

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.021288
COMMUNICATION
Electrochemical Identification of Yulania spp. by Fingerprinting of Leaves Using Glassy Carbon Electrode
1Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden, Mem. Sun Yat-Sen), Nanjing, 210014, China
2Hangzhou Botanical Garden (Hangzhou West Lake Academy of Landscape Science), Hangzhou, 310013, China
3College of Materials and Environmental Engineering, Hangzhou Dianzi University, Hangzhou, 310018, China
4Department of Chemistry and Biotechnology, Faculty of Science, Engineering and Technology, Swinburne University of Technology, Hawthorn, VIC 3122, Australia
*Corresponding Authors: Yuhong Zheng. Email: zhengyuhong@cnbg.net; Li Fu. Email: fuli@hdu.edu.cn
Received: 06 January 2022; Accepted: 15 March 2022
Abstract: In this communication, we used electrochemical sensor for recording the electrochemical profiles of eleven species of Yulania spp. from leaf extract. Two solvents and two buffer conditions were used for electrochemical fingerprints collection. Their electrochemical fingerprints can be converted to different patterns and consequently for species recognition. The results indicate the pattern recognition is much convenient than that of the recognition of species directly using voltammetric signal. The current information in electrochemical fingerprinting represents the type and amount of electrochemically active molecules, which linked to the genetic differences among the plants. Therefore, the electrochemical fingerprints were applied for further phylogenetic study. The phylogenetic tree deduced from voltametric curves is divided into three main groups. The first clade contains Y. denudate, Liriodendron chinense, Y. cylindrica, Y. biondii, Y. sprengeri. The second clade contains Y. zenii, Y. liliiflora, Y. kobus, and Y. amoena. The third clade contains Y. × soulangeana, Manglietia fordiana and Y. sinostellata. In addition, Y. salicifolia is not in these main clades. The results demonstrate that electrochemical fingerprinting can be used as a com-plementary tool in the study of phylogenetics.
Keywords: Phylogenetic; Yulania spp.; plant identification; fingerprints; chemotaxonomy
Yulania is a genus of deciduous trees or shrubs in the Magnoliaceae family, with about 25 species and varieties. They are mainly found in temperate and warm temperate zones, and are an intermittently distributed taxon in Asia and North America [1,2]. The leaves of Yulania spp. are very diverse in shape, including oval, obovoid, ovoid, sub orbicular and irregular. The buds are highly variable and can be divided into three categories: terminal buds, axillary buds and clustered buds [3]. The flower type is also diverse, and the number, shape and texture of single flowers with tepals are varied. Because of their excellent ornamental and application properties, Yulania spp. have a wide range of cultivation, thus leading to more natural hybridization [4]. Some Yulania spp. have small variation in traits and many crosses, and it is difficult to confirm their relatives correctly only by morphological characteristics such as leaf shape, flower type and fruit, thus leading to more phenomena of different names for the same species [5,6].
Plant taxonomy is the study of the origin, affinities and evolutionary development of different groups of plants by classifying the diverse and complex flora into precise species [7]. The oldest method of plant taxonomy is to observe the apparent morphology and internal anatomical features of the plants to be studied in combination with the growth environment and geographical features of the plants, and then to consider the evolution and affinities of the species [8,9]. This taxonomy method is influenced by external factors and has more uncertainties. In recent years, with the improvement of instrumentation and research methodology, many methods of taxonomy have been added, such as: molecular systematics, cellular taxonomy, quantitative taxonomy, chemical taxonomy, etc. Phytochemical taxonomy is an emerging discipline in which plant taxonomy and phytochemistry interpenetrate from each other [10,11]. It is based on classical taxonomy and phytochemistry to describe and classify plants, to study the interrelationship of phytochemical components in one taxon, and then to explore the evolutionary patterns [12,13]. The secondary metabolites of plants can be used as a basis for phytochemical taxonomy because they are also influenced by genetic factors and have genetic characteristics that can be used as taxonomic markers. However, traditional phytochemical taxonomy tracks only a limited number of secondary metabolites and therefore has a limited degree of interpretation [14–16]. This drawback can be overcome by electrochemical fingerprinting. Electrochemical fingerprinting can track all electrochemically active substances in plant tissues and can give comprehensive data on a large number of components. Electrochemical fingerprinting was proposed in 2015 and has been developed so far as a complementary tool for taxonomy [17]. Our group has also conducted extensive studies that have proven this technique to be a valuable one in the identification and phylogenetic studies of plants [18–25]. In this work, we tried to use this electro-phytochemotaxonomy for the identification and phylogenetic study of Yulania spp. Eleven species of Yulania and two exotaxa were collected as samples. The fingerprints of electrochemically active molecules in their leaf tissues were recorded. These data were used for pattern recognition and phylogenetic investigation.
Leaves of Yulania cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana were supplied by Nanjing Botanical Garden. All leaves were collected in May 2020. All leaves were kept frozen before extraction. 200 mg of leaves were added into 2 mL solvent. Then, 2 mill beads were added and the mixture were sonicated at a high throughput tissue grinding machine (MB-24S, Meibi Co., Ltd., China). Then, the extract was centrifuged at 8000 rmp for 3 min. The supernate was collected and added into electrolyte for electrochemical fingerprinting. The electrochemical recording process was according to previous report [26]. All electrochemical fingerprint were recorded using a CHI760 electrochemical workstation. A three-electrode system has been used for electrochemical fingerprint recording, where a commercial glassy carbon electrode (GCE), an Ag/AgCl electrode and a Pt electrode were used as the working electrode, reference electrode and counter electrode, respectively. Phosphate buffered solution (PBS, pH 7.0) and acetate buffered solution (ABS, pH 4.5) were used as electrolyte. For a typical electrochemical fingerprint recording process, 1 mL of plant extract was injected into a 4 mL electrolyte. Then, a differential pulse voltammetry (DPV) was recorded from −0.1 to 1.5 V. The experimental data was then normalized for further analysis.
A normalization treatment was carried out for fingerprints [23] before analysis. Principal component analysis (PCA) and cluster analysis were used for phylogenetic study.
Fig. 1 shows the electrochemical fingerprints of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana recorded after water extraction and under PBS condition. It can be observed that all three induvial recorded fingerprints were showed uniform profile, indicating excellent reproducibility of the electrochemical fingerprints recorded from the leaf extracts. Several oxidation peaks can be observed during the DPV curves. We can see that the fingerprints are highly variable between the majority of the species, representing different molecules involved in the electrochemical oxidation reaction. This is due to the differences in the type and content of electrochemically active molecules in different species. However, some of these species exhibit relatively similar profiles, such as Y. denudate, Y. × soulangeana and Y. biondii. In addition, Y. kobus and Y. liliiflora also showed similar profiles. These results indicate these species may have similar molecules underwent electrochemical oxidation at the scan window. This reflects the similarity of these species at the genetic level. For example, recent chloroplast genome-based phylogenetic studies of Magnoliaceae have confirmed that Y. kobus and Y. liliiflora are very closely related [27]. Similarity of electrochemical fingerprints is a common condition among species of the same genus. However, this does not necessarily mean that the types and contents of all electrochemically active substances are similar. However, the substances involved in the electrochemical reaction in these samples were relatively similar in the PBS environment. Therefore, it is difficult to identify different species directly with a single electrochemical fingerprint.
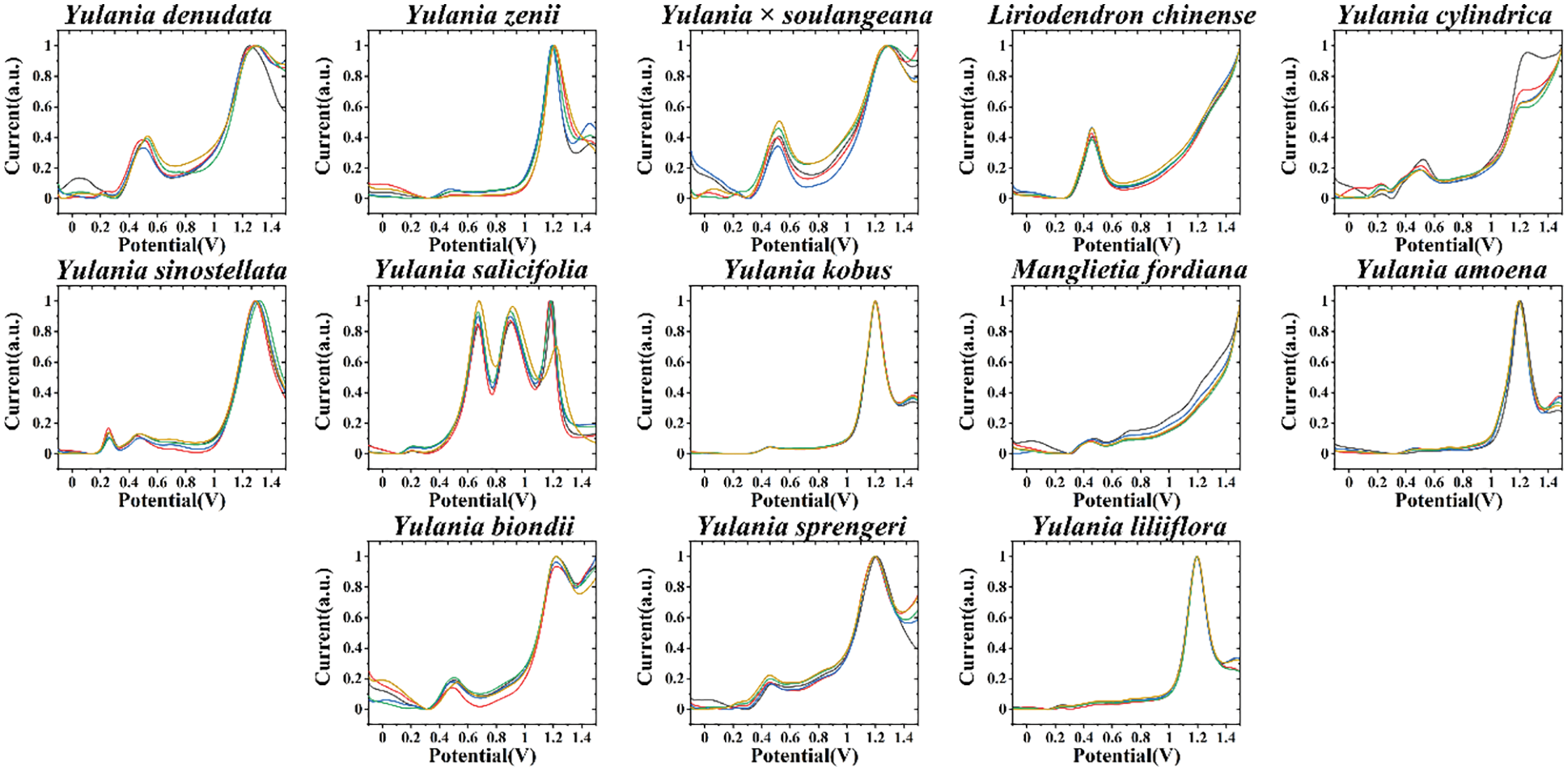
Figure 1: Electrochemical fingerprint of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana (solvent: water, electrolyte: PBS)
To amplify the variability between different species, we can perform fingerprinting of the samples under different conditions. This is because different solvents can extract different electrochemically active molecules, and also different electrolyte and pH environments can limit the electrochemical reactions of different molecules. This is mainly due to the difference of electrochemical behavior of electrochemically active substances in different pH environments. For organic small molecules, generally neutral pH environment can show more electrochemical correspondence. Therefore, we collected fingerprint of each species after ethanol extraction and under ABS as well (Fig. 2). The voltametric profile of each species showed different behavior compared with that of the fingerprint recorded in Fig. 1. The fact that Y. denudate and Y. biondii still have relatively similar electrochemical activities under this condition proves that these two species do have a high similarity in the molecules involved in electrochemical oxidation, potentially suggesting that they are more closely related at the genetic level. However, Y. × soulangeana exhibited a different electrochemical behavior, representing a greater difference between it and the other two species in terms of molecules involved in electrochemical oxidation under this condition. In cases of Y. kobus and Y. liliiflora, they still share a relatively similar electrochemical fingerprint profile, further representing their similarity at the genetic level.
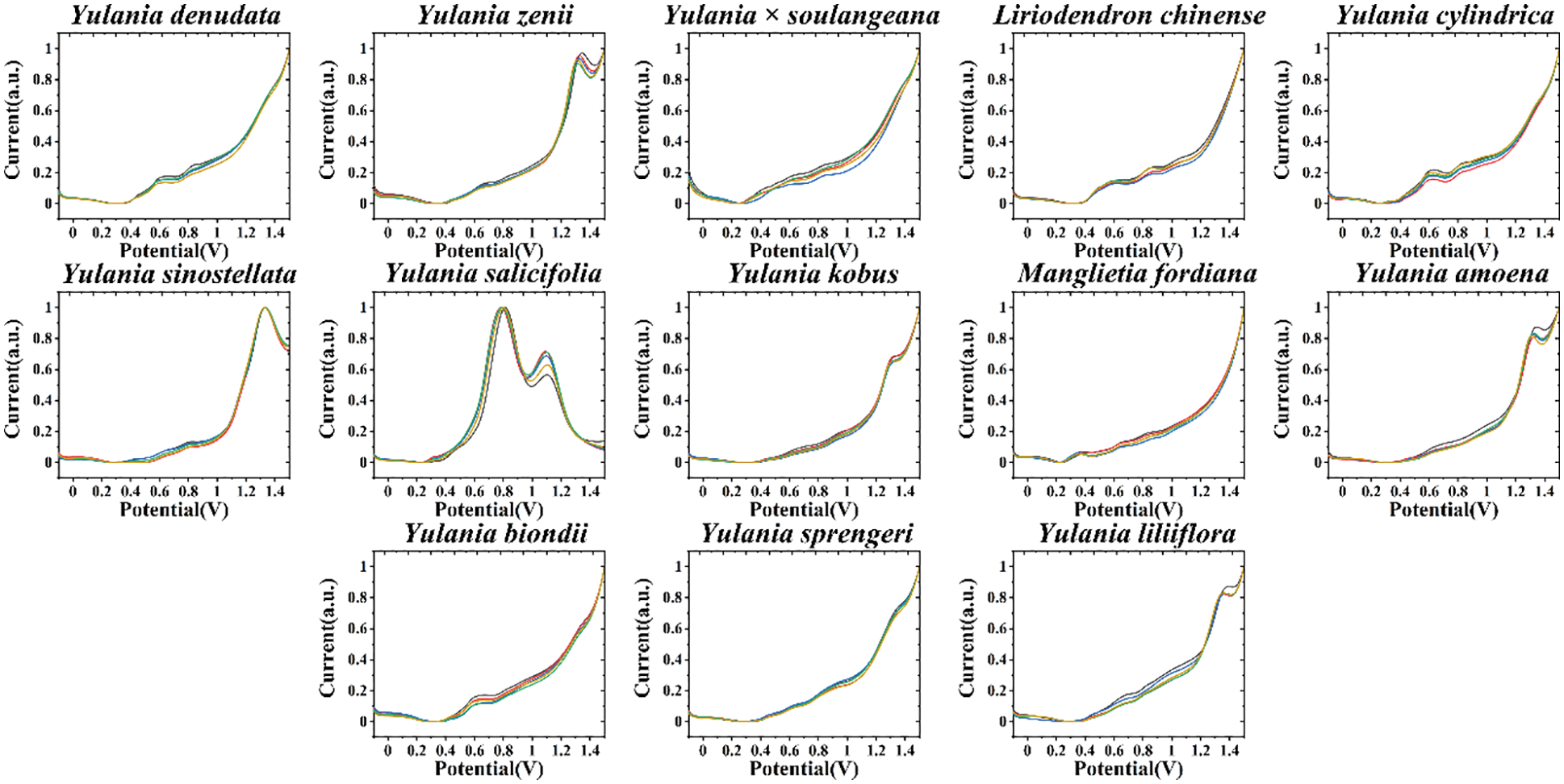
Figure 2: Electrochemical fingerprint of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana (solvent: ethanol, electrolyte: ABS)
The potential in electrochemical fingerprinting does not assume important information because the potential value is the same for each sample, so we can combine the current values of electrochemical fingerprints under different conditions to form different pattern recognition maps. For example, Fig. 3 shows scatter plots combining the currents recorded under PBS for the leaf water extract and under ABS for the leaf ethanol extract. The resolution of the pattern was improved by combining the two sets of fingerprint profiles. Comparing the fingerprint profiles of Figs. 1 and 2, this scatter plot better reflects the variability between different species. For example, the fingerprints of Y. denudate and Y. biondii were highly similar whether they were recorded from water extract under PBS or ethanol extract under ABS. However, their scatter plots showed some differences. In addition, electrochemical fingerprints of Y. kobus and Y. liliiflora were very similar in Figs. 1 and 2 as well, while showed clearly differences in the scatter plots.
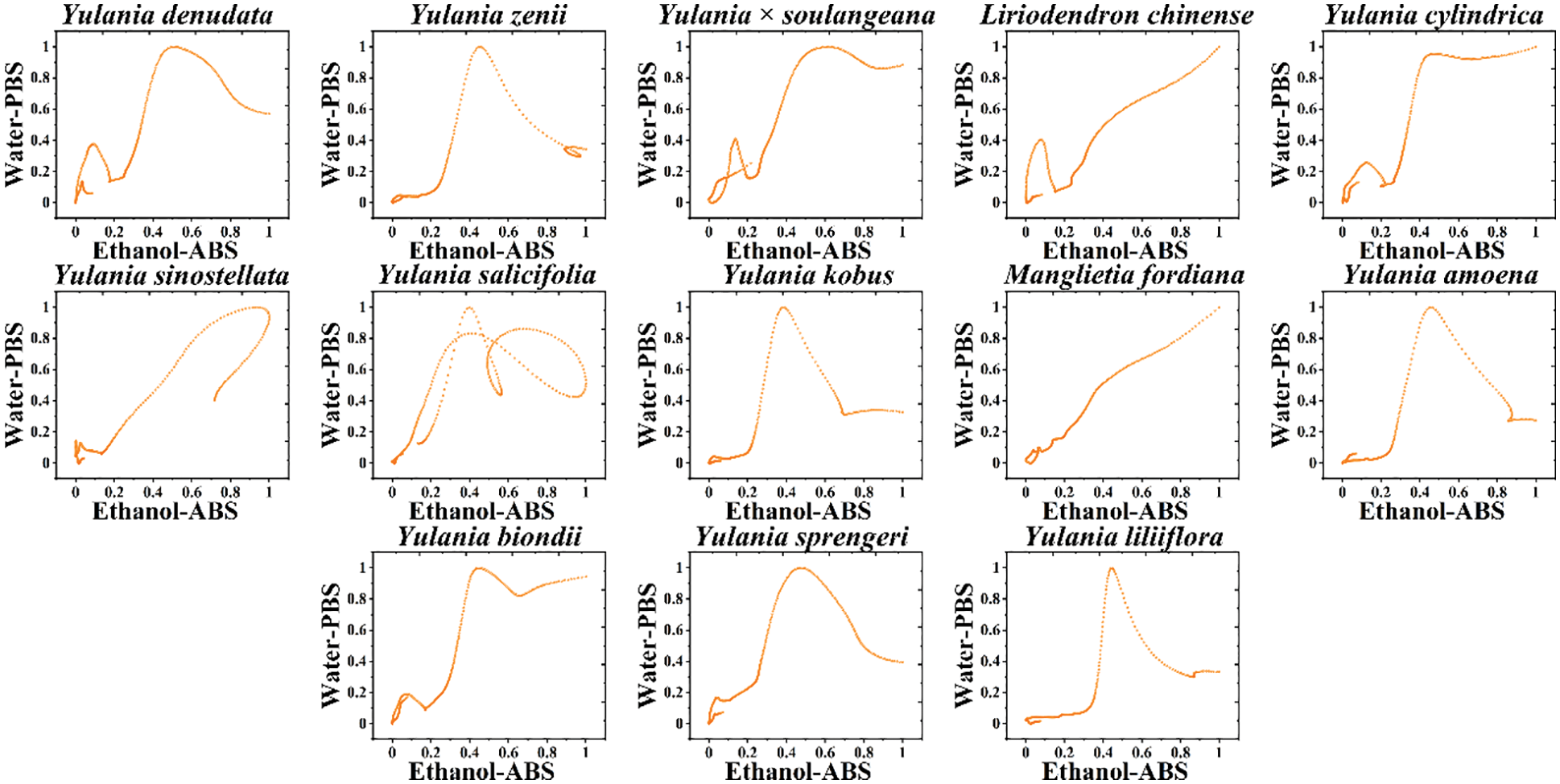
Figure 3: Scatter plots of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana combining the signals collected under PBS for the water extracts and under ABS for the ethanol extracts
Although scatter plots can clearly distinguish different species, it is not convenient to really use them as a database for identifying different species. Therefore, we believe that a two-dimensional density pattern is a more intuitive way for recognition. In a two-dimensional density pattern, data points appear in different colors depending on the density of the surrounding data. This pattern allows the identification of samples to be reached by locating areas of color. Fig. 4 shows the two-dimensional density pattern of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana combining the signals collected under PBS for the water extracts and under ABS for the ethanol extracts. We can see that some areas in the scatter plot become highlighted in this pattern, which is very conveniently for pattern recognition. Moreover, the heat map is able for the calculation of the similarity between different samples (Fig. 5). The heat map uses an alternative identification strategy. It can not only count data points in different regions like a scatter plot, but also locate different regions based on the density of the data like a two-dimensional density pattern. These pattern recognition strategies are more useful for plant species identification than direct fingerprinting.
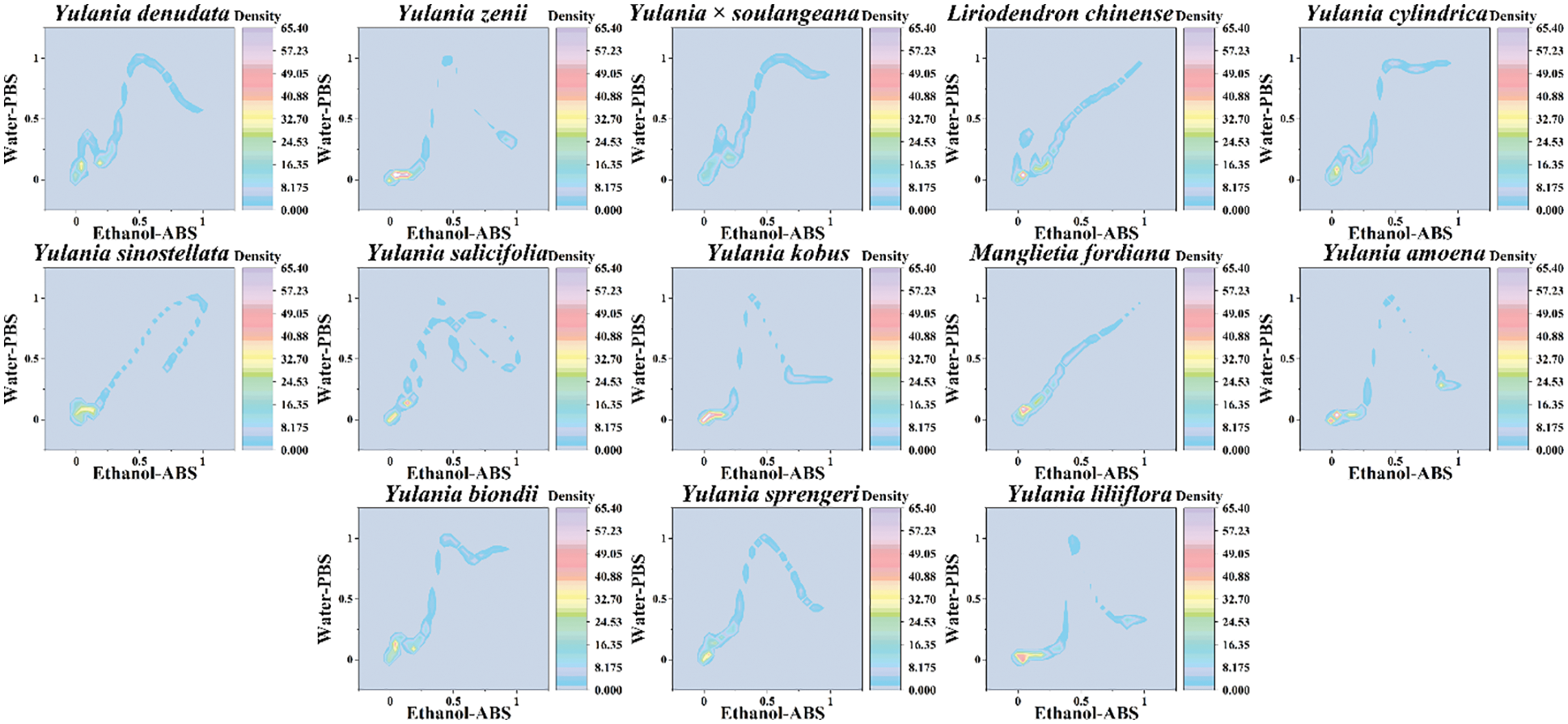
Figure 4: Two-dimensional density pattern of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana combining the signals collected under PBS for the water extracts and under ABS for the ethanol extracts
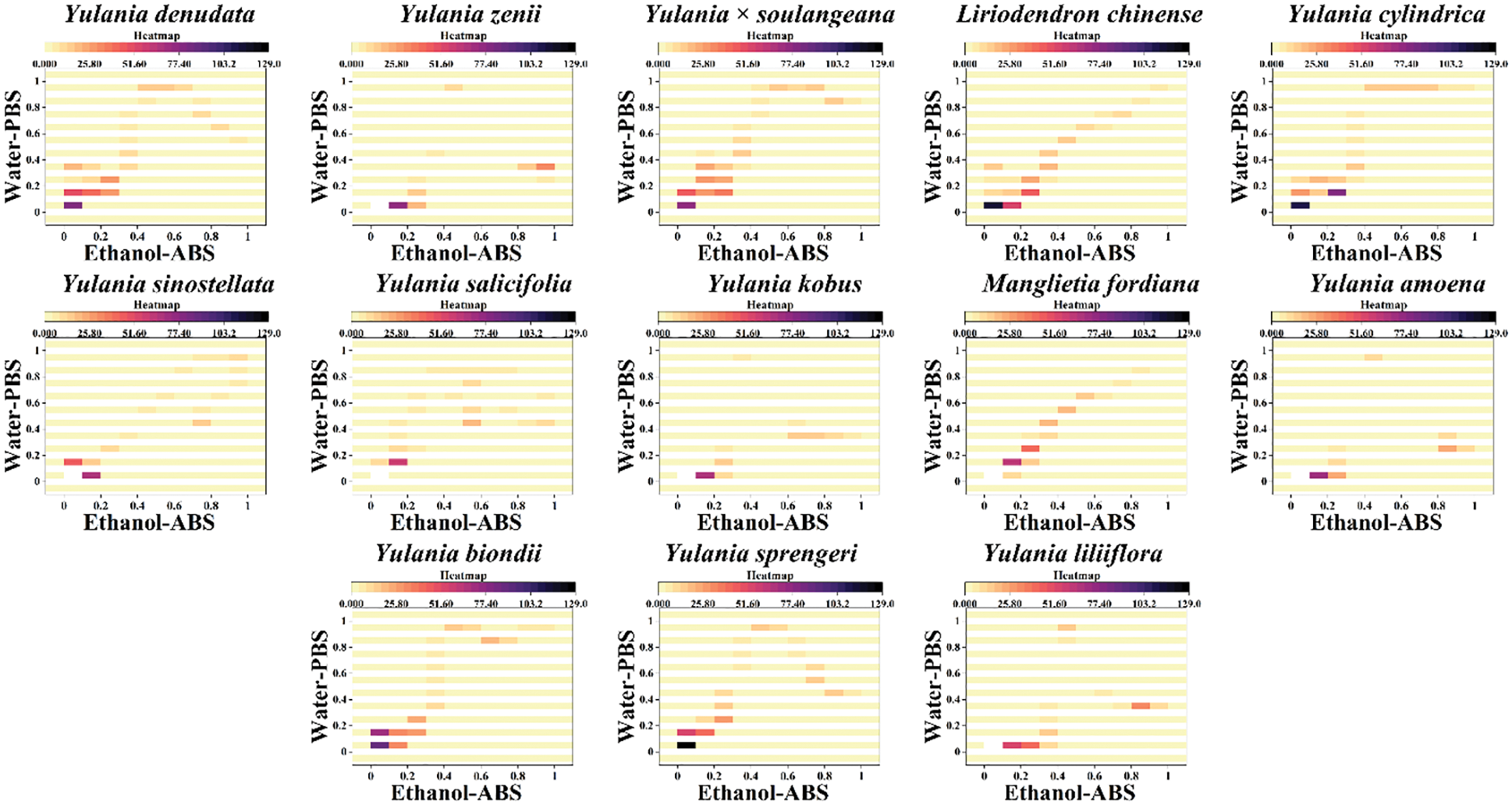
Figure 5: Heatmap of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana combining the signals collected under PBS for the water extracts and under ABS for the ethanol extracts
Since the current information in electrochemical fingerprinting represents the type and amount of electrochemically active molecules in plant tissues, these signals reflect the difference at genetic level. Therefore, the relationship of these species can be investigated using statistical methods. Fig. 6 shows the principal component analysis (PCA) of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana. PCA showed a 83.7% interpretation. This result suggested that electrochemical fingerprinting can be used for the identification of species of Yulania spp.
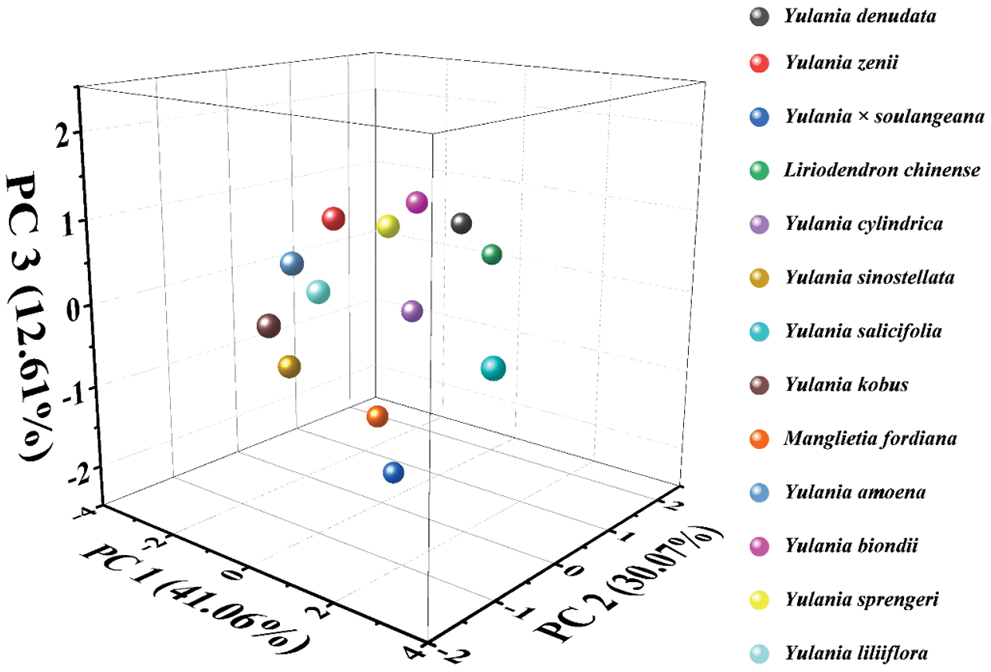
Figure 6: PCA analysis of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana
Yulania is one of the most important taxa in Magnoliaceae and is of great importance in the multidisciplinary theoretical studies of the taxonomic system of the family. The genus Yulania has a long history of cultivation with wide distribution. The species are easy natural hybridization with many types of morphological variation. It is difficult to find true wild populations of many species. There are many semi-wild and cultivated populations that have grown completely naturally. If only morphological variation is seen, without examining whether the species has a wild population, it will result in many species becoming endangered as soon as they are found. The question of whether such a single variant in the wild, semi-wild or cultivated wild state can be treated as a species or variety is a problem [28]. In recent years, molecular biology has been used with great success to analyze many controversial phylogenetic issues. Conventional morphological traits are easily influenced by environmental factors, and the variation types of some species in specific habitats are sometimes mistakenly used as a basis for discovering new species in taxonomy. In contrast, molecular data are heritable and stable. With the development of molecular biology, molecular biology tools such as Restriction Fragment Length Polymorphism (RFLP), Random Amplified Polymorphic DNA (RAPD), Internal Transcribed Spacer (ITS) and chloroplast DNA (cpDNA) sequencing have become more and more important in the systematics of Magnoliaceae [29–31]. While validating many anatomical and morphological views, many phylogenetic trees have been proposed that are very different from morphological taxonomy, making us question some traditional taxonomic bases [32–34]. Here, we performed a clustering analysis among different species using the electrochemical fingerprinting. As shown in Fig. 7, the dendrogram is divided into three groups. The first group contains Y. denudate, Liriodendron chinense, Y. cylindrica, Y. biondii, Y. sprengeri. The second group contains Y. zenii, Y. liliiflora, Y. kobus, and Y. amoena. The third group contains Y. × soulangeana, Manglietia fordiana and Y. sinostellata. In addition, Y. salicifolia is not in these main groups.
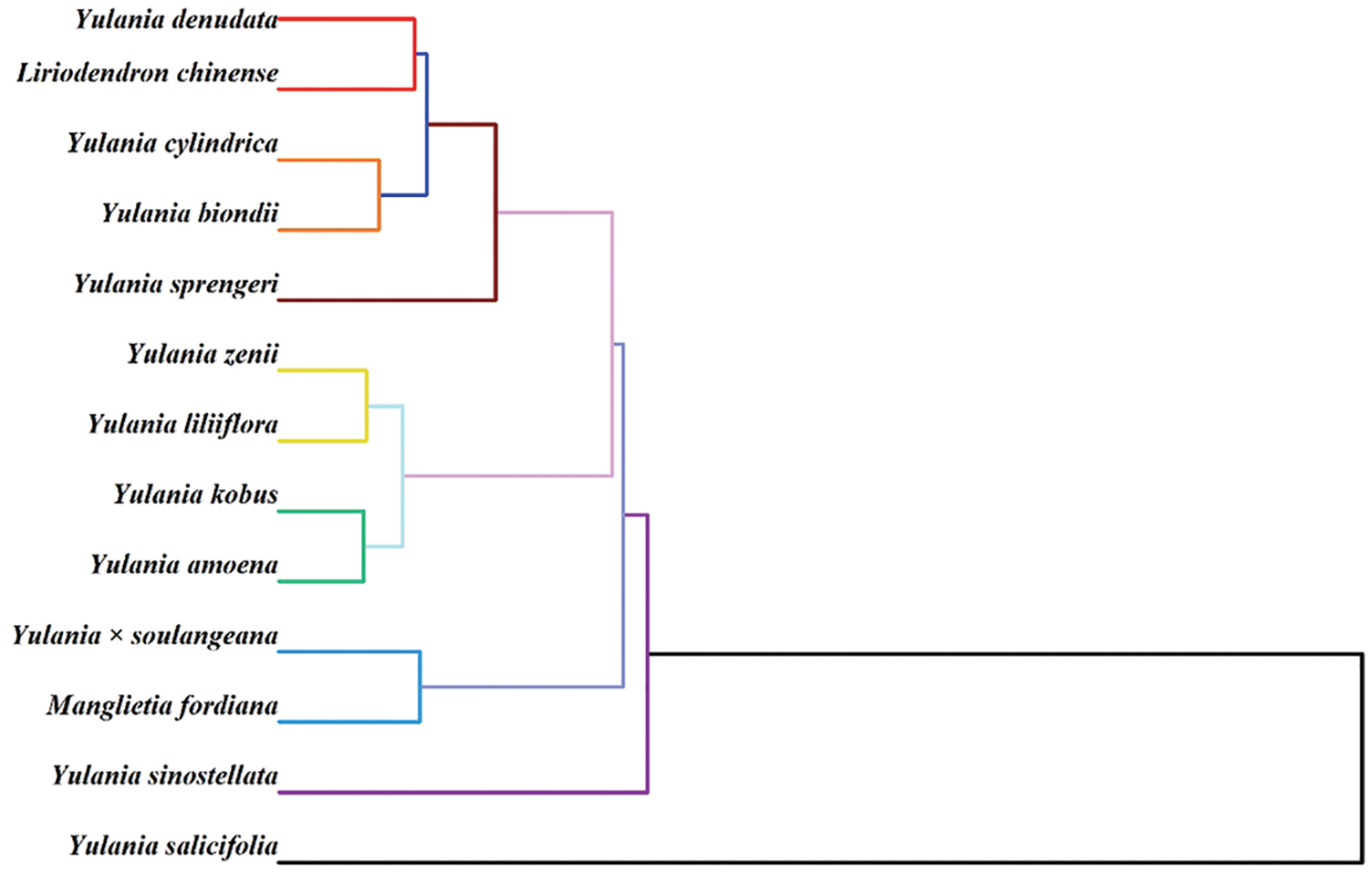
Figure 7: Dendrogram of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana based on electrochemical fingerprints
In the first cluster, the closer affinity between Y. denudata, Y. cylindrica and Y. sprengeri has been confirmed by the phylogenetic findings of RAPD [35]. It is not unexpected that Liriodendron chinense and these species are clustered in a cluster. Due to the primitive conserved DNA sequences of Magnoliaceae, the base variation sites are much less variable compared to other angiosperms [36]. However, the clustering of Y. biondii at this position is inconsistent with previously published results [37]. Y. biondii is generally considered to be closely related to Y. kobus and Y. liliiflora. In the second cluster, the positions of Y. liliiflora, Y. kobus and Y. amoena are very consistent with the previously published results. The relationship of Y. zenii is generally considered to be close to that of Y. denudate [38], but data based on electrochemical fingerprinting do not support this view. Previous studies did not include Y. fordiana, so this study gives its position in the phylogeny. A more distant relationship was shown between Y. sinostellata and other species in the RADP study [35]. Our results suggest that Y. salicifolia does not cluster with other species, probably because it is a specie of Japanese origin. The above results demonstrate that electrochemical fingerprinting can be used as a complementary tool in the study of phylogenetics.
The electrochemical fingerprint of Y. cylindrica, Y. denudata, Y. sprengeri, Y. kobus, Y. sinostellata, Y. × soulangeana, Y. zenii, Y. biondii, Y. amoena, Y. salicifolia, Y. liliiflora, Liriodendron chinense and Manglietia fordiana were recorded. These fingerprints are effective data for species identification via pattern recognition methods including two-dimensional density pattern and heatmap. The electrochemical fingerprints were used for further phylogenetic study. The dendrogram is divided into three groups. The first group contains Y. denudate, Liriodendron chinense, Y. cylindrica, Y. biondii, Y. sprengeri. The second group contains Y. zenii, Y. liliiflora, Y. kobus, and Y. amoena. The third group contains Y. × soulangeana, Manglietia fordiana and Y. sinostellata. In addition, Y. salicifolia is not in these main groups.
Author Contributions: Conceptualization, Y.Z. and L.F.; methodology, Y.Z., Z.L., A.Y. and P.Z.; investigation, Z.L., P.Z. and B.F.; writing—original draft preparation, Z.L. and B.F.; writing—review and editing, L.F., A.Y. and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding Statement: This research was supported by the Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF, No. CX (21)3044) and National Natural Science Foundation of China (22004026).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Fu, D. L., Fu, H., Qin, Y., Zhou, D. S., Duan, R. M. (2019). Analyses of chloroplast genomic and morphological evolutionomy of Yulania subsect. Cylindricae (Magnoliaceae). American Journal of Agriculture and Forestry, 7(5), 200–211. DOI 10.11648/j.ajaf.20190705.16. [Google Scholar] [CrossRef]
2. Fu, L., Mao, S., Chen, F., Zhao, S., Su, W. et al. (2022). Graphene-based electrochemical sensors for antibiotic detection in water, food and soil: A scientometric analysis in CiteSpace (2011–2021). Chemosphere, 297, 134127. DOI 10.1016/j.chemosphere.2022.134127. [Google Scholar] [CrossRef]
3. Sun, L., Jiang, Z., Wan, X., Zou, X., Yao, X. et al. (2020). The complete chloroplast genome of Magnolia polytepala: Comparative analyses offer implication for genetics and phylogeny of Yulania. Gene, 736, 144410. DOI 10.1016/j.gene.2020.144410. [Google Scholar] [CrossRef]
4. Fu, D. L., Zhang, Q., Xu, M., Zhou, D. S., Qin, Y. et al. (2019). Two particularly evolutionary loci of trnL-ndhJ of cpDNA of Yulania baotaina, a new species (Magnoliaceae) from China. American Journal of Agriculture and Forestry, 7(5), 229–233. DOI 10.11648/j.ajaf.20190705.19. [Google Scholar] [CrossRef]
5. Ye, K., Qin, J. (2019). Selection of photosynthesis light response model of Yulania stellata and its varieties. Journal of West China Forestry Science, 48(4), 132–136. [Google Scholar]
6. Xie, H., Wang, D., Li, M., Yang, Y., Yang, X. et al. (2019). The complete chloroplast genome of the endangered species Oyama sinensis (Magnoliaceae). Mitochondrial DNA Part B, 4(1), 830–831. DOI 10.1080/23802359.2019.1567286. [Google Scholar] [CrossRef]
7. Mannaa, M., Park, I., Seo, Y. S. (2019). Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated burkholderia species. International Journal of Molecular Sciences, 20(1), 121. DOI 10.3390/ijms20010121. [Google Scholar] [CrossRef]
8. Bonini, S. A., Premoli, M., Tambaro, S., Kumar, A., Maccarinelli, G. et al. (2018). Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. Journal of Ethnopharmacology, 227, 300–315. DOI 10.1016/j.jep.2018.09.004. [Google Scholar] [CrossRef]
9. Almerekova, S., Shchegoleva, N., Abugalieva, S., Turuspekov, Y. (2020). The molecular taxonomy of three endemic central asian species of Ranunculus(Ranunculaceae). PLoS One, 15(10), e0240121. DOI 10.1371/journal.pone.0240121. [Google Scholar] [CrossRef]
10. Leff, J. W., Bardgett, R. D., Wilkinson, A., Jackson, B. G., Pritchard, W. J. et al. (2018). Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. The ISME Journal, 2(7), 1794–1805. DOI 10.1038/s41396-018-0089-x. [Google Scholar] [CrossRef]
11. Koch, M. A., German, D. A., Kiefer, M., Franzke, A. (2018). Database taxonomics as key to modern plant biology. Trends in Plant Science, 23(1), 4–6. DOI 10.1016/j.tplants.2017.10.005. [Google Scholar] [CrossRef]
12. Cano-Ortiz, A., Spampinato, G., Piñar-Fuentes, J. C., Pinto-Gomes, C. J., Quinto-Canas, R. et al. (2021). Taxonomy, ecology and distribution of Juniperus oxycedrus L. group in the Mediterranean basin using bioclimatic, phytochemical and morphometric approaches, with special reference to the iberian peninsula. Forests, 12(6), 703. DOI 10.3390/f12060703. [Google Scholar] [CrossRef]
13. Yahyaabadi, Y., Mahmoudi-Otaghvari, A., Nazifi, E. (2020). Phytochemical and palynological study of several species of Mentha L. in North of Iran. Journal of Plant Research (Iranian Journal of Biology), 33(4), 843–853. [Google Scholar]
14. Arogundade, O. O., Adedeji, O. (2020). The importance of nutritive parameters in the taxonomy of some corm-producing members of the family araceae. Notulae Scientia Biologicae, 12(2), 318–333. DOI 10.15835/nsb12210537. [Google Scholar] [CrossRef]
15. Roma-Marzio, F., Najar, B., Alessandri, J., Pistelli, L., Peruzzi, L. (2017). Taxonomy of prickly juniper (Juniperus oxycedrus groupA phytochemical–morphometric combined approach at the contact zone of two cryptospecies. Phytochemistry, 141, 48–60. DOI 10.1016/j.phytochem.2017.05.008. [Google Scholar] [CrossRef]
16. Bilušić-Vundać, V. (2019). Taxonomical and phytochemical characterisation of 10 stachys taxa recorded in the Balkan peninsula flora: A review. Plants, 8(2), 32. DOI 10.3390/plants8020032. [Google Scholar] [CrossRef]
17. Domenech-Carbo, A., Ibars, A. M., Prieto-Mossi, J., Estrelles, E., Scholz, F. et al. (2015). Electrochemistry-based chemotaxonomy in plants using the voltammetry of microparticles methodology. New Journal of Chemistry, 39(9), 7421–7428. DOI 10.1039/C5NJ01233C. [Google Scholar] [CrossRef]
18. Fu, L., Su, W., Chen, F., Zhao, S., Zhang, H. et al. (2021). Early sex determination of ginkgo biloba based on the differences in the electrocatalytic performance of extracted peroxidase. Bioelectrochemistry, 140, 107829. DOI 10.1016/j.bioelechem.2021.107829. [Google Scholar] [CrossRef]
19. Fu, L., Zheng, Y., Zhang, P., Zhang, H., Wu, M. et al. (2019). An electrochemical method for plant species determination and classification based on fingerprinting petal tissue. Bioelectrochemistry, 129, 199–205. DOI 10.1016/j.bioelechem.2019.06.001. [Google Scholar] [CrossRef]
20. Xu, Y., Lu, Y., Zhang, P., Wang, Y., Zheng, Y. et al. (2020). Infrageneric phylogenetics investigation of chimonanthus based on electroactive compound profiles. Bioelectrochemistry, 133, 107455. DOI 10.1016/j.bioelechem.2020.107455. [Google Scholar] [CrossRef]
21. Fan, B., Wang, Q., Wu, W., Zhou, Q., Li, D. et al. (2021). Electrochemical fingerprint biosensor for natural indigo dye yielding plants analysis. Biosensors, 11(5), 155. DOI 10.3390/bios11050155. [Google Scholar] [CrossRef]
22. Zhou, J., Zheng, Y., Zhang, J., Karimi-Maleh, H., Xu, Y. et al. (2020). Characterization of the electrochemical profiles of lycoris seeds for species identification and infrageneric relationships. Analytical Letters, 53(15), 2517–2528. DOI 10.1080/00032719.2020.1746327. [Google Scholar] [CrossRef]
23. Fu, L., Zheng, Y., Zhang, P., Zhang, H., Zhuang, W. et al. (2018). Enhanced electrochemical voltammetric fingerprints for plant taxonomic sensing. Biosensors and Bioelectronics, 120, 102–107. DOI 10.1016/j.bios.2018.08.052. [Google Scholar] [CrossRef]
24. Fu, L., Zheng, Y., Zhang, P., Zhang, H., Xu, Y. et al. (2020). Development of an electrochemical biosensor for phylogenetic analysis of amaryllidaceae based on the enhanced electrochemical fingerprint recorded from plant tissue. Biosensors and Bioelectronics, 159, 112212. DOI 10.1016/j.bios.2020.112212. [Google Scholar] [CrossRef]
25. Zhang, M., Pan, B., Wang, Y., Du, X., Fu, L. et al. (2020). Recording the electrochemical profile of pueraria leaves for polyphyly analysis. ChemistrySelect, 5(17), 5035–5040. DOI 10.1002/slct.202001100. [Google Scholar] [CrossRef]
26. Fu, L., Zheng, Y., Wang, A., Zhang, P., Ding, S. et al. (2021). Identification of medicinal herbs in Asteraceae and Polygonaceae using an electrochemical fingerprint recorded using screen-printed electrode. Journal of Herbal Medicine, 30, 100512. DOI 10.1016/j.hermed.2021.100512. [Google Scholar] [CrossRef]
27. Zhu, B., Qian, F., Wang, X., Liu, Y. (2021). The phylogeny of magnoliaceae based on chloroplast genome. Journal of Biology, 1(1), 1–7. DOI 10.1007/s12374-013-0111-9. [Google Scholar] [CrossRef]
28. Dali, F. (2001). Notes on yulania spach. Wuhan Botanical Research, 19(3), 191–198. [Google Scholar]
29. Wang, Y. L., He, Z. C., Ejder, E., Yang, J. F., Zhang, S. Z. (2016). Cytological study of tetraploid species of Magnolia subgenus Yulania (Magnoliaceae). Cytologia, 81(2), 195–205. DOI 10.1508/cytologia.81.195. [Google Scholar] [CrossRef]
30. Zhao, D. X. (2008). Study on the classification characteristics of 35 species of Yulania spach. Journal of Anhui Agricultural Sciences, 35(36), 11811. [Google Scholar]
31. Lei, G., Mao, P., He, M., Wang, L., Liu, X. et al. (2016). Combination of column adsorption and supercritical fluid extraction for recovery of dissolved essential oil from distillation waste water of Yulania liliiflora. Journal of Chemical Technology & Biotechnology, 91(6), 1896–1904. DOI 10.1002/jctb.4785. [Google Scholar] [CrossRef]
32. Wang, Y. L., Li, Y., Zhang, S. Z., Yu, X. S. (2006). The utility of matK gene in the phylogenetic analysis of the genus magnolia. Journal of Systematics and Evolution, 44(2), 135–147. DOI 10.1360/aps040013. [Google Scholar] [CrossRef]
33. Si, H., Yu, J., Wu, X. (2019). Study on damage and regeneration of pine forest after fire caused by two types of fire. Journal of West China Forestry Science, 48(1), 7–11. [Google Scholar]
34. Sun, L., Jiang, Z., Liu, C., Yin, Z. (2018). Analysis of the adaptive and geographical distribution of Yulania liliiflora based on DIVA-GIS. Plant Science Journal, 36(6), 804–811. [Google Scholar]
35. Wang, Y., Yong, L., Zhang, S., Cui, A. (2003). Phylogenetic relationship among several subgenus Yulania species based onRAPD markers. Acta Horticulturae Sinica, 30(3), 299. [Google Scholar]
36. Kuang, D. Y., Wu, H., Wang, Y. L., Gao, L. M., Zhang, S. Z. et al. (2011). Complete chloroplast genome sequence of Magnolia kwangsiensis (MagnoliaceaeImplication for DNA barcoding and population genetics. Genome, 54(8), 663–673. DOI 10.1139/g11-026. [Google Scholar] [CrossRef]
37. Azuma, H., Chalermglin, P., Nooteboom, H. P. (2011). Molecular phylogeny of magnoliaceae based on plastid DNA sequences with special emphasis on some species from continental Southeast Asia. Thai Forest Bulletin, 39, 148–165. [Google Scholar]
38. Shi, S., Jin, H., Zhong, Y., He, X., Huang, Y. et al. (2000). Phylogenetic relationships of the magnoliaceae inferred from cpDNA matK sequences. Theoretical and Applied Genetics, 101(5), 925–930. DOI 10.1007/s001220051563. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |