 Open Access
Open Access
REVIEW
Adventitious Root Regeneration: Molecular Basis and Influencing Factors
Shanghai Key Laboratory of Bio-Energy Crops, Research Center for Natural Products, Plant Science Center, School of Life Sciences, Shanghai University, Shanghai, 200444, China
* Corresponding Author: Xiangyang Hu. Email:
Phyton-International Journal of Experimental Botany 2023, 92(10), 2825-2840. https://doi.org/10.32604/phyton.2023.030912
Received 02 May 2023; Accepted 25 June 2023; Issue published 15 September 2023
Abstract
Plant regeneration is a self-repair of the plant body in response to adverse conditions or damaged structures, and root regeneration allows the plant body to better adapt to its environment by supplementing the roots’ structure. Previous research has shown that adventitious roots can be made to occur from scratch in two ways. Studies that simulate adventitious root regeneration through natural conditions allow the regeneration process to be broadly divided into three stages: the perception of early signals, the massive accumulation of auxin, and the transformation of cell fate. The strength of regeneration, in turn, is influenced by wounding, stress, hormones, etc. This study mainly reviews the molecular mechanisms of de novo adventitious roots and discusses how the environment, hormones, and its own development in Arabidopsis thaliana.Keywords
Plant regeneration is the process by which the plant body repairs or replaces damaged structures [1,2]. In contrast to animals, plants cannot move to withstand damage from adverse environments, so regeneration is an important ability for plants to resist and adapt to their environment [3,4]. The essence of plant regeneration is the process by which cells transform their cell fate after adverse, environmental stress [5], and totipotency or pluripotency is the basis of regeneration [3].
De novo adventitious roots is the root organogenesis after tissue injury and is also a form of plant regeneration. Plants have a strong ability to perceive external stimuli. In response to biological stress (such as invasion of pathogenic bacteria) or abiotic stress (such as high temperature, light or flood), plants can activate the expression regulation of ARFs (AUXIN RESPONSE FACTORs), ERFs (ETHYLENE RESPONSE FACTOR) and other related regulations so as to regenerate adventitious roots (ARs) to repair damaged organs [6–8]. Most plants can regenerate ARs indirectly from extraneous tissues and organs, or directly from various airborne organs [9]. For a long time, the common regeneration method in agriculture is to heal tissues and cuttings, such as detached leaves, hypocotyls, cuttings, etc., to produce ARs [10].
ARs are widely used in cutting and tissue culture of plant organs because they enlarge the root system of plants and enable plants and cells to regenerate [11]. In rice and other grains, ARs represents a major component of the root system because the primary root RAM originating from the embryo is short-lived [12]. Some researchers prefer to refer to these types of ARs as dendritic crown roots because they are part of the normal developmental program of grains [12,13]. Compared with primary and lateral roots, ARs have no fixed location, are more influenced by environmental factors, and have a wider range of origins, such as isolated leaves, hypocotyls, and primary root wounds [14]. Although histological studies have shown that the formation of ARs begins in the primary cambium and its parenchyma cells, which are receptor cells for initiation of ARs [15], the lack of early molecular markers makes it much more difficult to identify protocells in ARs than in principal or lateral roots [16].
Phytohormone also play an important role in the regeneration of roots from nothing. The plant growth regulator auxin is a key regulator controlling rooting reaction, and the hormones cytokinin (CK), ethylene (ET), jasmonic acid (JA) and aurolactone generally reduce rooting ability [14,17–21]. As a result, a variety of hormones exist that tightly control and fine-tune the initiation and emergence of ARs. In tissue culture, the addition of growth hormone or CK can promote the regenerative ability of plant roots or shoots [22]. However, the key finding during tissue culture is that the ratio of added auxin and CK can determine cell fate, a high ratio of auxin promotes root regeneration, while a high ratio of CK promotes shoot regeneration [5]. Studies have confirmed that under natural conditions, plants can promote root regeneration from scratch by endogenous hormones on media without the addition of exogenous growth hormones [2,23].
According to the current research progress, the DNRR process can be roughly divided into three stages [2]: The first is the perception of early signals in isolated leaves, including damage signals and environmental signals such as light and temperature [24]. Accumulation of JA and ET caused by damage can stimulate the rooting of Arabidopsis explants, and dark treatment can promote the expression of YUCCA 5/8/9 (YUC 5/8/9) in leaf cells, resulting in the accumulation of auxin, thus promoting the formation of advents [2,25]. The second is the accumulation of auxin. Auxin, as a key plant hormone for de novo regeneration of ARs, will block rooting of leaf explants when its polar transport process is inhibited [26,27]. Finally, the fate of the cell changes. After auxin accumulation, ARF7/19 and other auxin response genes are activated to promote the transfer of auxin to regenerative cells and complete the transformation of cell fate [28,29]. With the participation of WUSCHEL-RELATED HOMEOBOX5/7 (WOX5/7) and LATERAL ORGAN BOUNDARIES DOMAIN16/59 (LBD16/29) genes, it promotes the regeneration of ARs [29,30].
Over the years, tissue culture techniques have been widely and well established in regeneration research, and some progress has been made in recent years in the cellular genealogy of the plant regeneration process and the molecular mechanisms controlling plant regeneration, Chen et al. studied the system of neoplastic root organogenesis and elucidated its regulatory mechanisms by culturing leaf explants from a plant, Arabidopsis thaliana, under model natural conditions [31]. Research on plant adventitious root regeneration has also significantly improved the efficiency of plant regeneration and transformation, and has been applied in the protection of endangered species and promotion of poplar clonal breeding program [32]. This paper reviews the molecular mechanisms of adventitious root formation, summarizing the process of cellular transition from response to a stimulus to fate. In addition, relevant factors that influence the presence or absence as well as the intensity of regeneration occurrences, such as wounding, phytohormones, and the developmental state of the plant itself, are summarized and discussed.
2 Adventitious Roots Regeneration
De novo organ regeneration is the process by which plants regenerate ARs or adventitious shoots from isolated tissues and organs, with cellular totipotency being the basis of regeneration [33], such as de novo root regeneration (DNRR). To gain a more comprehensive understanding of DNRR, the process of regenerating roots from damaged leaves or hypocotyls has been explored in a medium that simulates natural conditions [34]. The successful regeneration of ARs from isolated Arabidopsis leaves on a B5 medium without the addition of exogenous hormones confirmed the feasibility of this process occurring under natural conditions [15,27,31]. In addition, plant hormones play a key role in adventitious root regeneration, and growth hormone was found to accumulate rapidly near the wound after plant injury and is a key hormone affecting cell fate [35]. In addition, JA can also be activated rapidly after injury, mediating the activation of downstream signalling molecules, and is considered to be one of the response hormones [36].
2.2 De Novo Root Regeneration Pathway
There are two ways to regenerate adventitious rooting: one is the direct induction of adventitious root from isolated plant tissue or organs under suitable conditions, also known as the direct route. This process requires the involvement of endogenous plant Hormones, particularly the accumulation of growth hormones. Xu et al. demonstrated that the histological structure of Arabidopsis guaiac tissue is similar to that of the root primordium or root apical meristem in healing induction and that the intermediate cell layer is quiescent centre-like (QC-like), giving the plant organ the ability to regenerate ARs from scratch [37]. Root explants already have lateral root meristem tissue primordia along the somatic axis and can regenerate roots from the wound and near the primary root without callus inducing medium (CIM) pretreatment, whereas hypocotyl explants can only regenerate roots from the wound under these conditions [15,38]. ERF115 can promote the accumulation of auxin near damaged cells and promote regeneration and tissue repair after tissue damage [35]. The second is the generation of ARs by a two-step method, in which isolated organs are generated on a healing CIM with the addition of exogenous hormones, which are then transferred to a root induction medium (RIM) to generate ARs [27,39,40], also known as the indirect route. Probably because of the pluripotent nature of the CIM, which produces root meristem-like pluripotent cell masses, followed immediately by the development of these cell masses on the RIM into specific root meristematic tissues [37,41]. In the intermediate cell layer, WOX5 directly interacts with PLETHORA1 and 2 (PLT1/2) [37], Promoting the expression OF TRYPTOPHAN AMINOTRANSFERASE OF Arabidopsis and generating endogenous auxin. WOX5 also interacts with type b Arabidopsis RESPONSE REGULATOR12 (ARR12) to inhibit type-a ARRs and break the negative feedback loop of CK signaling [27,37]. That is, by enhancing the production of auxin and the sensitivity of CK to promote the acquisition of pluripotency in the intermediate cell layer, and then differentiate into ARs.
Isolated damaged leaves of Arabidopsis thaliana can sense external signals and changes in their endogenous hormones, and regulate and express relevant signaling molecules. Although the application of healing tissues has been relatively common, the perception of external signals by plants, and the mechanisms of self-regulation caused by them, have been subject to in-depth study. In recent years, Xu et al. have systematically studied DNRR under simulated natural conditions and divided the process into three stages: early signal perception, massive accumulation of growth hormone, and cell fate transformation [2,39,42].
In the first stage, a variety of signals are sensed by isolated leaf explants, such as trauma, abiotic stresses and the plant’s age. These stimuli influence the efficiency, speed and even the occurrence of regeneration of ARs in the plant. Early signals are sensed and can be transported to, for example, the chloroplast or some vascular cells [2]. After the leaf damage signal is sensed, it can cause rapid physicochemical changes and long-term hormonal signals to be activated [43]. First, Physical and chemical changes of intracellular Ca2+ concentration, membrane potential and reactive oxygen species [24,44]. After injury, the addition of a certain concentration of exogenous Ca2+ can promote the formation of adventitious roots, while the addition of broad-spectrum Ca2+ channel inhibitors and Ca2+ chelating agents can inhibit the formation of AR induced by indoleacetic acid (IAA) or NO [45]. Hydrogen, a bioinert gas, plays an active role in adventitious root regeneration of cucumber explants [46]. Then hormones in the plant are activated, causing intercellular signalling molecules such as auxin, CK, JA, etc. [18,36,47–49]. As a plant hormone that responds quickly to injury, JASMONATE binds to the F-box protein receptor CORONATINE INSENSITIVE1 (COI1) within a few hours of damage to plant tissues or organs, promoting the binding of Jasmonate-Zim-Domain (JAZ) protein to COI1. It leads to JAZ ubiquitination and degradation, and activates ja signaling pathway [36,50]. After a few hours of damage, long-term signalling is activated accordingly, causing long-term signalling. The first is the activation of regeneration-related genes, such as the expression of the YUC4 gene, which may be associated with the maintenance of plant growth hormone levels, in cells with a high regenerative capacity near plant injury [51,52]. After injury, (NAM/ATAF/CUC domain1) NAC1 transcription factor gene can be specifically expressed in leaf explants, and play a role in the degradation of cell wall expansion protein by regulating the expression of CEP gene, and then participate in adventitious root regeneration through an auxin independent pathway [31]. In addition, the expression of some genes that affect the cellular environment can also positively regulate root regeneration, such as the Arabidopsis ATAF1, ATAF2 and CUC2 genes [31].
Different environmental signals can also be sensed by the plant and affect the efficiency of adventitious root regeneration to a greater or lesser extent. Light is one of the most important factors influencing adventitious root regeneration. Hypocotyl under alternating light and dark conditions. can be promoted by the interaction of HY5 (LONG HYPOCOTYL) and AGO1 with growth hormone-related response factors to regenerate its ARs [52]. Another gene related to growth hormone synthesis, YUC5/8/9, is expressed during dark treatment and enhances growth hormone synthesis and thus adventitious root regeneration. In addition, drought, nutrition and temperature can also be perceived by plants to affect regeneration efficiency [20,51,53].
The developmental state of the explant, such as age, is also an important factor in regeneration. The regenerative capacity of older leaves is significantly reduced compared to younger leaves, and the phenomenon of growth hormone accumulation during leaf regeneration was found in the study, interestingly, there was a significant increase in regeneration capacity when exogenous growth hormone was added to mature leaves, which also suggests that auxin is one of the factors affecting regeneration capacity [2]. The response caused by changes in the plant’s state is complex, and the study of its regeneration mechanisms is important in breaking through the limits of regeneration. Early signals are sensed in time as the basis for regeneration and are coordinated by a variety of cellular tissues and organs, e.g., the cell wall can be sensed and transformed by molecules [54].
3.2 Phase II, Auxin Accumulation
Various plant hormones such as JA and abscisic acid (ABA) are involved in the regeneration of ARs from scratch, but the auxin is the key hormone in this process. Excessive amounts of growth hormone can lead to an increase in the number of ARs regenerated. In addition, the regeneration capacity of plants is significantly reduced when grown on media supplemented with the growth hormone biosynthesis inhibitors l-Kyn or yucasin or the growth hormone polar transport inhibitor naphthalic acid (NPA) [20,48]. Thus, growth hormone accumulation is a key step in the regulatory network of adventitious root regeneration signals, determining the presence or absence and intensity of regeneration.
Within hours of wound onset, plant hormones are activated for expression in response to stress signals. Growth hormone accumulates in transformed cells [55]. Tryptophan is a precursor substance for growth hormone biosynthesis, a process in which many genes are involved, such as those in the Arabidopsis tryptophan aminotransferase (TAA) family, the YUC family. YUC is highly sensitive to many early signals and the TAA gene can be continuously expressed before and after leaf isolation [26]. Thus, it has been shown that environmental factors and short-term trauma signals can activate the expression of YUC1/4 and YUC5/8/9 [26]. Hydrolysis of IAA coupled with amino acids also contributes to the rapid production of auxin [26]. The response of ARF to auxin can promote the activation and response of downstream genes. Overexpressed LkARF7 and LkARF19 in Larix kaempferi can positively regulate LkBBM1 and promote the formation of adventitic roots [28]. Li et al. confirmed that PHYB can interact with INDOLE-ACETIC ACID14 (IAA14), ARF7, and ARF9. This interaction stabilizes IAA14 and inhibits transcriptional activity of ARF7 and ARF19, thereby inhibiting dark-induced hypocotyl adventitious root biogenesis. The hydrolysis of IAA coupled with amino acids also contributes to the rapid formation of auxin [44].
Auxin is transported by polar transport from the chloroplast to the vicinity of the wound. Studies have shown that auxin accumulates rapidly near the wound at around 12 h of wound onset, mainly in the procambium layer and nearby parenchymal cells, a region of cells also known as cells with regenerative potential [20,43,56]. The addition of NPA retains growth hormone in transformed cells and prevents accumulation into regeneration-receptive cells [31], confirming that auxin from regeneration-receptive cells is transported by transformed cells. In addition, the polar transport process of auxin is also influenced by other hormones and can be inhibited by gibberellin, which in turn inhibits adventitious root regeneration. In the DNRR process, auxin accumulation is a pivotal influence in undertaking signal perception and cell fate transformation.
3.3 Stage III: Cell Fate Transformation
The accumulated growth hormone is transported to cells with regenerative potential for the cell fate transition to occur. The protoplast and its surrounding thin-walled cells have been shown to have regenerative potential [2,4]. The cells with the regenerative potential of leaf explants can transform their cell fate and regenerate adventitious rooting under natural conditions or with the addition of specific hormones. Xu proposed four steps for cells to undergo a fundamental shift in fate: initiation, initiation, pattern, and emergence [2].
The first step is ‘initiation’, a process that occurs within about 1–2 days of cell injury when the transition from regenerative cells to root-establishing cells occurs and does not involve cell division. The expression of the stem cell regulators WOX11 and WOX12 was identified by CHIP-seq and others, which are subfamilies of the WOX intermediate branch and are functionally redundant [2]. Furthermore, deletion of WOX11/12 leads to rooting defects, whereas overexpression of WOX11/12 enhances the rooting capacity [15]. The occurrence of this process also marks the transition of regeneration potential cells to ARs initiation cells. WOX11 is a direct target of the auxin signaling pathway [57]. During the regeneration of new roots, WOX11 expresses and activates the expression of its target genes, links the upstream auxin signal with the downstream cell fate transition, initiates root and callus primordials, and regulates the establishment of original cells [57]. In addition, WOX11/12 deficiency may lead to rooting defects, while overexpression of WOX11/12 may enhance rooting ability [27]. This process also marks the transition from regenerative potential cells to adventitious root initiation cells.
Next is ‘initiation’, a process that takes place within 2–4 days of injury and, in addition to a cell fate transition, cell division occurs in response to growth hormone, resulting in the formation of a dome-shaped root primordium with multiple cell layers. During cell division, many cell cycle-related genes are activated [58–61] Studies have shown that ERF115 acts as a CK signaling mechanism for AR initiation by activating CK, and ERF115 can be activated by JA transcription, inhibiting AR initiation in a ninja dependent and independent manner [19,62]. At the same time, cell division may be inhibited by solanum lactone. During a fate transition in the apical meristem, the transcription factor WOX11/12 activates the downstream LBD16 and WOX5/7 produces adventitious rooting primordia [15,27,63]. LBD16 is essential in the lateral root, ARs and healing tissue formation. lbd16 mutants showed significant defects in AR organogenesis in leaf explants, suggesting that lbd16 is required for AR initiation [29,64]. This study was proposed that WOX11/12 directly activated the expression of LBD16 in the process of adventitic root regeneration by explants of isolated leaves [15,64]. WOX5/7 can also be considered a key gene in the stem cell niche [63,65,66]. As for the upstream regulatory mechanism controlling WOX5 expression, Zhang et al. found that the suppressor of WUSCHEL1 (ROW1), also known as BARD1, directly binds to the WOX5 promoter region and restricts its transcription to QC [30]. It has been reported that WOX5/7 can interact with ARR12 and inhibit ARR5 to eliminate the negative feedback mechanism of CK signaling during callus induction, resulting in high CK sensitivity of explants. WOX5 may also maintain the pluripotency of the intermediate cell layer of callus on CIM and promote the adventitia formation of callus on root-inducing medium (RIM) through high levels of growth hormone [37].
This is followed by ‘moulding’ and ‘emergence’, in which the root primordial cells undergo successive cell divisions to form the root apical meristem (RAM). The genes involved in this process are SHOOT ROOT (SHR) and SCARECROW ROOT (SCR) [67]. The root tip meristem gradually matures to form a root tip, which grows rapidly to break through the explant thus forming ARs. In conclusion, adventitious root regeneration from the tip is a cell fate transformation process that undergoes complex, sequential signalling regulation.
The regeneration capacity of plants is closely related to their developmental state, with root regeneration capacity decreasing significantly as the plant ages. In general, the ability of mature plant explants to effectively regenerate branches is significantly lower than that of young branches [68]. The gradual maturation of plants reduces the regeneration capacity, which is a major obstacle to the asexual reproduction of good varieties, and part of this is related to phytohormone synthesis, with some studies showing that the conversion of peas from the nutritional mount state to the reproductive state is associated with reduced root regeneration, a corresponding reduction in auxin [69]. MiR156 is a factor associated with the regulation of plant development and can target the three SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors SPL2, SPL10 and SPL11 [62,68,70]. During the juvenile period, plants have a strong cell division and regeneration capacity. With age, miR156 synthesis decreases and inhibition of SPL targets is reduced [71,72]. SPL can directly inhibit Arabidopsis B-type response regulator activity and suppress shoot regeneration [68]. In addition, miR156 activity was reduced and the number of hypocotyls regenerated ARs was decreased, confirming that miR156 is also involved in adventitious rooting regeneration in Arabidopsis [73]. However, the mechanism of miR156/SPL in regulating adventitious rooting regeneration still needs to be studied in depth.
Damage is the main trigger for adventitious root regeneration, by reactivating cell proliferation and reprogramming cells. Two molecular modes associated with injury: the cell wall derivative oligogalacturonide and extracellular ATP are sensed to elicit a cellular defence response [74–76]. Such as the translocation of calcium signals and the burst of reactive oxygen species [41]. The damage signal will be rapidly responded to by translocation of electrical and chemical signals to relevant signalling receptors [8], which can promote high expression of the key transcription factor APETALA2/ET response factor (AP2/ERF) to induce healing tissue formation [62,77]. In addition, WOUND-INDUCED DEDIFFERENTIATION1 (WIND1) directly upregulates the expression of the enhancer of SHOOT REGENERATION1 (ESR1), an Arabidopsis AP2/ERF transcription factor, to promote healing tissue formation and specific transformation [78].
Recent studies have shown that genes related to signalling such as JA, ET and ROS are rapidly mobilized after leaf isolation, while stress response, subcellular regulation and metabolism-related genes are also mobilized to varying degrees, suggesting their involvement in cell fate transformation and root regeneration processes [39]. Analysis of trauma-induced transcriptome changes by RNA-seq data indicated that significant expression differences in 80% of gene expression profiles occur within 24 h after trauma [41]. Some genes encoding defence substance synthases, protein kinases that transduce calcium signals are more sensitive to trauma signals and activate expression rapidly within a short period of time [79]. After a period of time following the injury signal, genes associated with protein synthesis and cell proliferation are activated in response [39]. It is important to note that development, i.e., the environment, has a strong influence on the cellular protein synthesis capacity, and ribosomal transcript levels are significantly lower in mature organs than in areas of active cell proliferation [80]. These transcriptional dynamics may also affect endogenous hormone levels in traumatized organs. Hormone quantification and phenotypic analysis revealed that CK, a key hormone for healing tissue formation near the wound, is activated after injury and accumulates large amounts of tZ and cZ-type CKs [41]. Interestingly, the differences in the level of hormone activation response were highly significant under different physiological conditions. Wound-induced healing tissue formation strongly induced the CK synthesis-related genes LOG1, LOG4, LOG7, and CYP735A2, whereas there were no similar transcriptional changes in other wound inductions [81]. In addition, there is a significant accumulation of growth hormone during wound-induced ARs production, forming a growth hormone stream that promotes stem cell regeneration, whereas the role in healing tissue formation appears to be weak [35]. The coercive hormone ABA shows a significant inhibitory effect in both cell proliferation and healing tissue travel after injury [82].
AP2/ERFs are key transcriptional regulators of trauma induction, acting as receptors. Signals for trauma and activating the expression of downstream signals. Importantly, different groups of AP2/ERFs can trigger regenerative processes. Furthermore, in addition to trauma signalling plant hormones can also induce AP2/ERF promoters such as JA and salicylic acid (SA). GO analysis showed that SPL10 can induce AP2/ERF to inhibit root regeneration by regulating the synthesis of growth hormone. In addition, SPL10 can also affect the hormone-stimulated rooting action of JA and SA [62]. Under different circumstances, regulatory factors can play different roles. Overall, it appears that trauma has two main effects: firstly, it forms a barrier to the flow of growth hormone, leading to the accumulation of growth hormone near the wound and to the concentration required for adventitious root regeneration [35,50]; secondly, it indirectly increases the efficiency of adventitious root regeneration by promoting the synthesis of auxin [50].
Plant regeneration of ARs is influenced by various environmental conditions, such as nutrition, temperature and pH. Light is one of the most important factors affecting the regeneration of ARs from hypocotyls. In some cases, root or shoot regeneration can be inhibited by light [38]. In tissue culture experiments, light inhibited shoot regeneration in the first few hours after cotyledon tissue excision in Arabidopsis thaliana, while placing the explants in the dark promoted regeneration in only 2–6 h [38]. Light triggers the light signalling pathway and a key regulatory transcription factor downstream of light signalling, ELONGATED HYPOCOTYL5 (HY5) and AGO1, can induce ARs production in the hypocotyl under alternating light and dark conditions, and the number of ARs in the hypocotyl of the mutant ago1 is reduced and the bound growth hormone content in the mutant ago1 is decreased as revealed by proteomic analysis [53], ARF6 and ARF8 have been reported to be downstream targets of miR160, and the number of ARs was reduced in the hypocotyls of mutants arf6 and arf8, so ARF6 and ARF8 may be positive regulators of adventitious root regeneration [49]. Recent studies have shown that photoreceptors can negatively regulate hypocotyl adventitious root production under dark conditions [4,53,55]. Under dark conditions, hypocotyl adventitious root regeneration is completely dependent on ARF7, ARF19 and LBD16, and photoreceptor pigment B inhibits hypocotyl adventitious root formation by stabilizing IAA14 and inhibiting ARF7 and ARF19 [44,53,55].
ARs in plants can come from non-root organs. In Arabidopsis, ARs can be formed from hypocotyls that complement their root structures, and in addition, isolated plant leaves can also produce ARs. Plant hormones play a key role in the development of new roots. JA, a trauma-inducing hormone, plays a different role in the regeneration of ARs from hypocotyls and isolated organs, respectively [83].
JA plays a positive regulatory role in the regeneration of ARs from isolated organs in vitro [50,80,83]. As a wound hormone, JA is rapidly induced and activated within a short time of leaf damage. It then directly activates the downstream target site ERF 109 and the activated ERF109 can bind to the tryptophan biosynthesis precursor, ANTHRANILATE SYNTHASE α1 (ASA1) gene, promoting its expression [6,84]. The binding of ERF109 to the ASA1 promoter has been demonstrated by chromatin immunoprecipitation (ChIP) [50], a process that also involves the tri-methylation on Lys27 of histone H3 (H3K36me3) enzyme [50,83]. In contrast, persistently high JA levels were detrimental to adventitious root regeneration, and after two hours, JA levels decreased and the JAZ protein interacted with ERF109, leading to inhibition of ERF109 activity [50]. In addition, plant age also has an effect on adventitious root regeneration in isolated leaves. The miR156/SPL pathway in Arabidopsis plays a key role in senescence regulation [44,62,68,70,71]. SPL is a direct target site of miR156 and can be degraded upon binding by miR156. However, as leaves mature and senesce, miR156 content decreases and the ability to target SPL is diminished. The results of established studies confirm that SPL2, SPL10 and SPL11 are important regulatory genes in the regeneration of ARs from isolated leaves, negatively regulating ERF109 and limiting the synthesis of growth hormone thereby inhibiting rooting [78,85,86]. Therefore, the onset of the regeneration process is also age-sensitive. Thus, trauma-induced JA is dynamically variable and regulates the regeneration of ARs in conjunction with growth hormone and epigenetics.
In addition to promoting the regeneration of ARs in isolated leaves, JA also inhibits the regeneration of ARs in the hypocotyl. However, JA does not regulate regeneration alone but interacts with growth hormone and CK to regulate the regeneration signalling pathway of ARs [33,87]. Auxin has a positive regulatory role in promoting the regeneration of ARs. Three transcription factors, ARF6, ARF8 and ARF17, mediate auxin signalling and control the expression of the enzymes GRETCHEN HAGEN3.3 (GH3.3), GH3.5 and GH3.6 [88]. The enzymes GH3.3, GH3.5 and GH3.6 catalyse the binding of free JA and indole-3-acetic acid (IAA) into amino acids and maintain homeostasis [88]. It was found that DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) [86], an enzyme first identified in rice, controls a feedback pathway that stabilizes the crosstalk between growth hormone and JA during AR initiation [88]. CKs, on the other hand, inhibit ARs production and studies have found a decrease in CK content during the regeneration of ARs. CK oxidase/dehydrogenase 1 (CKX1) can degrade CK, and CKX1 activity is inhibited in the JA MYC2 signalling pathway. ERF115 can act as one of the downstream target genes of JA and promotes CK expression. In addition, JA and CK may synergistically activate APETALA2.6 LIKE (RAP2.6L) (also known as ERF 113) expression. It has been shown that JA can inhibit root regeneration in different ways, Lakehal et al. proposes that JA signalling inhibits adventitious root regeneration in both a ninja-dependent and ninja-independent manner and that JA-induced ERF115 transcription factor inhibits regeneration in a CK-dependent manner [19]. Therefore, JA, auxin and CK synergistically negatively regulate adventitious root regeneration in the hypocotyl.
As an important plant hormone, auxin plays a key role in the process of adventitious root regeneration. Feldmann et al. established a two-step method for indirect induction of Arabidopsis organ establishment by placing Arabidopsis explants in CIM and shoot-inducing medium (SIM) with different ratios of growth hormone and CKs [89], showing that organ regeneration from scratch requires the involvement of growth hormone. In medium-simulating natural conditions, there was a significant accumulation of growth hormone in isolated leaves of Arabidopsis with elevated levels, indicating the possible involvement of growth hormone in the process of adventitious root regeneration [90,91]. In addition, when exogenous auxin was added to the medium, adventitious root regeneration was significantly increased, whereas the addition of growth hormone inhibitors reduced growth hormone accumulation and inhibited adventitious root regeneration [31,55,92].
As mentioned earlier in the mechanism of adventitious root regeneration, during the regeneration of ARs from isolated organs, when growth hormone accumulates, it reaches the vicinity of cells with regenerative potential through polar transport and reaches maximum growth hormone concentration, which in turn induces high expression of WOX11 and WOX12 in the forming layer and nearby thin-walled cells, marking the completion of the first fate transition from potential to root cells in head regeneration [15,37]. WOX11 further activates the expression of LBD16/29 [87], a downstream gene involved in lateral root formation. which is directly regulated by the growth hormone-responsive genes AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 and controls cell proliferation and loose cell wall arrangement during lateral root initiation [52,87]. LBD promotes cell division through cell division promotes the transformation of root mother cells into root primordial cells labelled by WOX5/7, marking the completion of the second cell fate transition, after which the primordial cells continue to grow and differentiate into a complete ARs [44,63]. Thus, the LBD family of genes plays an important role in the generation of ARs in response to auxin.
In addition to auxin and JA, plant hormones such as CK and ET are also involved in the process of adventitious root regeneration in plants. Exogenous application of CKs results in larger root tip meristems and CKs contribute to the specialization of the QC of the primary root tip. In the CK-deficient mutant background, WOX5 is not specifically expressed in the resting centre of the primary root tip [93]. In Arabidopsis roots, there is an antagonistic effect of growth hormone and CK, whose minimal ratio is the boundary between cell division and differentiation and determines the fate of the cell [20,36]. The effect of CKs on regenerating roots is, on the one hand, through growth hormones, which affect their polar transport, and on the other hand, as downstream signalling molecules of the JA signalling pathway that inhibit root regeneration [20,36,93]. ET is also involved in the regeneration of ARs. When plant tissues are treated with ACC, a precursor substance of ET, the number of lateral roots is reduced and the transport of growth hormone is inhibited [94]. Moreover, the development of ARs is inhibited by the application of AVG or STS, inhibitors of ET synthesis [17].
The ability of plant regeneration reflects the high plasticity of plant cell fate. How to regenerate a single plant somatic cell into a complete plant has always been a very concerned problem in the field of plant research. It is the ability to regenerate ARs, adventitious buds, and even whole plants that enables tissue repair and survival in the face of harsh natural conditions or mechanical damage. Since the observation that plants have a strong regenerative ability, people began to develop and use them in agricultural production. Potatoes can be rapidly propagated by cutting, and fast-growing poplar trees commonly used in afforestation can be rapidly and economically planted by cuttings [32,95]. However, our understanding of the molecular mechanism is far from enough, and the technology and research method system are all obstacles for us to further explore the regeneration mechanism. For example, the mechanism of how plant cells respond to wounds and hormones to make the fate change, until the model plant Arabidopsis thaliana has the advantage of molecular genetic manipulation, we have entered a new era in the study of plant regeneration mechanism.
During regeneration, external stimuli such as wounds and stresses ectopically activate the intrinsic developmental program and initiate the ab initio occurrence of adventitious root in the hypocotyl or isolated leaf of the plant. This is an orderly process from cells with regenerative potentials, such as formative layer cells, to the formation of root meristems, which is regulated by a highly complex network of signals through at least three successive stages, culminating in the transformation of cell fate to produce ARs (summarized in Fig. 1). Through clonal propagation by in vitro organs, a large number of offspring can be efficiently generated to save endangered plants, or high-quality clones that are genetically stable can be produced to produce disease-resistant and pest-resistant plants, which can also be used for genetically improved trees in planting and breeding programs [69], through comparing the genotypes of different rooting abilities of cutting, Quantitative trait loci (QTLS) related to the number of cuttings in Cottonwood (Populus deltoides), eucalyptus (Eucalyptus spp.) and poplar were identified [69]. However, how exactly does a cell perceive a stimulus and then break through epigenetic constraints to reprogram the cell? How are the factors involved at different stages of the transition coordinated and how are resources allocated? Are the regenerative mechanisms consistent across sites? Answers to these questions will provide a deeper understanding of adventitious root regeneration from scratch, which will have important practical applications in the regeneration of cuttings and healing tissues.
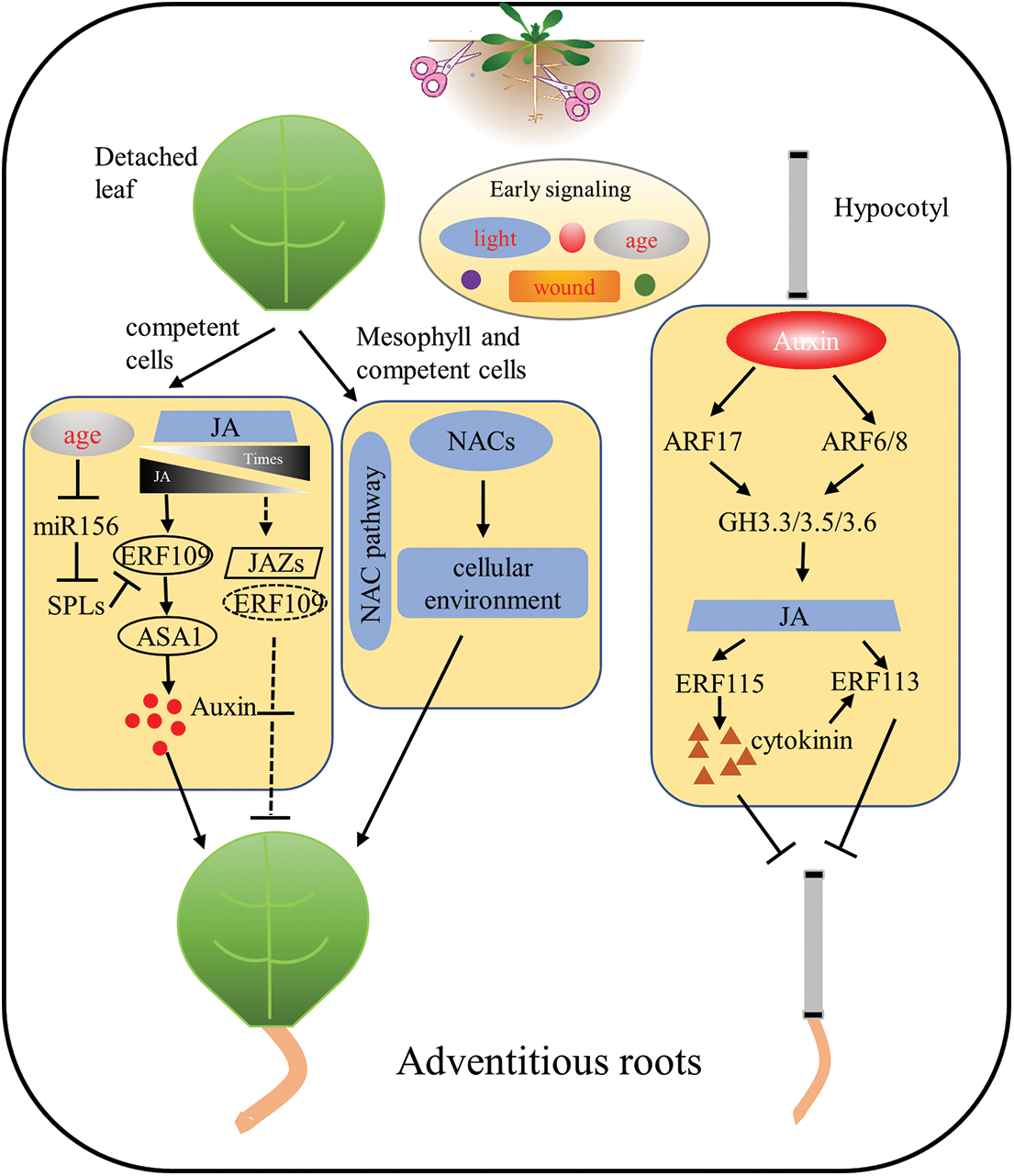
Figure 1: Molecular network of adventitious root regeneration in isolated organs of Arabidopsis thaliana. In the ARs process of isolated leaves, phytohormone (e.g., auxin, JA, CK) responses caused by wound signals and the age of the explant leaves simultaneously respond to regulate the ability of the explant to regenerate. In addition, NAC-related transcription factors are involved in adventitious root formation of detached leaves in a novel pathway. During hypocotyl adventitious root formation in Arabidopsis, auxin, JA and CK crosstalk with each other to form a molecular network for adventitious root formation in isolated organs
Much like animal regeneration, plant regeneration involves a variety of stem cells (or stem-like cells), Somatic embryogenesis is similar to dedifferentiation of animal somatic cells into induced pluripotent stem cells (iPS cells) or embryonic stem cells [13,96–99]. This dedifferentiation is easier in plant cells than in animal cells. The somatic cells of animals can achieve dedifferentiation by exogenous introduction of some specific genes or under certain conditions, but the frequency is very low. However, plant somatic cells can dedifferentiate efficiently under the induction of hormones or stress [13,96,97], so the research and development of plant regeneration will promote the progress in the field of regeneration. However, Among the factors affecting adventitious root development as well as efficiency, wounding, and state of its development, different phytohormones, and even different organs of the same hormone all have different effects. However, the regulation of adventitious root regeneration is more complex than we currently know, and there are many unanswered questions, such as the crosstalk between different signals, the critical points that trigger the response of different transcription factors, and how to balance the levels of different hormones to keep the plant in the most favourable state.
There are great differences in regeneration capacity among different species. The dicotyledonous model plant Arabidopsis thaliana is mostly used as the research object in the studies of adventitive root regeneration, and because the regenerative cells of Arabidopsis thaliana are all over the vascular system of leaves, it shows strong regenerative ability. However, for important food crops such as rice, wheat (Triticum aestivum) and corn (Zea mays), the model plants of monocotyledonous grasses, regenerative cells are limited to the base of immature leaves, so it is difficult to conduct tissue culture in mature tissues [12,45,100–102]. The explants usually used for monad cell regeneration are immature embryos, which are the main bottleneck for monad cell transformation, as these embryos need to be separated separately from the developing seed. As technology advances and develops, more research methods and tools are available for regeneration studies, such as single-cell RNA sequencing, which will give us more convenience and greater possibilities for future studies on adventitious root regeneration.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: Xiangyang Hu designed the research and editing the manuscript. Lulu Zhi obtained the related references and wrote the manuscript. Both authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ibáñez, S., Carneros, E., Testillano, P. S., Pérez-Pérez, J. M. (2020). Advances in plant regeneration: Shake, rattle and roll. Plants, 9(7), 897–908. [Google Scholar]
2. Xu, L. (2018). De novo root regeneration from leaf explants: Wounding, auxin, and cell fate transition. Current Opinion in Plant Biology, 41, 39–45. [Google Scholar] [PubMed]
3. Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell, 9, 841–857. [Google Scholar] [PubMed]
4. Ahkami, A. H., Lischewski, S., Haensch, K. T., Porfirova, S., Hofmann, J. et al. (2009). Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytologistogist, 181(3), 613–625. [Google Scholar]
5. Zhang, J., Long, X., Weng, Y., Cheng, T., Shi, J. et al. (2021). Efficient evergreen plant regeneration of Cinnamomum japonicum Sieb. through in vitro organogenesis. Phyton-International Journal of Experimental Botany, 90(2), 571–582. https://doi.org/10.32604/phyton.2021.014191 [Google Scholar] [CrossRef]
6. Cai, X. T., Xu, P., Zhao, P. X., Liu, R., Yu, L. et al. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nature Communications, 5, 5833. [Google Scholar] [PubMed]
7. Ye, B. B., Shang, G. D., Pan, Y., Xu, Z. G., Zhou, C. M. et al. (2020). AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell, 32(1), 226–241. [Google Scholar] [PubMed]
8. Omary, M., Matosevich, R., Efroni, I. (2023). Systemic control of plant regeneration and wound repair. New Phytologist, 237(2), 408–413. [Google Scholar] [PubMed]
9. Ikeuchi, M., Ogawa, Y., Iwase, A., Sugimoto, K. (2016). Plant regeneration: Cellular origins and molecular mechanisms. Development, 143(9), 1442–1451. [Google Scholar] [PubMed]
10. Lardon, R., Geelen, D. (2020). Natural variation in plant pluripotency and regeneration. Plants, 9(10), 1261–1288. [Google Scholar] [PubMed]
11. Jing, T., Ardiansyah, R., Xu, Q., Xing, Q., Müller-Xing, R. (2020). Reprogramming of cell fate during root regeneration by transcriptional and epigenetic networks. Frontiers in Plant Science, 11, 317. [Google Scholar] [PubMed]
12. Bhojwani, S. S., Evans, P. K., Cocking, E. C. (1977). Protoplast technology in relation to crop plants: Progress and problems. Euphytica, 26, 343–360. [Google Scholar]
13. Liao, R. Y., Wang, J. W. (2023). Analysis of meristems and plant regeneration at single-cell resolution. Current Opinion in Plant Biology, 74, 102378. [Google Scholar] [PubMed]
14. Ishida, S., Suzuki, H., Iwaki, A., Kawamura, S., Yamaoka, S. et al. (2022). Diminished auxin signaling triggers cellular reprogramming by inducing a regeneration factor in the liverwort marchantia polymorpha. Plant and Cell Physiology, 63(3), 384–400. [Google Scholar] [PubMed]
15. Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z. et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell, 26(3), 1081–1093. [Google Scholar] [PubMed]
16. Müller-Xing, R., Xing, Q. (2022). The plant stem-cell niche and pluripotency: 15 years of an epigenetic perspective. Frontiers in Plant Science, 13, 1018559. [Google Scholar]
17. Druege, U., Franken, P., Lischewski, S., Ahkami, A. H., Zerche, S. et al. (2014). Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in petunia cuttings. Frontiers in Plant Science, 5, 494. [Google Scholar] [PubMed]
18. Zhang, J., Mazur, E., Balla, J., Gallei, M., Kalousek, P. et al. (2020). Strigolactones inhibit auxin feedback on PIN-dependent auxin transport canalization. Nature Communications, 11(1), 3508. [Google Scholar] [PubMed]
19. Lakehal, A., Dob, A., Rahneshan, Z., Novák, O., Escamez, S. et al. (2020). ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. New Phytologist, 228(5), 1611–1626. [Google Scholar] [PubMed]
20. di Mambro, R., de Ruvo, M., Pacifici, E., Salvi, E., Sozzani, R. et al. (2017). Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Roceedings of the National Academy of Sciences, 114(36), E7641–E7649. [Google Scholar]
21. Hernández-Coronado, M., Araujo, P. C. D., Ip, P. L., Nunes, C. O., Rahni, R. et al. (2022). Plant glutamate receptors mediate a bet-hedging strategy between regeneration and defense. Developmental Cell, 57(4), 451–465.e6. [Google Scholar]
22. Cary, A. J., Che, P., Howell, S. H. (2002). Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant Journal, 32, 867–877. [Google Scholar]
23. Chen, X., Qu, Y., Sheng, L., Liu, J., Huang, H. et al. (2014). A simple method suitable to study de novo root organogenesis. Frontiers in Plant Science, 5, 208. [Google Scholar] [PubMed]
24. Wasternack, C. (2019). New light on local and systemic wound signaling. Trends in Plant Science, 24(2), 102–105. [Google Scholar] [PubMed]
25. Xu, D., Miao, J., Yumoto, E., Yokota, T., Asahina, M. et al. (2017). YUCCA9-mediated auxin biosynthesis and polar auxin transport synergistically regulate regeneration of root systems following root cutting. Plant and Cell Physiology, 58(10), 1710–1723. [Google Scholar] [PubMed]
26. Sun, B., Chen, L., Liu, J., Zhang, X., Yang, Z. et al. (2016). TAA family contributes to auxin production during de novo regeneration of adventitious roots from Arabidopsis leaf explants. Science Bulletin, 61(22), 1728–1731. [Google Scholar]
27. Hu, X., Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiology, 172(4), 2363–2373. [Google Scholar] [PubMed]
28. Tao, G. Y., Xie, Y. H., Li, W. F., Li, K. P., Sun, C. et al. (2023). LkARF7 and LkARF19 overexpression promote adventitious root formation in a heterologous poplar model by positively regulating LkBBM1. Communications Biology, 6(1), 372. [Google Scholar] [PubMed]
29. Lee, H. W., Cho, C., Pandey, S. K., Park, Y., Kim, M. J. et al. (2019). LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biology, 19(1), 46. [Google Scholar] [PubMed]
30. Zhang, Y., Jiao, Y., Liu, Z., Zhu, Y. X. (2015). ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nature Communications, 6, 6003. [Google Scholar] [PubMed]
31. Chen, X., Cheng, J., Chen, L., Zhang, G., Huang, H. et al. (2016). Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiology, 170(4), 2136–2145. [Google Scholar] [PubMed]
32. Xiong, Y., Chen, S., Wu, T., Wu, K., Li, Y. et al. (2022). Shoot organogenesis and somatic embryogenesis from leaf and petiole explants of endangered Euryodendron excelsum. Scientific Reports, 12(1), 20506. [Google Scholar] [PubMed]
33. Asahina, M., Azuma, K., Pitaksaringkarn, W., Yamazaki, T., Mitsuda, N. et al. (2011). Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proceedings of the National Academy of Sciences, 108(38), 16128–16132. [Google Scholar]
34. Falasca, G., Altamura, M. M. (2006). Histological analysis of adventitious rooting in Arabidopsis thaliana (L.) Heynh seedlings. Plant Biosystems, 137(3), 265–273. [Google Scholar]
35. Canher, B., Heyman, J., Savina, M., Devendran, A., Eekhout, T. et al. (2020). Rocks in the auxin stream: Wound-induced auxin accumulation and ERF115 expression synergistically drive stem cell regeneration. Proceedings of the National Academy of Sciences, 117(28), 16667–16677. [Google Scholar]
36. Zhang, G., Liu, W., Gu, Z., Wu, S., E, Y. et al. (2021). Roles of the wound hormone jasmonate in plant regeneration. Journal of Experimental Botany, 74(4), 1198–1206. [Google Scholar]
37. Zhai, N., Xu, L. (2021). Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nature Plants, 7(11), 1453–1460. [Google Scholar] [PubMed]
38. Nameth, B., Dinka, S. J., Chatfield, S. P., Morris, A., English, J. et al. (2013). The shoot regeneration capacity of excised Arabidopsis cotyledons is established during the initial hours after injury and is modulated by a complex genetic network of light signalling. Plant Cell and Environment, 36(1), 68–86. [Google Scholar]
39. Liu, W., Zhang, Y., Fang, X., Tran, S., Zhai, N. et al. (2022). Transcriptional landscapes of de novo root regeneration from detached Arabidopsis leaves revealed by time-lapse and single-cell RNA sequencing analyses. Plant Communications, 3(4), 100306. [Google Scholar] [PubMed]
40. Zeng, J., Dong, Z., Wu, H., Tian, Z., Zhao, Z. (2017). Redox regulation of plant stem cell fate. EMBO Journal, 36(19), 2844–2855. [Google Scholar] [PubMed]
41. Ikeuchi, M., Iwase, A., Rymen, B., Lambolez, A., Kojima, M. et al. (2017). Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiology, 175(3), 1158–1174. [Google Scholar] [PubMed]
42. Wang, S., Yan, J., Ha, B., Bai, Y. (2021). An efficient plant regeneration system of Hydrangea bretschneideri Dipp via stem segments as explants. Phyton-International Journal of Experimental Botany, 90(2), 595–604. https://doi.org/10.32604/phyton.2021.013693 [Google Scholar] [CrossRef]
43. Maffei, M. E., Mithofer, A., Boland, W. (2007). Before gene expression: Early events in plant-insect interaction. Trends in Plant Science, 12(7), 310–316. [Google Scholar] [PubMed]
44. Li, Q. Q., Zhang, Z., Wang, Y. L., Zhong, L. Y., Chao, Z. F. et al. (2021). Phytochrome B inhibits darkness-induced hypocotyl adventitious root formation by stabilizing IAA14 and suppressing ARF7 and ARF19. Plant Journal, 105(6), 1689–1702. [Google Scholar]
45. Lanteri, M. L., Pagnussat, G. C., Lamattina, L. (2006). Calcium and calcium-dependent protein kinases are involved in nitric oxide- and auxin-induced adventitious root formation in cucumber. Journal of Experimental Botany, 57(6), 1341–1351. [Google Scholar] [PubMed]
46. Lin, Y., Zhang, W., Qi, F., Cui, W., Xie, Y. et al. (2014). Hydrogen-rich water regulates cucumber adventitious root development in a heme oxygenase-1/carbon monoxide-dependent manner. Journal of Plant Physiology, 171(2), 1–8. [Google Scholar] [PubMed]
47. Aloni, R., Aloni, E., Langhans, M., Ullrich, C. I. (2006). Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Annals Botany, 97(5), 883–893. [Google Scholar]
48. Su, Y. H., Liu, Y. B., Zhang, X. S. (2011). Auxin-cytokinin interaction regulates meristem development. Molecular Plant, 4(4), 616–625. [Google Scholar] [PubMed]
49. Sorin, C., Bussell, J. D., Camus, I., Ljung, K., Kowalczyk, M. et al. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell, 17(5), 1343–1359. [Google Scholar] [PubMed]
50. Zhang, G., Zhao, F., Chen, L., Pan, Y., Sun, L. et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nature Plants, 5(5), 491–497. [Google Scholar] [PubMed]
51. Korasick, D. A., Enders, T. A., Strader, L. C. (2013). Auxin biosynthesis and storage forms. Journal of Experimental Botany, 64(9), 2541–2555. [Google Scholar] [PubMed]
52. Dharmasiri, N., Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends in Plant Science, 9(6), 302–308. [Google Scholar] [PubMed]
53. Nagpal, P., Ellis, C. M., Weber, H., Ploense, S. E., Barkawi, L. S. et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development, 132(18), 4107–4118. [Google Scholar] [PubMed]
54. Duman, Z., Eliyahu, A., Abu-Abied, M., Sadot, E. (2020). The contribution of cell wall remodeling and signaling to lateral organs formation. Israel Journal of Plant Sciences, 67(1–2), 110–127. [Google Scholar]
55. Gutierrez, L., Bussell, J. D., Pacurar, D. I., Schwambach, J., Pacurar, M. et al. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell, 21(10), 3119–3132. [Google Scholar] [PubMed]
56. Paul Chege, E. K., Lantos, C., Palágyi, A., Pauk, J. (2021). Doubled haploid production using an improved anther culture protocol for sorghum [Sorghum bicolor (L.) Moench]. Phyton-International Journal of Experimental Botany, 90(2), 475–487. https://doi.org/10.32604/phyton.2021.013557 [Google Scholar] [CrossRef]
57. Wan, Q., Zhai, N., Xie, D., Liu, W., Xu, L. (2023). WOX11: The founder of plant organ regeneration. Cell Regen, 12(1), 1. [Google Scholar] [PubMed]
58. Ahkami, A., Scholz, U., Steuernagel, B., Strickert, M., Haensch, K. T. et al. (2014). Comprehensive transcriptome analysis unravels the existence of crucial genes regulating primary metabolism during adventitious root formation in Petunia hybrida. PLoS One, 9(6), e100997. [Google Scholar] [PubMed]
59. Sukumar, P., Maloney, G. S., Muday, G. K. (2013). Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiology, 162(3), 1392–1405. [Google Scholar] [PubMed]
60. Neves, C., Hand, P., Amâncio, S. (2006). Patterns of B-type cyclin gene expression during adventitious rooting of micropropagated cork oak. Plant Cell, Tissue and Organ Culture, 86(3), 367–374. [Google Scholar]
61. Lindrotha, A. M., Kvarnhedenb, A., Arnolda, S. V. (2001). Isolation of a PSTAIRE CDC2 cDNA from Pinus contorta and its expression during adventitious root development. Plant Physiology and Biochemistry, 39(2001), 107–114. [Google Scholar]
62. Tian, J. J., Xing, Q., Jing, T. T., Fan, X., Zhang, Q. Z. et al. (2022). The epigenetic regulator ULTRAPETALA1 suppresses de novo root regeneration from Arabidopsis leaf explants. Plant Signaling and Behavior, 17(1), 2031784. [Google Scholar] [PubMed]
63. Liu, W., Yu, J., Ge, Y., Qin, P., Xu, L. (2018). Pivotal role of LBD16 in root and root-like organ initiation. Cellular and Molecular Life Sciences, 75(18), 3329–3338. [Google Scholar] [PubMed]
64. Liu, J., Hu, X., Qin, P., Prasad, K., Hu, Y. et al. (2018). The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant and Cell Physiology, 59(4), 734–743. [Google Scholar] [PubMed]
65. Cui, H., Hao, Y., Kovtun, M., Stolc, V., Deng, X. W. et al. (2011). Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiology, 157(3), 1221–1231. [Google Scholar] [PubMed]
66. Kong, X., Lu, S., Tian, H., Ding, Z. (2015). WOX5 is shining in the root stem cell niche. Trends in Plant Science, 20(10), 601–603. [Google Scholar] [PubMed]
67. Della Rovere, F., Fattorini, L., D’Angeli, S., Veloccia, A., Del Duca, S. et al. (2015). Arabidopsis SHR and SCR transcription factors and AUX1 auxin influx carrier control the switch between adventitious rooting and xylogenesis in planta and in in vitro cultured thin cell layers. Annals of Botany, 115(4), 617–628. [Google Scholar] [PubMed]
68. Zhang, T. Q., Lian, H., Tang, H. B., Karel, D., Zhou, C. M. et al. (2015). An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell, 27(2), 349–360. [Google Scholar] [PubMed]
69. Bellini, C., Pacurar, D. I., Perrone, I. (2014). Adventitious roots and lateral roots: Similarities and differences. Annual Review of Plant Biology, 65, 639–666. [Google Scholar] [PubMed]
70. Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D. et al. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138(4), 750–759. [Google Scholar] [PubMed]
71. Yamaguchi, A., Wu, M. F., Yang, L., Wu, G., Poethig, R. S. et al. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Developmental Cell, 17(2), 268–278. [Google Scholar] [PubMed]
72. Wang, J. W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 138(4), 738–749. [Google Scholar] [PubMed]
73. Xu, L., Huang, H. (2014). Genetic and epigenetic controls of plant regeneration. Current Topics in Developmental Biology, 108, 1–33. [Google Scholar] [PubMed]
74. Tanaka, K., Choi, J., Cao, Y., Stacey, G. (2014). Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Frontiers in Plant Science, 5, 446. [Google Scholar] [PubMed]
75. Choi, J., Tanaka, K., Cao, Y., Qi, Y., Qiu, J. et al. (2014). Identification of a plant receptor for extracellular ATP. Science, 343(6168), 290–294. [Google Scholar] [PubMed]
76. Bishop, P. D., Makus, D. J., Pearce, G., Ryan, C. A. (1981). Proteinase inhibitor-inducing factor activity in tomato leaves resides in oligosaccharides enzymically released from cell walls. Biochemistry, 78, 3536–3540. [Google Scholar]
77. Hilleary, R., Gilroy, S. (2018). Systemic signaling in response to wounding and pathogens. Current Opinion in Plant Biology, 43, 57–62. [Google Scholar] [PubMed]
78. Iwase, A., Harashima, H., Ikeuchi, M., Rymen, B., Ohnuma, M. et al. (2017). WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell, 29(1), 54–69. [Google Scholar] [PubMed]
79. Reymond, P., Weber, H., Damond, M., Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell, 12(5), 707–719. [Google Scholar] [PubMed]
80. Noir, S., Bomer, M., Takahashi, N., Ishida, T., Tsui, T. L. et al. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiology, 161(4), 1930–1951. [Google Scholar] [PubMed]
81. Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S. et al. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant Journal, 50(2), 347–363. [Google Scholar]
82. Wang, H., Qi, Q., Schorr, P., Cutler, A. J., Crosby, W. L. et al. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts. Plant Journal, 15(4), 501–510. [Google Scholar]
83. Pan, X., Yang, Z., Xu, L. (2021). Dual roles of jasmonate in adventitious rooting. Journal of Experimental Botany, 72(20), 6808–6810. [Google Scholar] [PubMed]
84. Wang, Z., Cao, G., Wang, X., Miao, J., Liu, X. et al. (2008). Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Reports, 27(1), 125–135. [Google Scholar] [PubMed]
85. Justamante, M. S., Ibanez, S., Peidro, A., Perez-Perez, J. M. (2019). A genome-wide association study identifies new loci involved in wound-induced lateral root formation in Arabidopsis thaliana. Frontiers in Plant Science, 10, 311. [Google Scholar] [PubMed]
86. Zhao, Z., Zhang, Y., Liu, X., Zhang, X., Liu, S. et al. (2013). A role for a dioxygenase in auxin metabolism and reproductive development in rice. Developmental Cell, 27(1), 113–122. [Google Scholar] [PubMed]
87. Schulze, A., Zimmer, M., Mielke, S., Stellmach, H., Melnyk, C. W. et al. (2019). Wound-induced shoot-to-root relocation of JA-Ile precursors coordinates Arabidopsis growth. Molecular Plant, 12(10), 1383–1394. [Google Scholar] [PubMed]
88. Lakehal, A., Dob, A., Novak, O., Bellini, C. (2019). A DAO1-mediated circuit controls auxin and jasmonate crosstalk robustness during adventitious root initiation in Arabidopsis. International Journal of Molecular Sciences, 20(18), 4428. [Google Scholar] [PubMed]
89. Feldmann, K. A., David Marks, M. (1986). Rapid and efficient regeneration of plants from explants of Arabidopsis thaliana. Plant Science, 47(1), 63–69. [Google Scholar]
90. Han, R., Khalid, M., Juan, J., Huang, D. (2018). Exogenous glycine inhibits root elongation and reduces nitrate-N uptake in pak choi (Brassica campestris ssp. Chinensis L.). PLoS One, 13(9), e0204488. [Google Scholar] [PubMed]
91. Nacry, P., Canivenc, G., Muller, B., Azmi, A., Van Onckelen, H. et al. (2005). A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiology, 138(4), 2061–2074. [Google Scholar] [PubMed]
92. Negi, S., Ivanchenko, M. G., Muday, G. K. (2008). Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant Journal, 55(2), 175–187. [Google Scholar]
93. Zhang, W., Swarup, R., Bennett, M., Schaller, G. E., Kieber, J. J. (2013). Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Current Biology, 23(20), 1979–1989. [Google Scholar] [PubMed]
94. Negi, S., Sukumar, P., Liu, X., Cohen, J. D., Muday, G. K. (2010). Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant Journal, 61(1), 3–15. [Google Scholar]
95. Bannoud, F., Bellini, C. (2021). Adventitious rooting in populus species: Update and perspectives. Frontiers in Plant Science, 12, 668837. [Google Scholar] [PubMed]
96. Ikeuchi, M., Favero, D. S., Sakamoto, Y., Iwase, A., Coleman, D. et al. (2019). Molecular mechanisms of plant regeneration. Annual Review of Plant Biology, 70, 377–406. [Google Scholar] [PubMed]
97. Druege, U., Hilo, A., Perez-Perez, J. M., Klopotek, Y., Acosta, M. et al. (2019). Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Annals of Botany, 123(6), 929–949. [Google Scholar] [PubMed]
98. Cao, X., He, Z., Guo, L., Liu, X. (2015). Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiology, 168(4), 1189–1196. [Google Scholar] [PubMed]
99. Verstraeten, I., Schotte, S., Geelen, D. (2014). Hypocotyl adventitious root organogenesis differs from lateral root development. Frontiers in Plant Science, 5(963525), 495. [Google Scholar] [PubMed]
100. Tiirid Becher, G. H., Koop, H. U. (1992). Callus formation and plant regeneration in standard and microexplants from seedlings of barley (Hordeum vulgare L.). Plant Cell Reports, 11, 39–43. [Google Scholar] [PubMed]
101. Meng, F., Xiang, D., Zhu, J. S., Li, Y., Mao, C. Z. (2012). Molecular mechanism of adventitious root formation in rice. Plant Growth Regulation, 68(3), 325–331. [Google Scholar]
102. Ahmadabadi, M., Ruf, S., Bock, R. (2007). A leaf-based regeneration and transformation system for maize (Zea mays L.). Transgenic Research, 16(4), 437–448. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools