

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018435
ARTICLE
Molecular Basis of Unique Branching Phenotypes in Salvia splendens and the Role of PSY
1College of Humanities and Urban-Rural Development, Beijing University ofAgriculture, Beijing, 102206, China
2College of Landscape Architecture, Beijing University of Agriculture, Beijing, 102206, China
3Beijing Collaborative Innovation Center for Eco-Environmental Improvement with Forestry and Fruit Trees, Beijing, 102206, China
4Beijing Engineering Research Center of Rural Landscape Planning and Design, Beijing, 102206, China
*Corresponding Author: Hongwei Chen. Email: chenhongwei2016@sina.com
#Co-first author
Received: 24 July 2021; Accepted: 29 November 2021
Abstract: The branching system of higher plants plays a very important role in plant morphogenesis, and the number of branches can directly affect crop yield and the ornamental value of plants. It is a complicated development process involving complex molecular mechanisms. The ‘Cailinghong’ variety of Salvia splendens is characterized by its great branching ability with the ability to grow into a spherical form naturally, without pinching. To gain insight into the molecular events during the branching development of S. splendens, suppressive subtractive hybridization (SSH) technology was used to screen differentially expressed genes between the erect plant type (strain 35) and the spherical plant type (‘Cailinghong’). In total, 96 and 116 unigenes were annotated. Four and eight unigenes up-regulated in ‘Cailinghong’ and strain 35, respectively, were associated with plant hormone anabolism and signal transduction, suggesting that they participate in the branching process. One of these genes, phytoene synthase (PSY), is a precursor of the new plant hormone group strigolactones. Using the PSY fragment (192 bp) as a template, the cDNA sequence of PSY in S. splendens was cloned and named SsPSY. A relative expression analysis and transgenic test results indicated that SsPSY plays an important role in lateral branch development in ‘Cailinghong’. These results provide new insight into the molecular mechanisms underlying branching in S. splendens.
Keywords: Lateral branch development; differential gene expression; Salvia splendens; suppression subtractive hybridization library; SsPSY
Resulting from the development and expansion of the terminal bud and axillary bud, branching is a complicated developmental trait in plants. Branching is a ubiquitous process in plant growth. The branching system of higher plants plays a very important role in plant morphogenesis. The number of branching events and tillering of food crops affect the seed setting rate and thus grain yield. In the process of maize cultivation, yield can be increased by reducing the number of branches. The tiller number of rice affects the yield. Branches of ornamental plants can be used as an important index to evaluate their ornamental value. In general, more branches are associated with more flowers, and a relatively compact plant form. However, in chrysanthemum, peonies and other ornamental plants, reduced branching can increase the flower size, and improve the ornamental value of flowers. Therefore, comprehensive experimental studies of the mechanism underly plant branching for controlling plant branching fundamentally, increase the yield of food crops and improve the value of ornamental plants.
Often, growing shoot tips influence shoot branching by inhibiting axillary bud formation, a phenomenon known as apical dominance, while the plant itself promotes axillary bud growth and new branches via complex signals involving hormones (auxin, strigolactones, and cytokinins), nutrients, and environmental factors [1]. Extensive research on plant physiology and molecular biology has improved our understanding of the mechanisms underlying plant branching based on in-depth studies of model wild-type and mutant plants.
In molecular biology, scientists have discovered a series of genes related to the growth and development of plant branching. These genes can be divided into five categories, CCD (carotenoid cleavage dioxygenase) family genes, GRAS (GAI (Gibberellin-insensitive), RGA (repressor of ga1–3), SCR (scarecrow)) family genes, bHLH (basic helix-loop-helix) transcription factor genes, cytochrome P450 family genes, and other genes that influence branching. Branched1 (BRC1), a member of the TCP (TB1 (TEOSINTE BRANCHED1), CYC (CYCLOIDEA), PCFs (PROLIFERATING CELL FACTORS)) transcription factor family, is a key factor in the regulation of axillary bud activity in the model plant Arabidopsis thaliana [2]. BRC1/TB1 regulates branching not only in A. thaliana, but also in many other important crops, such as tomato, pea, potato, and maize [3–6]. BRC1/TB1 is regulated at the transcriptional level by a variety of internal and external factors that affect branchings, such as phytochrome and cytokinins, and thus plays a central role in branching regulation. Despite research progress related to branching mutants, studies of additional plant mutants are needed owing to the complexity of the underlying molecular mechanism.
With respect to plant physiology, researchers initially thought that the synergistic effects of plant hormones co-regulate the development of plant branches, in which auxin and cytokinin play critical roles. Recently, a new class of hormones that inhibits plant branching, strigolactones (SLs), was discovered in 2008 [7,8]. Recent research suggests that plant branching is cooperatively controlled by auxin, cytokinin, and the newly discovered SLs. Subsequent research has clarified the biosynthesis, detection, and signal transduction pathways of SLs [9–11]. SLs are synthesized from β-carotene, which is converted into carlactone (CL) and transformed into a series of SLs [12,13].
Carotenoids are precursors of both SLs and abscisic acid by oxidase cleavage [14–16]. The main genes involved in the biosynthesis of carotenoids include DXS (1-deoxy-D-xylulose 5-phosphate synthase), DXR (1-deoxy-D-xylulose 5-phosphate reductoisomerase), and PSY (Phytoene synthase). PSY plays a key role in the first step in carotenoid biosynthesis and can control the flux of carotenoids [17,18]; it has become the primary gene target for improving the carotenoid content in plants. As a precursor for SL synthesis, PSY also plays a decisive role in the regulation of SL levels in plants [19].
In addition to hormones, nutrients and environmental factors are important determinants of the formation and development of plant branching. The growth relationship between shoot tips and axillary buds may be related to their competition for sugar and energy, and sucrose can induce branching by promoting cytokinin accumulation and vacuolar invertase activity [20,21]. Under dense planting conditions, the phototransduction transcription factors FHY3 and FAR1 synergically regulate branching formation by integrating the external light signaling pathway and the internal strigolactone signaling pathway [22].
Salvia splendens, also known as western red and scarlet sage, belongs to the family Lamiaceae and is characterized by bright color and a long bloom period. It is regarded as an essential plant material in flower decoration and parterre, with a very important role in Chinese landscaping. During its cultivation process, manual pinching is required to suppress the apical dominance and promote branching. Compared with the normal erect plant type, the ‘Cailinghong’ variety is a multi-branching type, with a natural spherical structure without pinching, early branching, and increased branch series.
In this study, candidate branching-related genes were identified by comparing the transcriptomes of the lateral buds between strain 35 (erect plant type) and ‘Cailinghong’ (spherical plant type, multi-branching), which further cloning and functional analyses of the PSY gene. These results provide a new perspective for studies of the molecular mechanism underlying the regulation of plant branching.
The ‘Cailinghong’ variety and strain 35 breeds by our research group were used as test materials. Seeds of the tested varieties were planted in a greenhouse. After about 80 days of sowing, elongation of the primary lateral branch of the seedling began, and the top bud of the primary lateral branch at the second or third nodes of the tested varieties was obtained as test materials for SSH library construction and the cloning of SsPSY. Stem nodes, root tips, and lateral buds before the formation of lateral branches (70 days of sowing, the lateral bud did not germinate) and after the formation of lateral branches (90 days of sowing, the lateral buds elongate to 5 cm) were used as experimental materials for qRT-PCR.
2.2 Morphological Characteristics of Strain 35 and ‘Cailinghong’
The tested varieties were grown in a greenhouse under a temperature of 25°C and 14 h of light a day. The morphology, plant height, and length of each branch were observed and measured regularly after sowing for 3 months. Measurements continued until 5 months after sowing.
Total RNA was extracted using the TRIzol Reagent Kit (Invitrogen, Carlsbad, USA). mRNA was separated from total RNA using a PolyA Tract mRNA Isolation System III (Promega, Madison, USA).
The purified mRNA was reverse transcribed into cDNA. Then cDNA of the tester and driver was purified and digested with RsaI. The digested tester cDNA was ligated with Adaptor 1 and Adaptor 2, which were provided in the kit. Subsequently, two rounds of hybridization were performed on the tester cDNA and excessive driver cDNA. After the hybridization product was diluted, two rounds of specific PCR were performed for the enrichment of differentially expressed sequences. All operations were performed according to the operation manual of the Clontech PCR-Select cDNA Subtraction Kit (Mountain View, CA, USA). The purified and concentrated SSH products were directly inserted into the PMD18-T Vector (TaKaRa, Dalian, China) and transformed into TOP10 competent cells. The competent cells were cultured in LB culture medium containing Amp/X2gal/IPTG, blue/white color screening was performed, and SSH libraries were constructed. The inserted element in each clone was confirmed by PCR. The positive clones were picked for sequencing. The sequencing was completed by Sangon Biotech Co., Ltd., Shanghai and M13 was used as the sequencing primer.
2.4 Clone Sequencing and Analysis
Briefly, the cDNA sequence was used for a search against the GenBank database using BLAST 2.2.26+ for a nucleotide sequence comparison (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The matching degree between the submitted sequence and target sequence was obtained by matrix scoring. The identity value was used to evaluate the degree of matching and similarity between the input sequence and its homolog in the database.
Based on the 192-bp sequence fragment of PSY obtained by SSH, two specific primers for 3′ RACE Outer and Inner were designed using Premier 5.0. The 3′ end of the cDNA sequence of SsPSY was obtained using a SMARTer® RACE 5′/3′ Kit (TaKaRa). According to the 3′ cDNA sequence, the specific primers for 5′ RACE Outer and Inner were designed, and the 5′ end of the cDNA sequence was obtained using the SMARTer® RACE 5′/3′ Kit (TaKaRa).
The 3′-end and 5′-end sequences were combined using DNAMAN 6.0 to obtain the full cDNA sequence of SsPSY. ORF Finder was used to search the largest open reading frame (ORF) online and to find the positions of the initiation codon and termination codon. Primers for full-length sequence amplification were designed with the initiation codon and the termination codon as the outer ends. Primers are shown in Table 1.
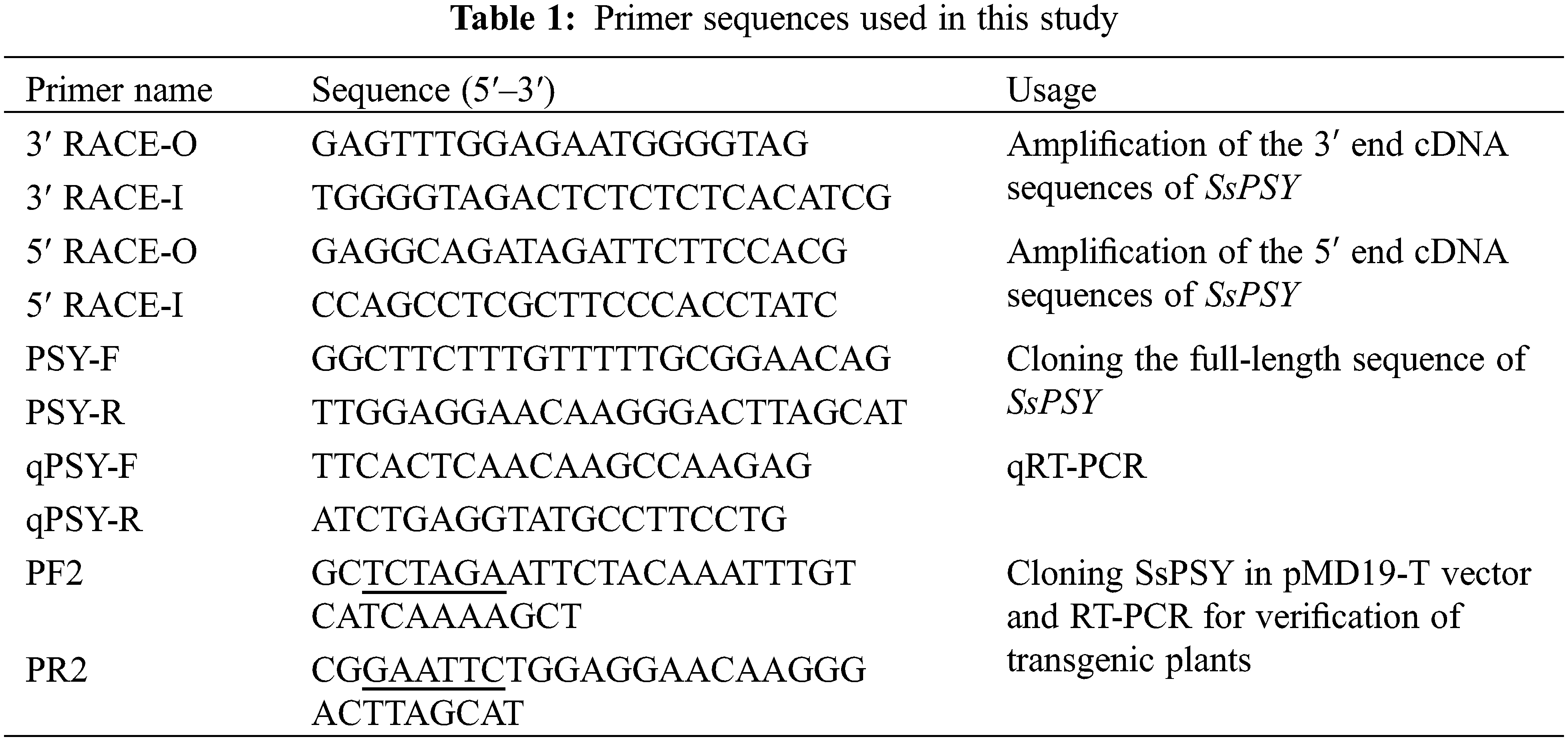
2.6 Bioinformatics Analysis of SsPSY
The full-length cDNA sequence obtained by cloning was used to infer the amino acid sequence, and a homology analysis was conducted using the BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST/). An ORF analysis was conducted using ORF Finder. DNAMAN 6.0 was used to generate a multiple sequence alignment and to perform a phylogenetic analysis.
2.7 Relative Expression Analysis of SsPSY
Total RNA was extracted from different tissues in the tested varieties at different periods using the SV Total RNA Isolation System Kit (Promega) according to the manufacturer’s instructions. Subsequently, 1 μg of RNA was transcribed into cDNA using the Reverse Transcription System Kit (Promega), and the quality of cDNA was verified using 18S rRNA-specific primers. Amplification was performed using the SYBR Premix Ex Taq II (Perfect Real Time) Kit (Takara), as per the manufacturer’s instructions. The qPCR data were obtained from three biological replicates. Three repetitions were prepared for each sample, and each experiment was conducted 4–6 times.
The expression of SsPSY in strain 35 was used as the test group, the expression of SsPSY in ‘Cailinghong’ was used as a control group, and 18S was used as an internal reference gene. The relative expression levels of target genes were determined using by the 2-ΔΔCT method. The primers pairs for qPCR are shown in Table 1.
2.8 Construction of SsPSY Plant Expression Vector and Plant Transformation
Primers containing the enzyme cleavage site sequence were redesigned to amplify the ORF of SsPSY. The primer pairs are listed in Table 1. The SsPSY ORF was introduced into the binary vector PMON721 using XbaI and EcoRI restriction sites. Next, the PMON-PSY recombinant vector was transformed into Agrobacterium strain EHA105 via the freeze-thaw method, and Agrobacterium-mediated transformation of Arabidopsis was performed by the floral dip method. Seeds harvested by the transformed Arabidopsis were disinfected with a 2% sodium hypochlorite solution and sown to 1/2MS medium supplemented with 120 mg/L kanamycin for positive plant screening. Positive seeds can germinate normally and grow into plants on medium. After the positive plants grew true leaves, total DNA was extracted from the leaves of the primary positive plants. PCR amplification was performed with specific full-length primers, and all transgenic plants were subjected to amplification. Positive plants were transferred into nutrient soil under normal conditions. One month after transplantation, branch phenotypes of T1 generation plants were observed.
3.1 Branch Growth in ‘Cailinghong’ and Strain 35
By observing the growth of the tested varieties (Figs. 1–3), we found that ‘Cailinghong’ was much shorter (mean ± S.D.: 15.944 ± 2.010 cm) than strain 35 (40.900 ± 9.795 cm), and the stem stretched to about 7–9 nodes in ‘Cailinghong,’ compared with 13–15 nodes in strain 35 after sowing for 5 months. During the growth process of the ‘Cailinghong’ variety, after the stem stretched to 2–3 nodes, lateral bud growth and elongation began from lower to upper nodes. The lateral buds at the upper nodes of the lateral branch exhibited continual growth and extension. In contrast, during the growth process of strain 35, the lower nodes started budding partly without elongation after the stem stretched to about 8–9 nodes.

Figure 1: Plant height of ‘Cailinghong’ and strain 35

Figure 2: Branching characteristics of ‘Cailinghong’ (breeding number 35–1) and strain 35 (A–B: 3 months after sowing; C–D: 4 months after sowing; E–F: 5 months after sowing)

Figure 3: Lateral branch growth of ‘Cailinghong’ and strain 35 (A: 3 months after sowing; B: 4 months after sowing; C: 5 months after sowing)
After sowing for 3 months, the ‘Cailinghong’ variety formed a spherical multi-branch structure. The longest lateral branch was at the first node, with a length of 3.480 ± 0.831 cm. With an increase in nodes, the length of the lateral branch successively decreased, allowing the plant to take a spherical form. However, the longest lateral branch of strain 35 was from the fourth node, at only 0.200 ± 0.082 cm, and the length of the lateral branch decreased gradually with an increase or decrease in the number of nodes from the fourth node (Figs. 2A, 2B and 3A).
After blooming (5 months after sowing, Figs. 2E, 2F and 3C), the growth of the lateral branches of ‘Cailinghong’ was similar to that observed at 3 months after sowing. Lateral branches at the first or second nodes grew the longest, i.e., up to 12.105 ± 1.455 cm, and the length of the lateral branches decreased as the number of nodes increased. The pattern of lateral growth of strain 35 was quite different; the lateral buds at nodes 3–6 under the flower showed elongation to form the lateral branch, with a length of 5.744 ± 3.918 to 8.328 ± 4.421 cm. However, the lateral buds at other nodes of the stem only stretched to 2–3 nodes and never showed elongation growth to form the lateral branch.
3.2 EST Sequence Analysis of the SSH Library
We compared the lateral bud transcriptomes of the erect plant type 35 (optional strain) and the ‘Cailinghong’ variety (spherical plant type, multi-branching) by SSH analyses and screened branching-related genes that were up-regulated in ‘Cailinghong’ (forward library) and strain 35 (reverse library).
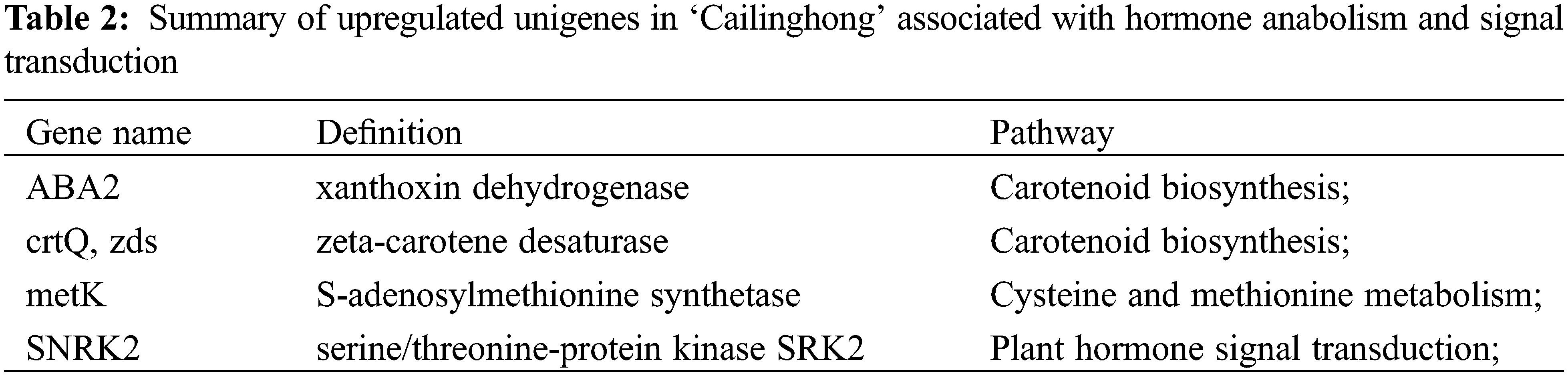
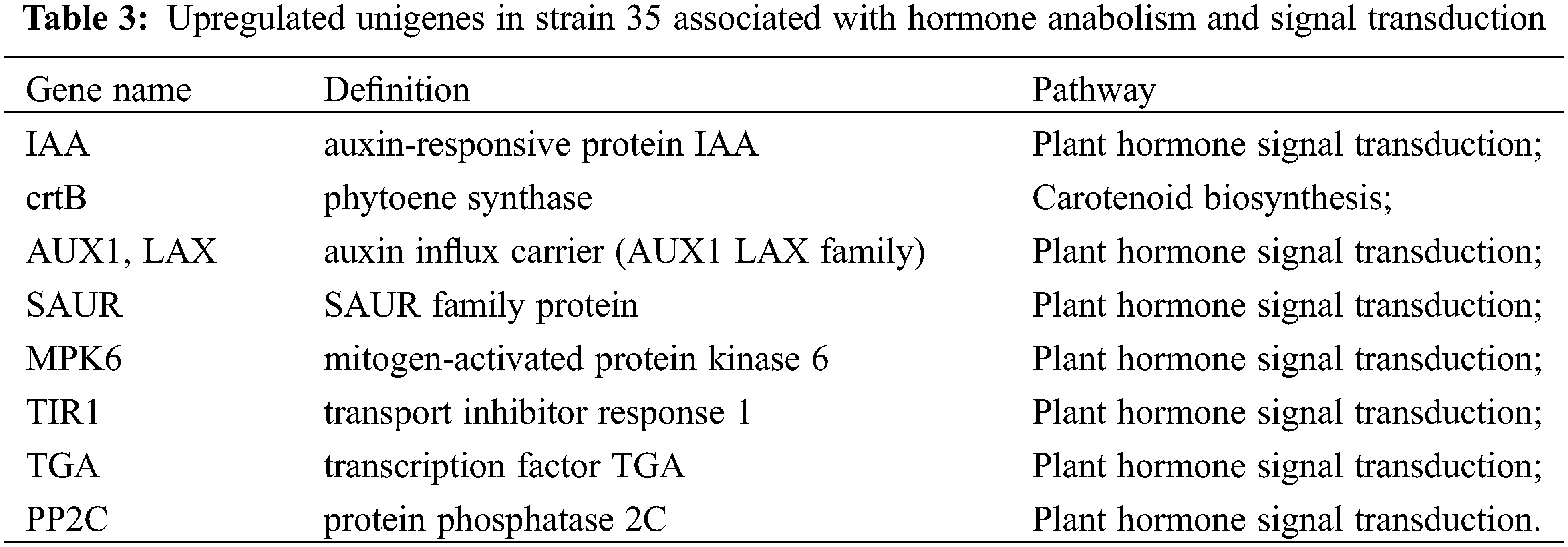
Using blue-white colony screening technology, we selected 570 clones and 645 clones from the forward and reverse libraries for sequencing. We obtained sequence data for 523 clones and 602 clones, accounting for 91.75% and 93.33% of all submitted clones. By Blast searches, 96 and 116 unigenes in the forward library (up-regulated in ‘Cailinghong’) and the reverse library (up-regulated in strain 35), respectively, matched to homologous sequences in the NCBI nr database. Among the up-regulated unigenes, four (forward library) and eight (reverse library) unigenes were closely related to plant hormone synthesis or signal transduction. Unigenes associated with hormone synthesis and signal transduction that were up-regulated in ‘Cailinghong’ were mainly related to abscisic acid and SL anabolism (ABA2 and crtQ, also named zds), ethylene anabolism (metK), and plant hormone signal transduction (SNRK2) (Table 2). Unigenes associated with hormone synthesis and signal transduction that were up-regulated in strain 35 were mainly associated with abscisic acid and SL anabolism (crtB , also named PSY) and plant hormone signal transduction (IAA, AUX1, SAUR, MPK6, TIR1, TGA, and PP2C) (Table 3).
3.3 Full-sequence Cloning and Bioinformatics Analysis of SsPSY
Among the 71 up-regulated unigenes obtained from the reverse library (up-regulated in strain 35) related to branching in the ‘Cailinghong’ variety, we obtained one gene sequence fragment encoding phytoene synthase (PSY), which contributes to the synthesis of carotenoids. The SL contents in the roots of dicotyledonous plants are determined by the PSY gene family, and extremely low levels of PSY result in a substantial reduction in SLs in plants [19]. It can be inferred that the PSY gene also plays an important role in the branching of plants. To determine if and how SsPSY regulates the development of the branching in ‘Cailinghong’, the full-length cDNA sequence was cloned by RACE and a bioinformatics analysis was performed.
The SsPSY (GenBank accession number MN548835) cDNA sequence was 1523 bp. It included an 86-bp 5′ untranslated region (UTR) and a 214-bp 3’ UTR. The ORF was 1245 bp. The deduced protein sequence was 414 amino acids. It had the highest homology with PSY from Pogostemon cablin (AHJ90431.1) based on a BLAST search, with an identity greater than 78.59%. SsPSY also had a high degree of homology with homologues in other common plant species, such as Sesamum indicum (77.68%), Osmanthus fragrans (75.8%), and Nicotiana tabacum (72.34%) (Fig. 4).
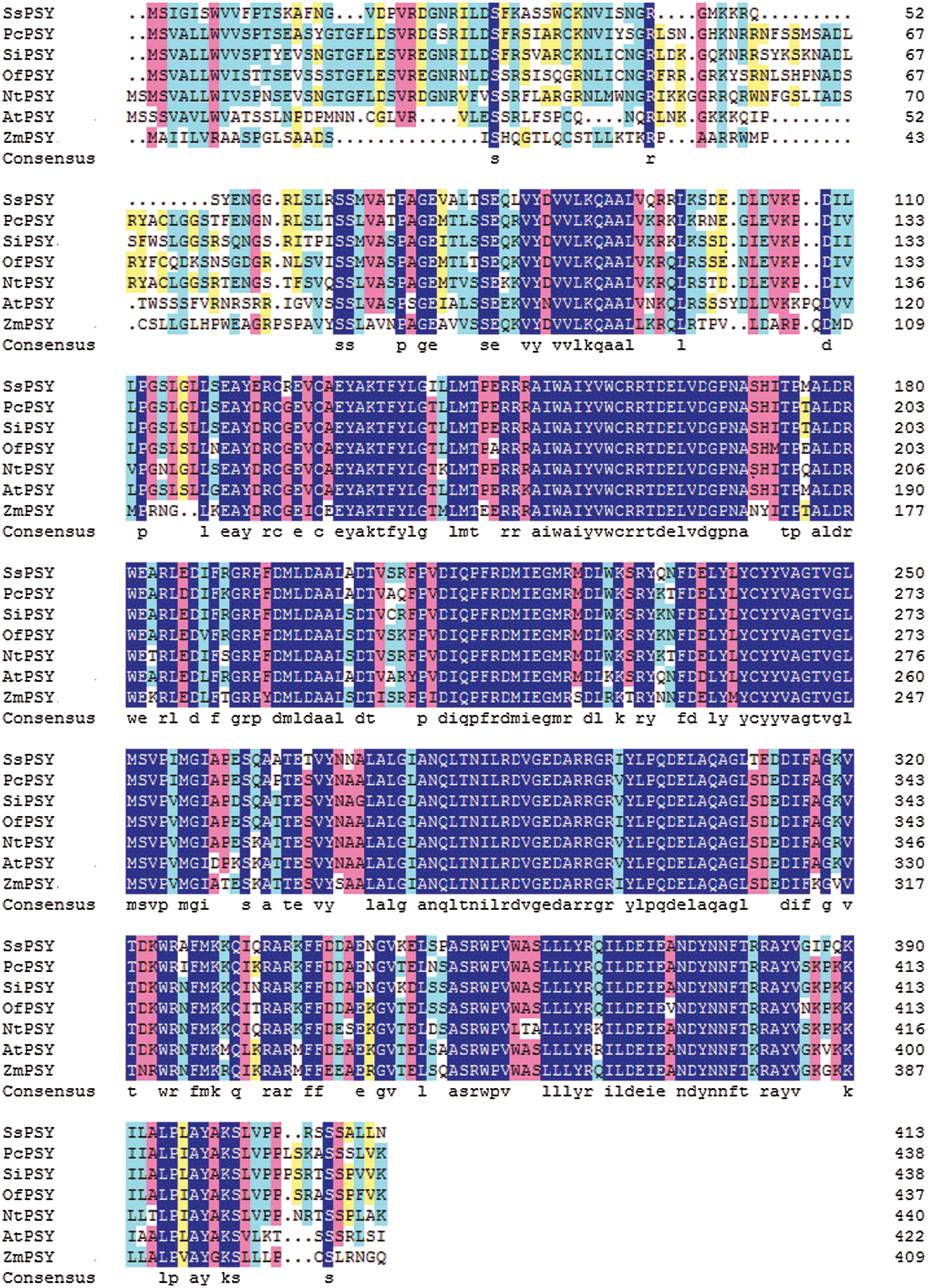
Figure 4: Comparison of deduced amino acid sequences of SsPSY with other phytoene synthases (deep blue, red, light blue, and yellow shading, respectively, represent homologies of 100%, ≤ 75%, ≥ 50%, and ≥ 30%)
SsPSY: Salvia splendens phytoene synthase; PcPSY: Pogostemon cablin phytoene synthase; SiPSY: Sesamum indicum phytoene synthase; OfPSY: Osmanthus fragrans phytoene synthase;
NtPSY: Nicotiana tabacum phytoene synthase; AtPSY: Arabidopsis thaliana phytoene synthase; ZmPSY: Zea mays phytoene synthase.
To determine the relationship between S. splendens PSY protein sequences and PSY protein sequences of other plants, we performed a phylogenetic analysis (Fig. 5). PSY isolated from S. splendens and Pogostemon cablin formed a group, indicating that the two proteins are highly homologous. PSY from Sesamum indicum also had high homology with SsPSY but formed a separate cluster. PSY from Osmanthus fragrans clustered separately from S. splendens, Pogostemon cablin, and Sesamum indicum.
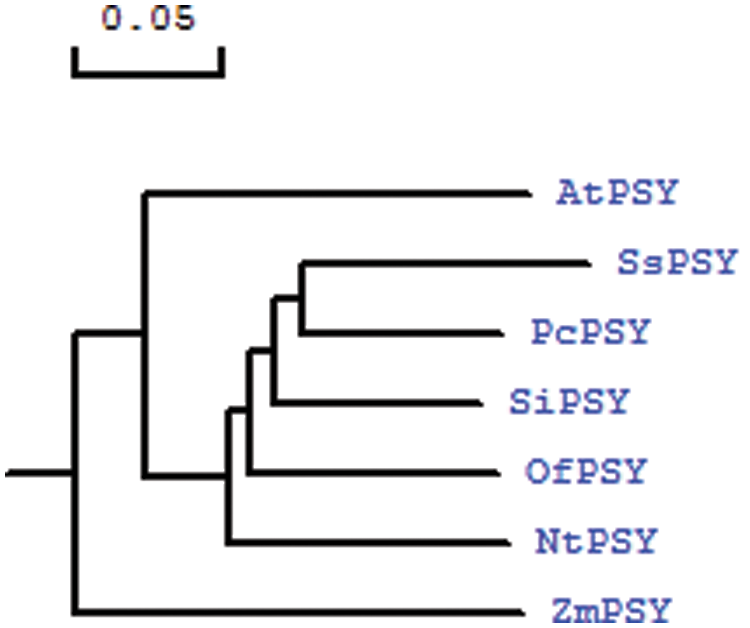
Figure 5: Phylogenetic tree based on the amino acid sequences of SsPSY and phytoene synthases from other species (scale bar represents 0.05 substitutions per site)
3.4 Relative Expression Analysis of SsPSY
Relative gene expression levels were examined in nodes of the stem, root tip, and lateral buds of the tested varieties to determine tissue-specific patterns of SsPSY expression before and after lateral branch formation (Fig. 6). The expression of SsPSY in ‘Cailinghong’ was used as the control group, and relationships between the expression of SsPSY in strain 35 and ‘Cailinghong’ were determined. Compared with the expression of SsPSY in ‘Cailinghong’, the relative expression level of SsPSY in nodes of the stem and root tip of strain 35 was significantly higher before and after the formation of lateral branches, with fold change values of 1.374 ± 0.096 and 1.845 ± 0.061. SsPSY gene expression levels in the lateral branches were the same as those in the nodes and root tip (Fig. 6B). The expression levels of SsPSY in strain 35 were significantly higher than those in ‘Cailinghong’ by 1.733 ± 0.145-fold.
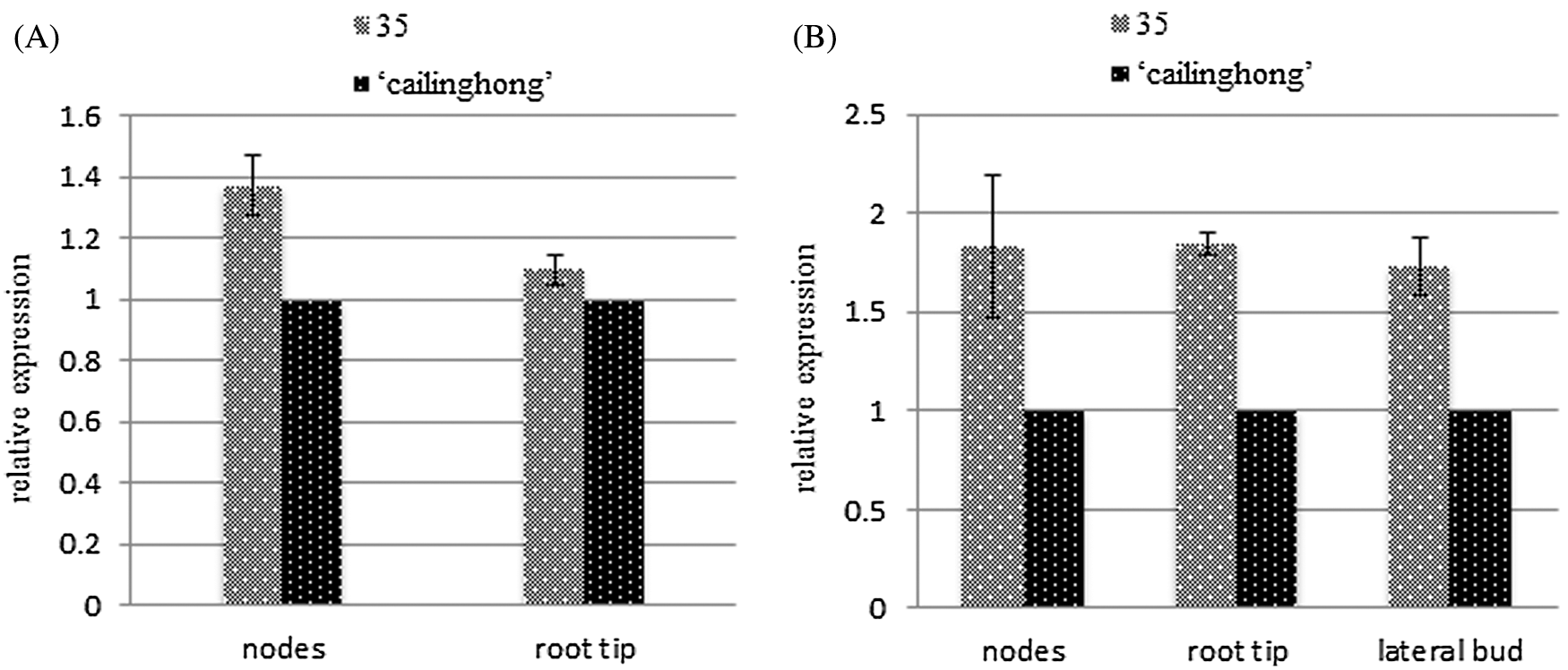
Figure 6: Relative expression analysis of SsPSY of different organs and growth stages. A: Relative expression of SsPSY in different organs of the tested varieties before the formation of lateral branches; B: Relative expression of SsPSY in different organs of the tested varieties after the formation of lateral branches
3.5 Phenotypic Analysis of PSY Transgenic Arabidopsis
To investigate the function of SsPSY in the regulation of branching, it was transferred into Arabidopsis, and the change of branching phenotype of transgenic plants was observed.
To screening the positive transgenic Arabidopsis plants, seeds harvested by the transformed Arabidopsis were sown to 1/2 MS medium supplemented with 120 mg/L kanamycin and seedling growth on medium was observed. Positive plants had a green leaf color and grew true leaves. Untransformed plants did not grow true leaves and roots and gradually withered and died (Fig. 7A). After the plants grew true leaves, the total leaf DNA of the primary positive plants was extracted and amplified by PCR using specific full-length primers (Table 1). The target bands were amplified in all the successful transgenic plants (Fig. 7B), and the PCR results for the transgenic plasmid were used as the control (CK). The positive plants were screened for a second time by this method.
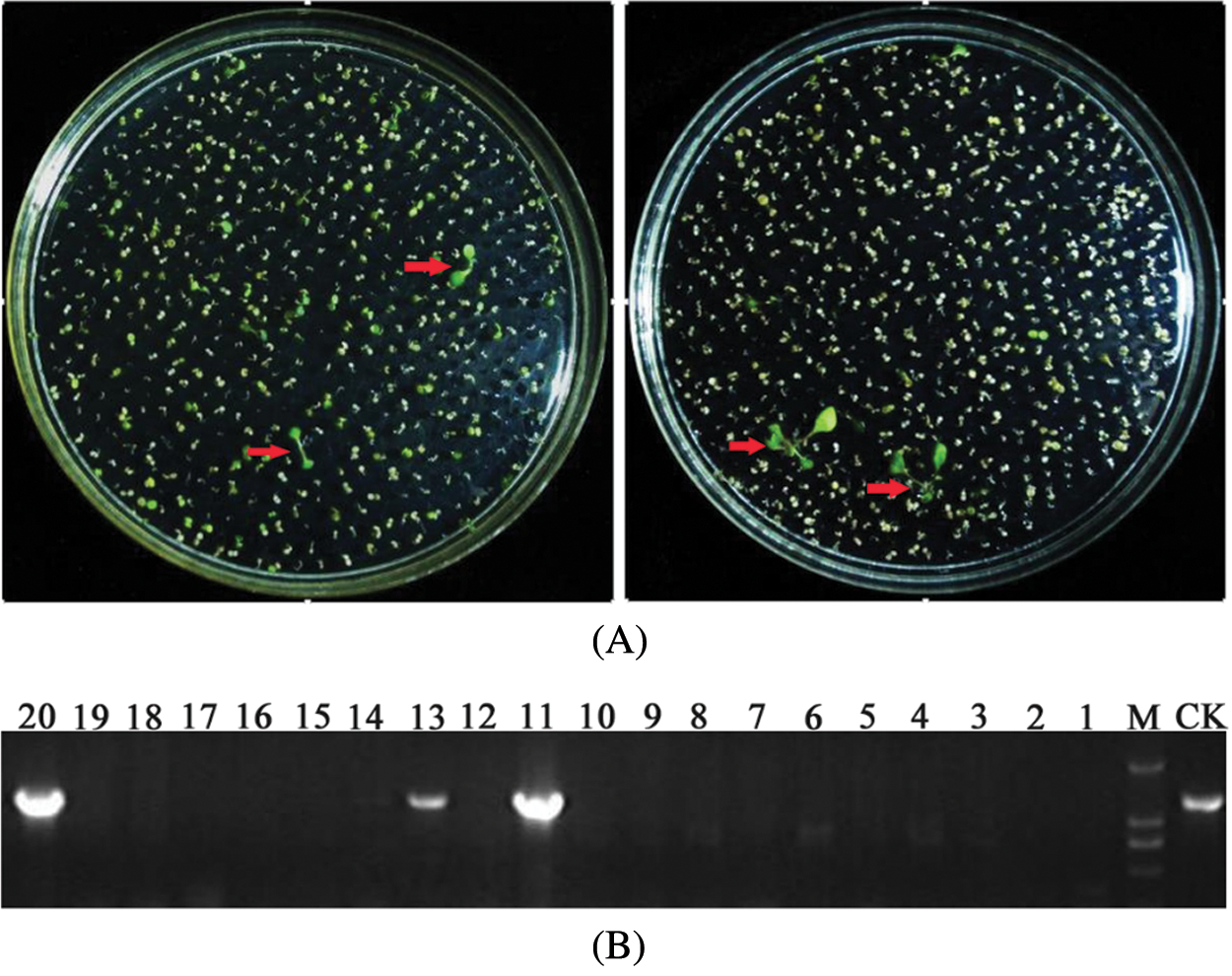
Figure 7: Screening of transgenic positive plants (A: T1 seeds of transgenic Arabidopsis on 1/2 MS; B: PCR verification of T1 seeds of transgenic Arabidopsis (M: Marker))
The positive plants screened by culture medium and detected by PCR were transplanted from the flat plate into soil for growth under natural conditions and observations of morphological changes in positive plants. SsPSY transgenic Arabidopsis (T1) was significantly different from wild-type plants (Col) in morphology. At the beginning of transplantation, transgenic plants had larger leaves with normal growth and no adverse reactions (Fig. 8A). Morphologically, the branching phenotype of transgenic plants differed significantly from that of wild-type plants and the branching of transgenic plants was significantly greater than that of wild-type plants after 1 month (Fig. 8B). The transgenic plants had 5–7 branches, while the wild-type plants had 2–4 branches.

Figure 8: Morphological characteristics of transgenic Arabidopsis (A: Growth of transgenic Arabidopsis after transplanting for 10 d; B: Branching characteristics of transgenic Arabidopsis after transplanting for 35 d)
Based on the results of the relative expression analysis of SsPSY, the SsPSY transgenic Arabidopsis was predicted to exhibit less branching than that in the wild-type plants. However, the experimental results were contrary to this expectation, and transgenic plants showed a multi-branching phenotype.
Branching is one of the basic features of plant growth and plays an important role in plant morphogenesis. The number of plant branches is directly related to the yield of food crops and the ornamental effect of ornamental plants; accordingly, the mechanism underlying plant branching is an important research topic. Researchers have identified a series of genes that regulate branching.
In this study, SSH technology was used to compare the ‘Cailinghong’ variety and the erect strain 35 and to screen up-regulated genes in each strain. A total of 212 up-regulated unigenes were obtained by SSH technology, including 96 unigenes in ‘Cailinghong’ and 116 unigenes in strain 35. In particular, these unigenes included genes related to plant hormone anabolism and signal transduction, including functions in abscisic acid and SL anabolism (ABA2, crtQ, and PSY), ethylene anabolism (metK), auxin signal transduction (IAA, AUX1, SAUR, and TIR1), and other hormone signal transduction (SNRK2, MPK6, TGA, and PP2C). Plant branching is controlled by the coordination of plant hormones; accordingly, the unigenes related to hormone synthesis and signal transduction that were up-regulated in ‘Cailinghong’ or strain 35 may contribute to the unique branching pattern found in ‘Cailinghong’.
SLs, involved in plant branching, are closely related to abscisic acid because they share a common synthetic substrate—carotenoids [23,24]. Carotenoid synthesis is carried out in plant plastids by several enzymes by a highly coordinated mechanism. Among these enzymes, phytoene synthase (PSY) is involved in the first step and is the most important rate-limiting enzyme [25–27]. Research on the PSY gene has mainly focused on color variation resulting from changes in the carotenoid content [28–30]. It is likely involved in the horizontal regulation of SL in plants and resistance to abscisic acid [19,31,32]. The regulatory effects of PSY on plant branching have not been evaluated.
Based on the important rate-limiting role of PSY in the biosynthesis of carotenoids, the role of PSY as a precursor of the synthesis of SLs and abscisic acid, and the identification of the PSY gene fragment by SSH and its role in the regulation of branching in the ‘Cailinghong’ variety in this study, PSY likely has a regulatory function in the development of branching of the ‘Cailinghong’ variety.
In this study, we cloned a PSY gene from S. splendens using RACE technology. The expression levels of this gene in different organs of the tested varieties before and after the occurrence of lateral branches were compared. In analyses of the nodes of the stem, root tip, and lateral branches before and after lateral branch formation, relative SsPSY expression levels in the erect plant type 35 were consistently significantly higher than those in ‘Cailinghong’ in the different plant parts. Zhang et al. [33] found that the overexpression of PSY can increase the carotenoid content in Hongkong kumquat. Lindgren et al. [34] also found that the overexpression of PSY in seeds results in higher carotenoid content in seeds produced by plants, and the abscisic acid content increased as the carotenoid content increased. Therefore, it is speculated that the high expression of SsPSY in strain 35 leads to a higher carotenoid content, and the increase of synthetic substrates indirectly leads to an increase in the synthesis of SLs, which inhibits the formation of branches in strain 35 and leads to a less branched phenotype. It has previously been shown that the down-regulation of PSY can reduce the levels of SLs in dicotyledonous plants [19], consistent with our results. Thus, we speculated that the low expression of SsPSY in S. splendens contributes to the increase in lateral branches.
To assess the function of the SsPSY gene, it was transformed into Arabidopsis to observe the phenotypic changes of positive strains. The branches of transgenic plants increased, contrary to our prediction. The expression of SsPSY was higher in strain 35 than in ‘Cailinghong’. Based on the results of Stauder et al. [19], we can infer that the strigolactone content in ‘Cailinghong’ is lower than that in strain 35. The over-expression of SsPSY gene in transgenic Arabidopsis is predicted to reduce plant branching. However, the transgenic assay showed that the branching phenotype of transgenic Arabidopsis increased, rather than decreased.
It is possible that this increase in branching can be explained by gene silencing, in which transgenic and endogenous genes are co-suppressed. This phenomenon was discovered by Napoli et al. in studies of transgenic petunia, in which the transgenes chs and dfr suppressed corresponding endogenous genes and their own expression simultaneously [35,36]. In many organisms, introducing a transgene can cause the co-suppression of the transgene and homologous endogenous genes [37–42].
Further studies are needed to determine whether the introduction of exogenous SsPSY triggered the gene silencing mechanism in transgenic plants in this study, explaining the multi-branching phenotype in transgenic plants. Although the experimental results for transgenic Arabidopsis were contrary to our predictions, the introduction of exogenous SsPSY altered the branching morphology of transgenic plants, suggesting that SsPSY plays an important role in plant branching.
In summary, genes related to the branching phenotype of ‘Cailinghong’ were screened by SSH technology. SsPSY, related to the synthesis of SLs, was cloned and analyzed. The results of a relative expression analysis suggested that the expression level of SsPSY was down-regulated in the ‘Cailinghong’ variety. However, the branching phenotype of transgenic Arabidopsis was contrary to the predictions, showing a multi-branching phenotype. This result may be explained by transgenic silencing. However, it can still be inferred that SsPSY plays an important role in the regulation of plant branching, as evidenced by changes in the branching phenotype in transgenic plants. Our future research will focus on the silencing of transgenic SsPSY, further plant transgenic experiments, verifying the function of SsPSY, and analyses of other candidates differentially expressed genes between varieties. These results provide a reference for understanding the molecular mechanism underlying plant branch development.
Funding Statement: This work was supported by grants from the Special Project of the University in 2019—Capacity Building of Science and Technology Innovation Service—Construction of Scientific Research—Beijing Collaborative Innovation Center for Eco-Environmental Improvement with Forestry and Fruit Trees (2011 Collaborative Innovation Center) (Project No. CEFF-PXM2019_014207_000099), the National Natural Fund (Project No. 31100509), Open Project of Beijing Engineering Research Center of Rural Landscape Planning and Design (KF2019065) and General Project of Science and Technology Plan of Beijing Education Commission (KM202010020006).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Le Moigne, M. A., Guérin, V., Furet, P. M., Billard, V., Lebrec, A. et al. (2018). Asparagine and sugars are both required to sustain secondary axis elongation after bud outgrowth in Rosa hybrida. Journal of Plant Physiology, 222, 17–27. DOI 10.1016/j.jplph.2017.12.013. [Google Scholar] [CrossRef]
2. Aguilar-Martínez, J. A., Poza-Carrión, C., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell, 19, 458–472. DOI 10.1105/tpc.106.048934. [Google Scholar] [CrossRef]
3. Martín-Trillo, M., Grandio, E. G., Serra, F., Marcel, F., Rodríguez-Buey, M. L. et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. The Plant Journal, 67, 701–714. DOI 10.1111/j.1365-313X.2011.04629.x. [Google Scholar] [CrossRef]
4. Braun, N., de Saint Germain, A., Pillot, J. P., Boutet-Mercey, S., Dalmais, M. et al. (2012). The Pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiology, 158, 225–238. DOI 10.1104/pp.111.182725. [Google Scholar] [CrossRef]
5. Finlayson, S. A. (2007). Arabidopsis TEOSINTE BRANCHED1-LIKE1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol, 48, 667–677. DOI 10.1093/pcp/pcm044. [Google Scholar] [CrossRef]
6. Nicolas, M., Rodríguez-Buey, M. L., Franco-Zorrilla, J. M., Cubas, P. (2015). A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Current Biology, 25, 1799–1809. DOI 10.1016/j.cub.2015.05.053. [Google Scholar] [CrossRef]
7. Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A. et al. (2008). Strigolactone inhibition of shoot branching. Nature, 455, 189–194. DOI 10.1038/nature07271. [Google Scholar] [CrossRef]
8. Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455(7210), 195–200. DOI 10.1038/nature07272. [Google Scholar] [CrossRef]
9. Al-Babili, S., Bouwmeester, H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology, 66, 161–186. DOI 10.1146/annurev-arplant-043014-114759. [Google Scholar] [CrossRef]
10. Borghi, L., Liu, G. W., Emonet, A., Kretzschmar, A., Martinoia, E. (2016). The importance of strigolactone transport regulation for symbiotic signaling and shoot branching. Planta, 243, 1351–1360. DOI 10.1007/s00425-016-2503-9. [Google Scholar] [CrossRef]
11. Waters, M. T., Gutjahr, C., Bennett, T., Nelson, D. C. (2017). Strigolactone signaling and evolution. Annual Review of Plant Biology, 68, 291–322. DOI 10.1146/annurev-arplant-042916-040925. [Google Scholar] [CrossRef]
12. Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M. et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science, 335, 1348–1351. DOI 10.1126/science.1218094. [Google Scholar] [CrossRef]
13. Seto, Y., Sado, A., Asami, K., Hanada, A., Umehara, M. et al. (2014). Carlactone is an endogenous biosynthetic precursor for strigolactones. Proceedings of the National Academy of Sciences of the United States of America, 111, 1640–1645. DOI 10.1073/pnas.1314805111. [Google Scholar] [CrossRef]
14. Walter, M. H., Strack, D. (2011). Carotenoids and their cleavage products: Biosynthesis and functions. Natural Product Reports, 28, 663–692. DOI 10.1039/c0np00036a. [Google Scholar] [CrossRef]
15. Mcquinn, R. P., Giovannoni, J. J., Pogson, B. J. (2015). More than meets the eye: Fromcarotenoid biosynthesis, to new insights into apocarotenoid signaling. Current Opinion in Plant Biology, 27, 172–179. DOI 10.1016/j.pbi.2015.06.020. [Google Scholar] [CrossRef]
16. Hou, X., Rivers, J., León, P., McQuinn, R. P., Pogson, B. J. (2016). Synthesis and function of apocarotenoid signals in plants. Trends in Plant Science, 21, 792–803. DOI 10.1016/j.tplants.2016.06.001. [Google Scholar] [CrossRef]
17. Gallagher, C. E., Matthews, P. D., Li, F., Wurtzel, E. T. (2004). Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiology, 135(3), 1776–1783. DOI 10.1104/pp.104.039818. [Google Scholar] [CrossRef]
18. Welsch, R., Arango, J., Bar, C., Salazar, B., Al-Babili, S. et al. (2010). Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell, 22, 3348–3356. DOI 10.1105/tpc.110.077560. [Google Scholar] [CrossRef]
19. Stauder, R., Welsch, R., Camagna, M., Kohlen, W., Walter, M. H. (2018). Strigolactone levels in dicot roots Are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE gene family. Frontiers in Plant Science, 9, 255. DOI 10.3389/fpls.2018.00255. [Google Scholar] [CrossRef]
20. Salam, B. B., Malka, S. K., Zhu, X., Gong, H., Ziv, C. et al. (2017). Etiolated stem branching is a result of systemic signaling associated with sucrose level. Plant Physiology, 175, 734–745. DOI 10.1104/pp.17.00995. [Google Scholar] [CrossRef]
21. Wingler, A. (2018). Transitioning to the next phase: The role of sugar signaling throughout the plant life cycle. Plant Physiology, 176(2), 1075–1084. DOI 10.1104/pp.17.01229. [Google Scholar] [CrossRef]
22. Xie, Y., Liu, Y., Ma, M., Zhou, Q., Zhao, Y. et al. (2020). FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nature Communications, 11, 1955. DOI 10.1038/s41467-020-15893-7. [Google Scholar] [CrossRef]
23. Crozier, A., Kamiya, Y., Bishop, G., Yolota, T. (2000). Biosynthesis of hormone and elicitor molecules. In: Buchanan, B., Gruissem, W., Jones, R. (Eds.Biochemistry and molecular biology of plants, pp. 865–872. Rockville: American Society of Plant Physiologists. [Google Scholar]
24. Moreno Beltran, J. C., Stange, C. (2016). Apocarotenoids: A new carotenoid-derived pathway. Subcellular Biochemistry, 79, 239–272. DOI 10.1007/978-3-319-39126-7. [Google Scholar] [CrossRef]
25. Li, F., Vallabhaneni, R., Wurtzel, E. T. (2008). PSY3, a New member of the phytoene synthase gene family conserved in the poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiology, 146, 1333–1345. DOI 10.1104/pp.107.111120. [Google Scholar] [CrossRef]
26. Rodríguez-Villalón, A., Gas, E., Rodríguez-Concepción, M. (2009). Colors in the dark: A model for the regulation of carotenoid biosynthesis in etioplasts. Plant Signaling & Behavior, 280, 699–700. DOI 10.4161/psb.4.10.9672. [Google Scholar] [CrossRef]
27. Rosas-Saavedra, C., Stange, C. (2016). Biosynthesis of carotenoidin plants: Enzymes and color. In: Stange, C. (Ed.Subcellular biochemistry, vol. 79. pp. 35–69. Switzerland: Springer International Publishing. [Google Scholar]
28. Zhang, X. J., Gao, Y., Wen, P. F., Hao, H. H., Chen, X. X. (2018). Cloning and expression analysis of the phytoene synthase gene in ‘Granny Smith’ apple (Malus × domestica Borkh). Biotechnology & Biotechnological Equipment, 32, 1105–1112. DOI 10.1080/13102818.2018.1512379. [Google Scholar] [CrossRef]
29. Andrew, J. P., Xue, G., Jonathan, R. P. (2019). A common phytoene synthase mutation underlies white petal varieties of the California poppy. Nature, 9, 11615. DOI 10.1038/s41598-019-48122-3. [Google Scholar] [CrossRef]
30. Wang, H., Ou, C. G., Zhuang, F. Y., Ma, Z. G. (2014). The dual role of phytoene synthase genes in carotenogenesis in carrot roots and leaves. Molecular Breeding, 34, 2065–2079. DOI 10.1007/s11032-014-0163-7. [Google Scholar] [CrossRef]
31. Ali, G. M. (2017). Novel phytoene synthase paralog from halophytic Salicornia plant confers salinity tolerance. Turkish Journal of Botany, 41, 117–126. DOI 10.3906/bot-1605-18. [Google Scholar] [CrossRef]
32. Simpson, K., Fuentes, P., Quiroz-Iturra, L. F., Flores-Ortiz, C., Contreras, R. et al. (2018). Unraveling the induction of phytoene synthase 2 (DcPSY2) expression by salt stress and abscisic acid in Daucus carota. Journal of Experimental Botany, 69, 4113–4126. DOI 10.1093/jxb/ery207. [Google Scholar] [CrossRef]
33. Zhang, J., Tao, N., Qiang, X., Zhou, W., Cao, H. et al. (2009). Functional characterization of Citrus PSY gene in Hongkong kumquat (Fortunella hindsii Swingle). Plant Cell Reports, 28, 1737–1746. DOI 10.1007/s00299-009-0774-3. [Google Scholar] [CrossRef]
34. Lindgren, L. O., Stålberg, K. G., Höglund, A. S. (2003). Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiology, 132, 779–785. DOI 10.1104/pp.102.017053. [Google Scholar] [CrossRef]
35. Napoli, C., Lemieux, C., Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. DOI 10.2307/3869076. [Google Scholar] [CrossRef]
36. Krol, A. R. V. D., Mur, L. A., Beld, M., Stuitje, M. A. R. (1990). Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. DOI 10.1105/tpc.2.4.291. [Google Scholar] [CrossRef]
37. Carlo, C. (2001). Homology-dependent gene silencing mechanisms in fungi. Annual Review of Microbiology, 55, 381–406. DOI 10.1146/annurev.micro.55.1.381. [Google Scholar] [CrossRef]
38. Dernburg, A. F., Zalevsky, J., Colaiácovo, M. P., Villeneuve, A. M. (2000). Transgene-mediated cosuppression in the C. elegans germ line. Genes & Development, 14, 1578–1583. DOI 10.1101/gad.14.13.1578. [Google Scholar] [CrossRef]
39. Kooter, J. M., Matzke, M. A., Meyer, P. (1999). Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends in Plant Science, 4, 340–347. DOI 10.1016/S1360-1385(99)01467-3. [Google Scholar] [CrossRef]
40. Vaucheret, H., Béclin, C., Elmayan, T., Feuerbach, F., Vernhettes, S. (1999). Transgene-induced gene silencing in plants. The Plant Journal, 16, 651–659. DOI 10.1046/j.1365-313x.1998.00337.x. [Google Scholar] [CrossRef]
41. Ruiz, F., Vayssie, L., Klotz, C., Sperling, L., Madeddu, L. (1998). Homology-dependent gene silencing in Paramecium. Molecular Biology of the Cell, 9, 931–943. DOI 10.1091/mbc.9.4.931. [Google Scholar] [CrossRef]
42. Sanders, M. (2002). An active role for endogenous beta-1, 3-glucanase genes in transgene-mediated co-suppression in tobacco. The European Molecular Biology Organization Journal, 21, 5824–5832. DOI 10.1093/emboj/cdf586. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |