

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018075
ARTICLE
Genome-Wide Analysis of Snapdragon WRKY and VQ Gene Families and Their Expression in Response to Drought and Cold Stresses
Zhejiang Institute of Landscape Plants and Flowers, Hangzhou, 311251, China
*Corresponding Authors: Qing Dong. Email: dqricezzh@163.com; Huaqiao Ding. Email: dhuaqiao@sina.com
Received: 27 June 2021; Accepted: 14 September 2021
Abstract: Snapdragon (Antirrhinum majus) is one of the most widely cultivated grass flowers in the world. WRKY transcription factors, VQ proteins and their interactions play crucial roles in plant response to abiotic stresses. However, little is known about WRKY and VQ gene families in snapdragon. In the present study, WRKY and VQ genes and their interactions were comprehensively analyzed in snapdragon using bioinformatics approaches, and their expression in response to drought and cold stresses was examined using real-time PCR. A total of 67 AmWRKY genes were identified in snapdragon, which were classified into different groups or subgroups based on phylogenetic analysis. Members in the same group or sub-groups exhibited similar exon-intron structure and conserved motifs distribution. Among these WRKY genes, 16 and 22 genes were found to differentially express under drought and cold stresses, respectively. A total of 32 AmVQ genes were identified, of which 10 and 18 were found to differentially express under drought and cold stresses, respectively. The WRKY-VQ or WRKY-WRKY interaction relationships were predicted for 11 cold-responsive genes, suggesting that they might exist in the same response pathway to cold stress. These results lay a foundation for further studies on roles of WRKY and VQ genes and their interactions in regulating abiotic responses in snapdragon.
Keywords: Snapdragon; WRKY genes family; VQ genes family; gene expression; abiotic stress
The WRKY gene family is one of the largest families of transcription factors, which have been identified throughout the plant kingdom. The name of the WRKY family is derived from the most prominent feature of these transcription factors, the WRKY core sequence, which generally is tryptophan-arginine-lysine-tyrosine-glycine-glutumine-lysine (WRKYGQK). Another defining feature of WRKY transcription factors is the zinc finger motif, which is either C2H2 or C2HC type. The core sequence and the zinc finger motif form the WRKY domain [1,2]. According to the number of WRKY domains and the type of zinc finger motif they contain, WRKY transcription factors are usually classified into three main groups. Group I contains two WRKY domains and a C2H2 zinc finger motif. Group II contains a single WRKY domain and a C2H2 zinc finger motif, which is further divided into five subgroups based on their primary sequences, including IIa, IIb, IIc, IId and IIe [1,2]. Group III contains a single WRKY domain and a C2HC zinc finger motif.
The WRKY transcription factors are essential regulatory components of plant response to abiotic stress. Numerus studies revealed that WRKY genes could rapidly differentially express under abiotic stresses, based on transcriptome analysis or real-time PCR [3–11]. Roles of WRKY genes in stress response have also been confirmed using mutant or transgenic plants. For instance, at least seven WRKY genes were validated to regulate abiotic stress response in Arabidopsis. Constitutive expressions of AtWRKY25, AtWRKY26 or AtWRKY33 enhance heat tolerance [12]. Loss function of AtWRKY34 improves cold tolerance of pollen [13]. Activating expression of AtWRKY30, AtWRKY53 or AtWRKY57 improve drought tolerance [14–16]. Overexpression of AtWRKY33 increases salt tolerance, while loss function of AtWRKY53 decreases salt tolerance [17,18]. The WRKY transcription factors also play important roles in many other biologic processes in plant, such as flowering time, pollen development, fruit ripening, seed size, leaf senescence and secondary metabolite biosynthesis [19–25].
In general, WRKY transcription factors performed their biologic function by interacting with various proteins [26]. The valine-glutamine (VQ) protein is one of the most important WRKY interactors identified in the past few years. The VQ proteins contained a conserved VQ motif structure FxxhVQxhTG (F, Phenylalanine; x, any amino acid; h, hydrophobic residue; V, Valine; Q, Glutamine; T, Tryptophan, G, Glycine) [27]. Like WRKY, VQ proteins also have significant effects on abiotic stress response in plants. In Arabidopsis, at least two VQ genes were reported to be involved in abiotic response. AtVQ9 was strongly induced by NaCl treatment. Mutation of AtVQ9 enhanced Arabidopsis tolerance to salt stress, while over-expression of this gene increased sensitivity to salt stress [28]. AtVQ15 was induced by dehydration, low temperature and high salinity. Knock-down of this gene enhances Arabidopsis tolerance to osmotic and salt stress. In contrast, over-expression of this gene showed increased sensitivity to these two abiotic stresses [28]. The role of VQ genes in plant response to abiotic stresses were also observed in other species, such as bamboo salt stress positive regulator PeVQ28 [29] and tomato heat negative regulator SlVQ6 [30].
It has been showed that VQ proteins could interact through their conserved V and Q residues with Group I and IIc WRKY transcription factors [31,32]). The two gene families jointly regulate plant development by forming complexes. For example, Arabidropsis AtVQ9 negatively regulates salt tolerance by interacting with AtWRKY8 [28]. AtVQ15 regulates osmotic stress tolerance, which interacts with AtWRKY25 and AtWRKY51 [32,33]. Banana MaWRKY26 interacts with MaVQ5 to regulate expression of JA biosynthetic genes involved in cold resistance [34]. Several WRKY-VQ interactions were found to regulate Arabidopsis resistance to Botrytis cinerea, including AtWRKY33/AtWRKY57-AtVQ16/AtVQ23, WRKY8-VQ10, and AtWRKY33-AtVQ23 [31,35,36]. In addition, co-expression between WRKY and VQ genes was observed in many plant species, such as rice, maize and melon [37–39].
Snapdragon (Antirrhinum majus) is a perennial herb of the Plantaginaceae. It originated in the Mediterranean coast, and has become one of the most widely cultivated grass flowers in the world. Snapdragon has a strong cold tolerance, which could keep blooming in early spring, late fall or even midwinter in some warm places. Snapdragon also has moderate drought tolerance, which could thrive in well-drained soils [40,41]. Several snapdragon genes have been found to enhance tolerance to cold or drought stresses [42,43]. The WRKY and VQ gene families have been systematically studied in many plant species [5,44,45], whereas there is little knowledge about WRKY genes in snapdragon. In this study, we performed a genome-wide analysis of WRKY and VQ genes in snapdragon, including gene classification, gene duplication investigation and protein-protein interaction prediction. The expression of these genes in response to cold and drought was further assayed using real-time PCR. Our study aimed to lay a foundation for further studies on the role of WRKY and VQ genes and their interactions in regulating snapdragon responses to abiotic stresses.
2.1 Plant Materials and Treatments
A snapdragon variety, Shiyun, was used in this study. Each snapdragon plant was grown in a plot with a diameter of 15 cm and peat moss-based soil. They were firstly planted in a plant growth chamber (MLR-351, PHCbi) for about four weeks at 15°C with a 16/8 h light/dark photoperiod and a relative humidity of 80%. The soil moisture was monitored by a soil moisture meter (L99, Luge), and was maintained at about 70%. Then these plants were divided equally into three groups. The first group was set up as a control. It was kept in the growth chamber, which was grown under 15°C and well-watered conditions. The second group was set up as a drought treatment. It was kept in the same growth chamber, which was grown under 15°C but received no irrigation for one week. The third group was set up as cold treatment. This group was kept at 15°C in the same growth chamber for six days, and then was transferred to a low-temperature light incubator (PRXD-300, Chuanyi) with 0°C for one day. Leaves were collected from five-week-old plants growing under the control, drought stress (after one week treatment) and cold stress (after one day treatment) treatments.
2.2 Identification of WRKY and VQ Genes in Snapdragon
The snapdragon genome and annotation data (version 2) were downloaded from the Snapdragon Genome Database (http://bioinfo.sibs.ac.cn/Am/index.php). The WRKY protein sequences of Arabidopsis were downloaded from the TAIR (http://www.arabidopsis.org/), and used as query sequences. A BLASTP search was performed to identify AmWRKY protein using TBtools with an E-value threshold of 10−5 [46]. The HMM of the VQ motif (PF05678) was downloaded from the Pfam database (http://pfam.sanger.ac.uk/), and was used to search AmVQ proteins with HMM 3.2 software [47]. All potential AmWRKY and AmVQ genes were assessed for presence of the WRKY domain or VQ motif with the Web CD-search Tool (http://www.ncbi.nlm.nih.gov/structure/cdd/). Only the genes with the complete WRKY domain or VQ motif were accepted.
2.3 Sequence Alignment and Phylogenetic Analysis
Sequence alignment was performed by MEGA7 software using the Muscle method with default parameters [48]. Phylogenetic tree was constructed by MEGA7 using Maximum Likelihood method with the following parameters: 1000 bootstrap replicates, Jones-Taylor-Thomton model, partial deletion and 80% site coverage cutoff.
2.4 Analysis of Gene Structure, Conserved Motif, Gene Duplication and Cis-Acting Regulatory Element, and Prediction of Protein-Protein Interaction
Molecular weights and isoelectric points were analyzed using the ExPASy website (http://web.expasy.org/protparam). Conserved motifs were identified using the MEME program (http://alternate.memesuite.org/tools/meme) with the following parameters: minimum motif width = 6; maximum motif width = 50; maximum number of motif = 10; minimum sites per motif = 2; maximum sites per motif = 600 [49]. Gene duplication and synteny analysis were performed using the MCScanX software with the default parameters [50]. The cis-regulatory elements were analyzed using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare). The protein-protein interactions were predicted by STRINGV11 online software with default score thresholds [51]. Only interactions predicted based on experimentally determined or from curated databases are presented.
2.5 RNA Extraction and Real-Time PCR
Total RNA was extracted using an RNeasy Plus Mini Kit (QIAGEN, German). First-strand cDNA was synthesized using a ReverTra Ace qPCR RT Master Mix with gDNA Remover kit (Toyobo, Japan). Real-time PCR was performed on an Applied Biosystems 7900HT using SYBR qPCR Mix kit (Toyobo, Japan). Ubiquitin was used as the endogenous control. Data were analyzed according to the 2−ΔΔCt method [52]. Three independent biological replicates and three technical replicates per biological replicate were used. Differences in gene expression under abiotic stress and normal conditions were compared using t-tests, with a threshold of P < 0.01. Primers were designed using Primer Premier 5.0 software (Supplementary Table 1).
3.1 Identification of WRKY Genes in Snapdragon
A total of 72 genes with WRKY domain were identified in snapdragon (Supplementary Table 2). Five of them were excluded from further study due to lack of the WRKYGQK sequence or zinc finger motif. The remaining 67 genes were designated from AmWRKY1 to AmWRKY67 based on their locations on the respective chromosome. The lengths of these AmWRKY proteins varied from 109 to 725 amino acids, the isoelectric points ranged from 4.90 to 10.13, and the molecular weights ranged from 12.9 to 78.3 kDa. Twelve of these 67 proteins contained two WRKY domains, and 45 of them contained one WRKY domain.
3.2 Phylogenetic Tree Analysis, Group Classification and Multiple Sequence Alignment of AmWRKY Genes
A phylogenetic tree was constructed based on the WRKY domain sequences of these AmWRKY proteins, and Arabidopsis WRKY domain sequences were included as references (Fig. 1). Sixty-six of 67 AmWRKY proteins were divided into three groups, according to the classification in Arabidopsis [1]. Twelve proteins harboring two WRKY domains were categorized into Group I, while 45 and nine proteins containing one WRKY domain were categorized into Group II and Group III, receptively. The remaining one protein, AmWRKY65, was not assigned to any group. The Group II was further divided into five subgroups. They were named IIa, IIb, IIc, IId and IIe, consisting of four, nine, eighteen, six and eight genes, respectively. This classification was consistent with the phylogenetic relationship base on full-length sequence of these AmWRKY proteins (Fig. 2a). Multiple sequence alignment was conducted for the WRKY domain of these proteins (Supplementary Fig. 1). All of them contained the conserved WRKYGQK sequence, except for AmWRKY42 belonging to Group IIc, which carried a variant of WRKYGKK. Members in Groups I and III contained the C2H2-type (C-X4-5-C-X22-23-H-X-H) and C2HC-type (C-X7-C-X23-H-X-C) zinc finger motif, respectively. Members in Group II contained C2H2-type zinc finger motif, except for AmWRKY8 and AmWRKY54 belonging to Group IIe. AmWRKY8 had a C-X5-C-X27-H-X-H type zinc finger motif, while AmWRKY54 carried a 14 amino acids deletion at the N terminus of zinc finger motif.
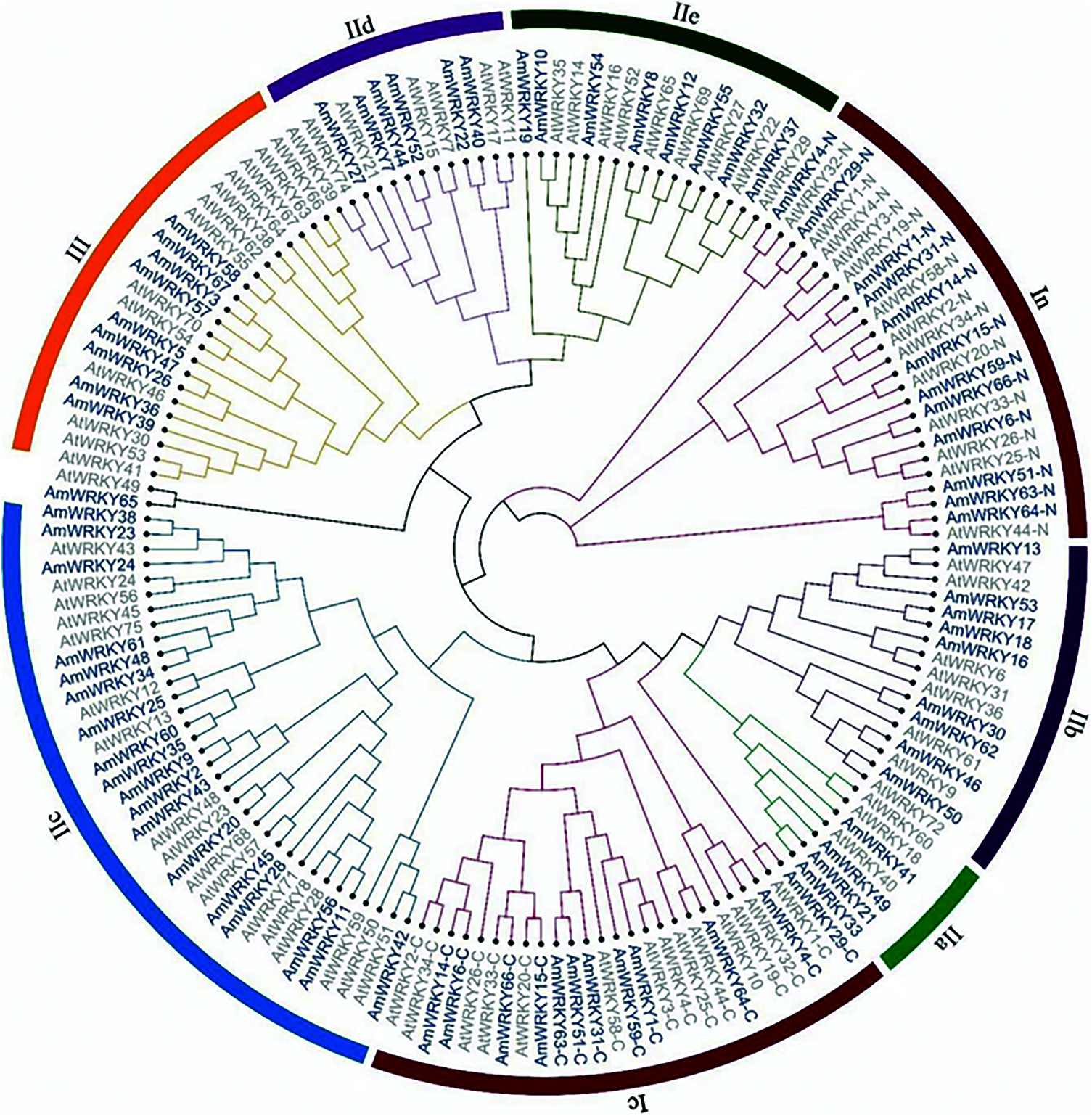
Figure 1: Phylogenetic tree of WRKY proteins among snapdragon and Arabidopsis. The phylogenetic tree of AmWRKY proteins was constructed using MEGA7.0 by the maximum likelihood method with 1,000 bootstrap replicates. AtWRKY proteins were used as references to classify AmWRKY proteins

Figure 2: Structure characterization of AmWRKY genes. (a) Phylogenetic tree of AmWRKY proteins. (b) The exon-intron structures of AmWRKY genes. The introns phases 0, 1 and 2 were indicated by numbers 0, 1 and 2, respectively. (c) The conserved motifs of AmWRKY proteins
3.3 Gene Structure, Motif Composition and Cis-Acting Regulatory Elements of AmWRKY Genes
The number of introns varied from two to seven for these AmWRKY genes (Fig. 2b). Most members of the same group had a similar intron number, except for Group IIb. All members of Groups IId, IIe and III contained two introns, most members of Groups I and IIa contained three or four introns, while most members of Group IIc contained one or two introns. In contrast, the intron number of Group IIb members was variable, including one, two, four, five or six introns.
Most members of the same group also shared similar exon-intron structures. An intron was located between two complete codons named phase 0, and after the first or second nucleotides in the codon defined as phases 1 and 2. All members of Groups IId, IIe and III were found in phase 0 at exon 1st and phase 2 at exon 2nd and 3rd (Fig. 2b). Members of Group IIa and IIb were only found in phase 0, except for AmWRKY49 and AmWRKY50. Phases 0 and 1 were widely distributed in Groups I and IIc with phase 2 having the least distribution.
The conserved motifs of AmWRKY proteins were further identified. As shown in Fig. 2c and Supplementary Fig. 2, the motifs 1, 2, 3, and 7 contained the WRKY core sequence and zinc finger motif. The remaining motifs exhibited a similar distribution in the same group. All members of Group I contained motifs 4 and 10. All members of Group IIa contained motifs 5 and 8. All members of Group IIb contained motifs 6 and 9. All members of Group IIc contained motif 4.
The potential cis-regulatory elements were investigated in the promoter region of AmWRKY genes. Five types of stress-related and six types of hormone-related elements were identified (Supplementary Table 3). Among the stress-related elements, anoxic-responsive element (ARE and GC-motif) was the most abundant, which was found in promoters of 55 of 67 AmWRKY genes. The remaining four types, including drought-responsive element (MBS), low temperature-responsive element (LTR), defense- and stress-responsive element (TC-rich repeats) and wound-responsive element (WUN-motif), were found in 30, 28, 23 and 3 AmWRKY genes, respectively. The hormone-related elements included abscisic acid (ABA)-responsive element (ABRE), gibberellin-responsive elements (GARE-motif, P-box and TATC-box), methyl jasmonate (MeJA)-responsive element (CGTCA-motif), ethylene-responsive element (ERE), auxin-responsive elements (AuxRR-core and TGA-element) and salicylic acid (SA)-responsive element (TCA-element), which were found in 60, 56, 52, 45, 31 and 28 AmWRKY genes, respectively.
3.4 Chromosome Location and Gene Duplication of AmWRKY Genes
The 67 AmWRKY genes were distributed across all the eight snapdragon chromosomes, but their distribution across the respective chromosomes was uneven (Fig. 3). Chromosome 2 contained the highest number of 18, followed by chromosomes 4 and 11. Chromosome 5 contained the lowest number of three. The number of WRKY genes on the remaining five chromosomes ranged from five to nine. In general, a chromosomal region within 200-kb containing multiple genes is defined as a gene cluster [53]. According to this criterion, three gene clusters were found, which were distributed on chromosomes 4 and 7 (Fig. 3).
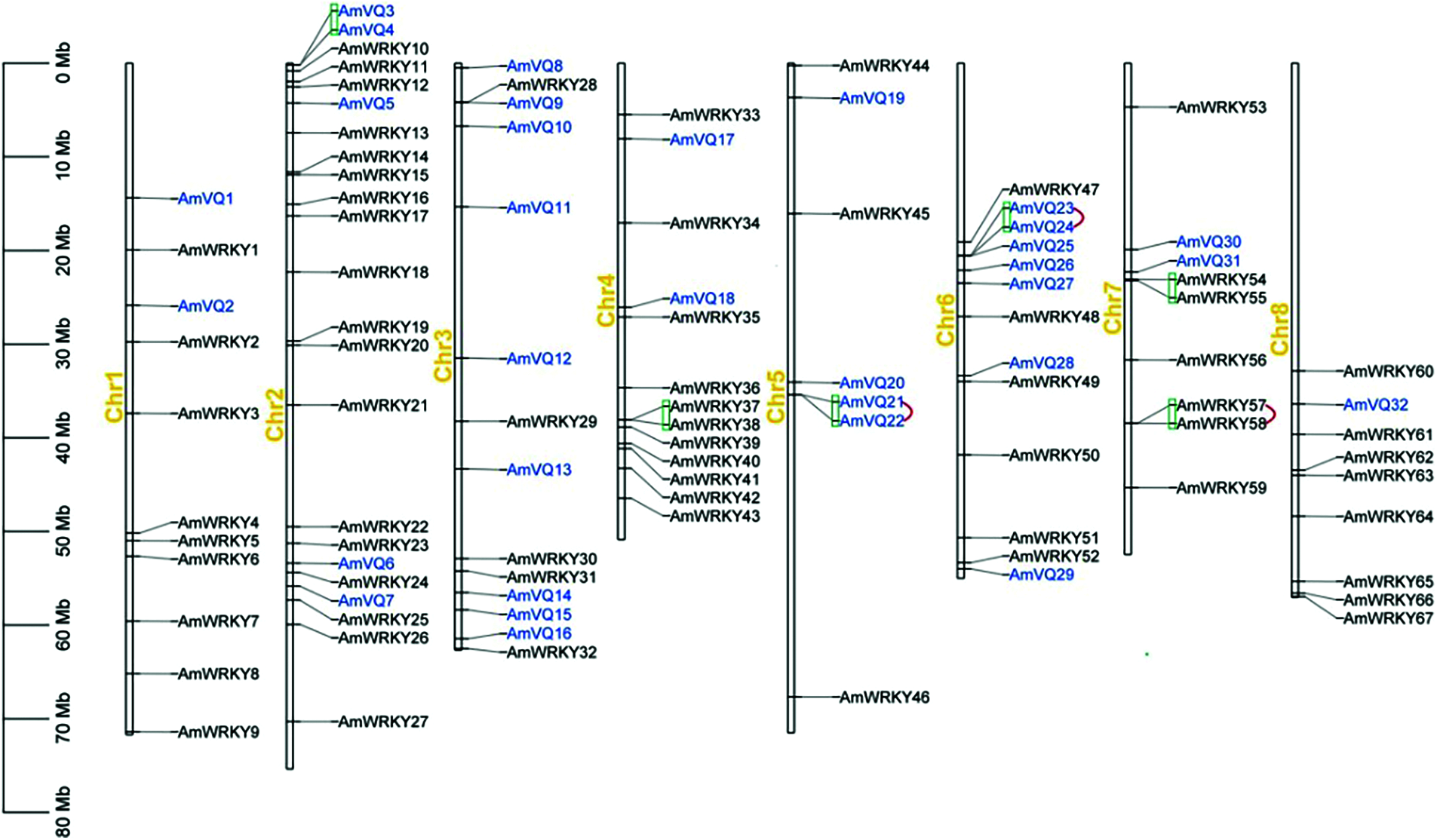
Figure 3: Locations of AmWRKY and AmVQ genes on snapdragon chromosomes. Gene clusters were marked in green line, and tandem duplication genes were marked in red line
Segmental and tandem duplication were investigated for these AmWRKY genes. A total of 74 segmental duplication events involving 44 AmWRKY genes were found, which were distributed on all eight chromosomes (Fig. 4a). In contrast, only one tandem repeat gene pair AmWRKY57/AmWRKY58 was identified, which was distributed on chromosome 7 (Fig. 3). The collinear relationship between snapdragon and Arabidopsis was further analyzed. A total of 34 orthologous WRKY gene pairs were identified, which involved in 34 AmWRKY genes and 29 AtWRKY genes (Supplementary Table 4).

Figure 4: Segmental duplications of AmWRKY (a) and AmVQ genes (b). The gray lines indicated segmentally duplicated gene pairs
3.5 Identification of VQ Genes in Snapdragon
A total of 32 AmVQ genes were identified, which were designated from AmVQ1 to AmVQ32 based on their locations on the respective chromosome (Supplementary Table 5). For these genes, the protein lengths varied from 102 to 396 amino acids, the isoelectric points ranged from 4.58 to 11.51, and the molecular weights ranged from 11.30 to 42.40 kDa. All of these VQ proteins contained the conserved FxxhVQxhTG motif, except for AmVQ24 which carried a variant of FxxhVQxhTC (Supplementary Fig. 3).
A phylogenetic tree was constructed based on the VQ-motif sequences of these AmVQ proteins (Fig. 5). They were cluster into three groups, according to the classification in Arabidopsis. Number of members ranged from one to five in these groups. All of AmVQ genes had no introns, except for AmVQ25 and AmVQ26 that contained two and one introns, respectively (Supplementary Fig. 4b). The conserved motifs exhibited a district distribution in different group, apart from that of the motif 1 corresponding to the VQ-motif presented in each of AmVQ protein (Supplementary Fig. 4c). For example, all members of Group I contained motifs 4, all members of Group IV contained motifs 1 and 2, and all members of Group VI contained motifs 3, 6 and 9. The cis-regulatory elements in the promoter were also investigated. Among the 32 AmVQ genes, 29 were found to carry at least one type of stress-related element, and 31 were found to carry at least one type of hormone-related element (Supplementary Table 6).
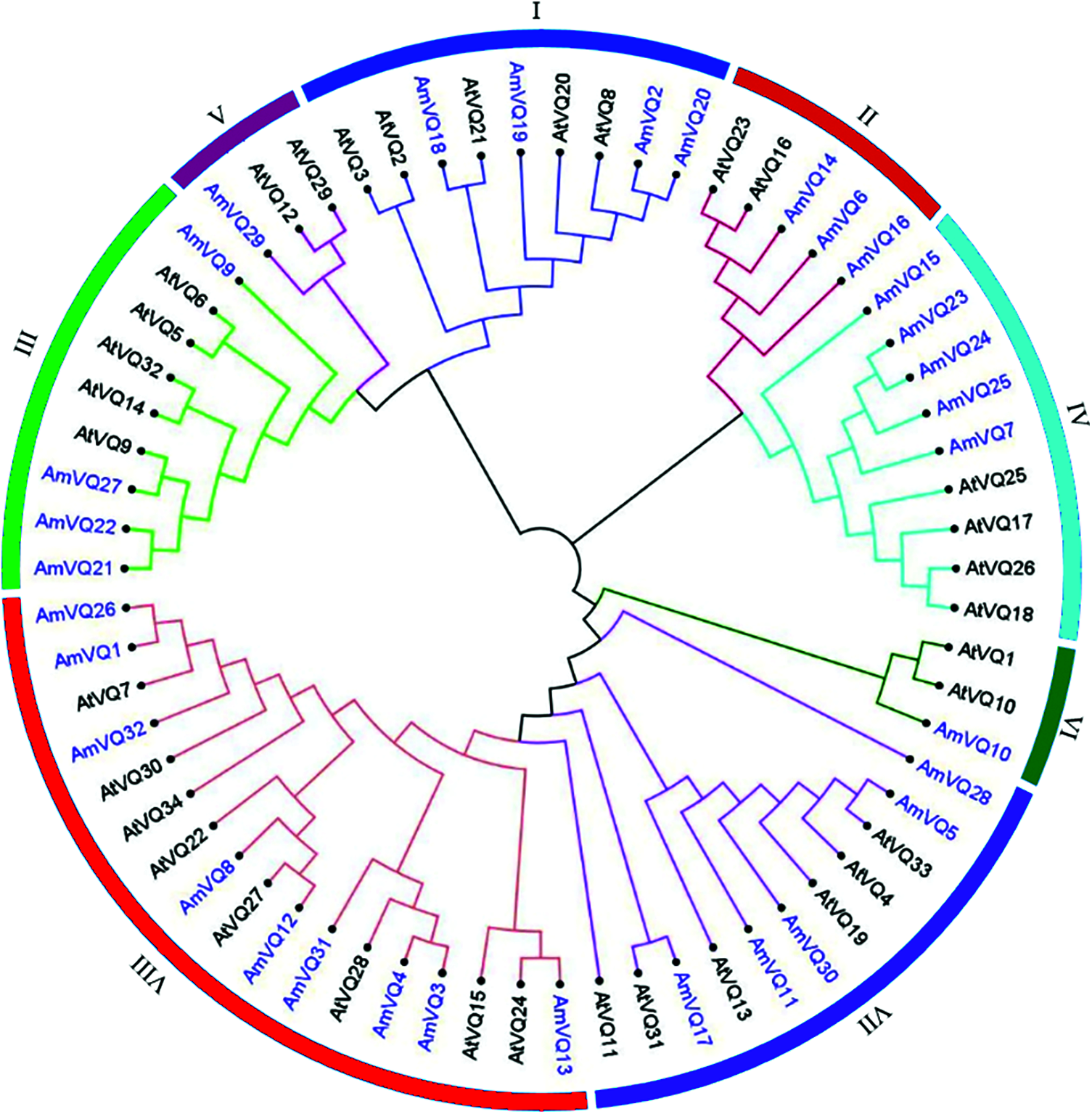
Figure 5: Phylogenetic tree of VQ proteins among snapdragon and Arabidopsis. The phylogenetic tree of AmVQ proteins was constructed using MEGA7.0 by the maximum likelihood method with 1,000 bootstrap replicates. AtVQ proteins were used as references to classify AmVQ proteins
The 32 AmVQ genes were distributed across all the eight snapdragon chromosomes, and their distribution across the respective chromosomes was also uneven (Fig. 3). Chromosome 3 contained the highest number of nine, followed by chromosomes 6, 2 and 5. Chromosomes 1, 4, 7 and 8 only contained one or two AmVQ genes. Three gene clusters were found, which were distributed on chromosomes 2, 5 and 6 (Fig. 3). Two of the clusters consisted of tandem repeat gene pair, including AmVQ21/AmVQ22 and AmVQ57/AmVQ58. A total of five segmental duplication events involving nine AmVQ genes were found, which were distributed on chromosomes 1, 2, 3, 4 and 6 (Fig. 4b). The orthologous VQ gene pairs between snapdragon and Arabidopsis were also analyzed. Ten orthologous gene pairs were identified, which involved in seven AmVQ genes and ten AtVQ genes (Supplementary Table 4).
3.6 Expression of AmWRKY and AmVQ Genes in Response to Drought and Cold Stresses
Expression levels of AmWRKY and AmVQ genes under drought and low temperature treatments were investigated using real-time PCR. Sixteen AmWRKY genes showed no detectable expression (Supplementary Table 1). Among the remaining 51 genes, 16 responded to drought and 22 responded to cold (Fig. 6). Compared with normal condition, eight AmWRKY genes were up-regulated and the other eight AmWRKY genes were down-regulated under the drought treatment. Under the low temperature treatment, eight AmWRKY genes were up-regulated and the remaining 14 genes were down-regulated.
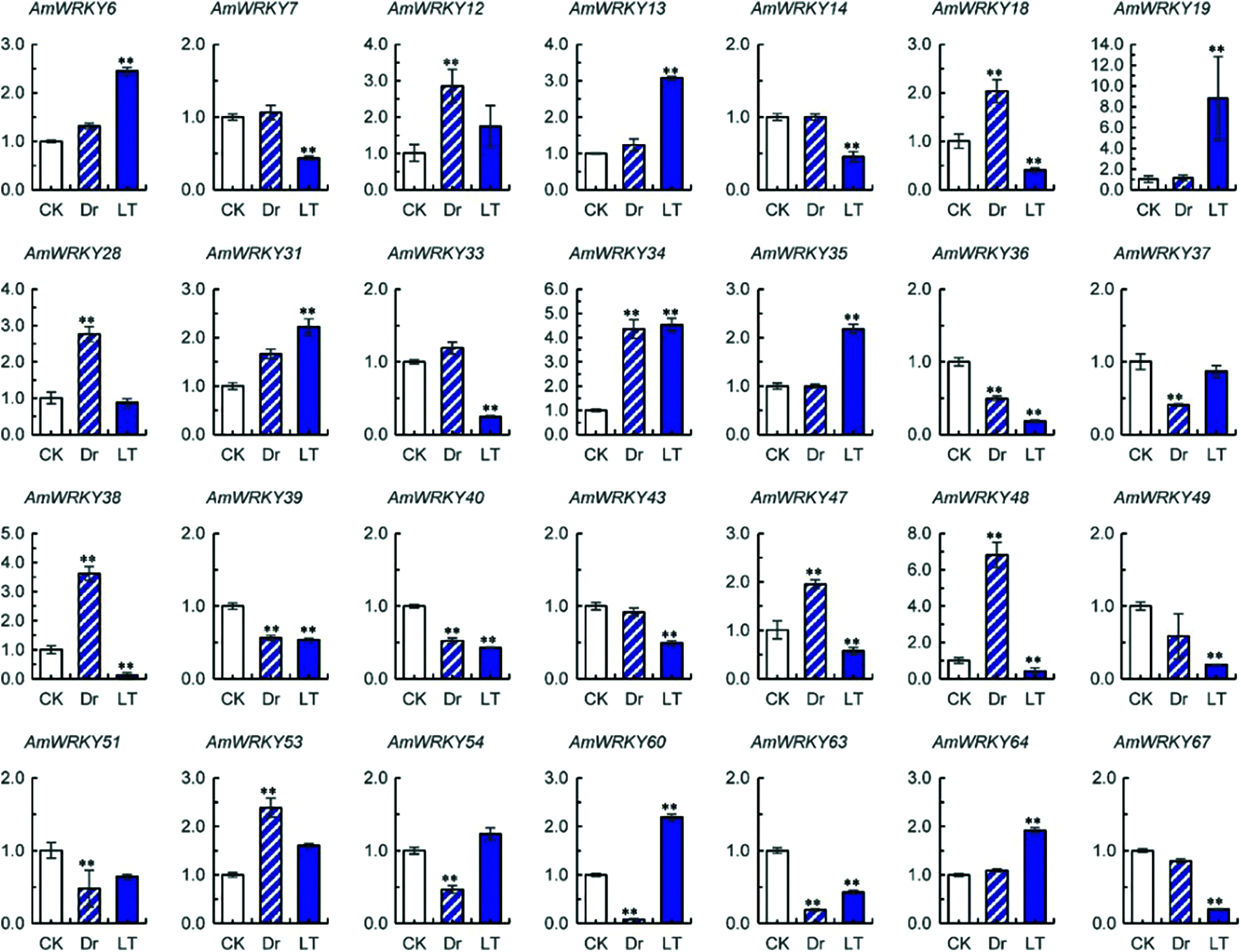
Figure 6: Expressions of AmWRKY genes under drought (Dr) and low temperature (LT) treatments. Values are means ± SD (n = 3). Significance was determined using t-test for comparison of gene expression under normal (CK) and abiotic conditions (Dr or LT) (** P < 0.01)
Ten of these AmWRKY genes simultaneously responded to drought and cold stresses. Five of them exhibited an opposite response pattern to different stresses. AmWRKY18, AmWRKY38, AmWRKY47 and AmWRKY48 were up-regulated under drought but down-regulated under cold, while AmWRKY60 was down-regulated under drought but up-regulated under cold. The remaining five genes exhibited a same response pattern to the different stresses. AmWRKY34 was up-regulated under both drought and cold stresses, while AmWRKY36, AmWRKY39, AmWRKY40 and AmWRKY63 were down-regulated under both stresses.
For the 32 AmVQ genes, two showed no detectable expression (Supplementary Table 1). Among the remaining 30 genes, 10 were found to response to drought stress and 18 were found to respond to cold stress (Fig. 7). In the drought treatment, six AmVQ genes were up-regulated and the other four genes were down-regulated. In the low temperature treatment, nine AmVQ genes were up-regulated and the other nine genes were down-regulated. Eight of these AmVQ genes simultaneously responded to drought and cold stresses, all of which showed a same response pattern to both stresses. AmVQ10, AmVQ15 and AmVQ31 were up-regulated under both stresses, while AmVQ12, AmVQ14, AmVQ16, AmVQ19 and AmVQ29 were down-regulated under both stresses.
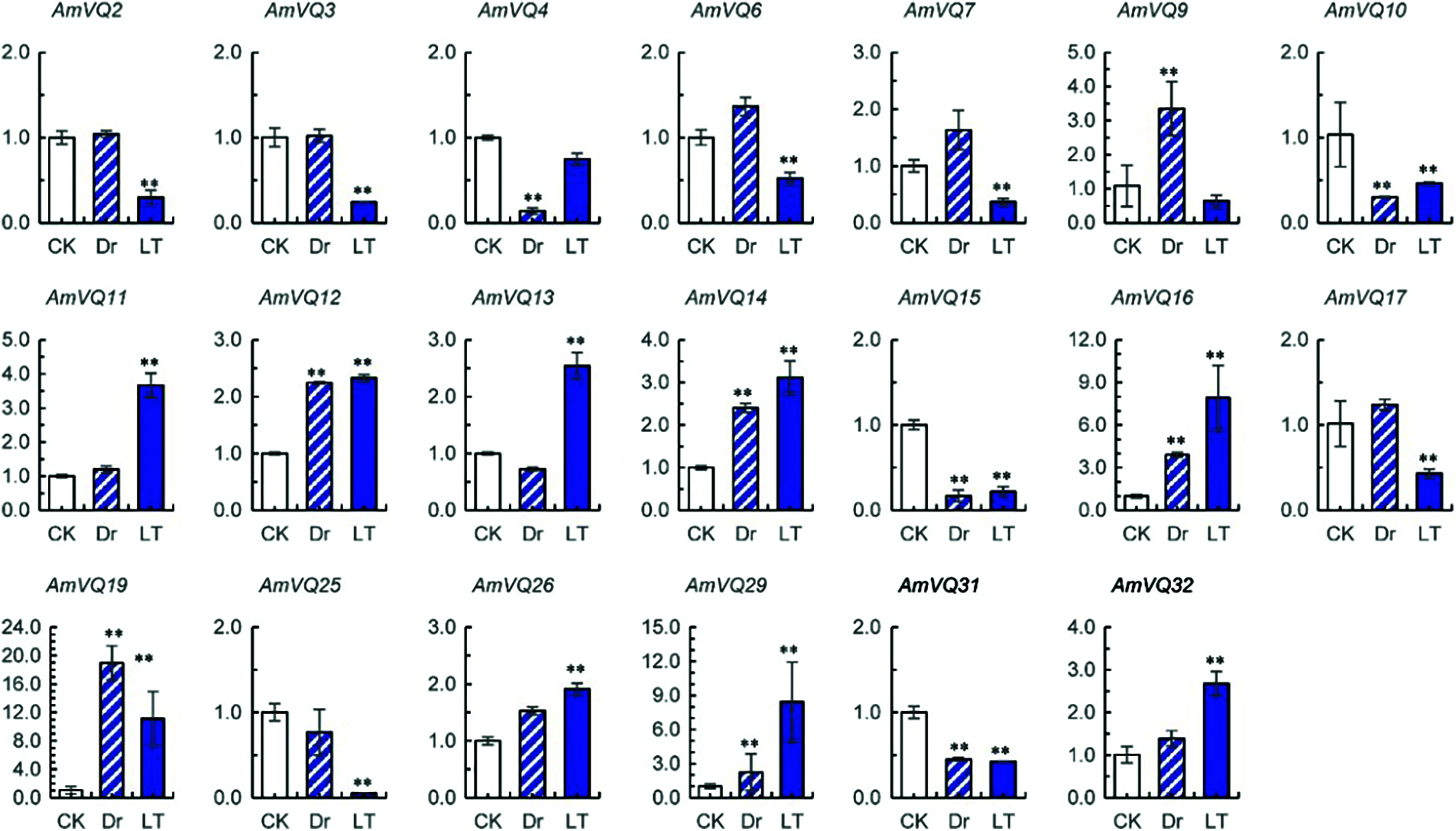
Figure 7: Expressions of AmVQ genes under drought (Dr) and low temperature (LT) treatments. Values are means ± SD (n = 3). Significance was determined using the t-test for comparison of gene expression under normal (CK) and abiotic conditions (Dr or LT) (** P < 0.01)
3.7 Protein-Protein Interaction of AmWRKY and AmVQ Proteins
Protein-protein interactions between AmWRKY and AmVQ proteins were predicted based on the Arabidopsis orthologs (Fig. 8). Three AmWRKY proteins belonging to Groups I or IIc were predicted to interact with four AmVQ proteins. AmWRKY6 (AtWRKY25 ortholog) was presumed to interact with four AmVQ proteins, including AmVQ7 (AtVQ8/18 ortholog), AmVQ11 (AtVQ19/33 ortholog), AmVQ15 (AtVQ25 ortholog) and AmVQ28 (AtVQ11 ortholog). AmWRKY14 (AtWRKY34 ortholog) and AmWRKY43 (AtWRKY24 ortholog) were also presumed to interact with AmVQ7 (AtVQ8/18 ortholog).
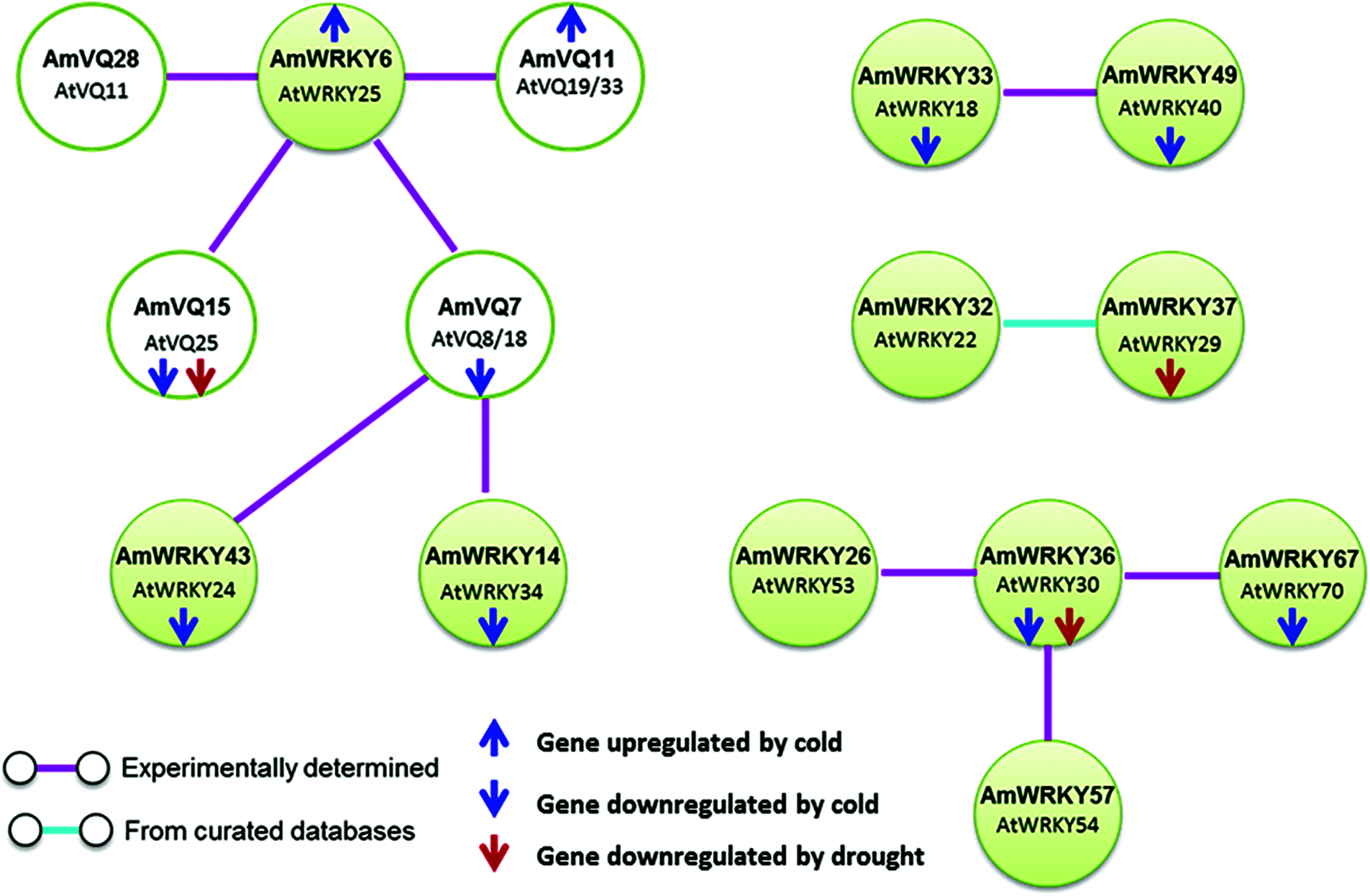
Figure 8: Protein-protein interactions of WRKY and VQ proteins predicted in snapdragon
Interactions among AmWRKY proteins were also identified. Two members of Group IIa, AmWRKY33 (AtWRKY18 ortholog) and AmWRKY49 (AtWRKY40 ortholog), were presumed to interact with each other. Two members of Group IIe, AmWRKY32 (AtWRKY29 ortholog) and AmWRKY37 (AtWRKY22 ortholog), were presumed to interact with each other. AmWRKY36 (AtWRKY30 ortholog) was predicted to interact with AmWRKY26 (AtWRKY53 ortholog), AmWRKY57 (AtWRKY54 ortholog) and AmWRKY67 (AtWRKY70 ortholog), all of which belonging to the Group III.
The WRKY gene family is one of the largest families of transcription factors. The VQ protein is the most important WRKY-interacting protein identified in the past few years [26]. The WRKY and VQ genes and their interactions are crucial for regulating life processes in plants [26,27,54]. In this study, we performed a genome-wide analysis of WRKY and VQ genes in snapdragon, including gene classification, gene duplication investigation, gene expression response to abiotic stresses and protein-protein interaction prediction. Our work lays a foundation for elucidating the structure, evolution and functional roles of WRKY and VQ genes in snapdragon, which generates new knowledge on essential and critical components of abiotic stress tolerance in this plant species.
A total of 67 AmWRKY genes were identified in this study (Supplementary Table 2). These genes were classified into the different groups or subgroups based on the phylogenetic tree, of which Group IIc possessed the most members (Fig. 1). This was similar to many other dicot plant species [6,7,9,11,55,56], suggesting that this subgroup was more active in the evolution of dicots. The WRKYGQK core sequence is a typical signature of WRKY proteins. Overwhelming majority of AmWRKY proteins had the highly conserved WRKYGQK core sequence. However, AmWRKY42 belonging to Group IIc was found to carry the WRKYGKK variant (Supplementary Fig. 2). This was consistent with that the WRKYGKK variant was frequently occurred in Group IIc of plant WRKY genes [4,7,9,55,57–59]. It has been showed that variation of the WRKYGQK core sequence might affect the binding ability of the WRKY transcription factor to the W-box motif [60,61]. In Arabidopsis, three WRKY proteins carring the variant WRKYGKK domain were identified, including AtWRKY50, AtWRKY51 and AtWRKY59. These three genes were clustered into a same clade with AmWRKY42, based on the WRKY domain sequence (Fig. 1). Among these three Arabidopsis genes, AtWRKY50 was reported to lack W-box binding activity [62], while AtWRKY59 was found to activate downstream genes mainly through a newly discovered WT-box motif rather than the W-box motif [63]. Therefore, AmWRKY42 might have different DNA binding specificity. A total of 32 AmVQ genes were found in this study (Supplementary Table 5). All of them contained the conserved FxxhVQxhTG motif, except for AmVQ24 which carried a variant of FxxhVQxhTC (Supplementary Fig. 3). The FxxhVQxhTG motif mediates protein localization and the interaction between VQ proteins and other regulators [64]. Thus, AmVQ24 may possess a different biological function.
Tandem duplication and segmental duplication play important roles in the amplification and evolution of the plant gene family. In the present study, 74 segmental duplication events involved 44 AmWRKY genes were identified (Fig. 4), whereas only one tandem duplication event was found with two AmWRKY genes (Fig. 3). Therefore, segmental duplication events might be the main driving force in the evolution of WRKY gene family in snapdragon. For AmVQ genes, five segmental duplication events and two tandem duplication events were found, implying that both duplication events contributed to the evolution of VQ gene family in snapdragon.
Gene expression reprogramming is one of the vital strategies for plants to adapt to diverse environmental stresses. In the present study, 28 AmWRKY genes and 21 AmVQ genes were found to respond to drought and cold stresses (Figs. 6 and 7), indicating they might participate in the regulation of snapdragon abiotic stress tolerances. Two of these WRKY genes, AmWRKY14 and AmWRKY36, were found to be the orthologs of Arabidopsis abiotic stress regulators AtWRKY34 and AtWRKY30, respectively (Supplementary Table 4). AtWRKY34 and AtWRKY30 were up-regulated under cold and drought stresses, respectively [13,14]. However, AmWRKY14 and AmWRKY36 were repressed by cold and drought in the present study, respectively (Fig. 6). These two AmWRKY genes might have different functioning ways compared with their orthologs in Arabidopsis. Among the AmVQ genes, eight were found to simultaneously respond to drought and cold stresses, all of which showed similar expression patterns under the two different stresses (Fig. 7). A similar phenomenon was also observed for rice VQ genes involved in drought and salinity responses [65], and for Eucalyptus grandis VQ genes involved in cold and heat responses [66]. These results suggested that VQ genes may exist in the same response pathway to different abiotic stresses.
Accumulated evidences suggest that WRKY-VQ interaction plays important roles in plant response to abiotic stresses. For instance, Arabidopsis AtWRKY8 and AtVQ9 could be strongly induced by salt stress, which antagonistically regulates salinity stress tolerance by forming a physical complex [28]. Banana MaWRKY26 and MaVQ5 could be rapidly induced by cold stress, and they positively regualtes cold stress tolerance by physical interacting with each other [34]. In the present study, WRKY-VQ interactions were predicted between three AmWRKY transcription factors and four AmVQ proteins (Fig. 7). All these AmWRKY genes and three AmVQ genes, including AmWRKY6, AmWRKY14, AmWRKY43, AmVQ7, AmVQ11 and AmVQ15 exhibited differentially expression under cold stresses (Figs. 6–8). These AmWRKY and AmVQ genes might regulate snapdragon cold response by forming a complicate network. AmWRKY6, AmWRKY14 and AmWRKY43 belonged to either Group I or Group IIc. This was consistent with the previous observation that VQ proteins interact only with Group I and IIc WRKY transcription factors, but failed to interact with Group IIa, IIb, IId, IIe, and III WRKY transcription factors [32,67].
Cooperation among WRKY proteins is another area of interest in the research community. WRKY transcription factors regulate downstream gene expression by binding conserved W-box cis-acting elements of the downstream gene. Some WRKY genes also have W-box elements in their promoters. Thus, these WRKY transcription factors could perform self-regulation or cross-regulation by binding with their own W-box elements [5]. Cooperation among WRKY proteins could also be achieved by protein-protein interactions. The WRKY-WRKY interactions have been previously observed within members of Group IIa, Group IIb and Group III, or between members of these groups in plants [26]. In the present study, a total of 41 AmWRKY genes were found to carry the W-box cis-acting elements in their 2-kb promoter region (Supplementary Tab. 3). The WRKY-WRKY interaction was also predicted within members of Group IIa and Group IIb (Fig. 8). These results indicated that the cooperation among WRKY transcription factors may also exist in snapdragon. Three WRKY genes with W-box elements, AmWRKY36, AmWRKY67 and AmWRKY49 were predicted to participate in WRKY-WRKY interactions. All of them were found to be downregulated under cold stress. These three genes could be good targets for studying the cooperation of WRKY genes in snapdragon response to abiotic stresses.
Funding Statement: This research was funded by the Key Research and Development Plan of Zhejiang Province (2019C02025) and the Youth Talent Program of Zhejiang Academy of Agricultural Sciences (2020R25R08E02).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends in Plant Science, 5(5), 199–206. DOI 10.1016/s1360-1385(00)01600-9. [Google Scholar] [CrossRef]
2. Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends in Plant Science, 15(5), 247–258. DOI 10.1016/j.tplants.2010.02.006. [Google Scholar] [CrossRef]
3. Chen, C., Chen, X., Han, J., Lu, W., Ren, Z. (2020). Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biology, 20(1), 443. DOI 10.1186/s12870-020-02625-8. [Google Scholar] [CrossRef]
4. Goyal, P., Manzoor, M. M., Vishwakarma, R. A., Sharma, D., Dhar, M. K. et al. (2020). A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Scientific Reports, 10(1), 373. DOI 10.1038/s41598-019-57232-x. [Google Scholar] [CrossRef]
5. Li, W., Pang, S., Lu, Z., Jin, B. (2020). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants, 9(11), 1515. DOI 10.3390/plants9111515. [Google Scholar] [CrossRef]
6. Li, Z., Hua, X., Zhong, W., Yuan, Y., Wang, Y. (2019). Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant and Cell Physiology, 61(3), 616–630. DOI 10.1093/pcp/pcz227. [Google Scholar] [CrossRef]
7. Liu, Z., Saiyinduleng, Chang, Q., Cheng, C., Zheng, Z. et al. (2020). Identification of yellowhorn (Xanthoceras sorbifolium) WRKY transcription factor family and analysis of abiotic stress response model. Journal of Forest Research, 32, 987–1004. DOI 10.1007/s11676-020-01134-6. [Google Scholar] [CrossRef]
8. Mao, P., Jin, X., Bao, Q., Mei, C., Zhou, Q. et al. (2020). WRKY transcription factors in Medicago sativa L.: Genome-wide identification and expression analysis under abiotic stress. DNA and Cell Biology, 39(12), 2212–2225. DOI 10.1089/dna.2020.5726. [Google Scholar] [CrossRef]
9. Song, Y., Cui, H., Shi, Y., Xue, J., Ji, C. et al. (2020). Genome-wide identification and functional characterization of the Camelina sativa WRKY gene family in response to abiotic stress. BMC Genomics, 21(1), 786. DOI 10.1186/s12864-020-07189-3. [Google Scholar] [CrossRef]
10. Yang, Y., Liu, J., Zhou, X., Liu, S., Zhuang, Y. (2020). Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ, 8(6), e8777. DOI 10.7717/peerj.8777. [Google Scholar] [CrossRef]
11. Zhao, N., He, M., Li, L., Cui, S., Hou, M. et al. (2020). Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS One, 15(4), e0231396. DOI 10.1371/journal.pone.0231396. [Google Scholar] [CrossRef]
12. Li, S., Fu, Q., Chen, L., Huang, W., Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta, 233(6), 1237–1252. DOI 10.1007/s00425-011-1375-2. [Google Scholar] [CrossRef]
13. Zou, C., Jiang, W., Yu, D. (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in arabidopsis. Journal of Experimental Botany, 61(14), 3901–3914. DOI 10.1093/jxb/erq204. [Google Scholar] [CrossRef]
14. El-Esawi, M. A., Al-Ghamdi, A. A., Ali, H. M., Ahmad, M. (2019). Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.). Genes, 10(2), 163. DOI 10.3390/genes10020163. [Google Scholar] [CrossRef]
15. Sun, Y., Yu, D. (2015). Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Reports, 34(8), 1295–1306. DOI 10.1007/s00299-015-1787-8. [Google Scholar] [CrossRef]
16. Jiang, Y., Qiu, Y., Hu, Y., Yu, D. (2016). Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Frontiers in Plant Science, 7, 145. DOI 10.3389/fpls.2016.00145. [Google Scholar] [CrossRef]
17. Jiang, Y., Deyholos, M. K. (2009). Functional characterization of arabidopsis naCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology, 69(1–2), 91–105. DOI 10.1007/s11103-008-9408-3. [Google Scholar] [CrossRef]
18. Zheng, Y., Ge, J., Bao, C., Chang, W., Liu, J. et al. (2020). Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Molecular Plant, 13(4), 598–611. DOI 10.1016/j.molp.2019.12.011. [Google Scholar] [CrossRef]
19. Yang, Y., Chi, Y., Wang, Z., Zhou, Y., Fan, B. et al. (2016). Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. Journal of Experimental. Botany, 67(15), 4727–4742. DOI 10.1093/jxb/erw252. [Google Scholar] [CrossRef]
20. Cheng, Y., Ahammed, G. J., Yu, J., Yao, Z., Ruan, M. et al. (2017). Corrigendum: Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Scientific Reports, 7(1), 43498. DOI 10.1038/srep43498. [Google Scholar] [CrossRef]
21. Lei, R., Li, X., Ma, Z., Lv, Y., Hu, Y. et al. (2017). Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant Journal, 91(6), 962–976. DOI 10.1111/tpj.13619. [Google Scholar] [CrossRef]
22. Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., Chaudhury, A. (2005). MINISEED3 (MINI3a WRKY family gene, and HAIKU2 (IKU2a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in arabidopsis. Proceedings of the National Academy of Science of the United States of America, 102(48), 17531–17536. DOI 10.1073/pnas.0508418102. [Google Scholar] [CrossRef]
23. Gu, Y., Li, W., Jiang, H., Wang, Y., Gao, H. et al. (2017). Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. Journal of Experimental Botany, 68(11), 2717–2729. DOI 10.1093/jxb/erx147. [Google Scholar] [CrossRef]
24. Ay, N., Irmler, K., Fischer, A., Uhlemann, R., Reuter, G. et al. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant Journal, 58(2), 333–346. DOI 10.1111/j.0960-7412.2009.03782.x. [Google Scholar] [CrossRef]
25. Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiology, 167(2), 295–306. DOI 10.1104/pp.114.251769. [Google Scholar] [CrossRef]
26. Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B. et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Molecular Plant, 6(2), 287–300. DOI 10.1093/mp/sst026. [Google Scholar] [CrossRef]
27. Yuan, G., Qian, Y., Ren, Y., Guan, Y., Wu, X. et al. (2021). The role of plant-specific VQ motif-containing proteins: An ever-thickening plot. Plant Physiology and Biochemistry, 159, 12–16. DOI 10.1016/j.plaphy.2020.12.005. [Google Scholar] [CrossRef]
28. Hu, Y. R., Chen, L. G., Wang, H. P., Zhang, L. P., Wang, F. et al. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. The Plant Journal, 74(5), 730–745. DOI 10.1111/tpj.12159. [Google Scholar] [CrossRef]
29. Cheng, X. R., Wang, Y. J., Xiong, R., Gao, Y. M., Yan, H. W. et al. (2020). A moso bamboo gene VQ28 confers salt tolerance to transgenic arabidopsis plants. Planta, 251(5), 99. DOI 10.1007/s00425-020-03391-5. [Google Scholar] [CrossRef]
30. Ding, H. D., Yuan, G. B., Mo, S. R., Qian, Y., Wu, Y. et al. (2019). Genome-wide analysis of the plant-specific VQ motif-containing proteins in tomato (Solanum lycopersicum) and characterization of SlVQ6 in thermotolerance. Plant Physiol. Biochem, 143, 29–39. DOI 10.1016/j.plaphy.2019.08.019. [Google Scholar] [CrossRef]
31. Lai, Z. B., Li, Y., Wang, F., Cheng, Y., Fan, B. F. et al. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell, 23(10), 3824–3841. DOI 10.1105/tpc.111.090571. [Google Scholar] [CrossRef]
32. Cheng, Y., Zhou, Y., Yang, Y., Chi, Y. J., Zhou, J. et al. (2012). Structural and functional analysis of VQ motif-containing proteins in arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiology, 159(2), 810–825. DOI 10.1104/pp.112.196816. [Google Scholar] [CrossRef]
33. Perruc, E., Charpenteau, M., Ramirez, B. C., Jauneau, A., Galaud, J. P. et al. (2004). A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant Journal, 38(3), 410–420. DOI 10.1111/j.1365-313X.2004.02062.x. [Google Scholar] [CrossRef]
34. Ye, Y. J., Xiao, Y. Y., Han, Y. C., Shan, W., Fan, Z. Q. et al. (2016). Banana fruit VQ motif-containing protein5 represses cold-responsive transcription factor MaWRKY26 involved in the regulation of JA biosynthetic genes. Scientific. Reports, 6(1), 23632. DOI 10.1038/srep23632. [Google Scholar] [CrossRef]
35. Jiang, Y., Yu, D. (2016). The WRKY57 transcription factor affects the expression of jasmonate ZIM-domain genes transcriptionally to compromise botrytis cinerea resistance. Plant Physiology, 171(4), 2771–2782. DOI 10.1104/pp.16.00747. [Google Scholar] [CrossRef]
36. Chen, J., Wang, H., Li, Y., Pan, J., Hu, Y. et al. (2018). Arabidopsis VQ10 interacts with WRKY8 to modulate basal defense against Botrytis cinerea. Journal of Integrative Plant Biology, 60, 956–969. DOI 10.1111/jipb.12664. [Google Scholar] [CrossRef]
37. Li, N., Li, X., Xiao, J., Wang, S. (2014). Comprehensive analysis of VQ motif-containing gene expression in rice defense responses to three pathogens. Plant Cell Reports, 33(9), 1493–1505. DOI 10.1007/s00299-014-1633-4. [Google Scholar] [CrossRef]
38. Jiao, Z., Sun, J., Wang, C., Dong, Y., Xiao, S. et al. (2018). Genome-wide characterization, evolutionary analysis of WRKY genes in cucurbitaceae species and assessment of its roles in resisting to powdery mildew disease. PLoS One, 13(12), e0199851. DOI 10.1371/journal.pone.0199851. [Google Scholar] [CrossRef]
39. Song, W., Zhao, H., Zhang, X., Lei, L., Lai, J. (2015). Genome-wide identification of VQ motif-containing proteins and their expression profiles under abiotic stresses in maize. Frontiers in Plant Science, 6, 1177. DOI 10.3389/fpls.2015.01177. [Google Scholar] [CrossRef]
40. Zeng, Z. (2010). Discussion on the perennial cultivation techniques of Antirrhinum majus. Heilongjiang Agricultural Sciences, 2, 49–51. DOI 10.3969/j.issn.1002-2767.2010.02.019. [Google Scholar] [CrossRef]
41. Zhu, G. H., Tang, R., Deng, B., Wang, C. Z., Gu, G. H. (2013). Physiological responses of different cultivars of cut-flowers Antirrhinum majus and its cold tolerance analysis under low temperature. Northerm Horticulture, 302, 100–102. DOI CNKI:SUN:BFYY.0.2013-23-031. [Google Scholar]
42. Naing, A. H., Park, K. I., Ai, T. N., Chung, M. Y., Han, J. S. et al. (2017). Overexpression of snapdragon delila (Del) gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance. BMC Plant Biology, 17(1), 65. DOI 10.1186/s12870-017-1015-5. [Google Scholar] [CrossRef]
43. Naing, A. H., Ai, T. N., Lim, K. B., Lee, I. J., Kim, C. K. (2018). Overexpression of rosea1 from snapdragon enhances anthocyanin accumulation and abiotic stress tolerance in transgenic tobacco. Frontiers in Plant Science, 9, 1070. DOI 10.3389/fpls.2018.01070. [Google Scholar] [CrossRef]
44. Finatto, T., Viana, V. E., Woyann, L. G., Busanello, C., Maia, L. C. et al. (2018). Can WRKY transcription factors help plants to overcome environmental challenges? Genetics and Molecular Biology, 41(3), 533–544. DOI 10.1590/1678-4685-gmb-2017-0232. [Google Scholar] [CrossRef]
45. Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J. et al. (2017). WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 59(2), 86–101. DOI 10.1111/jipb.12513. [Google Scholar] [CrossRef]
46. Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. DOI 10.1016/j.molp.2020.06.009. [Google Scholar] [CrossRef]
47. Wheeler, T. J., Eddy, S. R. (2013). Nhmmer: DNA homology search with profile HMMs. Bioinformatics, 29(19), 2487–2489. DOI 10.1093/bioinformatics/btt403. [Google Scholar] [CrossRef]
48. Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. DOI 10.1093/molbev/msw054. [Google Scholar] [CrossRef]
49. Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research, 34(Issue suppl_2), W369–W373. DOI 10.1093/nar/gkl198. [Google Scholar] [CrossRef]
50. Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J. et al. (2012). MCScanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40(7), e49. DOI 10.1093/nar/gkr1293. [Google Scholar] [CrossRef]
51. Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S. et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47, 607–613. DOI 10.1093/nar/gky1131. [Google Scholar] [CrossRef]
52. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
53. Holub, E. B. (2001). The arms race is ancient history in arabidopsis, the wildflower. Nature Reviews Genetics, 2(7), 516–527. DOI 10.1038/35080508. [Google Scholar] [CrossRef]
54. Bakshi, M., Oelmuller, R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signaling& Behavior, 9(2), e27700. DOI 10.4161/psb.27700. [Google Scholar] [CrossRef]
55. Yang, X., Zhou, Z., Fu, M., Han, M., Liu, Z. et al. (2020). Transcriptome-wide identification of WRKY family genes and their expression profiling toward salicylic acid in Camellia japonica. Plant Signaling & Behavior, 16(1), 1844508. DOI 10.1080/15592324.2020.1844508. [Google Scholar] [CrossRef]
56. Villano, C., Esposito, S., D’Amelia, V., Garramone, R., Alioto, D. et al. (2020). WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Scientific Reports, 10(1), 7196. DOI 10.1038/s41598-020-63823-w. [Google Scholar] [CrossRef]
57. Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology, 5, 1. DOI 10.1186/1471-2148-5-1. [Google Scholar] [CrossRef]
58. Tiika, R. J., Wei, J., Ma, R., Yang, H., Cui, G. et al. (2020). Identification and expression analysis of the WRKY gene family during different developmental stages in Lycium ruthenicum murr. fruit. PeerJ, 8(1), e10207. DOI 10.7717/peerj.10207. [Google Scholar] [CrossRef]
59. Sun, W., Ma, Z., Chen, H., Liu, M. (2020). Genome-wide investigation of WRKY transcription factors in tartary buckwheat (Fagopyrum tataricum) and their potential roles in regulating growth and development. PeerJ, 8(1), e8727. DOI 10.7717/peerj.8727. [Google Scholar] [CrossRef]
60. Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G. et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic arabidopsis plants. Plant Biotechnology Journal, 6(5), 486–503. DOI 10.1111/j.1467-7652.2008.00336.x. [Google Scholar] [CrossRef]
61. Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., Linthorst, H. J. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiology, 146(4), 1983–1995. DOI 10.1104/pp.107.112789. [Google Scholar] [CrossRef]
62. Dong, J., Chen, C., Chen, Z. (2003). Expression profiles of the arabidopsis WRKY gene superfamily during plant defense response. Plant Molecular Biology, 51, 21–37. DOI 10.1023/a:1020780022549. [Google Scholar] [CrossRef]
63. Kanofsky, K., Rusche, J., Eilert, L., Machens, F., Hehl, R. (2021). Unusual DNA-inding properties of the Arabidopsis thaliana WRKY50 transcription factor at target gene promoters. Plant Cell Reports, 40(1), 69–83. DOI 10.1007/s00299-020-02611-2. [Google Scholar] [CrossRef]
64. Jing, Y., Lin, R. (2015). The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiology, 169(1), 371–378. DOI 10.1104/pp.15.00788. [Google Scholar] [CrossRef]
65. Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology, 49(6), 865–879. DOI 10.1093/pcp/pcn061. [Google Scholar] [CrossRef]
66. Fan, C., Yao, H., Qiu, Z., Ma, H., Zeng, B. (2018). Genome-wide analysis of eucalyptus grandis WRKY genes family and their expression profiling in response to hormone and abiotic stress treatment. Gene, 678, 38–48. DOI 10.1016/j.gene.2018.08.003. [Google Scholar] [CrossRef]
67. Zhou, Y., Yang, Y., Zhou, X., Chi, Y., Fan, B. et al. (2016). Structural and functional characterization of the VQ protein family and VQ protein variants from soybean. Scientific Reports, 6(1), 34663. DOI 10.1038/srep34663. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |