 Open Access
Open Access
ARTICLE
Characterization and Pathogenicity of Pseudopestalotiopsis vietnamensis Causing Gray Blight of Wuyi Rock Tea (Camellia sinensis) in China and Specific Mechanisms of Disease Infection
1 Fujian Provincial Key Laboratory of Eco-Industrial Green Technology, College of Ecology and Resources Engineering, Wuyi University, Wuyishan, 354300, China
2 Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Plant Immunity Center, Fujian Agriculture and Forestry University, Fuzhou, 350002, China
3 Chongqing Engineering Laboratory of Green Planting and Deep Processing of Famous-Region Drug in the Three Gorges Reservoir Region, College of Biology and Food Engineering, Chongqing Three Gorges University, Chongqing, 404120, China
4 State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, 611130, China
5 College of Tea and Food Science, Wuyi University, Wuyishan, 354300, China
* Corresponding Author: Guangheng Wu. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Plant–Environment Interactions)
Phyton-International Journal of Experimental Botany 2023, 92(1), 131-147. https://doi.org/10.32604/phyton.2022.021919
Received 19 February 2022; Accepted 11 May 2022; Issue published 06 September 2022
Abstract
Gray blight disease (GBD) causes significant losses in tea production in China. Although genes and biological processes involved in resistance to fungal disease in tea plants have been identified, specific mechanisms of the GBD infection process remain unknown. In this study, morphological and multi-gene (TEF-TUB-ITS) phylogenetic characteristics were used to identify isolate CLBB1 of Pseudopestalotiopsis vietnamensis. Pathogenicity tests confirmed that isolate CLBB1 from tea leaves caused GBD in the susceptible tea cultivar Wuyi Rock (Camellia sinensis var. sinensis cv. Shuixian). Spores began to germinate 24 h after infection (hai), and after 48 h, elongated fungal hyphae formed from a single conidium. Transcriptome analysis revealed that 482, 517, and 369 genes were differentially expressed at 24, 48, and 72 hai, respectively, in Wuyi Rock tea leaves. Isolate CLBB1 infection elicited phenotype-related responses and activated defense-related pathways, including plant–pathogen interaction, MAPK signaling, and plant hormone signal transduction, suggesting a possible mechanism underlying phenotype-based susceptibility to CLBB1. Thus, a new Ps. vietnamensis strain causing GBD in the tea cultivar ‘Shuixian’ was discovered in this study. Transcriptome analysis indicated that pathogen invasion activated chitin-related MAPK pathways and that tea plants required a hormone to restrict CLBB1.Keywords
Tea, coffee, and cocoa are the three most popular nonalcoholic beverages worldwide, with tea being especially popular in Asia [1]. Tea contains a variety of secondary metabolites and aromas, such as polyphenols, polysaccharides, and γ-amino butyric acids, which may reduce the risk of diabetes, cardiovascular disease, Parkinson’s disease, and Alzheimer’s disease [2–4]. Polyphenols in tea can also help prevent a few types of cancer [5,6]. In addition, drinking tea can reduce mental stress and anxiety [7]. Tea is mainly produced from young leaves and buds of tea plants (Camellia sinensis (L.) O. Kuntze), and it is a valuable commercial crop in China, India, Sri Lanka, Vietnam, Indonesia, and Myanmar [8]. To improve tea quality, an effective approach is to select varieties of tea plants with good traits. Currently, tea is commercially cultivated in over 50 countries, including China, which has hundreds of tea germplasm resources from which to select tea cultivars [9]. In China, tea is further classified into different types on the basis of growing conditions, harvesting procedures, and processing methods, and types include black, green, yellow, white, oolong, and Pu-erh teas [10]. Oolong tea is a partially fermented tea with the fragrance of green tea and the smoothness of black tea, in addition to a distinct woody fragrance. Oolong tea has high levels of theanine, polyphenols, polysaccharides, and total catechins [10–12]. The anti-obesity components in oolong tea may reduce the risk of type II diabetes [13–15].
Because of the distinct flavor and excellent health benefits, oolong tea is popular among most Chinese. Traditional oolong tea is primarily grown in Fujian Province in southeastern China. Wuyi Rock tea is a well-known type of oolong tea and one of China’s top ten tea types [16]. Named after its natural habitat, Wuyi Rock tea grows from cracked rocks of Wuyi Mountain. The two main cultivars of Wuyi Rock tea in production are ‘Rougui’ and ‘Shuixian’ [12,16]. ‘Shuixian’ (C. sinensis var. sinensis cv. Shuixian) is an autotriploid (2n = 3x = 45) variety that was bred using asexual reproduction to maintain stable traits across generations. The variety was designated as a “national variety” in 1985.
Pathogenic microorganisms are a threat to tea plants, and warm and humid conditions promote the occurrence of most tea plant diseases. Pathogenic microorganisms such as anthracnose (Colletotrichum) typically cause lesions and disease spots on tea leaves [17]. Despite numerous studies on interactions between pathogens and tea plants, the dominant pathogens causing significant losses of Wuyi Rock tea remain unknown. Gray blight disease (GBD) is a common disease that causes significant yield losses in a variety of economically important crops, including apple, blueberry, coconut, grape, guava, hazelnut, lychee, mango, ginger, and tea [18]. It is also one of the most common tea diseases in China, with outbreaks in Jiangsu, Fujian, Anhui, Sichuan, Guizhou, Hunan, Yunnan, Zhejiang, and Guangdong [19]. Lesions are the most common symptom of GBD. At the beginning of infection, small yellow or green spots appear on tea leaves at leaf tips or margins that gradually develop into brown semicircular, circular, or irregular shapes and become grayish white as lesions become more visible [18].
Pestalotiopsis-like species can cause necrotic lesions on C. sinensis [20]. Gray blight lesion symptoms are also observed during the growing season of Wuyi Rock tea (C. sinensis var. sinensis cv. Shuixian) in Wuyi Mountain, causing yield losses. Furthermore, because ‘Shuixian’ is autotriploid, it is more susceptible to pathogens. However, the pathogen responsible for GBD in Wuyi Mountain has not yet been identified and characterized. The aim of this study was to identify the isolates causing GBD in Wuyi Rock tea cultivar ‘Shuixian’ based on morphological characteristics and multigene phylogenetic analysis, and reveal mechanisms of disease infection using RNA sequencing (RNA-seq) and reverse-transcription quantitative PCR.
2.1 Plant Materials, Fungal Isolates, and Treatments
The study was conducted at the Tea Garden of Wuyi University in Wuyishan, Fujian Province, China (27°72’67’’N, 118°00’47’’E; 130 m above sea level). Twenty tea leaves with disease symptoms were collected in June 2018. Leaves were surface-sterilized and incubated on potato dextrose agar (PDA, 200 g potato + 2% [w/v] dextrose + 2% [w/v] agar) for fungal strains. Petri dishes were incubated at 25°C ± 2°C under light of 3,000 to 4,000 lux until fungal colonies were observed. Pure cultures were obtained from hyphal tips collected at margins of colonies. In pathogenicity tests, mechanical inoculation of plants was performed in vitro based on methods described by Wang et al. [20]. Three-year-old tea plants (C. sinensis var. sinensis cv. Shuixian) were used as experimental materials. The third and fourth healthy mature leaves were sampled at 1, 2, 3, 4, and 10 days after inoculation (dai). Alcoholic trypan blue and 3, 3-diamino benzidine hydrochloride (DAB) staining assays were performed, and fungal structures were examined under a Leica DM2500 microscope (Germany) [21]. To observe the ultrastructure of CLBB1, gluta fixative (4%) was used to observe hyphae and conidia with a scanning electron microscope (TESCAN VEGA3, Czech).
2.2 DNA Extraction, PCR Amplification, and Sequencing
From mycelia grown on PDA plates, DNA was extracted with a PlantGen DNA Kit of CW Biotech (Beijing, China). The genes internal transcribed spacer (ITS) of rDNA, translation elongation factor−1α (TEF), and β-tubulin (TUB) were amplified and sequenced with primers ITS1/ITS4, EF-1α-F/R, and β-tub-F/R, respectively [22]. Primer sequences are listed in Table 1. The 2xTaq PCR Mix of LabLead were used for PCR amplifications (Fuzhou, China), and Sangon Biotech Co., Ltd. (Shanghai, China) conducted the sequencing.
Sequences of ITS, TEF, and TUB loci connected end to end were analyzed to investigate the phylogenetic relationships among Pseudopestalotiopsis and Neopestalotiopsis species using DNA sequences available from the GenBank (Table 2). DNASTAR (Lasergene 7.0) was used to obtain consensus sequences from sequences generated from ITS, EF-1α and β-tub forward and reverse primers. Combination sequence data obtained from the three gene regions were aligned using ClustalX (http://www.clustal.org/). Before phylogenetic analysis, ambiguous sequences at the start and the end were deleted, and gaps were manually adjusted to optimize the alignment. Maximum parsimony (MP) and maximum likelihood (ML) were conducted with branch-and-bound and heuristic searches as implemented in MEGA7.0 with the default options method. Clade stability was evaluated in a bootstrap analysis with 1,000 replicates, random sequence additions with maximum trees set at 10,000, and other default parameters in MEGA.
2.4 RNA Isolation, Library Construction, and Sequencing
To construct RNA-seq libraries, CLBB1 was cultured on PDA and incubated at 25°C ± 2°C under light of 3,000 to 4,000 lux for 10 days to promote sporulation. Conidia were harvested and suspended in sterile distilled water. Conidia concentration was determined using a hemocytometer and adjusted to 1 × 106 mL−1 for inoculation. The third and fourth healthy mature leaves of three-year-old tea plants were wounded on the upper surface with a sterile needle, and 20 μL of conidial suspension was applied to the wound. An equal volume of sterile water was applied as a mock inoculation. Inoculated and mock leaf samples were collected from tea plants at 0, 24, 48, and 72 h after inoculation (hai). Samples were immediately frozen in liquid nitrogen and then stored at −80°C until processed further. Three biological replicates were used in each experiment.
2.5 Library Preparation for Transcriptome Sequencing
To generate sequencing libraries, RNA of each leaf sample was extracted using a NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s protocol and guidelines. Concentration of RNA was measured on a Qubit2.0 Fluorometer, and RNA was diluted to 1.5 ng/μL for RNA sequencing. Library quality was assessed on an Agilent Bioanalyzer 2100 System. The Novogene Corporation (Beijing, China) performed library construction and RNA-seq, according to the manufacturer’s protocols. A total of 21 libraries (with three biological replicates per sample) were sequenced on an Illumina Novaseq platform, and 150-bp paired-end reads were generated.
2.6 Quality Control and Reads Mapping with the Reference Genome
Raw reads in fastq format were filtered using in-house perl scripts. Filtering primarily included removing reads containing an adapter or poly-N and those of low quality from raw data. To obtain high-quality clean reads, Q20 and Q30 values and GC contents were calculated before downstream analyses to ensure accuracy. The reference genome of ‘Shuchazao’ and gene model annotations were downloaded from the Tea Plant Information Archive database (http://tpia.teaplant.org/) [23–25], and paired-end reads were mapped to the tea plant reference genome sequence using Hisat2 v2.0.5 [26].
2.7 Quantification of Gene Expression Levels and Differential Expression Analysis
Fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) was used to determine gene expression levels. FeatureCounts v1.5.0-p3 was used to count numbers of reads mapped to each gene, following FPKM calculation for each gene based on its length. To compare differences in expression patterns among groups (with biological replications), the DESeq2 R package (1.16.1) was used [27]. Resulting p-values were adjusted using Benjamin and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value < 0.05 found by DESeq2 were assigned as differentially expressed.
2.8 GO and KEGG Enrichment Analyses of Differentially Expressed Genes
Gene ontology (GO) [28] and enrichment analysis and the statistical enrichment of differentially expressed genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were performed using the clusterProfiler R package.
2.9 Reverse-Transcription Quantitative PCR Verification
To verify results of RNA-seq data, leaf samples of H2O and CLBB1-inoculated Wuyi Rock tea ‘Shuixian’, ‘Rougui’, and ‘Shuchazao’ cultivars were collected at 0, 24, 48, and 72 hai. Total RNA was extracted with an RNAprep Pure Kit (Tiangen, China). First-strand cDNA from 2 μg of total RNA was synthesized using a NovoScript® Plus All-in-one 1st Strand cDNA Synthesis SuperMix Kit (NovoProtein, China). Accumulation of transcripts was examined using a NovoStart® SYBR qPCR SuperMix Plus Kit (NovoProtein) with a Bio-rad CFX96 (California, USA). Each reaction system contained 10 μL of SYBR Premix, 7.2 μL of ddH2O, 2 μL of diluted cDNA, and 0.4 μL of forward/reverse primer. Reverse-transcription quantitative PCR (RT-qPCR) conditions consisted of a preliminary step at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 20 s, and an extension step at 72°C for 30 s. Tea 18S rRNA was used to normalize transcript abundance. All gene-specific primers (RT-) are listed in Table 1 (CsCHITINASE, CsMPK3, CsPR, CsPR4, CsWIN1-L1, and Cs18S). Three biological replicates were used in each experiment. Statistical analysis of RT-qPCR data was performed with DPS7.05.
3.1 Identification of CLBB1, A New Isolate of a Pestalotiopsis-Like Genus
To identify the microorganism species causing GBD of Wuyi Rock tea in Fujian Province, fungal strain CLBB1 was screened and purified from leaves of Wuyi Rock tea (C. sinensis var. sinensis cv. Shuixian) exhibiting typical GBD symptoms in the Tea Garden of Wuyi University at Wuyi Mountain. Harvested diseased, susceptible leaves had large round lesions with dark brown discoloration at leaf edges.
Colony diameter was 52 mm at 3 dai under daylight at 25°C on PDA (Fig. 1A), and over the next three days, the colony spread covering large portions of the plate and produced conidia under aerial mycelium of raised curves (Fig. 1B). There were obvious dark dots on curves with abundant conidia on the plate at 9 dai (Fig. 1C). From 6 dai, observed from the bottom, the culture appeared yellowish white, with gradual accumulation of brown, most likely because CLBB1 produced pale brown pigments released into the medium (Figs. 1D–1F). Germination and developmental course of spores and hyphae in PDA culture were consistent. The white aerial mycelium produced many branches with smooth surfaces at 3 dai (Figs. 1G–1I). Conidia had the following characteristics: five cells, fusiform to ellipsoid, straight to slightly curved, four-septate, 22.4 to 31.7 μm × 5.6 to 7.3 μm (mean = 27.4 μm × 6.2 μm, n = 30), basal cell conic or obconic, hyaline, thin and smooth-walled, 5.5 to 7.1 μm long (mean = 6.5 μm, n = 30) slightly constricted at septa, three pale brown or olivaceous concolorous median cells, and three hyaline apical appendages arising from the apex (Figs. 1J–1L).
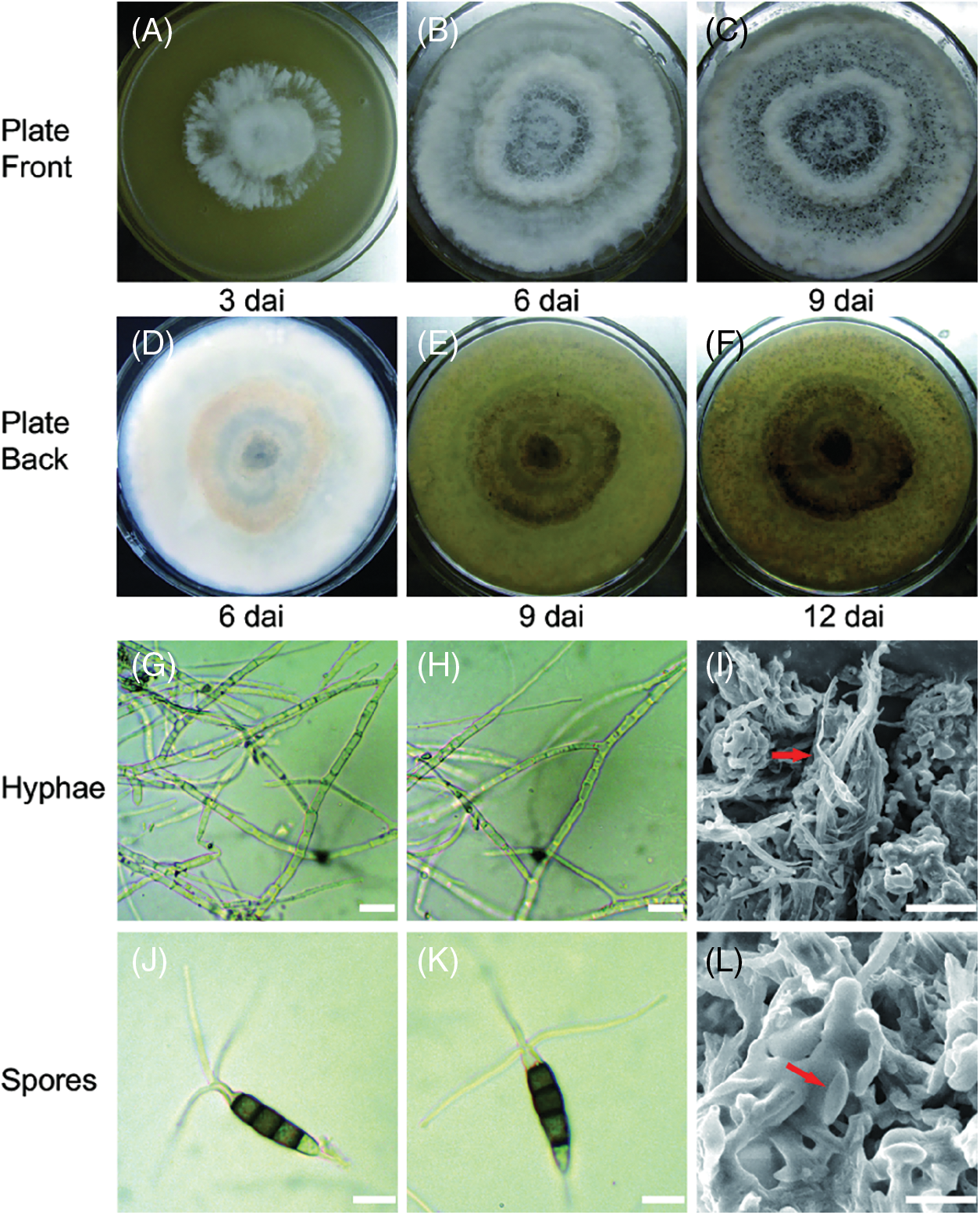
Figure 1: Culture traits and morphology of CLBB1 isolated from symptomatic tea leaves and grown on 90 mm PDA plates. (A–F) Representative culture traits of CLBB1 on PDA at 25°C at 3, 6, 9, and 12 days after inoculation (dai). (G–I) Images of hyphae of CLBB1. Bar = 25 μm. (J–L) Conidial morphology of CLBB1. Bar = 10 μm
Although Pestalotiopsis-like species on tea have been reported [18,19], the infection process on tea plants has not been described. In this study, three-year-old Wuyi Rock tea (C. sinensis var. sinensis cv. Shuixian) plants were infected with CLBB1 spores and examined for disease responses at 10 dai. Sterile water was used as the control. Leaves inoculated with CLBB1 had small brown necrotic spots on front and back sides at 10 dai. Symptoms were similar to those occurring in the field. There were no changes in sterile water-infected sites (Figs. 2A and 2B). Koch’s postulates were tested by re-isolation and morphological comparison.
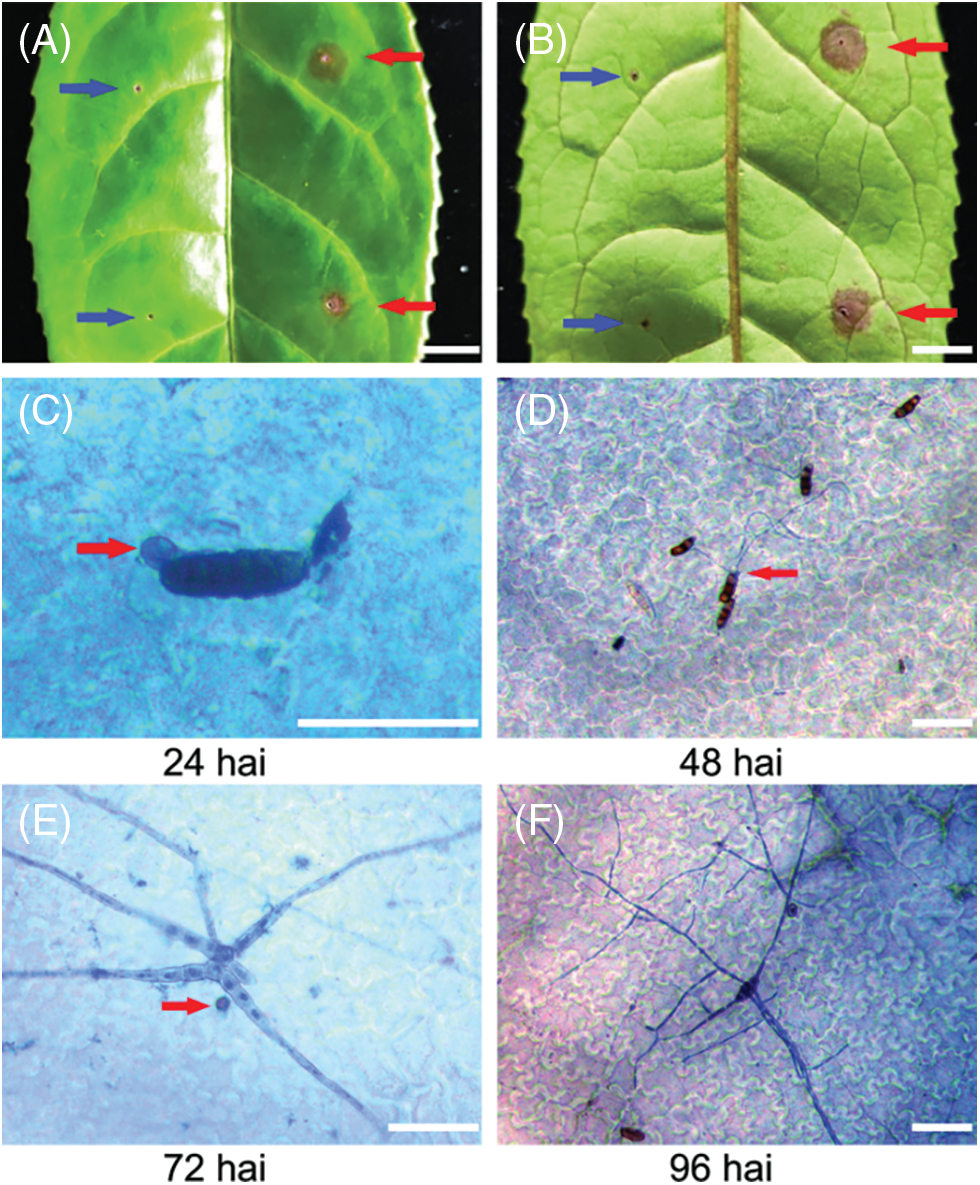
Figure 2: Infection progress of CLBB1 on host tea leaves at 24, 48, 72, and 96 h after infection (hai). (A, B) representative symptoms on leaves removed from three-year-old ‘Shuixian’ tea plants inoculated with CLBB1 at 10 days after inoculation. Red and blue arrows indicate CLBB1-infected and water-treated sites, respectively. Bar = 0.5 cm. (C–F) Trypan blue staining of representative leaves inoculated with CLBB1 at various times (hai). Red arrows indicate (C) penetration peg, (D) elongated fungal hyphae, and (E) H2O2 accumulation in infection site. Bar = 25 μm
To examine the early stage of fungal development on tea leaves, infected leaves at various times were stained with trypan blue to observe fungal structures. CLBB1 spore germination and penetration pegs at infection sites were observed in epidermal cells at 24 hai (Fig. 2C). At 48 hai, elongated fungal hyphae formed from a single conidium (Fig. 2D). At 72 hai, CLBB1 spores developed extensive branched hyphae with secondary germ tubes that invaded underlying epidermal cells (Figs. 2E and 2F). Infected leaves were also stained with 3, 3-DAB, and there were few accumulations of H2O2 in infection sites at 72 hai (Fig. 2E).
3.3 Phylogenetic Tree Analysis
The CLBB1 ITS sequence (MK909901) analyzed by BLASTn showed that most isolates of the pathogen had 99% sequence homology with strains of Pestalotiopsis-like species in the GenBank. To further analyze the specific species of Pseudopestalotiopsis and Neopestalotiopsis, phylogenetic tree analyses were conducted derived from the combined ITS, partial β-tubulin (TUB), and partial translation elongation factor 1α (TEF) gene regions. Sequence data of CLBB1 and reference strains of Pestalotiopsis-like species were obtained from the GenBank (Table 2).
From the varied lengths of sequences obtained from NCBI, regions containing many leading or trailing gaps were removed from the ITS, TUB, and TEF alignment. Sequences contained 500 nucleotides including gaps for ITS, 340 bp for TUB, and 905 bp for TEF. Phylogenetic trees were constructed using the MP method. Tree topology of the best tree revealed by ML analyses was identical to that of the MP tree (not shown). Species were separated into two clades including the genera Neopestalotiopsis and Pseudopestalotiopsis with high bootstrap support. Isolate CLBB1 was in the clade of Pseudopestalotiopsis (Fig. 3) and was most closely related to Ps. vietnamensis isolate 3-KW-2016–17 collected in Vietnam [29]. Thus, CLBB1 was considered as Ps. vietnamensis on the basis of morphological characteristics and multigene phylogenetic analyses of ITS, TUB, and TEF.
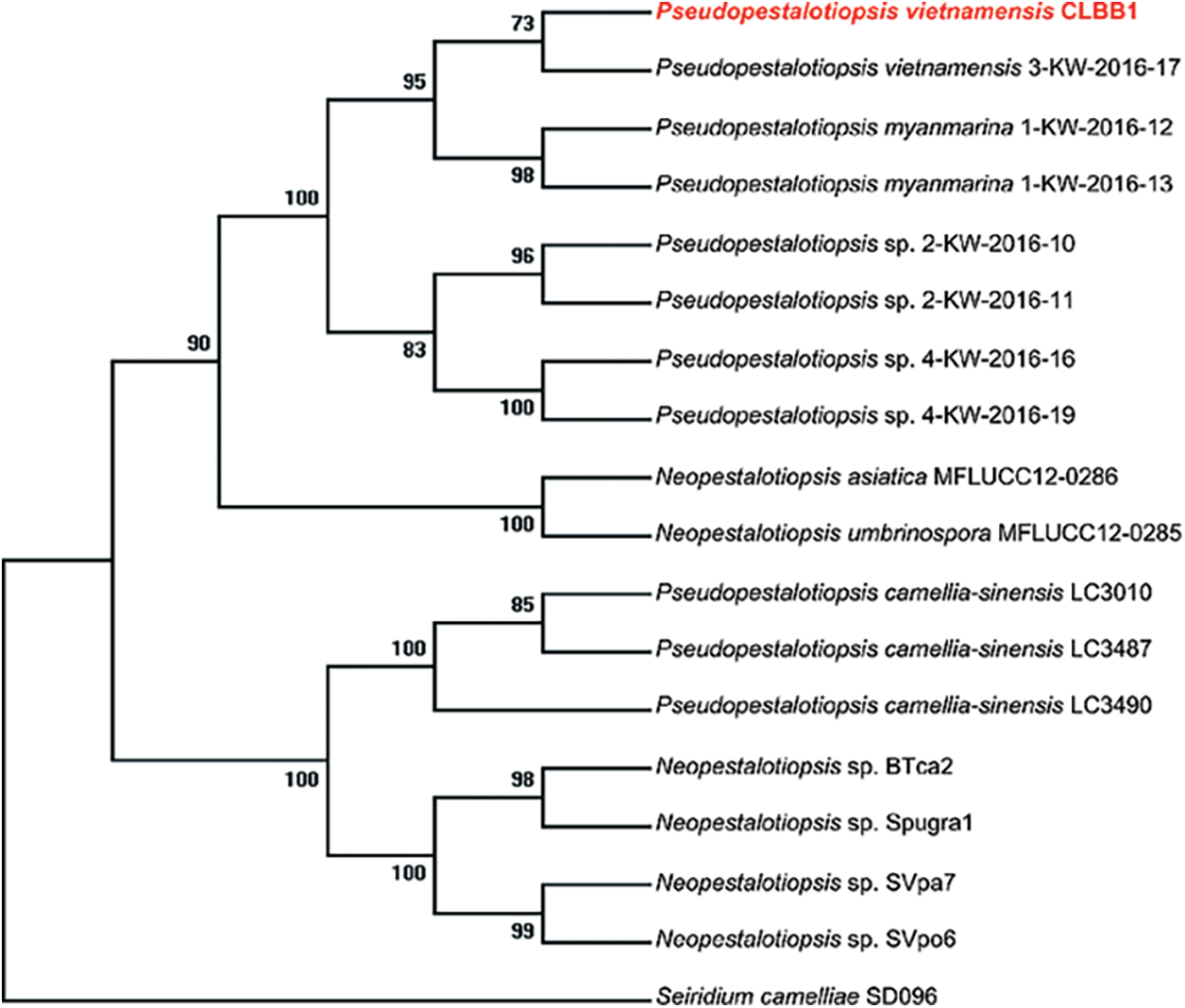
Figure 3: Molecular phylogenetic tree of Ps. vietnamensis isolate CLBB1 based on maximum parsimony generated from combined (TEF–TUB–ITS) alignment. The tree was rooted to Seiridium camelliae SD096. Ps. vietnamensis strain CLBB1 was in red and bold
3.4 CLBB1 Induced Extensive Immune Responses in Tea Plants
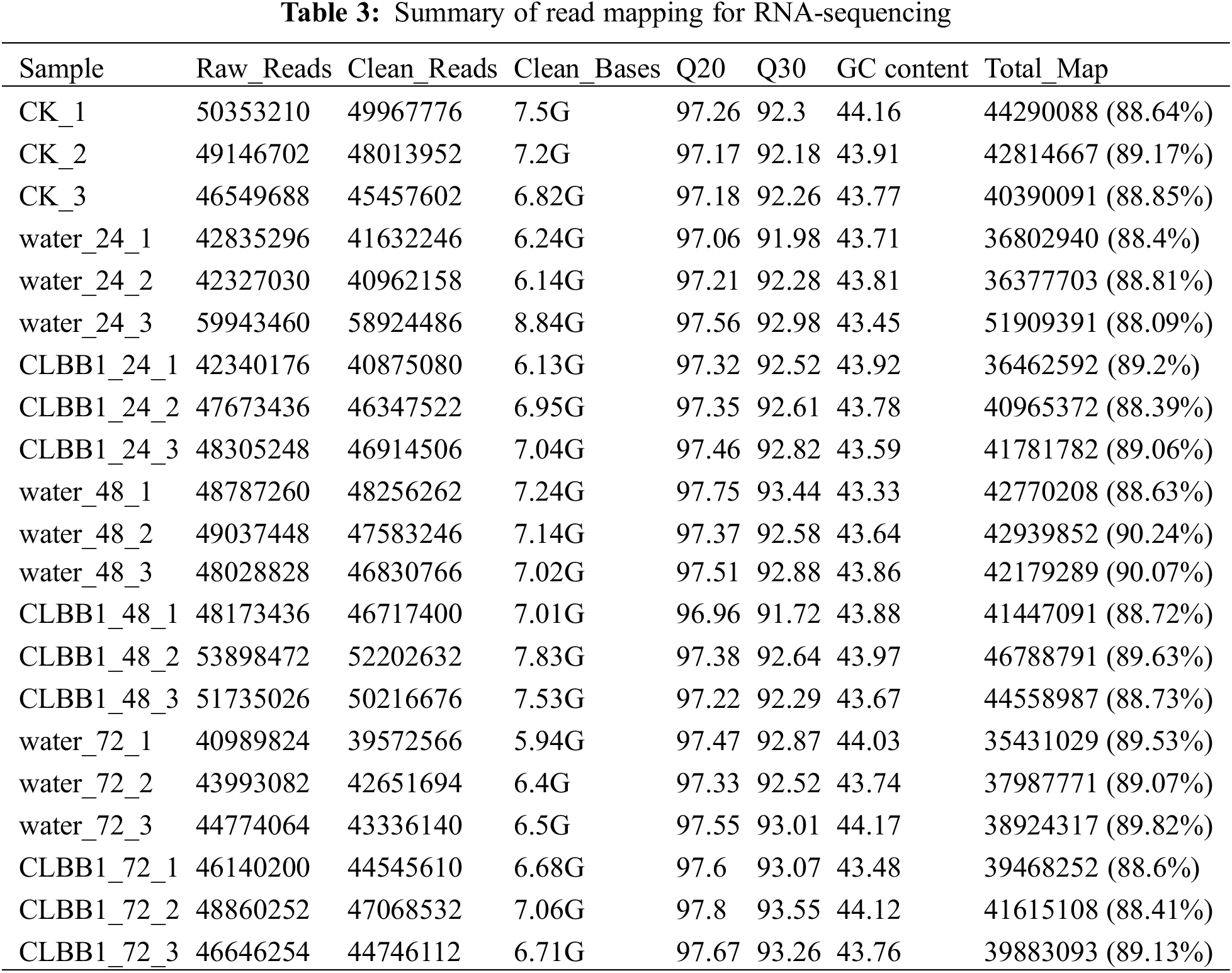
To further investigate the general immune response of tea against CLBB1 at different times after inoculation, RNA was extracted, and 21 sequencing libraries were generated from water (as the control) and CLBB1-infected samples at three different times, with three biological replicates per sample. In total, 145.92 G of high-quality clean data were produced by paired-end Illumina RNA-seq, and each library produced over 40,000,000 clean bases with an average of 97.42% at Q20 and a minimum of 91.72% at Q30. The percentage of clean reads in each library mapped either to a unique location or to multiple genomic locations was greater than 88.4% (Table 3).
Genes were globally differentially expressed, and more than 4,000 differentially expressed genes (DEGs) were identified in each sample. To reduce the deviation caused by negative factors such as environment, buffer, and wound by punch inoculation, sterile water was dropped on wound sites of tea leaves as an inoculation control. The DEGs found in pathogen-inoculated samples but not expressed in water-treated or uninoculated samples were analyzed at 24, 48, and 72 hai. As shown in the Venn diagram (Fig. 4A), 428, 517, and 369 genes were detected at 24, 48, and 72 hai, respectively. The largest number of DEGS at 48 hai was consistent with hyphal formation of CLBB1 fungi at 48 hai. Notably, only one DEG was expressed at all three times after inoculation, whereas 35 DEGs were co-expressed at 24 and 48 hai and also at 48 and 72 hai. In addition, 13 DEGs were co-expressed between 24 and 72 hai. Therefore, data suggested that CLBB1 activated expression of various genes at certain times after inoculation to increase pathogenicity or induce host defense.
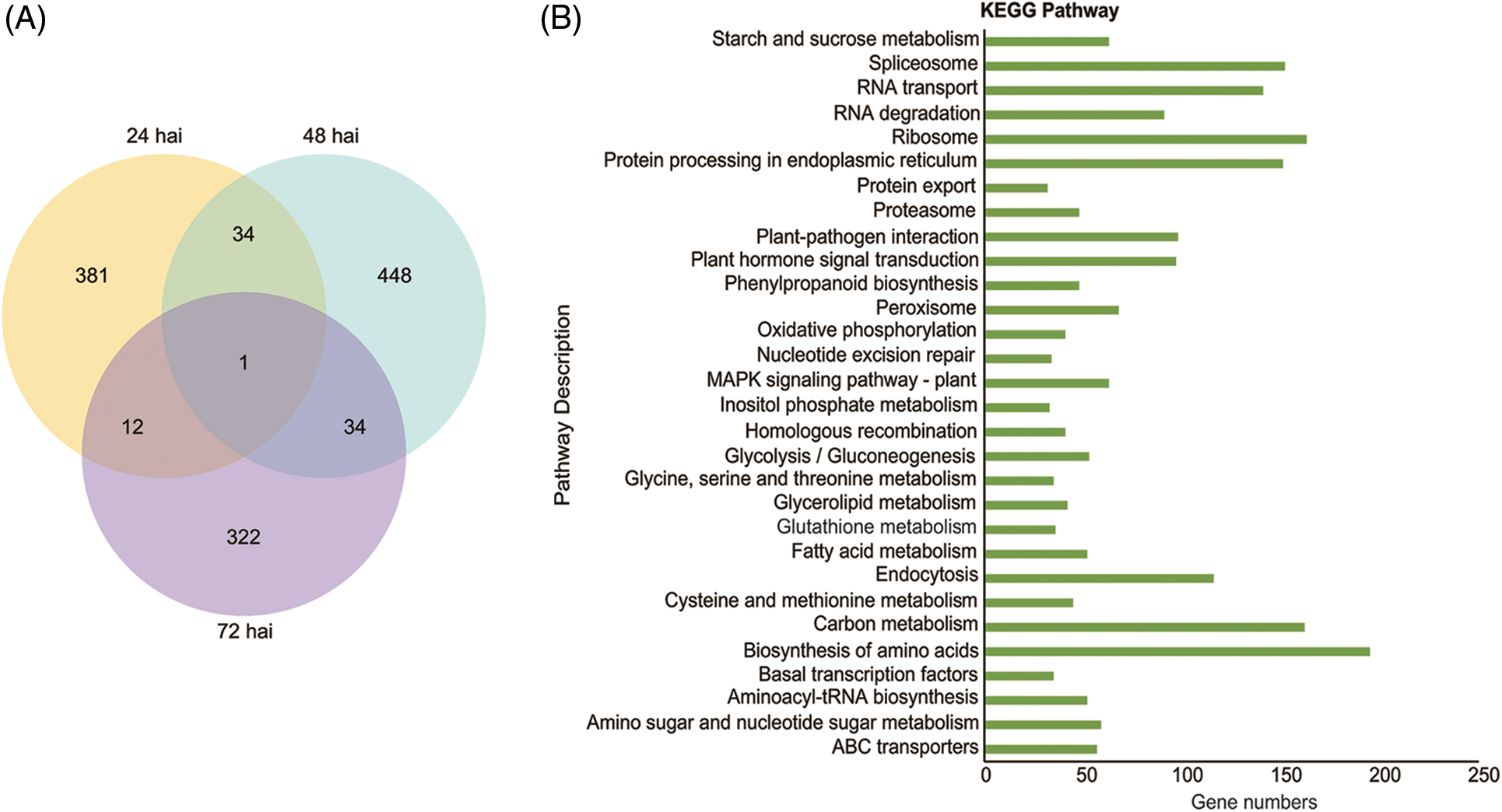
Figure 4: Transcripts induced by CLBB1 at three times after inoculation of tea leaves. (A) Venn diagram of numbers of differentially expressed genes (DEGs) with CLBB1 infection at 24, 48, and 72 h after inoculation (hai). (B) Significantly enriched KEGG pathways of DEGs after CLBB1 inoculation
Gene functions in higher-ordered pathways were investigated using KEGG analysis. The DEGs were highly enriched in RNA and protein biosynthesis, transportation, processing, and modification pathways, such as “spliceosome”, “RNA transport”, “ribosome”, “biosynthesis of amino acids”, and “protein processing in endoplasmic reticulum”. The enriched pathways indicated tea plants synthesized proteins in defense against CLBB1. Transcriptome data showed DEGs were also enriched in “plant-pathogen interaction”, “Mitogen Activated Protein Kinase (MAPK) signaling”, and “plant hormone signal transduction” pathways after CLBB1 infection (Fig. 4B). In addition, the endocytosis pathway was also enriched, which is involved in the immune response in Arabidopsis thaliana [21]. Collectively, the results revealed that CLBB1 induced extensive defense responses in tea plants, particularly by gene enrichment in protein kinase MAPK and plant resistance hormone transduction pathways.
According to GO analysis, many genes in tea plants were activated in resisting pathogen invasion at different stages. A total of 1,232 DEGs were classified into three GO categories, i.e., biological process, molecular function, and cellular component. Most DEGs involved in metabolic processes were enriched at the three times after inoculation (Fig. 5). Genes involved in chitin catabolic and metabolic processes and chitin-responsive (chitin binding, chitinase activity) genes were activated at 24 hai (Fig. 5A). In addition, many sequence-specific DNA-binding genes were expressed, which might induce early defense response gene expression against CLBB1 invasion (Fig. 5B). Genes responsive to pathogens and stress were highly expressed at 48 and 72 hai (Figs. 5B and 5C), which was consistent with growth of the pathogen.
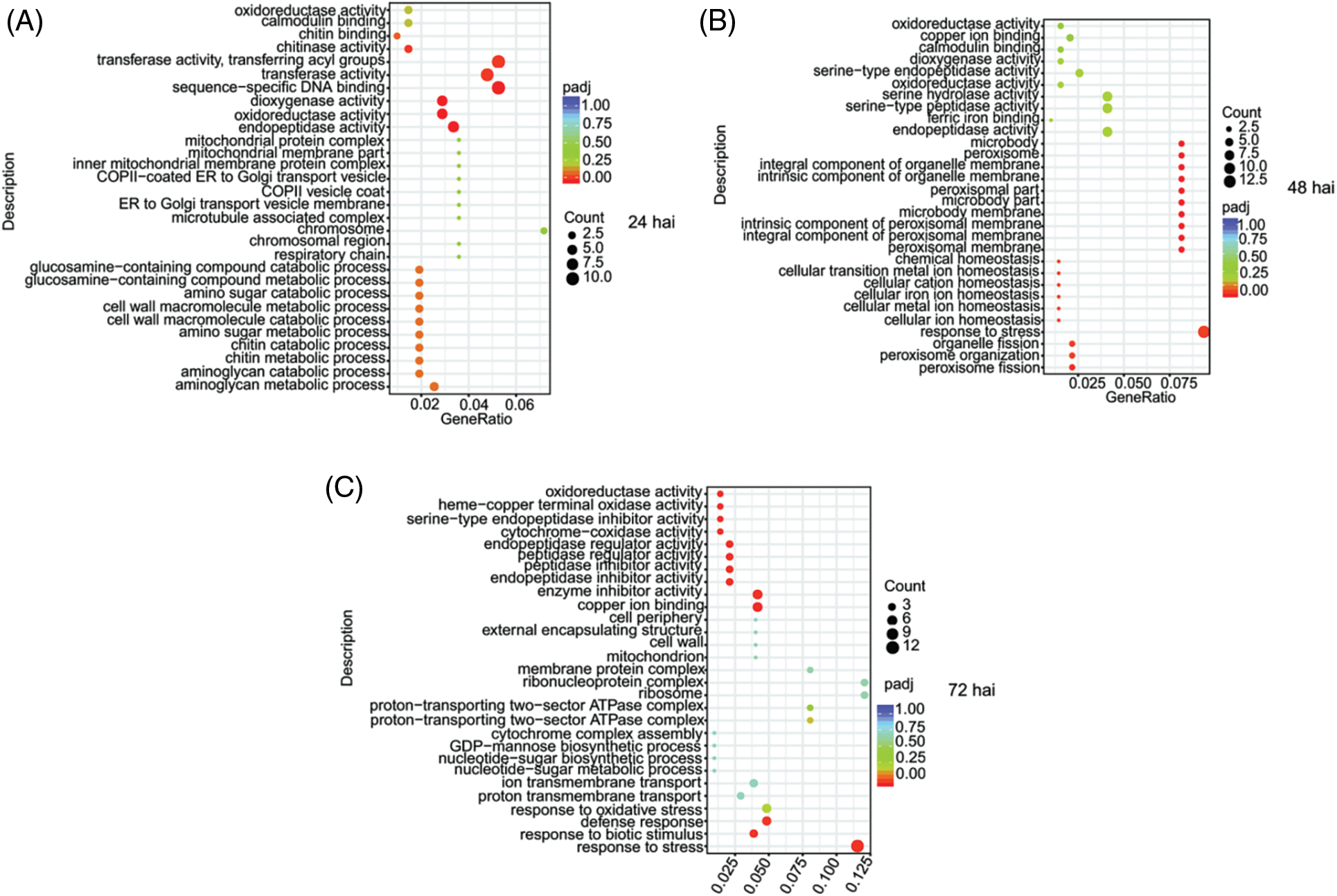
Figure 5: Gene ontology enrichment analysis of differentially expressed genes induced by CLBB1 at three times after inoculation (hai, hours after inoculation). (A) 24 hai, (B) 48 hai, (C) 72 hai
3.5 Reverse-Transcription Quantitative PCR Verification of Candidate Differentially Expressed Genes
To verify genes enriched in “chitin catabolic”, “MAPK signaling”, and “plant hormone signal transduction” pathways of plant immunity, RT-qPCR was performed to determine expression patterns of five candidate DEGs. Chitinases isolated from plants can limit the growth of chitin-containing fungi, and overexpressed chitinases in plants can resist infection of different fungal pathogens [30]. After CLBB1 infection, the expression level of CsCHITINASE III (TEA002526) was highly up-regulated at 48 hai in tea cultivar ‘Shuixian’ (Fig. 6A). To investigate expression levels of DEGs in other cultivars, transcripts of DEGs were also examined in commercial varieties ‘Shuchazao’ and ‘Rougui’. ‘Shuchazao’ represented Camellia sinensis var. sinensis, and its whole genome was sequenced. In addition to ‘Shuixian’, ‘Rougui’ is also a Wuyi Rock tea. With CLBB1 infection in ‘Shuchazao’, accumulation of CsCHITINASE III transcripts was highest at 24 hai. However, at 72 hai, CsCHITINASE III transcript expression levels in CLBB1-inoculated ‘Rougui’ plants were significantly higher than those in the control.
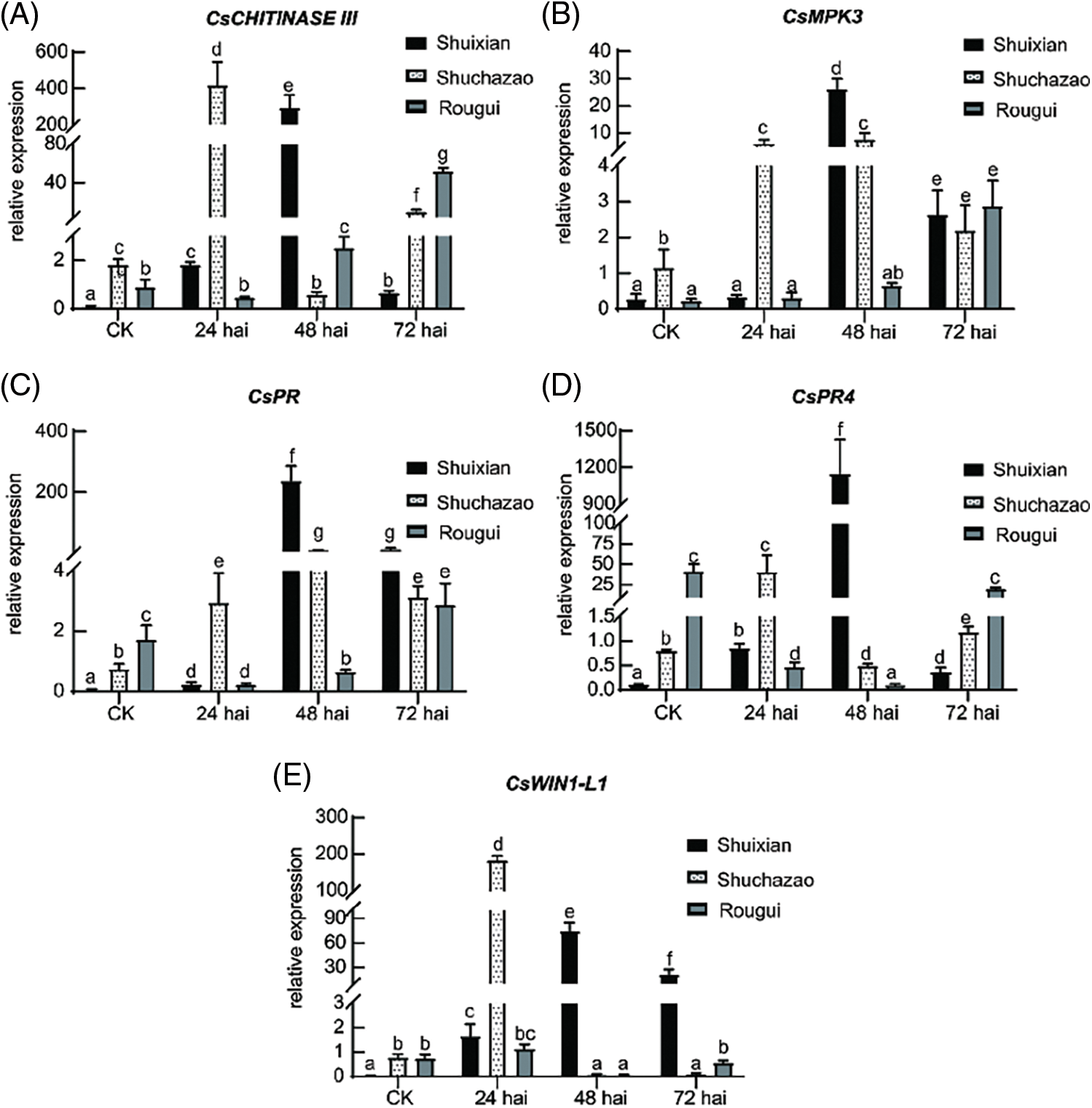
Figure 6: Relative expression levels of defense-related genes in ‘Shuixian’, ‘Shuchazao’, and ‘Rougui’ tea cultivars. Transcript accumulation of defense-related genes was examined by reverse-transcription quantitative PCR, with Cs18S used as the internal control. Relative transcriptional levels are shown for (A) CsCHITINASE III, (B) CsMPK3, (C) CsPR, (D) CsPR4, and (E) CsWIN1-L1. CK represents the uninoculated relative expression amount at 0 hai. Values are the mean and standard deviation obtained from three independent biological samples. Three technical replicates for each biological sample were examined in the experiment. Lowercase letters indicate significant differences (p < 0.05; one-way ANOVA). Experiments were repeated three times with similar results
After pathogen infection, plants activate extensive responses to pathogens. In particular, activation of MPKs and high expression of pathogenesis-related genes are typical responses against pathogen invasion in PAMP-triggered immunity (PTI) [31–33]. In this study, CsMPK3 (TEA026040) expression level was highly elevated by CLBB1 at 24, 48, and 72 hai in ‘Shuchazao’, ‘Shuixian’, and ‘Rougui’, respectively (Fig. 6B), which was consistent with expression levels of CsCHITINASE III (Fig. 6A). Transcript levels of CsPR (TEA004541) and CsPR4 (TEA013240) were also measured from 0 to 72 hai with CLBB1 infection. Before inoculation, CsPR and CsPR4 transcript levels were very low in all varieties, but basal expression levels of CsPR and CsPR4 were higher in ‘Rougui’ than in ‘Shuixian’ and ‘Shuchazao’. With CLBB1 infection, CsPR and CsPR4 transcripts accumulated at the highest levels at 48 hai in ‘Shuixian’ (Figs. 6C and 6D).
Plants biosynthesize plant resistance hormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) to defend against pathogens [32]. To determine whether defense-related hormones were induced by CLBB1, expression level of APETALA2/ethylene-responsive transcription factor CsWIN1-L1 (TEA001387, WIN1-like 1) was examined [34]. As shown in Fig. 6E, CsWIN1-L1 was highly expressed at 24 hai in ‘Shuchazao’ and at 48 hai in ‘Shuixian’, whereas its expression levels were relatively low and stable in ‘Rougui’.
In general, relative transcript levels of candidate DEGs were low in the control group, but expression was induced to relatively high levels after CLBB1 infection, especially at 48 hai in the cultivar ‘Shuixian’. Collectively, data indicated that tea plants activated immune responses with CLBB1 infection. Defense responses were activated at higher levels in ‘Shuixian’ at 48 hai than at other times, which was consistent with global changes in DEGs and growth of the pathogen.
Plant pathogens are typically divided into biotrophs and necrotrophs, with biotrophs consuming live plant tissues and necrotrophs killing their host and feeding on dead tissues [35]. In this study, CLBB1 was identified as a new strain of Ps. vietnamensis. It is a necrotrophic pathogen isolated from a necrotic lesion of GBD-infected leaves of Wuyi Rock tea C. sinensis var. sinensis cv. Shuixian in Wuyi Mountain, Fujian Province, a subtropical region of China. Because of the special “Danxia” landform and ecological environment [36], Wuyi Mountain is a unique region suitable for tea production, as well as proliferation of CLBB1. In addition, combined ITS, TUB, and TEF gene regions revealed that Ps. vietnamensis strain CLBB1 is different from strains previously identified, which might be related to the unique growing environment of Wuyi Rock tea (Fig. 3; Table 2).
Because infections are limited to tea plant surfaces, particularly those of leaves, early stages of CLBB1 infection in the epidermis were observed microscopically in this study. At 24 hai, CLBB1 conidia germinated, developed appressoria, formed penetration pegs, and penetrated outer epidermal cuticle and cell wall of the host to establish nutritional support for hyphal growth (Fig. 2C). From 48 hai (Figs. 2D–2F), elongated fungal hyphae extended across the leaf surface, leading to plant cell decay. Therefore, 24 and 48 hai were key times for development and spread, respectively, of CLBB1 pathogenicity. Those findings are consistent with growth behaviors of other strains of Pestalotiopsis in culture medium [37].
Pestalotiopsis-like species are common phytopathogens that cause GBD in tea plants; however, pathogens of other genera can also cause similar lesions [38,39]. Although other strains were not isolated in this study, it is uncertain whether the lesions observed were caused by a single strain or whether other pathogens, especially endophytes, caused synergistic effects. Germinating spores of the necrotrophs Botrytis cinerea and Alternaria brassicicola produce cell wall-degrading enzymes and toxins resulting in death and maceration of tissues [40]. Similarly, Pestalotiopsis produces phytotoxins and infects undamaged tea leaves [41]. In this study, whether CLBB1 could cause disease on undamaged tea leaves was not determined. On the basis of all data, it was postulated that CLBB1 conidia may germinate on tea leaves alone with wounds or surface breaks and activate enzymes and toxins to ensure infection or with other pathogens that produce enzymes and toxins to assist entry.
To examine the pathogenicity of CLBB1 in Wuyi Rock tea, RNA-seq was performed to analyze DEGs at different times after mechanical introduction of CLBB1 into tea leaves. Comparative analyses of transcriptome data revealed 428, 517, and 369 DEGs in leaves infected with CLBB1 at 24, 48, and 72 hai, respectively (Fig. 4A). The results indicated that the pathogen induced many genes to establish pathogenicity or induce host defense [42,43]. Spores of CLBB1 germinated at 24 hai, and elongated fungal hyphae formed at 48 hai, which developed further at 72 and 96 hai (Figs. 2C–2F). Many DEGs were identified at 24 and 48 hai, suggesting the transition stage from 24 to 48 h was key to germination, growth, and pathogenicity [44,45]. However, the specific genes that control pathogen growth and the associated mechanisms remain unknown.
Plants rely on a complicated innate system to counter invasions of diverse pathogens. Activation of MAPKs (consisting of MAPKKK, MAPKK, and MAPK) is one of the most crucial steps involved in PAMP-triggered immunity (PTI) [46–48]. In this study, CLBB1 activated the MAPK pathway at the transcriptional level, although specific components of the pathway in Wuyi Rock tea remain unclear (Fig. 4B). Expression levels of CsMAPK3 were highly elevated by CLBB1 in three different cultivars, suggesting the gene had a pivotal role against CLBB1 infection (Fig. 6B). In addition, many chitin-responsive and pathogenesis-related genes were differentially expressed after CLBB1 infection (Figs. 4B, 6A, 6C and 6D). Those results suggested that the MAPK signaling pathway was mediated by chitin recognition following CLBB1 infection and that MAPK cascades further induced extensive expression of PRs genes, which constitute the main defense-signaling pathway in PTI. However, the MAPKs and PRs involved in the CLBB1 and Wuyi Rock tea ‘Shuixian’ interaction still need to be identified and characterized.
Plant hormones also have essential roles in plant immunity, and SA, JA, ET, abscisic acid, auxins, brassinosteroids, gibberellic acid, and cytokinins are activated by pathogen invasion [49–51]. Salicyclic acid signaling pathways are activated as plant defense mechanisms against biotrophic pathogens, and both ET and JA signaling pathways are required to activate plant defense against necrotrophic pathogens [50,51]. In this study, CLBB1 infection activated the plant hormone signal transduction pathway, and many ET-related transcriptional factors were expressed. Results of KEGG analysis indicated that Wuyi Rock tea used the ET-signaling pathway to restrict CLBB1 invasion, a necrotrophic pathogen (Fig. 4B). In addition, ethylene-responsive factor CsERF96 was recently found to have a critical role in the response to GBD [52]. In this study, CsWIN1-L1 was identified as a potential candidate regulating plant defense-related ET pathways in tea (Fig. 6E).
In conclusion, spores on tea leaves began to germinate at 24 hai with CLBB1, and metabolism-related genes were activated to promote pathogen establishment. Tea plants recognized the pathogen via PAMPs, such as chitin, and activated MAPK pathways and expression of pathogenesis-related genes and transcription factors to resist effects of the pathogen. After 48 h, hyphae formed, and tea plants activated additional genes and pathways to restrict the invasion.
Author Contributions: Study concept and design: Guangheng Wu, Lu Rui, Xiang Lu. Acquisition of data: Guangheng Wu, Lu Rui, Gan Lv, Xianyu Fu, Jinxian Liu. Methodology: Libo Han, Nong Zhou, Chuanhai Zhang. Analysis and interpretation of data: Lu Rui, Xiang Lu. Draft manuscript preparation: Guangheng Wu, Lu Rui, Libo Han.
Acknowledgement: We thank Dr. Qing Xiong (Sichuan Agricultural University, Chengdu, China) and Charlesworth for linguistic assistance during the preparation of this manuscript.
Funding Statement: This work was financially supported by the Central Special Project for Fujian Local Science and Technology Development (2020L3025), the Fujian Natural Science Foundation (2020J01410), a Fujian Education and Scientific Research Project (JAT190789), the Talent-Recruiting Program of Wuyi University (YJ201503), the Open Fund of Fujian Provincial Key Laboratory of Eco-Industrial Green Technology (WYKF2019-2), a Chongqing Natural Science Foundation Project (cstc2021jcyj-msxmX0322), and the Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJQN202101246).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Xia, E. H., Zhang, H. B., Sheng, J., Li, K., Zhang, Q. J. et al. (2017). The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Molecular Plant, 10(6), 866–877. DOI 10.1016/j.molp.2017.04.002. [Google Scholar] [CrossRef]
2. Liu, C., Guo, Y., Sun, L., Lai, X., Li, Q. et al. (2019). Six types of tea reduce high-fat-diet-induced fat accumulation in mice by increasing lipid metabolism and suppressing inflammation. Food & Function, 10(4), 2061–2074. DOI 10.1039/c8fo02334d. [Google Scholar] [CrossRef]
3. Lim, H. J., Shim, S. B., Jee, S. W., Lee, S. H., Lim, C. J. et al. (2013). Green tea catechin leads to global improvement among Alzheimer’s disease-related phenotypes in NSE/hAPP-c105 Tg mice. Journal of Nutritional Biochemistry, 24(7), 1302–1313. DOI 10.1016/j.jnutbio.2012.10.005. [Google Scholar] [CrossRef]
4. Xu, Y., Zhang, Y., Quan, Z., Wong, W., Guo, J. et al. (2016). Epigallocatechin gallate (EGCG) inhibits alpha-synuclein aggregation: A potential agent for Parkinson’s disease. Neurochemical Research, 41(10), 2788–2796. DOI 10.1007/s11064-016-1995-9. [Google Scholar] [CrossRef]
5. Khan, N., Mukhtar, H. (2018). Tea polyphenols in promotion of human health. Nutrients, 11(1), 39. DOI 10.3390/nu11010039. [Google Scholar] [CrossRef]
6. Yang, C. S., Wang, X., Lu, G., Picinich, S. C. (2009). Cancer prevention by tea: Animal studies, molecular mechanisms and human relevance. Nature Reviews Cancer, 9(6), 429–439. DOI 10.1038/nrc2641. [Google Scholar] [CrossRef]
7. Unno, K., Yamada, H., Iguchi, K., Ishida, H., Iwao, Y. et al. (2017). Anti-stress effect of green tea with lowered caffeine on humans: A pilot study. Biological & Pharmaceutical Bulletin, 40(6), 902–909. DOI 10.1248/bpb.b17-00141. [Google Scholar] [CrossRef]
8. Zhou, Q., Chen, Z., Lee, J., Li, X., Sun, W. (2017). Proteomic analysis of tea plants (Camellia sinensis) with purple young shoots during leaf development. PLoS One, 12(5), e0177816. DOI 10.1371/journal.pone.0177816. [Google Scholar] [CrossRef]
9. Drew, L. (2019). The growth of tea. Nature, 566(7742), S2–S4. DOI 10.1038/d41586-019-00395-4. [Google Scholar] [CrossRef]
10. Ng, K. W., Cao, Z. J., Chen, H. B., Zhao, Z. Z., Zhu, L. et al. (2017). Oolong tea: A critical review of processing methods, chemical composition, health effects, and risk. Critical Reviews in Food Science and Nutrition, 58(17), 2957–2980. DOI 10.1080/10408398.2017.1347556. [Google Scholar] [CrossRef]
11. Chen, S., Li, M., Zheng, G., Wang, T., Lin, J. et al. (2018). Metabolite profiling of 14 Wuyi Rock tea cultivars using UPLC-QTOF MS and UPLC-QqQ MS combined with chemometrics. Molecules, 23(2), 104. DOI 10.3390/molecules23020104. [Google Scholar] [CrossRef]
12. Wang, C., Lv, S., Wu, Y., Gao, X., Li, J. et al. (2016). Oolong tea made from tea plants from different locations in Yunnan and Fujian, China showed similar aroma but different taste characteristics. SpringerPlus, 10(5), 576. DOI 10.1186/s40064-016-2229-y. [Google Scholar] [CrossRef]
13. Hayashino, Y., Fukuhara, S., Okamura, T., Tanaka, T., Ueshima, H. et al. (2011). High oolong tea consumption predicts future risk of diabetes among Japanese male workers: A prospective cohort study. Diabetic Medicine, 28(7), 805–810. DOI 10.1111/j.1464-5491.2011.03239.x. [Google Scholar] [CrossRef]
14. Iso, H., Date, C., Wakai, K., Fukui, M., Tamakoshi, A. et al. (2006). The relationship between green tea and total caffeine intake and risk for self-reported type 2 diabetes among Japanese adults. Annals of Internal Medicine, 144(8), 554–562. DOI 10.7326/0003-4819-144-8-200604180-00005. [Google Scholar] [CrossRef]
15. Wu, T., Xu, J., Chen, Y., Liu, R., Zhang, M. (2018). Oolong tea polysaccharide and polyphenols prevent obesity development in sprague-dawley rats. Food & Nutrition Research, 62, 1599. DOI 10.29219/fnr.v62.1599. [Google Scholar] [CrossRef]
16. Zhao, F., Lin, H. T., Zhang, S., Lin, Y. F., Yang, J. F. et al. (2014). Simultaneous determination of caffeine and some selected polyphenols in Wuyi Rock tea by high-performance liquid chromatography. Journal of Agricultural and Food Chemistry, 62(13), 2772–2781. DOI 10.1021/jf4056314. [Google Scholar] [CrossRef]
17. Wang, Y., Hao, X., Lu, Q., Wang, L., Qian, W. et al. (2018). Transcriptional analysis and histochemistry reveal that hypersensitive cell death and H2O2 have crucial roles in the resistance of tea plant (Camellia sinensis (L. ) O. kuntze) to anthracnose. Horticulture Research, 5, 18. DOI 10.1038/s41438-018-0025-2. [Google Scholar] [CrossRef]
18. Maharachchikumbura, S. S. N., Hyde, K. D., Groenewald, J. Z., Xu, J., Crous, P. W. (2014). Pestalotiopsis revisited. Studies in Mycology, 79, 121–186. DOI 10.1016/j.simyco.2014.09.005. [Google Scholar] [CrossRef]
19. Maharachchikumbura, S. S. N., Guo, L. D., Liu, Z. Y., Hyde, K. D. (2016). Pseudopestalotiopsis ignota and Ps. camelliae spp. nov. associated with grey blight disease of tea in China. Mycological Progress, 15, 22. DOI 10.1007/s11557-016-1162-3. [Google Scholar] [CrossRef]
20. Wang, S., Mi, X., Wu, Z., Wei, C., Zhang, L. (2019). Characterization and pathogenicity of Pestalotiopsis-like species associated with gray blight disease on Camellia sinensis in Anhui Province, China. Plant Disease, 103(11), 2786–2797. DOI 10.1094/PDIS-02-19-0412-RE. [Google Scholar] [CrossRef]
21. Wu, G., Liu, S., Zhao, Y., Wang, W., Kong, Z. et al. (2015). ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. The Plant Cell, 27(3), 857–873. DOI 10.1105/tpc.114.134668. [Google Scholar] [CrossRef]
22. Liu, F., Hou, L., Raza, M., Cai, L. (2017). Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Scientific Reports, 7(1), 866. DOI 10.1038/s41598-017-00972-5. [Google Scholar] [CrossRef]
23. Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C. et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. PNAS, 115(18), E4151–E4158. DOI 10.1073/pnas.1719622115. [Google Scholar] [CrossRef]
24. Xia, E. H., Li, F. D., Tong, W., Li, P. H., Wu, Q. et al. (2019). Tea plant information archive: A comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnology Journal, 17(10), 1938–1953. DOI 10.1111/pbi.13111. [Google Scholar] [CrossRef]
25. Xia, E. H., Tong, W., Hou, Y., An, Y., Chen, L. et al. (2020). The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into genome evolution and adaptation of tea plants. Molecular Plant, 13(7), 1013–1026. DOI 10.1016/j.molp.2020.04.010. [Google Scholar] [CrossRef]
26. Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods, 5(7), 621–628. DOI 10.1038/nmeth.1226. [Google Scholar] [CrossRef]
27. Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550. DOI 10.1186/s13059-014-0550-8. [Google Scholar] [CrossRef]
28. Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biology, 11(2), R14. DOI 10.1186/gb-2010-11-2-r14. [Google Scholar] [CrossRef]
29. Nozawa, S., Yamaguchi, K., Yen, L. H., Hop, D. V., Phay, N. et al. (2017). Identification of two new species and a sexual morph from the genus Pseudopestalotiopsis. Mycoscience, 58(5), 328–337. DOI 10.1016/j.myc.2017.02.008. [Google Scholar] [CrossRef]
30. Xin, Y., Wang, D., Han, S., Li, S., Gong, N. et al. (2021). Characterization of the chitinase gene family in mulberry (Morus notabilis) and MnChi18 involved in resistance to Botrytis cinerea. Genes, 13(1), 98. DOI 10.3390/genes13010098. [Google Scholar] [CrossRef]
31. Meng, X., Xu, J., He, Y., Yang, K. Y., Mordorski, B. et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. The Plant Cell, 25(3), 1126–1142. DOI 10.1105/tpc.112.109074. [Google Scholar] [CrossRef]
32. Tena, G., Boudsocq, M., Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Current Opinion in Plant Biology, 14(5), 519–529. DOI 10.1016/j.pbi.2011.05.006. [Google Scholar] [CrossRef]
33. Van Loon, L. C., Rep, M., Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology, 44(1), 135–162. DOI 10.1146/annurev.phyto.44.070505.143425. [Google Scholar] [CrossRef]
34. Djemal, R., Khoudi, H. (2021). The barley SHN1-type transcription factor HvSHN1 imparts heat, drought and salt tolerances in transgenic tobacco. Plant Physiology and Biochemistry, 164, 44–53. DOI 10.1016/j.plaphy.2021.04.018. [Google Scholar] [CrossRef]
35. Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology, 43, 205–227. DOI 10.1146/annurev.phyto.43.040204.135923. [Google Scholar] [CrossRef]
36. Sheng, C., Wang, Y., Ye, S. (2020). Discussion on the national park management mode under China’s national park system: A case of Wuyishan National Park System Pilot Area. International Journal of Geoheritage and Parks, 8(4), 230–234. DOI 10.1016/j.ijgeop.2020.11.004. [Google Scholar] [CrossRef]
37. Qian, Z., Oren, A., Wei, D., Chen, L., Liu, H. et al. (2018). A MYST histone acetyltransferase modulates conidia development and secondary metabolism in Pestalotiopsis microspora, a taxol producer. Scientific Reports, 8(1), 8199. DOI 10.1038/s41598-018-25983-8. [Google Scholar] [CrossRef]
38. Li, D., Bao, X., Yong, W., Ren, Y., Song, B. et al. (2019). First report of Lasiodiplodia theobromae causing leaf spot on tea plant in Guizhou Province of China. Plant Disease, 103(2), 374. DOI 10.1094/PDIS-06-18-1032-PDN. [Google Scholar] [CrossRef]
39. Thangaraj, K., Cheng, D., Cheng, L. L., Deng, W. W., Zhang, Z. (2018). Report of Phoma herbarum causing leaf spot disease of Camellia sinensis in China. Plant Disease, 102(11), 2373. DOI 10.1094/PDIS-01-18-0121-PDN. [Google Scholar] [CrossRef]
40. Otani, H., Kohnobe, A., Kodama, M., Kohmoto, K. (1998). Production of a host-specific toxin by germinating spores of Alternaria brassicicola. Physiological and Molecular Plant Pathology, 52(5), 285–295. DOI 10.1006/pmpp.1998.0147. [Google Scholar] [CrossRef]
41. Xie, J., Wei, J. G., Wang, K. W., Luo, J., Wu, Y. J. et al. (2020). Three phytotoxins produced by Neopestalotiopsis clavispora, the causal agent of ring spot on Kadsura coccinea. Microbiological Research, 238, 126531. DOI 10.1016/j.micres.2020.126531. [Google Scholar] [CrossRef]
42. Ke, X., Yin, Z., Song, N., Dai, Q., Voegele, R. T. et al. (2014). Transcriptome profiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genetics and Biology, 68, 31–38. DOI 10.1016/j.fgb.2014.04.004. [Google Scholar] [CrossRef]
43. Moore, J. W., Loake, G. J., Spoel, S. H. (2011). Transcription dynamics in plant immunity. The Plant Cell, 23(8), 2809–2820. DOI 10.1105/tpc.111.087346. [Google Scholar] [CrossRef]
44. Buscaill, P., Rivas, S. (2014). Transcriptional control of plant defence responses. Current Opinion in Plant Biology, 20, 35–46. DOI 10.1016/j.pbi.2014.04.004. [Google Scholar] [CrossRef]
45. de Souza, A. A., Takita, M. A., Coletta-Filho, H. D., Caldana, C., Goldman, G. H. et al. (2003). Analysis of gene expression in two growth states of Xylella fastidiosa and its relationship with pathogenicity. Molecular Plant-Microbe Interactions, 16(10), 867–875. DOI 10.1094/MPMI.2003.16.10.867. [Google Scholar] [CrossRef]
46. Macho, A. P., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Molecular Cell, 54(2), 263–272. DOI 10.1016/j.molcel.2014.03.028. [Google Scholar] [CrossRef]
47. Pitzschke, A., Schikora, A., Hirt, H. (2009). MAPK cascade signalling networks in plant defence. Current Opinion in Plant Biology, 12(4), 421–426. DOI 10.1016/j.pbi.2009.06.008. [Google Scholar] [CrossRef]
48. Wang, W., Feng, B., Zhou, J. M., Tang, D. (2020). Plant immune signaling: Advancing on two frontiers. Journal of Integrated Plant Biology, 62(1), 2–24. DOI 10.1111/jipb.12898. [Google Scholar] [CrossRef]
49. Asselbergh, B., de Vleesschauwer, D., Hofte, M. (2008). Global switches and fine-tuning-ABA modulates plant pathogen defense. Molecular Plant-Microbe Interactions, 21(9), 709–719. DOI 10.1094/MPMI-21-6-0709. [Google Scholar] [CrossRef]
50. Burger, M., Chory, J. (2019). Stressed out about hormones: How plants orchestrate immunity. Cell Host & Microbe, 26(2), 163–172. DOI 10.1016/j.chom.2019.07.006. [Google Scholar] [CrossRef]
51. Yang, D. L., Yao, J., Mei, C. S., Tong, X. H., Zeng, L. J. et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. PNAS, 109, E1192–E1200. DOI 10.1073/pnas.1201616109. [Google Scholar] [CrossRef]
52. Wang, S., Liu, L., Mi, X., Zhao, S., An, Y. (2021). Multi-omics analysis to visualize the dynamic roles of defense genes in the response of tea plants to gray blight. The Plant Journal, 106(3), 862–875. DOI 10.1111/tpj.15203. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools