

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.021530
ARTICLE
Genome-Wide Characterization of the Cellulose Synthase Gene Superfamily in Tea Plants (Camellia sinensis)
1College of Tea Science, Guizhou University, Guiyang, 550025, China
2Institute of Agro-Bioengineering, College of Life Sciences, The Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Guizhou University, Guiyang, 550025, China
*Corresponding Authors: Litang Lu. Email: ltlv@gzu.edu.cn; Qi Zhao. Email: zhaoqizq@yeah.net
Received: 20 January 2022; Accepted: 25 February 2022
Abstract: The cellulose synthase gene superfamily, including Cellulose synthase A (CesA) and cellulose synthase-like (Csl) gene families, is responsible for the synthesis of cellulose and hemicellulose, respectively. The CesA/Csl genes are vital for abiotic stress resistance and shoot tenderness regulation of tea plants (Camellia sinensis). However, the CesA/Csl gene family has not been extensively studied in tea plants. Here, we identified 53 CsCesA/Csl genes in tea plants. These genes were grouped into five subfamilies (CsCesA, CsCslB, CsCslD, CsCslE, CsCslG) based on the phylogenetic relationships with Arabidopsis and rice. The analysis of chromosome distribution, gene structure, protein domain and motif revealed that most genes in CsCesA, CsCslD and CsCslE subfamilies were conserved, whereas CsCslB and CsCslG subfamily members are highly diverged. The transcriptome analysis showed that most CsCesA/Csl genes displayed tissue-specific expression pattern. In addition, members of CsCslB4, CsCesA1/3/6, CsCslB3/4, CsCslD3, CsCslE1 and CsCslG2/3 subfamilies were up-regulated under drought and cold stresses, indicating their potential roles in regulating stress tolerance in tea plants. Furthermore, the expression levels of CsCslG2_6 and CsCslD3_5 in different tissues and cultivars, respectively, were positively correlated with the cellulose content that is negatively related with shoot tenderness. Thus, these two genes were speculated to be involved in the regulation of shoot tenderness in tea plants. Our findings may help elucidate the evolutionary relationships and expression patterns of the CsCesA/Csl genes in tea plants, and provide more candidate genes responsible for stress tolerance and tenderness regulation in tea plants for future functional research.
Keywords: Tea plant (Camellia sinensis); cellulose synthase superfamily; phylogeny; stress resistance; shoot tenderness regulation
The tea plant (Camellia sinensis (L.) O. Kuntze) is one of the most important economic crops in China. It has been cultivated in China for thousands of years [1], and now is planted in more than 50 countries worldwide [2]. Its buds and young leaves are generally used for manufacturing tea, which is one of the most widely consumed non-alcoholic health beverages in the world [3]. As a perennial evergreen woody crop, the tea plant is often exposed to various abiotic stresses such as drought, low temperature and high salt during its growth and development, which are the main environmental factors that significantly affect the growth, survival and geographical distribution of tea plants, and seriously constrain the yield and quality of tea products [4–8]. Additionally, the quality and economic value of tea products are also affected by a number of other factors such as the tea plant cultivars, the types and tenderness of leaves, as well as the processing methods used to make tea products [9].
The cell wall acts as the first line of defense when plants encounter various stresses [10]. Cellulose and hemicellulose are the main components of plant cell walls, and they were synthesized by the cellulose synthase A (CesA) and cellulose synthase-like (Csl) enzymes, respectively. Celluloses are the main constituents of both primary and secondary cell walls, and provide the major structural rigidity of the cell-wall matrix [11,12]. Hemicelluloses are the main components of the primary cell wall contributing to strengthening the cell wall by interaction with cellulose and/or lignin [13]. A number of studies have reported the involvement of cellulose and hemicellulose in plant defensing against environmental stresses. For instance, cellulose deficient mutant plants lesioned in CesA6, POM2/CSI1, or KOR1 led to enhanced sensitivity to salt stress [14–18]. Besides, mutation of hemicellulose biosynthesis-related genes such as CslD4, CslD5 and CslF6 significantly affect the abiotic stress-tolerance of plants [14,19,20]. These studies pointed out an important role of the cellulose and hemicellulose synthesis machinery in abiotic stress responses.
In addition to various environmental stresses, the tenderness of tea shoots is another important factor affecting the quality and economic value of tea products. The content of cellulose was considered to be negatively correlated with the tenderness of tea shoots [21,22]. The tender shoots generally contain lower amounts of cellulose than the older ones, and tea products made from tender shoots are usually more expensive than those made from older leaves [9,21]. Besides, the tenderness of new shoots declined gradually with the growth of tea plants, whereas the cellulose content increased during the maturity process of tea shoots [21,22]. Furthermore, 3 CsCesA genes involved in the second cell wall cellulose synthesis have been identified in the ‘Huangjinya’ and ‘Yujinxiang’ cultivars of tea plants. Their expression levels were positively correlated with the cellulose content in tea leaves and stems, as well as the thickness of leaf tissues, indicating their involvement in modifying the tenderness of new shoots via regulating cellulose biosynthesis [23].
The CesA and Csl genes belong to cellulose synthase superfamily which is classified as glycoside transferase gene family (GT2). Multiple isoforms of CesA enzymes form the cellulose synthase complex (CSC) and catalyze the assembly of β-(1,4)-glucan chains forming protofibrils, which are then transformed into cellulose microfibrils [24–26]. The Csl enzymes share sequence similarity with the CesA family [26]. The Arabidopsis genome encodes 40 AtCesA/Csl genes, including 10 AtCesA and 30 AtCsl genes [27–29]. The AtCsl genes are grouped into 6 subfamilies, including AtCslA, AtCslB, AtCslC, AtCslD, AtCslE, AtCslG. In addition, CslF, CslH, CslJ and CslM subfamilies exist in some plant genomes [30–32]. At present, the cellulose synthase superfamily has been extensively characterized in a variety of plant species, such as Arabidopsis [32], rice [33], maize [34], barley [35], cotton [36], pear [37], tomato [38], poplar [39,40], pine [41] and moss [42]. However, even after the whole-genome sequence for tea plants is available [43], the CesA/Csl family genes have not been fully characterized in tea plants.
In this work, we identified 53 CsCesA/Csl family members in tea plants at the whole-genome level and analyzed their evolutionary relationships with Arabidopsis and rice. Besides, we studied the expression patterns of CsCesA/Csl genes in different tissues and under abiotic stresses by analyzing the transcriptome data. Furthermore, we examined the distinctive expression patterns of several selected CsCesA/Csl genes in sampled tissues and cultivars. We also investigated the correlation between the expression patterns of these genes and cellulose content alterations in different tissues and cultivars. These results may present new insights into the evolution, expression profiles and functional divergence of CsCesA/Csl genes, as well as provide more candidate genes for further study to improve the economic value of tea plants.
2.1 Plant Growth and Tissue Sampling
The tea plants were grown under standard field conditions in the tea plant resource garden of Guizhou University. Three biological replicates of the first three leaves and the tender stems of ‘Fuding’ cultivar, as well as the tender stems of ‘Longjing’, ‘Wuniuzao’, ‘Zhonghuang’, ‘Naibai’ and ‘Shuijingbai’ were collected to determine the cellulose content and expression patterns of CsCesA/Csl genes. All samples were stored at −80°C for further use.
2.2 Identification of CsCesA/Csl Genes in the Tea Genome and Physicochemical Property Analysis
The genome data and protein sequence of tea plant, Arabidopsis and rice were obtained from Tea Plant Information Archive (TPIA) (http://tpia.teaplant.org/), The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/) and Rice Genome Annotation Project (RGAP) (http://rice.plantbiology.msu.edu/index.shtml) database, respectively. To identify the tea CsCesA/Csl genes, we used the Arabidopsis AtCesA/Csl amino acid sequences to search against tea proteome with Basic Local Alignment Search Tool (BLAST-P) (E-value < 10−5) and deleted the redundant sequences. All candidate CsCesA/Csl proteins were submitted to the online bioinformatics tool, the National Center of Biotechnology Information Conserved Domain-Search Tool (NCBI CD-Search Tool) (https://www.ncbi.nlm.nih.gov/Structure/index.shtml) (E-value < 10−2) [44] to confirm CsCesA/Csl proteins that contained the core domains. The resulting candidate CsCesA/Csl proteins were further confirmed through BLAST-P against the Universal Protein Resource (UniProt) proteome databases (https://www.uniprot.org/) (E-value < 10−10). The length of chromosome, exon and amino acid of CsCesA/Csl proteins were analyzed based on the annotated information downloaded from tea plant genome database. The molecular weight and isoelectric point of tea CsCesA/Csl amino acid sequences were predicted by using ExPASy (http://web.expasy.org/compute_pi/).
2.3 Phylogenetic Analysis of CsCesA/Csl Genes
DNAMAN software (Lynnon Biosoft, Quebec, Canada) was used to perform multiple sequence alignment with default settings for CesA/Csl protein sequences of tea plant, Arabidopsis and rice. The phylogenetic tree was constructed by MEGA X software (Mega Limited, Auckland, New Zealand) through the Neighbor joining (NJ) method with a bootstrap option of n = 1000.
2.4 Chromosomal Distribution and Gene Structure Analysis
The information of chromosome localization of CsCesA/Csl genes was downloaded from TPIA (http://tpia.teaplant.org/) and visualized by using TBtools (https://github.com/CJ-Chen/TBtools/releases), including the localization and length in corresponding chromosomes. The exon-intron structures about the quantity and distribution of the CsCesA/Csl genes based on the genome annotation were visualized by TBtools software.
2.5 Protein Domain and Conserved Motif Analysis
The conserved domains of CsCesA/Csl proteins were obtained by NCBI CD-Search Tool (https://www.ncbi.nlm.nih.gov/Structure/index.shtml) and visualized by TBtools. The conserved protein motifs of CsCesA/Csl proteins were analyzed through the Multiple Em for Motif Elicitation (MEME) program (http://meme-suite.org/) and visualized by TBtools.
2.6 Expression Pattern Analysis
The transcriptome data (TPM value) of CsCesA/Csl genes in different tissues (apical bud, young leaf, mature leaf, old leaf, stem, root, flower and fruit) of ‘Shuchazao’ cultivar, and transcriptome data in ‘Longjing43’ and ‘Tieguanyin’ in response to drought and cold stresses, respectively, were downloaded from TPIA (http://tpia.teaplant.org). The Log2-based-fold changes were used to create heatmaps via TBtools software.
2.7 RNA Extraction and qRT-PCR Analysis
Primers for the selected CsCesA/Csl genes were designed using the online program Integrated DNA Technologies (IDT) (https://sg.idtdna.com/pages) (Supplementary Table 1), the primer sequences were synthesized by Beijing Qingke Biotechnology Limited Company (China). Total RNA was extracted from the first, second and third leaves and tender stem segments of tea plants by cetyltrimethylammonium bromide (CTAB) method. The concentration of RNA was detected by a NanoPhotometer N50 Touch (Implen, Munich, Germany), and the RNA was then reversed transcribed into the first-strand cDNA by qRT-PCR kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The cDNA was subsequently used as a template for qRT-PCR analysis. SYBR Green qPCR Mix (Genenode, Wuhan, China) reagent was used for qRT-PCR amplification. Each reaction system contains 10 μL SYBR Green qPCR Mix, 0.8 μL forward and reverse primers, 1.5 μL cDNA template, 7.7 μL ddH2O. Reaction procedure was as follows: 95°C for 3 min; 40 cycles of 95°C for 10 s and 60°C for 20 s; 72°C for 30 s, CsGAPDH was used as the reference gene. The 2−ΔΔCT algorithm was used to calculate the relative expression of genes [45].
2.8 Determination of Cellulose Content
The tea samples were ground into fine powder and filtered through a 30-mesh sieve. The cell wall structural components were extracted with 70% ethyl alcohol and acetone reagent. The crystalline cellulose was hydrolyzed completely to glucose under highly acidic conditions. The glucose content in the supernatant was determined by anthrone-sulfuric colorimetry [46]. The crystallized cellulose was calculated according to the standard curve established by absorbance on the same 96-well polystyrene microtitration plate by the 1510 Multiskan GO spectrophotometer (Thermo Fisher Scientific, Vanta, Finland) under 620-nm wavelength [46]. All data were analyzed by Statistical Product and Service Solutions (SPSS) software (SPSS, Chicago, USA).
All results were given as means ± standard deviation of at least three biological replicates. The data were subjected to one-way analysis of variance using SPSS software. A p value smaller than 0.05 was considered to be statistically significant.
3.1 Identification and Phylogenetic Analysis of the CsCesA/Csl Genes in the Tea Genome
To identify the tea plant CsCesA/Csl genes, Arabidopsis AtCesA/Csl protein sequences were used to search against tea plant proteome with BLAST-P program (E-value < 10−5). To confirm CsCesA/Csl proteins that contained the core domains, all candidate CsCesA/Csl proteins were submitted to the online bioinformatics tool, NCBI CD-Search Tool (https://www.ncbi.nlm.nih.gov/Structure/index.shtml) (E-value < 10−2) [44]. The authenticity of the candidate CsCesA/Csl proteins was further confirmed through BLAST-P against the Universal Protein Resource (UniProt) proteome databases (https://www.uniprot.org/) (E-value < 10−10). A total of 53 candidate CsCesA/Csl genes were identified from tea plants, and they were named after their corresponding orthologs in Arabidopsis and classified into five subfamilies, including CsCesA, CsCslB, CsCslD, CsCslE, CsCslG based on the phylogenetic relationships with Arabidopsis and rice (Fig. 1, Supplementary Table 2).

Figure 1: Phylogenetic relationships and subfamily designations of CesA/Csl proteins from the tea plant (Camellia sinensis), Arabidopsis thaliana and rice (Oryza sativa). The phylogenetic tree was constructed with MEGA X using the neighbor-joining (NJ) method and 1000 bootstrap replicates. The superfamily was divided into 9 subfamilies, including CesA, CslA, CslB, CslC, CslD, CslE, CslF, CslH and CslG. Prefix ‘At’, ‘Os’ and ‘Cs’ indicate CesA/Csl proteins from Arabidopsis thaliana, Oryza sativa and Camellia sinensis
To investigate the functional associations and evolutionary relationships of CsCesA/Csl genes, a multi-species phylogenetic tree of CesA/Csl genes from Arabidopsis, rice and tea plant were constructed. This tree comprised 9 subfamilies, including CesA, CslA, CslB, CslC, CslD, CslE, CslF, CslG and CslH (Fig. 1, Supplementary Table 2). Among them, the CesA subfamily was the largest subfamily, containing 15 tea CsCesA genes, 10 Arabidopsis genes and 11 rice genes, accounting for 26.1% of the total CesA/Csl genes. The second largest subfamily was the CslD subfamily, with 9, 6 and 5 CslDs from tea plant, Arabidopsis and rice, respectively. Since rice lacks any CslG gene [47], the third largest family, CslG subfamily, involved 16 tea genes and 3 Arabidopsis genes. The CslH subfamily was the smallest one with only 3 rice OsCslH genes. No tea plant genes were found in the subfamilies of CslA, CslC, CslF and CslH, and there were 9 AtCslAs and 5 AtCslCs from Arabidopsis, and 9 OsCslAs and 6 OsCslCs from rice, respectively. The CslF and CslH subfamilies only contained 8 and 3 genes from rice, respectively. These results suggested evolutionary conservation and closer homology existed in closely related CesA/Csl gene subfamilies.
As shown in Table 1, the genomic DNA size of these genes varied from 448 bp (CsCslG2_4) to 27,703 bp (CsCslB3_1), and the average length was 9,512 bp. Among them, 18.9% CsCesA/Csl genes were <4000 bp, 24.5% of them were 4000–7000 bp, and 56.6% were >7000 bp. The encoded protein sequences consisted of 79 (CsCslG2_4) to 1,174 (CsCslD5_2) amino acids, with an average of 779 amino acids. Their corresponding molecular weights varied from 9 kDa (CsCslG2_4) to 132.1 kDa (CsCslD5_2), with the theoretical isoelectric points ranging from 6 (CsCesA1_2 and CsCesA1_3) to 9.51 (CsCslB4_1). The diverse physical and molecular properties of the 53 CsCesA/Csl genes might be resulted from the selection pressure during gene family evolution, as well as the genomic assembly and transcript annotation quality, indicating their different roles in various microenvironments.
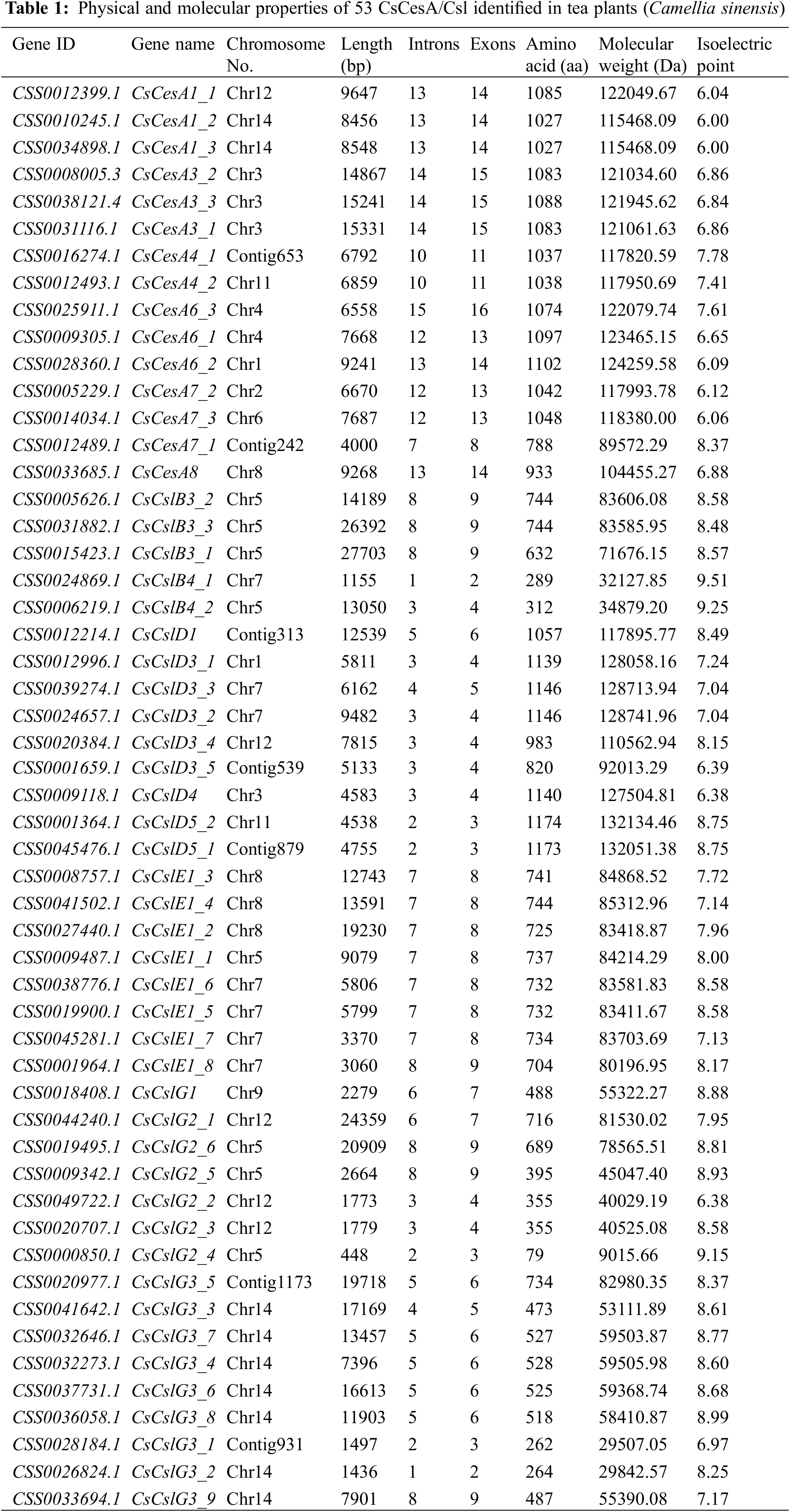
3.2 Genomic Distribution and Structural Feature of the CsCesA/Csl Genes
To clarify the genomic distribution of the CsCesA/Csl genes, a map of chromosomal locations was constructed based on the information annotated in the tea plant genome. A total of 46 CsCesA/Csl genes were unevenly distributed on 12 of 15 chromosomes, with the exception being Cs_chr10, Cs_chr13 and Cs_chr15 (Table 1, Fig. 2). Chromosome 14 contained the most CsCesA/Csl gene members, including 2 CsCesA1s and 7 CsCslG3s, and each subfamily was presented in the form of gene clusters. Besides, Chromosome 5 included 8 genes, followed by chromosome 7 containing 7 genes. In addition, only one gene was found on chromosome 2, 6 and 9, respectively. However, 7 out of 53 CsCesA/Csl genes were located on scaffolds, and were not presented in the figure (Table 1).

Figure 2: Distribution of CsCesA/Csl genes on the tea plant chromosomes. A total of 46 CsCes/Csl genes were localized to tea chromosomal regions, while the other genes were detected on scaffolds and were not presented in the figure
To provide more valuable information involved in evolutionary pattern and structural diversity of CsCesA/Csl genes, their exon-intron structures were analyzed. As shown in Fig. 3, most of the CsCesA/Csl genes in each of the CsCesA, CsCslE and CsCslD subfamilies showed similar exon-intron structure features. For instance, CsCesA genes had more than 10 exons (ranging from 11 to 16 exons), except for CsCesA7_1 with 8 exons. In contrast, all of the remaining CsCsl genes contained fewer than 10 exons. Specifically, CsCslE subfamily had 8 to 9 exons, and CsCslDs included 3 to 6 exons. However, the numbers of exons in both CsCslB and CsCslG subfamilies varied from 2 to 9, indicating a higher degree of divergence might exist in these two subfamilies than others.

Figure 3: Exon-intron structure of tea plant CsCesA/Csl genes. Green boxes indicate untranslated 5’- and 3-regions, yellow boxes represent exons, and black lines indicate introns
3.3 Protein Conserved Domains and Motifs Analysis of the CsCesA/Csl Family
All of the 53 identified CsCesA/Csl proteins contained one conserved cellulose synthase domain, most of which were located at the C-terminus (Fig. 4). In addition, several of the CsCesA/Csl proteins contained other domains. For instance, 13 CsCesA proteins included the Zinc finger-UDP domain located near the N-terminal, 5 CsCslD3 proteins and one CsCslD4 contained the Zinc finger-RING_4 domain, which existed upstream of the cellulose synthase domain, and 2 CsCslD5s and one CsCesA8 had the RING-Ubox superfamily domain, with the domain in CsCesA8 located at the N-terminal (Fig. 4).

Figure 4: Conserved domains of tea plant CsCesA/Csl proteins. Green, yellow, pink and blue boxes indicate cellulose synthase, Zinc finger-UDP, RING_Ubox superfamily and Zinc finger-RING_4 domains, respectively
To further explore the diversification of CsCesA/Csl gene family in tea plants, the putative conserved motifs of CsCesA/Csl proteins were predicted by the MEME program. A total of 15 distinct motifs were identified in the CsCesA/Csl gene family (Fig. 5). The number of different motifs was similar in each of the CsCesA, CsCslD, CsCslE subfamilies, but varied in the subfamilies of CsCslB and CsCslG. In the CsCesA subfamily, 14 out of the 15 (93.3%) members contained all of the 15 motifs, except for CsCesA7_1 lacking motif 10. Similarly, the 15 different motifs were also predicted in 6 out of the 9 (66.7%) CsCslD genes. For the CsCslE genes, 8 out of the 9 (87.5%) members had 12 motifs. In contrast, the number of distinct motifs varied from 2 to 13 and 2 to 12 in the CsCslB and CsCslG subfamilies, respectively. Motif 7 and 10 were not predicted in the CsCslB, CsCslE or CsCslG subfamilies, indicating these motifs might be key players during the functional differentiation of CsCesA/Csl genes during evolution.

Figure 5: Motif analysis of the tea plant CsCesA/Csl proteins. Motif 1 to motif 15 displayed by different colors represent different conserved motifs. The order of the motifs corresponds to their position within individual protein sequence
3.4 Expression Patterns of CsCesA/Csl Genes in Different Tissues
To further explore the particular function of the CsCesA/Csl genes in tea plants, we conducted a transcriptome analysis of the CsCesA/Csl genes in different tissues of the tea cultivar ‘Shuchazao’ of tea plant (apical bud, young leaf, mature leaf, old leaf, stem, root, flower and fruit) [43,48]. A heat map was used to display the CsCesA/Csls expression patterns in eight tissues (Fig. 6). A total of 17 genes (3 CsCesA1s, 3 CsCesA3s, 2 CsCesA4s, 3 CsCesA6s, a CsCslD3, and 5 CsCslG3s) were expressed in all sampled tissues at high levels, indicating that these genes might play crucial roles in the whole process of tea plant development. Besides, the tissue-specific expression patterns were identified in a number of tea CsCesA/Csl genes. For instance, 4 CsCslG3s (CsCslG3_1, CsCslG3_3, CsCslG3_5 and CsCslG3_8) and 2 CsCslE1s (CsCslE1_5 and CsCslE1_6) displayed moderate-to-high expression levels in 6 out of the 8 tissues, including mature leaves, old leaves, stems, roots, flowers and fruits, but expressed at low levels in the apical buds and young leaves, suggesting their potential tissue-specific functions in the corresponding developmental processes. In addition, 2 CsCesA7s (CsCesA7_1 and CsCesA7_2) and 2 CsCslG2s (CsCslG2_5 and CsCslG2_6) might be important for the development of the apical buds, young leaves, stems and roots because of their higher expression levels in these tissues than in others. Interestingly, it was found that several CsCesA/Csl gene family members displayed opposite expression patterns. For example, the CsCslD1 and CsCslD4 genes showed high expression levels in flowers and fruits and low expression in other tissues. In contrast, the expression levels of the CsCslE1_1 and 4 CsCslBs (CsCslB3_1, CsCslB3_2, CsCslB3_3 and CsCslB4_2) were low in flowers and fruits, but high in other sampled tissues. The contrary expression pattern was also found between 2 CsCslE1s (CsCslE1_7 and CsCslE1_8) and CsCslD3_1. The former genes expressed at higher levels in mature and old leaves than in other tissues, whereas the latter showed lower transcript abundances in mature and old leaves than in other sampled tissues. Furthermore, the expression levels of 2 CsCslE1s (CsCslE1_3 and CsCslE1_4) were lower in the root than in the other tissues, while CsCslE1_2 and 3 CsCslG2 genes (CsCslG2_1, CsCslG2_2 and CsCslG3) showed high root-specific expression. These results suggested that these genes, especially CsCslD and CsCslE1 subfamily members, might play antagonistic roles during tea plant development.
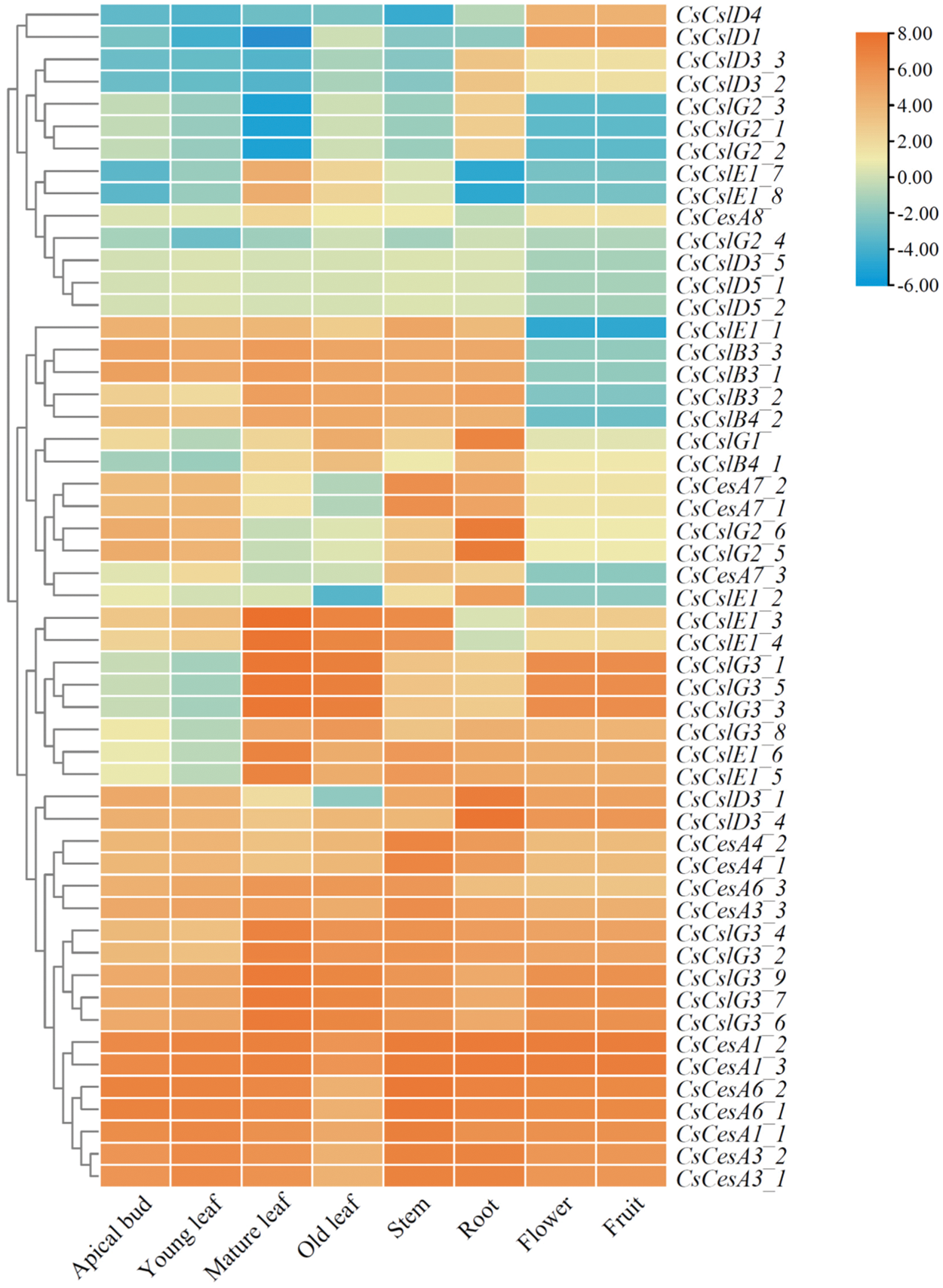
Figure 6: Expression profiles of tea plant CsCesA/Csl genes in different tissues. The heatmap was generated by TBtools software according to the RNA-seq database downloaded from TPIA (http://tpia.teaplant.org). Log2-based-fold changes were used to create a heatmap. The gene expression level is displayed in different colors on the map, as shown in the bar at the upper right corner. Three biological replicates of each tissue were used [3,43,48]
3.5 Expression Profiles of CsCesA/Csl Genes under Abiotic Stresses
To further describe the role of CsCesA/Csl genes, we performed a comparative transcriptome analysis of the CsCesA/Csl genes based on the downloaded abiotic (drought and cold) stress-responsive transcriptome data [3,48–50]. All of the 53 CsCesA/Csl genes were clustered into two main groups based on the gene expression pattern analysis of drought-responsive dataset (Fig. 7A). A total of 23 CsCesA/Csl genes, accounting for 56.6% of the whole CsCesA/Csl gene family, were clustered into Group I. Most of the genes in this group expressed at lower levels in the control and drought-stressed tea leaves than Group II members. In Group I, the expression level of CsCslB4_1 increased first but then decreased to low level, peaking at 24 h. The transcript abundances of CsCesA4_1 and 3 CsCslGs (CsCslG2_1, CsCslG2_4 and CsCslG3_5) were decreased in response to drought treatment for 24, 48 and 72 h compared with control plants. Besides, 2 CsCesAs (CsCesA1_2 and CsCesA7_1) showed down-regulated expression patterns under drought stress for 48 and 72 h. Besides, the expression levels of CsCesA7_2, CsCslB3_1 and 2 CsCslE1s (CsCslE1_2 and CsCslE1_6) were down-regulated in response to drought stress for 72 h. Other genes were not significantly affected by the drought stress. Among the CsCesA/Csl genes clustered in Group II, the expression level of CsCslB4_2 was gradually increased with the duration of drought stress (from 24 to 72 h), which was very different from other differentially expressed CsCesA/Csl genes in the Group II (Fig. 7A). The up-regulated expression of the 2 CsCslB4s in each subgroup suggested their potential roles in increasing tea plant defense against drought stress. Besides, the transcript abundances of 7 genes, including 3 CsCslG3s (CsCslG3_1, CsCslG3_4 and CsCslG3_7), 2 CsCesAs (CsCesA3_1 and CsCesA6_3), CsCslD5_2 and CsCslE1_4, were gradually decreased along with the duration of drought stress (from 24 to 72 h). Furthermore, drought stress for 72 h resulted in the reduced expression of 3 CsCslGs (CsCslG2_5, CsCslG3_2 and CsCslG3_6), CsCesA6_2 and CsCslD3_3.

Figure 7: Expression profiles of tea plant CsCesA/Csl genes under drought (A) and cold (B) stresses. The heatmap was generated by TBtools software according to the RNA-seq database downloaded from TPIA (http://tpia.teaplant.org). Log2-based-fold changes were used to create a heatmap. The gene expression level is displayed in different colors on the map, as shown in the bar at the upper right corner. Drought treatments was included 25% polyethylene glycol (PEG) treatment for 0, 24, 48 and 72 h [50]. Cold treatments included non-acclimated (CK), fully acclimated (CA1) and de-acclimated stages (CA3) [49]. Three biological replicates were used in each experiment [49,50]
The expression patterns of the 53 CsCesA/Csl genes under cold stress were also analyzed based on the cold stress-responsive transcriptome data during three stages of tea plant cold acclimation, including non-acclimated (CK), fully acclimated (CA1) and de-acclimated (CA3) stages (Fig. 7B) [3,48,49]. All of the CsCesA/Csl genes were clustered into two main groups with most of the Group I members showing relatively higher expression levels than those in Group II. Nine genes displayed the highest expression levels at the CA1 stage compared with other two stages, including 5 CsCesAs (CsCesA1_2, CsCesA1_3, CsCesA3_3, CsCesA6_1 and CsCesA6_2), 3 CsCslGs (CsCslG3_6, CsCslG3_7 and CsCslG3_9) and CsCslD3_4. The expression levels of 2 CsCslE1s (CsCslE1_5 and CsCslE1_6) peaked at the CA3 stage. Besides, 7 genes showed gradual up-regulated expression patterns across CA1 to CA3 stages compared with the control plants, including 3 CsCslBs (CsCslB3_1, CsCslB3_3 and CsCslB4_1), 2 CsCslGs (CsCslG2_5 and CsCslG2_6), CsCslD3_1 and CsCslE1_2. In contrast, the transcript level of CsCslE1_1 was gradually down-regulated at CA1 and CA3 stages compared with the CK stage. These results indicated that genes in tea plant CsCesA/Csl subfamilies might have distinct functions in response to different abiotic environments.
3.6 Analysis of Cellulose Content and Expression Patterns of the Selected CsCesA/Csl Genes in Different Tissues and Tea Plant Cultivars
Since the cellulose content is one of the major factors affecting the tenderness of tea plants, we measured the cellulose content in four tissues, as well as stems in five different cultivars of tea plants (Fig. 8). The cellulose content was gradually increased with the leaf maturity degree (Fig. 8A), and the third leaves from tea plants contained the highest amount of cellulose among the four tested tissues (Fig. 8A). The absolute amount of cellulose in stems was higher than those in the first and second leaves, and lower than that in the third leaves (Fig. 8A). The comparison of the cellulose content in stems of the five cultivars revealed that the ‘Zhonghuang’ had a higher cellulose content than other cultivars (Fig. 8B), indicating a cultivar-specific accumulation of cellulose might exist in certain tea plant cultivars, such as ‘Zhonghuang’.
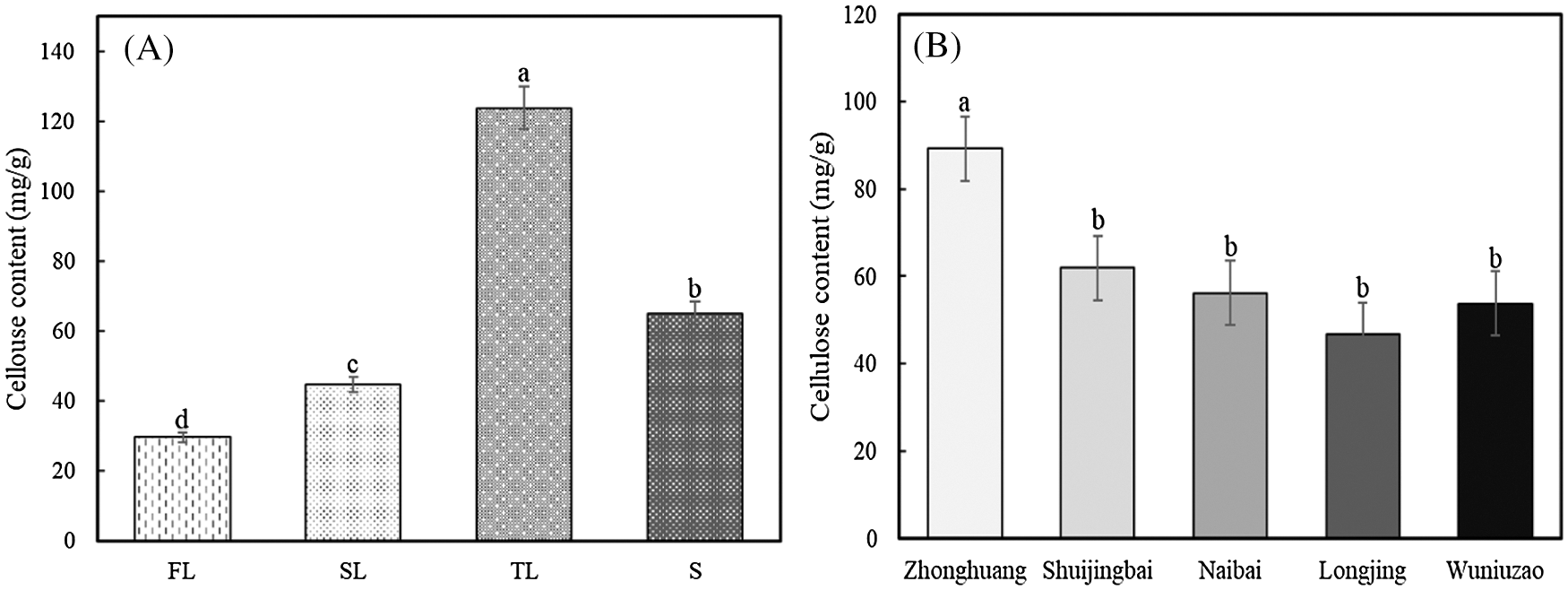
Figure 8: Cellulose content in different tissues of the ‘Fuding’ cultivar (A) and in stems of different tea cultivars (B). Error bars show the standard error among three replicates. Different letters above the columns indicate the significant differences calculated by one-way ANOVA method. L1: the first leaves; L2: the second leaves; L3: the third leaves; S: stems, respectively
To further validate the roles of CsCesA/Csl genes in the development of tea plants and the relationship between cellulose contents and expression patterns of the CsCesA/Csl genes, 6 CsCesA/Csl family members, including CsCesA6_2, CsCslD3_3, CsCslD3_5, CsCslE1_4, CsCslG2_5 and CsCslG2_6, were selected and their transcriptional activity in various tissue types and different tea plant cultivars were analyzed using qRT-PCR with gene-specific primers (Fig. 9). The expression level of the CsCesA6_2 in the third leaves was significantly lower than in other tissues, which is correlated well with the transcriptomic findings that the old leaves expressed the lowest level of the CsCesA6_2 compared with tested leaves and stems (Fig. 9). However, the expression patterns of other 5 genes were different from those in the transcriptomic results, suggesting these genes might adopt cultivar-specific expression strategies in different tea plant cultivars (Fig. 9). Besides, the CsCslG2_6 showed a gradual elevation of cellulose amount in the first leaves, the second leaves, and the stems, which is in accordance with the increased trend of cellulose content in the corresponding tissues (Fig. 9). The expression levels of the CsCesA6_2, CsCslD3_5, CsCslG2_5 and CsCslG2_6 genes were significantly increased in the second leaves than those in the first leaves, which is also well correlated with the change patterns of cellulose content in these two tissues (Fig. 9). Furthermore, the transcript level of the CsCslD3_5 in the stems of ‘Zhonghuang’ cultivar was the highest among all five cultivars. This result is in accordance with the highest cellulose contents in the same cultivar (Fig. 10). However, the remaining 5 CsCesA/Csl genes showed varied expression patterns in different cultivars (Fig. 10).

Figure 9: Expression profiles of selected CsCes/Csl genes in different tea tissues determined by qRT-PCR (A to F). Values represent the average ± standard error of three biological replicates with three technical replicates in each tissue. Different letters above the columns indicate the significant differences calculated by one-way ANOVA method. L1, L2, L3 and S represent the first leaves, the second leaves, the third leaves and stems, respectively

Figure 10: Expression profiles of selected CsCes/Csl genes in stems of different tea cultivars determined by qRT-PCR (A to F). Values represent the average ± standard error of three biological replicates with three technical replicates in each tissue. Different letters above the columns indicate the significant differences calculated by one-way ANOVA method. L1, L2, L3 and S represent the first leaves, the second leaves, the third leaves and stems, respectively
The cell wall is crucial for plants to adapt and survive through modulating plant growth and development, as well as by acting as the front line of plant immunity [51]. Cellulose and additional polysaccharides are the main components of plant cell walls. CesA and Csl enzymes are responsible for synthesizing cellulose and most hemicellulose polysaccharides, respectively, and they constitute the cellulose synthase superfamily that is classified as GT2 family [52]. Here, we conducted a genome-wide analysis and identified a total of 53 CsCesA/Csl genes with cellulose synthase domains in the tea plant genome database. Our results showed that the tea genome encodes 15 CsCesA genes, and they were classified into two distinct major groups based on the type of cell wall production and adaptive involvement in cellulose synthesis, with CsCesA1s, CsCesA3s and CsCesA6s in a group while CsCesA4s, CsCesA7s, and a CsCesA8 in another group (Fig. 1), which is similar with the classification of Arabidopsis AtCesA/Csl genes. In Arabidopsis, AtCesA1, AtCesA3 and AtCesA6-like proteins (AtCesA2, AtCesA5, AtCesA6 and AtCesA9) participate in the primary wall cellulose synthesis, whereas AtCesA4, AtCesA7 and AtCesA8 are components of the secondary wall cellulose synthase complex [27–29,53]. Thus, the molecular function of the CsCesAs in tea plants may be similar with their corresponding orthologs in Arabidopsis.
In addition, we found that the tea plant lacks CslA/C/F/H/J/M subfamilies based on the phylogenetic comparison with Arabidopsis and rice (Fig. 1). Similarly, it was reported that pear also lacks CslA genes [37] and there is only one SlCsl gene identified in the subgroup of SlCslA and SlCslC genes in tomato [38]. These results indicate that CsCslA and CsCslC subfamilies may have been lost during the evolution of the tea genome. Furthermore, the composition of hemicellulose between monocots and dicots is highly diverged, which partially resulted from the fact that some specific Csl classes exist only in monocots or dicots [54]. Specifically, the CslB and CslG classes were found exclusively in dicots. In contrast, the CslF and CslH classes were considered to be unique to monocots, thus they were not identified in both Arabidopsis and tea plants. The CslJ/M members were not identified in the genomes of Arabidopsis, rice and tea, but presented in a few monocot and eudicot species [31,33].
The gene structures and protein motif features are critical for elucidating the evolution, differentiation, or conservation of the function of gene family members. In the tea plant, most CsCesA, CsCslE and CsCslD genes have nearly the same gene structure and protein motifs in each subfamily, suggesting that these genes are conserved during evolution (Figs. 3 and 5). In contrast, CsCslB and CsCslG genes are highly diverged with different exon-intron structures and protein motifs existing in most family members (Figs. 3 and 5), probably due to chromosome fusion and/or rearrangement [55]. However, the gene structure of CslD was reported to be varied in other plant species, such as pear [37], pineapple [56] and tomato [38]. These results indicated that evolutionary mechanism of CsCslD subfamily genes might be special in tea plants and needs to be further elucidated.
Our analysis based on the transcriptomic data also provided valuable information on the potential function of CsCesA/Csl genes in tea plants. A number of CsCesA/Csl genes, including members of CsCesA1/3/4/6, CsCslD3 and CsCslG3s, were found to be expressed highly in all sampled tissues, indicating these genes were necessary for the whole processes of tea plant growth (Fig. 6). Besides, members from CsCesA7, CsCslE1, CsCslG2 and CsCslG3 subfamilies expressed highly in one or more specific tissues (Fig. 6). Additionally, the expression pattern of CsCesA6_2 verified by qRT-PCR displayed lower expression levels in the older leaves than in other tissues of the ‘Fuding’ cultivar (Fig. 9), which was in accordance with the changes of CsCesA6_2 in the ‘Shuchazao’ cultivar revealed by the transcriptomic data (Fig. 6). These tissue-specific expressed genes were considered to be closely related to the biosynthesis of cellulose and hemicellulose polysaccharides during the corresponding tissue development. Interestingly, CsCslD and CsCslE1 subfamily members displayed contrary tissue-specific expression patterns (Fig. 6), suggesting the potential antagonistic relationship between these two subfamilies. In Arabidopsis, the function of AtCesA/Csl genes has been extensively studied via gene mutation. For instance, null mutations in AtCesAs are usually lethal during embryogenesis or early seedling development [26]. Point mutations in AtCesAs often lead to abnormal phenotypes, such as a radial swelling phenotype caused by rsw1-1 mutation and an abnormal swollen cell phenotype resulted from rsw1-20 mutation [57,58], revealing the important roles of CesA/Csl genes in plant growth and development. Although the functional exploration of CsCesA/Csl genes in tea plants is still lacking, the analysis of their tissue expression patterns would provide more candidate genes for elucidating the specific and biologically important roles of CsCesA/Csl family members during tea plant growth and development.
The plant cell wall acts as the first line of defensing against a number of abiotic stresses, such as drought, low temperature and salinity [10]. Therefore, the cell wall-related genes are supposed to play important roles in regulating cell wall function under abiotic stresses. In tea plants, two members of CsCslB4 subfamily involved in cell wall hemicellulose biosynthesis displayed increased expression patterns under drought stress (Fig. 7A), and a number of genes were found to be up-regulated in response to different cold treatments, including members of CsCesA1/3/6, CsCslB3/4, CsCslD3, CsCslE1 and CsCslG2/3 classes (Fig. 7B), suggesting that these genes would be required for tea plants to survive under abiotic stresses. In other plant species, a series of studies have been conducted in order to elucidate the specific function of CesA/Csl genes when plants confront environmental stresses due to their involvement in cell wall biosynthesis. However, most of the research only focused on a few CesA/Csl subfamilies. For instance, OsCesA10 was found to exhibit a close correlation with rice drought tolerance [59]. Overexpression of OsCslD4 can enhance rice salt tolerance by elevating the ABA synthesis gene expression and increasing ABA content [20]. Additionally, AtCslD5 was confirmed to be required for Arabidopsis osmotic stress tolerance [60]. Our analysis of the expression pattern of tea CsCesA/Csl genes under drought and cold stresses provided more candidate stress-resistant genes, which would be valuable for a better understanding of tea plant stress-tolerant mechanism and the subsequent breeding of stress-tolerant tea plants.
The content of cellulose, one of the most important factors affecting the tenderness of new shoots, was considered to be positively correlated with the maturity of tea plant tissues [21–23]. This has been confirmed in our data that the cellulose content was gradually increased with the maturity of the leaves in the ‘Fuding’ cultivar (Fig. 8A), which was similar in both ‘Huangjinya’ and ‘Yujinxiang’ cultivars reported in previous studies [23]. However, the cellulose content in stems was not the highest among the four tested samples (Fig. 8A), probably due to that the stems of the first three internodes were mixed together as an integral stem sample, within which the first two internodes were tender than the third one [21]. Additionally, among the selected CsCesA/Csl genes, we found that the expression level of CsCslG2_6 in the ‘Fuding’ cultivar and CsCslD3_5 in the ‘Zhonghuang’ cultivar were positively correlated with the change pattern of the cellulose contents among different tissues and different cultivars, respectively (Figs. 9 and 10). A previous study has found that the expression levels of 3 CsCesAs involved in the secondary cell wall cellulose synthesis were positively related to the cellulose content alteration, showing their potential regulatory roles in modulating shoot tenderness [23]. Furthermore, several AtCsl genes were also proved to be directly involved in the biosynthesis of cellulose in Arabidopsis [61]. For instance, AtCslD1 and AtCslD4 mutation caused significantly reduced cellulose deposition in the cell wall of pollen tube [61]. Thus, we speculate that CsCslG2_6 and CsCslD3_5 in tea plants might also be involved in the cellulose synthesis to some extent, and thereby affect the tenderness of the new shoots. Taken together, these two genes would act as valuable targets for the further study on deciphering the molecular mechanism of the regulatory roles of CsCesA/Csl superfamily in modulating the tenderness of tea shoots.
The CsCesA/Csl gene family involved in the biosynthesis of cellulose and hemicellulose is critical for tea plant stress tolerance and shoot tenderness regulation. Although the whole-genome sequence for tea plants is available, information about the CsCesA/Csl gene family is still lacking. In this study, a total of 53 CsCesA/Csl genes were identified and classified into five subfamilies. Their phylogenetic relationships with CesA/Csl genes in Arabidopsis and rice, chromosome location, gene structure, expression patterns in different tissues and under drought and cold stresses were analyzed, providing a number of candidate genes probably involved in the regulation of tea plant growth, development and stress resistance. The correlation between selected CsCesA/Csl genes and shoot cellulose contents in different tissues and cultivars were also studied and revealed that two genes, CsCslG2_6 and CsCslD3_5, might be involved in regulating shoot tenderness by affecting the changes of cellulose content. Our results may be useful for further elucidating the function of candidate CsCesA/Csl genes in order to improve the stress-tolerance of tea plant, as well as the quality of tea products.
Authors’ Contributions: Conceive the study and revise the manuscript: Litang Lu and Qi Zhao. Perform the bioinformatic analysis, qRT-PCR validation mainly and draft the manuscript: Qianqian Li. Collect the materials and participate in the qRT-PCR validation: Xinzhuan Yao. Participate in the bioinformatic analysis: Baohui Zhang. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This work was supported by the Technology Creation Center of Guizhou Tea Industrialization (Qiankezhongyindi [2017]4005), Training Project for Guizhou Excellent Young Scientific and Technological Talents (Qiankehe Platform Talent [2019]5651), and Guizhou Science and Technology Planning Project (Qiankehe Support [2021] General 111) to Litang Lu, and Research Funds for Introduced Talents of Guizhou University to Qi Zhao.
Conflicts of Interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
1. Chen, Y., Yu, M., Xu, J., Chen, X., Shi, J. (2009). Differentiation of eight tea (Camellia sinensis) cultivars in China by elemental fingerprint of their leaves. Journal of the Science of Food and Agriculture, 89(14), 2350–2355. DOI 10.1002/jsfa.3716. [Google Scholar] [CrossRef]
2. Chen, L., Apostolides, Z., Chen, Z. M. (2012). Global tea breeding: Achievements, challenges and perspectives. Advanced Topics in Science & Technology in China, 378(378), 588. DOI 10.1007/978-3-642-31878-8. [Google Scholar] [CrossRef]
3. Xia, E. H., Tong, W., Wu, Q., Wei, S., Zhao, J. et al. (2020). Tea plant genomics: Achievements, challenges and perspectives. Horticulture Research, 7(1), 1–19. DOI 10.1038/s41438-019-0225-4. [Google Scholar] [CrossRef]
4. Li, J., Yang, Y., Sun, K., Chen, Y., Chen, X. et al. (2019). Exogenous melatonin enhances cold, salt and drought stress tolerance by improving antioxidant defense in tea plant (Camellia sinensis (L.) O. Kuntze). Molecules, 24(9), 1826. DOI 10.3390/molecules24091826. [Google Scholar] [CrossRef]
5. Liu, S., Yao, M., Ma, C., Jin, J., Ma, J. Q. et al. (2015). Physiological changes and differential gene expression of tea plant under dehydration and rehydration conditions. Scientia Horticulturae, 184, 129–141. DOI 10.1016/j.scienta.2014.12.036. [Google Scholar] [CrossRef]
6. Zhang, Y., Gao, W., Cui, C., Zhang, Z., He, L. et al. (2020). Development of a method to evaluate the tenderness of fresh tea leaves based on rapid, in-situ Raman spectroscopy scanning for carotenoids. Food Chemistry, 308, 125648. DOI 10.1016/j.foodchem.2019.125648. [Google Scholar] [CrossRef]
7. Zheng, C., Wang, Y., Ding, Z., Zhao, L. (2016). Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in camellia sinensis. Frontiers in Plant Science, 7, 1858. DOI 10.3389/fpls.2016.01858. [Google Scholar] [CrossRef]
8. Zhou, L., Xu, H., Mischke, S., Meinhardt, L. W., Zhang, D. et al. (2014). Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress. Horticulture Research, 1(1), 1–9. DOI 10.1038/hortres.2014.29. [Google Scholar] [CrossRef]
9. Chen, Y., Yu, Z., Zhang, Y., Zhou, J., Ni, D. et al. (2005). Effect of tea cultivars and tenderness on tea polysaccharide. Journal of Huazhong Agricultural, 24(4), 406–409. DOI 10.13300/j.cnki.hnlkxb.2005.04.021. [Google Scholar] [CrossRef]
10. Houston, K., Tucker, M. R., Chowdhury, J., Shirley, N., Little, A. (2016). The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Frontiers in Plant Science, 7, 984. DOI 10.3389/fpls.2016.00984. [Google Scholar] [CrossRef]
11. McFarlane, H., Döring, A., Persson, S. (2014). The cell biology of cellulose synthesis. Annual Review of Plant Biology, 65, 69–94. DOI 10.1146/annurev-arplant-050213-040240. [Google Scholar] [CrossRef]
12. Speicher, T., Li, P. Z., Wallace, I. (2018). Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants, 7(3), 52. DOI 10.3390/plants7030052. [Google Scholar] [CrossRef]
13. Scheller, H. V., Ulvskov, P. (2010). Hemicelluloses. Annual Review of Plant Biology, 61, 263–289. DOI 10.1146/annurev-arplant-042809-112315. [Google Scholar] [CrossRef]
14. Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A. et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell, 162(6), 1353–1364. DOI 10.1016/j.cell.2015.08.028. [Google Scholar] [CrossRef]
15. Kang, J. S., Frank, J., Kang, C. H., Kajiura, H., Vikram, M. et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proceedings of the National Academy of Sciences, 105(15), 5933–5938. DOI 10.1073/pnas.0800237105. [Google Scholar] [CrossRef]
16. Vain, T., Crowell, E. F., Timpano, H., Biot, E., Desprez, T. et al. (2014). The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiology, 165(4), 1521–1532. DOI 10.1104/pp.114.241216. [Google Scholar] [CrossRef]
17. Wang, T., McFarlane, H. E., Persson, S. (2016). The impact of abiotic factors on cellulose synthesis. Journal of Experimental Botany, 67(2), 543–552. DOI 10.1093/jxb/erv488. [Google Scholar] [CrossRef]
18. Zhang, S., Sun, L., Dong, X., Lu, S., Tian, W. et al. (2016). Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis. Journal of Integrative Plant Biology, 58(7), 623–626. DOI 10.1111/jipb.12442. [Google Scholar] [CrossRef]
19. Vega-Sánchez, M. E., Verhertbruggen, Y., Christensen, U., Chen, X., Sharma, V. et al. (2012). Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiology, 159(1), 56–69. DOI 10.1104/pp.112.195495. [Google Scholar] [CrossRef]
20. Zhao, H., Li, Z., Wang, Y., Wang, J., Xiao, M. et al. (2021). Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnology Journal, 20(3), 468–484. DOI 10.1111/pbi.13729. [Google Scholar] [CrossRef]
21. Huang, Y., Wei, K., Wang, L., Cheng, H., He, W. et al. (2012). Study of developmental pattern of tea shoot tenderness on the base of texture analyser. Journal of Tea Science, 32(2), 173–178. DOI 10.13305/j.cnki.jts.2012.02.014. [Google Scholar] [CrossRef]
22. Śmiechowska, M., Dmowski, P. (2006). Crude fibre as a parameter in the quality evaluation of tea. Food Chemistry, 94(3), 366–368. DOI 10.1016/j.foodchem.2004.11.026. [Google Scholar] [CrossRef]
23. Zhang, Z., Zhao, X., Zhang, Z., Huang, X., Zhang, L. et al. (2021). Cellulose accumulation in the secondary cell walls is negatively correlated with the tenderness of new shoots in Camellia sinensis. Physiologia Plantarum, 172(3), 1700–1710. DOI 10.1111/ppl.13376. [Google Scholar] [CrossRef]
24. Burton, R. A., Gidley, M. J., Fincher, G. B. (2010). Heterogeneity in the chemistry, structure and function of plant cell walls. Nature Chemical Biology, 6(10), 724–732. DOI 10.1038/nchembio.439. [Google Scholar] [CrossRef]
25. Cho, S. H., Purushotham, P., Fang, C., Maranas, C., Díaz-Moreno, S. M. et al. (2017). Synthesis and self-assembly of cellulose microfibrils from reconstituted cellulose synthase. Plant Physiology, 175(1), 146–156. DOI 10.1104/pp.17.00619. [Google Scholar] [CrossRef]
26. Daras, G., Templalexis, D., Avgeri, F., Tsitsekian, D., Karamanou, K. et al. (2021). Updating insights into the catalytic domain properties of plant cellulose synthase (CESA) and cellulose synthase-like (Csl) proteins. Molecules, 26(14), 4335. DOI 10.3390/molecules26144335. [Google Scholar] [CrossRef]
27. Desprez, T., Juraniec, M., Crowell, E. F., Jouy, H., Pochylova, Z. et al. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, 104(39), 15572–15577. DOI 10.1073/pnas.0706569104. [Google Scholar] [CrossRef]
28. Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M. et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proceedings of the National Academy of Sciences, 104(39), 15566–15571. DOI 10.1073/pnas.0706592104. [Google Scholar] [CrossRef]
29. Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, 100(3), 1450–1455. DOI 10.1073/pnas.0337628100. [Google Scholar] [CrossRef]
30. Fincher, G. B. (2009). Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiology, 149(1), 27–37. DOI 10.1104/pp.108.130096. [Google Scholar] [CrossRef]
31. Little, A., Schwerdt, J. G., Shirley, N. J., Khor, S. F., Neumann, K. et al. (2018). Revised phylogeny of the cellulose synthase gene superfamily: Insights into cell wall evolution. Plant Physiology, 177(3), 1124–1141. DOI 10.1104/pp.17.01718. [Google Scholar] [CrossRef]
32. Richmond, T. A., Somerville, C. R. (2000). The cellulose synthase superfamily. Plant Physiology, 124(2), 495–498. DOI 10.1104/pp.124.2.495. [Google Scholar] [CrossRef]
33. Yin, Y., Huang, J., Xu, Y. (2009). The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biology, 9(1), 1–14. DOI 10.1186/1471-2229-9-99. [Google Scholar] [CrossRef]
34. Appenzeller, L., Doblin, M., Barreiro, R., Wang, H., Niu, X. et al. (2004). Cellulose synthesis in maize: Isolation and expression analysis of the cellulose synthase (CESA) gene family. Cellulose, 11(3), 287–299. DOI 10.1186/s12870-017-1142-z. [Google Scholar] [CrossRef]
35. Burton, R. A., Shirley, N. J., King, B. J., Harvey, A. J., Fincher, G. B. (2004). The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiology, 134(1), 224–236. DOI 10.1104/pp.103.032904. [Google Scholar] [CrossRef]
36. Zou, X., Zhen, Z., Ge, Q., Fan, S., Liu, A. et al. (2018). Genome-wide identification and analysis of the evolution and expression patterns of the cellulose synthase gene superfamily in Gossypium species. Gene, 646, 28–38. DOI 10.1016/j.gene.2017.12.043. [Google Scholar] [CrossRef]
37. Li, G., Liu, X., Liang, Y., Zhang, Y., Cheng, X. et al. (2020). Genome-wide characterization of the cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Functional Integrative Genomics, 20(5), 723–738. DOI 10.1007/s10142-020-00747-8. [Google Scholar] [CrossRef]
38. Song, X., Xu, L., Yu, J., Tian, P., Hu, X. et al. (2019). Genome-wide characterization of the cellulose synthase gene superfamily in Solanum lycopersicum. Gene, 688, 71–83. DOI 10.1016/j.gene.2018.11.039. [Google Scholar] [CrossRef]
39. Djerbi, S., Lindskog, M., Arvestad, L., Sterky, F., Teeri, T. T. (2005). The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CESA) genes. Planta, 221(5), 739–746. DOI 10.1007/s00425-005-1498-4. [Google Scholar] [CrossRef]
40. Joshi, C. P., Bhandari, S., Ranjan, P., Kalluri, U. C., Liang, X. et al. (2004). Genomics of cellulose biosynthesis in poplars. New Phytologist, 164(1), 53–61. DOI 10.1111/j.1469-8137.2004.01155.x. [Google Scholar] [CrossRef]
41. Nairn, C. J., Haselkorn, T. (2005). Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary cell wall CesA genes of angiosperms. New Phytologist, 166(3), 907–915. DOI 10.1111/j.1469-8137.2005.01372.x. [Google Scholar] [CrossRef]
42. Roberts, A. W., Bushoven, J. T. (2007). The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Molecular Biology, 63(2), 207–219. DOI 10.1007/s11103-006-9083-1. [Google Scholar] [CrossRef]
43. Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C. et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proceedings of the National Academy of Sciences, 115(18), E4151–E4158. DOI 10.1073/pnas.1719622115. [Google Scholar] [CrossRef]
44. Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J. et al. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Research, 45(D1), D200–D203. DOI 10.1093/nar/gkw1129. [Google Scholar] [CrossRef]
45. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
46. Foster, C. E., Martin, T. M., Pauly, M. (2010). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: Carbohydrates. Journal of Visualized Experiments, 37, e1837. DOI 10.3791/1837. [Google Scholar] [CrossRef]
47. Hazen, S. P., Scott-Craig, J. S., Walton, J. D. (2002). Cellulose synthase-like genes of rice. Plant Physiology, 128(2), 336–340. DOI 10.1104/pp.010875. [Google Scholar] [CrossRef]
48. Xia, E., Li, F., Tong, W., Li, P., Wu, Q. et al. (2019). Tea plant information archive: A comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnology Journal, 17(10), 1938–1953. DOI 10.1111/pbi.13111. [Google Scholar] [CrossRef]
49. Wang, X., Zhao, Q., Ma, C., Zhang, Z., Cao, H. et al. (2013). Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics, 14(1), 1–15. DOI 10.1186/1471-2164-14-415. [Google Scholar] [CrossRef]
50. Zhang, Q., Cai, M., Yu, X., Wang, L., Guo, C. et al. (2017). Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genetics Genomes, 13(4), 1–17. DOI 10.1007/s11295-017-1161-9. [Google Scholar] [CrossRef]
51. Malinovsky, F. G., Fangel, J. U., Willats, W. G. (2014). The role of the cell wall in plant immunity. Frontiers in Plant Science, 5, 178. DOI 10.3389/fpls.2014.00178. [Google Scholar] [CrossRef]
52. Lairson, L., Henrissat, B., Davies, G., Withers, S. (2008). Glycosyltransferases: Structures, functions, and mechanisms. Annual Review of Biochemistry, 77, 521–555. DOI 10.1146/annurev.biochem.76.061005.092322. [Google Scholar] [CrossRef]
53. Somerville, C. (2006). Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology, 22, 53–78. DOI 10.1146/annurev.cellbio.22.022206.160206. [Google Scholar] [CrossRef]
54. Carpita, N. C. (1996). Structure and biogenesis of the cell walls of grasses. Annual Review of Plant Biology, 47(1), 445–476. DOI 10.1146/annurev.arplant.47.1.445. [Google Scholar] [CrossRef]
55. Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences, 109(4), 1187–1192. DOI 10.1073/pnas.1109047109. [Google Scholar] [CrossRef]
56. Cao, S., Cheng, H., Zhang, J., Aslam, M., Yan, M. et al. (2019). Genome-wide identification, expression pattern analysis and evolution of the Ces/Csl gene superfamily in pineapple (Ananas comosus). Plants, 8(8), 275. DOI 10.3390/plants8080275. [Google Scholar] [CrossRef]
57. Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W. et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science, 279(5351), 717–720. DOI 10.1126/science.279.5351.717. [Google Scholar] [CrossRef]
58. Beeckman, T., Przemeck, G. K., Stamatiou, G., Lau, R., Terryn, N. et al. (2002). Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiology, 130(4), 1883–1893. DOI 10.1104/pp.102.010603. [Google Scholar] [CrossRef]
59. Narciso, J., Oane, R. H., Kumar, A., Kohli, A. (2010). Cellulose synthase as a major candidate gene in the large effect QTL for rice yield under drought stress. Philippine Journal of Crop Science, 35, 7. [Google Scholar]
60. Zhu, J., Lee, B. H., Dellinger, M., Cui, X., Zhang, C. et al. (2010). A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. The Plant Journal, 63(1), 128–140. DOI 10.1111/j.1365-313X.2010.04227.x. [Google Scholar] [CrossRef]
61. Liu, X., Liu, L., Niu, Q., Xia, C., Yang, K. et al. (2011). MALE GAMETOPHYTE DEFECTIVE 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. The Plant Journal, 65(4), 647–660. DOI 10.1111/j.1365-313X.2010.04452.x. [Google Scholar] [CrossRef]

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |