 Open Access
Open Access
ARTICLE
The Cotton GhWRKY91 Gene Negatively Regulates Root Elongation in Overexpressed Transgenic Arabidopsis thaliana
College of Forestry, Hebei Agricultural University, Baoding, 071000, China
* Corresponding Author: Lijiao Gu. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Abiotic and Biotic Stress Tolerance in Crop)
Phyton-International Journal of Experimental Botany 2023, 92(11), 2937-2946. https://doi.org/10.32604/phyton.2023.043306
Received 28 June 2023; Accepted 06 August 2023; Issue published 24 October 2023
Abstract
WRKY transcription factors play important roles in plant growth, development, and stress responses. Our previous research has shown that the GhWRKY91 gene can delay age-, abscisic acid (ABA)-, and drought-induced leaf senescence when overexpressed in transgenic Arabidopsis plants. To explore in more depth the biological functions of the GhWRKY91 gene, we further observed the root growth of overexpressing transgenic Arabidopsis thaliana under ABA and drought treatment. In this study, we transplanted the germinated seeds of wild-type (WT) and three transgenic lines (OE-12, OE-13 and OE-20) to 1/2 MS solid medium containing ABA and different concentrations of mannitol (simulated drought treatment) for culturing. The results showed that the transgenic plants had dark green leaves and short root lengths when no stress treatment was added. After ABA and mannitol treatment, the root growth of the WT and transgenic Arabidopsis was inhibited to varying degrees, and the root length downregulation of the transgenic plants was higher than that of the WT, indicating that they were more sensitive to ABA and drought. A bimolecular fluorescence complementation (BiFC) assay showed that the GhWRKY91 and GhWRKY3 proteins interact and emit yellow fluorescence in tobacco leaf cells. These results indicate that the GhWRKY91 gene negatively regulates root elongation in transgenic Arabidopsis and provide a basis for further research on the molecular mechanism of its involvement in regulating cotton root development.Keywords
WRKY transcription factors constitute one of the largest transcription factor families in higher plants. The WRKY gene family contains a WRKY domain of approximately 60 amino acids, which contains a conserved WRKYGQK sequence and a zinc finger domain [1]. According to the number of WRKY domains and the characteristics of the zinc finger structure, the WRKY gene family is mainly divided into subfamilies I, II (IIa, IIb, IIc, IId and IIe) and III [1]. Since the first WRKY transcription factor SPF1 was identified from sweet potato [2], an increasing number of WRKY transcription factors have been identified from plants as the cost of genome sequencing has decreased. In recent years, 64 IiWRKY genes have been identified from Isatis indigotica [3], 62 PdWRKY genes from Prunus dulcis [4], 72 TkWRKY genes from Taraxacum kok saghyz Rodin [5], and 138 ClWRKY genes from Chrysanthemum lavandulifolium [6]. Clearly, the WRKY transcription factor is increasingly being extensively studied, laying the foundation for the further exploration of the function of the WRKY genes.
The orderly expression of plant genes is the foundation of plant growth and development. Numerous studies have shown that WRKY transcription factors play an important role in various physiological processes, such as plant growth, fruit ripening, aging, and stress response. For example, MdWRKY9 directly inhibits the transcription of the brassinosteroid (BR) restricted synthase MdDWF4, reduces the production of BRs, and positively regulates plant dwarfism [7]. Chinese flowering cabbage BrWRKY6 binds to the promoter of senescence-related genes BrSAG12, BrNYCl and BrSGR1 through W-box cis-elements, inhibits the expression of gibberellin (GA) biosynthetic genes BrKAO2 and BrGA20ox2, and accelerates leaf senescence [8]. The AcWRKY40 gene responds to ethylene treatment during the postharvest ripening of kiwifruit and may participate in the postharvest ripening of kiwifruit by regulating the expression of genes related to ethylene biosynthesis [9]. In Iris germanica, the expression of IgWRKY50 and IgWRKY32 can be induced by PEG6000, enhancing the drought resistance of overexpressing transgenic Arabidopsis through the abscisic acid (ABA) signaling pathway [10].
Some WRKY genes can affect root structure by influencing plant hormone signals or gene expression. Compared with the wild-type (WT), overexpression of OsWRKY31 in rice leads to reduced and shortened lateral roots, which may be achieved by interfering with the response or transport of auxin [11]. Ethylene (ETH) induces the formation of root hairs and adventitious roots during root development. In the wheat experiment, the 1-amino cyclopropene 1-carboxylate synthase gene for ethylene synthesis was downregulated in the TaWRKY51-OE overexpression lines and upregulated in the TaWRKY51-RNAi silenced lines. Further research found that TaWRKY51 is a negative regulator of ETH synthesis. TaWRKY51 inhibits the expression of the ETH synthesis gene ACS by combining with the W-box cis-element in the promoter region and coordinates ethylene synthesis and lateral root formation in wheat [12]. Soybean GmWRKY46 has a strong inducing effect under phosphate (Pi) starvation conditions, especially in soybean roots. Overexpression of GmWRKY46 significantly enhances the tolerance of transgenic Arabidopsis to Pi starvation and lateral root development [13]. These research results indicate that WRKY transcription factors play important roles in regulating plant root development.
Cotton is an important cash crop in China and the main source of natural fiber. WRKY transcription factors play an important role in the growth and development of plants and their response to stress. Previous studies have found that the GhWRKY91 gene can delay leaf senescence in transgenic Arabidopsis [14], but further research is needed to determine whether this gene has other functions. In this study, overexpression of the GhWRKY91 gene in Arabidopsis affected root development and increased sensitivity to ABA and drought stress. This provides a basis for further research on the molecular mechanism by which GhWRKY91 participates in regulating cotton root development and is important for stress resistance breeding in transgenic cotton.
2.1 Planting and Stress Management of GhWRKY91 Transgenic Arabidopsis
In a previous study, we constructed an overexpression vector of the GhWRKY91 gene driven by the 35S promoter and obtained three transgenic Arabidopsis thaliana positive lines, OE-12, OE-13 and OE-20, by floral dipping [14]. In this study, we further observed the phenotype of transgenic Arabidopsis under ABA and drought stress treatment. A. thaliana seeds of WT, OE-12, OE-13 and OE-20 were sterilized in 1.5 mL clean centrifuge tubes, planted on 1/2 MS solid medium (0.22 g 1/2 Murashige & Skoog medium, 3 g sucrose, 0.8 g agar powder, sterilized at 121°C for 15 min), and placed in a refrigerator at 4°C for 3 days for vernalization. Then, the solid plates were placed in a light incubator, the temperature was 22°C, and the photoperiod was 16 h of light/8 h of darkness until the seeds germinated. The germinated seeds of WT, OE-12, OE-13 and OE-20 with healthy and consistent growth were transplanted to 1/2 MS solid medium containing ABA (0.5 μM) and mannitol at different concentrations (50, 150 and 200 mM) and cultured in a light incubator. In the control group, the germinated seeds of WT, OE-12, OE-13 and OE-20 were planted on 1/2 solid medium without any stress treatment. The phenotypes were observed after approximately 12 days of cultivation. Three repetitions were carried out for each treatment.
2.2 Construction of BiFC Vector
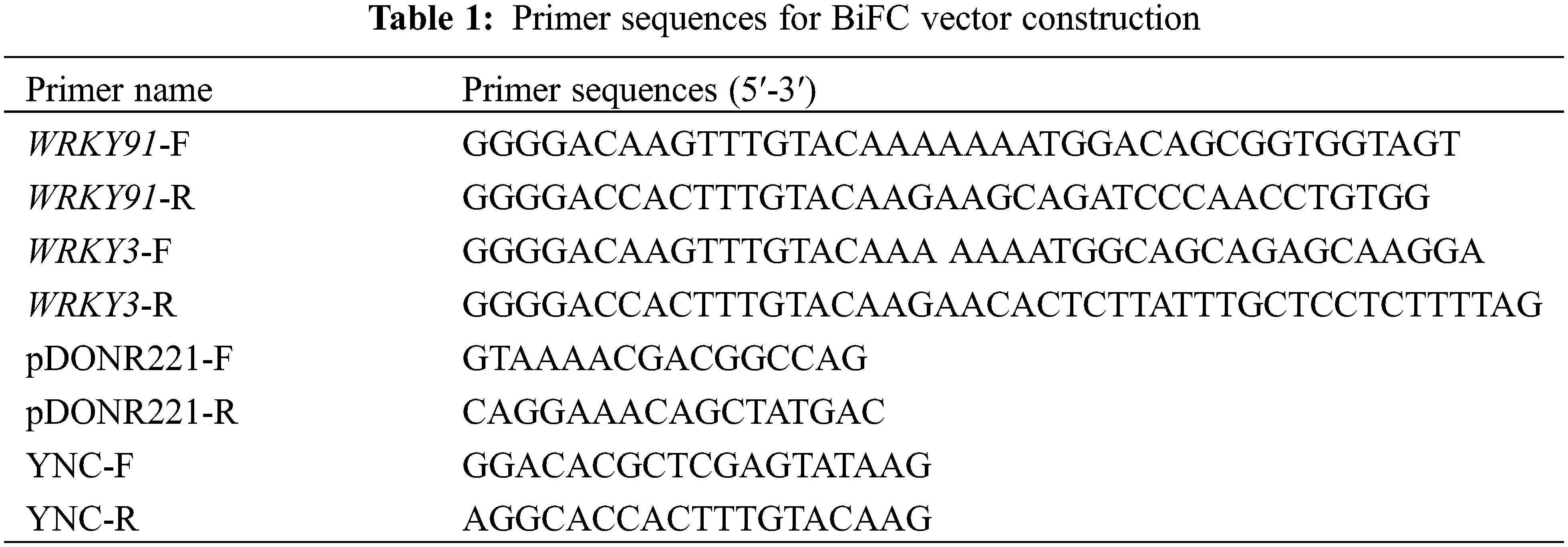
Using WRKY91-F/R and WRKY31-F/R primers, the GhWRKY91 and GhWRKY3 genes were cloned from the cDNA of CCRI10 cotton variety leaves, respectively. After the PCR product was purified and recovered, the purified product was connected to the pDONR221 entry vector by Gateway BP Clonase II Enzyme Mix (Invitrogen), and the recombinant plasmids pDONR221-WRKY91 and pDONR221-WRKY3 were transformed into TOP10 competent Escherichia coli cells. Positive clones were identified by pDONR221-F/R universal primer PCR and sent to the company for sequencing. The pDONR221-WRKY91 and pDONR221-WRKY3 plasmids with correct sequencing were extracted, and GhWRKY91 and GhWRKY3 were connected to the pEarleyGate201-YN and pEarleyGate202-YC vectors by Gateway LR Clonase II enzyme mix (Invitrogen), respectively. The connected products WRKY91-YN and WRKY3-YC were transformed into TOP10 competent E. coli cells. The positive clones were identified by universal primers YNC-F/R of pEarleyGate201-YN and pEarleyGate202-YC vectors using PCR. The empty vector pEarleyGate201-YN and pEarleyGate202-YC plasmids and the positive clones WRKY91-YN and WRKY3-YC plasmids were transformed into Agrobacterium GV3101 receptor cells and identified using YNC-F/R primers. All primers are listed in Table 1.
2.3 Tobacco Infection and Fluorescence Observation
Tobacco was grown in a greenhouse at 22°C under 16 h of light and 8 h of darkness. Tobacco that was approximately 3 weeks old was used for injection. The successful Agrobacterium solution was shaken overnight at 28°C and 200 rpm. pEarleyGate202-WRKY91-YN & pEarleyGate201-WRKY3-YC, pEarleyGate201-WRKY91-YN & pEarleyGate202-YC, pEarleyGate201-YN & pEarleyGate202-WRKY3-YC and pEarleyGate202-YC & pEarleyGate201-YN combinations were mixed at a ratio of 1:1. After centrifugation, four kinds of mixed bacterial solutions were resuspended in osmotic solution (10 mM MgCl2, 10 mM MES pH 5.7 and 0.2 mM acetosyringone) and adjusted to an OD600 ranging from 0.01 to 0.1. Before infection, the tobacco was placed under a white fluorescent lamp for 1 h to open its pores. Four kinds of suspensions were aspirated into a 1 mL syringe without the needle, injected into the back of the tobacco leaf and marked as the infected area. The infected tobacco was returned to the culture room, and fluorescence observation was performed by laser confocal scanning microscopy after 2–3 days.
3.1 GhWRKY91 Transgenic Arabidopsis Root Elongation Is Sensitive to ABA
In a preliminary study, we obtained three transgenic A. thaliana lines overexpressing the GhWRKY91 gene [14]. In this study, we further observed the effect of ABA on the root elongation of GhWRKY91 transgenic plants. Germinated seeds of WT, OE-12, OE-13, and OE-20 plants were placed on a 1/2 MS solid plate without ABA and with 0.5 μM ABA for approximately 12 days. From Fig. 1A, under normal growth conditions, the leaves of the seedlings of the three transgenic plants were darker green than those of WT, with shorter roots and overall smaller plants. After ABA treatment, the root growth of both WT and transgenic plants was inhibited, and the leaves of seedlings of the three transgenic plants showed varying degrees of yellowing. Fig. 1B shows that the root length of the three transgenic lines was consistently shorter than that of WT when ABA treatment was applied or not, which is consistent with the phenotype observation in Fig. 1A. The root length of the three transgenic lines was significantly lower than that of the WT before and after ABA treatment (Fig. 1B). We further analyzed the relative root length of WT and three transgenic strains under 0.5 μM ABA treatment relative to normal growth conditions using the data in Fig. 1B. The relative root length was 80.19% (WT), 46.22% (OE91-12), 54.43% (OE91-13) and 51.41% (OE91-20), respectively. The relative root length of the three transgenic lines was shorter than that of WT, indicating that the root length downregulation in the three transgenic lines was higher than that of WT, making them more sensitive to ABA treatment.
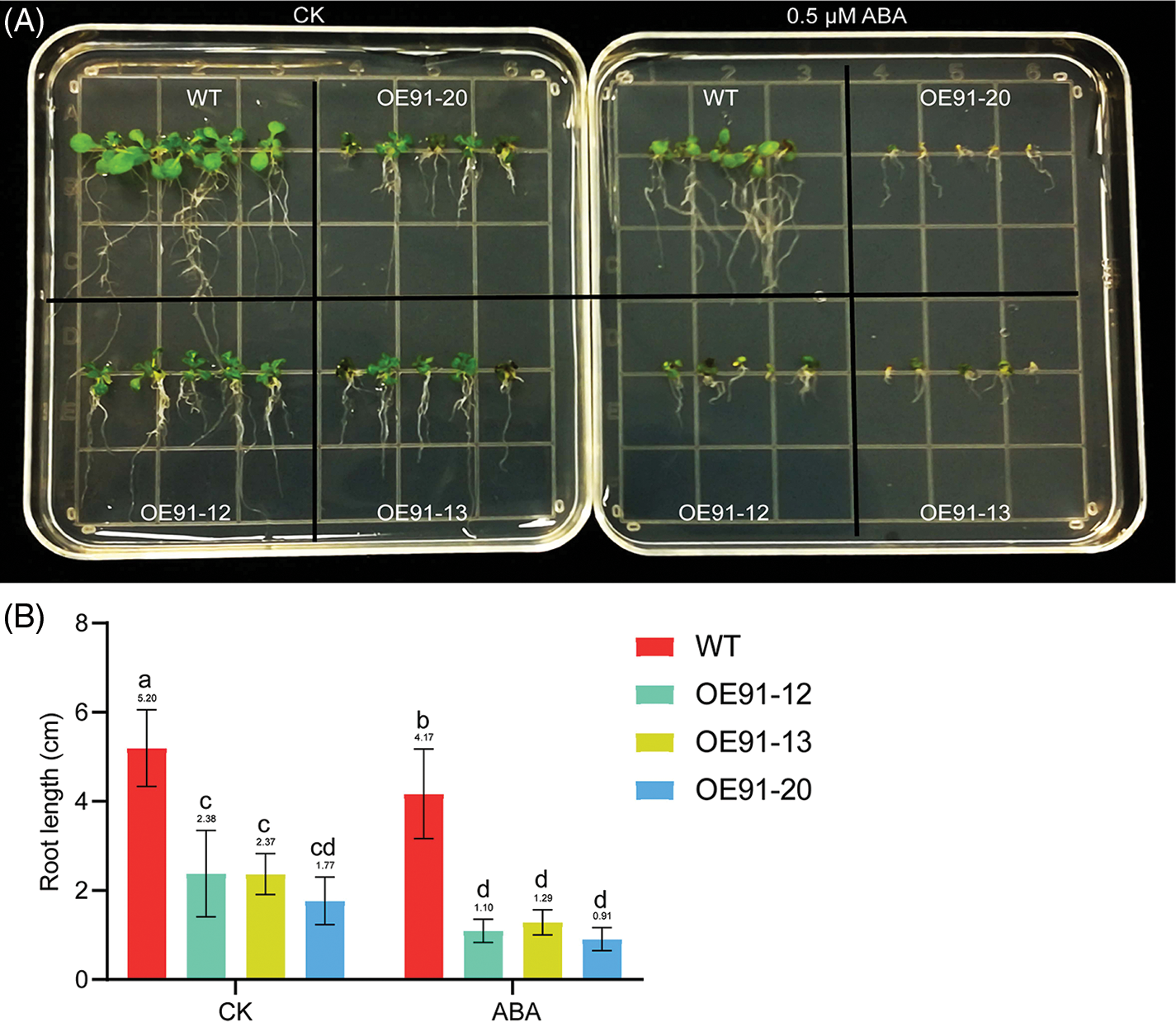
Figure 1: GhWRKY91 transgenic Arabidopsis root elongation is sensitive to ABA. (A) Phenotypes of the WT and three transgenic lines treated with 0 and 0.5 μM ABA. (B) Root length of the WT and three transgenic lines treated with 0 and 0.5 μM ABA
3.2 GhWRKY91 Transgenic Arabidopsis Root Elongation Is Sensitive to Drought
The germinated seeds of WT, OE-12, OE-13 and OE-20 were cultured on 1/2 MS solid plates containing 0, 50, 150 and 200 mM mannitol, and the effects of simulated drought treatment on root development were observed. When mannitol was not added, the leaves of the three transgenic lines were dark green, and the root length was shorter than that of the WT (Fig. 2A). After mannitol treatment, the root length of the WT and the three transgenic plants became shorter to varying degrees, the root length of the three transgenic lines was shorter than that of the WT, and the root length of the three transgenic lines decreased precipitously when treated with 150 mM mannitol (Fig. 2A). The statistical results for root length are consistent with the phenotypic observations of root length (Fig. 2B). The root length downregulation ratio of WT and three transgenic lines treated with mannitol at different concentrations compared with normal growth conditions was further analyzed based on the data in Fig. 2B. There was no significant difference in WT root length between the control and 50 mM mannitol treatment, nor was there any significant difference in the three transgenic lines. However, under 150 and 200 mM mannitol treatment, the WT and the three transgenic lines were significantly different from the untreated lines (Fig. 2B). With the increase in mannitol concentration, the root length of the WT and the three transgenic lines decreased continuously. Under 50 mM mannitol treatment, there was no significant difference in the root length decrease between the three transgenic lines and WT. Under 150 mM mannitol treatment, the root length of WT was decreased by 38.40%, and those of the three transgenic lines were decreased by 85.23%, 83.33% and 85.81%. Under 200 mM mannitol treatment, the root length of WT was decreased by 65.98%, and those of the three transgenic lines were decreased by 87.92%, 90.53% and 88.58%. These results showed that the root length reduction ratio of transgenic lines under drought treatment was higher than that of WT, and they were more sensitive to drought.
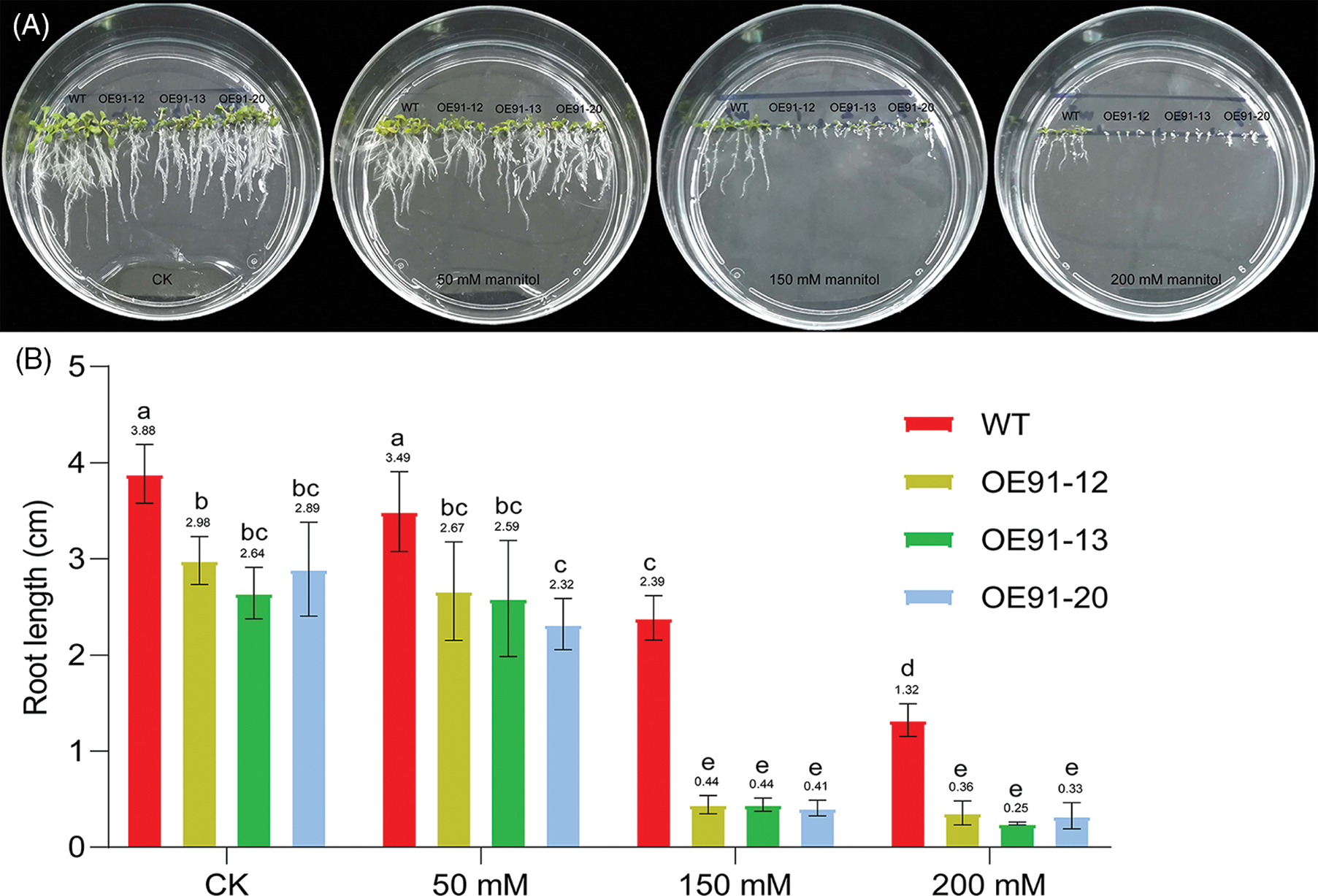
Figure 2: GhWRKY91 transgenic Arabidopsis root elongation is sensitive to drought. (A) Phenotypes of the WT and three transgenic lines under 0 (CK), 50, 150 and 200 mM mannitol treatment. (B) Root length of the WT and three transgenic lines under different concentrations of mannitol
3.3 Interaction between the GhWRKY91 and GhWRKY3 Proteins
Previous studies have shown that GhWRKY3 belongs to the group I WRKY transcription factor subfamily, is located in the nucleus and is induced by various hormones, biological stress and abiotic stress [15]. In addition, GhWRKY3 is constitutively expressed in roots [15], and its homolog in Arabidopsis thaliana, AtWRKY3 (NM_126385), regulates root elongation under stress treatments [16]. Therefore, GhWRKY3 was selected as a candidate gene to validate the interaction with GhWRK91, in order to preliminarily explore the regulatory mechanism of GhWRKY91 in regulating root growth. To verify whether the GhWRKY91 transcription factor interacts with GhWRKY3, we conducted BiFC experiments in tobacco leaf cells. The results showed that the three control combinations YN+YC, WRKY91-YN+YC, and YN+WRKY3-YC could not emit yellow fluorescence in tobacco leaf cells, and only WRKY91-YN+WRKY3-YC could emit yellow fluorescence in tobacco leaf cells (Fig. 3). This result indicates that GhWRKY91 and GhWRKY3 can interact and emit yellow fluorescence.
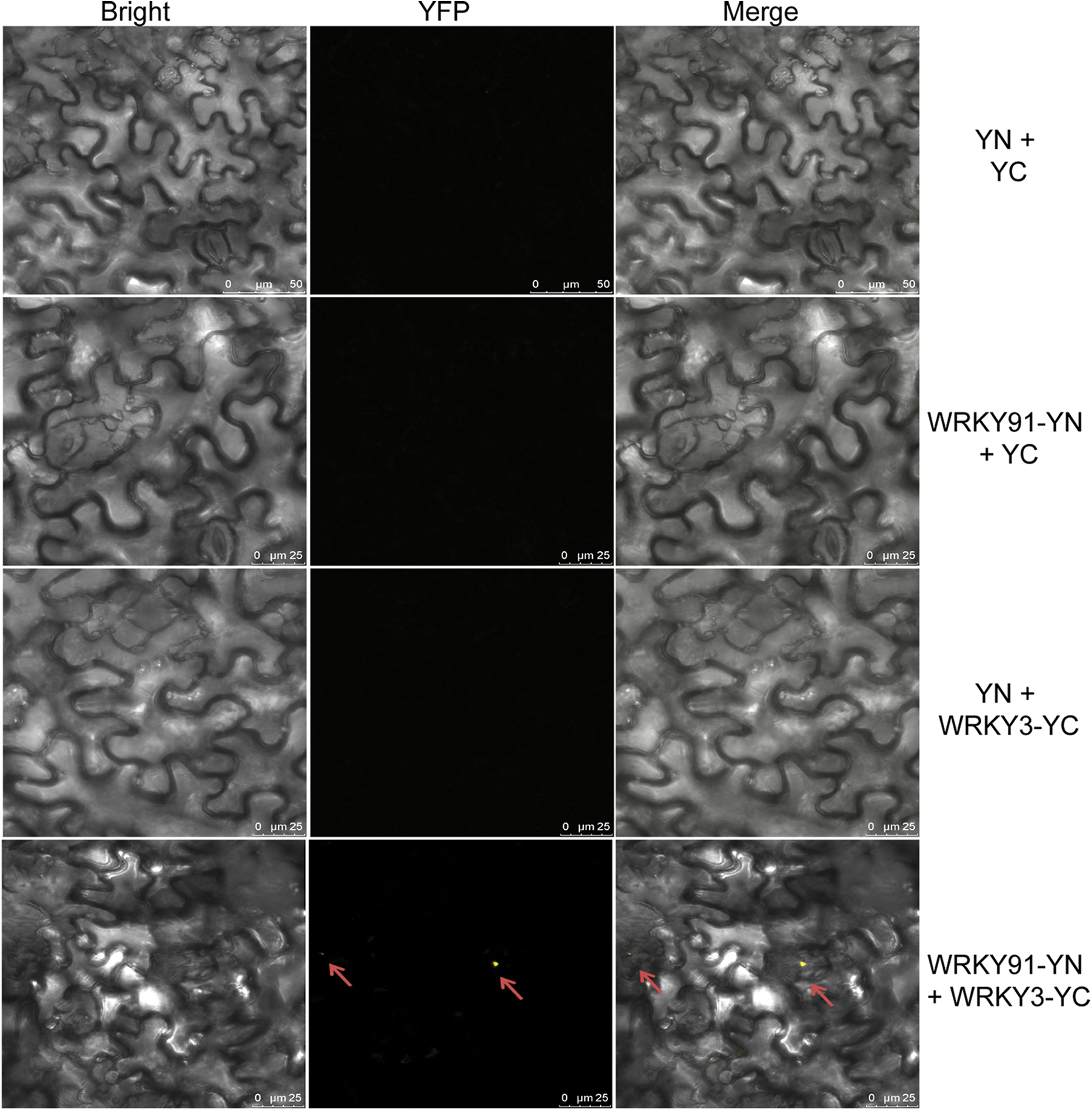
Figure 3: Interaction between GhWRKY91 and GhWRKY3 in tobacco leaf cells. The red arrow indicates the position of yellow fluorescence
In the process of plant growth, adverse environmental conditions such as drought, salinity, high temperature, low temperature, heavy metals and ultraviolet rays are often encountered, which affect the growth and development of plants and even lead to plant death in severe cases [17]. Plants have evolved a series of mechanisms to resist adverse environments after long-term adversity training so that they can better adapt to changes in the external environment [18–20]. As an important factor regulating the expression of stress-related genes, plant WRKY genes play an important role in plant adaptation and resistance to stress [5,21,22]. Cotton is the largest cash crop in China and the source of natural plant fiber. The function of WRKY genes in the cotton response to abiotic stress has been studied. For example, virus-induced GhWRKY5 gene silencing results in greater salt sensitivity in cotton than in WT plants, while overexpression of GarWRKY5 positively regulates salt tolerance in Arabidopsis seeds during germination and vegetative growth stages [23]. Under drought stress, GhWRKY33-overexpressing transgenic A. thaliana showed a higher water loss rate, higher wilting degree, and faster decay rate than the WT. GhWRKY33 may act as a negative regulator to mediate the plant response to drought stress [24]. Therefore, WRKY transcription factors play an important role in the regulation of cotton stress resistance, and it is very important to study the function of WRKY genes in cotton.
ABA is an important plant hormone involved in the stress response that can inhibit root elongation by delaying the initiation of cell elongation. Takatsuka et al. [25] showed that ABA inhibits root cell elongation by inhibiting the cytokinin signaling pathway. Qin et al. [26] showed that ABA can promote the biosynthesis of auxin and inhibit the elongation of primary roots in rice [27]. Drought stress can affect root development. For example, PEG simulation of drought stress can inhibit root hair elongation in 35S:ZmTIP1Mo17 and 35S:ZmTIP1CIMBL55 transgenic Arabidopsis [28]. In this study, root elongation of the WT and three transgenic lines was inhibited after ABA and drought treatment, which is consistent with previous research results. When no stress treatment was applied, the GhWRKY91 transgenic lines grown on 1/2 MS solid medium had a stronger green leaf color and smaller plant size than the WT, which is consistent with the results of our previous research on GhWRKY91 transgenic and WT plants in nutrient soil [14]. In addition, the root length of GhWRKY91 transgenic plants was shorter than that of WT in the absence of stress treatment. Dark green leaves can promote photosynthesis and effectively increase crop yield [29]. Root systems play a crucial role in soil nutrient absorption capacity and largely control plant productivity and adaptation to different soil conditions [30]. A greener leaf color indicates that GhWRKY91 transgenic plants may have high yield potential; however, yield potential may be limited by shorter root length. Our previous research found that transgenic Arabidopsis with GhWRKY91 can delay ABA- and drought-induced leaf senescence, exhibiting insensitivity to ABA and drought [14]. In this study, after ABA and drought treatment, the root length of GhWRKY91 transgenic A. thaliana decreased more than that of WT, indicating sensitivity to ABA and drought. This result is inconsistent with our previous research results, and there are three possible reasons for this discrepancy. First, different treatment periods were used. In this study, germinated seeds were used, while in the previous study, 3-week seedlings were used; second, the growth environment was different. In this study, plants were grown on 1/2 MS solid medium and cultured in a light incubator. In the previous study, plants were grown in nutrient soil and cultured in a greenhouse; third, different observation sites were observed. In this study, the root system was observed, while in previous studies, leaf senescence was observed. The GhWRKY91 transgenic seedlings had increased sensitivity to ABA and drought compared to the WT, indicating that the transgenic seedlings may be more susceptible to stress conditions at the seedling stage. However, the effects of ABA and drought treatment on growth and development as well as root elongation in the transgenic plants need to be further confirmed by measuring a series of morphological, physiological and biochemical indicators, including chlorophyll content, stomata, superoxide dismutase, peroxidase, and catalase activity. In addition, the molecular mechanism by which GhWRKY91 participates in regulating root elongation still needs further exploration.
BiFC is a technology that intuitively and quickly determines the location and interaction of target proteins in living cells and can utilize the characteristics of fluorescent proteins to study protein interactions [31,32]. For example, Gu et al. [33] conducted a BiFC experiment using Arabidopsis protoplasts to demonstrate that GhWRKY27 and GhTT2 interact to jointly regulate the expression of the cytochrome P450 gene GhCYP94C1 and ripen-related protein GhRipen2-2 in the process of leaf senescence. Li et al. [34] showed that AcMYB1 and AcbHLH1 of Hypericum perforatum interacted in the protoplast of A. thaliana and participated in the biosynthesis of anthocyanidin. Therefore, BiFC experiments are also an effective means of analyzing the molecular mechanisms of gene regulation. Previous studies have found that the GhWRKY3 gene can be transcriptionally upregulated by the plant hormones SA, ABA, MeJA, ET, GA and H2O2 and is also induced by injury and infection by three fungal pathogens, Rhizoctonia solani, Colletotrichum gossypii and Fusarium oxysporum f. sp. vasinfectum but is not induced by 6-BA, NAA, drought, salt and cold [15]. In addition, the GhWRKY3 gene is constitutively expressed in roots [15], and its homolog in Arabidopsis thaliana, AtWRKY3, regulates root elongation under stress treatments [16], suggesting that GhWRKY3 has potential functions in root growth and development. In this study, GhWRKY91 can interact with GhWRKY3, indicating that they may jointly participate in regulating multiple stress responses and root elongation regulation, which provides a preliminary exploration for the mechanism of GhWRKY91 in regulating root growth. The BiFC assay confirmed that the GhWRKY91 and GhWRKY3 interact in tobacco leaf cells; however, whether they interact in the nucleus requires further verification by DAPI staining. Furthermore, the interaction between GhWRKY91 and GhWRKY3 proteins requires further validation using techniques such as yeast two-hybrid, co-immunoprecipitation, and GST pull down. Whether GhWRKY3 is involved in the regulation of root growth and development needs to be further verified by obtaining transgenic plants through transgenic technology in the future.
In summary, using GhWRKY91 transgenic A. thaliana obtained previously, we further studied the effects of ABA and drought on the root growth of transgenic A. thaliana. ABA and drought negatively regulated the root elongation of GhWRKY91 transgenic A. thaliana, and GhWRKY91 and GhWRKY3 interacted. In our future research, transgenic cotton with GhWRKY91 gene overexpression, RNAi, gene editing deletion and gene editing transcription activation needs to be achieved for further functional verification to analyze the effect of GhWRKY91 on root development in cotton. In addition, we can use yeast one-hybrid, electrophoretic mobility shift assay (EMSA), chromatin immunoprecipitation sequencing (ChIP-Seq), dual luciferase assay and other technologies to further identify the downstream target genes of GhWRKY91 regulation of root development and explore the molecular mechanism of GhWRKY91 regulation of root development. Our findings provide a basis for the future development of resistant cotton materials.
Acknowledgement: Not applicable.
Funding Statement: This research was supported by the Hebei Agricultural University Introduced Talents Scientific Research Project (No. YJ2021011).
Author Contributions: Study conception and design: Lijiao Gu; data collection: Yuqing Wang; analysis and interpretation of results: Yueying Liu; draft manuscript preparation: Yueying Liu, Yuqing Wang and Lijiao Gu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analysed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends in Plant Science, 5(5), 199–206. [Google Scholar] [PubMed]
2. Ishiguro, S., Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Molecular Genetics and Genomics, 244(6), 563–571. [Google Scholar]
3. Qu, R. J., Cao, Y. W., Tang, X. Q., Sun, L. Q., Wei, L. et al. (2022). Identification and expression analysis of the WRKY gene family in Isatis indigotica. Gene, 783, 145561. [Google Scholar]
4. Yu, Z. F., Zhang, D. D., Zeng, B., Liu, X. Y., Yang, J. H. et al. (2022). Characterization of the WRKY gene family reveals its contribution to the adaptability of almond (Prunus dulcis). PeerJ, 10, e13491. [Google Scholar] [PubMed]
5. Cheng, Y. F., Lu, J. X., Li, H., Wei, F., Zhang, Y. Q. et al. (2022). Identification of the WRKY gene family and characterization of stress-responsive genes in Taraxacum kok-saghyz rodin. International Journal of Molecular Sciences, 23(18), 10270. [Google Scholar] [PubMed]
6. Ayoub, K. M., Kang, D. R., Wu, Y. F., Wang, Y., Ai, P. H. et al. (2022). Characterization of WRKY gene family in whole-genome and exploration of flowering improvement genes in Chrysanthemum lavandulifolium. Frontiers in Plant Science, 13, 861193. [Google Scholar]
7. Zheng, X. D., Zhao, Y., Shan, D. Q., Shi, K., Wang, L. et al. (2018). MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytologist, 217(3), 1086–1098. [Google Scholar] [PubMed]
8. Fan, Z. Q., Tan, X. L., Shan, W., Kuang, J. F., Lu, W. J. et al. (2018). Characterization of a transcriptional regulator, BrWRKY6, associated with Gibberellin-suppressed leaf senescence of chinese flowering cabbage. Journal of Agricultural and Food Chemistry, 66(8), 1791–1799. [Google Scholar] [PubMed]
9. Gan, Z. Y., Yuan, X., Shan, N., Wan, C. P., Chen, C. Y. et al. (2021). AcWRKY40 mediates ethylene biosynthesis during postharvest ripening in kiwifruit. Plant Science, 309, 110948. [Google Scholar] [PubMed]
10. Zhang, J. W., Huang, D. Z., Zhao, X. J., Zhang, M., Wang, Q. et al. (2022). Drought-responsive WRKY transcription factor genes IgWRKY50 and IgWRKY32 from Iris germanica enhance drought resistance in transgenic Arabidopsis. Frontiers in Plant Science, 13, 983600. [Google Scholar] [PubMed]
11. Zhang, J., Peng, Y. L., Guo, Z. J. (2008). Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Research, 18(4), 508–521. [Google Scholar] [PubMed]
12. Hu, Z. R., Wang, R., Zheng, M., Liu, X. B., Meng, F. et al. (2018). TaWRKY51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum L.). The Plant Journal, 96(2), 372–388. [Google Scholar] [PubMed]
13. Li, C., Li, K. N., Liu, X. Y., Ruan, H., Zheng, M. M. et al. (2021). Transcription factor GmWRKY46 enhanced phosphate starvation tolerance and root development in transgenic plants. Frontiers in Plant Science, 12, 700651. [Google Scholar] [PubMed]
14. Gu, L. J., Ma, Q., Zhang, C., Wang, C. C., Wei, H. L. et al. (2019). The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Frontiers in Plant Science, 10, 1352. [Google Scholar] [PubMed]
15. Guo, R. Y., Yu, F. F., Gao, Z., An, H. L., Cao, X. C. et al. (2011). GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Molecular Biology Reports, 38(1), 49–58. [Google Scholar] [PubMed]
16. Li, P., Li, X. W., Jiang, M. (2021). CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Molecular Biology Reports, 48(8), 5821–5832. [Google Scholar] [PubMed]
17. Gong, Z. Z., Xiong, L. M., Shi, H. Z., Yang, S. S., Herrera-Estrella, L. R. et al. (2020). Plant abiotic stress response and nutrient use efficiency. Science China-Life Sciences, 63(5), 635–674. [Google Scholar] [PubMed]
18. Paes de Melo, B., Carpinetti, P. A., Fraga, O. T., Rodrigues-Silva, P. L., Fioresi, V. S. et al. (2022). Abiotic stresses in plants and their markers: A practice view of plant stress responses and programmed cell death mechanisms. Plants, 11(9), 1100. [Google Scholar] [PubMed]
19. Zhang, H. M., Zhu, J. H., Gong, Z. Z., Zhu, J. K. (2022). Abiotic stress responses in plants. Nature Reviews Genetics, 23(2), 104–119. [Google Scholar] [PubMed]
20. Salvi, P., Agarrwal, R., Kajal., Gandass, N., Manna, M. et al. (2022). Sugar transporters and their molecular tradeoffs during abiotic stress responses in plants. Physiologia Plantarum, 174(2), e13652. [Google Scholar] [PubMed]
21. Yang, Y. L., Cushman, S. A., Wang, S. C., Wang, F., Li, Q. et al. (2013). Genome-wide investigation of the WRKY transcription factor gene family in weeping forsythia: Expression profile and cold and drought stress responses. Genetica, 151(2), 153–165. [Google Scholar]
22. Ehsan, A., Naqvi, R. Z., Azhar, M., Awan, M. J. A., Amin, I. et al. (2023). Genome-wide analysis of WRKY gene family and negative regulation of GhWRKY25 and GhWRKY33 reveal their role in whitefly and drought stress tolerance in cotton. Genes, 14(1), 171. [Google Scholar] [PubMed]
23. Guo, Q., Zhao, L., Fan, X. Q., Xu, P., Xu, Z. Z. et al. (2019). Transcription factor GarWRKY5 is involved in salt stress response in diploid cotton species (Gossypium aridum L.). International Journal of Molecular Sciences, 20(21), 5244. [Google Scholar] [PubMed]
24. Wang, N. N., Xu, S. W., Sun, Y. L., Liu, D., Zhou, L. et al. (2019). The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Scientific Reports, 9(1), 724. [Google Scholar] [PubMed]
25. Takatsuka, H., Umeda, M. (2019). ABA inhibits root cell elongation through repressing the cytokinin signaling. Plant Signaling & Behavior, 14(3), e1578632. [Google Scholar]
26. Qin, H., Wang, J., Zhou, J. H., Qiao, J. Z., Li, Y. X. et al. (2023). Abscisic acid promotes auxin biosynthesis to inhibit primary root elongation in rice. Plant Physiology, 191(3), 1953–1967. [Google Scholar]
27. Gupta, A., Rico-Medina, A., Cano-Delgado, A. I. (2020). The physiology of plant responses to drought. Science, 368(6488), 266–269. [Google Scholar] [PubMed]
28. Zhang, X. M., Mi, Y., Mao, H., Liu, S. X., Chen, L. M. et al. (2020). Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize. Plant Biotechnology Journal, 18(5), 1271–1283. [Google Scholar] [PubMed]
29. Su, X. J., Yue, X. N., Kong, M. Y., Xie, Z. W., Yan, J. H. et al. (2023). Leaf color classification and expression analysis of photosynthesis-related genes in inbred lines of Chinese cabbage displaying minor variations in dark-green leaves. Plants, 12(11), 2124. [Google Scholar] [PubMed]
30. Cholidah, L., Wang, L. C., Ma, S. Min (2018). Short term effect of biochar application on rapeseed root performance and nutrient content of purple soil: A greenhouse study. Summary and Summary of the 8th Member Representative Conference and Academic Annual Meeting of the Oil Crop Professional Committee of the Chinese Crop Society, Qingdao, Shandong, China. [Google Scholar]
31. Edgar, C. R., Dikeakos, J. D. (2022). Bimolecular fluorescence complementation to visualize protein-protein interactions in cells. Methods in Molecular Biology, 2440, 91–97. [Google Scholar] [PubMed]
32. Chen, J. J., Yu, Z. L., Unruh, J. R., Slaughter, B. D., Jaspersen, S. L. (2020). Super-resolution microscopy-based bimolecular fluorescence complementation to study protein complex assembly and co-localization. Bio-Protocol, 10(4), e3524. [Google Scholar] [PubMed]
33. Gu, L. J., Dou, L. L., Guo, Y. N., Wang, H. T., Li, L. B. et al. (2019). The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.). BMC Plant Biology, 19(1), 116. [Google Scholar] [PubMed]
34. Li, J., Wu, K. L., Li, L., Ma, G. H., Fang, L. et al. (2022). AcMYB1 interacts with AcbHLH1 to regulate anthocyanin biosynthesis in Aglaonema commutatum. Frontiers in Plant Science, 13, 886313. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools