

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.016951
ARTICLE
Genome-Wide Identification, Expression Profiling and Protein-Protein Interaction Properties of the BEL-Like Homeodomain Gene Family in Apple
1Shandong Institute of Pomology, Tai’an, 271000, China
2College of Horticulture, Qingdao Agricultural University, Qingdao, 266109, China
3College of Horticulture, Northwest A and F University, Yangling, 712100, China
4College of Plant Protection, Nanjing Agricultural University, Nanjing, 210095, China
*Corresponding Authors: Qinglong Dong. Email: dong19850412@163.com; Yi Xu. Email: xuyiqdpd@njau.edu.cn
Received: 13 April 2021; Accepted: 09 June 2021
Abstract: BEL1-like homeodomain (BLH) family proteins are homeodomain transcription factors, which are found ubiquitously in plants and play important roles in regulating meristem and flower development. Although BLH proteins have been reported in some plant species, there is very little information available for plants in the Malus genus (e.g., apple tree:Malus domestica). In the present study, we identified 19 apple MdBLH genes. Phylogenetic analysis revealed that the MdBLH genes could be divided into five groups. Analysis of gene structure showed that MdBLH gene has four exons, and the third exon was 61 bp in length. Chromosomal location analysis suggested that the MdBLH genes are not distributed uniformly on 12 chromosomes. Eleven MdBLH genes were cloned by RT-PCR, and their expression patterns were also determined. Among them, the expression levels of MdBLH4.1 and MdBLH9.1 could be induced by sodium chloride stress, while the expression levels of MdATH1.1, MdBLH8.1, MdBLH8.3, and MdBLH11.1 were down-regulated by such stress. Transcriptional levels of MdATH1.1 and MdBLH7.2 were down-regulated by mannitol stress. The result of yeast two-hybrid experiment showed that MdBEL1.1 interacted with apple ovate family proteins 6 (MdOFP6), and MdBLH3.1 interacted with the MdOFP4, MdOFP6, MdOFP13, and MdOFP16 proteins. Our results provide a strong theoretical basis and a valuable reference for analyzing of the biological functions of MdBLH proteins as transcription factors in apple growth, development, and stress and also for the construction of regulatory networks.
Keywords: Apple; BEL-like homeodomain; gene cloning; expression analysis; interaction analysis
The homeobox gene is a transcriptional regulator that encodes the homeodomain (HD), which is widely found in eukaryotes. The typical HD consists of 60 amino acids, which can form a 3-helical domain in space [1–3]. However, there are some exceptions, such as the TALE (Three Amino-acid Loop Extension) gene families, which encode an atypical DNA binding domain consisting of 63 amino acids, with three additional amino acid residues (P-Y-P) inserted between the first and second helices [3]. In plants, the TALE gene families include two gene subfamilies, which are known as homeodomain) and BLH, which is sometimes called BEL (BEL1-like homeodomain) [4]. The BLH protein consists of three domains: SKY, BEL, and homeobox KN. The SKY domain is a conserved region of about 17 amino acids at the N-terminal of the BEL protein [5]. The homeobox KN domain, which is located at the C-terminus of the BLH protein, is a typical homologous box region. It is a 60 amino acid helix-turn-helix DNA-binding domain involved in DNA binding and the formation of homologous protein dimers. Besides the 60 amino acids, there are three extra amino acids (P-Y-P) between the first and second helix [4]. The BEL domain is located between the SKY and the homeobox KN domains [4]. These three conserved domains are of great significance for the functioning of the BLH family proteins in plants. The SKY and BEL domains of the BLH proteins interact with the MEINOX domain of the KNOX proteins to form the BLH-KNOX heterodimer, which can be transferred from the cytoplasm to the nucleus. In the nucleus, their HD domains specifically bind to their respective target sequences to regulate the expression of downstream genes [4–7]. In addition, ovate family proteins (OFP) negatively regulate the complex formation of BLH-KNOX heterodimers, which results in relocating the heterodimers from the nucleus to the cytoplasm [8].
There are 13 BLH family genes in Arabidopsis thaliana, which play important roles in the development of plant meristems and flowers. For example, the BLH family members PENNYWISE (PNY/BLH9), POUNDFOOLISH (PNF/BLH8), and ARABIDOPSIS THALIANA HOMEOBOX 1 (ATH1) play important roles in the initiation, maintenance, and development of apical meristems and inflorescence formation [9–13]. SAWTOOTH1 (SAW1/BLH2) and SAWTOOTH2 (SAW2/BLH4) negatively regulate expression of the Class I KNOX gene BREVIRUELLUS (BP), which leads to leaf development defects [14]. The abnormal expression of BLH1 affects the establishment of mature embryo sac cells fate [15]. Integument was transformed into carpel structure in the bel1 mutant without BEL1 function [16]. In addition, SlBEL11 regulates the development of chloroplasts and chlorophyll synthesis in tomato fruits [17]. RI and RIL1 genes of rice BLH regulate the maintenance of the inflorescence structure and meristems [18].
At present, most of the studies on BLH family genes are focused on Arabidopsis, while the related researches on apple (Malus domestica) BLH have not been reported. As one of the most important economic fruits grown in temperate regions worldwide, apples are deeply loved by people and used for cooking, fresh eating, and cider production. The improved draft of apple genomes provides an excellent platform for identifying and analyzing the functions of the BLH transcription factor family [19–21]. In this study, the high-quality ‘GDDH13 v1.1’ apple reference genome database was used to investigate and identify the BLH gene family. We analyzed conserved domains, evolutionary relationships, subgroup classification, gene structures, and chromosomal locations of the MdBLH family. In addition, we also examined gene cloning, stress and tissue expression, and protein interaction. These results not only provide a strong theoretical basis and reference for analyzing the biological function of the MdBLH transcription factor in apple growth, development and stress, but also provide a strong theoretical basis for the construction of regulatory networks.
2.1 Plant Materials and Stress Treatments
‘Gala’ (Malus × domestica cv. Gala) tissue culture seedlings were used as test material in the expression analysis under stress treatments. Under 14-h light/10-h darkness (24 ± 2)°C, ‘Gala’ tissue culture seedlings were grown on a subculture medium (MS medium + 0.2 mg·L−1 IAA+ 0.8 mg·L−1 6-BA+30 g·L−1 sucrose + 7 g·L−1 agrose) and subcultured every 30 d; 150 mM·L−1 NaCl and 300 mM·L−1 mannitol were added to the subculture medium. ‘Gala’ tissue culture seedlings with the same growth status (about 20 d after the subculture) were inoculated in the subculture media with NaCl and mannitol as treatments, and the basic subculture medium was used as a control [22].
2.2 Identification of MdBLH Genes in Apple
Thirteen BLH amino acid sequences of Arabidopsis were downloaded from the Arabidopsis website [4,23]. We searched the apple genome functional database (https://www.rosaceae.org/species/malus/malus_x_domestica/genome_GDDH13_v1.1) using the BlastP program provided by the GDR database (https://www.rosaceae.org/blast/protein/protein); Protein BLAST Databases: [Apple Genome (GDDH13 V1.1) proteins], and then the nucleotide and amino acid sequence of the candidate genes were downloaded. The Pfam database (http://pfam.xfam.org/) and NCBI Conserved Domains (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to ensure that the above candidate genes contained the SKY, BEL and HD domains [22].
2.3 Analysis of Evolutionary Relationships, Gene Structure, Conserved Motifs, and Genome Location of MdBLH in Apple
Sequence analysis of MdBLH protein in apple was conducted by using the DNAMAN 6.0 software, and then evolution analysis was conducted by using the phylogenetic tree analysis software MEGA 6 (http://www.megasoftware.net) based on the NJ method (execution parameters: Poission correction, pairwise deletion and bootstrap (1000 repeats)) [24]. Structures of MdBLH and Arabidopsis BLH genes were downloaded from the PLAZA 3.0 comparative genome database (http://bioinformatics.psb.ugent.be/plaza/versions/plaza_v3_dicots/). The genomic location information for the MdBLH genes was obtained from the apple genome ‘GDDH13 V1.1’ database. The MdBLH genome localization was made using the MapInspect software (http://www.plant-breeding.wur.nl/UK/software_mapinspect.html). Tandem and fragment repeats were defined according to the method of Tian et al. [25]. MdBLH conserved motifs were identified using the online software MEME (http://meme-suite.org/tools/meme). Cis-elements in the MdBLH promoter were analyzed using the online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Supplementary Sequence: A1) [22].
RNA from fully expanded leaves of ‘Zihong Fuji’ (Malus domestica cv. Zihong Fuji) was extracted by a modified hot boric acid method, and then synthesized into cDNA using PrimeScriptTM1ST Strand cDNA Synthesis Kit (TaRaKa Company, Dalian, China) [22]. Specific primers (Tab. 1) were designed according to the MdBLH nucleotide sequence in the apple ‘GDDH13 V1.1’ genomic database for PCR amplification. The PCR reaction conditions were as follows: predenaturation at 94°C for 5 min; 94°C during 60 s, 56–62°C during 60 s, 72°C during 120 s; 35 cycles and elongation at 72°C for 10 min. PCR products were purified and cloned into the pMD18-T vector, recombinant plasmids were transformed into E. coli DH5α competent cells, and the positive clones were screened and sequenced (Supplementary Sequence: A2).
2.5 Expression Analysis of MdBLH Genes
The tissue expression data for MdBLH genes was obtained using the GEO database in NCBI (https://www.ncbi.nlm.nih.gov/) with GEO accession no. GSE42873 [26]. These data included a set of expression arrays from 16 different apple tissues, with two biological replicates for each tissue. RNA in leaves of ‘Gala’ apple was extracted by RNeasy Plant Mini Kit (QIAGEN, China, Item No. 74903) and then cDNA was synthesized by using the reverse transcription system PrimeScriptTM 1ST Strand cDNA Synthesis Kit. Primers for qRT-PCR analysis were designed based on the 3 ‘-UTR or 5’ -UTR of the MdBLH gene (Tab. 1). MdMDH was selected as the internal reference gene [27], and qRT-PCR was performed using the 3-step method in BIO-RAD IQ5 (USA). All qRT-PCR reactions were repeated three times. MdSOS2 and MdSUT2, as control genes, were demonstrated to respond to NaCl stress and osmotic stress, respectively [22,28,29]. The PCR reaction system was as follows: SYBR Green Master I 10 μL, upstream and downstream primers (5 μmol·L−1) 1 μL each, template 1 μL, and ddH2O was added to obtain a total of 20 μL. The qRT-PCR conditions were 95°C for 3 min, then 40 cycles at 95°C for 10 s, 58°C for 30 s, 72°C for 15 s; after annealing to 55°C, temperature was increased 0.5°C every 7 s until 95°C, 81 cycles in total [22]. The 2−ΔΔCT method was used to analyze the experimental data [30].
The Y2H experiment was carried out based on the Matchmaker® Gold Yeast Two-Hybrid System (Clontech, Code No. 630489). Full-length cDNAs of MdBLH were amplified with designed primers (Supplementary File 3) and recombined with the pGAD424 (GAL4 activation domain, AD) vector. The amplified full-length cDNAs of MdOFP were recombined with the pGBT9 (GAL4 DNA-binding domain, BD) vector [22]. The recombinant plasmids were transformed into the yeast strain Y2HGold in different combinations and then coated in the screening medium SD/-Trp/-Leu for inverted cultivation at 30°C. Three days later, screening was conducted on a SD/-Trp/-Leu/-His/-Ade+X-α-Gal medium. Finally, the β-galactosidase activity was surveyed to represent the expression of the LacZ reporter gene. The empty pGAD424 plasmid together with the pGBT9-MdOFP co-transformed yeast were used as the negative control, and the pGAD424-MdVQ10 and PGBT9-MDWRKY52 co-transformed yeasts were used as the positive control (Tab. 1; Supplementary Sequence: A3) [31].
3.1 Identification and Evolutionary Analysis of the Apple MdBLH Transcription Factor
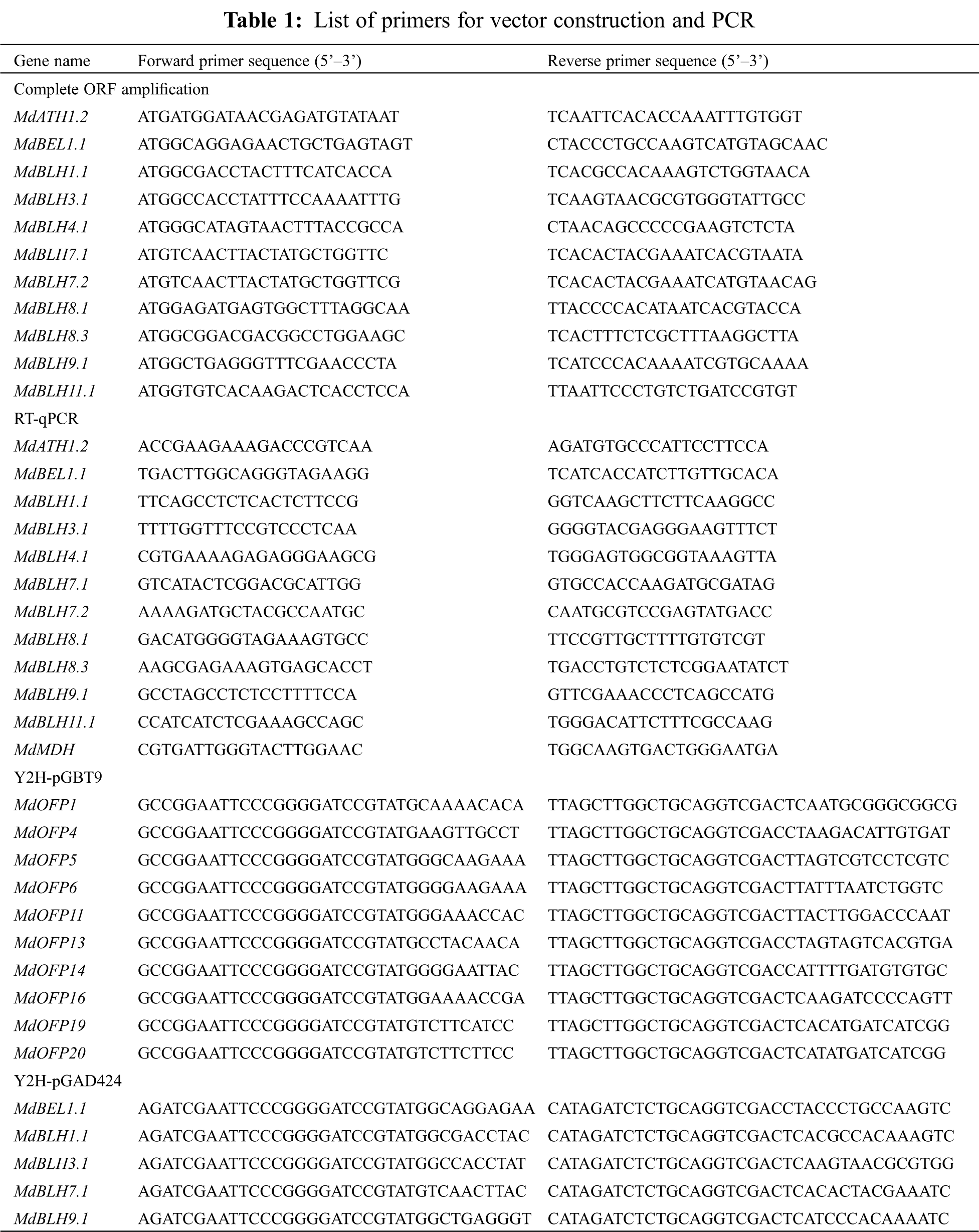
A total of 24 MdBLH-related genes were searched in the ‘GDDH13’ database for studying the apple genome (Tab. 2). The SKY, BEL, and HD domains of the candidate protein were predicted through using the Pfam database and the NCBI Conserved Domains. Five candidate MdBLH genes were found to be incomplete, and 19 candidate MdBLH genes were identified with full information for further investigation.
MEGA 6 software was used to analyze the evolution of the full-length sequences of the BLH proteins in apple (19 sequences) and Arabidopsis (13 known sequences). We found that BLH proteins from apple and Arabidopsis could be classified into five categories (Fig. 1A) [5].
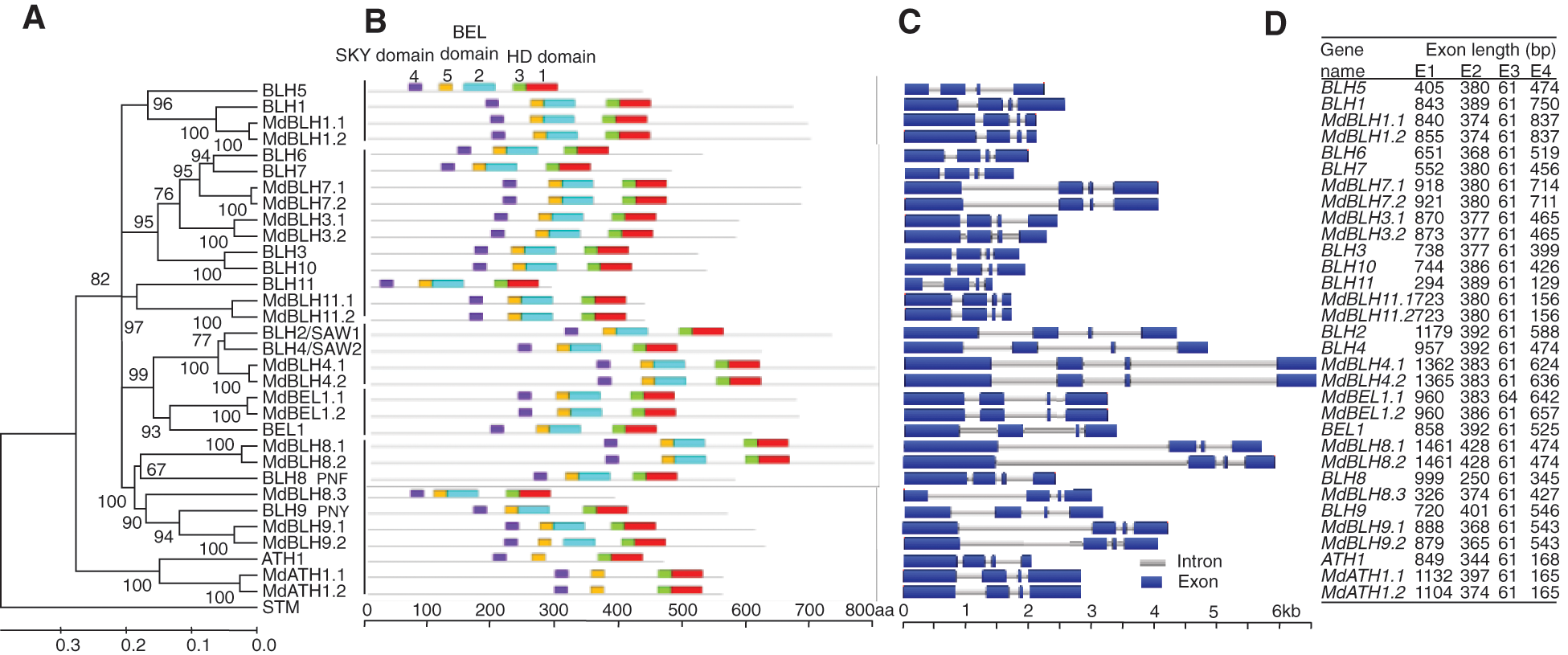
Figure 1: Analyses of phylogenetic relationships, conserved motifs, gene structures, and exon lengths of BLH genes in apple and Arabidopsis (A) The phylogenetic tree was constructed with MEGA 6 software using full-length amino acid sequences from the 33 BLH and STM proteins of apple and Arabidopsis. (B) Conserved motifs of apple and Arabidopsis BLH proteins that were identified using the MEME program. Motifs 1-5 are indicated by different colored boxes. Motifs 1 and 3: HD domain; Motifs 2 and 5: BEL domain; Motif 4: SKY domain. (C) The exon-intron structure of apple and Arabidopsis BLH genes. Introns and exons are represented by grey lines and blue boxes, respectively. (D) Analysis of exon length distribution of apple and Arabidopsis BLH genes
3.2 Analyses of Conserved Motifs and Gene Structure of the MdBLH Transcription Factor in Apple
Conserved motifs in the BLH gene family of apple and Arabidopsis were identified using online software MEME. Motif 4 represents the SKY domain, motifs 2 and 5 represent the BEL domains, and motifs 1 and 3 represent the HD domain (Fig. 1B). BLH gene family members in both apple and Arabidopsis contained the SKY, BEL, and HD conserved domains (Fig. 1B).
PLAZA3.0 comparative genome database was used to download structure maps of introns and exons of BLH genes in apple and Arabidopsis (Fig. 1C). BLH genes from both apple and Arabidopsis were composed of four exons, and the length of the third exon was 61 bp (31/32) (Figs. 1C and 1D).
(A) The phylogenetic tree was constructed with MEGA 6 software using full-length amino acid sequences from the 33 BLH and STM proteins of apple and Arabidopsis. (B) Conserved motifs of apple and Arabidopsis BLH proteins that were identified using the MEME program. Motifs 1–5 are indicated by different colored boxes. Motifs 1 and 3: HD domain; Motifs 2 and 5: BEL domain; Motif 4: SKY domain. (C) The exon-intron structure of apple and Arabidopsis BLH genes. Introns and exons are represented by grey lines and blue boxes, respectively. (D) Analysis of exon length distribution of apple and Arabidopsis BLH genes.
3.3 Analysis of Chromosome Localization of the Apple MdBLH Transcription Factor
The apple genome ‘GDDH13’ database was used to query related chromosome location information for the apple MdBLH gene family, and MapInspect software was used to map MdBLH genes on chromosomes. Nineteen apple MdBLH genes were mapped onto 12 chromosomes with inhomogeneity (Fig. 2). Chromosome 15 had the maximum with four MdBLH genes, Chromosome 8 followed with three, and Chromosomes 3, 4, 5, 6, 7, 10, 11, and 17 had the minimum of 1. Meanwhile, to elucidate the reasons for the expansion of MdBLH gene family, we examined gene replication events, which included tandem and segmental replication; MdATH1.1/MdATH1.2, MdBLH1.1/MdBLH1.2, MdBLH3.1/MdBLH3.2, MdBLH4.1/MdBLH4.2, MdBLH7.1/MdBLH7.2, MdBLH8.1/MdBLH8.2, MdBLH9.1/MdBLH9.2, and MdBLH11.1/MdBLH11.2 undergone segmental replication (Fig. 2).

Figure 2: Chromosomal locations of apple MdBLH genes
The scale is in megabases (Mb). The red font group represents segmental duplication. The grey area represents genome-wide duplications.
3.4 Cloning and Sequence Analysis of the Apple MdBLH Gene from Cultivar ‘Zihong Fuji’
Nineteen MdBLH genes were observed in apple based on genome-wide identification, and 11 of them were cloned using RT-PCR technology. Comparison of homologies showed that 11 deduced amino acid sequences of cloned MdBLH proteins contained SKY, BEL, and HD domains (Fig. 3).

Figure 3: Comparison of homologies of the deduced amino acid sequence alignment of cloned MdBLH proteins
Black-highlighted residues are identical, and light gray-highlighted residues are similar in all proteins. Three conserved domains and one box were marked as indicated as SKY domain, BEL domain, HD domain, and VSLTLGL box.
The promoter sequences of 11 MdBLH genes were obtained by downloading the upstream 1500 bp sequences to the transcriptional initiation site of the 11 cloned MdBLH genes. The cis-acting elements of promoter sequences were analyzed by the PlantCARE database. In addition to the light response element, there were 12 different types of cis-acting elements, such as defense and stress responsiveness element, hormone responsiveness element, and so on (Fig. 4). These cis-acting elements responded to stresses such as anaerobic induction, low temperature, heat, drought, pathogen infections and hormones (such as MeJA, salicylic acid, auxin, gibberellin, ethylene and abscisic acid) (Fig. 4). That is, these cis-acting elements in the MdBLH promoters might play important roles in apple growth and development and the response to stress.

Figure 4: The cis-acting elements of 11 promoters in MdBLH genes
Different colored triangles represent different cis-acting elements. Bright green: Defense and stress responsiveness; Red: Fungal elicitor responsive element; Water green: Auxin-responsive element; Blue: MYB binding site involved in drought-inducibility; Crimson: wound-responsive element; Light blue: MeJA-responsiveness; Yellow: Heat stress responsiveness; Gray: Anaerobic induction element; Orange: Abscisic acid responsiveness; Bright purple: Low-temperature responsiveness; Green: Gibberellin-responsive element; Purple: Salicylic acid responsiveness; Black: Ethylene-responsive element.
3.5 Expression Analysis of BLH Transcription Factor in Apple
The transcription profiles of 16 different apple tissues (GSE42873) were downloaded from the GEO database from the NCBI website to detect the expression of MdBLH in different tissues (Fig. 5). We found that the MdBLH genes were expressed in all the tested tissues, and MdBLH1.1 and MdBLH7.1 showed relatively high expression levels in Leaf M49, Flower M74, Fruit M20 (100DAM), and Fruit M74 (harvest) tissues (Fig. 5).
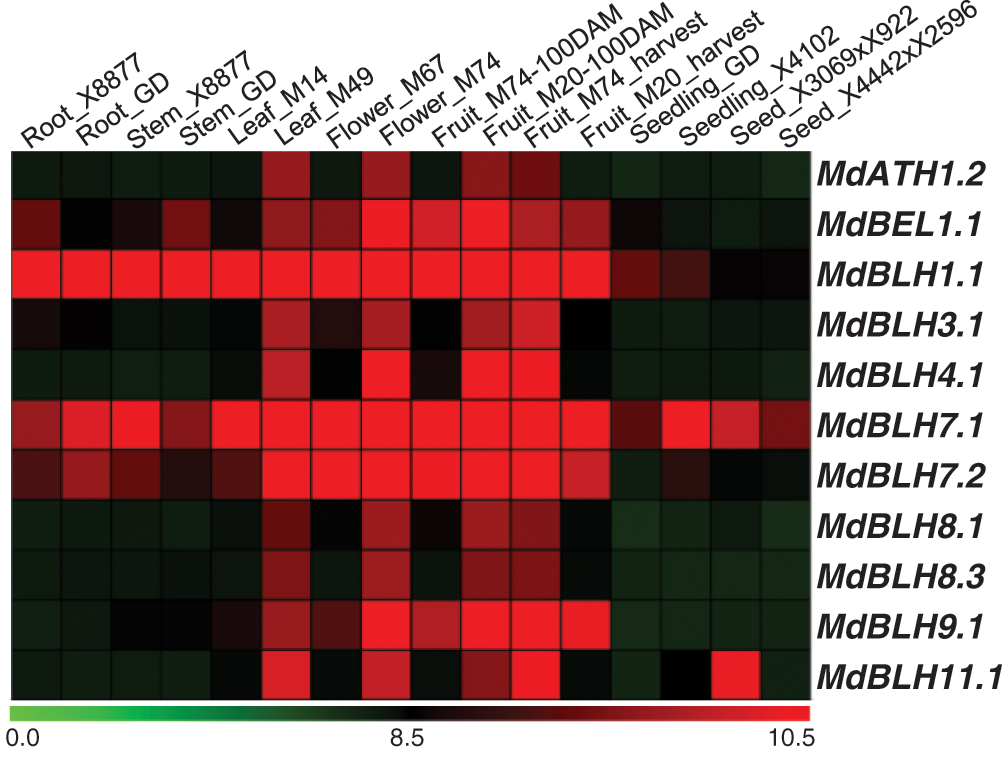
Figure 5: Expression profiles of MdBLH genes in various tissues in apple
The expression levels of the MdBLH genes in ‘Gala’ tissues from cultured seedlings under salt or mannitol treatments were analyzed further using qRT-PCR. Under the control treatment, the relative expression levels of MdBLH had no significant change within a 48 h period (Fig. 6A). Compared with the control group, the relative expression levels of MdBLH4.1 and MdBLH9.1 increased under sodium chloride treatment, reaching 3.5 and 2.3 times that of the control group at 48 h, respectively. Compared with the control group, the relative expression levels of MDBLH1.1, MdBLH8.1, MdBLH8.3, and MdBLH11.1 significantly decreased, and the relative expression level of MdBLH8.3 at 48 h was only 0.13 times of that of the control group (Fig. 6B). Compared with the control group, the relative expression levels of MdATH1.1 and MdBLH7.2 decreased after the mannitol treatment, and decreased to 0.41 and 0.31 times of the control group at 24 h, respectively (Fig. 6C).

Figure 6: Expression analysis of MdBLHs in apple under (A) normal growth, (B) salt, and (C) mannitol treatments. Three independent biological replicates were used for calculations. Error bars indicate standard deviation. * and ** indicate statistically significant differences, as determined by Student’s t-tests, at p < 0.05 and p < 0.01, respectively
3.6 Interactions between MdBLH and MdOFP Proteins
To investigate the interaction regulatory network of MdBLH with OFP proteins, a Y2H assay was performed to detect the interactions between MdBLH and MdOFP proteins. Five MdBLH full-length cDNA sequences were fused into the AD prey vector, and 10 MdOFP full-length cDNA sequences were fused into the BD bait vector. The fusion vectors of AD-MdBLH and BD-MdOFP were co-transformed into yeast cells, and the activity of β-galactosidase was tested by observing the expression of the LacZ reporter gene. The 10 BD-MdOFP fusion bait vectors, and the empty AD trap vectors, did not turn blue on a SD/-Trp/-Leu/-His/-Ade/X-α-gal minimal medium, which indicated that the 10 MdOFP proteins had no transcriptional self-activation activity (Fig. 7). But the co-transformed yeast cells of AD-MdBEL1.1 and BD-MdOFP6 and those of AD-MdBLH3.1 and BD-MdOFP4/MdOFP6/MdOFP13/MdOFP16 turned blue on the SD/-Trp/-Leu/-His/-Ade/X-α-gal minimal medium. These results indicated that the MdBEL1.1 protein interacts with the MdOFP6 protein, and the MdBLH3.1 protein interacts with the MdOFP4, MdOFP6, MdOFP13, and MdOFP16 proteins (Fig. 7).
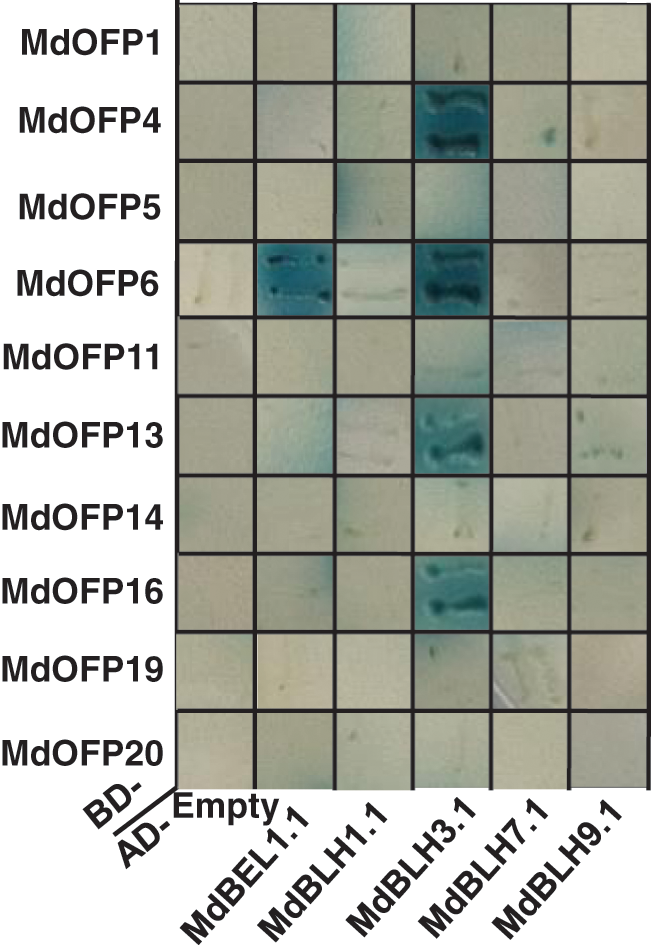
Figure 7: Interactions between MdBLH and MdOFP proteins in yeast cells
The AD-MdBLH fusion capture vectors were co-transformed with the BD-MdOFP fusion bait vectors into yeast cells. Positive interactions were indicated by the ability of cells to grow on synthetic dropout medium additive x-α-gal that lacked Leu, Trp, His, and Ade. The empty AD vector plus BD-MdOFP fusion vectors were used as negative controls.
In recent years, the technology of sequencing and sequence assembly has been continuously improved and innovated, and the cost of next generation sequencing has dropped rapidly. Genome maps of high quality can be drafted for a variety of plants, which is very convenient for investigating specific agronomic traits of plants, accelerating plant breeding, increasing production, and improving resistance to abiotic and biotic stresses. We have now a deeper understanding of the BLH gene accompanied by successive identification of the BLH transcription factor families in A. thaliana, Oryza sativa, Populus trichocarpa, Zea mays, Solanum tuberosum, Vandenboschia speciosa, Brassica rapa, Daucus carota, Phyllostachys edulis, Pisum sativum, Medicago truncatula, Camellia chekiangoleosa and five legume species [2–5,32–36]. Extensive studies have shown that gene replication, including tandem, fragment and genome-wide replication, not only plays an important role in genome rearrangement and amplification, but also in gene function diversification and gene families expression [37]. Velasco et al. [19] found that genome-wide duplication occurred in the apple genome 60–65 million years ago, which resulted in the formation of 17 new chromosomes from the original nine ancestral chromosomes. Recently, Daccord et al. [21] reported an apple genome map (apple ‘GDDH13 v1.1’ genome) of the highest quality at present by integrating three different techniques. In our study, 19 MdBLH genes were identified in the apple ‘GDDH13 v1.1’ genome (Tab. 2). Eighteen members of the apple MdBLH family were located in genome-wide replication regions and, of those, MdATH1.1/MdATH1.2, MdBLH1.1/MdBLH1.2, MdBLH3.1/MdBLH3.2, MdBLH4.1/MdBLH4.2, MdBLH7.1/MdBLH7.2, MdBLH8.1/MdBLH8.2, MdBLH9.1/MdBLH9.2, and MdBLH11.1/MdBLH11.2 (16/19; 84.2%) undergone segmental replication. Interestingly, members of the MdBLH family did not undergo tandem replication. Similar results were also found in soybeans, in which 89.1% of homeobox genes undergone segmental replication [32]. These results indicated that the event of segmental replication plays an important role in the extension of the BLH gene family in apple and soybean.
BLH transcription factors form either heterodimers with OFP proteins, and also interact with KNOX proteins, or form functional complexes with KNOX and OFP proteins, and then they participate in the regulation of plant growth and development [7,15,38–41]. For example, the heterodimers formed by BLH1 and KANT3 are involved in the normal development of embryo sacs and fate of cells in embryo sacs, and their activity is regulated by AtOFP5 [15]. Heterodimers that are formed by the interaction of BLH6 and KNAT7 proteins regulate secondary cell wall formation by inhibiting the expression of homeodomain-leucine zipper transcription factor REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1) [38]. Functional complexes formed by BLH6-KNAT7 heterodimers interact with AtOFP1 and AtOFP4 that were involved in the regulation of secondary cell wall formation by increasing the transcriptional inhibitory activity of BLH6 [39]. The BLH3 protein interacts with the AtOFP1 protein to regulate the conversion from vegetative to reproductive growth in Arabidopsis [40]; OsOFP2 in rice interact with OsKNAT7, BLH6-like, and BLH-Like2 to regulate vascular development [41]. BLH12 and BLH14 in maize interact with KNOTTED1 to play an important role in the structural development of internode patterning and vein anastomosis [7]. In addition, the BELL1 protein in Arabidopsis interact with the MADS-box protein and SPOROCYTELESS to participate in the regulation of ovule development [16,42]. Therefore, the BLH transcription factor acts as a focal point to interact with multiple proteins, and it constitutes a complex network to regulate plant growth and development accurately and efficiently. In the present study, we found that MdBEL1.1 could interact with the MdOFP6 protein, and MdBLH3.1 interacted with MdOFP4, MdOFP6, MdOFP13, and MdOFP16. This indicates that MdBEL1.1 and MdBEL3.1 may play important roles in regulating multiple aspects of plant growth and development, and are fine-tuned regulated by MdOFP proteins. Moreover, we speculated that the interaction between the MdBLH3.1 protein and MdOFP4, MdOFP6, MdOFP13, and MdOFP16 may affect the activity of the BLH-KNOX heterodimers. This in turn will change the expression level of the target genes, leading to regulation of growth and development of apples. However, further experiments are needed to analyze the relevant molecular mechanisms of the MdBLH-MdKNOX-MdOFP complexes involved in regulating apple growth and development.
At present, there are few studies on the response mechanism of the BLH protein to biotic and abiotic stresses. OsBIHD1, a member of the BEL family in rice, was rapidly up-regulated after 6 h from inoculation with Magnaporthe grisea and participated in the response of rice disease resistance [43]. Overexpression of OsBIHD1 in tobacco (Nicotiana tabacum) increases resistance to the tomato mosaic virus and to Phytophthora parasitica var. nicotianae; however, the transgenic tobacco plants overexpressing OsBIHD1 showed more sensitivity to salt and oxidative stresses [44]. Recent studies have shown that some BEL genes in chickpea roots and stems respond to stress treatment when plants are exposed to drought, salt, and cold conditions [32]. Under drought treatment, there are few BEL genes induced by stress in soybeans. A variety of BEL genes in soybeans were induced to different degrees by the pathogens Heterodera glycines, Phakopsora pachyrhizi, and P. sojae [32]. Overexpression of soybean GmBLH4 in Arabidopsis changes leaf morphology and fruit pod length, improving the response ability to high temperature and high humidity [45]. In the present study, we found that MdBLH4.1 and MdBLH9.1 were induced by salt stress; MdATH1.1, MdBLH8.1, MdBLH8.3, and MdBLH11.1 were inhibited by salt stress, and MdATH1.1 and MdBLH7.2 were inhibited by mannitol stress. These results indicated that MdBLH proteins also played a role in apple response to abiotic stress. The molecular mechanisms of BLH genes in regulating apple response to biotic and abiotic stresses should be investigated in future studies.
Acknowledgement: The authors are grateful to Professor Thomas Alan Gavin, Cornell University, for help in revising our English composition.
Author Contributions: HL and QD collected the public dataset, performed bioinformatics analysis, and drafted the manuscript. QZ contributed to the bioinformatics analysis and preparation of all Figures and tables. QZ and HW conducted the experiments. QD and YX conceived this study and reviewed the manuscript. All authors have read and approved the final manuscript.
Funding Statement: This study was supported by Shandong Provincial Natural Science Foundation, China (Grant No. ZR2019MC071).
Conflicts of Interest: The authors declare no conflict of interest.
1. Bürglin, T. R., Affolter, M. (2016). Homeodomain proteins: An update. Chromosoma, 125(3), 497–521. DOI 10.1007/s00412-015-0543-8. [Google Scholar] [CrossRef]
2. Ruiz-Estévez, M., Bakkali, M., Martín-Blázquez, R., Garrido-Ramos, M. A. (2017). Identification and characterization of TALE homeobox genes in the endangered fern Vandenboschia speciosa. Genes (Basel), 8(10), 275. DOI 10.3390/genes8100275. [Google Scholar] [CrossRef]
3. Dolgikh, A., Rudaya, E., Dolgikh, E. (2020). Identification of BELL transcription factors involved in nodule initiation and development in the legumes Pisum sativum and Medicago truncatula. Plants, 9(12), 1808. DOI 10.3390/plants9121808. [Google Scholar] [CrossRef]
4. Yan, C., Hu, Z., Nie, Z., Li, J., Yao, X. et al. (2021). CcBLH6, a bell-like homeodomain-containing transcription factor, regulates the fruit lignification pattern. Planta, 253, 90. DOI 10.1007/s00425-021-03610-7. [Google Scholar] [CrossRef]
5. Ghate, T. H., Sharma, P., Kondhare, K. R., Hannapel, D. J., Banerjee, A. K. (2017). The mobile RNAs, StBEL11 and StBEL29, suppress growth of tubers in potato. Plant Molecular Biology, 93, 563–578. DOI 10.1007/s11103-016-0582-4. [Google Scholar] [CrossRef]
6. Ikeda, T., Tanaka, W., Toriba, T., Suzuki, C., Maeno, A. et al. (2019). BELL1-like homeobox genes regulate inflorescence architecture and meristem maintenance in rice. The Plant Journal, 98(3), 465–478. DOI 10.1111/tpj.14230. [Google Scholar] [CrossRef]
7. Tsuda, K., Abraham-Juarez, M. J., Maeno, A., Dong, Z., Aromdee, D. et al. (2017). KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. The Plant Cell, 29(5), 1105–1118. DOI 10.1105/tpc.16.00967. [Google Scholar] [CrossRef]
8. Hackbusch, J., Richter, K., Muller, J., Salamini, F., Uhrig, J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proceedings of the National Academy of Sciences of the United States of America, 102(13), 4908–4912. DOI 10.1073/pnas.0501181102. [Google Scholar] [CrossRef]
9. Smith, H. M. S., Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis in florescence. The Plant Cell, 15(8), 1717–1727. DOI 10.1105/tpc.012856. [Google Scholar] [CrossRef]
10. Ragni, L., Belles-Boix, E., Günl, M., Pautot, V. (2008). Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. The Plant Cell, 20(4), 888–900. DOI 10.1105/tpc.108.058230. [Google Scholar] [CrossRef]
11. Ung, N., Lal, S., Smith, H. M. (2011). The role of PENNYWISE and POUND-FOOLISH in the maintenance of the shoot apical meristem in arabidopsis. Plant Physiology, 156(2), 605–614. DOI 10.1104/pp.110.171462. [Google Scholar] [CrossRef]
12. Khan, M., Tabb, P., Hepworth, S. R. (2012). BLADE-on-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signaling & Behavior, 7(7), 788–792. DOI 10.4161/psb.20599. [Google Scholar] [CrossRef]
13. Xu, Y., Wang, Y., Wang, X., Pei, S., Kong, Y. et al. (2020). Transcription factors BLH2 and BLH4 regulate demethylesterification of homogalacturonan in seed mucilage. Plant Physiology, 183, 96–111. DOI 10.1104/pp.20.00011. [Google Scholar] [CrossRef]
14. Kumar, R., Kushalappa, K., Godt, D., Pidkowich, M. S., Pastorelli, S. et al. (2007). The Arabidopsis BEL1LlIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. The Plant Cell, 19(9), 2719–2735. DOI 10.1105/tpc.106.048769. [Google Scholar] [CrossRef]
15. Pagnussat, G. C., Yu, H. J., Sundaresan, V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. The Plant Cell, 19(11), 3578–3592. DOI 10.1105/tpc.107.054890. [Google Scholar] [CrossRef]
16. Brambilla, V., Battaglia, R., Colombo, M., Masiero, S., Bencivenga, S. et al. (2007). Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. The Plant Cell, 19(8), 2544–2556. DOI 10.1105/tpc.107.051797. [Google Scholar] [CrossRef]
17. Meng, L., Fan, Z., Zhang, Q., Wang, C., Gao, Y. et al. (2018). BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. The Plant Journal, 94(6), 1126–1140. DOI 10.1111/tpj.13924. [Google Scholar] [CrossRef]
18. Ikeda, T., Tanaka, W., Toriba, T., Suzuki, C., Maeno, A. et al. (2019). BELL1-like homeobox genes regulate inflorescence architecture and meristem maintenance in rice. The Plant Journal, 98(3), 465–478. DOI 10.1111/tpj.14230. [Google Scholar] [CrossRef]
19. Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A. et al. (2010). The genome of the domesticated apple (Malus × domestica borkh.). Nature Genetics, 42(10), 833–839. DOI 10.1038/ng.654. [Google Scholar] [CrossRef]
20. Li, X., Ling, K., Zhang, J., Xie, Y., Wang, L. et al. (2016). Improved hybrid de novo genome assembly of domesticated apple (Malus x domestica). Gigascience, 5(1), 35. DOI 10.1186/s13742-016-0139-0. [Google Scholar] [CrossRef]
21. Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N. et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nature Genetics, 49(7), 1099–1106. DOI 10.1038/ng.3886. [Google Scholar] [CrossRef]
22. Li, H., Dong, Q., Zhao, Q., Ran, K. (2019). Genome-wide identification, expression profiling, and protein-protein interaction properties of ovate family proteins in apple. Tree Genetics & Genomes, 15, 45. DOI 10.1007/s11295-019-1354-5. [Google Scholar] [CrossRef]
23. Arnaud, N., Pautot, V. (2014). Ring the BELL and tie the KNOX: Roles for TALEs in gynoecium development. Frontiers in Plant Science, 5, 93. DOI 10.3389/fpls.2014.00093. [Google Scholar] [CrossRef]
24. Li, H., Ran, K., Dong, Q., Zhao, Q., Shi, S. (2020). Cloning, sequencing, and expression analysis of 32 NAC transcription factors (MdNAC) in apple. PeerJ, 8, e8249. DOI 10.7717/peerj.8249. [Google Scholar] [CrossRef]
25. Tian, Y., Dong, Q., Ji, Z., Chi, F., Cong, P. et al. (2015). Genome-wide identification and analysis of the MADS-box gene family in apple. Gene, 555(2), 277–290. DOI 10.1016/j.gene.2014.11.018. [Google Scholar] [CrossRef]
26. Li, H., Dong, Q., Zhao, Q., Shi, S., Ran, K. (2020). Isolation, sequencing, and expression analysis of 30 AP2/ERF transcription factors in apple. PeerJ, 8, e8391. DOI 10.7717/peerj.8391. [Google Scholar] [CrossRef]
27. Dong, Q., Duan, D., Zhao, S., Xu, B., Luo, J. et al. (2018). Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic arabidopsis. International Journal of Molecular Sciences, 19, 2478. DOI 10.3390/ijms19092478. [Google Scholar] [CrossRef]
28. Ma, Q. J., Sun, M. H., Liu, Y. J., Lu, J., Hu, D. G. et al. (2016). Molecular cloning and functional characterization of the apple sucrose transporter gene MdSUT2. Plant Physiology & Biochemistry, 109, 442–451. DOI 10.1016/j.plaphy.2016.10.026. [Google Scholar] [CrossRef]
29. Dong, Q., Zheng, W., Duan, D., Huang, D., Wang, Q. et al. (2020). MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Science, 299, 110611. DOI 10.1016/j.plantsci.2020.110611. [Google Scholar] [CrossRef]
30. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
31. Dong, Q., Zhao, S., Duan, D., Tian, Y., Wang, Y. et al. (2018). Structural and functional analyses of genes encoding VQ proteins in apple. Plant Science, 272, 208–219. DOI 10.1016/j.plantsci.2018.04.029. [Google Scholar] [CrossRef]
32. Bhattacharjee, A., Ghangal, R., Garg, R., Jain, M. (2015). Genome-wide analysis of homeobox gene family in legumes: Identification, gene duplication and expression profiling. PLoS One, 10(3), e0119198. DOI 10.1371/journal.pone.0119198. [Google Scholar] [CrossRef]
33. Que, F., Wang, G., Li, T., Wang, Y., Xu, Z. et al. (2018). Genome-wide identification, expansion, and evolution analysis of homeobox genes and their expression profiles during root development in carrot. Functional & Integrative Genomics, 18(6), 685–700. DOI 10.1007/s10142-018-0624-x. [Google Scholar] [CrossRef]
34. Ruiz-Estévez, M., Bakkali, M., Martín-Blázquez, R., Garrido-Ramos, M. A. (2017). Identification and characterization of TALE homeobox genes in the endangered fern Vandenboschia speciosa. Genes (Basel), 8(10), 275. DOI 10.3390/genes8100275. [Google Scholar] [CrossRef]
35. Khan, N., Hu, C., Khan, W. A., Wang, W., Han, K. et al. (2018). Genome-wide identification, classification, and expression pattern of homeobox gene family in Brassica rapa under various stresses. Scientific Reports, 8(1), 16265. DOI 10.1038/s41598-018-34448-x. [Google Scholar] [CrossRef]
36. Xu, X., Lou, Y., Yang, K., Shan, X., Zhu, C. et al. (2019). Identification of homeobox genes associated with lignification and their expression patterns in bamboo shoots. Biomolecules, 9(12), 862. DOI 10.3390/biom9120862. [Google Scholar] [CrossRef]
37. Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology, 4, 10. DOI 10.1186/1471-2229-4-10. [Google Scholar] [CrossRef]
38. Liu, Y., You, S., Taylor-Teeples, M., Li, W. L., Schuetz, M. et al. (2014). BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. The Plant Cell, 26(12), 4843–4861. DOI 10.1105/tpc.114.128322. [Google Scholar] [CrossRef]
39. Liu, Y., Douglas, C. J. (2015). A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in arabidopsis thaliana. Plant Signaling & Behavior, 10(7), e1033126. DOI 10.1080/15592324.2015.1033126. [Google Scholar] [CrossRef]
40. Zhang, L., Zhang, X., Ju, H., Chen, J., Wang, S. et al. (2016). Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochemical & Biophysical Research Communications, 470(3), 492–497. DOI 10.1016/j.bbrc.2016.01.135. [Google Scholar] [CrossRef]
41. Schmitz, A. J., Begcy, K., Sarath, G., Walia, H. (2015). Rice ovate family protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Science, 241, 177–188. DOI 10.1016/j.plantsci.2015.10.011. [Google Scholar] [CrossRef]
42. Bencivenga, S., Simonini, S., Benková, E., Colombo, L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. The Plant Cell, 24(7), 2886–2897. DOI 10.1105/tpc.112.100164. [Google Scholar] [CrossRef]
43. Luo, H., Song, F., Goodman, R. M., Zheng, Z. (2005). Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biology, 7(5), 459–468. DOI 10.1055/s-2005-865851. [Google Scholar] [CrossRef]
44. Luo, H., Song, F., Zheng, Z. (2005). Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. Journal of Experimental Botany, 56(420), 2673–2682. DOI 10.1093/jxb/eri260. [Google Scholar] [CrossRef]
45. Tao, Y., Chen, M., Shu, Y., Zhu, Y., Wang, S. et al. (2018). Identification and functional characterization of a novel BEL1-lIKE homeobox transcription factor GmBLH4 in soybean. Plant Cell Tissue & Organ Culture, 134, 331–344. DOI 10.1007/s11240-018-1419-4. [Google Scholar] [CrossRef]
Appendix A
Supplementary Sequence: A1. Prediction of cis-acting elements in MdBLH promoters
Supplementary Sequence: A2. Genomic information for 11 cloned MdBLH genes
Supplementary Sequence: A3. Interactions of MdWRKY52 protein with MdVQ10 protein in yeast cells
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |