

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.017629
ARTICLE
Proteomic Analysis of Chrysanthemum Lateral Buds after Removing Apical Dominance Based on Label-Free Technology
Department of Ornamental Horticulture, Sichuan Agricultural University, Chengdu, 611130, China
*Corresponding Author: Beibei Jiang. Email: 13786@sicau.edu.cn
#These authors contributed equally to this work
Received: 25 May 2021; Accepted: 13 August 2021
Abstract: Studying the genetic basis and regulatory mechanism of chrysanthemum lateral bud outgrowth is of great significance for reduction the production cost of cut chrysanthemum. To clarify the molecular basis of lateral bud elongation after removal of apical dominance in chrysanthemum, label-free quantification analysis was used to analyze the proteome changes after apical bud removal. Quantitative real-time PCR (qPCR) was used to analyze the changes in the expression of three plant hormone-related genes. A total of 440 differentially expressed proteins were successfully identified at three time points during the lateral bud elongation. The number of differentially expressed proteins in the three stages (24 h/0 h, 48 h/0 h, 48 h/24 h) were 219, 332, and 97, respectively. The difference in expressed proteins in the three comparison stages mainly involves RNA processing and modification; translation, ribosomal structure and biogenesis; Posttranslational modification, protein turnover, and chaperones. Path analysis showed that there was various physiological activities in the process of lateral bud dormancy breaking and elongation, which involved energy metabolism, biosynthesis, signal transduction and stress response in the growth process of lateral buds. qPCR indicated that the expression of cytokinin synthesis related gene was significantly increased after the removal of apical dominance, while the expression of strigolactones synthesis related gene experiences a dramatic fall to promote the development of the lateral buds. However, there was a drop before a slight increase in the expression of the auxin synthesis related gene, which was mainly due to the removal of apical dominance that led to the loss of indoleacetic acid in the main stem. However, with formation of the new apical source, indoleacetic acid can be released again.
Keywords: Branches; proteins; decapitation; proteome; 4D label-free quantification analysis; quantitative real-time PCR
Cut chrysanthemum is one of the four major cut flowers in the world, and it is also China’s main cut flower and an advantageous export flower product. Single-headed chrysanthemum is the main type of cut chrysanthemum [1]. Most single-headed cut chrysanthemum varieties have numerous lateral buds, and the manual removal method in production has led to increased labor costs and management difficulties in cut flower production. For example, the pinching cost of the main cut chrysanthemum variety ‘Jinba’ accounts for about 1/3 of its production cost, and it is increasing year by year [2,3]. Therefore, cultivating standard cut chrysanthemum varieties with few or no lateral branches is one of the goals of cut chrysanthemum breeding. The exploration of the regulation mechanism of chrysanthemum branching can provide a theoretical basis for the development of chrysanthemum branching control technology.
Plant branching can be divided into two stages: the formation of lateral buds and subsequent elongation of lateral buds [4]. The elongation of lateral buds is mainly inhibited by the top of the plant. This phenomenon is called apical dominance. When the top of the plant is removed, the lateral buds will be released from dormancy and elongate and develop into lateral branches [5]. Studies have shown that after pinching the cut chrysanthemum ‘Jinba’, the top three lateral buds were found to elongate significantly, indicating that apical dominance has an inhibitory effect on the lateral buds of chrysanthemum [6]. In the past few decades, great progress has been made in the regulation mechanism of plant branching. The auxin transportation channel model and the second messenger model have laid a good foundation for the study of plant apical advantages and branching regulation, but these two models still need to be developed and improved in future research [7]. At present, there are preliminary outlines for the development and regulation of chrysanthemum lateral branches, but many aspects still need to be further clarified. The molecular mechanism of plant top dominance is studied through proteomics strategy, and the screening of different proteins by molecular biology and bioinformatics technology is conducive to further revealing the regulatory network of chrysanthemum branches [8].
Proteomics is a new research field that emerged in the post-genome era, which aims to study the protein composition and dynamic changes of cells, tissues, or organisms [9]. It can be used to study the growth and development of plants, the response to biotic and abiotic stresses, post-translational modification, and protein-protein interactions [10]. As a functional component, a protein is a direct reflection of gene expression. Therefore, compared with the transcriptome, the proteome can more directly reflect the expression status of plant genes [11]. Liu et al. [12] analyzed the differentially expressed proteins in the process of tobacco senescence, in which most of the proteins related to anabolism processes such as photosynthesis were down regulated, while the proteins related to catabolism, such as stress and respiratory action, were mostly up regulated.
Xu et al. [13] studied the protein differences in the shoots and stem nodes of rice (Oryza sativa) released from dormancy after removing the apical dominance, and considered that several proteins involved in signal transduction, transporters, and cytokinin (CTK) biosynthesis may play an important role in the top dominance mechanism.
In this study, the proteome of Chrysanthemum morifolium ‘Jinba’ lateral buds was compared at three time points after the apical buds were removed, the differentially expressed proteins during the elongation of the lateral buds were screened, and functional annotation as well as statistical analysis of these proteins was performed. Meanwhile, qPCR was used to analyze the expression of three plant hormone-related genes. The metabolic pathways, key proteins and changes in the expression of three plant hormone-related genes during the process of lateral bud elongation of ‘Jinba’ were discussed, which provided a reference at the protein level for further revealing the elongation process of lateral buds as well as a theoretical basis for reducing the production cost of cut chrysanthemum.
The material used in the experiment was cut chrysanthemum ‘Jinba’ (same batch, consistent growth). Under aseptic conditions, chrysanthemum seedlings were propagated in a jar with MS (Murashige and Skoog 1962) agar medium. The following steps were performed. The bottle cap was opened when the seedlings grew to 8–10 cm. After four days, the seedlings were taken out and the roots were washed with deionized water. Then the seedlings were transplanted to flower pots of the same size (upper diameter of the flowerpot = 7 cm, lower diameter = 5 cm, and height = 10 cm). The seedlings grown in a mixture of perlite and turf soil (1:1) were placed in a light incubator with a temperature of 23°C and a light/dark photoperiod of 16 h/8 h for one week.
After removal of the apical buds, the first lateral bud was called Bud 1, and the second lateral bud was called Bud 2. Buds 1 and 2 (1 cm stems with buds) were collected at 0 h, 24 h, and 48 h after apical bud removal and were stored in a refrigerator at −80°C after rapid freezing in liquid nitrogen. There were six plants per treatment, and each treatment was replicated three times (Fig. 1).
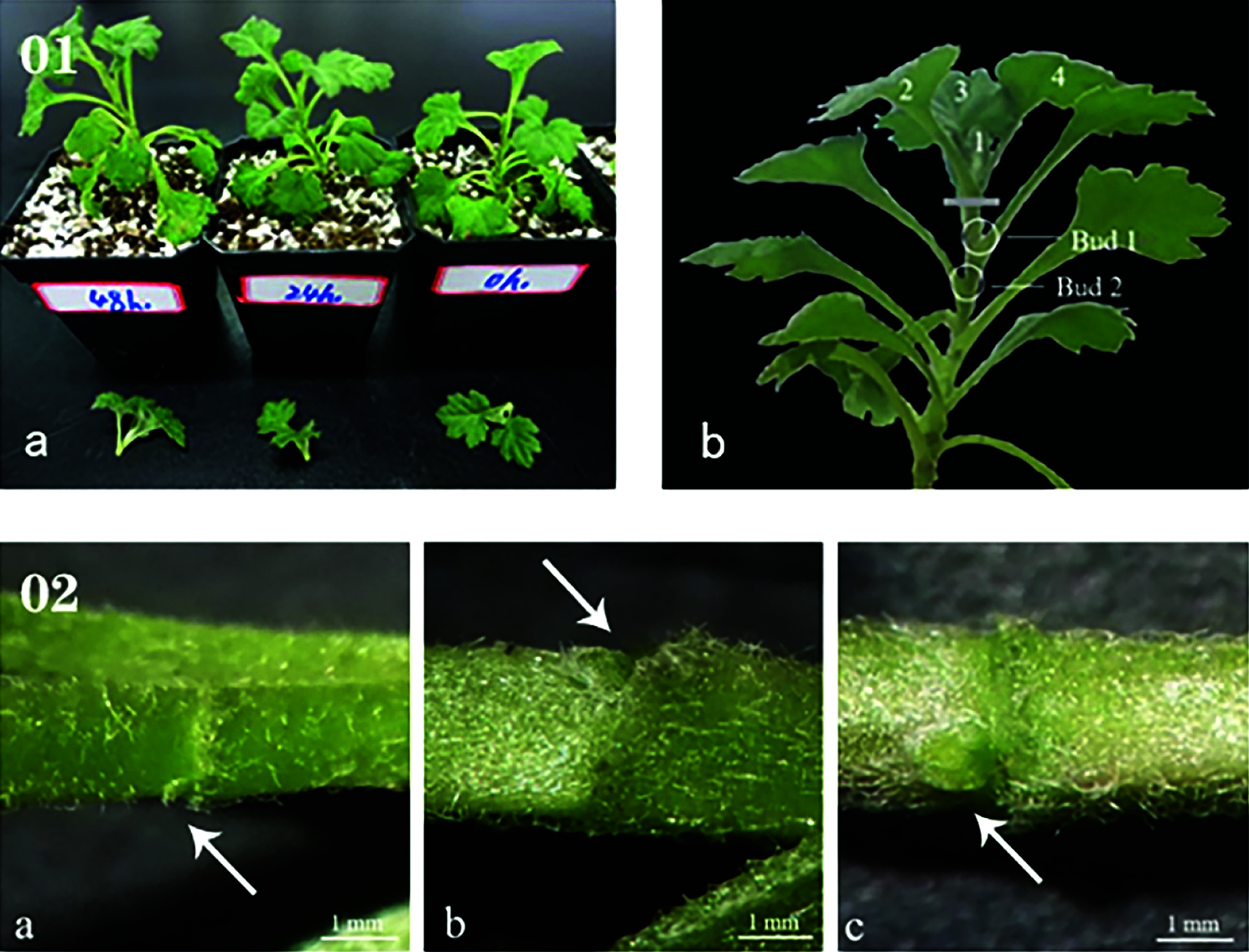
Figure 1: (01) Schematic of sampling. (a) Every plant underwent decapitation treatment at the same time, (b) Treatment method and sampling location of samples. (02) The state of lateral buds at 0 h (a), 24 h (b), and 48 h (c)
The sample was taken out from −80°C storage, an appropriate amount of the sample was weighed into a mortar pre-cooled with liquid nitrogen, and liquid nitrogen was added to fully grind the sample to a powder, which was transferred to a 5 mL centrifuge tube. Afterwards, four volumes of lysis buffer (containing 10 mM dithiothreitol, 1% protease inhibitor) were added to the cell powder and were ultrasonically lysed. An equal volume of Tris-Phenol was added, followed by centrifugation at 5500 g for 10 min at 4°C. Five volumes of 0.1 M ammonium acetate/methanol were added to the supernatant and allowed to precipitate overnight. Thereafter, the precipitate was washed with methanol and acetone sequentially. Finally, the precipitate was reconstituted with 8 M urea, and the protein concentration was determined using a bicinchoninic acid (BCA) kit.
For digestion, the protein was precipitated with cold 20% trichloroacetic acid (TCA) for 2 h on ice. After centrifugation at 4°C for 5 min, the supernatant was discarded. The remaining precipitate was washed with cold acetone three times. The protein was redissolved in buffer (200 mM TEAB, pH 8.0). Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight, and at a 1:100 trypsin-to-protein mass ratio for a second 4-h digestion. The protein solution was reduced with 5 mM dithiothreitol (DTT) for 30 min at 56°C and alkylated with 11 mM indoleacetic acid (IAA) for 15 min at room temperature in darkness.
2.4 LC-MS/MS Analysis and Database Search
The tryptic peptides were dissolved in solvent A (0.1% formic acid in water) and directly loaded onto a home-made reversed-phase analytical column (25-cm length, 75 μm i.d.). Peptides were separated with a gradient from 4% to 6% solvent B (0.1% formic acid in acetonitrile) during 2 min, 6% to 24% during 68 min, 24% to 32% during 14 min, increased to 80% during 3 min and then held at 80% for the last 3 min, all at a constant flow rate of 300 nL/min on a nanoElute UHPLC system (Bruker Daltonics Bremen, Germany).
The peptides were subjected to capillary source followed by the timsTOF Pro (Bruker Daltonics) mass spectrometry. The electrospray voltage applied was 1.60 kV. Precursors and fragments were analyzed by the TOF detector, with an MS/MS scan range from 100 to 1700 m/z. The timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states 0 to 5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set to 30 s.
Lateral bud proteins were then examined using the UniProt Database (http://www.uniprot.org/) for annotation and enrichment analysis. They were annotated by gene ontology (GO) based on molecular function, biological process, and cellular components. Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.kegg.jp/) annotation and enrichment were performed for pathway analysis. To understand the potential systemic roles of the identified proteins, clusters of orthologous groups (COG) of protein (http://www.ncbi.nlm.nih.gov/COG) annotation were also performed by a web server for metagenomic analysis from WebMGA (http://weizhong-lab.ucsd.edu/metagenomic-analysis/). The search tool for the STRING-DB (Retrieval of Interacting Genes/Proteins) (http://string-db.org/) physical and functional interactions was used to analyze the protein–protein interaction (PPI) of all the differentially expressed proteins.
Total RNA is extracted using the trizol method and less than 0.1 g of sample was used for RNA preparation per replicate. The reverse transcription reaction was performed using the TransScript® II All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) (TransGen Biotech, Beijing), according to the manufacturer’s protocol. Using qPCR, gene expression was detected using an ABI 7500 Fast Real-Time PCR system with SYBR Green Master Mix (Bio-Rad, CFX Connect). The Actin gene was used as internal control. Expression data of each target gene in each replicate were normalized with the reference gene β-actin expression level using the formula: expression data = 2Ct (reference gene) − Ct (target gene) (CT means cycle threshold). Three biological replicates were conducted. Quantitative real-time PCR primers are listed in Table 1.
3.1 Lateral Bud Development in Chrysanthemum morifolium ‘Jinba’
The formation and elongation of lateral branches are inhibited before the apical bud is removed due to the existence of apical dominance. When the apical bud is removed, the formation process of the side bud can be divided into four steps: 1) the establishment of the characteristics of the axillary part of the leaf; 2) the maintenance of the formation ability of the branched tissue in the axillary of the leaf; 3) the adjustment of the axillary branched tissue; 4) The formation of side buds. After the lateral bud is formed, it can be immediately extended to the side branch, or it can be kept in a dormant or semi-dormant state. In this experiment, after removing the apical buds, the side branches at the axillary of the first leaf do not go through dormancy and directly elongate to the side branches (Fig. 1).
3.2 Protein Identification and Quantification
Label-free nanoscale LC-MS quantitative proteomics was used to analyze proteins obtained from developing lateral buds of Chrysanthemum morifolium ‘Jinba’, which were collected at 0, 24 and 48 h after removing the apical buds. We identified 46,430 unique peptides, mapped to 6473 quantifiable proteins using a false discovery rate (FDR) parameter of 1%. Peptide fragments were predominantly in the range of 8 to 20 amino acids (AA) (Fig. 2). Filtering results with a 2-fold cut-off for significant temporal changes in abundance, we identified a total of 440 differentially expressed proteins (Fig. 3a). (see https://docs.qq.com/sheet/DR3d0UEt6SHpxSW9v?tab=BB08J2 for details of 440 proteins).
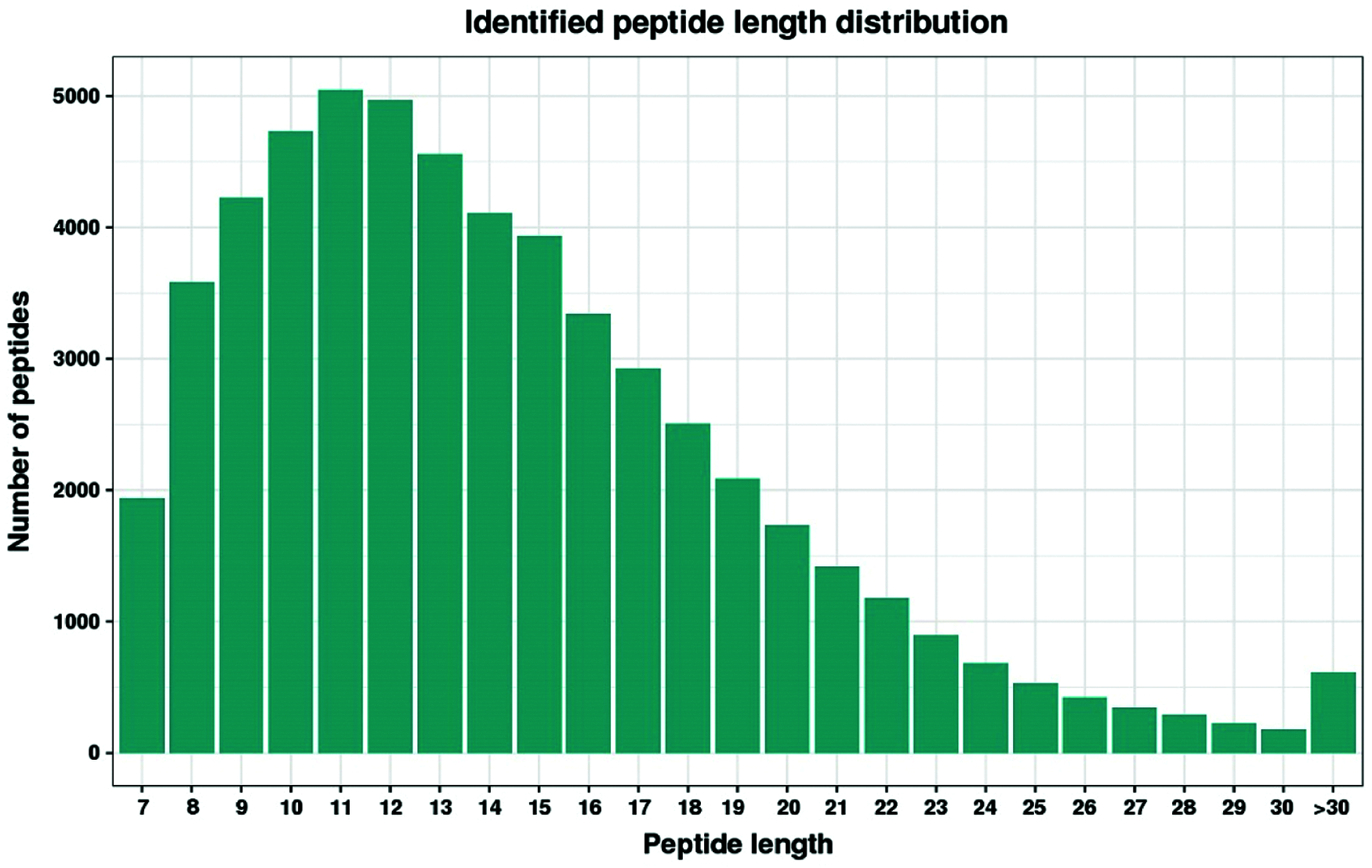
Figure 2: Distribution of peptides
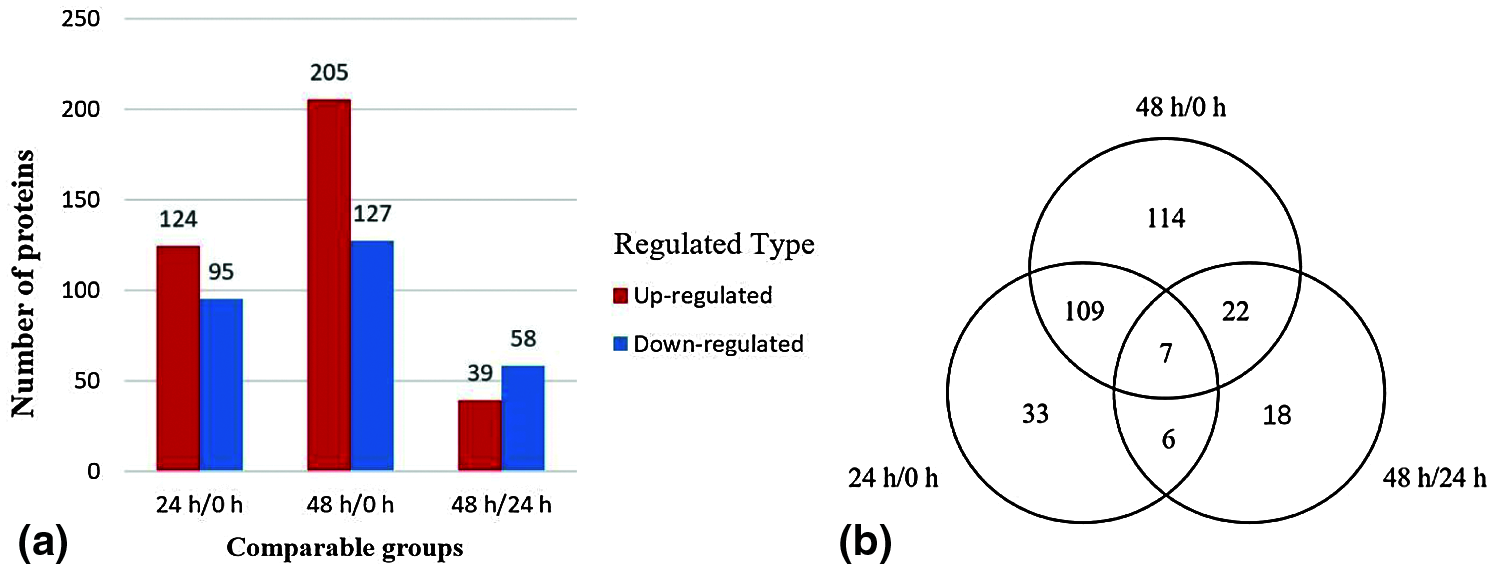
Figure 3: Histogram (a) of upregulated and downregulated differentially expressed proteins in three stages. Venn diagram (b) of differentially expressed proteins with KOG numbers (https://ftp://ftp.ncbi.nih.gov/pub/COG/KOG/kyva) at different stages
Compared with the lateral buds collected immediately after decapitation, the number of differentially expressed proteins of the lateral buds 24 h after decapitation was 219, of which 124 were upregulated and 95 were downregulated. Compared with the lateral buds collected immediately after decapitation, the number of differentially expressed proteins of the lateral buds that were grown for 48 h after decapitation was 332, of which 205 were upregulated and 127 were downregulated. Compared with the lateral buds collected 24 h after decapitation, the number of differentially expressed proteins in the lateral buds collected 48 h after decapitation was 97, of which 39 were upregulated and 58 were downregulated.
There were 7 proteins detected in all three stages, of which 3 proteins were upregulated and 4 proteins were downregulated. The 24 h/0 and 48 h/0 h stages shared 109 proteins, indicating that they were continuously involved in the process of lateral bud elongation within 48 h. There were 114 specific proteins identified in the 48 h/0 h stage, which implies that between 0 and 48 h there were many proteins that changed slowly, and the change was not obvious at 24 h, and it was not until 48 h that significant upregulation or downregulation was detected (Fig. 3b)
3.3 GO Annotation and GO Enrichment Analysis of Differential Proteins
Based on GO database, proteins were classified by function into three groups: cellular components, molecular functions, and biological processes. We found 28 biological processes involved in different proteins in the three stages, and the main trends were the same (Fig. 4). In the 28 biological processes, the number of differential proteins in the three stages increased during the 0–24 h stage and decreased at 24–48 h.
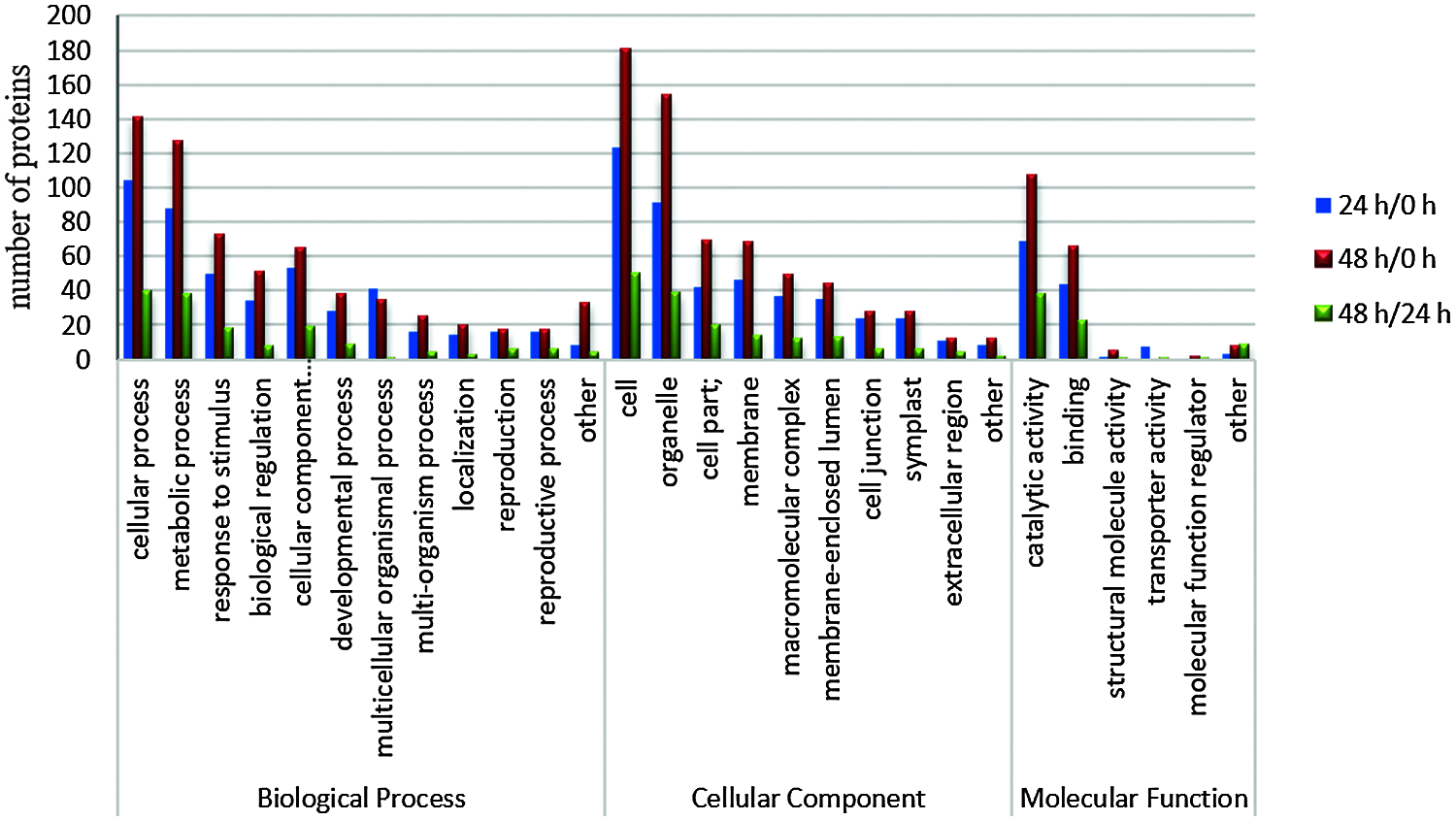
Figure 4: Functional categorization of differentially expressed proteins in different time periods after decapitation
Among all the 28 biological processes, the top three with a high proportion were cellular process (in the stages 24 h/0 h, 48 h/0 h, and 48 h/24 h, the percentages of differentially expressed proteins were 22.2%, 22%, and 25.6%, respectively), metabolic process (18.8%, 19.8%, 24.4%), and response to stimulus (10.7%, 11.4%, 11.5%). In the cell components, cell accounted for 27.9%, 28.1%, 32.1%, organelle accounted for 20.6%, 23.9%, 25%, and cell part accounted for 9.5%, 10.7%, and 12.8%; in the category of molecular function, catalytic activity, binding and structural molecule activity accounted for 55.6%, 54.6%, 52.1%; 35.5%, 33.7%, 31.5%; and 0.8%, 2.5%, 1.4%, respectively.
COG analysis represents a database of genes related to evolutionary generations, through which we can further understand the potential functions of differential proteins. The differentially expressed proteins identified in the three periods were classified into 23 types (Fig. 5). During the 24 h/0 and 48 h/0 h periods, most of the differentially expressed proteins were concentrated in four types, A, J, S, and O. With the elongation of lateral buds, the number of A, J, and O type proteins increased from 21, 17, and 17 to 28, 28, and 26, respectively. Among them, the J-type protein had the highest upregulation, and the O-type protein has the highest downregulation; the M- and W-type functions had only downregulated proteins. This shows that the process of lateral bud elongation after apical dominance removal is mainly affected by translation, ribosomal structure, and biogenesis (J), posttranslational modification, protein turnover, chaperones (O), and RNA processing and modification (A).
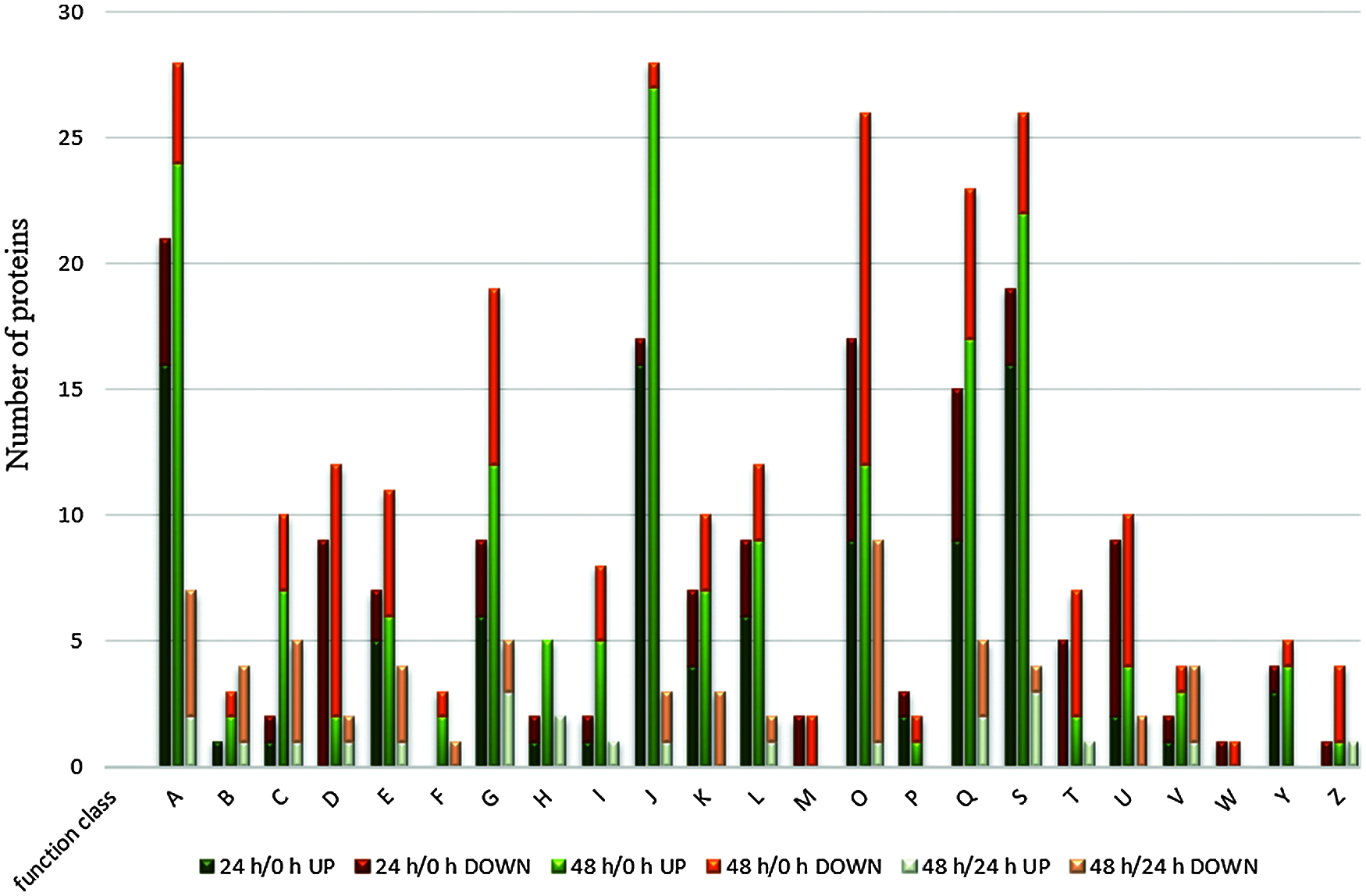
Figure 5: COG classification of differentially expressed proteins in different time periods after decapitation
To understand the metabolic processes involved in the three time periods, all differentially expressed proteins were analyzed for KEGG metabolic pathways, and 12 pathways were found to have significant changes (P1 < 0.05). The number of pathways with significant changes in the three periods were 8, 7, and 6, respectively (Fig. 6). In the 24 h/0 and 48 h/0 h stages, the protein involved in ribosome biogenesis in the eukaryotic pathway was abundant and accounted for a significant proportion of all pathways with significant changes, and all the involved proteins showed significant (P2 < 0.01) differences in both stages. In the 24 h/0 and 48 h/24 h stages, the number of proteins involved in the phenylpropanoid biosynthesis pathway was the highest, and the proteins involved showed significant differences in both stages (P3 < 0.01).

Figure 6: Bubble diagram of differential protein KEGG enrichment results. (a) 24 h/0 h (b) 48 h/0 h (c) 48 h/24 h
3.6 Gene Expression after Removal of the Apical Buds
IAA, CTK and strigolactones (SLs) are the three most vital hormones on the regulation of chrysanthemum lateral buds. Since these types of genes had not fully reflected in the protein pathway, we used qPCR to detect the gene expression that related to these three hormones after removing apical buds. (Fig. 7). The chrysanthemum samples collected at 0 h after removing the apical dominance were the control group, and those collected at 24 and 48 h were the treatment group. Obviously, it could be concluded from the results that the expression of DgD14 which belonged to SLs pathway continued to rise within 24 and 48 h, showing a significant higher level than that in the control group. In contrast, a continuous decrease in expression was observed on DgD27 which also belonged to the SLs pathway after removal of the apical buds. In the CTK pathway, the expression of CmERF053 saw a steady rise, while the DgIPT3 expression witnessed a slight declination at 24 h followed by a huge increase at 48 h after removal of the apical buds. Finally, in the auxin pathway, there was a fall in CmPIN1 expression after removing the apical dominance before increasing, but it was always lower than that of the control group; on the contrary, it was the expression level of DgBRC1 that significantly higher than that of the control group at 24 h, which accompanied by a decrease at 48 h but still higher than the control group.
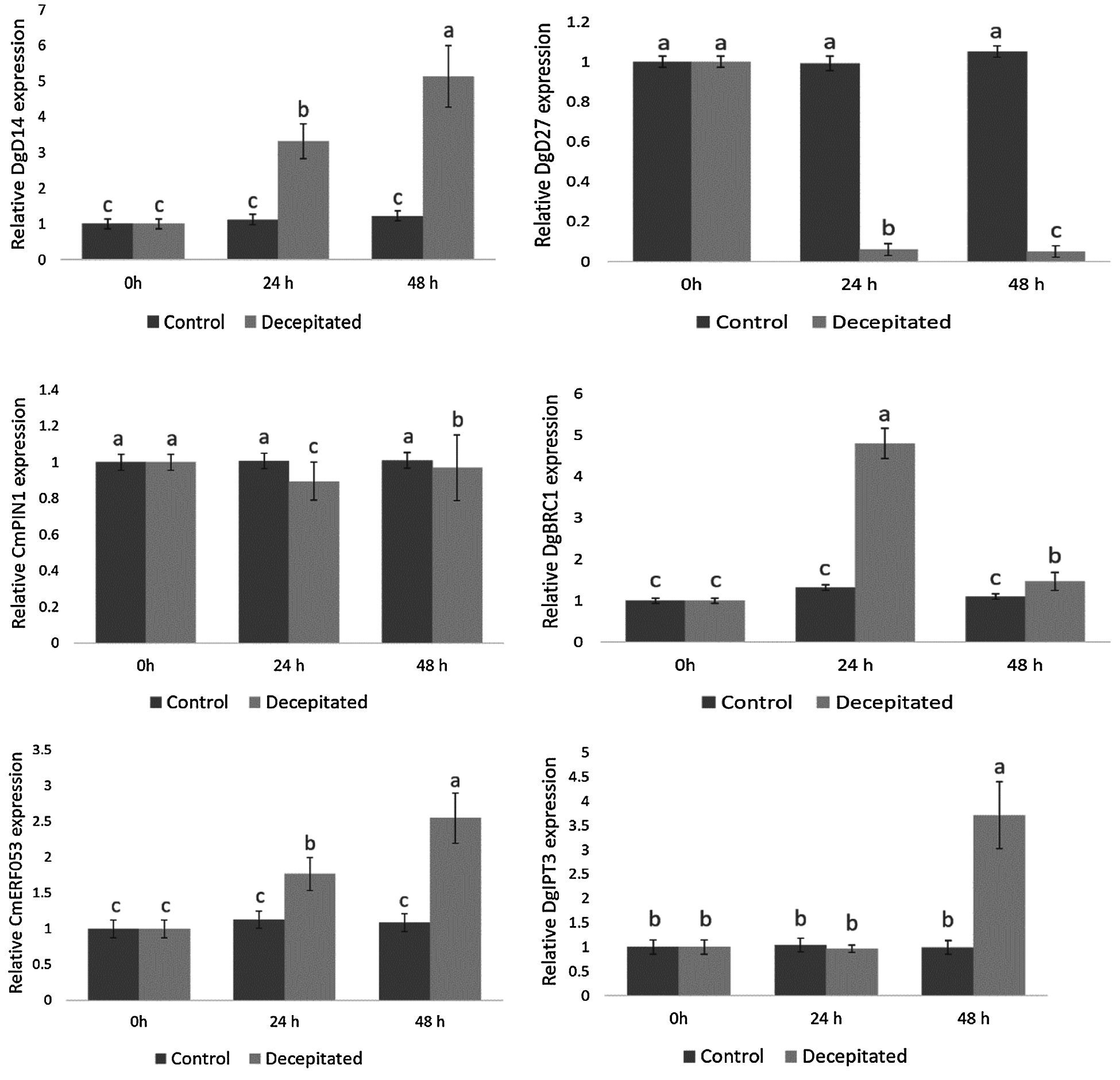
Figure 7: Expression of six related genes after removal of the apical buds
4.1 Apical Dominance in Chrysanthemum morifolium
Regulating the branches of chrysanthemum is of great significance to reducing the production cost of cut chrysanthemum. Research on the regulation mechanism of chrysanthemum branches can provide a theoretical basis for the development of technology to control chrysanthemum branches. Research based on plant apical dominance is an important aspect of the developmental regulation of chrysanthemum shoot branching. In Chrysanthemum morifolium ‘Jinba’, field observations have shown that the elongation of its lateral buds is related to apical dominance [13].
Traditional research on the mechanism of the elongation of the posterior bud of chrysanthemum after the breaking of apical dominance mainly focused on plant hormones and gene regulation [6]. However, the molecular properties and signal cellular basis for regulating the growth of lateral buds are still unclear. In this study, we performed extensive proteomic analysis on lateral buds collected at three time points (0, 24, and 48 h) after decapitation. These data will help us understand what biological processes occur in the lateral buds.
In our study, the length of lateral buds increased significantly during 0–48 h after removal of the top buds of chrysanthemum. The rate and process of lateral bud elongation are reflected by the executor of life function—the protein and the change of its expression is directly related to the cell function and metabolic activity of the tissue under specific physiological and ecological conditions.
4.2 Signal Transduction and Transport Across Membranes
After removal of the apical buds, some signal-related proteins are downregulated in the lateral buds, which means that signal transduction to maintain apical dominance is reduced. In this study, serine/threonine protein kinase was significantly downregulated from 0 to 24 h. This protein may prevent the production of auxin by regulating the PIN protein in the buds [14]. The transport carrier of auxin, PIN-FORMED (PIN), is a cell membrane protein. The regulation of its activity and location is very important for the transport direction of auxin and for plants to respond to environmental stimuli and to develop new organs. Serine/threonine protein kinase participates in the regulation of PIN phosphorylation and changes the polar positioning of PIN at the bottom and top plasma membranes of cells [15]. The vacuolar protein sorting-associated protein was significantly downregulated from 0–48 h; this protein plays an important role in intracellular vesicle transport, maintains cell polarity, and responds to the regulation of plant auxin [16].
The formation and growth process of lateral buds is a process of cell division, growth, and differentiation. This process requires the participation of energy. Recent studies have found that sucrose is an important signal molecule involved in the regulatory network of plant branch development. The absorption and accumulation of sugar by the terminal buds keep the lateral buds in a dormant state [17]. Beta-fructofuranosidase exists widely in plants and catalyzes the irreversible cleavage of sucrose to form glucose and fructose. Uridine−5′-diphosphate-glucose (UDP-glucose) pyrophosphorylase is significantly expressed within 0–48 h, and the expression level gradually increases. The enzyme uses glucose-1-phosphate and uridine triphosphate as substrates to catalyze the reaction to produce uridine diphosphate glucose and pyrophosphate, directly participating in the biosynthesis of plant sugar metabolism [18]. Changes in the expression of UDP-glucose pyrophosphorylase indicated that as the lateral buds elongated, the decomposition of polysaccharides gradually increased, and the synthesis and transport of monosaccharides increased.
The process of lateral bud outgrowth after the removal of apical dominance is a complex growth process of releasing dormancy and elongation. After decapitation, some proteins involved in ribosomal structure and translation function continued to be significantly upregulated from 0 to 48 h, which means that protein synthesis in lateral buds increased rapidly. GATA-4/5/6 transcription factors were significantly upregulated from 0 to 48 h, and their expression is induced by CTK. When the lateral buds of plants grow out, the CTK content in the lateral buds is significantly increased. GATA transcription factors play a unique role in the integration of CTK stimulation and the red light signal, thereby controlling the normal development of plants under these physiological stimuli [19].
The current study found that the synthesis of SLs starts from β-carotene, which is catalyzed by β-carotene isomerase, carotenoid dioxygenase 7, carotenoid dioxygenase 8, cytochrome P450 (CYP2 subfamily), and SLs molecules catalyzed by oxidoreductase. Alpha/beta hydrolaseD14 is the receptor for SLs in the cell, which can catalyze the hydrolysis of SLs and covalently bond with the active intermediate of SLs causing conformational changes. It forms a complex with the F-box protein DWARF3/MORE AXILLARY BRANCHES2 (D3/MAX2) to degrade the substrate protein [20,21]. Five cytochrome P450 (CYP2 subfamily) proteins and an alpha/beta hydrolase showed a continuous and significant up-regulation trend within 0–48 h, indicating that after decapitation, the auxin flow of the main stem of the plant weakened, and the IAA at the lateral buds could flow out, so the lateral buds would be after activation; the lateral buds can connect with the vascular bundles in the main stem to form a new auxin transport channel. At the same time, SLs inhibits the polar transport of auxin in the main stem and promotes the transfer of auxin between the lateral buds. The polar transport flow forms a competitive effect. The upper lateral buds are first activated to export the auxin and occupy the dominant position of the polar auxin transport flow in the main stem, while the lower lateral buds are in a disadvantageous position in the auxin transport flow competition, and the effect of auxin is difficult to overcome; therefore, these lateral buds will continue to be dormant [22–25].
When the apical buds are removed, the plant suffers mechanical damage. The serine carboxypeptidases content is increased and can participate in catalyzing proteolytic reactions, participate in injury response reactions, and can synthesize sinapoyl malate and brassinosteroid [26], which are rapidly and significantly upregulated during 0–48 h. Glutamate decarboxylase and related proteins are significantly upregulated in the 0–48 h stage. Glutamate decarboxylase is a type of intracellular enzyme that is ubiquitous in organisms. It is mainly distributed in the cytoplasm and is the key rate-limiting enzyme for catalyzing the decarboxylation of glutamate, the product being γ-aminobutyric acid (GABA). When subjected to abiotic stresses such as mechanical damage and tissue hypoxia, the activity of the GAD enzyme will change accordingly, and then GABA will be rapidly enriched in various plant tissues. In this way, it plays a role in such plant physiological processes as nitrogen metabolism, transportation and storage, and plant defense [27,28].
4.6 Regulation of Lateral Buds by Plant Hormones
Studies have revealed that the higher the content of IAA, the stronger the inhibitory effect on the development of lateral buds [29]. Instead of entering the lateral buds directly, IAA distributes according to a top-down gradient, which is related to PIN1 (PIN-FORMED), the transporter of auxin [30–32]. The expression of CmPIN1 in 24 h was slightly lower than that of the control group after removal of the apical buds, followed by a recovery in 48 h. This trend was the same as the result of Lv’s [5] experiment, supporting that removing apical dominance could lead to the loss of auxin transport in the main stem, thereby inhibiting the CmPIN1 expression. However, after which, the expression of CmPIN1 will be restored or increased, guiding the formation of a new apical auxin source and exporting IAA.
In our study, what was identical with Abdurazak’s [33] experimental result was that the expression level of CmERF053 at 24 h as well as 48 h was significantly higher than that of the control group, which proved that apical dominance removal could quickly promote CTK expression. Moreover, after removal of apical dominance in chrysanthemum, CmERF053 could respond to the dynamic changes of auxin in a rapid speed, and the overexpression of CmERF053 will lead to the increase of lateral buds in Arabidopsis. These findings suggest CmERF053 in its promotion to the development of lateral buds and its functional conservation.
In fact, SLs is a newly discovered plant hormone or its precursor, which can inhibit the growth of lateral buds as well as regulate the number of branches of plants together with auxin and CTK [6]. After removing the apical dominance, the expression of DgD27 which belongs to the SLs pathway continued to decrease, showing the same as Wen’s [4] experimental results. In addition, the number of lateral buds experienced a dramatic fall when DgD27 was used to transform Arabidopsis, indicating that DgD27 can inhibit the development of lateral buds and its function is conservative.
In summary, we identified 440 differentially expressed proteins. These proteins underwent significant changes during the formation of lateral buds after the breaking of apical dominance. Several of the proteins we have discussed may play an important role in the apical dominance mechanism, and they are involved in signal transduction, energy metabolism, biosynthesis, and resistance processes. This result shows that the signal transduction to maintain apical dominance after decapitation sharply declines, energy is generated through the accumulation of sugar during the initiation and elongation of lateral buds, and the synthesis of CTK increases rapidly. The rapid growth of SLs inhibits the formation of buds in the lower part of the plant. Meanwhile, changes in the expression of genes which related to IAA, CTK as well as SLs further proves the important regulatory effects of plant hormones on the development of chrysanthemum lateral buds. However, it still remains unclear about the specific mechanism of hormones regulation of chrysanthemum lateral buds, especially the regulation mechanism of SLs on lateral bud development, which needs further elucidation in the future.
Funding Statement: This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31800601).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Yu, J., Dong, L. L., Xi, L., Zhao, R. Y., Ma, N. et al. (2012). Isolation and characterization of cytokinin synthase gene DgIPT3 in chrysanthemum ‘Jinba’. Acta Horticulturae Sinica, 39, 721–728. DOI 10.16420/j.issn.0513-353x.2012.04.016. [Google Scholar] [CrossRef]
2. Dong, L. L., Ishak, A., Yu, J., Zhao, R. Y., Zhao, L. J. (2013). Identification and functional analysis of three MAX2 orthologs in chrysanthemum. Journal of Integrative Plant Biology, 55, 434–442. DOI 10.1111/jipb.12028. [Google Scholar] [CrossRef]
3. Liang, J. L., Zhao, L. J., Challis, R., Leyser, O. (2010). Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum). Journal of Experimental Botany, 61, 3069–3078. DOI 10.1093/jxb/erq133. [Google Scholar] [CrossRef]
4. Wen, C., Xi, L., Gao, B. (2015). Roles of DgD14 in regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum ‘Jinba’). Plant Physiology and Biochemistry, 96, 241–253. DOI 10.1016/j.plaphy.2015.07.030. [Google Scholar] [CrossRef]
5. Lv, S. H., Xi, L., Nie, J., Wen, C., Yuan, C. Q. et al. (2016). Isolation and expression analysis of auxin efflux transporter CmPIN1 in chrysanthemum (Chrysanthemum morifolium ‘Jinba’). Journal of China Agricultural University, 3, 58–66. DOI CNKI:SUN:NYDX.0.2016-03-009. [Google Scholar]
6. Wen, C., Liu, M. J., Shi, J. T., Ma, N., Zhao, L. J. (2017). Research progress on the formation and regulation of shoot branching in chrysanthemum. Journal of China Agricultural University, 22, 45–52. DOI CNKI:SUN:NYDX.0.2017-12-006. [Google Scholar]
7. Wang, W. F. (2019). Transcriptome analysis of axillary buds induced by tobacco topping and related gene functions (Ph.D. Thesis). Chinese Academy of Agricultural Sciences. [Google Scholar]
8. Lu, T. C. (2010). Proteomic study of mutants of pinus sylvestris var. sylvestris with loss of apical advantage and cloning of rad23 gene (Master Thesis). Northeast Forestry University. [Google Scholar]
9. Xie, Z. Q., Han, B. M., Wu, J. H. (2019). Proteome analysis of pericarp during grape coloring period. Acta Horticulturae Sinica, 46(4), 649–663. DOI 10.16420/j.issn.0513-353x.2018-0332. [Google Scholar] [CrossRef]
10. Tang, H. M., Zhang, X., Gong, B., Yan, Y. Y., Shi, Q. H. (2020). Proteomics and metabolomics analysis of tomato fruit at different maturity stages and under salt treatment. Food Chemistry, 311, 126009. DOI 10.1016/j.foodchem.2019.126009. [Google Scholar] [CrossRef]
11. Wei, X. D., Shi, D. W., Chen, G. X. (2013). Physiological, structural, and proteomic analysis of chloroplasts during natural senescence of Ginkgo leaves. Plant Growth Regulation, 69(2), 191–201. DOI 10.1007/s10725-012-9761-8. [Google Scholar] [CrossRef]
12. Liu, X. T., Zhai, R., Feng, W. T., Zhang, S. W., Wang, Z. G. et al. (2014). Proteomic analysis of ‘Zaosu’ pear (Pyrus bretschneideri Rehd.) and its early-maturing bud sport. Plant Science, 224, 120–135. DOI 10.1016/j.plantsci.2014.04.012. [Google Scholar] [CrossRef]
13. Xu, J. X., Ding, C. Q., Ding, Y. F., Tang, S., Zha, M. R. et al. (2015). A proteomic approach to analyze differential regulation of proteins during bud outgrowth under apical dominance based on the auxin transport canalization model in rice (Oryza sativa L.). Journal of Plant Growth Regulation, 34(1), 122–136. DOI 10.1007/s00344-014-9450-0. [Google Scholar] [CrossRef]
14. Jin, Y. D. (2013). Phosphoproteomic analysis of a Ser/Thr protein kinase PID mediated signaling in arabidopsis (Master Thesis). Northeast Forestry University. [Google Scholar]
15. Friml, J., Yang, X., Michniewicz, M., Ab, W. D. Q., Tietz, O. et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science,306(5697), 862–865. DOI 10.1126/science.1100618. [Google Scholar] [CrossRef]
16. Guo, M. M., Chen, M., Liu, R. B., Ma, Y. Z., Li, L. C. et al. (2014). Vacuolar protein sorting AtVPS25 regulates auxin responses in arabidopsis thaliana. Scientia Agricultura Sinica, 17, 3501–3512. DOI CNKI:SUN:ZNYK.0.2014-17-018. [Google Scholar]
17. Barbier, F. F., Dun, A. E., Kerr, C. S., Chabikwa, T. G., Beveridge, C. A. (2019). An update on the signals controlling shoot branching. Trends in Plant Science, 24, 220–236. DOI 10.1016/j.tplants.2018.12.001. [Google Scholar] [CrossRef]
18. Bai, B. B., Chen, Y. Y., Gai, J. T., Ye, X. X., Wang, P. (2019). Identification and sequence evolution analysis of plant UDP glucose pyrophosphorylase family genes. Molecular Plant Breeding, 17(4), 1186–1190. DOI 10.13271/j.mpb.017.001186. [Google Scholar] [CrossRef]
19. Naito, T., Kiba, T., Koizumi, N., Yamashino, T., Mizuno, T. (2007). Characterization of a unique GATA family gene that responds to both light and cytokinin. in Arabidopsis thaliana. Bioscience, Biotechnology, and Biochemistry, 71, 1557–1560. DOI 10.1271/bbb.60692. [Google Scholar] [CrossRef]
20. Zhao, L. H., Zhou, X. E., Wu, Z. S., Yi, W., Xu, Y. et al. (2013). Crystal structures of two phytohormone signal-transducing α/β hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Research, 23, 436–439. DOI 10.1038/cr.2013.19. [Google Scholar] [CrossRef]
21. Yao, R. F., Min, Z. H., Yan, L. M., Li, S. H., Wang, F. et al. (2016). DWARF14 is a non-canonical hormone receptor for strigolactone. Nature, 536(7617), 469–473. DOI 10.1038/nature19073. [Google Scholar] [CrossRef]
22. Crawford, S., Shinohara, N., Sieberer, T., Williamson, L., George, G. et al. (2010). Strigolactones enhance competition between shoot branches by dampening auxin transport. Development, 137, 2905–2913. DOI 10.1242/dev.051987. [Google Scholar] [CrossRef]
23. Domagalska, M. A., Leyser, O. (2011). Signal integration in the control of shoot branching. Nature Reviews Molecular Cell Biology, 12(4), 211–221. DOI 10.1038/nrm3088. [Google Scholar] [CrossRef]
24. Chabikwa, T. G., Brewer, P. B., Beveridge, C. A. (2019). Initial bud outgrowth occurs independent of auxin flow from out of buds. Plant Physiology, 179, 55–65. DOI 10.1104/PP.18.00519. [Google Scholar] [CrossRef]
25. Booker, J., Chatfield, S., Leyser, O. (2003). Auxin acts in xylen associated OT medullary cells to mediate apical dominance. Plant Cell, 15, 495–507. DOI 10.1105/tpc.007542. [Google Scholar] [CrossRef]
26. Li, C. (2015). Analysis of serine carboxypeptidase KL58 in arabidopsis and study on silencing NADK2 using CRISPR/cas9 technology (Master Thesis). Henan University. [Google Scholar]
27. Molina-Rueda, J. J., Pascual, M. B., Cánovas, F. M., Gallardo, F. (2010). Characterization and developmental expression of a glutamate decarboxylase from maritime pine. Planta, 232, 1471–1483. DOI 10.1007/s00425-010-1268-9. [Google Scholar] [CrossRef]
28. Bouché, N., Fait, A., Zik, M., Fromm, H. (2004). The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in arabidopsis. Plant Molecular Biology, 55, 315–325. DOI 10.1007/s11103-004-0650-z. [Google Scholar] [CrossRef]
29. Jiang, B., Chen, S., Jiang, J., Zhang, S., Chen, F. et al. (2012). Changes of endogenous hormones in lateral buds of chrysanthemum during their outgrowth. Russian Journal of Plant Physiology, 59, 356–363. DOI 10.1134/S1021443712020045. [Google Scholar] [CrossRef]
30. Booker, J., Chatfield, S., Leyser, O. (2003). Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell, 15, 495–507. DOI 10.1105/tpc.007542. [Google Scholar] [CrossRef]
31. Xi, L., Wen, C., Fang, S., Chen, X., Nie, J. et al. (2015). Impacts of strigolactone on shoot branching under phosphate starvation in chrysanthemum (Dendranthema grandiflorum cv. Jinba). Frontiers in Plant Science, 6, 694. DOI 10.3389/fpls.2015.00694. [Google Scholar] [CrossRef]
32. Wisniewska, J., Xu, J., Seifertová, D., Brewer, P. B., Ruzicka, K. et al. (2006). Polar PIN localization directs auxin flow in plants. Science, 312, 883. DOI 10.1126/science.1121356. [Google Scholar] [CrossRef]
33. Abdurazak, I. (2018). Cloning and functional analysis of the genes CmERF053 and CmIPT1 related to collateral development in chrysanthemum (Ph.D. Thesis). China Agricultural University. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |