

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.021199
ARTICLE
Identification and Characterization of a Novel Yellow Leaf Mutant yl1 in Rice
1Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region, Ministry of Education, Institute of Agro-Bioengineering and College of Life Sciences, Guizhou University, Guiyang, 550025, China
2Guizhou Plant Conservation Technology Center, Guizhou Key Laboratory of Agricultural Biotechnology, Guizhou Academy of Agricultural Sciences, Guiyang, 550006, China
*Corresponding Author: Degang Zhao. Email: dgzhao@gzu.edu.cn
#These authors contributed equally to this study
Received: 02 January 2022; Accepted: 03 March 2022
Abstract: Leaf-color mutants play an important role in the study of chlorophyll metabolism, chloroplast development, and photosynthesis system. In this study, the yellow leaf 1 (yl1) rice mutant was identified from the ethyl methane sulfonate-treated mutant progeny of Lailong, a glutinous japonica rice landrace cultivated in Guizhou Province, China. Results showed that yl1 exhibited yellow leaves with decreased chlorophyll content throughout the growth period. Chloroplast development in the yl1 mutant was disrupted, and the grana lamellae was loosely packed and disordered. RNA sequencing and real-time quantitative polymerase chain reaction (qRT-PCR) analysis revealed that the chlorophyll synthesis-related genes OsCHLH, OsCHLM, OsCHLG, PORB, and YGL8, as well as the chloroplast development-related genes FtsZ, OsRpoTp, and RbcL, were down-regulated in the yl1 mutant. Genetic analysis revealed that the yellow leaf phenotype of yl1 was controlled by recessive nuclear gene. By employing the MutMap method, the mutation responsible for the phenotype was mapped to a 6.17 Mb region between 17.34 and 23.51 Mb on chromosome 3. Two non-synonymous single-nucleotide polymorphisms (SNPs) located in the gene locus LOC_Os03g31210 and LOC_Os03g36760 were detected in this region. The two SNPs were further confirmed by PCR and Sanger sequencing. The expression patterns of the two candidate genes indicated that LOC_Os03g36760 showed greater potential for functional verification. Subcellular protein localization revealed that the encoded product of LOC_Os03g36760 was localized in the nucleus, cytoplasm, and plasma membrane. These results will be useful for further characterization and cloning of the yl1 gene, and for research on the molecular mechanisms controlling biogenesis and chloroplast biochemical processes.
Keywords: Rice; yl1; chlorophyll biosynthesis; map-based cloning
Rice (Oryza sativa L.) is an important crop that is a staple food for more than half of the world’s population [1]. It has been estimated that rice production must increase by 40% by 2030 to meet the demands of the growing population [2]. The pigment of leaves is normally primarily green, but abnormal biosynthesis and degradation of chlorophyll can change the pigment content and proportions, and lead to alteration of the leaf-color phenotype. Leaf color has an important influence on photosynthesis, growth, and development in plants [3]. To date, more than 200 leaf-color rice mutants have been reported and at least 50 leaf-color genes have been cloned, and are distributed among all 12 chromosomes in rice. Mutation of genes participating in chloroplast development, the chlorophyll metabolism pathway, and associated regulatory genes affect chlorophyll synthesis, thus resulting in leaf-color mutants. More than 20 chlorophyll synthesis-related genes and 14 genes involved in chloroplast development have been cloned in rice [4–10].
In rice, OsCHLH encodes the H subunit of magnesium chelatase (Mg-chelatase), a crucial enzyme for chlorophyll synthesis and chloroplast development [11]. The YGL7 and OsChl9 genes encode the CHLD and CHLI subunits of Mg-chelatase, respectively. Mutations in these subunits reduce the activity of Mg-chelatase, resulting in a yellow-green leaf phenotype [4–6]. Mutation of OsCRD1, a gene encoding Mg-protoporphyrin IX monomethyl ester cyclase, causes abnormal chlorophyll synthesis and chloroplast development, resulting in a yellow leaf and dwarf phenotype in rice [12]. The genes OsCAO1/PGL and OsCAO2 catalyze the synthesis of chlorophyll b from chlorophyll a. Of these two genes, OsCAO1 plays a major role in the synthesis of chlorophyll b, whereas OsCAO2 may play a less obvious role [7,13]. Protochlorophyllide oxidoreductase (POR) catalyzes the photoreduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide) during chlorophyll synthesis. The OsPORB gene is essential for maintenance of light-dependent chlorophyll synthesis, especially under high light intensity, whereas OsPORA functions mainly in the early stages of leaf development [14]. The OsPORB loss-of-function mutant ygl16 shows a decreased pigment concentration and photosynthetic capacity [15].
Abnormal chloroplast development causes changes in the normal proportion and content of each chloroplast pigment, which is manifested as diverse leaf-color mutant phenotypes. Therefore, mutations in genes associated with the regulation of chloroplast development, such as VYL (NAL9), RNRL1 (V3), OsNOA1/RIF1, OsGluRS, and ASL2, may also cause leaf-color mutations [16–22]. Most proteins localized in chloroplasts are the coding products of nuclear genes. Thus, the mutation of genes involved in the regulation of signal transduction between chloroplasts and the nucleus may also cause a change in leaf color. In rice, BGL encodes a guanine nucleotide exchange factor, OsRopGEF10, which activates Rop/Rac GTPases. OsRopGEF10 acts as a molecular switch in signal transduction by substituting GTP (the active form) for bound GDP (the inactive form) in eukaryotic responses to external or internal signals. Mutation of BGL results in a bright green leaf phenotype [23]. In addition, a number of other associated genes regulate leaf color. OsPDF1B encodes peptide deformylase, which acts to remove the N-terminal formyl group from methionine residues in protein synthesis. The Pdf1b/PDf1B homozygous mutant shows a degreening and growth-arrested phenotype with low chlorophyll content [24].
Studies of cloned rice leaf-color-associated genes have shown that such genes are involved in chlorophyll metabolism, chloroplast development, and chloroplast and nuclear signaling pathways. These results indicate that rice leaf color is regulated by a series of genes in multiple pathways. Gratifying progress has been made in elucidating the mechanism of rice leaf-color regulation, but leaf color development is a complex process, and the understanding of this regulatory network remains incomplete. Mutants are basic materials of genetic research and important materials for genomic research [25]. Leaf-color mutants are important in the study of photosynthesis, as well as morphogenesis of chloroplasts and other photosynthetic organs in response to environmental signals, and can possibly be used as screening markers in heterosis [26]. Therefore, the screening and identification of new leaf-color-related mutants, and gene mapping, cloning, and functional analysis of these mutants have important theoretical and practical applications in rice research.
In the current study, we isolated and characterized the yellow leaf 1 (yl1) rice mutant from the ethyl methane sulfonate (EMS)-treated mutants of Lailong, a glutinous rice landrace grown in Guizhou Province, China. The yl1 mutant exhibited a yellow leaf phenotype and reduced chlorophyll content throughout the growth period. Morphological observation and genetic analysis of the yl1 mutant, and fine mapping of the yl1 gene were performed, with the ultimate objective of cloning and functional analysis of the candidate gene.
2.1 Plant Materials and Growth Conditions
The yellow leaf 1 (yl1) rice mutant was isolated from the M2 generation raised from EMS-treated seeds of Lailong, a glutinous rice landrace from Guizhou Province, China. The Lailong wild type (WT) and yl1 seeds were sown in a seedbed for 20 days and seedlings were transplanted to a paddy field in Guiyang, Guizhou Province. The mutants were differentiated from normal segregants by the yellow leaf phenotype. Morphological traits of the WT and yl1 mutant were recorded in the mature stage.
2.2 Transmission Electron Microscopic Analyses
For transmission electron microscopy, leaves from 30-day-old seedings of the WT and yl1 mutant were cut into 5-mm-long pieces, fixed in 2.5% glutaraldehyde in phosphate buffer at 4°C for 4 h, rinsed, and incubated overnight in 1% OsO4 solution at 4°C. The samples were subsequently dehydrated, embedded, sectioned, and stained as described by Wang et al. [8]. The sections were observed with a Hitachi H-7650 transmission electron microscope (Hitachi, Tokyo, Japan).
2.3 Pigment and Chlorophyll Precursor Determination
Pigments were extracted from fresh leaf tissue with 80% acetone. The absorbance of the extract was measured at 470, 645, and 663 nm with a Varioskan Flash spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Total chlorophyll, chlorophyll a, chlorophyll b, and carotene contents were determined following the method described by Arnon [27]. The content of δ-aminolevulinic acid (ALA) was measured based on the method of Morton [28]. The chlorophyll precursors protoporphyrin IX (Proto IX), magnesium-protoporphyrin IX (Mg-Proto IX), Pchlide, and Chlide were assayed following the previously described methods [29,30]. The chlorophyll precursors in the acetone phase were quantified with a Varioskan Flash spectrophotometer (Thermo Fisher Scientific, Waltham City, MA, USA) using Ex400:Em632 for Proto IX, Ex440:Em633 for Pchlide, Ex440:Em672 for Chlide, and Ex420:Em595 for Mg-Proto IX.
2.4 RNA-Sequencing Analysis and Real-Time Quantitative RT-PCR
Total RNA was extracted from leaves of 4-week-old yl1 and WT plants using an RNA extraction kit (Omega Bio-Tek, Doraville, GA, USA) in accordance with the manufacturer’s instructions. The cDNA libraries were constructed and sequenced on a BGISEQ-500 platform by the Beijing Genomics Institute (Shenzhen, China; http://www.genomics.org.cn). Three biological replicates were prepared for each sequencing library. The DEGseq method was used to screen genes differentially expressed between groups [31]. The differentially expressed genes (DEGs) were selected according to a previously described method [32]. Clusters of orthologous groups (COG) functional classification, gene ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways annotation and enrichment analyses were conducted by accessing the NCBI COG, GO, and KEGG databases, respectively.
For real-time quantitative RT-PCR (qRT-PCR), cDNAs were generated by RT-PCR using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham City, MA, USA). The qRT-PCR analyses were performed using an qTower3G Real-time PCR System (Analytik Jena AG, Jena City, Germany) and the SYBR® Green PCR Master Mix Kit (Thermo Fisher Scientific, Waltham City, MA, USA). The OsActin1 gene was used as an internal reference to normalize the gene expression level. The relative gene expression levels were calculated using the 2−ΔΔCt method [33]. The primers used for real-time PCR are listed in Table S1.
2.5 Genetic Analysis and Map-Based Cloning of the yl1 Mutant
For genetic analysis, the yl1 mutant was crossed with two indica rice cultivars, ‘Yinguizhan’ and ‘9311’. The segregation of normal and yellow-leaved plants in the F2 populations was investigated at the tillering stage. The mapping population was generated by crossing the yl1 mutant and WT to map the yl1 gene using the MutMap approach described as Abe et al. [34]. Genomic DNA bulks were prepared from the yl1 mutant Lailong, 20 yellow-leaved individuals, and 20 green-leaved plants from among the F2 progeny. The DNA bulks were subjected to whole-genome sequencing using the Illumina HiSeq 2500 platform. The obtained reads were aligned to the reference Nipponbare genome (http://rice.uga.edu/) using SOAP2 software [35]. The single-nucleotide polymorphism (SNP) index was calculated as described by Abe et al. [34]. To identify the mutation sites of the yl1 alleles, we amplified the candidate genes using genomic DNA extracted from the WT and yl1 mutant. The amplified DNA fragment products were sequenced. The primers used for sequencing are listed in Table S1.
2.6 Protein Subcellular Localization
To determine the subcellular localization of the LOC_Os03g36760 protein, the pCambia-35S- LOC_Os03g36760:GFP vector was constructed. Agrobacterium tumefaciens strain EHA105 harboring this vector was introduced into rice seedling protoplasts for transient expression following the method of Li et al. [36]. Agrobacterium tumefaciens strain EHA105 harboring the pCambia-35S:GFP vector was used as the control. The green fluorescent protein (GFP) fluorescence signal was observed with a laser confocal microscope (Leica SP8 STED, Leica Biosystems, Wetzlar City, Germany) under excitation at 488 nm.
The statistical significance of differences in mean values of the examined parameters between the WT and yl1 mutant were determined using Student’s t-test as implemented in Microsoft Excel 2016 and PASW Statistics 18 (SPSS Inc., Chicago, IL, USA).
3.1 Phenotype of the yl1 Mutant
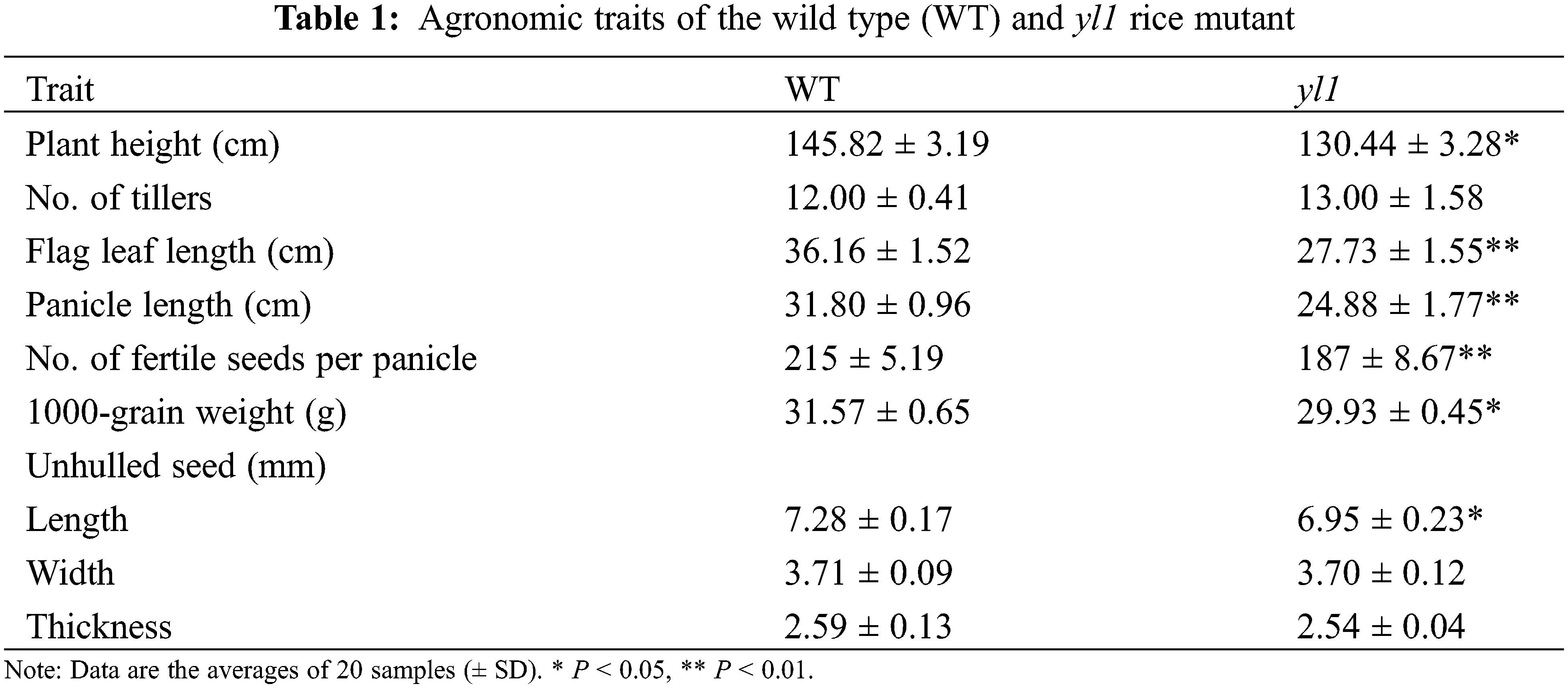
The yl1 mutant was isolated from a pool of EMS-mutagenized progeny of the japonica glutinous rice landrace Lailong. The yl1 mutant exhibited a yellow leaf phenotype throughout the growth period (Figs. 1A–1D). No significant differences in tiller number, grain width, and grain thickness were observed between the yl1 mutant and WT (Table 1). Plant height, panicle length, flag leaf length, grain number per panicle, and 1000-grain weight of the yl1 mutant were significantly decreased compared with those of the WT. The effective grain number per spike, grain length, and 1000-grain weight of the yl1 mutant were 14.97%, 4.75%, and 5.66% lower than those of the WT, respectively (Table 1). These results indicated that mutation of the yl1 gene affected the yield.
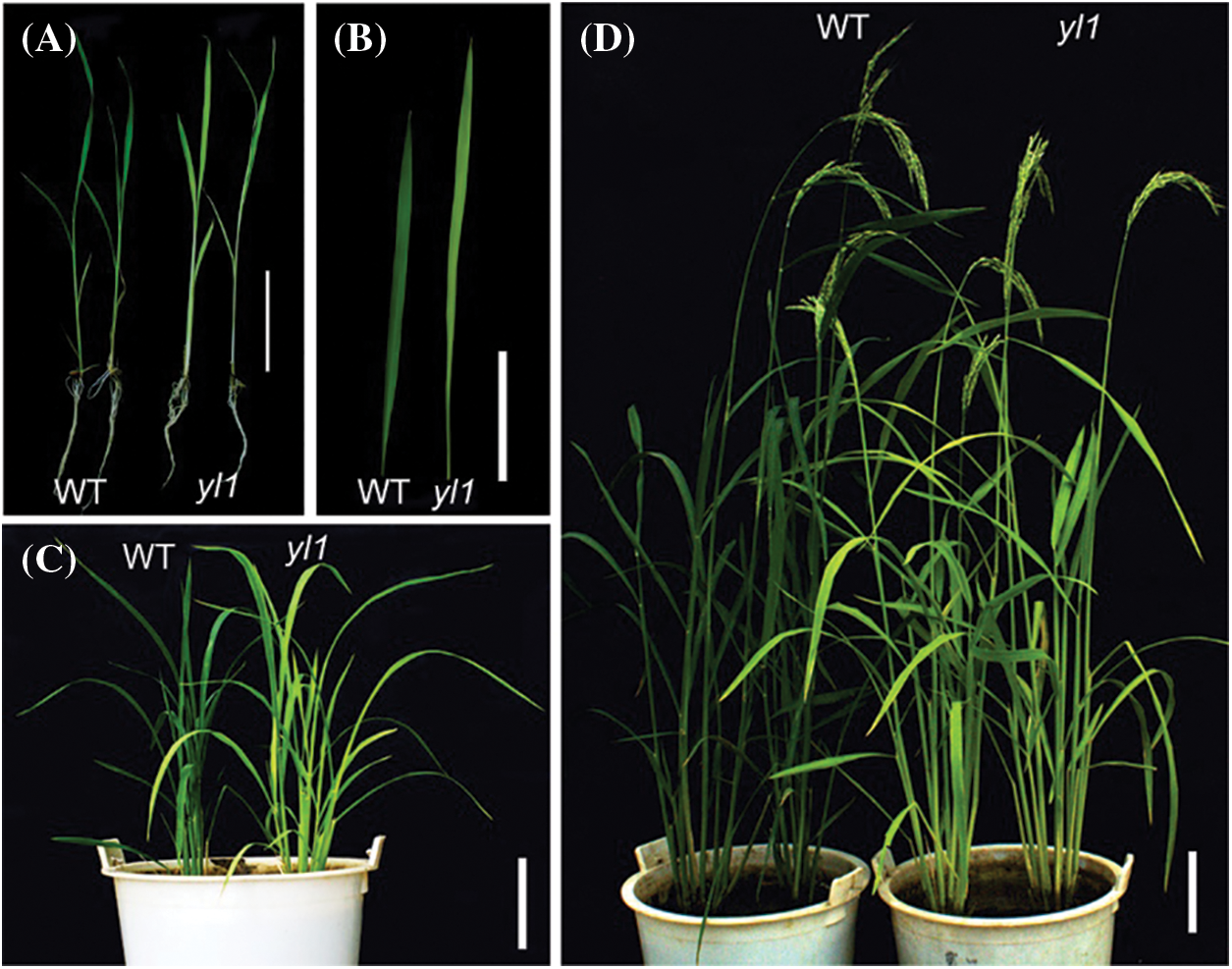
Figure 1: Rice leaf-color mutant yl1 and the wild type (WT) at different growth stages. A: Seedling stage, bar = 5 cm; B: seedling stage leaves, bar = 2 cm; C: tillering stage, bar = 10 cm; D: heading stage, bar = 10 cm
3.2 Determination of Photosynthetic Pigment Content in the yl1 Mutant
To characterize the yellow leaf phenotype of the yl1 mutant, we determined the pigment content of the yl1 and WT at the seedling, tillering, and grain filling stages. The contents of chlorophyll a, chlorophyll b, and total chlorophyll of yl1 were significantly reduced at each growth stage compared with those of the WT (Figs. 2A–2C). The content of carotene in the yl1 mutant was significantly decreased at the seedling and tillering stages, but markedly increased at the grain filling stage (Figs. 2A–2C). No significant difference in chlorophyll a to chlorophyll b (Chl a/b) ratio was observed between the yl1 mutant and WT leaves at the seedling and tillering stages (Fig. 2D). The Chl a/b ratio of the yl1 mutant was 6.67% lower than that of the WT at the grain filling stage. These data suggested that the yellow leaf phenotype of the yl1 mutant was the result of reduced photosynthetic pigment contents (Fig. 2D).
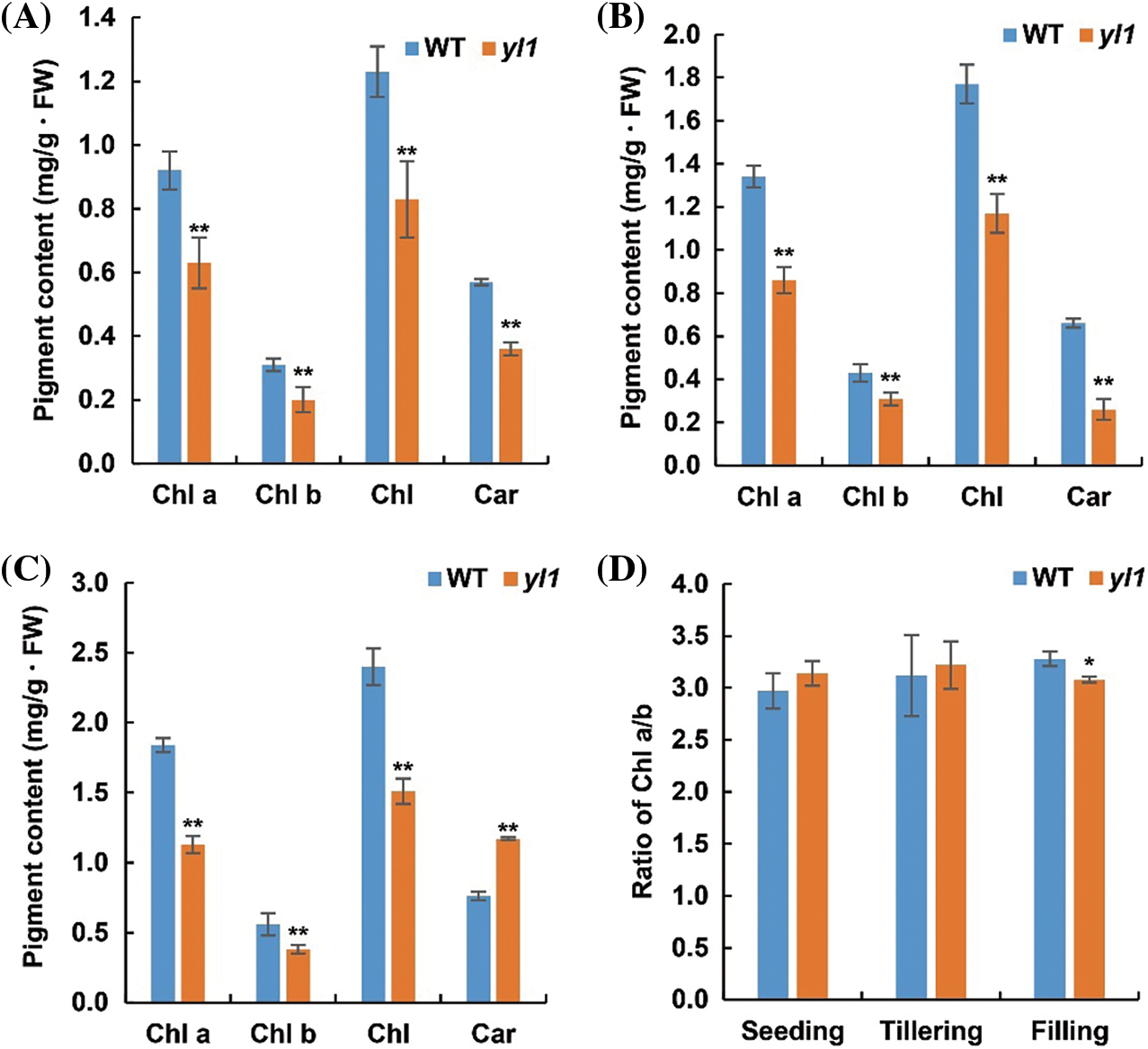
Figure 2: Photosynthetic pigment contents in the leaf of the yl1 mutant and wild type (WT) at three growth stages. A: Pigment contents at the seedling stage, B: pigment contents at the tillering stage, C: pigment contents at the grain filling stage, D: chlorophyll a to chlorophyll b ratio (Chl a/b) of the yl1 mutant and WT at the seedling, tillering, and grain filling stages. * P < 0.05, ** P < 0.01
To explore the affected pathway of chlorophyll synthesis in the yl1 mutant, the contents of chlorophyll intermediates were measured in the yl1 mutant and WT. No significant difference in the contents of ALA, porphobilinogen (PBG), uroporphyrinogen III (Urogen III), and coproporphyrinogen III (Coprogen III) were detected between the yl1 mutant and WT (Figs. 3A and 3B). However, the content of Proto IX in the yl1 mutant was significantly higher, and the contents of Mg-Proto IX and Pchlide were significantly decreased in the yl1 mutant, compared with those in the WT (Figs. 3B and 3C). These data indicated that the contents of Mg-Proto IX and subsequent steps of chlorophyll synthesis in the yl1 mutant were significantly lower than those of the WT. It was speculated that the yellow leaf phenotype of the yl1 mutant was due to the reduction of chlorophyll content caused by impaired synthesis of Mg-Proxo IX. This hypothesis suggested that the yl1 mutant may be involved in chlorophyll biosynthesis at the Mg-Proto IX synthesis step.
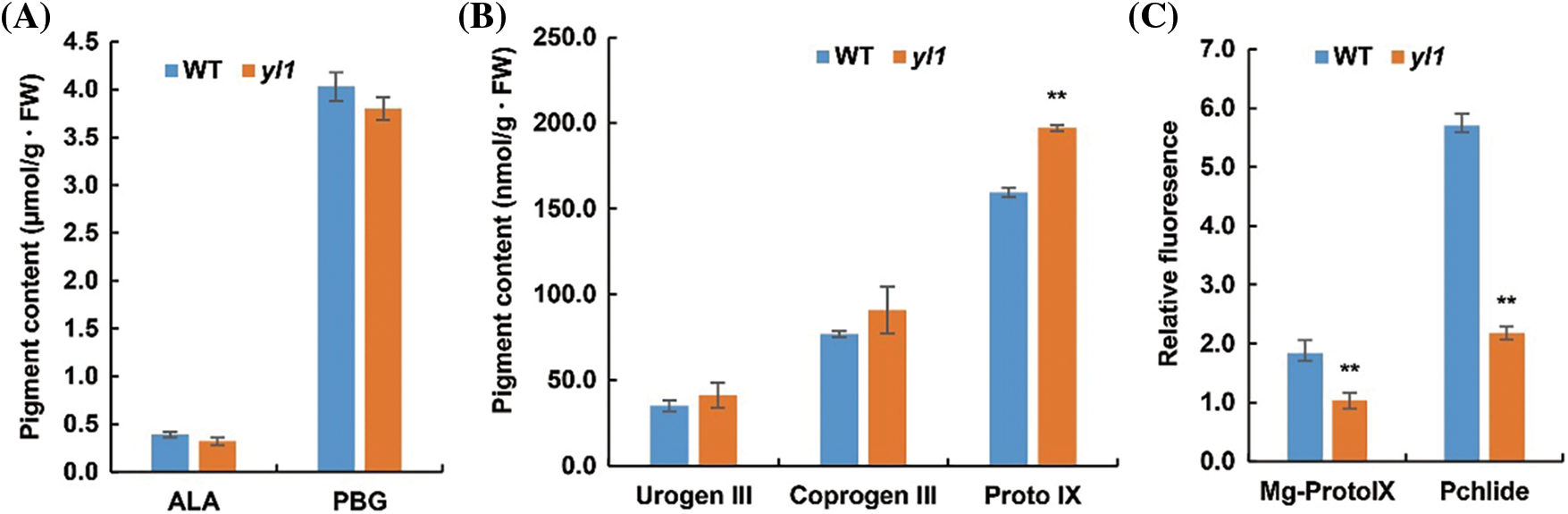
Figure 3: Contents of chlorophyll intermediates, and expression levels of chlorophyll biosynthetic genes in the wild type (WT) and yl1 mutant. A: Contents of δ-aminolevulinic acid (ALA) and porphobilinogen (PBG) in the second leaf of 14-day-old seedlings. B: Contents of uroporphyrinogen III (Urogen III), coproporphyrinogen III (Coprogen III), and protoporphyrin IX (Proto IX). C: Relative fluorescence of magnesium-protoporphyrin IX (Mg-Proto lX) and protochlorophyllide (Pchlide)
3.3 Disruption of Chloroplast Lamellae in the yl1 Mutant
To explore the effect of reduced photosynthetic pigment contents on the chloroplast structure of the yl1 mutant, the chloroplast ultrastructure was observed by transmission electron microscopy. Chloroplasts from WT leaves showed intact thylakoid and granum structure, with densely arranged grana lamellae (Figs. 4A and 4B). In contrast, chloroplast development was disrupted, and the grana lamellae were loose and disordered in the yl1 mutant (Figs. 4C and 4D). It was speculated that the yellow leaf phenotype of the yl1 mutant was caused by the decrease in photosynthetic pigment content resulting from impaired chloroplast development.
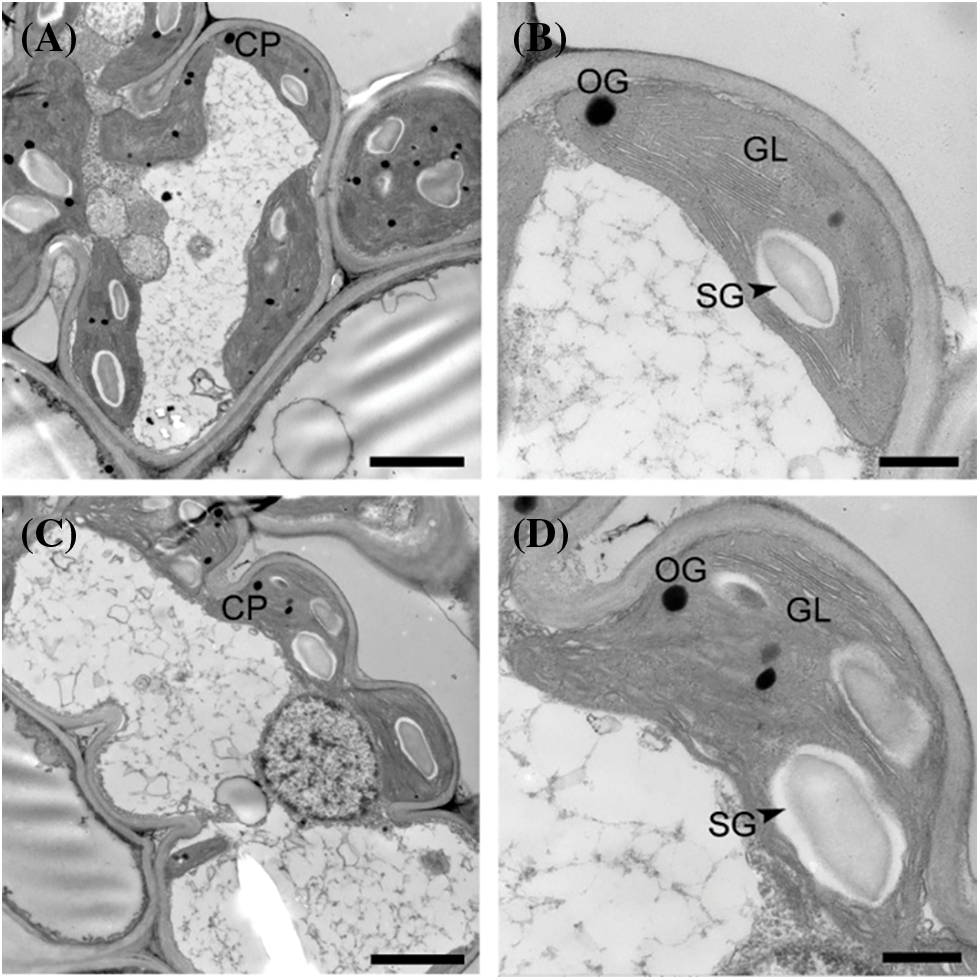
Figure 4: Ultrastructure of chloroplasts in the yl1 mutant and wild type at the seedling stage. A and B: Transmission electron micrographs of chloroplasts of the wild type. C and D: Transmission electron micrographs of chloroplasts of the yl1 mutant. A, C: bar = 2 µm; B, D: bar = 500 nm. CP: chloroplast, OG: osmiophilic granule, GL: grana lamellae, SG: starch granule
3.4 Differential Gene Expression between the yl1 Mutant and WT
To investigate the biological function of yl1, RNA-sequencing analysis was conducted to compare the transcriptome profiles of DEGs in the leaves of 4-week-old seedlings of the WT and yl1 mutant. A total of 1,329 DEGs were identified between the yl1 mutant and WT, of which 462 genes were up-regulated and 867 genes were down-regulated (Figs. 5A and 5C). These DEGs were distributed in all seven branches of KEGG metabolic pathways, such as metabolic pathways, genetic information processing pathways, and cell process pathways. All identified DEGs were clustered according to their biological roles. The GO enrichment of genes in the molecular function category were mainly significantly enriched for oxidoreductase activity and cofactor binding (Fig. 5B). The GO functional annotation was further divided into three components: molecular function, cellular component, and biological process. The highest number of genes was involved in the molecular function of catalytic activity, followed by an integral part of the cell and binding (Fig. 5D). KEGG pathway enrichment analysis revealed three significant KEGG pathways, comprising biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, and starch and sucrose metabolism (Fig. 6). The changes in transcript abundance of genes involved in the chlorophyll and carotene synthesis and metabolism pathway and in chloroplast development between the yl1 mutant and WT were further analyzed. The chlorophyll synthesis-related genes OsCHLH, OsCHLM, OsCHLG, PORB, and YGL8 were down-regulated in the yl1 mutant, especially OsCHLH, PORB, and YGL8 (Fig. 7A). These results indicated that the reduced chlorophyll content in the yl1 mutant was caused by down-regulation of chlorophyll synthesis genes.
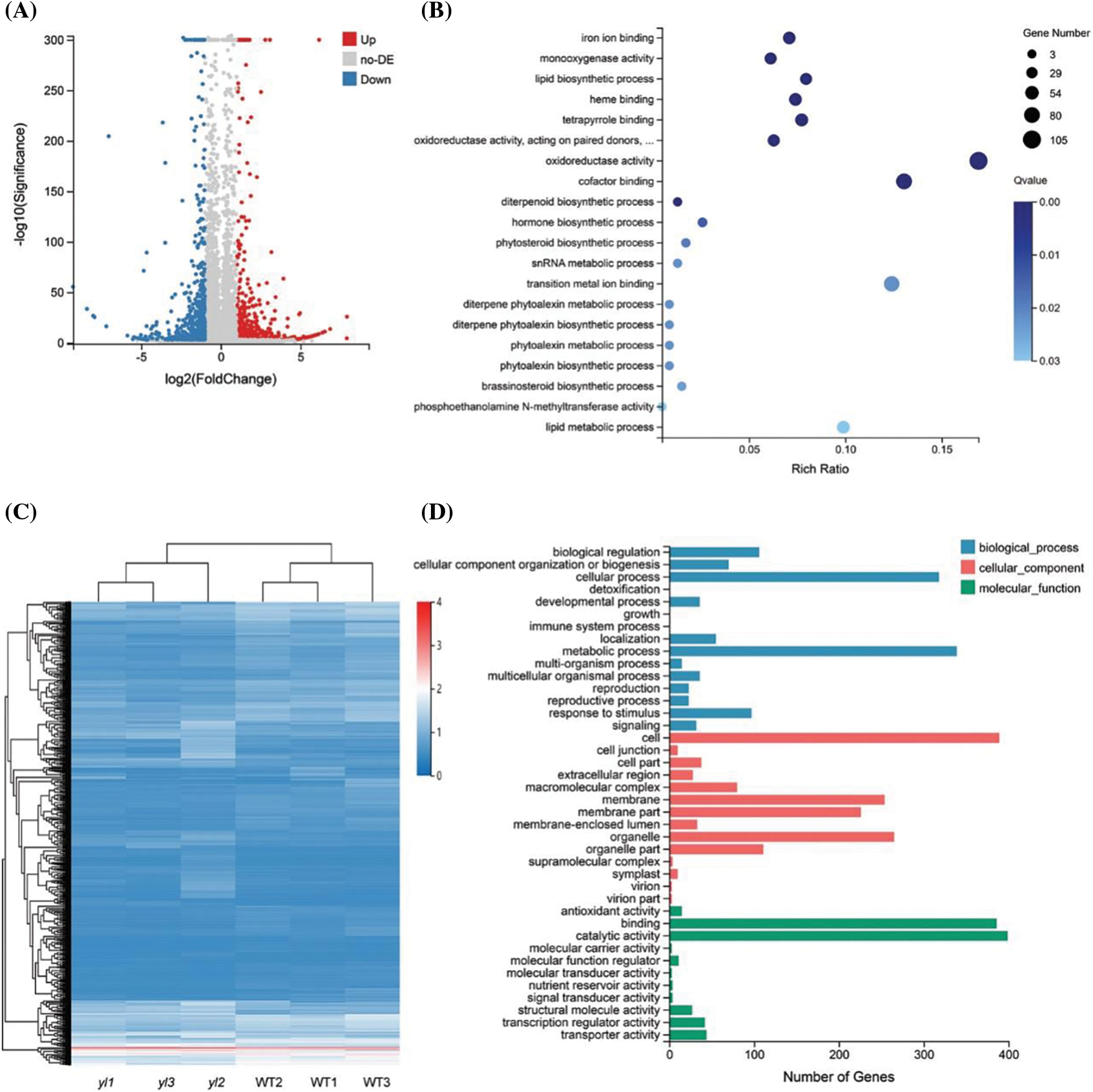
Figure 5: Transcriptional changes between the yl1 mutant and wild type (WT). A: Volcano map of differentially expressed genes (DEGs) between the yl1 mutant and WT. B: GO enrichment of DEGs between the yl1 mutant and WT. C: Hierarchical cluster analysis of DEGs between the yl1 mutant and WT. yl1–yl3, yl1 mutant; WT1–WT3, wild type. D: GO functional classification of DEGs between the yl1 mutant and WT
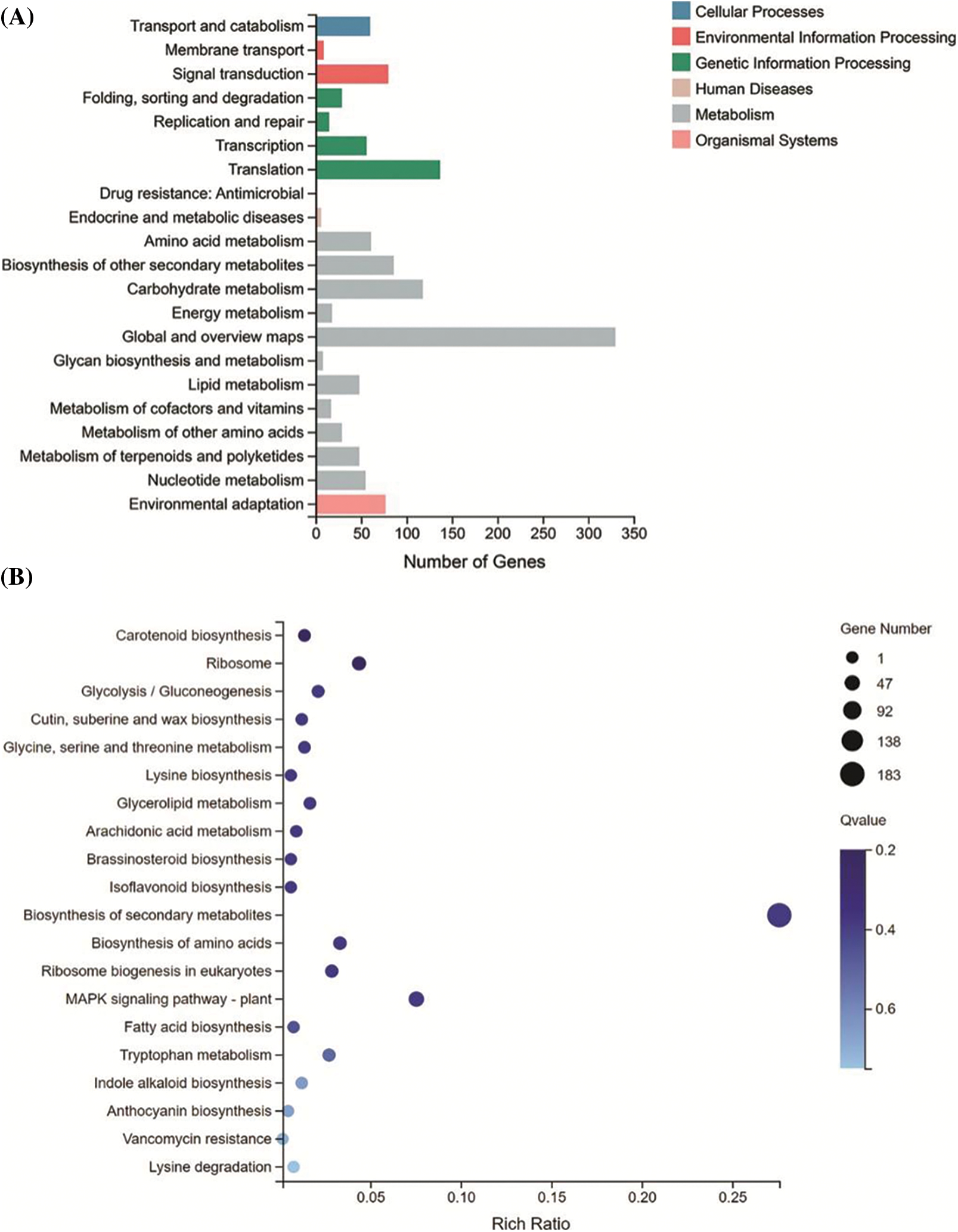
Figure 6: KEGG pathway functional classification and enrichment of differentially expressed genes (DEGs) between the wild type (WT) and yl1 mutant. A: KEGG functional classification of DEGs between the yl1 mutant and WT. B: KEGG enrichment of DEGs between the yl1 mutant and WT
To investigate further the effect of yl1 mutation on chloroplast development, we analyzed the expression of genes associated with chloroplast development in the WT and yl1 mutant. It was notable that the expression levels of the plastid division protein-encoding gene FtsZ and NEP core subunits-encoding gene OsRpoTp were significantly decreased in the yl1 mutant. The expression levels of the D1 protein of photosystem II (PsbA), PsaA (encoding the PsaA protein of photosystem I), and the large subunit of ribulose-bisphosphate carboxylase/oxygenase (RbcL) were also significantly reduced in the yl1 mutant (Fig. 7B). The expression of ASL3 and OsHAP3B was increased in the yl1 mutant (Fig. 7B). These results suggested that the mutation of yl1 affected the levels of a broad range of mRNAs of imported precursors targeted to the chloroplast in rice.
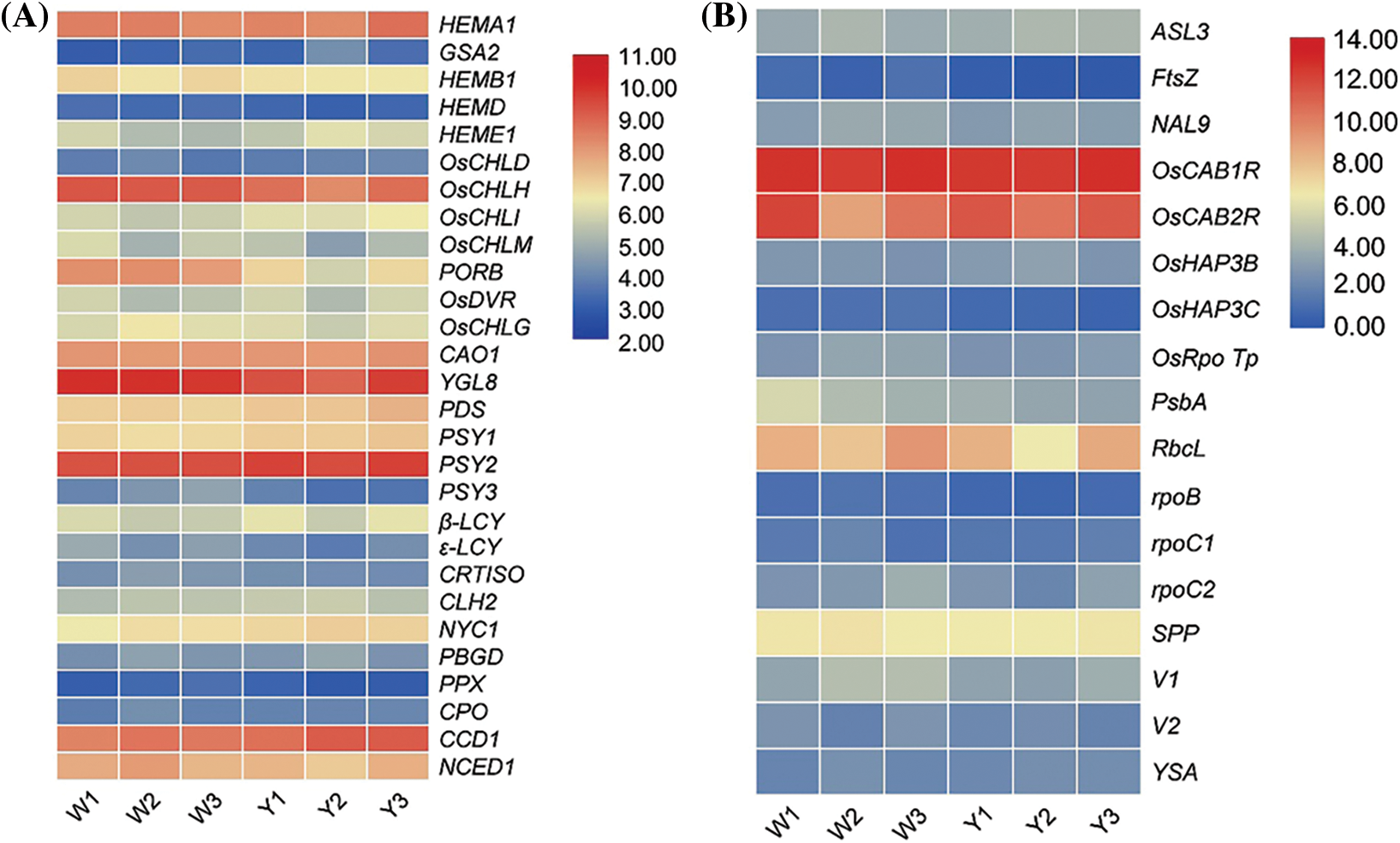
Figure 7: Relative expression of chlorophyll and carotene synthesis and metabolism-related genes (A) and chloroplast development-related genes (B) in the yl1 mutant and wild type as indicated by RNA-sequencing analysis. Different genes are indicated by different colors. Relative expression levels are shown as a color gradient from low (blue) to high (red). W1−W3, wild type; Y1−Y3, yl1 mutant
We further used qRT-PCR to analyze the expression of 10 genes involved in the chlorophyll biosynthetic pathway and chloroplast development pathway in rice. Compared with the WT, the yl1 mutant displayed no difference in the expression levels of OsCHLD and OsCHLI, which encode the CHLD and CHLI subunits of Mg-chelatase, respectively. OsCHLH, which encodes the Mg-chelatase CHLH subunit, was significantly down-regulated. Expression of the chlorophyll biosynthetic genes OsCHLG, OsCHLM, YGL8, and PORB was also significantly down-regulated in the yl1 mutant (Fig. 8). The chloroplast development-related genes FtsZ, OsRpoTp, and RbcL were also significantly down-regulated in the yl1 mutant (Fig. 8). Overall, the results indicated that mutation of yl1 affected the expression of genes associated with chloroplast development and resulted in the abnormal chloroplast structure observed in the yl1 mutant.
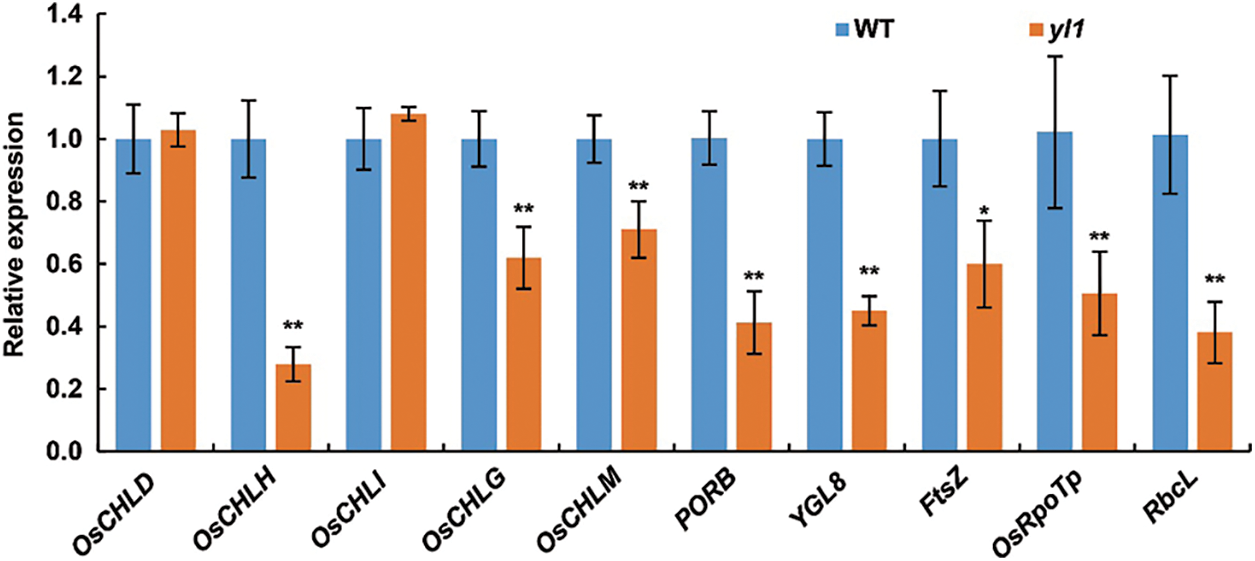
Figure 8: Relative expression levels of chlorophyll biosynthetic and chloroplast development genes. Data are means (n = 3), error bars indicate the SD. Asterisks indicate significant differences between the WT and yl1 mutant. * P < 0.05, ** P < 0.01
3.5 Genetic Analysis and Map-Based Cloning of yl1 Mutation Site
To analyze the genetic basis of the yl1 mutation, crosses were performed using the yl1 mutant as the female parent and two indica rice cultivars, ‘9311’ and ‘Yinguizhan’, as pollen donors. The leaf color of F1 individuals and segregation ratios were investigated in the F2 populations. All F1 individuals displayed a green leaf phenotype identical to that of WT. The F2 individuals derived from the crosses yl1 × 9311 and yl1 × Yinguizhan were separated into green leaf and yellow leaf phenotypes with a segregation ratio of 3:1 (χ2 < χ20.05 = 3.84, P > 0.05) (Table 2). These results indicated that the leaf-color mutation of yl1 was controlled by recessive nuclear gene.

To map the chromosomal site of yl1, another F2 population was constructed by crossing the yl1 mutant with WT. The yl1 mutant and WT parents, and 20 green-leaved individuals and 20 yellow-leaved plants from the F2 population were sequenced using an Illumina GAIIx DNA platform. We obtained 44.57 Gbp of short (75-bp) reads (Tables S2 and S3) that were aligned to the Nipponbare genome, resulting in identification of 2,759 SNP positions. For each SNP position, the value of the SNP index (the ratio of short reads harboring SNPs compared with the reference) was determined, and a graph relating SNP positions and the SNP index was generated for all 12 rice chromosomes. The causative SNP should be shared by all mutant F2 plants and, therefore, have a SNP index = 1, whereas SNPs unrelated to the mutant phenotype should segregate in a 1:1 ratio among the F2 progeny, resulting in a SNP index of 0.5. The MutMap approach applied to the F2 progeny revealed a cluster of SNPs with high SNP index values, and that showed statistically significant deviations from the SNP index of 0.5 (P < 0.01), in the region between 17.34 Mb and 23.51 Mb on chromosome 3 (Fig. 9A). Sixteen SNPs with an SNP index of 1 were identified in the candidate region (Table 3). Among these 16 SNPs, 14 SNPs were located in the spacer region and two SNPs were located in the exon region, resulting in nonsynonymous mutations. Two SNPs at nucleotide positions 17,776,204 and 20,394,504 corresponded to the second exon of the genes LOC_Os03g31210 and LOC_Os03g36760, and represented the nonsynonymous mutation of a proline (GCC) codon to a leucine (GCT) at residue 478 in LOC_Os03g31210 and a lysine (AAA) codon to a glutamic acid (GAA) at residue 906 in LOC_Os03g36760, respectively (Table 3, Figs. 9B and 9D). We further confirmed the SNP presence in the genome using PCR and Sanger sequencing (Figs. 9C and 9E). LOC_Os03g31210 encodes UDP-glucose 6-dehydrogenase, a crucial enzyme in the polysaccharide synthesis pathway, while LOC_Os03g36760 encodes an expressed protein of unknown function with a plant mobile domain (PMD). Both of these genes have not been reported previously. In the UniProt database (https://www.uniprot.org/uniprot/Q75GS4), the function of LOC_Os03g31210 is stated to produce UDP-glucuronic acid, which provides nucleotide sugars for cell-wall polymers. According to the Information Commons for Rice Database (IC4R) (http://ic4r.org/) and transcriptomic database Rice eFP (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi), LOC_Os03g31210 expresses in the ovary and root, and LOC_Os03g36760 widely expresses in various tissues (Fig. S1). Therefore, we concluded that LOC_Os03g36760 was the yl1 candidate gene.
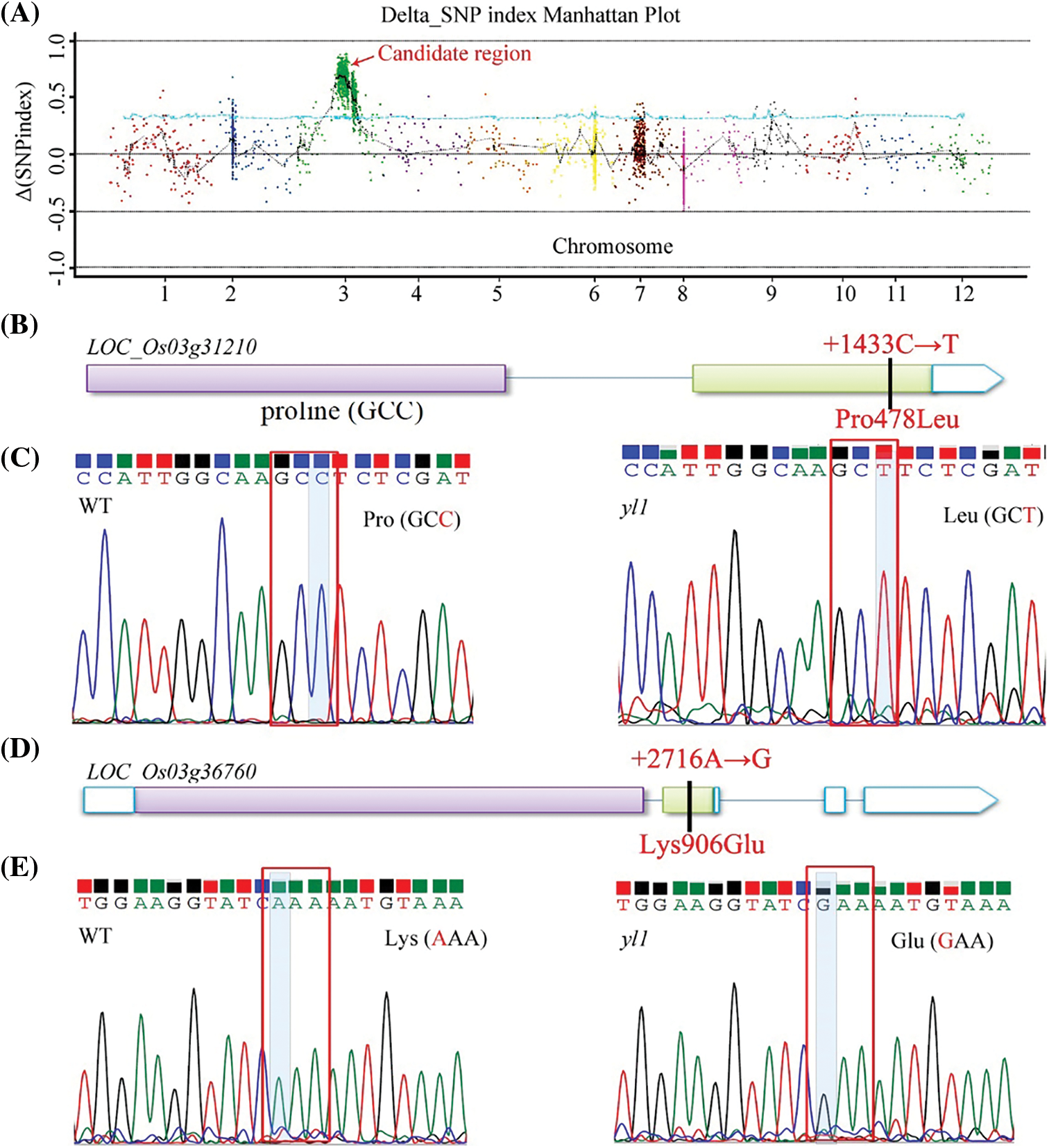
Figure 9: Identification of the yl1 mutation using the MutMap method. A: SNP index plot of rice chromosome 3 generated by the MutMap analysis, showing a genomic region in which the highest SNP index peak harbored the candidate mutation. B: Gene structure and mutation sites in LOC_Os03g31210. C: Confirmation of the yl1 mutation in LOC_Os03g31210 by Sanger sequencing. A red box indicates the C-to-T transition. D: Gene structure and mutation sites in LOC_Os03g36760. E: Confirmation of the yl1 mutation in LOC_Os03g36760 by Sanger sequencing. A red box indicates the A-to-G transition
3.6 Subcellular Localization of LOC_Os03g36760 Protein
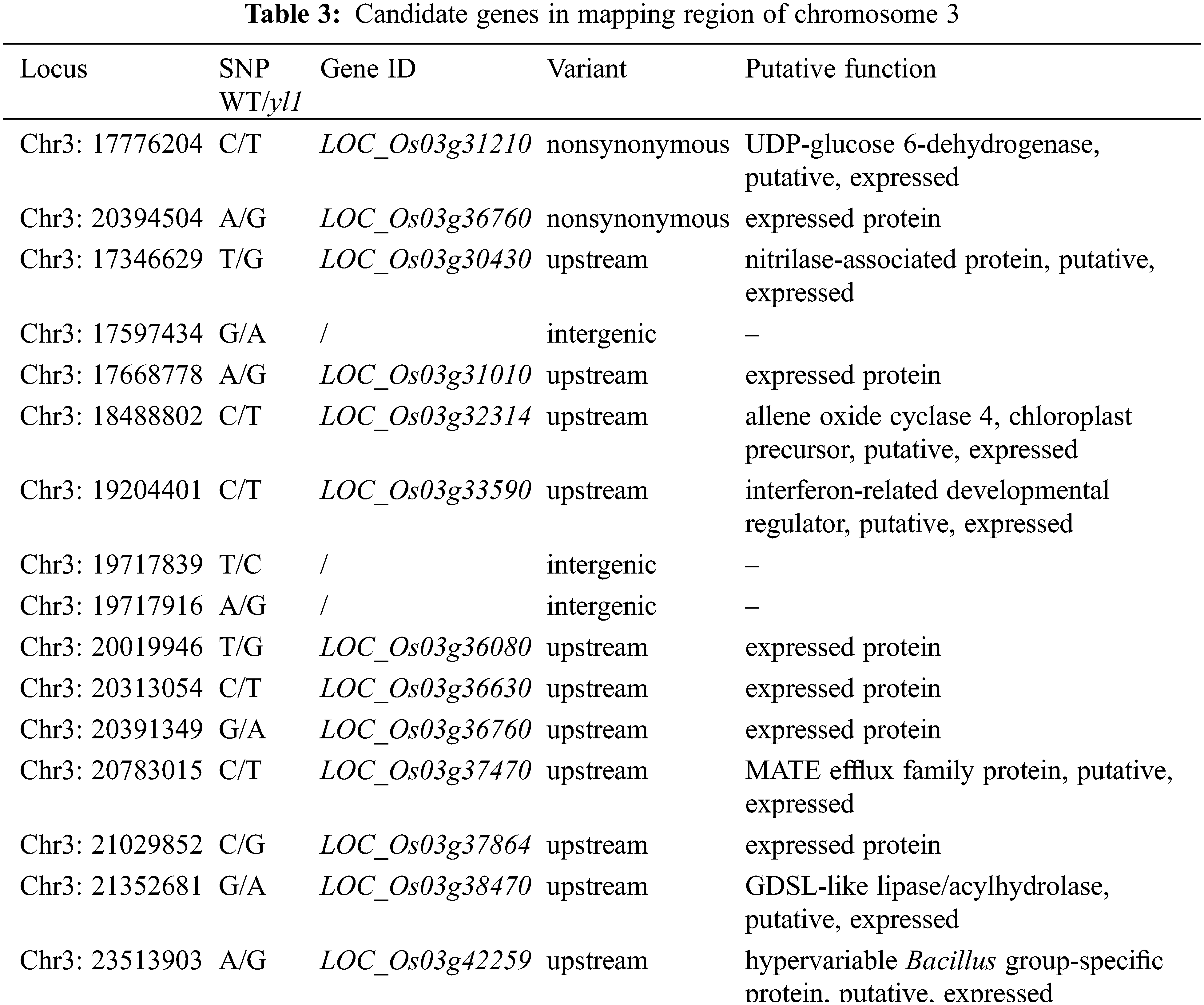
To visualize the subcellular localization of LOC_Os03g36760, we fused GFP to the C-terminus of LOC_Os03g36760. The fluorescent signal of the LOC_Os03g36760-GFP fusion protein was detected in the nucleus, cytoplasm, and plasma membrane (Fig. 10).
Figure 10: Subcellular localization of the LOC_Os03g36760 protein
The leaf is the main organ for photosynthesis in plants. Chlorophyll plays a vital role in plant development and crop yield. The regulatory mechanism of chlorophyll synthesis and chloroplast structure is an important research focus in plant physiology and molecular biology. In recent years, the chlorophyll biosynthetic and degradative pathways have received much attention from plant physiologists and breeders [21]. Leaf-color mutants are important materials for studying photosynthesis, chlorophyll synthesis and degradation, and the regulatory mechanisms of growth and development, and are of high utility in hybrid seed production and biomass improvement of rice. In plants, chlorophyll biosynthesis begins with the precursor L-glutamyl-tRNA and involves a total of 15 steps. The entire synthesis pathway involves the participation of 15 enzymes and the expression of 27 genes encoding these enzymes [37]. Any obstacle encountered in this process may impair chlorophyll biosynthesis and result in leaf-color mutations [38]. The enzyme of Mg-chelatase catalyzes the insertion of Mg2+ into Proto IX to produce Mg-Proto IX. Reduced activity of Mg-chelatase may ultimately cause a decrease in chlorophyll synthesis [5]. In the yl1 mutant, the yellow leaf phenotype expressed at all growth stages. Compared with those in Lailong, the contents of chlorophyll a and chlorophyll b were significantly decreased in the yl1 mutant at the mature stage. Analysis of the contents of chlorophyll precursors showed that Proto IX accumulated, whereas Mg-Proto IX and Pchlide contents were significantly decreased, in the yl1 mutant (Figs. 3B and 3C), indicating that the synthesis of Mg-Proto IX from Proto IX in the mutant was blocked. In rice, OsCHLH, OsCHLD, and OsCHLI encode the ChlH, ChlD, and ChlI subunits of Mg-chelatase, respectively [4,5]. Compared with the WT, the yl1 mutant displayed no difference in expression of OsCHLD and OsCHLI, whereas OsCHLH was significantly down-regulated (Fig. 8). Previous studies have shown that mutations and down-regulation of OsCHLH may cause a decrease in leaf chlorophyll content, resulting in yellow leaf phenotypes [4]. Therefore, we concluded that the decreased expression of OsCHLH resulted in the blocked synthesis of Mg-Proto IX, which caused the reduced chlorophyll contents in leaves of the yl1 mutant.
Chloroplast differentiation and development are controlled by the interaction and co-regulation of nuclear genes and plastid genes [39]. This differentiation and development process can be divided into seven steps, and any obstructed pathway leads to chloroplast hypoplasia, thus affecting plant chlorophyll content and causing leaf-color mutations [38,40]. FtsZ encodes a plastid division protein that plays an important role in chloroplast division [41–43]. In the yl1 mutant, the expression levels of FtsZ, RbcL, and OsRpoTp were significantly decreased (Fig. 8), whereas those of ASL3 and OsHAP3B were increased (Fig. 7). Overall, these results suggested that the mutation of yl1 affected the expression of genes associated with chloroplast development, and thereby caused the abnormal chloroplast structure in the yl1 mutant. Impairment of chloroplast development affects the photosynthetic efficiency of plants. This impairment then reduces the photosynthetic efficiency of chloroplast-related mutants and reduced plant yield. In addition, mutations causing deficiency in the capability to synthesize chlorophyll, including the genes OsCHLH, OsCHLD, OsCHLI, OsCHLG, and YGL8, also reduce yield to varying degrees [4,5,7,44,45]. In the yl1 mutant, the chlorophyll content and expression of chlorophyll synthesis-related genes were decreased, and the chloroplasts exhibited abnormal morphology, with loose thylakoid membranes and disordered grana lamellae. Compared with Lailong, grain number per spike, 1000-grain weight, and plant height in the yl1 mutant were all reduced. Therefore, we considered that the mutation of yl1 caused abnormal chloroplast development and blocked chlorophyll synthesis in rice, which ultimately affected yield-related traits of the yl1 mutant.
At present, more than 120 rice leaf color-related genes have been cloned and are distributed on the 12 chromosomes of rice, of which chromosome 3 harbors the highest number of cloned genes. Genes associated with chlorophyll synthesis, including OsCHLH, OsCHLD, OsCHLI, NOL, PAO, and OsDVR, are all located on chromosome 3. These results suggest that a mutation hotspot region for chlorophyll content may exist on chromosome 3 in rice. In this study, using the MutMap method, we mapped the yl1 gene located near the centromeric region of chromosome 3, and identified two previously unreported non-synonymous SNPs located in LOC_Os03g31210 and LOC_Os03g36760. LOC_Os03g36760 encodes a protein of unknown function with a PMD. The PMD proteins are ubiquitous in plants. A previous phylogenetic analysis of the PMD family indicated that LOC_Os03g36760, together with two other rice PMD proteins and eight Arabidopsis PMD proteins, was placed in the PMD-B clade and associated with PCGs [46]. The PMD-A clade proteins in Arabidopsis define an alternative silencing pathway independent of DNA methylation and short interfering RNAs [46]. However, the function of PMD-B clade proteins in Arabidopsis and rice has not been reported. Therefore, the function of LOC_Os03g36760 requires further study.
A yellow leaf rice mutant, yl1, was identified from the EMS-treated progeny of Lailong, a rice landrace cultivated in Guizhou. The yellow leaf phenotype of yl1 was caused by abnormal chloroplast development and inhibition of chlorophyll synthesis. Genetic analysis revealed that the yellow leaf phenotype of the yl1 mutant was controlled by a single recessive gene. By employing the MutMap method, two non-synonymous SNPs located in LOC_Os03g31210 and LOC_Os03g36760 were identified. LOC_Os03g36760 may be the candidate gene for the yl1 mutant. These results will be useful for further characterization and cloning of yl1, and for research on the molecular mechanisms controlling biogenesis and chloroplast biochemical processes.
Authorship: Z. X. F., L. G. Z., L. N. A., L. Y., H. X. Z. and L. J. R. assisted in the experiments. Z. X. F. wrote the manuscript. Z. X. F. and Z. D. G. supervised the research.
Funding Statement: This work was supported by grants from the Guizhou Province High-Level Innovative Talent Training Program Project ([2016]4003), the Guizhou Science and Technology Major Project [20126005] and the Guizhou Science and Technology Project (20171039).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Wang, W., Gao, H., Liang, Y., Li, J., Wang, Y. (2022). Molecular basis underlying rice tiller angle: Current progress and future perspectives. Molecular Plant, 15(1), 125–137. DOI 10.1016/j.molp.2021.12.002. [Google Scholar] [CrossRef]
2. Khush, G. S. (2005). What it will take to Feed 5.0 Billion Rice consumers in 2030. Plant Molecular Biology, 59(1), 1–6. DOI 10.1007/s11103-005-2159-5. [Google Scholar] [CrossRef]
3. Singh, U., Prithiviraj, B., Sarma, B. (2000). Development of Erysiphe pisi (powdery mildew) on normal and albino mutants of pea (Pisum sativum L.). Journal of Phytopathology, 148(11–12), 591–595. DOI 10.1046/j.1439-0434.2000.00558.x. [Google Scholar] [CrossRef]
4. Jung, K. H., Hur, J., Ryu, C. H., Choi, Y., Chung, Y. Y. et al. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant and Cell Physiology, 44(5), 463–472. DOI 10.1093/pcp/pcg064. [Google Scholar] [CrossRef]
5. Zhang, H., Li, J., Yoo, J. H., Yoo, S. C., Cho, S. H. et al. (2006). Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Molecular Biology, 62(3), 325–337. DOI 10.1007/s11103-006-9024-z. [Google Scholar] [CrossRef]
6. Deng, X. J., Zhang, H. Q., Wang, Y., He, F., Liu, J. L. et al. (2014). Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS One, 9(6), e99564. DOI 10.1371/journal.pone.0099564. [Google Scholar] [CrossRef]
7. Lee, S., Kim, J. H., Yoo, E. S., Lee, C. H., Hirochika, H. et al. (2005). Differential regulation of chlorophyll a oxygenase genes in rice. Plant Molecular Biology, 57(6), 805–818. DOI 10.1007/s11103-005-2066-9. [Google Scholar] [CrossRef]
8. Wang, P., Gao, J., Wan, C., Zhang, F., Xu, Z. et al. (2010). Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiology, 153(3), 994–1003. DOI 10.1104/pp.110.158477. [Google Scholar] [CrossRef]
9. Wang, Q., Zhu, B., Chen, C., Yuan, Z., Guo, J. et al. (2021). A single nucleotide substitution of GSAM gene causes massive accumulation of glutamate 1-semialdehyde and yellow leaf phenotype in rice. Rice, 14(1), 50. DOI 10.1186/s12284-021-00492-x. [Google Scholar] [CrossRef]
10. Liu, X., Huang, Q., Yang, Y., Tang, J., Zhao, Y. et al. (2021). Characterization and map-based cloning of the novel rice yellow leaf mutant yl3. Journal of Plant Biology, 64(1), 35–44. DOI 10.1007/s12374-020-09275-1. [Google Scholar] [CrossRef]
11. Goh, C. H., Satoh, K., Kikuchi, S., Kim, S. C., Ko, S. M. et al. (2010). Mitochondrial activity in illuminated leaves of chlorophyll-deficient mutant rice (OsCHLH) seedlings. Plant Biotechnology Reports, 4(4), 281–291. DOI 10.1007/s11816-010-0146-z. [Google Scholar] [CrossRef]
12. Wang, X., Huang, R., Quan, R. (2017). Mutation in Mg-Protoporphyrin IX monomethyl ester cyclase decreases photosynthesis capacity in rice. PLoS One, 12(1), e0171118. DOI 10.1371/journal.pone.0171118. [Google Scholar] [CrossRef]
13. Yang, Y., Xu, J., Huang, L., Leng, Y., Dai, L. et al. (2016). PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. Journal of Experimental Botany, 67(5), 1297–1310. DOI 10.1093/jxb/erv529. [Google Scholar] [CrossRef]
14. Sakuraba, Y., Rahman, M. L., Cho, S. H., Kim, Y. S., Koh, H. J. et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant Journal, 74(1), 122–133. DOI 10.1111/tpj.12110. [Google Scholar] [CrossRef]
15. Cai, L., Liu, J., Yun, H., Du, D., Zhong, X. et al. (2021). Characterization and candidate gene analysis of the yellow-green leaf mutant ygl16 in rice (Oryza sativa L.). Phyton-International Journal of Experimental Botany, 90(4), 1103–1117. DOI 10.32604/phyton.2021.015532. [Google Scholar] [CrossRef]
16. Dong, H., Fei, G. L., Wu, C. Y., Wu, F. Q., Sun, Y. Y. et al. (2013). A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiology, 162(4), 1867–1880. DOI 10.1104/pp.113.217604. [Google Scholar] [CrossRef]
17. Li, W., Wu, C., Hu, G., Xing, L., Qian, W. et al. (2013). Characterization and fine mapping of a novel rice narrow leaf mutant nal9. Journal of Integrative Plant Biology, 55(11), 1016–1025. DOI 10.1111/jipb.12098. [Google Scholar] [CrossRef]
18. Yoo, S. C., Cho, S. H., Sugimoto, H., Li, J., Kusumi, K. et al. (2009). Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiology, 150(1), 388–401. DOI 10.1104/pp.109.136648. [Google Scholar] [CrossRef]
19. Yang, Q., He, H., Li, H., Tian, H., Zhang, J. et al. (2011). NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice. PLoS One, 6(5), e20015. DOI 10.1371/journal.pone.0020015. [Google Scholar] [CrossRef]
20. Liu, H., Lau, E., Lam, M. P. Y., Chu, H., Li, S. et al. (2010). OsNOA1/RIF1 is a functional homolog of AtNOA1/RIF1: Implication for a highly conserved plant cGTPase essential for chloroplast function. New Phytologist, 187(1), 83–105. DOI 10.1111/j.1469-8137.2010.03264.x. [Google Scholar] [CrossRef]
21. Liu, W., Fu, Y., Hu, G., Si, H., Zhu, L. et al. (2007). Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta, 226(3), 785–795. DOI 10.1007/s00425-007-0525-z. [Google Scholar] [CrossRef]
22. Lin, D., Jiang, Q., Zheng, K., Chen, S., Zhou, H. et al. (2015). Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biology, 17(3), 599–607. DOI 10.1111/plb.12271. [Google Scholar] [CrossRef]
23. Yoo, J. H., Park, J. H., Cho, S. H., Yoo, S. C., Li, J. et al. (2011). The rice bright green leaf (bgl) locus encodes OsRopGEF10, which activates the development of small cuticular papillae on leaf surfaces. Plant Molecular Biology, 77(6), 631–641. DOI 10.1007/s11103-011-9839-0. [Google Scholar] [CrossRef]
24. Moon, S., Giglione, C., Lee, D. Y., An, S., Jeong, D. H. et al. (2008). Rice peptide deformylase PDF1B is crucial for development of chloroplasts. Plant and Cell Physiology, 49(10), 1536–1546. DOI 10.1093/pcp/pcn121. [Google Scholar] [CrossRef]
25. Krishnan, A., Guiderdoni, E., An, G., Yue-Ie, C. H., Han, C. D. et al. (2009). Mutant resources in rice for functional genomics of the grasses. Plant Physiology, 149(1), 165–170. DOI 10.1104/pp.108.128918. [Google Scholar] [CrossRef]
26. Zhu, Y., Yan, P., Dong, S., Hu, Z., Wang, Y. et al. (2021). Map-based cloning and characterization of YGL22, a new yellow-green leaf gene in rice (Oryza sativa). Crop Science, 61(1), 529–538. DOI 10.1002/csc2.20347. [Google Scholar] [CrossRef]
27. Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta Vulgaris. Plant Physiology, 24(1), 1–15. DOI 10.1104/pp.24.1.1. [Google Scholar] [CrossRef]
28. Morton, R. A. (1975). Biochemical spectroscopy. London: Adam Hilger. DOI 10.1016/0076-6879(95)46007-1. [Google Scholar] [CrossRef]
29. Santiago-Ong, M., Green, R. M., Tingay, S., Brusslan, J. A., Tobin, E. M. (2001). shygrl1 is a mutant affected in multiple aspects of photomorphogenesis. Plant Physiology, 126(2), 587–600. DOI 10.1104/pp.126.2.587. [Google Scholar] [CrossRef]
30. Masuda, T., Fusada, N., Oosawa, N., Takamatsu, K. I., Yamamoto, Y. Y. et al. (2003). Functional analysis of isoforms of NADPH: Protochlorophyllide OXIDOREDUCTASe (PORPORB and PORC, in Arabidopsis thaliana. Plant and Cell Physiology, 44(10), 963–974. DOI 10.1093/pcp/pcg128. [Google Scholar] [CrossRef]
31. Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2009). DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics, 26(1), 136–138. DOI 10.1093/bioinformatics/btp612. [Google Scholar] [CrossRef]
32. Li, Y., Huang, R., Li, J., Huang, X., Zeng, X. et al. (2020). DWARF and SMALL SEED1, a novel allele of OsDWARF, controls rice plant architecture, seed size, and chlorophyll biosynthesis. Phyton-International Journal of Experimental Botany, 90(1), 111–127. DOI 10.32604/phyton.2020.013933. [Google Scholar] [CrossRef]
33. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
34. Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H. et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology, 30(2), 174–178. DOI 10.1038/nbt.2095. [Google Scholar] [CrossRef]
35. Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M. et al. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics, 25(15), 1966–1967. DOI 10.1093/bioinformatics/btp336. [Google Scholar] [CrossRef]
36. Li, H., Sun, H., Jiang, J., Sun, X., Tan, L. et al. (2021). TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnology Journal, 19(1), 64–73. DOI 10.1111/pbi.13440. [Google Scholar] [CrossRef]
37. Beale, S. I. (2005). Green genes gleaned. Trends in Plant Science, 10(7), 309–312. DOI 10.1016/j.tplants.2005.05.005. [Google Scholar] [CrossRef]
38. Zhao, M. H., Li, X., Zhang, X. X., Zhang, H., Zhao, X. Y. (2020). Mutation mechanism of leaf color in plants: A review. Forests, 11(8), 851. DOI 10.3390/f11080851. [Google Scholar] [CrossRef]
39. Chan, K. X., Crisp, P. A., Estavillo, G. M., Pogson, B. J. (2010). Chloroplast-to-nucleus communication. Plant Signaling & Behavior, 5(12), 1575–1582. DOI 10.4161/psb.5.12.13758. [Google Scholar] [CrossRef]
40. Pogson, B. J., Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiology, 155(4), 1545–1551. DOI 10.1104/pp.110.170365. [Google Scholar] [CrossRef]
41. Vitha, S., Mcandrew, R. S., Osteryoung, K. W. (2001). FtsZ ring formation at the chloroplast division site in plants. Journal of Cell Biology, 153(1), 111–120. DOI 10.1083/jcb.153.1.111. [Google Scholar] [CrossRef]
42. Glynn, J. M., Yang, Y., Vitha, S., Schmitz, A. J., Hemmes, M. et al. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant Journal, 59(5), 700–711. DOI 10.1111/j.1365-313X.2009.03905.x. [Google Scholar] [CrossRef]
43. Liu, X., An, J., Wang, L., Sun, Q., An, C. et al. (2021). A novel amphiphilic motif at the C-terminus of FtsZ1 facilitates chloroplast division. Plant Cell, 34(1), 419–432. DOI 10.1093/plcell/koab272. [Google Scholar] [CrossRef]
44. Wu, Z., Zhang, X., He, B., Diao, L., Sheng, S. et al. (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiology, 145(1), 29–40. DOI 10.1104/pp.107.100321. [Google Scholar] [CrossRef]
45. Zhu, X., Guo, S., Wang, Z., Du, Q., Xing, Y. et al. (2016). Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biology, 16(1), 134. DOI 10.1186/s12870-016-0821-5. [Google Scholar] [CrossRef]
46. Ikeda, Y., Pélissier, T., Bourguet, P., Becker, C., Pouch-Pélissier, M. N. et al. (2017). Arabidopsis proteins with a transposon-related domain act in gene silencing. Nature Communications, 8(1), 15122. DOI 10.1038/ncomms15122. [Google Scholar] [CrossRef]
Supplementary Materials
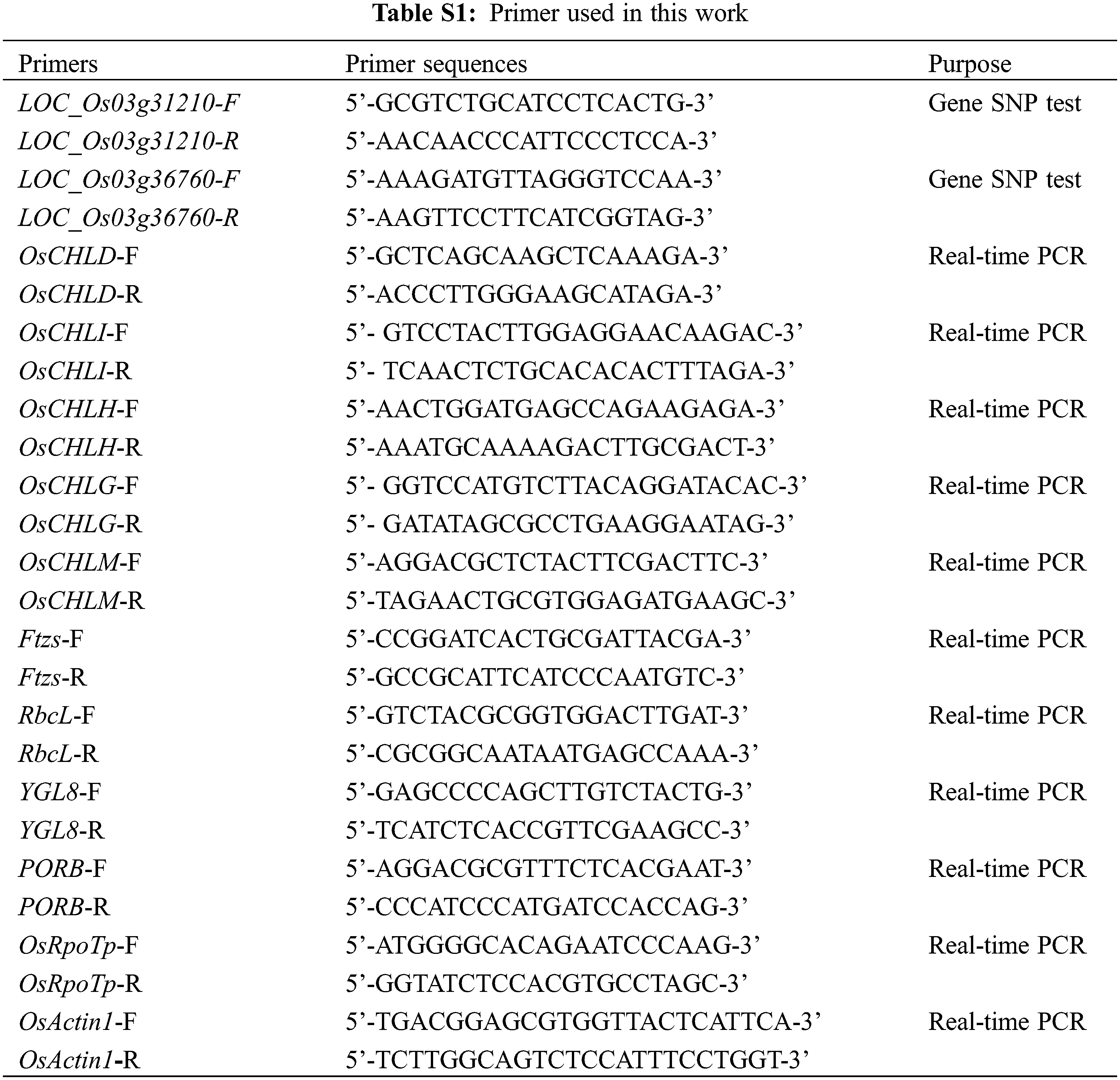
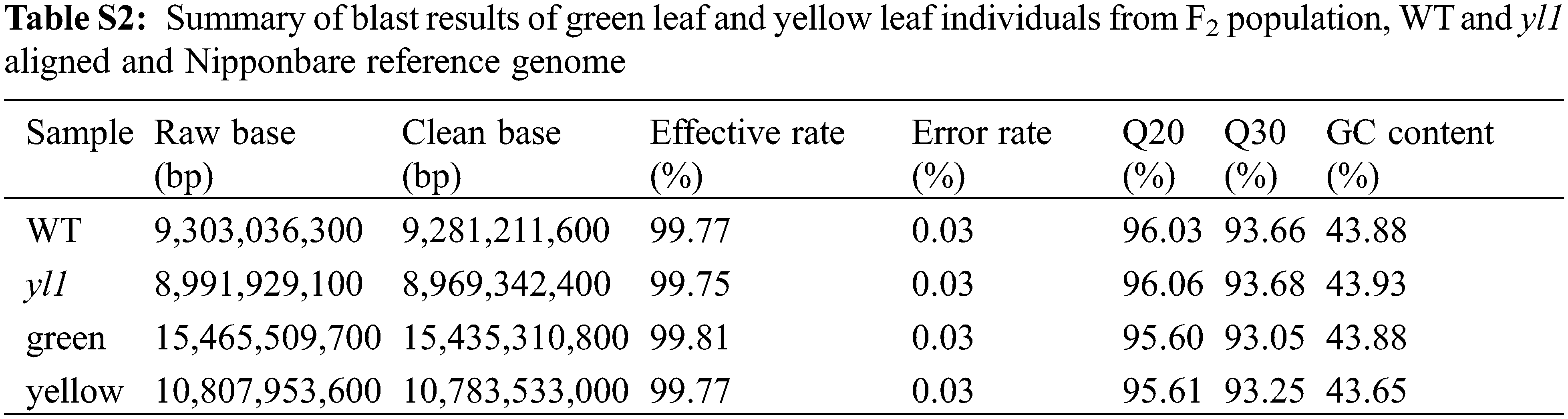
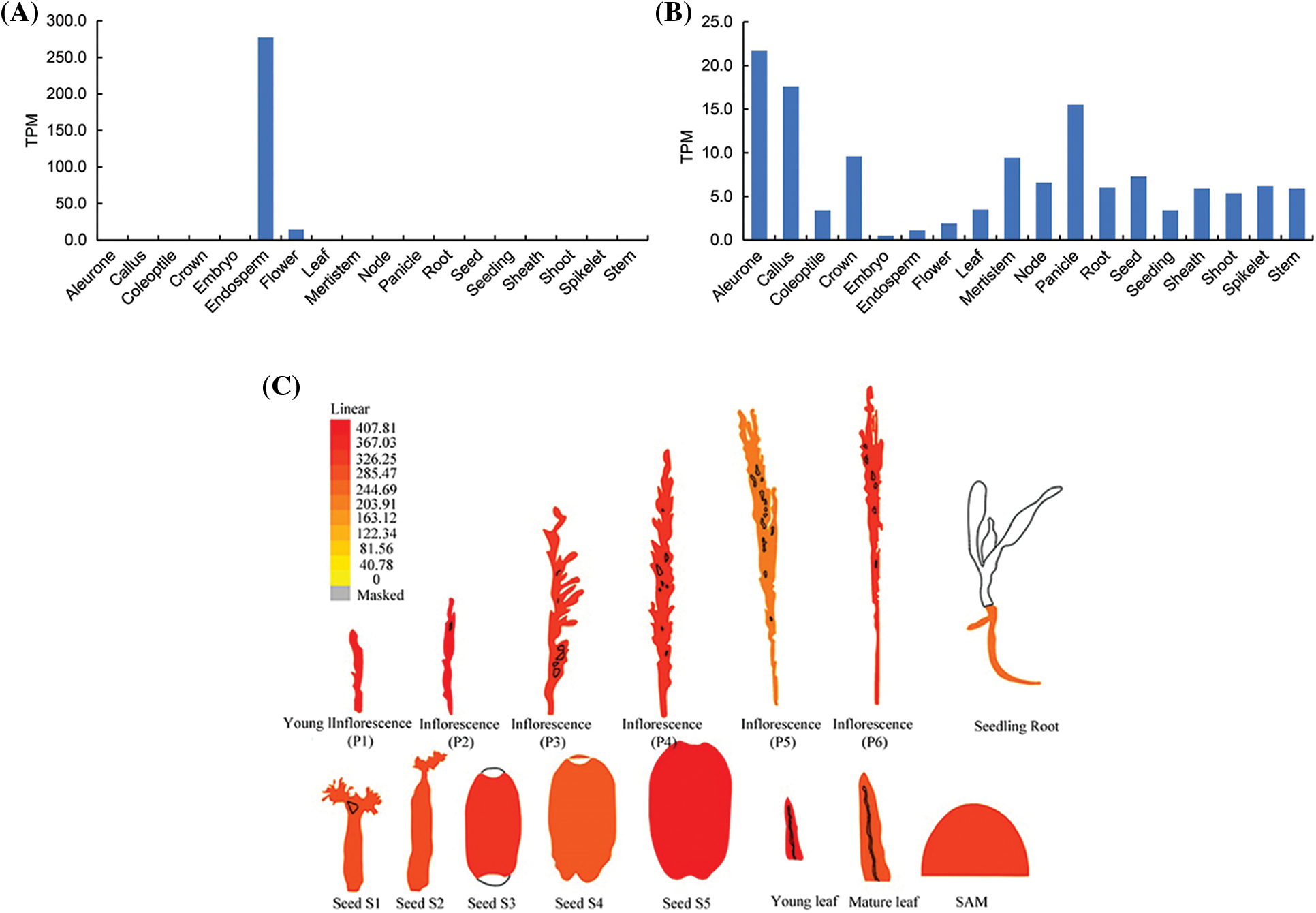
Figure S1: The expression levels in various tissues A: The gene expression levels of LOC_Os03g31210 in various tissues in Information Commons for Rice Database (IC4R); B: The gene expression levels of LOC_Os03g36370 in various tissues in Information Commons for Rice Database (IC4R); C: The gene expression levels of LOC_Os03g36370 in various tissues in transcriptomic database Rice eFP
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |