

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.017451
ARTICLE
Construction and Functional Analysis of CRISPR/Cas9 Vector of FAD2 Gene Family in Soybean
College of Agronomy, Jilin Agricultural University, Changchun, 130118, China
*Corresponding Author: Piwu Wang. Email: peiwuw@163.com
Received: 11 May 2021; Accepted: 18 June 2021
Abstract: Soybean oleic acid content is one of the important indexes to evaluate the quality of soybean oil. In the synthesis pathway of soybean fatty acids, the FAD2 gene family is the key gene that regulates the production of linoleic acid from soybean oleic acid. In this study, CRISPR/Cas9 gene editing technology was used to regulate FAD2 gene expression. Firstly, the CRISPR/Cas9 single knockout vectors GmFAD2-1B and GmFAD2-2C and double knockout vectors GmFAD2-2A-3 were constructed. Then, the three vectors were transferred into the recipient soybean variety Jinong 38 by Agrobacterium-mediated cotyledon node transformation, and the mutant plants were obtained. Functional analysis and comparison of the mutant plants of the T2 and T3 generations were carried out. The results showed that there was no significant difference in agronomic traits between the CRISPR/Cas9 single and double knockout vectors and the untransformed CRISPR/Cas9 receptor varieties. The oleic acid content of the plants that knocked out the CRISPR/Cas9 double gene vector was significantly higher than that of the single gene vector.
Keywords: Soybean; FAD2 gene family; CRISPR/Cas9; oleic acid content
Soybean oil is the main source of human edible vegetable oil and the largest vegetable oil for food in the world after palm oil [1]. It is also an indispensable part of people’s healthy daily life. The most important determinant of soybean oil quality is fatty acid composition and the proportion of soybean fatty acid determines the quality of soybean oil. Soybean fatty acid is mainly composed of palmitic (C16:0), stearic (C18:0), oleic (C18:1), linoleic (C18:2) and linolenic acids (C18:3) [2]. As a monounsaturated fatty acid, oleic acid has strong antioxidant activity and high stability [3]. It has also a health effect by lowering cholesterol, blood lipids and maintaining a high density of lipoprotein [4]. Soybean oil improvement is one of the important concerns of soybean chemical quality improvement, mainly by reducing the ratio of palmitic acid to linolenic to increase the ratio of linoleic acid to oleic acid [5]. Its high oleic acid content plays a significant role in improving the stability and nutritional quality of soybean edible oil [6]. This is because oleic acid concentration is only 20% and linoleic acid concentration is 55%. Increasing oleic acid content and reducing linoleic acid content is an important goal of soybean breeders.
Composition of soybean fatty acids is mainly regulated by the FAD2 gene family. In the metabolic pathway of soybean, saturated fatty acids are desaturated to form unsaturated fatty acids, such as oleic, linoleic and linolenic acids, under the catalysis of various desaturases. Among them, under the catalysis of the fatty acid dehydrogenase (FAD2 enzyme), and the oleic acid dehydrogenates to polyunsaturated linoleic fatty acid [7]. The activity of the FAD2 enzyme is the main factor affecting the synthesis of various unsaturated fatty acids in plants, and FAD2 gene is the coding gene of FAD2 enzyme. Therefore, the FAD2 gene is the key gene to control the conversion of oleic to linoleic acid [8]. It directly determines the proportion of fatty acid components and plays an important role in fatty acid metabolism. The regulation of the function of the FAD2 gene family can change the composition of soybean oleic acid. At present, FAD2 genes have been cloned on other studies in peanut, cotton, perilla, and Camellia oleifera plants [9–13]. Combs used RNAi technology to inhibit the expression of FAD2-2 genes in soybean seeds and obtained 81.9% transgenic soybean with oleic acid content [14,15]. Demorest and his collegues used TALENs technology to target GmFAD2-1A and GmFAD2-1B genes in soybean and knock out them. In the offspring of homozygous mutants, it was found that the fatty acid content of seeds changed, the oleic acid concentrations increased from 20% to 80%, and the linoleic acid concentration decreased from 50% to 4% [16]. Therefore, in the synthesis pathway of soybean fatty acids, the genes of the FAD2 gene family play an important role in reducing the formation of linoleic acid by inhibiting the expression of each gene in the FAD2 gene family, thus increasing the content of oleic acid. This provides a molecular basis for the cultivation of high oleic acid soybean.
CRISPR/Cas9 gene editing technology is a favorable tool to control gene expression. It has been applied to maize, rape, soybean, Arabidopsis, rice and other crops, which has brought new opportunities in the field of plant gene manipulation [17–21].
The plant material soybean Jinong 38 was provided by the Plant Biotechnology Center of Jilin Agricultural University. In the growth cycle, Jinong 38 has early maturity, good lodging resistance, higher oil content among all local varieties and good adaptability. Therefore, we chose this material to carry out our experiment.
2.2 Construction of CRISPR/Cas9 Carrier
The gRNA target sites of genes were designed by the CRISPR-P website (http://crispr.hzau.edu.cn/CRISPR2/), and the CRISPR/Cas9 vector was constructed in the Biogle Biotechnology Company by using the CRISPR/Cas vector.
We transferred the constructed CRISPR vector plasmid into the soybean Jinong 38 cotyledon node by using the Agrobacterium-mediated method. First, seeds with uniform size were selected and sterilized with 5 mL concentrated hydrochloric acid (12.27 mol/L) and 25 mL sodium hypochlorite (25.54 mol/L) for 16 h. Three days after germination, the cotyledon nodes of the germinated seeds were cut in half on the aseptic operating platform, pre-cultured for three days, and co-cultured with Agrobacterium tumefaciens for three days. The primary screening was performed after 15 days, and 1 mg/mL herbicide was used on the second screening for 15 days. The selected callus was transferred to an elongation medium and it was cultivated for 15 days. Then, it was transferred to a root induction medium until the root system was strong and well-developed. Thereafter, the seedlings were refined and transplanted, and the generation was increased indoors to obtain the gene-edited soybean offspring.
2.4 Genome Extraction and PCR Detection
The first group of three composite leaves of soybean V1 was selected as the material for extracting the soybean genome. The NuClean Plant Genomic DNA Kits were purchased from the Beijing ComWin Biotech Co., Ltd., China; we followed the company’s instructions for extraction. The primer 5.0 (Primer design software) software was used to mark the Bar specificity primer and Cas9 primer screening. The specific primers for the Bar gene were S: TCAAATCTCGGTGACGGGC, and AS: ATGAGCCCAGAACGACGC, the annealing temperature was 58°C. The specific primers for the Cas9 gene were S: CCCAAGAGGAACAGCGATAAG, and AS: GTCGATGGTGGTGTCAAAGT, and the annealing temperature was 64°C.
2.5 Detection of Target Mutations in Mutant Plants
From the T2 gene-edited offspring seeds obtained by the Agrobacterium-mediated cotyledon node method, 5 plants for each knockout vector were randomly selected for sequencing, and comparison with the target sequence of unmutated plants.
2.6 Fluorescence Quantitative PCR Detection in the Gene Editing Offspring
RNAiso Plus kits (TaKaRa) were used to extract the total RNA of soybean roots, stems, and leaves that were positive by PCR in the T2 and T3 generations. RNAlater® kit (Thermo Fisher, AM7020, China) was used to reverse transcribed into cDNA. Prime-ScriptTM RT Master Mix (Fermentas) reagent was used for qRT-PCR amplification. The amplification conditions were: 94°C pre-denaturation for 4 min; 94°C denaturation for 10 s; 60°C annealing and extension for 40 s; 40 cycles. The 2-ΔΔCT calculation method was used to analyze the expression and relative expression of the target gene and it was repeated twice, the average value was taken.
2.7 Determination of Fatty Acid in Gene Editor’s Fluxes
NIRSTM DS 2500 (Foss, Hillerod, Denmark) was used to determine the contents of oleic acid and four other fatty acids (stearic, palmitic, linoleic acid and linolenic acids) in transgenic soybean seeds after harvesting.
2.8 Analysis of Agronomic Characters in the Gene-Editing Generation
The agronomic traits and phenotype analyses of the gene-edited progeny plants were conducted. Investigate the leaf shape, flower color and growth period of the plant. After the plant matures, investigate and measure the plant height, number of main stem nodes, number of branches per plant, seed coat color and seed umbilicus color. Finally, SPSS 23.0 was used to analyze the data.
3.1 Construction of the CRISPR/Cas9 Carrier
CRISPR/Cas9 vectors of different targets were constructed to explore the differences of mutation efficiency and mutation types among different targets. According to the principle of target design, the target sequence was as Tab. 1.
The carrier structure was as Figs. 1–3.
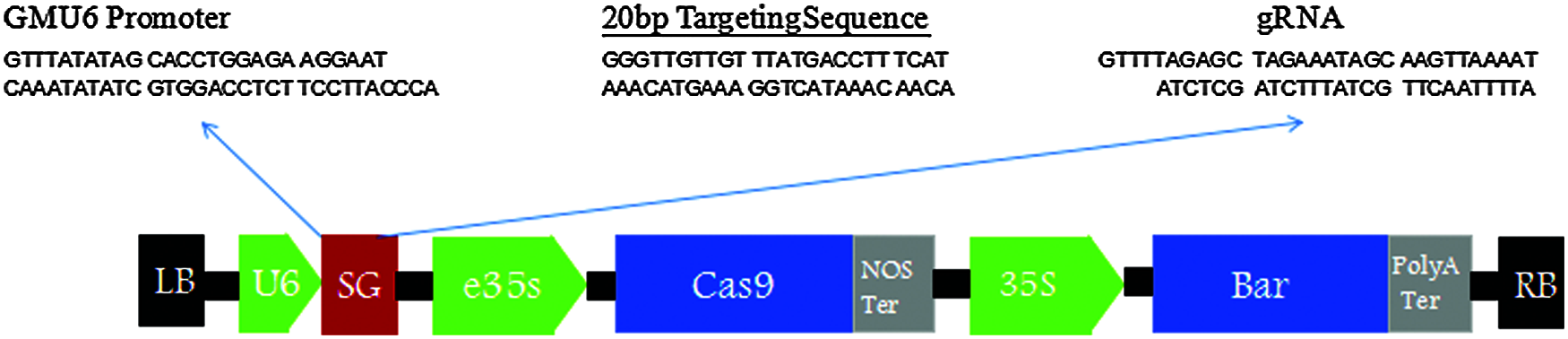
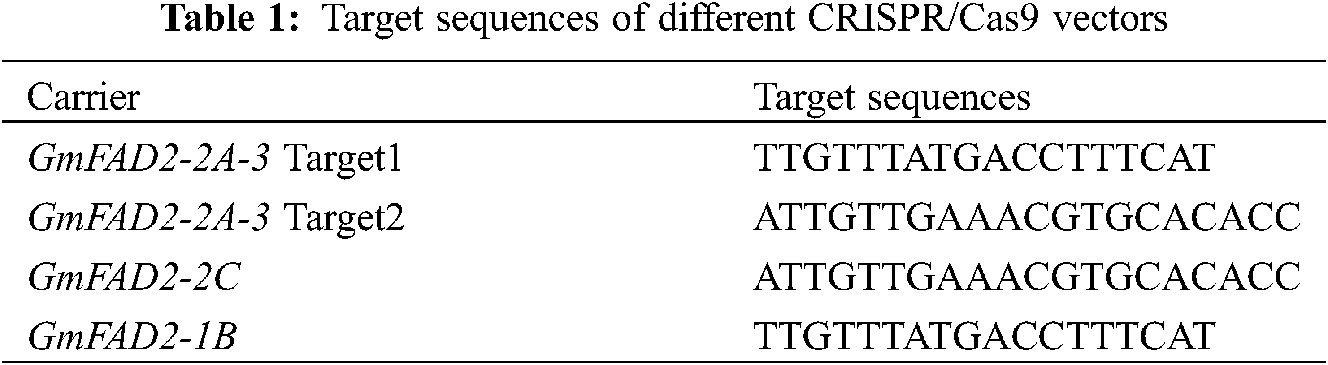
Figure 1: Vector structure diagram of CRISPR/Cas9-FAD2-1B
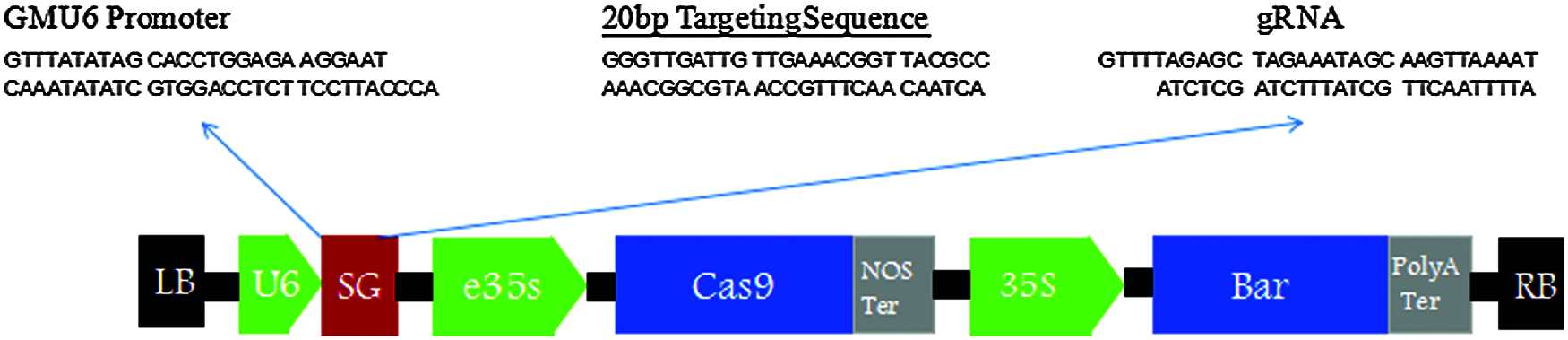
Figure 2: Vector structure diagram of CRISPR/Cas9-FAD2-2C
Figure 3: Vector structure diagram of CRISPR/Cas9-FAD2-2A&FAD2-3
Through Agrobacterium tumefaciens-mediated soybean cotyledon node, 9 transformed plants were obtained from the GmFAD2-1B single knockout vector, 7 transformed plants were found from the GmFAD2-2C single knockout vector, and 7 transformed plants were obtained from the GmFAD2-2A-3 double knockout vector (Fig. 4).

Figure 4: Agrobacterium-mediated soybean cotyledon node. (A) Germination culture, (B) Pre-culture, (C) Co-culture, (D) First screening culture, (E) Second screening culture, (F) Elongation culture, (G) Rooting culture, (H) Transplanting
3.3 Detection of Mutant Plants in the CRISPR/Cas9 System
The target fragments of 15 positive plants selected from different vectors of the T1 generation were sequenced randomly. The editing sites of the CRISPR/Cas9 system were detected and compared with the sequences of the unmodified plants. A total of 8 mutant plants (numbered L1–L8) were obtained (Fig. 5). All these plants were edited at the target, and the editing sites were different. The average mutation rate was 53.3%. The mutation type was base insertion, deletion and replacement (Fig. 5). The mutant plants of three double knockout vectors were edited at both targets, in which L1 inserted one base at both targets, L2 one base at target 1 and two bases at target 2. One base was missing at both targets in L3. Two single-gene GmFAD2-2C knockout vector mutant plants, L4 inserted a base; AA was replaced with TC in L5. Three mutant plants with single gene GmFAD2-1B knockout vector, two bases were replaced in the L6. Two bases were inserted in the L7, and one base in the L8 was missing (Fig. 5).
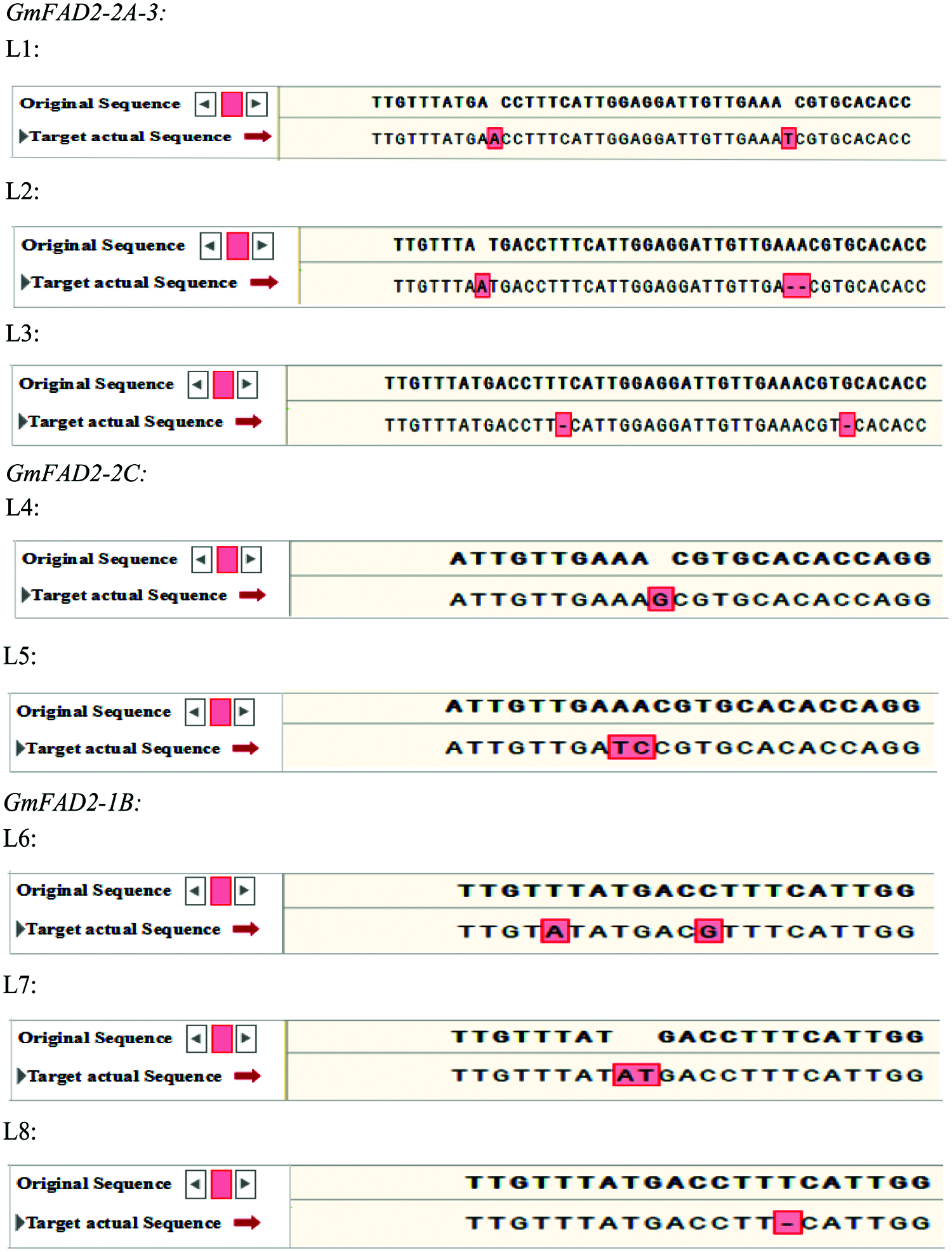
Figure 5: Sequencing results of each target
3.4 PCR Detection of Mutant Plants
To extract the genome of T2 and T3 mutants, untransformed strains, a DNA template was used based on the constructed CRISPR/Cas9 vector plasmid. The untransformed strain genome was controlled. The ddH2O negative was used to test for contamination. Bar screening marker genes and Cas9 genes were detected. PCR detection on Cas9 genes were done in T2 and T3 transformation plants. Specific bands were consistent with the location of the target fragment (screening marker Bar gene: 552 bp, Cas9 gene: 663 bp). There were no bands in the blank and negative water controls (Figs. 6 and 7). A transferred CRISPR/Cas9 vector was found in the transformed plants, and can stabilize heredity.
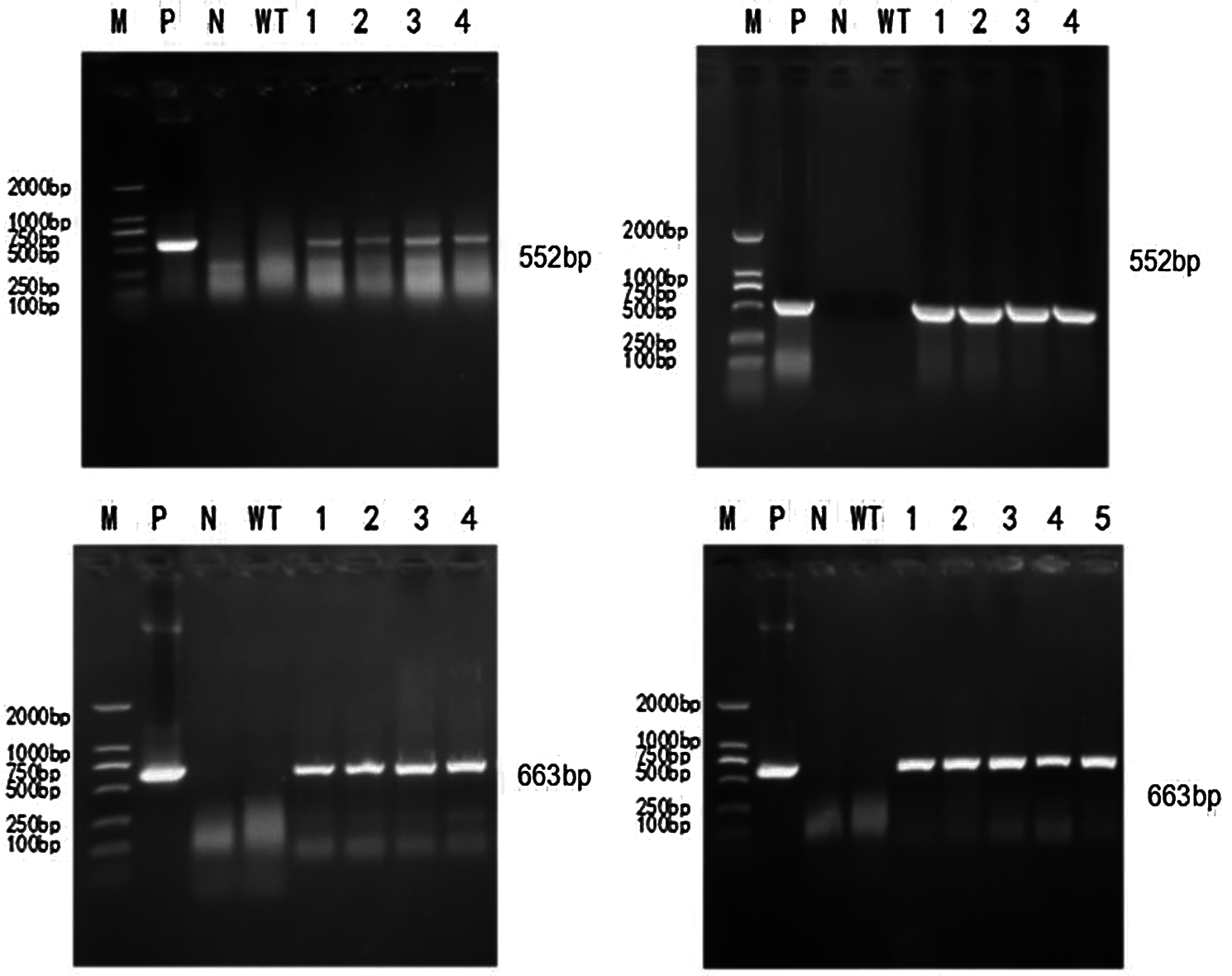
Figure 6: PCR detection of T2 generation transformed plants
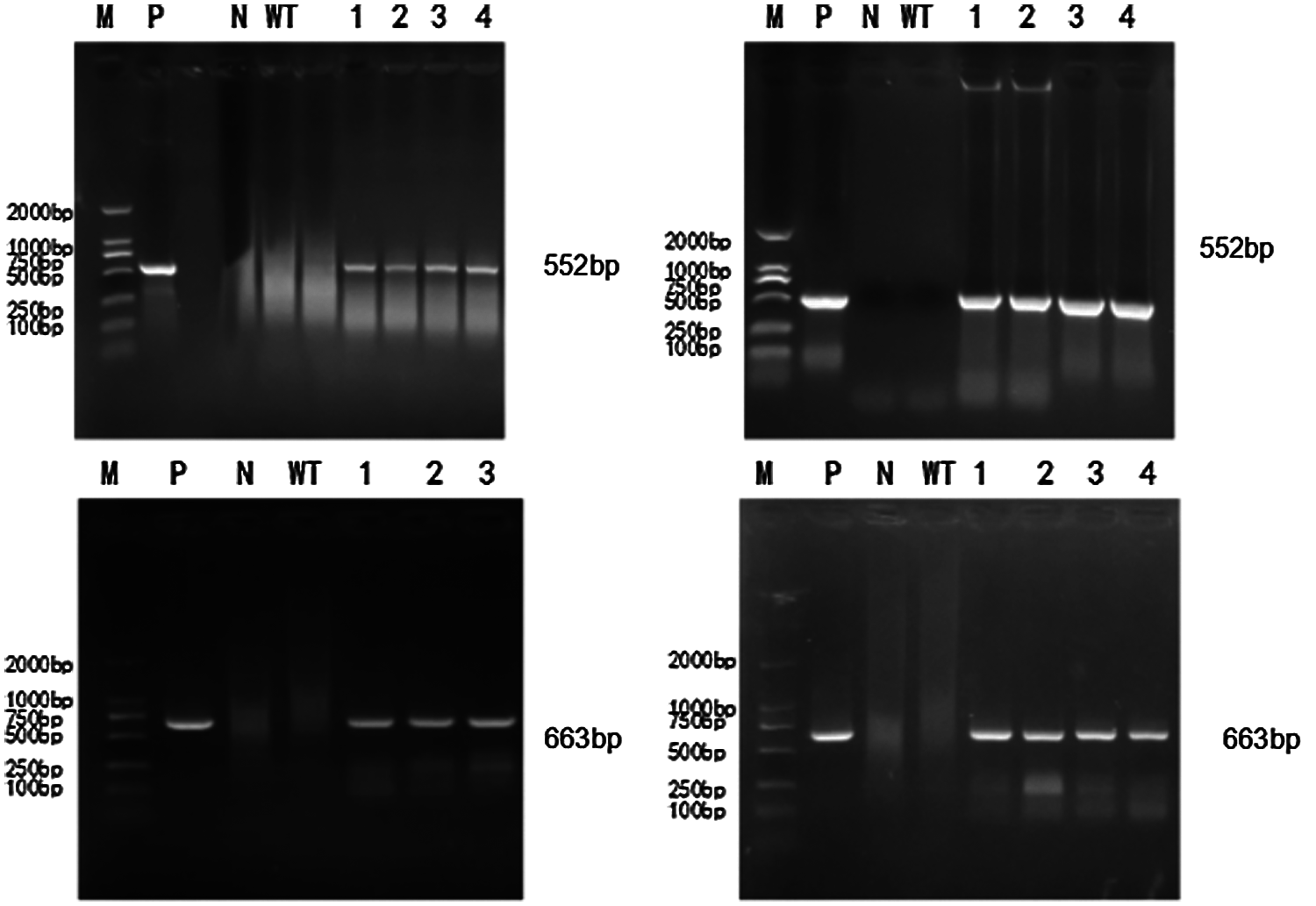
Figure 7: PCR detection of T3 generation transformed plants
In Figs. 6 and 7, M represents the DL2000 Marker, P represents the positive control, N represents water, WT represents wild-type soybean Jinong 38, 1–4 refers to all transformed plants, 552 bp refers to the screening Marker Bar gene detection of transformed plants, and 663 bp refers to the Cas9 gene detection of transformed plants.
3.5 Fluorescence Quantitative PCR Detection
The roots, stems and leaves of the T2 and T3 generation positive plants, and of the wild-type Jinong 38 soybean were obtained and transcribed into cDNA for fluorescence quantitative PCR detection. This was done to investigate the relative expression of GmFAD2-1B, GmFAD2-2C and GmFAD2-2A-3 genes in soybean receptors. The results showed that the T2 and T3 generation positive plants containing the GmFAD2-1B, GmFAD2-2C and GmFAD2-2A-3 genes were expressed in rhizomes and leaves. The T2 and T3 positive plants were significantly lower than those of the control varieties, and the highest expression was found in leaves(Figs. 8A and 8B). There were significant differences in the expression of different genes in the same tissues of soybean. In roots and stems, the expression of the GmFAD2-3 genes was the lowest. However, the expression of the GmFAD2-2C genes was the highest (Figs. 8A and 8B). In leaves, the expression of the GmFAD2-2A and GmFAD2-3 genes was the lowest whereas the expression of the GmFAD2-2C genes was the highest (Figs. 8A and 8B). The relative expression of the GmFAD2-3 gene was the lowest in rhizome and leaf, and that of the GmFAD2-2C gene in rhizome and leaf was the highest (Figs. 8A and 8B).

Figure 8: (A) T2 fluorescence quantification, (B) T3 fluorescence quantification
3.6 Determination of Fatty Acid Content in Gene Editors
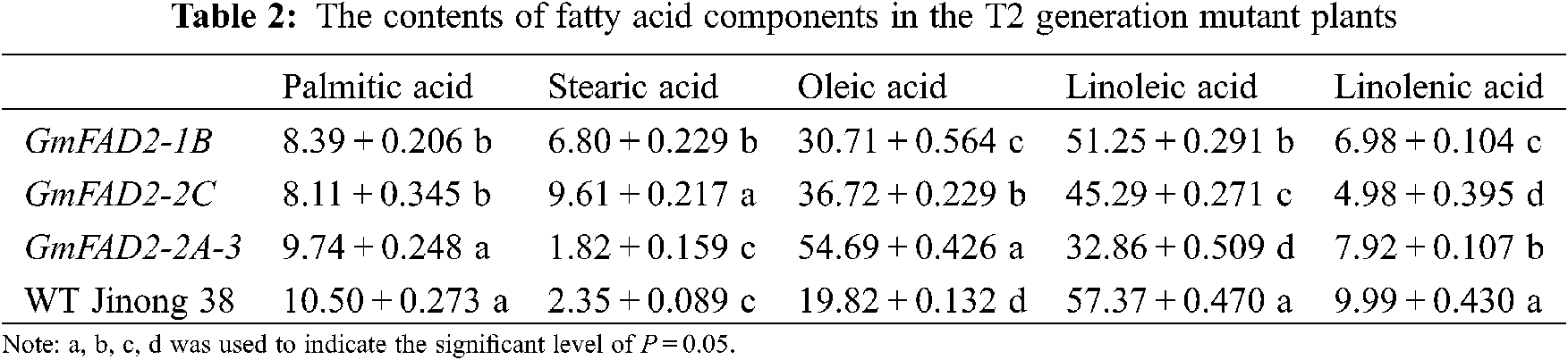
The T2 mutant plants showed significant differences in palmitic acid content between the knockout GmFAD2-1B and GmFAD2-2C genes and the knockout GmFAD2-2A-3 and wild-type Jinong 38 (Tab. 2). There were significant differences in stearic acid content among mutant GmFAD2-1B, GmFAD2-2C and knockout GmFAD2-2A-3 genes. There were significant differences between the knockout GmFAD2-2C genes and the wild-type Jinong 38. There were significant differences in oleic acid content among the mutants GmFAD2-1B, GmFAD2-2C, and the wild-type Jinong 38, and the knockout GmFAD2-2A-3 genes. There were significant differences in the content of linoleic acid in the mutant plants, GmFAD2-1B, GmFAD2-2C and the knockout GmFAD2-2A-3 genes. All of them were significantly different from the wild-type Jinong 38. There were significant differences in linolenic acid content among the mutant plants, GmFAD2-1B, GmFAD2-2C and the knockout GmFAD2-2A-3 genes, and all of them were significantly different from the wild-type Jinong 38.
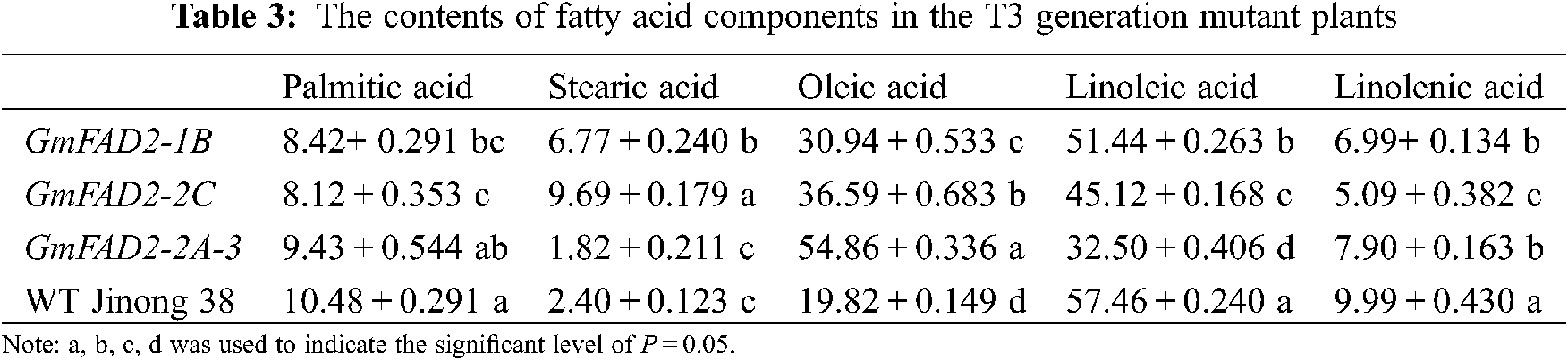
There were significant differences in the content of palmitic acid among the T3 mutant plants, GmFAD2-1B and GmFAD2-2C and the wild-type Jinong 38 (Tab. 3). There were significant differences in stearic acid content between the GmFAD2-1B and GmFAD2-2C genes in the mutant plants and the wild-type Jinong 38. There were significant differences in oleic acid content among the mutants GmFAD2-1B, GmFAD2-2C, wild-type Jinong 38, and the knockout GmFAD2-2A-3 genes. There were significant differences in the content of linoleic acid in the mutant plants, GmFAD2-1B and GmFAD2-2C and the knockout GmFAD2-2A-3 genes, all of them were significantly different from the wild-type Jinong 38. There were significant difference between the knockout GmFAD2-1B gene and GmFAD2-2A-3 gene and the knockout GmFAD2-2C gene in the content of linolenic acid in the mutant plants. All of them were significantly different from the wild-type Jinong 38.
3.7 Analysis of Agronomic Characters in the Gene-Editing Generation
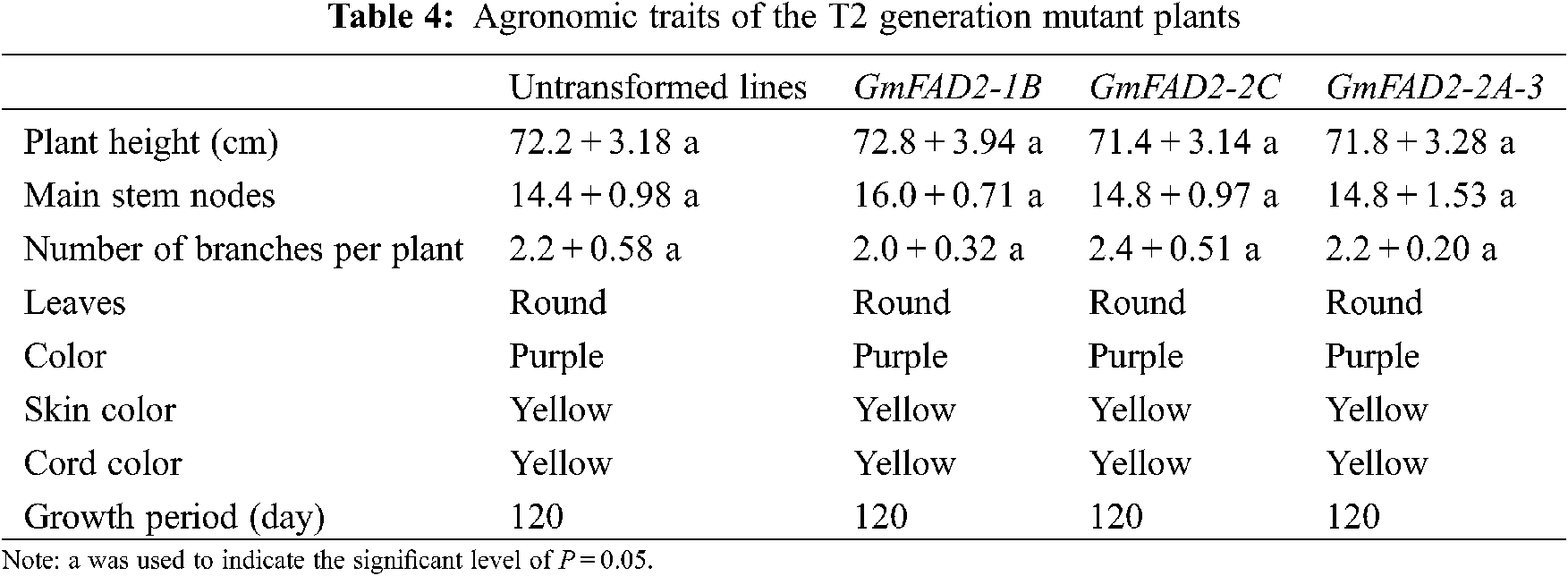
There was no significant differences in plant height, number of main stem nodes, number of branches, leaf shape, color, seed coat color, umbilical color and growth period between mutants GmFAD2-1B, GmFAD2-2C and GmFAD2-2A-3 genes (Tabs. 4 and 5). However; there was no significant difference between mutant lines. It was proved that the single target knockout and double target knockout of GmFAD2-1B and GmFAD2-2C genes by CRISPR/Cas9 gene editing technology did not affect the agronomic traits of mutant plants.

With the increase of population and improvement of living standard, the quality and demand of soybean products are increasing. So, improving soybean quality and increasing soybean oleic acid content is one of the main objectives of soybean breeding [22]. In a previous study William and his team knocked out two genes of the FAD2 gene family and reported genetically modified soybeans with an oleic acid content of up to 80% [16]. However, in recent years, using gene editing technology like CRISPR/Cas9 system, researchers are trying to enhance the content of oleic to linoleic acid ratio. Therefore, it is one of the hotspots in soybean research in China.
Wu and her research team in China constructed single and double knockout vectors of the FAD2 family functional genes using the CRISPR/Cas9 gene editing technology. They transferred the vector to the receptor Jinong 38 to determine its oleic acid content. They also reported that the oleic acid content was higher than that of the single gene vector after knockout of the double gene vector [23].
The soybean breeders mainly inhibited the expression of the FAD2 gene family by the RNAi and CRISPR/Cas9 gene editing technologies, and also reported the transfer of FAD2 antisense genes into oil crops through antisense inhibition technology to increase oleic acid content [8]. The antisense FAD2 gene was transferred into rape by the Agrobacterium tumefaciens mediated method and the obtained transgenic plants with the highest content of oleic acid (68.72%), which was 20% higher than that of the control. In contrast, the lowest content of linoleic acid (5.02%) was reported [24]. Liu and others used the RNAi technology to reduce the gene expression, oleic acid accumulation and linoleic acid content, and found increased oleic acid content from 18.18% to 58.92%, and linoleic acid content from 51.54% to 15.27% [25]. By using RNAi silencing the FAD2 gene in rape seed and reducing the expression of the FAD2 gene, the average oleic acid content in transgenic mature seeds (81.5%) higher than the control average (70.89%) [26]. The content of polyunsaturated fatty acids (linoleic and linolenic acids) were 5.25%, and 69.51%, respectively, that were lower than values on the control [26]. All these reports showed that, the degree of inhibition of gene expression of the FAD2 gene family was different and the increase of oleic acid content was also different, however, the content and the decrease of linoleic acid content were consistent. On the other hand, the genes of the FAD2 gene family can be expressed in many crops by inhibiting their expression. The oleic acid content can be increased and the linoleic acid content can be reduced.
In this study, 15 T1 positive plants were randomly selected to target mutation detection. The results showed that the average mutation rate was 53.3%, and the mutation types were base insertion, deletion and replacement (Fig. 5). The CRISPR/Cas9 gene editing techniques have been used in recent years to target genes of interest to increase the oleic acid content in soybean. A previous study in Jinong 38 cultivar by using Agrobacterium tumefaciens mediated-method to transfer the constructed FAD2-2 gene showed that the oleic acid concentration was as high as 65.9%, which was 48.56% higher than that on the control variety, and the linoleic acid concentration reported was decreased to 16.08% [27]. However, using the CRISPR/Cas9 technology to edit the soybean oleic acid gene GmFAD2-1A, stable homozygous soybean mutant materials were obtained [22]. Hagely et al. [22] reported that the oleic acid content of mutant plants was 23%, 15% higher than that of the control varieties, while the linoleic, palmitic and stearic acids were significantly reduced, and the linolenic acid did not change significantly. In the present study we found that the content of oleic acid in the T2 and T3 generations were different. The oleic acid concentrations that we obtained were 30.71%, 36.72%, 54.69% and 19.82% for GmFAD2-1B, GmFAD2-2C, GmFAD2-2A-3 and WT Jinong 38 respectively on the T2 generation (Tab. 2). The linoleic acid concentrations that we found were 51.25%, 45.29%, 32.86% and 57.37% for GmFAD2-1B, GmFAD2-2C, GmFAD2-2A-3 and WT Jinong 38 respectively for the T2 generation (Tab. 2). This Indicated that the oleic acid contents were increased as compared to the wild-type Jinong 38 and the linoleic acid was found to be decreased. Also was shown that CRISPR/Cas9 was the best tool to improve the oleic acid to linoleic acid ratio (O/L) in soybean. In addition, the present study found the oleic acid and the linoleic acid contents on the T3 generation. The T3 generation of oleic acid showed 30.94%, 36.59%, 54.86% and 19.82% for GmFAD2-1B, GmFAD2-2C, GmFAD2-2A-3 and WT Jinong 38, respectively (Tab. 3). The linoleic acid content we obtained was 51.44%, 45.12%, 32.50% and 57.46% for GmFAD2-1B, GmFAD2-2C, GmFAD2-2A-3 and WT Jinong 38 respectively on the T3 generation (Tab. 3). There were significant differences between the oleic acid content among the mutant GmFAD2-1B, GmFAD2-2C, wild-type Jinong 38, and knockout GmFAD2-2A-3 genes (Tabs. 2 and 3). There were significant differences in the content of linoleic acid in mutant plants, GmFAD2-1B, GmFAD2-2C and knockout GmFAD2-2A-3 genes (Tab. 2 and 3). This finding is in line with results in a previous study [23]. Agronomic traits were found consistent with those of control varieties, indicating that these traits on mutant plants were not affected by the site editing of FAD2 gene family genes. Other researchers [22,27] reported high oleic acid content and low linoleic acid content compared with the control varieties, which is in agreement with our findings (e.g., [22,27]). The morphological data obtained did not show any significant change. This might be due to the environmental factors or genetic factors. The oleic acid synthesis is greatly affected by the external environment or genetic factors, resulting in a hindered gene expression process and the oleic acid content is greatly affected [27,28].
The four genes in the FAD2 gene family were knocked out by single target and double target by the CRISPR/Cas9 gene editing technique, and the mutant plants were obtained by genetic transformation of Agrobacterium tumefaciens-mediated cotyledon node. The function of these four genes in the soybean FAD2 gene family was verified. By inhibiting the expression of genes in the FAD2 gene family, the contents of soybean oleic acid and linoleic acids could be increased. The expression of the GmFAD2-2C gene was stronger than that of the GmFAD2-1B gene. The expression of double gene was stronger than that of single gene. The FAD2 gene family genes can regulate the synthesis pathway of oleic acid, which provides a theoretical and practical basis for the cultivation of soybean with higher oleic acid contents in the future.
Acknowledgement: We would like to thank Jilin Agricultural University, Plant Biotechnology Center for facilitating the working environment to conduct the experiment.
Funding Statement: This work was supported by the National Natural Science Foundation of China Project Nos. [31771817, 31801381], National Key Research and Development Program [2019YFD1002603-1]. Furthermore, the funding body have no role in designing the study, collection and interpretation of data or writing up the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Silva, L., Matta, L., Pereira, G. R., Bueno, R. D., Dal-Bianco, M. (2021). Association studies and QTL mapping for soybean oil content and composition. Euphytica, 217(2). DOI 10.1007/s10681-020-02755-y. [Google Scholar] [CrossRef]
2. Sung, M., van, K., Lee, S., Nelson, R., Mian, M. (2021). Identification of SNP markers associated with soybean fatty acids contents by genome-wide association analyses. Molecular Breeding, 41(4). DOI 10.1007/s11032-021-01216-1. [Google Scholar] [CrossRef]
3. Kim, H. J., Ha, B. K., Ha, K. S., Chae, J. H., Park, J. H. et al. (2015). Comparison of a high oleic acid soybean line to cultivated cultivars for seed yield, protein and oil concentrations. Euphytica, 201(2285–292. DOI 10.1007/s10681-014-1210-5. [Google Scholar] [CrossRef]
4. Nan, H., Lu, S., Fang, C., Hou, Z., Yang, C. et al. (2020). Molecular breeding of a high oleic acid soybean line by integrating natural variations. Molecular Breeding, 40(9), 1–10. DOI 10.1007/s11032-020-01168-y. [Google Scholar] [CrossRef]
5. Bao, A., Zhang, C., Huang, Y., Chen, H., Cao, D. (2020). Genome editing technology and application in soybean improvement. Oil Crop Science, 5(1),31–40. DOI 10.1016/j.ocsci.2020.03.001. [Google Scholar] [CrossRef]
6. Lee, K. J., Lee, J. R., Shin, M. J., Cho, G. T., Ma, K. H. et al. (2019). Selection and molecular characterization of soybeans with high oleic acid from plant germplasm of genebank. Journal of Crop Science and Biotechnology, 22(4), 323–333. DOI 10.1007/s12892-018-0113-0. [Google Scholar] [CrossRef]
7. Xue, Y., Zhang, X., Wang, R., Chen, B., Jiang, J. et al. (2017). Cloning and expression of perilla frutescens FAD2 gene and polymorphism analysis among cultivars. Acta Physiologiae Plantarum, 39(3), 84. DOI 10.1007/s11738-017-2377-x. [Google Scholar] [CrossRef]
8. Ustun, R., Uzun, B. (2017). Breeding for introgression of FAD2-1A and FAD2-1B genes to local soybean cultivars of Turkey. Journal of Biotechnology, 256, S103. DOI 10.1016/j.jbiotec.2017.06.1151. [Google Scholar] [CrossRef]
9. Wang, L. M., Chen, C. Y., Tonnis, B., Pinnow, D., Davis, J. et al. (2018). Changes of seed weight, fatty acid composition, and oil and protein contents from different peanut FAD2 genotypes at different seed developmental and maturation stages. Journal of Agricultural and Food Chemistry, 66(14), 3658–3665. DOI 10.1021/acs.jafc.8b01238. [Google Scholar] [CrossRef]
10. Zhao, S., Li, A., Li, C., Xia, H., Zhao, C. et al. (2016). Development and application of KASP marker for high throughput detection of AHFAD2 mutation in peanut. Electronic Journal of Biotechnology, 25, 9–12. DOI 10.1016/j.ejbt.2016.10.010. [Google Scholar] [CrossRef]
11. Sturtevant, D., Horn, P., Kennedy, C., Hinze, L., Percy, R. et al. (2017). Lipid metabolites in seeds of diverse gossypium accessions: Molecular identification of a high oleic mutant allele. Planta, 245(3), 595–610. DOI 10.1007/s00425-016-2630-3. [Google Scholar] [CrossRef]
12. Lee, K. R., Lee, Y., Kim, E. H., Lee, S. B., Roh, K. H. et al. (2016). Functional identification of oleate 12-desaturase and ω-3 fatty acid desaturase genes from perilla frutescens var. frutescens. Plant Cell Reports, 35(12), 1–15. DOI 10.1007/s00299-016-2053-4. [Google Scholar] [CrossRef]
13. Ping, L., Wang, K., Zhou, C., Xie, Y., Yao, X. et al. (2018). Seed transcriptomics analysis in camellia oleifera uncovers genes associated with oil content and fatty acid composition. International Journal of Molecular Sciences, 19(1), 118. DOI 10.3390/ijms19010118. [Google Scholar] [CrossRef]
14. Combs, R., Bilyeu, K. (2019). Novel alleles of FAD2-1A induce high levels of oleic acid in soybean oil. Molecular Breeding, 39(6). DOI 10.1007/s11032-019-0972-9. [Google Scholar] [CrossRef]
15. Yang, J., Xing, G., Niu, L., He, H., Guo, D. et al. (2018). Improved oil quality in transgenic soybean seeds by RNAi-mediated knockdown of GmFAD2-1B. Transgenic Research, 27(2). DOI 10.1007/s11248-018-0063-4. [Google Scholar] [CrossRef]
16. Demorest, Z. L., Coffman, A., Baltes, N. J., Stoddard, T. J., Feng, Z. (2016). Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biology, 16(1), 225. DOI 10.1186/s12870-016-0906-1. [Google Scholar] [CrossRef]
17. Zhu, J., Ning, S., Sun, S., Yang, W., Zhao, H. et al. (2016). Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. Journal of Genetics and Genomics, 43(1), 25–36. DOI 10.1016/j.jgg.2015.10.006. [Google Scholar] [CrossRef]
18. Li, C., Hao, M., Wang, W., Wang, H., Fan, C. et al. (2018). An efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Frontiers in Plant Ence, 9, 442. DOI 10.3389/fpls.2018.00442. [Google Scholar] [CrossRef]
19. Du, H., Zeng, X., Meng, Z., Cui, X., Yu, D. (2016). Efficient targeted mutagenesis in soybean by talens and CRISPR/Cas9. Journal of Biotechnology, 217, 90–97. DOI 10.1016/j.jbiotec.2015.11.005. [Google Scholar] [CrossRef]
20. Beying, N., Schmidt, C., Pacher, M., Houben, A., Puchta, H. (2020). CRISPR–Cas9-mediated induction of heritable chromosomal translocations in arabidopsis. Nature Plants, 6(6), 1–8. DOI 10.1038/s41477-020-0663-x. [Google Scholar] [CrossRef]
21. Li, H., Li, X. F., Xu, Y., Liu, H. L. (2020). High-efficiency reduction of rice amylose content via CRISPR/Cas9-mediated base editing. Rice Science, 27(6), 4–7. DOI 10.1016/j.rsci.2020.09.001. [Google Scholar] [CrossRef]
22. Hagely, K., Konda, A. R., Kim, J. H., Cahoon, E. B., Bilyeu, K. (2021). Molecular-assisted breeding for soybean with high oleic/low linolenic acid and elevated vitamin e in the seed oil. Molecular Breeding, 41(1), 1–13. DOI 10.1007/s11032-020-01184-y. [Google Scholar] [CrossRef]
23. Wu, N., Lu, Q., Wang, P. W., Zhang, Q., Zhang, J. et al. (2020). Construction and analysis of GmFAD2-1A and GmFAD2-2A soybean fatty acid desaturase mutants based on CRISPR/Cas9 technology. International Journal of Molecular Sciences, 21(3), 1104. DOI 10.3390/ijms21031104. [Google Scholar] [CrossRef]
24. Lee, K. R., Kim, E. H., Roh, K. H., Kim, J. B., Kang, H. C. et al. (2016). High-oleic oilseed rapes developed with seed-specific suppression of FAD2 gene expression. Applied Biological Chemistry, 59(4), 669–676. DOI 10.1007/s13765-016-0208-1. [Google Scholar] [CrossRef]
25. Liu, F., Zhao, Y. P., Zhu, H. G., Zhu, Q. H., Sun, J. (2017). Simultaneous silencing of GhFAD2-1 and GhFATB enhances the quality of cottonseed oil with high oleic acid. Journal of Plant Physiology, 215, 132–139. DOI 10.1016/j.jplph.2017.06.001. [Google Scholar] [CrossRef]
26. Chen, Y., Zhou, X. R., Zhang, Z. J., Dribnenki, P., Singh, S. et al. (2015). Development of high oleic oil crop platform in flax through RNAi-mediated multiple FAD2 gene silencing. Plant Cell Reports, 34(4), 643–653. DOI 10.1007/s00299-015-1737-5. [Google Scholar] [CrossRef]
27. Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T. et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2–2 microsomal omega-6 desaturase in soybean (Glycine max. L). BMC Biotechnology, 19(1). DOI 10.1186/s12896-019-0501-2. [Google Scholar] [CrossRef]
28. Dar, A. A., Choudhury, A. R., Kancharla, P. K., Neelakantan, A. (2017). The FAD2 gene in plants: Occurrence, regulation, and role. Frontiers in Plant Science, 8, 1789. DOI 10.3389/fpls.2017.01789. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |