
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016947
ARTICLE
Villin Family Members Associated with Multiple Stress Responses in Cotton
1State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, 210095, China
2Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, 210014, China
3Institute of Food Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, 210014, China
*Corresponding Author: Kang Liu. Email: kangliu@njau.edu.cn
#These authors have contributed equally to this work
Received: 13 April 2021; Accepted: 31 May 2021
Abstract: Villin (VLN) is considered to be one of the most important actin-binding proteins, participates in modulating the actin cytoskeleton dynamics, plays essential role in plant development and resisting adverse environments. However, systematic studies of the VLN gene family have not been reported in cotton (Gossypium). In this study, 14 GhVLNs were identified in G. hirsutum. These GhVLN genes were distributed in 6 A-subgenome chromosomes and 6 D-subgenome chromosomes of the allotetraploid upland cotton and classified into three phylogenetical groups based on the classification model of AtVLNs. In addition, the 14 GhVLN genes have highly conserved gene structure and motif architecture. The number of introns was ranged from 18 to 22 and the length of protein sequences was varied from 901 to 1077. Six gelsolin homology domains, G1–G6, and villin headpiece domain, VHP, were existed in all GhVLNs with the exception of two VLNs (GhVLN6 and GhVLN13) which lacked VHP. Cis-elements analysis revealed that the promoter regions of GhVLNs contained various light related components and also elements responsible for phytohormones and stresses response, indicating that, when subjected to those adverse environments, cotton plants may activate the response system by targeting VLN genes to survive the crisis. Heatmaps showed that the GhVLN genes exhibited various expression patterns, some were accumulated in certain tissues, root, petal, stamen or elongating fibers, and some were obviously induced by environmental changes. Especially GhVLN3 and GhVLN10 were highly and preferentially expressed in elongating fibers and distinctly upregulated by abiotic (salt, PEG, cold and heat) and biotic (Verticillium dahliae V991) stresses. This study may provide useful information for biological function identification of GhVLN genes and gene resources for creating high-quality and various resistant cotton germplasms.
Keywords: Gossypium hirsutum; villin proteins; cis-elements; abiotic and biotic stresses
In the process of growth and development, plants are inevitably subjected to various types of abiotic and biotic stresses, whether long-or short-term. The biotic stresses including fungi, bacteria, oomycetes, nematodes, insects, and weeds cause diseases or physical damages to plants. On the other hand, effects by drought, salinity, floods, extreme temperatures, etc., are included in abiotic stresses. Fortunately, plants have developed various mechanisms in order to overcome these threats. In this series of response mechanisms, the actin cytoskeleton plays an extremely vital role.
The actin cytoskeleton is a highly organized and dynamic system that not only involves in direction of cell growth, maintenance of cell morphogenesis, but also participates in responses to numerous environmental stresses by actin filaments assembling, depolymerization, and bundling [1–6]. In the apical and subapical regions of extending pollen tubes, actin filaments would be persistently polymerized and bundled, and finally generated two structures, the one is the shank-localized bundles and the other one is the “apical actin structure” [6–8]. Low temperature induced the actin filaments depolymerization, moreover, the Ca2+ influx and the expression level of the cold acclimatization-specific gene (cas) could be decreased by stabilization of actin cytoskeleton, but increased by depolymerization of actin filaments [9]. Additionally, under salt and osmotic stresses, facilitation of the actin filaments polymerization rescued plants from death, however blocking the polymerizing process or depolymerizing the actin filaments obviously reduced the plant survival rate [4,10]. Moreover, actin filament arrays and actin bundles were increased around the pathogen infection sites and the changes were related to various plant immune defense response processes [11–13].
The dynamic rearrangement of actin cytoskeleton is directly modulated by actin-binding proteins (ABPs). Villin (VLN) participates in modulating the actin cytoskeleton dynamics, including nucleating, capping, severing, and bundling actin filaments, is considered to be one of the most important ABPs [14–23]. VLN proteins contain six gelsolin homology domains, G1–G6, and an extra villin headpiece domain, VHP, at C-terminus, belong to villin/gelsolin/fragmin superfamily [23]. Previous studies showed that VLNs play various important roles in many cellular processes [14–23]. In Arabidopsis thaliana genome, five villins were identified and classified into three phylogenetical groups [23]. AtVLN1 was classified into group I alone, AtVLN2 and AtVLN3 belonged to group II, and group III contained AtVLN4 and AtVLN5. AtVLN1 bundled and stabilized actin filaments, however, had no actin-nucleating,-severing or-capping activity [21,24]. The other four AtVLNs have been reported to participate in actin nucleation, bundling, severing, and capping actin filaments, in other words, AtVLN2, AtVLN3, AtVLN4 and AtVLN5 are all-rounders [18–21,25]. Additionally, biological function analysis discovered that AtVLN1-5 were related to plant cell polar growth or diffuse growth [18–21,25]. OsVLN2 was also detected to affect plant architecture through regulating actin cytoskeleton dynamics [17]. Moreover, in a previous study, we isolated a cotton villin gene (Gh_D08G0231 in this study) and demonstrated that this gene was involved in cell elongation and multiple biotic and abiotic stresses via regulation of actin organization [14,15].
For more information about the villins in cotton (Gossypium), the important economic crop that produces natural fibers, in this study, we genome-widely researched this important family based on the whole genome sequencing of the tetraploid cotton G. hirsutum and the release of publicly sequencing data [26,27]. We identified 14 GhVLN genes and further analyzed their physicochemical properties, chromosomal distribution, phylogenetic relationships, gene structures, and conserved motifs. Additionally, cis-elements analysis of the GhVLN promoters and expression profiling of GhVLNs in various cotton tissues and organs, as well as the different GhVLNs expression patterns under biotic (Verticillium dahliae V991) and abiotic (salt, PEG, cold and heat) stresses, revealed that the transcripts of GhVLNs differently accumulated in cotton tissues and GhVLNs may participate in responding to different environmental changes in cotton.
2.1 Identification and Sequence Retrieval of VLN Family Members
The HMM files (PF00626 and PF02209) of the conserved gelsolin domain and villin headpiece domain were downloaded from the Pfam database (https://pfam.xfam.org/) [28]. Putative VLNs identified from G. hirsutum (http://mascotton.njau.edu.cn/), G. arboretum (http://cgp.genomics.org.cn), G. herbaceum (https://www.cottongen.org/), G. raimondii (http://www.phytozome.net/) were verified by the HMMER software with the default parameters [29]. AtVLNs were also genome-widely searched against Arabidopsis thaliana genome sequencing data (https://www.arabidopsis.org/) and OsVLNs were obtained from Oryza sativa published genomes (https://phytozome.jgi.doe.gov/pz/portal.html). Furtherly, gene prediction programs SMART (http://smart.embl-heidelberg.de/) and FGENESH (http://www.softberry.com/berry.phtml) were used to correct and to detect the G1–G6 and VHP conserved motifs. Finally, the VLN genes were confirmed by BLASTp. Remove the VLNs that did not contain the gelsolin domain and retain proteins that contained the gelsolin domain but not the VHP domain. These VLN genes were named according the position on the chromosome. The amino acid length, isoelectric point (pI), theoretical molecular weight (Mw), and subcellular localization of the identified VLNs were predicted and calculated by submitting the protein sequences to the online program CELLO v2.5 (http://cello.life.nctu.edu.tw/) [30] and Pepstats (https://www.ebi.ac.uk/Tools/seqstats/emboss_pepstats/) [31].
2.2 Phylogenetic Analysis of VLN Proteins
The ClustalW program was used to perform the multiple sequence alignment of VLNs from G. hirsutum, G. arboretum, G. herbaceum, G. raimondii, A. thaliana, and Oryza sativa with the default parameters [32]. The unrooted phylogenetic tree was constructed by employing the alignment sequences using the neighbor-joining (NJ) method in MEGA 7.0 software, with 1000 replicates boot-strap test [33].
2.3 Gene Structure and Conserved Motif Analysis
The online program GSDS 2.0 [34] (http://gsds.cbi.pku.edu.cn/) was employed to draw the exon/intron structures of GhVLN genes. And the conserved motifs of GhVLNs were identified using one other online program MEME 5.3.3 [35] (http://meme-suite.org/tools/meme).
The promoter regions of 14 GhVLN genes were extracted from G. hirsutum genome database, which contains 2000 bp upstream of “ATG” of GhVLNs open reading frames. The online program PlantCARE [36] (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/search_CARE.html) was used to predict the cis-acting elements in the 14 GhVLNs promoter regions.
2.5 Expression Pattern Analysis
For analyzing the expression profiling of GhVLN genes, we downloaded the cotton RNA-seq data [26,37], including vegetative and reproductive tissues (stamen, root, stem, leaf, petal, −3, 0 or 3 DPA ovules, 5, 10, 20 and 25 DPA fibers), and responding to abiotic stresses (salt, PEG, heat and cold) and biotic stress (V. dahliae V991). Expression patterns were displayed in TBtools [38].
3.1 Identification of VLN Genes and Chromosomal Distribution Analysis
In this study, 14 VLN genes from upland cotton G. hirsutum, 7 from G. arboreum, 7 from G. herbaceum, 7 from G. raimondii, 5 from Arabidopsis, and 5 from rice were identified. 7 out of the 14 genes identified from upland cotton were distributed on the A subgenome, and the other 7 genes were mapped on the D subgenome. These 14 VLNs in G. hirsutum were named GhVLN1 to GhVLN14 in turn, according to the distribution of VLNs on chromosomes. The VLNs in G. arboreum, G. herbaceum and G. raimondii were named GaVLN1 to GaVLN7, GheVLN1 to GheVLN7, GrVLN8 to GrVLN14 in turn, according to their homology VLNs in G. hirsutum. The gene names and their corresponding gene ID numbers were listed in Tab. 1 and Tab. S1.
The length of GhVLNs protein sequences was varied from 901 to 1077 aa, and the average length was 963 aa. The other physicochemical parameters were also shown in Tab. 1, including the molecular weight (MW) of GhVLNs which was ranged from 100.73 to 120.56 kDa with an average value of 106.60 kDa, and the isoelectric point (pI) of GhVLNs which was ranged from 4.99 to 6.61 with an average value of 5.74. Additionally, the subcellular localization was predicted and the results demonstrated great majority of the GhVLN genes were located in outermembrane (Tab. 1). The chromosome localization analysis shown that the 14 GhVLNs were unevenly distributed on 12 chromosomes of G. hirsutum acc. TM-1. A01/D01 homologous chromosomes contained two GhVLN genes, the other chromosomes contained only one GhVLN gene (Fig. 1). Besides a pair of homoeologous genes on chromosome A2 and D3, the other gene pairs of the GhVLN gene family were exhibited to locate on the same numbered A and D chromosomes. In addition, notably, the distribution of GhVLN genes on chromosomes was scattered, three of them on the top ends, five on the opposite ends, four distributed in the middle of the chromosome and the other two was detected in the scaffold 2268 and scaffold 3706 (Fig. 1).
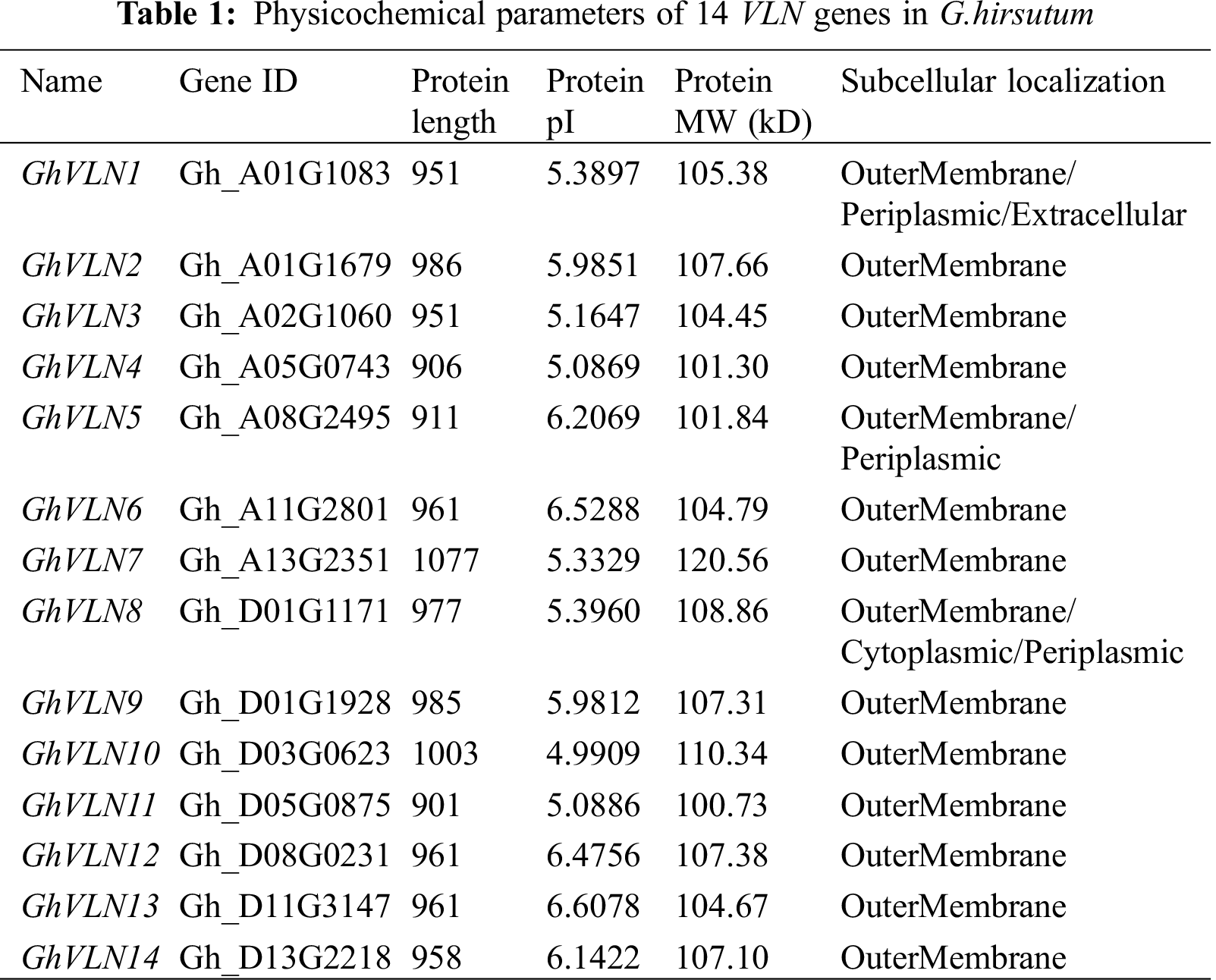
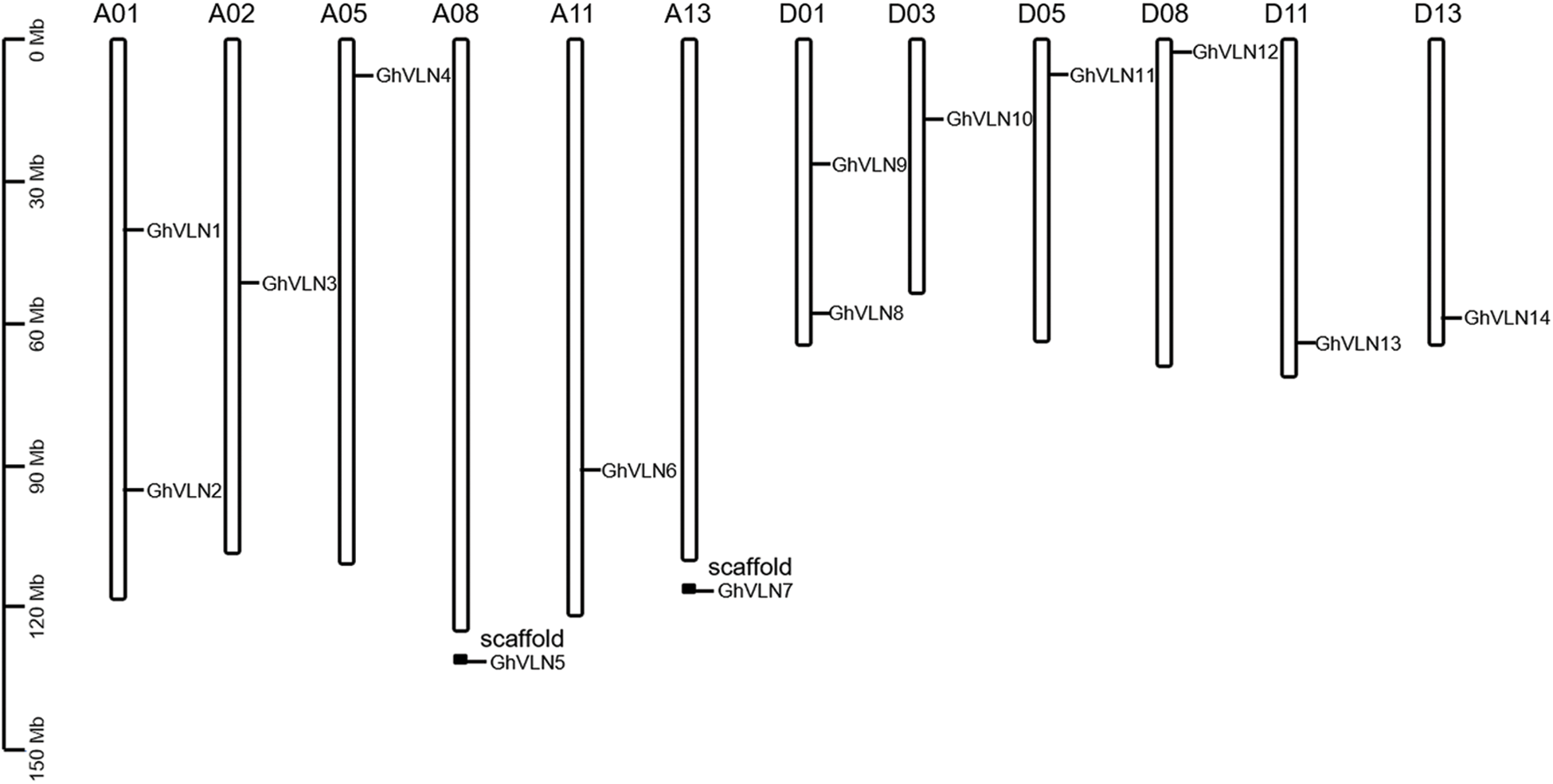
Figure 1: Chromosomal localization of VLN genes in Gossypium hirsutum. The scale bars represent the length in mega bases (Mb)
3.2 Phylogenetic Relationship, Gene Structure and Conserved Motif Analysis of VLNs
To investigate the evolutionary relationships of VLNs identified from G. hirsutum, G. arboreum, G. herbaceum, G. raimondii, Arabidopsis and rice, 14 GhVLNs, 7 GaVLNs, 7 GheVLNs, 7 GrVLNs, 5 AtVLNs and 5 OsVLNs protein sequences were used to construct an unrooted phylogenetic tree (Fig. 2). Based on the formulated subfamilies in Arabidopsis [19,21,23,24], the 45 VLNs were classified into 3 subfamilies (Groups I–III). The Group II containing 8 GhVLNs, 4 GaVLNs, 4 GheVLNs, 4 GrVLNs, 2 AtVLNs and 1 OsVLN, was the largest subfamily. However, the Group I held the fewest VLN members, only containing 2 GhVLNs, 1 GaVLN, 1 GheVLN, 1 GrVLN, 1 AtVLN and 1 OsVLN genes (Fig. 2). Specially, the Group III contained 4 GhVLNs, 2 GaVLNs, 2 GheVLNs, 2 GrVLNs and 2 AtVLN genes, whereas 3 OsVLN genes. Overall, in this phylogenetic tree, each group contained a fair number of VLN genes from the six tested species, suggesting that VLN genes were relatively conserved in different species during evolution, also exposing the difference between monocotyledonous and dicotyledonous plants. Additionally, all Ka/Ks values were less than 1 (ranging 0.09–0.83) (Tab. S1), indicating that, during the long-term evolutionary process, VLN gene family had subjected to purifying selection in cotton species.
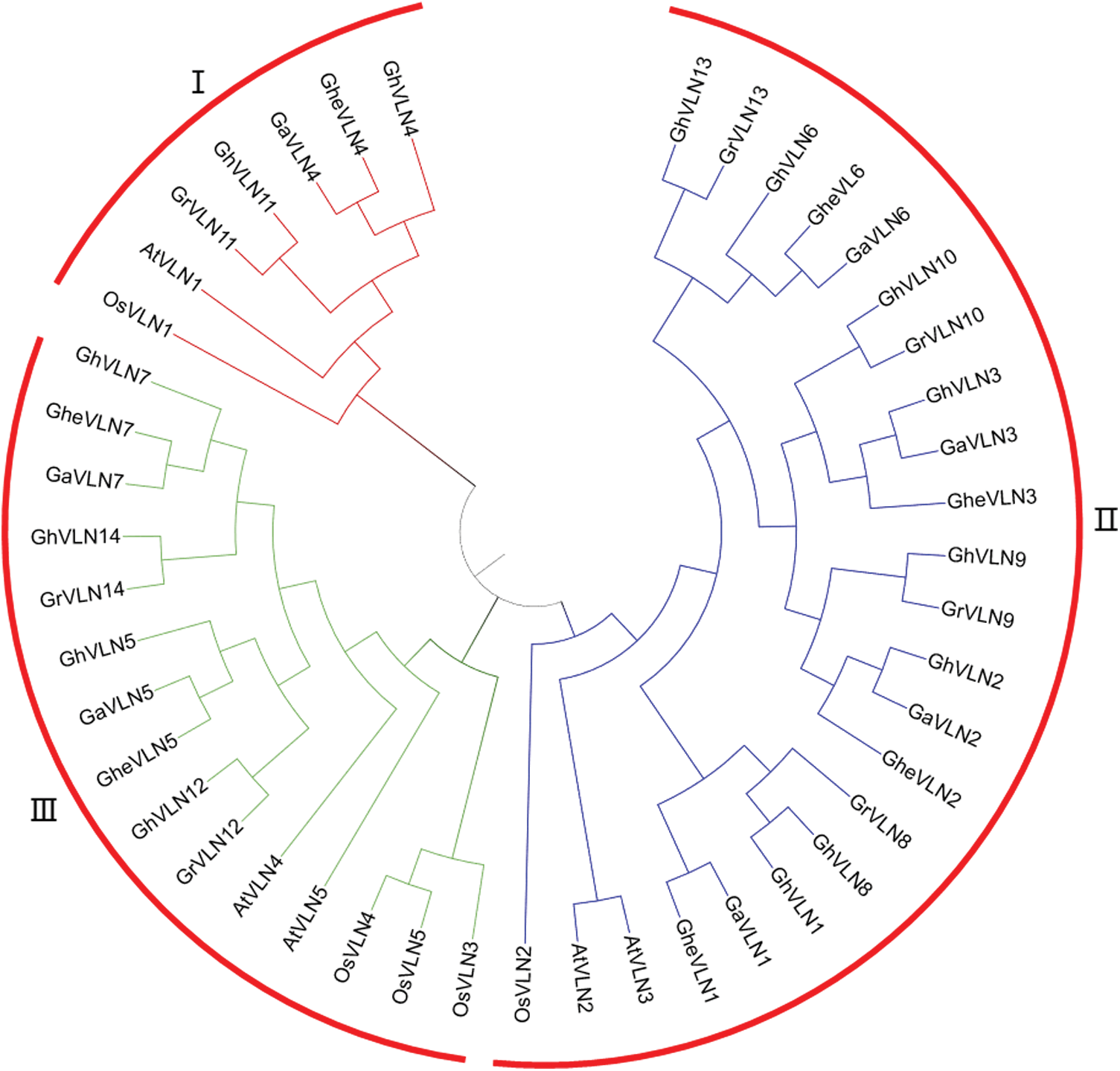
Figure 2: Phylogenetic relationships of VLN proteins from G. hirsutum, G. arboreum, G. herbaceum, G. raimondii, Arabidopsis thaliana and Oryza sativa. MEGA 7 was employed to construct the unrooted neighbor-joining (NJ) tree, and 1000 replicates were performed in the bootstrap testing. AtVLN1, At2g29890; AtVLN2, At2g41740; AtVLN3, At3g57410; AtVLN4, At4g30160; AtVLN5, At5g57320; OsVLN1, Os05g06110; OsVLN2, Os03g24220; OsVLN3, Os06g44890; OsVLN4, Os04g51440; and OsVLN5, Os08g14230
The GhVLN genes were also analyzed to investigate their exon/intron structure and conserved motif. As shown in Fig. 3, all of the GhVLNs contained a large number of exons and introns, showing the long genome sequences of GhVLNs. The number of introns of 14 GhVLNs was ranged from 18 to 22. In addition, to investigate whether the gene structure of GhVLNs is consistent with the evolutionary relationship, an unrooted phylogenetic tree was constructed employing the 14 GhVLN protein sequences. Combining the results of phylogenetic relationships and gene structure analysis, we found that evolutionarily closely related genes have similar exon/intron structures.
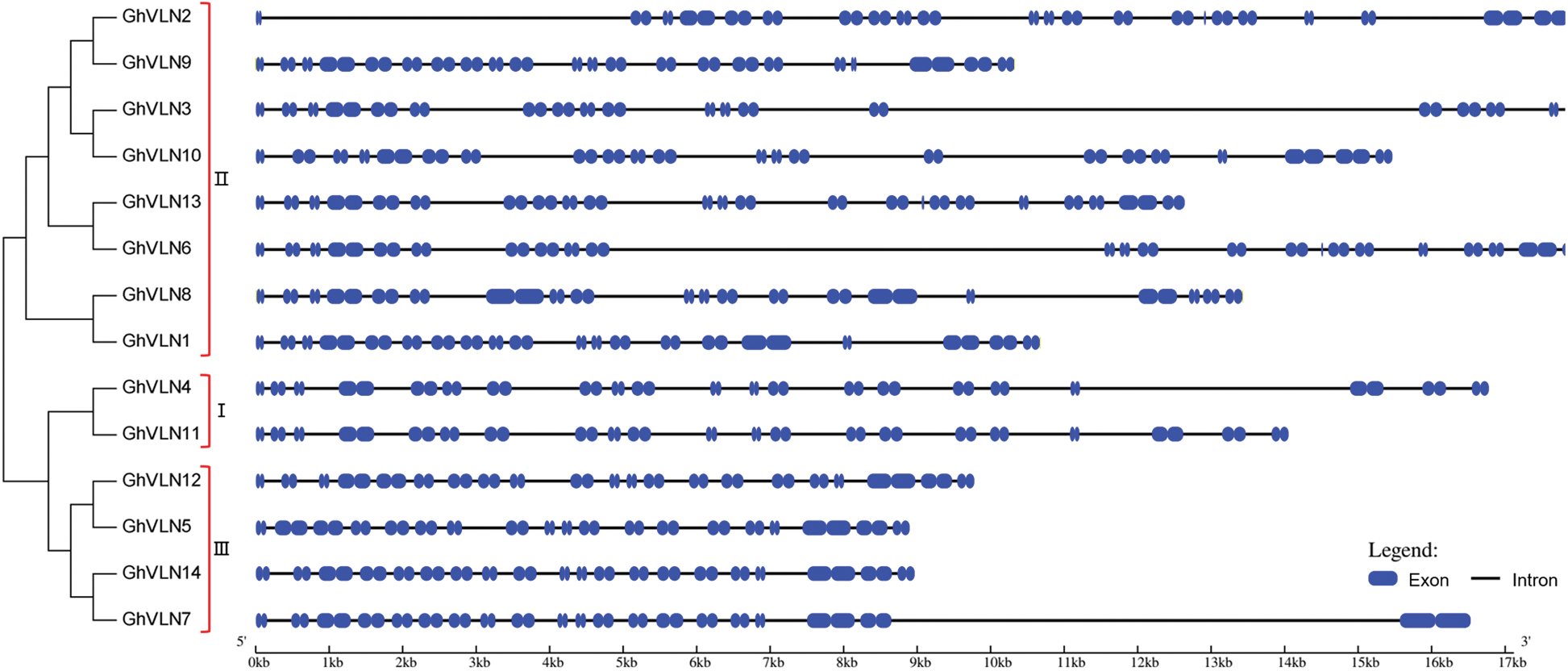
Figure 3: Phylogenetic relationships and gene structures of GhVLNs. MEGA 7 was employed to construct the unrooted NJ tree of 14 GhVLN genes (shown in left), and 1000 replicates were performed in the bootstrap testing. The exon (blue boxes)/intron (black lines) distribution of GhVLN genes (listed in right) was constructed using an online program GSDS 2.0
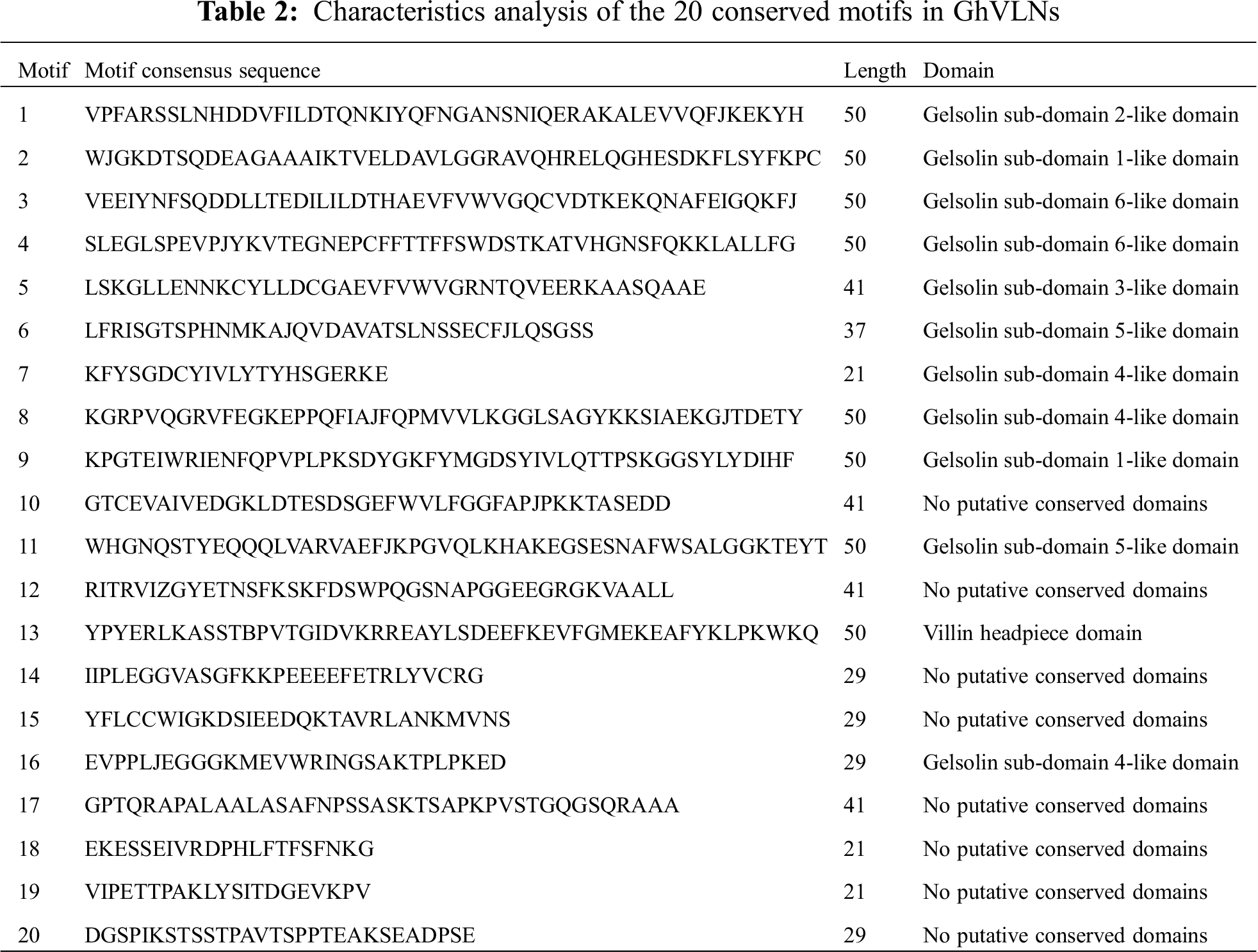
To discover the conserved motifs of GhVLNs, the MEME [35] was used by submitting the identified 14 GhVLN protein sequences to the online program. We detected 20 motifs and named them motif 1–20 accordingly (Fig. 4). The corresponding sequences and domain analysis of 20 identified motifs were shown in Tab. 2, and their motif logos were illustrated in Fig. S1. The length of the conserved motifs was ranged from 21 to 50 amino acids. The number of the conserved motifs in each GhVLN protein was varied from 17 to 20. All of the GhVLNs contained motif 1–16 and 18, while GhVLN4 and GhVLN11 lacked motif 17, 19 and 20, GhVLN6 and GhVLN13 lacked motif 13, GhVLN5 lacked motif 9 and 20, GhVLN7 and GhVLN14 lacked motif 9, 19 and 20. In other words, all GhVLN proteins contain six gelsolin homology domains, G1–G6, and villin headpiece domain, VHP, with the exception of two VLNs (GhVLN6 and GhVLN13) which lacked VHP. The sequence alignment of the conserved domains in GhVLNs and AtVLNs was shown in Fig. S2. Therefore, the gelsolin homology domains were highly conserved in cotton VLNs, moreover, there are similar types and number of motif architecture and exon/intron distribution pattern between GhVLN orthologs with close evolutionary relationship.
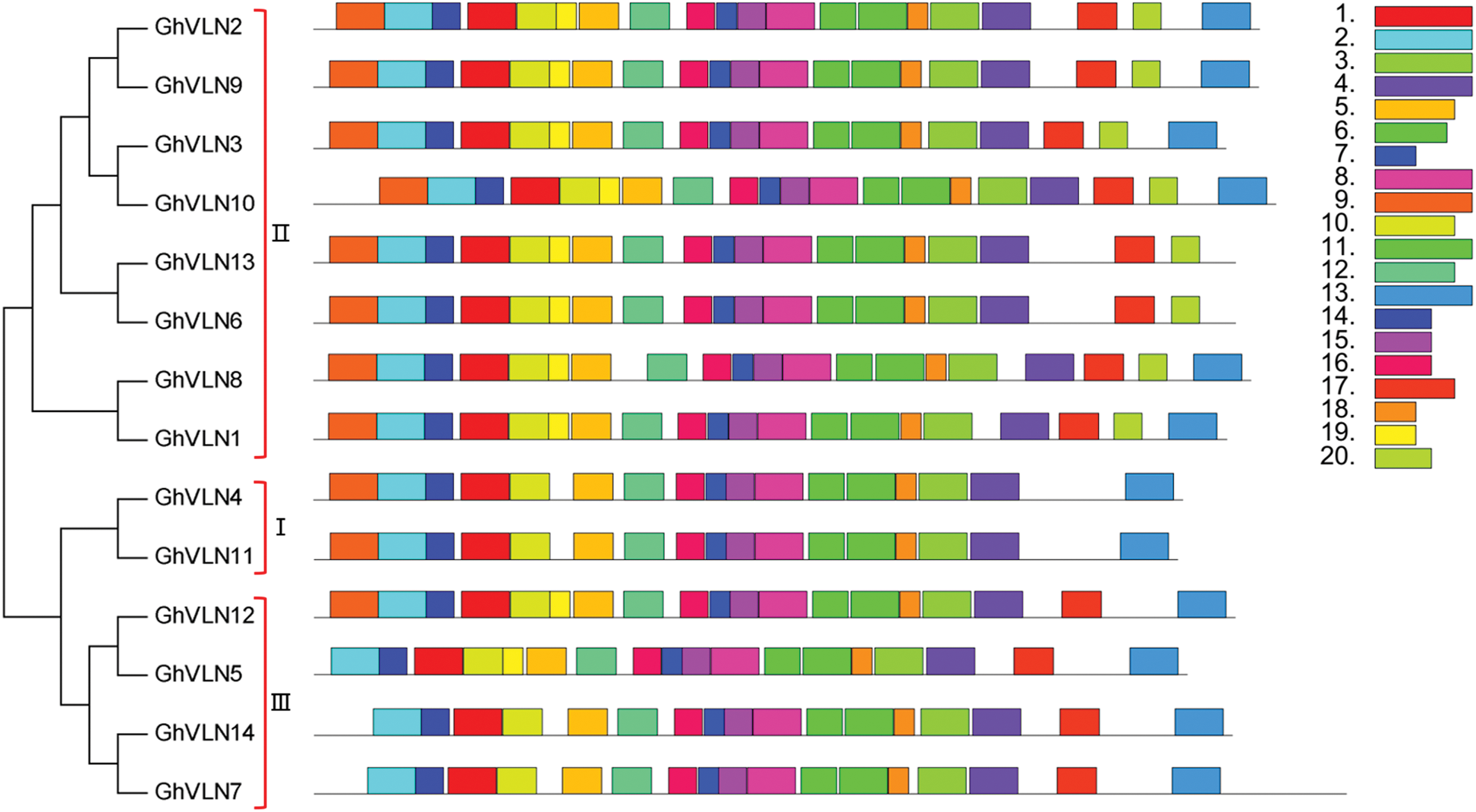
Figure 4: Conserved motifs of GhVLN proteins. The conserved motifs were identified using MEME (suite 5.3.3) by loading the GhVLN protein sequences. Each of the 20 identified motifs is indicated with a colored box (listed in right)
3.3 The Cis-Acting Elements Analysis of GhVLN Gene Promoters
When subjected to adverse environments or hormone stresses, what is the regulatory functions of VLN genes in cotton? In order to clarify this problem, we firstly extracted the 2000 bp upstream sequences of 14 GhVLN genes and submitted to the PlantCARE database for cis-acting element prediction. Besides 22 types of light response-related regulatory elements, there were several other cis-elements involved in stress response and phytohormone response in the promoter regions of GhVLNs. Stress response related cis-elements included defense and stress (TC-rich repeats), drought (MBS), WRKY binding site (W box), wound (WUN-motif), and low-temperature (LTR). While phytohormone response related cis-elements included abscisic acid (ABRE), ethylene (ERE), MeJA (CGTCA-motif and TGACG-motif), gibberellin (P-box, TATC-box and GARE-motif), salicylic acid (TCA-element), and auxin (TGA-element). As shown in Fig. 5, several promoter regions of GhVLNs contained a series of stress-responsive and hormone-responsive components, such as GhVLN6 (2 LTR, 3 WUN-motif, 1 MBS, 1 W box, 2 TC-rich repeats, 1 ERE, 1 P-box, 1 TCA-element, 2 TGACG-motif) and GhVLN10 (2 LTR, 1 MBS, 2 P-box, 3 TGACG-motif, 1 TC-rich repeats, 1 TCA-element, 3 CGTCA-motif). In addition, among the hormone-related elements, MeJA-responsive and SA-responsive elements appeared more frequently. These results indicated that GhVLNs possibly related to MeJA and SA hormone signaling pathways, and suggested the important roles of GhVLN genes in response to adverse environments stresses and phytohormone signals in cotton.
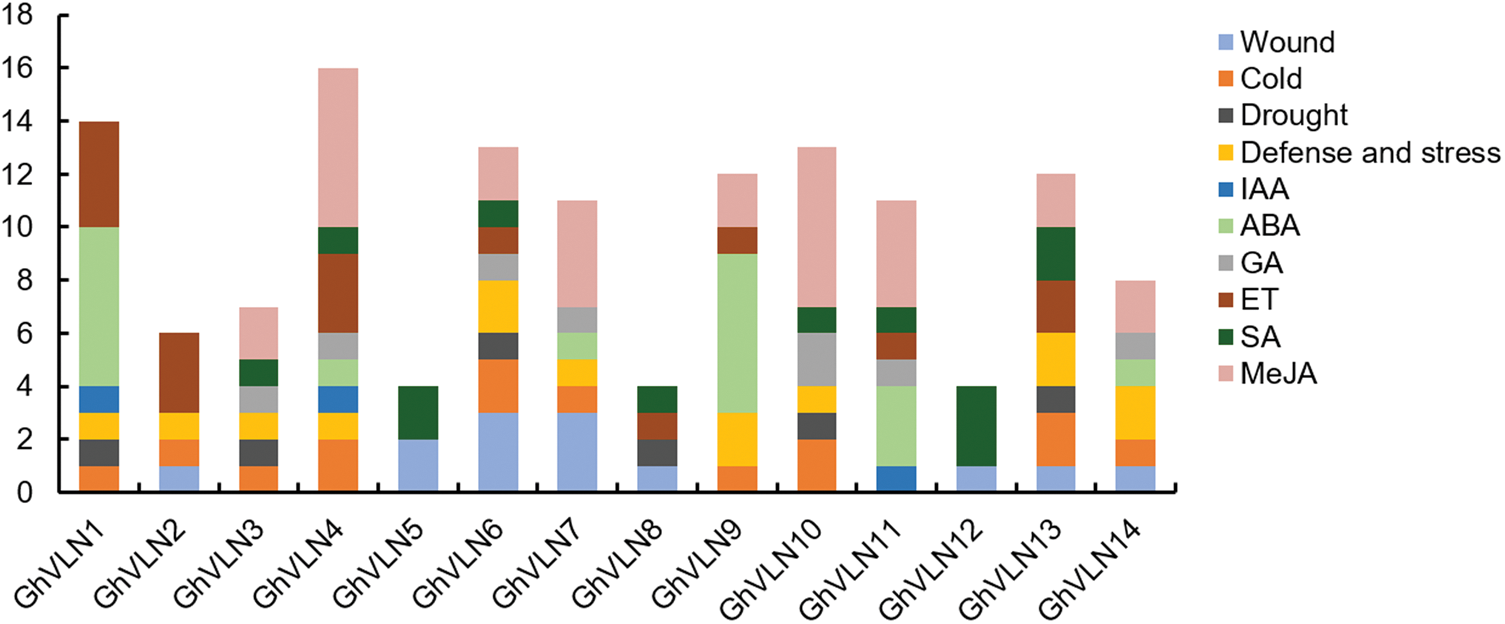
Figure 5: Cis-elements analysis of GhVLNs promoters. The numbers of different cis-elements are presented in the form of bar graphs, and similar cis-elements are exhibited with the same colors
3.4 Expression Patterns Analysis of GhVLN Genes in Various Tissues and under Different Biotic and Abiotic Stresses
To explore the potential functions of GhVLNs, transcription patterns of all 14 GhVLNs in various cotton tissues and organs were investigated by analyzing transcriptome data [26,37]. GhVLNs showed different expression patterns (Fig. 6). GhVLN4, GhVLN11, GhVLN3, GhVLN10, GhVLN5, GhVLN12, GhVLN2 and GhVLN9 were detected at a high level of transcripts in all tested tissues. Whereas, GhVLN4 and GhVLN11 had the most transcription in petal, while GhVLN3, GhVLN10, GhVLN2 and GhVLN9 in elongating fibers. And in particular, one pair of homologous genes (GhVLN5 and GhVLN12) exhibited a similar expression pattern with relative changeless expression level in the tested tissues. Furthermore, some GhVLNs transcripts were accumulated mainly in one specific tissue. GhVLN6, GhVLN8 and GhVLN13 were such genes, they were preferentially expressed in cotton stamen, implying that these genes are necessary for the growth and development of cotton stamens. Therefore, the expression profiles of GhVLN genes demonstrated that members of VLN family in cotton were widely expressed in both vegetative tissues and reproductive tissues and which might endow them multiple biological functions.
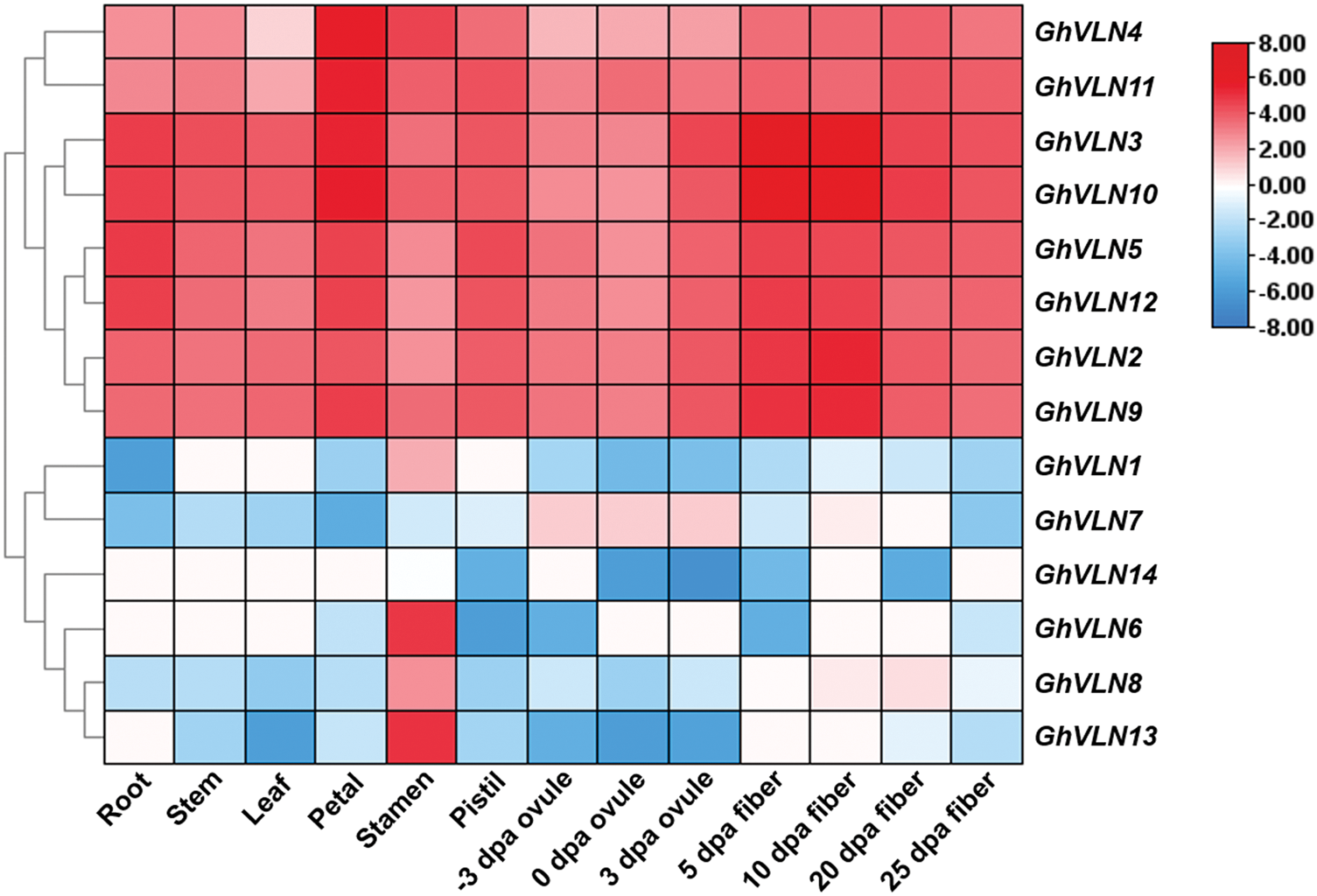
Figure 6: Expression patterns of GhVLNs in different tissues and organs in G. hirsutum. TBtools was used for generating the heatmaps
The analysis of cis-elements in promoter regions suggest that GhVLNs possibly respond to biotic and abiotic stresses. In order to test this hypothesis, we furtherly researched the expression patterns of 14 VLNs under salt, PEG, cold and heat stresses, as well as V. dahliae V991 biotic stress. As shown in Figs. 7 and 8, the expression of most GhVLN genes were induced under biotic and abiotic stresses in varying degrees. GhVLN3, GhVLN10, GhVLN11, GhVLN5, GhVLN12 were highly expressed before and after all the abiotic and biotic stress treatments, and had the similar expression patterns under these stresses. Moreover, GhVLN2 and GhVLN9 showed significantly upregulated expression under salt, PEG and heat stresses, however, under cold stress, the upregulated expression was not conspicuous. On the other hand, some genes (GhVLN8, GhVLN13 and GhVLN14) exhibited marked downregulation in response to the abiotic stresses. In addition, some genes can respond to one or numbers of specific abiotic stresses. For instance, the transcripts of GhVLN1 were significantly decreased under salt, PEG and heat, but not cold, and GhVLN6 was significantly decreased only under salt and heat. Importantly, it should be noticed that, when subjected to biotic stress, some VLN genes exhibited opposite expression patterns in G. barbadense and G. hirsutum. GhVLN14 and GhVLN6 were such genes. Under V991 biotic stress, the expression of GhVLN14 was upregulated in G. hirsutum but downregulated in G. barbadense. While the transcripts of GhVLN6 were decreased in G. hirsutum but increased in G. barbadense. To verify the results of transcriptome data, we analyzed the expression pattern of GhVLN12 gene in different cotton tissues and under abiotic and biotic stresses by comparing data of qRT-PCR and FPKM. As shown in Fig. S3, GhVLN12 exhibited the same expression trend in both transcriptome and qRT-PCR analysis.
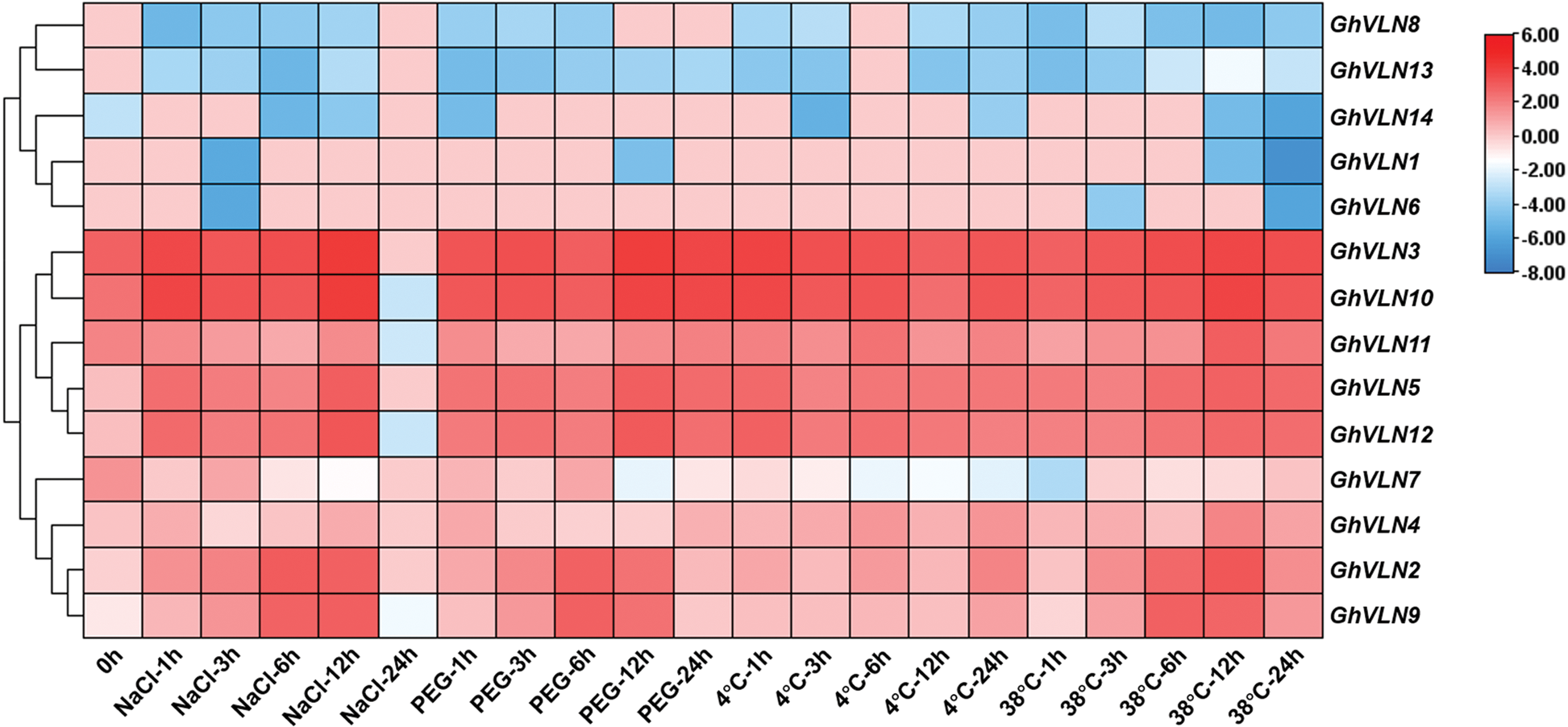
Figure 7: Expression patterns of GhVLNs in G. hirsutum under four different abiotic stresses. The normalization and visualization of abiotic stress related transcriptomic data were performed using TBtools
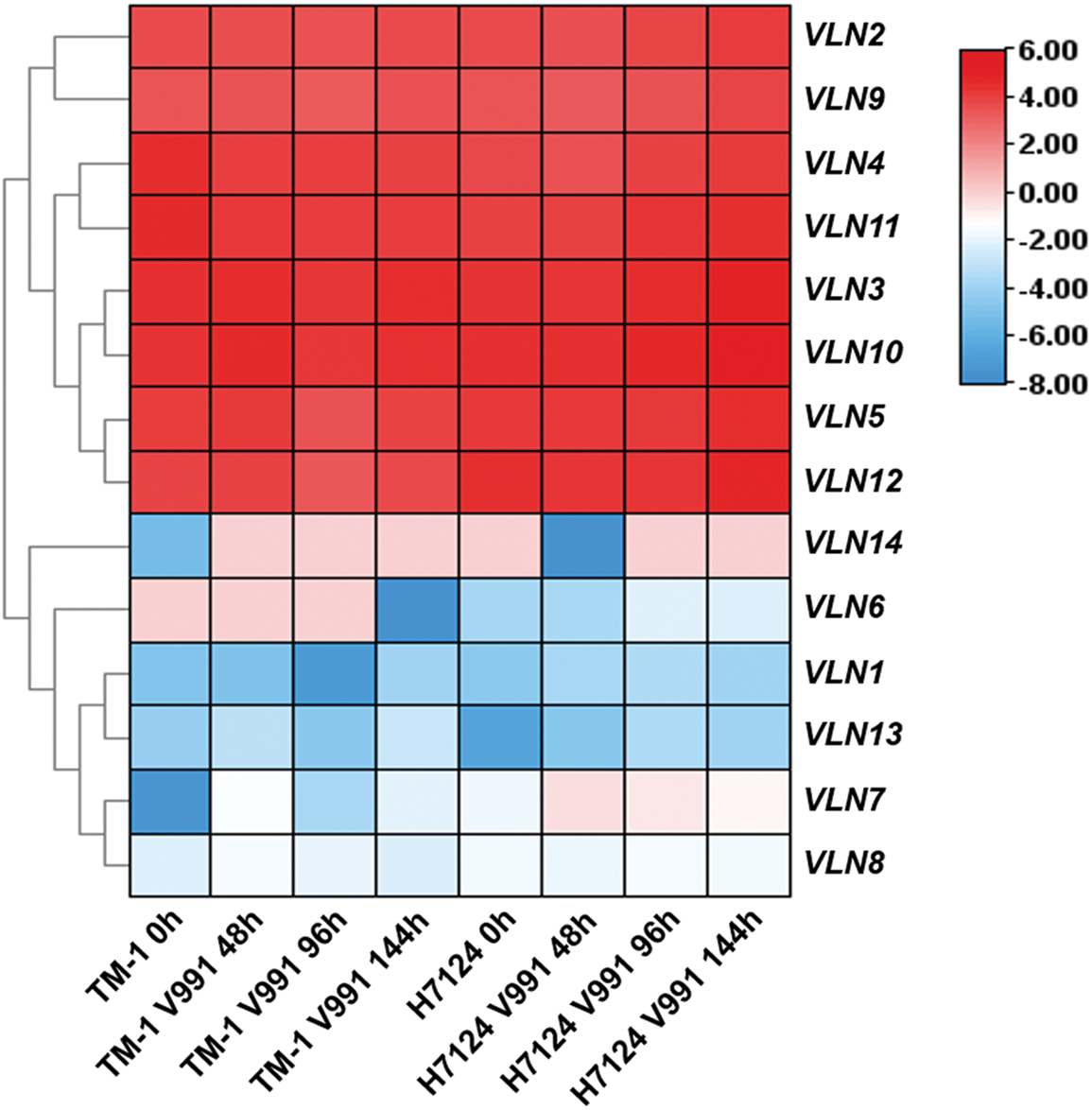
Figure 8: Expression patterns of VLNs in G. hirsutum and G. barbadense under Verticillium dahliae stress. The heatmap of biotic stress related transcriptomic data was generated using TBtools
Plant villins are widely detected, from algae to land plants [39]. Lots of VLN genes have been extensively analyzed in plant [14,15,17,23,40–43]. These studies demonstrated that VLNs play very important roles in plant growth and development and in resisting adverse environments. However, studies on cotton VLN gene family are relatively scarce so far.
In this study, we genome-widely identified for the first time the VLN family genes in cotton and systematically verified the chromosome distribution, phylogenetic relationship, gene structure and function of VLN genes. A total of 14 VLN genes were identified in G. hirsutum (Tab. 1). According to the classification model of AtVLN genes, 14 GhVLN genes were classified into three groups (Fig. 2), consistent with the previous classification of Arabidopsis [21,44]. The gene structure might decide its biological function [45]. Cotton VLN orthologs shared highly similar motif architectures (Fig. 4). Villin conserved domain, G1–G6 and VHP motifs, existed in all GhVLN proteins, with the exception of two VLNs (GhVLN6 and GhVLN13) which lacked VHP. Moreover, due to the GhVLNs motifs shared highly similarity with AtVLN1-5 [21], we speculate that VLN isoforms of cotton possibly play similar functions with Arabidopsis.
G1–G6, each G domain has a different biochemical function, including binding, capping, cutting, and nucleation of MFs [23]. The VHP domain at the C-terminal of villin proteins provides another microfilament binding site, which has microfilament bundle activity [46]. Since 1998, the plant villin P-115-ABP and P-135-ABP [47,48] have been identified from lily (Lilium longiflorum) for the first time. More and more studies about VLN genes have been reported in recent years, including AtVLNs [18–21,24,25] and OsVLN2 [17]. These studies have shown that plant villin has typical biochemical activities, which regulates the rearrangement of microfilaments and the formation of microfilament networks, exhibiting a significant impact on cell growth and cell morphology as well as responding to various abiotic and biotic stresses.
The expression pattern of one gene was determined in a certain degree by the cis-elements in its promoter regions [49]. In promoter sequences of GhVLNs, we investigated multiple types of cis-elements (Fig. 5), suggesting that expression of GhVLNs might be regulated by various stresses and phytohormones. Further expression patterns analysis verified this inference. The identified GhVLN genes showed different expression patterns (Fig. 6). The transcripts of some VLN orthologs were obviously detected in all tested tissues, some VLN genes were mainly accumulated in specific tissues, and others were low detected. This result is consistency with other studies, the five Arabidopsis villin-like genes (AtVLN1-5) were also reported to have different expression patterns at high expression levels with preferential expression in certain tissues [44]. In addition, when subjected to biotic and abiotic stresses, VLNs exhibited various expression patterns to response stresses (Figs. 7 and 8). These foundations suggest that VLN gene family is essential to cotton vegetative growth and fiber elongation, furthermore, there are functional differentiation, gained new functions, or lost functions probably presented in these cotton genes.
Additionally, a very interesting point caught our attention, that is the promoter regions of GhVLN5 and GhVLN12 containing only two types of cis-elements about SA and wound responses, however, the two genes induced highly in all the biotic and abiotic stress treatment. On the contrary, GhVLN6 and GhVLN13 have relatively more cis-elements, but expressed lowly in treatment. Therefore, we furtherly analyzed the promoter regions and found that in addition to cis-elements mentioned in Fig. 5, there are also MYB-and MYC-elements in all the VLN promoter regions. The MYB and MYC transcription factors have been reported to associated with drought, salt, heat, chilling stress responses and hormonal signaling pathways [50–55]. Transcription factor can activate or inhibit the expression of its target genes by combining with the cis-elements in the promoter regions of the genes. Perhaps, this is the reason for which the number of stress-related elements is inconsistent with the gene expression level in stress treatment. In our previous studies, GhVLN12 exhibited the actin-bundling activity, which induced cell extension by increasing the abundance of thick actin bundles [15], furthermore, this gene was reported to participate in salt and drought stress responses and required for tolerance to Verticillium Wilt in cotton [14]. To date, these are only two biological function studies of VLN in cotton, which consistence with the results in this study. Taken together, we speculate that VLN genes may involve in root growth, petal and stamen development, fiber elongation, response to stresses (salt, drought, cold, heat, V. dahliae) and yield in cotton. Even so, the detailed function of each VLN genes remains to be researched in cotton.
In this study, we genome-widely identified the villin family genes in cotton. A total of 14 GhVLN genes were identified. A series of systematic analysis of GhVLNs in physicochemical properties, chromosomal distribution, phylogenetic relationships, gene structures, conserved motifs, cis-elements in promoter regions, and expression patterns revealed that some of them involved in cotton fiber elongation, and some VLN orthologs are associated with all tested or specific stress responses. Collectively, the data here might provide useful information for well functional verification of GhVLN genes in cotton and gene resources for creating high-quality and various resistant cotton germplasms in future.
Acknowledgement: We especially thank Dr. Dayong Zhang (Nanjing Agricultural University, China) for providing cotton samples and figure production of chromosomal distribution of VLN genes.
Funding Statement: This work was financially supported by the National Natural Science Foundation of China (No. 31801408), the Natural Science Foundation of Jiangsu Province, China (No. BK20180517).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Jia, H. L., Chen, S. S., Wang, X. F., Shi, C., Liu, K. N. et al. (2020). Copper oxide nanoparticles alter cellular morphology via disturbing the actin cytoskeleton dynamics in Arabidopsis roots. Nanotoxicology, 14(1), 127–144. DOI 10.1080/17435390.2019.1678693. [Google Scholar] [CrossRef]
2. Szymanski, D., Staiger, C. J. (2018). The actin cytoskeleton: Functional arrays for cytoplasmic organization and cell shape control. Plant Physiology, 176(1), 106–118. DOI 10.1104/pp.17.01519. [Google Scholar] [CrossRef]
3. Sakai, Y., Takagi, S. (2017). Roles of actin cytoskeleton for regulation of chloroplast anchoring. Plant Signaling & Behavior, 12(10), e1370163. DOI 10.1080/15592324.2017.1370163. [Google Scholar] [CrossRef]
4. Wang, C., Zhang, L. J., Huang, R. D. (2011). Cytoskeleton and plant salt stress tolerance. Plant Signaling & Behavior, 6(1), 29–31. DOI 10.4161/psb.6.1.14202. [Google Scholar] [CrossRef]
5. Fu, Y. (2010). The actin cytoskeleton and signaling network during pollen tube tip growth. Journal of Integrative Plant Biology, 52(2), 131–137. DOI 10.1111/j.1744-7909.2010.00922.x. [Google Scholar] [CrossRef]
6. Xu, Y. N., Huang, S. J. (2020). Control of the actin cytoskeleton within apical and subapical regions of pollen tubes. Frontiers in Cell and Developmental Biology, 8, 614821. DOI 10.3389/fcell.2020.614821. [Google Scholar] [CrossRef]
7. Qu, X. L., Zhang, R. H., Zhang, M., Diao, M., Xue, Y. B. et al. (2017). Organizational innovation of apical actin filaments drives rapid pollen tube growth and turning. Molecular Plant, 10(7), 930–947. DOI 10.1016/j.molp.2017.05.002. [Google Scholar] [CrossRef]
8. Rounds, C. M., Hepler, P. K., Winship, L. J. (2014). The apical actin fringe contributes to localized cell wall deposition and polarized growth in the lily pollen tube. Plant Physiology, 166(1), 139–151. DOI 10.1104/pp.114.242974. [Google Scholar] [CrossRef]
9. Orvar, B. L., Sangwan, V., Omann, F., Dhindsa, R. S. (2000). Early steps in cold sensing by plant cells, the role of actin cytoskeleton and membrane fluidity. The Plant Journal, 23(6), 785–794. DOI 10.1046/j.1365-313x.2000.00845.x. [Google Scholar] [CrossRef]
10. Wang, C., Zhang, L., Yuan, M., Ge, Y., Liu, Y. et al. (2010). The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biology, 12(1), 70–78. DOI 10.1111/j.1438-8677.2009.00201.x. [Google Scholar] [CrossRef]
11. Wang, W. Y., Sun, Y. D., Han, L. B., Su, L., Xia, G. X. et al. (2017). Overexpression of GhPFN2 enhances protection against Verticillium dahliae invasion in cotton. Science China Life Sciences, 6(8), 861–867. DOI 10.1007/s11427-017-9067-2. [Google Scholar] [CrossRef]
12. Henty-Ridilla, J. L., Shimono, M., Li, J., Chang, J. H., Day, B. et al. (2013). The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathogens, 9(4), e1003290. DOI 10.1371/journal.ppat.1003290. [Google Scholar] [CrossRef]
13. Underwood, W., Somerville, S. C. (2008). Focal accumulation of defences at sites of fungal pathogen attack. Journal of Experimental Botany, 59(13), 3501–3508. DOI 10.1093/jxb/ern205. [Google Scholar] [CrossRef]
14. Ge, D. D., Pan, T., Zhang, P. D., Wang, L. J., Zhang, J. et al. (2021). GhVLN4 is involved in multiple stress responses and required for resistance to Verticillium wilt. Plant Science, 302(49), 110629. DOI 10.1016/j.plantsci.2020.110629. [Google Scholar] [CrossRef]
15. Lv, F. N., Han, M. Y., Ge, D. D., Dong, H., Zhang, X. T. et al. (2017). GhVLN4 is involved in cell elongation via regulation of actin organization. Planta, 246(4), 687–700. DOI 10.1007/s00425-017-2723-7. [Google Scholar] [CrossRef]
16. Tominaga, M., Yokota, E., Vidali, L., Sonobe, S., Hepler, P. K. et al. (2000). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta, 210(5), 836–843. DOI 10.1007/s004250050687. [Google Scholar] [CrossRef]
17. Wu, S. Y., Xie, Y. R., Zhang, J. J., Ren, Y. L., Zhang, X. et al. (2015). VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. The Plant Cell, 27(3), 2829–2845. DOI 10.1105/tpc.114.134668. [Google Scholar] [CrossRef]
18. van der Honing, H. S.,Kieft, H., Emons, A. M., Ketelaar, T. (2012). Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiology, 158(3), 1426–1438. DOI 10.1104/pp.111.192385. [Google Scholar] [CrossRef]
19. Zhang, Y., Xiao, Y. Y., Du, F., Cao, L. J., Dong, H. J. et al. (2011). Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytologist, 190(3), 667–682. DOI 10.1111/j.1469-8137.2010.03632.x. [Google Scholar] [CrossRef]
20. Zhang, H., Qu, X. L., Bao, C. C., Khurana, P., Wang, Q. N. et al. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. The Plant Cell, 22(8), 2749–2767. DOI 10.1105/tpc.110.076257. [Google Scholar] [CrossRef]
21. Khurana, P., Henty, J. L., Huang, S., Staiger, A. M., Blanchoin, L. et al. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. The Plant Cell, 22(8), 2727–2748. DOI 10.1105/tpc.110.076240. [Google Scholar] [CrossRef]
22. Thomas, C., Hoffmann, C., Dieterle, M., van Troys, M., Ampe, C. et al. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. The Plant Cell, 18(9), 2194–2206. DOI 10.1105/tpc.106.040956. [Google Scholar] [CrossRef]
23. Huang, S. J., Qu, X. L., Zhang, R. H. (2015). Plant villins, versatile actin regulatory proteins. Journal of Integrative Plant Biology, 57(1), 40–49. DOI 10.1111/jipb.12293. [Google Scholar] [CrossRef]
24. Huang, S. J., Robinson, R. C., Gao, L. Y., Matsumoto, T., Brunet, A. et al. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. The Plant Cell, 17(2), 486–501. DOI 10.1105/tpc.104.028555. [Google Scholar] [CrossRef]
25. Bao, C. C., Wang, J., Zhang, R. H., Zhang, B. C., Zhang, H. et al. (2012). Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. The Plant Journal, 71(6), 962–975. DOI 10.1111/j.1365-313X.2012.05044.x. [Google Scholar] [CrossRef]
26. Zhang, T. Z., Hu, Y., Jiang, W. K., Fang, L., Guan, X. Y. et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology, 33(5), 531–537. DOI 10.1038/nbt.3207. [Google Scholar] [CrossRef]
27. Li, F. G., Fan, G. Y., Lu, C. R., Xiao, G. H., Zou, C. S. et al. (2015). Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nature Biotechnology, 33(5), 524–530. DOI 10.1038/nbt.3208. [Google Scholar] [CrossRef]
28. El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A. et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Research, 47(D1), D427–D432. DOI 10.1093/nar/gky995. [Google Scholar] [CrossRef]
29. Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Computational Biology, 7(10), e1002195. DOI 10.1371/journal.pcbi.1002195. [Google Scholar] [CrossRef]
30. Wang, J. R., Sung, W. K., Krishnan, A., Li, K. B. (2005). Protein subcellular localization prediction for gram-negative bacteria using amino acid subalphabets and a combination of multiple support vector machines. BMC Bioinformatics, 6(1), 174. DOI 10.1186/1471-2105-6-174. [Google Scholar] [CrossRef]
31. Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T. et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research, 47(W1), W636–W641. DOI 10.1093/nar/gkz268. [Google Scholar] [CrossRef]
32. Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A. et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics, 23(21), 2947–2948. DOI 10.1093/bioinformatics/btm404. [Google Scholar] [CrossRef]
33. Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. DOI 10.1093/molbev/msw054. [Google Scholar] [CrossRef]
34. Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J. C. et al. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics, 31(8), 1296–1297. DOI 10.1093/bioinformatics/btu817. [Google Scholar] [CrossRef]
35. Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME, discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research, 34(suppl_2), W369–W373. DOI 10.1093/nar/gkl198. [Google Scholar] [CrossRef]
36. Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y. et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30(1), 325–327. DOI 10.1093/nar/30.1.325. [Google Scholar] [CrossRef]
37. Hu, Y., Chen, J. D., Fang, L., Zhang, Z. Y., Ma, W. et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nature Genetics, 51(4), 739–748. DOI 10.1038/s41588-019-0371-5. [Google Scholar] [CrossRef]
38. Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. DOI 10.1016/j.molp.2020.06.009. [Google Scholar] [CrossRef]
39. Yokota, E., Vidali, L., Tominaga, M., Tahara, H., Orii, H. et al. (2003). Plant 115-kDa actin-filament bundling protein, P-115-ABP, is a homologue of plant villin and is widely distributed in cells. Plant & Cell Physiology, 44(10), 1088–1099. DOI 10.1093/pcp/pcg132. [Google Scholar] [CrossRef]
40. Yokota, E., Tominaga, M., Mabuchi, I., Tsuji, Y., Staiger, C. J. et al. (2005). Plant villin, lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant & Cell Physiology, 46(10), 1690–1703. DOI 10.1093/pcp/pci185. [Google Scholar] [CrossRef]
41. Du, F., Zhang, Y., Ren, H. Y. (2011). The universal bundling activity of AtVLN4 in diffusely growing cells. Plant Signaling & Behavior, 6(9), 1290–1293. DOI 10.4161/psb.6.9.16501. [Google Scholar] [CrossRef]
42. Zou, M. X., Ren, H. Y., Li, J. J. (2019). An auxin transport inhibitor targets villin-mediated actin dynamics to regulate polar auxin transport. Plant Physiology, 181(1), 161–178. DOI 10.1104/pp.19.00064. [Google Scholar] [CrossRef]
43. Lu, L., Wu, G. W., Xu, X. M., Luan, H. X., Zhi, H. J. et al. (2015). Soybean actin-depolymerizing factor 2 interacts with Soybean mosaic virus-encoded P3 protein. Virus Genes, 50(2), 333–339. DOI 10.1007/s11262-014-1150-0. [Google Scholar] [CrossRef]
44. Klahre, U., Friederich, E., Kost, B., Louvard, D., Chua, N. H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiology, 122(1), 35–48. DOI 10.1104/pp.122.1.35. [Google Scholar] [CrossRef]
45. Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science, 290(5494), 1151–1155. DOI 10.1126/science.290.5494.1151. [Google Scholar] [CrossRef]
46. George, S. P., Wang, Y. H., Mathew, S., Srinivasan, K., Khurana, S. (2007). Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. The Journal of Biological Chemistry, 282(36), 26528–26541. DOI 10.1074/jbc.M703617200. [Google Scholar] [CrossRef]
47. Nakayasu, T., Yokota, E., Shimmen, T. (1998). Purification of an actin-binding protein composed of 115-kDa polypeptide from pollen tubes of lily. Biochemical and Biophysical Research Communications, 249(1), 61–65. DOI 10.1006/bbrc.1998.9088. [Google Scholar] [CrossRef]
48. Yokota, E., Takahara, K. I., Shimmen, T. (1998). Actin-bundling protein isolated from pollen tubes of lily. Biochemical and immunocytochemical characterization. Plant Physiology, 116(4), 1421–1429. DOI 10.1104/pp.116.4.1421. [Google Scholar] [CrossRef]
49. Bilas, R., Szafran, K., Hnatuszko-Konka, K., Kononowicz, A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue and Organ Cultur, 127(2), 269–287. DOI 10.1007/s11240-016-1057-7. [Google Scholar] [CrossRef]
50. Chen, H. H., Lai, L. Y., Li, L. X., Liu, L. P., Jakada, B. H. et al. (2020). AcoMYB4, an Ananas comosus L. MYB transcription factor, functions in osmotic stress through negative regulation of ABA signaling. International Journal of Molecular Sciences, 21(16), 5727. DOI 10.3390/ijms21165727. [Google Scholar] [CrossRef]
51. Zhao, Y. Y., Yang, Z. E., Ding, Y. P., Liu, L. S., Han, X. et al. (2019). Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Science: An International Journal of Experimental Plant Biology, 286, 28–36. DOI 10.1016/j.plantsci.2019.05.021. [Google Scholar] [CrossRef]
52. Li, M., Lin, L., Zhang, Y. H., Sui, N. (2019). ZmMYB31, a R2R3-MYB transcription factor in maize, positively regulates the expression of CBF genes and enhances resistance to chilling and oxidative stress. Molecular Biology Reports, 46(4), 3937–3944. DOI 10.1007/s11033-019-04840-5. [Google Scholar] [CrossRef]
53. Akhter, D., Qin, R., Nath, U. K., Eshag, J., Jin, X. L. et al. (2019). A rice gene, OsPL, encoding a MYB family transcription factor confers anthocyanin synthesis, heat stress response and hormonal signaling. Gene, 699, 62–72. DOI 10.1016/j.gene.2019.03.013. [Google Scholar] [CrossRef]
54. Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. The FEBS Journal, 287(12), 2560–2576. DOI 10.1111/febs.15157. [Google Scholar] [CrossRef]
55. Nakata, M., Mitsuda, N., Herde, M., Koo, A. J., Moreno, J. E. et al. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. The Plant Cell, 25(5), 1641–1656. DOI 10.1105/tpc.113.111112. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |