

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018984
ARTICLE
Cloning and Characterization of EuGID1 in Eucommia ulmoides Oliver
1The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Institute of Agro-Bioengineering and College of Life Sciences, Guizhou University, Guiyang, 550025, China
2College of Tea Sciences, Guizhou University, Guiyang, 550025, China
*Corresponding Author: Yan Li. Email: yli@gzu.edu.cn
Received: 27 August 2021; Accepted: 15 October 2021
Abstract: Gibberellic acid controlled the key developmental processes of the life cycle of landing plants, and regulated the growth and development of plants. In this study, a novel gibberellin receptor gene EuGID1 was obtained from Eucommia ulmoides Oliver. The cDNA of EuGID1 was 1556 bp, and the open reading frame was 1029 bp, which encoded 343 amino acids. EuGID1 had the homology sequence with the hormone-sensitive lipase family. Amino acid sequence alignment confirmed EuGID1 protein had the highest homology with the GID1 protein of Manihot esculenta. EuGID1 was located in the nucleus and cell membrane and had expression in four plant organs. Overexpression of EuGID1 in transgenic Arabidopsis plants promoted plant elongation and increased siliques yield.
Keywords: Eucommia ulmoides Oliver; EuGID1 gene; overexpression; subcellular localization; heterologous transformation
Gibberellin (GA) was a tetracyclic diterpenoid compound that controlled key developmental processes in the life cycle of landing plants, including seed germination, flower bud differentiation, leaf extension, rhizome elongation and xylem development [1,2]. When GA was sensed by the receptor, the signal pathway was activated and regulated downstream gene expression [3]. Gibberellin Insensitive Dwarf 1 (GID1) gene was a key receptor gene in this pathway. GID1 protein was a nucleocytoplasmic protein, which was a soluble GA-receptor, displayed high affinity for active GA [4]. DELLA protein acted as major growth repressors [5,6], it typically inhibited GA signaling [7–9]. Upon interaction with GA, GID1 underwent a conformational change that increased its affinity for DELLA. It was reported that GA first bound to the receptor protein GID1, then formed a GA-GID1-DELLA complex with DELLA protein, the formation of these stable complex helped F-box protein recognized the DELLA protein, the DELLA protein was further degraded by 26S protease ubiquitination [10,11].
GID1 was a key point in the GA signaling pathway, and GID1 gene loss-of-function mutants were usually expressed as dwarf plants. Conversely, overexpression of this gene might cause plants to be taller or larger [12,13]. The gibberellin receptor gene was cloned in rice, which was subsequently found in Arabidopsis [14], peach tree [15], dasypyrum villosum [16], wheat [17], barley [18], fern [19], tea tree [20] and other plants. There was only one gibberellin receptor in rice, and the gene in the rice mutant was first cloned and named OsGID1 by Ueguchi-Tanaka, this mutant showed severe plant dwarfing [21]. In the model plant Arabidopsis, it contained three GA receptor genes, namely AtGID1 a, AtGID1 b and AtGID1 c. Studies had found that they were orthologs to the GID1 gene in rice, and transformation experiments had also confirmed that AtGID1S could restore the dwarf phenotype of rice gidi-1 [22]. El-Sharkawy transformed the PslGID1 gene in the plum tree into the Arabidopsis gid1-ac double mutant, which was found that various growth defects in the mutant were inhibited by the presence of PslGID1, which showed that PslGID1 was similar to AtGID1 in function, and the higher the transgene level had the more active the growth performance [23]. The capsicum gibberellin receptor genes CaGID1b.1, CaGID1b.2 and CaGID1c were expressed into the Arabidopsis double mutant gid1a gid1c by CAO, among which, the effect of CaGID1b.2 was the most significant [24], the plant height and stem elongation were increased.
Eucommia ulmoides Oliver was a unique forest resource in China, whose wood yield was valued by more and more people [25]. This plant was a deciduous dioecious tree unique to the Eucommia genus in the family Eucommia [26]. The extracts obtained from different tissues of E. ulmoides showed a variety of pharmacological benefits [27–29], which was also the main source of separation and production of gutta-percha. Gene regulation of GA signaling pathway in forest trees had been widely reported [30,31], that was becoming particularly essential to dig out genes that regulated growth and development in woody plants [32,33]. GID1 gene from E. ulmoides was not been reported previously, which might play a crucial role in regulating its growth and development, and also had a guiding significance for its wood yield. This research cloned a GA receptor gene EuGID1 from E. ulmoides and constructed an overexpression vector to reveal its function, which committed to providing a theoretical basis for the study and application of EuGID1 gene function.
The age of E. ulmoides was more than 10 years old, which were planted in the Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region, Guiyang, Guizhou Province, China. The roots, stems, leaves and fruits of E. ulmoides were used for total RNA extraction. They were put into liquid nitrogen and stored at −80°C for subsequent RNA extraction. Nicotiana benthamiana seeds was provided by Key Laboratory of Plant Resources Conservation, Guiyang, Guizhou Province, China, the Arabidopsis seeds were provided by Germplasm Innovation in Mountainous Region, Guiyang, Guizhou Province, China.
2.2 5′-RACE and 3′-RACE Amplifications
The transcriptome and genome annotation database of E. ulmoides were compared to screen out homologous gene sequences. The experiment used the mRNA isolated and purified from the delicate leaves of E. ulmoides as a template, and the specific primers were synthesized by Sangon Biotech Company (Shanghai, China) (Table 1). Followed the instructions of Invitrogen 5′-RACE kit (Wuhan, China) and Clontech’s 3′-RACE kit (Dalian, China) to do the experiment, the amplification procedures used were as follows: pre-denaturation (94°C, 2 min); denaturation (94°C, 30 s), annealing (55°C, 30 s), extension (72°C, 1 min), 35 cycles; total extension (72°C, 10 min), the PCR product was ligated with pMD18T, then the positive clones were sent to Sangon Biotech Company (Shanghai, China) for direct sequencing after transformation.
2.3 Obtaining the Full-Length cDNA Sequence of the Target Gene
Two specific primers were designed, and synthesized by Sangon Biotech Company (Shanghai, China) (Table 1). The first strand of cDNA was used as a template for PCR amplification. The PCR conditions used were as follows: pre-denaturation (94°C, 2 min); denaturation (94°C, 30 s), annealing (55°C, 30 s), extension (72°C, 2 min 30 s), 35 cycles; total extension (72°C, 10 min). The PCR products were analyzed by 1.5% agarose gel electrophoresis. The experiment used the OMEGA kit to recover the agarose gel containing the target DNA (Shanghai, China). TaKaRa’s T4 ligase kit was used for ligation (Dalian, China). After transformation, the identification and screening of positive clones were carried out. The PCR procedures for identification of positive clones were as follows: pre-denaturation (94°C, 2 min); denaturation (94°C, 30 s), annealing (55°C, 30 s), extension (72°C, 1 min 40 s), 35 cycles; total extension (72°C, 10 min). After electrophoresis, the positive clones were picked out and sent to Sangon Biotech Company (Shanghai, China) for sequencing.
2.4 Bioinformatics Analysis of the EuGID1 Protein
Mega 6.0 was used for gene homology comparison; DNAMAN software was used to construct evolutionary trees; TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used to analyze protein transmembrane structure Domain; Prot Param (http://web.expasy.org/prot-param/) was used to analyze the composition and so on; and Compute pI tool (ExPASy-Compute pI/Mw tool/) was used to calculate the relative molecular weight and theoretical isoelectric point of EuGID1 protein. Prot Scale (ExPASy-ProtScale) was used for hydrophilicity and hydrophobicity analysis; Signal P 4.1 was used for signal peptide analysis; (https://NPS@:SOPM Secondary Structure Prediction (ibcp.fr)/) was used to predict protein secondary structure; and SMART (http://smart.embl-heidelberg.de/) was used to predict the functional domain of EuGID1 protein.
2.5 Construction of the Vector
Amplification primers were designed according to the full-length ORF sequence of E. ulmoides gene (Table 1). The PCR conditions used were as follows: 98°C pre-denaturation for 5 min; 98°C denaturation for 10 s, 55°C annealing for 30 s, 68°C extension for 1 min, 35 cycles; total Extension at 68°C for 5 min. The PCR products were analyzed after electrophoresis on a 1.5% agarose gel. The PCAMBIA-super1300 plasmid was subjected to double digestion with XbaI and PstI (provided by Prof. Qinglin Liu, Sichuan Agricultural University), then the PCR product and double digestion product were recovered. The target gene was ligated with the PCAMBIA-super1300 overexpression vector. The PCAMBIA-super1300-35S::EuGID1::EGFP fusion expression vector was transformed into E. coli DH5α competent cells, and then the positive clones were identified by PCR. After the plasmid containing the target gene was extracted, it was transformed into Agrobacterium GV3101 competent cells. The detection primers were the same as the amplification primers. The PCR conditions used were as follows: pre-denaturation (94°C, 2 min); denaturation (94°C, 30 s), annealing (55°C, 30 s), extension (72°C, 1 min), 35 cycles; total extension (72°C, 10 min). The positive monoclonal colonies were preserved.
2.6 Subcellular Localization of EuGID1 Protein
Nicotiana benthamiana seeds were sown and cultivated for 1 month under the conditions of light for 12 h, temperature of 25°C and humidity of 60%; 35S::EuGID1::EGFP overexpression vector and 35S-sGFP empty vector were amplified by Agrobacterium. The experiment used a 1 mL syringe with a pipette tip to inject the epidermis of Nicotiana benthamiana leaves and mark them: plants were cultured in low light for 2 days and observed using a laser confocal microscope (Nikon C2-ER). The excitation wavelength of chloroplast fluorescence signal was as follows: 640 nm, emission wavelength: 675 nm. Green fluorescent protein GFP: excitation light 488 nm, emission light 510 nm.
2.7 Tissue-Specific Expression Analysis of EuGID1
The CTAB method was used to extract RNA from roots, stems and leaves of E. ulmoides seedlings and RNA from E. ulmoides fruits, with 3 replicates in each tissue. Invitrogen’s superscript III first-strand synthesis system was used to synthesize the first strand of cDNA. The fluorescent quantitative PCR primers of EuGID1 gene were designed and synthesized according to the instrument operation guide (Table 1). Using EuActin as the internal standard (Table 1), the 7500 Real-time PCR system (ABI, USA) was used for amplification, and the relative gene expression was calculated according to the software provided by ABI. This experiment was analyzed and compared the specific expression of EuGID1 gene in different organs and tissues of various E. ulmoides plants.
2.8 Genetic Transformation of EuGID1
Arabidopsis seeds were sown and cultivated under the conditions of light for 12 h, temperature of 23°C. The Agrobacterium solution containing the overexpression vector was used as the working solution, and the flowering Arabidopsis plants were transformed by the floral dip method. The T0 generation seeds were collected and screened on the MS solid medium plate containing hygromycin. The total DNA of leaves was used CTAB Method extraction, and the hygromycin gene-specific primers were used for PCR identification and detection. T2 generation was screened by same method, and the gene-specific primers were used for PCR identification and detection (Table 1).
3.1 Obtaining of the Full-Length Sequence of EuGID1 Gene
Based on the data acquired from a previous transcriptomics experiment, the 5′-RACE, 3′-RACE and ORF cloning was used to obtain the full-length sequence of the gene (Fig. 1). After the sequence was spliced and the vector sequence was removed, the comparison was performed on NCBI to further confirmed the final result. The sequencing result was as follows: the EuGID1 was named according to its strong sequence similarity to the Arabidopsis gene AtGID1. The full-length EuGID1 cDNA was 1556 bp, the open reading frame (ORF) was 1029 bp in length. The 5′untranslated region (UTR) was 84 bp, 3′untranslated region (UTR) was 443 bp, and there was an obvious tailing signal downstream of its 3′end (Fig. 2).
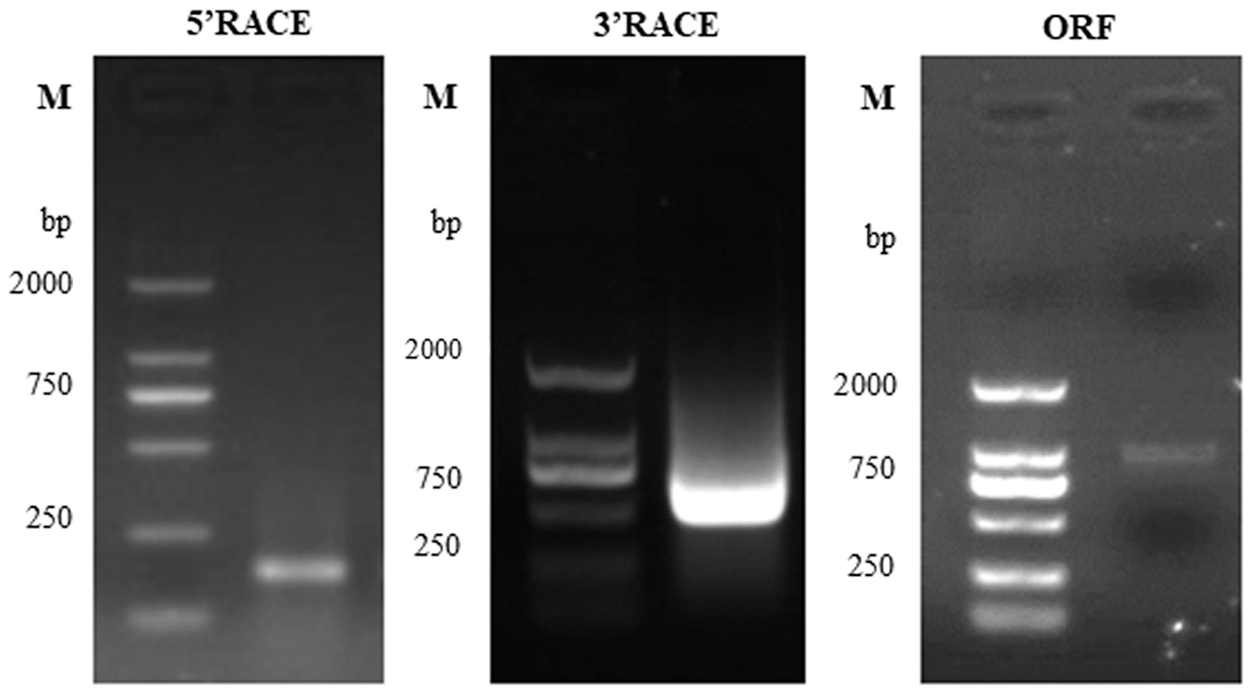
Figure 1: Electrophoresis results of EuGID1 cloning
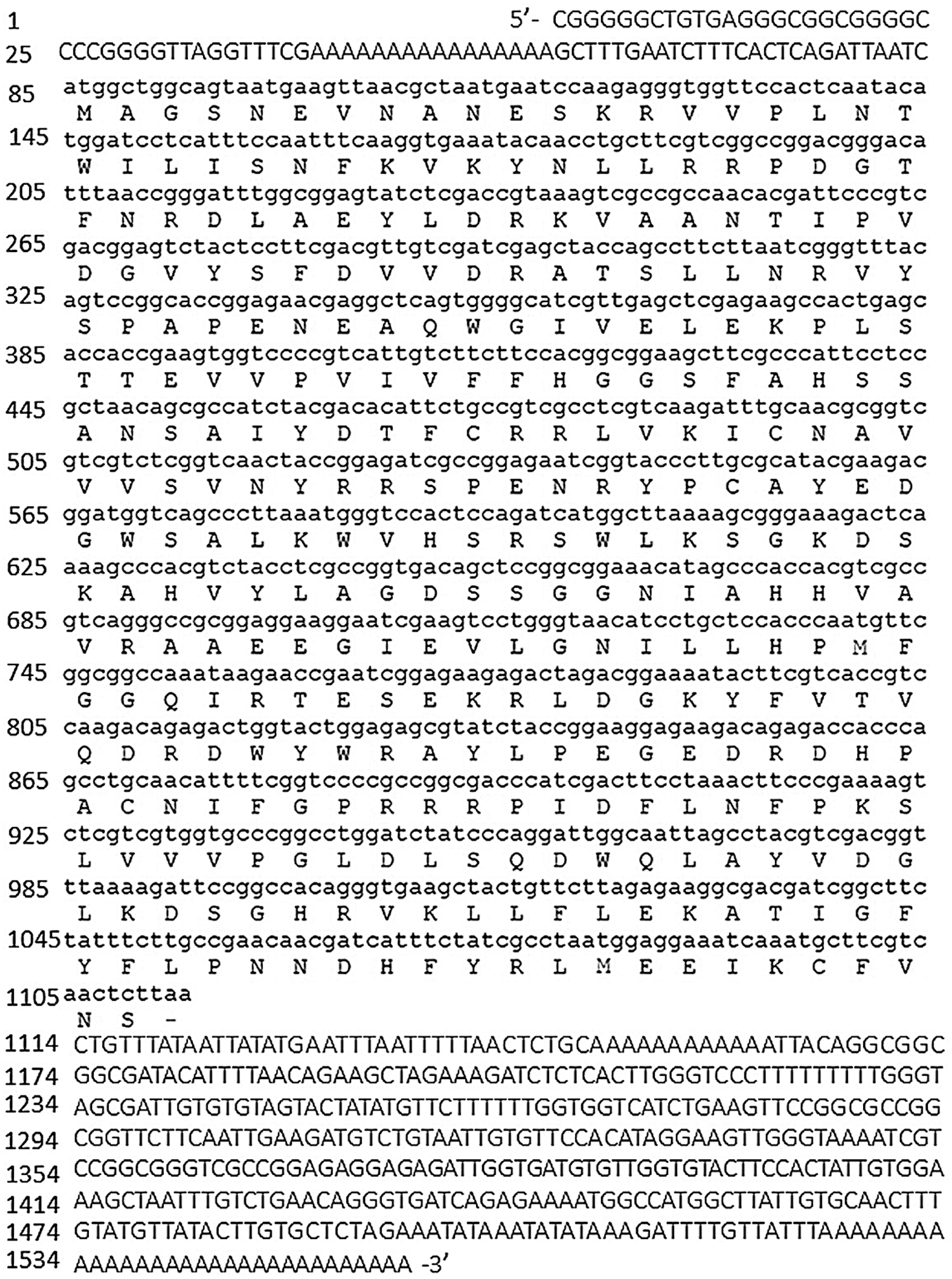
Figure 2: Full-length of EuGlD1 gene
3.2 Molecular Characteristics of the EuGID1 Protein
The gene encoded 342 amino acids, and the molecular formula was C1756H2687N485O501S8, Molecular weight was equal to 38.87 kDa, the theoretical isoelectric point was 7.15, the positive and negative charge residues were 41, and the fat coefficient was 86.32. Used NCBI BLAST to search, the protein ID number was XP_012092333.1. The protein sequence encoded by this gene had no transmembrane domain and no signal peptide, whose hydrophilicity was −0.296. The instability coefficient of the encoded protein sequence was 44.33, which was an unstable protein. The gene predicted protein secondary structure: there were more α-helixes and random coils, α-helix (α-helix) was 30.99%, random coil (random coil) was 34.50%, β-turn (β-turn) turn) was less at 10.23%.
3.3 Amino Acid Sequence Alignment of EuGID1 Protein
EuGID1 belonged to the α/β hydrolase superfamily. The protein sequence encoded by this gene contained the conserved sequence of the hormone-sensitive lipase family (HSL), which had 15 key sites for the binding of GID1 to active GA. In addition, GID1 protein bound GA and DELLA protein to a total of 13 functional domains. The amino acid sequence included TWVLIS, LDR, FFHGGSF, HS, IYD, YRR, DGW, GDSSGGNI, GNI, MF, LDGKYF, WYW and GFY functional domains. Among them, the V (valine) of the TWVLIS domain in EuGID1 was replaced by I (isoleucine) (Fig. 3), which showed that these domains play a crucial role in the process of GA and DELLA binding. Through comparison, it was found that EuGID1 protein sequence was similar to Jatropha curcas, Manihot esculenta, Camellia sinensis, Hevea brasiliensis, Populus trichocarpa, Capsicum annuum, and Durian. The similarity of was 82.32%∼85.17%. Among them, it had the highest homology with Manihot esculenta, up to 85.17%. It had the lowest homology with Camellia sinensis, only 82.32%.
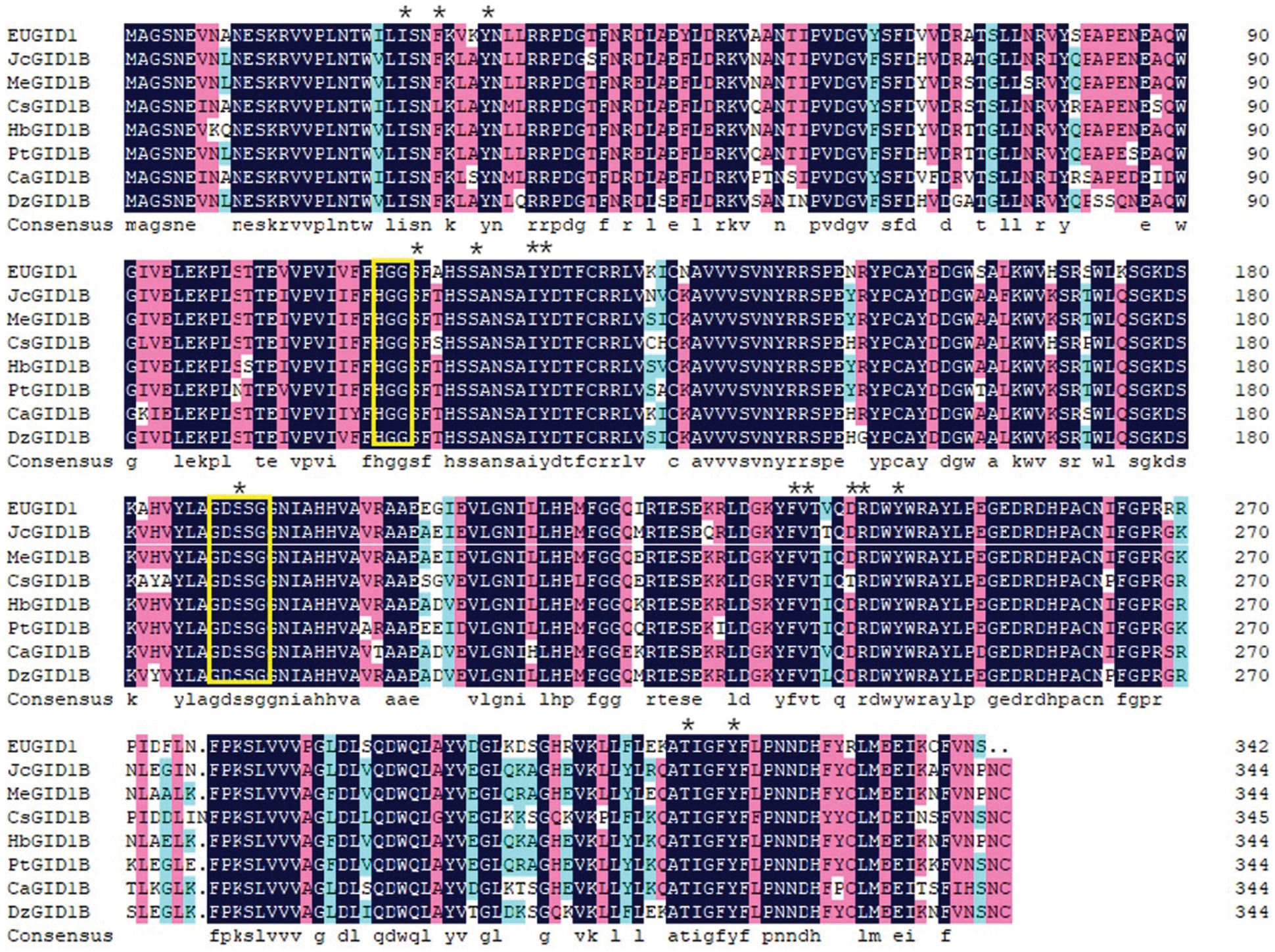
Figure 3: Amino acid sequence comparison between EuGID1 and other species
Jc: Jatropha curcas; Me: Manihot esculenta; Cs: Camellia sinensis; Hb: Hevea brasiliensis; Pt: Populus trichocarpa; Ca: Capsicum annuum; Dz: Durian. *Represented the key site where GID1 bind to active GA. The sequence in the yellow box was the conserved sequence of the hormone-sensitive lipase family (HSL).
3.4 Phylogenetic Analysis of EuGID1
A comparison of EuGID1 with the GID1 protein sequences of 28 representative plant species from NCBI (Fig. 4), which revealed that 29 species of plants were divided into 3 clusters: EuGID1 and Camellia sinensis, Nyssa sinensis, Panax ginseng, Coffea arabica, Ipomoea triloba, Ipomoea nil, Capsicum chinense, Petunia x hybrida, Nicotiana attenuata, Solanum lycopersicum, Solanum tuberosum were clustered into one branch; Jatropha curcas, Ricinus communis, Manihot esculenta, Hevea brasiliensis, Populus euphratica, Populus tomentosa, Populus alba, Castanea mollissima, Hibiscus syriacus, Durio zibethinus, Gossypium hirsutum, Gossypium arboreum, Gossypium barbadense and Gossypium mustelinum were the second cluster. Momordica charantia, Cucumis sativus, Cucumis melo were the third cluster.
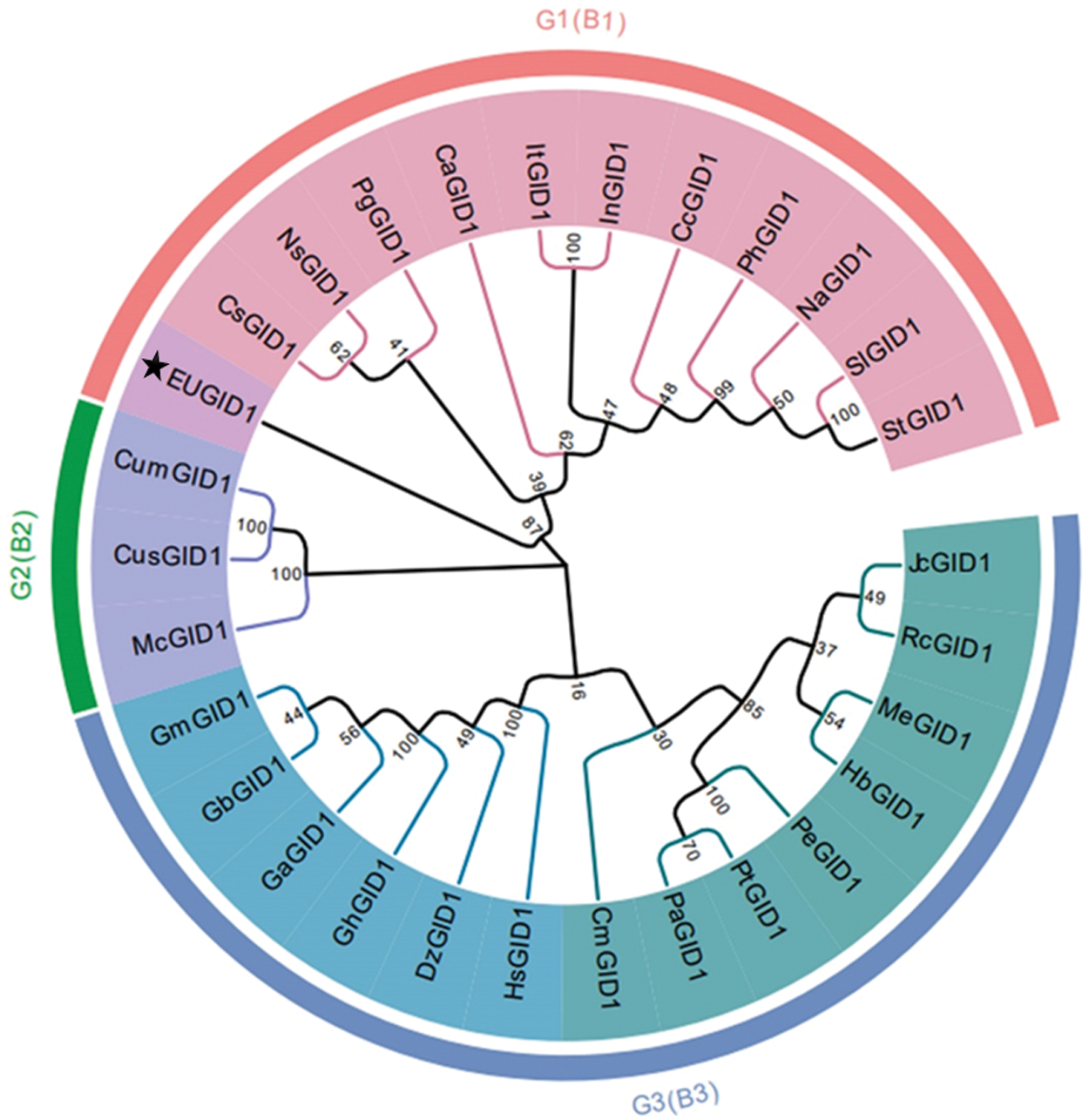
Figure 4: Phylogenetic tree of EuGID1
The following species were arranged into a phylogenetic tree using the neighbor connection method: Camellia sinensis KAF5930343.1 (CsGID1), Nyssa sinensis KAA8550923.1 (NsGID1), Panax ginseng QEX51270.1 (PgGID1), Coffea arabica XP_027097574.1 (CaGID1), Ipomoea triloba XP_031100613.1 (ItGID1), Ipomoea nil XP_019197492.1 (InGID1), Capsicum chinense PHU22188.1 (CcGID1), Petunia x hybrida AGN72649.1 (PhGID1), Nicotiana attenuate XP_019232495.1 (NaGID1), Solanum lycopersicum NP_001352577.1 (SlGID1), Solanum tuberosum XP_006362976.1 (StGID1), Jatropha curcas XP_012092333.1 (JcGID1), Ricinus communis XP_002524767.1 (RcGID1), Manihot esculenta XP_021613000.1 (MeGID1), Hevea brasiliensis XP_021681392.1 (HbGID1), Populus euphratica XP_011009471.1 (PeGID1), Populus topulus alba ANB66267.1 (PtGID1), Populus alba XP_034908255.1 (PaGID1), Castanea mollissima KAF3975015.1 (CmGID1), Hibiscus syriacus KAE8692101.1 (HsGID1), Durio zibethinus XP_022771057.1 (DzGID1), Gossypium hirsutum ABQ96123.1 (GhGID1), Gossypium arboretum XP_017627860.1 (GaGID1), Gossypium barbadense KAB2056790.1 (GbGID1), Gossypium mustelinum TYJ09289.1 (GmGID1), Momordica charantia XP_022150283.1 (McGID1), Cucumis sativus NP_001277149.1 (CusGID1), Cucumis melo XP_008451414.1 (CumGID1), ★ represented EuGID1 protein.
3.5 Overexpression Vector Construction and Subcellular Localization of EuGID1 Protein
An overexpression vector of 35S::EuGID1::EGFP was constructed (Fig. 5A). PCR verification and sequencing showed that EuGID1 had been inserted into the vector (Fig. 5B). The laser confocal microscope was used to determine the accurate localization of EuGID1 protein subcellular, EuGID1 and 35S-sEGFP enhanced green fluorescent protein were fused, and the transient expression of the epidermal cells of leaves was observed under a laser confocal microscope (Fig. 6). With 35S-sGFP empty as a control, a 35S::EuGID1::EGFP overexpression vector was constructed. The experimental results showed that the fluorescence distribution was determined on the nucleus and cell membrane.
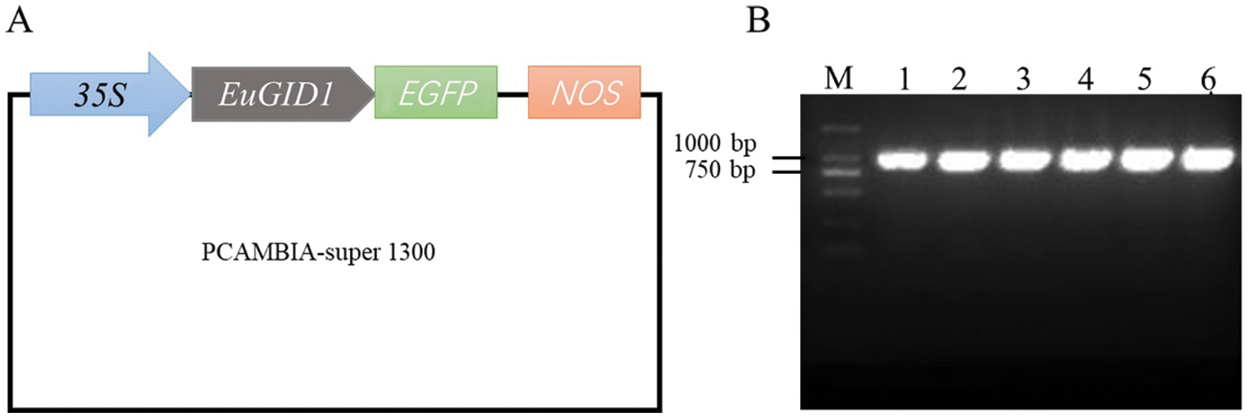
Figure 5: Construction of 35S::EuGID1::EGFP overexpression vector and PCR detection of agrobacterium liquid. (A) 35S::EuGID1::EGFP overexpression vector structure. In the PCAMBIA-super1300 vector, 35S was the strong promoter in the vector, EGFP was the green fluorescent protein signal, and NOS was the terminator. (B) Lanes 1–6 were PCR products of single colonies with 35S::EuGID1::EGFP, M was for DNA marker
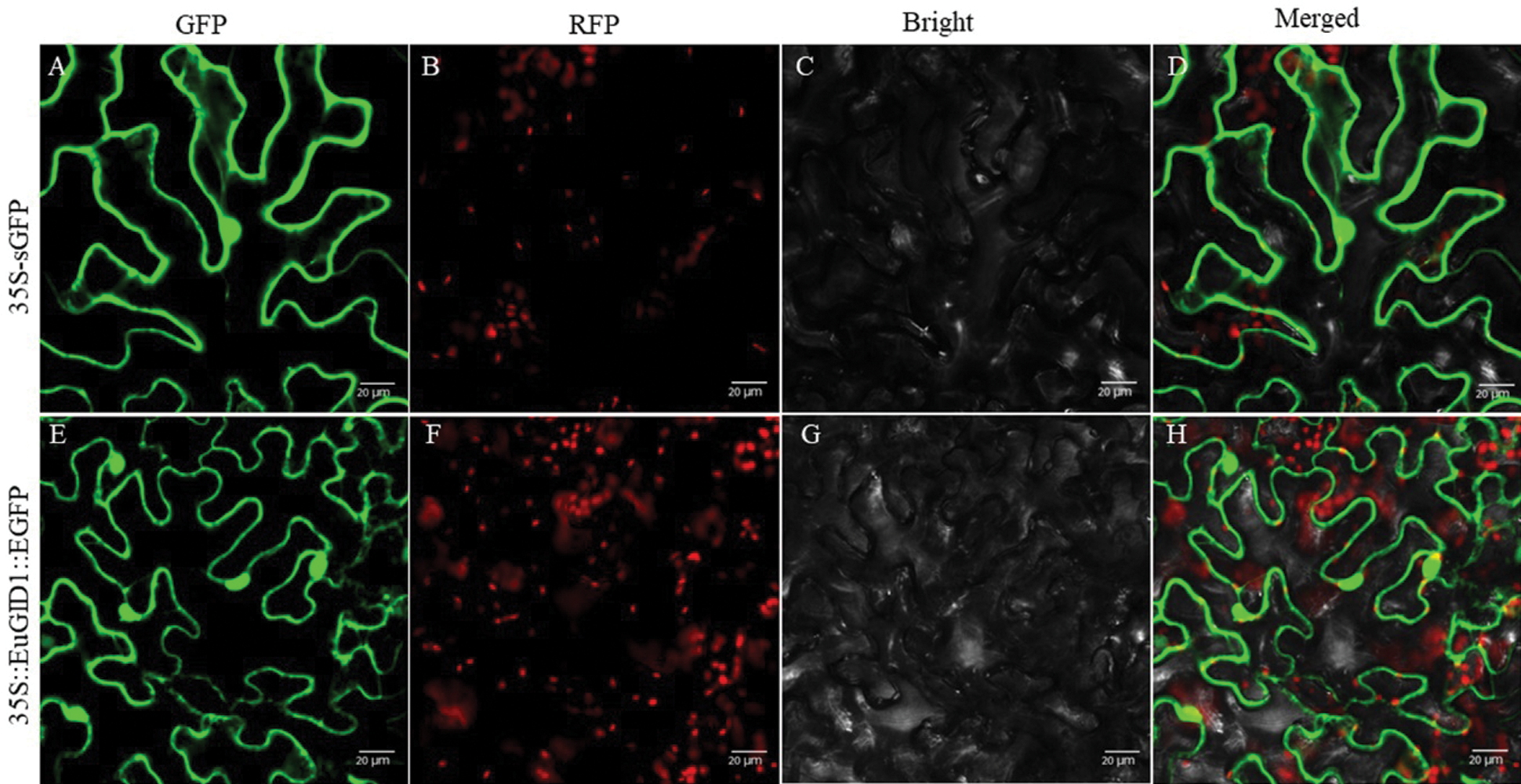
Figure 6: Analysis of subcellular localization of EuGID1 protein in Nicotiana benthamiana leaves. (A, E) GFP field; (B, F) RFP field; (C, G) Bright field; (D, H) Merged pictures. 35S-sGFP indicated the empty vector, and 35S::EuGID1::EGFP indicated the fusion protein with EuGID1. Bar: 20 μm
3.6 Relative Expression of GA Receptor Gene EuGID1
The expression of EuGID1 gene was detected in four organs, that implied this gene has a role in these organs. Real-time fluorescent quantitative PCR was used to analyze the expression pattern of this gene in the roots, stems, leaves and fruits of E. ulmoides (Fig. 7), The research found: with higher expression in roots and fruits, followed by stems and leaves, that the expression level in the root was 2.17 times the expression level in the stem, and the expression level in the root was 2.46 times the expression level in the leaf. It indicated that EuGID1 gene had the highest expression in roots in four organs, which might be an important effect gene in root development.
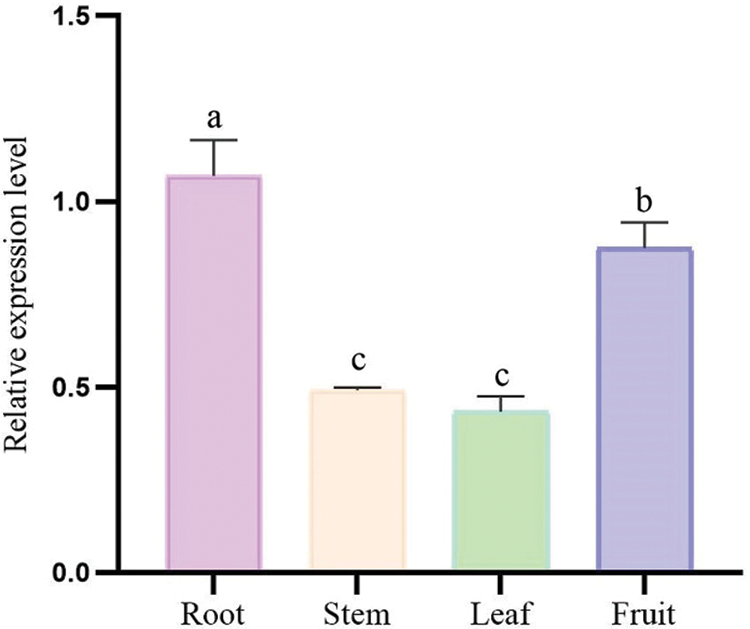
Figure 7: The relative expression of EuGID1 in the organs of E. ulmoides. Data are mean SDs (n = 3). Different letters indicate the difference significance at the 0.05 level, respectively.
3.7 Genetic Transformation and Phenotype Analysis of EuGID1
EuGID1 gene might have similar functions to AtGID1 in stem elongation and fruit development (Fig. 8B). Under the same growth conditions, the plant height and the number of siliques in the same period were measured (Fig. 8A). It was found that the average plant height of positive regenerated plants was 24.57 cm, and the average plant height of the wild-type plants in the same period was 19.37 cm (Fig. 8C), thus, the average plant height of the positive regenerated plants was 5.20 cm higher than the average plant height of the wild type. All transgenic types showed more significant elongation than the wild type, and positive regenerated plants had an average number of siliques of 171.80, while the average number of siliques of the wild type during the same period was 85.17 (Fig. 8C). The average number of siliques of the positive regenerated plants was about twice that of the wild type. All transgenic plants showed earlier silique maturation. There was no significant difference in the length of the siliques, the number of seeds in each silique and the size of the seeds. In Arabidopsis, heterologous transformation increased the number of gibberellin receptors and triggered the overall growth and development.
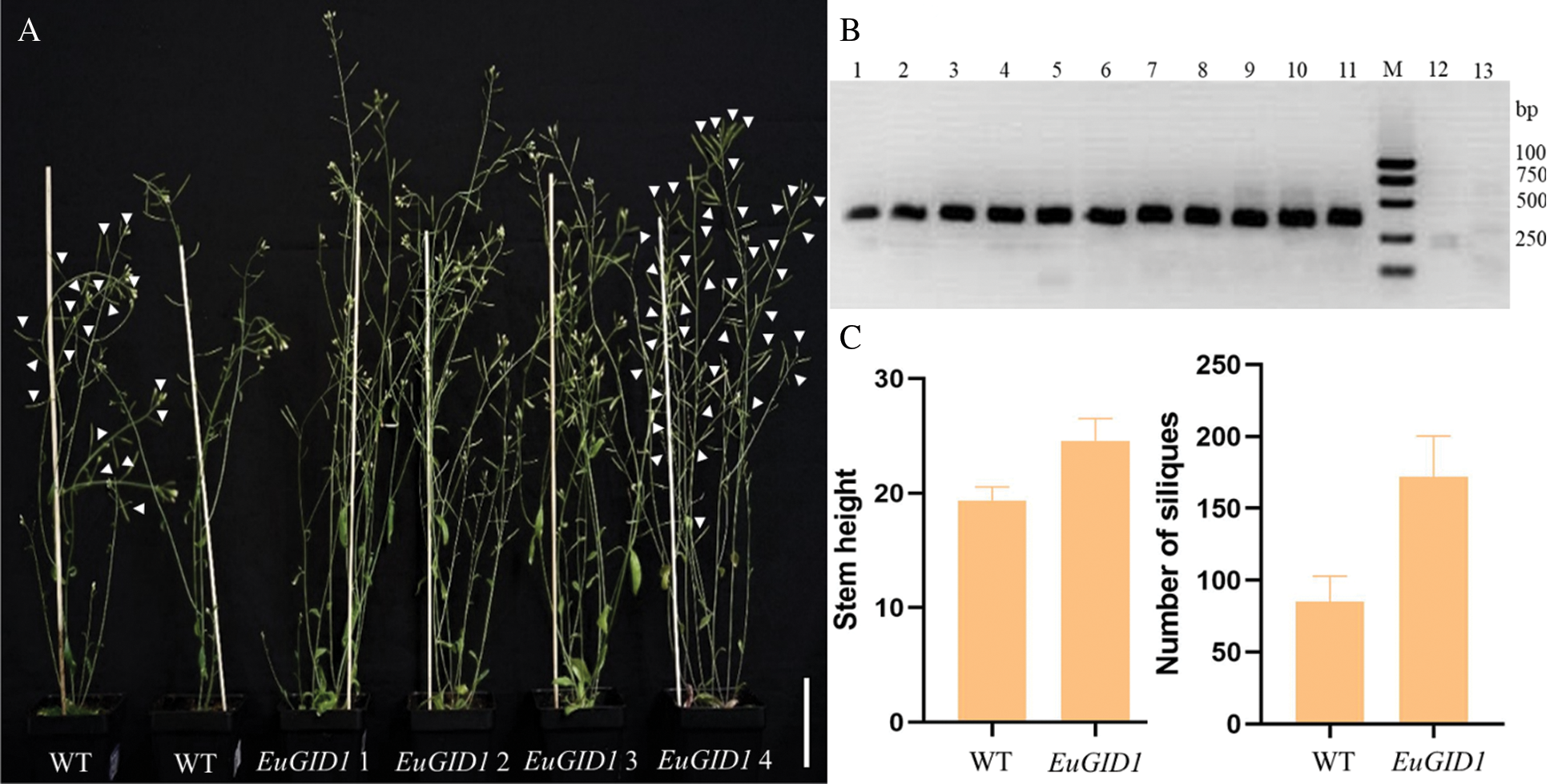
Figure 8: Phenotypic overexpression of EuGID1 gene in wild-type Arabidopsis. (A) The gross morphology of the wild-type plants and transgenic plants at the life stage; arrows indicate the siliques; Bar = 5 cm. (B) Detection map of the target gene intermediate fragment of Arabidopsis positive plants. Lanes 1–10 were transformed plants with EuGID1 gene, 11 was Plasmid control, M was DNA marker, 12–13 were wild type plants. (C) The comparison diagram of the stem height and the number of siliques between wild-type plants and positive plants
Gibberellin receptors were widely in plants, which was considered a key factor in the gibberellin signal transduction pathway. Since the gibberellin receptor gene was first positionally cloned in rice mutants [21], it was found that the GID1 gene played a significant role in regulating plant height, flower development and fruit length [34,35]. This study used E. ulmoides transcriptome and genome database as a reference to screen out homologous gene sequences [36], and RACE technology was used to amplify, sequence, and splice to obtain the full-length cDNA sequence of EuGID1. The full length of the amino acid sequence and the physical and chemical properties were similar to those of other species GID1 [37,38].
The EuGID1 gene might be the receptor gene of active GA in E. ulmoides, it was involved in the recognition and transduction of GA signals. EuGID1 protein belonged to the α/β hydrolase superfamily. The amino acid sequence of EuGID1 and other species were compared, the protein sequence encoded by this gene contained the conserved sequence of the hormone-sensitive lipase family (HSL), included the conserved HSL motifs HGG and GXSXG [39]. Among the three important amino acids in the HSL family that could form a catalytic triad (Ser, Asp and His), Asp and Ser were conserved in EuGID1 sequence, His was replaced by Ile, which might be the reason why they lost catalytic ability [14], which was consistent with the amino acid sequence of GID1 in poplar and Arabidopsis [22,40]. Compared the amino acid sequence, it was found that the homology was greater than 82%, indicated that the gene had a high degree of conservation in evolution. The construction of amino acid phylogenetic tree analysis showed that it was divided into 3 clusters.
GA synthesis was carried out in different area of the cell. In the relevant research on the GID1 gene report, the subcellular localization analysis found that the GID1 protein of rice, grape, and newhall navel orange were all expressed in the nucleus [21,41,42]. The localization results of this study indicated that EuGID1 protein was localized in the nucleus and cell membrane, result was for further study. Gibberellin was a hormone involved in multiple biological processes [43]. The results showed that EuGID1 was expressed in all organs of E. ulmoides, which was similar to the GID1 gene in Arabidopsis and grapes [35,41], this was consistent with GA being active during plant development [44]. Studies had found that the highest expression level in the roots, the maximum expression level of the MsGID1b gene in alfalfa and the PttGID1.4 gene in hybrid poplars also appeared in the roots [40,45]. The expression level in the roots and fruits of E. ulmoides was significantly higher than that in the stems and leaves, which was consistent with the conclusion that Arabidopsis AtGID1B was a minor effect gene in stem elongation, but AtGID1B was a major effect gene in root development [46]. Therefore, the expression of this gene was more advantageous in tissues with slow differentiation rate [47].
Gibberellin enhanced plant growth characteristics by stimulating the interaction between GID1 and DELLA, which triggered the degradation of DELLA and activated the overall GA response. Nakajima found three AtGID1 genes in Arabidopsis through homologous gene analysis [22]. Griffiths Jayne reported that the mutations of three AtGID1 genes in Arabidopsis resulted in significant dwarfing characteristics, including height reduction, slow growth and low seed setting rate [35]. When genes from other species were overexpressed in Arabidopsis, the phenotype of GID1 gene loss-of-function mutants might be appeared normal type or even larger type [46]. In this study, the average plant height of Arabidopsis positive regenerated plants was 5.20 cm higher than that of the wild type. The average siliques of the positive regenerated plants were about twice the average number of wild-type siliques, and all transgenic plants showed earlier silique maturation, which indicated that EuGID1 was similar to AtGID1 in function. This result was consistent with that El-Sharkawy transformed the PslGID1 gene from the plum tree into the gid1-ac double mutant of Arabidopsis, the resulting transgenic plants were taller than the control plants and the leaves of the transgenic plants were longer than the control group. This conclusion was also basically consistent with Li Jun’s conclusion: the GoGID gene from Galega orientalis Lam was transformed into the Arabidopsis gid1ac mutant, which showed increased plant height and increased germination rate [23,48]. Therefore, the overexpression of the EuGID1 gene had a significant effect on the growth and development of the gibberellin. It was reasonable to speculate that overexpression of the EuGID1 gibberellin receptor gene could speed up plant growth, increase plant height, and increase biomass.
In this work, a gibberellin receptor gene was obtained, EuGID1 had the homology sequence with the hormone-sensitive lipase family, it was located in the nucleus and cell membrane. EuGID1 had expression in four organs. Overexpression of EuGID1 in transgenic Arabidopsis plants promoted plant elongation and increased siliques yield. It provided excellent candidate genes for molecular breeding of E. ulmoides. In the future, we will continue to determine the relative expression level of transgenic Arabidopsis and the morphological phenotype of transgenic Arabidopsis after GA treatment.
Author Contributions: Yan Li designed the research and conducted the experiments. Yulu Chen collected and analyzed the data. All authors approved the final version of manuscript.
Funding Statement: This work was supported by grants from the Open Project of Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education) (MOELP-201801), the Young Scholars and Technology Talents Development Project of Guizhou Education Department KY ([2018]124) and the Introduction of Talent Project of Guizhou University ([2017]56).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Sun, T. P. (2011). The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Current Biology, 21(9), R338–345. DOI 10.1016/j.cub.2011.02.036. [Google Scholar] [CrossRef]
2. Yamamoto, Y., Hirai, T., Yamamoto, E., Kawamura, M., Sato, T. et al. (2010). A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell, 22(11), 3589. DOI 10.1105/tpc.110.074542. [Google Scholar] [CrossRef]
3. Gazara, R. K., Moharana, K. C., Bellieny-Rabelo, D., Venancio, T. M. (2017). Expansion and diversification of the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) family in land plants. Plant Molecular Biology, 97, 435–449. DOI 10.1007/s11103-018-0750-9. [Google Scholar] [CrossRef]
4. Livne, S., Weiss, D. (2014). Cytosolic activity of the gibberellin receptor GIBBERELLIN INSENSITIVE DW ARF1A. Plant Cell Physiology, 55(10), 1727–1733. DOI 10.1093/pcp/pcu104. [Google Scholar] [CrossRef]
5. Harberd, N. P., Belfield, E., Yasumura, Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. The Plant Cell, 21(5), 1328–1339. DOI 10.1105/tpc.109.066969. [Google Scholar] [CrossRef]
6. Sun, T. P. (2010). Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiology, 154(2), 567–570. DOI 10.1104/pp.110.161554. [Google Scholar] [CrossRef]
7. Dill, A., Jung, H. S., Sun, T. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proceedings of the National Academy of Sciences, 98(24), 14162–14167. DOI 10.1073/pnas.251534098. [Google Scholar] [CrossRef]
8. Silverstone, A. L., Jung, H. S., Dill, A., Kawaide, H., Sun, T. P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in arabidopsis. The Plant Cell, 13(7), 1555–1566. DOI 10.1105/TPC.010047. [Google Scholar] [CrossRef]
9. Nelson, S. K., Steber, C. M. (2016). Gibberellin hormone signal perception: Down-regulating DELLA repressors of plant growth and development. UK: John Wiley & Sons, Ltd. [Google Scholar]
10. Shimada, A., Ueguchi-Tanaka, M., Nakatsu, T., Nakajima, M., Naoe, Y. et al. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature, 456(7221), 520–523. DOI 10.1038/nature07546. [Google Scholar] [CrossRef]
11. Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K. et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science, 299(5614), 1896–1898. DOI 10.1126/science.1081077. [Google Scholar] [CrossRef]
12. Ariizumi, T., Murase, K., Steber, S. C. M. (2008). Proteolysis-independent downregulation of DELLA repression in arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. The Plant Cell, 20(9), 2447–2459. DOI 10.1105/tpc.108.058487. [Google Scholar] [CrossRef]
13. Li, J., Gao, H., Jiang, J., Dzyubenko, N., Chapurin, V. et al. (2013). Overexpression of the Galega orientalis gibberellin receptor improves biomass production in transgenic tobacco. Plant Physiology & Biochemistry, 73, 1–6. DOI 10.1016/j.plaphy.2013.07.015. [Google Scholar] [CrossRef]
14. Murase, K., Hlirano, Y., Sun, T. P., Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature, 456(7221), 459–63. DOI 10.1038/nature07519. [Google Scholar] [CrossRef]
15. Hollender, C. A., Hadiarto, T., Srinivasan, C., Scorza, R., Dardick, C. (2015). A brachytic dwarfism trait (dw) in peach trees is caused by a nonsense mutation within the gibberellic acid receptor PpeGID1c. New Phytologist, 210(1), 227–239. DOI 10.1111/nph.13772. [Google Scholar] [CrossRef]
16. Razumova, O. V., Bazhenov, M. S., Nikitina, E. A., Nazarova, L. A., Divashuk, M. G. (2020). Molecular analysis of gibberellin receptor gene GID1 in Dasypyrum villosum and development of DNA marker for its identification. RUDN Journal of Agronomy and Animal Industries, 15(1), 62–85. DOI 10.22363/2312-797X-2020-15-1-62-85. [Google Scholar] [CrossRef]
17. Li, A., Yang, W., Li, S. D., Guo, X., Sun, J. et al. (2013). Molecular characterization of three GIBBERELLIN-iNSENSITIVE DWARF1 homologous genes in hexaploid wheat. Journal of Plant Physiology, 170(4), 432–443. DOI 10.1016/j.jplph.2012.11.010. [Google Scholar] [CrossRef]
18. Chandler, P. M., Harding, C. A., Ashton, A. R., Mulcair, M. D., Dixon, N. E. et al. (2008). Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Molecular Plant, 1(2), 285–294. DOI 10.1093/mp/ssn002. [Google Scholar] [CrossRef]
19. Yasumura, Y., Crumpton-Taylor, M., Fuentes, S., Harberd, N. P. (2007). Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Current Biology, 17(14), 1225–1230. DOI 10.1016/j.cub.2007.06.037. [Google Scholar] [CrossRef]
20. Yue, C., Zeng, J., Cao, H., Hao, X., Yang, Y. (2013). Cloning and expression analysis of gibberellin receptor gene csgid1a in tea plant (Camellia sinensis). Acta Agronomica Sinica, 39(4), 599–608. DOI 10.3724/SP.J.1006.2013.00599. [Google Scholar] [CrossRef]
21. Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E. et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature, 437(7059), 693–8. DOI 10.1038/nature04028. [Google Scholar] [CrossRef]
22. Nakajima, M., Shimada, A., Takashi, Y., Kim, Y. C., Park, S. H. et al. (2010). Identification and characterization of Arabidopsis gibberellin receptors. Plant Journal, 46(5), 880–889. DOI 10.1111/j.1365-313X.2006.02748.x. [Google Scholar] [CrossRef]
23. El-Sharkawy, I., Sherif, S., Kayal, W. E., Mahboob, A., Abubaker, K. et al. (2014). Characterization of gibberellin-signalling elements during plum fruit ontogeny defines the essentiality of gibberellin in fruit development. Plant Molecular Biology, 84(4–5), 399. DOI 10.1007/s11103-013-0139-8. [Google Scholar] [CrossRef]
24. Cao, Y. C., Zhang, Z. H., Wang, L. H., Sui, X. L., Zhang, Z. X. et al. (2016). Cloning and characterization of CaGID1S and CAGAI in Capsicum annuum L. Journal of Integrative Agriculture, 15(4), 775–784. DOI 10.1016/S2095-3119(15)61275-8. [Google Scholar] [CrossRef]
25. Suzuki, N., Uefuji, H., Nishikawa, T., Mukai, Y., Nakazawa, Y. (2012). Construction and analysis of est libraries of the trans-polyisoprene producing plant, Eucommia ulmoides oliver. Planta, 236(5), 1405–1417. DOI 10.1007/s00425-012-1679-x. [Google Scholar] [CrossRef]
26. Zhu, M. Q., Sun, R. C. (2018). Eucommia ulmoides oliver: A potential feedstock for bioactive products. Journal of Agricultural & Food Chemistry, 66(22), 5433. DOI 10.1021/acs.jafc.8b01312. [Google Scholar] [CrossRef]
27. Cho, S., Hong, R., Yim, P., Yeom, M., Lee, B. et al. (2017). An herbal formula consisting of Schisandra chinensis (Turcz.) Baill, Lycium chinense Mill and Eucommia ulmoides Oliver alleviates disuse muscle atrophy in rats. Journal of Ethnopharmacology, 213, 328–339. DOI 10.1016/j.jep.2017.10.008. [Google Scholar] [CrossRef]
28. Fujiwara, A., Nishi, M., Yoshida, S., Hasegawa, M., Yasuma, C. et al. (2016). Eucommicin A, a beta-truxinate lignan from Eucommia ulmoides, is a selective inhibitor of cancer stem cells. Phytochemistry, 122, 139–145. DOI 10.1016/j.phytochem.2015.11.017. [Google Scholar] [CrossRef]
29. Xiang, C., Li, Y., Xiao, Y., Chen, Y., Li, W. et al. (2016). Chlorogenic acid-enriched extract from Eucommia ulmoides leaves inhibits hepatic lipid accumulation through regulation of cholesterol metabolism in hepg2 cells. Pharmaceutical Biology, 54(2), 251–259. DOI 10.3109/13880209.2015.1029054. [Google Scholar] [CrossRef]
30. Ni, J., Zhao, M. L., Chen, M. S., Pan, B. Z., Tao, Y. B. et al. (2017). Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellin A3 and 6-benzyladenine in Jatropha curcas. Scientific Reports, 7(1), 11417. DOI 10.1038/s41598-017-11588-0. [Google Scholar] [CrossRef]
31. Yang, H., Yang, X., Wang, L., Gong, C., Chen, B. et al. (2016). Single nucleotide polymorphisms in two GID1 orthologs associate with growth and wood property traits in Populus tomentosa. Tree Genetics & Genomes, 12(6), 109. DOI 10.1007/s11295-016-1070-3. [Google Scholar] [CrossRef]
32. Wisniewski, M., Norelli, J., Artlip, T. (2015). Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Frontiers in Plant Science, 85(6), 1–13. DOI 10.3389/fpls.2015.00085. [Google Scholar] [CrossRef]
33. Liu, Q., Ding, C., Chu, Y., Chen, J., Zhang, W. et al. (2016). Poplargene: Poplar gene network and resource for mining functional information for genes from woody plants. Scientific Reports, 6(1), 31356. DOI 10.1038/srep31356. [Google Scholar] [CrossRef]
34. Kwame, A. A., Hu, J., Ariel, R., Zheng, C., Tamar, H. et al. (2015). Functional characterization and developmental expression profiling of gibberellin signalling components in Vitis vinifera. Journal of Experimental Botany, 66(5), 1463–1476. DOI 10.1093/jxb/eru504. [Google Scholar] [CrossRef]
35. Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z. L. et al. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell, 18(12), 3399–3414. DOI 10.1105/tpc.106.047415. [Google Scholar] [CrossRef]
36. Zhao, D. G., Li, Y., Zhao, Y. C., Zhao, D., Lv, L. T. et al. (2015). Transcriptome data assembly and gene function annotation of female and male plants in Eucommia ulmoides. Journal of Mountain Agriculture and Biology, 34(01), 1–13. DOI 10.15958/j.cnki.sdnyswxb.2015.01.001. [Google Scholar] [CrossRef]
37. Aleman, L., Kitamura, J., Abdel-Mageed, H., Lee, J., Yan, S. et al. (2008). Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Molecular Biology, 68(1–2), 1. DOI 10.1007/s11103-008-9347-z. [Google Scholar] [CrossRef]
38. Zhou, T., Wang, P., Sun, J. K., Meng, L. I. (2017). Cloning of GID1-homologous genes and expression analysis during seed germination in Zanthoxylum dissitum Hemsl. Plant Physiology Journal, 53(8), 1499–1506. DOI 10.13592/j.cnki.ppj.2017.0078. [Google Scholar] [CrossRef]
39. Hirano, K., Ueguchi-Tanaka, M., Matsuoka, M. (2008). GID1-mediated gibberellin signaling in plants. Trends in Plant Science, 13(4), 192–199. DOI 10.1016/j.tplants.2008.02.005. [Google Scholar] [CrossRef]
40. Mauriat, M., Moritz, T. (2010). Analyses of GA20ox-and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant Journal, 58(6), 989–1003. DOI 10.1111/j.1365-313X.2009.03836.x. [Google Scholar] [CrossRef]
41. Wang, X. C., Weimin, W. U., Fang, J. G., Qian, Y. M., Wang, C. et al. (2013). Isolation, subcellular localization and expression analysis of gibberellin receptor gene VvGID1A from grapevine. Acta Horticulturae Sinica, 40(5), 839–848. DOI 10.16420/j.issn.0513-353x.2013.05.006. [Google Scholar] [CrossRef]
42. Shen, S. H. (2012). Functional analysis of gibberellin receptor CsGID1 from newhall navel orange. China: Huazhong Agricultural University. [Google Scholar]
43. Binenbaum, J., Weinstain, R., Shani, E. (2018). Gibberellin localization and transport in plants. Trends in Plant Science, 5(23), 410–421. DOI 10.1016/j.tplants.2018.02.005. [Google Scholar] [CrossRef]
44. Fleet, C. M., Sun, T. P. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Current Opinion in Plant Biology, 8(1), 77–85. DOI 10.1016/j.pbi.2004.11.015. [Google Scholar] [CrossRef]
45. Chen, M., Lin, M. A., Jia, C., Liu, X., Gong, P. et al. (2016). Cloning and expression analysis of gibberellin receptor gene in Medicago sativa L. Acta Botanica Boreali-Occidentalia Sinica, 36(11), 2159–2166. DOI 10.7606/j.issn.1000-4025.2016.11.2159. [Google Scholar] [CrossRef]
46. Iuchi, S., Suzuki, H., Kim, Y. C., Iuchi, A., Kuromori, T. et al. (2010). Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant Journal, 50(6), 958–966. DOI 10.1111/j.1365-313X.2007.03098.x. [Google Scholar] [CrossRef]
47. Ge, H., Zhao, X., Wang, Z., Chen, S. W., Ma, H. Q. (2011). Putative GA receptor gene family of Vitis vinifera L. and their differential expression under GA treatment. Journal of China Agricultural University, 16(2), 58–63. [Google Scholar]
48. Li, J. (2014). Characterization of gibberellin receptor and aquaporin genes in Galega orientalis. China: Chinese Academy of Agricultural Sciences. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |