

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.017625
ARTICLE
Identification of Genes Involved in Celastrol Biosynthesis by Comparative Transcriptome Analysis in Tripterygium wilfordii
1Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, 430074, China
2University of Chinese Academy of Sciences, Beijing, 100049, China
3Shanghai Key Laboratory of Bio-Energy Crops, Research Center for Natural Products, Plant Science Center, School of Life Sciences, Shanghai University, Shanghai, 200444, China
4State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, 430062, China
*Corresponding Authors: Shiyou Lü. Email: shiyoulu@wbgcas.cn; Changfu Li. Email: changfuli@shu.edu.cn
Received: 25 May 2021; Accepted: 07 July 2021
Abstract: Tripterygium wilfordii has been renowned mostly because of the anticancer effects of its root extracts, which is partly ascribed to the presence of celastrol, a pentacyclic triterpenoid, as one of the main active components. Celastrol also has recently been reported as an effective prodrug in the treatment of obesity. Despite the promising activities, the pathway leading to celastrol biosynthesis, especially cytochrome P450 (CYP) enzyme(s) that occur in its downstream steps, are largely unknown. This study conducted a comparative analysis of the T. wilfordii transcriptome derived from its root and leaf tissues. Differential gene expression analysis identified a number of root-specific CYP genes. Further phylogenetic analysis suggested specific family members of CYPs that may participate in the late steps during celastrol biosynthesis. Root-specific transcription factors (TFs) that may play regulatory roles in celastrol biosynthesis were also discussed. This genetic resource will aid in isolating the celastrol biosynthetic genes as well as engineering the celastrol biosynthesis pathway.
Keywords: Tripterygium wilfordii; celastrol biosynthesis; cytochrome P450
Tripterygium wilfordii hook F. (known as Lei Gong Teng in China) is a perennial woody vine of the Celastraceae family, which has been used as a Chinese traditional medicine in the treatment of arthritis for centuries [1]. The plant extracts of T. wilfordii also show activities toward other diseases, such as dermatosis, renal and bowel diseases [2,3]. Phytochemical studies showed that T. wilfordii produces diverse bioactive terpenoids with strong activities [4]. Among the T. wilfordii terpenoids, triptolide (a diterpenoid epoxide) and celastrol (a pentacyclic triterpenoid) have received the most interest due to their anticancer and anti-inflammatory activities [5,6]. This study focused only on celastrol. Recently, Liu et al. [7] have reported that celastrol was very effective in the treatment of obesity, a disease that has been a big problem accompanied with the development of modern societies. The anti-obesity effect of celastrol was believed to be mediated by an interleukin 1 receptor 1 (IL1R1) [8].
Currently, the common way to prepare celastrol is direct extraction of wild plant resources, which has endangered the continuation of the wild plant species and has brought adverse effects on the environment. The advent of synthetic biology opens an alternative avenue to produce celastrol with the prerequisite that genes coding for all the enzymes involved in celastrol biosynthesis are available. However, despite its biological importance, the pathway leading to celastrol biosynthesis is still poorly understood, especially the enzymes occurring on its downstream steps. Starting from 2,3-oxidosqualene, it is cyclized to form the friedelane backbone by friedelin synthase (FRS) [9] (Fig. 1). Genes encoding a monofunctional FRS have been isolated from Populus davidiana [10], Maytenus ilicifolia [11] and T. wilfordii [9]. In view of the molecular structure of celastrol, friedelin is subsequently converted to celastrol by multiple oxidations at the C-29, C-2, and C-24 positions, which are possibly mediated by specific cytochrome P450s (CYPs). The reaction order among these multiple oxidations is not clear yet. However, the occurrence of polpunonic acid (friedelin 29-carboxylic acid) in several plant species of Celastraceae (e.g., Maytenus senegalensis [12] and Tripterygium regelii [13]), may suggest that the 29-carboxylic acid might be formed prior to the other two oxidations. Indeed, the C29 methyl group of friedelin can be oxidized by an effective CYP enzyme, CYP712K1, to yield polpunonic acid [14] (Fig. 1). The corresponding enzymes for oxidations at the C-2 and C-24 positions have not yet been identified to date.
Comparative transcriptome analysis is an efficient methodology to identify genes involved in the biosynthesis of plant secondary metabolites. Given that the content of celastrol in roots is far higher than that in leaves [9,14,15], in this study we established the T. wilfordii transcriptome using raw RNA-sequencing datasets derived from its root and leaf tissues, which have been available online when we started this project. Through differential expression analysis, this study identified genes that were highly expressed in the T. wilfordii root with respect to its leaf tissue. We focused on the genes related to friedelin backbone biosynthesis and discovered new CYP enzymes that could be involved in the modification of friedelin leading to celastrol biosynthesis. Transcription factors (TFs) that might play regulatory effects on celastrol biosynthesis were also discussed.
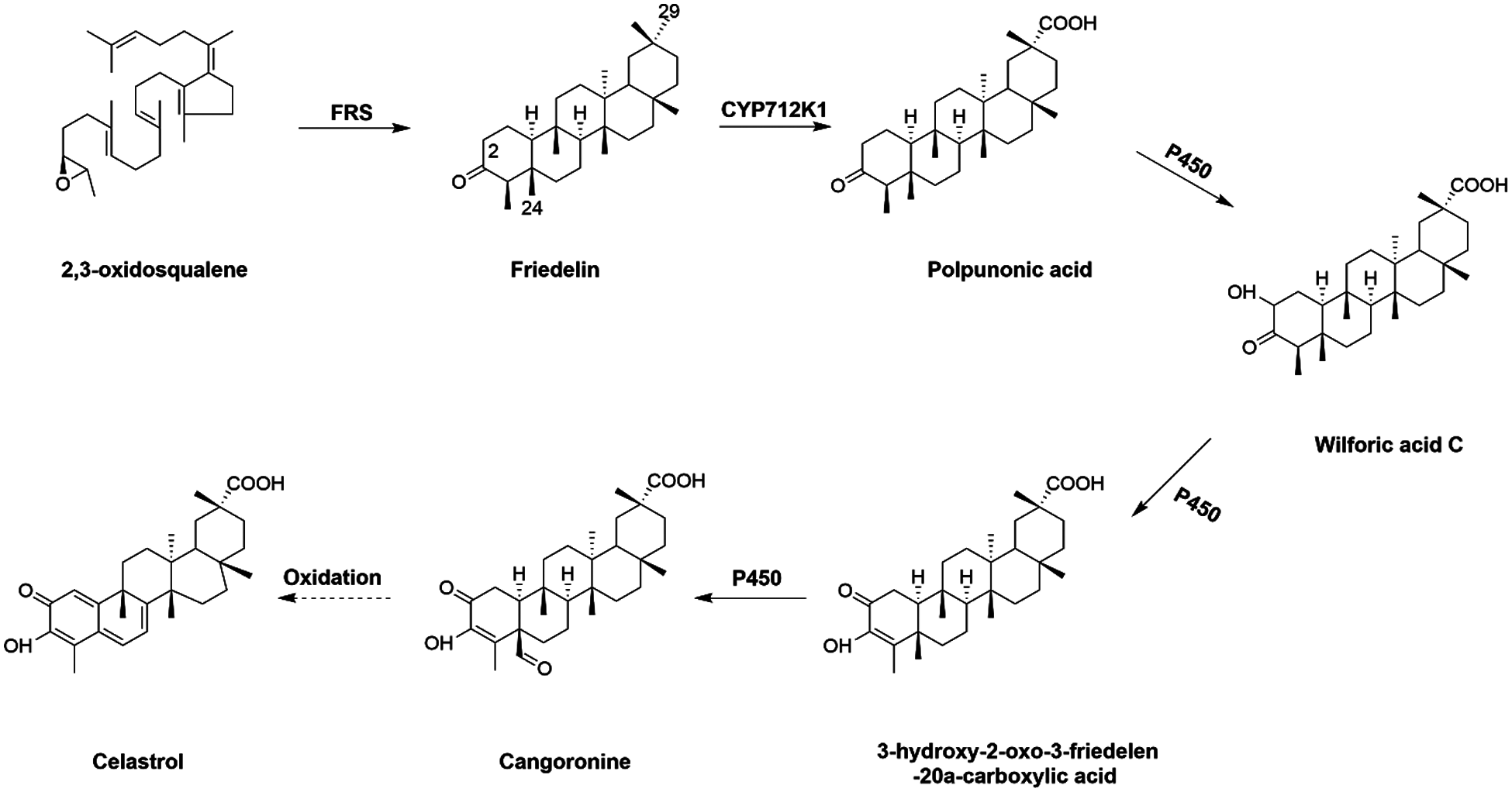
Figure 1: Proposed biosynthesis steps leading to celastrol from 2,3-oxidosqualene (the solid arrows indicate one-step reaction and the dashed arrow represents multiple step reactions)
Raw reads derived from the T. wilfordii root (deposit no. SRR708388 at NCBI) and leaf (SRR1171189 at NCBI) tissues were cleaned, and then assembled using the assembly program “Trinity”, following the protocol described by Grabherr et al. [16]. First, clean reads with certain length of overlap were combined to yield contigs. Then, the Trinity program (default set) was used to construct unigenes with the paired-end information.
2.2 Annotation and Classification of Unigenes
Using an E-value cut-off of 10–5, unigenes were searched against several resources, including Nr, UniProt, KEGG, GO, Pfam, eggNOG, and KOG databases, and their putative functions were annotated according to the highest similarity with the known sequences. If results of different databases conflicted, Nr then UniProt was set as the priority order of the database. For the unigenes that were not aligned to any databases described above, the ESTScan program [17] was then used to determine their sequence directions. The Blast2GO program [18] was employed to obtain GO annotations according to molecular function, biological process, and cellular component categories. Enzyme commission number was assigned based on the Blast2GO results.
2.3 Digital Differential Gene Expression Analysis
To determine the expression of each transcript, clean reads were mapped individually to the assembled transcriptome, and fragments per kb per million reads (FPKM) [19] were used to show the transcript abundance of each unigene. To obtain the differential expression genes (DEGs) between the root and leaf tissues, the DEGseq R package [20] was used. For DEG significance analysis, a threshold of |log2 (fold change)| > 1 and corrected p-value < 0.05 [21,22] were used.
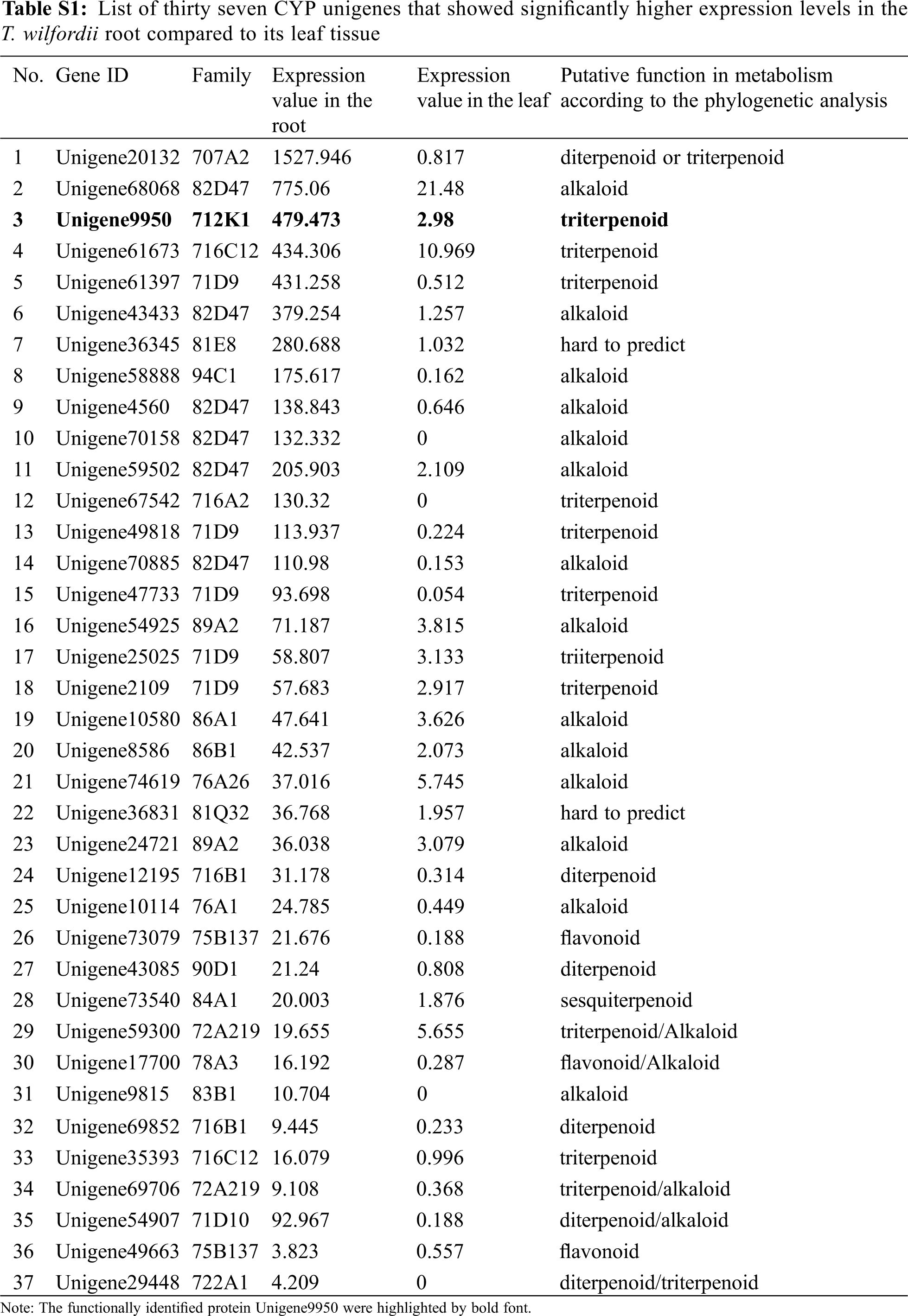
The open reading frames (ORFs) and amino acid sequences of the 37 T. wilfordii root-specifically expressed cyp450s were identified on ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder). The amino acid sequences of the other 211 published cyp450s involved in terpenoids and (iso)flavonoids biosynthesis were downloaded from the NCBI and Uniprot databases. All the amino acid sequences of the selected CYPs were aligned by ClustalW [23]. The best substitution model was selected using ModelFinder [24], and the maximum likelihood tree was inferred using IQ-TREE v1.6.12 [25]. The tree topology was then examined by 1000 ultrafast bootstrap sampling using UFBoot2 [26]. Finally, the phylogenetic tree was visualized on iTOL (https://itol.embl.de/).
3.1 De Novo Assembly and Functional Annotation
De novo assembly was performed to construct transcripts from the downloaded RNA-seq reads, resulting in a total of 75305 unigenes from the root and leaf tissues. The sequence length of the assembled unigenes ranged from 201 bp to 12932 bp, with an N50 value of 1073 bp (Fig. 2), indicating a high quality of the transcriptome assembly. Gene function was annotated by BLASTx (E-value < 1e–5) search against Nr (NCBI non-redundant sequence database), UniProt (Universal Protein), KEGG (Kyoto Encyclopedia of Genes and Genomes), GO (Gene Ontology), Pfam, and eggNOG (Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups) databases. In total, 38962 (51.74%) unigenes could be annotated to at least one database. According to the Nr annotation (Fig. 3), 8840 (11.94%) unigenes had the most hit from Theobroma cacao, followed by Picea sitchensis (7711, 10.24%), Jatropha curcas (7485, 9.94%), and Populus trichocarpa (6973, 9.26%).
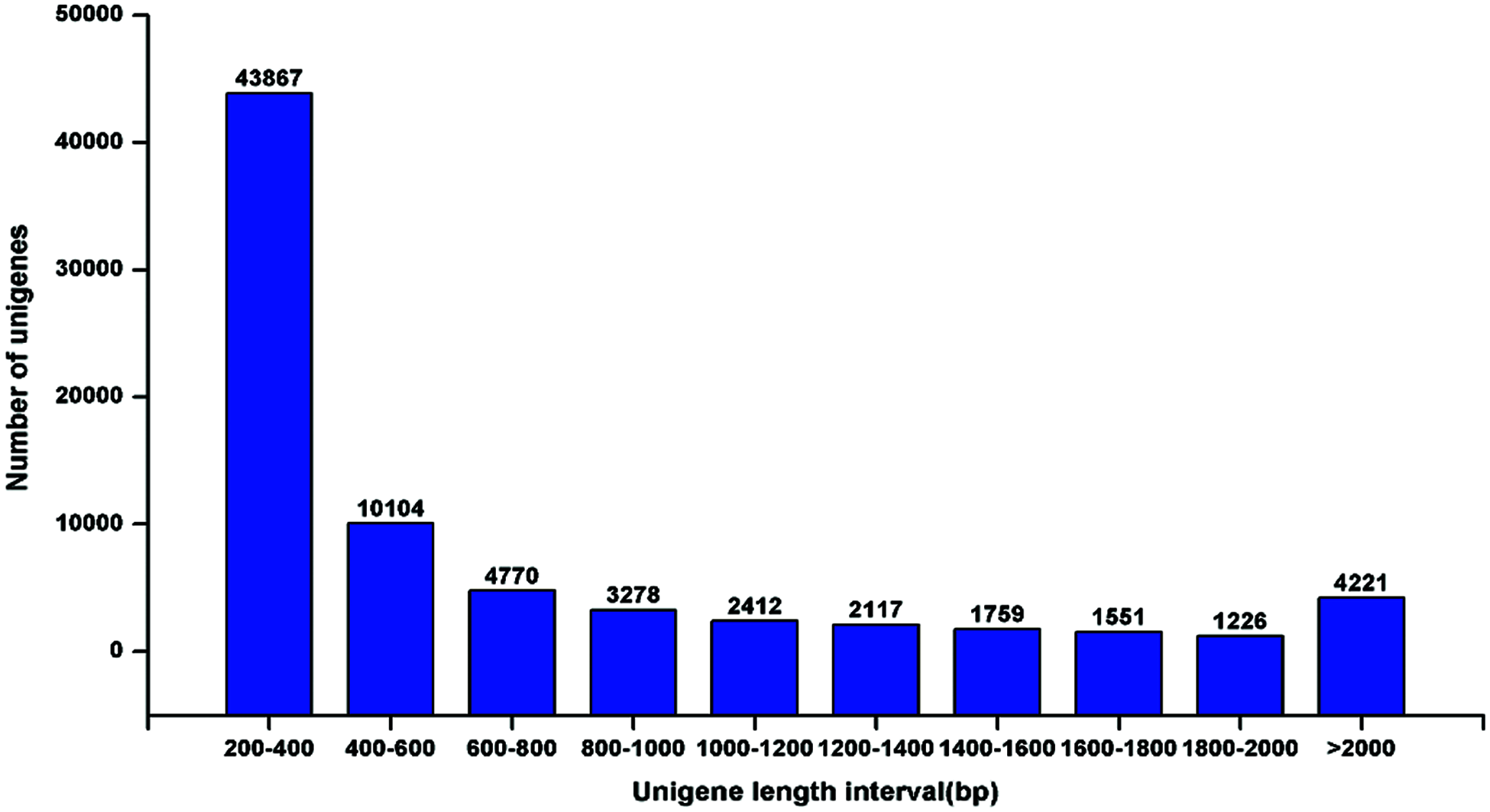
Figure 2: Assembled unigene length distribution of the T. wilfordii transcriptome
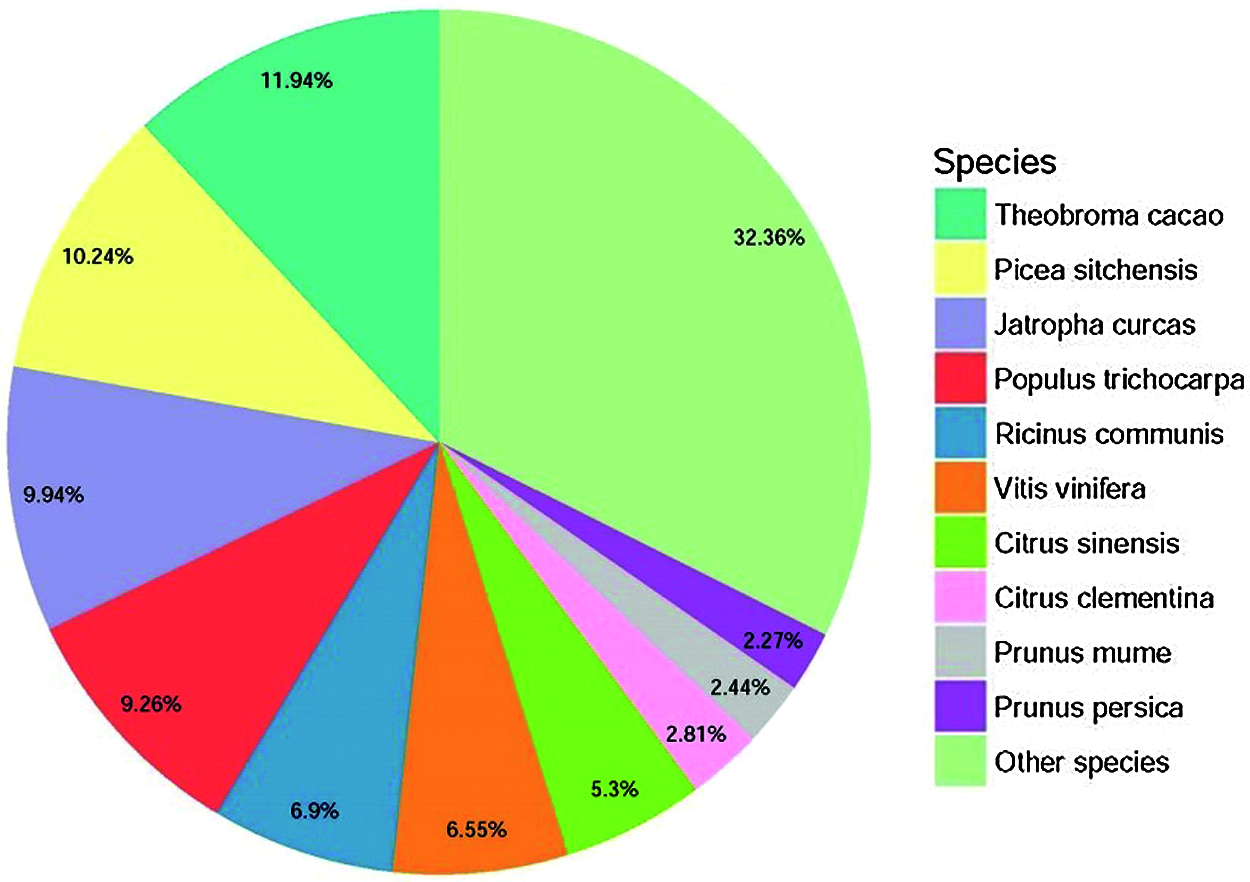
Figure 3: Species distribution of the homolog search of the unigenes against the Nr database
3.2 Identification of Genes Related to Friedelane Backbone Biosynthesis
Celastrol biosynthesis definitely proceeds via the isoprenoid unit, isopentenyl pyrophosphate (IPP), which is derived either from the MVA or from the MEP pathway. From the T. wilfordii transcriptome, a total of eight unigenes putatively encode four of the MVA pathway enzymes, including three for acetyl- CoA C-acetyltransferase (AACT), one for 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS), two for 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), one for mevalonate kinase (MK), and one for phosphomevalonate kinase (PMK). Meanwhile, eleven unigenes were annotated as molecules in the MEP pathway, and they included three unigenes for 1-deoxy-D-xylulose 5-phosphate synthase (DXS), three for 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), three for 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), one for 4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (HDS), and one for 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR). Based on the differential expression analysis, nearly all the MVA unigenes (AACT, HMGS, HMGR, and MK), except for PMK, were transcribed at a much higher level in the root than in the leaf (Tab. 1). On the contrary, the MEP pathway genes (DXS, DXR, CMK, HDS, and HDR) showed a similar or even a lower expression level in the root compared to the leaf tissues (Tab. 1).
Additionally, 10 unigenes were discovered as the candidates in the pathway beyond IPP toward the formation of the friedelane skeleton (Tab. 1), including two for isopentenyl diphosphate isomerase (IPPI), one for geranyl diphosphate synthase (GPPS), two for farnesyl diphosphate synthase (FPPS), one for squalene synthase (SS), three for squalene epoxidase (SE), and one for friedelin synthase (FRS, unigene11138, accession number: MZ488492). FRS catalyzes the first committed step to form the friedelane backbone as present in celastrol. Most of the unigenes in the steps from IPP to friedelin were up-regulated in the root compared to the leaf (Tab. 1). In particular, the FRS gene was root-specifically expressed with its FPKM value of 373.947 in the root while only 0.655 in the leaf.
3.3 Identification of the CYP-Encoding Genes Putatively Involved in Converting Friedelin to Celastrol
The subsequent steps of converting friedelin to celastrol are largely unknown. From a structural point of view, multiple oxidizations at the C-2, C-24, and C-29 positions of friedelin may be required, and these oxidizing steps were believed to be catalyzed by specific cytochrome P450 enzymes [14]. Given that the friedelane backbone genes were up-regulated in the root (Tab. 1), which is also the primary organ that accumulates celastrol in T. wilfordii, it is reasonable to speculate that the downstream P450 candidates involved in celastrol biosynthesis might be highly expressed in the root as well. For instance, from the T. wilfordii transcriptome of this study, CYP712K1, a known enzyme catalyzing the C29-oxidation of friedelin to form polpunonic acid [14], was present in the root by an FPKM value of 479.473 while it was expressed in the leaf only at an FPKM value of 2.98. This suggests that CYP712K1 is also specifically expressed in the root, as described above for FRS. Differential expression analysis led to the identification of thirty seven CYP unigenes that were specifically expressed in the root with respect to the leaf (Tab. S1). They belong to 21 different subfamilies of CYP707A, CYP82D, CYP712K, CYP716C, CYP71D, CYP81E, CYP94C, CYP716A, CYP89A, CYP86A, CYP86B, CYP76A, CYP81Q, CYP716B, CYP75B, CYP90D, CYP84A, CYP72A, CYP78A, CYP83B, and CYP722A. Among the root-specifically expressed CYPs, members from the clans of CYP82, CYP716 and CYP71 were the most abundant, including six (Unigene68068, Unigene43433, Unigene4560, Unigene70158, Unigene 59502 and Unigene70885) from the CYP82, six (Unigene61673, Unigene67542, Unigene12195, Unigene69852, Unigene35393, and Unigene54151) from the CYP716, and six (Unigene61397, Unigene49818, Unigene47733, Unigene25025, Unigene2109 and unigene54907) from the CYP71 class. When these root-highly expressing CYPs were subjected to a phylogenetic analysis (Fig. 4) together with the previously published CYPs with their known roles in biosynthesis of different plant secondary metabolites, the candidates that displayed a relatively closer relationship to the CYPs in triterpenoid metabolism were revealed; they included three candidates (Unigene 61673, Unigene67542 and Unigene35393) from the CYP716 clan, one (Unigene9950; CYP712K1) from the CYP712 clan, one (Unigene20132) from the CYP707 clan, and one (Unigene 29448) from the CYP722 clan. Based on their transcript abundances in the T. wilfordii root (see their FPKM values in Tab. S1), the order of Unigene20132 > Unigene9950 > Unigene61673 > Unigene67542 > Unigene35393 > Unigene29448 would be given precedence in considering them as potential candidates involved in celastrol biosynthesis. Except for the Unigene9950 that has recently been reported for a role in the C29-oxidation of friedelin [14], the remaining candidates identified by this study await further investigation to see whether they participate in oxidations at the C-2 and C-24 positions during celastrol biosynthesis. The accession numbers of the six CYP candidates can be found in the Genebank: Unigene20132(XM_038842675.1), Unigene9950(MN621243.1), Unigene61673(XM_038834352.1), Unigene67542(MZ488493), Unigene35393(XM_038866833.1), Unigene29448(XM_038848236.1).

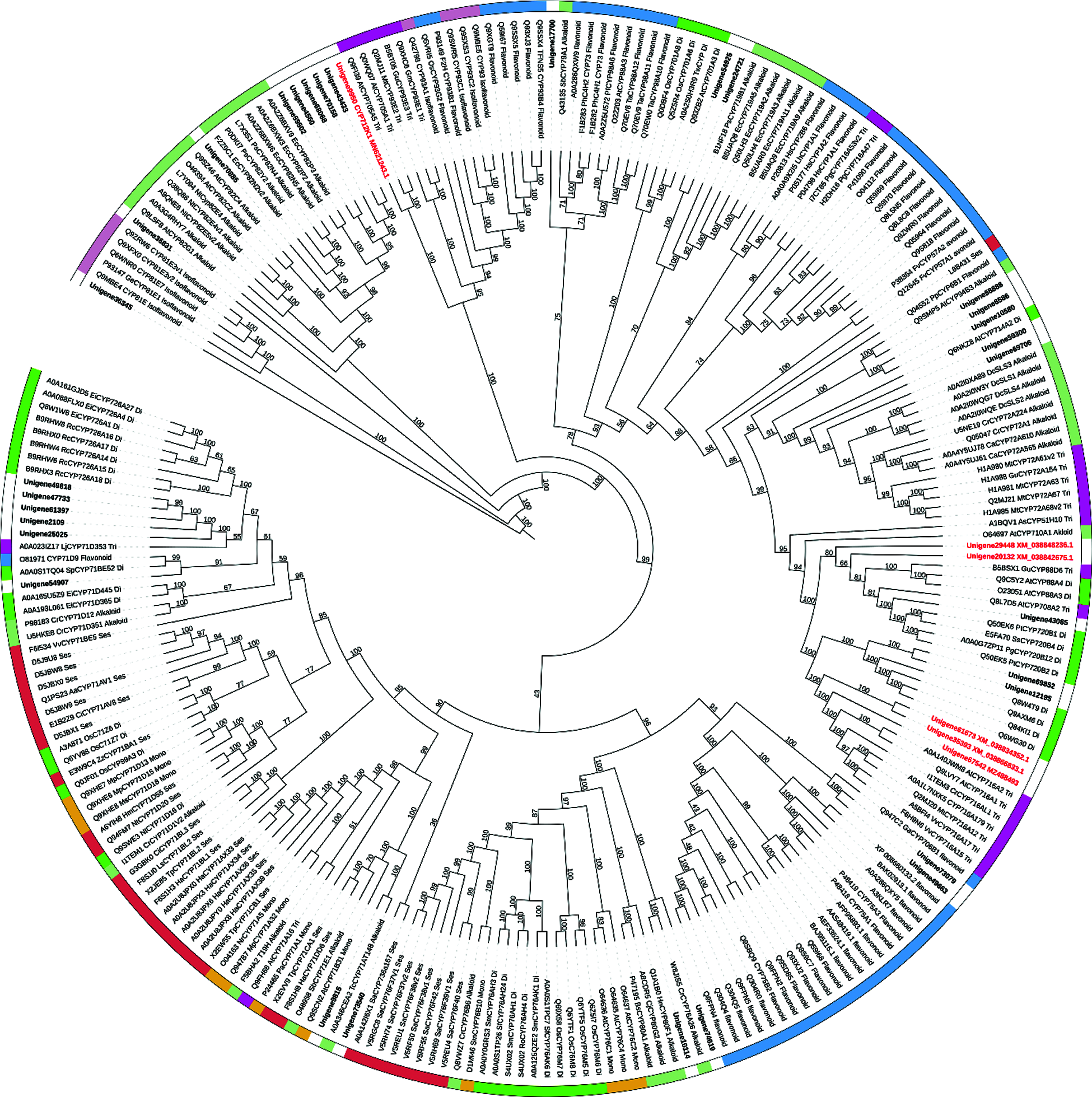
Figure 4: Phylogenetic analysis of the 37 T. wilfordii root-specifically expressed P450s (highlighted by bold font and with white bar) together with the 211 previously characterized P450s that function in the biosynthesis of sesquiterpenoid (dark red), monoterpenoid (brown), diterpenoid (Mustard green), triterpenoid (purple), alkaloid (celadon), flavonoid (blue), and isoflavonoid (grey purple). Six candidate CYPs that putatively involved in celastrol biosynthesis were marked in red
3.4 Identification of the Transcription Factors that May Play Regulatory Roles in Celastrol Biosynthesis
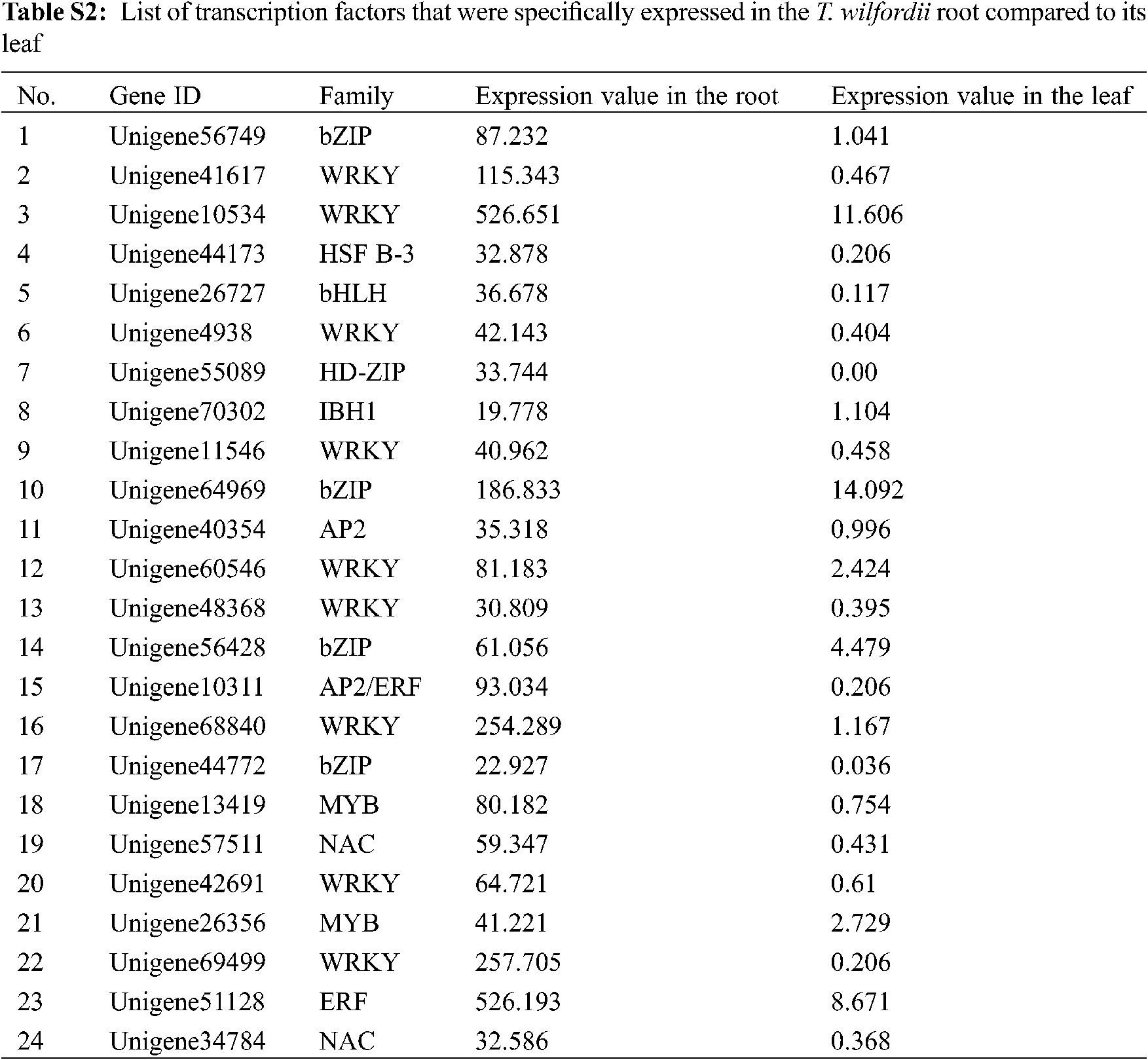
In total, 329 unigenes were annotated as transcription factors (TFs). There were 24 TFs that were specifically expressed in the T. wilfordii root compared to its leaf tissue (Tab. S2). These root highly expressed TFs belonged to the families of bZIP, WRKY, HSP B-3, bHLH, HD-ZIP, IBH1, AP2/ERF, MYB, and NAC, of which members of WRKY were the most abundant. It has been reported that members of WRKY participate in regulating the biosynthesis of triterpenoids [27]. They constitute a valuable gene resource for further studies of their regulatory functions in celastrol biosynthesis.
It is commonly accepted that triterpenoid stems from isoprenoid precursors, IPP and DMAPP, through the cytosolic MVA pathway [28]. With the construction of a T. wilfordii transcriptome derived from its root and leaf tissues, we have opportunities to know about the precursors originating for celastrol biosynthesis in this plant species. We have revealed that the gene transcript abundances of the MVA pathway were higher in its root than in the leaf (Tab. 1), consistent with the fact that T. wilfordii root is a known source of celastrol. On the contrary, the MEP pathway showed similar gene expression levels between the two tissues. This observation may indicate that celastrol, like other triterpenoids, is preferably biosynthesized from the MVA route.
Celastrol is biosynthesized from friedelin, which is formed by cyclization of 2,3-oxidosqualene catalyzed by a specific terpene synthase (friedelin synthase, FRS) [9]. Friedelin is then converted to celastrol by multiple oxidations at its C-29, C-2 and C-24 positions (Fig. 1). Very recently, a unique friedelin C-29 oxidase (CYP712K1) involved in celastrol biosynthesis has been characterized from T. wilfordii [14]. However, to date, there have been no reports regarding the C-2 and C-24 oxidases during celastrol biosynthesis. In this study, we performed a comparative transcriptome analysis of T. wilfordii root and leaf tissues. Predicted by FPKM values obtained from the RNA-sequencing analysis, both FRS and CYP712K1 transcripts in the roots were more than 200 times higher than in the leaves, suggesting that root is the predominant site for transcribing celastrol biosynthetic genes in T. wilfordii. To reveal the missing CYPs in the late steps toward celastrol biosynthesis, this study focused on CYP-encoding genes with high expression in the roots. A cluster of thirty-seven root-specifically expressed CYP-encoding unigenes, including the known C-29 oxidase CYP712K1, fell into our list. To identify the best candidate unigenes, these putative TwCYPs were subjected to a phylogenetic analysis using a variety of previously published P450s with known functions in different secondary metabolisms, including terpenoids (mono-, sesqui-, di-, and tri-terpenoids), alkaloids, and (iso) flavonoids. In stark contrast with relatively more strong clustering of (iso)flavonoid related CYPs on the tree, the selected terpenoid- and alkaloid-CYPs displayed a scattered clustering phenotype, indicating that across different plant species CYPs have independently evolved for those metabolisms. Closely examining the sub-clades of the tree allowed us to identify 6 CYP candidates (Unigene20132, Unigene9950, Unigene61673, Unigene67542, Unigene35393 and Unigene29448) that are most relevant to triterpenoid celastrol metabolism. Given that the Unigene9950 (CYP712K1) is an already known enzyme [14] in the steps toward celastrol biosynthesis, the identification of CYP712K1 within our list suggested that some of the other CYP candidates may play roles in celastrol biosynthesis. The candidates of Unigene61673, Unigene67542 and Unigene35393 all belong to the CYP716 clan. It has been reported that some of the CYP716 members participate in the biosynthesis of triterpenoids [29,30]. All the three CYP716 candidates were annotated as beta-amyrin 28-oxidase in our T. wilfordii transcriptome; however, blastp analysis of them against Nr database revealed that they all displayed less than 60% homology with the previously characterized beta-amyrin 28-oxidases. When searched the T. wilfordii transcriptome for beta-amyrin synthase, two putative beta-amyrin synthase genes (Unigene67989 and Unigene68089) stood up, and they both displayed a leaf-specific expression pattern with no or an extremely low expression in the root. Moreover, to the best of our knowledge, oleanolic acid (the direct product of beta-amyrin 28-oxidase [30] and its derivatives are rarely detected in the T. wilfordii roots. These observations may suggest that the three CYP716 candidates identified by this study are not the routine enzymes accepting beta-amyrin as a physiological substrate. It will be of interest to further test whether they catalyze the C2- and C24-oxidations for celastrol biosynthesis.
Our transcriptome data suggested that in T. wilfordii the genes in the late steps leading to celastrol biosynthesis are specifically expressed in the roots (Tab. 1). The regulatory mechanism that mediates this root-specific expression is not known; however, this pattern may be directed by specific transcription factors (TFs) that are specifically expressed in the roots. Based on the DGE data, we found a number of TFs (Tab. S2) that were specifically expressed in the T. wilfordii root compared to its leaf tissue. Among the root-specific TFs, members of the WRKY family were found to be the most abundant. It will be interesting to investigate whether some of these root-specific TFs up-regulate the expression of the late celastrol biosynthetic genes. These TFs may also be utilized as effective components through genetic engineering for enhancing celastrol content.
Author Contributions: Shiyou Lü and Changfu Li designed the project. Xiujun Zhang provided assistance in the bioinformatics analysis. Yaru Zhu analyzed the data and drafted the manuscript. Yansheng Zhang made discussions on the data. Changfu Li revised the manuscript.
Funding Statement: This work was supported in part by a grant from the National Key R&D Program of China (SQ2018YFC170017) and a grant from the National Natural Science Foundation of China (31670300).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Tao, X., Lipsky, P. E. (2000). The Chinese anti-inflammatory and immunosuppressive herbal remedy Tripterygium wilfordii Hook F. Rheumatic Diseases Clinics of North America, 26(1), 29–50. DOI 10.1016/s0889-857x(05)70118-6. [Google Scholar] [CrossRef]
2. Song, C. Y., Xu, Y. G., Lu, Y. Q. (2020). Use of Tripterygium wilfordii Hook F for immune-mediated inflammatory diseases: Progress and future prospects. Journal of Zhejiang University Science B, 21, 280–290. DOI 10.1631/jzus.B1900607. [Google Scholar] [CrossRef]
3. Wang, Y. D., Wang, Q., Zhang, J. B., Dai, Z., Lin, N. et al. (2019). Research progress on chemical constituents and quality control of Tripterygium wilfordii preparations. China Journal of Chinese Materia Medica, 44(16), 3368–3373. DOI 10.19540/j.cnki.cjcmm.20190606.501. [Google Scholar] [CrossRef]
4. He, M. F., Liu, L., Ge, W., Shaw, P. C., Jiang, R. et al. (2009). Antiangiogenic activity of Tripterygium wilfordii and its terpenoids. Journal of Ethnopharmacology, 121(1), 61–68. DOI 10.1016/j.jep.2008.09.033. [Google Scholar] [CrossRef]
5. Lee, D., Kim, I. Y., Saha, S., Choi, K. S. (2016). Paraptosis in the anti-cancer arsenal of natural products. Pharmacology & Therapeutics, 162, 120–133. DOI 10.1016/j.pharmthera.2016.01.003. [Google Scholar] [CrossRef]
6. Ziaei, S., Halaby, R. (2016). Immunosuppressive, anti-inflammatory and anti-cancer properties of triptolide: A mini review. Avicenna Journal of Phytomedicine, 6(62), 149–164. https://pubmed.ncbi.nlm.nih.gov/27222828/. [Google Scholar]
7. Liu, J., Lee, J., Salazar Hernandez, M. A., Mazitschek, R., Ozcan, U. (2015). Treatment of obesity with celastrol. Cell, 161(5), 999–1011. DOI 10.1016/j.cell.2015.05.011. [Google Scholar] [CrossRef]
8. Feng, X., Guan, D., Auen, T., Choi, J. W., Salazar Hernandez, M. A. et al. (2019). IL1R1 is required for celastrol’s leptin-sensitization and antiobesity effects. Nature Medicine, 25, 575–582. DOI 10.1038/s41591-019-0358-x. [Google Scholar] [CrossRef]
9. Zhou, J., Hu, T., Gao, L., Su, P., Zhang, Y. et al. (2019). Friedelane-type triterpene cyclase in celastrol biosynthesis from Tripterygium wilfordii and its application for triterpenes biosynthesis in yeast. New Phytologist, 223(2), 722–735. DOI 10.1111/nph.15809. [Google Scholar] [CrossRef]
10. Han, J. Y., Ahn, C. H., Adhikari, P. B., Kondeti, S., Choi, Y. E. (2019). Functional characterization of an oxidosqualene cyclase (PdFRS) encoding a monofunctional friedelin synthase in Populus davidiana. Planta, 249, 95–111. DOI 10.1007/s00425-018-2985-8. [Google Scholar] [CrossRef]
11. Souza-Moreira, T. M., Alves, T. B., Pinheiro, K. A., Felippe, L. G., de Lima, G. M. et al. (2016). Friedelin Synthase from Maytenus ilicifolia: Leucine 482 plays an essential role in the production of the most rearranged pentacyclic triterpene. Scientific Reports, 6, 36858. DOI 10.1038/srep36858. [Google Scholar] [CrossRef]
12. Sosa, S., Morelli, C. F., Tubaro, A., Cairoli, P., Speranza, G. et al. (2007). Anti-inflammatory activity of Maytenus senegalensis root extracts and of maytenoic acid. Phytomedicine: International Journal of Phytotherapy and Phytopharmacology, 14(2–3), 109–114. DOI 10.1016/j.phymed.2005.11.002. [Google Scholar] [CrossRef]
13. Fan, D., Parhira, S., Zhu, G. Y., Jiang, Z. H., Bai, L. P. (2016). Triterpenoids from the stems of Tripterygium regelii. Fitoterapia, 113, 69–73. DOI 10.1016/j.fitote.2016.07.006. [Google Scholar] [CrossRef]
14. Hansen, N. L., Miettinen, K., Zhao, Y., Ignea, C., Kampranis, S. C. (2020). Integrating pathway elucidation with yeast engineering to produce polpunonic acid the precursor of the anti-obesity agent celastrol. Microbial Cell Factories, 19(1), 1138. DOI 10.1186/s12934-020-1284-9. [Google Scholar] [CrossRef]
15. Su, P., Guan, H., Zhao, Y., Tong, Y., Xu, M. et al. (2018). Identification and functional characterization of diterpene synthases for triptolide biosynthesis from Tripterygium wilfordii. Plant Journal, 93(1), 50–65. DOI 10.1111/tpj.13756. [Google Scholar] [CrossRef]
16. Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A. et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29(7), 644–652. DOI 10.1038/nbt.1883. [Google Scholar] [CrossRef]
17. Iseli, C., Jongeneel, C. V., Bucher, P. (1999). ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings International Conference on Intelligent Systems for Molecular Biology, 99, 138–148. https://pubmed.ncbi.nlm.nih.gov/10786296/. [Google Scholar]
18. Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M. et al. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18), 3674–3676. DOI 10.1093/bioinformatics/bti610. [Google Scholar] [CrossRef]
19. Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G. et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 28(5), 511–515. DOI 10.1038/nbt.1621. [Google Scholar] [CrossRef]
20. Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics, 26(1), 136–138. DOI 10.1093/bioinformatics/btp612. [Google Scholar] [CrossRef]
21. Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biology, 11, R106. DOI 10.1186/gb-2010-11-10-r106. [Google Scholar] [CrossRef]
22. Storey, J. D., Tibshirani, R. (2003). Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America, 100(16), 9440–9445. DOI 10.1073/pnas.1530509100. [Google Scholar] [CrossRef]
23. Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A. et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948. DOI 10.1093/bioinformatics/btm404. [Google Scholar] [CrossRef]
24. Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14(6), 587–589. DOI 10.1038/nmeth.4285. [Google Scholar] [CrossRef]
25. Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D. et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37(5), 1530–1534. DOI 10.1093/molbev/msaa015. [Google Scholar] [CrossRef]
26. Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35(2), 518–522. DOI 10.1093/molbev/msx281. [Google Scholar] [CrossRef]
27. Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S. et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytologist, 215, 1115–1131. DOI 10.1111/nph.14663. [Google Scholar] [CrossRef]
28. Rodriguez-Concepcion, M., Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiology, 130(3), 1077–1089. DOI 10.1104/pp.007138. [Google Scholar] [CrossRef]
29. Fukushima, E. O., Seki, H., Ohyama, K., Ono, E., Umemoto, N. et al. (2011). CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant & Cell Physiology, 52(12), 2050–2061. DOI 10.1093/pcp/pcr146. [Google Scholar] [CrossRef]
30. Huang, L., Li, J., Ye, H., Li, C., Wang, H. et al. (2012). Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus. Planta, 236, 1571–1581. DOI 10.1007/s00425-012-1712-0. [Google Scholar] [CrossRef]
Appendixes
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |