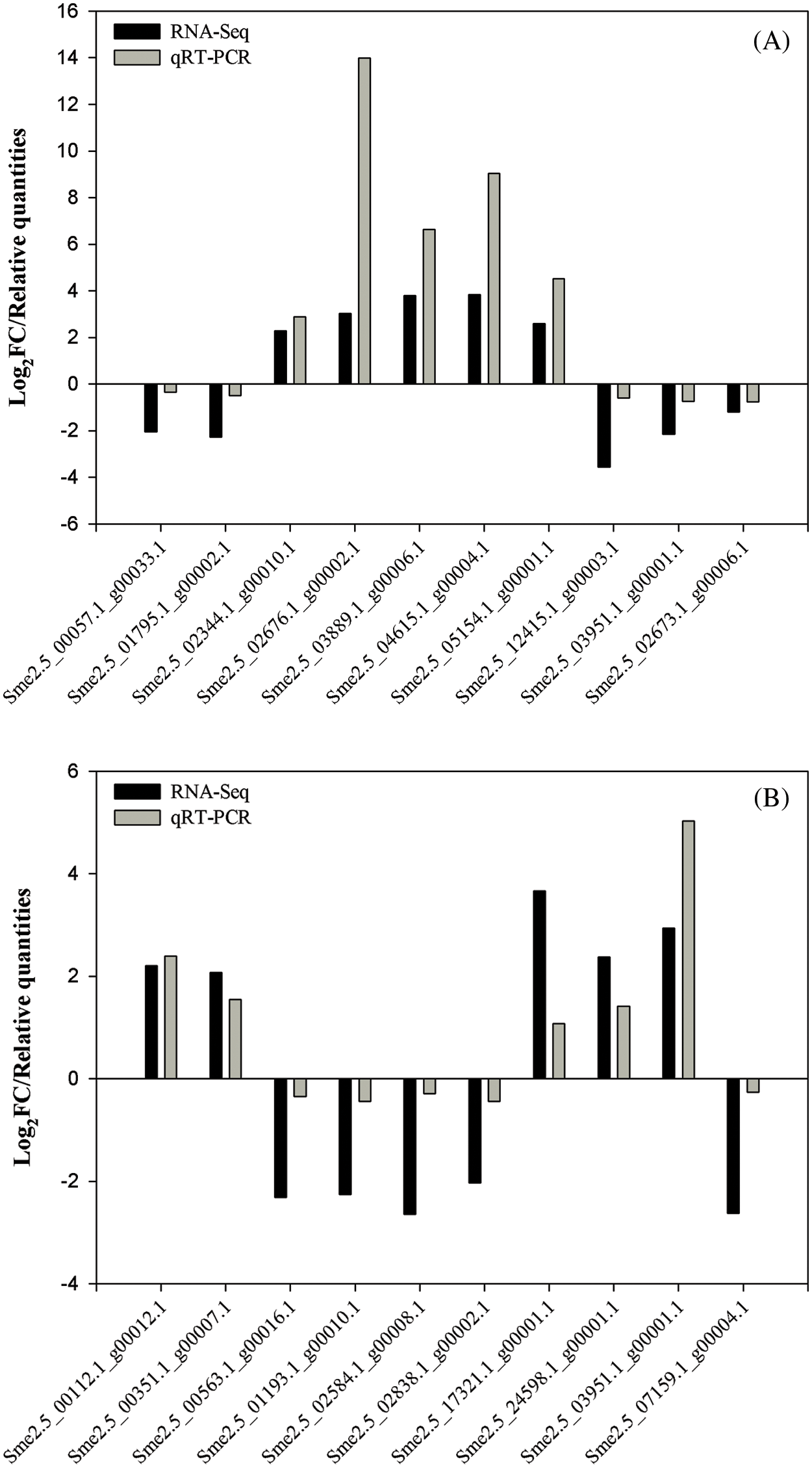
Figure 6: qRT –PCR validation for some differentially expressed genes (DEGs). (A) M_12 h vs. M_0 h; (B) M_48 h vs. M_12 h

Molecular research on plant resistance to Verticillium wilt has been carried out in tomato [25], potato [26] and tobacco [27]. However, the process of plant response to Verticillium wilt infection involves many factors and multiple resistance genes, and the information provided by these studies is insufficient to clarify the molecular mechanism of eggplant defence against V. dahliae infection. In addition, the current research mainly focuses on Verticillium wilt-resistant materials, such as S. torvum, S. sisymbriifolium, and other wild eggplant species, and there has been little research on Verticillium wilt susceptible materials. The significant differences in genetic background and growth environment between the wild and cultivated eggplants have greatly interfered with related research at the transcriptional level. Screening wild eggplant materials with similar growth environments and similar phenotypes vs. the cultivated eggplant can reduce the difference between them to a certain extent and build a bridge between the wild and cultivated eggplants.
In this study, we used M239, a material highly susceptible to Verticillium wilt, as the transcriptome material to analyse the DEGs, KEGG pathways and resistance related genes in its roots at different times within 48 h post-inoculation with V. dahliae, and compared the results with the reported disease-resistant materials S. aculeatissimum [17] and S. sisymbriifolium [18].
4.1 Analysis of DEGs and Their KEGG Enrichment Pathways
In their long-term coevolution with pathogens, plants have developed a series of disease resistance responses to inhibit the infection of pathogens. The resistance response is mainly composed of two innate immunity pathways, namely, PTI (pattern-associated molecular patterns, PAMP-triggered immunity) and ETI (effector-triggered immunity) [28]. From the analysis results of DEGs in this study, it can be seen that DEGs in each stage of M239 were mainly downregulated genes. In the study of Zhou et al. [17], 11,696 DEGs were upregulated and 5,949 were downregulated at 72 hpi. In a previous transcriptome study on S. sisymbriifolium inoculated with V. dahliae [18], there were significantly more upregulated (4,137) than downregulated DEGs (2,922). After the resistant wild eggplant material was inoculated with V. dahliae, more genes participated in the response, which showed that more genes were significantly upregulated, so as to prevent the invasion of pathogens. After the susceptible material M239 was inoculated with V. dahliae, due to its innate immune ability, some genes participated in the resistance response and showed differential expression. However, compared with the resistant material, M239 lacked the positive response of some genes, showing that most genes were downregulated.
In addition, in the plant disease resistance response, secondary metabolites can be used as biochemical barriers to prevent pathogen infection and as signal substances participating in disease resistance [29]. In the KEGG enrichment analysis of DEGs in this study, it was found that only a few metabolic pathways in M239 reached the enrichment level, among which ribosom-related metabolic pathways such as “ribosome”, “ribosome biogenesis in eukaryotes”, “glycolysis/gluconeogenesis”, and “ribosome biogenesis in eukaryotes” were the most enriched. In the resistant materials of S. aculeatissimum [17], “metabolic pathways”, “biosynthesis of secondary metabolites”, “plant pathogen interaction”, and “plant hormone signal transmission” were the four most enriched metabolic pathways. In S. sisymbriifolium [18], the most enriched pathways were “plant hormone signal transduction”, “phenylpropanoid biosynthesis”, “plant pathogen interaction”, and “taurine and taurine metabolism”.
Through the comparison of the above transcriptome data, it was found that after V. dahliae inoculation, many metabolic pathways were enriched in the Verticillium wilt-resistant materials S. aculeatissimum and S. sisymbriifolium; these mainly included the secondary metabolic pathways related to disease resistance, such as plant disease interaction, plant hormone signal transduction and secondary metabolite synthesis. In the susceptible material M239, the enriched metabolic pathways were increasingly less related to primary metabolism, such as sugar metabolism, and amino acid and ribosome formation.
It is known that plant disease interactions are one of the most important pathways in the defence response of plants to pathogens. Several studies have shown that many genes of the plant disease interaction pathway are involved in the interaction process between plants and V. dahliae [18,30–32]. In addition, plant hormones are important signalling molecules that regulate the development process of plants; they regulate plants in various biotic and abiotic stress responses by forming signal networks [33]. Studies have shown that jasmonic acid (JA)-mediated disease resistance signal transduction plays a key role in the interaction between cotton and V. dahliae [34,35]. Lignin and phytoalexin produced by the phenylpropanoid metabolic pathway also play key roles in the process of plant antibacterial activity. Lignin can enhance the ability of the host cell wall to resist pathogen penetration, while phytoalexin has a direct toxic effect on pathogens [36]. In this study, M239 may be unable to resist the further invasion of pathogens, and finally become susceptible, due to the lack of response of the above key metabolic pathways. The above results proved that plant-pathogen interaction, plant hormone signal transduction and secondary metabolite synthesis are the key disease resistance pathways in the interaction between eggplant and V. dahliae. In addition, the brassinolide synthesis pathway was significantly enriched at 12~48 h after M239 was inoculated with V. dahliae, indicating that this pathway may play an important role in plant innate immune resistance.
Plant resistance genes can mainly prevent pathogen invasion in plants, i.e., R genes, which include NB-LRR (nucleus-binding leucine rich repeat) genes, RLK (receptor-like kinases) genes, and RLP (receptor-like protein) genes [37,38]. Among them, plant receptor-like proteins (RLPs), as a class of cell surface receptors, widely exist in higher plants and play important roles in regulating plant growth and development and the stress response [39]. NB-LRR proteins can directly or indirectly recognize pathogen-specific effectors and stimulate specific immune responses [40,41].
Ve genes encode leucine-rich repeat proteins as cell surface receptors, namely, eLRR-RLPs (leucine-rich repeat receptor-like proteins), which are the most reported important genes related to Verticillium wilt resistance [4–6,9,11–13]. The Ve gene was first isolated and cloned from tomato (S. lycopersicum) and can effectively resist V. dahliae race 1. This gene locus includes two members, Ve1 and Ve2 [5]. Previous studies have shown that Ve1 rather than Ve2 plays a major role in resistance to V. dahliae race 1 in tomato [4–6]. In potato, Ve1 and Ve2 were both found to have Verticillium wilt resistance [42,43]. In addition, three homologous Ve genes, mVe1, Vr1, and Gbve1, were proven to play major roles in resistance to Verticillium wilt in mint, lettuce, and cotton, respectively [9,44,45]. However, it is not clear whether Ve1 or Ve2 is the main resistance gene in the resistance process of eggplant Verticillium wilt.
In this study, there was only a Ve2 homologous gene in M239. However, the Ve1 and Ve2 genes were both transcriptionally expressed in the resistant materials S. torvum [13] and S. sisymbriifolium [18]. Therefore, compared with Ve2, Ve1 may play a key role in wild eggplant Verticillium wilt resistance, though this speculation still needs further verification.
Plant pathogenesis-related proteins (PRs) are a class of proteins produced by plants induced by biotic or abiotic stress and are related to the response of plants to stress. Studies have shown that PR1, PR2, PR5 and PR10 play a wide range of roles in biotic stress [46–48] and are often used as indicator genes in plant disease resistance response and systemic acquired resistance (SAR) [49–51]. Among them, PR1 has been proven to have antiviral diffusion functions, limiting fungal invasion and protecting plants against stress [52]. PR5, also known as thaumatin-like protein, can destroy the integrity of the fungal cell wall by changing the permeability of the cell membrane and then inhibit fungal growth [53,54]. PR10 is a ribozyme, and studies have shown that it protects plants against fungi [48,55].
In this study, the differential expression of PR1 and PR5 was detected in M239. However, in the transcriptome sequencing of S. aculeatissimum [17] and S. sisymbriifolium [18], PR1, PR5 and PR10 were found to be simultaneously differentially expressed, and PR10 showed a significant upwards trend within 48 h after S. sisymbriifolium was inoculated with V. dahliae. The PR1, PR5 and PR10 proteins may all have certain functions in the interaction between the wild eggplant and V. dahliae, and the PR10 protein may play a more critical role in this process.
TFs, as important upstream regulatory proteins, play important roles in plant pathogen interactions [56]. Among them, bZIP, NAC, MYB, AP2-EREBPE and WRKY are common transcription factor families in plants. These transcription factors usually act as positive or negative regulatory factors in plant hormone-mediated disease resistance and stress signal regulation [3,57]. WRKY transcription factors are one of the largest transcription factor families in higher plants and play an important role in regulating the plant response to stress [31,58]. In addition, bHLH transcription factors often combine with the MBY family to form complexes and regulate the downstream expression of target genes together [59].
In this study, after M239 was inoculated with V. dahliae, six TF families MYB, AP2-EREBP, WRK, bHLH, NAC, and Orphans, were all detected at different time periods. In S. sisymbriifolium, MYB, AP2-EREBP, Orphan, and bHLH were the top six TF families with the largest numbers, indicating that these four types of transcription factors may play a basic regulatory role in the interaction between the wild eggplant and V. dahliae. In addition, the WRKY family had more DEGs within 48 h after S. sisymbriifolium was inoculated with V. dahliae, while in M239, WRKY genes were differentially expressed only at the initial stage of infection (0~12 h post-inoculation). It is suggested that WRKY transcription factors may also play an important role in transcriptional regulation of wild eggplant resistance to Verticillium wilt.
In this study, we screened and analyzed DEGs, KEGG enrichment pathways and key resistance genes in the roots of M239, a highly susceptible material to Verticillium wilt, within 48 h after inoculation with V. dahliae by transcriptome sequencing, and compared the results with the reportedly resistant wild eggplant materials (S. aculeatissimum and S. sisymbriifolium). It was found that in the process of responding to V. dahliae, the susceptible material M239 had fewer genes and metabolic pathways involved in the defence response, lacked the responses of some known key disease resistance genes and metabolic pathways, failed to form an effective defence network, and ultimately became susceptible to infection.
The above results are helpful to better understand the interaction process between the wild eggplant and V. dahliae, and provide some ideas for further understanding the molecular mechanism of plant resistance to Verticillium wilt. However, further research is needed due to the complex diversity of plant Verticillium wilt resistance mechanisms, and the significant difference in the genetic backgrounds of different materials.
Authorship: Liyan Wu and Guanghui Du designed the experiments. Jie Cheng, Yaju Gong, Rui Bao, Zhibin Li and Min Gui conducted the experiments and analyzed the data. Liyan Wu and Guanghui Du wrote the manuscript.
Availability of Data and Materials: The raw sequencing data in this study were deposited at the NCBI Sequence Read Archive (SRA) Database (Accession No. PRJNA495025).
Funding Statement: This work was supported by the National Natural Science Foundation of China (31960594), the Yunnan Fundamental Research Projects (202201AT070074, 2019FB059), and the Yunnan Technical Innovation Talent Training Project (202205AD160029).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Lian, Y., Liu, F. Z., Chen, Y. H. (2006). Chinese eggplant cultivars (Solanum melogena L.) distribution and germplasm resources research advancement. China Vegetables, 9–14 (in Chinese). [Google Scholar]
2. Wu, L. Y., Guo, Z. X., Zeng, L., Bao, R., Li, Z. B. et al. (2017). Resistance identification of Yunnan wild eggplant resources to Verticillium wilt. Journal of Plant Genetic Resources, 18(6), 1046–1054 (in Chinese). [Google Scholar]
3. Scholz, S. S., Schmidt-Heck, W., Guthke, R., Furch, A. C. U., Reichelt, M. et al. (2018). Verticillium dahliae-Arabidopsis interaction causes changes in gene expression profiles and jasmonate levels on different time scales. Frontiers in Microbiology, 9, 217. DOI 10.3389/fmicb.2018.00217. [Google Scholar] [CrossRef]
4. Diwan, N., Fluhr, R., Eshed, Y., Zamir, D., Tanksley, S. D. (1999). Mapping of Ve in tomato: A gene conferring resistance to the broad-spectrum pathogen, Verticillium dahliae race 1. Theoretical and Applied Genetics, 98(2), 315–319. DOI 10.1007/s001220051075. [Google Scholar] [CrossRef]
5. Kawchuk, L. M., Hachey, J., Lynch, D. R., Kulcsar, F., van Rooijen, G.et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proceedings of the National Academy of Sciences, 98(11), 6511–6515. DOI 10.1073/pnas.091114198. [Google Scholar] [CrossRef]
6. Fradin, E. F., Zhang, Z., Juarez Ayala, J. C., Castroverde, C. D. M., Nazar, R. N. et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiology, 150(1), 320–332. DOI 10.1104/pp.109.136762. [Google Scholar] [CrossRef]
7. Xu, L., Zhu, L., Tu, L., Liu, L., Yuan, D. et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. Journal of Experimental Botany, 62(15), 5607–5621. DOI 10.1093/jxb/err245. [Google Scholar] [CrossRef]
8. Sun, Q., Jiang, H., Zhu, X., Wang, W., He, X. et al. (2013). Analysis of sea-island cotton and upland cotton in response to Verticillium dahliae infection by RNA sequencing. BMC Genomics, 14(1), 852. DOI 10.1186/1471-2164-14-852. [Google Scholar] [CrossRef]
9. Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T. et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One, 7(12), e51091. DOI 10.1371/journal.pone.0051091. [Google Scholar] [CrossRef]
10. Zhang, W. W., Jiang, T. F., Cui, X., Qi, F. J., Jian, G. L. (2013). Colonization in cotton plants by a green fluorescent protein labelled strain of Verticillium dahliae. European Journal of Plant Pathology, 135(4), 867–876. DOI 10.1007/s10658-012-0131-1. [Google Scholar] [CrossRef]
11. Yang, Y., Ling, X., Chen, T., Cai, L., Liu, T. et al. (2015). A cotton Gbvdr5 gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Molecular Biology Reporter, 33(4), 987–1001. DOI 10.1007/s11105-014-0810-5. [Google Scholar] [CrossRef]
12. Chai, Y., Zhao, L., Liao, Z., Sun, X., Zuo, K. et al. (2003). Molecular cloning of a potential Verticillium dahliae resistance gene SlVe1 with multi-site polyadenylation from Solanum licopersicoides. DNA Sequence, 14(5), 375–384. DOI 10.1080/10425170310001605509. [Google Scholar] [CrossRef]
13. Fei, J., Chai, Y., Wang, J., Lin, J., Sun, X. et al. (2004). cDNA cloning and characterization of the Ve homologue gene StVe from Solanum torvum Swartz. DNA Sequence, 15(2), 88–95. DOI 10.1080/1042517042000199942. [Google Scholar] [CrossRef]
14. Wang, Z., Yang, Q., Wang, R., Zheng, Y. H. (2012). Optimization and establishment of cDNA-AFLP technical system in Solanum melongena. Northern Horticulture, (19), 108–112 (in Chinese). [Google Scholar]
15. Yang, L., Jue, D., Li, W., Zhang, R., Chen, M. et al. (2013). Identification of miRNA from eggplant (Solanum melongena L.) by small RNA deep sequencing and their response to Verticillium dahliae infection. PLoS One, 8(8), e72840. DOI 10.1371/journal.pone.0072840. [Google Scholar] [CrossRef]
16. Yang, X., Cheng, Y. F., Deng, C., Ma, Y., Wang, Z. W. et al. (2014). Comparative transcriptome analysis of eggplant (Solanum melongena L.) and turkey berry (Solanum torvum Sw.Phylogenomics and disease resistance analysis. BMC Genomics, 15(1), 412. DOI 10.1186/1471-2164-15-412. [Google Scholar] [CrossRef]
17. Zhou, X., Bao, S., Liu, J., Zhuang, Y. (2016). De novo sequencing and analysis of the transcriptome of the wild eggplant species Solanum aculeatissimum in response to Verticillium dahliae. Plant Molecular Biology Reporter, 34(6), 1193–1203. DOI 10.1007/s11105-016-0998-7. [Google Scholar] [CrossRef]
18. Wu, L., Du, G., Bao, R., Li, Z., Gong, Y. et al. (2019). De novo assembly and discovery of genes involved in the response of Solanum sisymbriifolium to Verticillium dahliae. Physiology & Molecular Biology of Plants, 25(4), 1009–1027. DOI 10.1007/s12298-019-00666-4. [Google Scholar] [CrossRef]
19. Zhuang, Y., Wang, S. B. (2009). Identification and inheritance of Verticillium wilt resistance in eggplant related species. Jiangsu Journal of Agricultural Sciences, 25(4), 847–850 (in Chinese). [Google Scholar]
20. Zheng, D. S., Gao, A. N., Li, L. H., Liu, X. (2013). Wild relatives of agricultural crop in Yunnan province and its peripheral area. Journal of Plant Genetic Resources, 14(2), 193–201 (in Chinese). [Google Scholar]
21. Wu, L. Y., Du, G. H., Bao, R., Li, Z. B., Gong, Y. J. (2018). Classification and genetic diversity of wild eggplant resources in Yunnan. Chinese Journal of Tropical Crops, 39(6), 275–280 (in Chinese). [Google Scholar]
22. Hirakawa, H., Shirasawa, K., Miyatake, K., Nunome, T., Negoro, S. et al. (2014). Draft genome sequence of eggplant (Solanum melongena L.The representative Solanum species indigenous to the old world. DNA Research, 21(6), 649–660. DOI 10.1093/dnares/dsu027. [Google Scholar] [CrossRef]
23. Mao, X., Cai, T., Olyarchuk, J. G., Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21(19), 3787–3793. DOI 10.1093/bioinformatics/bti430. [Google Scholar] [CrossRef]
24. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
25. Faino, L., de Jonge, R.,Thomma, B. P. H. J. (2012). The transcriptome of Verticillium dahliae-infected Nicotiana benthamiana determined by deep RNA sequencing. Plant Signaling & Behavior, 7(9), 1065–1069. DOI 10.4161/psb.21014. [Google Scholar] [CrossRef]
26. Derksen, H., Badawi, M., Henriquez, M. A., Yao, Z., El-Bebany, A. F. et al. (2013). Differential expression of potato defence genes associated with the salicylic acid defence signalling pathway in response to weakly and highly aggressive isolates of Verticillium dahliae. Journal of Phytopathology, 161(3), 142–153. DOI 10.1111/jph.12038. [Google Scholar] [CrossRef]
27. Chen, J. Y., Huang, J. Q., Li, N. Y., Ma, X. F., Wang, J. L. et al. (2015). Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biology, 15(1), 148. DOI 10.1186/s12870-015-0508-3. [Google Scholar] [CrossRef]
28. Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature, 444(7117), 323–329. DOI 10.1038/nature05286. [Google Scholar] [CrossRef]
29. Bednarek, P., Osbourn, A. (2009). Plant-microbe interactions: Chemical diversity in plant defense. Science, 324(5928), 746–748. DOI 10.1126/science.1171661. [Google Scholar] [CrossRef]
30. Ekengren, S. K., Liu, Y., Schiff, M., Dinesh-Kumar, S. P., Martin, G. B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. The Plant Journal, 36(6), 905–917. DOI 10.1046/j.1365-313x.2003.01944.x. [Google Scholar] [CrossRef]
31. Guo, S., Zuo, Y., Zhang, Y., Wu, C., Su, W. et al. (2017). Large-scale transcriptome comparison of sunflower genes responsive to Verticillium dahliae. BMC Genomics, 18(1), 42. DOI 10.1186/s12864-016-3386-7. [Google Scholar] [CrossRef]
32. Zhang, W., Zhang, H., Liu, K., Jian, G., Qi, F. et al. (2017). Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis. PLoS One, 12(8), e0181609. DOI 10.1371/journal.pone.0181609. [Google Scholar] [CrossRef]
33. Bari, R., Jones, J. D. G. (2009). Role of plant hormones in plant defence responses. Plant Molecular Biology, 69(4), 473–488. DOI 10.1007/s11103-008-9435-0. [Google Scholar] [CrossRef]
34. Pieterse, C. M. J., Leon-Reyes, A., van der Does, D., Verhage, A., Koornneef, A. et al. (2012). Networking by small-molecule hormones in plant immunity. IOBC-WPRS Bulletin, 83, 77–80. [Google Scholar]
35. Gao, W., Long, L., Zhu, L. F., Xu, L., Gao, W. H. et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Molecular & Cellular Proteomics, 12(12), 3690–3703. DOI 10.1074/mcp.M113.031013. [Google Scholar] [CrossRef]
36. Naoumkina, M. A., Zhao, Q., Gallego-Giraldo, L., Dai, X., Zhao, P. X. et al. (2010). Genome-wide analysis of phenylpropanoid defence pathways. Molecular Plant Pathology, 11(6), 829–846. DOI 10.1111/j.1364-3703.2010.00648.x. [Google Scholar] [CrossRef]
37. Kruijt, M., Brandwagt, B. F., de Wit, P. J. M. (2004). Rearrangements in the Cf-9 disease resistance gene cluster of wild tomato have resulted in three genes that mediate Avr9 responsiveness. Genetics, 168(3), 1655–1663. DOI 10.1534/genetics.104.028985. [Google Scholar] [CrossRef]
38. Yang, L., Mu, X., Liu, C., Cai, J., Shi, K. et al. (2015). Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. Journal of Integrative Plant Biology, 57(12), 1078–1088. DOI 10.1111/jipb.12348. [Google Scholar] [CrossRef]
39. Wu, J., Liu, Z., Zhang, Z., Lv, Y., Yang, N. et al. (2016). Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. Journal of Experimental Botany, 67(11), 3339–3351. DOI 10.1093/jxb/erw152. [Google Scholar] [CrossRef]
40. Nürnberger, T., Brunner, F., Kemmerling, B., Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunological Reviews, 198(1), 249–266. DOI 10.1111/j.0105-2896.2004.0119.x. [Google Scholar] [CrossRef]
41. Zhang, J., Zhou, J. M. (2010). Plant immunity triggered by microbial molecular signatures. Molecular Plant, 3(5), 783–793. DOI 10.1093/mp/ssq035. [Google Scholar] [CrossRef]
42. Bae, J. J., Halterman, D., Jansky, S. (2008). Development of a molecular marker associated with Verticillium wilt resistance in diploid interspecific potato hybrids. Molecular Breeding, 22(1), 61–69. DOI 10.1007/s11032-008-9156-8. [Google Scholar] [CrossRef]
43. Meier, A., Halterman, D. A. (2016). Structural variation within the potato Ve gene locus and correlation with molecular marker analysis. Crop Science, 56(6), 3133–3142. DOI 10.2135/cropsci2016.02.0137. [Google Scholar] [CrossRef]
44. Vining, K., Davis, T. (2009). Isolation of a Ve homolog, mVe1, and its relationship to verticillium wilt resistance in Mentha longifolia (L.) Huds. Molecular Genetics and Genomics, 282(2), 173–184. DOI 10.1007/s00438-009-0454-6. [Google Scholar] [CrossRef]
45. Hayes, R. J., McHale, L. K., Vallad, G. E., Jose Truco, M., Michelmore, R. W. et al. (2011). The inheritance of resistance to Verticillium wilt caused by race 1 isolates of Verticillium dahliae in the lettuce cultivar La Brillante. Theoretical and Applied Genetics, 123(4), 509–517. DOI 10.1007/s00122-011-1603-y. [Google Scholar] [CrossRef]
46. van Ooijen, G.,van den Burg, H. A., Cornelissen, B. J. C., Takken, F. L. W. (2007). Structure and function of resistance proteins in solanaceous plants. Annual Review of Phytopathology, 45(1), 43–72. DOI 10.1146/annurev.phyto.45.062806.094430. [Google Scholar] [CrossRef]
47. van der Hoorn, R. A. L., Kamoun, S. (2008). From guard to decoy: A new model for perception of plant pathogen effectors. The Plant Cell, 20(8), 2009–2017. DOI 10.1105/tpc.108.060194. [Google Scholar] [CrossRef]
48. Agarwal, P., Agarwal, P. K. (2014). Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Molecular Biology Reports, 41(2), 599–611. DOI 10.1007/s11033-013-2897-4. [Google Scholar] [CrossRef]
49. van Loon, L. C.,Rep, M., Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology, 44(1), 135–162. DOI 10.1146/annurev.phyto.44.070505.143425. [Google Scholar] [CrossRef]
50. Wang, F. X., Ma, Y. P., Yang, C. L., Zhao, P. M., Yao, Y. (2011). Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics, 11(22), 4296–4309. DOI 10.1002/pmic.201100062. [Google Scholar] [CrossRef]
51. Zhang, W., Zhang, H., Qi, F., Jian, G. (2016). Generation of transcriptome profiling and gene functional analysis in Gossypium hirsutum upon Verticillium dahliae infection. Biochemical and Biophysical Research Communications, 473(4), 879–885. DOI 10.1016/j.bbrc.2016.03.143. [Google Scholar] [CrossRef]
52. Rauscher, M., Adám, A. L., Wirtz, S., Guggenheim, R., Mendgen, K. et al. (1999). PR-1 protein inhibits the differentiation of rust infection hyphae in leaves of acquired resistant broadbean. The Plant Journal, 19(6), 625–633. DOI 10.1046/j.1365-313x.1999.00545.x. [Google Scholar] [CrossRef]
53. van Loon, L. C.,van Strien, E. A. (1999). The families of pathogen-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiology & Molecular Biology of Plants, 55(2), 85–97. DOI 10.1006/pmpp.1999.0213. [Google Scholar] [CrossRef]
54. Liu, J. J., Sturrock, R., Ekramoddoullah, A. K. M. (2010). The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Reports, 29(5), 419–436. DOI 10.1007/s00299-010-0826-8. [Google Scholar] [CrossRef]
55. Park, C. J., Kim, K. J., Shin, R., Park, J. M., Shin, Y. C. (2004). Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant Journal, 37(2), 186–198. DOI 10.1046/j.1365-313x.2003.01951.x. [Google Scholar] [CrossRef]
56. Singh, K. B., Foley, R. C., Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology, 5(5), 430–436. DOI 10.1016/S1369-5266(02)00289-3. [Google Scholar] [CrossRef]
57. Gutterson, N., Reuber, T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology, 7(4), 465–471. DOI 10.1016/j.pbi.2004.04.007. [Google Scholar] [CrossRef]
58. Yang, C. Q., Fang, X., Wu, X. M., Mao, Y. B., Wang, L. J. et al. (2012). Transcriptional regulation of plant secondary metabolism. Journal of Integrative Plant Biology, 54(10), 703–712. DOI 10.1111/j.1744-7909.2012.01161.x. [Google Scholar] [CrossRef]
59. Zhang, Y., Wang, X. F., Ding, Z. G., Ma, Q., Zhang, G. R. et al. (2013). Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics, 14(1), 637. DOI 10.1186/1471-2164-14-637. [Google Scholar] [CrossRef]
Table S1: Transcription factors at M_12 h vs. M_0 h in M239
Table S2: Transcription factors at M_48 h vs. M_12 h in M239
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools