

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.021889
ARTICLE
Selection and Verification of Reference Genes for qRT-PCR Analysis in Iris domestica under Drought
College of Chinese Medicinal Materials, Jilin Agricultural University, Changchun, 130118, China
*Corresponding Authors: Cuijing Liu. Email: mary1011@126.com; Mei Han. Email: hanmei77@sohu.com
Received: 11 February 2022; Accepted: 28 March 2022
Abstract: Iris domestica is a plant of the Iridaceae family and is drought-tolerant, but its drought-resistance mechanism is not yet clear. Analysing the gene expression changes of I. domestica by qRT-PCR is an important mean to understand its drought resistance characteristics. Nevertheless, a lack of reference genes greatly hinders investigation and research on the adaptation of I. domestica to drought at the molecular and genetic levels. In this study, we assessed the expression stability of 11 candidate gene in I. domestica under drought stress conditions and different tissues using geNorm, NormFinder, BestKeeper and RefFinder tools. The results showed that EF1β was the most stable reference genes under drought stress and in different tissues. To validate further the stability of the identified reference genes, the expression patterns of VP gene in I. domestica was analysed. These results will be conducive to more accurate quantification of gene expression levels in I. domestica.
Keywords: Iris domestica; gene expression; qRT-PCR; reference genes; drought
Iris domestica (L.) Goldblatt et Amberley [syn. Belamcanda chinensis (L.) DC.] is a perennial herb of the Iridaceae family. Due to its strong stress tolerance (such as drought), I. domestica is a good material for studying the drought resistance mechanisms of plants [1]. The qRT-PCR technology has high specificity, strong sensitivity and good repeatability [2,3], which plays an important role in quantifying the expression of drought resistance genes in plants under drought [4]. However, no relative researches of suitable reference genes on I. domestica have been reported, which greatly limits the research on the drought resistance mechanism in I. domestica. Therefore, evaluation of the stability of candidate reference genes in I. domestica under drought and different tissues is necessary.
Stable reference genes play an important role in the accuracy and reliability of qRT-PCR analysis, especially when plants are under environment stress. Typically, the ideal reference genes for qRT-PCR are usually housekeeping genes, including β-tubulin (β-TUB), Elongation factor 1 alpha (EF1α), Elongation factor 1 beta (EF1β), Ubiquitin (UBQ), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and Actin (ACT) genes [5–8]. Nevertheless, the stability of these genes differs with tissues, developmental stages, environmental conditions and plant species [9–12]. Therefore, it is necessary to select reference genes suitable for plants. In the present study, 11 candidate reference genes were selected to perform qRT-PCR tests in I. domestica under drought and different tissues. The four tools of geNorm [13], NormFinder [14], BestKeeper [15], and RefFinder (https://github.com/fulxie/RefFinder) were applied to assess the identified reference genes and to select the most stably expressed genes. The expression levels of vacuolar proton pyrophosphatase gene (VP) from previous study were quantified to validate the chosen best-ranked reference genes. The results will contribute to provide reliable reference genes during qRT-PCR for gene expression studies in I. domestica.
2.1 Plant Materials and Treatment
The experiment was carried at the agricultural base of Jilin Agricultural University, China (43°48′N, 125°25′E) with an annual average temperature of 4.8°C, a maximum temperature of 39.5°C, and a minimum temperature of −39.8°C. The average annual amount of precipitation is 568.5 mm, mainly concentrated on June–August. The annual average sunshine hours are 2690 h.
Fresh dried mature seeds were collected from the Medicinal Botanical Garden of Jilin Agricultural University in October 2019, appraised by Professor Mei Han of Jilin Agricultural University. Those seeds were sprinkled on breeding trays with one seed per tray, which was placed in the greenhouse of Jilin Agricultural University. The about 5 cm height and one-year-old seedings were selected, transplanted and planted in plastic pots which had an inner diameter of 20.5 cm and a pot depth of 14.5 cm, and were put in a rain-proof shed for normal field management. The properties of all soils tested were pH 7.187 and contained 248 mg·kg−1 available nitrogen, 17 mg·kg−1 available phosphorus, and 140 mg·kg−1 available potassium.
The materials tested for this experiment were 2-year-old plants. Before drought stress, the plants were irrigated with 500 mL water every three days. Soil water content data were recorded after no irrigation with a HH2 Soil Moisture Meter (Delta-T Devices Ltd., UK). A total of 5 samples were taken, and the soil water content was 22.83% (1 d), 14.30% (13 d), 7.23% (27 d), 5.57% (42 d), and 2.13% (61 d), respectively. The rhizome samples under drought and untreated samples of the leaves, buds, roots and rhizomes of I. domestica were collected and immediately frozen in liquid nitrogen, and stored at −80°C. Among them, the rhizome samples of 1 d, 13 d, 27 d and 61 d were used for mRNA sequencing.
2.2 Total RNA Extraction and cDNA Synthesis
Total RNA was extracted with liquid nitrogen from the frozen different samples by the Plant RNA Extration Kit (Solarbio, Beijing, China). The RNA concentration and purity were assessed by nucleic acid/protein detector (Nanodrop2000, USA). A total of 1ug RNA was used to synthesize first-strand cDNA with a reverse transcription kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
2.3 Reference Gene Selection and Primer Design
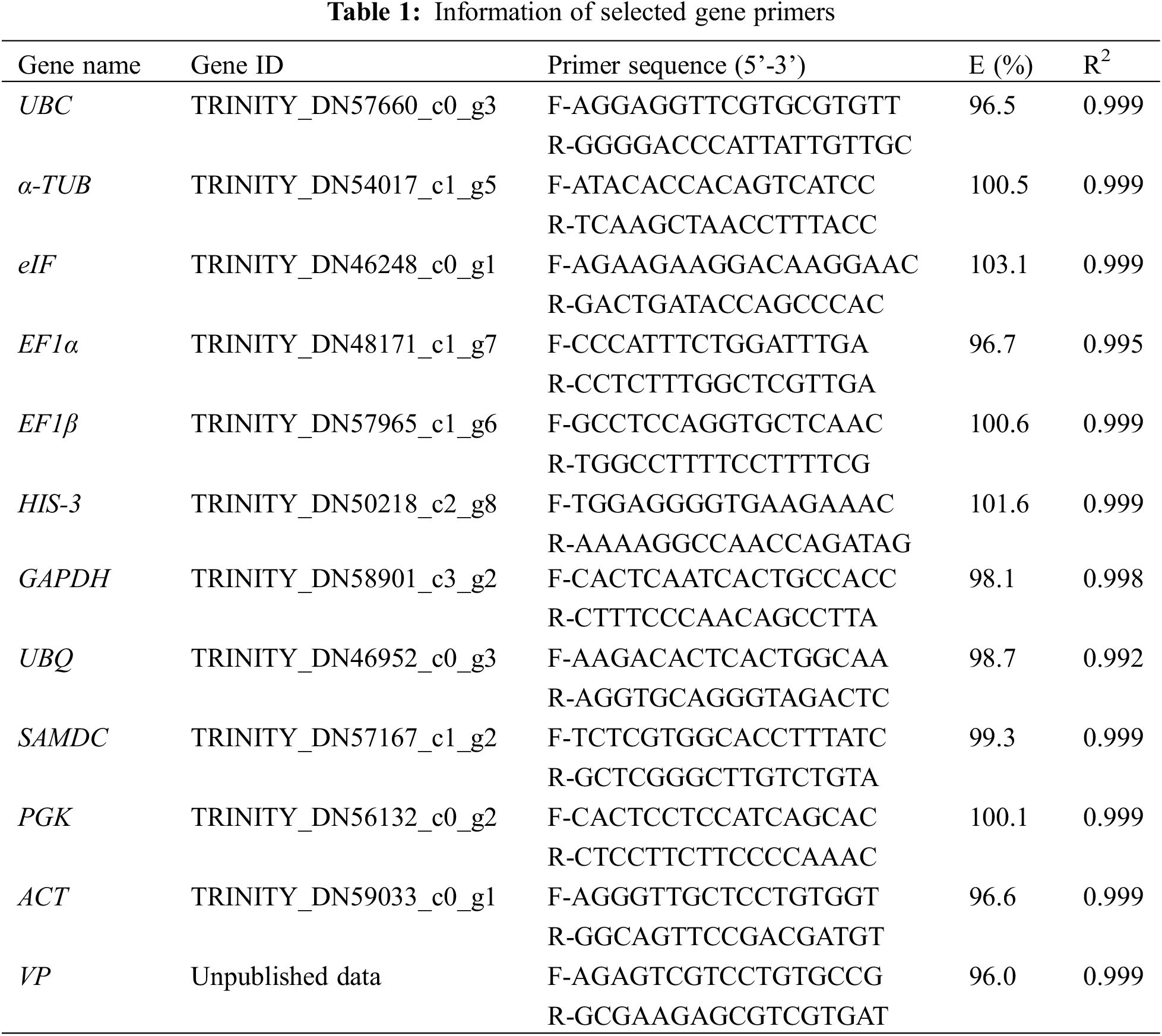
Based on the transcriptome of I. domestica under different drought conditions and the filter conditions of TPM values ≥0.2, 11 candidate reference genes were selected including Ubiquitin-protein ligase UBC9 (UBC), Tubulin alpha-5 (α-TUB), Eukaryotic translation initiation factor (eIF), Translation elongation factor EF1α (EF1α), Translation elongation factor EF1β (EF1β), Histone H3 (HIS-3), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Ubiquitin (UBQ), S-Adenosylmethionine decarboxylase (SAMDC), Phosphoglycerate kinase (PGK), and Actin (ACT). These genes are often used as housekeeping genes [5], and some are also used in the Iris family plant I. germanica and I. lactea [16,17]. The VP gene was selected as the verification gene. The primers for each gene were designed by Primer Premier 5 software, and all primers were synthesized by Sangon Biotech (Changchun, China). The PCR amplification efficiency (E) and correlation coefficient (R2) for each primer pair were calculated with a fourfold dilution series cDNA.
The qRT-PCR was performed on the Mx3000 P instrument (Agilent, SantaClara, CA, USA) using SYBR Premix Ex TaqTM II (Tli RNaseH Plus). The 20 μL qRT-PCR reaction system contained 1 μL template, 1 μL of upstream and downstream primers (10 μM) , 7 μL RNase Free d2H2O, and 10 μL of SYBR Premix Ex TaqII (TaKaRa, Dalian, China). The PCR reaction program was as follows: 95°C/30 s and 40 cycles of 95°C/5 s, 55°C/30 s, and 72°C/30 s and then a final extension at 72°C for 10 min. Three independent replicates were used for each treatment. Relative gene expression levels were calculated using the 2−ΔΔCt method [18].
The cycle threshold (Ct) value was recorded for each qRT-PCR analysis of gene expression. Box plot was drawn to visualize the reference gene expression levels and variations. Next, the four software tools of geNorm, NormFinder, BestKeeper, and RefFinder were used to assess the stability of the 11 selected reference genes. The geNorm selected the stable reference gene by calculating the M value of each reference gene and determined the number of optimal reference genes according to the Vn/Vn+1 value [19]. NormFinder calculated a stability value for each gene based on the variance analysis. The gene with the lowest value was identified as the most stably expressed gene [14]. For both algorithms, the Ct values should first be transformed by 2−ΔΔCt. The standard deviation (SD), and coefficient of variation (CV) of pairing between each gene can be calculated by BestKeeper, with smaller SD and CV values denoting better stability of the reference gene [15]. Finally, the reference genes were ranked based on the geometric mean (GM) values calculated with RefFinder.
2.6 Validation of Reference Genes
In order to verify the analysis results of the candidate genes, the expression profiles of the VP gene responding to drought were determined and normalized using the two most stable and the least stable reference genes [20]. The qRT-PCR amplification conditions were as described above. Statistical analyses were performed with the SPSS 19 (SPSS Inc., Chicago, IL, USA).
3.1 Primer Specificity and Amplification Efficiency Verification
Fifteen genes were selected to verify the transcriptome results, and the strong positive correlation was determined between the results of RNA-seq and qRT-PCR. The correlation coefficients R2 at 13 d vs. 1 d, 27 d vs. 1 d and 61 d vs. 1 d were 0.88, 0.74, and 0.77, respectively, which meant the reliability of the transcriptome sequencing data (Figs. 1a–1c). The TPM-based clustering heatmap of the 11 genes under different drought conditions was built. The results showed that the gene with large changes expression levels were clustered together, and the rest of the genes were clustered together, which indicated that some genes exhibited relatively stable expression except EF1α and UBC, and needed a further analysis (Fig. 1d).
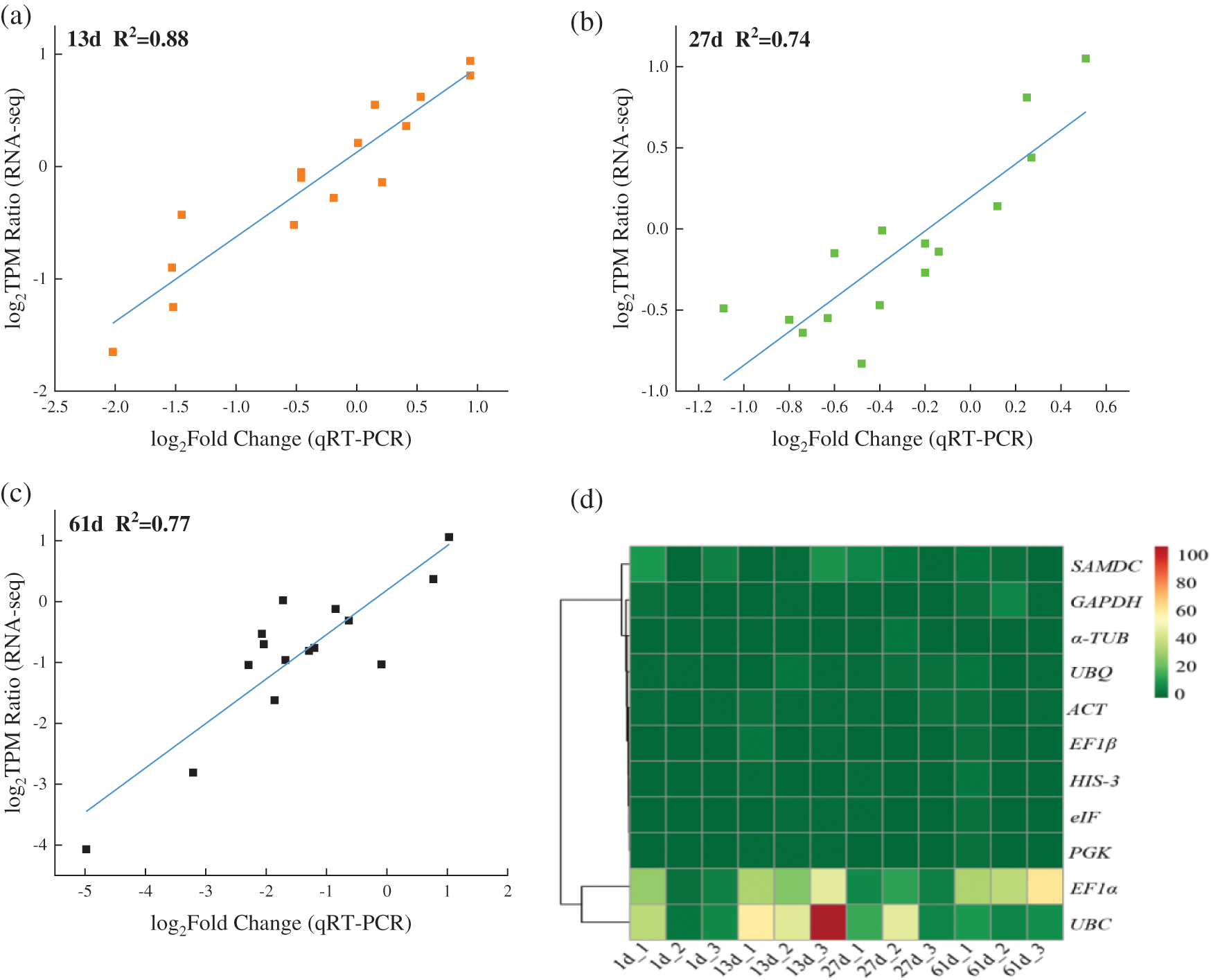
Figure 1: (a–c) Correlation between RNA-seq and qRT-PCR results. X-axis denoted Log2 (the ratio of qRT-PCR) using the method 2−ΔΔCt to calculate the relative expression level. Y-axis represented the TPM values in RNA-Seq data. (d) Heatmap of 11 candidate reference genes
12 genes were selected as candidate genes including a validation gene-VP gene. Detailed information was listed in Table 1. These genes were specific as their melting curves had a single peak for selected primer pairs (Supplementary File). According to the slope of the standard curve, the amplification efficiency and R2 of the qRT-PCR assays were calculated, indicating that the amplification efficiency ranged from 96% to 103.1%, and the R2 value ranged from 0.992 to 0.999. These results indicated that all the primers of the 12 genes had high specificity and amplification efficiency and were thus suitable for further analysis.
3.2 Expression Profiles of Reference Genes in I. domestica
qRT-PCR assays were performed with the designed specific primers for the 11 candidate reference genes using the cDNA as template for drought-treated materials and different tissues of I. domestica. Based on the Ct values obtained from qRT-PCR, a box diagram was done to reflect the differences in the expression levels of the 11 candidate genes (Fig. 2). The average Ct values of all samples varied from 17.83 to 28.21, reflecting the diversity of expression levels. The results showed that SAMDC had the smallest variation, followed by HIS-3, EF1β, and ACT, while UBC and PGK had the largest variation. Therefore, SAMDC, HIS-3, EF1β and ACT were identified as relatively stable genes in comparison to the other reference genes.
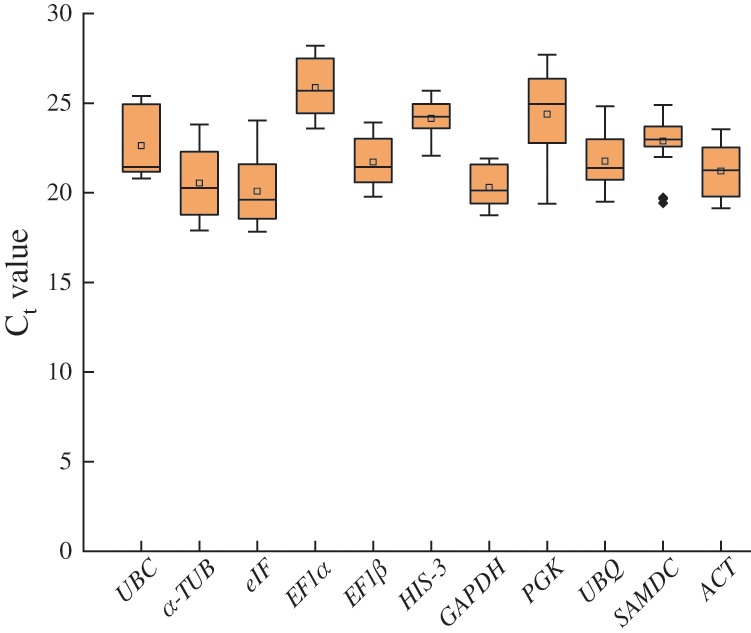
Figure 2: Distribution of qRT-PCR Ct values for the 11 candidate reference genes across all I. domestica samples
The geNorm evaluates the stability of reference gene expression by calculating the average M values, with a low value representing high stability. For the drought treatment, EF1α and ACT were the most stably expressed genes, with an M value of 0.96, whereas UBQ was the least stably expressed gene, with an M value of 4.55 (Fig. 3a). For the different tissues, EF1β and GAPDH (M = 0.33) were the most stably expressed genes, whereas PGK (M = 6.84) was the least stably expressed gene (Fig. 3b). Overall, EF1β and ACT (M = 3.28) were the more stably expressed genes in all samples, whereas PGK (M = 21.22) was the least stably expressed gene (Fig. 3c). The geNorm was also used to calculate the paired variation value (Vn/Vn+1) to determine the optimal number of reference genes for qRT-PCR standardization [19]. Considering that values are all above 0.15, the optimal number of reference genes was not determined in I. domestica (Fig. 3d). However, 0.15 is just a theoretical value, and higher cutoff values of Vn/n+1 have been used in other reports [21,22].
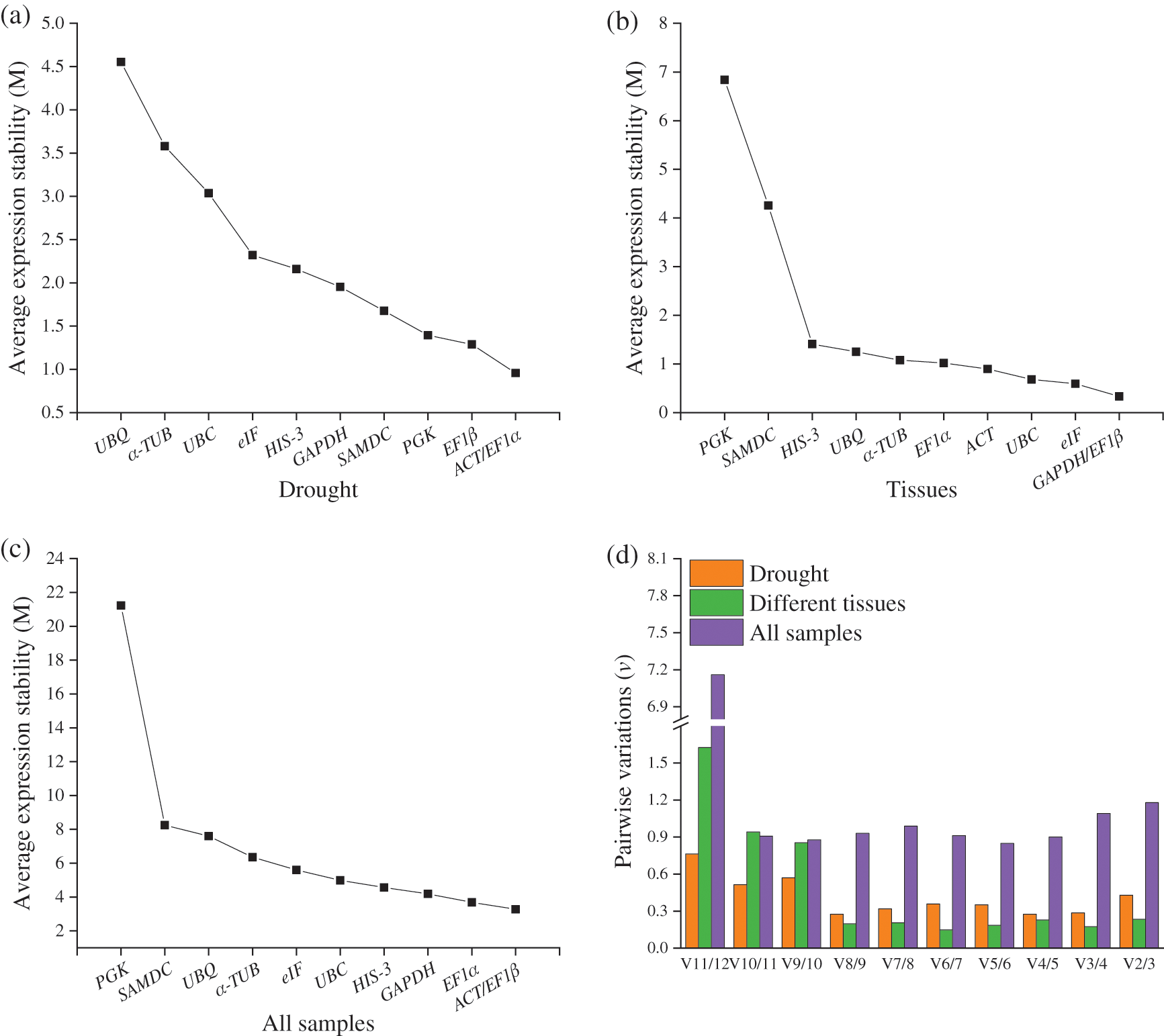
Figure 3: (a–c) The geNorm analysis of expression stability and (d) the variation values for the 11 reference genes under drought, different tissues and all samples
For the drought treatment, different tissues and all samples, NormFinder suggested that EF1β were the most expressed stably reference gene, with an stability value of 0.129, 0.235 and 0.198, respectively, whereas UBC was the least expressed stably reference gene with the value of 0.575 for drought condition, and PGK was the least expressed stably reference gene for both tissues and all samples with the value of 0.881 and 1.576, respectively (Fig. 4).

Figure 4: Robustness of candidate genes by NormFinder analysis
As shown in Table 2, BestKeeper manifested that HIS-3 gene was identified as the most stable reference genes under drought (CV ± SD = 1.83 ± 0.46), and in all samples (CV ± SD = 3.71 ± 0.90). For the different tissues, eIF gene (CV ± SD = 2.37 ± 0.44) was the most expressed stably. The most stable reference genes produced by the above three programs are not the same, a comprehensive ranking of the reference genes was needed with RefFinder. The results showed that ACT and EF1β under drought stress, EF1β and UBC in different tissues were the most stable genes. For all samples, EF1β and ACT were the two most stable reference genes (Fig. 5).
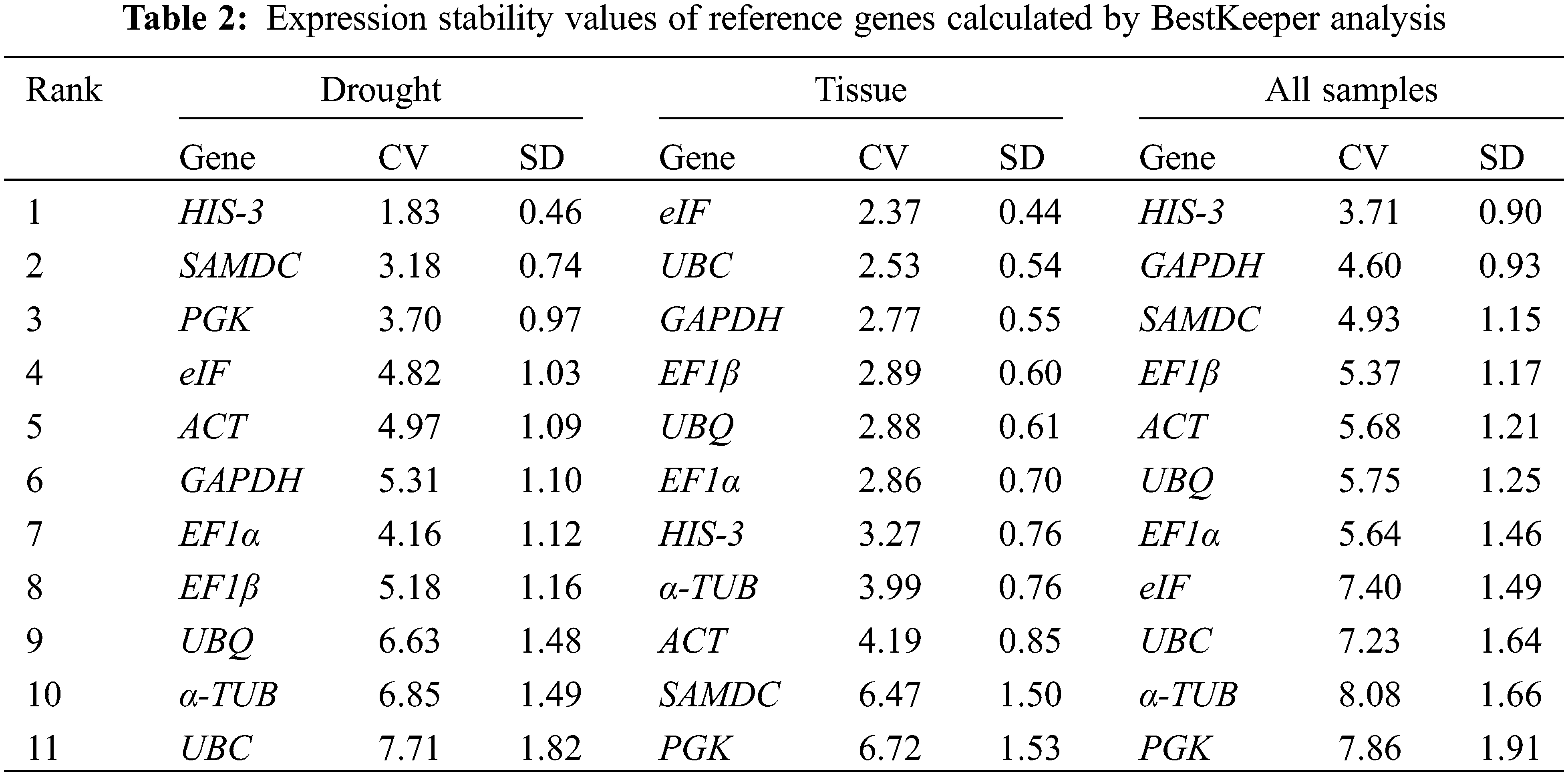

Figure 5: Comprehensive ranking of stability with RefFinder analysis (a, b and c). The lower the column, the more stable the gene was
To verify target gene expression stability of the identified reference genes, the two stable relatively reference genes and one unstable reference gene were selected to normalize gene expression of VP gene. The two more stable reference genes under drought according to RefFinder analysis were ACT and EF1β, and the most unstable was UBC, while the two more stable genes in different tissues were EF1β and UBC, and the most unstable was PGK. When the more ideal reference genes were used alone or combined as the internal reference control, the expression of VP exhibited little changes, whereas the VP gene expression showed large changes with the unstable reference gene to normalize data. Besides, the expression of VP gene was significantly higher than that of other reference genes when using UBC as a correction gene in the early stage of drought. In extreme drought (soil moisture content below 5.57%), the expression level of VP was significantly lower than other reference genes. The unstable internal reference gene had different effects on the expression of the target gene under different drought conditions. And compared with leaves, the expression level of VP gene was lower in other tissues. However, its expression level was higher, even higher than that in leaves when the unstable reference gene UBC was used (Fig. 6).

Figure 6: (a) Expression of VP gene with different validated reference genes for data normalization. (b) VP expression under drought condition. VP expression in different tissues. Data (means ± SD, N = 3) with different letter above bars indicated significant differences between treatments at the 5% level of probability
Because qRT-PCR technology can help scientists save experimental time and improve experimental efficiency, it is widely used in various fields including basic science, biotechnology, medical research, forensic medicine, and diagnostics [4,23,24]. However, regarding the expression levels of these reference genes exist variability in different plants, tissues, developmental stages, and environmental conditions [16,25,26]. The stable internal reference gene is a reliable guarantee for accurate qRT-PCR results in plants. So far, there is no research on the systematic verification of internal reference genes for I. domestica. In this paper, we aimed to select ideal reference genes in I. domestica under drought and among tissues. A total of 11 candidate reference genes, including UBC, α-TUB, eIF, EF1α, EF1β, HIS-3, GAPDH, UBQ, SAMDC, PGK were screened, and were evaluated with geNorm, NormFinder, BestKeeper, and RefFinder. The evaluation results of different algorithms for the reference genes were inconsistent, in which the most expressed stably genes in this study included EF1α by geNorm under drought, EF1β by geNorm and NormFinder in tissues and all samples, EF1β by NormFinder under drought, HIS-3 by BestKeeper under drought and in all samples, eIF by BestKeeper in tissues. In addition, the expression level of UBC under drought fluctuated greatly, which was similar to our transcriptome results (Fig. 1d). Throughout comprehension ranking of RefFinder, the most ideal reference genes in drought, tissues and all samples were ACT, EF1β, EF1β, respectively. However, we can consider using EF1β as the internal reference gene in all samples, because ACT (2.2) and EF1β (2.3) have similar comprehensive ranking scores. EF1β gene catalyzes the extension of the amino acid chain on the ribosome in RNA transcription and regulates the synthesis of related proteins. This gene is usually used as an internal reference gene in many plants to standardize target genes under plants [27], tissues [28] and different conditions [29,30]. Nevertheless, there is also an opposite example that the expression of EF1α is the most stable and the expression of EF1β is the most unstable in the process of Phytophthora Nicotiana infection and methyl jasmonate treatment [31]. These results indicate that the stability of the same reference gene in different plants is inconsistent.
Stability of reference genes identified in this article was further validated with a target gene VP, in different drought samples and different tissues. For drought samples, regardless of whether it is normalized with one internal reference gene (ACT or EF1β) or two internal reference genes (ACT & EF1β), the expression level of VP fluctuated a little. With the aggravation of drought, the expression of VP gene changes greatly. When the unstable internal reference gene was used, the variation error of the VP gene was greater. For the tissues, the expression patterns of VP gene showed similar trends when the two most stable reference genes (EF1β & UBC) or a single gene (EF1β or UBC) was used as internal controls, while the expression level of VP was higher than that of other groups, when the most unstable gene was used as an internal reference gene. Interestingly, we also found that UBC was the most unstable reference gene under drought, but was stably expressed in different tissues similarly to the same results in other reports [16,32,33]. These results indicate that stable internal reference genes are very important for the standardization of target genes in plants. In this article, the screened internal reference genes are only suitable for gene quantitative experiments under drought stress except extreme drought stress and in different tissues.
Acknowledgement: The authors thank Zeliang Lü and Linlin Yang for their help on the manuscript and experiment.
Availability of Data and Materials: The data that support the findings of this study are available on request to the first author, and will be made available upon reasonable request. Other data see Supplementary File.
Funding Statement: This study was funded by the Modern Agricultural Industrial Technology System Project (CARS-21).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Anderson, N. O. (2019). Breeding for dwarf, winter-hardy Iris domestica, blackberry lily (Iridaceae). Acta Horticulturae, 1263, 275–282. DOI 10.17660/ActaHortic.2019.1263.36. [Google Scholar] [CrossRef]
2. Yan, J., Yuan, F., Long, G., Qin, L., Deng, Z. (2012). Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Molecular Biology Reports, 39(2), 1831–1838. DOI 10.1007/s11033-011-0925-9. [Google Scholar] [CrossRef]
3. Zhang, Y. F., Zhao, L. J., Zeng, Y. L. (2014). Selection and application of reference genes for gene expression studies. Plant Physiology Journal, 50(8), 1119–1125. DOI 10.13592/j.cnki.ppj.2014.0201. [Google Scholar] [CrossRef]
4. Guénin, S., Mauriat, M., Pelloux, J., van Wuytswinkel, O., Bellini, C. et al. (2009). Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of Experimental Botany, 60(2), 487–493. DOI 10.1093/jxb/ern305. [Google Scholar] [CrossRef]
5. Gantasala, N. P., Papolu, P. K., Thakur, P. K., Kamaraju, D., Sreevathsa, R. et al. (2013). Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Research Notes, 6(1), 1–11. DOI 10.1186/1756-0500-6-312. [Google Scholar] [CrossRef]
6. Janská, A., Hodek, J., Svoboda, P., Zámečník, J., Prášil, I. T. et al. (2013). The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Molecular Genetics and Genomics, 288(11), 639–649. DOI 10.1007/s00438-013-0774-4. [Google Scholar] [CrossRef]
7. Gutierrez, L., Mauriat, M., Guénin, S., Pelloux, J., Lefebvre, J. F. et al. (2008). The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal, 6(6), 609–618. DOI 10.1111/j.1467-7652.2008.00346.x. [Google Scholar] [CrossRef]
8. Zheng, T., Chen, Z., Ju, Y., Zhang, H., Cai, M. et al. (2018). Reference gene selection for qRT-PCR analysis of flower development in Lagerstroemia indica and L. speciosa. PLoS One, 13(3), e0195004. DOI 10.1371/journal.pone.0195004. [Google Scholar] [CrossRef]
9. Carvalho, K., de Campos, M., Pereira, L., Vieira, L. (2010). Reference gene selection for real-time quantitative polymerase chain reaction normalization in “Swingle” citrumelo under drought stress. Analytical Biochemistry, 402(2), 197–199. DOI 10.1016/j.ab.2010.03.038. [Google Scholar] [CrossRef]
10. Pabuayon, I. M., Yamamoto, N., Trinidad, J. L., Longkumer, T., Raorane, M. L. et al. (2016). Reference genes for accurate gene expression analyses across different tissues, developmental stages and genotypes in rice for drought tolerance. Rice, 9(1), 1–8. DOI 10.1186/s12284-016-0104-7. [Google Scholar] [CrossRef]
11. Guo, J., Ling, H., Wu, Q., Xu, L., Que, Y. (2014). The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Scientific Reports, 4(1), 1–10. DOI 10.1038/srep07042. [Google Scholar] [CrossRef]
12. Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F., Park, C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biology, 8(1), 1–11. DOI 10.1186/1471-2229-8-112. [Google Scholar] [CrossRef]
13. Etschmann, B., Wilcken, B., Stoevesand, K., von Der Schulenburg, A., Sterner-Kock, A. (2006). Selection of reference genes for quantitative real-time PCR analysis in canine mammary tumors using the GeNorm algorithm. Veterinary Pathology, 43(6), 934–942. DOI 10.1354/vp.43-6-934. [Google Scholar] [CrossRef]
14. Andersen, C. L., Jensen, J. L., Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64(15), 5245–5250. DOI 10.1158/0008-5472.CAN-04-0496. [Google Scholar] [CrossRef]
15. Pfaffl, M. W., Tichopad, A., Prgomet, C., Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnology Letters, 26(6), 509–515. DOI 10.1023/B:BILE.0000019559.84305.47. [Google Scholar] [CrossRef]
16. Wang, Y., Zhang, Y., Liu, Q., Liu, L., Huang, S. et al. (2021). Reference gene selection for qRT-PCR normalization in Iris germanica L. Phyton-International Journal of Experimental Botany, 90(1), 277–290. DOI 10.32604/phyton.2020.011545. [Google Scholar] [CrossRef]
17. Gu, C. S., Liu, L. Q., Xu, C., Zhao, Y. H., Zhu, X. D. et al. (2014). Reference gene selection for quantitative real-time RT-PCR normalization in Iris. lactea var. chinensis roots under cadmium, lead, and salt stress conditions. Scientific World Journal, 2014, 7. DOI 10.1155/2014/532713. [Google Scholar] [CrossRef]
18. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
19. Vandesompele, J., de Preter, K., Pattyn, F., Poppe, B., van Roy, N. et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7), 1–12. DOI 10.1186/gb-2002-3-7-research0034. [Google Scholar] [CrossRef]
20. Park, S., Li, J., Pittman, J. K., Berkowitz, G. A., Yang, H. et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proceedings of the National Academy of Sciences, 102(52), 18830–18835. DOI 10.1073/pnas.0509512102. [Google Scholar] [CrossRef]
21. Chen, Y., Tan, Z., Hu, B., Yang, Z., Xu, B. et al. (2015). Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiologia Plantarum, 155(2), 138–148. DOI 10.1111/ppl.12302. [Google Scholar] [CrossRef]
22. Chen, M., Wang, B., Li, Y., Zeng, M., Liu, J. et al. (2021). Reference gene selection for qRT-PCR analyses of luffa (Luffa cylindrica) plants under abiotic stress conditions. Scientific Reports, 11(1), 1–12. DOI 10.21203/rs.3.rs-65316/v1. [Google Scholar] [CrossRef]
23. Bustin, S. A., Mueller, R. (2005). Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clinical Science, 109(4), 365–379. DOI 10.1042/CS20050086. [Google Scholar] [CrossRef]
24. Bollmann, F., Casper, I., Henke, J., Pautz, A. (2012). qRT-PCR: A method and its difficulties. Naunyn-Schmiedeberg’s Arch Pharmacol, 385(10), 949–951. DOI 10.1007/s00210-012-0786-3. [Google Scholar] [CrossRef]
25. Li, R., Cui, K., Xie, Q., Xie, S., Chen, X. et al. (2021). Selection of the reference genes for quantitative gene expression by RT-qPCR in the desert plant Stipagrostis pennata. Scientific Reports, 11(1), 1–11. DOI 10.1038/s41598-021-00833-2. [Google Scholar] [CrossRef]
26. Wei, M., Chen, Y., Zhang, M., Yang, J., Lu, H. et al. (2020). Selection and validation of reference genes for the qRT-PCR assays of Populus ussuriensis gene expression under abiotic stresses and related ABA treatment. Forests, 11(4), 476. DOI 10.3390/f11040476. [Google Scholar] [CrossRef]
27. Liu, X., Zhang, Z., Bian, W., Duan, A., Zhang, H. (2020). Enhancing the expression of ARK1 genes in poplar leads to multiple branches and transcriptomic changes. Royal Society Open Science, 7(9), 201201. DOI 10.1098/rsos.201201. [Google Scholar] [CrossRef]
28. Narancio, R., John, U., Mason, J., Spangenberg, G. (2018). Selection of optimal reference genes for quantitative RT-PCR transcript abundance analysis in white clover (Trifolium repens L.). Functional Plant Biology, 45(7), 737–744. DOI 10.1071/FP17304. [Google Scholar] [CrossRef]
29. Bertini, L., Proietti, S., Focaracci, F., Canini, F., Bravo, L. A. et al. (2021). Identification and validation of new reference genes for accurate quantitative reverse transcriptase-PCR normalization in the Antarctic plant Colobanthus quitensis under abiotic stress conditions. Polar Biology, 44(2), 389–405. DOI 10.1007/s00300-021-02801-y. [Google Scholar] [CrossRef]
30. Li, D., Hu, B., Wang, Q., Liu, H., Pan, F. et al. (2015). Identification and evaluation of reference genes for accurate transcription normalization in safflower under different experimental conditions. PLoS One, 10(10), e0140218. DOI 10.1371/journal.pone.0140218. [Google Scholar] [CrossRef]
31. Zhang, Y., Wang, R., Yang, Z., Lu, Z., Li, J. et al. (2019). Screening of suitable reference genes for qRT-PCR normalization in Sisal. Chinese Journal of Tropical Crops, 40(11), 2166–2173. DOI 10.3969/j.issn.1000-2561.2019.11.010. [Google Scholar] [CrossRef]
32. Hu, X., Zhang, L., Nan, S., Miao, X., Yang, P. et al. (2018). Selection and validation of reference genes for quantitative real-time PCR in Artemisia sphaerocephala based on transcriptome sequence data. Gene, 657, 39–49. DOI 10.1016/j.gene.2018.03.004. [Google Scholar] [CrossRef]
33. Han, X., Lu, M., Chen, Y., Zhan, Z., Cui, Q. et al. (2012). Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS One, 7(8), e43084. DOI 10.1371/journal.pone.0043084. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |