 Open Access
Open Access
ARTICLE
Silencing of the Nonspecific Phospholipase C6 (NPC6) Gene Induces Ricinoleic Acid Accumulation in Castor Seeds
1 College of Life Science and Food, Inner Mongolia Minzu University, Tongliao, 028000, China
2 College of Life Science, Baicheng Normal University, Baicheng, 137000, China
3 College of Life Science, Northeast Forestry University, Harbin, 150040, China
4 Key Laboratory of Castor Breeding of the State Ethnic Affairs Commission, Inner Mongolia Minzu University, Tongliao, 028043, China
5 Inner Mongolia Industrial Engineering Research Center of Universities for Castor, Inner Mongolia Minzu University, Tongliao, 028043, China
6 Inner Mongolia Key Laboratory of Castor Breeding and Comprehensive Utilization, Inner Mongolia Minzu University, Tongliao, 028000, China
7 Inner Mongolia Engineering Research Center of Industrial Technology Innovation of Castor, Inner Mongolia Minzu University, Tongliao, 028000, China
* Corresponding Authors: Fenglan Huang. Email: ; Fanjuan Meng. Email:
# Yong Zhao and Lili Li contributed equally to this work and shared co-first authorship
Phyton-International Journal of Experimental Botany 2023, 92(12), 3237-3250. https://doi.org/10.32604/phyton.2023.031495
Received 23 May 2023; Accepted 12 September 2023; Issue published 28 December 2023
Abstract
Castor, scientifically known as Ricinus communis L., is among the top ten oil crops globally. It is considered a renewable resource and is commonly referred to as ‘green oil’. Castor seeds contain castor oil as their main component, which is predominantly composed of ricinoleic acid. This study utilized RNAi technology to silence the NPC6 gene in NO.2129 castor, resulting in the creation of mutant plants L1 and L2. The weight of 100 dry seed kernels from L1 and L2 exceeds that from NO.2129. The crude fat and ricinoleic acid levels of L1 and L2 were higher than those of NO.2129 at various developmental stages. In the proteomics analysis of 60-day-old castor seeds, a total of 21 differentially expressed proteins were identified, out of which 19 were successfully recognized. Eleven of the differentially expressed proteins identified were legumins, which play a crucial role in nutrient storage within the seed. Silencing the NPC6 gene results in the accumulation of ricinoleic acid in castor seeds. The findings of this study not only enhance our knowledge of NPC6’s role in regulating castor seed oil synthesis but also offer fresh perspectives for investigating oil synthesis and accumulation in other plant species.Keywords
Supplementary Material
Supplementary Material FileCastor (Ricinus communis L.) belongs to the Euphorbiaceae family and is primarily cultivated in China, India, and Brazil. Castor seeds have high economic value because they contain a high-value oil. The ricinoleic acid content in castor oil can reach 89%–92% [1]. Ricinoleic acid is an unusual unsaturated fatty acid found in castor oil, with a hydroxyl group on the C12 atom. Ricinoleic acid is a crucial raw material in the biodiesel, cosmetics, and other industries. Previous studies have shown that ricinoleic acid possesses bactericidal, anti-inflammatory, and anti-herpes properties attributed to the presence of functional groups such as -COOH, -OH, and -C=C [1]. In addition, it can be used to generate the important industrial raw material sebacic acid through a series of chemical reactions [2,3].
Phospholipase C (PLC) is a crucial enzyme that plays a significant role in regulating various plant cell functions. It is responsible for controlling signal transduction, lipid metabolism, growth, and development in plants. The process of PLC hydrolysis plays a significant role in various physiological functions within cells. It involves the hydrolysis of phospholipid molecules, resulting in the formation of sn-1,2-diacylglycerol (DAG) [4,5]. PLC can be divided into two subtypes, namely, phosphoinositide-specific PLC (PI-PLC) and nonspecific PLC (NPC). PI-PLC is evolutionarily conserved and specifically targets the hydrolysis of phosphatidylinositol phosphate (PIP). On the other hand, the hydrolysis of phospholipids by NPC is not specific [6,7]. PI-PLC contains a PH domain, catalytic X-Y domain, and C2 domain, while NPC only contains one phosphatase domain [8]. These differences indicate that NPC may function using different structures. Previous studies have shown that NPC is a calcium-independent phospholipase. Six and five NPC genes were found in Arabidopsis and rice, respectively. In Arabidopsis, NPC plays a crucial role in multiple biological processes [9], such as root development [10], flower development [11], and seed oil production [12]. NPC1 is responsible for the plant response to heat [13], NPC2 plays a role in gametophyte development and glycerol metabolism [14], and NPC3 is involved in root development and brassinolide signal transduction [15]. The study found that Arabidopsis NPC4 is highly effective in breaking down PC and PG, indicating that its acts preferentially on substrates that have these specific acyl chain components [16,17]. Additionally, research has revealed that NPC4 and NPC5 mutants exhibit a decrease in diacylglycerol (DAG) species [17,18]. The NPC6 gene plays a crucial role in various biological processes, such as gametophyte development, glycerolipid metabolism, root development, seed yield, and oil production [19–21]. Overexpression of NPC6 in oilseed Brassicaceae plants results in an increase in seed oil content, seed weight, and yield. Conversely, knockout of NPC6 leads to a decrease in seed oil content and seed size [21].
This study found that NCP6 plays a vital role in regulating the metabolic pathways of oil in seeds. Despite progress made in researching the oil synthesis pathway, the molecular mechanism regulating ricinoleic acid synthesis in castor seeds remains largely unknown. This study investigated the impact of silencing the NPC6 gene on the accumulation of ricinoleic acid in castor seeds. In this study, we used RNAi technology to silence the NPC6 gene in castor plants. The silenced plants were then subjected to biochemical testing to assess the accumulation of fatty acids. To evaluate proteomic differences between mutant and wild-type mature seeds, two-dimensional gel electrophoresis was utilized. The findings of this research will enhance our understanding of the role of NPC6 in regulating castor seed oil synthesis.
Castor NO.2129 was provided by Tongliao Academy of Agricultural Sciences, Inner Mongolia, China. It is widely planted in the Inner Mongolia and Jilin regions of China.
2.1 Construction of the RNAi Vector for the NPC6 Gene
The C1 fragment of the sense orientation of NPC6 (ID: 8269216) was PCR-amplified from castor variety NO.2129 using the primer pair C1-S and C1-X (Supplementary Table 1). The primer C1-S was used to introduce a SpeI restriction site at one end of the fragment. The C2 fragment of NPC6 in antisense orientation was PCR-amplified from castor variety NO.2129 using the primer pair C2-S and C2-X (Supplementary Table 1). To introduce a SmaI restriction site at one end of the fragment, the primer C2-S was utilized. The intron fragment was acquired through double digestion of pHANNIBAL using the HindIII and KpnI restriction enzymes (Supplementary Fig. 1). Similarly, the pBI-121 vector was linearized through the use of the XbaI and SmaI restriction enzymes. The hairpin RNAi vector pBI-C1-IN-C2 was obtained by cloning C1, the intron, and C2 into the pBI-121 vector using T4 DNA ligase.
Agrobacterium GV3101 containing the plasmid pBI-C1-IN-C2 was utilized for the transformation of the cotyledon nodes of castor plant NO.2129. NO.2129 castor seeds were inoculated in germination medium (30 g/L sucrose, 5.6 g/L agar, pH 5.6–5.8). After the seeds had germinated, the cotyledon nodes were cut off and inoculated into culture medium (1/8 MS, 10 g/L agar, 20 g/L saccharose, 8.0 mg/L ZT, 1.0 mg/L NAA, 100 μm/L AS, pH 5.6–5.8) and cultivated for 2 days. The castor bean cotyledon nodes were soaked in Agrobacterium GV3101 for 20 min and then inoculated in screening medium (1/8 MS, 10 g/L agar, 20 g/L saccharose, 8.0 mg/L ZT, 1.0 mg/L NAA, 25 mg/L Kan, pH 5.6–5.8) for 30 days. The surviving castor plants in Kan medium were transplanted into rooting medium (1/16 MS, 10 g/L agar, 20 g/L saccharose, 0.4 mg/L NAA, pH 5.6–5.8). Once the roots grew 2–3 cm, they were into soil. Castor plants were grown in greenhouses under natural sunlight at 28°C–35°C.
2.3 PCR Analysis of Transgenic Castor Plants
The molecular characterization of putative transgenic plants involved PCR analyses. PCR amplification was performed using genomic DNA and the template primers CT-S and CT-X (Supplementary Table 1). The expected product was a 1595 bp fragment from the pBI-C1-IN-C2 vector. In this study, DNA was isolated from the leaves of putative transgenic plants using a genomic DNA extraction kit (Beijing Zoman Biotechnology Co., Ltd., Beijing, China). The PCR (50 μL) mixture contained 200 ng of template DNA, 10 pmol of each primer, and 25 μL of TaKaRa Taq Version 2.0 plus dye (TaKaRa, Beijing, China). The PCR was set up as follows: initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, elongation at 72°C for 60 s, and a final extension step at 72°C for 10 min.
A total plant RNA extraction kit (Beijing Zoman Biotechnology Co., Ltd., Beijing, China) was used to separate RNA from transgenic and wild-type seeds, and the mRNA was purified with an mRNA kit (Qiagen). Determination of the concentration and quality of the mRNA was conducted using a microplate reader (Bio-Rad Microplate Reader 550). A cDNA reverse transcription kit (Takara) was used to reverse transcribe 1000 ng of mRNA to cDNA. The qRT–PCR (50 μL) contained 100 ng of template cDNA, 10 μL of SYBR Premix Ex Taq (Takara), 10 pmol of each primer, and 0.4 μL of ROX Reference Dye II. P-S and P-X were the primers used for the NPC6 gene, and 18s-s and 18s-x were the primers for the ribosomal 18S reference gene (Supplementary Table 1). The 2−ΔΔCT method was used for relative quantification. All qRT–PCR experiments were performed with four biological replicates.
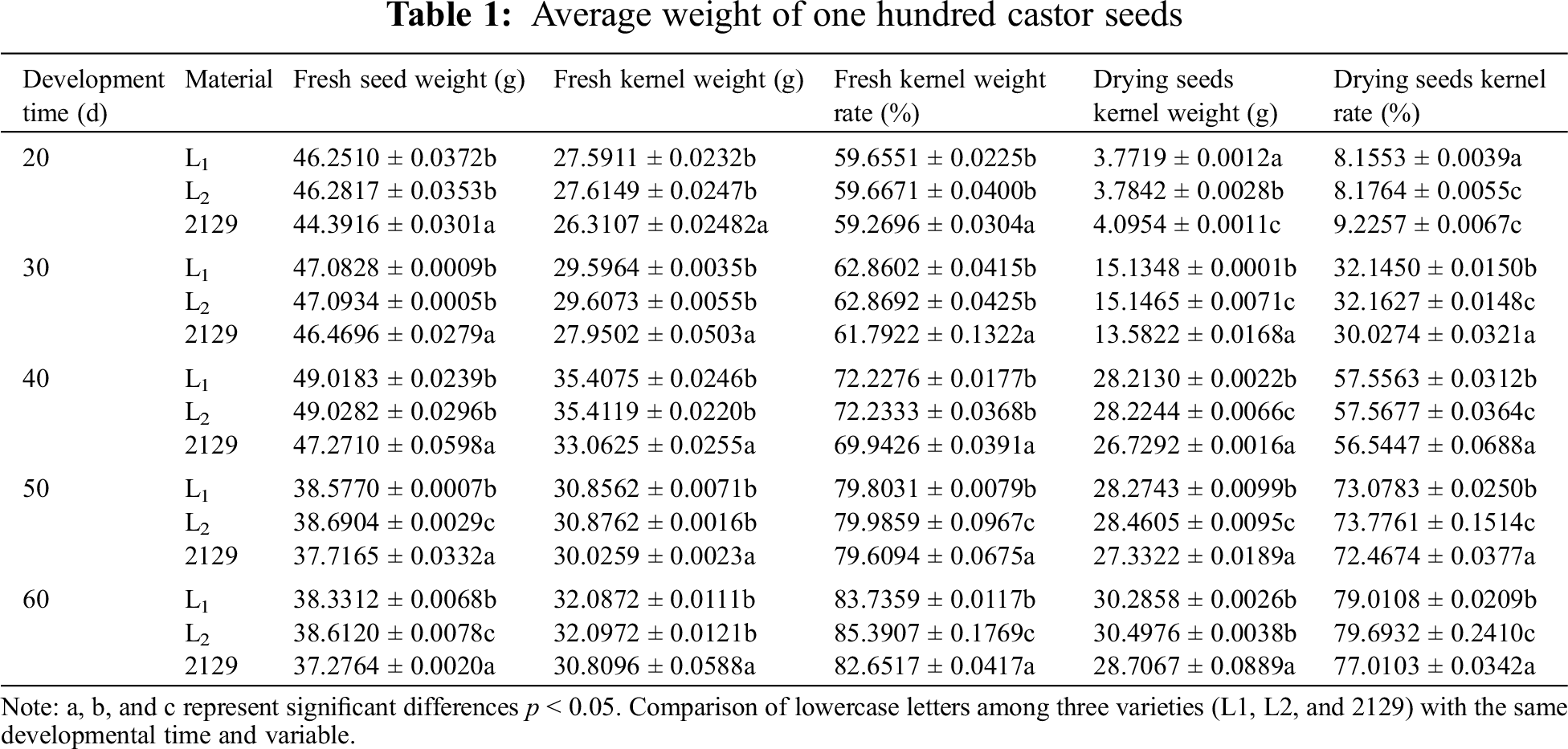
To further measure the weight of castor seeds produced by the plants, 100 seeds were randomly selected in each developmental period to measure the average weight. The seed coat was removed, and the weight of 100 fresh kernels (g) was measured. This process was repeated three times to determine the average value. Fresh kernel percentage (%) = weight of 100 fresh kernels (g)/weight of 100 fresh seeds (g) × 100%. Then, the fresh kernels were dried in a 65°C constant temperature oven, the weight of 100 dried kernels was measured, and this process was repeated three times to obtain the average value. Dried kernel rate (%) = dried kernel weight (g)/fresh seed weight (g) × 100%.
2.6 Determination of Crude Fat and Fatty Acid Content in Castor Seeds
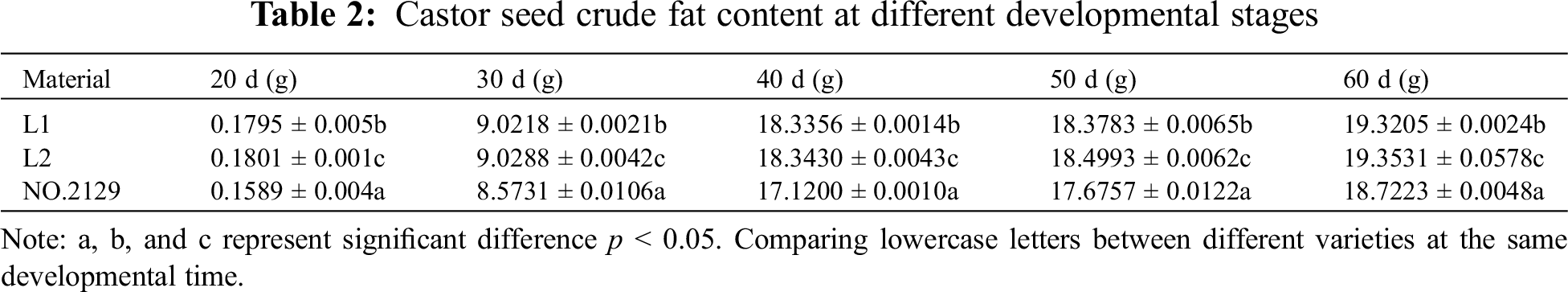
The determination of crude fat in castor seeds was performed according to the Chinese standard GB/T2906-82. The Soxhlet extraction method was used to determine the crude fat content of seeds [22]. Dried seed kernels (5 g) were ground to powder, put into a dry filter paper cylinder, and extracted with petroleum ether for 8 h in a Soxhlet extractor. The extracted crude fat was dried in an oven and weighed.
The determination of fatty acids in castor seeds was performed according to the Chinese standard GB5009.168-2016. The method for determining fatty acids involved saponification and methyl-esterification of castor kernel crude fat under alkaline conditions, resulting in the production of fatty acid methyl ester. The fatty acid methyl ester content was quantitatively determined by capillary column gas chromatography. The chromatographic column was an HP-INNOWAX (15.0 m × 250 μm × 0.25 μm) column. The gas chromatography conditions were as follows: injection temperature of 250°C, column flow rate of 1.0 mL/min, column box temperature: 180°C retention for 5 min, 180°C~230°C 3°C/min.
2.7 Proteomics Analysis of Castor Seeds
The total protein from 60-day-old castor seeds was separated by the phenol extraction method [23]. Proteins (1 g) were dissolved in the lysate (7 M urea, 2 M thiourea, 4% CHAPS (W/V), 40 mM DTT, 2% pharmalyte 3–10, 4% protease inhibitor). The dissolved protein was subjected to one-dimensional isoelectric focusing gel electrophoresis (IEFGE) at the following voltages: 30 V 8 h, 50 V 4 h, 100 V 1 h, 300 V 1 h, 600 V 1 h, 1000 V 2 h, 8000 V 12 h, and 1000 V 10 h. After one-dimensional electrophoresis, a 12% polyacrylamide gel was used for two-dimensional electrophoresis [24]. The gel was stained with Coomassie brilliant blue R250 after electrophoresis. The stained gel was photographed with a white light scanner. Image Master 2D Platinum 7.0 software (GE Healthcare) was used to analyze the gel (fold change ≥ 2-fold and p < 0.05), and differential protein spots were identified for mass spectrometry analysis.
3.1 L1 and L2 Castor Generated by Silencing NPC6
The RNAi vector pBI-C1-IN-C2 was transformed into castor cotyledon nodes using Agrobacterium tumefaciens (Fig. 1). PCR amplification from genomic DNA of T2 generation plants subjected to NPC6 gene silencing yielded 1595 bp bands for only the L1 and L2 plants (Fig. 2B). The expression level of the NPC6 gene in L1 and L2 was found to be significantly lower than that in wild-type NO.2129 through qRT–PCR analysis (Fig. 2C). In summary, L1 and L2 are NPC6 gene mutant plants.
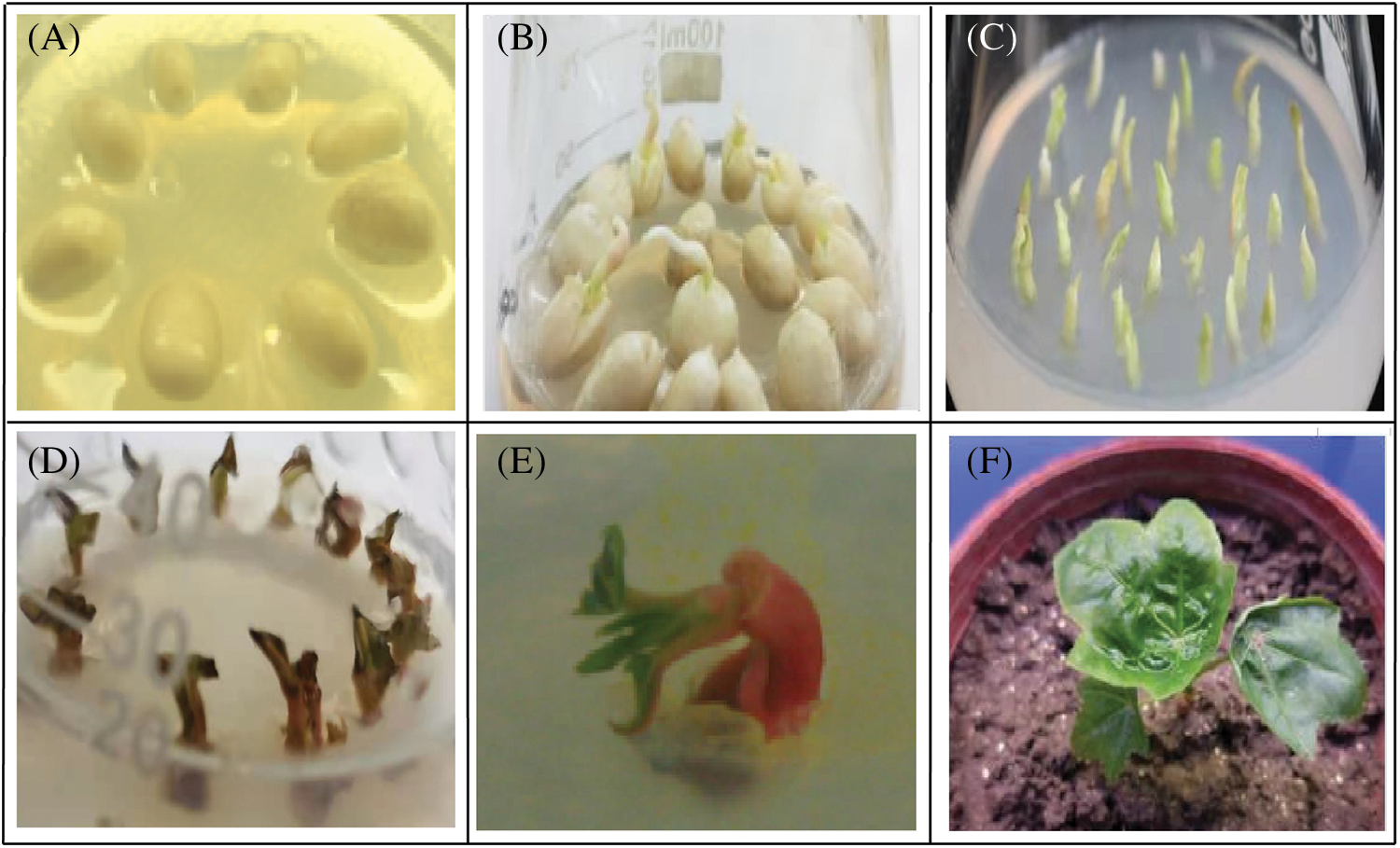
Figure 1: Genetic transformation of castor cotyledon nodes with the pBI-C1-IN-C2 vector. (A) NO.2129 seeds were inoculated into germination medium after removing the seed coat. (B) Seed germination. (C) The cotyledon nodes of castor seeds were cut and inoculated in medium. (D) Castor cotyledon nodes were inoculated into a screening medium containing Kan after being impregnated with Agrobacterium tumefaciens. (E) Kan-resistant seedlings. (F) Putative transgenic plant culture
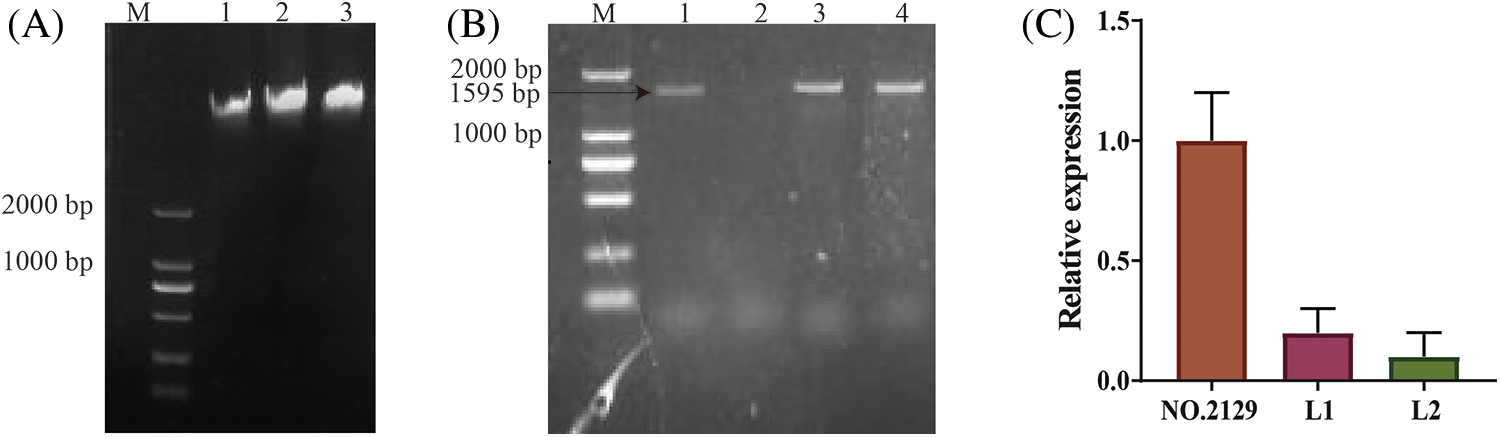
Figure 2: PCR-based identification of transgenic plants. (A) Genomic DNA electrophoresis results for castor seeds; M: DL2000 DNA marker, 1: L1 genomic DNA, 2: L2 genomic DNA, 3: NO.2129 genomic DNA. (B) PCR amplification of partial fragments of the pBI-C1-IN-C2 vector using genomic DNA as a template; M: DL2000 Marker, 1: positive control, 2: negative control, 3: L1 seed DNA PCR, 4: L2 seed DNA PCR. (C) qRT–PCR analysis of the relative expression levels of the NCP6 gene in L1, L2, and NO.2129 seeds
Meng-Yuan et al. found that seed weight was significantly positively correlated with seed oil content [25]. In this study, the 100-seed weight of castor seeds at different developmental stages was determined (Table 1). Table 1 shows that the fresh weight of seeds reaches its maximum on the 40th day of development, and gradually decreases from 50 to 60 days, with significant differences between the three varieties (p < 0.05). The weight of fresh kernel increases with the development of seeds but not significantly. The weight of dry kernel also increased with the development of seeds, and the difference was significant (p < 0.05). Interestingly, the fresh and dry kernel weights of L1 and L2 were both greater than those of NO.2129, and there is a significant difference between L1 and L2 compared to NO.2129 (p < 0.05) The above results indicate that silencing the NPC6 gene promotes dry matter accumulation in castor seeds.
3.2 Crude Fat and Fatty Acids in Seeds
In this study, the crude fat content of castor seeds at different developmental stages was determined (Table 2). Crude fat accumulated with the growth of castor seeds. Seed crude fat accumulated rapidly during the 30 to 40 days of development. The determination of the crude fat content of L1, L2, and NO.2129 showed that the crude fat content of L1 and L2 seeds from 20 to 60 days of development was higher than that of NO.2129, but the difference between L1 and L2 was not significant. According to the above results, NPC6 silencing promotes the accumulation of crude fat in castor seeds.
For further investigation, we examined the levels of different types of fatty acids in crude fat (Table 3), including palmitic acid, stearic acid, oleic acid, linoleic acid, linolenic acid, arachidic acid, arachidonic acid, behenic acid, and ricinoleic acid. The most abundant fatty acid was ricinoleic acid. Table 3 shows that fatty acids accumulate rapidly in seeds from 30 to 40 days of development, while slowly accumulating from 40 to 60 days. This is consistent with the crude fat determination results. In addition, the levels of ricinoleic acid, arachidonic acid, oleic acid, and palmitic acid in L1 and L2 seeds were higher than those in NO.2129, but the difference in ricinoleic acid was the largest. These results confirmed that NPC6 gene silencing promoted ricinoleic acid accumulation in castor seed crude fat.
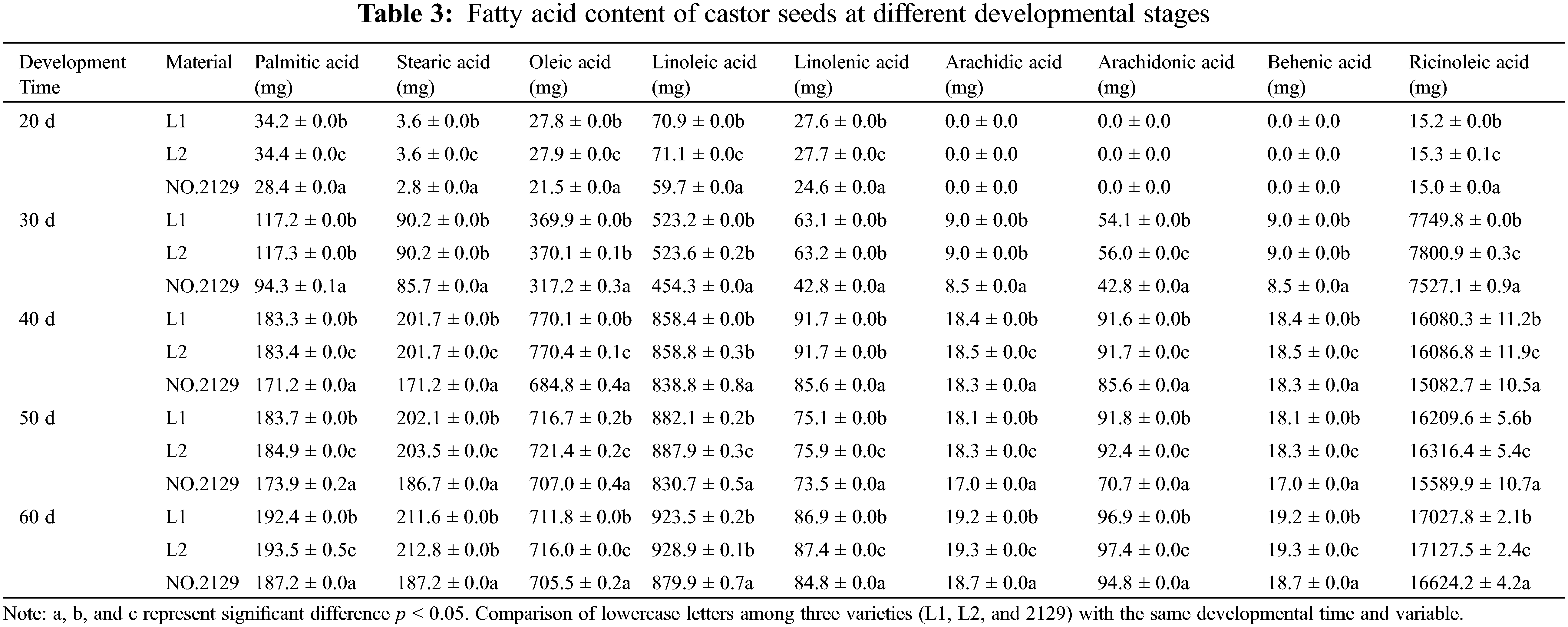
3.3 Castor Seed Proteomic Analysis
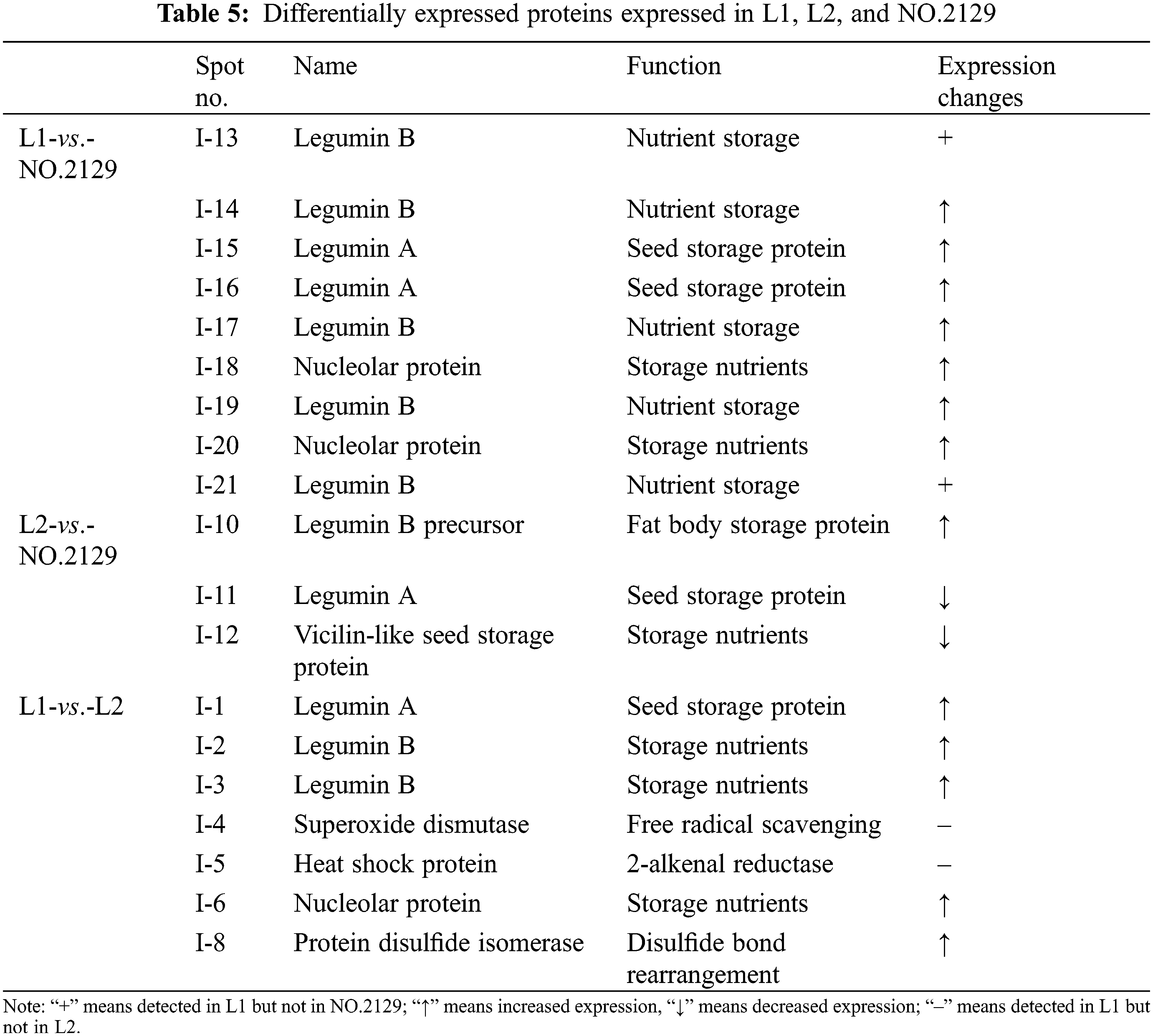
We performed proteomic analysis on L1, L2, and NO.2129 seeds developed for 60 days (Fig. 3). To assess variations in protein amounts, we performed pairwise alignment based on fold change ≥ 2 and p < 0.05. In all, 21 differentially expressed proteins were detected: nine in L1-vs.-NO.2129, four in L2-vs.-NO.2129, and eight in L1-vs.-L2. Mass spectrometry analysis of 21 differentially expressed proteins successfully identified 19 proteins (Table 4 and Supplementary Fig. 2). A heatmap was drawn based on the expression levels of these 19 proteins (Fig. 4). Of the 12 proteins, 7 were found in L1-vs.-NO.2129, 2 were found in L2-vs.-NO.2129, and the remaining 3 were found in L1-vs.-L2 (Table 5). The expression levels of legumins in L1 and L2 were found to be higher than those in NO.2129. Furthermore, upregulation of nucleolar proteins was also observed in L1 and L2.
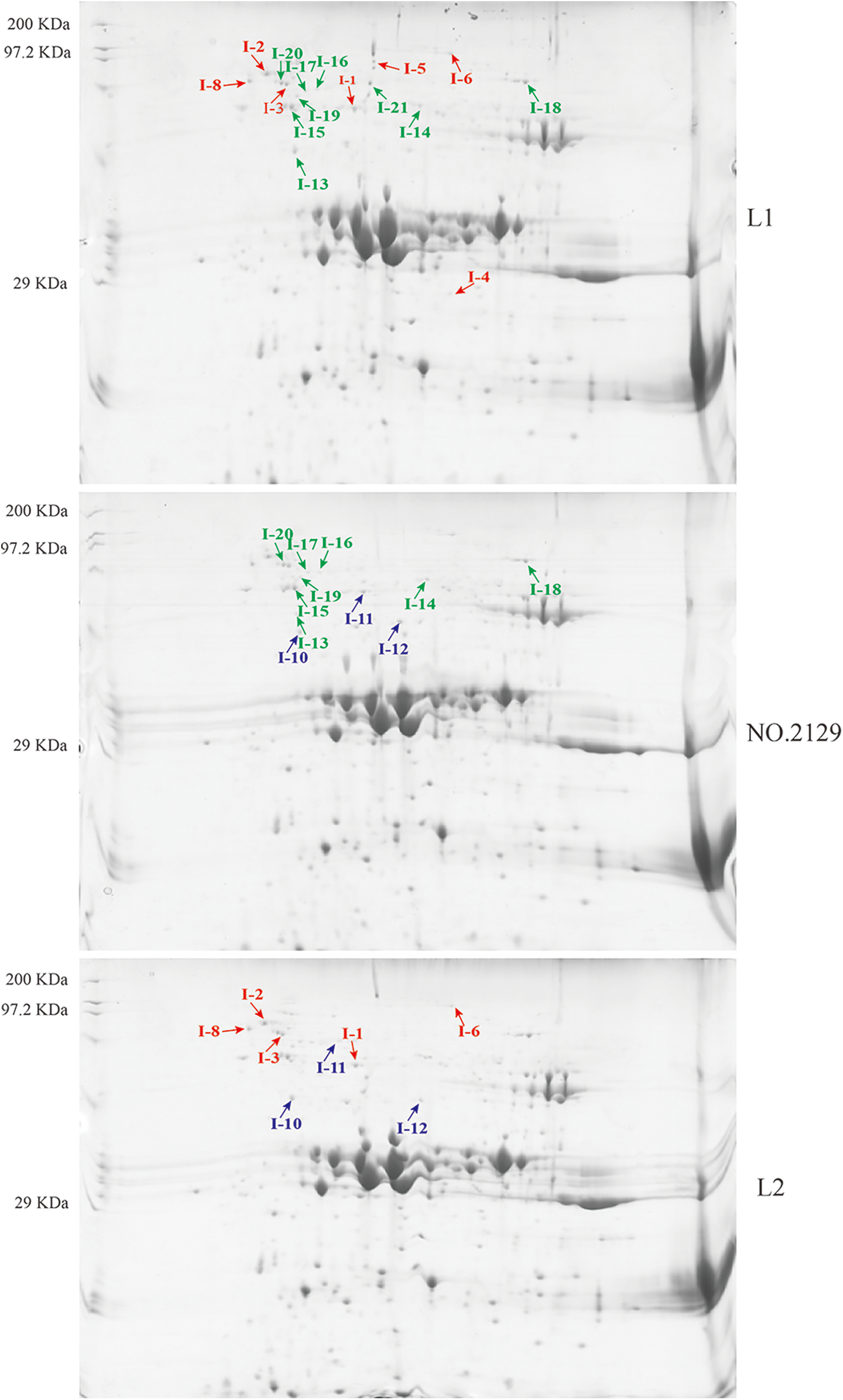
Figure 3: Two-dimensional electrophoresis analysis of castor seeds grown for 60 days. “ : L1-vs.-L2 differentially expressed proteins.” “
: L1-vs.-L2 differentially expressed proteins.” “ : L1 vs. NO.2129 differentially expressed proteins.” “
: L1 vs. NO.2129 differentially expressed proteins.” “ : L2-vs.-NO.2129 differentially expressed proteins.”
: L2-vs.-NO.2129 differentially expressed proteins.”
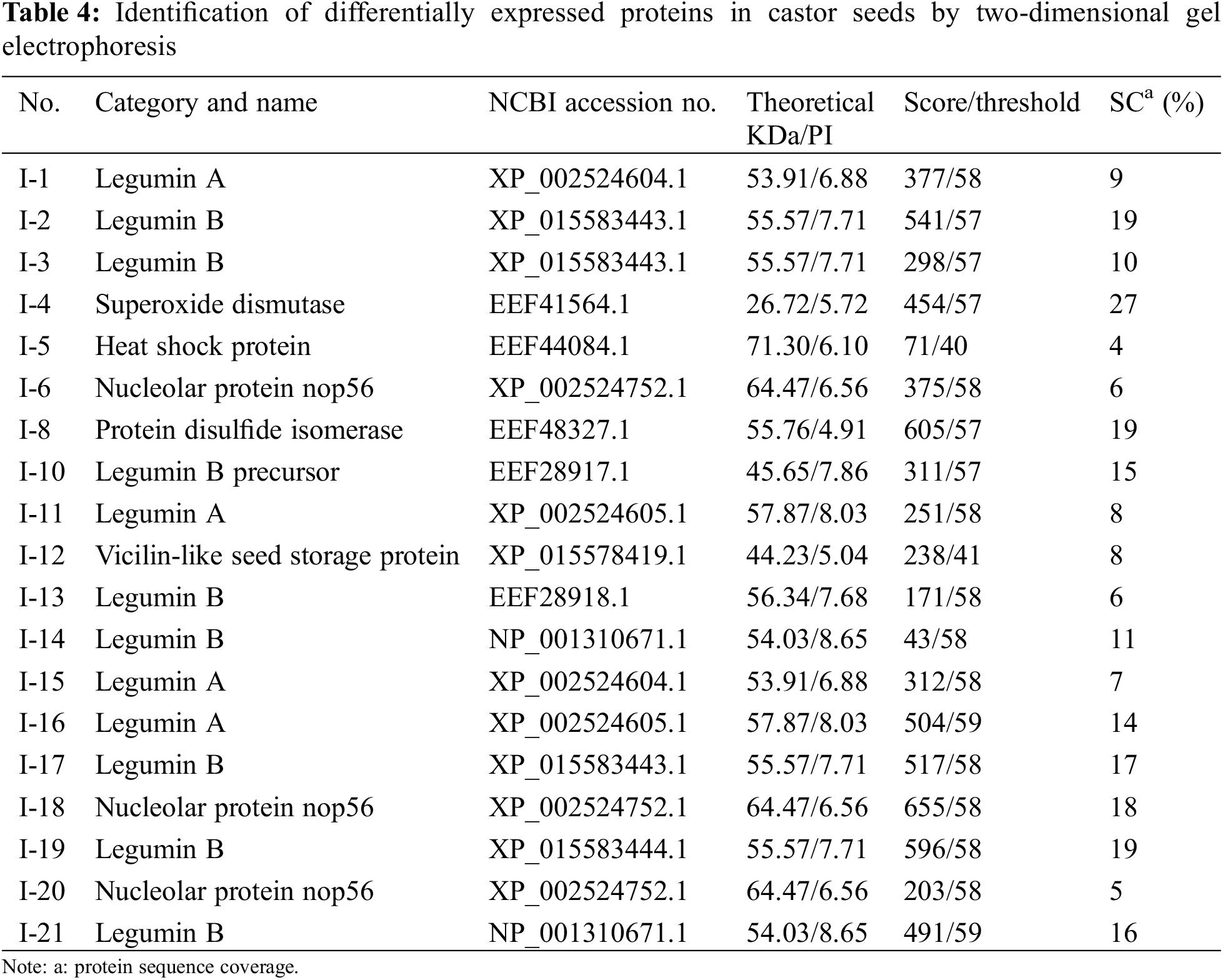
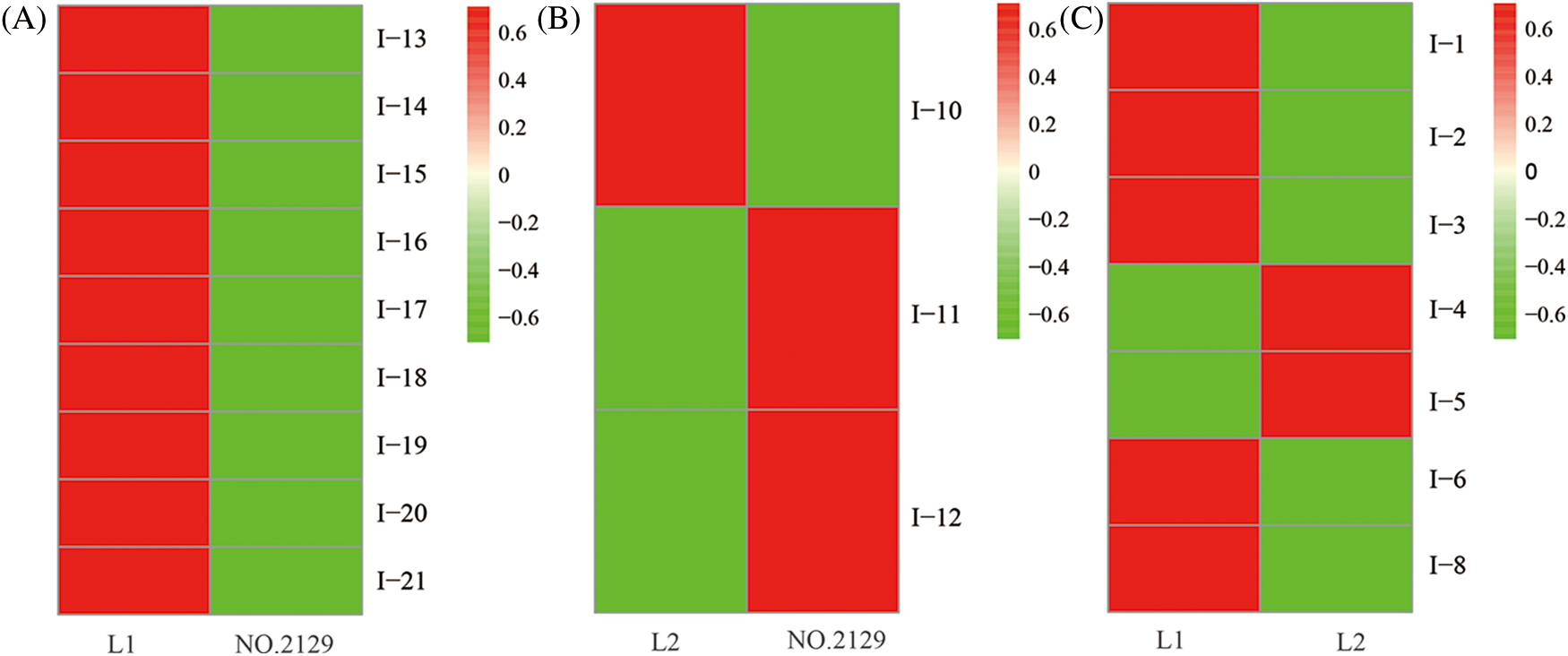
Figure 4: Heatmap of 19 differentially expressed proteins. (A) The differentially expressed proteins between L1 and NO.2129; (B) The differentially expressed proteins between L2 and NO.2129; (C) The differentially expressed proteins between L1 and L2
NPC is a type of nonspecific PLC that plays an important role in many biological processes, including in plant photosynthesis, root growth, response to heat stress, phosphorus deficiency, and salt stress [26–30]. This paper reports a new function of castor NPC6, which regulates the biosynthesis of ricinoleic acid. This work contributes to a better understanding of the function of the NPC6 gene and provides new mechanistic insights into castor oil biosynthesis.
RNAi was utilized to silence the castor NPC6 gene, resulting in two mutant plants, L1 and L2, which exhibited higher crude fat content than NO.2129. The analysis of the castor seeds fatty acid determination results revealed that L1 and L2 had higher ricinoleic acid levels than NO.2129. Studies on oilseed Brassicaceae plants showed that knockout of NPC6 reduced the production of fatty acids from phospholipids and galactolipids to triglycerides, but the result of overexpression of the NPC6 gene was the opposite [21]. This is consistent with our research results. It is well known that 2-oleoyl PC is the substrate for ricinoleic acid synthesis, but NPC6 can promote the conversion of 2-oleoyl-PC to triacylglycerol [31]. Therefore, NPC6 gene silencing may increase the content of ricinoleic acid in seeds by preventing nonhydroxy fatty acids from entering triacylglycerol synthesis (Fig. 5).
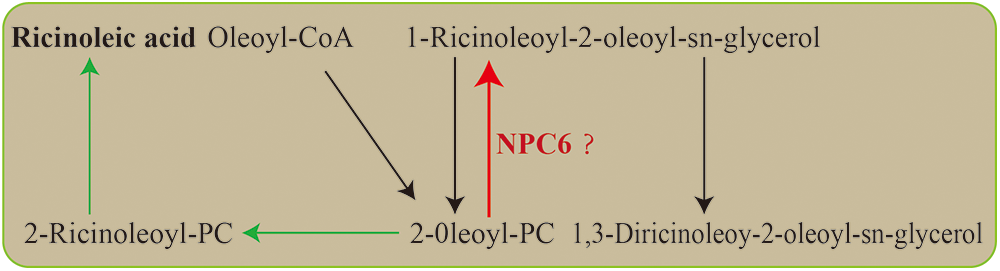
Figure 5: Ricinoleic acid biosynthesis pathway. The green arrow represents the synthesis pathway of ricinoleic acid. The red arrow represents the metabolic processes that NCP6 may regulate
This study analyzed the proteome of castor seeds from the L1, L2, and NO.2129 varieties. Twelve legumins were found to have differential expression, with higher expression in L1 and L2 compared to NO.2129. The synthesis and accumulation of storage proteins require a large amount of energy and substances, and lipid metabolism can provide energy and substances for their synthesis [32]. NPC not only produces energy in the process of hydrolyzing membrane phospholipids but also stimulates the release of Ca2+ stored in the endoplasmic reticulum (ER), which increases the concentration of cytoplasmic Ca2+ [33,34]. Therefore, NCP6 gene silencing may increase the concentration of Ca2+ in the ER, thereby promoting the expression of storage proteins and processing enzymes in seeds, but the specific regulatory mechanism still needs to be further studied.
The proteomics research found that legumin expression was upregulated in L1 and L2. Nucleolar proteins also exhibited the same trend. Nucleolar proteins are a class of proteins that exist in the nucleus and play a crucial role in the formation and functional regulation of nucleoli. In addition, nucleolar proteins play an important role in ribosome biosynthesis, including in transcription, processing, and assembly of rRNA (ribosome RNA) [35,36]. Therefore, it is speculated that nucleolar proteins regulate the expression of legumin by participating in ribosome biosynthesis. However, further in-depth research is needed on how NPC6 regulates nucleolar proteins and legumin.
In this study, RNAi was used to silence the NPC6 gene and increase the content of ricinolic acid in castor seeds. NPC6 gene silencing may not only increase the content of ricinolic acid in seeds but also promote the accumulation of nutrients by preventing nonhydroxy fatty acids from entering triacylglycerol synthesis.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by the following agencies: Natural Science Foundation of Jilin Province (YDZJ202201ZYTS453), Scientific Research Project of the Jilin Provincial Department of Education (JJKH20220010KJ), supported by Program for Innovative Research Team of Baicheng Normal University, National Natural Science Foundation of China (31860071), Inner Mongolia Autonomous Region Natural Science Foundation Project (2021MS03008), Inner Mongolia Autonomous Region Grassland Talent Innovation Team (2022), 2022 Basic Scientific Research Business Cost Project of Universities Directly under the Autonomous Region (237), Open Fund Project of Inner Mongolia Castor Industry Collaborative Innovation Center (MDK2021011, MDK2022014, MDK2022008, MDK2021008, MDK2022009), and Fundamental Research Funds for Universities Directly under the Autonomous Region in 2023 of Inner Mongolia University for Nationalities (225, 227, 243, 244), New Agricultural Science Research and Reform Practice Project of the Ministry of Education (2020114). In 2023, the Science and Technology Department of Inner Mongolia Autonomous Region approved the Construction Project of the Key Laboratory for Castor Breeding and Comprehensive Utilization in Inner Mongolia Autonomous Region.
Author Contributions: Yong Zhao, Lili Li, Mu Peng, and Tongtong Jiang performed the experiments. Rui Luo and Tongtong Jiang guided the experimental process. Lili Li, Yong Zhao, Rui Luo, Mingda Yin, Yanpeng Wen, and Zhiyan Wang completed the data analysis. Yong Zhao completed the writing of his paper. All the authors have read and agreed to the final text. Fenglan Huang and Fanjuan Meng designed the experiment and provided funding.
Availability of Data and Materials: All data are incorporated into the article and its supplementary material.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.031495.
References
1. Nitbani, F. O., Tjitda, P. J. P., Wogo, H. E., Detha, A. I. R. (2022). Preparation of ricinoleic acid from castor oil: A review. Journal of Oleo Science, 71(6), 781–793. [Google Scholar] [PubMed]
2. Ogunniyi, D. S. (2006). Castor oil: A vital industrial raw material. Bioresource Technology, 97(9), 1086–1091. [Google Scholar] [PubMed]
3. Goswami, D., Basu, J. K., De, S. (2012). Lipase applications in oil hydrolysis with a case study on castor oil: A review. Critical Reviews in Biotechnology, 33(1), 81–96. [Google Scholar] [PubMed]
4. Carrasco, S., Mérida, I. (2007). Diacylglycerol, when simplicity becomes complex. Trends in Biochemical Sciences, 32(1), 27–36. [Google Scholar] [PubMed]
5. Goni, F. M., Montes, L. R., Alonso, A. (2012). Phospholipases C and sphingomyelinases: Lipids as substrates and modulators of enzyme activity. Progress in Lipid Research, 51(3), 238–266. [Google Scholar] [PubMed]
6. Ali, U., Lu, S., Fadlalla, T., Iqbal, S., Yue, H. et al. (2022). The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses. Progress in Lipid Research, 86, 101158. [Google Scholar] [PubMed]
7. Yang, B., Li, M., Phillips, A., Li, L., Guo, L. (2021). Nonspecific phospholipase C4 hydrolyzes phosphosphingolipids and sustains plant root growth during phosphate deficiency. The Plant Cell, 33, 766–780. [Google Scholar] [PubMed]
8. Fan, R., Zhao, F., Gong, Z., Chen, Y., Yang, B. et al. (2023). Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C. Nature Communications, 14(1), 1–11. [Google Scholar]
9. Pokotylo, I., Pejchar, P., Potocky, M., Kocourkova, D., Krckova, Z. et al. (2013). The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Progress in Lipid Research, 52(1), 62–79. [Google Scholar] [PubMed]
10. Wimalasekera, R., Pejchar, P., Holk, A., Martinec, J., Scherer, G. F. (2010). Plant phosphatidylcholine-hydrolyzing phospholipase C NPC3 and NPC4 with roles in root development and brassinolide signaling in Arabidopsis thaliana RID A-7853-201. Molecular Plant, 3(3), 610–625. [Google Scholar] [PubMed]
11. Nakamura, Y., Liu, Y. C., Lin, Y. C. (2014). Floral glycerolipid profiles in homeotic mutants of Arabidopsis thaliana. Biochemical and Biophysical Research Communications, 450(4), 1272–1275. [Google Scholar] [PubMed]
12. Cai, G. Q., Fan, C. C., Liu, S., Yang, Q. Y., Liu, D. X. et al. (2020). Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants. New Phytologist, 226(4), 1055–1073. [Google Scholar] [PubMed]
13. Zuzana, K. K., Jitka, B., Michal, D., Daniela, K., Dominique, R. et al. (2015). Arabidopsis non-specific phospholipase C1: Characterization and its involvement in response to heat stress. Frontiers in Plant Science, 6(17), 928. [Google Scholar]
14. Ngo, A. H., Lin, Y. C., Liu, Y. C., Gutbrod, K., Peisker, H. et al. (2018). A pair of nonspecific phospholipase C, NPC2 and NPC6, are involved in gametophyte development and glycerolipid metabolism in Arabidopsis. New Phytologist, 219(1), 163–175. [Google Scholar] [PubMed]
15. Wimalasekera, R., Pejchar, P., Holk, A., Martinec, J., Scherer, G. F. E. (2010). Plant Phosphatidylcholine-Hydrolyzing Phospholipases C NPC3 and NPC4 with Roles in Root Development and Brassinolide Signaling in Arabidopsis thaliana. Molecular Plant, 3(3), 610–625. [Google Scholar] [PubMed]
16. Nakamura, Y., Awai, K., Masuda, T., Yoshioka, Y., Takamiya, K. I. et al. (2005). A Novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. Journal of Biological Chemistry, 280(9), 7469–7476. [Google Scholar] [PubMed]
17. Peters, C., Li, M., Narasimhan, R., Roth, M., Welti, R. et al. (2010). Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. The Plant Cell, 22(8), 2642–2659. [Google Scholar] [PubMed]
18. Peters, C., Kim, S. C., Devaiah, S., Li, M., Wang, X. (2014). Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell & Environment, 37(9), 2002–2013. [Google Scholar]
19. Ngo, A. H., Lin, Y. C., Liu, Y. C., Gutbrod, K., Peisker, H. et al. (2018). A pair of nonspecific phospholipases C, NPC2 and NPC6, are involved in gametophyte development and glycerolipid metabolism in Arabidopsis. New Phytologist, 219(1), 163–175. [Google Scholar] [PubMed]
20. Ngo, A. H., Kanehara, K., Nakamura, Y. (2019). Non-specific phospholipases C, NPC2 and NPC6, are required for root growth in Arabidopsis. The Plant Journal, 100(4), 825–835. [Google Scholar] [PubMed]
21. Cai, G., Fan, C., Liu, S., Yang, Q., Liu, D. et al. (2020). Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants. New Phytologist, 226(4), 1055–1073. [Google Scholar] [PubMed]
22. Gu, Z. X., Wu, B. C., Wu, L. Y., Hang, Y. Y. (2009). Analysis of total lipid contents and fatty acids composition of three species of Euphorbia in Jiangsu Province. Chemistry and Industry of Forest Products, 29(4), 63–66. [Google Scholar]
23. Paul, S., Gayen, D., Datta, S. K., Datta, K. (2015). Dissecting root proteome of transgenic rice cultivars unravels metabolic alterations and accumulation of novel stress responsive proteins under drought stress. Plant Science, 234, 133–143. [Google Scholar] [PubMed]
24. Cho, S. H., Lee, K. M., Kim, C. H., Kim, S. S. (2020). Construction of a Lectin-Glycan interaction network from enterohemorrhagic Escherichia coli strains by multi-omics analysis. International Journal of Molecular Sciences, 21(8), 2681. [Google Scholar] [PubMed]
25. Dai, M. Y., Cui, C. S., Gao, M., Li, W. C. (2019). Correlation analysis of oil content, fatty acid content and seed phenotypic traits of Ricinus Communis L. Seed. Southwest China Journal of Agricultural Sciences, 32(6), 55–62. [Google Scholar]
26. Wei, Y. L., Liu, X. Y., Ge, S. N., Zhang, H. Y., Che, X. Y. et al. (2022). Involvement of phospholipase C in photosynthesis and growth of maize seedlings. Genes, 13(6), 1011. [Google Scholar] [PubMed]
27. Ngo, A. H., Kanehara, K., Nakamura, Y. (2019). Non-specific phospholipases C, NPC2 and NPC6, are required for root growth in Arabidopsis. Plant Journal, 100(4), 825–835. [Google Scholar]
28. Krckova, Z., Brouzdova, J., Danek, M., Kocourkova, D., Rainteau, D. et al. (2015). Arabidopsis non-specific phospholipase C1: Characterization and its involvement in response to heat stress. Frontiers in Plant Science, 6, 928. [Google Scholar] [PubMed]
29. Yang, B., Zhang, K., Jin, X., Yan, J. Y., Lu, S. P. et al. (2021). Acylation of non-specific phospholipase C4 determines its function in plant response to phosphate deficiency. Plant Journal, 106(6), 1647–1659. [Google Scholar]
30. Kocourkova, D., Krckova, Z., Pejchar, P., Veselkova, S., Valentova, O. et al. (2011). The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. Journal of Experimental Botany, 62(11), 3753–3763. [Google Scholar] [PubMed]
31. Pokotylo, I., Pejchar, P., Potocký, M., Kocourková, D., Krčková, Z. et al. (2013). The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Progress in Lipid Research, 52(1), 62–79. [Google Scholar] [PubMed]
32. Wang, X., Du, G., Wu, Y., Zhang, Y., Wu, R. (2019). Association between different levels of lipid metabolismrelated enzymes and fatty acid synthase in Wilms’ tumor. International Journal of Oncology, 56(2), 568–580. [Google Scholar] [PubMed]
33. Wu, Q. W., Kapfhammer, J. P. (2021). Modulation of increased mGluR1 signaling by RGS8 protects Purkinje cells from dendritic reduction and could be a common mechanism in diverse forms of spinocerebellar ataxia. Frontiers in Cell and Developmental Biology, 8, 569889. [Google Scholar] [PubMed]
34. Tang, Y., Rijal, R., Zimmerhanzel, D. E., Mccullough, J. R., Gomer, R. H. (2021). An autocrine negative feedback loop inhibits dictyostelium discoideum proliferation through pathways including IP3/Ca2+. mBio, 12(3), e0134721. [Google Scholar] [PubMed]
35. Boisvert, F. O. M., van Koningsbruggen, S., Navascués, J., Lamond, A. I. (2007). The multifunctional nucleolus. Nature Reviews Molecular Cell Biology, 8(7), 574–585. [Google Scholar] [PubMed]
36. Tiku, V., Antebi, A. (2018). Nucleolar function in lifespan regulation. Trends in Cell Biology, 28(8), 662–672. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools