 Open Access
Open Access
ARTICLE
Characterization of Endophytic Microorganisms of Rice (Oryza sativa L.) Potentials for Blast Disease Biocontrol and Plant Growth Promoting Agents
1 Division of Plant Pathology, Faculty of Agriculture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Wadura, India
2 Mountain Research Centre for Field Crops, SKUAST-Kashmir, Khudwani, India
3 Division of Genetics and Plant Breeding, Faculty of Agriculture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Wadura, India
4 Division of Microbiology, Faculty of Agriculture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Wadura, India
5 Division of Statistics, Faculty of Agriculture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Wadura, India
6 Division of Agronomy, Faculty of Agriculture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Wadura, India
7 Indian Council of Agricultural Research-Central institute of Temperate Horticulture (ICAR-CITH), Old Airport, Rangreth, Srinagar, India
8 Division of Plant Pathology, Faculty of Horticulture, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Shalimar, India
9 Directorate of Extension, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Shalimar, India
10 Department of Plant Production, College of Food and Agriculture Sciences, King Saud University, Riyadh, Saudi Arabia
11 College of Food, Agricultural and Environmental Sciences, The Ohio State University, Columbus, OH, USA
12 Department of Agronomy, Faculty of Agriculture, University of Kafrelsheikh, Kafrelsheikh, Egypt
* Corresponding Authors: Fayaz A. Mohiddin. Email: ; Ibrahim Al-Ashkar. Email:
Phyton-International Journal of Experimental Botany 2023, 92(11), 3021-3041. https://doi.org/10.32604/phyton.2023.030921
Received 03 May 2023; Accepted 14 August 2023; Issue published 24 October 2023
Abstract
One hundred twenty-five endophytic microorganisms were isolated from the roots, stems, and leaves of four prominent rice cultivars growing in temperate regions. Their potential to combat rice blast disease and promote plant growth was investigated. The dual culture tests highlighted the strong antagonistic activity of five fungal (ranging from 89%–70%) and five bacterial (72%–61%) endophytes. Subsequent examination focused on volatile compounds produced by selected isolates to counter the blast pathogen. Among these, the highest chitinase (13.76 µg mL−1) and siderophore (56.64%), was exhibited by Aspergillus flavus, and the highest HCN production was shown by Stenotrophomonas maltophilia (36.15 µM mL−1). In terms of growth promotion traits, Aspergillus flavus and Enterobacter cloacae excelled in activities viz, phosphorous solubilization, ammonia production, auxin and gibberellic acid production, and nitrogen fixation. The Identity of these endophytes was confirmed through molecular analysis as Trichoderma afroharzianum, Trichoderma harzianum, Penicillium rubens, Aspergillus flavus, Stenotrophomonas rhizophila, Stenotrophomonas maltophilia, Bacillus cereus, Enterobacter cloacae, and Bacillus licheniformis. Under greenhouse conditions, the highest disease control was shown by isolate Bacillus licheniformis and A. flavus with an inhibition of 79%, followed by S. rhizophila (77%) and T. afroharzianum (73%). The overall results of this study showed that Bacillus licheniformis and Stenotrophomonas rhizophila have great potential to be used as bio-stimulant and biocontrol agents to manage rice blast disease.Keywords
Supplementary Material
Supplementary Material FileRice (Oryza sativa L.) is one of the most frequently consumed cereal crops in the world which is consumed as a staple diet by 50% of the world’s population [1]. By 2025, the worldwide demand for rice is anticipated to be over 800 metric million tons [2]. Rice production is becoming increasingly challenging due to severe yield losses caused by biotic and abiotic stress factors [3]. Among the biotic stresses, rice blast can cause yield losses of 65% [4] and during epidemic years, they can reach to 100% [5]. In field conditions, rice blast can attack all stages of rice development [6]. Crop rotation, fertilizer management, blast-resistant variety planting, and fungicide administration are the common management strategies used to fight against this disease [7]. However, each of these methods has its own set of disadvantages. Chemical fungicides are often used, resulting in the development of resistance to pathogens, environmental contamination, and effects on non-target microorganisms [8]. Biocontrol using endophytic microorganisms is an alternative management strategy and is being used throughout the world for the management of rice blast [9–11]. An endophyte is a fungal or bacterial microbe that spends all or part of its life cycle inhabiting the host plant’s healthy tissues, usually without persuading disease symptoms [12]. Rice contains a diversity of endophytes, with some strains having a better influence on disease prevention and control [13]. Therefore, the objective of the present study was to characterize and explain the effect of isolated endophytes on rice growth promotion by producing phytohormones, ammonia, phosphorous solubilization and to look into the potential application of rice endophytes against blast diseases of rice.
Sample collection for the isolation of endophytic microorganisms was done at an altitude ranging from 1500 to 2200 meters above mean sea level. The roots, stems and leaves of disease-free and healthy rice plants (10 from each variety) in disease-affected fields were randomly selected at the panicle initiation stage (active phase) from four varieties of rice (C1039, SR3, SR4 and SR5). Paper bags were used to transport samples to the laboratory.
2.2 Sample Preparation for Isolation of Endophytic Microorganisms
To isolate endophytic microorganisms, plant tissues were washed under running tap water to remove soil residues and then surface sterilized with 70% alcohol and 5% sodium hypochlorite for 5 min each, followed by three washes with sterile distilled water. For purification, well-established endophytic fungal and bacterial colonies were removed from plates and sub-cultured in test tubes [14].
2.3 In vitro Screening for Antagonistic Activity by Diffusible Compounds
Dual culture [15] was used to test the antagonism of endophytic bacterial and fungal isolates with diffusible compound(s) against Pyricularia oryzae. In the case of endophytic fungi, a 5 mm mycelial disc of pathogenic fungi was put on one side of the potato dextrose agar (PDA) plate, and an endophytic fungal disc was put on the other side. In the case of bacterial endophytes, one loopful of 48-hour-old culture was streaked on one side of the plate, and a 5 mm mycelial disc of pathogenic fungi was placed on the other side of the plate with PDA and nutrient agar (NA) in a 2:1 ratio [8]. The plates were put in a biological oxygen demand (BOD) incubator for 20 days at 251°C for fungi and 281°C for bacteria. The experiment was done using a complete randomised design with three replications. The percentage of inhibition of mycelial growth over control was calculated using the formula below. The isolates showing potent antifungal activity were selected for further studies.
Percent inhibition = (C − T/C) 100, where C represents the radial growth of the test pathogen in the control plates (measured in mm), and T is the radial growth of the test pathogen in the test plates (mm) [16].
2.4 In vitro Screening for Antagonistic Activity of Volatile Compounds
The volatile compound(s) (VOCs) production of the superior antagonists was screened according to Dennis and Webster (16). A 5 mm disc of P. oryzae and the fungal endophyte were placed at the centre of two different Petri plates containing PDA. In the case of bacterial endophytes, on one plate, a 5 mm disc of P. oryzae was placed at the centre of the Petri plate, and the bacterial endophytes were streaked on another Petri plate containing NA. The lids were then removed and replaced by the bottom of the Petri plate containing the testing endophytic isolates. The two dishes were then taped with parafilm. On the control plate, no bioagent was inoculated but taped in the same manner. The plates were incubated at 251°C in a BOD incubator for 20 days. The percent inhibition was measured by the formula mentioned above.
2.5 Assay for Phosphate Solubilization
A loopful of endophytic bacteria and a mycelial disc of endophytic fungus were added to 3.9 mL of Pikovskaya’s broth medium [17]. The slants were kept in an incubator shaker and incubated at 28°C with shaking at 200 rpm for 3 days for bacterial endophytes and 5 days for fungal endophytes. After that, 1 mL of culture was centrifuged at 14,000 rpm for 10 min, and the soluble P in the supernatant was determined with the method of Bray et al. [18]. The amount of soluble P was calculated from the standard curve of P concentration using KH2PO4.
2.6 Indole Acetic Acid (IAA) Determination
The production of IAA was demonstrated by the method of Bano et al. [19]. The fungal endophytes were inoculated in the PDA amended with 2 mg mL−1 of L-tryptophan (Trp) and incubated at 120 rpm, 25°C, for 5 to 7 days, whereas the bacterial endophytes were grown in Luria broth amended with 2 mg mL−1 of L-tryptophan (Trp) and incubated at 120 rpm, 25°C, for 3 days. The broth was then centrifugated at 10,000 rpm for 10 min, and 1 mL of the supernatant was mixed with 2 mL of Salkowski reagent. Pink colour production indicated IAA production. For the quantitative assay, the above mixture was incubated for 20 min, and a reading was taken at 530 nm.
2.7 Gibberellic Acid Estimation
In a 250-mL Erlenmeyer flask, 100 mL of nutrient broth (NB) and potato dextrose broth (PDB) were sterilised, and 1 mL of standard inoculum (109 cells mL−1) of endophytic isolates was added, which was shaken at 30°C. The experiment was conducted according to standard techniques [20]. The absorbance at 254 nm was measured using a spectrophotometer.
The fungal and bacterial endophytes were grown in 10 mL of peptone broth and kept in an incubator shaker (120 rpm, 25°C ± 1°C) for 4–5 days. A volume of 0.5 mL of Nessler’s reagent was added to each tube. The development of a brown-to-yellow color indicated ammonia production. The liberated ammonia was determined spectrophotometrically at 450 nm [21].
Colloidal chitin was prepared by the method adopted by Prabavathy et al. [22]. The bacterial and fungal endophytes were grown in the slants containing colloidal chitin media for 3 days and 5 days, respectively. A colorimetric chitinase enzymatic assay was conducted according to Rojas-Avelizapain triplicates [23]. The absorbance was measured at 535 nm. One unit (U) of chitinase activity was expressed as the amount of enzyme required to produce 1 mol of N-acetyl glucosamine (Sigma assay kit) per hour under the above conditions [23].
2.10 Screening for Siderophore Production
Siderophore production was assayed by the method given by Schwyn et al. [24]. In this assay, CAS media was prepared by dissolving 60.5 mg CAS (Sigma Aldrich, USA) in 50 mL of water and mixing it with 10 mL of iron (III) solution (1 mM FeCl3•6H2O and 10 mM HCl). This solution was slowly added to 72.9 mg of HDTMA dissolved in 40 mL of water. Also, minimal media were prepared, and both were autoclaved separately. The colonies of bacterial and fungal endophytes were then added to the broth. The reference was also made to the reference, which had exactly the same components but lacked the endophyte. The cultures were incubated in an incubator shaker at 28°C for 3–5 days. After that, the absorbance was measured at 630 nm. The experiment was carried out in three replications.
2.11 Hydrogen Cyanide (HCN) Production
The bacterial endophytic cultures were streaked on pre-poured plates of King’s medium B amended with 4.4 gL−1 glycine, while the fungal endophytic cultures were grown on PDA amended with 4.4 gL−1 glycine. The filter paper (Whatman No. 1) strips were saturated in 0.5% picric acid and 0.2% sodium carbonate. Now the strips were placed in the center of the Petri plates. The parafilm-sealed Petri plates were transferred into an incubator at 28°C for 1–5 days. The control (uninoculated plate) was used for comparison. The experimental plates were analyzed through observation of the change in color of the filter paper from yellow to orange-brown or dark brown [25].
2.12 Morphological Identification of Endophytic Microorganisms
The cultures of endophytic fungi were morphologically identified by observing their morphology, both macro- and micro- characteristics. The macro morphological characteristics tested included the colour of the colony, the texture, and the surface of the colony. The microscopic morphological observations were made by forming a smear of the different isolates of endophytic fungi on the glass slide cover and staining it with lactophenol cotton blue. The isolates were examined under a light microscope. The result of morphological identification (macro and micromorphological characteristics) was compared with the reference available [26]. The colony morphology of bacterial endophytes was studied on plates after streaking a loopful of the isolated colony, and different morphological characteristics like shape, size, texture, margin, and colour were observed. The shape of a single colony was observed under a light microscope. The isolates of Bacteria were gramme-stained. The bacterial smear was prepared using isolated colonies and stained with Gramme’s stain.
2.13 Isolation and Amplification of Genomic DNA
The isolates of endophytes that showed antifungal properties of the tested pathogens were cultured and selected at 27°C for 8 days using PDB (Himedia). The mycelial mat was separated from the PDB and freeze-dried. The mats were quenched in liquid nitrogen, and the genomic DNA was isolated by the Cetyl Trimethyl Ammonium Bromide (CTAB) procedure with some modifications following the protocol of Sambrook et al. [27]. Similarly, for bacterial endophytes, selected endophytic bacterial isolates were cultured for 24 h at 28°C in 10 mL of Luria broth medium, and DNA was extracted following the standard protocol [28]. The DNA extracted was separated on a 0.8 percent (w/v) agarose gel in 1 TAE buffer for 30 min and visualised under UV light by staining with ethidium bromide. The extracted DNA was amplified using ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primers for fungi and 27F (5′-AGAGTTTGATCTTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) for bacteria [29,30] in a thermocycler. The PCR products were run on electrophoresis along with a molecular ladder in a 1.5% agarose gel.
2.14 Sequencing and Sequence Submission
The PCR products of partial 16S rRNA and ITS regions of the gene were custom sequenced by Biokart, Bangalore, India. Consensus sequences were constructed in DNA Baser version 5.15. Before, manual editing was conducted, and then pairwise alignment was accomplished by BioEdit software version 7.2. The similarity of these sequences was checked with the already present sequences in the National Centre for Biotechnology Information (NCBI) gene bank database using the Basic Local Alignment Search Tool (BLASTn) program. The isolates were identified through the observation of a percentage of sequence identity (90%) with that of a prototype strain sequence in the GenBank [31]. The consensus sequences were deposited in GenBank using the BankIt tool of the NCBI.
2.15 Sequence Identity Matrix and Phylogenetic Analysis
The retrieved sequences were subjected to BLAST analysis after sequencing to check for the best hits. The consensus sequences were submitted to GenBank. A neighbour-joining phylogenetic study was performed by MEGA v. 7 using generated sequences [32]. Bootstrap analysis was done with 10,000 replicate run data sets, and the sequence identity matrix was done using BioEdit version 7 software.
2.16 Evaluation under Greenhouse Conditions
A greenhouse experiment was performed to investigate the efficacy of endophytes against P. oryzae using a completely randomised design. The experiment was performed in pots containing sterile soil. The soil and vermicompost were mixed in a ratio of 2:1 (w/w) and sterilised by autoclaving at 121°C at 15 psi for two hours for three days. The susceptible variety of P. oryzae Mushkbudji was used. The seeds were soaked in the 30 mL spore suspension of potential bacterial (108 cfu mL−1) and fungal (106 spores mL−1) endophytes for 24 h. At the planting stage, additional 5 mL inoculums were added to the inoculated seeds in the pots. The pathogen was added 7 days after the inoculation of endophytes. Untreated rice seeds were planted in control pots. After germination, each container had five plants. The containers were watered with sterile water to keep the soil wet. Samples from rice plants were obtained 30 days after sowing (DAS) for assessing various plant biometric characteristics. Disease incidence and disease intensity (%) were evaluated using the Standard Evaluation System (1–9 scale) (IRRI, 1980). The lengths of the shoot and root, as well as the fresh and dry weights, were measured [33].
The data from the experiments were subjected to statistical analysis as per the methods detailed by Gomez et al. [34]. A one-way analysis of variance (ANOVA) was carried out using OPSTAT online software. Molecular data analysis was carried out by BioEdit software version 7.2 and MEGA software version 11.0 [32].
3.1 Isolation of Rice Endophytic Microorganisms
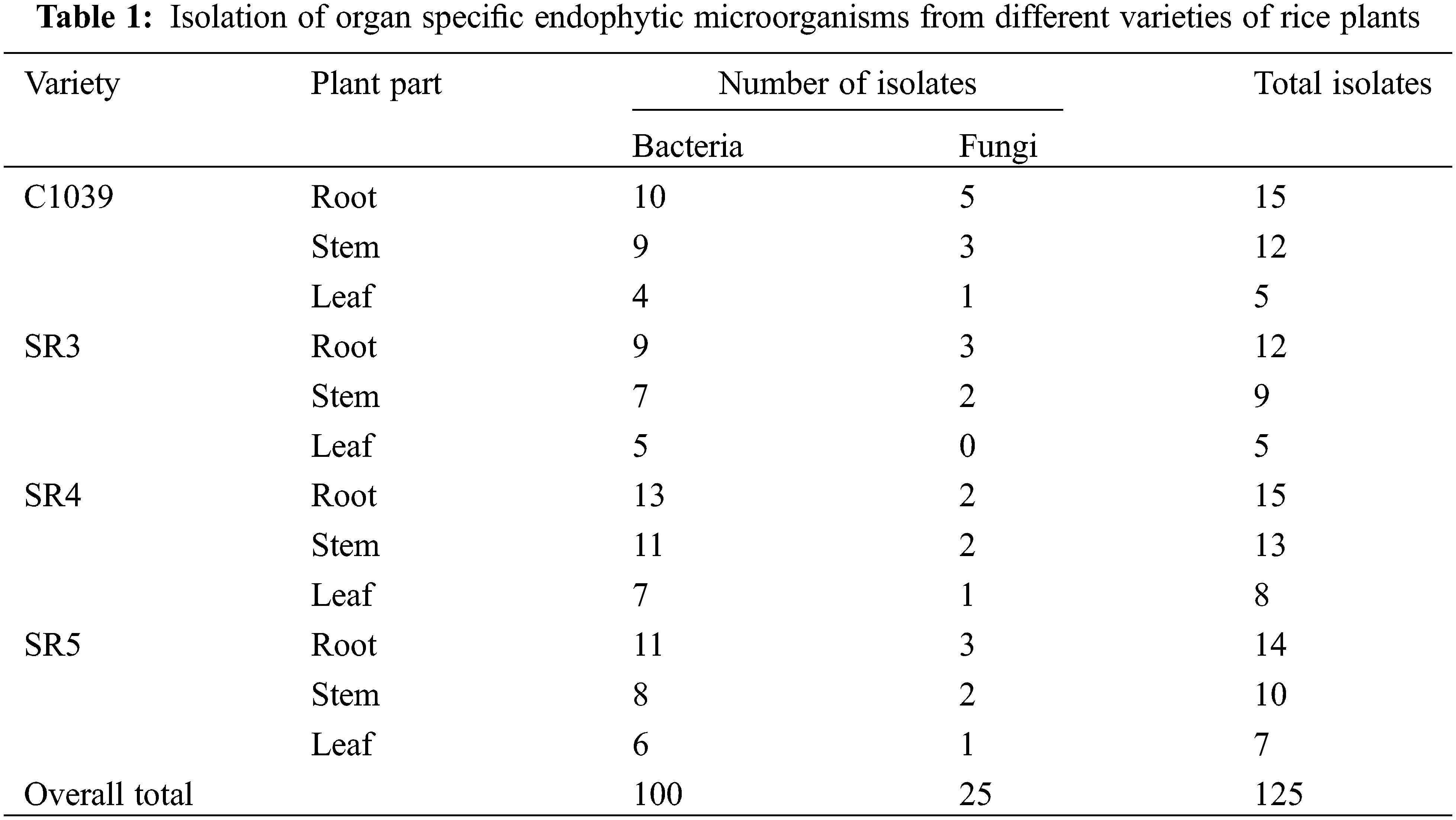
A total of 125 endophytic microorganisms were isolated from the leaves, stems, and roots of the four different varieties of rice grown in the Kashmir Himalayas, out of which 100 were bacterial endophytes and 25 were fungal endophytes. It was found that the highest number of endophytes were isolated from roots, followed by stems and leaves (Table 1). In particular, 48% of bacterial endophytes were isolated from the root. Whereas the endophytes isolated from stem and leaf were 35% and 20% of the total number of isolated endophytes, respectively. The highest number of 14 endophytes was isolated from SR4 (36) followed by C1039 (32) and SR5 (31) while the fewest endophytes were isolated from the SR3 variety of rice.
3.2 In vitro Test for Antagonism by Dual Culture of Endophytes
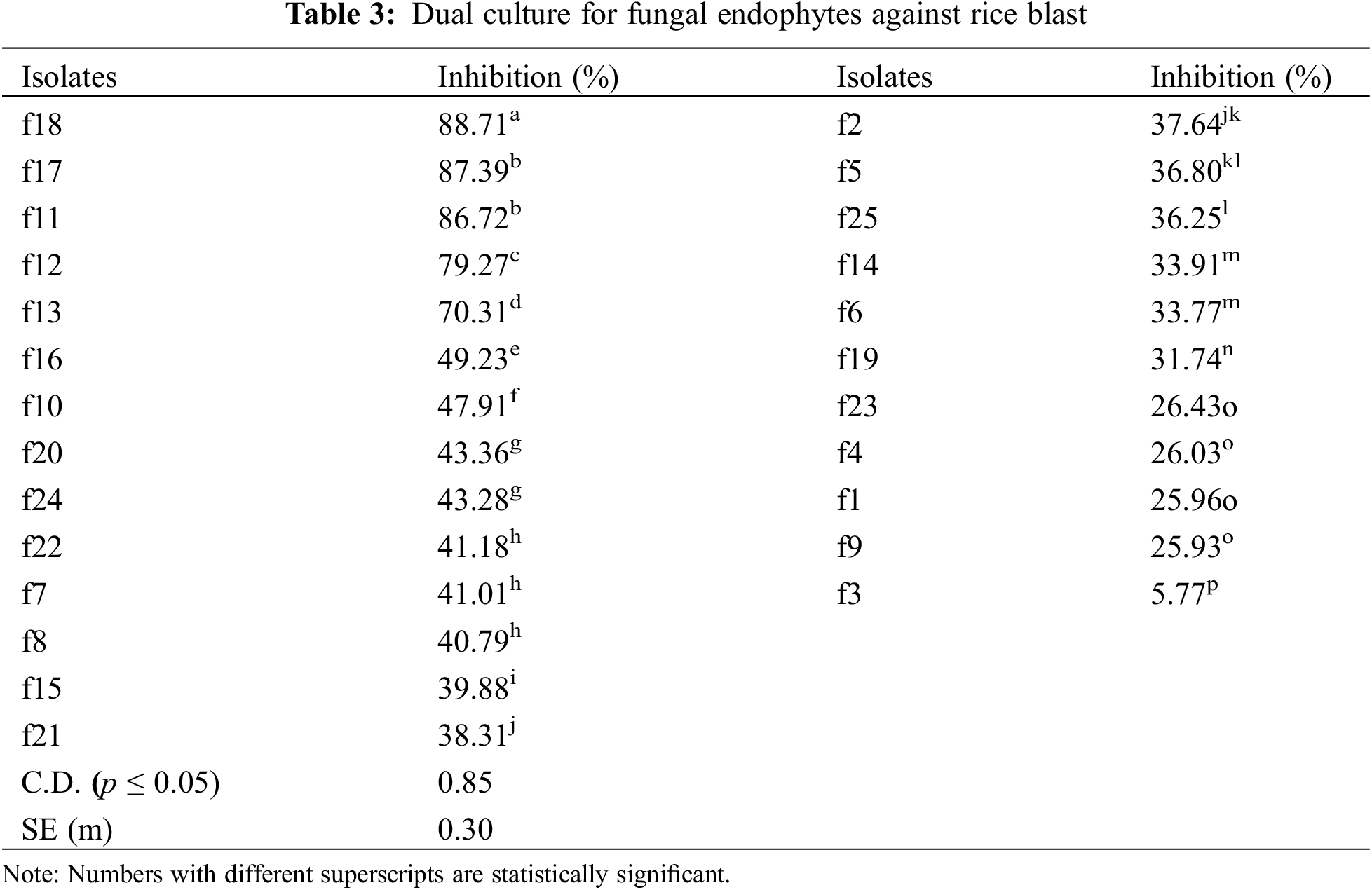
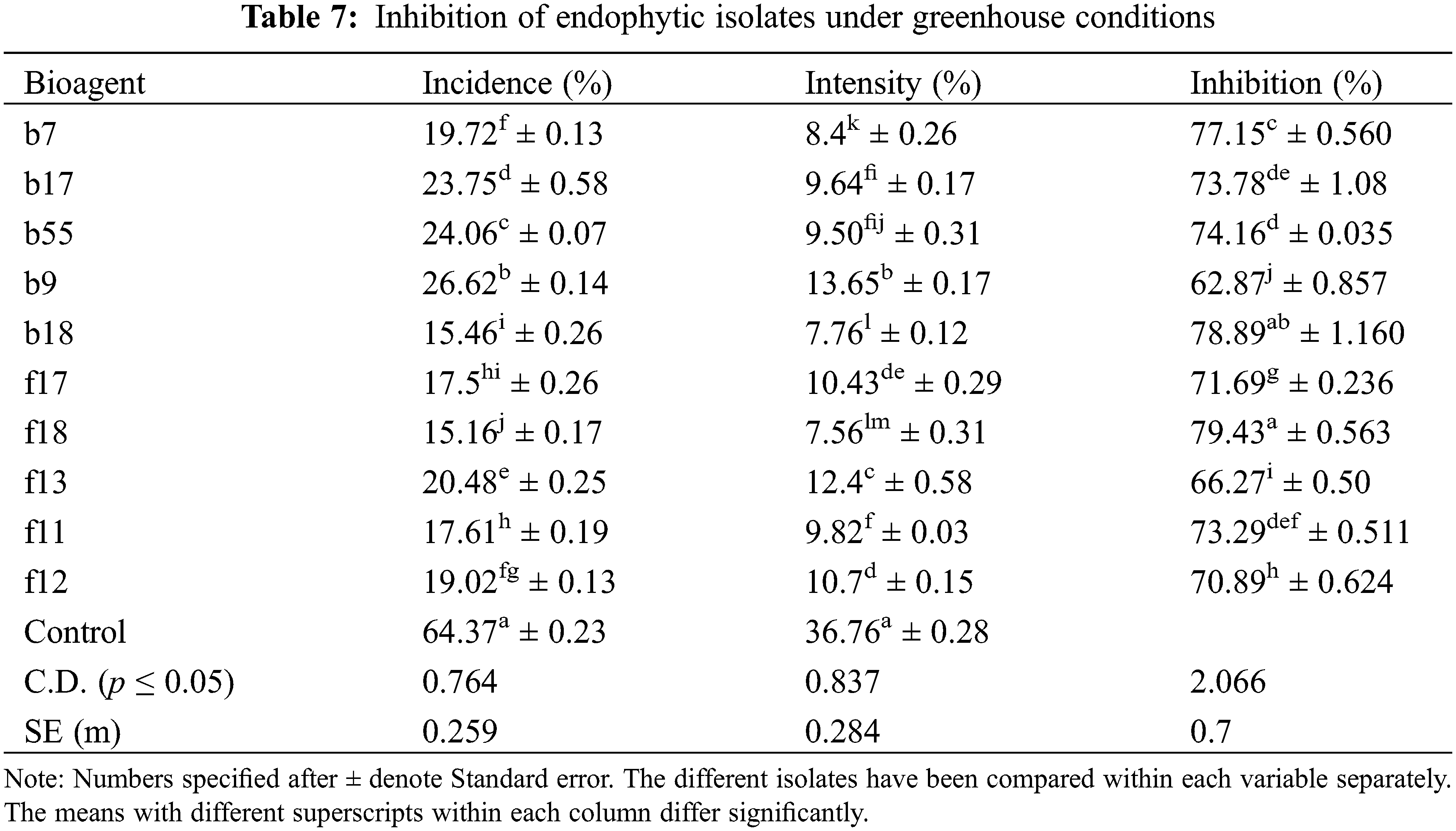
The isolates were evaluated for antagonistic activity against P. oryzae using the dual culture method. Five isolates among bacterial endophytes and five isolates among fungal endophytes showed antagonistic activity against pathogens with percent inhibition greater than 50%. It was found that in the case of bacterial endophytes, isolates b18 and b7 had the highest percent inhibition of 71.56% and 70.43% respectively, which were statistically significant (p < 0.05), followed by b17 (67.83%), b55 (62.93%), and b9 (61.23%). However, the lowest inhibition was found by the isolate b45 (8.46%) (Table 2; Figs. 1A, 1B). In the case of fungal endophytes, the highest inhibition was found by f18 with a percent inhibition of 90.46% which was significant at p < 0.05 followed by f17 (87.39%). However, the lowest inhibition was found by f3 (5.77%) (Table 3; Figs. 1A, 1C). The endophytes that showed greater inhibition in dual culture tests were used for further studies.
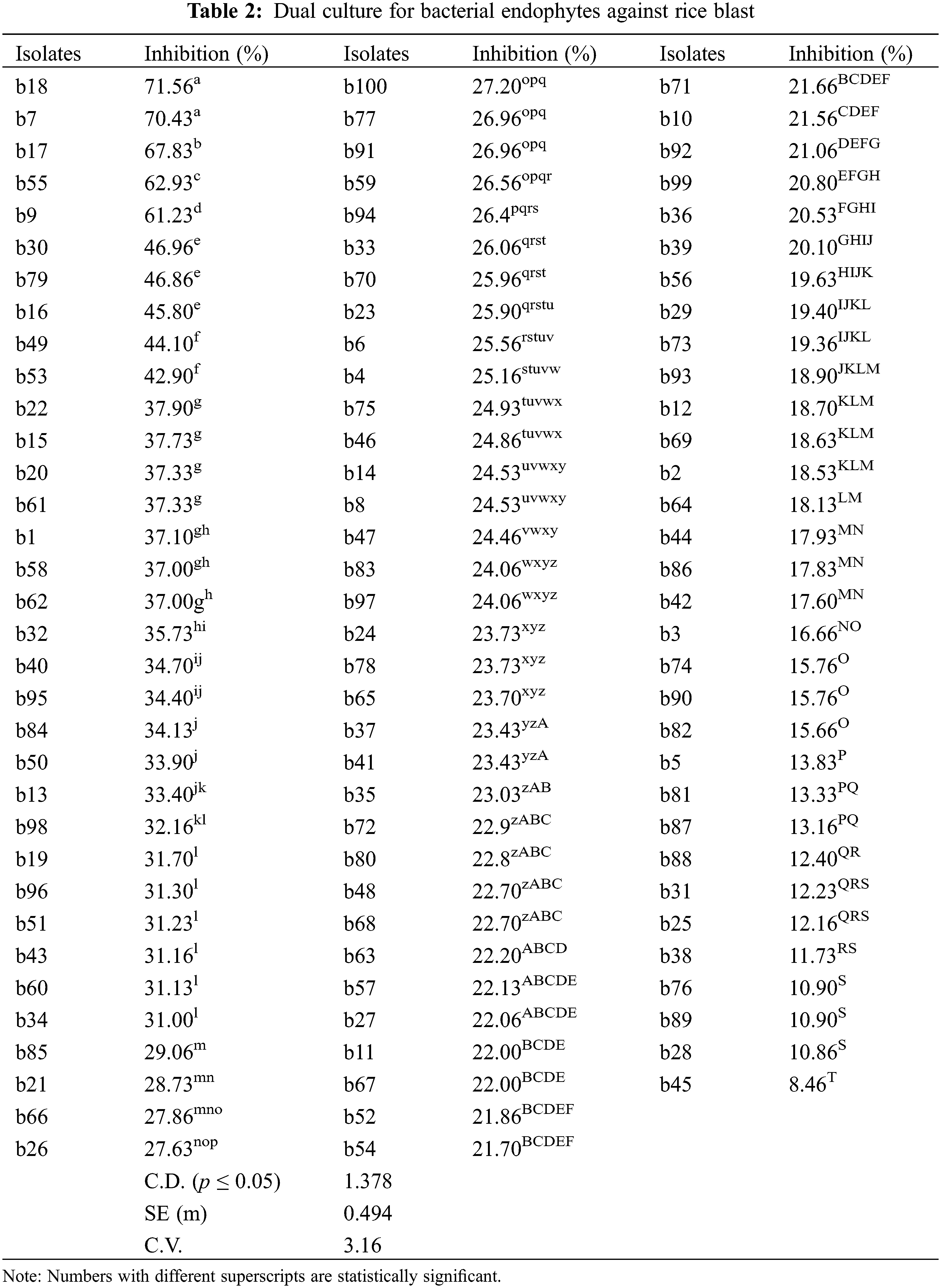

Figure 1: Dual culture of bacterial and fungal endophytes using potato dextrose agar plate against Pyricularia oryzae. (A) Selection of appropriate strains of bacterial and fungal isolates for good endophytes potential; (B) Five isolates among bacterial endophytes were showing antagonistic activity. (C) Five isolates among fungal endophytes were showing antagonistic activity
3.3 In vitro Screening for Antagonistic Activity of Volatile Compounds
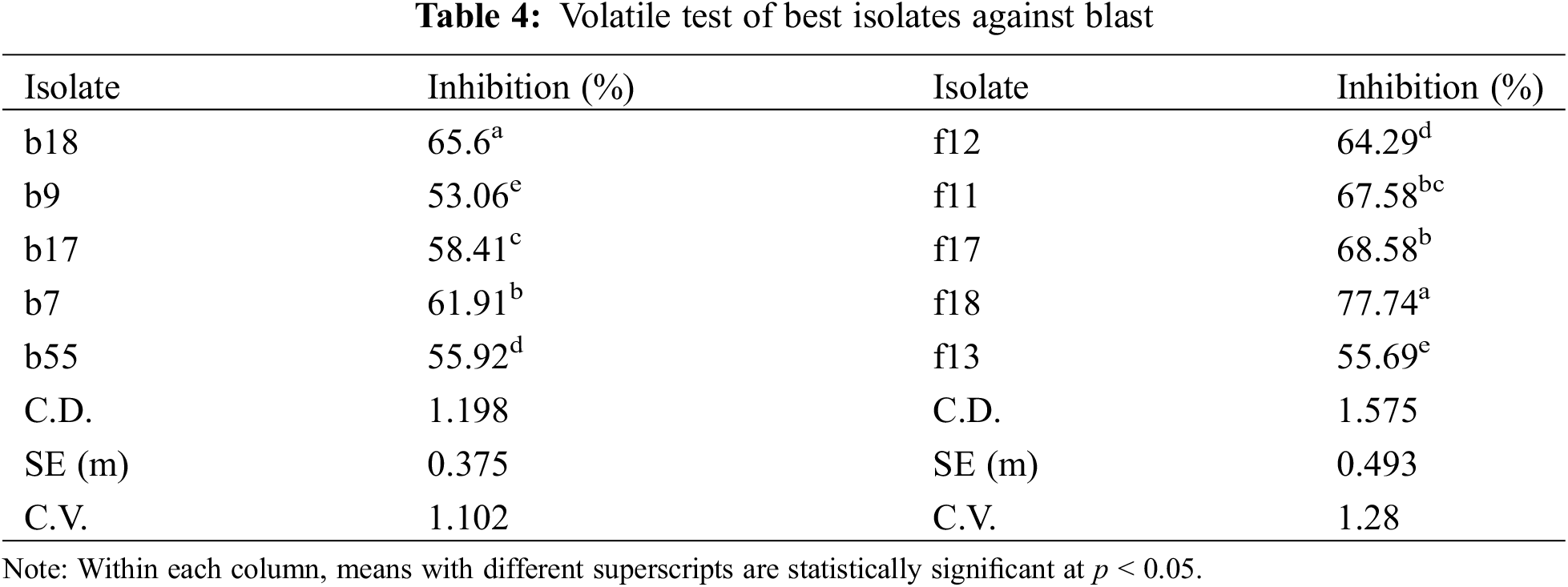
All the isolates produced volatile compounds that inhibit the growth of the pathogen. Among the bacterial isolates, b18 showed the highest inhibition of 65.6% followed by b7 and b17 with percent inhibitions of 61.91% and 58.41%, respectively (Table 4). Among the fungal isolates, the highest inhibition was shown by f18 (77.74%) followed by f17 (68.58%), and f11 (67.58%) (Table 4).
3.4 Phosphorus Solubilization Test of Effective Biocontrol Agents
The phosphorus solubilization test revealed that in the case of fungal endophytes, phosphorous solubilization varied from 34.28 (f12) to 88.44 g mL−1 (f18). On the other hand, in the case of bacterial isolates, it varied from 48.24 (b7) to 70.73 g mL−1 (b55) (Tables 5, 6).
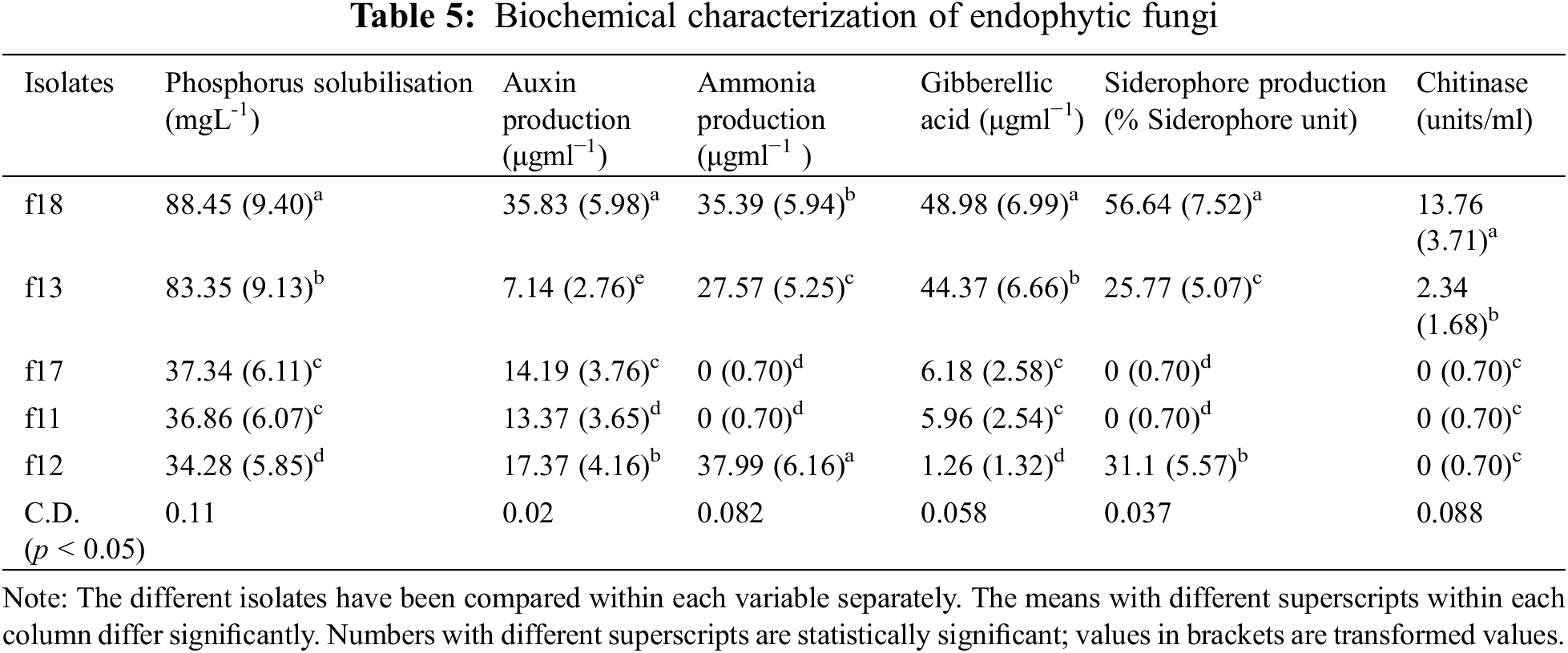
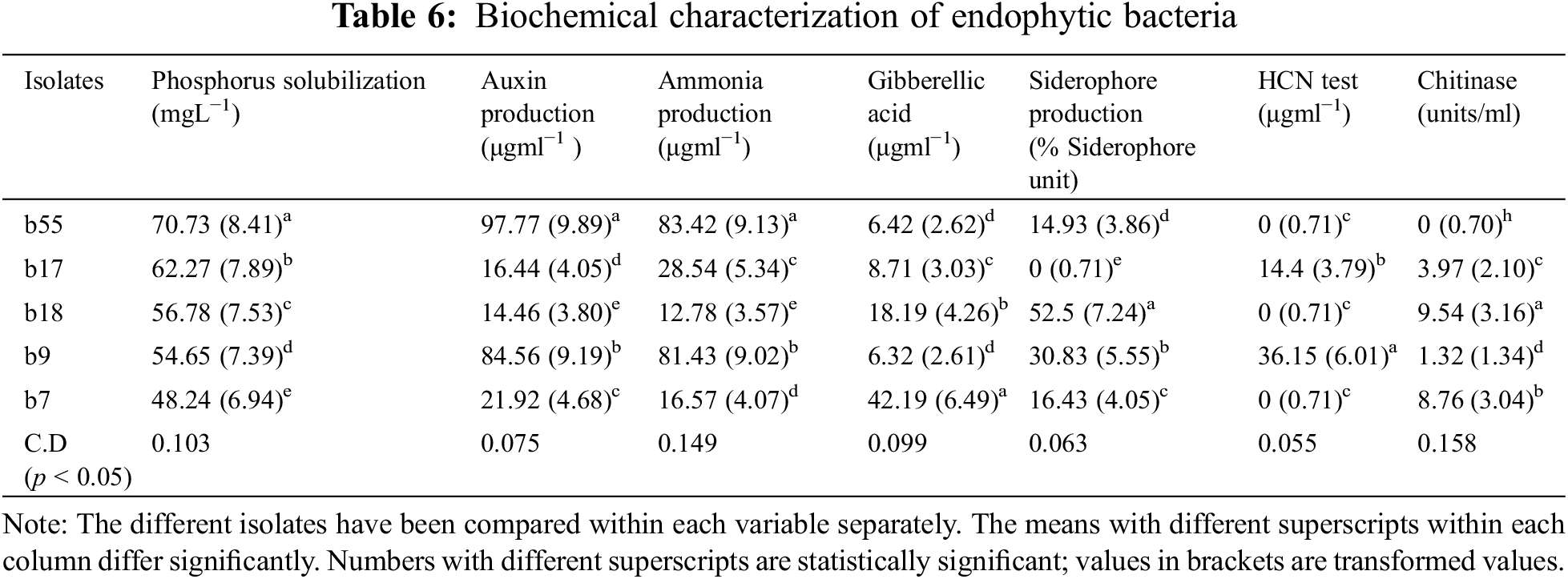
3.5 Assay for in vitro Auxin and Gibberellic Acid Production
All the bacterial and fungal isolates were able to produce auxin as well as gibberellic acid. The auxin production varied from 7.14 (g mL−1) to 97.77 (g mL−1). Among the bacterial endophytes, the highest auxin production was shown by isolate b55 (97.77 µgml−1) (significant at p < 0.05), followed by isolate b17 (84.56 µgml−1). The lowest auxin production was shown by isolate b18 (14.46 µgml−1). Among the fungal isolates, the highest production was shown by f18 (significant at p < 0.05), which showed auxin production of 35.83 µgml−1 followed by f12 (17.37 µgml−1), whereas the lowest auxin production was exhibited by f13 (7.14 µgml−1). In the case of gibberellic acid (GA) production, isolate b7 showed the highest GA production of 42.19 µgml−1 (significant at p < 0.05), and the lowest production was shown by isolate b9 (6.32 µgml−1) among bacterial isolates. Among fungal isolates, the highest production was shown by isolate f18 (48.98 µgml−1), followed by isolate f13 (44.37 µgml−1), whereas the lowest production was shown by isolate f12 (1.26 µgml−1) (Tables 5, 6).
3.6 Detection of Siderophore and HCN Production
Among the five bacterial isolates, four were confirmed to produce siderophores by CAS assay, while, only three were able to produce siderophores among the fungal isolates. The isolates that produce siderophores change the colour of the media from blue to orange. In the case of bacteria, the isolate b18 showed the highest siderophore production of 52.5 M mL−1, whereas in the case of fungi, the isolate f18 produced 56.64 M mL−1 of siderophore (Tables 5, 6). Among all the ten isolates tested for HCN production, only two bacterial isolates were able to produce HCN. The isolate b9 produced HCN of 36.15 µgml−1, while b17 produced HCN of 14.4 µgml−1 (Tables 5, 6).
3.7 Detection of Ammonia Production and Chitinase Production
All the bacterial isolates were able to produce ammonia, varying from 83.42 µgml−1 (b55) to 12.78 µgml−1 (b18). However, only three fungal isolates were able to produce ammonia, where the highest production was found by isolate f12 (37.99 µgml−1) followed by f18 (35.39 µgml−1) and f13 (27.57 µgml−1) (Tables 5, 6). Also, some isolates were able to produce chitinase. The isolate b18 had the highest chitinase activity of 9.54 units/ml, followed by b7 (8.76 units/ml) and b17 (3.97 units/ml). In the case of fungi, only two fungi were able to produce the chitinase enzyme. The isolate f18 showed chitinase production of 13.76 units/ml, and f13 showed a production of 2.34 units/ml (Tables 5, 6). The isolates were also tested for nitrogen fixation by using Jensen’s media, and it was confirmed that all the bacterial isolates were able to fix the atmospheric nitrogen by forming glistering colonies, while none of the fungal isolates were able to fix the atmospheric nitrogen.
3.8 Morphological Characterization of Endophytic Microorganisms
The morphological characteristics of bacterial endophytes revealed that b7 and b9 were gram-negative, while b18, b55, and b17 were gramme-positive in gramme staining. The colony morphology of the bacterial isolates varied from smooth and round in the case of b7 to irregular and rough in b18, while b17 has granular and wavey margins. The colony’s colour varied from white to greyish white. All the bacterial endophytes were rod-shaped (Supplementary Table 1). In the case of fungal isolates, the colour of the colony was initially white, which later turned dark olivaceous green in the cases of f11 and f17 (Supplementary Table 2). The mycelium is abundant, cottony, dark olive green in colour, and aerial. The conidia are globose to subglobose with an average size of 5.21-3.14.13-2.17 m (av. 4.033.11 m) and 4.27-2.783.87-2.17 m (av. 3.783.18 m), respectively. The average width of mycelium was found to be 5.524.3 m (av. 4.96 m) and 5.633.66 m (av. 4.85 m), respectively. In f12, the mycelium is floccose, tufted green, fast-growing, and sporulates abundantly. The edge of the colony was smooth and spreading. The conidia were light green in colour and globose to subglobose in shape, with an average size of 3.88–2.7–2.97–1.61 (av. 2.97–2.51) m and an average mycelial width of 4.94–3.64 (av. 4.59) m. In the case of f18, the colony was a powdery mass with a yellow-green colour and abundant sporulation. Usually, the margins of the colony were white. The conidia were globose in shape, with an average size of 4.67-3.85.61-4.00 (av. 4.873.92) m and a mycelial width of 8.644.74 (av. 6.53) m. In the case of f13, the colonies were bluish-green in colour with a white periphery and were radially sulcate. The globose to subglobose with an average size of 4.89–3.95–4.45–3.18 (av. 3.84–3.78) m and mycelial width of about 4.97–3.26 (av. 3.74) m. The phialides were flask-shaped, and their size was also measured, which ranged from 4.3-9.53.1-4.5 m (av. 5.83.2) m in the case of f12 to 5.2-7.84.6-8.2 m (av. 6.87.5 m) in f13 (Supplementary Table 2).
To confirm the identity of potential bacterial and fungal endophytes, the 16S rRNA gene in the case of bacteria and the ITS region in the case of fungal endophytes were amplified using PCR. The 16S rRNA gene was amplified from all five bacterial endophytes, and an amplicon of 1200 bp was obtained after PCR amplification. Similarly, the ITS region was amplified from all five fungal endophytes, and an amplicon of 750 bp was obtained.
Both the fragments from bacteria and fungi were sequenced and analysed. All the sequences generated were submitted to NCBI GenBank, and accession numbers were received. Detailed illustrations of these bacterial and fungal endophytes and their accession numbers, percentage similarity to the closest species, and genetic identity based on BLASTn searches are given in Supplementary Table 3. The results revealed that the endophytes were identified as: Trichoderma afroharzianum (f11 and f17), Trichoderma harzianum (f12), Penicillium rubens (f13), Aspergillus flavus (f18), Stenotrophomonas rhizophila (b7), Stenotrophomonas maltophilia (b9), Bacillus cereus (b17), Enterobacter cloacae (b55), and Bacillus licheniformis (b18). The percent similarity of endophytes varied from 93 percent in the case of Stenotrophomonas maltophilia to 100 percent in the cases of Trichoderma harzianum and Aspergillus flavus. The sequences of isolated bacterial and fungal endophytes were compared with sequences of other bacterial and fungal species, respectively, using a similarity of above 99% retrieved from GenBank. MEGA 11 software was used to analyse and construct phylogenetic relationships between the endophytes and their related fungi and bacteria. The evolutionary history was inferred using the neighbour-joining method. In the bacterial phylogenetic tree, it was evident that two broad clusters were formed, with Bacillus licheniformis (B. licheniformis), Bacillus cereus (B. cereus), and Stenotrophomonas rhizophila (S. rhizophila), S. maltophilia, and E. coaleae in another cluster (Supplementary Fig. 1). All the isolates were grouped into two phyla, viz., Firmicutes and Proteobacteria. In the case of a fungal phylogenetic tree, the tree divides into two broad clusters. T. afroharzianum and T. harzianum were in one cluster, and P. crysogenum and A. flavus were in a separate cluster. Both clusters further subdivide into two more clusters. All the fungal endophytes fall under the division Ascomycota.
The sequence identity matrix of our endophytes with other endophytes in the NCBI gene bank revealed that similarity ranged between 23% and 100%. Most of the bacterial endophytes were similar to endophytes from India and China. However, Stentrophomonas rhizopila was 93.5% similar to isolates from Iran and China (MZ895036 and EU135790). Similarly, Bacillus licheniformis was 96% similar to isolates from Denmark and South Korea (CP045814 and CP042252). S. maltophila was 93% similar to Indian isolates (MW282918 and HE963840). Furthermore, Enterobacter cloacae were 96% similar to isolates obtained from China and France (KX431213 and KJ49995). Bacillus cereus was 99% similar to isolates from India (KC68396 and HQ388817) (Supplementary Fig. 1). In the case of fungal endophytes, the sequence identity matrix revealed that T. harzianum showed 100% similarity with the isolates from China (MT102390) and Hungary (MN516457), and T. afroharzianum showed 100% similarity with isolates from Brazil (KC576738) and the USA (KC007211). P. chrysogenum showed 99% similarity with isolates from the USA (MN644620) and South Africa (MN413181). Similarly, A. flavus was showing 100% similarity with the isolates of Cassava (KC994648) and France (KC964101) (Supplementary Fig. 2).
3.12 Efficacy of Fungal Endophytes under Greenhouse Conditions
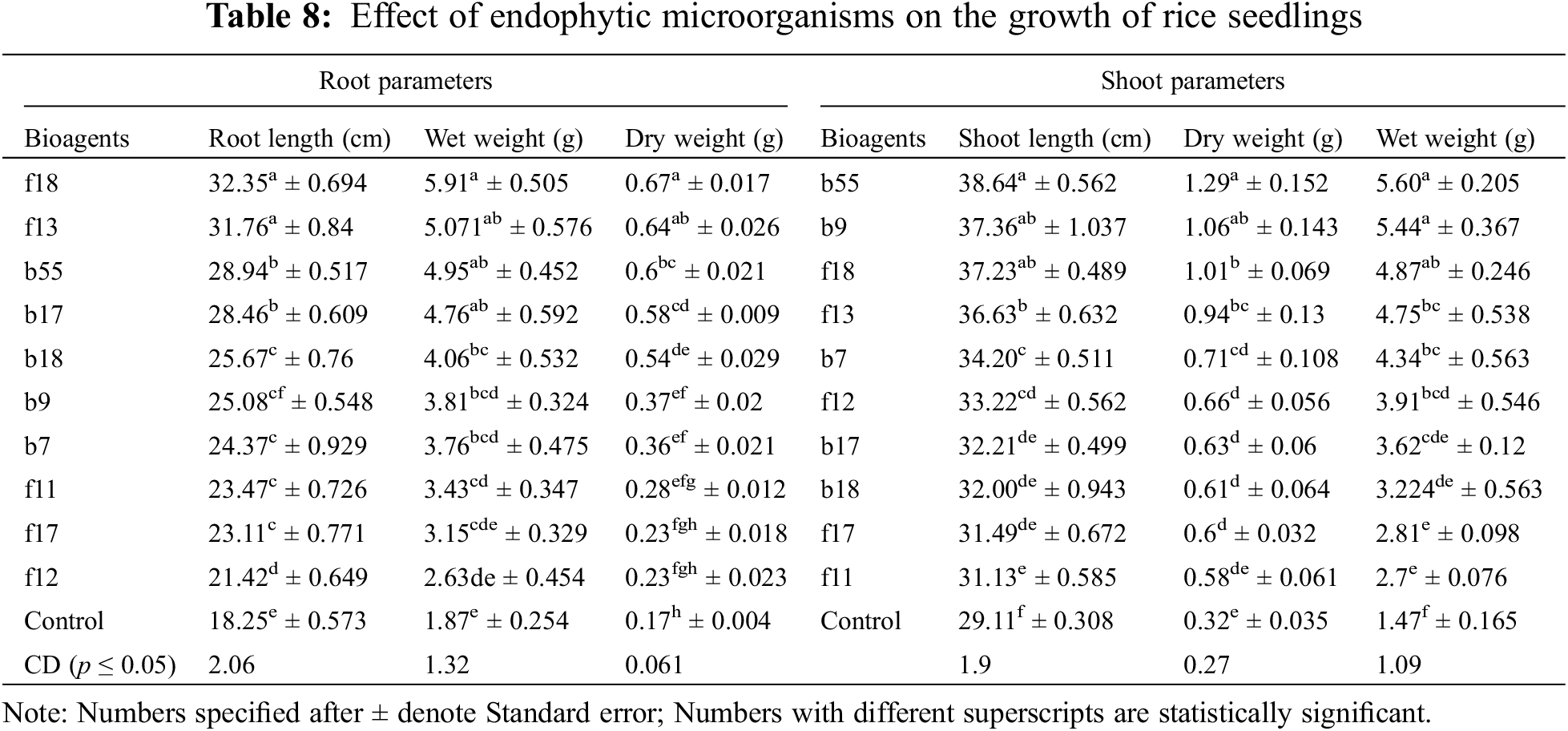
The highest disease control was observed by A. flavus with a percent inhibition of 79.43%, followed by T. afroharzianum with an inhibition of 71.63% (Table 7). Among bacterial endophytes, the highest inhibition was found by B. licheniformis with an inhibition of 78.89% and was statistically significant at p < 0.05, followed by S. rhizophila with an inhibition of 77% (Table 7). The plants were also evaluated for growth parameters, and the results indicate that all the isolates had a positive impact on considered plant growth parameters like elongation of the shoot (38.64–31.13 cm) and root (32.35–21.42 cm) when compared with the control seedling treated with only distilled water. Among bacterial isolates, the isolate E. cloacae showed a significant increase in root length (28.94 cm), shoot length (38.64 cm), fresh root weight (4.95 g), fresh shoot weight (5.60 g), dry root weight (0.6 g), and dry shoot weight (1.29 g) over the control at p < 0.05 (Table 8). In the case of fungal isolates, the f18 showed the highest root growth (32.35 cm), shoot growth (37.23 g), dry root weight (0.67 g), dry shoot weight (1.01 g), fresh root weight (5.91 g), and fresh shoot weight (4.87 g) at p < 0.05 (Table 8).
Endophytes are microorganisms that live within plant tissues and are beneficial for the growth of plants [35]. Apart from being a notable option for endosymbiosis, endophytes have the ability in terms of influencing the global climatic scenario. These features have made this one of the most cost-effective and environmentally friendly techniques in the integrated crop disease control strategy. In our study, a total of 125 endophytic microorganisms (100 bacterial and 25 fungal) were isolated from roots, stems and leaves of four varieties of rice collected from three different districts of Kashmir namely; Anantnag, Budgam and Baramullah. In our study, the majority of endophytes were isolated from roots followed by the stem and leaves. This is Ji et al. [36] who also isolated the highest number of endophytes from roots. The difference in endophytic presence in tissue and organs might be the ability of the endophytes to use the substrates in addition to the physiochemical properties of the tissue. Furthermore, the variance in endophytic colonization might be due to the environmental circumstances in which the rice plants thrive. The highest number of endophytes was isolated from SR4 followed by C1039 and SR3 variety of rice.
Endophytes can prevent or mitigate the harmful effects of certain pathogenic microorganisms. They benefit their host plant through various mechanisms such as antibiosis, growth promotion, induced resistance, competition, and parasitism [37]. Our results suggest that from 125 endophytic microorganisms isolated, ten endophytes (five bacterial and five fungal) were effective in controlling the growth of P. oryzae under in vitro conditions. In the case of bacterial endophytes, B. licheniformis showed the highest inhibition, followed by S. rhizophila. Among fungal endophytes, A. flavus showed the highest inhibition, followed by T. afroharzianum and T. harzianum. Similar results have been also reported by several workers where they reported endophytes to inhibit the growth of disease-causing pathogens, including P. oryzae in Suwandel and Kaluheenati varieties of paddy [38]. A previous study showed that B. licheniformis inhibits the development of Rhizoctonia solani in maize [39]. However, the effectiveness of B. licheniformis and S. rhizophila in controlling P. oryzae has not been reported yet. Moreover, Aspergillus and Penicillium were shown to inhibit the mycelial growth of Magnaporthe oryzae [40]. Trichoderma sp. was also shown to limit the development of Rhizoctonia solani, according to Sharma et al. [41].
VOCs are known to serve as stimuli, activating a variety of signals that govern physiological processes and cellular impacts [42]. These compounds might have a variety of chemical configurations and could help plants defend themselves against fungal infections [43]. A volatile test was conducted and it was found that all the selected endophytes produced volatile compounds that were able to inhibit the disease. The highest inhibition was exhibited by B. licheniformis, followed by S. rhizophila. In the case of fungal endophytes, the highest inhibition was shown by A. flavus, followed by T. afroharzianum in the volatile test. Similar results were also reported by Strobel [44] who observed VOCs are being emitted by endophytic Bacillus and Enterobacter strains. Aspergillus niger and T. harzianum volatile-mediated antagonistic action against F. oxysporum f. sp. lycopersici was mentioned by Suciatmih et al. [45]. Production of volatile compounds by Trichoderma longibrachiatum was also reported by Sornakili et al. [46] against phytopathogens P. oryzae.
The bacteria used for biocontrol produce siderophores with a greater affinity for iron than fungal pathogens, allowing antagonistic bacteria to scavenge the majority of the available iron and restrict fungal pathogen multiplication [36]. In the present study, all the bacterial isolates produced siderophores except Bacillus cereus, whereas, in the case of fungus, only Trichoderma afroharzianum did not produce siderophores. The highest siderophore production was found by Bacillus. licheniformis, followed by Stenotrophomonas maltophilia in the case of bacterial endophyte. In the case of fungal endophytes, the highest siderophore production was found in Aspergillus flavus. Siderophore-producing bacteria and fungi have been reported to be found in rice, with the main genera being Pantoea, Bacillus, Pseudomonas, and Burkholderia [47,48]. Regarding hydrogen cyanide production, it was found that only two bacterial endophytes were able to produce HCN with the highest production by Stenotrophomonas maltophilia, followed by Bacillus cereus. Ammonia production provides a variety of roles in plant physiology and growth promotion and is mostly formed by diazotrophic bacteria by nitrite ammonification [49]. In our study among bacterial endophytes highest ammonia production was found by Enterobacter cloacae, followed by Stenotrophomonas maltophilia. In the case of fungal endophytes, the highest ammonia production was found by T. harzianum followed by Aspergillus flavus. Ammonia in association with HCN and siderophore synthesis, works as an antagonistic factor against phytopathogens [50]. Production of siderophores, ammonia, and HCN were also earlier reported [51,52]. In the context of chitinase activity, all isolates except Enterobacter cloacae were shown to be capable of producing this enzyme. The highest chitinase activity was shown by Stenotrophomonas rhizophila, followed by Bacillus cereus and B. licheniformis. In the case of fungal isolates, only Aspergillus flavus and Penicillium rubens showed chitinase activity. Also, all five bacterial isolates were able to fix nitrogen by forming glistering colonies on Jensen’s media; however, no fungal endophyte was able to fix atmospheric nitrogen. The results are in agreement with previous reports, where Enterobacter has been reported to have the capability of IAA, siderophores, HCN, ammonia, and phosphate solubilization [53]. Nitrogenase activity, IAA production, and phosphate solubilization were also reported by Mehnaz et al. [54] while working on Stenotrophomonas maltophilia which was isolated from sugarcane plants. Stenotrophomonas. rhizophila has been reported to possess plant growth-promoting activity as reported by Wei et al. [55]. It was also reported that Trichoderma isolates have been found to possess disease suppression activities and growth-promoting properties like phosphate solubilization, secretion of indole acetic acid, gibberellins, and siderophores activity [56,57].
Plant hormones are signalling molecules that have important roles in the growth and development of plants [58]. Endophytes have been shown to boost plant development through a variety of processes, including the production of phytohormones such as IAA and GA [59]. In the present, all the isolates were able to produce pink colour in Luria broth supplemented with tryptophan. Among bacterial endophytes, the highest IAA was produced by isolate Enterobacter Cloacae, whereas, in the case of fungal endophytes, the highest IAA production was found by isolate A. flavus. In case the of GA production isolate B. subtilis showed the highest GA production. Among fungal isolates, the highest production was shown by isolate A. flavus. Plant growth-promoting fungi and bacteria have been shown to boost crop growth and production in several studies [60]. There have also been reports of filamentous fungi from the genera Aspergillus, Penicillium, and bacteria synthesizing phytohormones [61]. In our study, Enterobacter cloacae produces more IAA than other bacteria which is also reported by Coulson and Patten [62]. Endophytic fungus such as Aspergillus niger, Aspergillus flavus, Penicillium funiculosum, and Penicillium corylophilum, have been shown to produce phytohormones such as GAs and IAA [63].
Phosphorous is an essential macronutrient for plant growth and development. Endophytes are known to enhance plant growth by phosphate [64]. Insoluble forms of phosphates are converted by endophytes into soluble and plant-available forms. In our case it was found that among bacterial endophytes, all the isolates were able to solubilise phosphorous. It was revealed that isolate Enterobacter cloacae have highest phosphorous solubilisation, followed by Bacillus cereus and Bacillus licheniformis. In case of fungal endophytes Aspergillus flavus and Penicillium chrysogenum has highest phosphorous solubilisation, followed by Trichoderma afroharzianum and Trichoderma harzianum. These findings are consistent with those of Kumar et al. [65,66], who found that endophytic diazotrophic bacteria were able to solubilize calcium phosphate and Sharma et al. also reported Aspergillus, Penicillium and Trichoderma were most prevalent P-solubilizing strains [67].
The amplification and comparison of 16S rRNA and 18S rRNA is a sensitive tool for the identification of bacteria and fungi [68] and provided highly similar sequence identities to known species for the isolates studied. Molecular identification revealed that the entophytes were identified as Trichoderma afroharzianum (f11 and f17), Trichoderma harzianum (f12), Penicillium rubens (f13), Aspergillus flavus (f18), Stenotrophomonas rhizophila (b7), Stenotrophomonas maltophilia (b9), Bacillus cereus (b17), Enterobacter cloacae (b55) and Bacillus licheniformis (b18). The greenhouse experiment results emphasized the superiority of endophytic microbial application over harmful chemical fungicides. The study’s statistically significant findings suggest that chemical disease control can be replaced by the environmentally friendly and cost-effective usage of potential endophytes isolated from rice. All the treatments with endophytes were found statistically similar, however, the highest disease control was observed by Aspergillus flavus, followed by T. afroharzianum. Among bacterial endophytes, the highest inhibition was found by B. licheniformis, followed by Stenotrophomonas rhizophila. Some of the known biocontrol agents namely Trichoderma species, Bacillus subtilis and Pseudomonas fluorescens have shown good results in managing rice blast [9–11]. The identified endophytes used in the study were found as good as other reported biocontrol agents for managing rice blasts. The possible reason for their management is either due to the production of various types of volatile and non-volatile compounds or the development of the chitinase enzyme, siderophore production, HCN production, and ammonia production [69,70]. In the case of fungal endophytes like Trichoderma, where chitinase activity was not detected, processes such as mycoparasitism and competition, may play a role in disease suppression. All these factors might have contributed to effective disease suppression. Endophytic bacteria are known to control plant pathogenic fungi [14]. Endophytic bacteria’s antifungal processes are strain-specific, according to Hallmann et al. [14]. Similar results were also reported by several workers who use Bacillus subtilis as a foliar treatment which is efficient in controlling R. solani [71]. Martinez-Absalón et al. [72] used Bacillus licheniformis as a bio-control agent for suppressing phytopathogens. Bacillus sp. has been identified as a biocontrol agents against R. solani. Stenotrophomonas rhizophila has disease-suppression properties [73]. Khalili et al. [74] reported that Trichoderma was able to manage the brown spot disease in greenhouse conditions when used as seed treatment and foliar spray. Waqas et al. [75] also found that Aspergillus terreus was able to control stem rot caused by Sclerotium rolfsii in sunflowers. High seedling vigour plays a major role in plant growth promotion and biocontrol activities in endophytic bacteria mostly by Bacillus species [76]. Rice seeds when inoculated with the endophytes resulted in a significant increase in seedling height and dry weight compared to uninoculated plants [35,38]. Trichoderma isolate has the ability to promote the growth of rice seedlings [77].
Most of the isolated endophytes showed plant growth-promoting activity through the production of IAA, GA, and ammonia. Out of 125 endophytes, ten (10) showed promising results in dual culture experiments and were further assessed for volatile compound production tests. Also, the isolates had chitinase activity and produced siderophores and HCN which may be one of their mechanisms of disease control. Morphological and molecular characterization of endophytes were done to ascertain their identity. Differences among treated rice plants with these endophytes under greenhouse conditions demonstrated their positive effects on rice blast disease management and plant growth and development. In this study two novel bacterial endophytes were identified viz, Bacillus licheniformis and Stenotrophomonas rhizophila which were showing promising results both for growth promotion of rice and for managing blast disease caused by Pyricularia oryzae. Pot trial results also validated our results as all selected endophytes showed good results. Further research is needed for making formulations of both bacterial and fungal endophytes and using them in field conditions in the future.
Acknowledgement: The authors extend their appreciation to the Researchers Supporting Project number (RSP2023R298), King Saud University, Riyadh, Saudi Arabia. The authors are highly thankful to Associate Director Research, MRCFC, Khudwani SKUAST-K for providing space for lab as well as field experiments.
Funding Statement: The authors extend their appreciation to the Researchers Supporting Project Number (RSP2023R298), King Saud University, Riyadh, Saudi Arabia.
Author Contributions: The Author confirm contribution to the paper as follows: Study Conception and design: Guoqiang Fan; data collection: Shugufta Parveen, Fayaz A. Mohiddin, M. Ashraf Bhat, Zahoor Ahmed Baba, ;analysis and interpretation of results: Fehim Jeelani, M. Anwar Bhat, Sajad Un Nabi, Burhan Hamid, Saba Bandey; draft manuscript preparation: Farhanaz Rasool, Zakir Amin, Ibrahim Al-Ashkar, Muhammad Adnan and Ayman El Sabagh. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this article.
Ethics Approval: None.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.030921.
References
1. Parajuli, M., Khadka, G. B., Chaurasia, J. (2022). A review on comparative effect of chemicals and botanicals in management of brown spot diseases of rice (Oryza sativa L.). Archives of Agriculture and Environmental Science, 7, 127–131. [Google Scholar]
2. Nawaz, A., Rehman, A. U., Rehman, A., Ahmad, S., Siddique, K. M. et al. (2022). Increasing sustainability for rice production systems. Journal of Cereal Science, 103, 103400. [Google Scholar]
3. Pérez-Méndez, N., Miguel-Rojas, C., Jimenez-Berni, J. A., Gomez-Candon, D., Pérez-de-Luque, A. et al. (2021). Plant breeding and management strategies to minimize the impact of water scarcity and biotic stress in cereal crops under Mediterranean conditions. Agronomy, 12, 75. [Google Scholar]
4. Nalley, L., Tsiboe, F., Durand-Morat, A., Shew, A., Thoma, G. (2016). Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLoS One, 11, e0167295. [Google Scholar] [PubMed]
5. Dean, R., van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., di Pietro, A. et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. [Google Scholar] [PubMed]
6. Shahriar, S. A., Imtiaz, A. A., Hossain, M. B., Husna, A., Eaty, M. N. K. (2020). Review: Rice blast disease. Annual Research and Review in Biology, 35(1), 50–64. [Google Scholar]
7. Hamid, M. I., Ghazanfar, M. U. (2022). Integrated management of rice diseases. In: Sarwar, N., Rehman, A., Ahmad, S., Hasanuzzaman, M. (Eds.Modern techniques of rice crop production, pp. 401–421. Singapore: Springer. [Google Scholar]
8. Shabanamol, S., Jisha, M. S. (2014). Assessment of rice endophytic diazotrophic bacteria for biocontrol of rice sheath blight. International Multidisciplinary Research Journal, 3, 1–6. [Google Scholar]
9. Zeng, Q., Man, X., Huang, Z., Zhuang, L., Yang, H. et al. (2023). Effects of rice blast biocontrol strain Pseudomonas alcaliphila Ej2 on the endophytic microbiome and proteome of rice under salt stress. Frontiers in Microbiology, 14, 1129614. https://doi.org/10.3389/fmicb.2023.1129614 [Google Scholar] [PubMed] [CrossRef]
10. Law, J., Ser, W., Khan, H., Chuah, L., Pusparajah, T. M. et al. (2017). The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Frontiers in Microbiology, 8, 3. https://doi.org/10.3389/fmicb.2017.00003 [Google Scholar] [PubMed] [CrossRef]
11. Murunde, R., Ringo, G., Robinson-Boyer, L., Xu, X. (2022). Effective biocontrol of rice blast through dipping transplants and foliar applications. Agronomy, 12, 592. https://doi.org/10.3390/agronomy12030592 [Google Scholar] [CrossRef]
12. Mishra, S., Bhattacharjee, A., Sharma, S. (2021). An ecological insight into the multifaceted world of plant-endophyte association. Critical Reviews in Plant Sciences, 40(2), 127–146. [Google Scholar]
13. Ali, M., Ali, Q., Sohail, M. A., Ashraf, M. F., Saleem, M. H. et al. (2021). Diversity and taxonomic distribution of endophytic bacterial community in the rice plant and its prospective. International Journal of Molecular Sciences, 22, 10165. [Google Scholar] [PubMed]
14. Hallmann, J., Berg, G. (2006). Spectrum and population dynamics of bacterial root endophytes. In: Schulz, B. J. E., Boyle, C. J. C., Sieber, T. N. (Eds.Microbial root endophytes, vol. 9, pp. 15–31. Berlin, Heidelberg: Springer. [Google Scholar]
15. Naik, B. S., Shashikala, J., Krishnamurthy, Y. L. (2009). Study on the diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Microbiological Research, 164, 290–296. [Google Scholar] [PubMed]
16. Dennis, C., Webster (1971). Antagonistic properties of species-groups of Trichoderma: II. Production of volatile antibiotics. Transactions of the British Mycological Society, 57, 41–48. [Google Scholar]
17. Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiology, 17, 362–370. [Google Scholar]
18. Bray, R. H., Kurtz, L. T. (1945). Determination of total, organic, and available forms of phosphorus in soils. Soil Science, 59, 39–46. [Google Scholar]
19. Bano, N., Musarrat, J. (2003). Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential biocontrol agent. Current Microbiology, 46, 324–328. [Google Scholar] [PubMed]
20. Paleg, L. G. (1965). Physiological effects of gibberellins. Annual Review of Plant Physiology and Plant Molecular Biology, 16, 291–322. [Google Scholar]
21. Cappuccino, J. G., Sherman, N. (2002). Techniques for isolation of pure culture. Cultural and morphological characteristics of microorganisms. In: Welsh, C. T. (Ed.Microbiology: A laboratory manual, vol. 6, pp. 13–23. New York: Pearson Education. [Google Scholar]
22. Prabavathy, V. R., Mathivanan, N., Sagadevan, E., Murugesan, K., Lalithakumari, D. (2006). Self-fusion of protoplasts enhances chitinase production and biocontrol activity in Trichoderma harzianum. Bioresource Technology, 97, 2330–2334. [Google Scholar] [PubMed]
23. Rojas-Avelizapa, L. I., Cruz-Camarillo, R., Guerrero, M. I., RodríguezVázquez, R., Ibarra, J. E. (1999). Selection and characterization of a proteo-chitinolytic strain of Bacillus thuringiensis, able to grow in shrimp waste media. World Journal of Microbiology and Biotechnology, 15, 299–308. [Google Scholar]
24. Schwyn, B., Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry, 160, 47–56. [Google Scholar] [PubMed]
25. Baker, A. W., Schippers, B. (1987). Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp. mediated plant growth stimulation. Soil Biology and Biochemistry, 19, 51–457. [Google Scholar]
26. Boone, D. R., Castenholz, R. W., Garrity, G. M., Brenner, D. J., Krieg, N. R. et al. (2001). Bergey’s manual of systematic bacteriology, vol. 2. Springer Science and Business Media. [Google Scholar]
27. Sambrook, J., Fritsch, E. F., Maniatis, T. (1989). Molecular cloning: A laboratory manual, 2nd EditionNew York: Cold Spring Harbor Press. [Google Scholar]
28. Sun, L., Qiu, F. B., Zhang, X. X., Dai, X., Dong, X. Z. et al. (2008). Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microbial Ecology, 55, 415–424. [Google Scholar] [PubMed]
29. Khaskheli, M. A., Wu, L., Chen, G., Chen, L., Hussain, S. et al. (2020). Isolation and characterization of root-associated bacterial endophytes and their biocontrol potential against major fungal phytopathogens of rice (Oryza sativa L.). Pathogens, 9, 172. [Google Scholar] [PubMed]
30. Etesami, H., Emami, S., Alikhani, H. A. (2017). Potassium solubilizing bacteria (KSB) mechanisms, promotion of plant growth, and future prospects. A Review Journal of Soil Science and Plant Nutrition, 17, 897–911. [Google Scholar]
31. Tamura, K., Dudley, J., Nei, M., Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599. [Google Scholar] [PubMed]
32. González-Teuber, M., Vilo, C., Bascuñán-Godoy, L. (2017). Molecular characterization of endophytic fungi associated with the roots of Chenopodium quinoa inhabiting the Atacama Desert, Chile. Genomics Data, 11, 109–112. [Google Scholar]
33. Mingma, R., Duangmal, K. (2018). Characterization, antifungal activity and plant growth promoting potential of endophytic actinomycetes isolated from rice (Oryza sativa L.). Chiang Mai Journal of Science, 45, 2652–2665. [Google Scholar]
34. Gomez, K. A., Gomez, A. A. (1984). Statistical procedures for agricultural research. New York: John Wiley & Sons. [Google Scholar]
35. Fadiji, A. E., Babalola, O. O. (2020). Exploring the potentialities of beneficial endophytes for improved plant growth. Saudi Journal of Biological Sciences, 27, 3622–3633. [Google Scholar] [PubMed]
36. Ji, S. H., Gururani, M. A., Chun, S. C. (2014). Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiological Research, 169, 83–98. [Google Scholar] [PubMed]
37. Adeleke, B. S., Ayilara, M. S., Akinola, S. A., Babalola, O. O. (2022). Biocontrol mechanisms of endophytic fungi. Egyptian Journal of Biological Pest Control, 32(1), 1–17. [Google Scholar]
38. Atugala, D. M., Deshappriya, N. (2015). Effect of endophytic fungi on plant growth and blast disease incidence of two traditional rice varieties. Journal of the National Science Foundation of Sri Lanka, 43(2), 173–187. [Google Scholar]
39. Wang, H., Wen, K., Zhao, X., Wang, X., Li, A. et al. (2009). The inhibitory activity of endophytic Bacillus sp. strain CHM1 against plant pathogenic fungi and its plant growth-promoting effect. Crop Protection, 28(8), 634–639. [Google Scholar]
40. Schulz-Bohm, K., Martín-Sánchez, L., Garbeva, P. (2017). Microbial volatiles: Small molecules with an important role in intra and inter-kingdom interactions. Frontiers in Microbiology, 8, 2484. [Google Scholar] [PubMed]
41. Sharma, P., Leon, V. C., Raja, M., Pandian, R. T. P., Javeria, S. et al. (2016). Opportunistic endophytism of Trichoderma spp. and its biocontrol activity against Rhizoctonia solani causing sheath blight in rice. Preceedings of the Meeting of Biocontrol and Microbial Ecology, 116, 117–121. [Google Scholar]
42. Kai, M., Effmert, U., Piechulla, B. (2016). Bacterial-plant-interactions: Approaches to unravel the biological function of bacterial volatiles in the rhizosphere. Frontiers in Microbiology, 7(289), 108. [Google Scholar] [PubMed]
43. Li, T., Li, L., Du, F., Sun, L., Shi, J. et al. (2021). Activity and mechanism of action of antifungal peptides from microorganisms: A review. Molecules, 26, 3438. [Google Scholar] [PubMed]
44. Strobel, G. (2006). Muscodor albus and its biological promise. Journal of Industrial Microbiology and Biotechnology, 33, 514–522. [Google Scholar] [PubMed]
45. Suciatmih, M., Rahmansyah. (2013). Endophytic fungi isolated from mangrove plant and have antagonism role against Fusarium wilt. American Journal of Agricultural and Biological Science, 8, 251–257. [Google Scholar]
46. Sornakili, A., Thankappan, S., Sridharan, A. P., Nithya, P., Uthandi, S. (2020). Antagonistic fungal endophytes and their metabolite-mediated interactions against phytopathogens in rice. Physiological and Molecular Plant Pathology, 112, 101525. [Google Scholar]
47. Zhang, Z., Liu, T., Zhang, X., Xie, J., Wang, Y. et al. (2021). Cultivable endophytic bacteria in seeds of Dongxiang wild rice and their role in plant-growth promotion. Diversity, 13(12), 665. [Google Scholar]
48. Borah, M., Das, S., Bora, S. S., Boro, R. C., Barooah, M. (2021). Comparative assessment of multi-trait plant growth-promoting endophytes associated with cultivated and wild Oryza germplasm of Assam, India. Archives of Microbiology, 203(5), 2007–2028. [Google Scholar] [PubMed]
49. Geetha, K., Venkatesham, E., Hindumathi, A., Bhadraiah, B. (2014). Isolation, screening and characterization of plant growth promoting bacteria and their effect on Vigna radiata (L.) R. Wilczek. International Journal of Current Microbiology and Applied Sciences, 3, 799–803. [Google Scholar]
50. Allu, S., Pradeep, K. N., Amrutha (2014). Isolation, biochemical and PGP characterization of endophytic Pseudomonas aeruginosa isolated from chilli red fruit antagonistic against chilli anthracnose disease. International Journal of Current Microbiology and Applied Sciences, 3, 318–329. [Google Scholar]
51. Maheshwari, R., Bhutani, N., Suneja, P. (2019). Screening and characterization of siderophore producing endophytic bacteria from Cicer arietinum and Pisum sativum plants. Journal of Applied Biology and Biotechnology, 7, 7–14. [Google Scholar]
52. Abdenaceur, R., Farida, B. T., Mourad, D., Rima, H., Zahia, O. et al. (2022). Effective biofertilizer Trichoderma spp. isolates with enzymatic activity and metabolites enhancing plant growth. International Microbiology, 25(4), 817–829. [Google Scholar] [PubMed]
53. Ahemad, M., Khan, M. S. (2010). Influence of selective herbicides on plant growth promoting traits of phosphate solubilizing Enterobacter asburiae strain PS2. Research Journal of Microbiology, 5, 849–857. [Google Scholar]
54. Mehnaz, S., Baig, D. N., Lazarovits, G. (2010). Genetic and phenotypic diversity of plant growth promoting rhizobacteria isolated from sugarcane plants growing in Pakistan. Journal of Microbiology and Biotechnology, 20, 1614–1623. [Google Scholar] [PubMed]
55. Wei, Y., Li, L., Hu, W., Ju, H., Zhang, M. et al. (2020). Suppression of rice blast by bacterial strains isolated from cultivated soda saline-sodic soils. International Journal of Environmental Research and Public Health, 17, 5248. [Google Scholar] [PubMed]
56. Joo, J. H., Hussein, K. A. (2022). Biological control and plant growth promotion properties of volatile organic compound-producing antagonistic Trichoderma spp. Frontiers in Plant Science, 13, 897668. [Google Scholar] [PubMed]
57. Safarimotlagh, M. R., Mohammadian, S. (2016). Biological control of rice brown spot disease caused by Bipolaris victoriae by some fungal isolates in the greenhouse and in vitro conditions. Biocontrol in Plant Protection, 4, 11–25. [Google Scholar]
58. Poveda, J., Eugui, D., Abril-Urías, P., Velasco, P. (2021). Endophytic fungi as direct plant growth promoters for sustainable agricultural production. Symbiosis, 85, 1–19. [Google Scholar]
59. Woźniak, M., Gałązka, A., Tyśkiewicz, R., Jaroszuk-Ściseł, J. (2019). Endophytic bacteria potentially promote plant growth by synthesizing different metabolites and their phenotypic/physiological profiles in the Biolog GEN III MicroPlateTM test. International Journal of Molecular Sciences, 20, 5283. [Google Scholar] [PubMed]
60. Wani, Z. A., Kumar, A., Sultan, P., Bindu, K., Riyaz-Ul Hassan, S. et al. (2017). Mortierella alpina CS10E4, an oleaginous fungal endophyte of Crocus sativus L. enhances apocarotenoid biosynthesis and stress tolerance in the host plant. Scientific Reports, 7(1), 8598. [Google Scholar] [PubMed]
61. Egamberdieva, D., Wirth, S. J., Alqarawi, A. A., Abd Allah, E. F., Hashem, A. (2017). Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Frontiers in Microbiology, 8, 2104. [Google Scholar] [PubMed]
62. Coulson, T. J., Patten, C. L. (2015). The TyrR transcription factor regulates the divergent akr-ipd Coperons of Enterobacter cloacae UW5. PLoS One, 10, e0121241. [Google Scholar] [PubMed]
63. Khan, A. L., Hussain, J., Al-Harrasi, A., Al-Rawahi, A., Lee, I. J. (2015). Review endophytic fungi: Resource for gibberellins and crop abiotic stress resistance. Critical Reviews in Biotechnology, 35, 62–74. [Google Scholar] [PubMed]
64. Varga, T., Hixson, K. K., Ahkami, A. H., Sher, A. W., Barnes, M. E. et al. (2020). Endophyte-promoted phosphorus solubilization in Populus. Frontiers in Plant Science, 11, 567918. [Google Scholar] [PubMed]
65. Kumar, V., Jain, L., Jain, S. K., Chaturvedi, S., Kaushal, P. (2020). Bacterial endophytes of rice (Oryza sativa L.) and their potential for plant growth promotion and antagonistic activities. South African Journal of Botany, 134, 50–63. [Google Scholar]
66. Aarab, S., Addas, F. E. H., Laglaoui, A., Bakkali, M., Arakrak, A. (2015). Solubilization of inorganic phosphate by Pseudomonas strains isolated from rice rhizosphere. International Journal of Biosciences, 6, 116–124. [Google Scholar]
67. Sharma, P., Leon, V. C., Raja, M., Pandian, R. T. P., Javeria, S. et al. (2016). Opportunistic endophytism of Trichoderma spp. and its biocontrol activity against Rhizoctonia solani causing sheath blight in rice. Proceedings of the Meeting of Biocontrol and Microbial Ecology, 116, 117–121. [Google Scholar]
68. Weisburg, W. G., Barns, S. M., Pelletier, D. A., Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173, 697–703. [Google Scholar] [PubMed]
69. Khan, M. R., Mohiddin, F. A. (2018). Trichoderma: Its multifarious utility in crop improvement crop improvement through microbial biotechnology. In: Prasad, R., Gill, S. S., Tuteja, N. (Eds.New and future developments in microbial biotechnology and bioengineering, pp. 263–291. Amsterdam, Netherlands: Elsevier. [Google Scholar]
70. Mohiddin, F. A., Padder, S. A., Bhat, A. H., Ahanger, M. A., Shikari, A. B. et al. (2021). Phylogeny and optimization of Trichoderma harzianum for chitinase production: Evaluation of their antifungal behaviour against the prominent soil borne phyto-pathogens of temperate India. Microorganisms, 9, 1962. https://doi.org/10.3390/microorganisms [Google Scholar] [CrossRef]
71. Liu, B., Qiao, H., Huang, L., Buchenauer, H., Han, Q. et al. (2009). Biological control of take in all wheat by endophytic Bacillus subtilis E1R-j and potential mode of action. Biological Control, 49, 277–285. [Google Scholar]
72. Martinez-Absalón, S. C., Orozco-Mosqueda, M., del, C., Martínez-Pacheco, M. M., Farías Rodríguez, R. et al. (2012). Isolation and molecular characterization of a novel strain of Bacillus with antifungal activity from the sorghum rhizosphere. Genetics and Molecular Research, 11, 1–9. [Google Scholar]
73. Ulrich, K., Kube, M., Becker, R., Schneck, V., Ulrich, A. (2021). Genomic analysis of the endophytic Stenotrophomonas strain 169 reveals features related to plant-growth promotion and stress tolerance. Frontiers in Microbiology, 12, 1542. [Google Scholar]
74. Khalili, E., Sadravi, M., Naeimi, S., Khosravi, V. (2012). Biological control of rice brown spot with native isolates of three Trichoderma species. Brazilian Journal of Microbiology, 43, 297–305. [Google Scholar] [PubMed]
75. Waqas, M., Khan, A. L., Hamayun, M., Shahzad, R., Kang, S. M. et al. (2015). Endophytic fungi promote plant growth and mitigate the adverse effects of stem rot: An example of Penicillium citrinum and Aspergillus terreus. Journal of Plant Interactions, 10, 280–287. [Google Scholar]
76. Ansary, W. R., Prince, F. R. K., Haque, E., Sultana, F., West, H. M. et al. (2018). Endophytic Bacillus spp. from medicinal plants inhibit mycelial growth of Sclerotinia sclerotiorum and promote plant growth. Journal for Nature Research, 73, 247–256. [Google Scholar]
77. Bilal, L., Asaf, S., Hamayun, M., Gul, H., Iqbal, A. et al. (2018). Plant growth promoting endophytic fungi Aspergillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis, 76, 117–127. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools