 Open Access
Open Access
ARTICLE
The Endosperm-Specific Expression of YUCCA Genes Enhances Rice Grain Filling
1
College of Life Sciences, Shaoxing University, Shaoxing, 312000, China
2
College of Chemistry and Life Sciences, Zhejiang Normal University, Jinhua, 321004, China
3
School of Life Sciences, Lanzhou University, Lanzhou, 730000, China
4
School of Life Science, South China Normal University, Guangzhou, 510631, China
* Corresponding Author: Jianxin Shou. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: High-Yield Rice Physiology & Genetics)
Phyton-International Journal of Experimental Botany 2022, 91(12), 2633-2648. https://doi.org/10.32604/phyton.2022.021474
Received 16 January 2022; Accepted 11 April 2022; Issue published 29 August 2022
Abstract
Grain filling is a crucial process that affects yield in rice (Oryza sativa L.). Auxin biosynthesis and signaling are closely related to rice yield; therefore, it is important to understand the effects of auxin biosynthesis on rice grain filling to improve crop yield. In this study, we used physiological and molecular strategies to identify the roles of auxin in rice grain filling. Exogenous application of auxin (IAA) or auxin analogues (2, 4-D) to young spikelets and flag leaves improved the seed-setting rate and yield per spike. Furthermore, real-time quantitative PCR assays confirmed that nine members of the OsYUCCA family of auxin biosynthetic genes were upregulated during grain filling, implication that auxin biosynthesis plays a major role in grain development. The specific expression of either Arabidopsis AtYUCCA1 or OsYUCCA2 in the endosperm or leaves resulted in increased expression of OsIAA genes and auxin content of seeds, as well as increased grain filling and seed-setting rate. This result establishes that the auxin content in grains and leaves is important for grain development. Our findings further highlight the potential applications for improving rice yield by elevating targeted gene expression in specific tissues.Keywords
Rice (Oryza sativa) is one of the most important food crops globally, and increasing its production has long been the primary goal of rice breeding. Grain filling and seed-setting rate are the most basic traits that determine rice yield [1–5]. Many factors affect these traits, including transcription factors, plant hormones, and signaling proteins [6–9]. Auxin was identified about 80 years ago to be an important regulator of seed development and seed weight [10,11]. Understanding the roles of auxin in determining rice yield is of great biological relevance.
Auxin is mainly biosynthesized by the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA)/YUCCA flavin monooxygenase (YUC) pathway, which is highly conserved throughout the plant kingdom [12]. TAA1 catalyzes the conversion of tryptophan to indole-3-pyruvate (IPA), and then the flavin monooxygenase-like enzyme YUCCA converts IPA to indole-3-acetic acid (IAA) [13,14]. Auxin biosynthesized by the TAA/YUC pathway plays a critical role in determining rice yield, and auxin biosynthesis mediated by OsYUCCA9 and OsYUCCA11 regulates grain filling via the rice endosperm [6,15]. TILLERING AND SMALL GRAIN1 (TSG1) encodes a tryptophan aminotransferase in rice that promotes endogenous auxin levels, and loss of TSG1 function decreases yield [16]. Furthermore, OsYUCCA12 and OsIAA29 are expressed transiently during early grain development, which suggests that OsYUCCA12 may regulate grain filling in rice [15,17]. The fruit set of plants is highly dependent on auxin metabolism [18]. The gene TILLER ANGLE CONTROL4 (TAC4) regulates rice plant architecture and yield-related traits by affecting the endogenous auxin content and its asymmetrical distribution [19]. The AUXIN RESPONSE FACTOR (ARF)–mediated activation of NO3-transporter and N-metabolism genes in response to auxin increases grain yield [20]. Moreover, OsPIN5b influences auxin levels, and its overexpression alters plant architecture and yield, leading to changes that include decreases in plant height, tiller number, leaf number, seed-setting rate, and the number of full grains per plant [8]. Collectively, these observations indicate that regulators of auxin signaling mediate rice yield via several different mechanisms.
Treatment with exogeneous auxin can improve the seed-setting rate in plants [21,22]. For example, treatment with the synthetic auxin 2, 4-dichlorophenoxyacetic acid (2, 4-D) or 1-naphthaleneacetic acid (NAA) or with IAA reverses male sterility and restores the seed-setting rate in barley [23]. Moreover, NAA significantly increases the yield of sweet potato under drought stress [24]. Application of exogenous IAA partially rescues the seed-setting rate defects in the Arabidopsis ADAPTOR PROTEIN COMPLEX 2 (AP2) mutant [25]. To date, more than 1,000 synthetic plant growth regulators have been used to improve food production. However, synthetic reagents may be harmful to the environment and human health; therefore, increasing the level of endogenous auxin is a more desirable option. Previous work suggested that transgenic expression of an auxin biosynthesis gene, iaaM, can increase the total IAA content in young flower buds of strawberry (Fragaria × ananassa) and raspberry (Rubus idaeus), as well as plant fecundity and fruit production [26,27]. Here, we explored the potential to increase rice yield via similar molecular approaches.
We determined that the application of exogenous auxin or elevation of the endogenous auxin level via overexpression of native or heterologous YUCCA genes in rice dramatically increased yield. We also established that treatment of spikelets and flag leaves of rice with IAA, 2, 4-D, or NAA increased the seed-setting rate and yield per spike, dependent on the concentration of auxin. Quantitative reverse transcription-PCR (qRT-PCR) assays revealed that nine OsYUCCA genes were upregulated and four OsYUCCA genes were downregulated, pointing to the importance of the spatiotemporal regulation of YUCCA expression during rice grain filling. Furthermore, expressing AtYUCCA1 (as an alternative to OsYUCCA1 because overexpression of the latter severely compromises viability) or OsYUCCA2 under the control of endosperm-and leaf-specific promoters resulted in increased expression of OsIAA genes accompanied by an increased seed-setting rate. These results further support the notion that increasing in specific tissues of auxin concentrations is a promising method to increase rice yield.
2.1 Plant Materials and Growth Conditions
Grains of wild-type rice (Oryza sativa L. ssp. japonica cv. Nipponbare) and of transgenic plants carrying pOsGt1::AtYUCCA1:GUS, pOsGluB::OsYUCCA2:GUS, pRubisco::AtYUCCA1:GUS, and pRubisco::OsYUCCA2:GUS were surface sterilized with 70% ethanol for 10 min followed by 10% NaClO for 30 min and then rinsed eight times with water. Wild-type and transgenic plants were grown in the field under natural conditions in Jinhua and Sanya, China.
The chemicals 2, 4-D, IAA, 1-NAA, and Tween-20 were obtained from Sigma-Aldrich. Stock solutions were prepared as follows: 10 mM stock solutions of 2, 4-D, IAA, and 1-NAA were made by dissolving each reagent in a few drops of 1 M KOH and diluting with ddH2O. The stock solutions were diluted to 50, 100, and 200 μM with ddH2O and 0.5% (v/v) Tween-20. Distilled water with 0.5% (v/v) Tween-20 was used as a control.
Spikelets and flag leaves were treated at 15:00 every other day, and the treatment was performed three times in total. The seed-setting rate, yield per spike, and 1,000-grain weight were measured after harvest for three seasons, from 2012 to 2014, as follows: seed setting rate (%) = (total grain number – shell number)/total grain number; yield per spike = the weight of the full seed per spike; yield per plant = the weight of the full seed per plant; and 1,000-grain weight = 100-grain weight × 10. Statistical analyses were performed using two-tailed Student’s t-tests for data from three independent experiments.
2.3 Quantitative Reverse Transcription-PCR (qRT-PCR)
To quantify the expression levels of endogenous OsYUCCA genes in wild-type grains and of OsIAA genes in the grains of transgenic plants, total RNA was isolated from grains using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized with a SuperScript III First-Strand Synthesis System (Invitrogen). qRT-PCR was performed with Thunderbird SYBR qPCR mix (Toyobo) and a StepOnePlus Real-Time PCR System (Applied Biosystems). The reactions were performed in a 20 μL volume containing 10 μL 2 × SYBR qPCR mix (Toyobo), 10 ng cDNA, and 1 μM of each gene-specific primer (Table S1). The PCR cycles were performed as follows: one cycle of 95°C for 3 min and 40 cycles of 95°C for 5 s and 60°C for 50 s. The resulting data were analyzed using StepOne Software v2.1. The transcript levels were normalized to that of the housekeeping gene OsACTIN2 (Table S1). For statistical analysis, the transcript levels from three independent experiments were analyzed with two-tailed Student’s t-tests.
2.4 Construct Generation, Transformation, and Molecular Identification in Rice
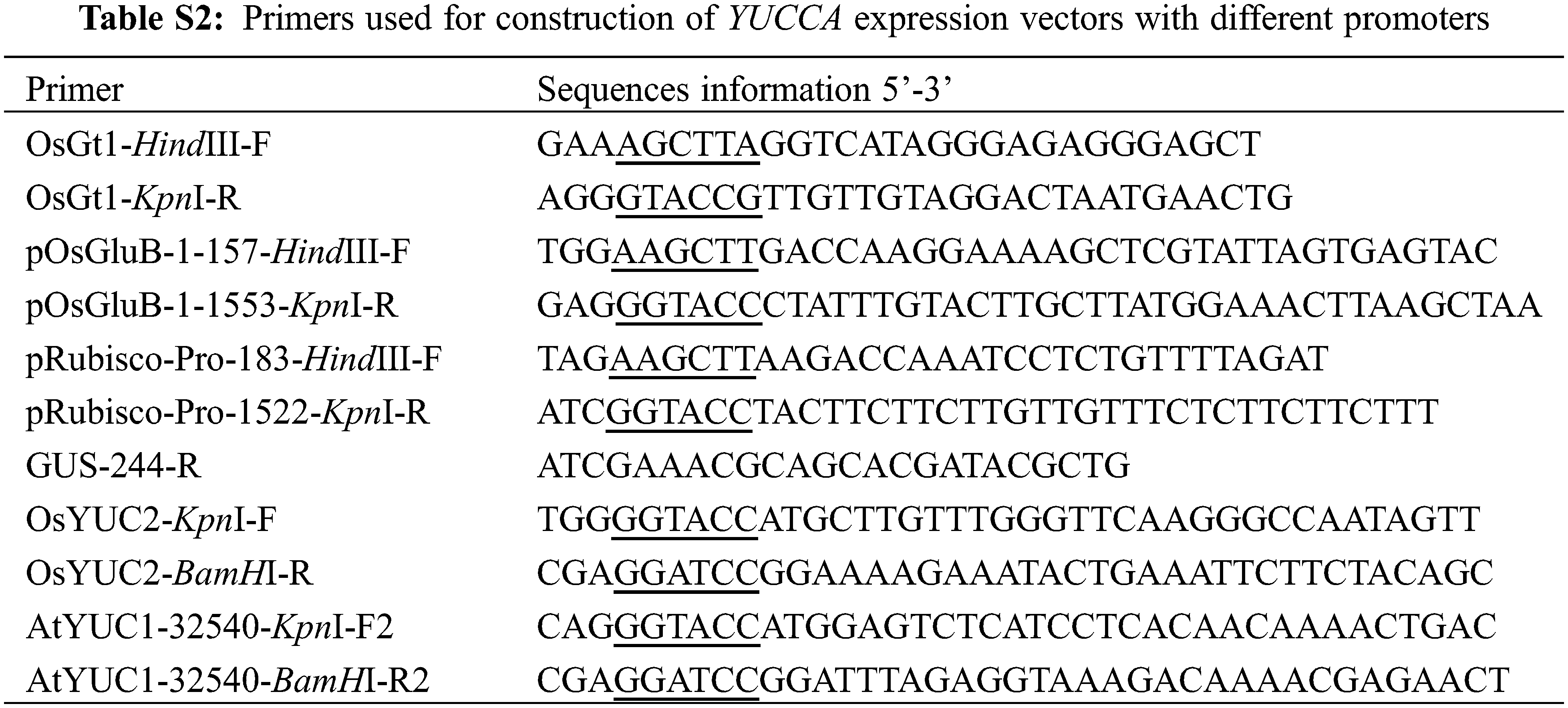
To specifically overexpress the AtYUCCA1 and OsYUCCA2 genes in rice, constructs were generated using PCR and restriction digestion and were ligated with the plant transformation vector pCAMBIA2300S-GUS containing the OsGt1, OsGluB, and Rubisco promoters. The resulting constructs were confirmed by sequencing. All primer sequences for the constructs are indicated in Table S2.
Rice transformation was conducted as described previously [28]. To determine whether the transgenic plants harboring the constructs expressed the constructs, RT-PCR and GUS staining were performed in T1 plants. RNA isolation and cDNA synthesis were conducted as described above. RT-PCR primers were designed to amplify the coding sequences of AtYUCCA1and OsYUCCA2. Expression of rice OsACTIN2, a housekeeping gene, was used as an internal control and wild-type Nipponbare cDNA served as a negative control (NC). Twenty-seven cycles of PCR were used to amplify AtYUCCA1, OsYUCCA2, and OsACTIN2 from the rice transgenic lines. Homozygous transgenic lines that overexpressed the AtYUCCA1 or OsYUCCA2 constructs were recovered in the T3 generation via selection for G418 resistance. To stain for GUS, samples were immersed in GUS staining solution, placed under vacuum for 30 min, incubated at 37°C overnight, and destained with solution (3:1 ethyl alcohol:acetic acid [v/v]) for 16 h.
3.1 Treating Rice with Exogenous Auxin Increases Yield
Auxin affects grain filling, photosynthesis, and photosynthate distribution in rice and consequently can enhance seed-setting rate and yield. To further demonstrate the function of auxin in rice, we treated young spikelets and flag leaves with exogenous IAA or its analogs 2, 4-D and NAA and analyzed how auxin affected yield. Young spikelets (2 days after full heading) were treated with different concentrations of IAA, 2, 4-D, and NAA (0, 50, 100, or 200 μM). Analysis of the seed-setting rate, yield per spike, and 1,000-grain weight showed that the 50 and 100 μM IAA treatments caused a significant increase in yield per spike, and 50, 100, and 200 μM IAA treatments increased the seed-setting rate, compared with those of the mock-treated control (0 μM) (Fig. 1A). Similarly, treatment with 50, 100, or 200 μM 2, 4-D improved the seed-setting rate, yield per spike, and 1,000-grain weight relative to the control values (Fig. 1B). Treatment with NAA caused an increase in seed-setting rate, although it did not significantly change the yield per spike or the 1,000-grain weight (Fig. 1C). These results demonstrate that exogenous auxin effectively increases rice yield.
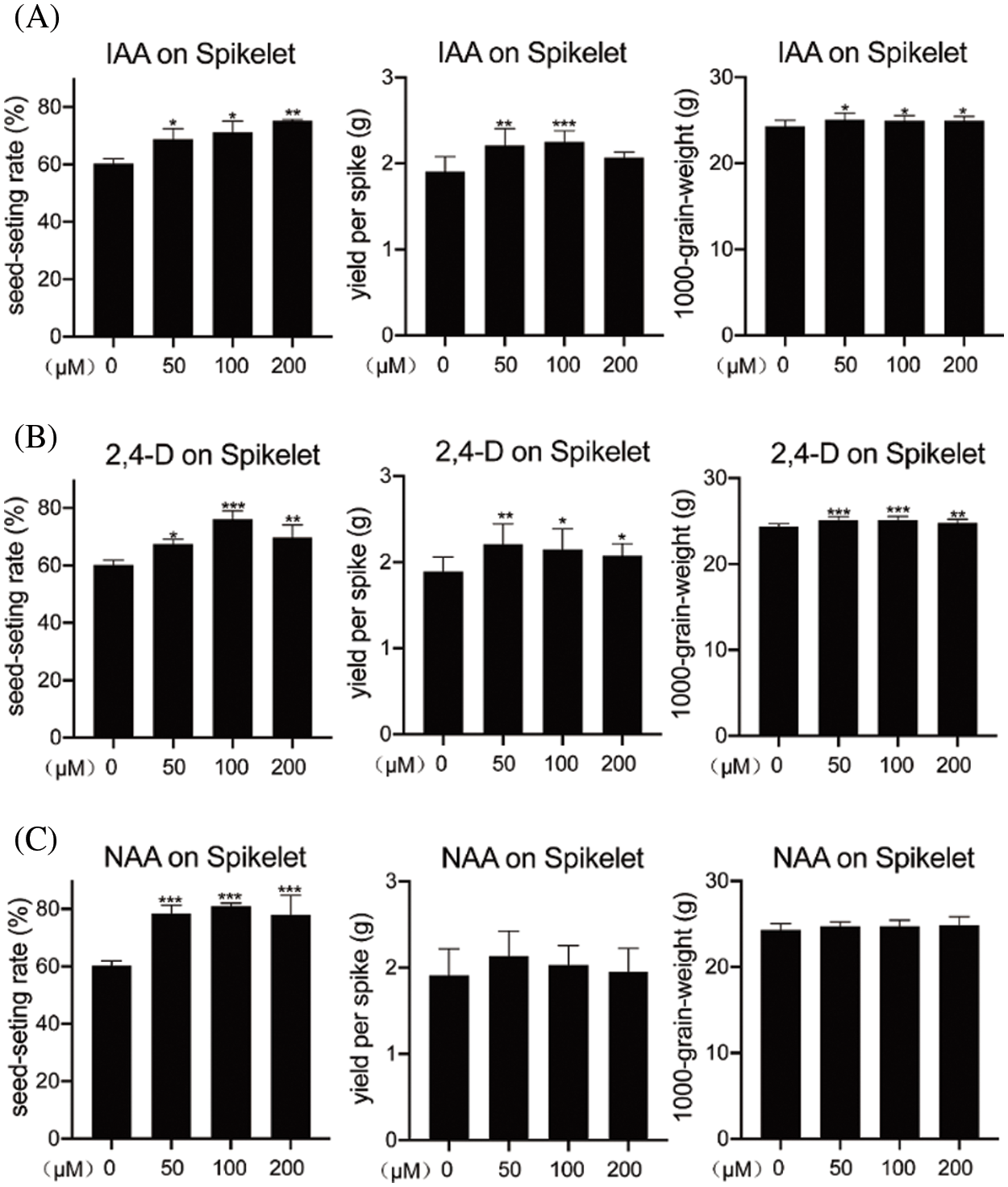
Figure 1: Effect of treating spikelets with exogenous auxin on rice yield traits. Effect of IAA (A), 2, 4-D (B), and NAA (C) treatment on seed-setting rate (%), yield per spike (gram), and 1,000-grain weight (gram) in rice spikelets. The concentrations of 2, 4-D, IAA, and NAA were 0 (mock), 50,100, and 200 μM. Values are means ± SD. Statistically significant differences at P < 0.05, 0.01, and 0.001 are indicated by *, **, and ***, respectively (Student’s t-test; compared with the corresponding mock control)
Flag leaves provide the greatest nutrition for the spikelets during grain development; therefore, we also treated flag leaves with 50, 100, or 200 μM of 2, 4-D, IAA, and NAA. Similar to the results for spikelets, treatment of flag leaves with 50,100, or 200 μM auxin dramatically increased the seed-setting rate (by 12.4%, 11.3%, and 10.8% with IAA; 18.1%, 17.2%, and 17.4% with 2, 4-D; and 12.3%, 10.5%, and 9.6% with NAA) compared with the control values (Figs. S1A–S1C). Additionally, the yield per spike increased following treatment with 50 or 100 μM IAA as well as with 50, 100, or 200 μM 2, 4-D (Figs. S1A and S1B). However, 1,000-grain weight was not affected by auxin treatment (Figs. S1A–S1C). Collectively, these results confirm that treating different rice tissues with exogenous auxin effectively increases yield in a concentration-dependent manner.
3.2 OsYUCCA Genes are Differentially Expressed during Grain Filling
OsYUCCA genes are involved in IAA biosynthesis and are expressed in almost all organs, including roots and leaves, and in vascular tissues [29]. Rice contains 14 OsYUCCA homologs [22]. Our finding that auxin promotes rice grain filling prompted us to test whether OsYUCCA genes are also involved in rice grain filling. To this end, we quantified the expression levels of OsYUCCA1–OsYUCCA14 in rice kernels during grain filling at 0, 5, 10, 15, and 20 days after flowering by qRT-PCR. The time-course analysis revealed that the expression of nine genes (OsYUCCA1, OsYUCCA2, OsYUCCA3, OsYUCCA5, OsYUCCA7, OsYUCCA8, OsYUCCA9, OsYUCCA11, and OsYUCCA12) increased, particularly starting from day 10, whereas the expression of three genes (OsYUCCA4, OsYUCCA6, and OsYUCCA10) decreased as grain filling progressed, and OsYUCCA13 and OsYUCCA14 were not detected (Fig. 2). Among the upregulated genes, OsYUCCA5, OsYUCCA7, OsYUCCA8, and OsYUCCA12 were primarily expressed in the early stage of grain filling (by day 5), and the other five genes were upregulated in the middle or late stages. Similarly, OsYUCCA9 and OsYUCCA11 were previously reported to play important roles in rice grain filling [6]. These results suggest that YUCCA-dependent auxin biosynthesis is involved in rice grain filling, which prompted us to continue to analyze the potential of YUCCA genes to improve rice yield.
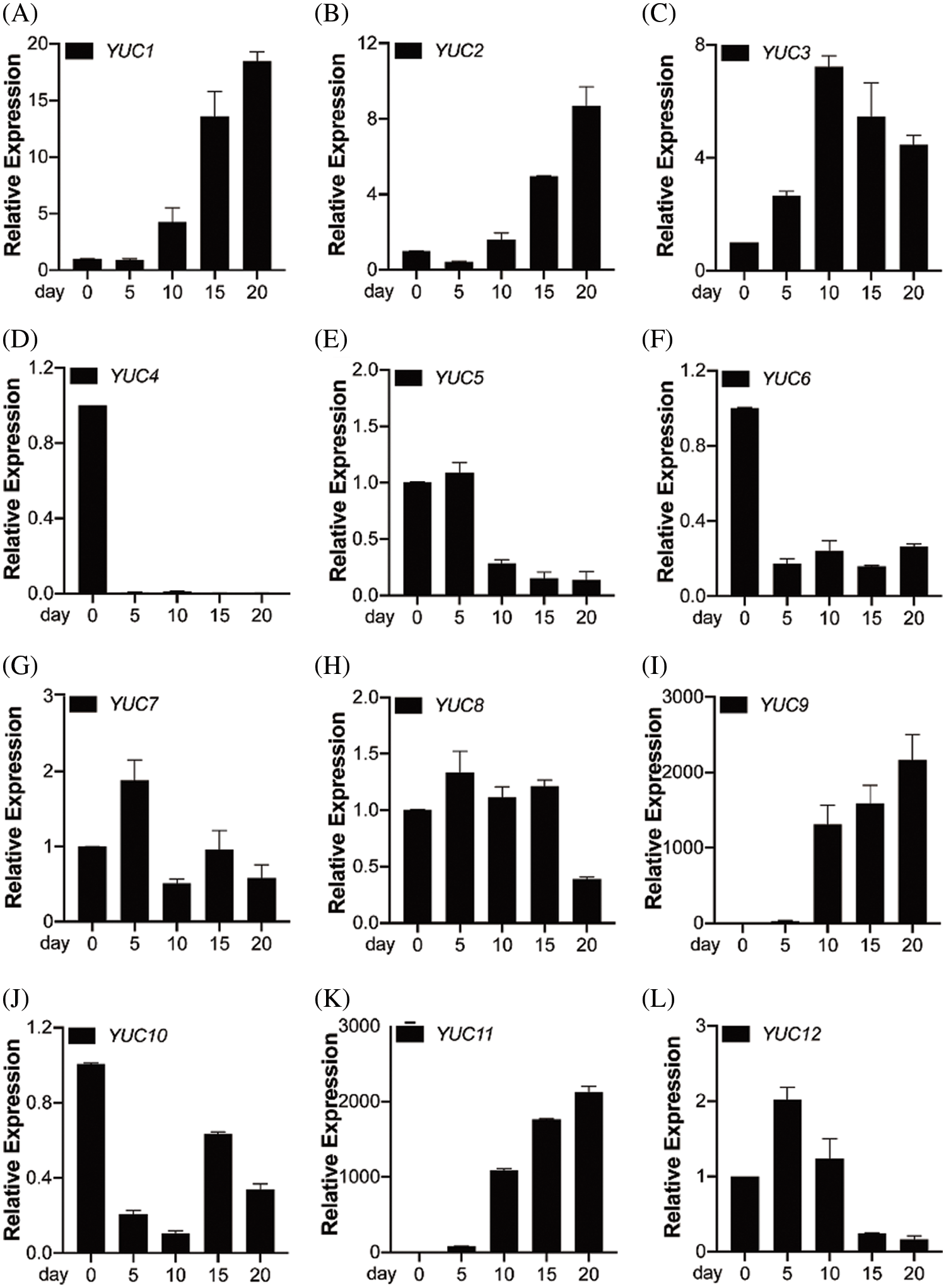
Figure 2: qRT-PCR analysis of transcript levels of OsYUCCA genes during grain filling. Expression of OsYUCCA genes in rice grains 0, 5, 10, 15, and 20 days after flowering. OsACTIN was used as an internal control. For each gene, the transcript level was normalized to the level at 0 day after flowering. Values shown are means ± SD
3.3 Confirmation of the Tissue-Specific Overexpression of YUCCA Genes in Transgenic Rice
The YUCCA enzymes represent the rate-limiting step auxin biosynthesis, and in Arabidopsis, their overexpression dramatically increases the concentration of IAA [30]. Plants expressing 35S:OsYUCCA1 accumulate high levels of auxin but are very difficult to regenerate due to abnormal organ development [29]. Therefore, we used the Arabidopsis homolog AtYUCCA1 in place of OsYUCCA1. We selected the promoters of Glutelin 1 (OsGt1) and OsGluB from rice and of the ribulose 1, 5-bisphosphate carboxylase/oxygenase small subunit promoter from Arabidopsis (Rubisco) to drive the tissue-specific expression of the YUCCA genes. OsGt1 and OsGluB are specifically expressed in the endosperm [31,32], and Rubisco is specifically expressed in leaves [33]. To further investigate the role of auxin biosynthesis genes in rice grain development, we generated transgenic plants that expressed AtYUCCA1 or OsYUCCA2 fused to the β-glucuronidase (GUS) reporter gene and driven by the tissue-specific OsGt1 and OsGluB promoters from rice and the Rubisco promoter from Arabidopsis (Fig. S2A).
We performed GUS staining and RT-PCR assays to examine the expression patterns and levels of the AtYUCCA1 and OsYUCCA2 transgenes in the endosperm and leaves of T1 plants. GUS staining was observed in the endosperm of pOsGt1::AtYUCCA1:GUS plants (lines #1, #2, #3, and #4) and pOsGluB::OsYUCCA2:GUS plants (lines #1, #2, #3, and #4) (Fig. 3A), whereas pRubisco::AtYUCCA1:GUS #1 and pRubisco::OsYUCCA2:GUS lines #1/#2 showed GUS staining in the leaves (Fig. 3C). RT-PCR assays confirmed that these lines showed higher tissue-specific expression of YUCCA1 and YUCCA2 than the non-transgenic plants (Fig. 3B). In the T3 generation, we selected homozygous transgenic lines according to their segregation for G418 resistance. The homozygous lines grew on agar supplemented with G418, whereas the root growth of wild-type plants was inhibited (Fig. S2B).
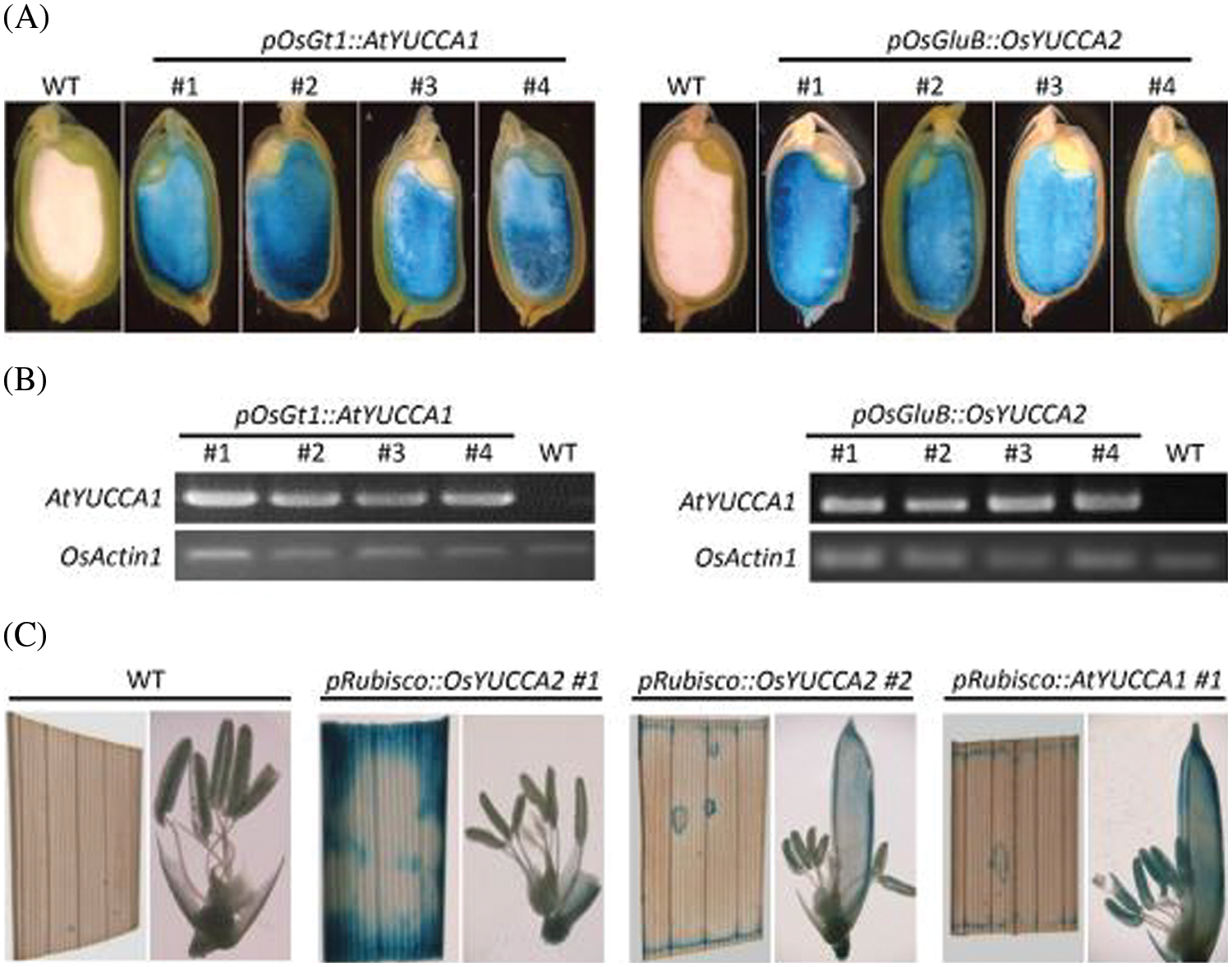
Figure 3: Tissue-specific expression of AtYUCCA1 or OsYUCCA2 in transgenic expression lines. (A) β-Glucuronidase (GUS) staining of transgenic lines expressing AtYUCCA1 or OsYUCCA2 driven by the endosperm-specific promoter pOsGt1::AtYUCCA1:GUS or pOsGluB::OsYUCCA2:GUS. Bar = 4 mm. (B) RT-PCR assays in individual T1 G418-resistant transgenic lines expressing AtYUCCA1 or OsYUCCA2 under the control of endosperm-specific promoters. Numbers indicate different transgenic lines. OsACTIN1 was used as an internal control. (C) GUS staining of transgenic lines expressing OsYUCCA2 and AtYUCCA1 driven by the leaf-specific promoter pRubisco
3.4 Tissue-Specific Expression of AtYUCCA1 and OsYUCCA2 Increases OsIAA Transcript Levels
Overexpression of YUCCA genes in Arabidopsis resulted in altered auxin phenotypes [13,30]. Rice contains 31 OsIAA homologs, which show different spatiotemporal expression patterns [34]. Treatment with exogenous IAA promotes the expression of OsIAA9 and OsIAA30 in young panicles and OsIAA1, OsIAA6, and OsIAA10 in leaves [34]. To determine the effect of tissue-specific AtYUCCA1 and OsYUCCA2 expression on the expression levels of endogenous OsIAA genes, we analyzed OsIAA gene expression in the young panicles of transgenic plants using qRT-PCR. The expression of both OsIAA9 and OsIAA30 was higher in caryopses of pOsGt1::AtYUCCA1:GUS and pOsGluB::OsYUCCA2:GUS transgenic lines compared to the wild type (Fig. 4A). These results demonstrate that the endosperm-specific expression of either AtYUCCA1 or OsYUCCA2 increases the expression of OsIAA9 and OsIAA30. We also analyzed the levels of OsIAA1, OsIAA6, and OsIAA10 expression in transgenic rice plants that carried the leaf-specific pRubisco::OsYUCCA2:GUS construct and observed that all three OsIAA genes were dramatically upregulated in lines #1 and #2 (Fig. 4B). In addition, IAA content determination of seeds indicated that pOsGt1::AtYUCCA1:GUS and pOsGluB::OsYUCCA2:GUS material content was increased compared to the wild type (Fig. 4C). These results further confirmed that the transgenic expression of AtYUCCA1 and OsYUCCA2 increased the expression of OsIAA genes in rice.
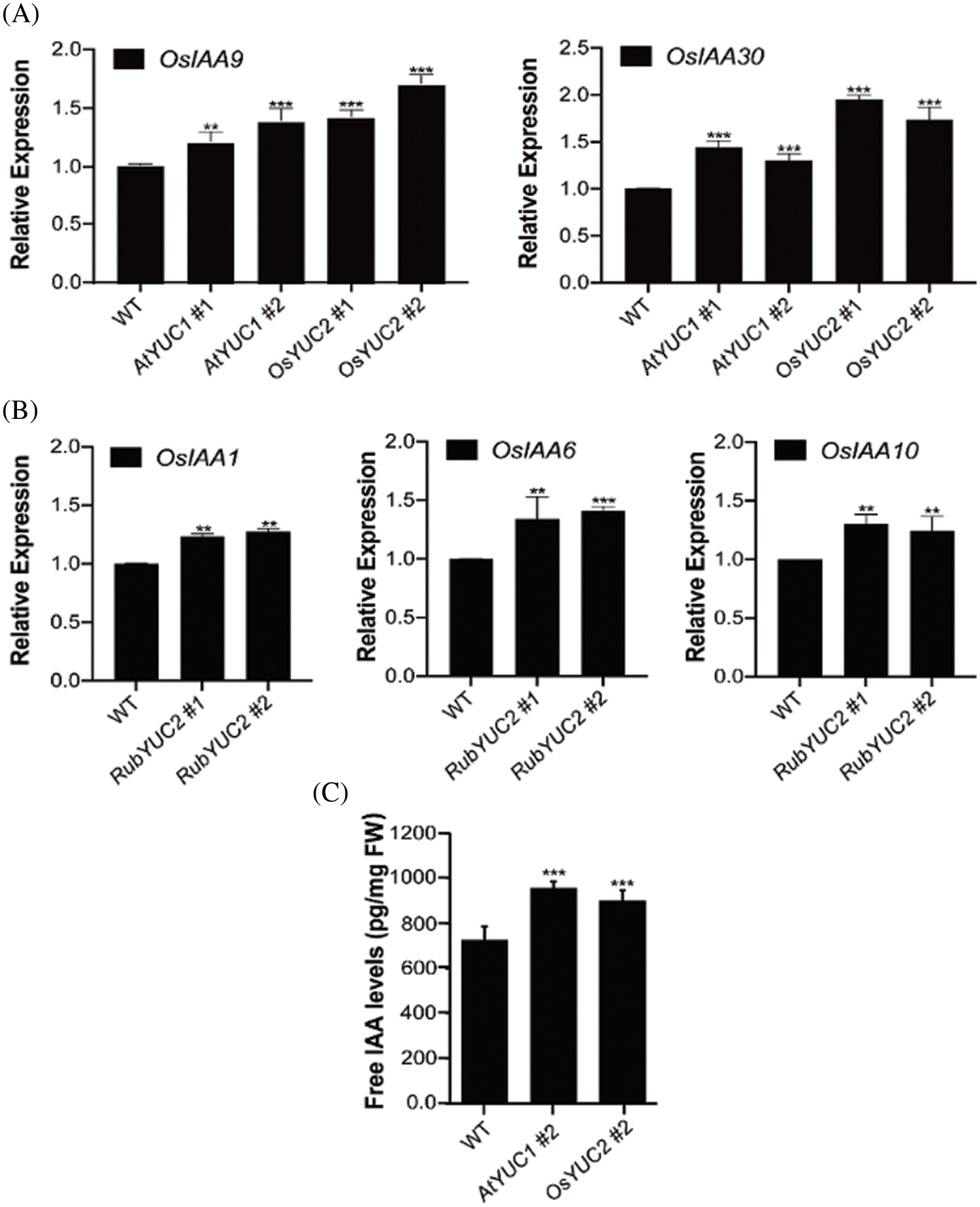
Figure 4: Tissue-specific overexpression of YUCCA1 or YUCCA2 increases OsIAA transcript levels. (A) Transcript levels of OsIAA9 and OsIAA30 determined by qRT-PCR in pOsGt1::AtYUCCA1:GUS (AtYUC1) and pOsGluB::OsYUCCA2:GUS (OsYUC2) transgenic lines. (B) qRT-PCR assays in transcript levels of OsIAA1 and OsIAA6 in pRubisco::OsYUCCA2:GUS (RubYUC2) transgenic lines. Wild-type Nipponbare was used as the negative control. Numbers indicate different transgenic lines. OsACTIN was used as an internal control. (C) Measurement of auxin transport in wild-type, pOsGt1::AtYUCCA1:GUS (AtYUC1) and pOsGluB::OsYUCCA2:GUS (OsYUC2) plants. Values are means ± SD. Statistically significant differences at P < 0.01 and 0.001 are indicated by ** and ***, respectively (Student’s t-test; compared with the wild type)
3.5 Transgenic Expression of AtYUCCA1 or OsYUCCA2 Improves Grain Yield
Next, we quantified grain yield traits in the transgenic expression lines to confirm the effect of endogenous auxin on rice yield. In the endosperm-specific expression lines pOsGt1::AtYUCCA1:GUS and pOsGluB::OsYUCCA2:GUS, the seed-setting rate was notably elevated (Figs. 5A, 5D), and the yield per spike was increased relative to the wild type (Figs. 5B, 5E). However, no significant differences in 1,000-grain weight were observed between the wild type and the endoderm-specific transgenic lines (Figs. 5C, 5F). Similarly, seed-setting rate and yield per plant, but not 1,000-grain weight, were increased in lines that expressed pRubisco::OsYUCCA2:GUS specifically in the leaves (Figs. 5G–5I). These results agree with previous findings that exogenous auxin promotes grain yield. Collectively, these results supported the notion that YUCCAs play a critical role in rice grain filling.
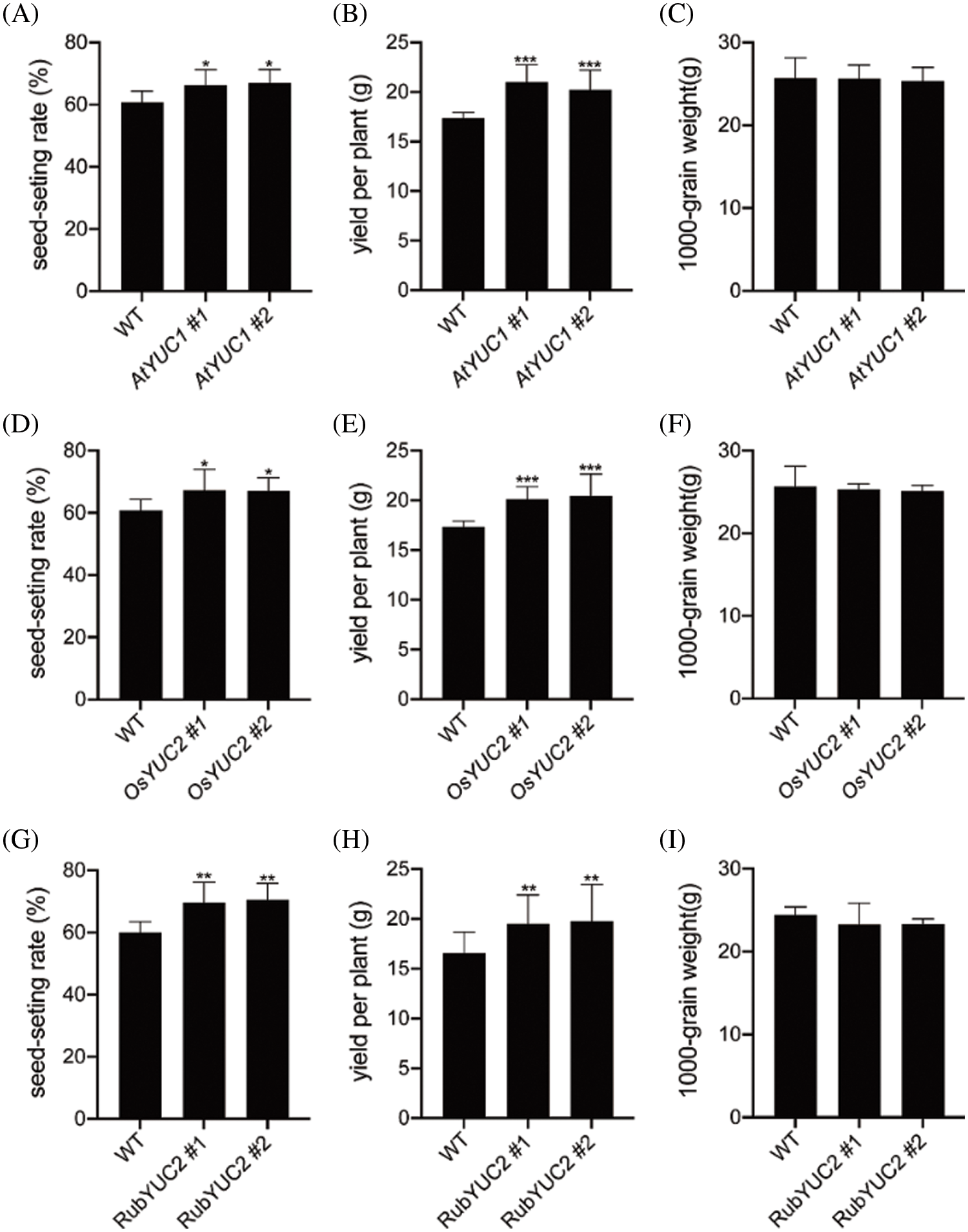
Figure 5: Analysis of yield traits in rice transgenic lines transgenically expressing AtYUCCA1 or OsYUCCA2. Seed-setting rate (%), yield per plant, and 1,000-grain weight in pOsGt1::AtYUCCA1:GUS (AtYUC1) (A–C), pOsGluB::OsYUCCA2:GUS (OsYUC2) (D–F), and pRubisco::OsYUCCA2:GUS (RubYUC2) (G–I) transgenic lines. CK, non-transgenic lines. Values are means ± SD. Statistically significant differences at P < 0.05, 0.01, and 0.001 are indicated by *, **, and ***, respectively (Student’s t-test; compared with the non-transgenic lines)
Plant hormones are critical in the regulation of seed set, and studies suggest that auxin can substitute for pollination and fertilization signals to promote seed growth [35]. In rice, the TAA/YUCCA pathway is essential for IAA biosynthesis during grain filling, and the IAA concentration is strongly correlated with the expression of IAA biosynthesis genes, including OsYUCCA9, OsYUCC11, and OsTAR1 [36]. Here, we confirmed that increasing the auxin level via both exogenous and endogenous methods improves grain yield in rice.
4.1 Exogenous Auxin Treatments Increase Rice Yield
Exogenous plant growth regulators are commonly applied to crops: For example, exogenous application of the gibberellin GA3 affects the deposition of storage compounds in seeds during seed filling in oilseed rape [37]. Auxins such as IAA repress the germination of soybean seeds by mediating the synthesis of abscisic acid and GA [21]. Consistent with this, our results indicated that exogenous auxin treatment promoted the seed-setting rate and yield per spike in rice (Fig. 1). When we treated young spikelets and flag leaves with 2, 4-D, IAA, or NAA, treatment of the flag leaf was more effective in promoting yield (Fig. S1). In particular, the seed-setting rates were substantially higher when the flag leaf, compared with the spikelet, was treated with different concentrations of 2, 4-D, IAA, or NAA. Notably, treating flag leaves with exogenous auxin mainly promotes photosynthesis in those leaves (Watson, 1952); by contrast, treating the young spikelet promotes the transport of photosynthetic products to the grain. This is why improving the photosynthetic efficiency of rice leaves is the main factor involved in increasing rice yield under conditions that limit photosynthetic capacity. Moreover, the effect of different exogenous auxins on crop yield is variable and may depend on environmental conditions, genotype, or the properties of the auxins. Collectively, these results confirm the important roles of exogenous auxins in promoting seed yield in rice.
4.2 Endosperm-Specific Expression of AtYUCCA1 and OsYUCCA2 Increases Rice Yield
Grain filling is the critical period that determines rice yield and quality [38]. The main component of rice grains is starch, and starch content and composition directly influence rice yield and quality. IAA affects the activities of enzymes involved in the conversion of sugar into starch. The protein products of TAA1 and the YUCCA genes are required to catalyze the biosynthesis of IAA from tryptophan. A recent study suggested that loss of OsYUCCA11 function reduces the seed weight and decreases the size of starch granules compared with wild-type plants [6], indicating that auxin biosynthesis genes in rice critically affect yield. Therefore, we investigated the expression of 14 OsYUCCA genes in kernels during grain filling and observed differential expression patterns. The expression of nine OsYUCCA genes was upregulated during grain filling, with two showing expression peaks at day 5 and the others late in grain filling (Fig. 2). By contrast, three OsYUCCA genes showed decreasing expression throughout grain filling, and two showed no expression during this process, implying that different OsYUCCA genes have stage-or tissue-specific functional roles and may be functionally redundant. This is consistent with previous findings that OsYUCCA7 is highly expressed early in grain development (2 days after pollination, DAP) and OsYUCCA9 and OsYUCCA11 are highly expressed at 7 DAP, whereas OsYUCCA12 shows a transient peak in expression at 3–4 DAP [36]. Furthermore, recent studies have reported that OsYUCCA1, OsYUCCA9, and OsYUCCA11 expression gradually increases during endosperm development, whereas OsYUCCA12 is specifically expressed in the endosperm at 2 DAP, and its expression then decreases [39]. These results suggest that the YUCCA pathway is critical for auxin biosynthesis and, therefore, for rice grain development.
Exogenous plant growth regulators can be expensive or harmful to the environment, and genetic modification technologies have been widely used in agriculture to improve crop performance (Phillips, 2010). The epidermis-specific promoter of FLORAL BINDING PROTEIN 7 (FBP7) from petunia has been used to drive the tissue-specific expression of an IAA biosynthesis gene, iaaM, in ovule epidermal cells, and this specifically increased IAA levels and substantially increased the number of lint fibers in cotton [40]. To further dissect the role of the auxin biosynthesis genes AtYUCCA1 and OsYUCCA2, we expressed them from the promoters of endosperm-and leaf-specific genes. We detected GUS signals in the endosperm and leaves of the transgenic plants (Fig. 3) and observed increased levels of OsIAA transcripts compared with those in wild-type plants (Fig. 4). The seed-setting rate and yield per spike were notably greater in the transgenic plants, whereas 1,000-grain weight was not affected (Fig. 5). The absence of an effect on 1,000-grain weight might arise mainly because an increased concentration of IAA causes more photosynthates to be transported to the smaller grains and less to the larger grains. Grain quality was improved by the endosperm-specific overexpression of iaaM in rice [39]. We also observed that in some plants with a high seed-setting rate, the yield per spike did not significantly increase because the yield per plant was also affected by the panicle number and total grain number per plant. It will be important to determine which other factors regulate crop yield, as well as to determine the most appropriate IAA concentration to increase yield.
Overexpression of AtYUCCA1 and iaaM leads to dramatic auxin overproduction phenotypes in Arabidopsis [41]; similarly, overexpression of OsYUCCA1 causes abnormal leaf, root, and stem development in rice [29]. Furthermore, rice lines constitutively expressing 35S::OsYUCCA1 are extremely difficult to regenerate and grow to seed set because they have defects in organ development. Therefore, by contrast with constitutive expression, the elevation of phytohormone levels at specific developmental stages or in specific tissues from tissue-specific promoters represents a potential and feasible method to improve crop yield. Here, we used endosperm-and leaf-specific promoters to express AtYUCCA1 and OsYUCCA2 to increase the IAA level, and subsequently grain yield, without adversely affecting plant development.
Acknowledgement: We thank Prof. Weihuai Pan for revising the manuscript.
Funding Statement: This work was supported by the National Natural Science Foundation of China (Grant Nos. 31801193, 31820103008, 91754104, and 31670283) and the Fundamental Research Funds for the Central Universities (No. lzujbky-2020-it13).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Zuo, J., Li, J. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annual Review of Genetics, 48(1), 99–118. DOI 10.1146/annurev-genet-120213-092138. [Google Scholar] [CrossRef]
2. Xu, Y., Yang, J., Wang, Y., Wang, J., Yu, Y. et al. (2017). OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genetics, 13(7), e1006906. DOI 10.1371/journal.pgen.1006906. [Google Scholar] [CrossRef]
3. Li, N., Xu, R., Duan, P., Li, Y. (2018). Control of grain size in rice. Plant Reproduction, 31(3), 237–251. DOI 10.1007/s00497-018-0333-6. [Google Scholar] [CrossRef]
4. Li, Q. P., Deng, F., Chen, H., Zeng, Y. L., Li, B. et al. (2020). Shading decreases rice yield by impeding grain-filling progress after heading. Agronomy Journal, 112(5), 4018–4030. DOI 10.1002/agj2.20372. [Google Scholar] [CrossRef]
5. Wang, T., Li, Y., Song, S., Qiu, M., Zhang, L. et al. (2021). Embryo sac development 1 affects seed setting rate in rice by controlling embryo sac development. Plant Physiology, 186(2), 1060–1073. DOI 10.1093/plphys/kiab106. [Google Scholar] [CrossRef]
6. Xu, X. Y., Zhiguo, E., Zhang, D. P., Yun, Q. B., Zhou, Y. et al. (2021). OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiology, 185(3), 934–950. DOI 10.1093/plphys/kiaa057. [Google Scholar] [CrossRef]
7. Liang, W. H., Shang, F., Lin, Q. T., Lou, C., Zhang, J. (2014). Tillering and panicle branching genes in rice. Gene, 537(1), 1–5. DOI 10.1016/j.gene.2013.11.058. [Google Scholar] [CrossRef]
8. Lu, G., Coneva, V., Casaretto, J. A., Ying, S., Mahmood, K. et al. (2015). OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant Journal, 83(5), 913–925. DOI 10.1111/tpj.12939. [Google Scholar] [CrossRef]
9. An, J., Almasaud, R. A., Bouzayen, M., Zouine, M., Chervin, C. (2020). Auxin and ethylene regulation of fruit set. Plant Science, 292, 110381. DOI 10.1016/j.plantsci.2019.110381. [Google Scholar] [CrossRef]
10. Gustafson, F. G. (1936). Inducement of fruit development by growth-promoting chemicals. PNAS, 22(11), 628–636. DOI 10.1073/pnas.22.11.628. [Google Scholar] [CrossRef]
11. Cao, J., Li, G., Qu, D., Li, X., Wang, Y. (2020). Into the seed: Auxin controls deed development and grain yield. International Journal of Molecular Sciences, 21(5), 1662. DOI 10.3390/ijms21051662. [Google Scholar] [CrossRef]
12. Zhao, Y. (2012). Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Molecular Plant, 5(2), 334–338. DOI 10.1093/mp/ssr104. [Google Scholar] [CrossRef]
13. Kakei, Y., Nakamura, A., Yamamoto, M., Ishida, Y., Yamazaki, C. et al. (2017). Biochemical and chemical biology study of rice OsTAR1 revealed that tryptophan aminotransferase is involved in auxin biosynthesis: Identification of a potent OsTAR1 inhibitor, pyruvamine2031. Plant and Cell Physiology, 58(3), 598–606. DOI 10.1093/pcp/pcx007. [Google Scholar] [CrossRef]
14. Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H. et al. (2011). The main auxin biosynthesis pathway in arabidopsis. PNAS, 108(45), 18512–18517. DOI 10.1073/pnas.1108434108. [Google Scholar] [CrossRef]
15. Nonhebel, H. M., Griffin, K. (2020). Production and roles of IAA and ABA during development of superior and inferior rice grains. Functional Plant Biology, 47(8), 716–726. DOI 10.1071/FP19291. [Google Scholar] [CrossRef]
16. Guo, T., Chen, K., Dong, N. Q., Ye, W. W., Shan, J. X. et al. (2020). Tillering and small grain 1 dominates the tryptophan aminotransferase family required for local auxin biosynthesis in rice. Journal of Integrative Plant Biology, 62(5), 581–600. DOI 10.1111/jipb.12820. [Google Scholar] [CrossRef]
17. French, S. R., Abu-Zaitoon, Y., Uddin, M. M., Bennett, K., Nonhebel, H. M. (2014). Auxin and cell wall invertase related signaling during rice grain development. Plants, 3(1), 95–112. DOI 10.3390/plants3010095. [Google Scholar] [CrossRef]
18. Zhang, S., Gu, X., Shao, J., Hu, Z., Yang, W. et al. (2021). Auxin metabolism is involved in fruit set and early fruit development in the parthenocarpic tomato “R35-P”. Frontiers in Plant Science, 12, 671713. DOI 10.3389/fpls.2021.671713. [Google Scholar] [CrossRef]
19. Li, H., Sun, H., Jiang, J., Sun, X., Tan, L. et al. (2021). TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnology Journal, 19(1), 64–73. DOI 10.1111/pbi.13440. [Google Scholar] [CrossRef]
20. Zhang, S., Zhu, L., Shen, C., Ji, Z., Zhang, H. et al. (2021). Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell, 33(3), 566–580. DOI 10.1093/plcell/koaa037. [Google Scholar] [CrossRef]
21. Shuai, H., Meng, Y., Luo, X., Chen, F., Zhou, W. et al. (2017). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Scientific Reports, 7(1), 12620. DOI 10.1038/s41598-017-13093-w. [Google Scholar] [CrossRef]
22. Cao, X., Yang, H., Shang, C., Ma, S., Liu, L. et al. (2019). The roles of auxin biosynthesis YUCCA gene family in plants. International Journal of Molecular Sciences, 20(24). DOI 10.3390/ijms20246343. [Google Scholar] [CrossRef]
23. Sakata, T., Oshino, T., Miura, S., Tomabechi, M., Tsunaga, Y. et al. (2010). Auxins reverse plant male sterility caused by high temperatures. PNAS, 107(19), 8569–8574. DOI 10.1073/pnas.1000869107. [Google Scholar] [CrossRef]
24. Wang, J. Q., Li, H., Liu, Q., Zeng, L. S. (2020). Effects of exogenous plant hormones on physiological characteristics and yield of sweet potato under drought stress. Ying Yong Sheng tai xue bao = The Journal of Applied Ecology, 31(1), 189–198. DOI 10.13287/j.1001-9332.202001.026. [Google Scholar] [CrossRef]
25. Kim, S. Y., Xu, Z. Y., Song, K., Kim, D. H., Kang, H. et al. (2013). Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell, 25(8), 2970–2985. DOI 10.1105/tpc.113.114264. [Google Scholar] [CrossRef]
26. Mezzetti, B., Landi, L., Pandolfini, T., Spena, A. (2004). The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnology, 4(1), 4. DOI 10.1186/1472-6750-4-4. [Google Scholar] [CrossRef]
27. Yin, Z., Malinowski, R., Ziolkowska, A., Sommer, H., Plcader, W. et al. (2006). The defh9-iaaM-containing construct efficiently induces parthenocarpy in cucumber. Cellular & Molecular Biology Letters, 11(2), 279–290. DOI 10.2478/s11658-006-0024-4. [Google Scholar] [CrossRef]
28. Pan, W., Shen, J., Zheng, Z., Yan, X., Shou, J. et al. (2018). Overexpression of the Tibetan plateau annual wild barley (Hordeum spontaneum) HsCIPKs enhances rice tolerance to heavy metal toxicities and other abiotic stresses. Rice, 11(1), 51. DOI 10.1186/s12284-018-0242-1. [Google Scholar] [CrossRef]
29. Yamamoto, Y., Kamiya, N., Morinaka, Y., Matsuoka, M., Sazuka, T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiology, 143(3), 1362–1371. DOI 10.1104/pp.106.091561. [Google Scholar] [CrossRef]
30. Cheng, Y., Dai, X., Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes and Development, 20(13), 1790–1799. DOI 10.1101/gad.1415106. [Google Scholar] [CrossRef]
31. Wu, C., Washida, H., Onodera, Y., Harada, K., Takaiwa, F. (2000). Quantitative nature of the prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: Minimal cis-element requirements for endosperm-specific gene expression. Plant Journal, 23(3), 415–421. DOI 10.1046/j.1365-313x.2000.00797.x. [Google Scholar] [CrossRef]
32. Russell, D. A., Fromm, M. E. (1997). Tissue-specific expression in transgenic maize of four endosperm promoters from maize and rice. Transgenic Research, 6(2), 157–168. DOI 10.1023/a:1018429821858. [Google Scholar] [CrossRef]
33. Yamakawa, S., Ando, K., Chisaka, A., Yoshida, K., Shinmyo, A. et al. (2004). Systematic transient assays of promoter activities for leaf-specific genes identified by gene-expression profiling with cDNA microarrays in Arabidopsis thaliana. Journal of Bioscience and Bioengineering, 98(2), 140–143. DOI 10.1016/S1389-1723(04)70257-1. [Google Scholar] [CrossRef]
34. Song, Y., Wang, L., Xiong, L. (2009). Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta, 229(3), 577–591. DOI 10.1007/s00425-008-0853-7. [Google Scholar] [CrossRef]
35. Uchiumi, T., Okamoto, T. (2010). Rice fruit development is associated with an increased IAA content in pollinated ovaries. Planta, 232(3), 579–592. DOI 10.1007/s00425-010-1197-7. [Google Scholar] [CrossRef]
36. Abu-Zaitoon, Y. M., Bennett, K., Normanly, J., Nonhebel, H. M. (2012). A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiologia Plantarum, 146(4), 487–499. DOI 10.1111/j.1399-3054.2012.01649.x. [Google Scholar] [CrossRef]
37. Huang, X. Q., He, R. Q., Liao, X. Y., Zhou, B., Peng, W. S. et al. (2014). Effect of exogenous gibberellin on reserve accumulation during the seed filling stage of oilseed rape. Genetics and Molecular Research, 13 (2), 2827–2839. DOI 10.4238/2014.January.22.7. [Google Scholar] [CrossRef]
38. Zhang, X., Rerksiri, W., Liu, A., Zhou, X., Xiong, H. et al. (2013). Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene, 530(2), 185–192. DOI 10.1016/j.gene.2013.08.048. [Google Scholar] [CrossRef]
39. Zhang, X. F., Tong, J. H., Bai, A. N., Liu, C. M., Xiao, L. T. et al. (2020). Phytohormone dynamics in developing endosperm influence rice grain shape and quality. Journal of Integrative Plant Biology, 62(10), 1625–1637. DOI 10.1111/jipb.12927. [Google Scholar] [CrossRef]
40. Zhang, M., Zheng, X., Song, S., Zeng, Q., Hou, L. et al. (2011). Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nature Biotechnology, 29(5), 453–458. DOI 10.1038/nbt.1843. [Google Scholar] [CrossRef]
41. Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X. et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in arabidopsis. PNAS, 108(45), 18518–18523. DOI 10.1073/pnas.1108436108. [Google Scholar] [CrossRef]
Appendix

Figure S1: Effect of auxins on rice yield traits in the flag leaf. Effects of IAA (A), 2, 4-D (B), and NAA (C) treatments on seed-setting rate (%), yield per spike (gram), and 1,000-grain weight (gram) in the flag leaf. Concentrations of 2, 4-D, IAA, and NAA were 0 (mock), 50, 100, and 200 μM. Values shown are means ± SD. Statistically significant differences at P < 0.05 and 0.01 are indicated by * and **, respectively (Student’s t-test; compared with the corresponding mock control)
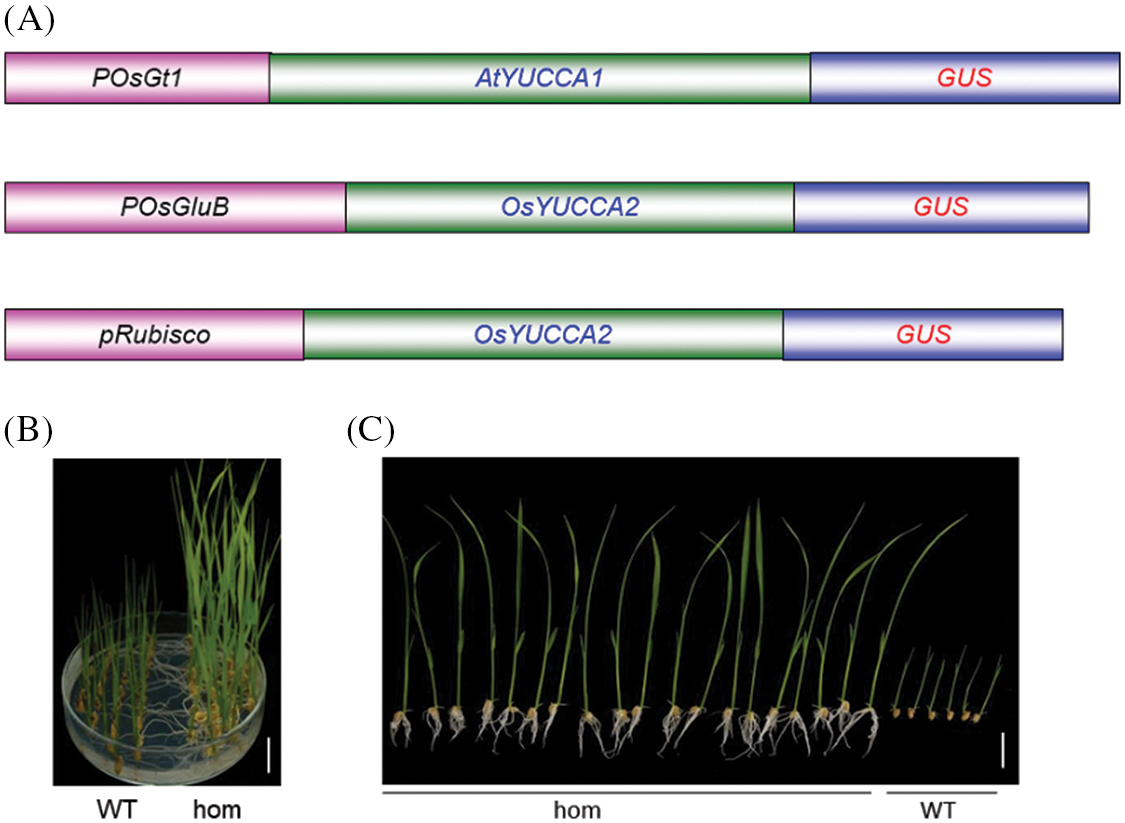
Figure S2: Molecular cloning and identification of homozygous transgenes. (A) Constructs used for the tissue-specific expression of AtYUCCA1/OsYUCCA2. The promoters pOsGt1 and pOsGluB from rice were used to drive endosperm-specific expression, and Arabidopsis pRubisco was used for leaf-specific expression. (B) and (C) Identification of homozygous lines in the T3 generation. B: Left, wild-type, and right, homozygous lines, Bar = 1 cm; C: Left, homozygous, and right, wild-type lines, bar = 2 cm

Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools