

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018600
ARTICLE
Vital Parameters Assessments of Starvation Tolerance of in vitro Populus alba Culture
1Horticulture Department, Faculty of Agriculture, Ain Shams University, Cairo, 11566, Egypt
2Botany and Microbiology Department, Faculty of Science, Helwan University, Cairo, 11795, Egypt
3Department of Biotechnology, Faculty of Sciences, Taif University, Taif, 21944, Saudi Arabia
*Corresponding Author: Eman Tawfik Hussien. Email: emantawfik@science.helwan.edu.eg
Received: 05 August 2021; Accepted: 18 September 2021
Abstract: Populus alba is a large woody deciduous plant. The plant has been introduced to shooting, then multiplication of rooting on Murashige and Skoog (MS) medium. This work was designed to estimate the effect of two factors (low levels of 1-Naphthaleneacetic acid NAA and sucrose) on P. alba response resulting in 6 treatments compared to the control, with twelve measured responses. There was a significant difference in some measurements in morphology, like plantlets fresh-weight, shoot-, root-length, and leaf number. In the physiological measurements, there were significant differences in all the measured parameters. The low concentrations of sucrose and media composition/power (MS grams/L) led to starvation in plants; however, these conditions led to enhancement in some morphological and physiological parameters to overcome the starvation effect, compared to the control. The RAPD-PCR molecular marker (four decamers) was used to evaluate the new individuals’ genetic variation (instability), resulting in a total polymorphism percentage of 50.83%. It was formerly known that the plantlets were identical to each other and to the mother plant. In this study, however, the use of distinct media power, hormonal and sucrose levels resulted in molecular variation reflected in P. alba’s morphological and physiological responses.
Keywords: Genetic stability; Populus alba; RAPD-PCR; somaclonal variations; starvation
The genus Populus is widely distributed in the whole world, especially in the Northern hemisphere. It belongs to the family Salicaceae [1,2]. The primary model system for genomic, genetic and physiological studies on trees was for Populus [3]. It is an essential model for perennial wood species biotechnology because it can be exposed to in vitro culture and genetic engineering through Agrobacterium-mediated transformation [4]. It was the first tree to be sequenced from the genome [5]. A native to the Mediterranean region is Populus alba (White poplar). It is a deciduous, fast-growing tree. The leaves of white poplar may be used as soil pollution bio-monitors [6]. Poplar trees and their rapid growth spread through sucker shoots arising from horizontal roots [7]. Aspen (Populus tremula and P. tremuloides) from woody cuttings are, however, rare to root [8]. An effective in vitro propagation system for aspen is therefore highly requested.
Populus alba is a common model for in vitro culture studies. Previous research has found the vegetative spread of a single bud and different plant regeneration based on a single bud callus. Initially, there were problems with the establishment of cultures and genetically determined differences between the species. The age of the mother plants controls the culture establishment. Recently, media optimization has been used for commercial purposes to develop micropropagation methods for poplars. Breeding work based on in vitro explants has initiated almost simultaneously with the development of an in vitro mass-propagation procedure for poplars. This protocol is based on protoplasts and cell sus-pension production, followed by plant regeneration [9].
In vitro culture studies integrated some molecular markers to estimate variable changes in the obtained plantlets. There are many molecular markers like random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and inter simple sequence repeats (ISSR). They were applied to estimate genomic polymorphism among different transformed plant lines and their non-transgenic ones [10,11]. Also, they were used at the DNA level for the detection of polymorphism. For example, RAPD-PCR gained much popularity because it is simple, and does not require prior information on nucleotide sequence. RAPD-PCR can be performed with a minimal amount of genomic DNA. The technique of RAPD is a simple, efficient, reliable, and economical mean of identification and analysis of cultivar diversity [12].
In various studies, some plants’ genetic diversity has been investigated using different molecular markers [13]. The most significant uses have been the study of molecular variability and phylogenetic relationships, the marker-assisted selection, varietal identification, quantitative trait loci (QTLs), or the map-based cloning of genes [14]. Despite using different molecular markers to examine genetic diversity in cultivated plant species, many of them identify a limited level of polymorphism. Thus, the identification of more polymorphic molecular markers is essential for research [15].
Phenotypic, physiological and genetic variations occur due to the propagation process with distinct media constituents under starvation conditions. Starvation conditions include the low components and concentrations of different medium compositions; however, the plant can survive and tolerate these conditions. The genetic stability of P. alba is therefore essential to assess. This study monitored that genetic stability using the RAPD-PCR molecular technique on long-term micro-propagated shoots of P. alba. None of these studies have been investigated previously for the in vitro propagation of P. alba. Our main purpose was to show the variation in the genetic content which was reflected on morphological and physiological responses on the new resulted plantlets. This variation came from the different constituents of the media compositions. Also, we indicated how the plant can survive and grow in such deficient conditions and leakage of media contents. P. alba is an essential plant source of wood and it is used in many fabrics industries. So, we seek to produce more plantlet amounts with low cost without decreasing the wood quality.
2.1 Plant Materials and Explants Establishment
Explants of P. alba nodal segments were brought from the Horticulture Research Station of the Governorate of Al-Gharbia. Stem segments were then taken to the ACGEB (Agriculture Center for Genetic Engineering and Biotechnology) tissue culture laboratory at the Faculty of Agriculture, University of Ain Shams. P. alba stem nodes were sterilized and cultured for 2 months on a free plant growth regulators MS medium (pH 5.6). The standard sterilization conditions were the elimination of stacked dust/soil practices. The explants were defoliated and washed thoroughly in tap water. They were then exposed to 20% Clorox + 0.1% HgCl2 for 20 min, and washed 4 to 5 times with sterile ddH2O for surface sterilization of the explants. Under the sterilized conditions, excision and culture procedures for stem nodal ex-plants were performed. The MS medium [16] contained the necessary macro- and micro-element nutrients for the in vitro plants. The medium was allocated into incubation jars containing 50 mL in each jar. Stem nodal cultures were incubated under cool, white, fluorescent lamps at 25 ± 2°C, and a satisfactory fluorescent light of 60 μmol m−2 s−1 using a 16-h photoperiod.
Resulting shoots from establishment were excised and transferred into a multiplication medium of MS supplemented with 0.075 mg/L of 6-Benzylaminopurine (BAP). This was to elicit plant growth and development responses, and obtain micro-shoots required for the rooting experiment.
2.2 In vitro Rooting Experiment
Stem node segments of Populus alba were sterilized and cultured for 2 months on free plant growth regulators MS media. According to Murashige et al. [16], P. alba was retained on the MS tissue culture media for full growth of plantlets. Shoots developed on free sterilized MS multiplication medium with 0.5 g/L activated charcoal for root formation. They were then transferred to culture jars containing 50 mL solidified MS medium and cultured.
This study was designed based on two main factors (different sucrose and NAA concentrations). NAA was selected as an essential auxin for root formation and enhancement of nutrient absorption. Four nodal segments/jar (about 10–15 mm long) were cultured on 1⁄4 MS (1.1 g/L), with different sucrose concentrations (0.0 and 5 g/L) supplemented with MS basic medium and NAA (0.0, 0.1 and 0.25 g/L). This experiment was compared with the control (3/4 MS, 15 g sucrose and 0.25 g/L NAA). The shoots grew during 4 weeks under the same normal growing conditions (16 h light/8 h dark at 25 ± 2°C and a light intensity of 38 μmol m−2 s−1) under white fluorescent tubes. Five replicates were performed for each treatment.
2.4 Morphological Measurements
The following seven morphological biometric parameters were measured on all treatments: plantlet fresh-weight (g), shoot-, root-length (cm), leaf length, leaf number and number of roots.
Physiological parameters included measurements of photosynthetic pigments (chlorophyll a, chlorophyll b, carotenoids and total pigmentation), total proteins and total soluble sugars at the various rooting starvation treatments on P. alba individuals.
2.5.1 Determination of Photosynthetic Pigments
The plantlets’ fresh leaves were used for chlorophyll a, b, carotenoids and total pigmentation measurements. They were determined following Metzener et al. [17]: A known weight of these leaves (0.5 g) was homogenized in 85% acetone, then centrifuged and transfer into a new tube. After that, the extract was measured using a colorimeter against a blank of 85% pure acetone at three wavelengths (i.e., 452, 645 and 664 nm). The concentrations of chlorophyll a, b and carotenoids were calculated as μg/ml using the following equations after Metzener et al. [17]:
where A = absorbance at different wave lengths.
After that, the fractions were calculated as mg/g fresh weight:
Fraction: results from Eqs. (1)–(3). Dilution: how many times solution is diluted from stock.
The total protein content was extracted according to Bradford [18]. Briefly, 0.5 g of leaves were weighted and grind well with 0.5 mL of [2x] Bradford reagent. Samples were then vortexed during 10 min and centrifuged for 15 min at 24,104 g at 4°C. The supernatant contained the total protein content on the studied leaves. Finally, the protein concentration was estimated according to Bradford [18] as follows: 0.1 mL of supernatant was placed into a test tube using a pipette. Thereafter, 5 mL of protein reagent was added, mixed and measured using a spectrophotometer at a 595 nm wavelength. The concentration of protein was determined from the protein standard curve (using bovine serum albumin), and calculated according to the following equation as indicated by Bradford (#):
where X was the protein concentration (mg/g), and Y the absorbance (nm).
2.5.3 Total Soluble Sugars (TSS)
A known leaf weight (0.5 g) was ground in 5 mL of 70% ethanol; the supernatant was completed with distilled water to a known volume (10 ml) after centrifugation. As described by Umbriet et al. [19], total sugars were determined using the anthrone technique. In a 3 ml sample, 6 ml anthrone solution (2 g/L H2SO4 95%) was added and kept for 3 min in a boiling water bath. The developed color was measured spectrophotometrically at 620 nm following cooling. The following equation was then used to compute the concentration of the total soluble sugars according to the following equation:
*R: samples reading (nm), B blank reading (nm). Dilution factor (how many times the solution was diluted from stock), and factor was fixed number from previous standard curve).
2.6 DNA Isolation and RAPD-PCR Bioassay
According to Doyle et al. [20], Populus alba’s total genomic DNA was extracted using the CTAB method as follow: Half gram of leaves was pulverized with 700 μL of 2% CTAB buffer and incubated at 65°C for 30 min (vortexed each 10 min). At 17,709 g for 10 min, the Eppendorf tubes were centrifuged, and the supernatant was transferred into new tubes. After that, equal chloroform volume [Isoamyl alcohol (24:1)] was added to each tube and set at room temperature for 2 min; then it was centrifuged at 17,709 g at 4°C for 10 min. The upper aqueous layer was transferred to new tubes, and 800 μL of absolute ice-cold ethanol were added, which were left at −20°C for about 2 h. Tubes were then centrifuged to precipitate DNA pellets, and then washed with 70% ice-cold ethanol. The TE buffer pellets were resuspended to 50 μL and kept to −20°C until the RAPD-PCR reaction was performed.
In this bioassay, eight RAPD decamer were applied, only 4 of which gave scorable clear bands. Table 3 listed these primers. The RAPD-PCR reaction in the Biometra thermocycler was performed. The reaction mixture had a total volume of 25 μL, with 12.5 μL of Taq master mix (COSMO PCR RED M. Mix, W1020300x), 2 μl of genomic DNA, 1 μL for each primer (Willowfort), and 9.5 μL dH2O for each primer (Willowfort). The program of reaction included 35 cycles of the following steps: 30-sec denaturation at 94°C, 30-sec annealing at different degrees for each primer, as shown in Table 4, and 1-min extension at 72°C. This was followed by one phase of final extension for 10 min at 72°C, then cooling at 4°C. Compared to the ladder (New England Biolab, #N3232S), the amplified PCR product was run on agarose gel (1.2%).
The images resulting from gel electrophoresis were analyzed and the presence of a band was scored as 1, while the absence was 0. Using Jaccard’s coefficient of similarity, a pairwise similarity matrix was generated and cluster analysis was performed to develop a dendrogram using the unweighted pair group method with the arithmetic averaging algorithm (UPGMA). These computations were performed by Bio-Rad Quantity one (4.6.2) [21].
Descriptive analysis of the variance test and one way ANOVA in Minitab 19 was performed on the data collected. The mean average was estimated, with standard deviations and correlations. Besides, comparison test (ANOVA) was performed to estimate significant differences among the means within each of the studied parameters. Community Package Analysis (CPA, 1.2) software was used to assess the relationship between all Populus treatments based on morphological, physiological and molecular data. CAP tools were complete linkage clustering and PCA blot of PCA covariance ordination.
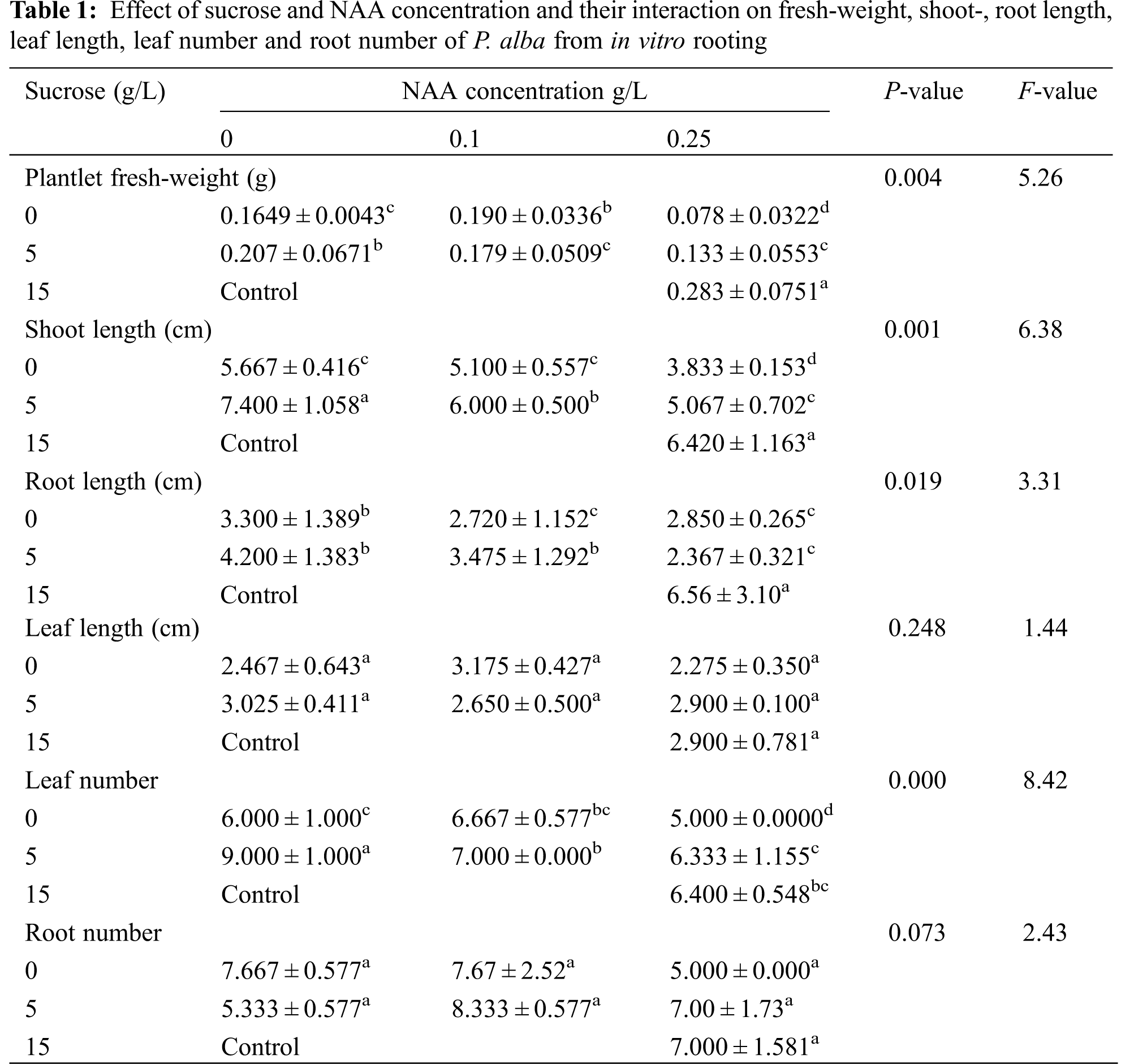
3.1 Morphological and Physiological Parameters
Result data laid out in Table 1 and Fig. 1 show the morphological responses of P. alba to the different sucrose and NAA hormone concentrations. The results had high significant differences on effect of fresh weight, shoot length, root length and leaf number. Some morphological parameters were higher in starvation treatments than control, such as shoot length, leaf length, leaf number and root number. The control treatment was used under normal growth conditions to show that P. alba can grow under normal and starvation treatment conditions.

Figure 1: Different starvation treatments of Populus alba. (1) 0 g/L sucrose, 0 g/L hormone, (2) 0 g/L sucrose, 0.1 g/L NAA; (3) 0 g/L sucrose, 0.25 g/L NAA; (4) 5 g/L sucrose, 0 g/L hormone; (5) 5 g/L sucrose, 0.1 g/L NAA; (6) 5 g/L sucrose, 0.25 g/L NAA; (C) control: 15 g/L sucrose, 0.25 g/L NAA
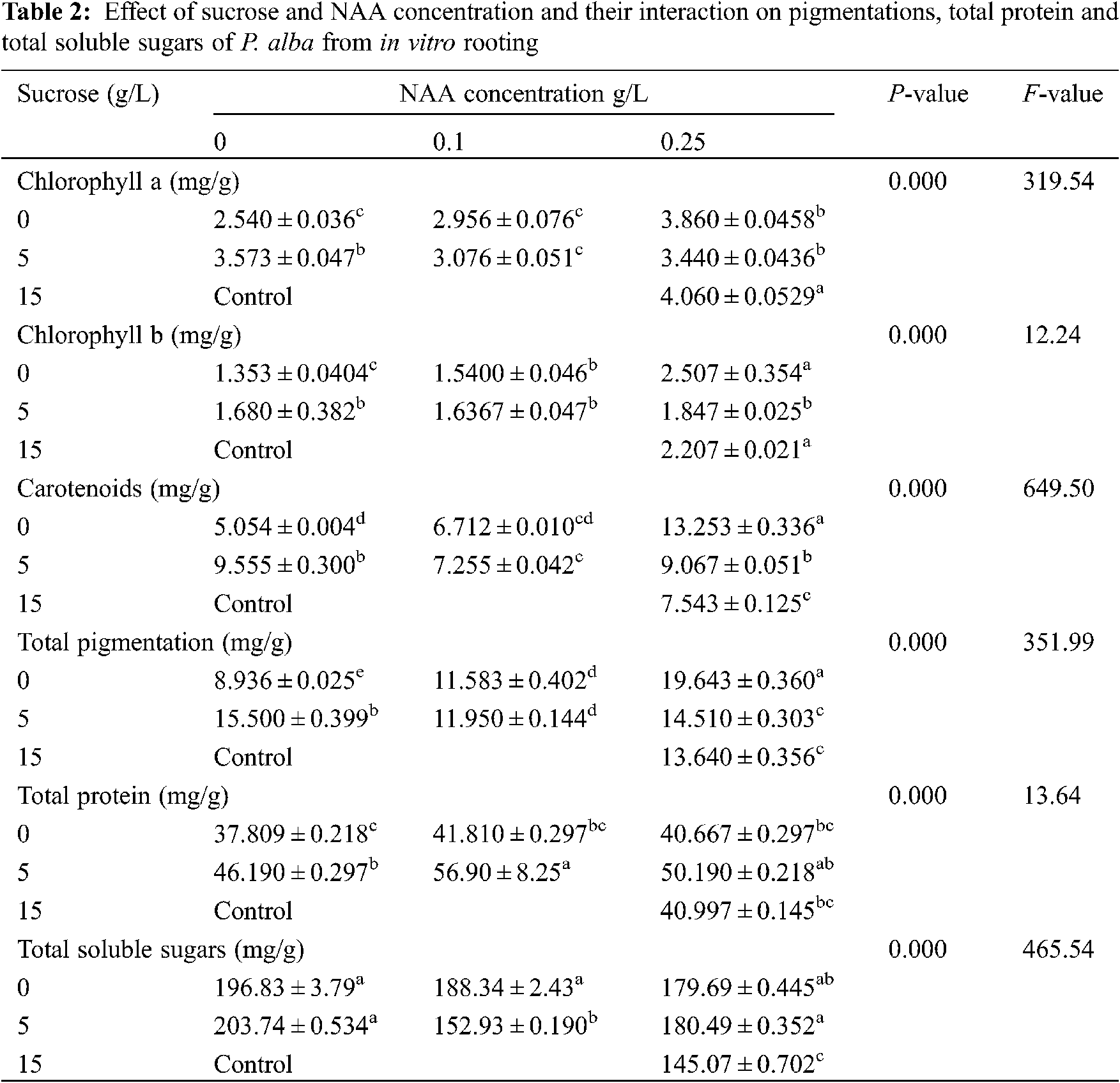
The response of the different physiological measured parameters was estimated and the average was recorded in Table 2. The measured pigmentation in leaves tissues (chlorophyll a, chlorophyll b, carotenoids and Total pigmentation), total protein and total soluble sugars showed a high significance difference. All parameters had a significant difference. There were some physiological responses raised more in response to starvation comparing to control, like carotenoids, total pigmentation, total proteins and total soluble sugars.
3.2 Molecular Marker (RAPD-PCR)
Genetic variation has practical utility and commercial implications for micro-propagated Populus alba plants. In this study, we evaluated fingerprinting profiles using RAPD markers for the different treatments of culture regenerants to confirm whether or not the plantlets were genetically stable. For initial screening, a total of 8 random RAPD primers were tested, of which only 4 primers gave reproducible and clear bands. The number of scorable loci ranged from 3 (Deca-10) to 8 (Deca-4) for each RAPD primer (Table 3). Twenty-two distinct and scorable bands ranged from 202 to 3048 bp from the 4 RAPD primers produced. The polymorphic bands ranged from 1 band (with Deca-11) to 4 bands (with Deca-4 and Deca-13) with a total polymorphism percentage of 50.83% from all primers. During the RAPD analysis of in vitro raised plants, polymorphism was detected (Figs. 2a–2d). The regenerated plants’ uniformity was proven to be maintained, indicating variation in genetic stability between the clones.
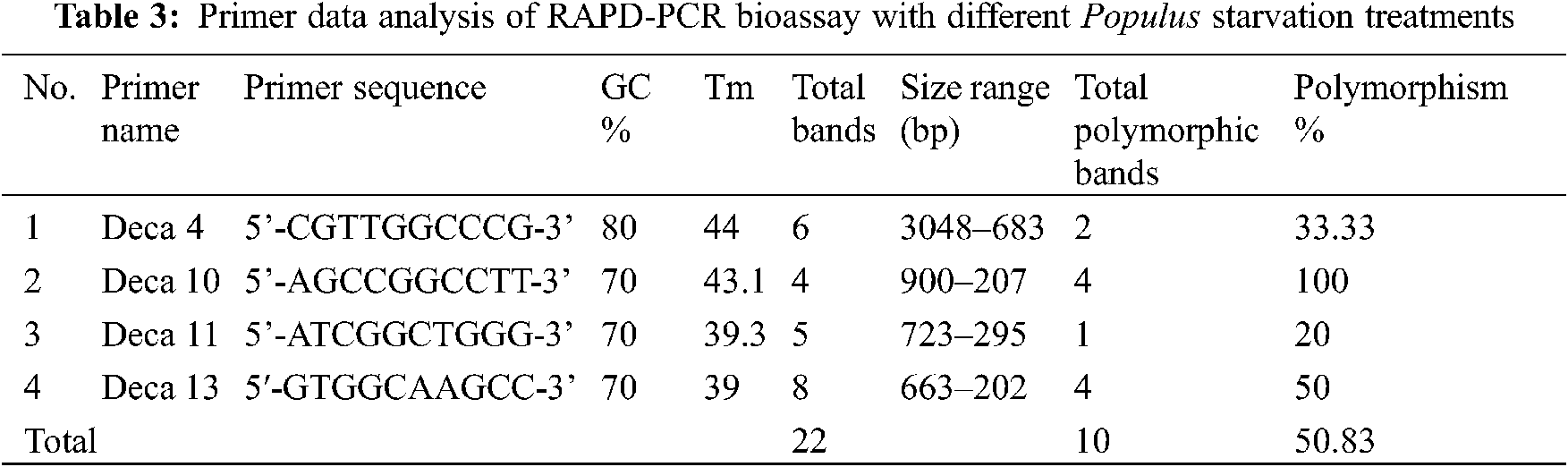
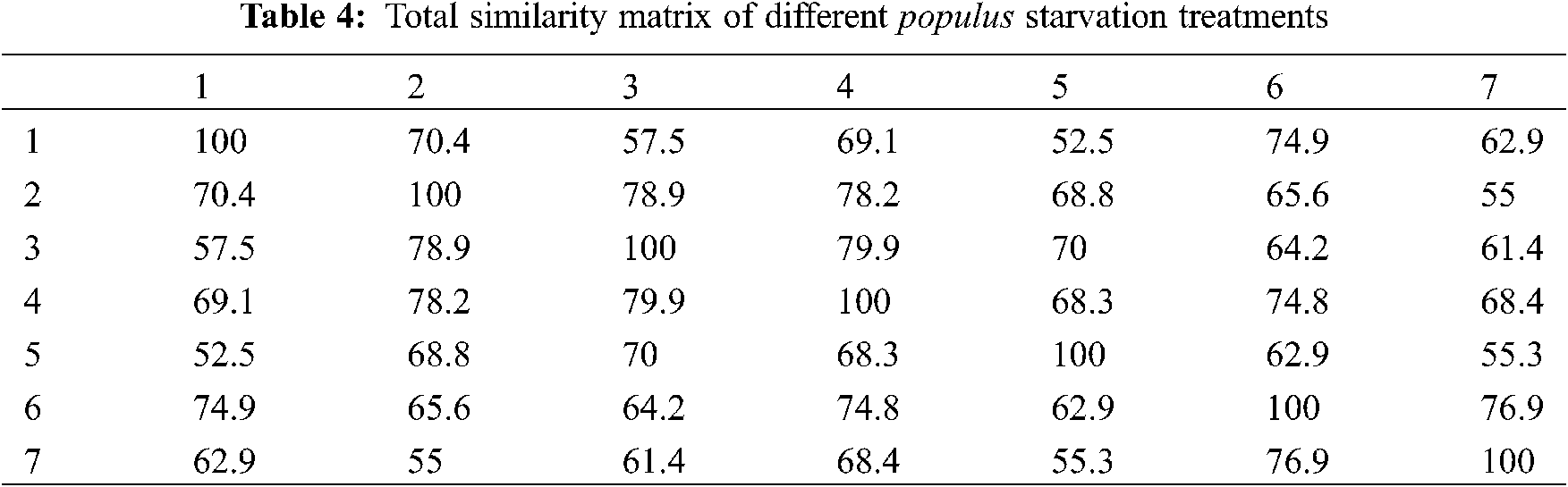

Figure 2: Gel banding pattern of RAPD-PCR for populus alba starvation treatments. (a) Deca-4 primer, (b) deca-10 primer, (c) Deca-11 primer and (d) Deca-13 primer. Specimens: (C) control: 15 g/L sucrose, 0.25 g/L NAA (1) 0 g/L sucrose, 0 g/L hormone, (2) 0 g/L sucrose, 0.1 g/L NAA; (3) 0 g/L sucrose, 0.25 g/L NAA; (4) 5 g/L sucrose, 0 g/L hormone; (5) 5 g/L sucrose, 0.1 g/L NAA; (6) 5 g/L sucrose, 0.25 g/L NAA
A dendrogram of 7 somaclones (Fig. 3) was constructed based on the genetic similarity matrix from Table 4. It was found that more or less, all the somaclones were near to each other, especially the following somaclones treatments (2, 3), (6, 7). In comparison, the dendrogram of these 7 somaclones (Fig. 4) was constructed based on the morphological, physiological and molecular results. It was found that only control, 5 and 6, were grouped, while 1, 2, 3 and 4 starvation treatments were grouped. These findings agreed with results obtained from CAP covariance ordination (Fig. 5).

Figure 3: UPGAMA phylogenetic tree of different populus starved somaclones treatments based on RAPD-PCR data. (1) 15 g/L sucrose, 0.25 g/L NAA; (2) 0 g/L sucrose, 0 g/L hormone, (3) 0 g/L sucrose, 0.1 g/L NAA; (4) 0 g/L sucrose, 0.25 g/L NAA; (5) 5 g/L sucrose, 0 g/L hormone; (6) 5 g/L sucrose, 0.1 g/L NAA; (7) 5 g/L sucrose, 0.25 g/L NAA

Figure 4: The total complete linkage clustering analysis of combined morphology, physiology and RAPD-PCR responses resulted from different treatments of P. alba. (C) control: 15 g/L sucrose, 0.25 g/L NAA (1) 0 g/L sucrose, 0 g/L NAA, (2) 0 g/L sucrose, 0.1 g/L NAA; (3) 0 g/L sucrose, 0.25 g/L NAA; (4) 5 g/L sucrose, 0 g/L NAA; (5) 5 g/L sucrose, 0.1 g/L NAA; (6) 5 g/L sucrose, 0.25 g/L NAA

Figure 5: PCA blot of PCA covariance ordination based on morphology, physiology and RAPD-PCR between different starvation treatments of P. alba
Starvation conditions mean that plant is exposed to nutrient deficiency supplemented in growth media, and also deficient in carbohydrates (i.e., sucrose). The ability of the plant to control and resist low sucrose levels may act as a controlling mechanism, integrating the influence of environmental conditions (e.g., biotic and abiotic stress factors) with internal developmental programs which could be controlled by hormones.
Some morphological and physiological responses were more elevated in starvation treatments than control, such as carotenoids, total pigmentation, total proteins and total soluble sugars. This may be a response to deficient in nutrients in supplemented media and as a defense mechanism.
Elazab et al. [22] applied different sucrose concentrations on the in vitro culture of Ficus carica. They reported that those concentrations enhance the rooting of Ficus. Also, Martins et al. [23] showed that different sucrose concentrations enhance shooting on in vitro cultures of Billbergia zebrina.
The composition of the culture medium is one of the most important factors affecting plant growth in vitro. The main components of in vitro culture media are mineral salts and carbohydrates as carbon sources. Carbohydrates are an essential component for plant development and in vitro growth when conditions are not conducive to photosynthesis; as a result, carbohydrates are supplemented as a carbon source to maintain carbon supply as well as cell osmotic potential. Sucrose, fructose, and glucose are common carbohydrate types. Sucrose is widely used in plant tissue culture because of its favorable growth properties and low cost [24]. Sucrose is essential in the growth medium for the in vitro regeneration process in many plants [23,25]. Nutrient amounts (represented as media powder or how many grams of ready MS medium/L dH2O) may become unavailable to the explants and lead to a restriction in their growth and explant mass. Fresh and dry mass accumulations are nutritionally related to the amounts of supplemented grams in the medium [26]. Adventitious root induction is closely linked to the concentration and endogenous balance of the nutrient composition and plant hormones such as Indole-3-acetic acid (IAA) [27]. NAA is a synthetic auxin plant hormone that is a rooting agent, and it is used for the vegetative propagation of plants from stem and leaf cuttings. It is also used for different plant tissue culture purposes. Yan et al. [28] investigated the significant role of NAA in the adventitious rooting in Hemarthria compressa.
The application of molecular analysis on in vitro regenerated plants has been well documented by many workers [29–32]. Molecular analysis is an efficient and reliable technique for screening tissue culture-derived plants’ types [33,34]. Using several PCR-based molecular markers such as RAPD, ISSR, SSR and AFLP, reliable monitoring of the variability in DNA sequences of plants has been achieved. In several cases, the absence of genetic variation using RAPD has been reported, such as on the axillary bud proliferation of chestnut rootstock hybrids [35] and almond plantlets [36,37].
Such differences could be attributed to the effects of media components on cellular behavior which was affected by the sucrose and hormone concentrations supplemented in the poor MS culture. Application of dendrograms to estimate the genetic stability of plants agreed with Kasim et al. [38]. These authors applied dendrograms to estimate the variation on Cyclanthus due to different hormonal and media compositions.
The in vitro culture may induce loss of cellular control, resulting in somaclonal variations [39]. These variations could be due to factors such as explant type, culture medium composition, culture duration, phyto-regulators, genotype, and number of subcultures or transfers. All these conditions are considered capable of inducing variability on in vitro cultures [40–44].
The present study described an efficient protocol to control Populus alba micropropagation. Shoot induction, and leaf and root numbers were affected mainly by the interaction between sucrose and hormone levels (in poor MS medium “1/4 MS”). Each medium composition was found to be effective in the micro propagation of each study parameter. Populus alba can lead to somaclonal variation among the micro-propagated plants responding to the different media compositions. The genetic variation resulting from these media was not so high (50.83%), which ensures some genetic instability within the somaclones of P. alba. Future studies in this field should focus on making plants more tolerant to extreme conditions, and study the stress effects on P. alba responses.
Acknowledgement: The authors thank the Agricultural Center for Genetic Engineering and Biotechnology “ACGEB”, Faculty of Agriculture, Ain Shams University, for facilitating working in tissue culture labs, and using their facilities and materials.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Bueno, M. A., Gomez, A., Manzanera, J. A. (2003). Micropropagation of woody trees and fruits. Netherlands: Kluwer Academic Publishers. [Google Scholar]
2. Wühlisch, G. (2009). Euforgen technical guidelines for genetic conservation and use of Eurasian Aspen (Populus tremula). Rome, Italy: Biodiversity International. [Google Scholar]
3. Luquez, V., Hall, D., Albrectsen, B. R., Karlsson, J., Ingvarsson, P. et al. (2008). Natural phenological variation in aspen (Populus tremulaThe SwAsp collection. Tree Genetics and Genomes, 4, 279–292. DOI 10.1007/s11295-007-0108-y. [Google Scholar] [CrossRef]
4. Soliman, M. H., Hussein, M. H. A., Gad, M. M. A., Mohamed, A. S. (2017). Genetic transformation of white poplar (Populus alba L.) with glutaredoxin 2 gene. Bioscience Research, 14(3), 464–472. [Google Scholar]
5. Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I. et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & gray). Science, 313, 1596–1604. DOI 10.1126/science.1128691. [Google Scholar] [CrossRef]
6. Madejón, P., Madejón, T., Murilloa, J. M., Robinson, B. (2004). White poplar (Populus alba) as a biomonitor of trace elements in contaminated riparian forests. Environmental Pollution, 132, 145–155. DOI 10.1016/j.envpol.2004.03.015. [Google Scholar] [CrossRef]
7. Bradshaw, H. D., Ceulemans, R., Davis, J., Stettler, R. (2000). Emerging model systems in plant biology: Poplar (Populus) as a model forest tree. Journal of Plant Growth Regulation, 19, 306–313. DOI 10.1007/s003440000030. [Google Scholar] [CrossRef]
8. Ahuja, M. R. (1983). Somatic cell differentiation and rapid clonal propagation of aspen. Silvae Genetica, 32, 131–135. [Google Scholar]
9. Keserű, Z., Balla, I., Antal, B., Rédei, K. (2015). Micropropagation of leuce-poplars and evaluation of their development under sandy site conditions in Hungary. Acta Silvatica et Lignaria Hungarica, 11(2), 139–152. DOI 10.1515/aslh-2015-0011. [Google Scholar] [CrossRef]
10. El-Khishin, D. A., Abdul Hamid, A., El Moghazy, G., Metry, E. A. (2009). Assessment of genetically modified potato lines resistant to potato virus Y using compositional analysis and molecular markers. Research Journal of Agriculture and Biological Sciences, 5, 261–271. [Google Scholar]
11. Saker, M. M., Mohamed, A. A., Aly, A. A. (2011). Comparative analysis of transformed potato microtubers and its non-transformed counterpart using some biochemical analysis along with inter simple sequence repeat (ISSR) marker. African Journal of Biotechnology, 10(34), 6401–6410. [Google Scholar]
12. Li, F., Zuo, R., Abad, J., Xu, D., Bao, G. et al. (2012). Simultaneous detection and differentiation of four closely related sweet potato potyviruses by a multiplex one-step RT-PCR. Journal of Virological Methods, 186(1–2), 161–166. DOI 10.1016/j.jviromet.2012.07.021. [Google Scholar] [CrossRef]
13. Ahmed, M. F., Hosni, A. M., Hewidy, M., Abd El razik, A. B., Bahnasy, M. I. (2019). Morphological, chemical characters and genetic analysis discrimination of five naturalized populus species inhabiting four governorate territories of Egypt. Arab Universities Journal of Agricultural Sciences, 27(4), 2273–2290. DOI 10.21608/ajs.2019.17898.1099. [Google Scholar] [CrossRef]
14. Ahmad, A., Wang, J. D., Pan, Y. B., Sharif, R., Gao, S. J. (2018). Development and use of simple sequence repeats (SSRs) markers for sugarcane breeding and genetic studies. Agronomy, 8(11), 260. DOI 10.3390/agronomy8110260. [Google Scholar] [CrossRef]
15. Abdein, M. A., Abd El-Moneim, D., Taha, S. S., Al-Juhani, W. S. M., Mohamed, S. E. (2018). Molecular characterization and genetic relationships among some tomato genotypes as revealed by ISSR and SCOT markers. Egyptian Journal of Genetics and Cytology, 47, 139–159. [Google Scholar]
16. Murashige, T., Skoog, F. K. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497. DOI 10.1111/j.1399-3054.1962.tb08052.x. [Google Scholar] [CrossRef]
17. Metzner, H., Rau, H., Senger, H. (1965). Studies on synchronization of some pigment-deficient Chlorella mutants. Planta, 65, 186–194. DOI 10.1007/BF00384998. [Google Scholar] [CrossRef]
18. Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochemical Journal, 72, 248–254. DOI 10.1016/0003-2697(76)90527-3. [Google Scholar] [CrossRef]
19. Umbriet, W. W., Burris, R. H., Stauffer, J. F., Cohen, P. P., Johanse, W. J. et al. (1959). Monometric technique: A manual description method applicable to study of describing metabolism. Minneapolis: Burgess Publishing Company. [Google Scholar]
20. Doyle, J. J., Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12, 13–15. [Google Scholar]
21. Shuaib, M., Zeb, A., Ali, Z., Ali, W., Ahmad, T. et al. (2007). Characterization of wheat varieties by seed storage protein electrophoresis. African Journal of Biotechnology, 6, 497–500. DOI 10.4314/ajb.v6i5.56863. [Google Scholar] [CrossRef]
22. Elazab, D. S., Shaaban, M. M. (2015). The impact of sucrose concentration on root growth and development in fig (Ficus carica L.) in vitro. Assiut Journal of Agricultural Sciences, 46(6), 67–75. DOI 10.21608/ajas.2015.521. [Google Scholar] [CrossRef]
23. Martins, J. P. R., Pasqual, M., Martins, A. D., Ribeira, S. F. (2015). Effects of salts and sucrose concentrations on in vitro propagation of Billbergia zebrina (Herbert) lindley (Bromeliaceae). Australian Journal of Crop Science, 9(1), 85–91. DOI 10.1590/2447-536X.v26i1.2092. [Google Scholar] [CrossRef]
24. Sumaryonoï Muslihatin, W., Ratnadewi, D. (2012). Effect of carbohydrate source on growth and performance of in vitro sago palm (Metroxylon sagu rottb.) plantlets. HAYATI Journal of Biosciences, 19(2), 88–92. DOI 10.4308/hjb.19.2.88. [Google Scholar] [CrossRef]
25. Williams, R. R. (1993). Mineral nutrition in vitro–A mechanistic approach. Australian Journal of Botany, 41(2), 237–251. DOI 10.1071/BT9930237. [Google Scholar] [CrossRef]
26. Huang, Z. C., Zeng, F. H., Lu, X. Y. (2010). Efficient regeneration of Eucalyptus urophylla from seedling-derived hypocotyls. Biologia Plantarum, 54(1), 131–134. DOI 10.1007/s10535-010-0020-4. [Google Scholar] [CrossRef]
27. Dong, N., Pei, D., Yin, W. (2012). Tissue-specific localization and dynamic changes of endogenous IAA during poplar leaf rhizogenesis revealed by in situ immunohistochemistry. Plant Biotechnology Reports, 6(2), 165–174 DOI 10.1007/s11816-011-0209-9. [Google Scholar] [CrossRef]
28. Yan, Y. H., Li, J. L., Zhang, X. Q., Yang, W. Y., Wan, Y. et al. (2014). Effect of naphthalene acetic acid on adventitious root development and associated physiological changes in stem cutting of hemarthria compressa. PLoS One, 2014(9), e90700. DOI 10.1371/journal.pone.0090700. [Google Scholar] [CrossRef]
29. Saha, S., Dey, T., Ghosh, P. D. (2010). Micropropagation of Ocimum kilimandscharicum guerke (Labiatae). Acta biologica Cracoviensia. Series botanica, 52, 50–58. DOI 10.2478/v10182-010-0023-7. [Google Scholar] [CrossRef]
30. Bhatia, R., Singh, K. P., Sharma, T. R., Jhang, T. (2011). Evaluation of the genetic fidelity of in vitro propagated gerbera (Gerbera jamesonii bolus) using DNA based markers. Plant Cell, Tissue and Organ Culture, 104, 131–135. DOI 10.1007/s11240-010-9806-5. [Google Scholar] [CrossRef]
31. Saha, S., Kader, A., Sengupta, C., Ghosh, P. D. (2012). In vitro propagation of Ocimum gratissimum L. (Lamiaceae) and its evaluation of genetic fidelity using RAPD markers. American Journal of Plant Sciences, 3, 64–74. DOI 10.4236/ajps.2012.31006. [Google Scholar] [CrossRef]
32. Saha, S., Adhikari, S., Dey, T., Ghosh, P. (2016). RAPD and ISSR based evaluation of genetic stability of micropropagated plantlets of Morus alba L. variety S-1. Meta Gene, 7, 7–15. DOI 10.1016/j.mgene.2015.10.004. [Google Scholar] [CrossRef]
33. Damasco, O. P., Graham, G. C., Henry, R. J., Adkins, S. W., Smiths, M. K. et al. (1996). Random amplified polymorphic DNA (RAPD) detection of dwarf off-types in micropropagated cavendish (Musa spp. AAA) bananas. Plant Cell Reports, 16(1–2), 118–123. DOI 10.1007/BF01275464. [Google Scholar] [CrossRef]
34. Khatab, I. A., Youssef, M. S. (2018). Micropropagation and assessment of genetic stability of musa sp. cv. williams using RAPD and SRAP markers. Egyptian Journal of Botany, 58(3), 1–10. DOI 10.21608/ejbo.2018.3199.1161. [Google Scholar] [CrossRef]
35. Carvalho Luísa, C., Luís, G., Cristina, O., Carlos, G. J., Amancio, S. (2004). RAPD assessment for identification of clonal identity and genetic stability of in vitro propagated chestnut hybrids. Plant Cell, Tissue and Organ Culture, 77(1), 23–27. DOI 10.1023/B:TICU.0000016482.54896.54. [Google Scholar] [CrossRef]
36. Martins, M., Sarmento, D., Oliveira, M. M. (2004). Genetic stability of micropropagated almond plantlets as assessed by RAPD and ISSR markers. Plant Cell Reports, 23(7), 492–496. DOI 10.1007/s00299-004-0870-3. [Google Scholar] [CrossRef]
37. Lakshmanan, V., Venkataramareddy, S. R., Neelwarne, B. (2007). Molecular analysis of genetic stability in long-term micropropagated shoots of banana using RAPD and ISSR markers. Electronic Journal of Biotechnology, 10(1), 106–113. DOI 10.2225/vol10-issue1-fulltext-12. [Google Scholar] [CrossRef]
38. Kasim, N. F. M., Yahya, H. N., Kadzimin, S., Awang, Y. (2018). Micropropagation and assessment of genetic variability of Cyclanthus bipartitus. Asian Journal of Plant Sciences, 17(1), 19–26. DOI 10.3923/ajps.2018.19.26. [Google Scholar] [CrossRef]
39. Pathak, H., Dhawan, V. (2012). Evaluation of genetic fidelity of in vitro propagated apple (Malus × domestica borkh.) rootstock MM 106 using ISSR markers. Acta Horticulturae, 961, 303–310. DOI 10.17660/ActaHortic.2012.961.40. [Google Scholar] [CrossRef]
40. Biswas, M. K., Dutt, M., Roy, U. K., Islam, R. (2009). Development and evaluation of in vitro somaclonal variation in strawberry for improved horticultural traits. Scientia Horticulturae, 122, 409–416. DOI 10.1016/j.scienta.2009.06.002. [Google Scholar] [CrossRef]
41. Chuang, S. J., Chen, C. L., Chen, J. J., Chou, W. Y. (2009). Detection of somaclonal variation in micro-propagated Echinacea purpurea using AFLP marker. Scientia Horticulturae, 120, 121–126. DOI 10.1016/j.scienta.2008.09.020. [Google Scholar] [CrossRef]
42. Khan, E. U., Fu, X. Z., Wang, J., Fan, Q. J. (2009). Regeneration and characterization of plants derived from leaf in vitro culture of two sweet orange (Citrus sinensis (L.) osbeck) cultivars. Scientia Horticulturae, 120, 70–76. DOI 10.1016/j.scienta.2008.10.004. [Google Scholar] [CrossRef]
43. Borse, N., Chimote, V. P., Jadhav, A. S. (2011). Stability of micropropagated Musa acuminate cv. grand naine over clonal generations: A molecular assessment. Scientia Horticulturae, 129, 390–395. DOI 10.1016/j.scienta.2011.04.001. [Google Scholar] [CrossRef]
44. Werner, E. T., Soares, T. C. B., Gontijo, A. B. P. L., Souza Neto, J. D., do Amaral, J. A. T. (2015). Genetic stability of micropropagated plants of Crambe abyssinica hochst using ISSR markers. Genetics and Molecular Research, 14(4), 16450–16460. DOI 10.4238/2015.December.9.16. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |