

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018767
ARTICLE
Selection of Stable Reference Genes for Quantitative Real-Time PCR on Paeonia ostii T. Hong et J. X. Zhang Leaves Exposed to Different Drought Stress Conditions
1College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, 225009, China
2Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou, 225009, China
*Corresponding Author: Jun Tao. Email: taojun@yzu.edu.cn
Received: 16 August 2021; Accepted: 27 October 2021
Abstract: The definition of relatively stable expressed internal reference genes is essential in both traditional blotting quantification and as a modern data quantitative strategy. Appropriate internal reference genes can accurately standardize the expression abundance of target genes to avoid serious experimental errors. In this study, the expression profiles of ten candidate genes, ACT1, ACT2, GAPDH, eIF1, eIF2, α-TUB, β-TUB, TBP, RNA Pol II and RP II, were calculated for a suitable reference gene selection in Paeonia ostii T. Hong et J. X. Zhang leaves under various drought stress conditions. Data were processed by the four regularly used evaluation software. A comprehensive analysis revealed that RNA Pol II was the most stable gene and eIF2 was the least stable one. In addition, the geNorm program provided the optimal choice of two reference gene combination, RNA Pol II and β-TUB, for qRT-PCR normalization in P. ostii subjected to different drought stress levels. Our research provided convenience for gene expression analysis in P. ostii under drought stress and promoted research of effective methods to alleviate P. ostii drought stress in the future.
Keywords: Paeonia ostii T. Hong et J. X. Zhang; reference gene; qRT-PCR; drought stress
The gene expression level analysis has been widely used in different research areas and species [1–6] to screen key genes, confirm specific regulatory mechanisms, as well as to study new gene functions. Quantitative real–time PCR (qRT–PCR) is predominant in different research fields and has been widely applied due to its high accuracy, good specificity, and high throughput at the mRNA level [7,8]. However, many restricting factors influenced the accuracy of results analyzed by qRT-PCR, including quality and integrity of RNA samples, efficiency in complementary DNA (cDNA) synthesis, primer specificity, and overall transcriptional activity of the tissue or cells [9–11]. Thus, with the aim to acquire precise experimental results, the target gene’s expression abundance must be corrected with reliable internal reference genes (housekeeping genes) to acquire normalized expression levels [9,12,13].
Some traditional housekeeping genes, such as actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 18S ribosomal RNA (18S rRNA), elongation factor 1α (EF-1α), α-tubulin (α-TUB) and β-tubulin (β-TUB), etc., have often been used as expression correction tools due to their stable expressions in previous studies [14,15]. Theoretically, these traditional reference genes maintain the most basic cellular activities and their expression levels are not affected by the external environment. However, many researches revealed that the common housekeeping genes presented different expression modes in different species as well as under different conditions [16,17]. The RPS2 (ribosomal protein S) and UBQ2 (ubiquitin) genes were validated as the two most suitable reference genes in various tissues, across different developmental stages and under different stresses in banana (Musa acuminata AAA Group, cv. Carven-dish) fruits [18]. In Eremosparton songoricum (Litv.) Vass., EsUBQ could only be used as a reference gene across multiple adult tissue samples, and EsEF and EsACT were the most stable reference genes for samples of differing germination stages [19]. The ACT and CACs (clathrin adaptor complexes medium subunit family protein) genes were identified as the most suitable combination for gene expression correction in several tissues of different collection dates in cork oak (Quercus suber L.) [20]. Similarly, common reference genes may display various expression patterns under different abiotic stresses [21]. Also, using unstable internal reference genes will have great impacts on the target gene expression level analysis and cause erroneous experimental results. In Kosteletzkya virginica (L.) Presl., 18S rRNA was identified to be the most stable reference gene under salt stress [22]. In herbaceous peony (Paeonia lactiflora Pall.) under light stress, ETI (eukaryotic translation initiation factor 5A-2) was recommended to be the most stable gene in the flowers, and ACT and pp2A (serine/threonine protein phosphatase 2A) showed the least expression variations in stems [23]. Therefore, it is very important to choose relatively stable housekeeping genes according to specific experimental conditions, especially under specific abiotic stresses, before their use in qRT-PCR normalization. Up to now, several housekeeping genes were identified in different species under various abiotic stresses, such as in goosegrass (Eieusine indica (L.) Gaertn.) under herbicide stress [24], in cotton (Gossypium hirsutum L.) under drought stress [25], in Moringa oleifera Lam. under water stress [26] and in Panax ginseng C. A. Meyer under heat stress [27].
Paeonia ostii T. Hong et J. X. Zhang is a famous traditional ornamental flower in China, which is internationally popular due to its large, showy, and aromatic flower. Studies have proved that its seed oil enriches a large amount of α-linolenic acid (ALA), which may be beneficial as a nutraceutical/pharmaceutical candidate and is good for human health [28]. Additionally, as an emerging woody oil crop, it has a lot of advantages such as strong adaption and low planting investment for large-scale cultivation promotion in many areas [29]. However, as a common abiotic stress, drought stress has greatly restricted the ornamental characteristics and economic values of P. ostii, especially in arid areas [30]. Therefore, it is urgent to find feasible ways to alleviate this damage caused by drought stress. At present, there are few reports in the literature on P. ostii internal reference gene selection; some scholars have studied the reference genes selection of P. ostii in different tissues and across different developmental stages [31,32]. However, there is no literature report on internal reference genes verification under drought conditions in P. ostii. Thus, when clarifying the underlying molecular mechanism of P. ostii under drought stress and finding effective mitigation routes, there is a lack of stable internal reference genes for qRT-PCR analysis.
In this study, we selected 10 commonly used candidate reference genes and used the geNorm, NormFinder, BestKeeper, and RefFinder to test their expression stability under different drought stress conditions in P. ostii. The identified internal reference genes provide valuable information for further qRT-PCR analysis and drought stress alleviation methods of P. ostii.
2.1 Plant Materials and Treatments
The potted seedlings (top diameter × height × bottom diameter = 20 cm × 25 cm × 12 cm) of Paeonia ostii (three-year-old) were used as plant materials. P. ostii plants with uniform traits were cultivated in a greenhouse maintained at 25–30°C under natural light in Yangzhou University (Yangzhou, Jiangsu Province, China) by regular agronomic practices. Before the drought treatment, watering was normally conducted for three consecutive days. Then, 12 potted seedlings were subjected to the natural drought treatment. According to our previous study, with the increase of drought treatment days, the damages caused by the drought stresses in P. ostii plants were gradually severe [33], and on the 0, 4, 8, and 12 d after the drought stresses, the soil water content was 32.23%, 18.31%, 11.68%, and 4.02%, respectively [34]. In this experiment, leaves were randomly sampled every 4 d on the 0, 4, 8, and 12 d after the drought treatment as described earlier [34], and leaves were collected from 2 to 4 pairs of leaves from top to bottom with three biological repeats. Then, they were quickly frozen in liquid nitrogen and stored at −80°C until further analysis.
2.2 RNA Extraction and cDNA Synthesis
Total RNA was extracted using TRIzol reagent (Takara, Dalian, China). The RNA integrity was analyzed by 1.0% agarose gels, and the RNA purity was assessed using BioPhotometer D30 (Eppendorf AG, Hamburg, Germany). Only qualified RNA samples (260/280 ratio ranging from 1.8 to 2.0; 260/280 ratio > 2.0) were used for subsequent experiments. The first-strand cDNA was synthesized using PrimeScript RT reagent Kit (Takara, Dalian, China).
2.3 Potential Reference Gene Selection and qRT-PCR Primer Design
Based on the RNA-seq database obtained from the transcriptome sequencing using P. ostii drought treatment (DT) and control (CK) leaves on the 12th day, a total of 78,392 unigenes’ fragments per kilobase million (FPKM) values were obtained. According to the differential expressions of genes in different samples, log2 (DT/CK) was set as a threshold between −0.1 to 0.1 to find stable expressed internal reference genes. We initially obtained 10 unigenes with the most stable expressions distributed in 8 gene families as candidate internal reference genes for further expression profiles analysis (Table 1). All of these candidate genes were chosen for their essential functions in maintaining the most basic cellular activities.
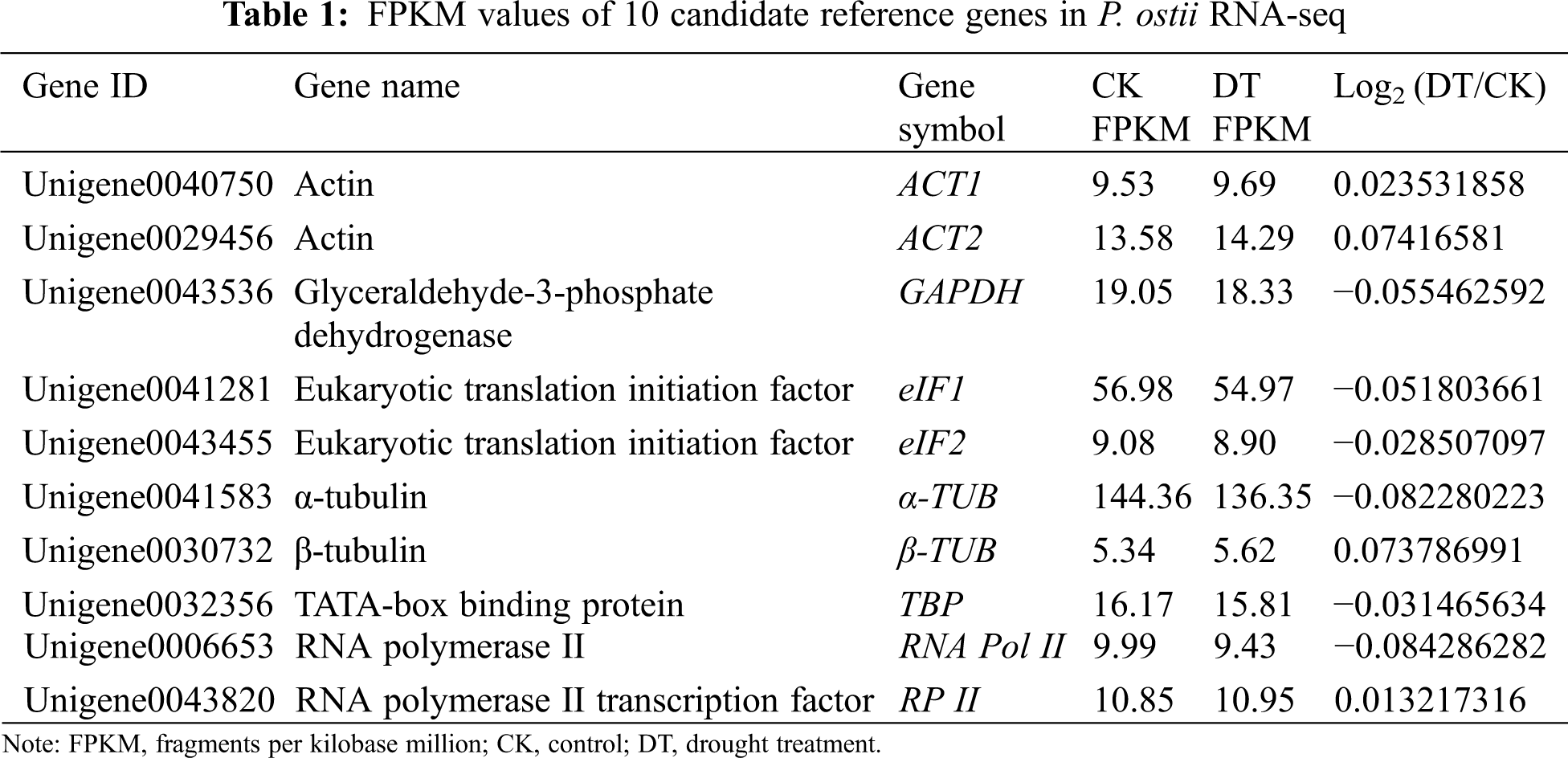
Using the Primer Premier 5 software, we designed qRT-PCR primers for each of the 10 reference genes from P. ostii. The specific sequence of each primer was shown in Table 2 and the design standard rules are as follows: the amplification product length: 100 to 200 bp; primer length: 18 to 22 bp; melting temperature (TM): 45 to 55°C; GC content: 40% to 60%. Additionally, we used the mixed cDNA sample of each sample as the cDNA template, and the diluted cDNA template was used to calculate PCR efficiency (E) and determination coefficient (R2). Finally, the amplified products of each reference gene were visualized on 1.0% agarose gels.
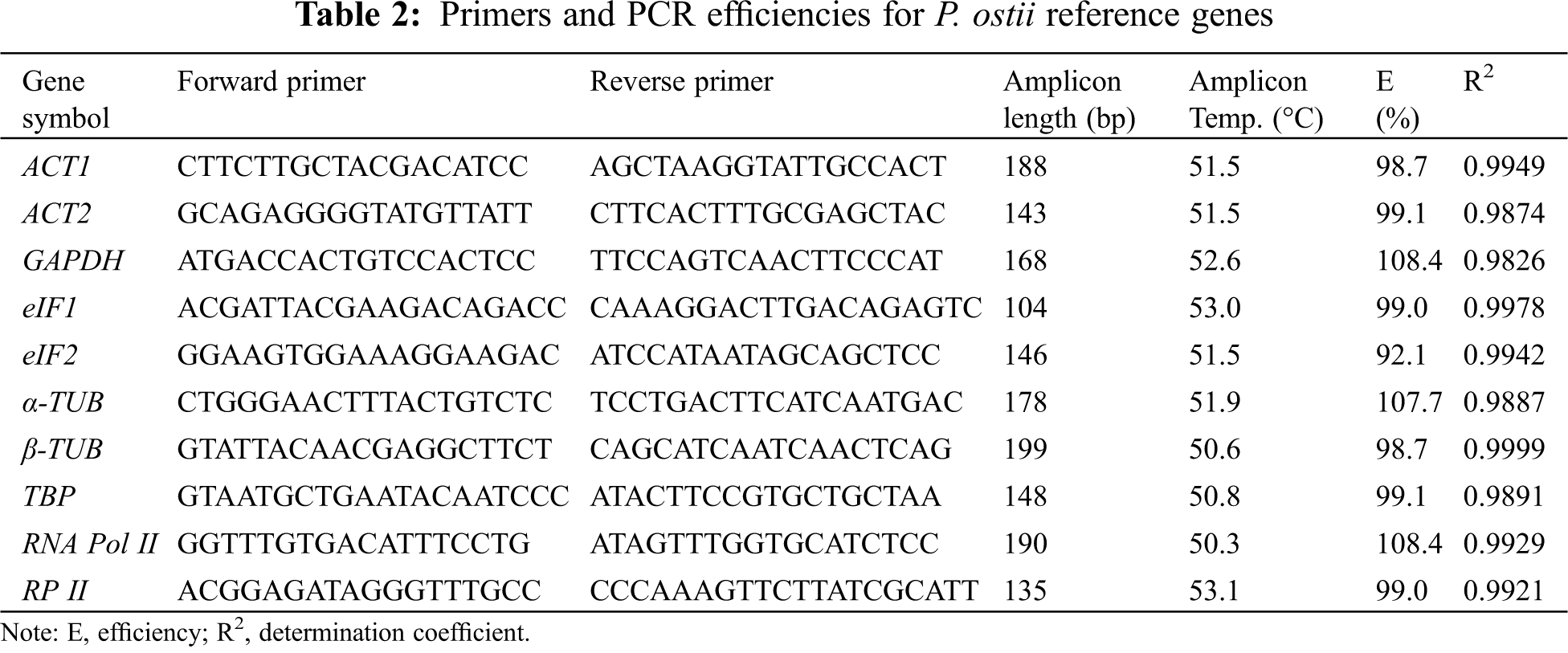
qRT-PCR reactions were conducted with a Bio-Rad PCR CFX96 system (Bio-Rad Laboratories, Hercules, CA, USA). The 10 candidate genes were amplified by qRT-PCR from different drought stress conditions using SYBR Premix Ex Taq (Takara, Dalian, China). Meanwhile, the reactions without cDNA were generated as the negative controls. The qRT-PCR system was conducted as follows: 95°C for 30 s, followed by 5 s at 95°C, 30 s at 55°C, and 30 s at 72°C (40 cycles). We repeated each reaction in triplicate.
The Data were processed by Bio-Rad CFX Manager software. Four different methods based on statistical algorithms including the geNorm [13], NormFinder [10], BestKeeper [35], and RefFinder were used to evaluate gene expression stability and availability in various drought stresses.
The geNorm program calculates the expression stability value (M) of each gene. The final criterion defines the gene with the lowest M value as the most stable one. Moreover, geNorm can also calculate the pair-wise variation (Vn/Vn+1) to determine the optimal numbers of reference genes combination used for qRT-PCR analysis.
The NormFinder’s reference gene definition principles are consistent with geNorm which is based on the variance between samples. It uses the M value to rank candidate reference genes for the most suitable reference gene selection.
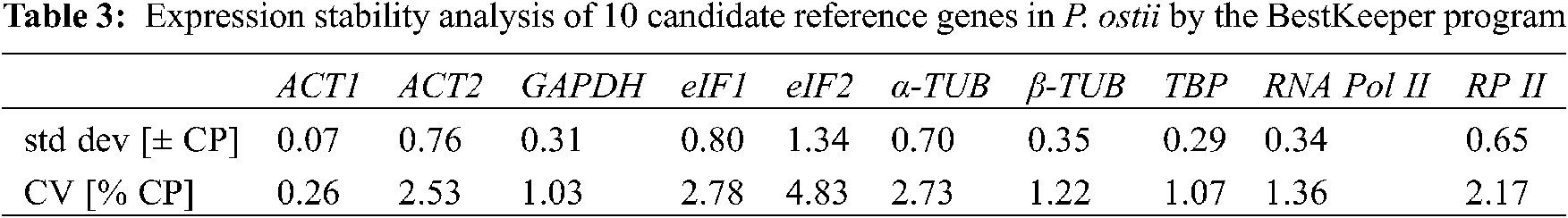
The BestKeeper program generates the coefficient of variation (CV) and standard deviation (SD) by comparing Ct values. Genes with lower CV and SD values imply more stability; conversely, they imply lower stability.
The RefFinder program (http://www.leonxie.com/referencegene.php) is an online website tool that integrates the ranking results of the Delta Ct, geNorm, NormFinder, and BestKeeper; it obtains an integrated result by combining the results of these four statistical algorithms and recommends the most suitable reference gene.
The primers of all candidate genes were designed based on the gene sequences obtained from P. ostii RNA-seq data [33]. The fragment length of PCR amplifications ranged from 104 to 199 bp (Fig. 1), and the PCR amplification results were verified to be consistent with the original RNA-seq. In addition, quality analysis results of each primer showed that the melting curve of each candidate gene revealed a single sharp peak, indicating good specificity (Fig. 2). The PCR efficiencies (E) ranged from 92.1% for eIF2% to 108.4% for RNA Pol II, and the determination coefficients (R2) varied from 0.9826 for GAPDH to 0.9999 for β-TUB (Table 2).
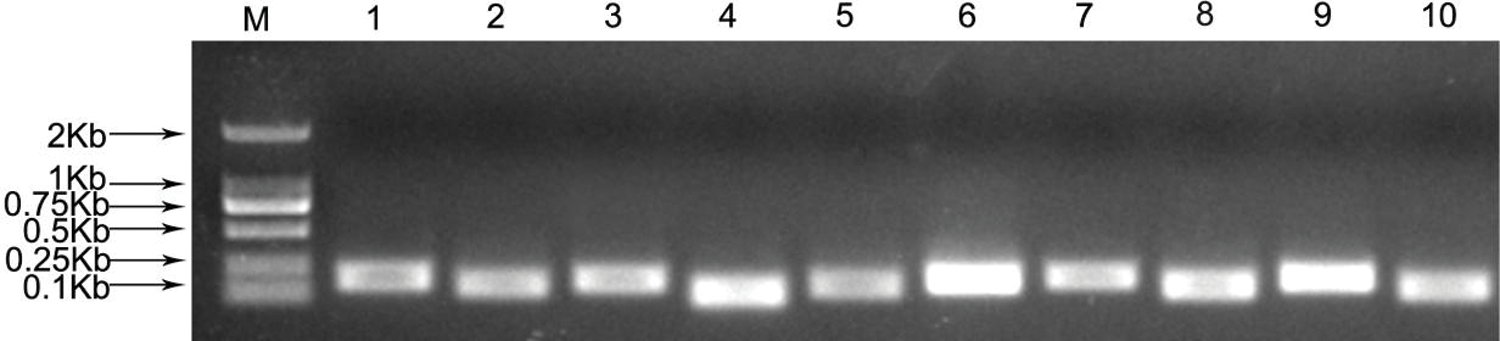
Figure 1: PCR amplifications of 10 candidate reference genes of P. ostii
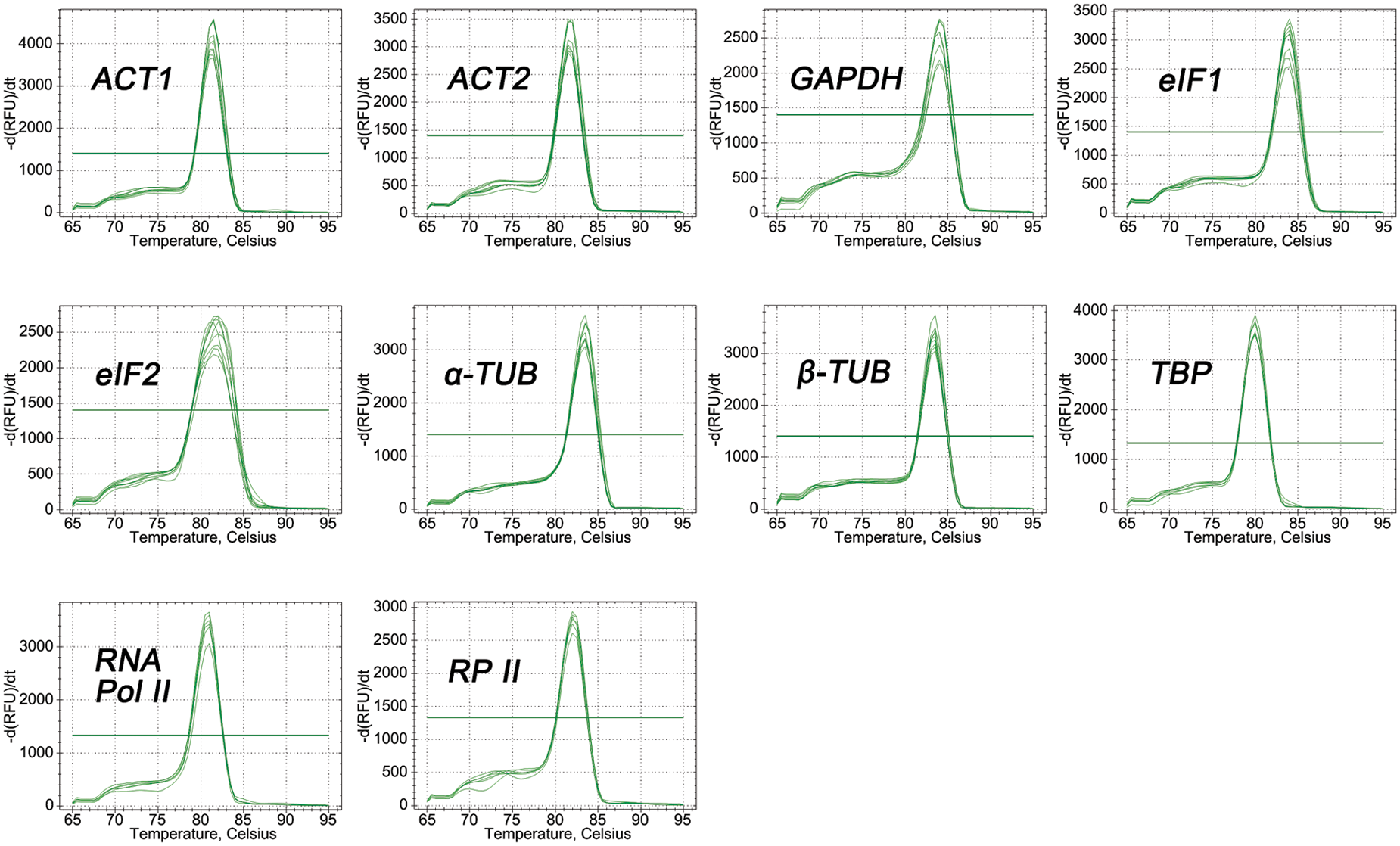
Figure 2: Melting curves of 10 candidate reference genes of P. ostii
M, 2000 marker; 1, ACT1; 2, ACT2; 3, GAPDH; 4, eIF1; 5, eIF2; 6, α-TUB; 7, β-TUB; 8, TBP; 9, RNA Pol II; 10, RP II
3.2 Expression Characteristics of the Candidate Reference Genes
The Ct values of 10 candidate genes under different drought conditions were summarized in Fig. 3. It showed that the Ct values of the 10 candidate genes ranged from 24.43 to 31.47. Among them, RNA Pol II had the lowest average Ct value (25.30); it indicated that RNA Pol II had the highest expression abundance at the mRNA level. Meanwhile, ACT2 had the highest average Ct value (29.85), indicating it had the lowest expression level. The CV value defined the stability of gene expression under the different drought conditions. Among these ten genes, ACT1 showed the smallest variation span value of 0.66%, which meant it had the highest expression stability, while eIF2 showed the highest variation span value of 5.08%, indicating that its expression level varied intensely under the diverse drought stresses. The line across the boxplot reflected the median value. The box showed the percentile range from 25% to 75%. Vertical lines indicate maximum and minimum values.
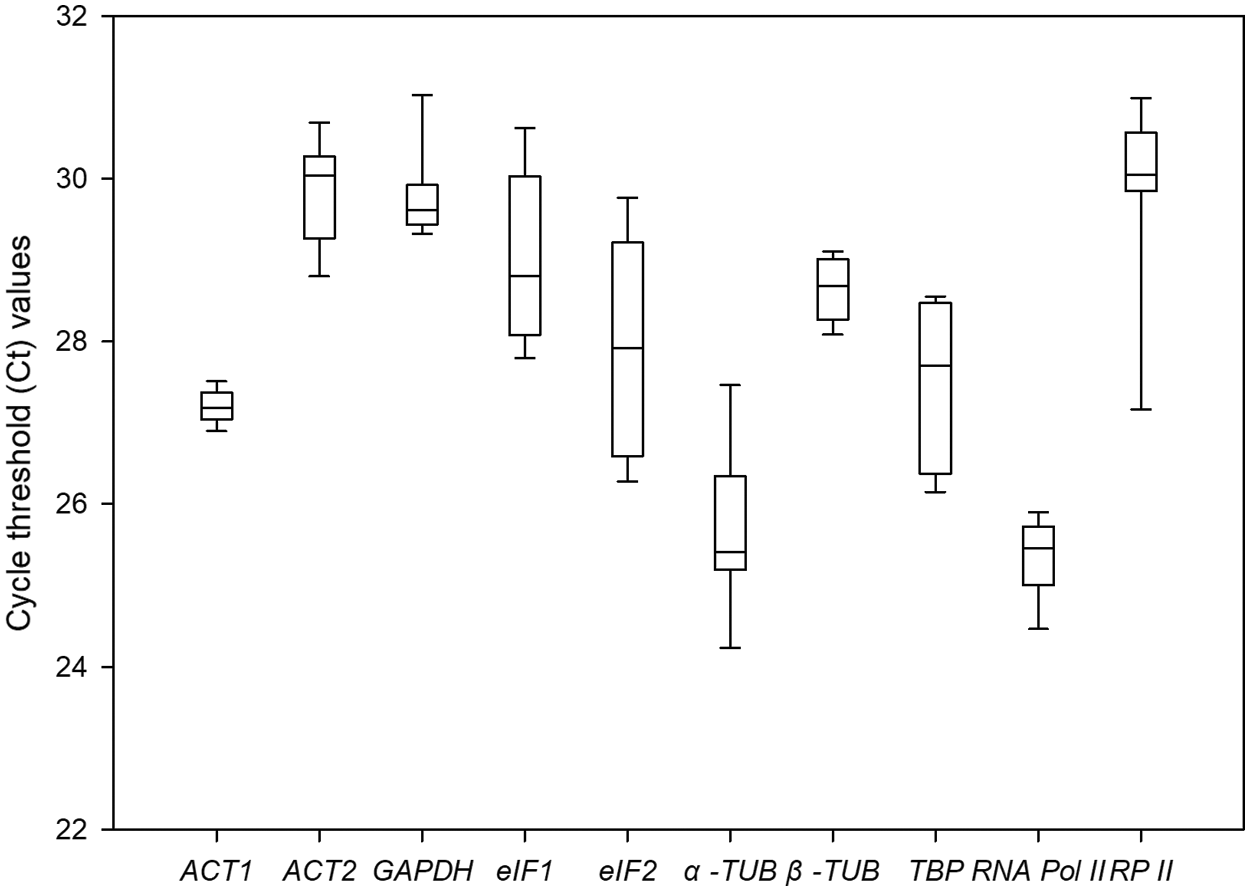
Figure 3: Comparative expression profiles of 10 candidate reference genes in P. ostii
GeNorm measures the stability of genes based on the average variation degree of the M value calculated by Ct-converted values. The default value of this software is 1.5. It was not recommended to use genes with M values higher than 1.5 as internal reference genes. Moreover, the lower the M value is, the more stable the gene is. In this study, the M values of the 10 candidate genes were all lower than 1.5, ranging from 0.14 to 0.96. Among them, eIF2 had the least stable expression, while RNA Pol II and β-TUB ranked first. These latter two genes were stably expressed despite changes in the external environment. The expression stability ranking of other candidate genes is as follows: TBP > ACT1 > α-TUB > eIF1 > GAPDH > RP II > ACT2 (Fig. 4).
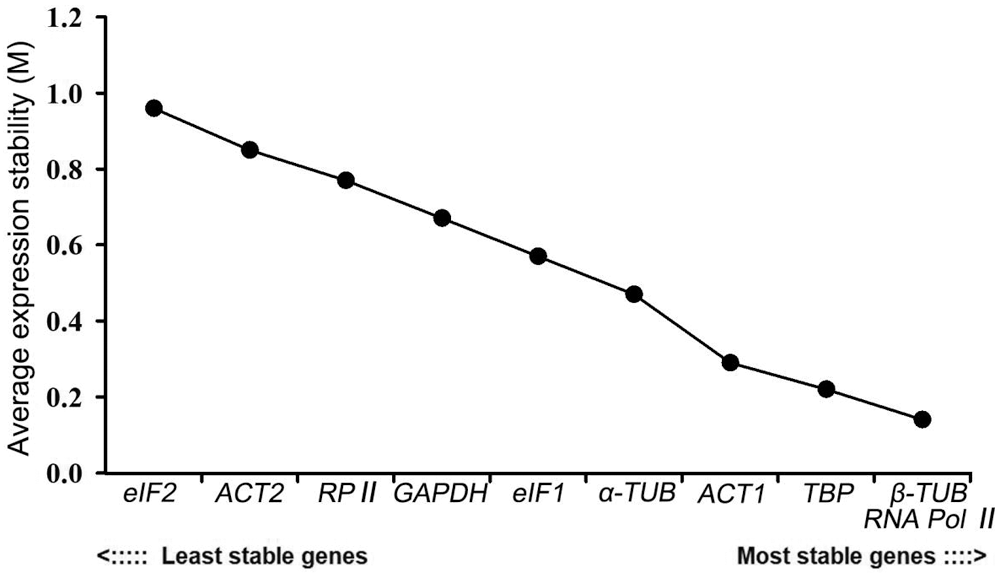
Figure 4: Expression stability analysis of 10 candidate reference genes in P. ostii by the geNorm program
Two or more internal reference gene combinations can be used for correcting target gene expressions under certain conditions to obtain more accurate experimental results. The geNorm software can also determine the appropriate number of internal reference genes according to the paired difference value Vn/Vn+1 of the candidate internal reference genes. The 0.15 threshold was proposed to determine optimal reference gene numbers, and the standard was as follows: if Vn/Vn+1 value < 0.15, the optimal number was n; if Vn/Vn+1 value > 0.15, the optimal number was n + 1. In this study, RNA Pol II and β-TUB were the optimal combinations for accurate qRT-PCR analysis in P. ostii with a V2/3 value of 0.085 under various drought stress conditions (Fig. 5). Thus, there was no need to introduce more genes for qRT-PCR correction.
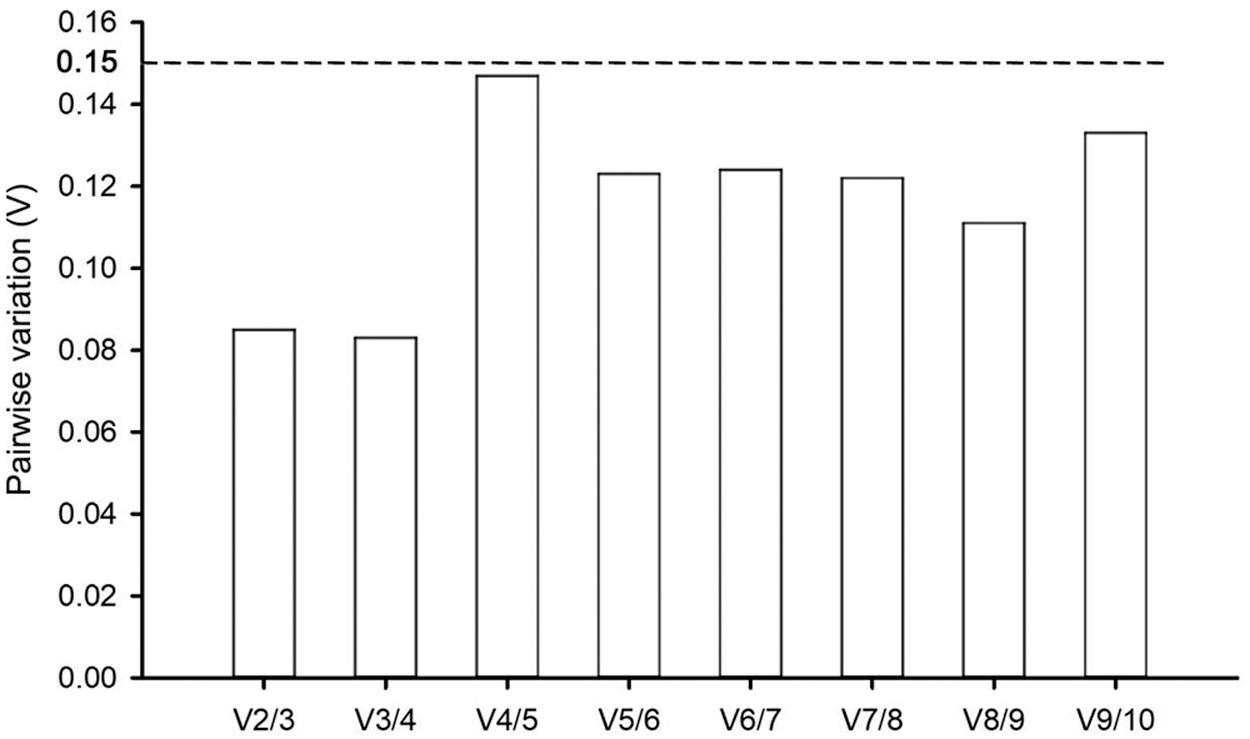
Figure 5: Pairwise variation (V) for the optimal number of candidate genes in P. ostii by the geNorm program
NormFinder combines the within-group variation and the between-group variation to calculate the stable value of each candidate reference gene. NormFinder considers the gene with the lowest M value as the most stable one. NormFinder only determines one gene as the most stable reference gene, which is different from geNorm. Among these ten genes, TBP had the lowest M value of 0.021, indicating that it was the most stable reference gene. RNA Pol II ranked second with an M value of 0.048, followed by β-TUB (0.203), ACT1 (0.322), α-TUB (0.381), eIF1 (0.539), GAPDH (0.566), RP II (0.626) and ACT2 (0.734). eIF2 was the most unstable gene with the largest M value of 0.909, which revealed the same result with geNorm (Fig. 6).
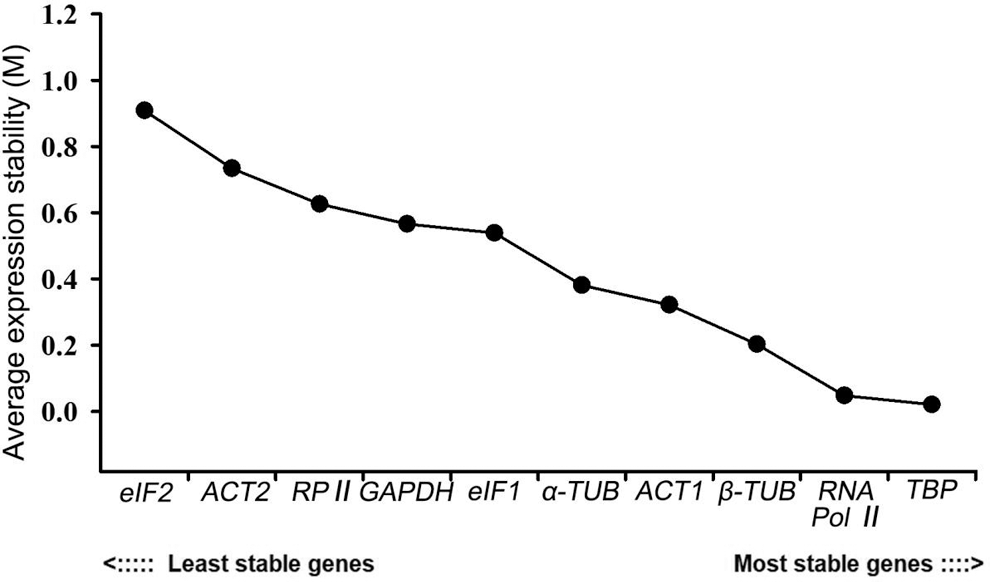
Figure 6: Expression stability analysis of 10 candidate reference genes in P. ostii by the NormFinder program
BestKeeper judges the gene expression stability based on the SD and the CV values by directly analyzing the Ct values. The program default threshold value is 1. When the SD value is greater than 1, the gene is considered to be unstable; conversely, a gene with an SD value less than 1 implies high expression stability. In this study, the SD values of all genes were less than 1, except eIF2. Thus, they were all suitable as internal reference genes, but eIF2. ACT1 was the most stable with an SD value of 0.07, which was the most suitable choice for an internal reference gene (Table 3).
RefFinder is an online analysis website that integrates the analysis methods of the Delta Ct, geNorm, NormFinder, and BestKeeper. It calculates the weight of the analysis results of each software to obtain a comprehensive ranking result. The re-rank order analysis result was as follows: RNA Pol II > RP II > β-TUB > ACT1 > GAPDH > α-TUB > ACT2 > eIF1 > TBP > eIF2. Thus, RNA Pol II was the most stable candidate reference gene which was consistent with the result of geNorm. eIF2 ranked last, indicating that it had the most unstable expression stability (Fig. 7).
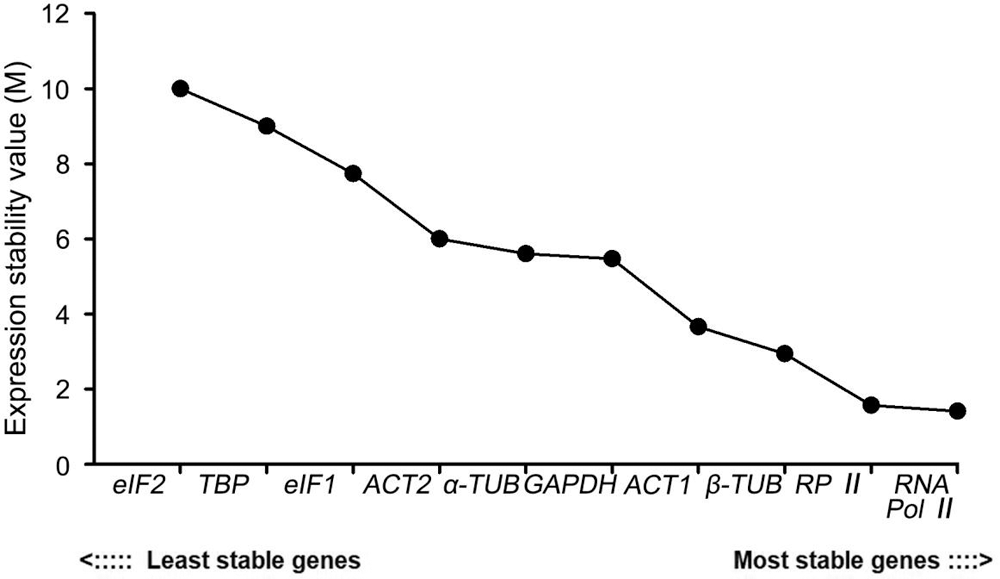
Figure 7: Expression stability analysis of 10 candidate reference genes in P. ostii by RefFinder
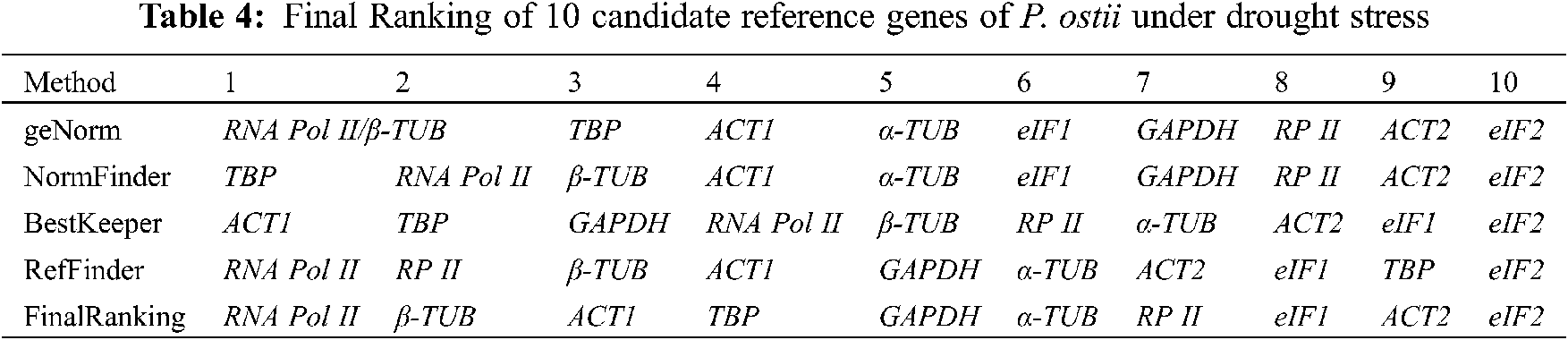
Based on the ranking results of the geNorm, NormFinder, BestKeeper, and RefFinder, a comprehensive analysis revealed that RNA Pol II was better than the other 9 candidate reference genes and ranked first due to its stable expression under various drought stress conditions (Table 4). The final ranking result was as follows: RNA Pol II > β-TUB > ACT1 > TBP > GAPDH > α-TUB > RP II > eIF1 > ACT2 > eIF2. Finally, we recommended RNA Pol II as the most suitable reference gene; and this result was the same as the geNorm and RefFinder programs.
Compared with conventional quantitative methods, qRT-PCR is superior in terms of its high sensitivity and good specificity contributing to its wide application. However, studies have shown that no gene expresses in the same abundance under any condition, and a single gene may have different expression levels in different tissues, growth stages, and experimental treatments [36–38]. Therefore, it is critically necessary to choose suitable reference genes according to specific experimental treatments before qRT-PCR.
At present, with the wide application of qRT-PCR in many areas [39] such as molecular biology [40], modern medicine [41,42], food detection [43], and environmental monitoring [44], some evaluation methods for judging gene stability have gradually emerged simultaneously. The geNorm, NormFinder, BestKeeper, and RefFinder are among these methods, which improved the efficiency of gene stability analysis and accelerated the pace of suitable reference genes selection. However, the results of each algorithm are different because of the unique algorithm of each program. Therefore, a comprehensive analysis needs to be synthesized according to the analysis results of each statistical algorithm.
P. ostii produces a traditional famous flower in China. Research on P. ostii is very rich from multiple viewpoints. It includes research on its antioxidant activity [45], seed dormancy [46], oil resource promotion [47], ornamental traits regulation [48], and growth resistance and ecological adaptation [49]. In recent years, as the medicinal values and economic benefits of P. ostii have become increasingly prominent, the planting scale has continued to expand in many areas. However, in arid and semi-arid areas, due to the lack of water resources, the use of P. ostii and the orderly development of a P. ostii industry have been greatly restricted. Therefore, many scholars have conducted numerous drought-related studies on P. ostii, such as on water physiology [34], cell membrane system [50], osmotic system [51], and photosynthetic mechanisms [34].
In our previous research, some drought stress-relevant indicators were measured, and we have tried to find useful ways to alleviate the drought stress of P. ostii [52,53]. At the same time, with the development of transcriptome sequencing, research on drought stress has gradually advanced and deepened from the physiological to the molecular level [54]. Therefore, it is essential to verify the expression of key genes involved in enhancing P. ostii drought stress tolerance.
To date, the evaluation of internal reference genes in P. ostii was only reported in different tissues across different developmental stages without environmental stresses [31,32]. As a result, different reference gene combinations [GAPDH/UBC (ubiquitin conjugating enzyme) for ‘Feng Dan’ and ‘Xi Shi’; EF-1α/UBC for ‘Que Hao.’] were identified in P. ostii flowers [31]. This indicates that it is essential to choose reference genes in different cultivars, even though they pertain to the same species. Some scholars screened the reference genes under different abiotic stresses such as plant hormone stress [55], light stress [23], cold or high-temperature stress [56,57], and salt stress [58], based on the special experimental needs to solve specific scientific problems. Similarly, due to the high industrial values of P. ostii, research on P. ostii drought stress and its mitigation methods is also very significant. Screening the key genes, and explaining the key mechanisms of drought stress, require suitable reference genes which can correct the expression levels of drought-related genes.
Leaves are the main organ of plants for photosynthesis and transpiration and are also the most sensitive plant part to drought stress [59]. Drought stress can cause leaf wilting and withering, and reduce leaf water content in P. ostii. In the drought research of plants, leaves are often used to detect drought-related physical and chemical indicators to measure the overall plant drought degree. Therefore, in this study, we used leaves as the experimental material to screen stable reference genes of P. ostii under different drought conditions to provide reference and help for P. ostii drought research.
In this study, we selected appropriate internal reference genes based on the unigenes’ FPKM values in the transcriptome sequencing database of P. ostii leaves under drought stress obtained in the previous stage. Then, we set a strict threshold (log2 (DT/CK) value ranging from −0.1 to 0.1) to initially obtain 10 stable expressed candidate reference genes. The qRT-PCR results indicated that α-TUB had the lowest average Ct value; this was consistent with the transcriptome data, while other candidate genes showed variable expression patterns. Then, a comprehensive evaluation was performed using the geNorm, NormFinder, BestKeeper, and RefFinder programs. Based on the geNorm analysis results, RNA Pol II and β-TUB were stably expressed under different drought stress conditions, which were identified as the most stable reference genes. RNA Pol II and β-TUB were also chosen for internal controls in foxtail millet (Setaria italica L.) [60] and sugarcane (Saccharum officinarum L.) [61] under dehydration stress and drought stress. NormFinder recommended TBP as the most stable reference gene, and RNA Pol II ranked second. At the same time, the ranking results of NormFinder from the fourth to the seventh-place were the same as those of geNorm. The BestKeeper analysis results revealed that ACT1 was the most stable gene in P. ostii, which ranked fourth in both geNorm and NormFinder; the remaining ranking results were almost different from these two programs. RefFinder determined RNA Pol II as the most stable one, which was consistent with the geNorm analysis results. Due to the different ranking results of the four programs, RNA Pol II was considered to be the most suitable reference gene because of its persistent stable expression under various drought stress conditions (Table 4). Additionally, RNA Pol II had high expression abundance among these ten candidate genes and could be used to standardize the expressions of highly expressed target genes. MIQE guideline for qPCR methods suggested that there is no perfect gene that is constantly expressed regardless of the external environment under any condition [9]. geNorm program showed that using two genes as internal reference corrections was more reliable than using only one gene. Due to the pairwise variation calculated by geNorm, a combination of two internal reference genes (RNA Pol II and β-TUB) could be used as internal controls for qRT-PCR analysis in P. ostii under drought stress conditions. This study provided convenience for qRT-PCR analysis and provided useful reference data for screening key genes to alleviate drought stress of P. ostii in the future.
We evaluated 10 candidate reference genes in the leaves of P. ostii under different drought stress conditions. These genes were ACT1, ACT2, GAPDH, eIF1, eIF2, α-TUB, β-TUB, TBP, RNA Pol II, and RP II. The final comprehensive ranking results analyzed by four programs recommended RNA Pol II as the most stable reference gene, and that it could be used to relatively quantify the expression levels of target genes. geNorm program defined RNA Pol II and β-TUB as the best combination for qRT-PCR normalization in P. ostii exposed to drought conditions. Our research facilitated the expression analysis of target genes in P. ostii under drought stress, and also promoted the establishment of an optional internal reference gene library in P. ostii.
Author Contributions: J. T. and D. Z. designed the experiments. Y. L., C. X., and X. W. performed the experiments. Y. L., C. X., and X. W. analyzed the data and wrote the manuscript. All authors have carefully read and approved the final manuscript.
Acknowledgement: The authors wish to thank three anonymous reviewers for their input and constructive criticisms.
Funding Statement: This work was supported by the National Key R&D Program of China (2018YFD1000405), and Jiangsu Modern Agricultural Industrial Technology System (JATS [2020]436).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Martinez-Beamonte, R., Navarro, M. A., Larraga, A., Strunk, M., Barranquero, C. et al. (2011). Selection of reference genes for gene expression studies in rats. Journal of Biotechnology, 151(4), 325–334. DOI 10.1016/j.jbiotec.2010.12.017. [Google Scholar] [CrossRef]
2. Li, H., Ni, D. H., Duan, Y. B., Chen, Y., Li, J. et al. (2013). Quantitative detection of the rice false smut pathogen Ustilaginoidea virens by real-time PCR. Genetics & Molecular Research, 12(4), 6433–6441. DOI 10.4238/2013.December.10.4. [Google Scholar] [CrossRef]
3. Yu, S. H., Yang, P., Sun, T., Qi, Q., Wang, X. Q. et al. (2016). Identification and evaluation of reference genes in the Chinese white wax scale insect Ericerus pela. SpringerPlus, 5(1), 791. DOI 10.1186/s40064-016-2548-z. [Google Scholar] [CrossRef]
4. Tan, Y., Zhou, X. R., Pang, B. P. (2017). Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bulletin of Entomological Research, 107(3), 359–368. DOI 10.1017/S0007485316000948. [Google Scholar] [CrossRef]
5. Dong, Z. D., Chen, P. S., Zhang, N., Huang, S. K., Zhang, H. R. et al. (2018). Evaluation of reference genes for quantitative real-time PCR analysis of gene expression in Hainan medaka (Oryzias curvinotus). Gene Reports, 14, 94–99. DOI 10.1016/j.genrep.2018.11.008. [Google Scholar] [CrossRef]
6. Chen, C. B., Wu, J. Y., Hua, Q. Z., Tel-Zur, N., Xie, F. F. et al. (2019). Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods, 15, 70. DOI 10.1186/s13007-019-0455-3. [Google Scholar] [CrossRef]
7. Heid, C. A., Stevens, J., Livak, K. J. (1996). Williams PM. Real time quantitative PCR. Genome Research, 6(10), 986–994. DOI 10.1101/gr.6.10.986. [Google Scholar] [CrossRef]
8. Ginzinger, D. G. (2002). Gene quantification using Real-time quantitative PCR: An emerging technology hits the mainstream. Experimental Hematology, 30, 503–512. DOI 10.1016/s0301-472x(02)00806-8. [Google Scholar] [CrossRef]
9. Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J. et al. (2008). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622. DOI 10.1373/clinchem.2008.112797. [Google Scholar] [CrossRef]
10. Andersen, C. L., Jensen, J. L., Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-pCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250. DOI 10.1158/0008-5472.CAN-04-0496. [Google Scholar] [CrossRef]
11. Expósito-Rodríguez, M., Borges, A. A., Borges-Pérez, A., Pérez, J. A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology, 8, 131. DOI 10.1186/1471-2229-8-131. [Google Scholar] [CrossRef]
12. Richards, M. P., Poch, S. M. (2002). Quantitative analysis of gene expression by reverse transcription polymerase chain reaction and capillary electrophoresis with laser-induced fluorescence detection. Molecular Biotechnology, 21(1), 19–37. DOI 10.1385/MB:21:1:019. [Google Scholar] [CrossRef]
13. Vandesompele, J., de Preter, K., Pattyn, F., Poppe, B., van Roy, N. et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, 1–12. DOI 10.1186/gb-2002-3-7-research0034. [Google Scholar] [CrossRef]
14. Aoki, T., Naka, H., Katagiri, T., Hirono, I. (2000). Cloning and characterization of glyceraldehyde-3-phosphate dehydrogenase cDNA of Japanese flounder Paralichthys olivaceus. Fisheries Science, 66, 737–742. DOI 10.1046/j.1444-2906.2000.00120. x. [Google Scholar] [CrossRef]
15. Zhang, Y. F., Zhao, L. J., Zeng, Y. L. (2014). Selection and application of reference genes for gene expression studies. Plant Physiology Journal, 50(8), 1119–1125 (in Chinese). DOI 10.13592/j.cnki.ppj.2014.0201. [Google Scholar] [CrossRef]
16. Wang, X. W., Wu, J., Bao, W. Q., Hu, H. Y., Chen, M. et al. (2019). Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biology, 19(1), 498. DOI 10.1186/s12870-019-2108-0. [Google Scholar] [CrossRef]
17. Yang, X. J., Zheng, H. L., Liu, Y. Y., Li, H. W., Jiang, Y. H. et al. (2020). Selection of reference genes for quantitative real-time PCR in Aquatica leii (Coleoptera: Lampyridae) under five different experimental conditions. Frontiers in Physiology, 11, 555233. DOI 10.3389/fphys.2020.555233. [Google Scholar] [CrossRef]
18. Chen, L., Zhong, H. Y., Kuang, J. F., Li, J. G., Lu, W. J. et al. (2011). Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta, 234(2), 377–390. DOI 10.1007/s00425-011-1410-3. [Google Scholar] [CrossRef]
19. Li, X. S., Yang, H. L., Zhang, D. Y., Zhang, Y. M., Wood, A. J. (2012). Reference gene selection in the desert plant Eremosparton songoricum. International Journal of Molecular Sciences, 13(12), 6944–6963. DOI 10.3390/ijms13066944. [Google Scholar] [CrossRef]
20. Marum, L., Miguel, A., Ricardo, C. P., Miguel, C. (2012). Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS One, 7(4), e35113. DOI 10.1371/journal.pone.0035113. [Google Scholar] [CrossRef]
21. Liu, Y., Liu, J., Lei, X., Lai, H., Chen, Y. et al. (2017). Identification and validation of reference genes for seashore paspalum response to abiotic stresses. International Journal of Molecular Sciences, 18(6), 1322. DOI 10.3390/ijms18061322. [Google Scholar] [CrossRef]
22. Tang, X. L., Wang, H. Y., Shao, C. Y., Shao, H. B. (2015). Reference gene selection for qPCR normalization of Kosteletzkya virginica under salt stress. BioMed Research International, 823806, 1–8. DOI 10.1155/2015/823806. [Google Scholar] [CrossRef]
23. Wan, Y. L., Hong, A. Y., Zhang, Y. X., Liu, Y. (2019). Selection and validation of reference genes of Paeonia lactiflora in growth development and light stress. Physiology and Molecular Biology of Plants, 25(4), 1097–1105. DOI 10.1007/s12298-019-00684-2. [Google Scholar] [CrossRef]
24. Chen, J. C., Huang, Z. F., Huang, H. J., Wei, S. H., Liu, Y. (2017). Selection of relatively exact reference genes for gene expression studies in goosegrass (Eleusine indica) under herbicide stress. Scientific Reports, 7(1), 46494. DOI 10.1038/srep46494. [Google Scholar] [CrossRef]
25. Wang, M., Wang, Q. L., Zhang, B. H. (2013). Evaluation and selection of reliable reference genes for gene expression under abiotic stress in cotton (Gossypium hirsutum L.). Gene, 530(1), 44–50. DOI 10.1016/j.gene.2013.07.084. [Google Scholar] [CrossRef]
26. Deng, L. T., Wu, Y. L., Li, J. C., Ouyang, K. X., Ding, M. M. (2016). Screening reliable reference genes for RT-qPCR analysis of gene expression in Moringa oleifera. PLoS One, 11(8), e0159458. DOI 10.1371/journal.pone.0159458. [Google Scholar] [CrossRef]
27. Wang, M. Z., Lu, S. F. (2016). Validation of suitable reference genes for quantitative gene expression analysis in Panax ginseng. Frontiers in Plant Science, 6, 696. DOI 10.3389/fpls.2015.01259. [Google Scholar] [CrossRef]
28. Kim, K. B., Nam, Y. A., Kim, H. S., Hayes, A. W., Lee, B. M. (2014). Alpha-linolenic acid: Nutraceutical, pharmacological and toxicological evaluation. Food and Chemical Toxicology, 70, 163–178. DOI 10.1016/j.fct.2014.05.009. [Google Scholar] [CrossRef]
29. Peng, L. P., Men, S. Q., Liu, Z. A., Tong, N. N., Imran, M. et al. (2019). Fatty acid composition, phytochemistry, antioxidant activity on seed coat and kernel of Paeonia ostii from main geographic production areas. Foods, 9(1), 30. DOI 10.3390/foods9010030. [Google Scholar] [CrossRef]
30. Ma, Y., Li, J. X., Zhang, Y., Zhang, G. (2018). Research progress on drought resistance in Paeonia suffruticosa. Journal of Anhui Agricultural Sciences, 46(13), 27–29, 32 (in Chinese). DOI 10.13989/j.cnki.0517-6611.2018.13.008. [Google Scholar] [CrossRef]
31. Li, J., Han, J. G., Hu, Y. H., Yang, J. (2016). Selection of reference genes for quantitative real-time PCR during flower development in tree peony (Paeonia suffruticosa Andr.). Frontiers in Plant Science, 7, 516. DOI 10.3389/fpls.2016.00516. [Google Scholar] [CrossRef]
32. Li, C. Q., Hu, L. Z., Wang, X. Q., Liu, H. Z., Tian, H. H. et al. (2019). Selection of reliable reference genes for gene expression analysis in seeds at different developmental stages and across various tissues in Paeonia ostii. Molecular Biology Reports, 46(6), 6003–6011. DOI 10.1007/s11033-019-05036-7. [Google Scholar] [CrossRef]
33. Zhao, D. Q., Zhang, X. Y., Fang, Z. W., Wu, Y. Q., Tao, J. (2019). Physiological and transcriptomic analysis of tree peony (Paeonia section Moutan DC.) in response to drought stress. Forests, 10(2), 135. DOI 10.3390/f10020135. [Google Scholar] [CrossRef]
34. Fang, Z. W., Wang, X. X., Zhang, X. Y., Zhao, D. Q., Tao, J. (2020). Effects of fulvic acid on the photosynthetic and physiological characteristics of Paeonia ostii under drought stress. Plant Signaling & Behavior, 15(7), 1774714. DOI 10.1080/15592324.2020.1774714. [Google Scholar] [CrossRef]
35. Pfaffl, M. W., Tichopad, A., Prgomet, C., Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnology Letters, 26(6), 509–515. DOI 10.1023/B: BILE.0000019559.84305.47. [Google Scholar] [CrossRef]
36. Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCRTrends and problems. Journal of Molecular Endocrinology, 29, 23–39. DOI 10.1677/jme.0.0290023. [Google Scholar] [CrossRef]
37. Bowen, J., Ireland, H. S., Crowhurst, R., Luo, Z., Watson, A. E. et al. (2014). Selection of low-variance expressed Malus x domestica (apple) genes for use as quantitative PCR reference genes (housekeepers). Tree Genetics & Genomes, 10(3), 751–759. DOI 10.1007/s11295-014-0720-6. [Google Scholar] [CrossRef]
38. Li, J. T., Han, X. P., Wang, C., Qi, W., Zhang, W. Z. et al. (2017). Validation of suitable reference genes for RT-qPCR data in Achyranthes bidentata blume under different experimental conditions. Frontiers in Plant Science, 8, 776. DOI 10.3389/fpls.2017.00776. [Google Scholar] [CrossRef]
39. An, G. L. (2018). The principle and application of real-time fluorescent quantitative PCR. China Modern Educational Equipment, 301, 19–21 (in Chinese). DOI 10.13492/j.cnki.cmee.20181120.002. [Google Scholar] [CrossRef]
40. Pabuayon, I. M., Yamamoto, N., Trinidad, J. L., Longkumer, T., Raorane, M. L. et al. (2016). Reference genes for accurate gene expression analyses across different tissues, developmental stages and genotypes in rice for drought tolerance. Rice, 9(1), 32. DOI 10.1186/s12284-016-0104-7. [Google Scholar] [CrossRef]
41. Gebeh, A. K., Marczylo, E. L., Amoako, A. A., Willets, J. M., Konje, J. C. (2012). Variation in stability of endogenous reference genes in fallopian tubes and endometrium from healthy and ectopic pregnant women. International Journal of Molecular Sciences, 13(3), 2810–2826. DOI 10.3390/ijms13032810. [Google Scholar] [CrossRef]
42. Sahoo, P. K., Parida, S., Mohapatra, A., Mohanty, J. (2019). Selection of candidate reference genes for RT-qPCR analysis in Argulus siamensis and their validation through screening of drugs and drug targets. Scientific Reports, 9(1), 18365. DOI 10.1038/s41598-019-54881-w. [Google Scholar] [CrossRef]
43. Ding, B. Q., Liu, S. N. (2020). Application of real-time quantitative PCR technology in rapid food detection. Science and Technology of Food Industry, 42(7), 366–373 (in Chinese). DOI 10.13386/j.issn1002-0306.2020050185. [Google Scholar] [CrossRef]
44. Chen, G. X., Lu, Z. X., Li, K. X., Wang, J. W., Wang, G. A. (2020). Effect of real-time quantitative PCR on abundance of nitrifying bacteria in different processes under temperature changes. Water Purification Technology, 39(1), 106–109, 226 (in Chinese). DOI 10.15890/j.cnki.jsjs.2020.s1.022. [Google Scholar] [CrossRef]
45. Zhang, H. F., Li, X. F., Wu, K., Wang, M. K., Liu, P. et al. (2016). Antioxidant activities and chemical constituents of flavonoids from the flower of Paeonia ostii. Molecules, 22(5), 1–15. DOI 10.3390/molecules22010005. [Google Scholar] [CrossRef]
46. Xue, J. Q., Wang, S. L., Zhang, P., Zhu, F. Y., Ren, X. X. et al. (2015). On the role of physiological substances, abscisic acid and its biosynthetic genes in seed maturation and dormancy of tree peony (Paeonia ostii ‘Feng dan’). Scientia Horticulturae, 182, 92–101. DOI 10.1016/j.scienta.2014.11.021. [Google Scholar] [CrossRef]
47. Li, S. S., Yuan, R. Y., Chen, L. G., Wang, L. S., Hao, X. H. et al. (2015). Systematic qualitative and quantitative assessment of fatty acids in the seeds of 60 tree peony (Paeonia section Moutan DC.) cultivars by GC-mS. Food Chemistry, 173, 133–140. DOI 10.1016/j.foodchem.2014.10.017. [Google Scholar] [CrossRef]
48. Gao, L. X., Yang, H. X., Liu, H. F., Yang, J., Hu, Y. H. (2015). Extensive transcriptome changes underlying the flower color intensity variation in Paeonia ostii. Frontiers in Plant Science, 6, 1205. DOI 10.3389/fpls.2015.01205. [Google Scholar] [CrossRef]
49. Peng, L. P., Cheng, F. Y., Hu, X. G., Mao, J. F., Xu, X. X. et al. (2019). Modelling environmentally suitable areas for the potential introduction and cultivation of the emerging oil crop Paeonia ostii in China. Scientific Reports, 9(1), 3213. DOI 10.1038/s41598-019-39449-y. [Google Scholar] [CrossRef]
50. Li, J. X., Zhang, Y., Bi, J. T., Yang, Y. (2019). Study on drought resistance of three varieties of oil peony. Journal of Anhui Agricultural Sciences, 47(21), 124–128 (in Chinese). DOI 10.3969/j.issn.0517-6611.2019.21.037. [Google Scholar] [CrossRef]
51. Li, J., Kong, X. S., Li, J. H., Liu, G. X., Guo, L. L. (2014). Effect of gradual drought stress on physiological indexes of Paeonia suffruticosa Andr. Northern Horticulture, 16, 50–53 (in Chinese). [Google Scholar]
52. Zhao, D. Q., Fang, Z. W., Tang, Y. H., Tao, J. (2020). Graphene oxide as an effective soil water retention agent can confer drought stress tolerance to Paeonia ostii without toxicity. Environmental Science and Technology, 54(13), 8269–8279. DOI 10.1021/acs.est.0c02040. [Google Scholar] [CrossRef]
53. Zhang, X. Y., Fang, Z. W., Liu, H. N., Zhao, D. Q., Tao, J. (2019). Exogenous calcium-induced physiological and biochemical changes in tree peony (Paeonia section Moutan DC.) under drought stress. Photosynthetica, 57(4), 904–911. DOI 10.32615/ps.2019.108. [Google Scholar] [CrossRef]
54. Guo, L. L., Guo, D. L., Yin, W. L., Hou, X. G. (2018). Tolerance strategies revealed in tree peony (Paeonia suffruticosa; Paeoniaceae) ecotypes differentially adapted to desiccation. Applications in Plant Sciences, 6(10), e01191. DOI 10.1002/aps3.1191. [Google Scholar] [CrossRef]
55. Qu, R. J., Miao, Y. J., Cui, Y. J., Cao, Y. W., Zhou, Y. et al. (2019). Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune. BMC Molecular Biology, 20(1), 9. DOI 10.1186/s12867-019-0126-y. [Google Scholar] [CrossRef]
56. Li, Q. Y., Li, Z. L., Lu, M. X., Cao, S. S., Du, Y. Z. (2019). Selection of valid reference genes for quantitative real-time PCR in Cotesia chilonis (Hymenoptera: Braconidae) exposed to different temperatures. PLoS One, 14(12), e0226139. DOI 10.1371/journal.pone.0226139. [Google Scholar] [CrossRef]
57. Yu, J. J., Su, Y., Sun, J. Y., Liu, J. R., Li, Z. Y. et al. (2020). Selection of stable reference genes for gene expression analysis in sweet potato (Ipomoea batatas L.). Molecular and Cellular Probes, 53, 101610. DOI 10.1016/j.mcp.2020.101610. [Google Scholar] [CrossRef]
58. Ma, L., Wu, J. Y., Qi, W. L., Coulter, J. A., Fang, Y. et al. (2020). Screening and verification of reference genes for analysis of gene expression in winter rapeseed (Brassica rapa L.) under abiotic stress. PLoS One, 15(9), e0236577. DOI 10.1371/journal.pone.0236577. [Google Scholar] [CrossRef]
59. Otto, M. S. G., Francisco, J. G., Gonsalez, B. T., de Almeida Calvo, L., de Mattos, E. M. et al. (2017). Changes in γ-aminobutyric acid concentration, gas exchange, and leaf anatomy in Eucalyptus clones under drought stress and rewatering. Acta Physiologiae Plantarum, 39(9), 208. DOI 10.1007/s11738-017-2507-5. [Google Scholar] [CrossRef]
60. Kumar, K., Muthamilarasan, M., Prasad, M. (2013). Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell, Tissue & Organ Culture, 115(1), 13–22. DOI 10.1007/s11240-013-0335-x. [Google Scholar] [CrossRef]
61. Crystian, D., Terto, J., Silva, J. V., Almeida, C. (2018). Validation of reference genes for accurate normalization by quantitative polymerase chain reaction in sugarcane drought stress studies using two cultivars. Ciência Rural, 48(11), e20180152. DOI 10.1590/0103-8478cr20180152. [Google Scholar] [CrossRef]

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |