

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.022013
REVIEW
Papaya Ring Spot Virus: An Understanding of a Severe Positive-Sense Single Stranded RNA Viral Disease and Its Management
1Institute for Forest Resources and Environment of Guizhou and Forestry College, Research Centre of Forest Ecology, Guizhou University, Guiyang, 550025, China
2Department of Plant Pathology, College of Agriculture, University of Sargodha, Sargodha, 40100, Pakistan
3College of Bioscience, University of Birmingham, Birmingham, B152TT, UK
4Department of Soil Science, Faculty of Agricultural Sciences and Technology, Bahauddin Zakariya University, Multan, 60800, Pakistan
5Department of Plant Pathology, PMAS University of Arid Agriculture, Rawalpindi, 43490, Pakistan
6Department of Agriculture and Agribusiness Management University of Karachi, Karachi, 75270, Pakistan
7Citrus Research Institute, Sargodha, 40100, Pakistan
8Department of Plant Production, Faculty of Agronomy, Universidad de Concepción, Chillán, 3780000, Chile
*Corresponding Authors: Yuejun He. Email: hyj1358@163.com; Yasir Iftikhar. Email: yasir.iftikhar@uos.edu.pk
Received: 16 February 2022; Accepted: 30 March 2022
Abstract: Viral diseases have been studied in-depth for reducing quality, yield, health and longevity of the fruit, to highlight the economic losses. Positive-sense single-stranded RNA viruses are more devastating among all viruses that infect fruit trees. One of the best examples is papaya ringspot virus (PRSV). It belongs to the genus Potyvirus and it is limited to cause diseases on the family Chenopodiaceae, Cucurbitaceae and Caricaceae. This virus has a serious threat to the production of papaya, which is famous for its high nutritional and pharmaceutical values. The plant parts such as leaves, latex, seeds, fruits, bark, peel and roots may contain the biological compound that can be isolated and used in pharmaceutical industries as a disease control. Viral disease symptoms consist of vein clearing and yellowing of young leaves. Distinctive ring spot patterns with concentric rings and spots on fruit reduce its quality and taste. The virus has two major strains P and W. The former cause disease in papaya and cucurbits while the later one in papaya. Virion comprises 94.4% protein, including a 36 kDa coat protein which is a component responsible for a non-persistent transmission through aphids, and 5.5% nucleic acid. Cross protection, development of transgenic crops, exploring the resistant sources and induction of pathogen derived resistance have been recorded as effective management of PRSV. Along with these practices reduced aphid population through insecticides and plant extracts have been found ecofriendly approaches to minimize the disease incidence. Adoption of transgenic crops is a big challenge for the success of disease resistant papaya crops. The aim of this review is to understand the genomic nature of PRSV, detection methods and the different advanced control methods. This review article will be helpful in developing the best management strategies for controlling PRSV.
Keywords: Papaya; obligate parasite; +ssRNA; PRSV; aphid; genomic characterization and functions
Plant virus diseases are ubiquitous. Apparently, one or more viruses probably affect all wild and domesticated plants grown for food, feed, esthetic and fiber. These diseases have been shown to be very important agents in reducing yield, quality, health, longevity and cause significant economic losses in many crops, which have pushed more attention to viruses of cultivated crops. Plant viruses have not been reported to have direct impact on humans. However, the damage they do to the food chain might have a significant indirect effect and must not be underestimated. Among these plants, fruit trees are a suitable host for a wide range of viral pathogens owing to their vegetative mode of propagation and being perennial plants [1]. A number of viruses have been detected in several cultivated species from a range of fruit trees. Some of the viruses may be considered less economically important but provide significant information concerning viruses and their possible capacity of infection in the continuously changing genotype of plants. This review on Papaya ringspot virus (PRSV), a positive sense ssRNA virus, has been written with the aim to understand the role of PRSV in causing disease in Papaya and other cucurbits. The present review highlights the economic importance, genomic organization and future prospect in research related to the PRSV. Carica papaya is an important tropical and sub-tropical fruit tree, belonging to the family Caricacea, due to its large nutritional and pharmaceutical values, and it gained great popularity among researchers due to its high health benefits. In 1955, the production of papaya fruits was estimated to be 5.7 million metric tons, and more or less this amount was double in 1980 which indicated the importance of papaya at the economic level. Phenotypically papaya tree has a single stem with a large crown having palmate like shaped leaves, and have male or female parts or may be hermaphrodite due to being polygamous. Nutritionally, the fresh fruit has low calories and it is rich in vitamins and minerals such as iron, thiamine, fiber, vitamin A and C. Some biological active chemicals, known as phytochemicals, have been isolated from papaya to control pathogens [2]. Papain and chymopapain are examples of such active compounds present in papaya that may help to cure cancer, diabetes, and heart diseases. Ripe fruit of papaya is consumed fresh or can be processed in the production of jam, jelly, marmalade, candies and vegetable if unripe [3]. Southern Mexico and Costa Rica are the origin of papaya and it is native to central and southern American states. Currently, it has been adapted in USA, India, South Africa, Indonesia, Malaysia, Australia and Philippines. Cultivation of papaya trees is expanding in tropical and sub-tropical countries because of its fast-growing nature and fruiting within 3 years. A Venn diagram showing different regions with papaya production, is displayed in Fig. 1. A leading country is India which share 43.7% on the world’s total production of papaya. Brazil and Indonesia share of 11.8% and 7%, respectively, of the total production of papaya [4,5]. Papaya production faced major challenges for the last few years due to diseases and environmental changes. Among them, viral diseases are destructive for papaya crops that cause significant yield losses all over the world. Some important viral diseases of papaya are reported in the literature (Table 1) such as Papaya ring spot virus (PRSV) [6], papaya mosaic virus (PapMV) [7], papaya leaf distortion mosaic virus (PLDMV) [8], papaya lethal yellowing virus (PLYV) [9], papaya leaf curl virus (PaLCuV) [10] and papaya meleira virus (PMeV). The literary on PRSV showed that it causes ring spot diseases. The characteristic symptoms of the disease are vein clearing and yellowing of young leaves, Symptoms on fruit are distinctive ring spot patterns, consisting of concentric rings, and spots lead to deterioration on fruit quality and its taste. Symptoms caused by PRSV other than ringspots are stunting of infected plants, water-soaked oily streaks on the petiole and the upper part of the trunk, banding of young leaves, leaf mosaic, chlorosis and flower abortion. PRSV is not only the cause of low fruit production, but also has an effect on the level of fruit sugar, which can be reduced by 50% or some time more. PRSV infected a number of host species belonging to the family Chenopodiaceae such as Chenopodium quinoa and Chenopodium amaranti but symptoms of the disease were restricted to local lesions of leaves [11]. Preliminary information regarding pathogenicity of the PRSV is purely based on infection assays on these local lesions of leaves. On the basis of infectivity [12], the Papaya ring spot virus (PRSV) was classified into two types; (i) papaya-infecting Type-P (PRSV-P) that infected the plant species belonging to the families Caricaceae, Cucurbitaceae and Chenopodiaceae, and (ii) non-papaya-infecting Type-W (PRSV-W) infected plant species that belong to the families Cucurbitaceae and Chenopodiaceae. PRSV-P type affects papaya and cucurbits, and PRSV-W type affects only cucurbits. Different aphid species were responsible for the transmission of both types of strains in a non-persistent manner. The comparative consequences of mutation and the movement of the virus around the world are unclear. They may affect the control of the virus and the economic constraints for the production of papaya and cucurbits worldwide [13,14].
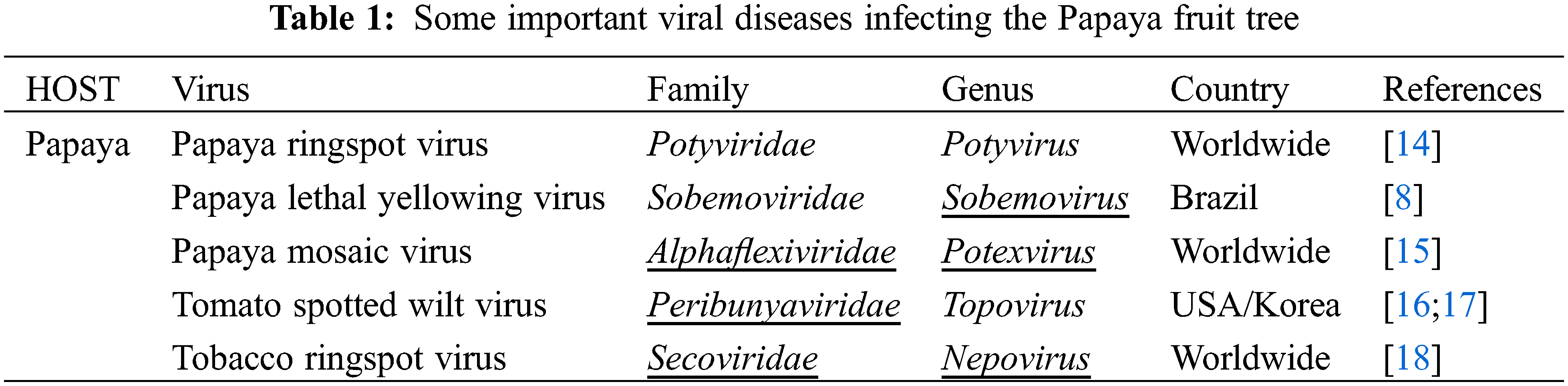
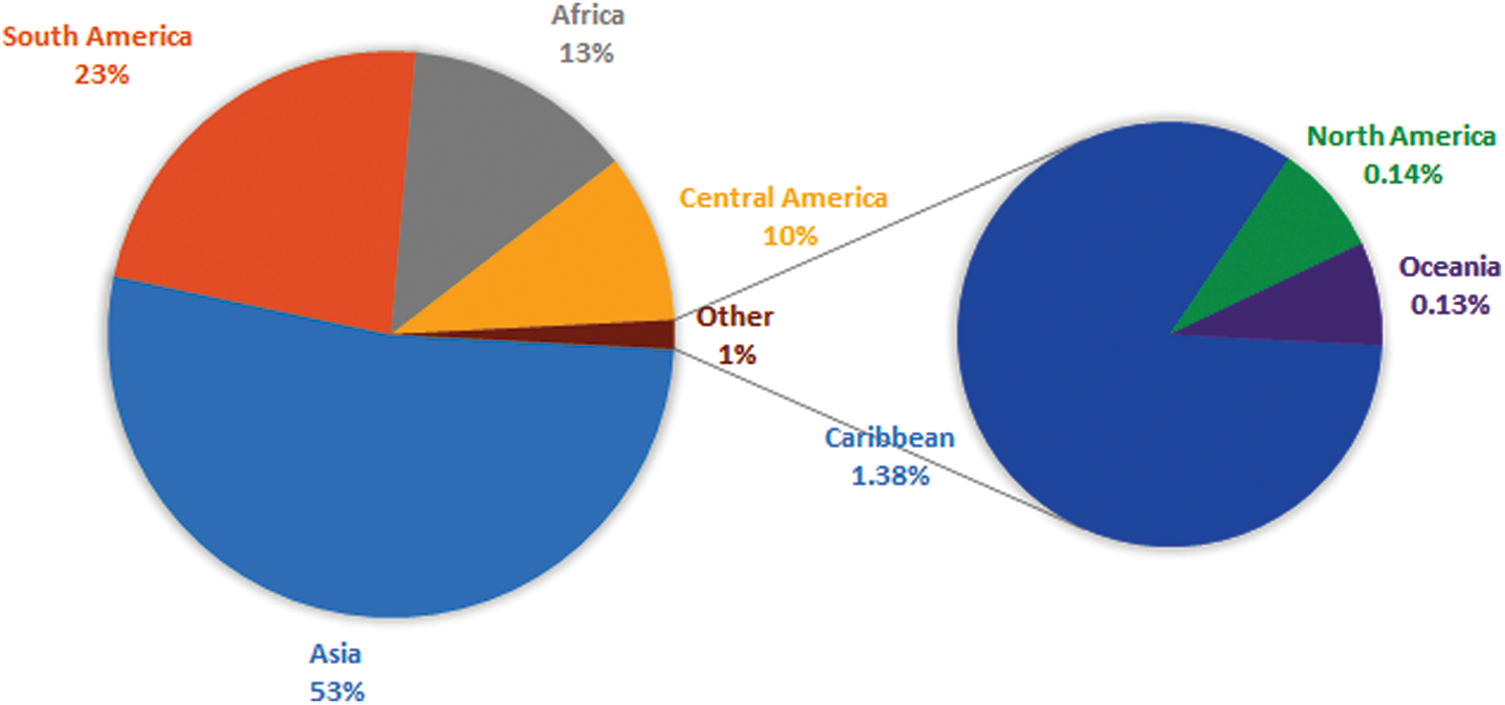
Figure 1: Venn diagram showing the share of different papaya growing regions of the world. Asia is a leading continent with 53% of the total world production of papaya
2 Geographical Distribution and Economic Losses
In 1949, Jenson documented the exceptional yield losses due to PRSV in papaya crop cultivated in areas of Hawaii. The disease had been reported in many papaya-growing tropical and subtropical countries including USA, Japan, Mexico, and India [19]. PRSV was first reported in China in 1959 and it turned into a devastating disease which completely limited the growth of the papaya industry [20]. In 1970, PRSV-W (known as watermelon mosaic virus-1) was reported in Australia, but there was no recorded data of PRSV-P, so it was reported as papaya dieback disease [21]. In 1990, the PRSV caused demolition of the Hawaiian papaya industry and commercial crops [22,23]. In 1992, PRSV disease in Hawaii, severely affected nearly 100% of the papaya plants within 3 years. PRSV developed as a severe problem in different areas of the world, like Brazil, Hawaii, Thailand and Taiwan [24]. The studies of the development and epidemiology of PRSV has been made difficult. (1) Initially due to the misperception on the appropriate identification of PRSV, particularly in cucurbits, and (2) the lack of satisfactory records from many countries.
3 Genomic Components and Their Function
The genome of PRSV is in a positive sense single stranded RNA, comprised of 10,324 nucleotides with a particle size of 800 to 900 nm long. It is non-enveloped flexuous filamentous. The virion comprised of protein (94.4%) and nucleic acid 5.5% [20,25]. Different PRSV isolates were recorded from different regions of the world, and their phylogenetic analysis showed their relatedness among them (Fig. 2).
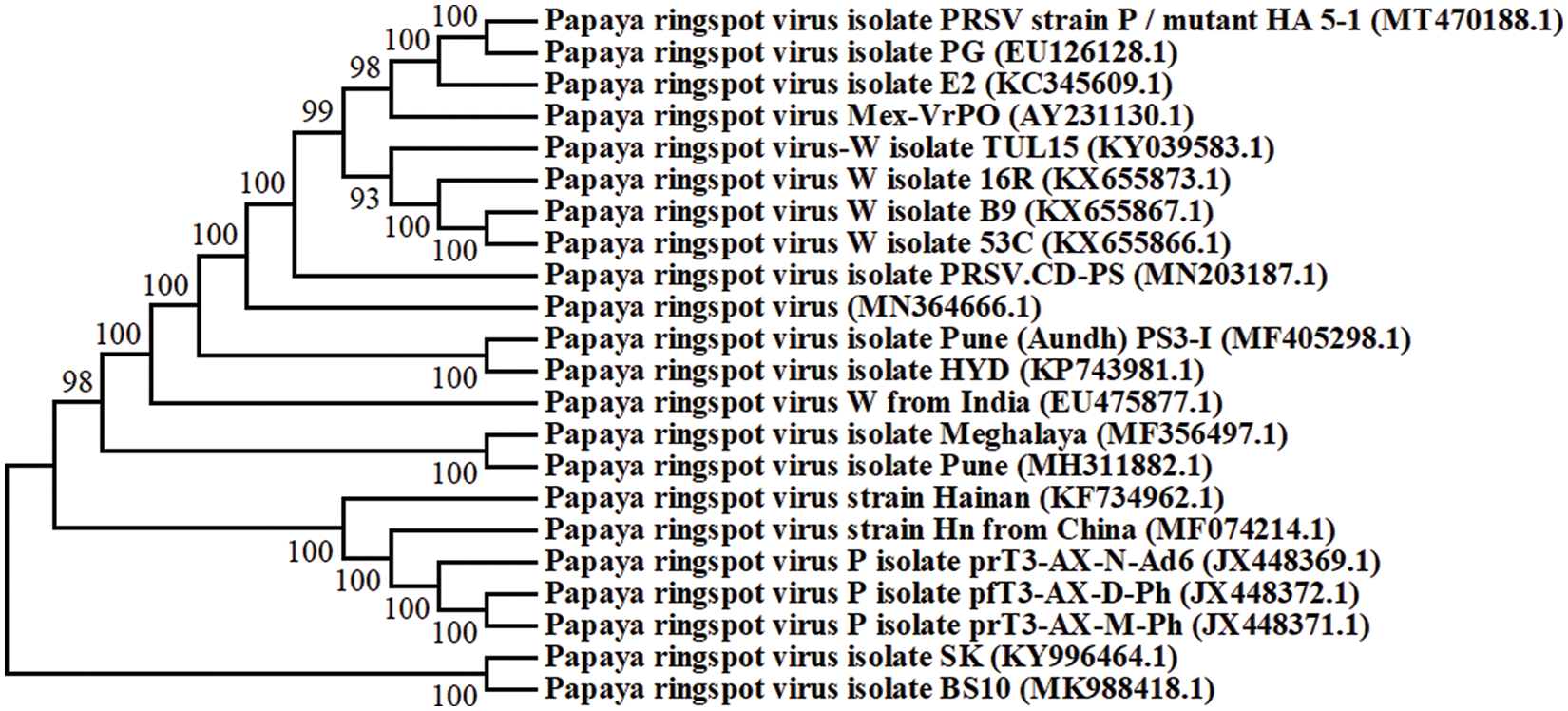
Figure 2: Phylogenetic analysis of 22 full-length genome sequences of different PSRV isolates strains from different regions of the world. The accession number of each isolate is shown in brackets and the full name is indicated with regions of identification. Each branch has a bootstraps number that was obtained from 1000 replicates
The PRSV genome was found as a single large protein of 3,344 amino acids, and it cleaves into small protein units. These small protein units have various functions. The recommended position of the cleavage sites in a single large protein of 3,344 amino acids produce 8 to 9 proteins. They include a helper component (HC-Pro, 52 KDa), coat protein (CP, 35 KDa), cylindrical inclusion protein (C1, 72 KDa), nuclear inclusion protein a (NIa, 48KDa), nuclear inclusion protein b (NIb, 59 KDa), P1 (63 KDa) and P3 (46 KDa) [25]. Each protein unit has its own specific function. The C terminus of HC-Pro cleaves catalytically [26] and it is responsible for a proteolytic activity either cis or trans to generate the CP, NIa, NIb and CI proteins [27–29]. Recently, the NIa protein of TEV and PSbMV showed delimitation of proteinase domains and genome-linked proteins (VPg) due to internal cleavage sites [30]. Sequencing results showed that the N terminus of HC-Pro cleaves catalytically in tobacco vein mottle virus [31]. HC-Pro a multifunctional protein, facilitated the transmission of the disease via aphids, the appearance of symptoms, amplification of the genome, and suppression of host resistance with post transcriptional gene silencing (PTGS) and effective RNA silencing suppressor [32]. HC-Pro affected the microRNA-mediated development pathways that resulted in a heterologous viral formation. The PTGS suppression mechanism, triggers genome replication and long-distance movement of HC-Pro [33]. It was also responsible for the synergistic relationship among potyviruses and unrelated viruses that could cause severe symptoms and an accumulation of virus in infected leaves.
Coat protein (CP) of PRSV have a molecular weight of 36 KDa [10]. CP induce amorphous inclusions (AIs) and cylindrical inclusions (CIs) in host cell cytoplasm [14]. The molecular weight of CIs protein is 70 KDa and AIs protein molecular weight is 51 KDa [34] from the total molecular weight of CP. Half of the coding capacity of the virus genome for CP is done by one reading frame [25]. The CP not only performs a duty of encapsidation of the viral genome, but also is involved in the systemic movement and transmission by aphids. Geographical location affects the genetic diversity of PRSV (e.g., the CP gene incorporated in the transgenic papaya variety (HA 5-1) showed resistance against the pathogenic strain (HA) from USA but was susceptible against the Thai and Australian strains [35]. The CI protein carried out different activities such as RNA binding, RNA helicase activity, and NTP binding and NTPase [36]. N and C-terminal genome-linked protein (VPg) has two domains of NIa protein that play an important role in RNA synthesis by codependent RNA polymerase triggered by the NIb protein replicase activity. The P1 protein was cleaved auto catalytically and it was the least conserved protein. The P1 protein moves systemically in infected plants [37].
3.2 Host Specificity Determining Factor and Vector Transmission
Aphid transmits the PRSV virion from infected to healthy plants when they use their stylet to puncture the leaf epidermal layer for suctioning of food. Viral encoded proteins such as HC-Pro and CP have been reported as vital for spreading of the disease [38]. Host and viral interaction help virion intercellular movement in plants. Viruses transfer among the host plants via mechanical injuries or vectors. Cucurbita pepo, Cucumis metuliferus and C. papaya have been reported as host plants of PRSV. The lesion assay hosts of PRSV were Chenopodium quinoa and Chenopodium amaranticolor [14]. Virion causes infection in papaya [39]. In papaya, the genes responsible for infection were NIa and some portions of NIb genes. Any abrupt change in HC-Pro and P1 genes may result in the weakening of symptoms in papaya and local lesions in Chenopodium quinoa [40]. To develop a management strategy against PRSV it is important to study the interaction between the host and the pathogen [41].
Correct identification of PRSV is the first and most important step to control the virus because PRSV exists in different strains [42]. The virus is uneven and has the ability to combine with the plant debris. Primarily, it can be identified by visual symptoms; visual identification is quickly but unreliable because symptoms caused by micronutrient deficiency are similar to PRSV symptoms. This is why molecular diagnosis is necessary for the correct identification of PRSV. Different molecular diagnostic tools are in use, such as the Enzyme-linked immunosorbent assay (ELISA), RT-PCR, Dot immune binding assay (DIBA) and Immuno-capture RT-PCR. ELISA is a quick and reliable technique used in different PRSV affected areas of the world [35,41]. RT-PCR is also a quick and reliable technique and can determine even very low concentrations of the virus. More diseased samples are used for the indexing of the virus; then, DIBA is a simple, easy, economical and time saving method [43].
5 Disease Management Strategies
PRSV can be managed by the individual or a combined use of different disease control mechanisms such as:
Mild strains of PRSV used to induce immunity in papaya plants against severe strains of the PRSV virus cause huge economic losses [44]. In Taiwan, the PRSV was controlled with a mild strain of PRSV by providing resistance against PRSV infection [45]. Availability of mild strains is a key factor to control the target virus. This method of protection needs extra care and agriculture practices. The limitation of this control practice is the availability of specific and pure mild strains [46]. It is a slightly effective method against PRSV approved during field experiments [14]. Even though we know little about cross protection at the molecular level, post transcriptional gene silencing (PTGS) is effective to manage PRSV [41,47].
5.2 Pathogen Derived Resistance
Sanford and Johnston suggested pathogen-derived resistance (PDR) as an ideal strategy [48] for developing resistance against pathogens. RNA-mediated, protein-mediated and RNA-mediated gene silencing were the methods of PDR. The last one was triggered due to expression of viral genes in host plants [43]. In spite of attainment of this PRSV technique, resistance levels varied during developmental stages of plants and environmental factors. If viral targeted genes had homology with transgenes and with divergent strains of PRSV based on geographical distribution, they were used to develop resistance against a wide range of PRSV strains. The transgenic papaya varieties must establish separation in each papaya-growing region. The best way to control the PRSV disease for long-time protection is the development of PRSV resistance lines [49].
5.3 Development of Resistant Varieties of Papaya
Transgenic crop developed through PDR derived genes from a virus sequence or other sources could interfere with natural resistance and targeted viral genes. Viral replication was being affected or altered due to PDR in the host plant. Gene technology was used by the researchers of the University of Hawaii and Cornell University for the development of resistant cultivars. The transgenic papaya variety was developed against PRSV using viral coat proteins, replicas gene technology and RNA silencing. Resistant varieties of papaya were developed, but these varieties did not grow at a commercial scale due to poor quality of fruit and vigor [50]. PRSV resistant genes were available in some wild varieties related to the Carica species. However, cultivated papaya and wild species were sexually incompatible. This was the biggest drawback to the use of conventional breeding methods. This disease control approach is limited due to the use of backcrosses with commercial papaya to develop disease tolerance [22].
5.4 Use of Coat Proteins for the Development of Transgenic Crops
The transgenic papaya was developed against PRSV after successful transformation and expression of the CP gene in transgenic tobacco plants against the tobacco mosaic virus [51]. The first demonstration of resistance-mediated CPs against PRSV was when CP genes were transferred in transgenic papaya via the immature zygotic embryo transformation system along with the neomycin phosphotransferase II (nptII) gene [52]. Resistant transgenic papaya was also developed from CP genes of PRSV (Taiwanese strain) pBGCP vector induced via Agrobacterium transformation. Different explants with nptII gene containing plasmids were used to develop many resistant varieties of papaya around the papaya-cultivating world [53]. CP mediated resistance depends upon the origin of viral isolates [54] even though it was excessively used to protect papaya against PRSV in the world [20]. Untranslatable CP gene and transferred with a gene gun for the development of a resistant variety of papaya that showed resistance against homologous isolates discorded from Australia, Mexico, Taiwan, Bahamas, Hawaii, Jamaica and Brazil. In Hawaii, scientists developed SunUp (transgenic variety of papaya) with the transformation of CP gene and somatic embryos of a Hawaiian strain. In 1994, researchers developed transgenic papaya expressing the CP gene of the mild PRSV strain from Hawaii (PRV HA 5-1) highly resistant to the PRV-HA strain [35]. Expression of CP genes in transgenic crops triggered vertical resistance against different strains which were geologically inhabitant throughout Taiwan in 2004 [55]. The CP gene of the PRSV Philippine strain was used to develop genetically modified papaya plants. Transgenic plants were moderately susceptible but their first generation was resistant.
RNA interference (RNAi) was used to develop resistance in transgenic tobacco plants against the Potato virus Y. RNA mediated gene resistance has been established as a new tool and a platform for functional studies of genes in the field of improvement and development of transgenic crops [56]. Gene silencing mediated by RNA played an important role in both biotic and abiotic biotic stress on plant defense against insects and pathogens. It helped the humanity to combat with the challenges arose in sustainable agriculture due to environmental conditions and climate change. This technology involved in suppression of the specific gene related to pathogenesis, and resulted in resistance development in plants [57]. PRSV is a +ssRNA virus containing only one open reading frame, it produces a large polyprotein that cleaved itself in final protein products during the phenomena of translation [58]. RNA-mediated interference would only be effective if the transgene is homologous to the pathogenic viruses, but if transgenes are heterologous and their origin is geologically distinct, it would be difficult to develop transgenic cultivars. Viral origin suppressor proteins mediated RNA silencing involved in the failure of resistant cultivars [59] but HC-Pro could counter the RNA silencing mechanism. Thus, HC-Pro must be considered for the development of resistant cultivars. Small-interfering RNAs were 21–25 base pairs long nucleotide sequences that mediated degradation and methylation of specific sequences of targeted mRNAs and targeted genes. This mechanism is a homology based resistance mechanism of PTGS [60]. Untranslatable CP genes involved in the PTGS mechanism against homologous viral strains.
5.6 Replicase Gene Mediated Resistance
The resistance mechanism is based on a replicase protein because mutation influenced the resistance phenotype by affecting the protein’s primary structure that transgenes encoded. Replicase genes were different in structure within various genera. Replicase gene mediated resistance was first demonstrated against the tobacco mosaic virus in tobacco plants by the introduction of the replicase gene [61]. The function of mutation was to confirm a mutation in the gene [62]. Reports showed that transgenic papaya plants with replicase gene conferred resistance against PRSV, and conformed high resistance to PRSV to mutate replicase genes in transgenic papaya [63].
Use of transgenic crops, cross protection and exploration of resistance sources are the effective management practices of PRSV. Other than these, insecticides, used for the control of aphid species, also played a vital role to reduce the disease incidence and yield loss. Although, Papaya has not been recorded as a main host, aphids were effective in transmission of PRSV. Therefore, reduction in the PRSV incidence was achieved by the application of insecticides alone and in combination with bio rationals. The efficacy of different insecticides, bio rationals and their combinations were evaluated, and found that foliar application of different insecticides (Tolfenpyrad 15% EC at 1 ml/lt, imidacloprid 17.8% SL at 0.2 ml/lt, thiacloprid 21.7 SC at 1 ml/lt and dinotefuran 20% SG at 0.5 g/lt) mixed with micronutrients with an interval of 30 days was found to be the most effective [64]. The PRSV is one of the major threats for papaya production. Different integrated approaches were carried out to combat the disease. Raising of Papaya seedlings under insect-free controlled conditions, and spray of Dimethoate (1.05%) alone and in combination with neem oil 1% were the different treatments to minimize the disease incidence during the field experiment. Among the different treatments, application of neem oil 1% + Dimethoate 1.05% was the most effective [65]. Therefore, control of the aphid population in papaya under field conditions always reduced the disease incidence caused by PRSV.
6 Adoption of Transgenic Crops
Transgenic cultivars of crops play an important role in the management of viral diseases, but its acceptance rate is very low. Some of the key challenges faced by adoption of transgenic crops were the use of biotechnological procedures for its development and sustainability as a marketable product. There were concerns related to trade and biosafety regulations. It was investigated that the transgenic crop had no impact on its adjacent atmosphere; for example, microbial flora in the soil adjacent to transgenic trees [66]. With reference to safety, assessments by the scientists had the recommendation of no harmful effects on nutritional and toxicological parameters [67]. Farmers were influenced by anti-genetic engineering. NGO networks were not easily adaptable to new technologies. This was one of the reasons for the less use of transgenic crops. Greenpeace International NGO stood against the use of transgenic crops in some countries. Developing countries faced problems in the adaptation of transgenic crops due to lack of training sessions for farming communities, infrastructure, biosafety laws and commercialization of transgenic crops. Markets of many developing countries were fully dependent on importing countries rules, political influences and consumer’s demands. Furthermore, traditional awareness, and social, political and lucrative factors also had an impact on the adoption of transgenic crops.
6.1 Adaptation of Transgenic Papaya Crop
The fruitful approval of transgenic papaya depends on regulatory issues regarding biosafety and the social acceptance of the technology. The resistant transgenic papaya had only partial possessions on the microbial life within the soil [68]. Resistant transgenic papaya varieties were released and adopted in the USA and China. Reports showed that the rate of adoption of resistant papaya was higher among farmers in Hawaii [23]. There was a variable adoption rate of transgenic papaya in Hawaii, Jamaica, and Venezuela, which was influenced by the demand for papaya, biosafety regulations, and the social acceptance of the technology [69].
7 Conclusion and Future Prospects
The key risk in the production of papaya is PRSV. There were different methods to control or manage plant diseases, but resistant varieties and application of bio rational along with nutrients and insecticides are the effective management of PRSV. These were no or least harmful to the environment and human health. The modern plant disease mechanisms of PRSV depended on the development of transgenic papaya crops. Socioeconomically, Hawaiian papaya industry is highly influenced by transgenic papaya. The gene technology used CP genes of RNAi to develop transgenic varieties of papaya. Transgenic crop adaptability depends on the resistance levels and durable horticultural characteristics. PRSV isolates geographically distinct and genetically diversifying is why the resistance in papaya is not sustainable against different isolates. If having a keen check on the population of diversifying PRSV, the disease can be managed successfully. PTGS-mediated RNA silencing mechanisms may be an effective and durable method to develop resistant varieties against geographically distinct isolates. This review recommends that papaya-producing countries are supposed to develop resistant transgenic varieties by using local isolates through PTGS technology.
Compliance with Ethical Standards: The authors declare that the review is in compliance with ethical standards of the journal.
Competing Financial Interests: The authors declare no competing financial interests.
Research Involving Human Participants and/or Animals: The authors declare that the manuscript does not contain research involving Human Participants and/or Animals.
Author’s Contribution: All the authors have equally contributed in gathering literature and manuscript write up and formatting.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare no conflict of interests and give their consent for publishing the material.
1. Reed, P. J., Foster, J. A. (2011). Exclusion of pome and stone fruit viruses, viroids and phytoplasmas by certification and quarantine. In: Hadidi, A., Barba, M., Candresses, T., Jelkmann, W. (Eds.Virus and virus-like diseases of pome and stone fruits, pp. 381–388, St. Paul, MN: APS Press. [Google Scholar]
2. Boshra, V., Tajul, A. Y. (2013). Papaya–An innovative raw material for food and pharmaceutical processing industry. Health Environmental Journal, 4, 68–75. [Google Scholar]
3. Aravind, G., Bhowmik, D., Duraivel, S., Harish, G. (2013). Traditional and medicinal uses of Carica papaya. Journal of Medicinal Plants Studies, 1(1), 7–15. [Google Scholar]
4. Varun, P., Saxena, S. (2017). Leaf curl disease of Carica papaya. In: Begomoviruses: Occurrence and management in Asia and Africa, pp. 111–135. Singapore: Springer. [Google Scholar]
5. Kebede, G., Kabeto, E. G., Dagnew, A. (2021). Distribution and Identification of anthracnose of papaya caused by Collectotrichum gloeosporioides in the central rift valley of Ethiopia. Plant Protection, 5(3), 139–147. DOI 10.33804/pp.005.03.3917. [Google Scholar] [CrossRef]
6. Jensen, D. (1949). Papaya ringspot virus and its insect vector relationships. Phytopathology, 39, 212–220. [Google Scholar]
7. Thérien, A., Bédard, M., Carignan, D., Rioux, G., Gauthier-Landry, L. et al. (2017). A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling. Journal of Nanobiotechnology, 15(1), 54. DOI 10.1186/s12951-017-0289-y. [Google Scholar] [CrossRef]
8. Tuo, D., Yan, P., Zhao, G., Cui, H., Zhu, G. et al. (2021). An efficient papaya leaf distortion mosaic potyvirus vector for virus-induced gene silencing in papaya. Horticulture Research, 8(1), 144. DOI 10.1038/s41438-021-00579-y. [Google Scholar] [CrossRef]
9. Pereira, A. J., Alfenas-Zerbini, P., Cascardo, R. S., Andrade, E. C., Murilo, Z. F. (2012). Analysis of the full-length genome sequence of papaya lethal yellowing virus (PLYVdetermined by deep sequencing, confirms its classification in the genus Sobemovirus. Archives of Virology, 157(10), 2009–2011. DOI 10.1007/s00705-012-1384-x. [Google Scholar] [CrossRef]
10. Lal, A., Kil, E. J., Rauf, K., Ali, M., Lee, S. (2020). First report of papaya leaf curl virus associated with leaf curl disease in Cestrum nocturnum in Pakistan. Plant Disease, 104(11), 3089. DOI 10.1094/PDIS-12-19-2681-PDN. [Google Scholar] [CrossRef]
11. Purcifull, D. E., Hiebert, E. (1979). Serological distinction of watermelon mosaic virus isolates. Phytopathology, 69, 112–116. DOI 00031-949X/79/000018$03.00/0. [Google Scholar]
12. Sultana, S., Roy, B., Sherpa, A. R. (2021). The complete sequence of a papaya ringspot virus (PRSV) isolate from West Bengal, India infecting papaya and study of genetic variation. European Journal of Plant Pathology, 159(1), 203–210. DOI 10.1007/s10658-020-02124-4. [Google Scholar] [CrossRef]
13. Suzuki, J. Y., Tripathi, S., Gonsalves, D. (2007). Virus-resistant transgenic papaya: Commercial development and regulatory and environmental issues. Biotechnology and Plant Disease Management, pp. 436–461. CABI Publishing Series. [Google Scholar]
14. Tripathi, S., Suzuki, J. Y., Ferreira, S. A., Gonsalves, D. (2008). Papaya ringspot virus-P: Characteristics, pathogenicity, sequence variability and control. Molecular Plant Pathology, 9(3), 269–280. DOI 10.1111/j.1364-3703.2008.00467.x. [Google Scholar] [CrossRef]
15. Abou Haidar, M. G., Erickson, J. W. (2018). Structure and in vitro assembly of papaya mosaic virus. In: Molecular plant virology, pp. 85–121. CRC Press. [Google Scholar]
16. Batuman, O., Turini, T. A., Oliveira, P. V., Rojas, M. R., Macedo, M. et al. (2017). First report of a resistance-breaking strain of Tomato spotted wilt virus infecting tomatoes with the Sw-5 tospovirus-resistance gene in California. Plant Disease, 101(4), 637. DOI 10.1094/PDIS-09-16-1371-PDN. [Google Scholar] [CrossRef]
17. Cho, S. Y., Kim, S. M., Kim, S., Lee, B. C. (2020). First report of tomato spotted wilt virus infecting Arachis hypogaea in Korea. Journal of Plant Pathology, 102(1), 271. DOI 10.1007/s42161-019-00410-7. [Google Scholar] [CrossRef]
18. Tabara, M., Nagashima, Y., He, K., Qian, X., Crosby, K. M. et al. (2021). Frequent asymptomatic infection with tobacco ringspot virus on melon fruit. Virus Research, 293, 198266. DOI 10.1016/j.virusres.2020.198266. [Google Scholar] [CrossRef]
19. Azad, M., Kalam, A., Amin, L., Sidik, N. M. (2014). Gene technology for papaya ringspot virus disease management. The Scientific World Journal, 2014(1), 1–11. DOI 10.1155/2014/768038. [Google Scholar] [CrossRef]
20. Ventura, J. A., Costa, H., da Silva Tatagiba, J. (2004). Papaya diseases and integrated control. In: Diseases of fruits and vegetables, vol. 2, pp. 201–268. Dordrecht: Springer. [Google Scholar]
21. Gonsalves, D., Vegas, A., Prasartsee, V., Drew, R., Suzuki, J. et al. (2006). Developing papaya to control papaya ringspot virus by transgenic resistance, intergeneric hybridization, and tolerance breeding. In: Plant breeding reviews, vol. 26, pp. 35–73. John Wiley & Sons, Inc. [Google Scholar]
22. Gonsalves, C., Lee, D. R., Gonsalves, D. (2007). The adoption of genetically modified papaya in Hawaii and its implications for developing countries. The Journal of Development Studies, 43(1), 177–191. DOI 10.1080/00220380601055650. [Google Scholar] [CrossRef]
23. Davidson, S. N. (2008). Forbidden fruit: Transgenic papaya in Thailand. Plant Physiology, 147(2), 487–493. DOI 10.1104/pp.108.116913. [Google Scholar] [CrossRef]
24. Yeh, S. D., Gonsalves, D. (1985). Translation of papaya ringspot virus RNA in vitro: Detection of a possible polyprotein that is processed for capsid protein, cylindrical-inclusion protein, and amorphous-inclusion protein. Virology, 143(1), 260–271. DOI 10.1016/0042-6822(85)90113-8. [Google Scholar] [CrossRef]
25. Carrington, J. C., Cary, S. M., Parks, T. D., Dougherty, W. G. (1989). A second proteinase encoded by a plant potyvirus genome. The EMBO Journal, 8(2), 365–370. DOI 10.1002/j.1460-2075.1989.tb03386.x. [Google Scholar] [CrossRef]
26. Carrington, J. C., Dougherty, W. G. (1987). Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. Journal of Virology, 61(8), 2540–2548. DOI 10.1128/jvi.61.8.2540-2548.1987. [Google Scholar] [CrossRef]
27. Carrington, J. C., Dougherty, W. G. (1987b). Processing of the tobacco etch virus 49K protease requires autoproteolysis. Virology, 160(2), 355–362. DOI 10.1016/0042-6822(87)90006-7. [Google Scholar] [CrossRef]
28. Dougherty, W. G., Carrington, J. C. (1988). Expression and function of potyviral gene products. Annual Review of Phytopathology, 26(1), 123–143. DOI 10.1146/annurev.py.26.090188.001011. [Google Scholar] [CrossRef]
29. Johansen, E., Rasmussen, O. F., Heide, M., Borkhardt, B. (1991). The complete nucleotide sequence of pea seed-borne mosaic virus RNA. Journal of General Virology, 72(11), 2625–2632. DOI 10.1099/0022-1317-72-11-2625. [Google Scholar] [CrossRef]
30. Mavankal, G., Rhoads, R. E. (1991). In vitro cleavage at or near the N-terminus of the helper component protein in the tobacco vein mottling virus polyprotein. Virology, 185(2), 721–731. DOI 10.1016/0042-6822(91)90543-K. [Google Scholar] [CrossRef]
31. Anandalakshmi, R., Pruss, G. J., Ge, X., Marathe, R., Mallory, A. C. et al. (1998). A viral suppressor of gene silencing in plants. Proceedings of the National Academy of Sciences, 95(22), 13079–13084. DOI 10.1073/pnas.95.22.13079. [Google Scholar] [CrossRef]
32. Kasschau, K. D., Carrington, J. C. (2001). Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology, 285(1), 71–81. DOI 10.1006/viro.2001.0901. [Google Scholar] [CrossRef]
33. Purcifull, D. E., Edwardson, J. R. (1967). Watermelon mosaic virus: Tubular inclusions in pumpkin leaves and aggregates in leaf extracts. Virology, 32(3), 393–401. DOI 10.1016/0042-6822(67)90289-9. [Google Scholar] [CrossRef]
34. de Mejia, M. V. G., Hiebert, E., Purcifull, D. E. (1985). Isolation and partial characterization of the amorphous cytoplasmic inclusions associated with infections caused by two potyviruses. Virology, 142(1), 24–33. DOI 10.1016/0042-6822(85)90419-2. [Google Scholar] [CrossRef]
35. Tennant, P. F., Gonsalves, C., Ling, K. S., Fitch, M., Manshardt, R. et al. (1994). Differential protection against papaya ringspot virus isolates in coat protein gene transgenic papaya and classically cross-protected Papaya. Phytopathology, 84, 1359–1365. [Google Scholar]
36. Fernandez, A., Lain, S., Carcia, J. A. (1995). RNA helicase activity of the plum pox potyvirus Cl protein expressed in Escherichia coli mapping of an RNA binding domain. Nucleic Acids Research, 23(8), 1327–1332. DOI 10.1093/nar/23.8.1327. [Google Scholar] [CrossRef]
37. Urcuqui-Inchima, S. (2001). Potyvirus proteins: A wealth of functions. Virus Research, 74(1–2), 157–175. DOI 10.1016/S0168-1702(01)00220-9. [Google Scholar] [CrossRef]
38. Peng, Y., Kadoury, D., Gal-On, A., Huet, H., Wang, Y. et al. (1998). Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: Effects on aphid transmission and binding to purified virions. Journal of General Virology, 79(4), 897–904. DOI 10.1099/0022-1317-79-4-897. [Google Scholar] [CrossRef]
39. Chen, G., Ye, C., Huang, J., Yu, M., Li, B. (2001). Cloning of the papaya ringspot virus (PRSV) replicase gene and generation of PRSV-resistant papayas through the introduction of the PRSV replicase gene. Plant Cell Reports, 20(3), 272–277. DOI 10.1007/s002990000311. [Google Scholar] [CrossRef]
40. Chiang, C. H., Lee, C. Y., Wang, C. H., Jan, F. J., Lin, S. S. et al. (2007). Genetic analysis of an attenuated Papaya ringspot virus strain applied for cross-protection. European Journal of Plant Pathology, 118(4), 333–348. DOI 10.1007/s10658-007-9130-z. [Google Scholar] [CrossRef]
41. Yeh, S. D., Gonsalves, D. (1994). Practices and perspective of control of papaya ringspot virus by cross protection. In: Advances in disease vector research, pp. 237–257. New York, NY: Springer. [Google Scholar]
42. Chiang, C. H., Wang, J. J., Jan, F. J., Yeh, S. D., Gonsalves, D. (2001). Comparative reactions of recombinant papaya ringspot viruses with chimeric coat protein (CP) genes and wild-type viruses on CP-transgenic papaya. Journal of General Virology, 82(11), 2827–2836. DOI 10.1099/0022-1317-82-11-2827. [Google Scholar] [CrossRef]
43. Smith, F. D., Banttari, E. E. (1987). Dot-ELISA on nitrocellulose membranes for detection of potato leafroll virus. Plant Disease, 69(9), 202–205. DOI 10.1094/PD-71-0795. [Google Scholar] [CrossRef]
44. Yeh, S. D., Gonsalves, D. (1984). Evaluation of induced mutants of papaya ringspot virus for control by cross protection. Phytopathology, 74(9), 1086–1091. DOI 10.1094/Phyto-74-1086. [Google Scholar] [CrossRef]
45. Yeh, S. D., Gonsalves, D., Wang, H. L. (1988). Control of papaya ringspot virus by cross protection. Plant Disease, 72(5), 375–380. DOI 10.1094/PD-72-0375. [Google Scholar] [CrossRef]
46. Yeh, S. D., Cheng, Y. H. (1989). Use of resistant Cucumis metuliferus for selection of nitrous-acid induced attenuated strains of papaya ringspot virus. Phytopathology, 79, 1257–1261. [Google Scholar]
47. Gonsalves, D., Garnsey, S. M. (1989). Cross-protection techniques for control of plant virus diseases in the tropics. Plant Disease, 73, 592–597. DOI 10.1094/PD-73-0592A. [Google Scholar] [CrossRef]
48. Sanford, J. C., Johnston, S. A. (1985). The concept of parasite-derived resistance-deriving resistance genes from the parasite’s own genome. Journal of Theoretical Biology, 113(2), 395–405. DOI 10.1016/S0022-5193(85)80234-4. [Google Scholar] [CrossRef]
49. Fermin, G. A., Castro, L. T., Tennant, P. F. (2010). CP-transgenic and non-transgenic approaches for the control of papaya ringspot: Current situation and challenges. Transgenic Plant Journal, 4(S1), 1–15. [Google Scholar]
50. Dillon, S., Ramage, C., Ashmore, S., Drew, R. A. (2006). Development of a codominant CAPS marker linked to PRSV-P resistance in highland papaya. Theoretical and Applied Genetics, 113(6), 1159–1169. DOI 10.1007/s00122-006-0375-2. [Google Scholar] [CrossRef]
51. Fitch, M. M., Manshardt, R. M., Gonsalves, D., Slightom, J. L., Sanford, J. C. (1992). Virus resistant papaya plants derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Nature Biotechnology, 10(11), 1466–1472. DOI 10.1038/nbt1192-1466. [Google Scholar] [CrossRef]
52. Cheng, Y. H., Yang, J. S., Yeh, S. D. (1996). Efficient transformation of papaya by coat protein gene of papaya ringspot virus mediated by Agrobacterium following liquid-phase wounding of embryogenic tissues with caborundum. Plant Cell Reports, 16(3), 127–132. DOI 10.1007/BF01890852. [Google Scholar] [CrossRef]
53. Azad, M., Kalam, A., Rabbani, M., Amin, L., Sidik, N. M. (2013). Development of transgenic papaya through agrobacterium-mediated transformation. International Journal of Genomics, 2013(5), 1–16. DOI 10.1155/2013/235487. [Google Scholar] [CrossRef]
54. Sajid, Q. A., Elçi, E. (2019). Investigation of virus diseases and molecular detection of little cherry virus 1 on cherry plants at Niğde Province. Turkish Journal of Agriculture-Food Science and Technology, 7(7), 1008–1013. DOI 10.24925/turjaf.v7i7.1008-1013.2490. [Google Scholar] [CrossRef]
55. Tripathi, S., Bau, H. J., Chen, L. F., Yeh, S. D. (2004). The ability of Papaya ringspot virus strains overcoming the transgenic resistance of papaya conferred by the coat protein gene is not correlated with higher degrees of sequence divergence from the transgene. European Journal of Plant Pathology, 110(9), 871–882. DOI 10.1007/s10658-004-0607-8. [Google Scholar] [CrossRef]
56. Eamens, A., Wang, M. B., Smith, N. A., Waterhouse, P. M. (2008). RNA silencing in plants: Yesterday, today, and tomorrow. Plant Physiology, 147(2), 456–468. DOI 10.1104/pp.108.117275. [Google Scholar] [CrossRef]
57. Ramesh, S. V., Mishra, A. K., Praveen, S. (2007). Hairpin RNA-mediated strategies for silencing of tomato leaf curl virus AC1 and AC4 genes for effective resistance in plants. Oligonucleotides, 17(2), 251–257. DOI 10.1089/oli.2006.0063. [Google Scholar] [CrossRef]
58. Yeh, S. D., Jan, F. J., Chiang, C. H., Doong, T. J., Chen, M. C. et al. (1992). Complete nucleotide sequence and genetic organization of papaya ringspot virus RNA. Journal of General Virology, 73(10), 2531–2541. DOI 10.1099/0022-1317-73-10-2531. [Google Scholar] [CrossRef]
59. Ruanjan, P., Kertbundit, S., Juříček, M. (2007). Post-transcriptional gene silencing is involved in resistance of transgenic papayas to papaya ringspot virus. Biologia Plantarum, 51(3), 517–520. DOI 10.1007/s10535-007-0110-0. [Google Scholar] [CrossRef]
60. Meins, F. (2000). RNA degradation and models for post-transcriptional gene silencing. Plant Molecular Biology, 43(2), 261–273. DOI 10.1023/A:1006443731515. [Google Scholar] [CrossRef]
61. Golemboski, D. B., Lomonossoff, G. P., Zaitlin, M. (1990). Plants transformed with a tobacco mosaic virus nonstructural gene sequence are resistant to the virus. Proceedings of the National Academy of Sciences, 87(16), 6311–6315. DOI 10.1073/pnas.87.16.6311. [Google Scholar] [CrossRef]
62. Nunome, T., Fukumoto, F., Terami, F., Hanada, K., Hirai, M. (2002). Development of breeding materials of transgenic tomato plants with a truncated replicase gene of cucumber mosaic virus for resistance to the virus. Breeding Science, 52(3), 219–223. DOI 10.1270/jsbbs.52.219. [Google Scholar] [CrossRef]
63. Wei, X., Lan, C., Lu, Z., Ye, C. (2007). Analysis on virus resistance and fruit quality for T4 generation of transgenic papaya. Frontiers of Biology in China, 2(3), 284–290. DOI 10.1007/s11515-007-0041-3. [Google Scholar] [CrossRef]
64. Hsieh, Y. T., Pan, T. M. (2006). Influence of planting papaya ringspot virus resistant transgenic papaya on soil microbial biodiversity. Journal of Agricultural and Food Chemistry, 54(1), 130–137. DOI 10.1021/jf051999i. [Google Scholar] [CrossRef]
65. Fermin, G., Inglessis, V., Garboza, C., Rangel, S., Dagert, M. et al. (2004). Engineered resistance against Papaya ringspot virus in Venezuelan transgenic papayas. Plant Disease, 88(5), 516–522. DOI 10.1094/PDIS.2004.88.5.516. [Google Scholar] [CrossRef]
66. Premchand, U., Mesta, R. K., Basavarajappa, M. P., Waseem, M. A., Mahesh, Y. S. et al. (2021). Management of Papaya ringspot virus (PRSV) using insecticides and bio rationals under field conditions. Biological Forum, 13(3a), 743–748. [Google Scholar]
67. Singh, S. K., Jha, P. K., Ray, P. K. (2010). Integrated management of Papaya Ring Spot Virus (PRSV) in agro ecological conditions of Bihar. Acta Horticulturea, 851, 487–494. DOI 10.17660/ActaHortic.2010.851.75. [Google Scholar] [CrossRef]
68. Sakuanrungsirikul, S., Sarindu, N., Prasartsee, V., Chaikiatiyos, S., Siriyan, R. et al. (2005). Update on the development of virus-resistant papaya: virus-resistant transgenic papaya for people in rural communities of Thailand. Food and Nutrition Bulletin, 26(4_Suppl3), S307–S311. DOI 10.1177/15648265050264S310. [Google Scholar] [CrossRef]
69. Roberts, M., Minott, D. A., Tennant, P. F., Jackson, J. C. (2008). Assessment of compositional changes during ripening of transgenic papaya modified for protection against papaya ringspot virus. Journal of the Science of Food and Agriculture, 88(11), 1911–1920. DOI 10.1002/jsfa.3295. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |