 Open Access
Open Access
ARTICLE
Ectopic Overexpression of EuCHIT30.7 Improves Nicotiana tabacum Resistance to Powdery Mildew
1 Key Laboratory of Mountain Plant Resources Protection and Germplasm Innovation of the Ministry of Education, College of Life Sciences/Institute of Agricultural Bioengineering, Guizhou University, Guiyang, 550025, China
2 Plant Conservation Technology Center, Guizhou Key Laboratory of Agricultural Biotechnology, Biotechnology Institute of Guizhou, Guizhou Academy of Agricultural Sciences, Guiyang, 550006, China
* Corresponding Author: Degang Zhao. Email:
Phyton-International Journal of Experimental Botany 2023, 92(11), 3043-3061. https://doi.org/10.32604/phyton.2023.031175
Received 18 May 2023; Accepted 08 August 2023; Issue published 24 October 2023
Abstract
Various strains of powdery mildew (PM), a notorious plant fungal disease, are prevalent and pose a significant threat to plant health. To control PM, transgenic technology can be used to cultivate more resistant plant varieties. In the present study, we utilized the rapid amplification of cDNA ends (RACE) technique to clone the full-length cDNA sequence of the EuCHIT30.7 gene to explore plant genes with disease resistance functions. Bioinformatics analysis revealed that this gene belongs to the GH18 family and is classified as a class III chitinase. The EuCHIT30.7 gene is expressed throughout the Eucommia ulmoides plant, with the most abundant expression in male flowers. Subcellular localization analysis indicated that the protein encoded by this gene was detected within both the cell membrane and cytoplasm. Upon PM inoculation, overexpression of EuCHIT30.7 in tobacco plants led to a significantly reduced relative lesion area and a decreased spore count compared to both wild-type and empty vector control plants. Activities of the protective enzymes, namely, peroxidase (POD), superoxide dismutase (SOD), catalase (CAT), and phenylalaninammo-nialyase (PAL), in tobacco plants overexpressing EuCHIT30.7 were significantly greater than those in wild-type and empty vector tobacco plants. Furthermore, the rate of increase in malondialdehyde (MDA) content was significantly lower in tobacco plants expressing EuCHIT30.7 compared to control tobacco plants. In EuCHIT30.7 transgenic tobacco, the expression of pathogen-related protein genes, namely, PR2, PR5, PR1a, PDF1.2, and MLP423, along with the tobacco PM negative regulatory gene, MLO2, were significantly higher compared to control tobacco plants. These findings suggested that EuCHIT30.7 significantly enhances the resistance of tobacco to PM.Keywords
Powdery mildew (PM) is a widespread and serious fungal disease of plants, and PM of different plants is usually caused by the infestation of different species of PM fungi [1]. Currently, the control of PM primarily relies on the use of chemical agents. However, excessive use of these chemicals may have a negative impact on the effectiveness of PM control because it not only increases the pathogen’s resistance to chemical agents but may also be harmful to human health and the environment. Therefore, an alternative strategy to control PM is to breed resistant plant varieties using transgenic technology.
Plant chitinases are a group of pathogenesis-related (PR) proteins. Based on phylogeny, plant chitinases are classified into classes I to VII [2]. The GH18 family contains both class III and V chitinases. Class III chitinases are mainly endochitinases, which randomly cleave chitin into chitin oligosaccharides of different sizes [3]. Amid pathogenic incursions, plant chitinases decompose chitin present in the cell walls of pathogenic fungi, and the resultant chitin oligosaccharides are identified by pathogen-associated molecular pattern (PAMP) recognition receptors, thereby inducing plant immunological responses [4]. Simultaneously, chitinases act as effectors of the immune response to disrupt fungal cell wall integrity, thereby resisting pathogen invasion [5].
Eucommia ulmoides Oliv. is the only genus and species in its family. This plant, a nationally protected species of the second class, is important in traditional Chinese medicine, with a plethora of medicinal applications [6,7]. E. ulmoides contains various antifungal proteins, such as chitinase and antimicrobial peptides. We have previously cloned and studied multiple chitinase genes. We transferred the EuCHIT1 gene into Micro-Tom tomato to improve resistance to Botrytis cinerea [8] and into wheat to improve resistance to stripe rust [9]. Dong et al. (2017) successfully cloned EuCHIT2, an E. ulmoides chitinase gene, and confirmed its resistance against PM in tobacco (Nicotiana tabacum). The antimicrobial peptide gene, EuAFP1.2, of E. ulmoides has been transferred into tobacco, resulting in inhibitory effects against Rhizopus microsporus [10]. However, because EuCHIT2 is part of the GH19 family and we have yet to clone and functionally analyze the E. ulmoides GH18 family genes, we utilized the E. ulmoides genome and transcriptome databases to screen for a new E. ulmoides chitinase gene belonging to the GH18 family, namely EuCHIT30.7. In the present study, we cloned this gene and genetically transformed tobacco to study its physiological and molecular mechanisms in the resistance to PM disease. In addition, we comprehensively evaluated the role of the chitinase 18 family within E. ulmoides, thereby providing a theoretical foundation for the molecular cultivation of disease-resistant crops.
All seeds were picked from E. ulmoides Oliver that were cultivated in the experimental field of our laboratory, and the Sanxing tobacco seeds used for transformation were preserved in the laboratory (Guiyang, China).
Escherichia coli strain DH5α was maintained by the Guizhou Institute of Agricultural Biology (Guiyang, China). Agrobacterium tumefaciens strain GV3101 and plant expression vectors were purchased from Wuhan Transduction Biology Laboratory Co., Ltd. (Wuhan, China).
2.2 Rapid Amplification of cDNA Ends (RACE) Cloning of the EuCHIT30.7 Gene from E. ulmoides
An Ultrapure RNA Kit (Kangwei Century, Beijing, China) was used to extract the total RNA of E. ulmoides seedling leaves according to the manufacturer’s instructions. RACE cloning was performed using a SMARTer RACE 5′/3′ Kit (TaKaRa, Shiga, Japan) to convert RNA into 5′/3′-RACE-Ready cDNA according to the manufacturer’s instructions. The EuCHIT30.7 reference sequence was obtained from a previously established E. ulmoides genome and transcriptome database [11]. The GSP1 and GSP2 specific primers were designed according to the requirements of the SMARTer RACE 5′/3′ Kit (TaKaRa) (Table 1). The following amplification protocol was utilized: denaturation at 94°C for 30 s; annealing at 65°C for 30 s; extension at 72°C for 3 min; and storage at 12°C. After electrophoresis using a 1.2% agarose gel, the single target band was recovered using a gel recovery kit (TaKaRa) and then recombined with PUC19-T by homologous recombination. After transforming DH5α competent cells, blue and white spots were screened on Luria-Bertani (LB) medium containing 100 mg·L−1 ampicillin (Amp). Positive single colonies were verified by colony PCR and then sent to Beijing Qingke Biotechnology Co., Ltd. (Wuhan, China) for sequencing.
2.3 Coding Sequence (CDS) Cloning of the EuCHIT30.7 Gene from E. ulmoides
An Ultrapure RNA Kit (Kangwei Century) was used to extract total RNA from the stem bark of E. ulmoides, and RNA was reverse-transcribed into cDNA by a ReverAid First Strand cDNA Synthesis Kit (Thermo Scientific, Vilnius, Lithuania). The EuCHIT30.7-F and EuCHIT30.7-R primers were designed according to the full-length cDNA sequence of EuCHIT30.7 (Table 1). The CDS was amplified by high-fidelity DNA polymerase using cDNA as template. After double enzyme digestion, the CDS was ligated into PUC19-T vectors by T4 DNA ligase. After transformation of DH5α competent cells, selective culture was conducted on LB medium supplemented with 100 mg·L−1 Amp. Positive colonies with Amp resistance were identified by colony PCR, and the bacterial liquid was sent to Beijing Qingke Biotechnology Co., Ltd. for sequencing.
2.4 Bioinformatics Analysis of the Protein Encoded by EuCHIT30.7
The physical properties, chemical properties, and hydrophobicity of the protein encoded by EuCHIT30.7 were predicted by the ProtParam ExPASy program (https://web.expasy.org/protparam/) and ProteinScale online software (https://web.expasy.Org/protscale/). The conserved domain was predicted using the Simple Modular Architecture Research (SMART) Tool (http://smart.embl.de/), and the signal peptide sequence was predicted using Signal P online software (https://services.healthtech.dtu.dk/services/SignalP-5.0/). The amino acid sequences of the Arabidopsis chitinase gene family were obtained from the Arabidopsis Information Resource site (https://www.arabidopsis.org/). To construct a systematic phylogenetic tree using the neighbor-joining method, Mega 11 software (Auckland, New Zealand) was utilized, and the resulting tree was enhanced using the iTOL online website (https://itol.embl.de/) for aesthetic purposes.
2.5 EuCHIT30.7 Spatial Expression Pattern Analysis
An Ultrapure RNA Kit (Kangwei Century) was used to extract total RNA from young leaves, stem bark, pericarp, root bark, and male flowers of 18-year-old E. ulmoides male and female plants according to the manufacturer’s instructions. RNA was reverse-transcribed into cDNA by a Reverse Transcriptase M-MLV Kit (TaKaRa). The primers designed for reverse-transcription quantitative PCR (RT-qPCR) were EuCHIT30.7-qF and EuCHIT30.7-qR (Table 1). The E. ulmoides EuActin [12] gene was used as the internal reference gene (Table 1). RT-qPCR analysis (BIO-RAD, Beijing, China) of EuCHIT30.7 was performed using a RT-qPCR kit (Novoprotein, Suzhou, China).
2.6 EuCHIT30.7 Subcellular Localization Analysis
Subcellular localization of the EuCHIT30.7 protein was determined by transient expression in Nicotiana benthamiana leaf epidermis. A. tumefaciens GV3101 containing pCAMBIA-1300-35S-EuCHIT30.7-enhanced green fluorescent protein (EGFP) plasmids were activated and cultured, and the cells were collected and resuspended in a heavy suspension [10 mmol·L−1 MgCl2, 10 mmol·L−1 2-morpholinoethanesulfonic acid (MES), and 0.1 mmol·L−1 acetosyringone (AS)]. To achieve an optical density (OD600) of approximately 0.5, the suspension was agitated and left undisturbed for 2–3 h. The suspension was then injected into the lower epidermis of 3- to 4-week-old tobacco leaves. After a 3-day transformation period, observations were conducted [13]. Leaves transformed with no-load pCAMBIA1300-35s-EGFP were used as the control. The EGFP fluorescence was observed by a laser confocal microscope, with an excitation wavelength of 488 nm and an emission wavelength of 507 nm.
2.7 Agrobacterium-Mediated Genetic Transformation of Tobacco
Tobacco was transformed by a leaf disk method mediated by A. tumefaciens [14]. Leaves of young sterile seedlings were cut into 1 cm2 squares, and the leaf squares were then put into A. tumefaciens culture carrying the pCAMBIA1301-35S-EuCHIT30.7 expression vector to infect for 5 min. During the infection period, the bacterial solution was continuously shaken to ensure leaves were in full contact with bacteria. The solution was then removed, and the leaf squares were dried with absorbent paper. The leaf squares were inoculated on co-culture medium and cultured in the dark for 48 h, and the leaf squares were then transferred to screening medium with 50 mg·L−1 hygromycin at 23°C. The photoperiod was 16 h, and culture medium was changed every 14 days. When the resistant buds grew to 1 to 2 cm in length, they were cut with sterilized scissors and inserted into the rooting medium for rooting culture.
2.8 Identification of EuCHIT30.7 Transgenic Tobacco Plants
Newly emerged leaves of resistant tobacco plants were used for anti-β-glucuronidase (GUS) tissue staining. Leaves were reacted at 37°C for 12 h and then decolorized at 37°C with 10%, 25%, 50%, 75%, and 95% ethanol [15]. Total DNA was extracted from resistant tobacco plants stained blue by GUS using a plant total DNA extraction kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Verification was performed by PCR amplification using the VF and VR primers designed to amplify the specific verification sequence (Table 1), including the 35s to EuCHIT30.7 common 205 bp sequence to verify the resistant plants. The GUS-F and GUS-R GUS primers (Table 1) were designed to verify empty vector plants.
2.9 Identification of Transgenic Tobacco Plant Resistance to PM
Nine leaves in a similar physiological state were selected from wild-type, empty vector, and transgenic tobacco lines at the 6- to 8-leaf stage and placed in a Petri dish containing moist absorbent paper. Petioles were wrapped in wet, aseptic, defatted cotton. Inoculation was conducted according to the methods of Miclot and Moyer [16–18]. After repeatedly washing 6 to 7 tobacco leaves infected with PM with 15 mL of sterile water and 1 drop of Tween-20, the spores were filtered with five layers of fine gauze. The spore suspension was collected and then counted using a blood cell count plate after centrifugation. The concentration of the spore suspension was adjusted to 106 per milliliter. Then, 10 drops (approximately 5 μL) of spore suspension was inoculated and evenly spread on the whole leaf surface with a glass rod. After drying for 25 min at room temperature, inoculated tobacco leaves were enclosed in a Petri dish and cultured at 25°C under a 16-h light/8-h dark cycle. Observations were recorded daily after inoculation. After inoculation, three 1 cm2 diseased sections were cut from each leaf, and the spores on the leaves were repeatedly washed with 2 mL of 0.85% NaCl isotonic solution. Then, 20 μL of the suspended spore suspension was used for blood cell plate counting, and the average value was obtained by repeating counts three times. The tobacco used for expression analysis and physiological index determination was inoculated as a whole plant. All experiments were repeated three times.
2.10 Determination of Physiological Indicators
At the 6- to 8-leaf stage, wild-type, empty vector, transgenic tobacco leaves, and homolateral leaves were inoculated with PM for 0, 12, 24, 48, and 72 h. The activities of the chitinase (EC 3.2.1.14) [19,20], catalase (CAT; EC 1.1.3.10) [21], superoxide dismutase (SOD; EC 1.1.3.9) [22,23], peroxidase (POD; EC 1.3.3.6) [24,25], and phenylalanine ammonia lyase (PAL; EC 4.3.1.24) [26] enzymes, as well as the content of malondialdehyde (MDA; EC 1.1.1.2) [22,23], were determined using kits from Solarbio Biological Co., Ltd. (Beijing, China) according to the manufacturer’s instructions. Wild-type and empty vector tobacco plants were used as the controls. All experiments were repeated three times.
2.11 Expression Analysis of Tobacco Resistance Genes and Mildew Resistance Locus O (MLO) Genes Related to PM Resistance
Sequences of the tobacco internal reference gene, namely, guanine-N(7)-methyltransferase (18s), and the PR genes, namely, pathogenesis-related protein 1a (PR1a), pathogenesis-related protein 2 (PR2), pathogenesis related protein 5 (PR5), plant defensin 1.2 (PDF1.2) [27], major latex protein (MLP)-like protein 423 (MLP423) [28], MLO2, and XTH [29], were obtained from GenBank, and RT-qPCR primers were designed (Table 1). Wild-type, empty vector, and transgenic tobacco lines at the 6- to 8-leaf stage were inoculated with PM at 0, 12, 24, 48, and 72 h. Total RNA was extracted using TRIzol and reverse-transcribed into cDNA. To measure gene expression levels, a Novoprotein SYBR Green dye kit (Suzhou, China) was used according to the manufacturer’s instructions, and the gene expression levels were calculated as previously described [30] using SPSS 22.0 (Chicago, IL, USA) and GraphPad Prism 8.0. (CA, USA). All experiments were repeated three times.
All data are presented as the mean ± standard error. Data were analyzed using one-way analysis of variance (ANOVA), followed by Duncan’s test, using IBM SPSS Statistics 27 software (IBM Corporation, Chicago, IL, USA). Different letters indicate significant differences at p < 0.05.
3.1 RRACE Cloning and Sequence Analysis of the EuCHIT30.7 Gene from E. ulmoides
Using reverse-transcribed 3′ and 5′-RACE-Ready cDNA as templates, PCR amplification was performed with the GSP1- and GSP2-specific primers, in addition to the UPM RACE primer. The electrophoresis results showed that 3′ and 5′ end fragments of EuCHIT30.7 were obtained by RACE amplification (Fig. 1). The full-length sequence of the EuCHIT30.7 chitinase gene of E. ulmoides was obtained by sequencing and splicing, and it was 1069 bp in length and included an 867-bp open reading frame. The start codon (ATG) was at positions 44 to 46, and the stop codon (TGA) was at positions 908 to 910. In addition, the terminal tailing signal sequence (AATAAA) was at positions 1010 to 1015. The sequence encoding a protein containing 288 amino acid residues included a 43-bp 5′ untranslated region (5′-UTR), a 158-bp 3′ untranslated region (3′-UTR), and a 33-bp poly (A) tail (Fig. 2).
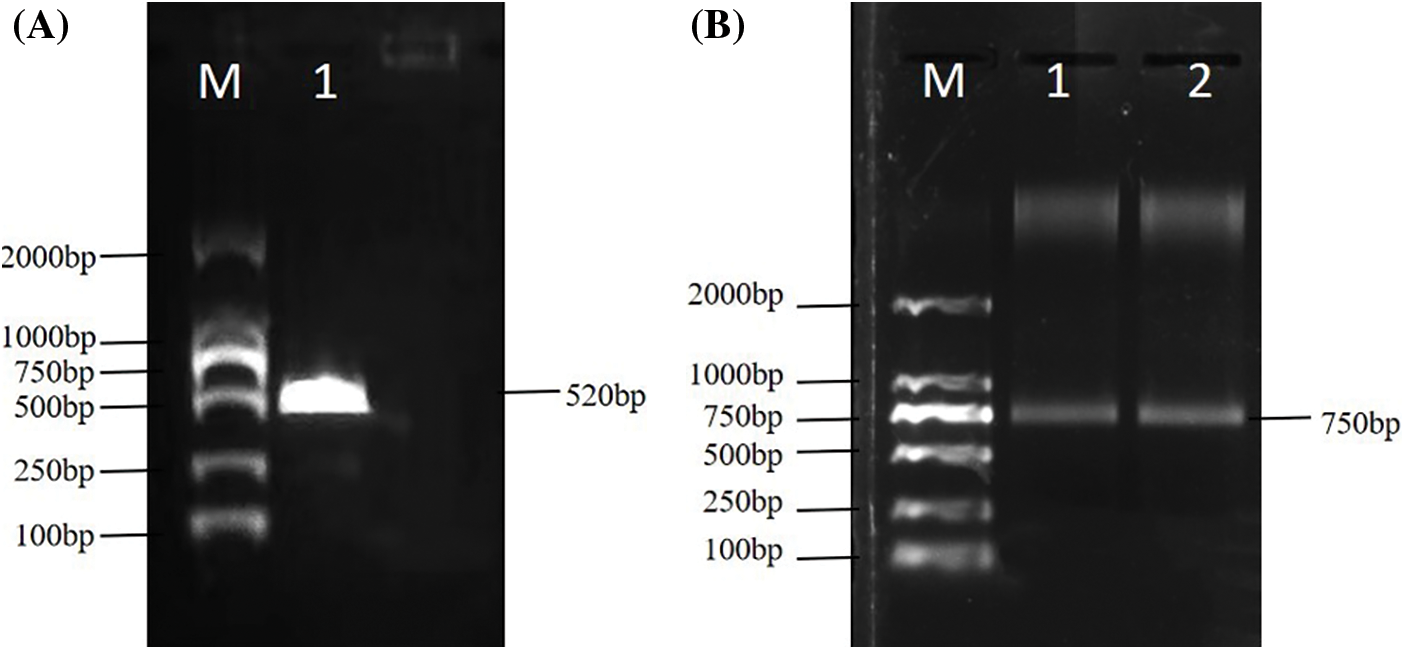
Figure 1: (A) RACE electrophoresis of the 5′ end of the EuCHIT30.7 gene. M: DNA marker; 1: 5′-end amplified fragment. (B) RACE electrophoresis at the 3′-end of the EuCHIT30.7 gene. M: DNA marker; 1 and 2: 3′-end amplified fragment

Figure 2: Base sequence of the EuCHIT30.7 chitinase gene of E. ulmoides and its encoded amino acid sequence. The sequence total length is 1069 bp. The nontranslated 5′-UTR and 3′-UTR ends are marked with gray shading, and the poly (A) tail signal is marked with blue shading. The termination codon is marked with an asterisk (*)
3.2 CDS Cloning and Bioinformatics Analysis of the EuCHIT30.7 Gene
For cloning, reverse-transcribed E. ulmoides stem bark cDNA was used as the template. According to the mRNA sequence of EuCHIT30.7 amplified by RACE, primers were designed to amplify the EuCHIT30.7 gene, and a specific band of expected size was obtained (Fig. 3). The CDS of the E. ulmoides EuCHIT30.7 chitinase gene was obtained by sequencing. After blast alignment, the sequence was the same as that of the transcript, with a size of 867 bp, encoding a protein of 288 amino acid residues. The physicochemical properties of the protein encoded by the gene were predicted. Because the molecular mass of the protein was predicted to be 30.7 kDa, the gene was named EuCHIT30.7. The protein encoded by the gene was acidic, stable, and hydrophobic, and it contained a signal peptide (sequence: -MAIHSLLSLLLLLIALFKSTESA-) at the N-terminal end. Subcellular localization of the protein was predicted to be in vacuoles. In the phylogenetic tree of chitinase family genes in Arabidopsis thaliana, the gene was a class III chitinase in the chitinase 18 family (Fig. 4). Similarly, through conservative domain analysis, we predicted that the amino acid sequence encoded by this gene contains a complete chitinase 18 family domain (Fig. 5).
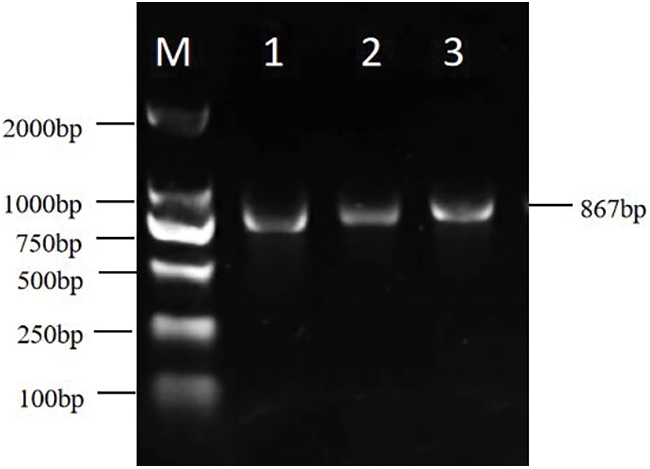
Figure 3: Electrophoresis detection of the amplified product of the EuCHIT30.7 coding sequence. M: DNA marker; 1 to 3: amplification products
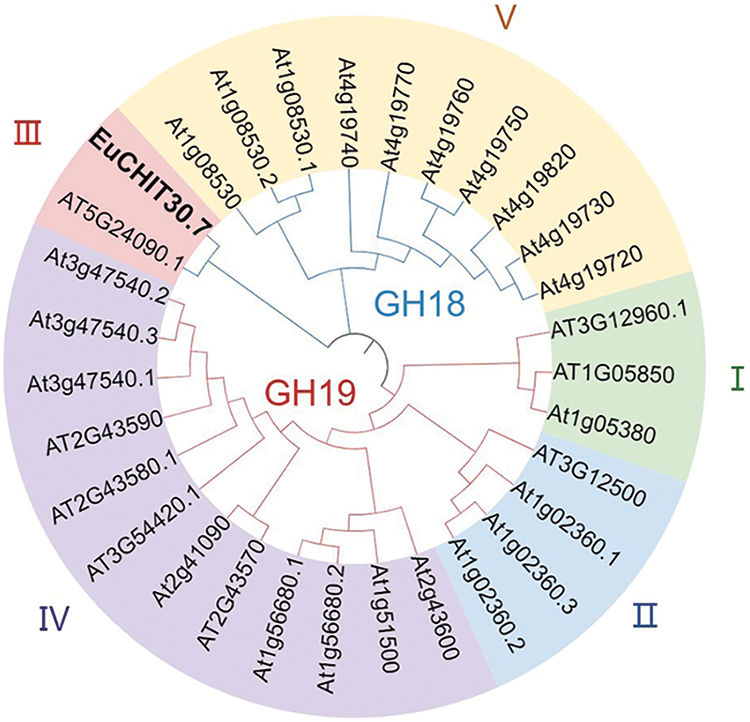
Figure 4: Phylogenetic tree of EuCHIT30.7. The protein was a class III chitinase in the GH18 chitinase family
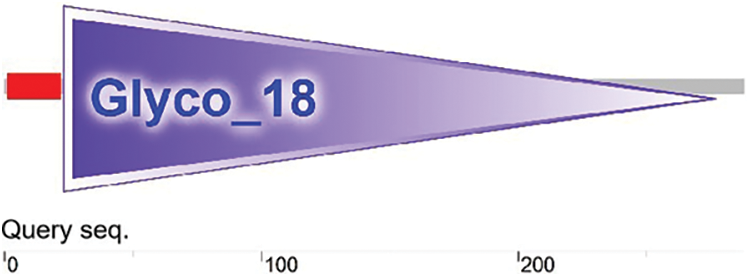
Figure 5: Prediction of the conserved domain in the EuCHIT30.7 protein
3.3 Analysis of Spatial Expression Pattern and Subcellular Localization of EuCHIT30.7
Expression of the EuCHIT30.7 gene in the root bark, tender leaf, tender stem bark, male flower, female young leaf, tender stem bark, and pericarp of 18-year-old E. ulmoides male and female plants was quantitatively analyzed by RT-qPCR. The EuCHIT30.7 gene was expressed in all parts of the plant, with its expression levels in the following order (from highest to lowest): male flower > pericarp > male leaf > female stem bark > female leaf > male stem bark > root bark (Fig. 6). The pCAMBIA1300-35S-EuCHIT30.7-EGFP fusion protein vector (Fig. 7) was constructed and transiently transformed in N. benthamiana leaves, and the empty EGFP vector was used as the control. Leaves containing the EGFP empty vector had strong green fluorescence in the nucleus, cytoplasm, and cell membrane, whereas leaves containing EuCHIT30.7-EGFP had strong green fluorescence in the cell membrane, cytoplasm, and around the nuclear membrane. Thus, EuCHIT30.7 was mainly localized in the cell membrane and cytoplasm, and may also be located in the nuclear membrane (Fig. 8).
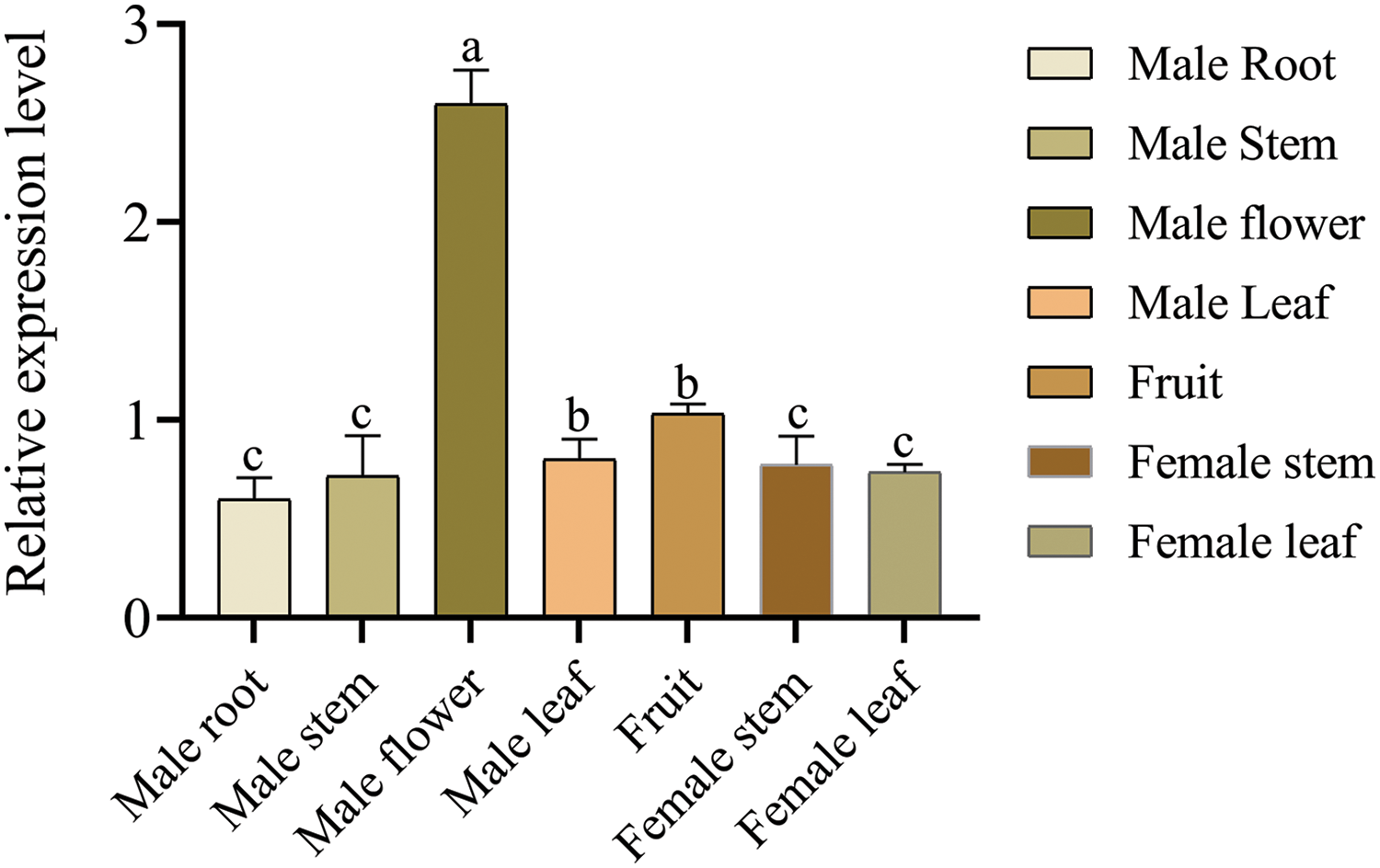
Figure 6: Expression pattern of the E. ulmoides EuCHIT30.7 gene. Different letters indicate significant differences at p < 0.05
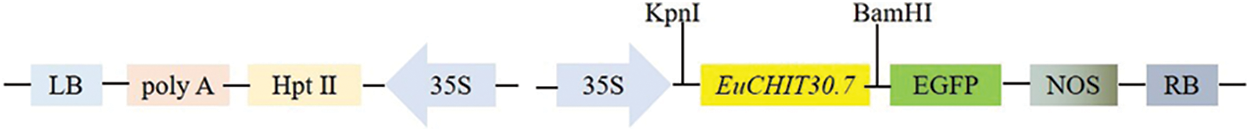
Figure 7: T-DNA structure of the pCAMBIA1301-35S-EuCHIT30.7-EGFP vector
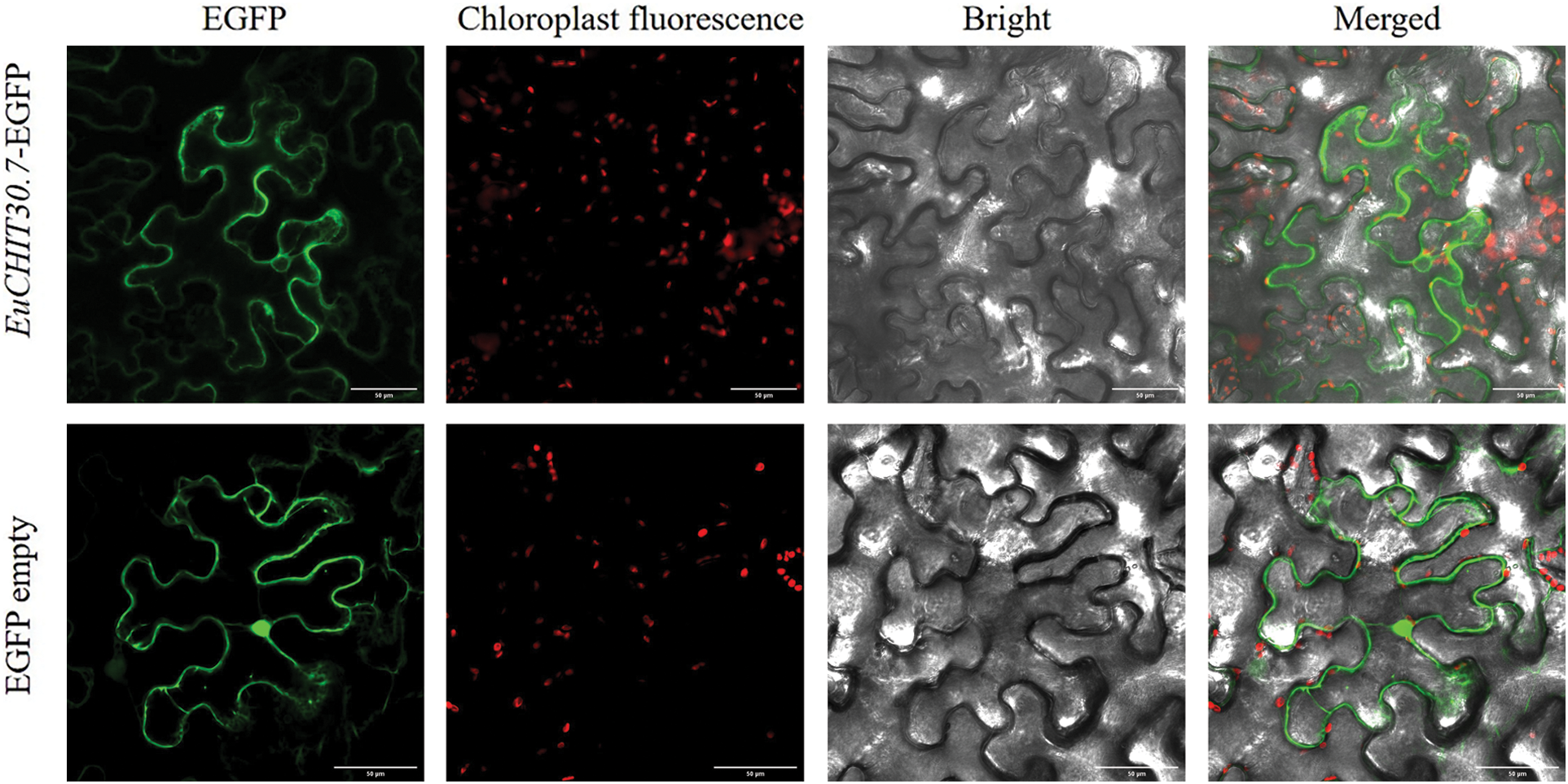
Figure 8: Subcellular localization of EuCHIT30.7. Bar = 50 μm. EGFP: Enhanced green fluorescent protein
3.4 Screening of Transgenic Tobacco Plants with the E. ulmoides chitinase EuCHIT30.7 Gene and Analysis of Resistance to PM
The pCAMBIA1301-35S-EuCHIT30.7 plant expression vector was constructed using a homologous recombination method (Fig. 9) and then transformed into tobacco by an A. tumefaciens-mediated leaf disk method (Figs. 10A–10D). Resistant plants were verified by GUS staining and PCR amplification. In total, 32 EuCHIT30.7 transgenic plant lines and 18 empty transgenic plant lines were obtained (Figs. 10E and 10F). Additionally, we assessed the expression levels of the EuCHIT30.7 gene in 17 lines, and we selected the TP8, TP15, and TP18 lines with relatively high levels of gene expression for subsequent experiments (Fig. 10G). The results of the chitinase enzyme activity assay indicate that the chitinase activity of the wild-type tobacco is 10.370 U·g−1, while the chitinase activity of the empty vector control EV is 10.740 U·g−1. On the other hand, TP8, TP15, and TP18 have chitinase activities of 20.741, 17.778, and 15.556 U·g−1, respectively. The chitinase activity in the overexpressing tobacco plants is 1.73 times and 1.67 times higher than that of the wild-type and empty vector plants, respectively (Fig. 10H). This demonstrates that the overexpression of EuCHIT30.7 in the transgenic tobacco plants enhances the activity of chitinase enzyme in tobacco plants.
Figure 9: T-DNA structure of vector pCAMBIA1301-35S-EuCHIT30.7
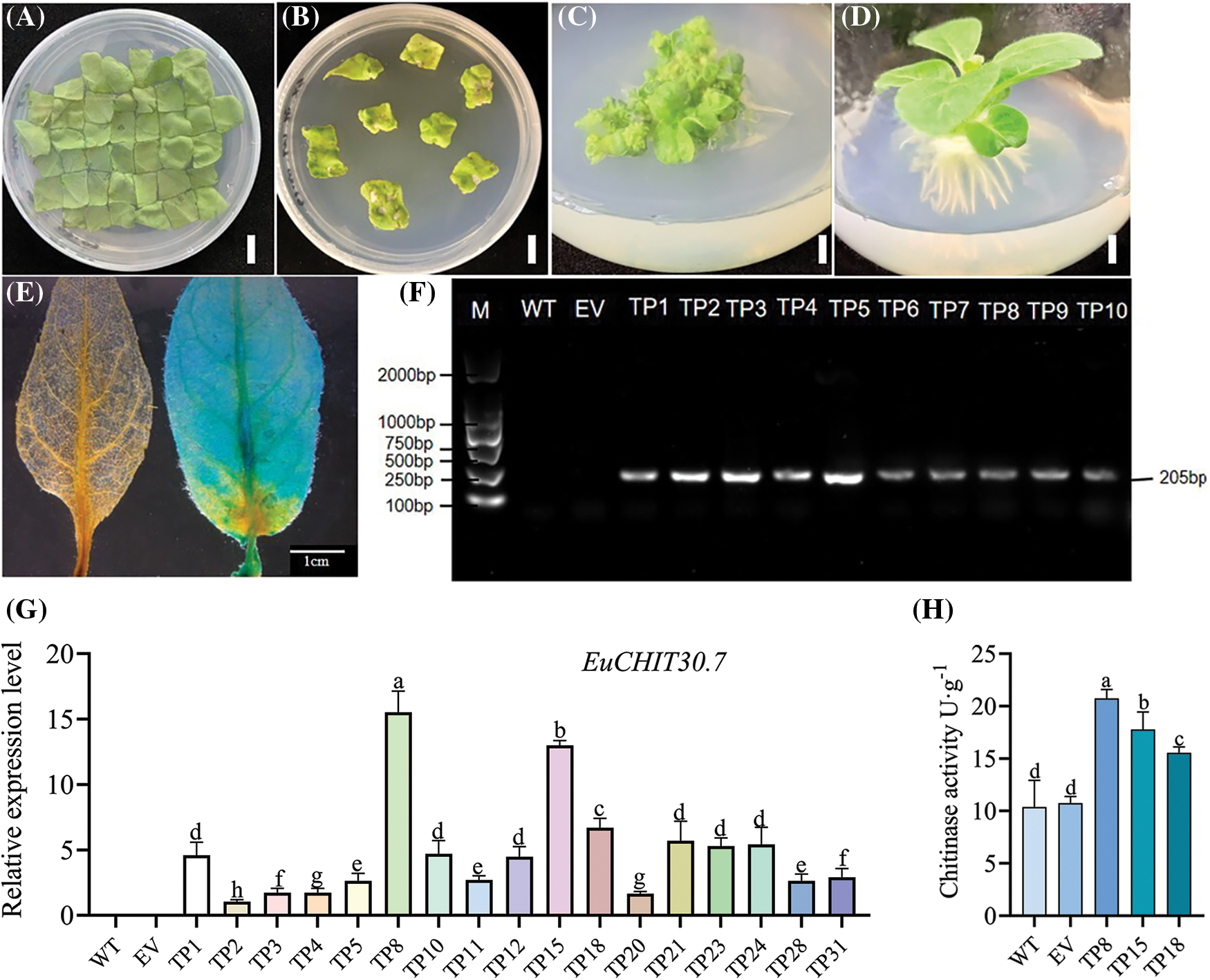
Figure 10: Genetic transformation process of tobacco and identification of transformed plants. (A) Coculture of transgenic tobacco; (B) Screening of transgenic tobacco; (C) Callus induction of transgenic tobacco; (D) Rooting of transgenic tobacco; (E) GUS staining verification of transgenic tobacco (left: wild type; right: transgenic); (F) PCR identification of transgenic plants (partial); (G) Eexpression level of EuCHIT30.7 in WT, EV and transgenic lines (partial); (H) Determination of chitinase activity of EuCHIT30.7 transgenic lines (TP8, TP15, and TP18). M: DNA marker; WT: wild-type plant; EV: empty-vector plant; TP1 to TP10: transgenic plants. Bar = 1 cm
The pathological transformations in tobacco leaves in vitro post-inoculation with PM fungus were comprehensively evaluated. There were no disease spots in wild-type, empty vector, and transgenic tobacco plants before inoculation, whereas disease spots appeared on leaves of the three types of tobacco plants Day 3 post-inoculation. Moreover, the disease spots in wild-type and empty vector tobacco plants were significantly larger than those in transgenic tobacco plants, and the leaf disease spots gradually expanded with time (Figs. 11A–11D). At Day 12 post-inoculation, the leaves of wild-type and empty vector tobacco plants were covered with many PM hyphae and spores, and the area of disease spots on the surface of transgenic tobacco leaves was relatively small, with mild symptoms (Fig. 11D). The relative disease spot area of transgenic tobacco leaves at Day 12 post-inoculation and the number of conidia per 1 cm2 of tobacco leaves at Days 3, 5, 8, and 12 post-inoculation were analyzed. The relative disease spot area of transgenic tobacco plants was approximately 24%, which was 2.54 times smaller than that of wild-type plants (61%) and 2.71 times smaller than that of empty vector plants (64%) (Fig. 12A). After inoculation, the number of PM spores on the leaves of the three transgenic tobacco lines (TP8, TP15, and TP18) was significantly lower than that of the wild-type tobacco plants. On Day 12, the wild-type, empty vector, and transgenic tobacco plants had 91,010, 88,907, and 25,353 spores/cm2, respectively (Fig. 12B). PM induced yellowing and wilting of tobacco leaves along with pathological alterations. However, EuCHIT30.7 transgenic tobacco plants effectively suppressed the proliferation of PM spores, thereby mitigating the symptoms of the disease.

Figure 11: Phenotypic observation of wild-type, empty-vector, and EuCHIT30.7 transgenic tobacco leaves inoculated with PM. (A) Leaves inoculated with PM for 3 days; (B) Leaves inoculated with PM for 5 days; (C) Leaves inoculated with PM for 8 days; (D) Leaves inoculated with PM for 12 days. WT: wild-type tobacco leaves; EV: empty-vector tobacco leaves; TP8, TP15, TP18: transgenic tobacco leaves. Bar = 1 cm
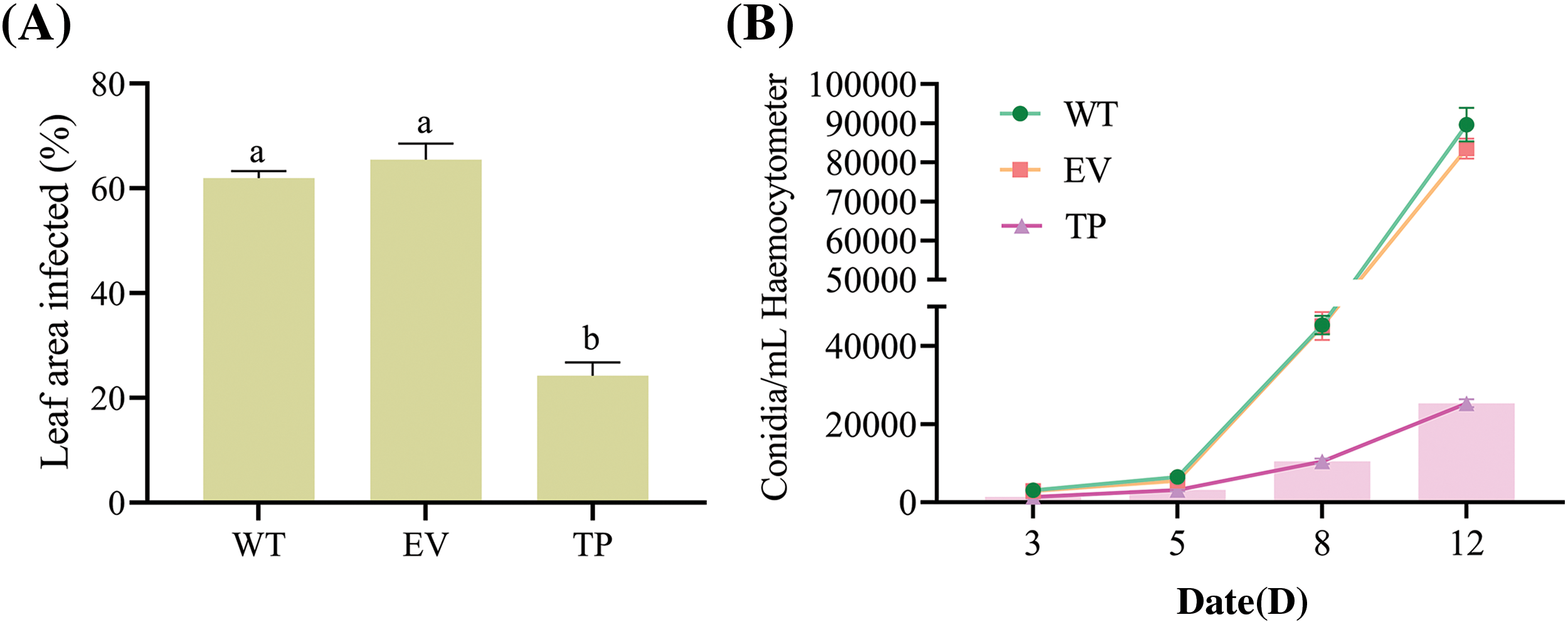
Figure 12: Quantification of infected leaf area and number of conidia. (A) Diseased spot area of tobacco leaves at Day 12 post-inoculation with PM; (B) Number of PM conidia per 1 cm2 on tobacco leaves at Days 3, 5, 8, and 12 post-inoculation. WT: wild-type tobacco leaves; EV: empty vector tobacco leaves; TP: transgenic tobacco leaves. Different letters indicate significant differences at p < 0.05
3.5 Effects of EuCHIT30.7 Gene Transfer on the Activities of Protective Enzymes and MDA Content in Tobacco
The CAT activity, SOD activity, POD activity, PAL activity and MDA content in wild-type, empty vector, and transgenic tobacco plants were determined at 0 h before inoculation, as well as at 12, 24, 48, and 72 h after inoculation with PM. The activities of POD, SOD, and CAT in wild-type, empty vector, and transgenic tobacco plants first increased and then decreased. After inoculation with PM, the activities of SOD, POD, and CAT were higher in transgenic tobacco leaves compared to wild-type and empty vector plant leaves (Figs. 13A–13C). At 24 h post-inoculation, the SOD activity in transgenic tobacco significantly increased to 471.3 U·g−1 (Fig. 13B), and the activities of POD and CAT significantly increased to 5,466.8 and 554.5 U·g−1 respectively at 48 h post-inoculation. The SOD, POD, and CAT activities in transgenic tobacco plants were significantly higher than those in wild-type and empty vector plants (Figs. 13A and 13C). After inoculation, the PAL activity increased with time, reaching the highest level of 48.7 U·g−1 at 72 h in transgenic tobacco plants, which was significantly higher than that in wild-type and empty vector plants (Fig. 13D). However, the MDA content first decreased and then increased. The MDA content sharpy decreased at 48 h post-inoculation to 12.8 nmol·g−1 in transgenic tobacco plants, which was significantly lower than that in wild-type and empty vector plants (Fig. 13E). Thus, these findings suggested that the introduction of the EuCHIT30.7 gene enhances the capacity of tobacco plants to quench reactive oxygen species (ROS), augment the accumulation of antifungal compounds, and enhance osmotic regulation, thereby increasing the resistance of tobacco plants to disease.
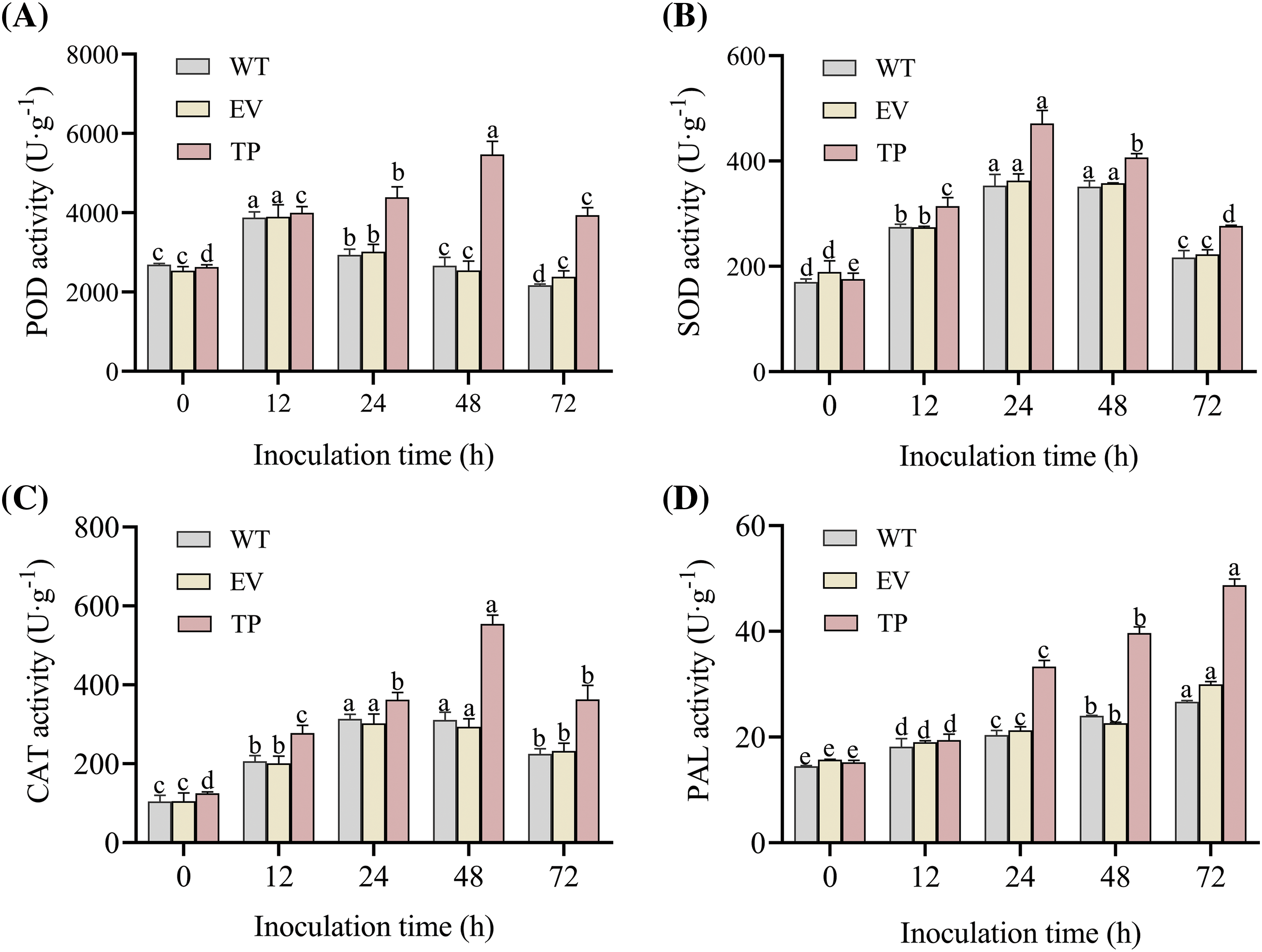
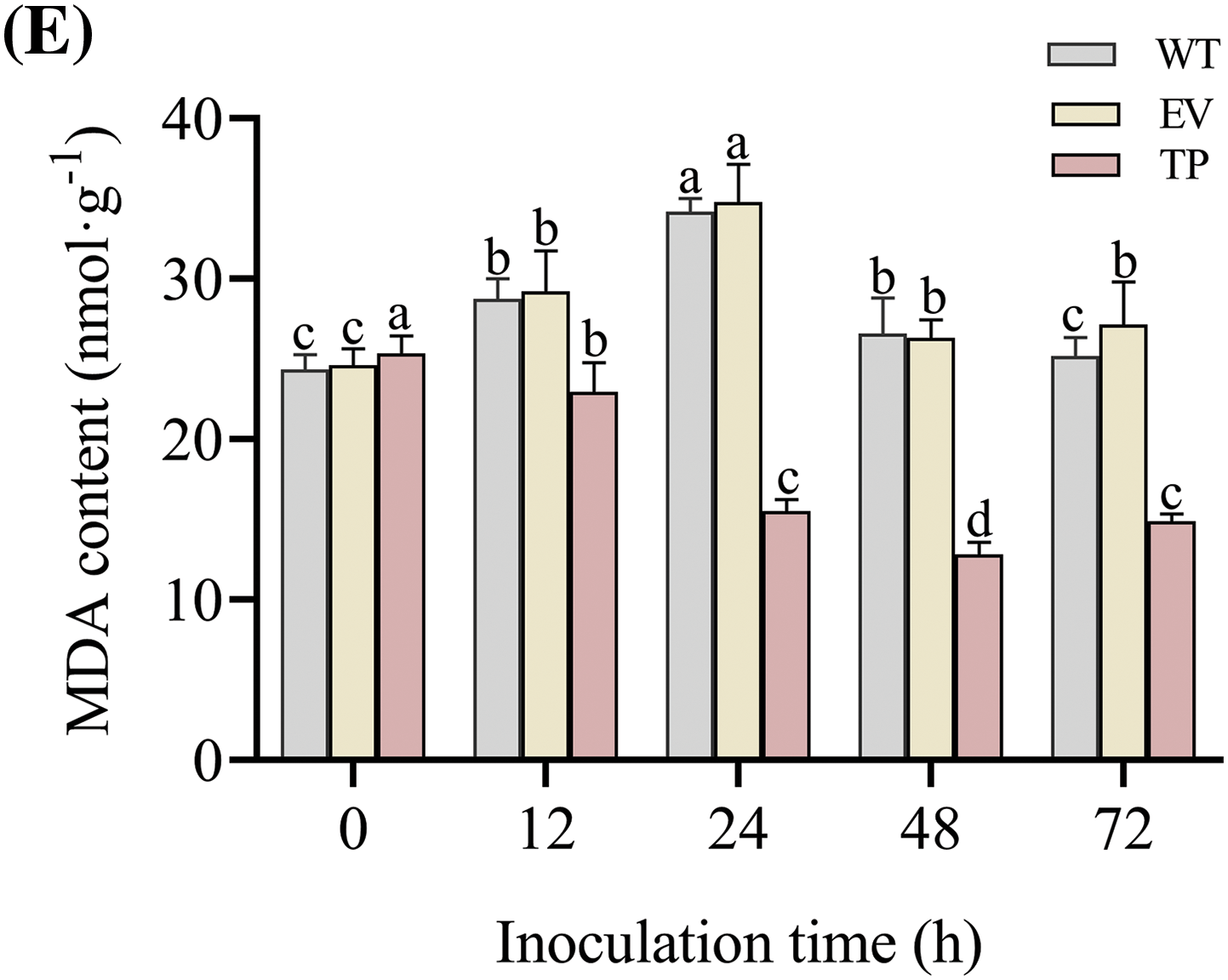
Figure 13: Effect of PM on physiological indices of wild-type, empty-vector, and EuCHIT30.7 transgenic tobacco. (A) POD activity; (B) SOD activity; (C) CAT activity; (D) PAL activity; (E) MDA content. WT: wild-type tobacco; EV: empty vector tobacco; TP: transgenic tobacco. Different letters indicate significant differences at p < 0.05
3.6 Effect of EuCHIT30.7 Gene Expression on the Expression of PR Genes in Tobacco
The relative expression of tobacco PR genes in wild-type, empty vector, and transgenic tobacco plants was analyzed at 0 h before inoculation, as well as 12, 24, 48, and 72 h after inoculation with PM. The relative expression of the PR5, PR1a, and PR2 genes in EuCHIT30.7 transgenic tobacco plants was significantly higher than that in wild-type and empty vector plants (1.70, 1.76, and 2.43 times higher than that in the wild-type plants, respectively; and 1.41, 1.79, and 2.67 times higher than that in empty-vector plants, respectively) (Figs. 14A–14C). At 12, 24, 48, and 72 h post-inoculation, the relative expression of PR1a (p < 0.05), PR5 (p < 0.05), PR2 (p < 0.05), PDF1.2 (p < 0.05), and MLP423 (p < 0.01) in transgenic plants was significantly higher than that in wild-type and empty vector plants (Fig. 14). At 48 h post-inoculation, the relative expression of PR5, PDF1.2, and MLP423 reached the highest level in transgenic plants (14.19, 17.15, and 20.35 times higher than that in wild-type plants, respectively; and 14.43, 16.9, and 23.27 times higher than that in empty vector plants, respectively) (Figs. 14C–14E). The relative expression of PR1a and PR2 reached the highest level at 72 h post-inoculation in transgenic plants (122.82 and 9.96 times higher than that in wild-type plants, respectively; and 115.61 and 9.78 times higher than that in empty vector plants, respectively) (Figs. 14A and 14B). Thus, these findings indicated that overexpression of the EuCHIT30.7 gene in tobacco increases the expression of PR genes and key genes of the jasmonic acid pathway to increase the resistance of tobacco plants to PM.
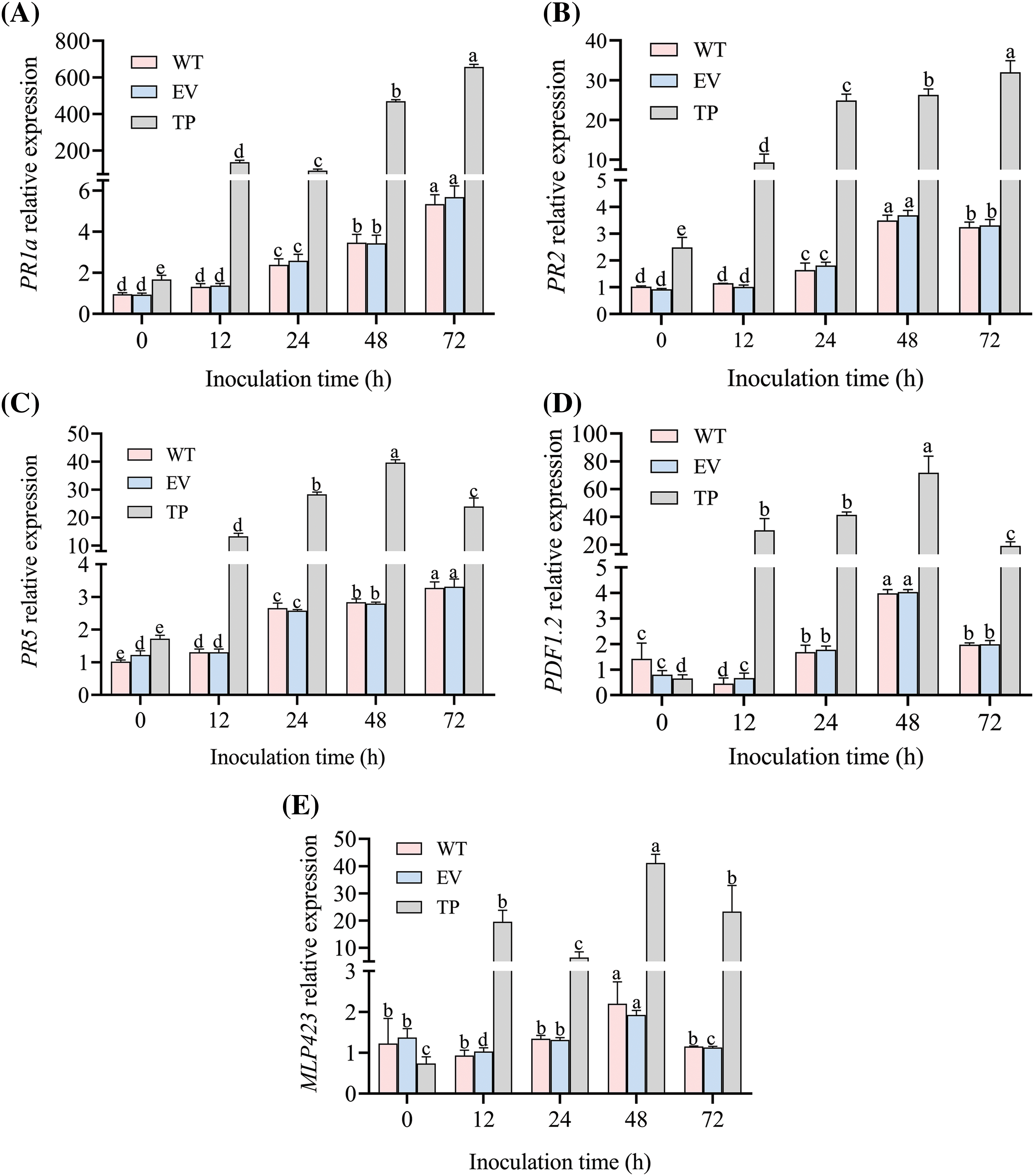
Figure 14: Relative expression of tobacco PR genes before and after inoculation with PM. Gene expression of (A) PR1a; (B) PR2; (C) PR5; (D) PDF1.2; and (E) MLP423. WT: wild-type tobacco; EV: empty-vector tobacco; TP: transgenic tobacco. Different letters indicate significant differences at p < 0.05
3.7 Effect of EuCHIT30.7 Gene Expression on MLO Gene Expression Related to Tobacco PM Resistance
To explore whether EuCHIT30.7 is involved in other mechanisms of tobacco resistance to PM, we analyzed the relative expression of NtMLO2 (PM phase-related gene) and NtXTH1 (downstream gene) in wild-type, empty vector, and transgenic tobacco plants at 0 h before inoculation, as well as at 12, 24, 48, and 72 h after inoculation with the PM pathogen. There was no significant difference in NtMLO2 expression among the wild-type, empty vector, and transgenic tobacco plants before inoculation, but the expression of NtXTH1 in transgenic tobacco plants was significantly lower than that in wild-type and empty vector tobacco plants after inoculation (Fig. 15). The expression of NtMLO2 and NtXTH1 in wild-type, empty vector, and transgenic tobacco plants gradually increased at 12, 24, 48, and 72 h post-inoculation. The expression level of NtMLO2 and NtXTH1 in transgenic tobacco plants was significantly higher than that in wild-type and empty-vector tobacco plants (p < 0.05). At 72 h post-inoculation, the expression of NtMLO2 and NtXTH1 was 10.08 and 10.11 times higher than that in wild-type plants, respectively, while the expression of NtMLO2 and NtXTH1 was 52.29 and 59.68 times higher than that in empty vector tobacco plants, respectively (Fig. 15). The transgenic EuCHIT30.7 gene increased the expression of MLO, providing an opportunity and the conditions for PM infection, but it still provided resistance to PM. Therefore, these results suggested that the ability of transgenic plants to improve resistance to PM is not achieved by regulating the expression of resistance-related genes to PM but by the gene products directly inhibiting pathogen growth.
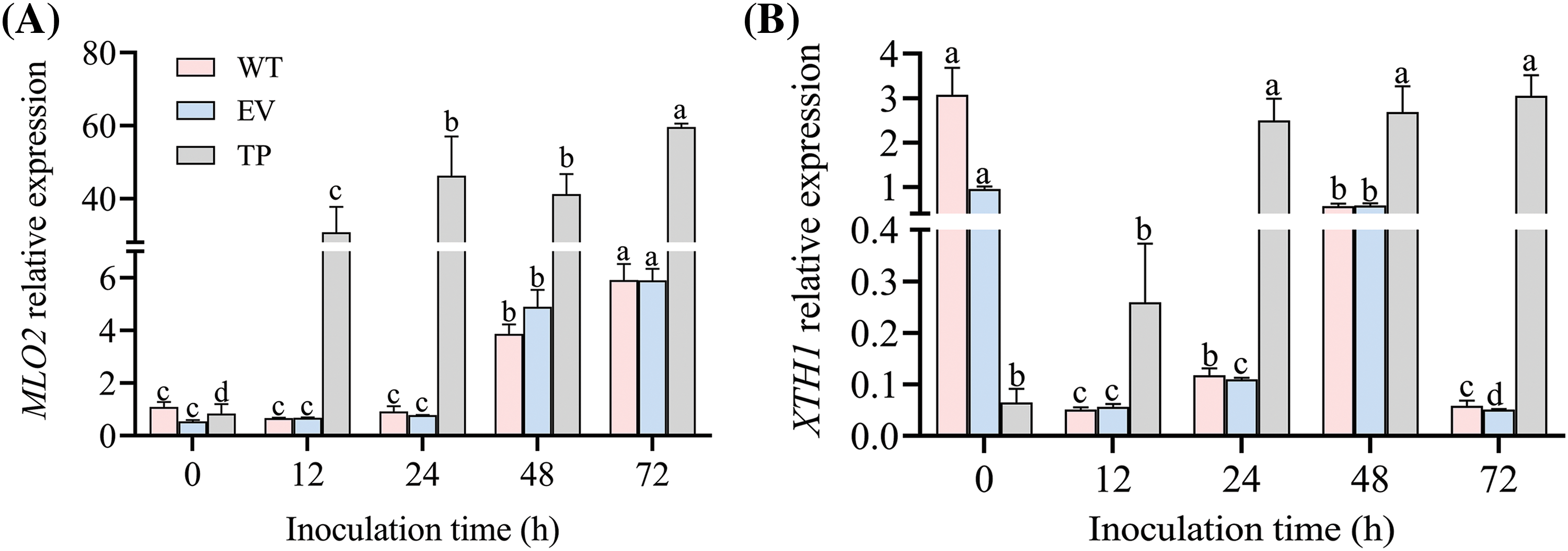
Figure 15: Relative expression of PM resistance-related genes in tobacco before and after inoculation with PM. Gene expression of (A) MLO2 and (B) XTH1. WT: wild-type tobacco; EV: empty-vector tobacco; TP: transgenic tobacco. Different letters indicate significant differences at p < 0.05
Plant diseases caused by a variety of pathogens are important causes of reductions in crop yields. Therefore, to further improve plant defense responses, it is important to identify additional enzymes that can degrade cell walls of invasive plant pathogenic fungi [31]. In the present study, a novel chitinase gene from Eucommia, named EuCHIT30.7, was successfully isolated. The phylogenetic position of EuCHIT30.7 among the chitinase family genes of Arabidopsis was elucidated, and its conserved domain was determined. The EuCHIT30.7 gene was identified as a constituent of the chitinase 18 family, and it encoded a stable hydrophobic acidic protein. The EuCHIT30.7 protein was localized in the cell membrane and cytoplasm in tobacco leaves. The location of the EuCHIT30.7 protein was not consistent with the predicted location of vacuoles, but it was consistent with observations of the chitinase 18 family gene, PbChia1, of Rhizobium brassicae [32]. After inoculation with PM, the wild-type, empty vector, and EuCHIT30.7 transgenic plants all showed symptoms of disease. However, the onset time in transgenic tobacco plants was later than that in wild-type and empty vector tobacco plants. Furthermore, the transgenic tobacco plants exhibited a significant inhibitory effect on PM spore count, indicating that EuCHIT30.7 tobacco plants have the ability to resist PM disease.
Many previous studies have reported excessive generation of ROS following the invasion of a pathogen, which disrupts standard plant physiological processes [33–35]. Consequently, the defensive enzymatic system of the plant is triggered to neutralize reactive oxygen free radicals and peroxides [36]. In the present study, the activities of SOD, POD, and CAT in EuCHIT30.7 transgenic tobacco plants first increased and then decreased after infection with PM, and the activities of these enzymes were significantly higher than those in wild-type and empty vector tobacco plants, indicating that EuCHIT30.7 enhances the capacity of tobacco plants to neutralize ROS. Plants possess the inherent capability to synthesize a diverse array of substances in response to stress or pathogenic incursion, encompassing plant defense compounds, lignin, and other phenolic compounds that are deposited in the cell wall, as well as chitinases and PR proteins. Nearly all these compounds are synthesized via the phenylpropanoid metabolic pathway. PAL is a pivotal enzyme in this metabolic pathway and also functions as a rate-limiting enzyme. Upon pathogenic attack, the overall activity of PAL in the plant is rapidly increased, inducing a series of reactions in phenylpropanoid metabolism and enhancing the accumulation of antimicrobial substances [37–39]. In the present study, PAL activity in transgenic plants was significantly higher than that in wild-type and empty vector plants following inoculation, suggesting that EuCHIT30.7 overexpression increases the biosynthesis of resistance-related metabolites in tobacco, thereby increasing plant defense ability. After plants are invaded by pathogens, MDA accumulates in plants, and excessive MDA content indicates damage to the membrane system of plant cells [40]. In the present study, the MDA content in transgenic plants decreased after inoculation, suggesting that EuCHIT30.7 overexpression reduces the damage to cells by membrane peroxidation, thereby improving plant resistance to PM disease.
PR proteins in plants resist fungal infection by invasive pathogens [41]. The PDF1.2 gene is specifically induced by jasmonic acid and can be used as a marker gene [42] in the jasmonic acid-dependent signaling pathway. MLP is a type of latex protein that shares a similar three-dimensional structure with the PR10 protein [43]. In the present study, the expression levels of PR1a, PR2, PR5, and MLP423, as well as PDF1.2 (key gene of the jasmonic acid signaling pathway) were significantly higher in EuCHIT30.7 transgenic tobacco plants than in wild-type and empty vector tobacco plants after inoculation with PM. Thus, overexpression of EuCHIT30.7 in carrier plants increased the expression of key genes in the salicylic acid and jasmonic acid signaling pathways in tobacco, as well as the expression of genes related to the disease process, thereby increasing the immunity of plants and resistance to fungal invasion. The MLO gene is one of the susceptibility genes present in plants, and it is associated with susceptibility to PM disease. In addition, the MLO gene negatively regulates the disease resistance process of plants [44]. NtXTH is mainly responsible for the degradation of the cell wall in plants, making it easier for pathogens to penetrate the first line of defense of plants, namely, the cell wall [45,46]. In the present study, the expression of NtMLO2 and NtXTH1 in EuCHIT30.7 transgenic tobacco plants significantly increased after inoculation with PM, improving the conditions and opportunities for PM infection. However, the transgenic plants still showed increased disease resistance.
In the present study, we successfully cloned the EuCHIT30.7 gene originating from E. ulmoides, a member of the chitinase 18 family. Overexpression of EuCHIT30.7 significantly increased the resistance of the tobacco plant against PM. Previous studies have suggested that in circumstances where plants are afflicted by fungal diseases, the EuCHIT30.7 chitinase decomposes chitin present in fungal cell walls, generating a multitude of chitooligosaccharides that resemble PAMPs. These fragments of chitin assume the role of signaling molecules, thereby triggering the immune response of plants, which enhances the expression of disease PR genes and the activities of protective enzymes, ultimately improving the disease resistance in transgenic plants.
Acknowledgement: Thank other teachers or members of the laboratory for their help in this study.
Funding Statement: This study was funded by the NSCF (Nos. 31360272 & 31870285) and Talent Special Project of Guizhou Academy of Agricultural Sciences (No. 2022-02) and Talent Base for Germplasm Resources Utilization and Innovation of Characteristic Plant in Guizhou (Grant No. RCJD2018-14).
Availability of Data and Materials: The datasets used and/or analysed during the current study are available from the author on reasonable request.
Ethics Approval: We do not contain any studies with human or animal subjects.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Takamatsu, S. (2004). Phylogeny and evolution of the powdery mildew fungi (Erysiphales, Ascomycota) inferred from nuclear ribosomal DNA sequences. Mycoscience, 45(2), 147–157. https://doi.org/10.1007/S10267-003-0159-3 [Google Scholar] [CrossRef]
2. Melchers, L. S., Groot, A. D., Knaap, J., Ponstein, A. P., Sela-Buurlage, M. B. et al. (1994). A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant Journal, 5(4), 469–480. [Google Scholar]
3. Sahai, A. S., Manocha, M. S. (1993). Chitinases of fungi and plants: Their involvement in morphogenesis and host-parasite interaction. FEMS Microbiology Reviews, (4), 317–338. https://doi.org/10.1111/j.1574-6976.1993.tb00004.x [Google Scholar] [CrossRef]
4. Nagpure, A., Choudhary, B., Gupta, R. K. (2014). Chitinases: In agriculture and human healthcare. Critical Reviews in Biotechnology, 34(3), 215–232. https://doi.org/10.3109/07388551.2013.790874 [Google Scholar] [PubMed] [CrossRef]
5. Ohnuma, T., Numata, T., Osawa, T., Mizuhara, M., Vrum, K. M. et al. (2011). Crystal structure and mode of action of a class V chitinase from Nicotiana tabacum. Plant Molecular Biology, 75(3), 291–304. https://doi.org/10.1007/s11103-010-9727-z [Google Scholar] [PubMed] [CrossRef]
6. He, X., Wang, J., Li, M., Hao, D., Yang, Y. et al. (2014). Eucommia ulmoides Oliv.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. Journal of Ethnopharmacology, 151(1), 78–92. https://doi.org/10.1016/j.jep.2013.11.023 [Google Scholar] [PubMed] [CrossRef]
7. Huang, L., Lyu, Q., Zheng, W., Yang, Q., Cao, G. (2021). Traditional application and modern pharmacological research of Eucommia ulmoides Oliv. Chinese Medicine, 16(1), 73. https://doi.org/10.1186/s13020-021-00482-7 [Google Scholar] [PubMed] [CrossRef]
8. Guo, L. X., Dong, X., Zhao, D. G. (2016). Transgenic tomato plants expressing a Eucommia ulmoides chitinase gene EuCHIT1 and their resistance to Botrytis cinerea. Plant Physiology Journal, 52(5), 703–714. https://doi.org/10.13592/j.cnki.ppj.2016.0066 [Google Scholar] [CrossRef]
9. Ding, Y. Q., Dong, X., Zhao, D. G. (2017). Transformed EuCHIT1 gene in wheat (Triticum aestivum) enhances resistance to stripe rust (Puccinia striiformis f. sp tritici). Journal of Agricultural Biotechnology, 25(6), 1013–1022. [Google Scholar]
10. Liu, Y., Ran, X., Zhao, D. G. (2022). Transgenic tobacco with EuAFP1. 2 gene improves fungal disease resistance. Genomics and Applied Biology, 41(8), 1767–1778. https://doi.org/10.13417/j.gab.041.001767 [Google Scholar] [CrossRef]
11. Zhao, D. G., Li, Y., Zhao, Y. C., Zhao, D., Lv, L. T. et al. (2015). Transcriptome data assembly and gene function annotation of female and male plants in Eucommia ulmoides. Journal of Mountain Agriculture and Biology, 34(1), 1–13. https://doi.org/10.15958/j.cnki.sdnyswxb.2015.01.001 [Google Scholar] [CrossRef]
12. Ye, J., Jin, C. F., Li, N., Li, Z. Q. (2018). Selection of suitable reference genes for RT-qPCR normalisation under different experimental conditions in Eucommia ulmoides Oliv. Scientific Reports, 8(1), 15043. https://doi.org/10.1038/s41598-018-33342-w [Google Scholar] [CrossRef]
13. Li, X. (2011). Infiltration of Nicotiana benthamiana protocol for transient expression via agrobacterium. Bio-Protocol, 1(14), e95. https://doi.org/10.21769/BioProtoc.95 [Google Scholar] [CrossRef]
14. Horsch, R. (1985). A simple and general method for transferring genes into plants. Science, 227(1), 331–336. https://doi.org/10.1016/0003-2697(88)90554-4 [Google Scholar] [CrossRef]
15. Jefferson, R. A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Molecular Biology Reporter, 5(4), 387–405. https://doi.org/10.1007/BF02667740 [Google Scholar] [CrossRef]
16. Miclot, A. S., Wiedemann-Merdinoglu, S., Duchêne, E., Merdinoglu, D., Mestre, P. (2012). A standardised method for the quantitative analysis of resistance to grapevine powdery mildew. European Journal of Plant Pathology, 133(2), 483–495. https://doi.org/10.1007/s10658-011-9922-z [Google Scholar] [CrossRef]
17. Moyer, M. M., Gadoury, D. M., Cadle-Davidson, L., Dry, I. B., Magarey, P. A. et al. (2010). Effects of acute low-temperature events on development of erysiphe necator and susceptibility of Vitis vinifera. Phytopathology, 100(11), 1240–1249. https://doi.org/10.1094/PHYTO-01-10-0012 [Google Scholar] [PubMed] [CrossRef]
18. Dong, X., Zhao, Y. C., Ran, X., Guo, L. X., Zhao, D. G. (2017). Overexpression of a new chitinase gene EuCHIT2 enhances resistance to Erysiphe cichoracearum DC. in tobacco plants. International Journal of Molecular Sciences, 18(11), 2361–2374. https://doi.org/10.3390/ijms18112361 [Google Scholar] [CrossRef]
19. Sela-Buurlage, M. B. (1993). Only specific tobacco (Nicotiana tabacum) chitinases and β-1,3-glucanases exhibit antifungal activity. Plant Physiology, 101(3), 857–863. https://doi.org/10.1104/pp.101.3.857 [Google Scholar] [PubMed] [CrossRef]
20. Loon, L. C. V., Strien, E. A. V. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiological & Molecular Plant Pathology, 55(2), 85–97. https://doi.org/10.1006/pmpp.1999.0213 [Google Scholar] [CrossRef]
21. Johansson, L. H., Håkan Borg, L. A. (1988). A spectrophotometric method for determination of catalase activity in small tissue samples. Analytical Biochemistry, 174(1), 331–336. https://doi.org/10.1016/0003-2697(88)90554-4 [Google Scholar] [PubMed] [CrossRef]
22. Masayasu, M., Hiroshi, Y. (1979). A simplified assay method of superoxide dismutase activity for clinical use. Clinica Chimica Acta, 92(3), 337–342. https://doi.org/10.1016/0009-8981(79)90211-0 [Google Scholar] [PubMed] [CrossRef]
23. Spitz, D. R., Oberley, L. W. (1989). An assay for superoxide dismutase activity in mammalian tissue homogenates. Analytical Biochemistry, 179(1), 8–18. https://doi.org/10.1016/0003-2697(89)90192-9 [Google Scholar] [PubMed] [CrossRef]
24. Reuveni, R., Shimoni, M., Karchi, Z., Kuc, J. (1992). Peroxidase activity as a biochemical marker for resistance of muskmelon (Cumcumis melo) to Pseudoperonospora cubensis. Phytopathology, 82(7), 749–753. https://doi.org/10.1094/Phyto-82-749 [Google Scholar] [CrossRef]
25. Doerge, D. R., Divi, R. L., Churchwell, M. I. (1997). Identification of the colored guaiacol oxidation product produced by peroxidases. Analytical Biochemistry, 250(1), 10–17. https://doi.org/10.1006/abio.1997.2191 [Google Scholar] [PubMed] [CrossRef]
26. Zhang, J., Lv, J., Xie, J., Gan, Y., Coulter, J. A. et al. (2020). Nitrogen source affects the composition of metabolites in pepper (Capsicum annuum L.) and regulates the synthesis of capsaicinoids through the GOGAT-GS pathway. Foods, 9(2), 150. https://doi.org/10.3390/foods9020150 [Google Scholar] [PubMed] [CrossRef]
27. Guo, W. L., Chen, B. H., Guo, Y. Y., Chen, X. J., Wang, G. Y. (2020). Expression of pumpkin CmbHLH87 gene improves powdery mildew resistance in tobacco. Frontiers in Plant Science, 11, 163–173. https://doi.org/10.3389/fpls.2020.00163 [Google Scholar] [CrossRef]
28. Liu, H., Ma, X., Liu, S., Du, B., Cheng, N. et al. (2020). The Nicotiana tabacum L. major latex protein-like protein 423 (NtMLP423) positively regulates drought tolerance by ABA-dependent pathway. BMC Plant Biology, 20(1), 475–488. https://doi.org/10.1186/s12870-020-02690-z [Google Scholar] [PubMed] [CrossRef]
29. Zhang, X. L. (2022). Study on the mechanism of MLO-based resistant to powdery mildew in Nicotiana tabacum L. (Ph.D. Thesis). Guizhou University, Guiyang, China. [Google Scholar]
30. Hellemans, J., Mortier, G., Paepe, A. D., Speleman, F., Vandesompele, J. (2007). QBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology, 8(2), 19–32. https://doi.org/10.1186/gb-2007-8-2-r19 [Google Scholar] [PubMed] [CrossRef]
31. Antony Ceasar, S., Ignacimuthu, S. (2012). Genetic engineering of crop plants for fungal resistance: Role of antifungal genes. Biotechnology Letters, 34(6), 995–1002. https://doi.org/10.1007/s10529-012-0871-1 [Google Scholar] [PubMed] [CrossRef]
32. Zhao, Y. L. (2022). Biological function of chitinase PbChia1 in Plasmodiophora brassicae (Ph.D. Thesis). Huazhong Agricultural University, Wuhan, China. [Google Scholar]
33. Heller, J., Tudzynski, P. (2011). Reactive oxygen species in phytopathogenic fungi: Signaling, development, and disease. Annual Review of Phytopathology, 49(1), 369–390. https://doi.org/10.1146/annurev-phyto-072910-095355 [Google Scholar] [PubMed] [CrossRef]
34. Sang, Y., Macho, A. P. (2017). Analysis of PAMP-triggered ROS burst in plant immunity. Methods in Molecular Biology, 1578, 143–153. https://doi.org/10.1007/978-1-4939-6859-6 [Google Scholar] [CrossRef]
35. Castro, B., Citterico, M., Kimura, S., Stevens, D. M., Coaker, G. L. (2021). Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nature Plants, 7(4), 403–412. https://doi.org/10.1038/s41477-021-00887-0 [Google Scholar] [PubMed] [CrossRef]
36. Tian, M., Rao, L. B., Li, G. (2005). Reactive oxygen species (ROS) and its physiological functions in plant cells. Plant Physiology Journal, (2), 235–241. [Google Scholar]
37. Chavez, E., Franco, M., Reyes-Vivas, H., Zazueta, C., Saltveit, M. E. (1998). Phenylalanine ammonia lyase inhibitors control browning of cut lettuce. Postharvest Biology & Technology, 14(2), 171–177. https://doi.org/10.1016/S0925-5214(98)00048-9 [Google Scholar] [CrossRef]
38. Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R. et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Scientific Reports, 10(1), 4818–4835. https://doi.org/10.1038/s41598-020-61140-w [Google Scholar] [PubMed] [CrossRef]
39. Pellegrini, L., Rohfritsch, O., Legrand, F. M. (1994). Phenylalanine ammonia-lyase in tobacco. Molecular cloning and gene expression during the hypersensitive reaction to tobacco mosaic virus and the response to a fungal elicitor. Plant Physiology, 106(3), 877–886. https://doi.org/10.1104/pp.106.3.877 [Google Scholar] [PubMed] [CrossRef]
40. Gawe, S., Wardas, M., Niedworok, E., Wardas, P. (2004). Malondialdehyde (MDA) as a lipid peroxidation marker. Wiad Lek, 57(9–10), 453–455. https://pubmed.ncbi.nlm.nih.gov/15765761/ [Google Scholar]
41. Chhikara, S., Chaudhury, D., Dhankher, O. P., Jaiwal, P. K. (2012). Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae. Plant Cell, Tissue and Organ Culture, 108(1), 83–89. https://doi.org/10.1007/s11240-011-0015-7 [Google Scholar] [CrossRef]
42. Penninckx, I. A. M. A., Thomma, B. P. H. J., Buchala, A., Métraux, J. P., Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. The Plant Cell, 10(12), 2103–2113. https://doi.org/10.1105/tpc.10.12.2103 [Google Scholar] [PubMed] [CrossRef]
43. Radauer, C., Lackner, P., Breiteneder, H. (2008). The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evolutionary Biology, 8(1), 286–304. https://doi.org/10.1186/1471-2148-8-286 [Google Scholar] [PubMed] [CrossRef]
44. Jrgensen, I. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica, 63(1), 141–152. https://doi.org/10.1007/BF00023919 [Google Scholar] [CrossRef]
45. Liu, R., Weng, K., Dou, M., Chen, T., Xu, Y. (2019). Transcriptomic analysis of Chinese wild Vitis pseudoreticulata in response to Plasmopara viticola. Protoplasma, 256(5), 1409–1424. https://doi.org/10.1007/s00709-019-01387-x [Google Scholar] [PubMed] [CrossRef]
46. Katarzyna, O. K., Edmund, K., Józef, B. (2018). Spatiotemporal changes in Xylan-1/Xyloglucan and Xyloglucan Xyloglucosyl Transferase (XTH-Xet5) as a step-in of ultrastructural cell wall remodelling in potato-potato virus Y (PVYNTN) hypersensitive and susceptible reaction. International Journal of Molecular Sciences, 19(8), 2287–2310. https://doi.org/10.3390/ijms19082287 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools