

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.020372
REVIEW
Genetic Improvement of Betula platyphylla Suk. in China: A Review
1College of Forestry and Grassland, Jilin Agricultural University, Changchun, 130118, China
2State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, 150040, China
3Qiqihar Institute of Heilongjiang Academy of Forestry, Qiqihar, 161021, China
4Sanchazi Forestry Bureau of Jilin Province, Baishan, 134600, China
*Corresponding Authors: Xiaoqing Hu. Email: huxiaoqing@jlau.edu.cn; Xiyang Zhao. Email: zhaoxyphd@163.com
#This symbol represents that these three authors have contributed equally to this work and have been added to the manuscript
Received: 19 November 2021; Accepted: 18 January 2022
Abstract: Birch (Betula platyphylla Suk.), distributed in Eurasia, North America, and Australia, is a kind of cold-resistant, fast-growing, and vital pulpwood tree species. It is also one of the most important ecological restoration tree species with high values of economic benefits in Northeast China. To improve the genetic gain and expand the economic benefit of B. platyphylla, many genetic improvements have been carried out. In China, B. platyphylla is widely distributed and varied, and there are many varieties with excellent genetic characteristics. In this paper, the genetic improvement of B. platyphylla was reviewed, and the previous research results were discussed from two aspects: conventional breeding and molecular breeding. Some problems and corresponding solutions in the genetic improvement were put forward to provide ideas for B. platyphylla breeding in the future.
Keywords: Betula platyphylla; conventional breeding; molecular breeding; genetic improvement
Betula platyphylla Suk., one of the most extensively distributed broadleaf trees in northern and southwestern forest areas of China [1], has a high reproduction ability of seeds and a strong adaptability of seedlings. It is an important component species of natural secondary forest and nearly one-third of mountain areas are covered by it in Northeast China [2,3]. Because of high wood quality and fast growth rate, birch is also an important commercial tree species for papermaking, furniture making, construction, plywood, and handicrafts production [4].
The genetic improvement of B. platyphylla had been investigated for more than 40 years, which included elite materials selection [5], physiological index evaluation [6], wood properties evaluation [7], stress resistance evaluation [8], population genetic structure [9], and transgenic breeding [10]. This review covered the recent research progresses in genetic improvement of B. platyphylla in China, specifically focused on conventional breeding and molecular breeding. Meanwhile, new research directions were devised with different breeding goals for further research.
2 Research Progress on Conventional Breeding of B. Platyphylla
2.1 Germplasm Resources of B. Platyphylla
In order to save, produce and utilize comprehensively the species, the germplasm resources of B. platyphylla have been investigated and collected by the Northeast Forestry University (NEFU) over the past two decades. More than 30 provenances and 400 full-sibling, half-sibling families and clones of birch were collected from China, Finland, Canada, Russia, Kazakhstan and Japan, including B. pendula, B. kirghisorum, B. pendula ‘Purple Rain’, B. pendula ‘Dalecarlica’, B. populifolia and Japanese birch. In addition, 13 germplasm conservation test forests have been established in Northeast and North China. Based on the materials, the mechanism of marginal cleavage in B. pendula ‘Dalecarlica’ [11,12] and the variation of leaf colour in B. pendula ‘Purple Rain’ [13–15] were studied.
2.2 Elite Provenances Selection of B. Platyphylla
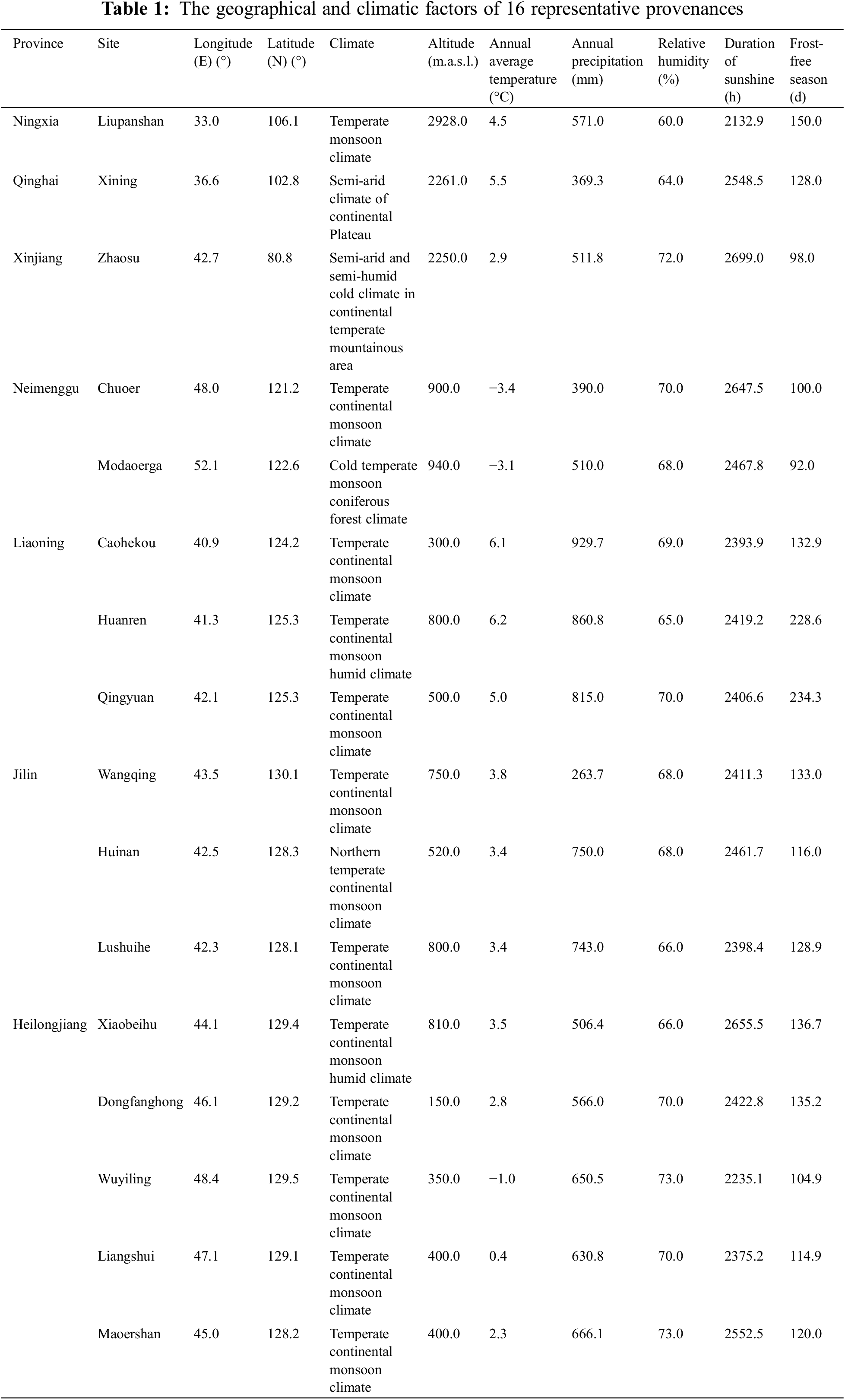
Due to the wide distribution of birch and environmental differences caused by geographic area, there were great differences between and within varieties. The longitude distribution range of birch in China is 42°07′(Zhaosu)∼130°01′(Wangqing), and latitude distribution range is 33°00′(Liupanshan)∼52°10′(Moerdaoga), where are mainly temperate continental monsoon climates (Table 1 and Fig. 1). Exploration and evaluation of 16 provenances in B. platyphylla were based on multiple breeding traits, and then Wangqing and Liangshui were preliminarily considered the optimal provenances [16]. Provenance tests were established in three different sites (Caohekou in Jilin Province, Maoershan in Heilongjiang Province and Jinhe in Mongolia autonomous region) in Northeast China in 1997 (Fig. 1). After that, further studies of provenance were conducted based on the above materials. For example, the height of B. platyphylla from different provenances was negatively correlated with latitude and positively correlated with longitude when the trees were 4-year-old [17]. Three excellent provenances for cellulose timber (Dongfanghong, Maoershan and Wuyiling) were identified from fourteen provenances of 10-year-old B. platyphylla in the Maoershan test site [18]. Growth traits and wood properties of eighteen B. platyphylla provenances in three trial sites were also investigated and analyzed, which suggested that there existed significant differences among different sites and also in site × provenance interaction. Ultimately, twelve elite provenances were selected based on growth traits including 6 provenances for Maoershan site, 3 for Caohekou site and 3 for Jinhe site, respectively [19].
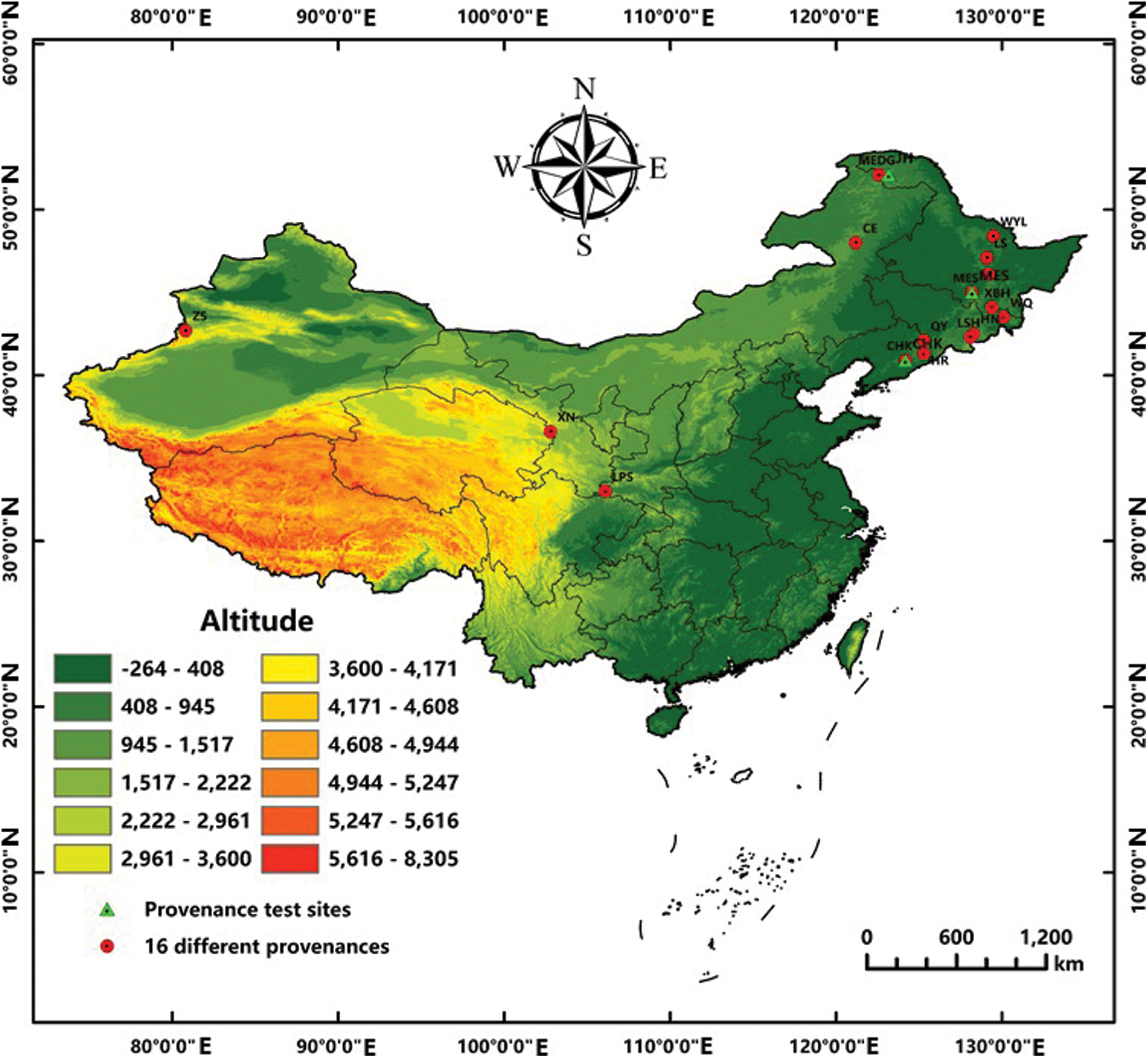
Figure 1: Distribution of 16 representative provenances of B. platyphylla in china and the provenance trial sites in Northeast China
Note: Red dots represent 16 representative provenance areas and the green triangles denote the provenance trial sites. LPS: Liupsan; XN: Xining; ZS: Zhaosu; CE: Chaoer; MEDG: Moerdaoga; CHK: Caohekou; HR: Huanren; QY: Qingyuan; WQ: Wangqing; HN: Huinan; LSH: Lushuihe; XBH: Xiaobeihu; DFH: Dongfanghong; WYL: Wuyiling; LS: Liangshui; MES: Maoershan; JH: Jinhe.
2.3 Elite Family Selection of B. Platyphylla
Selective breeding is usually required for tree genetic improvement to obtain superior clones and families. Due to the low survival rate of cutting and grafting in B. platyphylla, families and clones were important materials in the breeding research. Researchers made studies on the selection of elite families, which included higher yield, wood characteristics, and stress resistance of forest trees [5,20–22]. For instance, tree height and ground diameter were investigated among 21 half-sib families of 2-year-old B. platyphylla, and 8 elite families with higher tree height and 7 families with wider ground diameter were selected [21]. Based on 11 growth characters, 17 half-sib families of B. platyphylla were evaluated, and two families were selected as excellent families [5]. Growth traits of 53 half-sibling families in B. platyphylla at three different sites were investigated, including the height, timber volume and diameter at breast height. Thereafter, based on the breeding values, 11 families (B34, B15, B28, B16, B51, B40, B42, B45, B48, B35 and B19) were rated as superior families, whose mean timber volumes were 8.29%, 9.80% and 13.60% higher than the average of all the families planted in Langxiang, Maoershan and Jilin [22]. In addition, the growth traits of 37 B. platyphylla clones were evaluated; subsequently, seven superior clones were identified, whose mean height, ground diameter and number of lateral branches were 11.31%, 9.91% and 8.93% higher than the average of all the clones, respectively [23]. In another study, five superior clones of B. platyphylla were selected according to plant height, ground diameter and volume after fertilization [24].
Wood is one of the most dominant renewable resources for human beings [25], and its properties could be roughly divided into five categories: (1) Mechanical properties (e.g., longitudinal growth strain, modulus of elasticity, strength), (2) Technological characteristics (e.g., splitting index), (3) Physical properties (e.g., wood density, shrinkage), (4) Anatomical properties (e.g., fiber length, fiber thickness, coarseness), (5) Chemical properties (e.g., cellulose and lignin content, lignin composition) [26]. The estimated values for some wood properties of B. platyphylla are shown in Table 2 [7,27–29]. Wood properties (chemical properties of wood, fiber length and width, content of lignin, cellulose, and holocellulose) among 31 seed trees of tetraploid B. platyphylla were measured, which suggested that 19 tetraploid individuals with excellent wood properties can be used as the preferred variety of pulpwood [29]. In addition, other properties of wood such as the wood moisture content, air-dry density, oven-dry density, flexural strength, elastic modulus, shear strength, compressive strength parallel to the grain, and compressive strength stripes of B. platyphylla were also investigated, and 6 elite half-sib families in northeast China were deemed superior based on wood properties [7]. Regarding growth and wood properties, tree height, diameter at breast height, fiber aspect ratio, and hemicellulose content of 36 half-sib B. platyphylla families were researched, and the results indicated that 13 excellent families were identified [30].

2.4 Resistance Breeding of B. Platyphylla
Forest trees are vulnerable to natural and unnatural factors due to the complex climate, soil types, and complicated geographical environments. It is necessary to strengthen resistance breeding [31], including cold resistance, drought resistance, and saline-alkali resistance. Temperature is an essential environmental factor that seriously affects the growth, development, and geographical distribution of plants [32]. The enzyme recovery system of B. platyphylla was affected by low temperature; for example, some birches from warm climates in Europe were introduced and planted in higher latitudes/colder areas in China, and the growth and physiological characters were slightly lower than those of native B. platyphylla [33]. Promoted lignification and enhanced cold resistance of the seedlings were observed when the leaves of B. platyphylla were sprayed with potassium chloride, potassium dihydrogen phosphate, borax, and boric acid [34]. Drought is another important abiotic stress during tree growth and development [35] The drought tolerance of forest species has been investigated earlier. In 1973, Hsiao studied plants’ adaptation mechanisms to drought stress and put forward a specific standard for the water stress gradient in mesophytes. For B. platyphylla seedlings, the accumulation of malondialdehyde (MDA) decreased and the soluble sugar content significantly increased with the deepening of drought stress [36]. The leaves of 2-year-old B. platyphylla were sprayed with different concentrations of paclobutrazol (a kind of plant growth retardant), and then the drought resistances were evaluated by the membership function method. It was found that the drought resistance of seedlings treated with paclobutrazol was improved when the solution concentration was 750 mg L−1 [37].
Salt stress could inhibit plant growth and affect plant photosynthesis, protein synthesis, energy and fat metabolism [38]. There have been increasing studies on salt resistance of trees in recent years. Compared with other broadleaf species (such as Vlmus pumila, Fraxinus mandshurica and Tilia amurensis), the salt resistance of B. platyphylla is relatively weak by evaluating the plasma membrane permeabilities, which is an ideal indicator for studying salt resistance [39]. Introducing plants is also a good way to resistance breeding, and the salt-resistance of one-year seedlings of B. kirghisorum, B. pendula and B. pubescens brought from Kazakhstan, and the local B. platyphylla, were studied. The results showed that B. kirghisorum had the strongest resistance to neutral salt and B. kirghisorum and B. pubescens had more resistance to alkaline salt at high concentrations [40]. Moreover, growth traits and physiological characters from diverse B. platyphylla families showed significantly differences under salt stress, and the nine families with elite salt resistance were selected [41].
2.5 Ploidy Breeding of B. Platyphylla
Polyploidy is a ubiquitous phenomenon in higher plants [42,43]. The increase in chromosomes could result in increased gene dosages and cell volumes [44]. Therefore, polyploid plants usually have larger leaves, greater height and diameter, and increased ability to adapt to environment [45]. In order to obtain various ploidy materials, seeds of B. platyphylla were soaked in 0.1% colchicine, and 101 tetraploid individuals in total were obtained in the year of 2004 and 2009. Compared with diploids, these tetraploids showed larger leaf areas but shorter tree height during the seedling stages. Triploids were obtained by hybridization between diploid and tetraploid in B. platyphylla, and germination rate of the hybrid seeds were 3.33–45.33%; growth traits (tree height, leaf area and photosynthetic rate) of triploids were superior to those on diploids in current-year seedlings [46]. However, the growth traits (including tree height, diameter at breast height and volume) of 21 three-year-old B. platyphylla families from different ploidy level were investigated, which suggested that most diploid families were better than tetraploid and triploid families [47].
3 Research Progress on Molecular Breeding of B. Platyphylla
With the rapid development of modern molecular biotechnology and information technology, multiple breakthroughs have been made in plant genetic engineering and molecular breeding. Advanced breeding technologies have been increasingly implemented to improve critical commercial characteristics of plants (yield and resistance) [48]. Plant breeding in the world has gradually entered the molecular level, and the studies of genetics, genomics, molecular biology, and bioinformatics have been widely implemented. The conventional breeding methods are gradually transforming to molecular breeding methods. Molecular breeding represented by genetic engineering and molecular markers is gradually playing an essential role in plant breeding [49–51].
The Human Genome Project (HGP) [52] promoted the development of genome sequencing technology, which plays a crucial role in the field of modern genetic research. In 2000, the whole genome sequencing of Arabidopsis thaliana was accomplished by using the first generation of sequencing technology [53], which opened the door to the whole genome of plants. Afterward, the draft genome of Populus trichocarpa had been reported, which was the first woody plant to have its genome fully sequenced. There were important guiding significance for exploring the origin and evolution of woody plants, mapping and cloning of essential functional genes, and molecular marker-assisted selection. Until now, the whole genome sequencing has been completed on more than 40 woody plant species [54]. A whole-genome reference sequence for the diploid species B. nana (dwarf birch) was generated, which is the keystone woody species of subarctic scrub communities [55]. Then, silver birch (B. pendula) was sequenced and a reference genome from a fourth-generation inbred line was assembled, which showed high similarity with B. platyphylla [56]. Recently, researchers sequenced the B. platyphylla genome and assembled the sequences into 14 chromosomes, which facilitated the identification of important and essential genes governing important traits of trees and genetic improvement of B. platyphylla [57].
3.2 Molecular Marker-Assisted Breeding
Molecular markers are essential tools for genetic improvement, which can be applied to germplasm evaluation, genetic analysis and maker-assisted breeding. Multiple molecular markers were effectively used for genetic diversity, population structure and marker-trait association analysis of B. platyphylla. Random amplified polymorphic DNA (RAPD) is a convenient method to detect genetic diversity, which is based on the polymerase chain reaction (PCR) using randomly synthesized oligonucleotides (10 bp) as primers. The provenance division and genetic variation among 13 provenances of B. platyphylla in China were studied by using RAPD. The results showed that the differentiation of the percentage of polymorphic loci (PPL) among different provenances was evident, ranging from 20.17% to 32.19%. Among them, Maoershan provenance and Qingyuan provenance had the higher percentages of PPL; in contrast, Chuoer provenance had the lowest PPL percentage [58]. The fragment BFL significantly related to fiber length was selected from 100 B. platyphylla individuals with long-fiber using the RAPD technology. Then, BFL was successfully transformed into a sequence characterized amplified region (SCAR) marker by which the identification rate of long fiber B. platyphylla was more than 70% [59]. Using simple sequence repeat (SSR), four amplified fragments related to fiber length were identified in B. platyphylla [60]. In addition, genetic diversities of 41 white birch genotypes collected from 6 different geographical regions were analyzed using SSR markers. The result indicated that 111 selected SSR loci showed low to moderate similarity (0.025–0.610); by UPGMA-based clustering analysis of the allelic constitution, the six different geographical regions were further separated into four clusters: Cluster I, Huanren and Liangshui provenances; Cluster II, Xiaobeihu and Qingyuan provenances; Cluster III, Finland provenance; Cluster IV, Maoershan provenances [61]. Using ISSR (inter-simple sequence repeat) markers, 15 provenances from the provenance trial in Maoershan, and the result showed the range of the percentages of polymorphic loci was 48.03%∼62.20% [62]. Furthermore, some studies about genetic linkage maps of B. platyphylla were also based on molecular markers, such as ISSR and AFLP (Amplified fragment length polymorphism) [63].
3.3 Genetic Engineering Breeding
Genetic engineering has emerged as a method with high potential to modify traits more precisely and made functional genomics studies more efficiently. In the last decade, improving plant traits through genetic engineering has become an area of increased focus. Tree genetic engineering has advanced to the point at which genes for desirable traits can now be introduced and expressed efficiently [64,65]. Here, we describe major scientific discoveries on genetic engineering in terms of insect resistant, abiotic stress, wood property modification and flowering in B. platyphylla.
3.3.1 Insect-Resistant and Stress-Resistant Genetic Engineering
Unfortunately, B. platyphylla forests always were invaded by insects due to the abundance of secondary metabolites. It is an effectively pollution-free way to control plant pests by introducing insect-resistance genes. In 2001, a Chinese investigator obtained the first transgenic insect-resistant B. platyphylla by overpressing chimeric sequences of C peptide of Bt gene and spider insecticide peptide [66]; the transgenic birch could restrain the development of Lymantia dispar [67].
Researchers have found that there are many stress resistance genes in plants, and their functions could be studied in detail to understand the stress-resistant mechanism. In recent years, some resistance genes of B. platyphylla have been identified consecutively. The transgenic B. platyphylla plants with overexpressing BplMYB46 improved salt and osmotic tolerance by affecting gene expressions included SOD, POD and P5CS to increase reactive oxygen species scavenging and proline levels [68]. BpNAC012 positively activated the core sequence CGT(G/A) to induce SOD and POD genes, and the overexpression lines showed increased SOD and POD activities under salt and osmotic stress [69]. Moreover, the expression of BpARF1 was significantly up-regulated by drought stress, and the silencing of BpARF1 could improve the drought tolerance of B. platyphylla [70]. Compared with the wild type and BpERF11-overexpression lines, BpERF11 RNAi-silence in B. platyphylla showed increased ROS scavenging capability, reduced proline accumulation and enhanced water loss rate under salt and severe osmotic stress [71].
3.3.2 Genes Involved in Wood Formation
The properties of wood are determined by the composition and characteristics of the xylem secondary cell wall. B. platyphylla is one of the main pulpwood species; however, pulping yields for pulping industry were affected due to the high lignin content. Therefore, it is of great significance to study the lignin and cellulose biosynthesis for the genetic improvement of forest trees. The main mechanism of cellulose biosynthesis is that the thousands of glucose residues are combined into a full length chain by the glycosyltransferase. In the cDNA library of the B. platyphylla cambium, the gene for the glycosyltransferase was highly expressed during the important periods for wood formation [72]. Four Cellulose Synthase (CESA) genes from B. platyphylla were identified. BplCESA7 and BplCESA4 may be related to the formation of a cellulose synthase complex and participate mainly in secondary cell wall biosynthesis. BplCESA3 was possibly involved in primary cell wall biosynthesis and homogalacturonan synthesis [73]. Expression analysis of the cellulose synthase gene promoter in B. platyphylla has also been conducted. The promoter of BpCESA7 gene was cloned and the histochemical assays indicated that BpCESA7 may play an important role in growth and development of B. platyphylla [74]. Furthermore, studies of lignin synthesis genes have made a great progress recently. The Cinnamoyl-CoA reductase (CCR) gene was the key enzyme of the lignin-specific pathway, and its homologous gene BpCCR1 was isolated from B. platyphylla. Compared with WT, the higher lignin contents in overpressed BpCCR1 B. platyphylla seedlings, indicated that the BpCCR1 gene was related to the lignin synthesis [75]. BpCCoAOMT, isolated from B. platyphylla, played an important role in the precursor synthesis of G-lignin units. Compared with WT, the antisense BpCCoAOMT transgenic tobaccos showed the characteristics of decreased lignin content and reduced S-lignin content [76]. Besides enzyme genes, some transcription factors were reported to be involved in lignin synthesis. Ectopic expression of BpMYB4 in A. thaliana conferred lower lignin deposition and increased cellulose content [77]. Compared with the nontransgenic plants, the BpMADS12-overexpressing lines had higher lignin levels. There were differentially expressed genes involved in lignin and brassinosteroid biosynthesis, suggesting that BpMADS12 promoted the expression of lignin synthesis enzyme genes in response to brassinosteroid signaling [78]. In addition, the above-mentioned BpNAC012 and BplMYB46 were involved in the abiotic resistance of the regulated secondary wall biosynthesis as well [68,69]. For instance, BpNAC012 activated the expression of secondary wall-associated downstream genes (such as MYB46, MYB54, CCR1, 4CL1 and CesA3), resulting in ectopic secondary wall deposition in the transgenic stem epidermis.
Flowering is the transformation process from vegetative to reproductive growth and plays a vital role in life cycle of plants. However, B. platyphylla had a long juvenile period, which constrained breeding programs, and a few studies have focused on their floral development. The first reported case in birch flower development suggested that BpSPL1 (a SBP-box gene from B. pendula) was expressed in inflorescences as well as in shoots and leaves [79]. Subsequently, BplSPL1, the homologous gene of BpSPL1, was identified in B. platyphylla. Further studies showed that 35S::BplSPL1 transgenic A. thaliana had the early-flowering phenotype and the expression levels of flowering time genes and flower meristem identity genes had changed [80]. Early-flowering birches were generated by genetic transformation of 35S::BpAP1, and the inflorescences of the transgenic lines emerged miraculously beginning 2 months after transplanting. The male inflorescences rarely produced pollen, whereas the female inflorescences developed normally. The hybrid progeny by using the transgenic birch as female parent also completed flowering within only one year [81]. A similar early-flowering phenotype was observed in 35S::BpSEP4 transgenic A. thaliana, which changed the expression of flowering time genes and flower meristem identity genes. Moreover, the ectopic expression of BpSEP4 in A. thaliana caused aberrant floral organ development and delayed flower abscission [82].
In summary, a great deal of work has been done in China on the genetic improvement of B. platyphylla (Fig. 2), and some achievements have been made: (1) Many planting varieties with elite characteristics were cultivated through conventional breeding methods such as selection and hybridization. (2) Modern biotechnology has been widely used in the genetic improvement of B. platyphylla, and many transgenic B. platyphylla plants with stress resistance or economic traits have been bred. Nevertheless, there also have the following shortcomings: (1) Not enough attention has been paid to the utilization of B. platyphylla resources in many areas, and the B. platyphylla forest has been severely damaged. (2) The research on B. platyphylla in China started relatively late, most of the research results have not been widely used in production practice. (3) Breeding and spreading of excellent varieties are still at an early stage, and the economic benefits they reap have not been enough to produce a virtuous circle. (4) Despite the whole-genome sequencing of B. platyphylla has already been completed, transcriptome and metabonomics analyses for specific traits are still lacking.
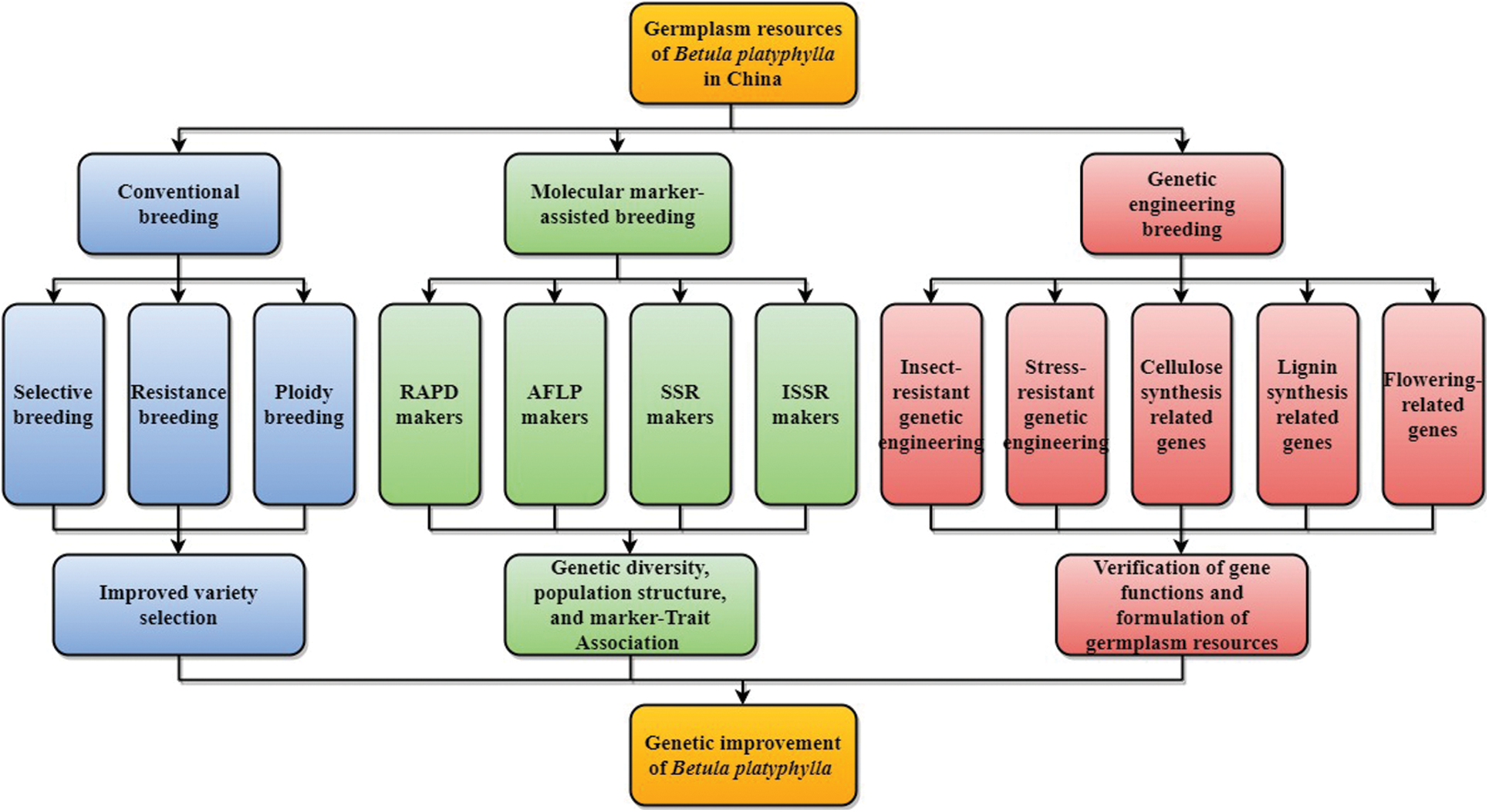
Figure 2: Genetic improvement progress of B. platyphylla in China
Given the above problems, further active measures should be taken to protect and utilize B. platyphylla resources in China (Fig. 3). (1) The genetic diversity of B. platyphylla should be protected, asafe utilization plan should be formulated, and the collection of germplasm resources should be strengthened. (2) The construction of intensive seed orchards for B. platyphylla should be strengthened for growth-promoting and shortening the generational cycle. (3) The breeding and promotion of B. platyphylla elite varieties should be strengthened, and attention should be paid to the cultivation of wood and greening varieties. (4) Further research involving multi-omics such as genomics, transcriptomics, and metabonomics should be developed to accelerate the breeding process of B. platyphylla.
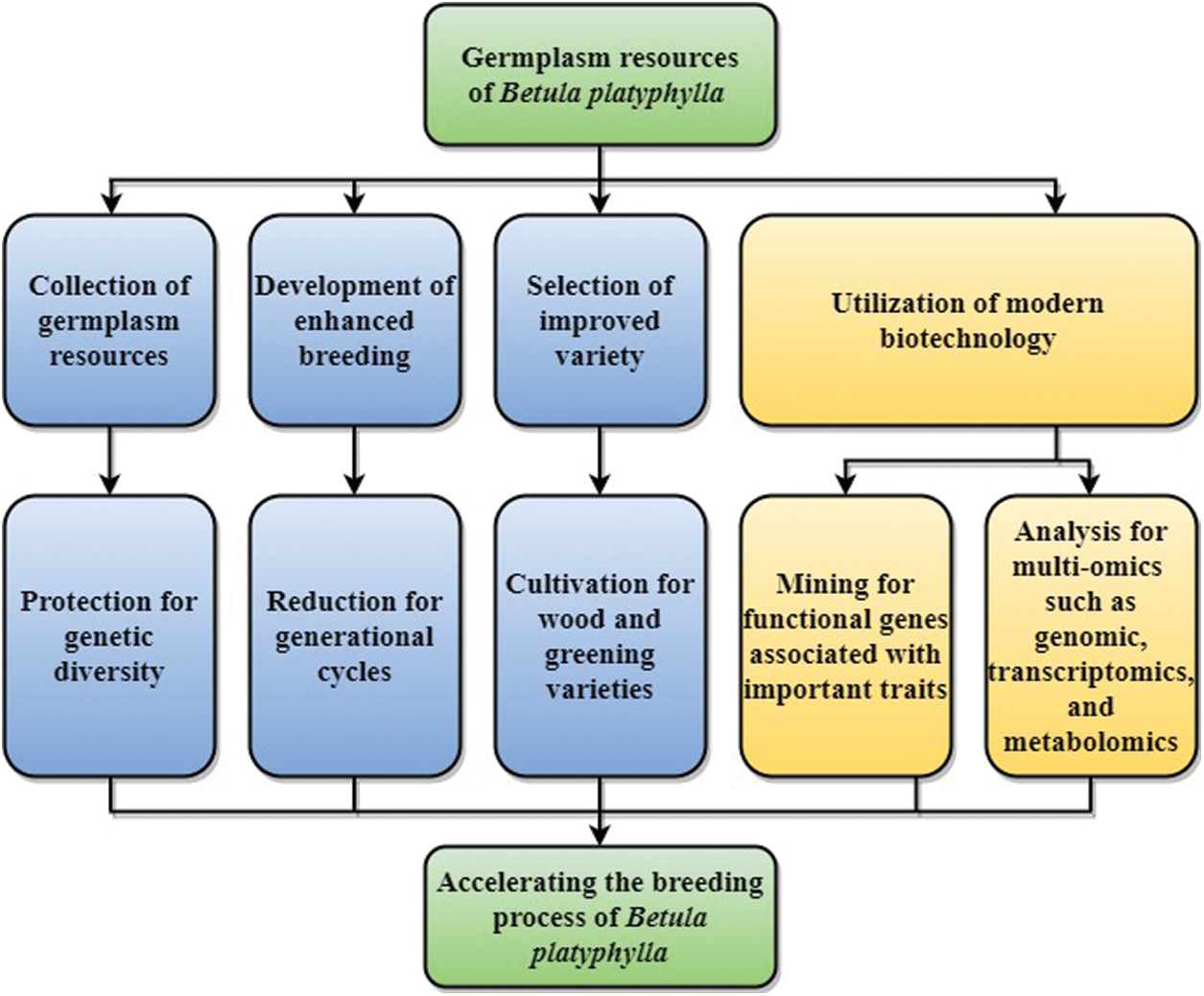
Figure 3: Genetic improvement strategies of B. platyphylla
Funding Statement: This research was found by the Scientific Research Start-Up Funds of Jilin Agricultural University (No. 2021002).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Jiang, J., Yang, G., Zhu, Z. B., Yang, Y. L., Yang, S. Z. (2011). Family selection from intensive seed orchard of Betula platyphylla. Journal of Northeast Forestry University, 39(1), 1–4. DOI 10.13759/j.cnki.dlxb.2011.01.018. [Google Scholar] [CrossRef]
2. Kitao, M., Lei, T. T., Nakamura, T., Koike, T. (2001). Manganese toxicity as indicated by visible foliar symptoms of Japanese birch (Betula platyphylla var. japonica). Environmental Pollution, 111(1), 89–94. DOI 10.1016/S0269-7491(99)00332-2. [Google Scholar] [CrossRef]
3. Yang, C. P. (2017). Study on the molecular mechanism of wood property variation and wood decay in Betula platyphylla population. China: Science Press. [Google Scholar]
4. Zhao, X. Y., Bian, X. Y., Liu, M. R., Li, Z. X., Zheng, M. et al. (2014). Analysis of genetic effects on a complete diallel cross test of Betula platyphylla. Euphytica, 200(2), 221–229. DOI 10.1007/s10681-014-1147-8. [Google Scholar] [CrossRef]
5. Li, Z. X., Li, R., Jiang, G. Y., Li, F., Liang, J. X. et al. (2019). Comprehensive evaluation of multiple-traits of Betula platyphylla half-sib family seedlings. Molecular Plant Breeding, 17(8), 2667–2674. DOI 10.13271/j.mpb.017.002667. [Google Scholar] [CrossRef]
6. Mijiti, M., Zhang, Y. M., Zhang, C. R., Wang, Y. C. (2017). Physiological and molecular responses of Betula platyphylla Suk to salt stress. Trees, 31(16), 1–13. DOI 10.1007/s00468-017-1576-9. [Google Scholar] [CrossRef]
7. Liang, D. Y., Zhang, X. X., Wang, C., Wang, X. W., Li, K. L. et al. (2018). Evaluation of Betula platyphylla families based on growth and wood property traits. Forest Science, 64(6), 663–670. DOI 10.1093/forsci/fxy027. [Google Scholar] [CrossRef]
8. Hoshika, Y., Watanabe, M., Inada, N., Mao, Q., Koike, T. (2013). Photosynthetic response of early and late leaves of birch (Betula platyphylla var. japonica) grown under free-air ozone exposure. Environmental Pollution, 182, 242–247. DOI 10.1016/j.envpol.2013.07.033. [Google Scholar] [CrossRef]
9. Jiang, T. B., Zhou, B. R., Gao, F. L., Guo, B. L. (2009). Genetic linkage maps of white birches (Betula platyphylla Suk. and B. pendula roth) based on RAPD and AFLP markers. Molecular Breeding, 27(3), 347–356. DOI 10.1007/s11032-010-9436-y. [Google Scholar] [CrossRef]
10. Zhang, W. B., Wei, R., Chen, S., Jiang, J., Li, H. Y. et al. (2015). Functional characterization of CCR in birch (Betula platyphylla × Betula pendula) through overexpression and suppression analysis. Physiologia Plantarum, 154(2), 283–296. DOI 10.1111/ppl.12306. [Google Scholar] [CrossRef]
11. Tian, S. L., Ma, Q., Wang, Y., Lin, X., Yang, Y. L. et al. (2019). Segregation of seed vigor and leaf traits in hybrid progenies of Betula pendula ‘Purple rain’ and Betula pendula ‘Dplecprlicp’. Forest Research, 32(3), 40–48. DOI 10.13275/j.cnki.lykxyj.2019.03.006. [Google Scholar] [CrossRef]
12. Qu, C., Bian, X. Y., Jiang, J., Chen, S., Liu, G. F. (2017). Leaf morphological characteristics and related gene expression characteristic analysis in Betula pendula ‘Dalecarlica’ and Betula pendula. Journal of Beijing Forestry University, 39(8), 9–16. DOI 10.13332/j.1000-1522.20160200. [Google Scholar] [CrossRef]
13. Lv, D. L., Lin, L., Guo, Y. W., Han, R., Jiang, J. (2018). Characterization of gene expression in anthocyanin synthesis and salt tolerance of Betula pendula ‘Purple rain’. Journal of Nanjing Forestry University (Natural Sciences Edition), 42(2), 25–32. DOI 10.3969/j.issn.1000-2006.201704048. [Google Scholar] [CrossRef]
14. Jiang, J., Wang, F., Lu, F., Jiang, J. (2017). Spatial and temporal content levels of anthocyanin and the expression characters of some related genes in Betula pendula. Journal of Southwest Forestry University (Natural Sciences), 37(2), 53–59. DOI 10.11929/j.issn.2095-1914.2017.02.009. [Google Scholar] [CrossRef]
15. Lin, L., Mu, H. Z., Jiang, J., Liu, G. F. (2013). Transcriptomic analysis of purple leaf determination in birch. Gene, 526(2), 251–258. DOI 10.1016/j.gene.2013.05.038. [Google Scholar] [CrossRef]
16. Jiang, J., Yang, C. P., Liu, G. F., You, X. L., Wang, Y. C. (1999). Provenance trial of Betula platyphylla Suk. in seedling stage. Journal of Northeast Forestry University, 27(6), 1–3. DOI 10.13759/j.cnki.dlxb.1999.06.001. [Google Scholar] [CrossRef]
17. Zhu, X., Yang, C. P., Zhu, Z., Pang, Z. H., Zhao, Y. et al. (2001). The geographic variance of provenance for Betula platyphylla in two years old. Journal of Northeast Forestry University, 29(6), 7–10. DOI 10.13759/j.cnki.dlxb.2001.06.003. [Google Scholar] [CrossRef]
18. Gao, Y. C., Wei, Z. G., Yang, C. P., Liu, G. F., Liu, G. J. (2009). A 10-year-old provenance trial of Betula platyphylla in the maoershan area of heilongjiang province. Journal of Zhejiang Forestry College, 26(6), 784–791. DOI 10.3969/j.issn.2095-0756.2009.06.004. [Google Scholar] [CrossRef]
19. Liu, Y., Xu, H. W., Shang, F. Q., Jiao, H., Zhang, L. M. et al. (2016). Variation and zoning of 16-year-old Betula platyphylla provenance. Scientia Silvae Sinicae, 52(9), 48–56. DOI 10.11707/j.1001-7488.20160906. [Google Scholar] [CrossRef]
20. Liu, Y., Xu, H. W., Bian, X. Y., Liu, G. F., Zhao, X. Y. (2013). Selection of seedling growth and photosynthetic traits assistant evaluated factors in Betula platyphylla half-sib families. Acta Botanica Boreali-Occidentalia Sinica, 33(5), 963–969. DOI 10.3969/j.issn.1000-4025.2013.05.015. [Google Scholar] [CrossRef]
21. Shang, F. Q. (2019). Estimation of genetic parameters of family for Betula platyphylla in two years old. Journal of Liaoning Forestry Science & Technology, 46(1), 32–33+66. DOI 10.3969/j.issn.1001-1714.2019.01.009. [Google Scholar] [CrossRef]
22. Liu, Y., Xu, H. W., Zhang, G. B., Wang, Y. J., Teng, W. H. et al. (2017). Multipoint growth trait test of half-sibling offspring and excellent family selection of Betula platyphylla. Journal of Beijing Forestry University, 39(3), 7–15. DOI 10.13332/j.1000-1522.20160154. [Google Scholar] [CrossRef]
23. Li, C. X., Liu, G. F., Liu, Y., Xu, H. W., Jiang, J. (2017). Preliminary seedling selection of superior clones of pot culture Betula platyphylla. Journal of Beijing Forestry University, 39(2), 16–23. DOI 10.13332/j.1000-1522.20160064. [Google Scholar] [CrossRef]
24. Song, S. Y., Cheng, S. Q., Jia, Z., Li, M. H., Ji, J. B. et al. (2020). Selection and optimum fertilization of Betula platyphylla hybrid clones for growth. Trees, 35(2), 469–478. DOI 10.1007/s00468-020-02049-9. [Google Scholar] [CrossRef]
25. Wildhagen, H., Paul, S., Allwright, M., Smith, H. K., Malinowska, M. et al. (2018). Genes and gene clusters related to genotype and drought-induced variation in saccharification potential, lignin content and wood anatomical traits in Populus nigra. Tree Physiology, 38(3), 320–339. DOI 10.1093/treephys/tpx054. [Google Scholar] [CrossRef]
26. Gion, J. M., Carouché, A., Deweer, S., Bedon, F., Pichavant, F. et al. (2011). Comprehensive genetic dissection of wood properties in a widely-grown tropical tree: Eucalyptus. BMC Genomics, 12(1), 1–19. DOI 10.1186/1471-2164-12-301. [Google Scholar] [CrossRef]
27. Wang, Q. Y., Qu, L. N., Jia, H. B. (2007). Variation of wood fiber characteristics microfibril angle and basic density of Betula platyphylla in natural populations. Journal of Northeast Forestry University, 35(2), 1–3+6. DOI 10.3969/j.issn.1000-5382.2007.02.001. [Google Scholar] [CrossRef]
28. Guo, M. H., Lu, Y., Wang, W. J., Cui, Y. Z. (1999). The radial variation pattens of wood density and ring width in different Betula platyphylla provenances. Journal of Northeast Forestry University, 27(4), 29–32. DOI 10.13759/j.cnki.dlxb.1999.04.007. [Google Scholar] [CrossRef]
29. Liu, C. Y., Liu, G. F., Fang, G. G., Jiang, C. M., Jiang, J. (2017). Comparison of tetraploid Betula platyphylla wood fiber traits and selection of superior seed trees. Journal of Beijing Forestry University, 39(2), 9–15. DOI 10.13332/j.1000-1522.20160091. [Google Scholar] [CrossRef]
30. Mu, H. Z., Liu, G. F., Jiang, J., Li, K. L., Zhu, Z. B. et al. (2009). Variations of growth and fiber properties of half-sib family progeny of Betula platyphylla. Journal of Northeast Forestry University, 37(3), 1–3+8. DOI 10.13759/j.cnki.dlxb.2009.03.027. [Google Scholar] [CrossRef]
31. Zhang, H. J., Zhao, Z. L. (2013). Innovative development of forest tree breeding resistance in the development of forestry. China Venture Capital, 16(30), 425–425. DOI 10.3969/j.issn.1673-5811.2013.30.397. [Google Scholar] [CrossRef]
32. Allen, D. J., Ort, D. R. (2001). Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Science, 6(1), 36–42. DOI 10.1016/S1360-1385(00)01808-2. [Google Scholar] [CrossRef]
33. Yang, D. H., Yang, M. S., Wang, J. M., Wang, S. L., Li, S. Y. et al. (2004). Europe birch’s membrane system changes under low temperature menace in the period of seedling. Journal of Northeast Forestry University, 32(6), 13–15. DOI 10.13759/j.cnki.dlxb.2004.06.005. [Google Scholar] [CrossRef]
34. Peng, X. Y., Suo, S. Q., Wang, W. D., Wang, H. W., Yan, H. Y. et al. (1996). Antifreeze effect of chemical drugs on Betula platyphylla seedlings. Science and Technology of Qinghai Agriculture and Forestry, (2), 60–62. [Google Scholar]
35. Wang, K. Y., Chen, F. Q., Huang, W. X. (2019). Research advance on drought stress response mechanism in plants. Journal of Agricultural Science and Technology, 21(2), 19–25. DOI 10.13304/j.nykjdb.2018.0115. [Google Scholar] [CrossRef]
36. Xue, J. P., Wang, Q. Y., Ren, H. (2007). Drought stress-induced responses of physiological indices of Betula platyphylla. Journal of Northeast Forestry University, 35(8), 12–15. DOI 10.3969/j.issn.1000-5382.2007.08.004. [Google Scholar] [CrossRef]
37. Jiang, Z. Z., Chen, X. W. (2006). Effect of paclobutrazol on drought resistance of Populusalba × Populus berolinensis, Ulmus pumila, and Betula platyphylla. Scientia Silvae Sinicae, 42(8), 130–134. DOI 10.3321/j.issn:1001-7488.2006.08.021. [Google Scholar] [CrossRef]
38. Serrano, R. (2002). Plants, genes and ions: Workshop on the molecular basis of ionic homeostasis and salt tolerance in plants. Embo Reports, 3(2), 116–119. DOI 10.1093/embo-reports/kvf030. [Google Scholar] [CrossRef]
39. Yang, C. P., Jiao, X. C., Liu, W. X., Liang, J. Y., Sheng, H. et al. (1997). The relationship of permeability of plasma membrane and salt resistance of wood plants. Journal of Northeast Forestry University, 25(1), 2–4. DOI 10.13759/j.cnki.dlxb.1997.01.001. [Google Scholar] [CrossRef]
40. Na, X. Y., Wang, X. W., Xu, H. Y., Qiao, L. N., Liu, G. F. et al. (2015). Analyzing and evaluating the salt tolerance of four kinds of birch seedlings. Bulletin of Botanical Research, 35(6), 873–882. DOI 10.7525/j.issn.1673-5102.2015.06.014. [Google Scholar] [CrossRef]
41. Yang, C. J., Liu, G. F., Zhang, X. H., Chen, J. Y., Liu, Y. Y. et al. (2013). Selection of salt-tolerant Betula platyphylla families under salt stress. Jiangsu Agricultural Sciences, 41(6), 139–142. DOI 10.15889/j.issn.1002-1302.2013.06.135. [Google Scholar] [CrossRef]
42. Masterson, J. (1994). Stomatal size in fossil plants: Evidence for polyploid in majority of angiosperms. Science, 264(5157), 421–424. DOI 10.1126/science.264.5157.421. [Google Scholar] [CrossRef]
43. Wendel, J. F. (2000). Genome evolution in polyploids. Plant Molecular Evolution, 42, 225–249. DOI 10.1023/a:1006392424384. [Google Scholar] [CrossRef]
44. Kang, X. Y. (2003). Advances in researches on polyploid breeding of forest trees. Journal of Beijing Forestry University, 25(4), 70–74. DOI 10.13332/j.1000-1522.2003.04.015. [Google Scholar] [CrossRef]
45. Qi, C. L., Jin, C. L., Li, K. L., Li, Z. X., Zhao, H. (2010). Comparison of photosynthetic characteristics and leaf anatomy structure of different ploidy Populus ussuriensis Kom. Plant Physiology Journal, 46(9), 917–922. DOI 10.13592/j.cnki.ppj.2010.09.022. [Google Scholar] [CrossRef]
46. Mu, H. Z. (1010). Inducement, growth and photosynthetic character study on polyploidy betula platyphylla (Master Thesis). Northeast Forestry University, Harbin. [Google Scholar]
47. Huang, H. J., Peng, R. S., Liu, Y. Y., Jiang, J. (2017). Growth traits variation analysis and family selection of 3-year-old various ploidy Betula platyphylla. Bulletin of Botanical Research, 37(2), 274–280. DOI 10.7525/j.issn.1673-5102.2017.02.016. [Google Scholar] [CrossRef]
48. Moose, S. P., Mumm, R. H. (2008). Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiology, 147(3), 969–977. DOI 10.1104/pp.108.118232. [Google Scholar] [CrossRef]
49. Baenziger, P. S., Russell, W. K., Graef, G. L., Campbell, B. T. (2006). Improving lives: 50 years of crop breeding, genetics, and cytology (C-1). Crop Science, 46(5), 2230–2244. DOI 10.2135/cropsci2005.11.0404gas. [Google Scholar] [CrossRef]
50. Varshney, R. K., Hoisington, D. A., Tyagi, A. K. (2006). Advances in cereal genomics and applications in crop breeding. Trends in Biotechnology, 24(11), 490–499. DOI 10.1016/j.tibtech.2006.08.006. [Google Scholar] [CrossRef]
51. Shao, L. T., Yin, S. P., Yan, D., Zhao, Y., Liu, T. T. et al. (2015). Transgenic research progress on genetic improvement in Betula platyphlla. World Forestry Research, 28(6), 29–33. DOI 10.13348/j.cnki.sjlyyj.2015.0020.y. [Google Scholar] [CrossRef]
52. Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J. et al. (2001). The sequence of the human genome. Science, 291(5507), 1304–1351. DOI 10.1126/science.1058040. [Google Scholar] [CrossRef]
53. Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M., Rooney, T. et al. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408(6814), 796–815. DOI 10.1038/35048692. [Google Scholar] [CrossRef]
54. Liu, H. L., Yin, T. M. (2018). Progress on the whole genome sequencing and the application in woody plants. Journal of Nanjing Forestry University (Natural Sciences Edition), 42(5), 172–178. DOI 10.3969/j.issn.1000-2006.201709020. [Google Scholar] [CrossRef]
55. Wang, N., Thomson, M., Bodles, W. J., Crawford, R. M. M., Hunt, H. V. et al. (2013). Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers. Molecular Ecology, 22(11), 3098–3111. DOI 10.1111/mec.12131. [Google Scholar] [CrossRef]
56. Salojarvi, J., Smolander, O., Nieminen, K., Rajaraman, S., Safronov, O. et al. (2017). Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nature Genetics, 49(6), 904–912. DOI 10.1038/ng.3862. [Google Scholar] [CrossRef]
57. Chen, S., Wang, Y. C., Yu, L. L., Zheng, T., Wang, S. et al. (2021). Genome sequence and evolution of Betula platyphylla. Horticulture Research, 8(1), 1–12. DOI 10.1038/s41438-021-00481-7. [Google Scholar] [CrossRef]
58. Jiang, J., Yang, C. P., Liu, G. F., Liu, Y. X., Ren, X. Q. (2001). Analysis of genetic variation within and among Betula platyphylla provenances in Northeast China using RAPD makers. Journal of Northeast Forestry University, 29(2), 30–34. DOI 10.3969/j.issn.1673-5102.2001.01.031. [Google Scholar] [CrossRef]
59. Wei, Z. G., Yang, C. P., Pan, H. (2006). Identification of molecular markers associated with birch fiber length trait by multiple regression analysis. Molecular Plant Breeding, 4(6), 835–840. DOI 10.3969/j.issn.1672-416X.2006.06.013. [Google Scholar] [CrossRef]
60. Wang, Y. M. (2007). Study on betula platyphylla long fiber trait by SSR maker technology (Master Thesis). Northeast Forestry University, Harbin. [Google Scholar]
61. Wei, H., Wang, S. J., Liu, H. J., Zhou, B. R., Wang, X. W. et al. (2015). Development of SSR markers and genetic diversity in white birch (Betula platyphylla). PLoS One, 10(4), e0125235. DOI 10.1371/journal.pone.0125235. [Google Scholar] [CrossRef]
62. Gao, Y. C. (2010). Genetic diversity of provenances of betula platyphylla (Master Thesis). Northeast Forestry University, Harbin. [Google Scholar]
63. Wei, Z. G., Zhang, K. X., Yang, C. P., Liu, G. F., Liu, G. J. et al. (2010). Genetic linkage maps of Betula platyphylla Suk. based on ISSR and AFLP markers. Plant Molecular Biology Reporter, 28(1), 169–175. DOI 10.1007/s11105-009-0138-8. [Google Scholar] [CrossRef]
64. Harfouche, A., Meilan, R., Altman, A. (2011). Tree genetic engineering and applications to sustainable forestry and biomass production. Trends in Biotechnology, 29(1), 9–17. DOI 10.1016/j.tibtech.2010.09.003. [Google Scholar] [CrossRef]
65. Lebedev, V. G., Shestibratov, K. A. (2021). Genetic engineering of lignin biosynthesis in trees: Compromise between wood properties and plant viability. Russian Journal of Plant Physiology, 68(4), 596–612. DOI 10.1134/S1021443721030109. [Google Scholar] [CrossRef]
66. Zhan, Y. G., Liu, Z. H., Wang, Y. C., Wang, Z. Y., Yang, C. P. et al. (2001). Transformation of insect resistant gene into birch. Journal of Northeast Forestry University, 29(6), 4–6. DOI 10.13759/j.cnki.dlxb.2001.06.002. [Google Scholar] [CrossRef]
67. Wang, Z. Y., Xue, Z., Fan, H. J., Zhan, Y. G. (2007). Resistance of transgenic Betula platyphylla to the defoliator Lymantria dispar. Scientia Silvae Sinicae, 43(1), 116–120. DOI 10.3321/j.issn:1001-7488.2007.01.020. [Google Scholar] [CrossRef]
68. Guo, H. Y., Wang, Y. C., Wang, L. Q., Hu, P., Wang, Y. M. et al. (2017). Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnology Journal, 15(1), 107–121. DOI 10.1111/pbi.12595. [Google Scholar] [CrossRef]
69. Hu, P., Zhang, K., Yang, C. (2019). BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiology, 179(2), 700–717. DOI 10.1104/pp.18.01167. [Google Scholar] [CrossRef]
70. Li, H. Y., Zhang, X., Tong, B. T., Wang, Y. C., Yang, C. P. (2020). Expression analysis of the BpARF genes in Betula platyphylla under drought stress. Plant Physiology and Biochemistry, 148, 273–281. DOI 10.1016/j.plaphy.2020.01.028. [Google Scholar] [CrossRef]
71. Zhang, W. H., Yang, G. Y., Mu, D., Li, H. Y., Zang, D. D. et al. (2016). An ethylene-responsive factor BpERF11 negatively modulates salt and osmotic tolerance in Betula platyphylla. Scientific Reports, 6, 1–13. DOI 10.1038/srep23085. [Google Scholar] [CrossRef]
72. Wang, C., Wang, Y. C., Diao, G. P., Jiang, J., Yang, C. P. (2010). Isolation and characterization of expressed sequence tags (ESTs) from cambium tissue of birch (Betula platyphylla Suk). Plant Molecular Biology Reporter, 28, 438–449. DOI 10.1007/s11105-009-0172-6. [Google Scholar] [CrossRef]
73. Liu, X. M., Wang, Q. Y., Chen, P. F., Song, F. N., Guan, M. X. et al. (2012). Four novel cellulose synthase (CESA) genes from birch (Betula platyphylla Suk.) involved in primary and secondary cell wall biosynthesis. International Journal of Molecular Sciences, 13(10), 12195–12212. DOI 10.3390/ijms131012195. [Google Scholar] [CrossRef]
74. Zhu, D. C., Hu, X. Q., Zhao, T., Tian, J., Zhang, Y. et al. (2018). Cloning and expression analysis of promoter of CESA7 gene in Betula platyphylla. Bulletin Botanical Research, 38(4), 551–558. DOI 10.7525/j.issn.1673-5102.2018.04.009. [Google Scholar] [CrossRef]
75. Wei, R. (2012). Gene clone and genetic transformation of cinnamoy1-CoA reductase gene 1 in betula platyphylla (Master Thesis). Northeast Forestry University, Harbin. [Google Scholar]
76. Zhang, Y., Hu, X. Q., Zheng, Y. Q., Liu, X. M. (2020). Ectopic expression of an antisense BpCCoAOMT gene from Betula platyphylla Suk. affects growth and development of tobacco due to lignin content reduction. Journal of Plant Biochemistry and Biotechnology, 29(2), 266–275. DOI 10.1007/s13562-019-00533-z. [Google Scholar] [CrossRef]
77. Yu, Y., Liu, H. Z., Zhang, N., Gao, C. Q., Qi, L. W. et al. (2021). The BpMYB4 transcription factor from Betula platyphylla contributes toward abiotic stress resistance and secondary cell wall biosynthesis. Frontiers in Plant Science, 11, 1–14. DOI 10.3389/fpls.2020.606062. [Google Scholar] [CrossRef]
78. Li, H. Y., Yang, Y., Wang, Z. J., Guo, X. H., Liu, F. F. et al. (2016). BpMADS12 gene role in lignin biosynthesis of Betula platyphylla Suk. by transcriptome analysis. Journal of Forestry Research, 27(5), 1111–1120. DOI 10.1007/s11676-016-0229-y. [Google Scholar] [CrossRef]
79. Lännenpää, M., Jänönen, I., Hölttä-Vuori, M., Gardemeister, M., Porali, I. et al. (2004). A new SBP-box gene BpSPL1 in silver birch (Betula pendula). Physiologia Plantarum, 120(3), 491–500. DOI 10.1111/j.0031-9317.2004.00254.x. [Google Scholar] [CrossRef]
80. Tian, J., Hu, X. Q., Zhang, Y., Xin, Q. Q., Li, D. et al. (2020). Molecular cloning and functional analysis of the BplSPL1 gene from Betula platyphylla Suk. Trees, 34(3), 801–811. DOI 10.1007/s00468-020-01959-y. [Google Scholar] [CrossRef]
81. Huang, H. J., Wang, S., Jiang, J., Liu, G. F., Li, H. Y. et al. (2013). Overexpression of BpAP1 induces early flowering and produces dwarfism in Betula platyphylla × Betula pendula. Physiologia Plantarum, 151(4), 495–506. DOI 10.1111/ppl.12123. [Google Scholar] [CrossRef]
82. Hu, X. Q., Tian, J., Xin, Q. Q., Li, D., Yao, L. M. et al. (2019). Cloning and functional characterization of a novel BpSEP4 gene from Betula platyphylla Suk. Tree Genetics & Genomes, 16(1), 1–11. DOI 10.1007/s11295-019-1405-y. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |