
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016362
ARTICLE
The BHLH Transcriptional Factor PIF4 Competes with the R2R3-MYB Transcriptional Factor MYB75 to Fine-Tune Seeds Germination under High Glucose Stress
Shanghai Key Laboratory of Bio-Energy Crops, School of Life Sciences, Shanghai University, Shanghai, 200444, China
*Corresponding Authors: Ping Li. Email: liping80@shu.edu.cn; Xiaojun Yuan. Email: yuanxiaojun@shu.edu.cn
Received: 01 March 2021; Accepted: 24 March 2021
Abstract: It is known that the high level of sugar including glucose suppresses seed germination through ABA signal. ABI5 is an essential component to mediate ABA-dependent seed germination inhibition, but underlying mechanism needs more investigation. Previous study demonstrated the PIF4 activated the expression of ABI5 to suppress seed germination in darkness. Here we reported that PIF4 also mediated the seed germination inhibition through ABI5 under high concentration of glucose treatment. Furthermore, we found that PIF4 interacted with PAP1, the central factor to control anthocyanin biosynthesis. Such interaction was confirmed in vitro and in planta. Biochemical and physiological analysis revealed that PAP1 bond the promoter of ABI5 to suppress its expression, thus enhanced seed germination under high concentration of glucose treatment. Specially, PAP1 competed with PIF4 to antagonize the activation of PIF4 on ABI5 expression, thus promoted seed germination under high glucose treatment. Given these, we uncover a novel role for PIF4 and PAP1 in controlling seed germination under high glucose treatment, and reveal their antagonistic mechanism by which coordinates ABI5 expression to control seed germination in response to the glucose signal.
Keywords: Seeds germination; glucose; MYB75; PIF4; ABI5
Sugar, including sucrose and glucose, not only act as the main product of photosynthesis to affect cellular metabolism, they serve also as signals to coordinate gene expression and enzyme activity in both source and sink tissue [1–3]. Glucose as one of the hexose hydrolytic products of sucrose is associated with various physiological processes, such as seed germination, cotyledon opening, leave greening, root growth and anthocyanin biosynthesis. It is reported that plant exists three glucose signal transduction pathways, the first one is the hexokinase 1 (AtHXK1)-dependent pathway, in which AtHXK1 as the glucose sensor to perceive glucose stimulation and then activate downstream gene expression, the secondary pathway is the glycolysis-dependent pathway that need AtHXK1-catalitic activity to activate the expression of pathogenesis-related genes [4,5]. The last one is the AtHXK1-independent pathway that requires cell wall Cytosolic invertase1 (CINV1) and CINV2 [1,6], genetic experiments also demonstrate that AtHXK1-independent glucose-sensing and signal process is also related to G-protein coupled receptor system in plant [7]. Some evidence reveals that high concentration of sugar induces the biosynthesis of anthocyanin, which can protect plants from excessive environmental stress, or defense from pathogen and herbivores [8,9]. Anthocyanin is biosynthesized from flavonoid, and catalyze by the enzyme encoded by early biosynthesis genes (EBGs) including chalcone synthase (CHS), chalcone isomerase (CHI), and flavonol 3-hydroxylase (F3H). A series of anthocyanin specific biosynthetic gene encoding dihydroflavonol-4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glucose: flavonoid-3-O-glycosyl-transferase (UF3GT) is also reported. This gene is controlled by the ternary MYB-bHLH-WD40 (MBW) protein complex containing R2R3-MYB, basic helix-loop-helix (bHLH), and WD40-repeat proteins. Among MBW, the PRODUCTION OF ANTHOCYANIN PIGMENTS 1 (PAP1) is the core component that activates the expressions of DFR, LDOX and UF3GF for anthocyanin biosynthesis [9,10]. The protein stability of PAP1 is degraded by E3 ubiquitin ligase COP1 in the dark [11], while light irradiation stabilizes PAP1 through MPK4 mediated phosphorylation modification [12]. Beside light or sugar induced anthocyanin biosynthesis, the transcriptional level of PAP1 is also regulated by PHR1 under phosphate deficiency stress to induce anthocyanin biosynthesis, during this process, SPX1 can sequestrate the activity of PHR1 to control PAP1 expression in response to phosphate-deficiency signal [13]. However, its detail mechanism by which PAP1 subtly controls anthocyanin biosynthesis to adapt environment remains unknown.
The phytochrome interaction proteins (PIFs), including PIF1, PIF3, PIF4 and PIF5 were firstly identified as the core component to perceive the light signal and regulate multiple physiological responses [14,15]. For example, PIF1 negatively control seed germination under red light irradiation [16], PIF1 and PIF3 also affect chlorophyll biosynthesis from dark to light shift [17,18]. PIF4 and PIF5 negatively control phytochrome-dependent inhibition of shade avoidance and dark-mediated leave senescence [19,20]. All of these PIFs also control hypocotyl elongation in an additive pattern as the single mutant presented weak hypocotyl elongation deficiency, but its quadruple mutant showed longer hypocotyl [21]. Exogenous sucrose treatment significantly induced hypocotyl elongation in wild-type, but not in pifs mutant, when the seedling was transferred from light to darkness [22,23]. What is more, sucrose-dependent promotion of hypocotyl elongation requires multiple PIFs, and overexpressing PIF5 shows the similar growth dynamics to those plants exposed to sucrose [22], suggesting the possible link between PIFs and sugar signal.
In this study, we investigated the effect of glucose on seed germination, and found glucose induced the accumulation of PIF4, which bond to the promoter of ABI5 to activate ABI5 expression, subsequently suppress seed germination, furthermore, we found PIF4 interacts with PAP1, and overexpressing PAP1 enhanced seed germination under high level of glucose stress. Biochemical analysis revealed that PIF4 and PAP1 antagonistically regulate ABI5 expression to fine-tune seed germination under glucose stress. Thus, our finding uncovers a novel mechanism by which PFI4 and PAP1 completely control ABI5 expression to sophistically control seed germination under high glucose stress.
2.1 Arabidopsis Seeds and Growth
All Arabidopsis (Arabidopsis thaliana) mutants, including pif4-1 and PAP1-D were obtained from the Arabidopsis Biological Resource Center. The transgenic PIF4-TAP line was provided by Professor Michael F Thomashow in Michigan State University [24]. Double mutants or lines carrying two different transgenes were generated by crossing individual lines and selecting homozygous progeny. Seeds were surface-sterilized and sown on 0.8% agar (pH 5.7) plates under white light (50 μmol−2s−1). Adult plants were grown in soil with vermiculite (3:1) at 22°C under long day (16-h light /8-h darkness) conditions for 6–8 weeks, and seeds were harvested at the same time in each batch for germination or dormancy assays.
Seeds were harvested and dried for 3−5 weeks at room temperature, and seed germination assays were performed as previously described [25,26]. In brief, seeds were surface-sterilized in a 5% hypochlorite and 0.02% Triton-X100 solution for about 10 min and then rinsed several times with sterile water before being plated on germination medium consisting of half-strength Murashige and Skoog salts with 1% sucrose and different concentration of glucose. After stratification at 4°C for 3 d, plates were placed in constant light to initiate germination, at a constant temperature at 22°C for 5 d. A seed was considered to have germinated when its radicle protruded from the seed coat. For each germination assay, at least three biological replicate experiments were performed.
2.3 Protein Extraction and Immunoblots
We extracted total protein from hydrated seeds using extraction buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA pH 8.0, 0.1% [v/v] Triton X-100, 10 mM NaF, 5% [v/v] glycerol) supplemented with phosphatase inhibitor cocktail (Roche) and 1 mM PMSF (Sigma) as previous method [26]. Proteins were cleared by centrifugation at 14,000 g for 10 min at 4°C. Protein concentration was measured using Bradford Quantitative Reagent (Invitrogen). The extracted protein (15 μg aliquot) was separated by electrophoresis on a 12% SDS-polyacrylamide gel and blotted onto polyvinylidene difluoride membranes, which were then probed with the appropriate primary anti-FLAG (1:3,000; Sigma-Aldrich), anti-MYC (1:3,000; Clontech), or anti-tubulin (1:1,000; Sigma-Aldrich) antibody and horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:3,000; Promega). Signals were observed using a ONE-HOUR IP-Western Kit (Genescript).
For Y2H, the full-length region of PIF4 and PAP1 was cloned into the pGBKT7 bait vector and the pGADT7 prey vector, respectively, using the In-Fusion cloning system (Clontech). Two-hybrid screening was performed using the mating protocol described in the Matchmaker Gold Yeast Two-Hybrid User Manual (Clontech).
The coding regions for PIF4 and PAP1 were cloned into the pGEX-4T-1 (Pharmacia) and pET28a (Merck) vectors to generate the pGST-PIF4 and pHis-PAP1 constructs, respectively. The primers used for constructions are listed in Supplemental Table S1. For prokaryotic protein expression, the constructs were transformed into E.coli Rosetta strain, and protein accumulation was induced by Isopropyl β-d-1-thiogalactopyranoside (IPTG). Soluble GST-PIF4 protein was extracted and immobilized onto Glutathione Sepharose Beads (GE healthcare), while the soluble His-PAP1 was extracted and immobilized onto Ni-NTA agarose beads (QIAGEN). For pull-down assays, 2 μg His-PAP1 was incubated with GST alone or GST-PIF4 in binding buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA) at 4°C overnight. Pulled-down proteins were extensively washed with buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 0.6% Triton X-100) before the samples were resolved on 8% SDS-PAGE gels and analyzed by immunoblot analysis using anti-His antibody (Abmart) or anti-GST antibody (Abmart) followed by a mouse secondary antibody (1:5,000; Promega).
For the in vivo Co-IP using PIF4-TAP as bait, the 1-week-old PIF4-TAP seedling was ground to a fine powder in liquid nitrogen as previous method [27]. Total proteins were extracted in MOPS buffer (100 mM MOPS, pH 7.6, 150 mM NaCl, 0.1% Nonidet P-40%, 1% Triton X-100, 20 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, 2 μg/L aprotinin, 5 μg/L leupeptin, 1 μg/L pepstatin, 2 × Complete Protease Inhibitor Cocktail, and PhosStop Cocktail from Roche), centrifuged at 13,000 rpm at 4°C for 10 min, and filtered through two layers of Miracloth. Supernatant (1.0 mL) was incubated with anti-Flag resin (Sigma Chemical) overnight under gentle rotation at 4°C. Beads were washed four times with wash buffer (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% [v/v] Triton X-100), and the proteins were eluted at 95°C for 10 min in 2× loading buffer (100 mM Tris-HCl, pH 6.8, 200 mM DTT, 2% SDS, 20% glycerol, and 0.2% bromophenol blue) and analyzed by immunoblotting with anti-FLAG (1:3,000; Sigma-Aldrich) or anti-PAP1 (1:1,000; PhytoAb) antibodies.
Chromatin affinity purification was performed as described previously [28]. Seeds were cross-linked with a 1% formaldehyde solution under a vacuum for 1 h. The chromatin was extracted and sheared to an average length of 300–500 bp by sonication and then immunoprecipitated with anti-PAP1 (Catalog no PHY1193S, PhytoAb). The crosslinking was then reversed, and the amount of each immunoprecipitated DNA fragment was determined by quantitative PCR using gene-specific primers (Supplemental Tab. S1).
The coding regions of PIF4 and PAP1 were cloned into pGreen binary vectors to add each half of the YFP coding sequence (nYFP and cYFP) upstream of and in-frame with PIF4 or PAP1, to generate nYFP-PIF4 and cYFP-PAP1. Combinations of nYFP-PIF4/cYFP-PAP1 or their corresponding control combinations were co-infiltrated into Nicotiana benthamiana leaves by Agrobacterium-mediated transient transfection. After 48 h, YFP fluorescence in N. benthamiana leaf cells was observed with a Zeiss LSM710 confocal microscope as previously.
Total RNA was extracted from hydrated seeds using TRIzol reagent (Invitrogen) as previous method [26]. RT-qPCR was performed as described. Briefly, first-strand cDNA was synthesized from 1.5 μg DNase-treated RNA in a 20 μl reaction volume using M-MuLV reverse transcriptase (Fermentas) with oligo (dT) 18 primer. For qPCR, cDNA samples were diluted to 2 to 10 ng/mL, and PCR was performed in the presence of SYBR Green I Master Mix on a Roche LightCycler 480 real-time PCR machine according to the manufacturer's instructions. All RT-qPCR experiments were independently performed in triplicate, and representative results are shown. PP2A was used as an internal control. The primer pairs for quantitative RT-qPCR are listed in Supplemental Tab. S1.
2.10 Protoplast Transient Expression Assay
For the transient expression assay, a 3-kbp ABI5 promoter fragment was inserted into the pGreenII 0800-LUC vector to generate a series of ABI5pro:LUC reporter constructs. The coding sequences of PIF4 and PAP1 were inserted into the pGreenII 62-SK vector and placed under the control of the 35S promoter. All primers used for these constructs are listed in Supplemental Tab. S1. After protoplast preparation and subsequent transfection, firefly LUC and renilla luciferase (REN) activities were measured using the Dual-Luciferase Reporter Assay System (Promega), and the LUC/REN ratio was presented.
3.1 High Glucose Induces the Accumulation of PIF4 to Suppress Seed Germination
It is reported that ABA suppresses the hypocotyl elongation through PIF4 under dark, and several ABA insensitive mutants also show tolerance to high glucose stress [29]. Thus we wonder whether the possible function of PIF4 during seed germination under high glucose. As shown in Figs. 1A and 1B, the MS medium containing 3% glucose partially reduced seed germination of wild-type Col, and the glucose at 5% suppressed the seed germination. In contrast to Col, the pif4-1 mutant showed more seed germination percentage, but the transgenic line overexpressing PIF4 (expressing PIF4-TAP fusion under the control of constitutive 35S promoter) showed lower seed germination. These data suggest that PIF4 negatively control seed germination in response to high glucose stress. Thus, these data suggest the necessary role of PIF4 in controlling seed germination under high glucose stress. We further tested the effect of high glucose on PIF4 expression. Using the total protein extracted from the imbibition seeds from wild-type Col, we found high glucose did not obviously affect the protein accumulation of endogenous PIF4 protein by anti-PIF4 antibody (Fig. 1C). Meanwhile, our RT-qPCR result also showed that high glucose did not obviously change the transcriptional level of PIF4 (Fig. 1D), these data indicate the new mechanism for PIF4 in controlling seed germination independent on its expression level under high glucose stress.
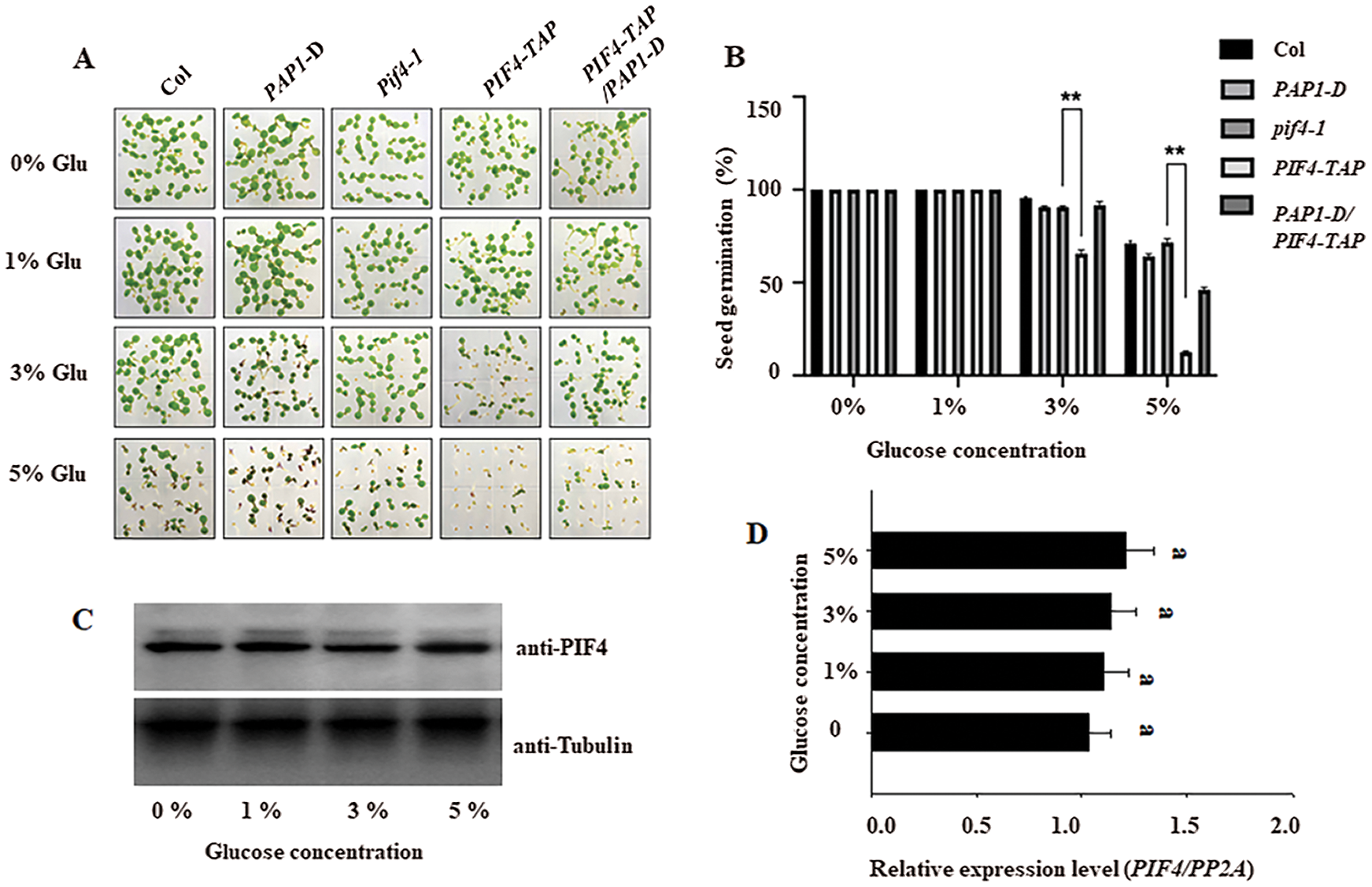
Figure 1: The different seed germination percentage among wild type Col, PAP1-D, pif4-1, PIF-TAP and the crossed pap1-D/PIF4-TAP under gradient glucose stress These seeds were cold stratificated for 3 days at 4°C and then placed at 22°C on the medium containing indicated concentration of glucose, the photo was taken after 6 days of germination (A), and the seed germination among Col, pif4-1 and PIF4-TAP on the gradient glucose treatment was calculated after 5 day of germination (B). The values are shown as means ±SD of triplicate experiments. Asterisks indicate significant difference by Student’s t-test (**P < 0.01). Protein accumulation of PIF4 (C) and the transcriptional level of PIF4 (D) and in the seedling after gradient glucose treatment for 3 days was also measured by RT-qPCR and western blotting analysis, respectively. For RT-qPCR analysis, the PP2A was used as the internal control. For western blotting analysis, anti-Tubulin was used as the loading control. Bars with different letters are significantly different at P < 0.05 (Tukey’s test)
3.2 PIF4 Activates ABI5 Expression to Suppress Seed Germination under High Glucose Stress
ABI5 acts as a negative regulator to suppress seed germination in response to ABA stress [30,31], and PIF4 can bind to the ABI5 promoter to activate ABI5 expression [29]. To determine the genetic relationship between PIF4 and ABI5 during seed germination after high glucose stress, we introduce PIF4-TAP into abi5-7 mutant to obtain PIF4-TAP/abi5-7, and introduced pif4-1 into abi5-7 to obtain pif4-1/abi5-7 line. Similar to abi5-7, both of the PIF4-TAP/abi5-7 and pif4-1/abi5-7 showed similarly higher seed germination. On the contrary, both of ABI5-MYC and ABI5-MYC/pif4-1 showed lower seed germination, though the seed germination of pif4-1 was higher under high glucose stress (Fig. 2A). Based on this genetic analysis, we propose that ABI5 is epistatic to PIF4 to control seed germination under high glucose stress.
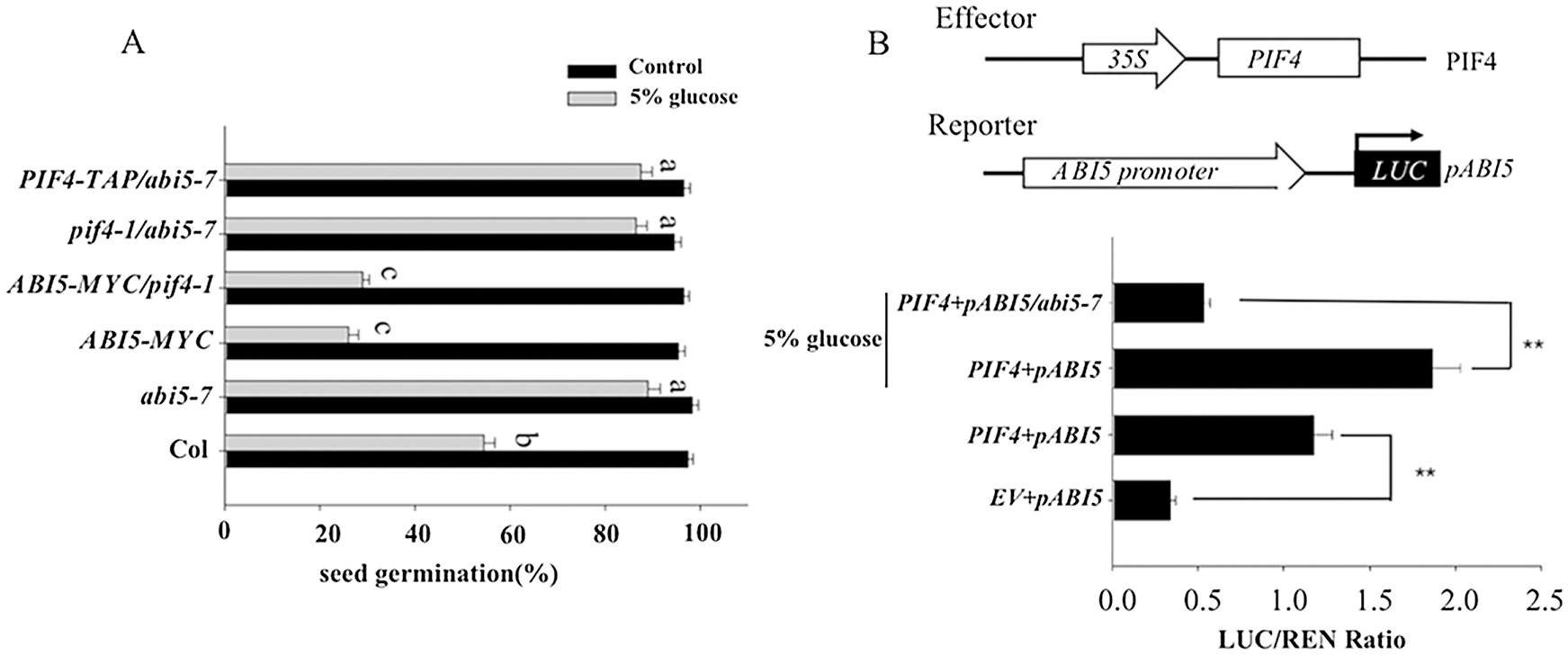
Figure 2: PIF4 suppresses seeds germination on the medium containing 5% glucose through activating ABI5 expression A) The seed germination percentage of Col, abi5-7, pif4-1, PIF4-TAP, pif4-1/abi5-7 and PIF4-TAP/abi5-7 on the medium containing 5% glucose were calculated. The values are shown as means ±SD of triplicate experiments. Bars labelled with different letters are significantly different at P < 0.05 (Tukey’s test). B) PIF4 activates the expression of ABI5 by transient protoplast analysis. Upper panel: Schematic diagram of the PIF4 effector and the ABI5pro: LUC reporter constructs used in the transient transactivation assay. Lower panel: The ABI5pro:LUC reporter was coexpressed with PIF4 effectors for 24 h; the firefly luciferase and Renilla luciferase (LUC/REN) ratio represents ABI5pro:LUC activity relative to the internal control (35Spro:REN). Data is meant SD of three biological replicates. Asterisks indicate significant difference by Student’s t-test (**P < 0.01)
To confirm ABI5 is the target gene for PIF4-dependent seed germination under high glucose treatment. We performed the transient protoplast transformation analysis to test the effect of PIF4 on ABI5 expression with or without high glucose treatment. As shown in Fig. 2B, overexpressing PIF4 indeed increased the transcriptional level of ABI5, and high glucose further increased the activation effect of PIF4 on ABI5 expression. However, PIF4 did not efficiently induce the expression of ABI5 in the protoplast from abi5-7 mutant line, suggest the synergistic effect of PIF4 and ABI5 on ABI5 expression under high glucose treatment. This data suggests that high glucose dependents on PIF4 to induce ABI5 expression.
3.3 PIF4 Physically Interacts with PAP1
To understand the underlying mechanism by which PIF4 control seed germination under high glucose stress, we performed yeast two hybrid and searched the PIF4-interaction protein from the normalized Arabidopsis cDNA library. After two rounds of screening, we obtained the several positive clones. We focus on one clone encoding PAP1 that is pivotal for anthocyanin biosynthesis. Interaction between PIF4 and PAP1 was confirmed by yeast two-hybrid experiment (Fig. 3A). Pull-down analysis using these purified GST-PIF4 and HIS-PAP1 showed that PIF4-GST, but not GST alone as the control, could be immunoprecipitated by HIS-PAP1 (Fig. 3B). These data suggest that PIF4 interacts with PAP1 in vitro. Furthermore, we performed Co-IP analysis to test the interaction of PAP1 and PIF4 in vivo. Using the transgenic PIF4-TAP and anti-PAP1 antibody, we extracted the total protein from the transgenic PIF4-TAP line and found PAP1 protein could be co-immunoprecipitated with anti-FLAG resin (Fig. 3C). As the control, we extracted the total protein from wild type Col seedling and the PAP1 could be co-immunoprecipitated by anti-FLAG resin, suggesting the interaction of PIF4 and PAP1 in planta. We subsequently adopted BiFC to check the interaction of PIF4 and PAP1 in vivo. We fused PIF4 with nYFP (PIF4-nYFP) and PAP1 with cYFP (PAP1-cYFP), the strong YFP fluorescence could be observed in the tobacco leave co-expressing PIF4-nYFP and PAP1-cYFP, as the control we did not the YFP fluorescence in the tobacco leave co-expressing the recombine of PIF4-nYFP and cYFP, or nYFP with PAP1-cYFP (Fig. 3D). At last, we transiently expressed PIF4-mCherry and PAP1-GFP in the tobacco leaves, and found both of them localized in the nucleus, and also co-colocalized with the nucleus marker H2B-CFP (Fig. 3E), suggesting both of them co-localized in the nucleus. Together, this evidence supports the interaction of PIF4 and PAP1 in planta.
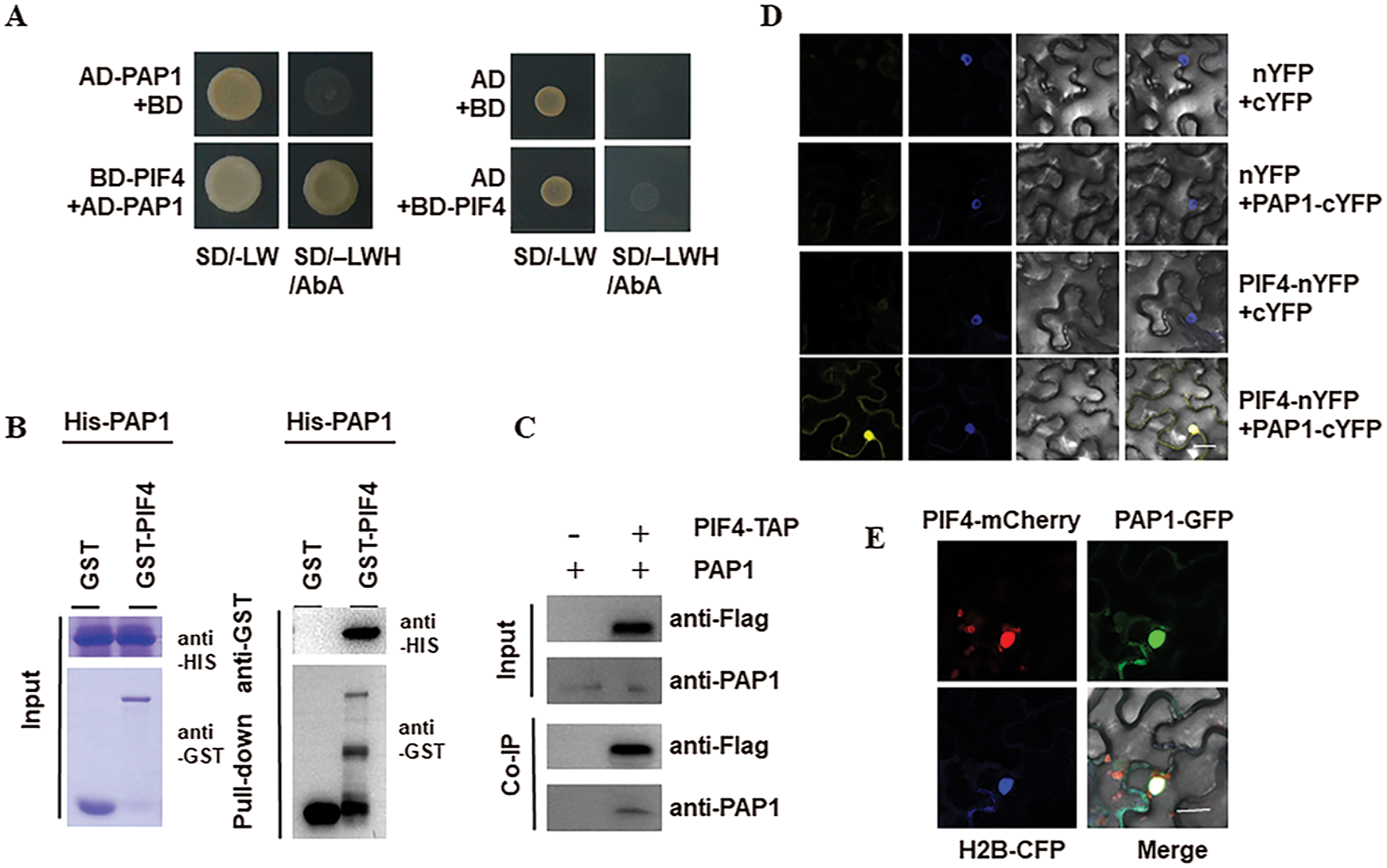
Figure 3: PIF4 interacts with PAP1 A) The interaction of PIF4 and PAP1 by Y2H analysis. Yeast cells co-transformed with the indicated construct combinations were grown on SD medium lacking Trp/Leu (-LW) or Trp/Leu/His (-LWH) with 100 ng mL−1 Aureobasidin A (AbA). AD: DNA-activation domain of GAL4; BD: DNA-binding domain of GAL4. B) Pull-down assay showing direct interaction between His-PAP1 and GST-PIF4 fusion proteins in vitro. His-PAP1 proteins were incubated with immobilized GST or GST-PIF4 proteins. Immunoprecipitated fractions were observed by anti-His or anti-GST antibody, respectively. C) Co-IP showing the interaction of PIF4 with PAP1 in Arabidopsis. Proteins extracted from hydrated seeds from wild type Col or PIF4-TAP plants were immunoprecipitated by anti-Flag resin beads. Coimmunoprecipitated proteins were detected by anti-PAP1 anti-Flag antibody. Immunoblots show the presence of proteins in total protein extracts from plants (input) and fractions after immunoprecipitation by anti-PAP1 or anti-Flag antibodies. D) The interaction between PIF4 and PAP1 by BIFC analysis BiFC assay showed that PIF4-cYFP interacts with PAP1-nYFP in the nuclei of N. benthamiana epidermal leaf cells. PIF4 was fused to the C-terminal fragment of YFP (cYFP) to form PIF4-cYFP. Full-length PAP1 was fused with the N-terminal fragment of YFP (nYFP) to generate PAP1-nYFP. YFP fluorescence was detected in N. benthamiana leaves co-infiltrated with the indicated constructs. Nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI). Bar, 50 mm. E) The co-localization of PIF4 and PAP1 in the nucleus Analysis of the colocalization of PIF4 and PAP1 in N. benthamiana leaves. PAP1-GFP and PIF4-mCherry colocalize in the nuclei of N. benthamiana epidermal leaf cells. Bar = 50 μm
3.4 PAP1 Enhances Seed Germination Tolerance to High Glucose Stress
It is well known that high glucose induces the accumulation of anthocyanin and PAP1 is the critical regulator for anthocyanin biosynthesis [32]. As PIF4 interacted with PAP1, and PIF4 also regulates seed germination under high glucose treatment [22,33], hinting the probable role of PAP1 in controlling seed germination under high glucose treatment. To test such possibility, we check the germination percentage of PAP1-D line, which is the dominant mutant with strong PAP1 expression [34]. As shown in Fig. 1A, the PAP1-D mutant still showed higher germination on the high glucose treatment in contrast to wild type line. Genetic analysis showed that the crossed pap1-D/ABI5-MYC line showed the lower germination under high glucose treatment, similar to ABI5-MYC line. These data suggest that ABI5 is epistatic to PAP1 to control seed germination after high glucose treatment.
PAP1 acts as the MYB transcriptional factor that recognizes the 7-bp MYB-recognizing element (MRE) [13,35]. We searched the promoter of ABI5 and found 6 regions containing the MRE element, we named these regions as P1 to P4. Among them, P1 and P4 contained two motifs, respectively. To test which region could be specially recognized by PAP1, we used anti-PAP1 antibody and ChIP analysis to check the interaction of these region with PAP1 protein. As shown in Figs. 4A and 4B, we found P3 region could be specially enriched by anti-PAP1 antibody, and 5% glucose treatment intensified such binding. Thus, these results suggest that high glucose adds the binding of PAP1 to the ABI5 promoter, subsequently suppress ABI5 expression, thereby reducing the inhibitory effect of high glucose on seed germination.
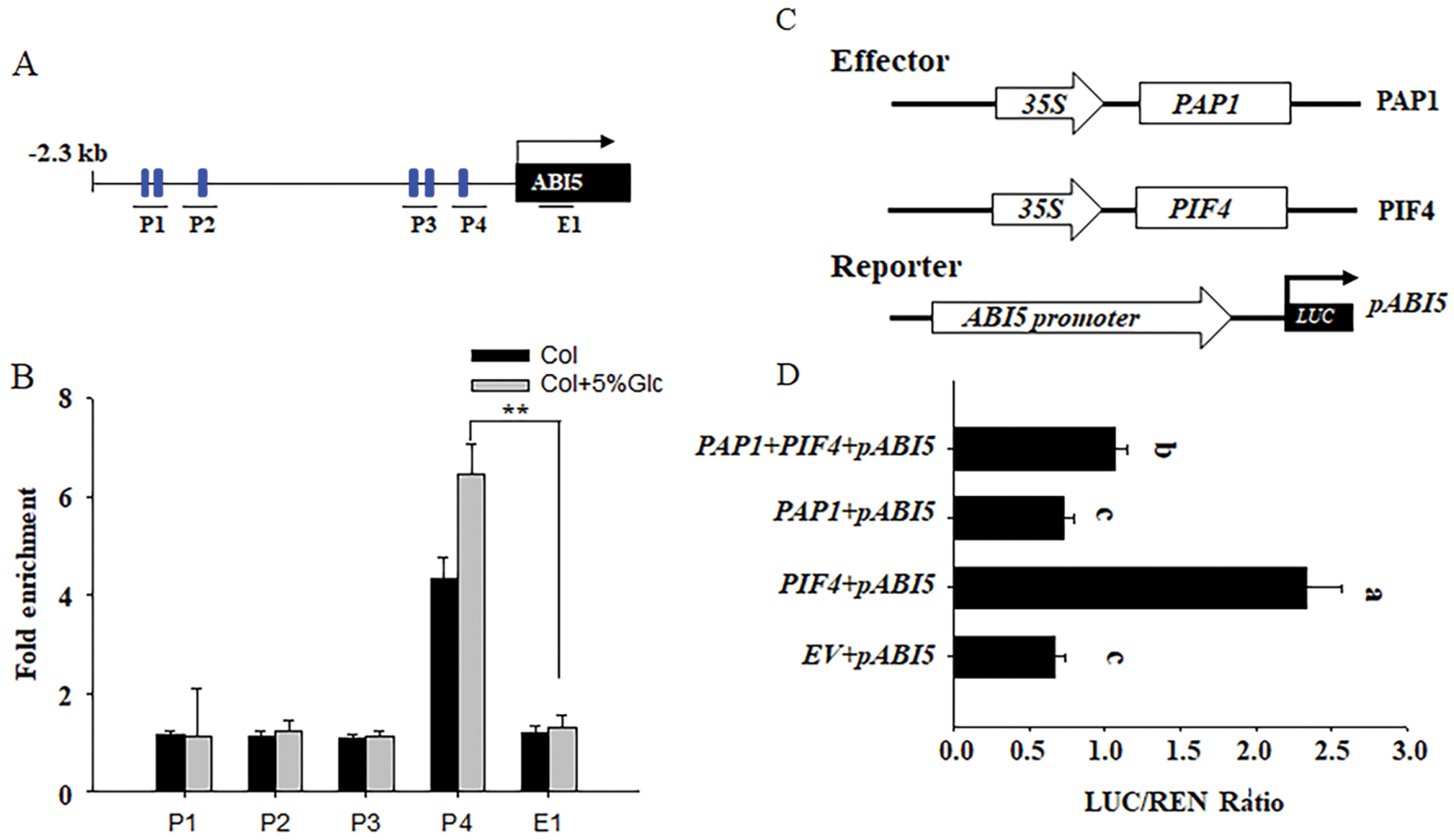
Figure 4: PAP1 binds to the promoter of ABI5 to antagonize the activation effect of PIF4 on ABI5 expression A&B) ChIP-qPCR assay of the association of PAP1 with the ABI5 promoter in vivo. Hydrated Col-0 with or without 5% glucose treatment were used (B). The diagram of the ABI5 promoter region showing the position of MRE boxes (blue rectangles), and four regions (P1 to P4) for ChIP-qPCR amplification, as indicated by black lines under the MRE boxes. The E1 box was used as the internal control. Anti-PAP1 antibody was used for ChIP. ACTIN2 served as an internal control, and enrichment values were normalized to the level of input DNA. Values are shown as the means ± SD (t-test, **P < 0.01). C&D) PAP1 and PIF4 antagonistically activate the expression of ABI5 by transient protoplast analysis. The ABI5pro: LUC reporter was coexpressed with PIF4 and PAP1 effectors for 24 h; the firefly luciferase and Renilla luciferase (LUC/REN) ratio represents ABI5pro:LUC activity relative to the internal control (35Spro:REN). Data are means ± SD of three biological replicates. Bars labelled with different letters are significantly different at P < 0.05 (Tukey’s test). The schematic diagram of the PIF4 and PAP1 effector, and the ABI5pro: LUC reporter constructs used in the transient transactivation assay (D). Bars with different letters are significant different at P < 0.05
3.5 PIF4 and PAP1 Antagonistically Control Seed Germination through ABI5
As PIF4 can activate the expression of ABI5 [28], but PAP1 suppresses the expression of ABI5 as described above, we suppose there is an antagonistic effect between PIF4 and PAP1 on ABI5 expression. To test such possibility, we crossed PIF4-TAP with PAP1-D, and compared the expression of ABI5 after high glucose stress. As shown in Fig. 5A, the crossed PIF4-TAP/PAP1-D showed relatively lower ABI5 expression than PIF4-TAP alone under high glucose stress. Similarly, the expression of ABI5 was also lower in PAP1-D mutant, but high in the PIF4-TAP seed, after high glucose treatment. Environment stress, such as strong light, increased the accumulation of PAP1. Thus, this genetic data suggests that PAP1 attenuates the inhibitory effect of PIF4 on seed germination through ABI5 under high glucose stress. Here we also measured the protein abundance of PAP1 after high glucose treatment. As shown in Fig. 5B, we found that high glucose gradually increased the protein accumulation of PAP1. As we described above, high glucose induced, but PAP1 suppressed the expression of ABI5, these data hint possible feedback regulation of PAP1 during seed germination under high glucose stress.
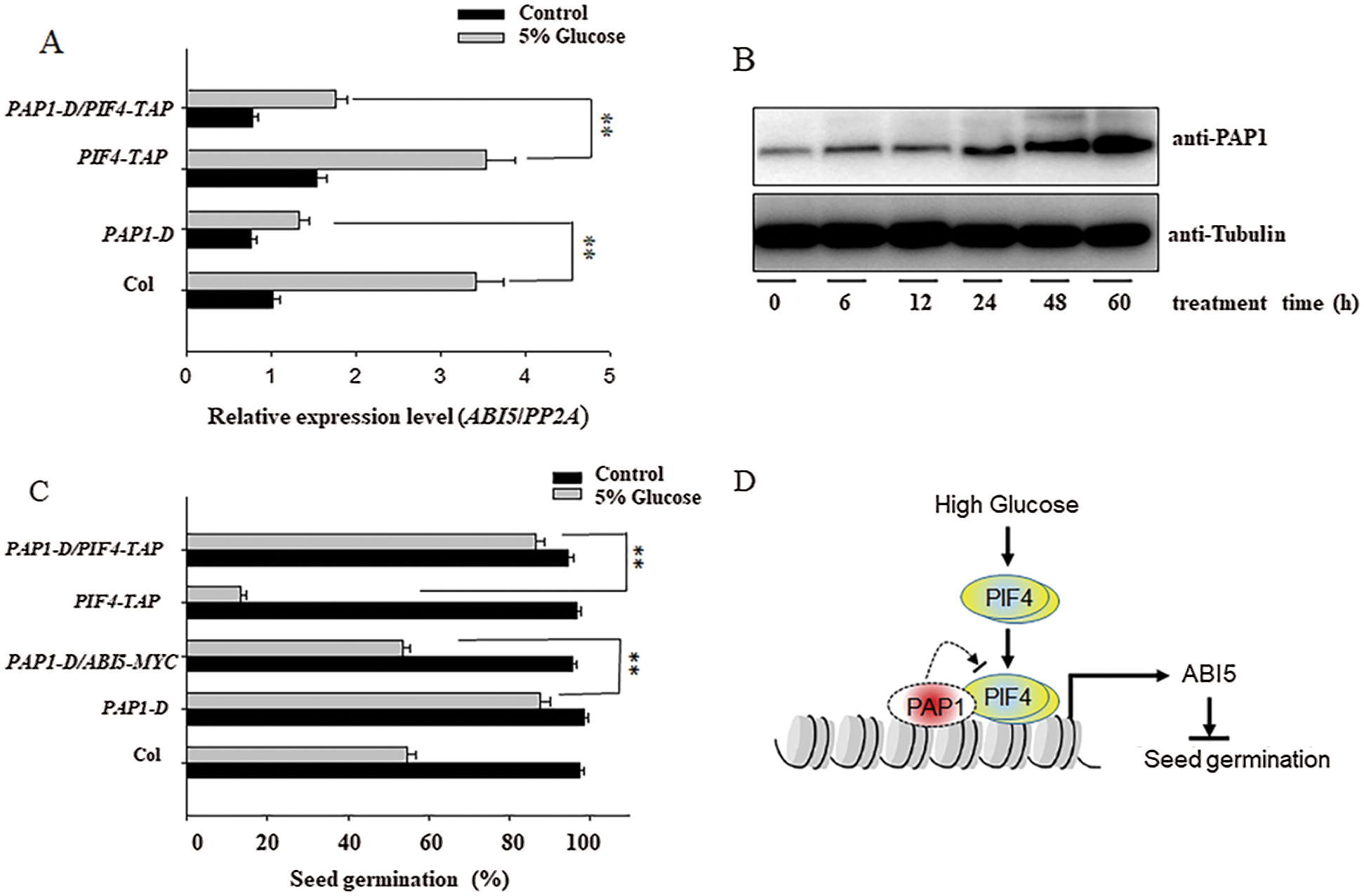
Figure 5: PAP1and PIF4 antagonistically control the seeds germination under high glucose treatment A) PAP1 and PIF4 antagonistically modulate glucose-induced ABI5 expression by RT-qPCR analysis. The Col, pap1-D, PIF4-TAP and their crossed seeds were incubated on the MS medium with or without 5% glucose treatment for 48 h, and the ABI5 expression level was measured by RT-qPCR analysis. The PP2A was used as the internal control. Values are shown as means ± SD from three biological replicates. Asterisks indicate significant difference by Student’s t-test (**P < 0.01). B) High glucose gradually induced the accumulation of PAP1 protein. The hydrated Col-0 seeds were incubated with 5% glucose for indicated time, and the total protein was extracted for western blotting analysis. Endogenous PAP1 protein was used by anti-PAP1 antibody, and the anti-Tubulin antibody was used for internal loading control. C) PAP1 and PIF4 antagonistically controlled the seed germination. The Col-0, PAP1-D, PIF4-TAP, the crossed PAP1-D/ABI5-MYC and PAP1-D/PIF4-TAP seeds were sowed on the MS with or without 5% glucose for 5 days, and the seed germination percentage was calculated. Values are shown as means ± SD from three biological replicates. Asterisks indicate significant difference by Student’s t-test (**P < 0.01). D) The propose model to demonstrate the antagonistic effect between PIF4 and PAP1 on ABI5 expression and seed germination under high glucose treatment. High glucose activates the expression of ABI5 through PIF4, thus suppresses seeds germination, meanwhile, high glucose also induces the accumulation of PAP1, which antagonizes the activation effect of PIF4 on ABI5 expression, ultimately coordinate the transcriptional level of ABI5 at the appropriate level to fine-tune seeds germination in response to high glucose stress
Furthermore, we utilized the transient protoplast transformation to check the opposition effect between PIF4 and PAP1 on ABI5 expression. The reporter construct contained the LUC driven by ABI5 promoter, while the effect construct expressed PIF4 or PAP1 under the control of the 35S promoter. As shown in Figs. 4C and 4D, solely expressing PIF4 obviously upregulated the LUC activity, suggesting activate the expression of ABI5. Conversely, solely expressing PAP1 suppressed the LUC activity and ABI5 expression. However, co-expressing PIF4 and PAP1 simultaneously attenuated the PIF4 activated LUC expression, confirming our above genetic conclusion that PIF4 and PAP1 antagonistically control ABI5 expression and seed germination under high glucose stress.
It is known that PIFs including PIF1/3/4/5 accumulate in the darkness to promote skotomorphogenic development [36,37], however, once light irradiation, the light receptor phytochrome interact with PIFs to induce their degradation through the 26S proteasome pathway. PIF can directly regulate the expression of downstream gene by binding to either G-box (CACGTG) or E-box (CANNTG) motifs in their promoter [37]. ABI5 as the important component of ABA signal pathway mediates seed germination [30,31]. Previous studies have demonstrated the crosstalk between PIFs and ABA signal [38,39]. For instance, PIF1 controls seed germination through ABA and GA pathway [16], ABA suppresses the hypocotyl elongation of wild-type seedling but not for pif4 pif5 mutants [29], indicating that ABA may suppress PIF4/PIF5 function during shade response. PIF1/3/4/5 can also directly bind to the G-box motifs in the ABI5 promoter to activate ABI5 expression in response to ABA [31]. In study we firstly confirmed that high concentration of glucose inhibited seed germination in the wild type Col seeds. However, the pif4-1 mutant seed showed higher germination percentage, but overexpressing PIF4 reduced seed germination under high concentration of glucose stress, suggesting that PIF4 negatively regulates seed germination under high glucose treatment. However, we did not observe the obvious effect of high glucose on the transcriptional level of PIF4 and its protein accumulation (Figs. 1C and 1D), this data suggests the post-translational mechanism of PIF4 in controlling seed germination under high glucose stress. Previous study showed that PIF4 can bind the G-box motif in the promoter region of ABI5 to activate ABI5 expression [31]. We thus compared the ABI5 level before and after glucose treatment. In line with the seed germination phenotype, glucose obviously stimulates the expression of ABI5 in the wild-type Col seed, such activation effect was compromised in the pif4-1 mutant seeds, but aggravated in the PIF4-TAP seeds, suggesting glucose required PIF4 to activate ABI5 expression. Our genetic experiment showed that the PIF4-TAP/abi5-7 and abi5-7 seeds showed relatively higher seed germination under glucose medium, though PIF4-TAP seeds germination were sensitive to glucose stress (Figs. 1A and 1B). We also observed that ABI5-MYC and ABI5-MYC/pif4-1 also show sensitive to high glucose stress, though pif4-1 seeds germination show insensitive to high glucose stress (Fig. 2A). Based on these phenotypes, we propose that ABI5 is genetically epistatic to PIF4 to control seed germination under high glucose stress.
In this study, we further found that PAP1 acts as the interaction protein of PIF4, such interaction was confirmed by several methods, including yeast two hybrid experiment, in vitro pull-down analysis and BiFC analysis. PAP1 is a pivotal regulator for anthocyanin biosynthesis under environmental stress. Previous study pointed out the PIF4 mediates the anthocyanin biosynthesis under red light irradiation [22,33], hinting the possible relationship between PIF4 and PAP1. Here we also check the seed germination of PAP1-D, the dominant mutant with high PAP1 expression and found that PAP1-D showed higher germination than that of wild-type Col seed under glucose stress. In according with it, the ABI5 transcripts in PAP1-D were also lower (Fig. 5A). As the PAP1 as the MYB transcriptional factor can bind the MYE motif to activate its target gene expression [13], there exist one MYE motif in the ABI5 promoter, and the ChIP analysis showed that PAP1 was specially enriched in such MYE motif, RT-qPCR analysis showed that PAP1 repressed the expression of ABI5 (Fig. 4B), these results propose that PAP1 as the negatively regulator of ABI5 to enhance seed germination under high concentration of glucose stress (Fig. 5C). Consistent with this conclusion, our genetic analysis showed that the crossed ABI5-MYC/PAP1-D seed showed relatively lower seed germination than PAP1-D under high glucose treatment, suggest that PAP1 enhances seed germination under high concentration of glucose through suppressing ABI5 (Fig. 5C).
As PIF4 activates the expression of ABI5 to suppress seed germination [28], but PAP1 suppresses the expression of ABI5 to enhance seed germination, under high concentration of glucose stress. These data suggest the antagonistic mechanism between PAP1 and PIF4 in controlling seed germination. Transient transformation analysis showed that PIF4 activated the expression of ABI5, while PAP1 assuaged the activation effect of PIF4 on ABI5 expression (Fig. 4D). Genetic analysis also showed that overexpressing PAP1 in the PIF4-TAP background partially improved the seed germination subjected to high glucose stress, compared with relatively lower ABI5 expression (Fig. 5C). Thereby these data support the antagonistic relationship between PAP1 and PIF4 on ABI5 expression and seed germination.
In summary, our study revealed the novel of PIF4 and PAP1 in controlling seed germination under high concentration of glucose stress. Under the same high concentration of glucose stress, we found PIF4 activated the ABI5 expression to suppress seed germination, reversely, PAP1 suppressed ABI5 to enhance seed germination, furthermore, the genetic and physiological analysis revealed the antagonistically mechanism between PAP1 and PIF4 in controlling seed germination through ABI5. Thus, these data demonstrate that PAP1 interacts with PIF4 to coordinate ABA signal by controlling ABI5 expression, and provide a new mechanism by which PAP1 and PIF4 integrate ABA signal to fine-tune seed germination in response to high glucose stimulation (Fig. 5D).
Funding Statement: This work was funded by the National Natural Science Foundation of China (Grant No. 31970289).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Roitsch, T. (1999). Source-sink regulation by sugar and stress. Current Opinion in Plant Biology, 2(3), 198–206. DOI 10.1016/S1369-5266(99)80036-3. [Google Scholar] [CrossRef]
2. Sergeeva, L. I., Keurentjes, J. J. B., Bentsink, L., Vonk, J., van der Plas, L. H. W. et al. (2006). Vacuolar invertase regulates elongation of arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proceedings of the National Academy of Sciences of the United States of America, 103(8), 2994–2999. DOI 10.1073/pnas.0511015103. [Google Scholar] [CrossRef]
3. Barratt, D. H., Derbyshire, P., Findlay, K., Pike, M., Wellner, N. et al. (2009). Normal growth of arabidopsis requires cytosolic invertase but not sucrose synthase. Proceedings of the National Academy of Sciences of the United States of America, 106(31), 13124–13129. DOI 10.1073/pnas.0900689106. [Google Scholar] [CrossRef]
4. Cho, Y. H., Yoo, S. D., Sheen, J. (2007). Glucose signaling through nuclear hexokinase1 complex in arabidopsis. Plant Signaling & Behavior, 2(2), 123–124. DOI 10.4161/psb.2.2.3894. [Google Scholar] [CrossRef]
5. Sheen, J. (2014). Master regulators in plant glucose signaling networks. Journal of Plant Biology, 57(2), 67–79. DOI 10.1007/s12374-014-0902-7. [Google Scholar] [CrossRef]
6. Lou, Y., Gou, J. Y., Xue, H. W. (2007). PIP5K9, an arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell, 19(1), 163–181. DOI 10.1105/tpc.106.045658. [Google Scholar] [CrossRef]
7. Xiao, W., Sheen, J., Jang, J. C. (2000). The role of hexokinase in plant sugar signal transduction and growth and development. Plant Molecular Biology, 44(4), 451–461. DOI 10.1023/A:1026501430422. [Google Scholar] [CrossRef]
8. Yoon, J., Cho, L. H., Tun, W., Jeon, J. S., An, G (2021). Sucrose signaling in higher plants. Plant Science, 302, 110703. DOI 10.1016/j.plantsci.2020.110703. [Google Scholar] [CrossRef]
9. Kreynes, A. E., Yong, Z., Liu, X. M., Wong, D. C. J., Castellarin, S. D. et al. (2020). Biological impacts of phosphomimic AtMYB75. Planta, 251(3), 60. DOI 10.1007/s00425-020-03350-0. [Google Scholar] [CrossRef]
10. Shi, M. Z., Xie, D. Y. (2014). Biosynthesis and metabolic engineering of anthocyanins in arabidopsis thaliana. Recent Patents on Biotechnology, 8(1), 47–60. DOI 10.2174/1872208307666131218123538. [Google Scholar] [CrossRef]
11. Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B. et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in arabidopsis. Plant Journal, 74(4), 638–651. DOI 10.1111/tpj.12153. [Google Scholar] [CrossRef]
12. Li, S., Wang, W., Gao, J., Yin, K., Wang, R. et al. (2016). MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in arabidopsis. Plant Cell, 28(11), 2866–2883. DOI 10.1105/tpc.16.00130. [Google Scholar] [CrossRef]
13. He, Y. Q., Zhang, X. Y., Li, L. Y., Sun, Z. T., Li, J. M. et al. (2021). SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytologist, 230(1), 205–217. DOI 10.1111/nph.17139. [Google Scholar] [CrossRef]
14. Pham, V. N., Kathare, P. K., Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiology, 176(2), 1025–1038. DOI 10.1104/pp.17.01384. [Google Scholar] [CrossRef]
15. Leivar, P., Monte, E. (2014). PIFs: Systems integrators in plant development. Plant Cell, 26(1), 56–78. DOI 10.1105/tpc.113.120857. [Google Scholar] [CrossRef]
16. Yang, L., Jiang, Z., Jing, Y., Lin, R. (2020). PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in arabidopsis. Journal of Integrative Plant Biology, 62(9), 1372–1384. DOI 10.1111/jipb.12938. [Google Scholar] [CrossRef]
17. Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G. et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proceedings of the National Academy of Sciences of the United States of America, 106(18), 7660–7665. DOI 10.1073/pnas.0812219106. [Google Scholar] [CrossRef]
18. Stephenson, P. G., Fankhauser, C., Terry, M. J. (2009). PIF3 is a repressor of chloroplast development. Proceedings of the National Academy of Sciences of the United States of America, 106(18), 7654–7659. DOI 10.1073/pnas.0811684106. [Google Scholar] [CrossRef]
19. Holalu, S. V., Reddy, S. K., Blackman, B. K., Finlayson, S. A. (2020). Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant Cell Environment, 43(9), 2224–2238. DOI 10.1111/pce.13824. [Google Scholar] [CrossRef]
20. Ueda, H., Ito, T., Inoue, R., Masuda, Y., Nagashima, Y. et al. (2020). Genetic interaction among phytochrome, ethylene and abscisic acid signaling during dark-induced senescence in arabidopsis thaliana. Frontiers in Plant Science, 11, 564. DOI 10.3389/fpls.2020.00564. [Google Scholar] [CrossRef]
21. Kupers, J. J., Oskam, L., Pierik, R. (2020). Photoreceptors regulate plant developmental plasticity through auxin. Plants (Basel Switzerland), 9(8), 940. DOI 10.3390/plants9080940. [Google Scholar] [CrossRef]
22. Liu, Z., Zhang, Y., Liu, R., Hao, H., Wang, Z. et al. (2011). Phytochrome interacting factors (PIFs) are essential regulators for sucrose-induced hypocotyl elongation in arabidopsis. Journal of Plant Physiology, 168(15), 1771–1779. DOI 10.1016/j.jplph.2011.04.009. [Google Scholar] [CrossRef]
23. Stewart, J. L., Maloof, J. N., Nemhauser, J. L. (2011). PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS One, 6(5), e19894. DOI 10.1371/journal.pone.0019894. [Google Scholar] [CrossRef]
24. Lee, C. M., Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 109(37), 15054–15059. DOI 10.1073/pnas.1211295109. [Google Scholar] [CrossRef]
25. Chang, G., Wang, C., Kong, X., Chen, Q., Yang, Y. et al. (2018). AFP2 as the novel regulator breaks high-temperature-induced seeds secondary dormancy through ABI5 and SOM in arabidopsis thaliana. Biochemical and Biophysical Research Communications, 501(1), 232–238. DOI 10.1016/j.bbrc.2018.04.222. [Google Scholar] [CrossRef]
26. Zhang, Q., He, D., Ying, S., Lu, S., Wei, J. et al. (2020). GABA enhances thermotolerance of seeds germination by attenuating the ROS damage in arabidopsis. Phyton-International Journal of Experimental Botany, 89(3), 619–631. [Google Scholar]
27. Li, P., Zhang, Q., He, D., Zhou, Y., Ni, H. et al. (2020). AGAMOUS-Like67 cooperates with the histone mark reader EBS to modulate seed germination under high temperature. Plant Physiology, 184(1), 529–545. DOI 10.1104/pp.20.00056. [Google Scholar] [CrossRef]
28. Hu, X., Kong, X., Wang, C., Ma, L., Zhao, J. et al. (2014). Proteasome-mediated degradation of FRIGIDA modulates flowering time in arabidopsis during vernalization. Plant Cell, 26(12), 4763–4781. DOI 10.1105/tpc.114.132738. [Google Scholar] [CrossRef]
29. Qi, L., Liu, S., Li, C., Fu, J., Jing, Y. et al. (2020). PHYTOCHROME-Interacting FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Molecular Plant, 13(3), 14–430. DOI 10.1016/j.molp.2020.02.001. [Google Scholar] [CrossRef]
30. Finkelstein, R. R., Lynch, T. J. (2000). The arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell, 12(4), 599–609. DOI 10.1105/tpc.12.4.599. [Google Scholar] [CrossRef]
31. Yu, F., Wu, Y., Xie, Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends in Plant Science, 20(9), 569–575. DOI 10.1016/j.tplants.2015.05.004. [Google Scholar] [CrossRef]
32. Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in arabidopsis requires the MYB75/PAP1 gene. Plant Physiology, 139(4), 1840–1852. DOI 10.1104/pp.105.066688. [Google Scholar] [CrossRef]
33. Liu, Z., Wang, Y., Fan, K., Li, Z., Jia, Q. et al. (2021). PHYTOCHROME-Interacting FACTOR 4 (PIF4) negatively regulates anthocyanin accumulation by inhibiting PAP1 transcription in arabidopsis seedlings. Plant Science, 303, 110788. DOI 10.1016/j.plantsci.2020.110788. [Google Scholar] [CrossRef]
34. Tohge, T., Matsui, K., Ohme-Takagi, M., Yamazaki, M., Saito, K. (2005). Enhanced radical scavenging activity of genetically modified arabidopsis seeds. Biotechnology Letters, 27(5), 297–303. DOI 10.1007/s10529-005-0683-7. [Google Scholar] [CrossRef]
35. Sablowski, R. W., Moyano, E., Culianez-Macia, F. A., Schuch, W., Martin, C. et al. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO Journal, 13(1), 128–137. DOI 10.1002/j.1460-2075.1994.tb06242.x. [Google Scholar] [CrossRef]
36. Shi, H., Lyu, M., Luo,Y., Liu, S., Li, Y. et al. (2018). Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proceedings of the National Academy of Sciences of the United States of America, 115(25), 6482–6487. DOI 10.1073/pnas.1803861115. [Google Scholar] [CrossRef]
37. Pham, V. N., Xu, X., Huq, E. (2018). Molecular bases for the constitutive photomorphogenic phenotypes in arabidopsis. Development, 145(23DOI 10.1242/dev.169870. [Google Scholar] [CrossRef]
38. Yadukrishnan, P., Datta, S. (2021). Light and abscisic acid interplay in early seedling development. New Phytologist, 229(2), 763–769. DOI 10.1111/nph.16963. [Google Scholar] [CrossRef]
39. Liang, S., Gao, X., Wang, Y., Zhang, H., Yin, K. et al. (2020). Phytochrome-interacting factors regulate seedling growth through ABA signaling. Biochemical and Biophysical Research Communications, 526(4), 1100–1105. DOI 10.1016/j.bbrc.2020.04.011. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |