
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016154
ARTICLE
Identification, Isolation and Characterization of GaCyPI Gene in Gossypium arboreum under Cotton Leaf Curl Virus Disease Stress
1Centre of Excellence in Molecular Biology, University of the Punjab, Lahore, 53700, Pakistan
2Department of Biological Sciences, Faculty of Fisheries & Wild Life, University of Veterinary and Animal Sciences-Ravi Campus, Pattoki, 55300, Pakistan
*Corresponding Author: Bushra Rashid. Email: bushra.cemb@pu.edu.pk
Received: 10 February 2021; Accepted: 14 April 2021
Abstract: Pakistan is facing the threat of Cotton Leaf Curl Virus (CLCuV) which is transmitted through whitefly to cotton crop. Molecular mechanism of leaf epicuticular wax protects the plants from different pathogens including insect attack and disease transmission. Objective of current study is the isolation and characterization of a wax related gene GaCyPI from Gossypium arboreum under CLCuV infection. A fragment of 475 bp was isolated from the total RNA and 3’ and 5’ RACE-PCR products were arranged by overlapping the extended sequences at both the ends. Deduced protein sequence of GaCyPI showed homology with Cyclophilin cis-trans isomerase gene of Gossypium ramondii and Gossypium barbadanse. Multiple sequence alignment also revealed homology among the coding sequences of same gene. GaCyPI protein comprised of 173 amino acids and ORF finder revealed the 69 bases upstream at 5’ while 350 bp at 3’UTR. InterProScan revealed that it belongs to Cyclophilin-type peptidyl prolyl cis/trans isomerase (PPIase) family. Active sites are visible at specific amino acid positions and 3D structure was stable in Ramachandran plot. Prosa server showed protein residues have average 3D-1D score >= 0.2 and Z-Score was −6.74. Phylogenetic analysis revealed that G. raimondii is the closest species that shares the same sequence. Hence, GaCyPI has strong role in plants’ epicuticular wax and its genetic transformation may protect the cotton from whitefly which transmits CLCuV.
Keywords: Cotton; plant wax; virus transmission; cyclophilin gene family; expressed sequence tag
Cotton, the backbone of Pakistan’s economy, is under the threat of Cotton Leaf Curl Virus transmitted by whitefly. This is the cause of Cotton leaf curl disease (CLCuD) which is one of the main limitations to cotton yield since the past two decades. The most distinguishing symptoms of CLCuD are veins darkening and thickening, upward or downward leaf curling, enation and stunted plant growth. This results in poor boll formation and ultimately leads to poor yield and fiber quality [1,2]. Gossypium hirsutum, the American cotton/Mexican cotton, is the most extensively cultivated species because of its yield and fiber quality. It contributes overall 90% of world’s cultivated cotton [3,4]. Gossypium arboreum also known as tree cotton is native to the old world [5] and contributes less than 1% of world’s total cotton production [4,6]. Although its yield is low but it has many agronomical and stress tolerance related characteristics such as high fiber strength, insect pest, virus and disease resistance as well as requires less inputs for its growth [7,8]. These characteristics reveals it as stress related gene pool against biotic and abiotic stresses [9,10] which makes it suitable for the study of stress related genes [11].
Plants have developed various defense mechanism or physical barriers that protect them from various biotic and abiotic stresses [12]. Epicuticular wax is also a similar defense barrier that protects the plants from the insects, pathogens and other environmental factors [13]. Leaf epicuticular wax acts as a first line of defense in plants and protect them from non-stomatal destructive water loss, ultraviolet (UV) radiations and microbial attack such as viruses, bacteria and fungi [14,15].
Differential display (dd) PCR is a sensitive technique that is employed to access the changes in gene expression at mRNA level. Basic approach of dd-PCR is to use multiple combinations of different primers to generate population of products that are further analyzed on gel [16–18]. The comparison of amplified products in test and control samples results in the identification of differentially expressed mRNA [19].
Cyclophilin proteins are found to be involved in diverse cellular activities and responses. These are among the most ancient and conserved proteins present in both animals and plants. These are also among the three subfamilies of immunophillins superfamily that trigger the organism’s metabolism to response in different ways under biotic and abiotic stresses [20]. A number of cyclophilin proteins have been identified in plants like 31 in Arabidopsis thaliana, 29 in Oryzae sativa, 62 in Glycine max and 94 in Brassica napus [21]. The mutation studies of various cyclophilin in Arabidopsis and rice revealed the loss or over expression of a particular function [22,23]. Moreover, cyclophilin also act as host restriction factors to inhibit the replication of viruses [24]. The current study is carried out to isolate and characterize the GaCyPI gene from local variety of Gossypium arboreum by differential display under the infection of CLCuV. Preliminary studies of the identified gene will lead to the insights into its structure and protein expression and will be helpful to correlate the role of this gene and CLCuV transmission through whitefly in cotton plants.
2.1 Seed Source, Growing of Plants and Experimental Design
Seeds of 33 local varieties of Gossypium were grown in greenhouse and field conditions. Among those 33, seeds of 30 CLCuV resistant varieties; (2 of G. arboreum and 28 of G. hirsutum) were obtained from Central Cotton Research Institute (CCRI) Multan Pakistan. Seeds of two highly susceptible varieties of G. hirsutum (S-12 and NIAB-78) were obtained from Department of Plant Breeding & Genetics, University of Agriculture Faisalabad, Pakistan. Seeds of CA-12, i.e., CLCuV resistant variety of G. hirsutum were obtained from Centre of Excellence in Molecular Biology (CEMB), University of the Punjab, Lahore Pakistan (Tab. 1). The behavior of varieties for CLCuD infection was observed for two years.

The seeds were sown in soil pots of 30 cm diameter in three replications in greenhouse whereas; the seeds were sown on ridges in the field. Viral inoculum (CLCuV) was maintained in both field and green house through naturally infected plants of S-12 and NIAB-78 by placing them along with the other genotypes for the transmission of CLCuV to the healthy plants. On an average, four whiteflies per leaf were infested to transmit the infection under greenhouse conditions. In field, natural infestation of whiteflies was observed. Normal agronomical practices such as irrigation and fertilizers (NPK) were applied to plants of both the groups and no pesticide was sprayed to maintain the whitefly infection and viral inoculum pressure. The data for the presence of whitefly and CLCuV disease symptoms was recorded daily in the early morning (after sunrise), noon (mid-day) and evening (just before sunset).
2.2 Selection of Stress Responsive Gene from Data Bank
A total 16 stress related ESTs accession number (GE654038, GE653432, GE654065, GE654072, GE653455, GE654052, GE654076, GE654064, GE654066, GE653951, GE653633, GE653410, GE653418, GE653776, GE653998 and GE653487) were selected from the literature (previous work of lab and NCBI data base (https://www.ncbi.nlm.nih.gov) (These ESTs were found effected (upregulated) in microarray analysis of wax mutants [25]. Degenerate arbitrary (stress genes sequence specific) and anchoring primers were used listed in Tab. 2.

2.3 Variety Selection, Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from fresh leaves of 2 month old plants of G. arboreum RAVI (control variety) and G. hirsutum S-12 (highly susceptible variety) as described by Muoki et al. [26]. Then total RNA was purified by using GeneJET Plant RNA Purification Mini Kit (Thermo Scientific cat #K0802 Lithuania EU). RNA integrity was visualized on 1% agarose gel and its quality and concentration was observed by nanodrop (version ND-1000 V3.3.0 USA). 1 μg of total RNA was taken for reverse transcription (cDNA synthesis) by using oligo (dT)18 primer of Thermo Scientific Revert-Aid First Strand cDNA Synthesis Kit (#K1621) according to manufacturer protocol.
Differential display PCR was performed according to the method of Liang et al. [27] with some modifications. Target specific and anchored primers were used to amplify the differentially displayed products from RAVI and S-12. PCR reaction mixture (25 μl) contained 500 ng of cDNA; 20 μM of forward (ESTs specific/target specific) and reverse primers (anchoring), 2.5 μl of 10X PCR buffer, 2 μl of 25 mM dNTPs, 2 units of Taq DNA polymerase. The cycling program was: initial denaturation at 94°C for 3 min, annealing at 40°C–55°C for up to 40 sec (according to primer) and final extension at 72°C for 40 sec for 30 cycles. Multiple combinations of primers were adopted. The amplified products were confirmed by using same primer pair to reduce false positive amplification. The differentially expressed transcript (DET2) was further characterized.
2.5 Sequence Analysis and Verification of Transcript
The transcript DET2 was TA cloned in PCR 2.1 vector (Invitrogen, Cat # K 4500–01) for sequencing. The ligation mixture for cloning contained 50 ng of vector, 1 μL of 10X ligation buffer, 1 μL of ligase and 25 ng of transcript template DNA in sterile water making the final volume up to 10 μL and incubated at 14°C overnight. The ligated product was transformed to competent cells of E.coli strain DH5α by heat shock method as described by Inoue et al. [28]. The transformants were screened on LB agar plates supplemented with Kanamycin (25 μg/mL) and tetracycline (12.5 μg/mL) at 37°C after overnight growth. The plasmid DNA was isolated from single colonies grown overnight by using GeneJET Plasmid Miniprep Kit (#K0503). The positive clones were confirmed through PCR by using same primers (A1B3) and restriction digestion with EcoR1. The confirmed positive clones were sequenced by Sanger’s sequencing by using M13 primers in 10 μL reaction mixture containing 2 μL DNA (100 ng); 1 μL of forward/reverse 10 μM M13 primers, 2 μL of 5X sequencing buffer, 1 μL of BigDyeTM and 4 μL deionized water. Thermal cycling conditions used for sequencing PCR were: an initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 94°C for 15 s, annealing at 52°C for 25 s, extension at 60°C for 4 min and a final extension at 60°C for 4 min. The sequencing product was precipitated by adding 2 μL of 3 M sodium acetate, 2 μL of 125 mM EDTA and 25 μL of absolute ethanol. The contents were vortexed briefly, spun shortly and incubated at room temperature for 20 min. The samples were then centrifuged at 3000 xg for 20 min at room temperature followed by the washing with 30 μL of 75% ethanol and air dried the pellet. Then the pellet was re-suspended in 13 μL of sequencing grade formamide, vortexed and then spun-out shortly. The samples were shifted to 96-well plate and denatured at 95°C for 5 min and quick chilled by placing in ice for 2 min before loading onto ABI PRISM 3100 genetic analyzer. Sequence was analyzed by using Chromas software version (v 1.45) and was submitted to NCBI GenBank. Once the sequence was retrieved, sequence specific primers were designed by using primers 3 database (http://bioinfo.ut.ee/primer3-0.4.0/) listed in Tab. 2.
2.6 Rapid Amplification of cDNA Ends (RACE)
The sequenced transcript DET2 was subjected to RACE PCR to find out the 5’ upstream and 3’downstream nucleotide bases by designing specific primers according to sequence information and parameters described in the protocol (Invitrogen GeneRacer RACE Ready cDNA kit version B101810, 25–0530). 1 μL of 3’ and 5’ GeneRacer cDNA was amplified by using 10x high fidelity buffer (10 mM), high fidelity platinum Taq DNA polymerase (5 U/μL), 50 mM MgSO4, 10 μM of each reverse and forward primers (Tab. 2) in sterile water. Touchdown PCR cycling parameters were used for RACE product amplification as described by Don et al. [29]. Amplification at 3’-end of 3’ cDNA was carried out by a forward (upstream) gene specific primer (GSP) GRF2 and a (reverse) downstream primer (GeneRacer 3’ primer). For 3’ RACE initial denaturation was conducted at 94°C for 2 min. Step 2 consisted of denaturation at 94°C for 30 s and annealing/extension at 72°C for 1 min. Step 3 consisted of denaturation at 94°C for 30 s and annealing/extension at 70°C for 1 min. 5 cycles of both the step 2 and 3 were followed by step 3 consisted of denaturation at 94°C for 30 s and final annealing at 65°C for 40 s and extension at 72°C for 1 min. After 10 cycles of annealing at 65°C, the temperature was decreased by 0.5°C after every 3 subsequent cycles, until 60°C was reached. The additional 30 cycles of annealing at 65°C were also performed. To perform 5’-end amplification, 5’cDNA was amplified by using an upstream GeneRacer 5’primer (Forward) and a downstream GSP (reverse primer) GRR2. The annealing temperature for 5’ RACE product was initially kept 67°C for 40 s and after 10 cycles, the temperature was decreased by 0.5°C after every 3 subsequent cycles, until 60°C was reached. All other steps were kept same as mentioned above for 3’ RACE PCR.
The 3’ and 5’ RACE PCR amplified products were resolved on 0.8% and 1.5% agarose gel, respectively, excised, eluted by using GeneJet Gel Extraction Kit (#K0691) and sequenced. A full length sequence was retrieved by joining 5’ and 3’ RACE sequenced fragments. Primers for full length gene specific sequence and coding sequence were also designed by primer 3’ software (Tab. 2). The full sequence of gene was amplified by PCR at 55°C annealing temperature while all other conditions were kept same as discussed above in Section 2.4. Different bioinformatics tools were applied to characterize the amplified gene as mentioned below.
The full length amplified nucleotide sequence (DET2) of Gossypium arboreum stress related transcript was submitted to ITesser server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) for homology modeling of identified gene with relevant protein. Sequence of the amplified transcript was analyzed for nucleotide and protein homology by using BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) as described by Altschul et al. [30]. The homology analyses for conserved sequences were obtained from NCBI and aligned by ClustalW T-coffee tool (https://www.ebi.ac.uk/Tools/msa/tcoffee/). Translation of the sequence was accomplished by using an open reading frame finder program (https://www.ncbi.nlm.nih.gov/orffinder/).
2.8 Molecular Weight, Stability Index and Localization of Full Length DET2 Protein
Subcellular localization and molecular weight of the transcribed protein was determined via plant-mploc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) and Expasy ProtParam server (https://web.expasy.org/cgi-bin/protparam/protparam), respectively. Protein super family and domains’ analysis was carried out by interPro scan (https://www.ebi.ac.uk/interpro/). PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) was used to determine the secondary structure of the sequence and its components with default parameters.
2.9 Structure and Quality of Protein and Gene Ontology
The proSa-web server (https://prosa.services.came.sbg.ac.at/prosa.php) was used to access the insight of protein structure of the transcript under study. Overall and local model quality was determined by calculating the Z score. Validation of 3D structure was done by verifying 3D, by plotting Ramachandran plot (http://services.mbi.ucla.edu/SAVES/Ramachandran) and calculation assessed by a versatile protein structure analysis PROCHECK statistics. Ramachandran plot analysis was used to draw between Phi (φ) and Psi (ψ) torsion angles of amino acids of protein to validate GaCyPI protein structure. Gene ontology (biological and molecular functions and cellular location) was determined by protein predict (https://www.predictprotein.org) and PSIPRED (http://bioinf.cs.ucl.ac.uk/).
Phylogenetic analysis was performed with the sequences having homology with the under study transcript through the sequences publicly accessible on NCBI. CLC sequence viewer software package 8.0.0 was used to generate a neighbor-joining phylogenetic tree as reported by Gregoretti et al. [31]. Bootstrap values from 1050 replicates were generated to verify the relative support level for tree topology.
2.11 Real Time Expression Analysis of GaCyPI Gene in Selected Cotton Varieties
The relative expression of GaCyPI in both resistant (RAVI) and susceptible (S-12) selected cotton varieties was observed by real time PCR studies. The RNA was extracted from leaf tissues of both the varieties, before and after the exposure of plants to CLCuV(2 months) and cDNA was synthesized. The primers for real time PCR were designed by using Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/) with expected product size of 120 bp. The sequence of primers is mentioned in Tab. 2. The cDNA was amplified by using SYBER green PCR master mix (Fermentas) according to manufacturer’s instructions. The reaction was carried out in ABI 7500 real time PCR machine (Applied Biosystem, USA). 1 μL of cDNA was amplified by using 0.2 μL of each forward (RGr-F) and reverse (RGr-R) primers (Tab. 2) with 7.5 μL of syber green master mix, making the volume 15 μL by adding up 6.1 μL of water. The cycling program was: initial denaturation at 94°C for 10 min, then 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and final extension at 72°C for 30 s. Primers of GAPDH sequence was selected as internal control for data normalization as mentioned in Tab. 2. The data was analyzed in GraphPad Prism 7 by plotting relative fold change expression.
3.1 Selection of Plants and Wax Related Transcript Identified from Gossypium arboreum
The highly susceptible G. hirsutum variety S-12 plants showed the symptoms of disease like thickening of veins, upward and downward curling of leaves with a large number of whiteflies on upper and lower surface of leaf while CLCuV resistant control plants (G. arboreum RAVI and FDH-170) did not show any symptoms of disease and none of the whiteflies were observed on lower or upper surface of leaves (Fig. 1). The selected varieties showing resistant and susceptible behavior against CLCuV are discussed here while the data of remaining varieties against CLCuV disease is already published by our group [32].

Figure 1: Leaves from control (RAVI) and stressed (S-12) cotton plants. (a) CLCuV symptoms: downward curling of leaf, thickening and darkening of veins and enation are visible. (b) Healthy leaves of Gossypium arboreum (RAVI)
The combination of primers in dd-PCR produced 6 DETs (Fig. 2) and DET2 was further characterized.

Figure 2: Amplification of DETs by DD-PCR. The amplified transcripts are tagged as DETs 1–6. The odd sample number represents healthy plant samples (G. arboreum) while even number are susceptible plant (G. hirsutum) samples. Lane L: 50 bp ladder, Lane 1: amplification of DET1(200+ bp). Lane 3: amplification of DET2 and DET3 (around 475 and 150 bp respectively, Lane 5: amplification of DET4 (around 300 bp), Lane 6: amplification of DET5 and 6 (around 400 and 200+ bp)
3.2 Identification and Amplification of Wax Related Transcript through Rapid Amplification of cDNA Ends (RACE)
Initially DET2 amplified product was of 475 bp in RAVI G. arboreum (Fig. 3a) which was confirmed by PCR amplification and restriction digestion after TA cloning and generated a fragment of 475 bp in Figs. 3b and 3c, respectively. The cloned transcript was sequenced and submitted to NCBI with accession number KY368095. The transcript was extended to 804 bp after 3’ and 5’ RACE (Figs. 3d–3f).

Figure 3: Amplification of DET2 from G. arboreum, cloning confirmation by restriction digestion and PCR and Rapid Amplification of cDNA Ends (RACE) PCR of GaCyPI (a) Fragment amplification of DET1; Lane L: 1 kb ladder, Lane 1: Fragment around 475 bp. (b) Confirmation of positive clones by digestion; Lane L: 1 kb ladder, Lane 1: undigested vector, Lanes 2–4: digested vector with a fragment of 475 bp, Lane A: 100 bp ladder. (c) Confirmation of positive clones by PCR; Lanes 1–6: Fragment of 475 bp, Lane L: 1 kb ladder. (d) 3’ RACE; Lane L:100 bp ladder, Lane 1: Fragment amplification of around 350 bp at 3’ end. (e) 5’ RACE; Lane 1: Fragment amplification of around 150 bp at 5’ end, Lane L:50 bp ladder. (f) Complete RACE fragment; Lane 1: Amplification of 475 bp transcript (before RACE), Lane 2: Amplification of full length around 804 bp transcript with extended basis on both ends after 3’ and 5’ RACE, Lane L:1 kb ladder
The products were arranged by overlapping the bases to form a full length sequence (Fig. 4). The sequence was named as ‘Gossypium arboreum Cyclophilin cis trans Isomerase’ (GaCyPI) due to its homology with this gene family and complete sequence was submitted to NCBI GenBank BankIt 2323393 Seq1 MT199216 (waiting for accession number).

Figure 4: Schematic representation of complete gene sequence of GaCyPI. (a) Complete 5’ to 3’sequence of GaCyPI. (b) Coding region of GaCyPI with 5’and 3’ UTRs. Solid black bars indicate exons and green dashed lines indicated un-translated regions. Number of nucleotides is indicated in each UTR
3.3 Homology Analysis of Gossypium arboreum Cyclophilin Cis Trans Isomerase (GaCyPI) Protein
Analysis of ORF finder revealed that the fragment of 69 bases upstream to the start codon (ATG) correspond to 5’ UTR while 350 bp right after the stop codon correspond to 3’ UTR along with a poly A tail (Fig. 5a) and protein of GaCyPI is found to be comprised of 173 amino acids. The deduced protein sequence of GaCyPI was observed to have 100% and 99% homology with Cyclophilin cis trans isomerase gene of G. ramondii (XP_012459367.1) and G. barbadanse (KAB2035964.1), respectively. Multiple sequence alignment with related gene sequences from different organism revealed the good homology (up to 99%) among coding sequences of the same gene (Fig. 5b), while the nucleotide sequences of these related sequences showed gaps and differences in nucleotide basis upstream to start codon as well as after the stop codon.

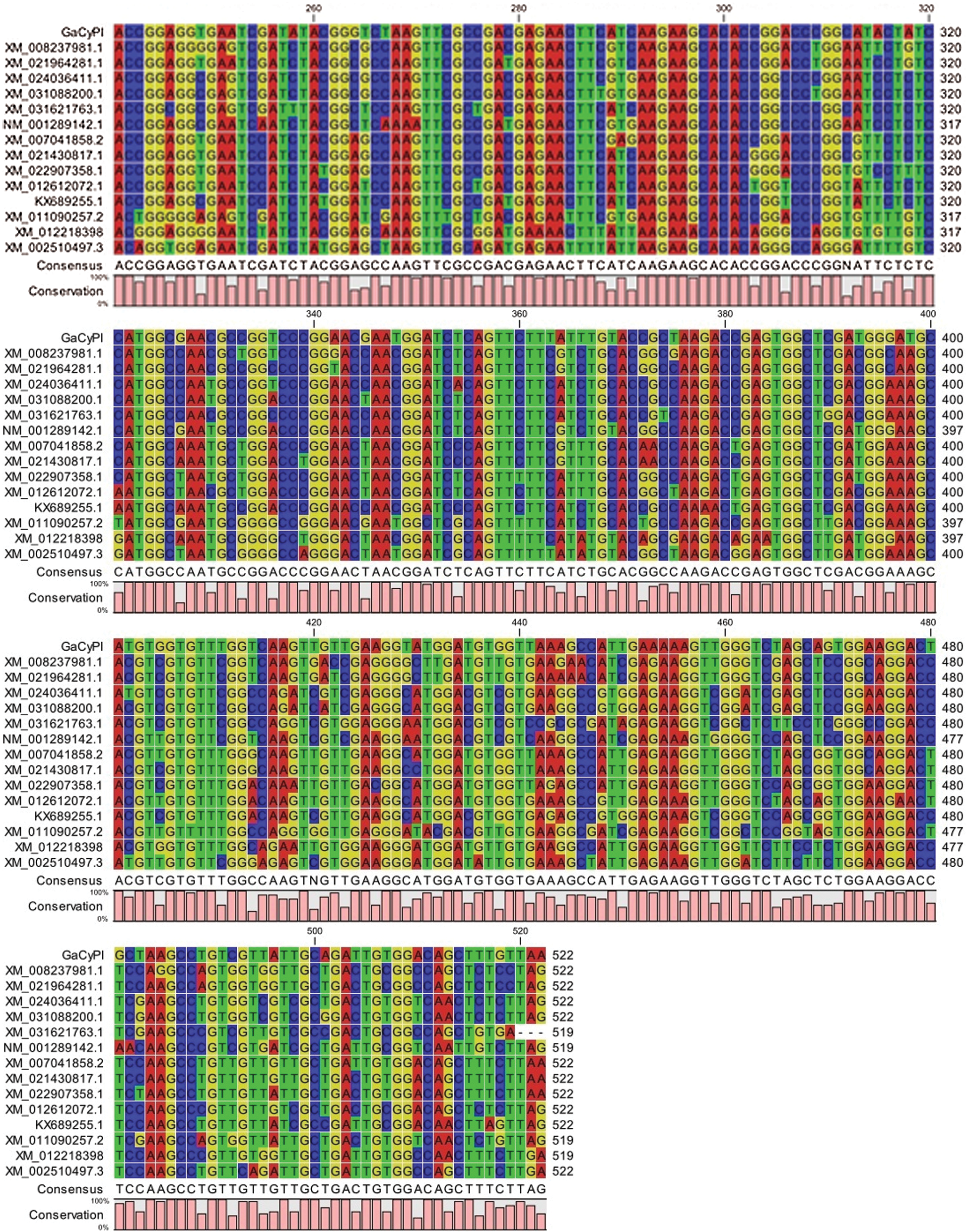
Figure 5: Schematic presentation of open reading frame of GaCyPI gene and multiple sequence alignment. (a) Horizontal bar ORF1 showing the full GaCyPI protein starting from first ATG at bases 70 to 590 producing a protein of 173 amino acids. (b) Multiple sequence alignment of coding sequence of GaCyPI with related genes
3.4 Annotation of GaCyPI Protein
GaCyPI protein was annotated by using InterProScan that revealed it belongs to cyclophilin-type peptidyl-prolyl cis-trans isomerase (Cyclophilin-type_PPIase) protein super family. A major cyclophilin-type peptidyl-prolyl cis-trans isomerase domain (IPR002130) is clearly detected in this protein. Other domains are PF00160 (Cyclophilin type peptidyl-prolyl cis trans isomerase/CLD also known as Pro_isomerase), PS50072 (Cyclophilin-type peptidyl-prolyl cis-trans isomerase domain profile, also known as CSA_PPIASE_2) and PR00153 (CSAPPISMRASE) (Fig. 6a). Six active sites are also found at position (amino acid) 62, 63, 68, 119, 121 and 129 (His-62, Arg-63, Phe-68, Gln-119, Phe-121, Trp-129) and the TM- score was found to be 0.942. Position of the specific amino acid with reference to structural elements is indicated in predicted secondary structure map obtained by PSIPRED (Fig. 6b).
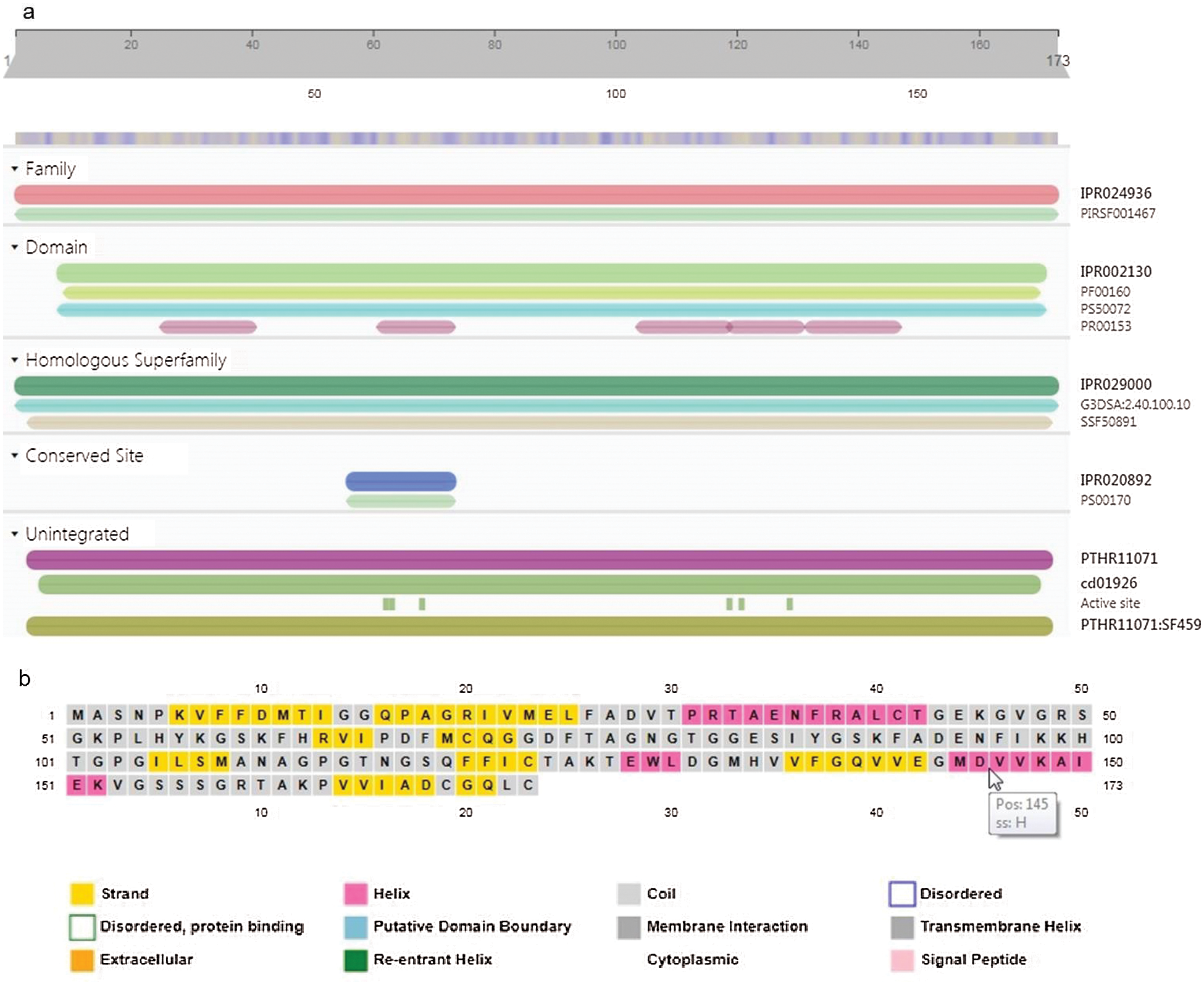
Figure 6: Inter Pro scan analysis for conserved domains and active sites and Sequence plot of secondary structure of GaCyPI protein by PSIPRED. (a) Inter Pro scan analysis detected the cyclophillin-type peptidyl-prolyl cis-trans isomerase conserved domain (IPR002130) in GaCyPI protein under IPR029000 homologous super family. (b) Secondary structure of GaCyPI protein showing helix, strand and coiled motifs in pink, yellow and light grey colors, respectively
3.5 3D Structure Prediction of GaCyPI Protein
The 3D structure of GaCyPI protein was predicted by I-TASSER modeling which made the use of templates of highest significance in multiple threading alignments identified from the PDB library (Fig. 7a). A PDB file was generated by I-TASSER for GaCyPI and a total of 10 templates that matched the target sequence were selected to predict the structure and function of protein. Among those, 5 models (5yb9A, 1e8kA, 2hqjA, 1ihgA1 and 2pluA) have been taken as the guide based on their similarity and identity to the target sequence. Active sites found in GaCyPI protein are also mentioned in Fig. 7b.

Figure 7: 3D structural model of GaCyPI protein predicted by Itesser and active sites in GaCyPI. (a) Predicted 3D Structure of protein (b) 3D structure of protein with active sites mentioned
3D structure of GaCyPI protein was found to be stable as observed by drawing the Ramachandran plot. The number of residues in favored and allowed region was observed to be 97.7%, while only 2.3% were located in outlier region (Fig. 8).
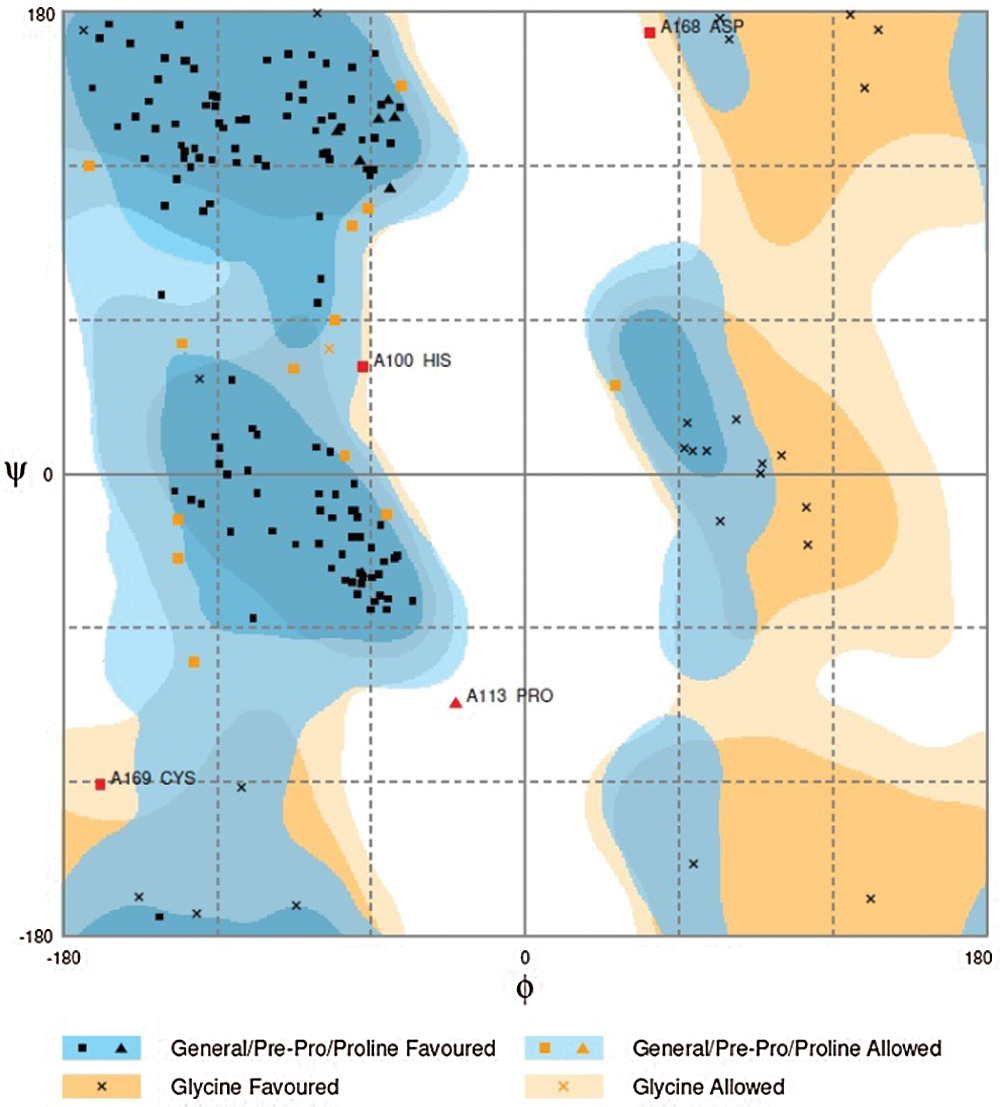
Figure 8: Ramachandran plot analysis of GaCyPI protein
Protein model evaluation was carried out by Prosa server and 100% of the residues have average 3D-1D score >= 0.2. At least 80% of the amino acids’ scored >= 0.2 in 3D/1D profile. Z-Score was found to be −6.74 that indicated the overall quality of protein. Z-Score of the structure was within the range of scores typically found for native proteins of similar size. Estimated value of RMSD of predicted protein structure (measure of the average distance between the atoms) was found to be 1.5 ± 1.4 Å (Figs. 9a and 9b).

Figure 9: Predicted local similarity of GaCYPI Protein. (a) Quality estimate of GaCYPI Protein, (b) Z-score indicated overall model quality of GaCyPI protein
3.6 Molecular Weight, Stability Index and Localization of GaCyPI Protein
The GaCyPI protein is predicted to be localized in cytoplasm (97%) and nucleus. The molecular weight was found to be 18285.92 and instability index of protein was 18.24. The aliphatic index was 68.80 which have been considered as a positive factor for the increased thermo-stability of globular proteins. Thus the analyses from different databases revealed that the GaCyPI protein is highly stable.
Phylogenetic tree has generated and compared the wax related sequences and determined the link in the evolution of GaCyPI related sequences (Fig. 10). On the basis of homology of GaCyPI, all selected species are divided into families, subfamilies and further to groups. The analysis revealed that the G. raimondii is the closest species that shares almost the same sequence of GaCyPI. Glycine max is found to be the most distant organism in terms of evolution of GaCyPI. Sequence gaps were considered as missing data.

Figure 10: Phylogenetic tree using CLC sequence viewer. A distance based neighborhood joining phylogenetic tree was created by aligning the most related sequences with GaCyPI sequence. A bootstrap analysis 1050 replicates was performed
3.8 Molecular and Biological Functions of GaCyPI
Prediction of molecular functions and biological processes by different functions’ prediction tools revealed that the GaCyPI gene is involved in various molecular and biological activities including protein binding, peptidyle-prolyl cis trans Isomerase activity, protein folding and transport, protein metabolic processes, regulation of protein expression and mRNA splicing (Tab. 3). Only functions with high reliability showing the major roles that can be helpful in understanding the further studies related to gene functionality are included in the table.

3.9 Expression Analysis of GaCyPI
The expression analysis of GaCyPI revealed the relative fold increase in expression of RAVI and S-12 after the exposure to CLCuV. The expression of GaCyPI was very low (0.29) in S-12 as compared to RAVI (6.78 fold) after the exposure to CLCuV. The expression of GaCyPI in control and susceptible varieties before the exposure to CLCuV is also shown in the Fig. 11 such as fold change is 1 in RAVI while the expression in S-12 was observed to be 0.07 fold.

Figure 11: Real time analysis of GaCyPI gene in RAVI and S-12 varieties. After the exposure to CLCuV infection, the expression of GaCyPI was upregulated under CLCuV stress in both varieties (grey bars) but the relative expression was much higher in RAVI (6.78) as compared to S-12 (0.29) while the fold change was 1 in control group
Cotton is the backbone of Pakistan’s economy but presently it is under stress of CLCuV which is transmitted through whitefly. Wax is a protective layer that plays an important role as first line of defense in plants against various biotic and abiotic stresses [33]. The study has been carried out to identify and characterize a wax related gene isolated from CLCuV stressed plants of local variety of G. arboreum (Ravi). Leaf epicuticular wax content is found to be linked with resistance towards CLCuV. A negative correlation between wax content and CLCuV is previously observed [32,34]. Difference in the response towards CLCuV is observed in G. arboreum varieties (RAVI and FDH-170) and G. hirsutum variety (S-12). In previously conducted experiments [32], the higher wax content (185 μg/cm2) was found in Ravi as compared to wax load 47 μg/cm2 in S-12 as reported in other studies [15,35]. As previously reported [15] that CLCuV resistant G. arboreum has higher leaf epicuticular wax as compared to G. hirsutum, makes the G. arboreum best candidate for the isolation of wax related genes.
The GaCyPI was amplified through dd-PCR by using target specific degenerate arbitrary and anchoring primers as described by [16,36] and RACE-PCR provided the extended sequences (at 3’ and 5’ ends) on either side of isolated transcript and the full length sequence was obtained. The in-silico analyses are always necessary to learn about the functionality and working of a protein, and to have the basic knowledge regarding its active sites, structure and nature. GaCyPI has homology with the cyclophilin cis-trans isomerase gene of G. raimondii. The homology analysis revealed the presence of gene in other higher organisms as well. A number of plant species were found to exhibit the same gene in their genome such as G. raimondii, Glycine max, Jatropha curcas etc. The GaCyPI has close resemblance (99%) with AT2g16600.1 found in Arabidopsis (arabidopsis.org) which is involved in cis-trans isomerization of proteins (NCBI GenBank accession No. NM_001335481). GaCyPI protein also has 95% homology with Cydophilin (ACT63839.1) of G. hirsutum which is a fiber preferential gene. In-silico analysis of GaCyPI revealed its localization in cytoplasm (97%). The structure and conformation analysis of GaCyPI protein is found to be stable by Ramachandran plot analysis. Most of the dihedral angles (89.5%) of GaCyPI protein were observed in favored (general) regions that validated its structure. In poor quality of protein structure, the dihedral angles are found in forbidden or non-favored regions of Ramachandran plot that indicate the problems in structure and conformation of protein structure [37]. The GaCyPI was found to be closely related to cyclophilin gene of G. raimondii in phylogenetic analysis. Analysis of molecular and biological functions by various tools also revealed that GaCyPI has role in stress related responses. Literature also provided the basis for its role in many biotic and abiotic stresses [38].
Cyclophilins (CyPs) are protein foldases found to be directly linked with various stress responses in higher organisms. The overexpression of cyclophilins have enhanced or induced the tolerance against various stresses including heat shock, low temperature, salt stress, light, wounding, chemical elicitors and pathogens [39]. The exact mechanism of cyclophilins, their physiological relevance and molecular basis is still unknown or not clear. However, their wide distribution in organisms and ubiquitous nature signifies their fundamental importance in plant’s survival under various stressful conditions [40].
GaCyPI sequence includes EST sequence (GE653410) that was found to be upregulated in wax deficient mutants as previously generated [25]. The G. arboreum Wax Mutant1 (GaWM1), GaWM2 and GaWM3 showed an altered wax pattern. The wax morphology was also changed from smooth strips to embedded tubules/fibers, irregular patches and non-stripy smooth layers respectively. In this study, the relative expression analysis of GaCyPI before and after the exposure to whitefly and transmission of CLCuV also showed the involvement of GaCyPI gene in plant response towards CLCuD. The expression of GaCyPI was increased in the cotton variety RAVI as compared to variety S-12. In response to various stresses, up-regulation of the cyclophilin gene expression has also been reported as [41–43]. Higher expression of potato CyPs mRNA was found in response to salicylic acid, P. infestans elicitor and P. infestans infection [44]. Overexpression studies of cyclophilin in diverse organisms also enhanced the tolerance towards viral infections [45]. Santos et al. [46] also described the role of cyclophilin in immune responses. Experiments proved a wide range of novel functions of cyclophilins but yet to clear to fully understand the unexplored mechanisms. It is described that cyclophilins present in diverse organisms act by assisting the folding and assembly of newly synthesized proteins [47]. These are involved in maturation and conformational modifications of different proteins by isomerization of peptidyl-prolyl bonds and also affect the protein trafficking. Moreover, the sequences of different forms of cyclophilins in different organisms are found to be conserved. All members of cyclophilin family share a 109-amino-acid-long cyclophilin-like domain that performs the PPIase activity, while additional unique sequences or domains are also there in cyclophilin protein that are important for the selection of particular protein substrates and for subcellular compartmentalization and other specialized functions [48]. These have been implicated in diverse cellular mechanisms such as signaling, transfer of reducing power [49] and preservation of protein structure [41].
This study provides a comprehensive knowledge about GaCyPI protein and presents a positive correlation for stress protection. Effect on the expression of this gene in wax mutants generated by Barozai et al. [25,35] provide a link of this gene with wax. But the underlying mechanism which is targeted by GaCyPI protein to be involved in stress protection either by affecting the wax load is in progress. Further studies of this gene will certainly be helpful to elucidate the mechanism of CLCuV resistance/tolerance and its transmission in cotton through whitefly as well as other economically important crops. It is a unique study of its type which will further provide the insight of environment related adaptations and genetic transformation of this gene in cotton will improve the wax load which would help to combat the environmental challenges and CLCuV related problems.
Acknowledgement: Authors are thankful to Central Cotton Research Institute (CCRI) Multan and Department of Plant Breeding & Genetics, University of Agriculture Faisalabad, Pakistan for provision of seeds of cotton varieties.
Availability of Data: All the data is available within the manuscript or the link for submission to NCBI repository has been mentioned in the text.
Funding Statement: Funds are provided by Higher Education Commission (HEC), Pakistan to Dr. Bushra Rashid; Principal Investigator of the Project No. 20-3692/NRPU/R&D/HEC/14 to complete this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Nawaz, B., Naeem, M., Malik, T. A., Muhae-Ud-Din, G., Ahmad, Q. et al. (2019). A review about cotton leaf curl viral disease and its control strategies in Pakistan. International Journal of Innovative Approaches in Agricultural Research, 3(1), 132–147. DOI 10.29329/ijiaar. [Google Scholar] [CrossRef]
2. Rahman, M., Khan, A. Q., Rahmat, Z., Iqbal, M. A., Zafar, Y. (2017). Genetics and genomics of cotton leaf curl disease, its viral causal agents and whitefly vector: A way forward to sustain cotton fiber security. Frontiers in Plant Science, 8, 1157. DOI 10.3389/fpls.2017.01157. [Google Scholar] [CrossRef]
3. Zhao, Y., Wang, H., Chen, W., Li, Y., Gong, H. et al. (2015). Genetic diversity and population structure of elite cotton (Gossypium hirsutum L.) germplasm revealed by SSR markers. Plant System and Evolution, 301(1), 327–336. DOI 10.1007/s00606-014-1075-z. [Google Scholar] [CrossRef]
4. Shim, J., Mangat, P. K., Angeles-Shim, R. B. (2018). Natural variation in wild gossypium species as a tool to broaden the genetic base of cultivated cotton. Journal of Plant Science Current Research, 2, 5. DOI 10.24966/PSCR-3743/100005. [Google Scholar] [CrossRef]
5. Viot, C. (2019). Domestication and varietal diversification of Old world cultivated cottons (Gossypium sp.) in the antiquity. Revue D’ethnoécologie, 15. DOI 10.4000/ethnoecologie. [Google Scholar] [CrossRef]
6. Li, R., Erpelding, J. E. (2016). Genetic diversity analysis of gossypium arboreum germplasm accessions using genotyping-by-sequencing. Genetica, 144, 535–545. DOI 10.1007/s10709-016-9921-2. [Google Scholar] [CrossRef]
7. Iqbal, M., Khan, M. A., Chattha, W. S., Abdullah, K., Majeed, A. (2019). Comparative evaluation of gossypium arboreum L. and gossypium hirsutum L. genotypes for drought tolerance. Plant Genetic Resources: Characterization and Utilization, 11(6), 1–8. DOI 10.1017/S1479262119000340. [Google Scholar] [CrossRef]
8. Mushtaq, R., Shahzad, K., Shah, Z. H., Alsamadany, H., Alzahrani, H. A. S. et al. (2020). Isolation of biotic stress resistance genes from cotton (Gossypium arboreum) and their analysis in model plant tobacco (Nicotiana tabacum) for resistance against cotton leaf curl disease complex. Journal of Virology Methods, 276, 113760. DOI 10.1016/j.jviromet.2019.113760. [Google Scholar] [CrossRef]
9. Naqvi, R. Z., Shan-e-Ali Zaidi, S., Akhtar, K. P., Strickler, S., Woldemariam, M. et al. (2017). Transcriptomics reveals multiple resistance mechanisms against cotton leaf curl disease in a naturally immune cotton species. gossypium arboreum. Scientific Reports, 7, 15880. DOI 10.1038/s41598-017-15963-9. [Google Scholar] [CrossRef]
10. Mushtaq, R., Shahzad, K., Shah, Z. H., Alsamadany, H., Alzahrani, H. A. S. et al. (2020). Isolation of biotic stress resistance genes from cotton (Gossypium arboreum) and their analysis in model plant tobacco (Nicotiana tabacum) for resistance against cotton leaf curl disease complex. Journal of Virological Methods, 276, 113760. DOI 10.1016/j.jviromet.2019.113760. [Google Scholar] [CrossRef]
11. Jia, Y., Pan, Z., He, S., Gong, W., Geng, X. et al. (2018). Genetic diversity and population structure of gossypium arboreum L. collected in China. Journal of Cotton Research, 1, 11. DOI 10.1186/s42397-018-0011-0. [Google Scholar] [CrossRef]
12. Abid Ullah, H., Hussain, A., Shaban, M., Khan, A. H., Alariqi, M. et al. (2018). Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiology and Biochemistry, 123, 149–159. DOI 10.1016/j.plaphy.2017.12.012. [Google Scholar] [CrossRef]
13. Xue, D., Zhang, X., Lu, X., Chen, G., Chen, Z. H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Frontiers in Plant Science, 8, 621. DOI 10.3389/fpls.2017.00621. [Google Scholar] [CrossRef]
14. Sharma, P., Kothari, S. L., Rathore, M., Gour, V. (2018). Properties, variations, roles, and potential applications of cuticular wax: A review. Turkish Journal of Botany, 42(2), 135–149. DOI 10.3906/bot-1702-25. [Google Scholar] [CrossRef]
15. Khan, M. A. U., Shahid, A. A., Rao, A. Q., Bajwa, K. S., Samiullah, T. R. et al. (2015). Molecular and biochemical characterization of cotton epicuticular wax in defense against cotton leaf curl disease. Iranian Journal of Biotechnology, 13(4), 1234–1242. DOI 10.15171/ijb.1234. [Google Scholar] [CrossRef]
16. Crawford, D. R., Kochheiser, J. C., Schools, G. P., Salmon, S. L., Davies, K. J. A. (2002). Differential display: A critical analysis. Gene Expression, 10, 101–107. [Google Scholar]
17. Saakre, M., Baburao, T. M., Salim, A. P., Ffancies, R. M., Achuthan, Y. P. et al. (2017). Identification and characterization of genes responsible for drought tolerance in rice mediated by pseudomonas fluorescens. Rice Science, 24(5), 291–298. DOI 10.1016/j.rsci.2017.04.005. [Google Scholar] [CrossRef]
18. Moustafa, M. F., Taha, T. H., Helal, M., Alrumman, S. A. (2016). Differential-display reverse transcription-pCR (DDRT-pCRA new technology for molecular detection and studying one of the antagonistic factors of bacillus endophyticus strain SA against staphylococcus aureus (MRSA). 3. Biotech, 6(2), 121. DOI 10.1007/s13205-016-0439-1. [Google Scholar] [CrossRef]
19. Abdelkhalek, A., Elmorsi, A., Alshehaby, O., Sanan-mishra, N., Hafez, E. (2018). Identification of genes differentially expressed in onion infected with iris yellow spot virus. Phytopathologia Mediterranea, 57(2), 334–340. DOI 10.14601/Phytopathol_Mediterr-21877. [Google Scholar] [CrossRef]
20. Singh, K., Winter, M., Zouhar, M., Rysanek, P. (2018). Cyclophilins: Less studied proteins with critical roles in pathogenesis. Phytopathology, 108, 6–14. DOI 10.1094/PHYTO-05-17-0167-RVW. [Google Scholar] [CrossRef]
21. Singh, H., Kaur, K., Singh, M., Kaur, G., Singh, P. (2020). Plant cyclophilins: Multifaceted proteins with versatile roles. Frontiers in Plant Science, 11, 585212. DOI 10.3389/fpls.2020.585212. [Google Scholar] [CrossRef]
22. Gochez, A. M., Shantharaj, D., Potnis, N., Zhou, X., Minsavage, G. V. et al. (2017). Molecular characterization of XopAG effector avrgf2 from xanthomonas fuscans ssp. Aurantifolii in Grapefruit Molecular Plant Pathology, 18(3), 405–419. DOI 10.1111/mpp.12408. [Google Scholar] [CrossRef]
23. Kong, G., Zhao, Y., Jing, M., Huang, J., Yang, J. et al. (2015). The activation of phytophthora effector avr3b by plant cyclophilin is required for the nudix hydrolase activity of avr3b. PLoS Pathogens, 11(8), e1005139. DOI 10.1371/journal.ppat.1005139. [Google Scholar] [CrossRef]
24. Kovalev, N., Nagy, P. D. (2013). Cyclophilin a binds to the viral RNA and replication proteins, resulting in inhibition of tombusviral replicase assembly. Journal of Virology, 87, 13330–13342. DOI 10.1128/JVI.02101-13. [Google Scholar] [CrossRef]
25. Barozai, M. Y. K., Husnain, T. (2012). Identification of biotic and abiotic stress up-regulated ESTs in gossypium arboreum. Molecular Biological Reports, 39, 1011–18. DOI 10.1007/s11033-011-0826-y. [Google Scholar] [CrossRef]
26. Muoki, R. C., Paul, A., Kumari, A., Singh, K., Kumar, S. (2012). An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. kuntze). Molecular Biotechnology, 52, 82–88. DOI 10.1007/s12033-011-9476-5. [Google Scholar] [CrossRef]
27. Liang, P., Pardee, A. B. (1992). Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. DOI 10.1126/science.1354393. [Google Scholar] [CrossRef]
28. Inoue, H., Nojima, H., Okayama, H. (1990). High efficiency transformation of escherichia coli with plasmids. Gene, 96(1), 23–28. DOI 10.1016/0378-1119(90)90336-P. [Google Scholar] [CrossRef]
29. Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K., Mattick, J. S. (1991). Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Research, 19(14), 4008. DOI 10.1093/nar/19.14.4008. [Google Scholar] [CrossRef]
30. Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. DOI 10.1016/S0022-2836(05)80360-2. [Google Scholar] [CrossRef]
31. Gregoretti, I., Lee, Y. M., Goodson, H. V. (2004). Molecular evolution of the histone deacetylase family. functional implications of phylogenetic analysis. Journal of Molecular Biology, 338, 17–31. DOI 10.1016/j.jmb.2004.02.006. [Google Scholar] [CrossRef]
32. Majid, M. U., Sher, Z., Rashid, B., Ali, Q., Sarwar, M. B. et al. (2020). Role of leaf epicuticular wax load and composition against whitefly population and cotton leaf curl virus in different cotton varieties. Cytology and Genetics, 54(5), 472–486. DOI 10.3103/S009545272005014X. [Google Scholar] [CrossRef]
33. Dhanyalakshmi, K. H., Soolanayakanahally, R. Y., Rahman, T., Tanino, K. T., Nataraja, K. N. (2019). Leaf cuticular wax, a trait for multiple stress resistance in crop plants. Abiotic Biotic Stress Plants, London UK, IntechOpen. [Google Scholar]
34. Muhammad, S., Song, X. L., Sun, X. Z., Muhammad, R. (2018). Leaf cuticular wax content is involved in cotton leaf curl virus disease resistance in cotton (Gossypium hirsutum L.). Spanish Journal of Agricultural Research, 16(4), 705–713. DOI 10.5424/sjar/2018164-13085. [Google Scholar] [CrossRef]
35. Barozai, M. Y. K., Husnain, T. (2014). Development and characterization of the asiatic desi cotton (Gossypium arboreum L.) leaf epicuticular wax mutants. Pakistan Journal of Botany, 46(2), 639–43. [Google Scholar]
36. Sturtevant, J. (2000). Applications of differential-display reverse transcription-pCR to molecular pathogenesis and medical mycology. Clinical Microbiology Reviews, 13(3), 408–427. DOI 10.1128/CMR.13.3.408. [Google Scholar] [CrossRef]
37. Wiltgen, M. (2019). Encyclopedia of bioinformatics and computational biology. Reference module in life sciences, Academic Press, USA. [Google Scholar]
38. Singh, K., Tzelepis, G., Zouhar, M., Ryšánek, P., Dixelius, C. (2018). The immunophilin repertoire of plasmodiophorabrassicae and functional analysis of PbCYP3 cyclophilin. Molecular Genetics Genomics, 293, 381–390. DOI 10.1007/s00438-017-1395-0. [Google Scholar] [CrossRef]
39. Chen, Q., Chen, Q. J., Sun, G. Q., Zheng, K., Yao, Z. P. et al. (2019). Genome-wide identification of cyclophilin gene family in cotton and expression analysis of the fibre development in gossypium barbadense. International Journal of Molecular Sciences, 20(2), 349. DOI 10.3390/ijms20020349. [Google Scholar] [CrossRef]
40. Kumari, S., Roy, S., Singh, P., Singla-Pareek, S., Pareek, A. (2013). Cyclophilins: Proteins in search of function. Plant Signaling and Behaviour, 8(1), e22734. DOI 10.4161/psb.22734. [Google Scholar] [CrossRef]
41. Ricarova, V., Kazda, J., Singh, K., Ryšánek, P. (2016). Club root caused by plasmodiophora brassicae Wor: A review of emerging serious disease of oilseed rape in the Czech Republic. Plant Protection Science, 52, 71–86. DOI 10.17221/87/2015-PPS. [Google Scholar] [CrossRef]
42. Wang, Q., Wang, Y., Chai, W., Song, N., Wang, J. et al. (2017). Systematic analysis of the maize cyclophilin gene family reveals ZmCYP15 involved in abiotic stress response. Plant Cell Tissue and Organ Culture, 128, 543–561. DOI 10.1007/s11240-016-1132-0. [Google Scholar] [CrossRef]
43. Yan, H., Zhou, B., He, W., Nie, Y., Li, Y. (2018). Expression characterisation of cyclophilin BrROC1 during light treatment and abiotic stresses response in brassica rapa subsp. rapa ‘Tsuda’. Functional Plant Biology, 45(12), 1223–1232. DOI 10.1071/FP18029. [Google Scholar] [CrossRef]
44. Dubery, I. A. (2007). An elicitor and pathogen-induced cDNA from potato encodes a stress-responsive cyclophilin. Biologia Plantarum, 51(2), 327–332. DOI 10.1007/s10535-007-0063-3. [Google Scholar] [CrossRef]
45. Mittapelly, P., Rajarapu, S. P. (2020). Applications of proteomic tools to study insect vector-plant virus interactions. Life, 10(8), 143. DOI 10.3390/life10080143. [Google Scholar] [CrossRef]
46. Santos, I. B. D., Park, S. W. (2019). Versatility of cyclophilins in plant growth and survival: A case study in arabidopsis. Biomolecules, 9(1), 20. DOI 10.3390/biom9010020. [Google Scholar] [CrossRef]
47. Zhu, C., Wang, Y., Li, Y., Bhatti, K. H., Tian, Y. et al. (2011). Overexpression of a cotton cyclophilin gene (ghcyp1) in transgenic tobacco plants confers dual tolerance to salt stress and pseudomonas syringae pv. Tabaci infection. Plant Physiology and Biochemistry, 49, 1264–1271. DOI 10.1016/j.plaphy.2011.09.001. [Google Scholar] [CrossRef]
48. Idris, M., Idris, M., Adeola, F., Sedzro, D. M. (2019). Cyclophilins: The structure and functions of an important peptidyl-prolyl isomerase. International Journal of Biochemistry, Biophysics & Molecular Biology, 4(1), 1–6. DOI 10.11648/j.ijbbmb.20190401.11. [Google Scholar] [CrossRef]
49. Kalinina, A. A., Khromykh, L. M., Kazansky, D. B. (2017). Cyclophilin A: Structure and functions. Advances in Molecular Oncology, 4(4), 17–23. DOI 10.17650/2313-805X-2017-4-4-17-23. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |