
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.015239
REVIEW
Functional Role of miRNAs: Key Players in Soybean Improvement
1CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow, 226015, India
2Department of Zoology, School of Applied Sciences, Centurion University of Technology and Management, Bhubaneswar, 751009, India
3Department of Biotechnology, Central University of Rajasthan, Ajmer, 305817, India
4Plant Molecular Biology, International Center for Genetic Engineering & Biotechnology, New Delhi, 110067, India
5Department of Biosciences and Bioinformatics, Khallikote University, Berhampur, 761008, India
6Mountain Research Center for Field Crops, Khudwani, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Anantnag, 192101, India
*Corresponding Authors: Shabir Hussain Wani. Email: shabirhussainwani@gmail.com; Jogeswar Panigrahi. Email: drjpanigrahi@gmail.com
Received: 03 December 2020; Accepted: 27 February 2021
Abstract: Soybean (Glycine max (L.) Merr) is an agro-economic crop growing across the world to cater nutrition for both human and animal feed due to the high oil and protein content in its edible seeds. The genes and QTLs associated with important agronomic traits in this crop have already been identified and validated for soybean cyst nematode (SCN), Phytophthora root and stem rot, Pythium root rot and aphid resistance, seed quality, nutrient values, and also employed for genetic improvement in soybean. In the last decade, micro RNAs (miRNAs) have been considered the effector molecules, so the detection and characterization of novel miRNAs in soybean have been taken up by several workers. The advancement in the strategy of sequencing and tools of bioinformatics during last decade has contributed to the discovery of many soybean miRNAs, thus miRNA can be used as a tool in molecular breeding studies, and this has opened new vistas for miRNA mediated genetic improvement of soybean to augment crop productivity as well as nutritional quality. This review addresses the current state of understanding of miRNA-mediated stress responses, nutrient acquisition, plant development and crop production processes in soybean.
Keywords: Soybean; miRNA; quality improvement; plant development; genetic engineering; molecular breeding
The legumes are major source of edible proteins in the vegetarian diets, and accommodate the need of protein supplement to the world. Among the legumes, soybean [Glycine max (L.) Merr.] is a major crop with a rich source of protein (40%) and oil (20%). Soybean is ranked number one (53%) among the major oil crops, such as rapeseed, groundnut, cottonseed, sunflower seed, linseed, etc. Soybean domestication is estimated to be about 2500 BC in Northeastern China and then spread to Southern China, Korea, Japan, and other South-Eastern Asian countries. Being a legume, soybean is predominantly self-pollinated crop with somatic chromosome complement of 2n = 4x = 40. The genome sequence of this crop has also been decoded, and there are 56,044 protein-coding loci and 88,647 mRNA transcripts in the present Williams 82 soybean reference genome (Wm82.a2.v1) assembly [1]. Non-coding RNAs, including microRNAs (miRNAs) and phased small interfering RNAs (phasiRNAs), are also present in the soybean genome in addition to coding genes. MicroRNAs (miRNAs) are 20–24 nucleotide long non-coding RNAs that repress the expression of target genes in eukaryotes at the post-transcriptional level [2]. miRNAs are small RNAs like the siRNAs, but they are transcribed from miRNA genes (MIR) by RNA polymerase II (Pol II). The primary miRNAs (pri-miRNAs) are then folded into stem-loop structures recognized by Dicer-like 1 enzyme (DCL1) that cleaves the stems from base leading to the formation of pre-miRNAs. DCL1 further cleaves at the loop regions of the pre-miRNA to liberate the mature miRNA/miRNA* duplexes from the hairpins. miRNA is then incorporated into the RNA-induced silencing complex (RISC) containing ARGONAUTE 1 (AGO1) for downregulation of target gene [3] (Fig. 1).
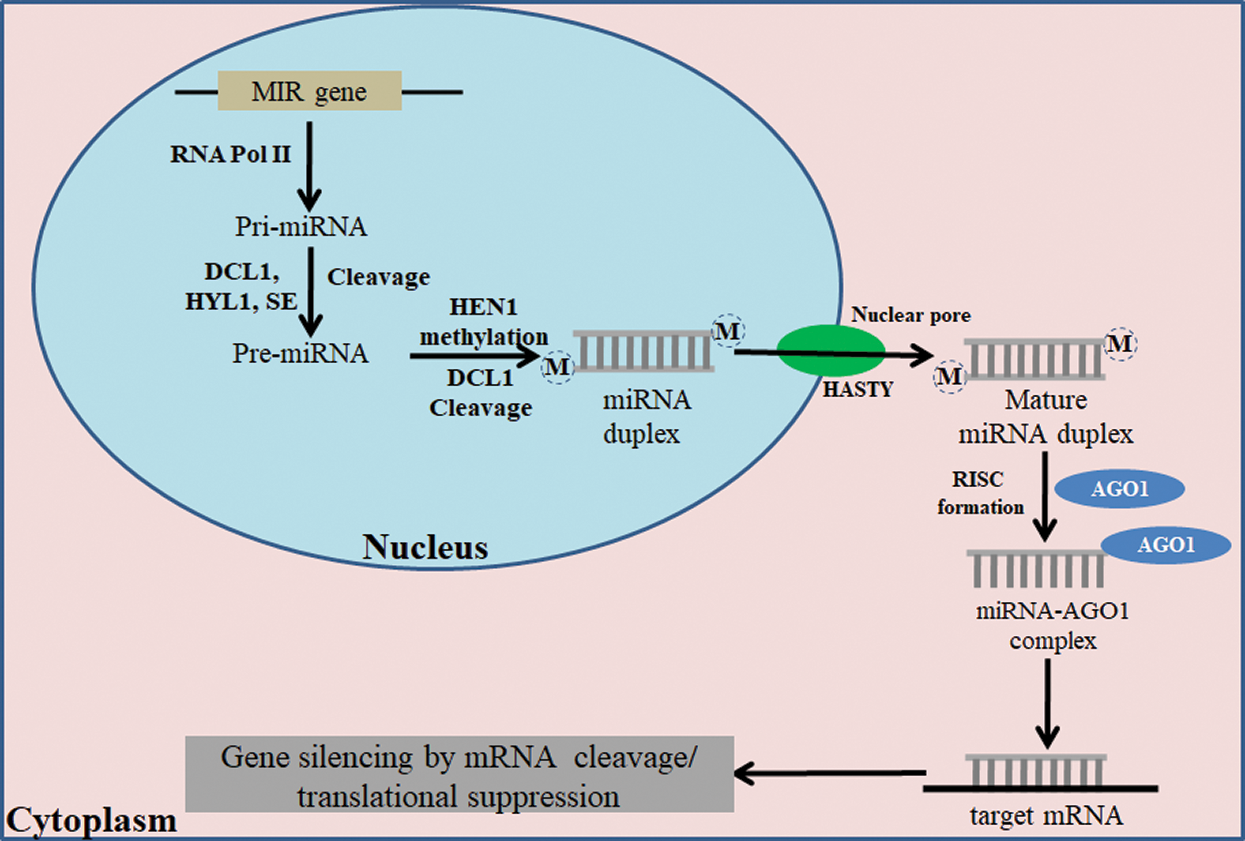
Figure 1: Mechanism of miRNA biogenesis in plants and mature miRNA generation in the nucleus. The mature miRNA is transported to the cytoplasm, and it further mediates post-translation repression and gene silencing with help of other co-factors. RNA pol II-RNA polymerase II; DCL1-Dicer-like 1; HYL1-hyponastic leaves 1; HEN1-HUA ENHANCER 1; AGO1-Argonaute1; RISC-RNA-induced silencing complex
In plants, most miRNAs bind and cleave to either the coding regions or the mRNA targets in the 5’ and 3’ UTR region, contributing to translational inhibition of the target mRNA. Plant miRNAs are known to regulate numerous biological functions as main regulators, including growth, organ recognition, metabolism, stress response, and physiological processes [4–6]. The miRNAs of plants control genes involved in different biological functions. However, their main targets are the transcription factor (TF) which modulates gene expression according to its growing habitat and biotic and abiotic factors ypresent during plant system growth and development. TFs control significant developmental processes such as cell division/expansion/differentiation, determination of organ recognition, phase transition, and nutrient homeostasis, while function as regulators of plant growth and production via the miRNAs-TF interplay [4,7]. Among plants, miRNAs are conserved and represent large families with a wide variety of targets, and their expression varies depending on the form of tissue, the type of organ, its developmental phases, and environmental biotic and abiotic conditions.
Subramanian et al. [8] identified 35 novel miRNA families and Zhang et al. [9] later identified sixty-nine miRNAs together with their targets and divided them into 33 families. From soybean nodules, a small RNA library was built and 32 small RNAs were reported from 11 miRNA families. Speech review and miRNA target prediction revealed that these miRNAs were active in nutrient acquisition and plant development [10]. Wild soybeans have registered 15 retained miRNAs and 9 new miRNAs, most of which have tissue-specific expression patterns [11]. Transgenic studies of miR482, miR1512, and miR1515 showed that their over-expression could lead to major increases in soybean nodule numbers [12].
129 miRNAs were identified from four small RNA libraries from root, seed, flower, and nodules in soybean; out of these, 42 miRNAs matched with known miRNAs in soybean or other species, while 87 novel miRNAs were identified and putative target were predicted [13]. Via small RNA and degradome-associated deep sequencing in soybean seeds, 26 new miRNAs were identified and their predicted targets included genes from the MYB, ARF, NAC, GRF, and TCP, TFs families as well as non-conserved genes, such as F-box protein, G-proteins, and gene silencing suppressor protein [14]. Deep-sequencing of degradome libraries from various soybean tissues, such as root, leaf, stem, inflorescence, and leaves collected at flowering stage led to the identification of sixty-nine novel miRNA from twenty-five novel miRNA families and two hundred eleven potential miRNA targets. Twenty-three miRNA targets were found to be guided by five miRNAs (GMA-miR393, gmamiR1508, GMA-miR1510, GMA-miR1514, and a novel miRNA-11) were also predicted that could produce secondary siRNAs [15]. Turner et al. [16] identified novel miRNAs and previously unknown family members for conserved miRNAs and classified all known soybean miRNAs based on their phylogenetic (conserved, legume- and soybean- specific miRNAs). Degradome data analysis revealed expression of miRNA-and/or phase RNA in lipid metabolism suggested that lipid metabolism in soybean is potentially regulated by a complex non-coding network, including miRNAs and phase RNAs [17]. Another study performed on root-, cotyledon-, leaf- and seed led to identification of miRNA regulatory networks with identification of six conserved GMA-miRNA families and three hundred sixty-five tissue-specific miRNA-target interactions (MTIs) containing two hundred and five conserved suggesting MTIs roles and mechanisms of miRNA-mediated regulation of tissue-specific growth and development in soybean [18]. Several miRNAs in soybeans have been revealed in recent years to play key regulatory roles in organ development [19–21], modulation [22]; nutrient signaling [23–25], seed development [26]; biotic and abiotic stress responses [27–29]. The characterization of novel miRNAs in soybeans is gaining more interest with the discovery of miRNAs as ultimate gene effector molecules. In addition, the rapid growth of soybean genomics and transcriptomics has provided an abundance of data and a considerable opportunity to delineate soybean genetic enhancement using miRNAs as the candidate molecule.
2 Functional Aspects of miRNAs in Soybean
Throughout its life cycle, soybean is exposed to many abiotic and biotic stresses. Drought, salinity, cold and heavy metals are the main abiotic stresses. Biotic stresses include infections caused by nematodes and insect pests caused by bacteria, viruses, fungi, and infestations. These stresses ultimately affect the crop yield. Exploration of miRNAs in soybean revealed many miRNAs with regulatory roles in different biological processes modulating stress responses, crop yield and nutritional quality [4,7]. These roles of miRNAs have been discussed in this review, and summarized in Figs. 2 and 3 and Tab. 1.
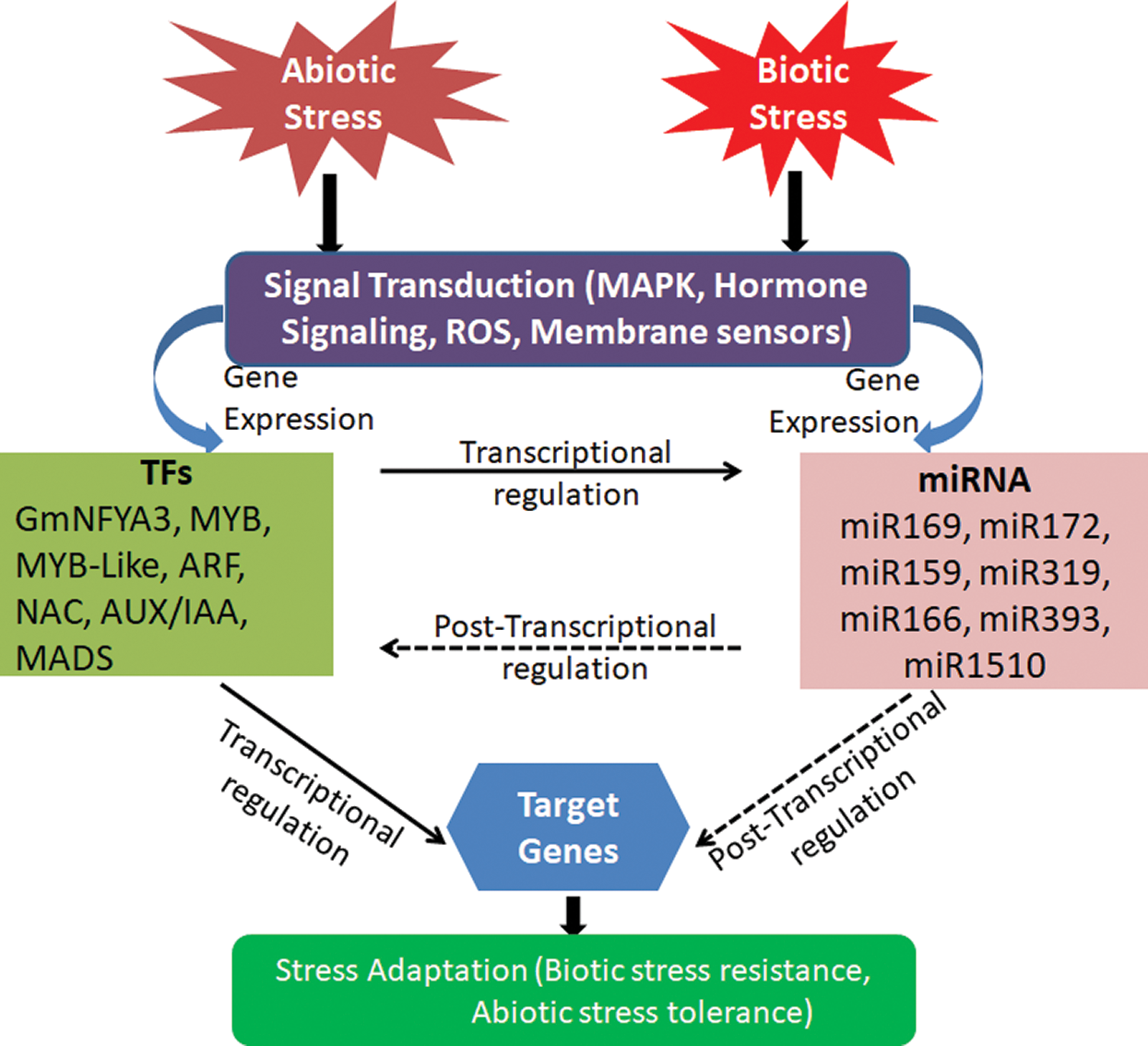
Figure 2: Interaction between various miRNAs and TFs in stress. Signal transduction initiated by abiotic and biotic stress and further contributes to activation of stress-tolerant miRNAs and TFs that control the target gene for stress adaptation
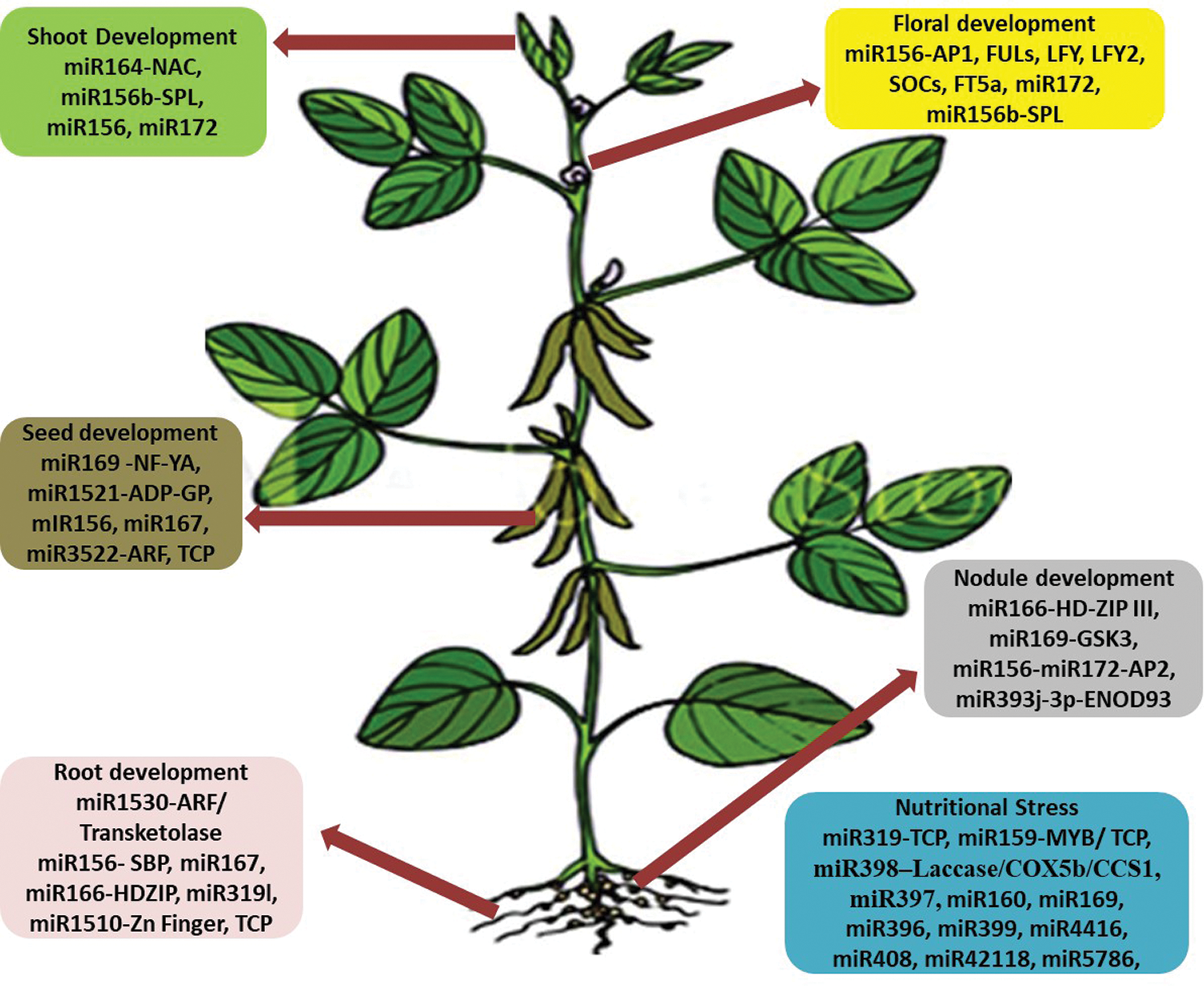
Figure 3: The role of numerous microRNAs and their target genes/TFs in the growth/development of soybean
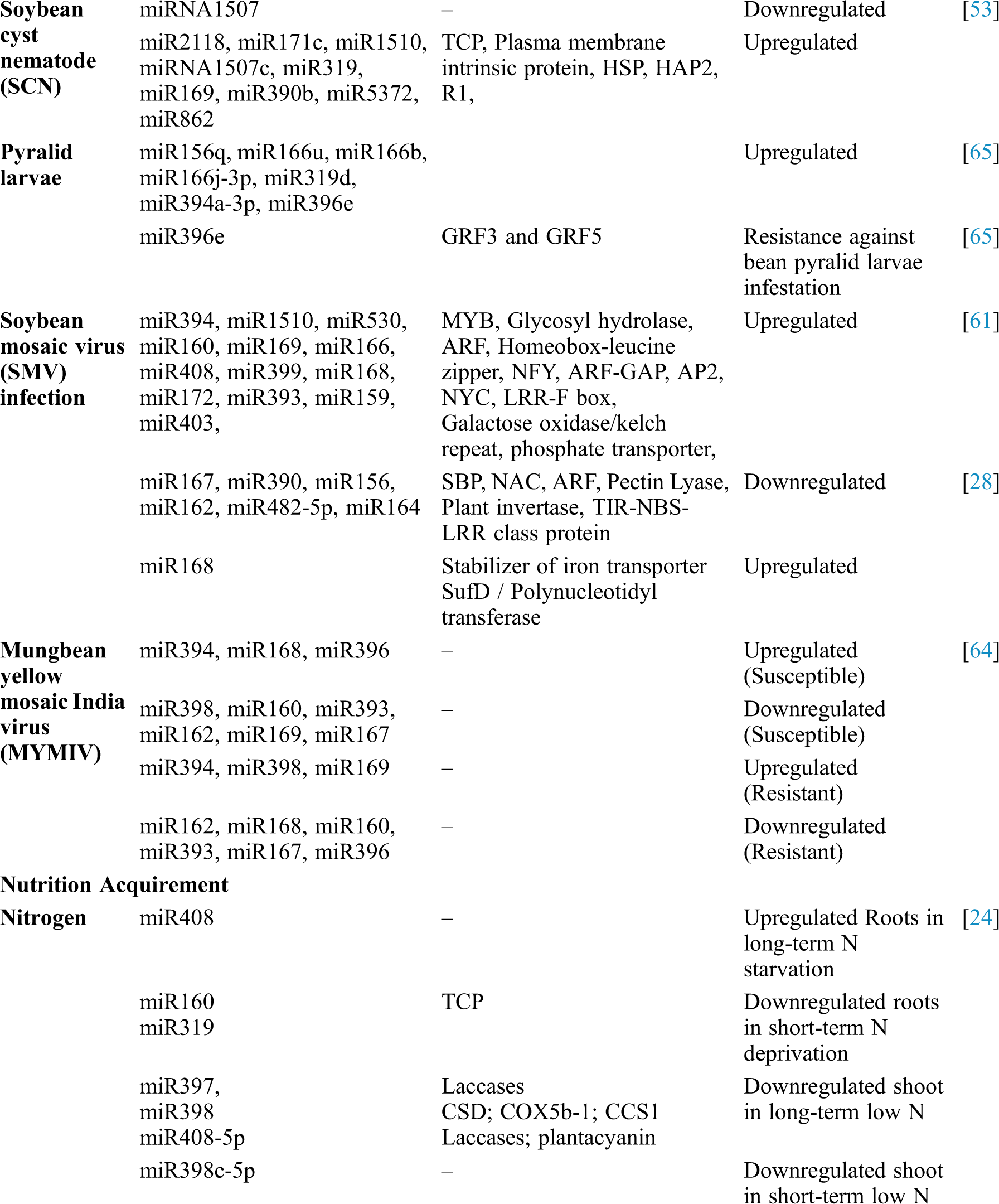
Table 1: Identification of soybean miRNAs showing differential expression and their role in different processes


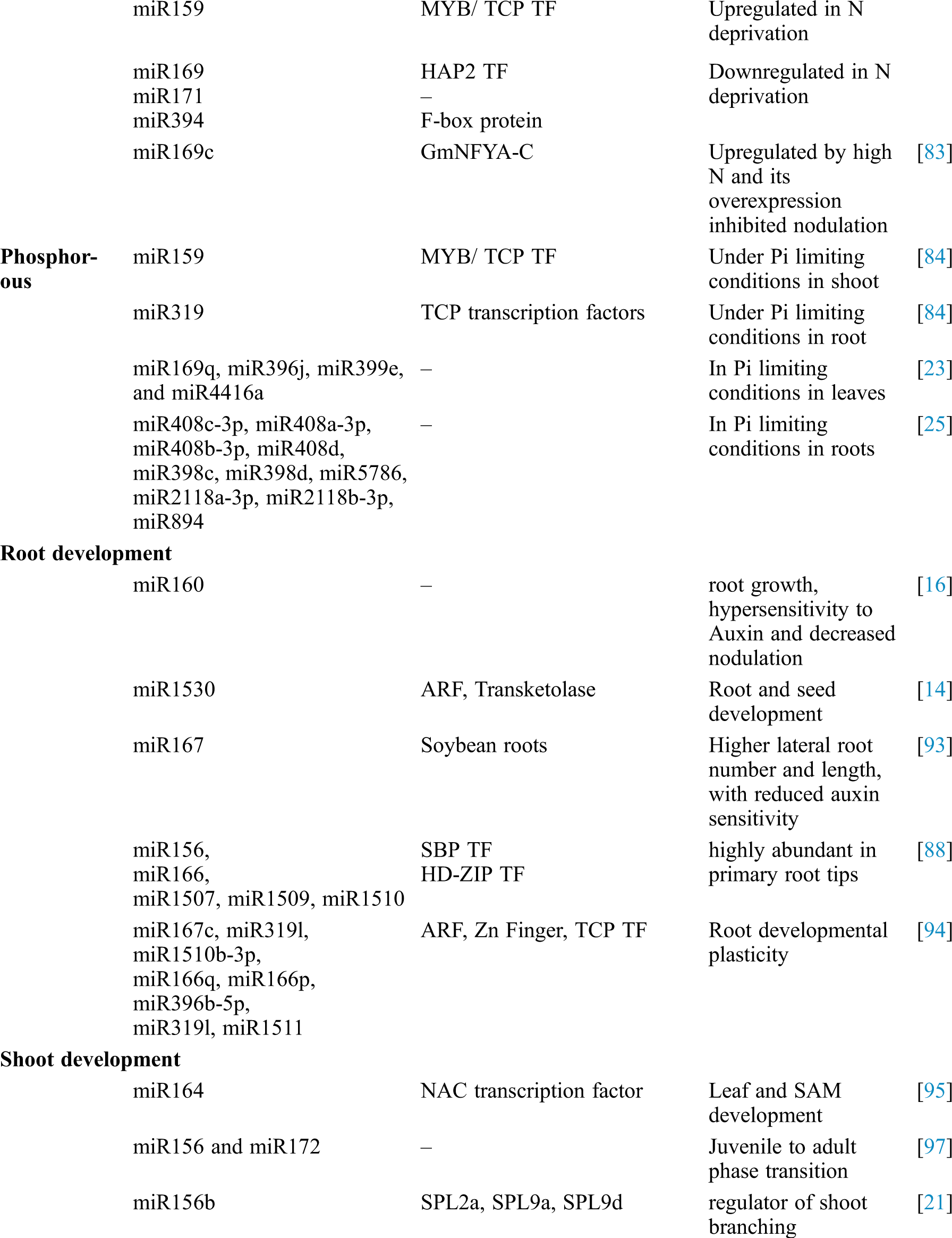

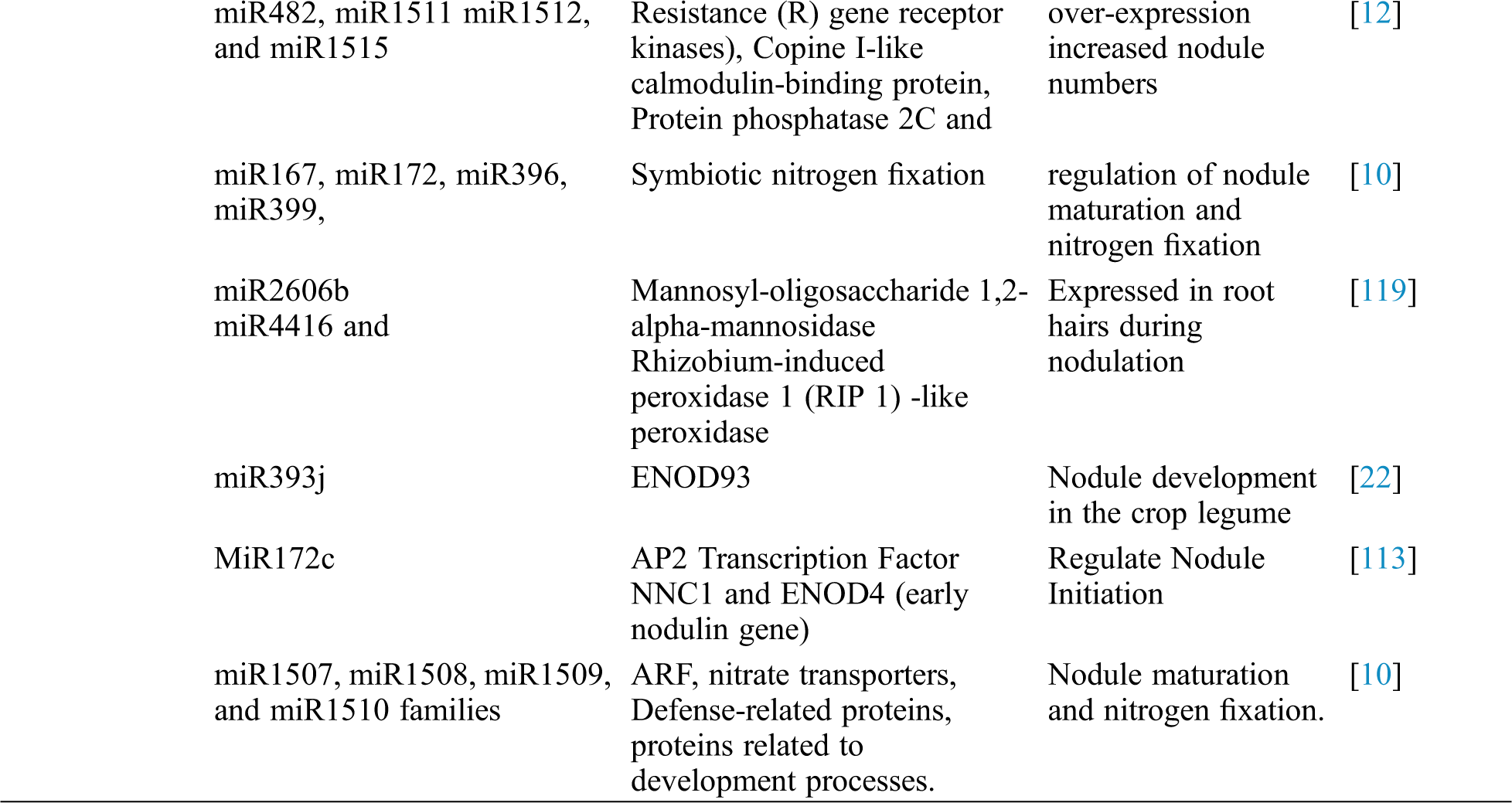
Abiotic stresses like drought stress, cold stress, heat stress, salt stress, and stresses laid upon by heavy metals are the major challenges to enhance the crop productivity in many crops [30–32]. However, plants usually develop some adoptive mechanisms to counteract these challenges. Micro-RNAs (miRNAs) are identified to have an important role in counteracting the effects of abiotic stresses. Like other crop plants, soybean also faces different abiotic stresses during different development stages, and diverse kinds of miRNAs have been characterized and well documented with their functional roles in soybean.
Drought, characterized by the water deficit, is considered one of the important environmental constraints affecting overall growth and development in many crops. The effects caused due to drought are apparent at all the stages of growth and development of plants and could be evident from morphological to molecular level [33]. In soybean, drought or water deficit causes severe yield losses by affecting nutrient uptake, carbon assimilation, stomatal movement and photosynthesis [32]. About 40% of yield loss has been estimated in soybean due to drought [34]. To understand the potentialities of miRNAs in soybean to drought tolerance, two genotypes with the contrasting response to drought (sensitive, tolerant) were exposed to drought stress and the resulted differential expressions of 11 miRNAs in both the genotypes were characterized [35]. Five miRNAs (miR169f-3p, miR166-5p, miR397ab, miR-Seq13, miR1513c) were upregulated in the sensitive genotype, and were downregulated in the tolerant genotype when exposed to drought stress [35]. Another target gene of miR169, GmNFYA3, was also characterized for its potential role in drought tolerance and found that the GmNFYA3 functions in positive modulation of drought stress [36]. Over-expression of miR172c from soybean was also found to be conferring tolerance to water deficit in transgenic Arabidopsis thaliana [37].
Like drought, salinity has also a profound effect on growth and yield of many crop plants, including the soybean. About 6% of the agricultural land over the world is exposed to excessive salinity stress [38]. Due to salinity stress, the soil rhizosphere impedes the roots and prevent uptake of water and essential nutrients. However, a number of miRNAs have been reported to express in crop plants to respond to the salinity stress [39–41]. Some candidate miRNAs were also identified in soybean with salinity responsiveness. Li et al. [29] analyzed the expressions of fifty miRNAs under different saline conditions, and among them, 46 miRNAs were found to be upregulated and four were downregulated in response to salt stress. Further, to understand the expression profile of soybean miRNAs under normal and salt-stressed conditions, deep sequencing of the miRNAs was carried out and was resulted in identification of two hundred thirty-eight miRNAs belongs to one hundred twenty-five miRNA families. Among them, one hundred four miRNAs have expressed differentially; four miRNAs (miR159c, gmamiR159b, miR169c and miR319a) showed more than tenfold decreased expression; 34 miRNAs were repressed; twelve miRNAs were induced due to salt stress in the matured root nodules of soybean [42]. Further exploration of these miRNAs identified 770 mRNA targets; among the majority are the transcription factors. The targets genes are found to perform various functions like ubiquitin-conjugating enzymes and Ca2+/calmodulin-dependent protein kinase [42].
Another study revealed miR172c as a salt responsive miRNA. Increased sensitivity to salinity stress was observed in a transgenic plant over-expressing the miR172c whereas reduced sensitivity to salt stress was observed when the miR172c was knocked down. The miR172c target gene, Nodule Number Control 1 (NNC1), was also downregulated in response to salt stress [43]. miR390-directed TAS3 cleavage was only found under long-term salt stress in soybean roots. Considering the fact that miR390-TAS3 pathway is highly conserved among plants, it might play an universal role when plants facing long-term abiotic stress [44]. GMA-miR169c role as a negative regulator in drought role has been described in gma-miR169c overexpressing Arabidopsis plants that showed higher sensitivity to drought stress and lower survival than WT plants. AtNFYA1 and AtNFYA5 TFs were predicted to be regulated by GMA-miR169c; this was also correlated with the reduction in stress-responsive genes RD29A, RD22, GSTU25 and COR15A expression [45]. Salt stress-induced expression of fifty-four miRNAs in Soybean drought-tolerant cultivar (Conquista) in comparison to drought-sensitive cultivar (C08); target genes of these miRNAs correspond to stress adaptation mechanism [46]. Expression of GMA-miR1508a induced in cold but was suppressed in drought stress; its overexpression in soybean resulted in cold resistance during germination and seedling phases but dwarfing and thickening of the cell wall were shown by plants [47]. Cold stress, which is characterized by either freezing (<0°C) or chilling (<20°C) is another detrimental abiotic stress affect the growth of plants and agricultural productivity [33]. Cold stress reduces the nitrogen fixation rate and ultimately causes yield loss in soybean. Eight numbers of miRNAs responsive to cold stress have been identified in nitrogen-fixing soybean root nodules [30]. In chilling condition, three of these miRNAs (miR397a, miR166u, miR171p) were upregulated while the expression of the rest.
(miR169c, miR159b, miR319a/b, miR5559) was significantly downregulated. Similarly, 51 cold responsiveness miRNAs with 898 mRNAs target transcripts have also been identified in vegetable soybean. The mRNAs target transcripts were associated with redox chemical reactions and different signaling pathways [48].
Heavy metal stress is another major environmental stress which is increasing due to heavy industrial activities which affects normal plant growth and productivity in plants. These include aluminium, arsenic, cadmium, lead, and mercury. Treatment of 50 μM AlCl3; among two soybean genotypes, BX10 (Al-tolerant) and BD2 (Al-sensitive), led to differential expression of 32 miRNAs. miRNAs, associated with root elongation, such as GMA-miR166k/o, GMA-miR390g, and gma-miR396c/k, DE in BX10 can help in Al tolerance in BX10 compared to BD2 [31]. Zeng et al. [49] identified thirty Al-responsive miRNAs in wild soybean. Out of those, ten miRNAs belong to seven conserved miRNA families, thirteen un-conserved miRNAs and seven novel miRNAs. Among DE miRNAs, miR396 and miR390 related to heavy metal stress were up-regulated in response to Al-stress and can be linked to metal tolerance in soybean. Another study reported differential expression of twenty-six miRNA in response to cadmium stress between two genotypes, Huaxia3 (HX3, Cd-tolerant) and Zhonghuang24 (ZH24, Cd-sensitive). Among them, nine were detected in both cultivars, while five were expressed only in HX3 and twelve only in ZH24 [50].
In addition to abiotic stresses, soybean is also affected by various biotic stresses such as insect-pests, and diseases caused by bacteria, virus, fungi, and nematodes [27,51]. The miRNAs’ role in mitigating biotic stresses has been characterized well in soybean.
The Heterodera glycines (soybean cyst nematode), is a major pest of soybean causes tremendous damage to soybean [52]. Various studies found responsive miRNAs acts against soybean cyst nematode (SCN) [53]. Comparative miRNAs profiling between a SCN resistant cultivar (Hairbinxiaoheidou) and a SCN susceptible cultivar (Liaodou 10) of soybean revealed one hundred one numbers of SCN-responsive miRNAs belongs to fourty families. Among the one hundred one SCN-responsive miRNAs, twenty were expressed differentially between the cultivars. In another study, siRNA of two soybean cultivars, SCN susceptible (KS4607) and another SCN resistant (KS4313N) were sequenced and sixty miRNAs (responsive to SCN) belong to twenty-five miRNA families were identified [54]. During SCN infection a number of legume-specifc miRNAs including miR2109, miR4996, miR1509, miR2118 and miR1510 were abundantly found along with the conserved miRNAs in soybean.
In addition to cyst nematode, fungal infection also severely damages the soybean crop. A major fungal pathogen of soybean, Phakopsora pachyrhizi, is the causal agent of foliar disease, the Asian soybean rust (ASR) which causes 10% to 90% damage in soybean [55,56]. In an expression study, miRNAs such as miR166f, miR166a-5p, miR397ab, miR-seq13 and miR169-3p were found to be down-regulated in a susceptible genotype (Embrapa 48) but no any expression was observed in the resistant genotype (PI561356) upon ASR infection. Again, another miRNA, miR4415b, showed reduced expression in the susceptible genotype (PI561356) when infected with ASR [35].
Phytophthora sojae is another devastating pathogen of soybean accounting for worldwide losses of about 2 billion US dollars of revenue each year [57,58]. Wong et al. [19] identified two miRNAs, miR393 and miR166, responsive to P. sojae infection and indicated their soybean basal defense involvement. They observed increased susceptibility of soybean to P. sojae infection when the level of mature miR393 was knocked down. Again, expression of isoflavonoid synthesizing genes was also radically decreased in the knockdown miR393 roots. Cui et al. [59] revealed that the over-expression of a miRNA, miR1510a/b, associated with susceptibility of soybean hairy roots to P. sojae infection.
Soybean mosaic virus (SMV) infection is also another devastating viral infection causes major productivity loss in soybean. During the early stages of infection, some strains of SMV causes downregulation of numerous defensive genes [60]. Profiling of SMV-inoculated soybean plants identified one hundred and seventy-nine miRNAs responsive to viral infection. miRNAs such as miR393, miR160 and miR1510 found to be resistant to SMV [61]. In addition to SMV infection, soybean is also affected by DNA viruses like mungbean yellow mosaic India virus (MYMIV) and Mungbean yellow mosaic virus (MYMV). Down-regulation of some miRNAs was observed in soybean when infected with MYMIV and MYMV transcripts [62,63]. Changes in the expression patterns of conserved miRNAs, putative miRNAs and their target transcripts were observed in two genotypes of soybean [susceptible (JS335), resistant (UPSM534)] when infected with MYMIV [64]. Moreover, the pattern of expressions of miRNAs resulting from soybean advocates the importance of argonaute (AGO) homeostasis and regulatory hormonal signaling pathways for controlling virus resistance [64].
Involvement of Gma-miR156q, Gma-miR166u, Gma-miR166b, Gma-miR166j-3p, Gma-miR319d, Gma-miR394a-3p and Gma-miR396e have been predicted to contribute resistance against soybean pyralid larvae. Gma-miR396e mainly target the GRF genes; GRF3 and GRF5 to enhance the resistance against bean pyralid larvae infestation [65].
2.3 Plant Growth and Development
Plant growth and development require optimal amount of macro and micronutrients from the soil to drive several metabolic processes. These nutrients serve both structural and catalytic roles, either by acting as structural component of several proteins and enzymes or catalyzing of plant metabolism. These nutrients play crucial role in the vital processes of plant system, such as photosynthesis, cellular respiration, growth and differentiation, etc. [66–68].
miRNAs are involved in organizing the nutrient signal, physiological processes that help plants adapt to nutrient stresses and toxicity and survive. Studies have shown that plants have developed advanced mechanisms for detecting their nutrient levels and responding to changes in nutrient availability [69,70]. The completion of genome sequences of several plant species and the development of modern genomics approaches have established a large number of regulatory components involved in reacting to nutritional stress [24,71,72]. By mediating post-transcriptional modulation of multiple effector genes, miRNAs have been active in plant responses to nutritional stresses [73–75].
Nitrogen (N) is an important plant macronutrient, which greatly influences seed development and nutrition in plants [76] and thus required by high-yielding crops insignificant amount [77]. During long-term N starvation conditions in soybean roots, miR408 family members were up-regulated and in short-term N starvation, miR160 and miR319 families were down-regulated [24]. Some species of the miR397, miR398 and miR408-5p family are down-regulated in response to long term N starvation in soybean shoots, and miR398c5p has been found to be downregulated in response to short-term starvation indicates conserved role of these miRNAs in nitrogen acquisition among plants like maize [78]. miR169 in maize and soybean has also been identified as N sensitive [24,78,79] and suggests its role in adaptation to N-deficiency in soybean. Changes in miRNA expression have been observed and linked to adaptive responses to N supply and transportation modification in root architecture, metabolite accumulation and free radical scavenging machinery, growth and flowering [80–82]. miR169cexpression is induced by high Nitrogen, its overexpression targeted 3’-UTR of GmNFYA-C and inhibited nodulation while CRISPR-cas9 mediated knockout promoted nodulation in soybean mediated through expression of gene GmENOD40 [83].
Phosphorus (Pi), an essential plant macronutrient plays a vital role in plant growth and productivity. In soybean nodule under Pi-deficiency miRNAs were detected to be responsive in Pi-deficiency. Few miRNAs have been reported in soybean to response to the Pi deficiency using Microarray and Next-Generation Sequencing [23,84,85]. miR399 expression was induced in soybean root similar to Arabidopsis, under Pi starvation, which suggests the essential role of miR399 in Pi starvation [86,87]. Under + Pi conditions, twelve and two miRNAs were detected in leaf and root respectively in Pi deprived conditions, out of which ten miRNAs were unique to the leaf and four to root. Under Pi limiting conditions miR169q, miR396j, miR399e, and miR4416a, Pi limits were strongly induced in leaves of strived plants but not detected in control plants [23]. Pi limitation induced expression of eight (miR156/157, miR159, miR1507, miR167, miR168, miR319, miR1509, and miR894) and seven miRNAs (miR1507, miR1509, miR159, miR396, miR474, miR482, and miR894) in soybean leaves and roots [88], respectively. Upregulation of miR5373 and ten miRNAs (miR408c-3p, miR408a-3p, miR408b-3p, miR408d, miR398c, miR398d, miR5786, miR2118a-3p, miR2118b-3p, and miR894) were down-regulated in roots under Pi starvation [25] suggest involvement of miRNAs in response to root Pi starvation in soybean. A total of seven hundred seventy-seven miRNAs were differentially expressed (DE) in response to different Pi levels. DE miRNAs’ putative targets mainly included ERF, ARF, zinc finger protein, MYB, and NAC domain TFs [89].
Roots are responsible for providing support to aerial part of the plant and are responsible for water uptake, nutrients translocation, hormone signaling, secondary metabolites biosynthesis, and protection of plant against stress. Root development occurs at embryonic and postembryonic stages with the formation of plant root system from primary and lateral roots [90]. miRNAs are important players in root initiation, development, root growth and root morphogenesis [91]. miRNAs are reported to regulate a zipper transcription factor and a nucleocytoplasmic transporter known for cell organization in root apical meristem leading to root length reduction, and also to xylem overgrowth in the stem [92]. Soybean miR167-ARF8 interaction was found to be a key regulator for root nodule and lateral root development. miR167c overexpression in roots exhibited significantly different root architecture with higher lateral root number and the lateral root length in miR167c plants compared to control plants [93].
Role of miRNAs has been observed in the root apical meristem of control as well as water deficit conditions. Five miRNA families namely miR156, miR166, miR1507, miR1509 and miR1510 were found to be highly abundant in primary root tips of which miR1507 is the most abundant miRNA [20]. Overexpression of miR172c promoted root formation during the stage of hairy root initiation and the number of lateral roots was increased in miR172c overexpressing plants [43]. Five miRNAs were specifically expressed in soybean roots treated without or with salt stress miR167c, miR319l, miR1510b-3p, miR166q, miR166p were found in the soybean. The abundance of miR396b-5p, miR319l, and miR1511 was observed very high in the control and salt-stressed root tissues, suggesting their involvement in the RAM of soybean [94]. These examples support soybean miRNAs to regulate the root meristem management process, primary root, and lateral root biogenesis.
The shoot apical meristem (SAM) is a region that contains stem cells that give rise to the above-ground organs of the plants, such as leaves and stem. A microarray study in soybean reported 31 and 42 miRNAs expressed in the SAM or leaf, respectively, and six of them were found to be legume specific miRNAs, suggesting their crucial roles in mediating SAM development. The higher number of miRNAs expression in the leaf than SAM can be related to the structural complexity, metabolic and developmental networks in leaves. The identified potential miRNAs can be key regulators and help understand miRNAs’ regulatory role in the soybean SAM during shoot development [95].
miR159 expression was observed in the primordial SAM and leaf and its pattern of expression is close to that recorded in Arabidopsis for miR159 [96], indicating a conserved role of miR159. The expression of miR166 was found beneath and on the abaxial side of the incipient leaf in soybean, while its star strand miR166* was found to be weak at the tip of leaf primordia but stronger at the lateral portion of the SAM with stronger in older leaf primordia indicates their involvement in controlling leaf development and suggests diversification of miRNA function [95]. The leaf’s different position can also show the difference in expression behavior of miRNAs in soybean seedlings from two leaf stages up to 6 leaf stages [97].
The expression of miR156 was high in the two-leaf stage and decreased moderately in the third leaf, which down-regulated in stages of fourth to sixth leaves that decreased the expression of miR156 expression with development. In case of miR172 inverse expression pattern, low in first and second leaves; increase in the third and fourth leaves and ultimately reached to highest in fifth and sixth leaves. This antagonistic expression pattern of miR156 and miR172 genes is associated with juvenile-adult transition and suggest miR156 and miR172 role in vegetative phase transition in soybean [97].
Two SPL13 genes (SPL13Ba and SPL13Bb) were predicted as target of miR156b at translational level; the rest of the SPL mRNAs are likely cleaved by miR156b. In the shoot apex and axillary buds of miR156bOE-5 plants, SPL2a, SPL9a and (particularly) SPL9d were significantly down-regulated, while the expression of other target genes was marginally down-regulated or not altered. Number of branches were found to increase in soybean miR15*6b overexpressing plants, which supports miR156 as a regulator of shoot branching [21].
The transformation from vegetative to reproductive stages is known as floral transformation, which comprises of vast changes during a plant’s life cycle. The conversion of shoot meristem into the reproductive organ is due to the interplay of several miRNAs and their target genes. MIR156 and MIR172 facilitate reproductive stage transformation in the vegetative tissue, caused by age or environmental signals. miR156b can negatively target SPL orthologs SPLs, and thus delays flowering. miR156b down-regulates flower time regulators like AP1, FULs, LFY, LFY2, SOCs, FT5a, and miR172 [98]. a-miR172a has role in a diurnal rhythm its abundance increased rapidly as the plants grew until the initiation of the soybean flowering phase. One of the a-miR172a target gene, AP2 like gene is involved in flowering. Over-expression of miR172a produced early flowering and increased expression of FT, AP1 and LFY was observed. miR156 and 172 play an important role in flowering by suppressing an AP2 TF which increased expression of LFY, FT1 and AP1 to encourage flowering [99].
A total of 200 mature miRNAs were identified by Kulcheski et al. [100] in soybean flower, of which 41 belonged to 32 new novel miRNA families. Expression of the flower specific and novel miRNAs in the different flower whorls suggests their role to be important in soybean flower regulatory network. The example of Cytoplasmic male sterile (CMS) plants puts a light on the flowering regulation. Two small RNA and two degrading libraries were constructed from the flower buds of the soybean CMS line NJCMS1A, and its restorer line NJCMS1C reported five hundred and fifty-eight known miRNAs, one hundred and three novel miRNAs, and some base-edited miRNAs [101]. Seventy-six differentially expressed miRNAs were discovered among the identified miRNAs between NJCMS1A and NJCMS1C. miR4393a and miR5667 were represented only in NJCMS1A and miR4346, miR4372a, miR4400, miR4994-5p, and miR5785 in NJCMS1C (restorer line) suggests involvement of these miRNAs in flower bud development [102]. Importantly, nearly half of the miR156 family members showed higher expression in the soybean CMS line’s flower buds than its maintainer line and GmSPL9 is negative regulated by miR156b during soybean flowering. Some important flowering-related gene like LBD22, LBD36, AGL30 and AGL104 which highly express in soybean flower were down-regulated in CMS NJCMS1A flower buds compared with NJCMS1B. Homologous genes of these four members were all down-regulated in flower buds of Arabidopsis: MIR156b transgenic lines compared to WT conclude that the miR156-targeted SPLs regulate these genes and play an essential role in the development of the flower bud [102].
In soybean seeds, during growth, a lot of nutritional compounds are accumulated. Soybean is the world’s most significant legume, commercially and essential double-purpose crop with seeds enriched with proteins and oils that provide food for livestock and human consumption. Soybean cotyledons have formed as complex sink tissue for protein and oil accumulation that directly affects the yield and quality of soybean seeds. The storage of proteins and oil in soybean cotyledons directly affects the yield and consistency of soybean seeds. Too little is known about soybean cotyledon miRNAs and their regulatory networks. Discovery of novel miRNA involved in soybean seed development will facilitate understanding of miRNA regulatory network in improving soybean seed quality. miRNA family members miR156, 159, 160, 164, 166, and 167 were reported from seed coats and cotyledons [103].
Shamimuzzaman et al. [104] studied miRNAs in soybean seed production to address miRNA-directed gene regulation in soybean seed development. They established a total of one hundred and eighty-three separate targets of 53 recognized miRNAs of soybean. A total of three hundred four miRNA genes were expressed in soybean cotyledons and one thousand nine hundred and ten targeted genes were predicted by Goettel et al. [105], among which one hundred thirty novel miRNA genes and seventy-two novel miRNA families were reported to be expressed in cotyledons. miRNAs (miR156, miR166, miR319, miR159, miR164, miR167, miR482, miR1508, miR1510 and miR3522) were highly expressed in seed development stages and TFs such as ARF and TCP containing a CCAAT box were predicted as their target.
A total of one hundred and fourty five genes were identified as miRNA targets and twenty-five as target for novel miRNAs [14]. TFs from the ARF, GRF NAC, MYB, and TCP families were found to be the majority of miRNA targets. Shamimuzzaman et al. [104] reported seven NF-YA TFs to be target in seed coats and cotyledons, miR169 family members will control NF-YA TFs during the production of soybean seeds. The majority of the known TF targets for soybean miRNA are ARFs, MYBs, TCPs, NACs, HD-ZIPs and NF-Y subunits with explained role in plant development. Strong expression of miR167 and miR1512 in seed coats and miR156 and miR3522 in cotyledons were observed [106]. Five of the MIR166 family members showed increment in expression during seed development reaching highest at the later stage of soybean seed development may be involved in the seed maturation program [107]. Seventeen novel miRNAs and fourteen isoforms of miRNA (isomiRs) related to seed development were identified. These novel miRNAs and isomiRs that represented tissue-specific expression and the isomiRs showed significantly higher abundance in different tissues indicating their regulatory roles in soybean seed development [108].
Soybean oil flavor and stability can be improved by reducing the content of alpha-linolenic acid (18:3). To down-regulate omega-3 fatty acid desaturase (enzyme catalyze linoleic acid (18:2) to alpha-linolenic acid (18:3), Flores et al. [109] used hairpin RNA strategy under seed-specific glycinin promoter with decreased alpha-linoleic acid content (1–3%) were observed (7–10%) in transgenic soybean seeds compared to non-transgenic seeds.
Legume plants in association with nitrogen-fixing soil bacteria, form a specialized organ, the nodule. The molecular mechanisms of signaling that regulate the formation of nodules have been extensively characterized, and microRNAs (miRNAs) have been reported in different legumes nodulation processes [8,13,110]. Subramanian et al. [8] identified involvement of several miRNAs in soybean like miR172, miR166, miR396 during nodulation. Inoculation of soybean roots with Bradyrhizobium japonicum, resulted in up-regulation of miR159 and miR393, downregulation of miR160 and miR1693 3 h after infection. miR168 and miR172 were found to up-regulated at 3 but down-regulated at 12 h and returned to basal level. MiR166 was reported to regulate the expression of class III homeodomain-leucine zipper (HD-ZIP III) genes in both Medicago truncatula and soybean, thereby modulating root and nodule development [8,111]. Similarly, in the later stages of the soybean-Rhizobium nodulation process, thirty-two separate miRNAs belonging to eleven miRNA families (for example, miR167, miR172, miR396, and miR399 families) were accused [10]. miR169 and miR482, miR1512 are involved in nodule numbers by regulation of target like GSK3-like kinase and putative Dicer-like protein [12]. Transgenic studies found over-expression of microRNAs; miR482, miR1512, and miR1515 to increase soybean nodule numbers [12]. MiR172 has retained a role in transforming the developmental phase, the identity of the floral organ, and the flowering period in plants; yet miR172’s particular role was found to be in the nodulation process. Over-expression of soybean miR172 culminated in a rise in nodule numbers in transgenic roots and in nodules of symbiotic leghemoglobin and nonsymbiotic hemoglobin, indicating higher levels of nitrogenase activity. miR156 mediated regulation of miR172 expression was reported which regulate the transcript level of an AP2 TF linked to nonsymbiotic hemoglobin, responsible for levels of nodulation [112]. One study on soybean reported that miRNA172 modulate both Rhizobium infection and nodule organogenesis. Inoculated soybean roots with either B. japonicum or lipooligosaccharide Nod factor strongly upregulated miR172c during nodule growth [113].
Symbiotic relationship with nitrogen-fixing bacteria (Rhizobia) to receive nitrogen during nodulation of plant roots [114,115]. Bean rhizobial infection increased the level of miR172c expression to rise until the stage of nodule growth, while uninfected nodules displayed low miR172c and high AP2 levels. Increased rhizobial infection, root growth, and higher expression level of nodulation genes resulted from over-expression of miR172c, which improved nodulation and nitrogen uptake [116]. miR393j-3p control ENOD93 mRNA levels, affects nodule meristematic activity and nitrogen-use efficiency during early nodule development stages; which can be an important control point in soybean nodule formation [22]. miR160 mediated targeting of ARF10/16/17 helps fine-tune auxin and cytokinin action in nodulation [117].
miR171o and miR171q expression are negatively associated with that of their target genes. These miRNAs in transgenic hairy roots resulted in a substantial decrease in nodule formation targeting GRAS TFs members. GRAS TFs SCL-6 and NSP2 are known to influence expression of the genes Nodule Inception (NIN), Early Nodulin 40 (ENOD40) and Ethylene Response Factor Required for Nodulation (ERN) during the process of Bradyrhizobium japonicum-soybean nodulation [118].
Soybean is a legume crop model that is used to explain plant morphology, adaptation, and domestication in practical genomics. A selection of data sets, such as the abundance of mRNA transcripts, small RNA communities and methylation status, offer unprecedented insight into gene regulation during developmental and environmental responses. There are also undefined and little-known features of many miRNAs as well as their operating mechanisms. miRNAs preserved in broad families include representatives with anonymous functions; exploring the involvement of different family members in relation to tissue/organ-specific tasks is of great importance. High-performance techniques of sequencing have assisted researchers with miRNA genome-wide identification and characterization supported by different computational tools and public web services. This has allowed the effective and quick identification of defining the targets of miRNAs in degradome sequencing with some limitations. A reliable method for improving crops is the ability of miRNA modification, but unintentional side effects may also be recorded since adverse improvements in plant growth and morphology may be seen separately from favorable and beneficial ones [120]. In the in-silico tools and databases developed in recent years, modifications are required to provide excellent help for the identification and prediction of the target gene networks of candidate miRNAs involved in plant development, growth and stress responses.
By modifying the target miRNA that controls the number of plant genetic traits [121,122], efficient bioengineering and endogenous RNA regulatory network modification are feasible, so miRNA offers an important basis for the use of quantitative and qualitative features for bioengineering. In addition to the essential role of miRNAs in the suppression of a target gene expression, more dynamic and diverse roles can be performed by miRNAs in crops. The elucidation of the roles of the miRNA will extend our comprehension and help to identify breeding strategies by following suitable goals for agronomic significance characteristics [123]. Through collecting observations of miRNA characteristics, innovations or the creation of new miRNA target prediction and validation methods, increased plant transformation techniques and more powerful gene-manipulating techniques such as short tandem target mimic (STTM) and CRISPR/Cas9 are used. An inexpensive and appealing genome-editing technique has been shown to be CRISPR/Cas9 and can be used for crop enhancement technology. It is therefore more essential for the creation of CRISPR/Cas9 controlled editing systems for targeting miRNA locus and miRNA binding sites.
This will allow researchers and breeders to understand the function of miRNA in improving plants by incorporating modern biotechnological techniques. With the combined efforts of various omics technologies and post-genomics resources, expanding awareness of miRNA regulatory roles will provide better molecular breeding options to speed up the selection process, and significantly shorten the time necessary for improved variety development. This modification of miRNA genes and miRNA tolerant target genes will help to enhance the engineering of desirable features in agricultural products.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no interest in reporting regarding the present study.
1. Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T. et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature, 463(7278), 178–183. DOI 10.1038/nature08670. [Google Scholar] [CrossRef]
2. Rogers, K., Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell, 25(7), 2383–2399. DOI 10.1105/tpc.113.113159. [Google Scholar] [CrossRef]
3. Chapman, E. J., Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nature Reviews Genetics, 8(11), 884–896. DOI 10.1038/nrg2179. [Google Scholar] [CrossRef]
4. Jones-Rhoades, M. W., Bartel, D. P., Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology, 57(1), 19–53. DOI 10.1146/annurev.arplant.57.032905.105218. [Google Scholar] [CrossRef]
5. Willmann, M. R., Poethig, R. S. (2007). Conservation and evolution of miRNA regulatory programs in plant development. Current Opinion in Plant Biology, 10(5), 503–511. DOI 10.1016/j.pbi.2007.07.004. [Google Scholar] [CrossRef]
6. Sunkar, R., Li, Y. F., Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends in Plant, Science,17(4), 196–203. DOI 10.1016/j.tplants.2012.01.010. [Google Scholar] [CrossRef]
7. Bartel, B., Bartel, D. P. (2003). MicroRNAs: At the root of plant development? Plant Physiology, 132(2), 709–717. DOI 10.1104/pp.103.023630. [Google Scholar] [CrossRef]
8. Subramanian, S., Fu, Y., Sunkar, R., Barbazuk, W. B., Zhu, J. K. et al. (2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics, 9(1), 160. DOI 10.1186/1471-2164-9-160. [Google Scholar] [CrossRef]
9. Zhang, B., Pan, X., Stellwag, E. J. (2008). Identification of soybean microRNAs and their targets. Planta, 229(1), 161–182. DOI 10.1007/s00425-008-0818-x. [Google Scholar] [CrossRef]
10. Wang, Y., Li, P., Cao, X., Wang, X., Zhang, A. et al. (2009). Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochemical and Biophysical Research Communications, 378(4), 799–803. DOI 10.1016/j.bbrc.2008.11.140. [Google Scholar] [CrossRef]
11. Chen, R., Hu, Z., Zhang, H. (2009). Identification of microRNAs in wild soybean (Glycine soja). Journal of Integrative Plant Biology, 51(12), 1071–1079. DOI 10.1111/j.1744-7909.2009.00887.x. [Google Scholar] [CrossRef]
12. Li, H., Deng, Y., Wu, T., Subramanian, S., Yu, O. (2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiology, 153(4), 1759–1770. DOI 10.1104/pp.110.156950. [Google Scholar] [CrossRef]
13. Joshi, T., Yan, Z., Libault, M., Jeong, D. H., Park, S. et al. (2010). Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinformatics, 11(S1), S14. DOI 10.1186/1471-2105-11-S1-S14. [Google Scholar] [CrossRef]
14. Song, Q. X., Liu, Y. F., Hu, X. Y., Zhang, W. K., Ma, B. et al. (2011). Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biology, 11(1), 5. DOI 10.1186/1471-2229-11-5. [Google Scholar] [CrossRef]
15. Hu, Z., Jiang, Q., Ni, Z., Chen, R., Xu, S. et al. (2013). Analyses of a Glycine max degradome library identify microRNA targets and microRNAs that trigger secondary siRNA biogenesis. Journal of Integrative Plant Biology, 55(2), 160–176. DOI 10.1111/jipb.12002. [Google Scholar] [CrossRef]
16. Turner, M., Nizampatnam, N. R., Baron, M., Coppin, S., Damodaran, S. et al. (2013). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiology, 162(4), 2042–2055. DOI 10.1104/pp.113.220699. [Google Scholar] [CrossRef]
17. Ye, C. Y., Xu, H., Shen, E., Liu, Y., Wang, Y. et al. (2014). Genome-wide identification of non-coding RNAs interacted with microRNAs in soybean. Frontiers in Plant Science, 5, 743. DOI 10.3389/fpls.2014.00743. [Google Scholar] [CrossRef]
18. Wang, R., Yang, Z., Fei, Y., Feng, J., Zhu, H. et al. (2019). Construction and analysis of degradome-dependent microRNA regulatory networks in soybean. BMC Genomics, 20(1), 534. DOI 10.1186/s12864-019-5879-7. [Google Scholar] [CrossRef]
19. Wong, J., Gao, L., Yang, Y., Zhai, J., Arikit, S. et al. (2014). Roles of small RNA s in soybean defense against Phytophthora sojae infection. Plant Journal, 79(6), 928–940. DOI 10.1111/tpj.12590. [Google Scholar] [CrossRef]
20. Zheng, Y., Hivrale, V., Zhang, X., Valliyodan, B., Lelandais-Brière, C. et al. (2016). Small RNA profiles in soybean primary root tips under water deficit. BMC Systems Biology, 10(S5), 126. DOI 10.1186/s12918-016-0374-0. [Google Scholar] [CrossRef]
21. Sun, Z., Su, C., Yun, J., Jiang, Q., Wang, L. et al. (2019). Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnology Journal, 17(1), 50–62. DOI 10.1111/pbi.12946. [Google Scholar] [CrossRef]
22. Yan, Z., Hossain, M. S., Arikit, S., Valdés-López, O., Zhai, J. et al. (2015). Identification of micro RNA s and their mRNA targets during soybean nodule development: Functional analysis of the role of miR393j-3p in soybean nodulation. New Phytologist, 207(3), 748–759. DOI 10.1111/nph.13365. [Google Scholar] [CrossRef]
23. Xu, F., Liu, Q., Chen, L., Kuang, J., Walk, T. et al. (2013). Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics, 14(1), 66. DOI 10.1186/1471-2164-14-66. [Google Scholar] [CrossRef]
24. Wang, Y., Zhang, C., Hao, Q., Sha, A., Zhou, R. et al. (2013). Elucidation of miRNAs-mediated responses to low nitrogen stress by deep sequencing of two soybean genotypes. PLoS One, 8(7), e67423. DOI 10.1371/journal.pone.0067423. [Google Scholar] [CrossRef]
25. Ning, L. H., Du, W. K., Song, H. N., Shao, H. B., Qi, W. C. et al. (2019). Identification of responsive miRNAs involved in combination stresses of phosphate starvation and salt stress in soybean root. Environmental and Experimental Botany, 167(2), 103823. DOI 10.1016/j.envexpbot.2019.103823. [Google Scholar] [CrossRef]
26. Yu, J. Y., Zhang, Z. G., Huang, S. Y., Han, X., Wang, X. Y. et al. (2019). Analysis of miRNAs targeted storage regulatory genes during soybean seed development based on transcriptome sequencing. Genes, 10(6), 408. DOI 10.3390/genes10060408. [Google Scholar] [CrossRef]
27. Ramesh, S. V., Ratnaparkhe, M. B., Husain, S. M., Bhatia, V. S. (2015). Viral micro RNA transcriptomics (miRNAomics). Transcriptomics, 3, 108. [Google Scholar]
28. Chen, H., Arsovski, A. A., Yu, K., Wang, A. (2016). Genome-wide investigation using sRNA-Seq, degradome-Seq and transcriptome-Seq reveals regulatory networks of microRNAs and their target genes in soybean during soybean mosaic virus infection. PLoS One, 11(3), e0150582. DOI 10.1371/journal.pone.0150582. [Google Scholar] [CrossRef]
29. Li, H., Dong, Y., Yin, H., Wang, N., Yang, J. et al. (2011). Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biology, 11(1), 170. DOI 10.1186/1471-2229-11-170. [Google Scholar] [CrossRef]
30. Zhang, S., Wang, Y., Li, K., Zou, Y., Chen, L. et al. (2014). Identification of cold-responsive miRNAs and their target genes in nitrogen-fixing nodules of soybean. International Journal of Molecular Sciences, 15(8), 13596–13614. DOI 10.3390/ijms150813596. [Google Scholar] [CrossRef]
31. Huang, S. C., Lu, G. H., Tang, C. Y., Ji, Y. J., Tan, G. S. et al. (2018). Identification and comparative analysis of aluminum-induced microRNAs conferring plant tolerance to aluminum stress in soybean. Biologia Plantarum, 62(1), 97–108. DOI 10.1007/s10535-017-0752-5. [Google Scholar] [CrossRef]
32. Singh, L., Mehta, B. K., Kumar, S., Tiwari, S., Kushwaha, A. K. (2017). Role of miRNAs in Legume crops in response to biotic and abiotic stress. Journal of Pure and Applied Microbiology, 11(1), 605–614. DOI 10.22207/JPAM.11.1.79. [Google Scholar] [CrossRef]
33. Tyczewska, A., Gracz, J., Kuczyński, J., Twardowski, T. (2016). Deciphering soybean molecular stress response via high-throughput approach. Acta Biochimica Polonica, 63, 631–643. [Google Scholar]
34. Thao, N. P., Tran, L. S. P. (2012). Potentials toward genetic engineering of drought-tolerant soybean. Critical Reviews in Biotechnology, 32(4), 349–362. DOI 10.3109/07388551.2011.643463. [Google Scholar] [CrossRef]
35. Kulcheski, F. R., de Oliveira, L. F.,Molina, L. G., Almerão, M. P., Rodrigues, F. A. et al. (2011). Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics, 12(1), 307. DOI 10.1186/1471-2164-12-307. [Google Scholar] [CrossRef]
36. Ni, Z., Hu, Z., Jiang, Q., Zhang, H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Molecular Biology, 82(1-2), 113–129. DOI 10.1007/s11103-013-0040-5. [Google Scholar] [CrossRef]
37. Li, W., Wang, T., Zhang, Y., Li, Y. (2016). Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. Journal of Experimental Botany, 67(1), 175–194. DOI 10.1093/jxb/erv450. [Google Scholar] [CrossRef]
38. Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review Plant Biology, 59(1), 651–681. DOI 10.1146/annurev.arplant.59.032607.092911. [Google Scholar] [CrossRef]
39. Sunkar, R., Zhu, J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell, 16(8), 2001–2019. DOI 10.1105/tpc.104.022830. [Google Scholar] [CrossRef]
40. Lu, X. Y., Huang, X. L. (2008). Plant miRNAs and abiotic stress responses. Biochemical and Biophysical Research Communications, 368(3), 458–462. DOI 10.1016/j.bbrc.2008.02.007. [Google Scholar] [CrossRef]
41. Arenas-Huertero, C., Pérez, B., Rabanal, F., Blanco-Melo, D., de la Rosa, C. et al. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Molecular Biology, 70(4), 385–401. DOI 10.1007/s11103-009-9480-3. [Google Scholar] [CrossRef]
42. Dong, Z., Shi, L., Wang, Y., Chen, L., Cai, Z. et al. (2013). Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. International Journal of Molecular Sciences, 14(2), 2717–2738. DOI 10.3390/ijms14022717. [Google Scholar] [CrossRef]
43. Sahito, Z. A., Wang, L., Sun, Z., Yan, Q., Zhang, X. et al. (2017). The miR172c-NNC1 module modulates root plastic development in response to salt in soybean. BMC Plant Biology, 17(1), 1–12. DOI 10.1186/s12870-017-1161-9. [Google Scholar] [CrossRef]
44. Wang, Q., Yang, Y., Lu, G., Sun, X., Feng, Y. et al. (2020). Genome-wide identification of microRNAs and phased siRNAs in soybean roots under long-term salt stress. Genes & Genomics, 42(11), 1239–1249. DOI 10.1007/s13258-020-00990-0. [Google Scholar] [CrossRef]
45. Yu, Y., Ni, Z., Wang, Y., Wan, H., Hu, Z. et al. (2019). Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Science, 285, 68–78. DOI 10.1016/j.plantsci.2019.05.003. [Google Scholar] [CrossRef]
46. Cadavid, I. C., Guzman, F., de Oliveira-Busatto, L., de Almeida, R. M., Margis, R. (2020). Transcriptional analyses of two soybean cultivars under salt stress. Molecular Biology Reports, 47(4), 1–18. DOI 10.1007/s11033-020-05398-3. [Google Scholar] [CrossRef]
47. Sun, M., Jin, Y., Wang, X., Zhang, Y., Zhang, Y. et al. (2020). Gma-miR1508a confers dwarfing, cold tolerance, and drought sensitivity in soybean. Molecular Breeding, 40(1), 1–13. DOI 10.1007/s11032-019-1080-6. [Google Scholar] [CrossRef]
48. Xu, S., Liu, N., Mao, W., Hu, Q., Wang, G. et al. (2016). Identification of chilling-responsive microRNAs and their targets in vegetable soybean (Glycine max L.). Scientific Reports, 6(1), 26619. DOI 10.1038/srep26619. [Google Scholar] [CrossRef]
49. Zeng, Q. Y., Yang, C. Y., Ma, Q. B., Li, X. P., Dong, W. W. et al. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biology, 12(1), 182. DOI 10.1186/1471-2229-12-182. [Google Scholar] [CrossRef]
50. Fang, X., Zhao, Y., Ma, Q., Huang, Y., Wang, P. et al. (2013). Identification and comparative analysis of cadmium tolerance-associated miRNAs and their targets in two soybean genotypes. PLoS One, 8(12), p.e81471. DOI 10.1371/journal.pone.0081471. [Google Scholar] [CrossRef]
51. Miransari, M. (2015). Abiotic and biotic stresses in soybean production. Soybean production. vol. 1. Oxford: Academic Press. [Google Scholar]
52. Wrather, A., Koenning, S. (2009). Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Progress, 10(1), 24. DOI 10.1094/PHP-2009-0401-01-RS. [Google Scholar] [CrossRef]
53. Li, X., Wang, X., Zhang, S., Liu, D., Duan, Y. et al. (2012). Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS One, 7(6), e39650. DOI 10.1371/journal.pone.0039650. [Google Scholar] [CrossRef]
54. Tian, B., Li, J., Oakley, T. R., Todd, T. C., Trick, H. N. (2016). Host-derived artificial microRNA as an alternative method to improve soybean resistance to soybean cyst nematode. Genes, 7(12), 122. DOI 10.3390/genes7120122. [Google Scholar] [CrossRef]
55. Yorinori, J. T., Paiva, W. M., Frederick, R. D., Costamilan, L. M., Bertagnolli, P. F. et al. (2005). Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Disease, 89(6), 675–677. DOI 10.1094/PD-89-0675. [Google Scholar] [CrossRef]
56. Sinclair, J. B. (1999). Soybean rust. Compendium of soybean disease, pp. 25–26. St. Paul, MN. [Google Scholar]
57. Tyler, B. M. (2007). Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Molecular Plant Pathology, 8(1), 1–8. DOI 10.1111/j.1364-3703.2006.00373.x. [Google Scholar] [CrossRef]
58. Ye, W., Wang, X., Tao, K., Lu, Y., Dai, T. et al. (2011). Digital gene expression profiling of the Phytophthora sojae transcriptome. Molecular Plant-Microbe Interactions, 24(12), 1530–1539. DOI 10.1094/MPMI-05-11-0106. [Google Scholar] [CrossRef]
59. Cui, X., Yan, Q., Gan, S., Xue, D., Dou, D. et al. (2017). Overexpression of GMA-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene, 621, 32–39. DOI 10.1016/j.gene.2017.04.015. [Google Scholar] [CrossRef]
60. Babu, M., Gagarinova, A. G., Brandle, J. E., Wang, A. (2008). Association of the transcriptional response of soybean plants with soybean mosaic virus systemic infection. Journal of General Virology, 89(4), 1069–1080. DOI 10.1099/vir.0.83531-0. [Google Scholar] [CrossRef]
61. Yin, X., Wang, J., Cheng, H., Wang, X., Yu, D. (2013). Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta, 237(5), 1213–1225. DOI 10.1007/s00425-012-1835-3. [Google Scholar] [CrossRef]
62. Ramesh, S. V., Govindasamy, V., Rajesh, M. K., Sabana, A. A., Praveen, S. (2019). Stress-responsive miRNAome of Glycine max (L.) Merrill: Molecular insights and way forward. Planta, 249(5), 1267–1284. DOI 10.1007/s00425-019-03114-5. [Google Scholar] [CrossRef]
63. Ramesh, S. V., Gupta, G. K., Husain, S. M. (2016). Soybean (Glycine max) microRNAs display proclivity to repress begomovirus genomes. Current Science, 110(3), 424–428. DOI 10.18520/cs/v110/i3/424-428. [Google Scholar] [CrossRef]
64. Ramesh, S. V., Chouhan, B. S., Kumar, G., Praveen, S., Chand, S. (2017). Expression dynamics of Glycine max (L.) Merrill microRNAs (miRNAs) and their targets during Mungbean yellow mosaic India virus (MYMIV) infection. Physiological and Molecular Plant Pathology, 100, 13–22. DOI 10.1016/j.pmpp.2017.05.001. [Google Scholar] [CrossRef]
65. Zeng, W., Sun, Z., Lai, Z., Yang, S., Chen, H. et al. (2019). Determination of the miRNAs related to bean pyralid larvae resistance in soybean using small RNA and transcriptome sequencing. International Journal of molecular Sciences, 20(12), 2966. DOI 10.3390/ijms20122966. [Google Scholar] [CrossRef]
66. Jagtap, U. B., Gurav, R. G., Bapat, V. A. (2011). Role of RNA interference in plant improvement. Naturwissenschaften, 98(6), 473–492. DOI 10.1007/s00114-011-0798-8. [Google Scholar] [CrossRef]
67. Zhang, Y. C., Chen, Y. Q. (2017). Epigenetic regulation by noncoding RNAs in plant development. Plant epigenetics, pp. 183–198. Cham: Springer. [Google Scholar]
68. Islam, W., Adnan, M., Huang, Z., Lu, G. D., Chen, H. Y. (2019). Small RNAs from seed to mature plant. Critical Reviews in Plant Sciences, 38(2), 117–139. DOI 10.1080/07352689.2019.1608404. [Google Scholar] [CrossRef]
69. Schachtman, D. P., Shin, R. (2007). Nutrient sensing and signaling: NPCS. Annual Review Plant Biology, 58(1), 47–69. DOI 10.1146/annurev.arplant.58.032806.103750. [Google Scholar] [CrossRef]
70. Ohkama-Ohtsu, N., Wasaki, J. (2010). Recent progress in plant nutrition research: Cross-talk between nutrients, plant physiology and soil microorganisms. Plant and Cell Physiology, 51(8), 1255–1264. DOI 10.1093/pcp/pcq095. [Google Scholar] [CrossRef]
71. Kobayashi, T., Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology, 63(1), 131–152. DOI 10.1146/annurev-arplant-042811-105522. [Google Scholar] [CrossRef]
72. Rubio, V., Bustos, R., Irigoyen, M. L., Cardona-López, X., Rojas-Triana, M. et al. (2009). Plant hormones and nutrient signaling. Plant Molecular Biology, 69(4), 361–373. DOI 10.1007/s11103-008-9380-y. [Google Scholar] [CrossRef]
73. Chiou, T. J. (2007). The role of microRNAs in sensing nutrient stress. Plant, Cell & Environment, 30(3), 323–332. DOI 10.1111/j.1365-3040.2007.01643.x. [Google Scholar] [CrossRef]
74. Kuo, H. F., Chiou, T. J. (2011). The role of microRNAs in phosphorus deficiency signaling. Plant Physiology, 156(3), 1016–1024. DOI 10.1104/pp.111.175265. [Google Scholar] [CrossRef]
75. Scheible, W. R., Pant, B. D., Musialak-Lange, M., Nuc, P. (2011). Nutrient-responsive plant microRNAs. Non-coding RNAs in plants, pp. 313–337. Berlin, Heidelberg: Springer. [Google Scholar]
76. Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J. et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the united States of America, 107(9), 4477–4482. DOI 10.1073/pnas.0909571107. [Google Scholar] [CrossRef]
77. Frink, C. R., Waggoner, P. E., Ausubel, J. H. (1999). Nitrogen fertilizer: Retrospect and prospect. Proceedings of The National Academy of Sciences of the United States of America, 96(4), 1175–1180. DOI 10.1073/pnas.96.4.1175. [Google Scholar] [CrossRef]
78. Xu, Z., Zhong, S., Li, X., Li, W., Rothstein, S. J. et al. (2011). Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS One, 6(11), e28009. DOI 10.1371/journal.pone.0028009. [Google Scholar] [CrossRef]
79. Trevisan, S., Nonis, A., Begheldo, M., Manoli, A., Palme, K. et al. (2012). Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant, Cell & Environment, 35(6), 1137–1155. DOI 10.1111/j.1365-3040.2011.02478.x. [Google Scholar] [CrossRef]
80. Fischer, J. J., Beatty, P. H., Good, A. G., Muench, D. G. (2013). Manipulation of microRNA expression to improve nitrogen use efficiency. Plant Science, 210, 70–81. DOI 10.1016/j.plantsci.2013.05.009. [Google Scholar] [CrossRef]
81. Zeng, H., Wang, G., Hu, X., Wang, H., Du, L. et al. (2014). Role of microRNAs in plant responses to nutrient stress. Plant and Soil, 374(1–2), 1005–1021. DOI 10.1007/s11104-013-1907-6. [Google Scholar] [CrossRef]
82. Sinha, S. K., Srinivasan, R., Mandal, P. K. (2015). MicroRNA-based approach to improve nitrogen use efficiency in crop plants. Nutrient use efficiency: From basics to advances, pp. 221–235. New Delhi: Springer. [Google Scholar]
83. Xu, H., Li, Y., Zhang, K., Li, M., Fu, S. et al. (2020). miR169c-NFYA-C-ENOD40 modulates nitrogen inhibitory effects in soybean nodulation. New Phytologist, 229(6), 3377–3392. DOI 10.1111/nph.17115.2. [Google Scholar] [CrossRef]
84. Zeng, H. Q., Zhu, Y. Y., Huang, S. Q., Yang, Z. M. (2010). Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). Journal of Plant Physiology, 167(15), 1289–1297. DOI 10.1016/j.jplph.2010.04.017. [Google Scholar] [CrossRef]
85. Sha, A., Chen, Y., Bai, H., Shan, Z., Zhang, X. et al. (2012). Identification of Glycine max MicroRNAs in response to phosphorus deficiency. Journal of Plant Biology, 55(4), 268–280. DOI 10.1007/s12374-011-0255-4. [Google Scholar] [CrossRef]
86. Bari, R., Pant, B. D., Stitt, M., Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology, 141(3), 988–999. DOI 10.1104/pp.106.079707. [Google Scholar] [CrossRef]
87. Hsieh, L. C., Lin, S. I., Shih, A. C. C., Chen, J. W., Lin, W. Y. et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology, 151(4), 2120–2132. DOI 10.1104/pp.109.147280. [Google Scholar] [CrossRef]
88. Zeng, H., Wang, G., Zhang, Y., Hu, X., Pi, E. et al. (2016). Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant and Soil, 398(1–2), 207–227. DOI 10.1007/s11104-015-2657-4. [Google Scholar] [CrossRef]
89. Liu, X., Chu, S., Sun, C., Xu, H., Zhang, J. et al. (2020). Genome-wide identification of low phosphorus responsive microRNAs in two soybean genotypes by high-throughput sequencing. Functional & Integrative Genomics, 20(6), 825–838. DOI 10.1007/s10142-020-00754-9. [Google Scholar] [CrossRef]
90. Rogers, E. D., Benfey, P. N. (2015). Regulation of plant root system architecture: Implications for crop advancement. Current Opinion in Biotechnology, 32(2), 93–98. DOI 10.1016/j.copbio.2014.11.015. [Google Scholar] [CrossRef]
91. Meng, Y., Ma, X., Chen, D., Wu, P., Chen, M. (2010). MicroRNA-mediated signaling involved in plant root development. Biochemical and Biophysical Research Communications, 393(3), 345–349. DOI 10.1016/j.bbrc.2010.01.129. [Google Scholar] [CrossRef]
92. Ochando, I., Jover-Gil, S., Ripoll, J. J., Candela, H., Vera, A. et al. (2006). Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiology, 141(2), 607–619. DOI 10.1104/pp.106.077149. [Google Scholar] [CrossRef]
93. Wang, Y., Li, K., Chen, L., Zou, Y., Liu, H. et al. (2015). MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiology, 168(3), 984–999. DOI 10.1104/pp.15.00265. [Google Scholar] [CrossRef]
94. Sun, Z., Wang, Y., Mou, F., Tian, Y., Chen, L. et al. (2016). Genome-wide small RNA analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Frontiers in Plant Science, 6, 1273. DOI 10.3389/fpls.2015.01273. [Google Scholar] [CrossRef]
95. Wong, C. E., Zhao, Y. T., Wang, X. J., Croft, L., Wang, Z. H. et al. (2011). MicroRNAs in the shoot apical meristem of soybean. Journal of Experimental Botany, 62(8), 2495–2506. DOI 10.1093/jxb/erq437. [Google Scholar] [CrossRef]
96. Allen, R. S., Li, J., Stahle, M. I., Dubroué, A., Gubler, F. et al. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proceedings of the National Academy of Sciences of the United States of America, 104(41), 16371–16376. DOI 10.1073/pnas.0707653104. [Google Scholar] [CrossRef]
97. Yoshikawa, T., Ozawa, S., Sentoku, N., Itoh, J. I., Nagato, Y. et al. (2013). Change of shoot architecture during juvenile-to-adult phase transition in soybean. Planta, 238(1), 229–237. DOI 10.1007/s00425-013-1895-z. [Google Scholar] [CrossRef]
98. Cao, D., Li, Y., Wang, J., Nan, H., Wang, Y. et al. (2015). GmmiR156b overexpression delays flowering time in soybean. Plant Molecular Biology, 89(4–5), 353–363. DOI 10.1007/s11103-015-0371-5. [Google Scholar] [CrossRef]
99. Wang, T., Sun, M. Y., Wang, X. S., Li, W. B., Li, Y. G. (2016). Over-expression of GmGIa-regulated soybean miR172a confers early flowering in transgenic Arabidopsis thaliana. International Journal of Molecular Sciences, 17(5), 645. DOI 10.3390/ijms17050645. [Google Scholar] [CrossRef]
100. Kulcheski, F. R., Molina, L. G., da Fonseca, G. C., de Morais, G. L.,de Oliveira, L. F. V. et al. (2016). Novel and conserved microRNAs in soybean floral whorls. Gene, 575(2), 213–223. DOI 10.1016/j.gene.2015.08.061. [Google Scholar] [CrossRef]
101. Ding, X., Zhang, H., Ruan, H., Li, Y., Chen, L. et al. (2019). Exploration of miRNA-mediated fertility regulation network of cytoplasmic male sterility during flower bud development in soybean. 3 Biotech, 9(1), 22. [Google Scholar]
102. Ding, X., Ruan, H., Yu, L., Li, Q., Song, Q. et al. (2020). miR156b from soybean CMS line modulates floral organ development. Journal of Plant Biology, 63(2), 1–13. DOI 10.1007/s12374-020-09237-7. [Google Scholar] [CrossRef]
103. Tuteja, J. H., Zabala, G., Varala, K., Hudson, M., Vodkin, L. O. (2009). Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell, 21(10), 3063–3077. DOI 10.1105/tpc.109.069856. [Google Scholar] [CrossRef]
104. Shamimuzzaman, M., Vodkin, L. (2012). Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics, 13(1), 310. DOI 10.1186/1471-2164-13-310. [Google Scholar] [CrossRef]
105. Goettel, W., Liu, Z., Xia, J., Zhang, W., Zhao, P. X. et al. (2014). Systems and evolutionary characterization of microRNAs and their underlying regulatory networks in soybean cotyledons. PLoS One, 9(1), e86153. DOI 10.1371/journal.pone.0086153. [Google Scholar] [CrossRef]
106. Zabala, G., Campos, E., Varala, K. K., Bloomfield, S., Jones, S. I. et al. (2012). Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC Plant Biology, 12(1), 177. DOI 10.1186/1471-2229-12-177. [Google Scholar] [CrossRef]
107. Li, X., Xie, X., Li, J., Cui, Y., Hou, Y. et al. (2017). Conservation and diversification of the miR166 family in soybean and potential roles of newly identified miR166s. BMC Plant Biology, 17(1), 1–18. DOI 10.1186/s12870-016-0951-9. [Google Scholar] [CrossRef]
108. Yu, L., Guo, R., Jiang, Y., Ye, X., Yang, Z. et al. (2019). Genome-wide identification and characterization of novel microRNAs in seed development of soybean. Bioscience, Biotechnology, and Biochemistry, 83(2), 233–242. DOI 10.1080/09168451.2018.1536513. [Google Scholar] [CrossRef]
109. Flores, T., Karpova, O., Su, X., Zeng, P., Bilyeu, K. et al. (2008). Silencing of GmFAD3 gene by siRNA leads to low α-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Research, 17(5), 839–850. DOI 10.1007/s11248-008-9167-6. [Google Scholar] [CrossRef]
110. Lelandais-Brière, C., Naya, L., Sallet, E., Calenge, F., Frugier, F. et al. (2009). Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell, 21(9), 2780–2796. DOI 10.1105/tpc.109.068130. [Google Scholar] [CrossRef]
111. Boualem, A., Laporte, P., Jovanovic, M., Laffont, C., Plet, J. et al. (2008). MicroRNA166 controls root and nodule development in Medicago truncatula. Plant Journal, 54(5), 876–887. DOI 10.1111/j.1365-313X.2008.03448.x. [Google Scholar] [CrossRef]
112. Yan, Z., Hossain, M. S., Wang, J., Valdés-López, O., Liang, Y. et al. (2013). miR172 regulates soybean nodulation. Molecular Plant-Microbe Interactions, 26(12), 1371–1377. DOI 10.1094/MPMI-04-13-0111-R. [Google Scholar] [CrossRef]
113. Wang, Y., Wang, L., Zou, Y., Chen, L., Cai, Z. et al. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell, 26(12), 4782–4801. DOI 10.1105/tpc.114.131607. [Google Scholar] [CrossRef]
114. Sasaki, T., Suzaki, T., Soyano, T., Kojima, M., Sakakibara, H. et al. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nature Communications, 5, 1–9. [Google Scholar]
115. Suzaki, T., Yoro, E., Kawaguchi, M. (2015). Leguminous plants: Inventors of root nodules to accommodate symbiotic bacteria. International review of cell and molecular biology, pp. 111–158. Cambridge, MA: Academic Press. [Google Scholar]
116. Nova-Franco, B., Íñiguez, L. P., Valdés-López, O., Alvarado-Affantranger, X., Leija, A. et al. (2015). The micro-RNA172c-APETALA2-1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiology, 168(1), 273–291. DOI 10.1104/pp.114.255547. [Google Scholar] [CrossRef]
117. Nizampatnam, N. R., Schreier, S. J., Damodaran, S., Adhikari, S., Subramanian, S. (2015). micro RNA 160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant Journal, 84(1), 140–153. DOI 10.1111/tpj.12965. [Google Scholar] [CrossRef]
118. Hossain, M. S., Hoang, N. T., Yan, Z., Tóth, K., Meyers, B. C. et al. (2019). Characterization of the spatial and temporal expression of two soybean miRNAs identifies SCL6 as a novel regulator of soybean nodulation. Frontiers in Plant Science, 10, 475. DOI 10.3389/fpls.2019.00475. [Google Scholar] [CrossRef]
119. Yan, Z., Hossain, M. S., Valdés-López, O., Hoang, N. T., Zhai, J. et al. (2016). Identification and functional characterization of soybean root hair micro RNA s expressed in response to Bradyrhizobium japonicum infection. Plant Biotechnology Journal, 14(1), 332–341. DOI 10.1111/pbi.12387. [Google Scholar] [CrossRef]
120. Kamthan, A., Chaudhuri, A., Kamthan, M., Datta, A. (2015). Small RNAs in plants: Recent development and application for crop improvement. Frontiers in Plant Science, 6, 208. DOI 10.3389/fpls.2015.00208. [Google Scholar] [CrossRef]
121. Ivashuta, S., Banks, I. R., Wiggins, B. E., Zhang, Y., Ziegler, T. E. et al. (2011). Regulation of gene expression in plants through miRNA inactivation. PLoS One, 6(6), e21330. DOI 10.1371/journal.pone.0021330. [Google Scholar] [CrossRef]
122. Yasin, J. K., Mishra, B. K., Pillai, M. A., Verma, N., Wani, S. H. et al. (2020). Genome-wide in-silico miRNA and target network prediction from stress-responsive Horsegram (Macrotyloma uniflorum) accessions. Scientific Reports, 10(1), 1–23. DOI 10.1038/s41598-020-73140-x. [Google Scholar] [CrossRef]
123. Wani, S. H., Kumar, V., Khare, T., Tripathi, P., Shah, T. et al. (2020). miRNA applications for engineering abiotic stress tolerance in plants. Biologia, 75(7), 1–19. DOI 10.2478/s11756-019-00397-7. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |