

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.019219
ARTICLE
Identification of a Unique Germacrene A Oxidase from Xanthium strumarium
1Shanghai Key Laboratory of Bio-Energy Crops, Research Center for Natural Products, Plant Science Center, School of Life Sciences, Shanghai University, Shanghai, 200444, China
2Songjiang Yunjian School Affiliated to Shanghai Foreign Language School, Shanghai, 201600, China
3College of Medicine, Henan University of Chinese Medicine, Zhengzhou, 450046, China
*Corresponding Author: Changfu Li. Email: changfuli@shu.edu.cn
Received: 09 September 2021; Accepted: 09 October 2021
Abstract: 8,12-sesquiterpene lactones (STLs) are an important class of natural products with unique pharmaceutical activities. For years the pathway leading to 8,12-STLs remains an enigma. Xanthium strumarium accumulates abundant 8,12-STLs, and xanthatin is a characteristic 8,12-STL in it. Xanthatin has been previously postulated to be derived from germacrene A, but the steps from germacrene A to xanthatin are unknown. As part of an effort to understand the xanthatin biosynthetic pathway. This study reports the characterization of a unique germacrene A oxidase (XsGAO) from X. strumarium. Unlike a classical GAO enzyme, which is known to catalyze a three-step oxidation of germarene A to yield germacrene A acid (GAA), surprisingly, XsGAO catalyzed only one-step conversion of germacrene A to germacrene A alcohol. We discussed that GAO may be pressured to acquire a novel activity during the evolutionary path for the biosynthesis of 8,12-STLs.
Keywords: Xanthium strumarium; 8,12-sesquiterpene lactones; germacrene A oxidase
Xanthium strumarium is native to North America, and is now widely distributed in China, Europe and other countries [1]. The fruit of this plant, known as Cang-Er-Zi in traditional Chinese medicine, has been prescribed for the treatment of rhinitis and headache for thousands of years [2]. Phytochemical studies of X. strumarium fruits and other tissues have revealed the presence of sesquiterpenes and phenylpropanoids as the most abundant constituents [3]. Among the X. strumarium sesquiterpenes, xanthatin is an important active compound [4]. The biological role of xanthatin is currently unclear, but it is implicated as an antifeedant in plant herbivore resistance [5]. In addition, it has shown inhibitory activities against several tumor cells, including breast cancer [6], colorectal cancer [7], and glioma cells [8]. In spite of many studies concerning the medical activities of xanthatin, the genes and enzymes involved in its biosynthesis are largely unknown [9].
Xanthatin belongs to the 8,12-seco-guaianolide, one subfamily of sesquiterpene lactones with a C-8-C-12 conjugated ring (Fig. 1). Concerning the lactone-ring position, guaianolides can be classified as 6,12- or 8,12-guaianolides (Fig. 1). To date, the biosynthetic mechanism of 8,12-guaianolides remains largely unknown, whereas some progresses have been achieved for biosynthesis of 6,12-guaianolides [10,11]. The biosynthesis of the 6,12-guaianolides are believed to be derived from costunolide [12], a simplest member of germacranolide. Costunolide is synthesized from FPP (farnesyl diphosphate) under the successive activities of a sesquiterpene synthase (germacrene A synthase) and two cytochrome P450s (germacrene A oxidase and costunolide synthase) (Fig. 1) [11,13]. Li et al. [14] reported the identification and characterization of a germacrene A synthase from X. strumarium (XsGAS), and have demonstrated that XsGAS is involved in xanthatin biosynthesis. Therefore, it appears that the germacrene A generally serves as an entry intermediate for biosynthesis of either 6,12- or 8,12-guaianolide. Nothing is known about the later stages of xanthatin biosynthesis after the formation of germacrene A. However, judging from the structure of xanthatin, the steps from germacrene A to xanthatin presumably include multiple oxidations, and enzymes of cytochrome P450s and alcohol and aldehyde dehydrogenases may be involved in these oxidation steps [9]. The specific accumulation of xanthatin in X. strumarium trichomes indicates that the trichome may be site for transcribing the xanthatin biosynthetic genes [15]. Indeed, the transcripts for a germacrene A synthase and some putative cytochrome P450s have proven to be trichome-specifically expressed in X. strumarium [9]. Therefore, it would be possible to elucidate the xanthatin biosynthesis mechanism by characterizing the biochemical functions of the trichome-dependent gene products.
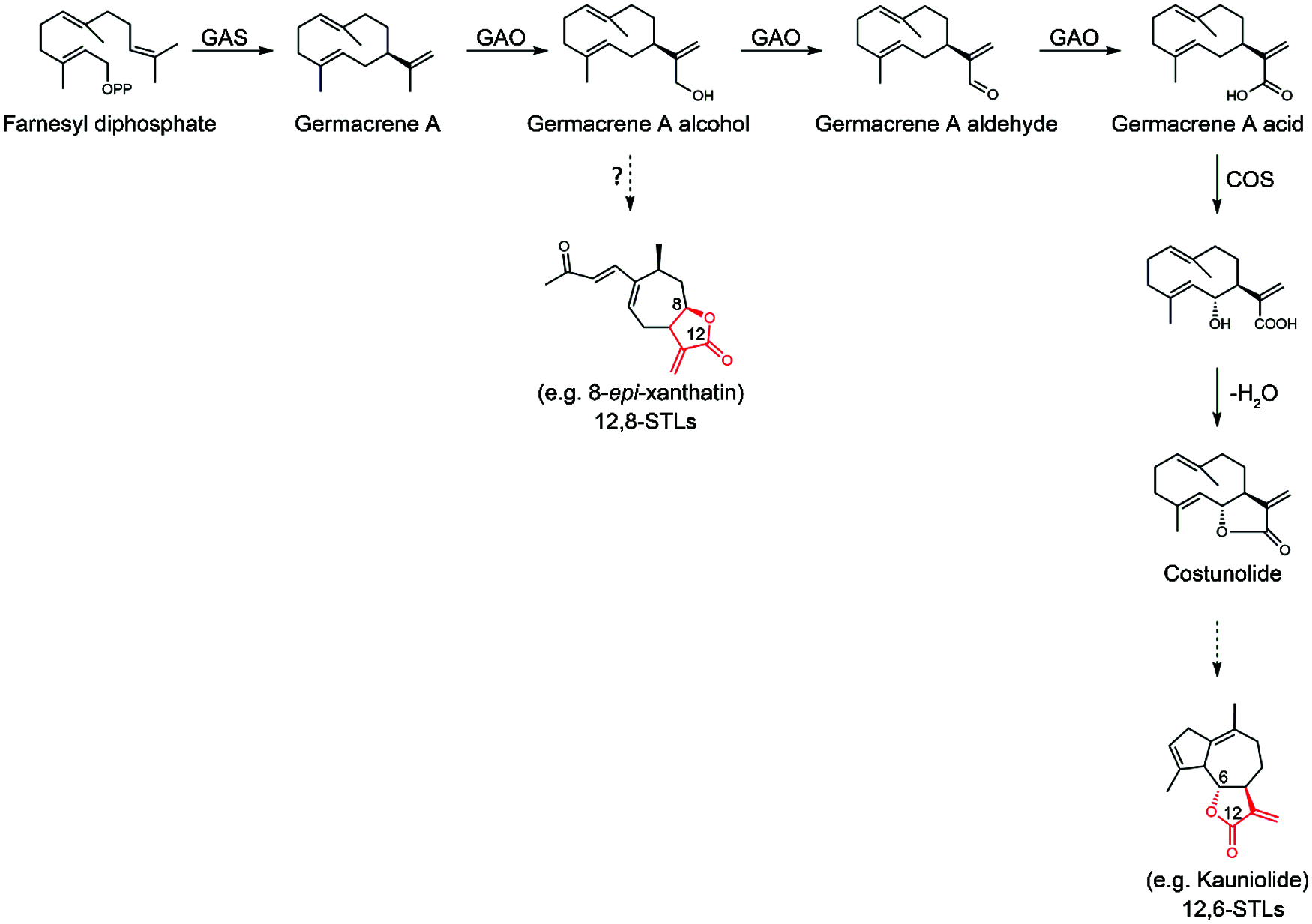
Figure 1: Proposed biosynthesis steps leading to 12,6- or 12,8-STLs starting from FPP (farnesyl pyrophosphate). FPP is cyclized by germacrene A synthase (GAS) to germacrene A, which is then oxidized three times at C12 by germacrene A oxidase (GAO) to form germacrene A acid via alcohol and aldehyde intermediates. Then, costunolide synthase (COS) catalyzes 6α-hydroxylation of germacrene A acid to yield costunolide. Costunolide serves as a central substrate leading to 12,6-sesquiterpene lactones (12,6-STLs). From this study, we proposed that the germacrene A alcohol may be the branch point for biosynthesis of 12,8-STLs
As part of a broader study of the elucidation of xanthatin biosynthesis mechanism in X. strumarium, we report the biochemical characterization of the trichome-specifically expressed P450 enzyme CYP71AV14 from X. strumarium. This study demonstrated that CYP71AV14 is a unique germacrene A oxidase (designated XsGAO) in catalyzing only one-step C12-oxidation of germacrene A to form germacrene A alcohol, which activity is in contrast from that catalyzed by a convenient GAO. A classical GAO activity is involved in a three-step oxidation of germacrene A at C12 to yield germacrene A acid (GAA). The novel activity of XsGAO important for xanthatin biosynthesis is discussed.
2.1 Identification and Cloning of the Germacrene A Oxidase from X. strumarium
The X. strumarium glandular trichomes were isolated and purified as previously described [16]. In short, the young leaves from three month-old plants were dissected by glass beads in an isolation buffer [25 mM MOPSO, pH 6.6, 200 mM sorbitol, 10 mM sucrose, 5 mM thiourea, 2 mM dithiothreitol, 5 mM MgCl2, 0.5 mM sodium phosphate, 0.6% (w/v) methylcellulose and 1% (w/v) polyvinylpyrrolidone (Mr 40000)]. The disrupted extracts were successively passed through a series of nylon mesh with different sizes, and the isolated glandular trichomes were retained on the 42 μm mesh screen. The total RNA was then isolated from the purified trichomes using Trizol reagent (Invitrogen) according to the protocol provided. Our previously constructed transcriptome [9] from the X. strumarium trichome allowed us to identify one putative GAO (i.e., CYP71AV14 or XsGAO) in X. strumarium. The XsGAO was amplified from the X. strumarium trichome cDNA using the primers 5′-atggaagtctccctcaccacttcc-3′ and 5′-ctaataacttggtaccaatatc-3′. The amplicon was cloned into pMD18-T vector and sequenced.
2.2 Phylogenetic Analysis of the GAOs from Different Plant Species
The phylogeny was constructed using the deduced amino acid sequences of the GAO sequences from different plant species. Sequences were aligned by ClustalW [17], and the alignment was used to construct a phylogenetic tree with MEGA 6.0 software.
2.3 Expression of XsGAO in Yeast
The ORF (open reading frame) of XsGAO was PCR-amplified, and the amplicons were subsequently digested with NotI/PacI, and cloned into the respective sites in pESC-leu2d-CPR [18]. To supply the germacrene A substrate for XsGAO, the germacrene A synthase gene (LsGAS; AF489965) from Lactuca sativa was amplified from a previously made plasmid pESC-leu2d-CPR-LsGAS-LsGAO supplied by the Ro’s group [19], and inserted into a yeast expression vector pESC-URA under BamHI/XhoI sites. The LsGAS was either expressed alone or co-expressed with XsGAO in the Saccharomyces cerevisiae EPY300 strain [20]. As a comparison, a classical GAO from L. sativa (LsGAO; KF981867), which is known to oxidize a three-step oxidation of germacrene A to yield germacrene A acid [19], was co-expressed with LsGAS by transferring the plasmid pESC-leu2d-CPR-LsGAS-LsGAO to the EPY300 strain. The transgenic yeast cells were standardly cultivated in appropriate dropout media, and the expression of the transferred genes was induced by 2% galactose.
To extract the germacrene A alcohol product, the yeast culture was overlaid with dodecane as previously described [21], and the trapped dodecane layer was used for GC-MS analysis. To extract the germacrene A acid product, the yeast culture was directly extracted three times with ethyl acetate, and the ethyl acetate extract was subjected to GC-MS analysis.
All GC-MS analyses were performed on Shimadzu GC-MS HP 2010 equipment. A sample volume of 1 μL was injected at an inlet temperature of 250°C, and compounds were separated with a HP-5MS column [15 m (length) × 250 μm (i.d.) × 0.1 μm (film thickness)] using helium as a carrier gas at a flow rate of 2.0 mL/min. The GC oven temperature program was as follows: 40°C for 3 min and then increased by 5 °C/min to 240°C. The electron impact ionization of the mass spectrometric detector was tuned at 70 eV, and operated at 50–400 Da in full scan mode.
LC-MS/MS analysis was performed using a Q-Exactive Focus system (Thermo Fisher Scientific, MA, USA) equipped with a Hypersil Gold column (particle size, 1.9 μm; pore size, 175 Å; 2.1 × 50 mm; Thermo Fisher Scientific, MA, USA). Samples were eluted with a mixture of A (HPLC grade water/5 mM ammonium acetate) and B (methanol/5 mM ammonium acetate) at a flow rate of 0.25 mL/min under the following program: 0–2 min 65% B; 2–27 min 65–99% B; 27–37 min 99% B; 37–37.5 min 99–65% B; 37.5–39 min 65% B. The injection volume was 3 μL.
3.1 Sequence Analysis of XsGAO from X. strumarium
Our previously transcriptomic analysis of the X. strumarium trichomes revealed one putative germacrene A oxidase (GAO) that is specifically expressed in the trichomes [9]. This putative GAO was designated XsGAO here, and this sequence was also assigned the name CYP71AV14 in the David Nelson’s cytochrome P450 database (http://drnelson.uthsc.edu/cytochromeP450.html). The XsGAO sequence has been deposited under an accession number of OK315336 in the NCBI GenBank. The full length open reading frame of XsGAO was amplified from the previously prepared X. strumarium trichome cDNA [9], and the resulting 1467 bp putatively encoded for a polypeptide with a molecular mass of 54.9 kDa. XsGAO shows 78%–94% amino acid identities with the previously characterized GAOs of other plant species [19], including Lactuca sativa (LsGAO, KF981867), Saussurea costus (ScGAO, D5JBW9), Cichorium intybus (CiGAO1, GU256644), Barnadesia spinosa (BsGAO, GU256647), and Helianthus annuus (HaGAO, GU256646). For cytochrome P450s, the overall structure is generally conserved and six substrate recognition sites have been identified [22]. Amino acid alignment of XsGAO with other GAOs revealed eight residue variations within the substrate recognition sites among the GAOs (Fig. 2).
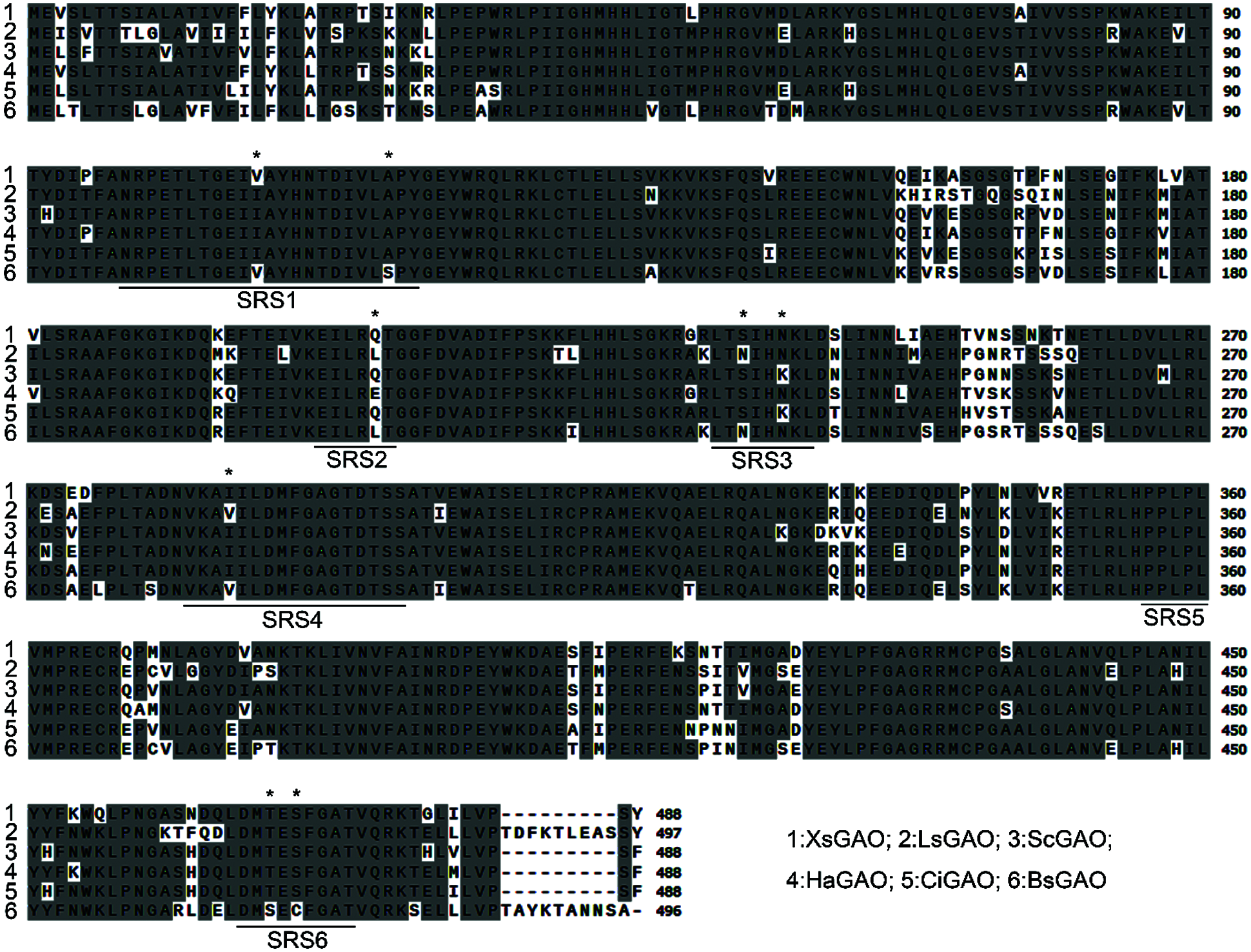
Figure 2: Amino acid alignment of XsGAO with several previously characterized GAOs. Identical residues are highlighted. Six conserved substrate recognition sites (SRSs) are underlined, and the residue variation within the SRSs are labeled with asterisks. Abbreviations and GenBank accession numbers of GAOs are as follows: Lactuca sativa (LsGAO, KF981867), Helianthus annuus (HaGAO, GU256646), Saussurea costus (ScGAO, D5JBW9), Barnadesia spinosa (BsGAO, GU256647), Cichorium intybus (CiGAO, GU256644)
A phylogenetic tree of GAOs (Fig. 3) indicated that XsGAO shows a close relationship with the sunflower GAO (i.e., HaGAO), but being distinct from other GAOs.
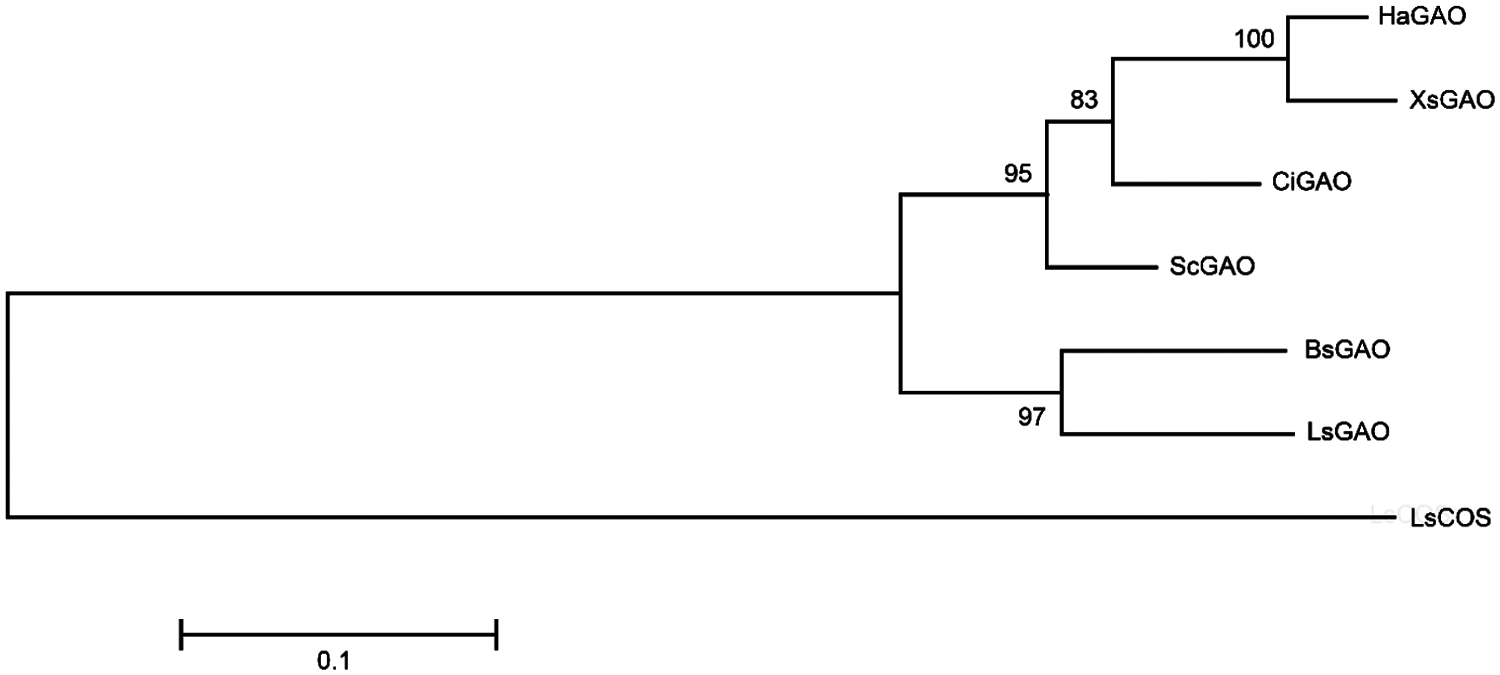
Figure 3: Molecular phylogenetic analysis of XsGAO and several previously characterized GAOs. The scale bar indicates 0.1 amino acid substitutions per site. LsCOS (Lactuca sativa costunolide synthase, HQ439599) serves an outgroup enzyme. The phylogenetic tree was constructed using the maximum-likelihood method in MEGA6.0 software
3.2 Biochemical Function of XsGAO Revealed by Expression in Yeast
To examine the germacrene A oxidizing activity, the full-length XsGAO cDNA was cloned into the yeast expression vector pESC-leu2d-CPR, and it was co-transferred into the EPY300 yeast strain with the germacrene A-producing plasmid pESC-URA-LsGAS. Compared to the yeast expressing the LsGAS alone or the control yeast bearing the empty vector, two new products (peaks 1 and 2) were formed in the cultures co-expressing LsGAS and XsGAO (Fig. 4a). Based on their mass fragmented patterns (Fig. 4b) compared with those previously reported by others [23], they were identified as the heat-and/or acid-induced rearrangement products of germacrene A alcohol. Surprisingly, no GAA was produced in the LsGAS/XsGAO-expressing cultures (Fig. 5), whereas it was clearly synthesized when XsGAO was replaced with LsGAO as a co-expression partner with LsGAS (Fig. 5). LsGAO is a classical GAO enzyme and is known to catalyze a three-step oxidation of germacrene A at C12 position to yield GAA [19]. Thus, not like a convenient GAO, it appears that XsGAO is a unique GAO enzyme, which catalyzes only one-step oxidation of germacrene A at C12 to give germacrene A alcohol.

Figure 4: GC-MS analysis of the products extracted from the transgenic yeasts. (a) Selective ion-chromatograms at m/z of 220, 202 and 184; (b) Mass fragmented patterns of the products. Two new products (Peaks 1 and 2) were formed upon the co-expression of LsGAS and XsGAO, compared to the empty vector control or the expression of LsGAS alone. The products of 1 and 2 were identified as the heat or acid-induced rearrangement products of germacrene A alcohol

Figure 5: LC-MS analysis of the products extracted from the yeast co-expressing LsGAS-LsGAO, LsGAS-XsGAO, or the empty vector-transferred yeast. Germacrene A acid (GAA) was produced by the LsGAS/LsGAO whereas not by the LsGAS/XsGAO expression or the yeast carrying the empty vector
The STLs are likely to be associated with the evolutionary success of Asteraceae plants [19]. Despite the structural diversity of STLs, their core structures can be traced to only several backbones, such as germacranolide, eudesmanolide and guaianolide [12]. This may suggest that STLs have risen from a limited number of ancestor compounds. Germacrene A could be one of such ancestors, as functional germacrene A synthases exist in Barnadesia spinosa [24], which is placed at the phylogenetic base of the Asteraceae [25]. Indeed, our recent phytochemical, biochemical and genetic analyses of the X. strumarium trichomes have revealed that germacrene A is relevant to the biosynthesis of xanthatin [9,14], a 12,8-seco-guaianolide.
After formation of germacrene A, the germacrene A oxidase (GAO) may represent a regulatory point for the divergence of the STL metabolic pathways. The classical GAO activity is involved in a three-step oxidation of germacrene A at its C12-position to yield germacrene A acid (GAA), and this activity has been reported for a number of plant species, such as lettuce [19], chicory [13] and costus [11], which produce 12,6-guaianolides. It is conceivable that 12,6-guaianolides would likely accumulate once GAA is formed in vivo, because that the C6 in GAA is allylic and, therefore is relatively reactive for C6 hydroxylation, catalyzed by a P450 like costunolide synthase (COS) (Fig. 1) [11]. Given that costunolide serves as a gateway intermediate of 12, 6-guaianolides [12], to ensure the success of accumulating the 12, 6-guaianolides, the three sequential oxidative reactions by GAO need to be strictly retained for properly supplying GAA to the COS. Indeed, this conventionally three-step oxidizing activity of GAO is conserved in lettuce [19], sunflower [19], costus [11], and chicory [13], all of which are known to accumulate 12, 6-guaianolides. When GAO meets a new substrate armorphadiene, that is restricted to a single species Artemisia annua, it has evolved to be a more specialized P450 AMO (amorphadiene oxidase) for the biosynthesis of the anti-malaria compound artemisinin [26]. Our study clearly showed that facing the ancestor substrate germacrene A, the XsGAO has lost the multiple oxidizing activity while only catalyzing the first step oxidation in converting germacrene A to germacrene A alcohol (Fig. 4). Given that GAA is the central and immediate precursor to the 12, 6-guaianolides (Fig. 1), X. strumarium might be pressured to evolve this unique single-step oxidizing activity of GAO to prevent the formation of GAA, consequently channeling the metabolic flux toward the 12, 8β-lactones, such as xanthatin. Apparently, GAO would be a branch point of enzyme functional adaptions for the lactone diversification in the Asteraceae. Of course, if a widely known phytochemical knowledge of more of the 12, 8-guaianolide type plant species is available in the future, a systematical analysis of the GAO biochemical function would help to examine whether the acquisition of this new mono-functional GAO activity is truly widespread and conserved in the 12, 8-guaianolide-producing plants [27].
The substrate-interacting residues (SIRs) of GAO, which fall into the general substrate recognition regions for P450s [28], have previously been identified based on a structural modeling analysis [26]. With a focus on the SIRs, XsGAO was revealed to have eight residue variants compared to the other classical GAOs (Fig. 2); however, each single variant could also be found within the selected classical GAOs. This finding suggested that the unique activity of XsGAO was not evolved by a single residue variation; it may be acquired by two or more or simultaneously eight mutations of the residues. This hypothesis could be inspected in the future by site-mutagenesis.
Author Contributions: Changfu Li designed the project and revised the manuscript. Lifen Zheng performed the biochemical assays and drafted the manuscript. Fangfang Chen cloned the full-length XsGAO gene. Yuanjun Li isolated the X. strumarium trichomes and prepared the X. strumarium cDNA.
Funding Statement: This work was supported by a grant from the National Natural Science Foundation of China (31370339).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Iqbal, M. F., Liu, M. C., Iram, A., Feng, Y. L. (2020). Effects of the invasive plant Xanthium strumarium on diversity of native plant species: A competitive analysis approach in North and Northeast China. PLoS One, 15, e0228476. DOI 10.1371/journal.pone.0228476. [Google Scholar] [CrossRef]
2. Fan, W., Fan, L., Peng, C., Zhang, Q., Wang, L. et al. (2019). Traditional uses, botany, phytochemistry, pharmacology, pharmacokinetics and toxicology of Xanthium strumarium L.: A review. Molecules, 24, 359. DOI 10.3390/molecules24020359. [Google Scholar] [CrossRef]
3. Khafagy, S. M., Sabry, N. N., Metwally, A. M., El-Naggar, S. F. (1974). Phytochemical investigation of Xanthium strumarium. Planta Medica, 26, 75–78. DOI 10.1055/s-0028-1097971. [Google Scholar] [CrossRef]
4. Nibret, E., Youns, M., Krauth-Siegel, R. L., Wink, M. (2011). Biological activities of xanthatin from Xanthium strumarium leaves. Phytotherapy Research, 25, 1883–1890. DOI 10.1002/ptr.3651. [Google Scholar] [CrossRef]
5. Ahern, J. R., Whitney, K. D. (2014). Sesquiterpene lactone stereochemistry influences herbivore resistance and plant fitness in the field. Annals of Botany, 113, 731–740. DOI 10.1093/aob/mct297. [Google Scholar] [CrossRef]
6. Takeda, S., Noguchi, M., Matsuo, K., Yamaguchi, Y., Kudo, T. et al. (2013). (-)-Xanthatin up-regulation of the GADD45 gamma tumor suppressor gene in MDA-MB-231 breast cancer cells: Role of topoisomerase II alpha inhibition and reactive oxygen species. Toxicology, 305, 1–9. DOI 10.1016/j.tox.2012.12.019. [Google Scholar] [CrossRef]
7. Piloto-Ferrer, J., Sanchez-Lamar, A., Francisco, M., Gonzalez, M. L., Merino, N. et al. (2019). Xanthium strumarium’s xanthatins induces mitotic arrest and apoptosis in CT26WT colon carcinoma cells. Phytomedicine, 57, 236–244. DOI 10.1016/j.phymed.2018.12.019. [Google Scholar] [CrossRef]
8. Ma, Y. Y., Di, Z. M., Cao, Q., Xu, W. S., Bi, S. X. et al. (2020). Xanthatin induces glioma cell apoptosis and inhibits tumor growth via activating endoplasmic reticulum stress-dependent CHOP pathway. Acta Pharmacologica Sinica, 41, 404–414. DOI 10.1038/s41401-019-0318-5. [Google Scholar] [CrossRef]
9. Li, Y., Gou, J., Chen, F., Li, C., Zhang, Y. (2016). Comparative transcriptome analysis identifies putative genes involved in the biosynthesis of xanthanolides in Xanthium strumarium L. Frontiers in Plant Science, 7, 1317. DOI 10.3389/fpls.2016.01317. [Google Scholar] [CrossRef]
10. Liu, Q., Kashkooli, A. B., Manzano, D., Pateraki, I., Richard, L. et al. (2018). Kauniolide synthase is a P450 with unusual hydroxylation and cyclization-elimination activity. Nature Communications, 9, 4657. DOI 10.1038/s41467-018-06565-8. [Google Scholar] [CrossRef]
11. Ikezawa, N., Göpfert, J. C., Nguyen, D. T., Kim, S. U., O’maille, P. E. et al. (2011). Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. Journal of Biological Chemistry, 286, 21601–21611. DOI 10.1074/jbc.M110.216804. [Google Scholar] [CrossRef]
12. De Kraker, J. W., Franssen, M. C., Joerink, M., De Groot, A., Bouwmeester, H. J. (2002). Biosynthesis of costunolide, dihydrocostunolide, and leucodin. demonstration of cytochrome P450-catalyzed formation of the lactone ring present in sesquiterpene lactones of chicory. Plant Physiology, 129, 257–268. DOI 10.1104/pp.010957. [Google Scholar] [CrossRef]
13. Liu, Q., Majdi, M., Cankar, K., Goedbloed, M., Charnikhova, T. et al. (2011). Reconstitution of the costunolide biosynthetic pathway in yeast and Nicotiana benthamiana. PLoS One, 6, e23255. DOI 10.1371/journal.pone.0023255. [Google Scholar] [CrossRef]
14. Li, Y. J., Chen, F. F., Li, Z. Q., Li, C. F., Zhang, Y. S. (2016). Identification and functional characterization of sesquiterpene synthases from Xanthium strumarium. Plant and Cell Physiology, 57, 630–641. DOI 10.1093/pcp/pcw019. [Google Scholar] [CrossRef]
15. Li, C., Chen, F., Zhang, Y. (2014). GA3 and other signal regulators (MeJA and IAA) improve xanthumin biosynthesis in different manners in Xanthium strumarium L. Molecules, 19, 12898–12908. DOI 10.3390/molecules190912898. [Google Scholar] [CrossRef]
16. Chen, F., Hao, F., Li, C., Gou, J., Lu, D. et al. (2013). Identifying three ecological chemotypes of Xanthium strumarium glandular trichomes using a combined NMR and LC-MS method. PLoS One, 8, e76621. DOI 10.1371/journal.pone.0076621. [Google Scholar] [CrossRef]
17. Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A. et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics, 23, 2947–2948. DOI 10.1093/bioinformatics/btm404. [Google Scholar] [CrossRef]
18. Ro, D. K., Ouellet, M., Paradise, E. M., Burd, H., Eng, D. et al. (2008). Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnology, 8, 83. DOI 10.1186/1472-6750-8-83. [Google Scholar] [CrossRef]
19. Nguyen, D. T., Göpfert, J. C., Ikezawa, N., Macnevin, G., Kathiresan, M. et al. (2010). Biochemical conservation and evolution of germacrene a oxidase in asteraceae. Journal of Biological Chemistry, 285, 16588–16598. DOI 10.1074/jbc.M110.111757. [Google Scholar] [CrossRef]
20. Ro, D. K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L. et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature, 440, 940–943. DOI 10.1038/nature04640. [Google Scholar] [CrossRef]
21. Newman, J. D., Marshall, J., Chang, M., Nowroozi, F., Paradise, E. et al. (2006). High-level production of amorpha-4, 11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnology Bioengineering, 95, 684–691. DOI 10.1002/(ISSN)1097-0290. [Google Scholar] [CrossRef]
22. Hamberger, B., Bak, S. (2013). Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philosophical Transactions of the Royal Society B, 368, 20120426. DOI 10.1098/rstb.2012.0426. [Google Scholar] [CrossRef]
23. De Kraker, J. W., Franssen, M. C., de Groot, A., Shibata, T., Bouwmeester, H. J. (2001). Germacrenes from fresh costus roots. Phytochemistry, 58, 481–487. DOI 10.1016/S0031-9422(01)00291-6. [Google Scholar] [CrossRef]
24. Nguyen, T. D., Faraldos, J. A., Vardakou, M., Salmon, M., O’maille, P. E. et al. (2016). Discovery of germacrene a synthases in Barnadesia spinosa: The first committed step in sesquiterpene lactone biosynthesis in the basal member of the asteraceae. Biochemical and Biophysical Research Communications, 479, 622–627. DOI 10.1016/j.bbrc.2016.09.165. [Google Scholar] [CrossRef]
25. Panero, J. L., Funk, V. A. (2008). The value of sampling anomalous taxa in phylogenetic studies: Major clades of the asteraceae revealed. Molecular Phylogenetics and Evolution, 47, 757–782. DOI 10.1016/j.ympev.2008.02.011. [Google Scholar] [CrossRef]
26. Nguyen, T. D., Kwon, M., Kim, S. U., Fischer, C., Ro, D. K. (2019). Catalytic plasticity of germacrene a oxidase underlies sesquiterpene lactone diversification. Plant Physiology, 181, 945–960. DOI 10.1104/pp.19.00629. [Google Scholar] [CrossRef]
27. Kebbi, S., Ciavatta, M. L., Mahmoud, A. M., Carbone, M., Ligresti, A. et al. (2021). Sesquiterpene lactones with the 12, 8-guaianolide skeleton from Algerian Centaurea omphalotricha. Biomolecules, 11, 1053. DOI 10.3390/biom11071053. [Google Scholar] [CrossRef]
28. Gotoh, O. (1992). Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. Journal of Biological Chemistry, 267, 83–90. DOI 10.1016/S0021-9258(18)48462-1. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |