
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016174
REVIEW
Pathways of Phosphorus Absorption and Early Signaling between the Mycorrhizal Fungi and Plants
1Facultad de Ciencias Agrotecnológicas, Universidad Autónoma de Chihuahua (UACH), Chihuahua, 31000, México
2Innovak Global, Chihuahua, 31375, México
*Corresponding Author: Graciela Dolores Avila-Quezada. Email: gdavila@uach.mx; gavilaq@gmail.com
Received: 13 February 2021; Accepted: 18 March 2021
#These authors contributed equally to this manuscript
Abstract: This review highlights the key role that mycorrhizal fungi play in making phosphorus (Pi) more available to plants, including pathways of phosphorus absorption, phosphate transporters and plant-mycorrhizal fungus symbiosis, especially in conditions where the level of inorganic phosphorus (Pi) in the soil is low. Mycorrhizal fungi colonization involves a series of signaling where the plant root exudates strigolactones, while the mycorrhizal fungi release a mixture of chito-oligosaccharides and liposaccharides, that activate the symbiosis process through gene signaling pathways, and contact between the hyphae and the root. Once the symbiosis is established, the extraradical mycelium acts as an extension of the roots and increases the absorption of nutrients, particularly phosphorus by the phosphate transporters. Pi then moves along the hyphae to the plant root/fungus interface. The transfer of Pi occurs in the apoplectic space; in the case of arbuscular mycorrhizal fungi, Pi is discharged from the arbuscular to the plant’s root symplasm, in the membrane that surrounds the arbuscule. Pi is then absorbed through the plant periarbuscular membrane by plant phosphate transporters. Furthermore, plants can acquire Pi from soil as a direct absorption pathway. As a result of this review, several genes that codify for high-affinity Pi transporters were identified. In plants, the main family is Pht1 although it is possible to find others such as Pht2, Pht3, Pho1 and Pho2. As in plants, mycorrhizal fungi have genes belonging to the Pht1 subfamily. In arbuscular mycorrhizal fungi we found L1PT1, GiPT, MtPT1, MtPT2, MtPT4, HvPT8, ZmPht1, TaPTH1.2, GmosPT and LYCes. HcPT1, HcPT2 and BePT have been characterized in ectomycorrhizal fungi. Each gene has a different way of expressing itself. In this review, we present diagrams of the symbiotic relationship between mycorrhizal fungi and the plant. This knowledge allows us to design solutions to regional problems such as food production in soils with low levels of Pi.
Keywords: Interaction; mycorrhiza establishment; symbiosis; phosphate uptake
The United Nations General Assembly designated 2021 as the International Year of Fruits and Vegetables. This celebration represents an opportunity to share information on the importance of knowing how plants are nourished, and produce quality fruits, for the consumption of the human population. Plants require essential elements to growth and phosphorus (Pi) is one of them [1]. It is involved in multiple biochemical processes such as energy storage, membrane integrity, photosynthesis, glycolysis, respiration, enzyme activation/inactivation, redox reactions, signaling, carbohydrate metabolism, nitrogen fixation, as well as the structure and function of nucleic acids [2]. Phosphorus is the basis of many compounds, of which the most important for plant nutrition are phosphates. Microorganisms form an integral part of the soil phosphorus cycle and mediate the availability of Pi to plants [3–5].
This element can be mobilized by different microorganisms [3,6] through its primary metabolism, particularly with the production of low molecular weight organic acids. These have a negative charge and create chelates with metal ions such as Ca2+, Mg2+, Fe2+ and Al3+, that are generally associated with phosphorus in an insoluble way, transforming it into soluble phosphorus for the plant [5,7] (Fig. 1).
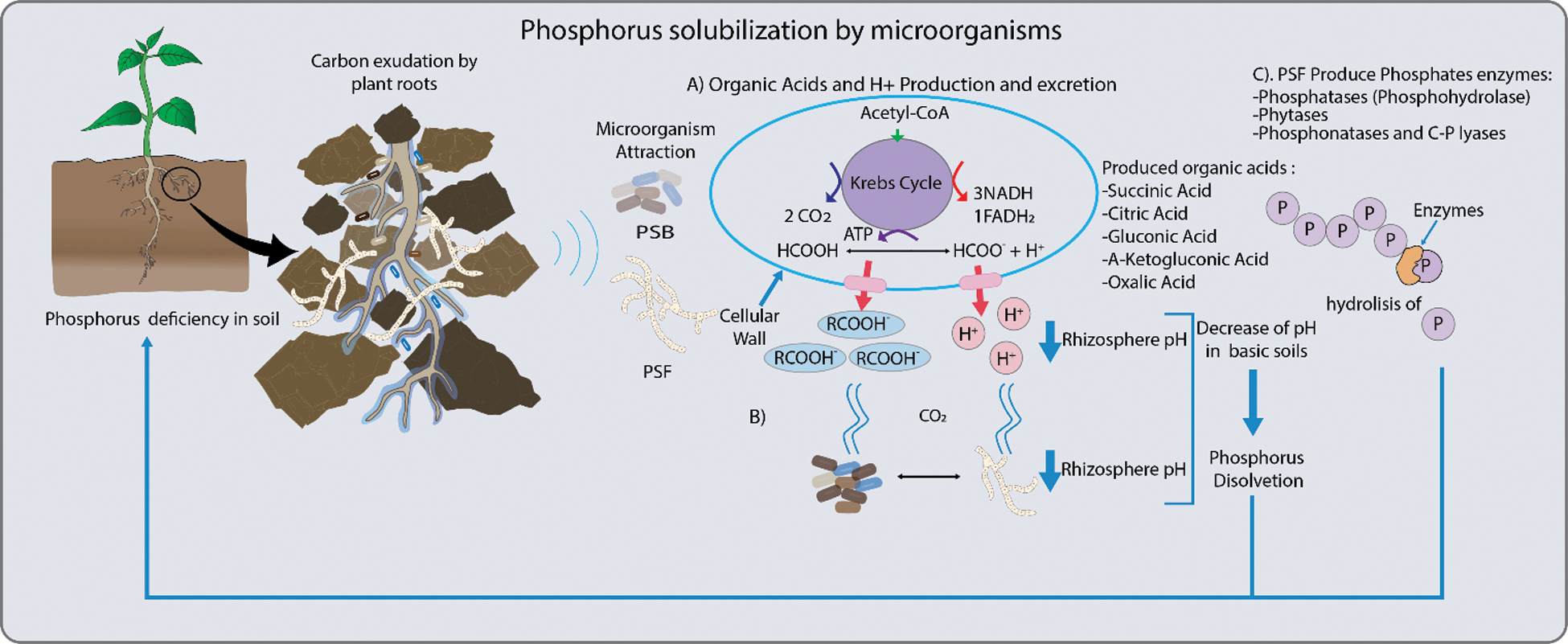
Figure 1: Mechanisms employed by phosphorus solubilizing bacteria (PSB) and fungi (PSF). These include: A. The production of organic acid and secretion of H+ ions, and the synthesis of enzymes to release phosphorus (Pi). Organic acids, such as citrate, malate, oxalate, succinate and fumarate increase Pi availability to roots. Another mechanism related to organic acid production is the release of H+ which is translocated to the rhizosphere by ATPase. This decreases pH and promotes Pi solubilization. B. Also, carbon dioxide (CO2) produced by microbial respiration dissolves in the soil water to form carbonic acid and this solubilizes Pi by the associated decrease in pH. C. In general, Pi is released from organic compounds in the soil by enzymes, such as phosphatases that release Pi from nucleotides and sugar phosphates, phytases degrade phytate to solubilize Pi and phosphonatases and carbon-phosphorus (C-Pi) lyases that cleave the C-Pi in organophosphonates [3–5]
In addition, certain microorganisms can promote an increment in the volume of the root, which leads to a raise in the absorption of phosphorus by the increase in the contact surface of the root with the soil. This increased area by microbial action can be for at least three types; (1) the association of mycorrhizal fungi, (2) by indoleacetic acid-producing bacteria, and (3) by ACC deaminase [8–10].
About the first type, pre-symbiotic molecular communication involves the reciprocal perception of diffusible molecules between plant and fungi [11,12]. Once symbiosis is established near the root apex, fungal hyphae increases the degree of exploration and absorption of nutrients [13–17]. Most plants experience greater growth and production when colonized by mycorrhizal fungi.
2 Plant-Mycorrhizal Fungus Symbiosis
In the establishment of a symbiotic association, plants can interact with many mycorrhizal fungi simultaneously, in the same way, each of these mycorrhizal fungi can potentially interact with many host plants [18]. Until now it is believed that most mycorrhizal fungi are unspecific with respect to their ability to colonize different species of plants. Mycorrhizal fungi perform vital functions for the productivity of cultivated plants, for example, the extraradical mycelium acts as an extension of the roots and increases the absorption of nutrients, particularly phosphorus. The compensation of fungi is the supply of carbohydrates derived from the host [19,20].
2.1 Mechanism of Colonization by Ectomycorrhizal Fungi (EMF)
The association of EMF corresponds preferably to tree species from boreal forests, temperate, subtropical and mountainous forests [21,22]. The first approach between both symbionts begins with the release of root exudates (sugars, amino acids, hormones, enzymes, flavonoids, organic acids, etc.) recognized by basidiomycete and ascomycete spores that stimulate their germination and consequent radial growth of their hyphae through the soil towards the roots to form a mantle. This recognition occurs through an exchange of molecular signals and point receptors, which promote the early stages of the establishment of EMF, with the subsequent proliferation of host hyphae and roots [19].
During colonization, the mycelium occupies the apoplastic spaces forming the Hartig network, defined as an interface of molecular communications and where the greatest exchange of nutrients takes place. Colonized roots undergo a series of morphological changes, such as the arrest of cell growth, rearrangement of microtubules and cellular microfibrils, changes in transmembrane transporters and thickening of walls in epidermal, and cortical cells to increase the surface of nutrient exchange [19,23,24].
The interaction of the ectomycorrhizal fungus (EMF) Laccaria bicolor with the plant involves the activity of the LbMiSSP7 gene of the fungus, which produces a protein in response to the detection of flavonoids exuded by the root (rutin and quercetin). This protein is brought to the nucleus, where it comes into contact with the PtJAZ6 protein, to avoid the formation of the PtJAZ6–PtCOI1 complex (inducer of a systemic response induced by the plant through jasmonic acid). Therefore, the defense signal is suppressed in the plant to begin the colonization of the fungus [25,26]. As a result of colonization, a mycelial mantle grows around the epidermal and cortical cells of the short lateral roots to form the Hartig network [27].
This mutualistic association causes modifications in the root structure with the formation of swollen lateral roots that are enveloped by a fungal mantle. This phenomenon is given by regulatory mechanisms during the colonization of EMF producing analogous auxins concentrated in the root tips, causing an increase in the number of short lateral roots [28]. Vayssières et al. [29] report that the fungus Laccaria bicolor releases high concentrations of indole-3-acetic acid (IAA) as an active signaling pathway for root growth and overall increased colonization of the fungus.
Another form of stimulation is by an increase in the auxin synthesis through the action of hyphaporin, secreted by the fungus that internalizes itself in the root cells and competes with the plant’s auxin production by binding to proteins and auxin receptors [30]. Other hormones involved are cytokinins (zeatin and riboside zeatin), gibberellins, abscisic acid, salicylic acid, jasmonic acid and ethylene that play a role in the stimulation and regulation of colonization [31–33].
2.2 Mechanism of Colonization of Endomycorrhizal Fungi
The symbiosis between two eukaryotes such fungi and plants is interesting, where fungal metabolites and plant exudates are the early signaling for a successful symbiosis [34–38]. Fungi in general have a cycle that includes spore germination, germ tube development, and contact with the host and penetration [39,40]. This process triggers resistance or defense mechanisms in the plant. In the specific case of the obligate symbiotic mycorrhizal fungi, during the "infection" process, it should suppress plants resistance to colonization [41].
The molecular signals released by the plant to promote the germination and growth of Arbuscular mycorrhizal fungi (AM) fungal hyphae are strigolactones and flavonoids [42]. AM perceive the signals and produce a mixture of chitooligosaccharides and liposaccharides (Myc Factors). These function as fungal molecules recognized in the plasma membrane of the plant [43], causing a stimulation on the signaling pathways of associated symbiosis. It causes an increase in nuclear and perinuclear calcium oscillations, with activation of calcium and calmodulin dependent on a serine/threonine protein kinase (CCAMK), with an extensive change on transcriptional reprogramming, to promote fungal colonization in the root [44].
The increase in the level of calcium facilitates the appressorial penetration to the root. These events are coordinated by the nucleus, beginning with structural changes in the cell wall, membrane, and cytoplasm, leading in the host cell, the development of the hypha. Once the hyphae enter the cell, form arbuscules and initiate a nutrient exchange [45–47].
The formation of arbuscules and hyphae in plant tissue causes morphological changes to improve the efficiency of the carbon and nutrient absorption system [34–38]. Both, plant and fungus induce physiological changes in the other organism to obtain reciprocal nutritional benefits [34,48]. This symbiosis also has the advantage of increasing the systemic resistance of the plant [49] and, therefore, it can provide protection to plant roots from attack by pathogens [50–52].
3 Plant-Mycorrhizal Fungus Communication; Warning Signs for Low Phosphorus in Soil
All interactions begin with a series of signals between two organisms. On the one hand, the plant emits a signaling related to low concentration of Pi controlled by a gene signaling network coupled to hormonal responses (such as auxins and ethylene), sugars, among other molecules such as flavonoids, amino acids, vitamins, etc. [53–55]. Among the molecules found in root exudates, the strigolactones released into the rhizosphere by plants.
According to Siddiqi et al. [56] and Visentin et al. [57] plants respond to Pi deficiency by exuding strigolactones such as: orobanchol, solanacol and didehydro-orobanchol (s). Yoneyama et al. [58] reported that the secretion of sorgomol and 5-deoxystrigol in Chinese pea is promoted under conditions of Pi deficiency. In alfalfa and red clover, Pi deficiency increases secretions of orobanchol and orobankyl acetate [58], while, in lettuce, wheat and calendula, Pi deficiency increases the exudation of strigolactones [59].
Strigolactones released by plants, act as early signals of symbiosis to attract AMF stimulate their growth and genetic expression and consequently the release of Myc factors. These activate the process of symbiosis through signaling pathways gene (SYM genes). Once symbiosis is established, lateral roots are grown [59,60–63].
Several Pi transporters have been identified in the mycelium of some AMFs, and their expression has been observed to increase under low Pi conditions [64–66]. Once the fungal structures are established in the plant cells, the hyphae branch and form arbuscules. Pi in polyphosphate granules is transported from the extraradical mycelium to the arbuscules [67].
Interestingly, the genes that encode proteins for the transport of mineral nutrients, such as phosphorus, are expressed in the hyphae of extraradial mycelial networks [68,69]. In addition, ectomycorrhizal fungi (EMF) root tips produce phosphomonoesterases when symbiosis occurs [70], as well as phytases and acid phosphatase [71]. This synthesis is related to low levels of Pi in the soil [72]. EMF also produces organic acids, which solubilize Pi from the soil; in their anionic form and with the protons release [73].
4 Two Pathways of Phosphorus Absorption
Phosphate is absorbed by plants from the soil as free ions of hydrogen phosphate (HPO42−) and dihydrogen phosphate (H2PO4−). There are two strategies for the acquisition of Pi in soil: (a) the direct absorption pathway by the plant (DUP) [74] and (b) the Mycorrhizal phosphorus uptake pathway (MUP) [2,8,60,75].
Plants have developed strategies such as root morphological modifications, physiological, biochemical and molecular responses to obtain Pi when the element is scarce [76]. The morphological modifications that plants undergo are the increase the root-soil interface. Root hairs grow to cover more surface in a lateral direction to maximize access to Pi [77]. In a biochemical way, the roots activate enzymes, make changes in protein phosphorylation, activate glycolytic bypass pathway, secrete RNases, organic acids and phosphatases to solubize the Pi from organic compounds [76,78].
In DUP inorganic phosphate (Pi) is absorbed from the rhizosphere into the root epidermis and root hairs of the plant, through Pi transporters located in the cells of the root apex and in the root hairs (Fig. 2A) [8,75,79].
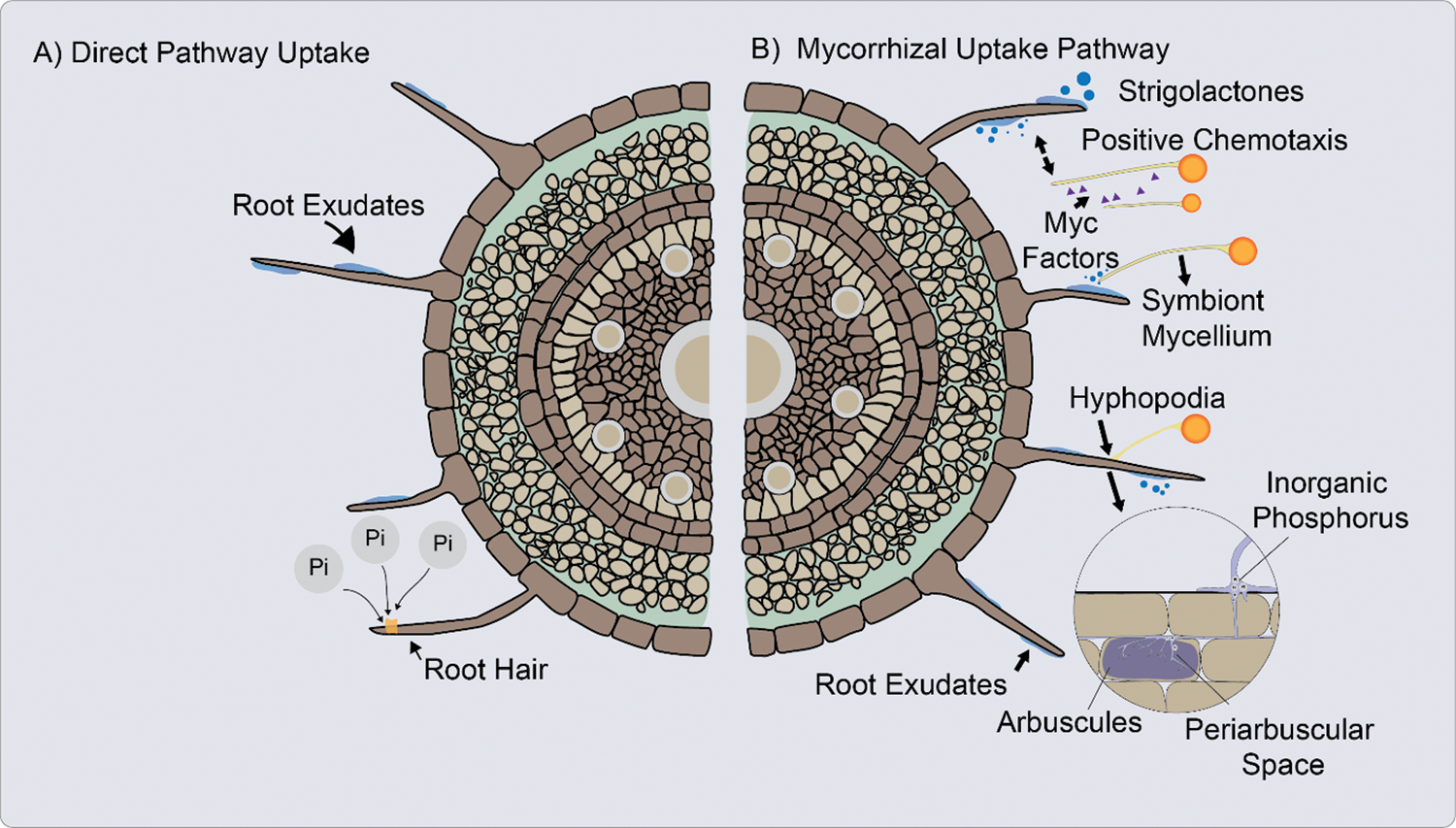
Figure 2: A. Direct uptake pathway (DUP). The inorganic phosphate (Pi) is absorbed from the rhizosphere into the plant through inorganic phosphate (Pi) transporters. B. Mycorrhizal uptake pathway (MUP), the Pi is uptake from the soil solution through fungal Pi transporters. Absorption of Pi in plants and fungi against a gradient of electrochemical potential [60,74,87]
When the AM fungus hyphae penetrate the cortical cells of the root, form an arbuscula that can fill most of the cell space, while the plasma membrane remains structured. Cortical cells form a periarbuscular membrane to surround the arbuscule [80].
Once established AM symbiosis, absorption through mycorrhizae occurs primarily because of the increased soil volume explored by the external hyphae and possibly by increasing the solubility of phosphorus in the soil due to the possible acidification produced by the release of organic acids by AM (Fig. 2B) [81]. This absorption of Pi is mediated by H+/Pi or Na+/Pi symporters located in the extraradical mycelium [8]. The hyphae absorb Pi, translocate it to AM structures such arbuscules, from where it is released to the interfacial apoplast space [75].
Studies of Chiou et al. [82] demostrated that in roots colonized by AM fungi, the DUP may be absent, due to the plant genes that encode the Pi transporters expressed in the root, may be negatively regulated in response to colonization by AM. Furthermore, the DUP may be affected by the low concentration of Pi left available in the soil solution at the root/soil interface [83].
In both types of pathways (DUP and MUP), Pi uptake occurs against an electrochemical potential gradient where an ATPase, which acts at the expense of ATP (adenosine triphosphate), exudes protons (H+). The proton concentration gradient and the generated membrane potential produce an electrochemical potential across the membrane. Proton mobilization along electrical and concentration gradients allows Pi to be distributed through Pht1 phosphate transporters [84,85]. The transmembrane topology of the Pht1 transporters indicates that the domains are arranged in two halves with amino (N–) and carboxyl (C–) terminals connected by a hydrophilic circuit, with possible sites for phosphorylation, myristoylation and glycosylation [84,86,87].
5.1 Phosphate Transport in Plants
The five families of Pi transporter genes in plants are Pht1, Pht2, Pht3, Pho1 and Pho2 [88]. The Pht1 family is found mainly in the root cell membrane and its expression depends on Pi levels [89]. High-affinity Pi transporters are integral membrane proteins that transport Pi from an external site to the cytoplasm. They consist of 12 regions that span the membrane, separated by a large hydrophilic region into two groups of six [76].
Phosphate transporters are mainly expressed in roots when Pi is low [90,91]. An example of this are the tomato (Lycopersicon esculentum) Pi transporters (LePT1 and LePT2), which are induced by the low concentration of Pi in roots. Faced with Pi starvation, there is a rapid activation of the expression of Pi transporters as indicated by studies in barley and corn [92,93].
5.1.1 Conditions of Apoplasmic Space
Although up to now it has not been elucidated how Pi is transported from the epidermis or cortex during asymbiosis and symbiosis, respectively, to the endodermis, it has been suggested that it takes the same pathway that nutrients take: apoplastic, simplistic or transcellular pathway [8]. The mucilage excreted by the cell walls makes the apoplasm to contain negatively charged hydroxyl groups serving as a substrate for the microorganisms in the rhizosphere. This negative charge causes some anions to be repelled and restricted to large pores in the apoplasm, reducing the concentration of Pi on the outer surface of the plasma membrane (Fig. 3). Once the ions are in the cytosol, within the symplasm, they can easily move into the stele via the plasmodesmata [94].
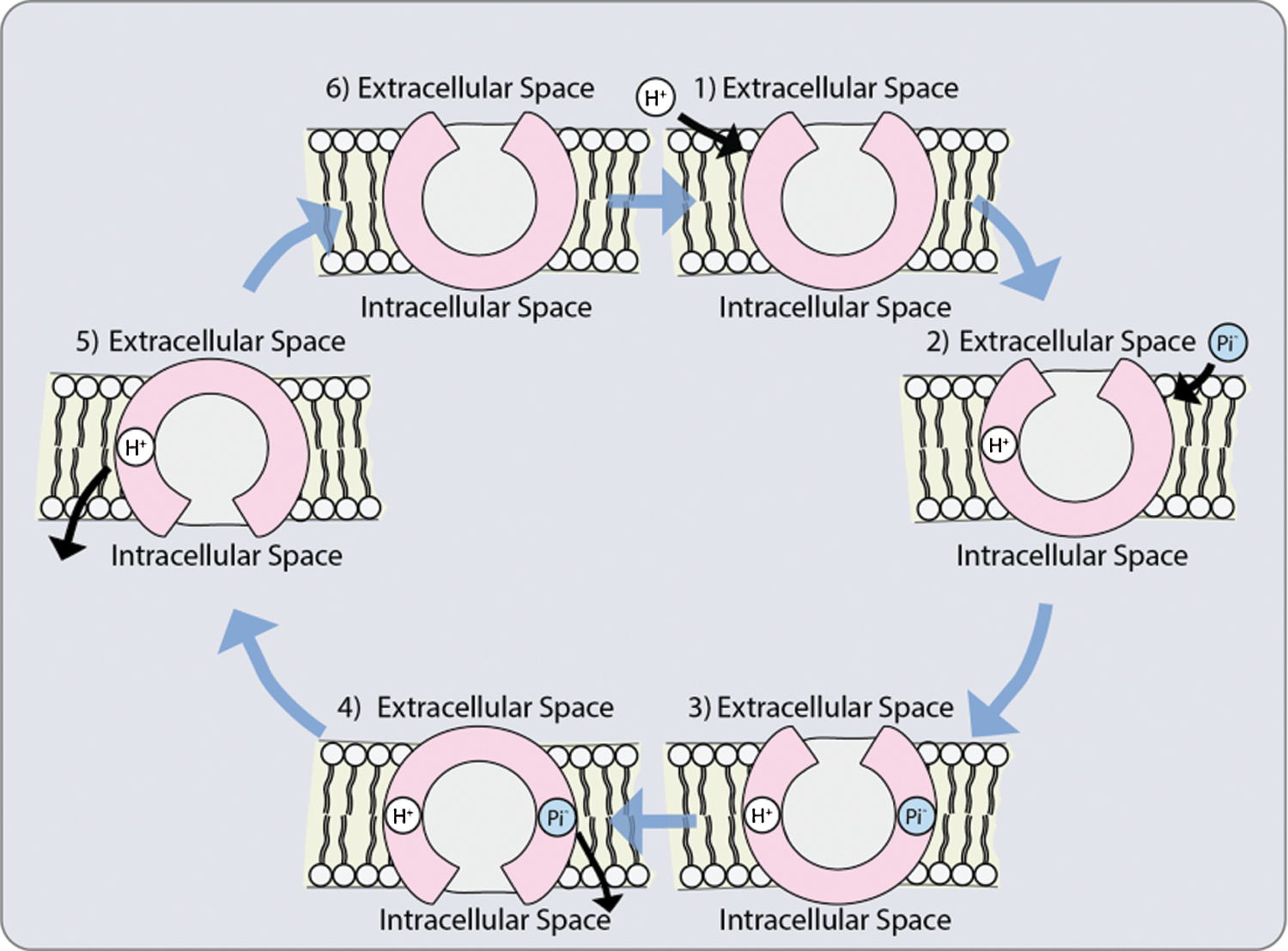
Figure 3: Possible co-transporter mechanism: 1. The protonation of a transporter with a hydrophilic pore open towards the outside; 2. Phosphate anion binding; 3. Conformational change that allows the opening of the pores towards the interior; 4. Phosphate release; 5. Deprotonation of the transporter; 6. Return of the initial conformation towards the outside [118]
Therefore, the transport of nutrients to the plant depends on the movement of ions across the plasma membrane of the epidermal and cortical cells of the root [95,96]. The phosphate concentration in the apoplasm is very low and it is higher in the cytosol [97,98], thus, strong electrochemical gradients must be overcome for the transport of phosphate anions to the root cells. This is the function of the specialized proteins of the Pht1 family of phosphate transporters in plants.
5.1.2 Studies of Pht1 in Plants
The genes related to the transport of Pi induce a change in the functional relationships of the root types when entering into symbiosis [99]. The assimilation of Pi is regulated by phosphate transporter proteins through two affinity systems. Low-affinity transporters have a role in Pi translocation from the root to the shoot, whereas high-affinity transporters has a role in Pi uptake capacity in Pi homeostasis responses [100].
Because sympathetic connections are lost in the membranes, proteins of the Pht1 family and at least one member of Pht2 family are those that transport Pi within the plant through the membrane, [101,102]. Some plants induce Pht1 phosphate transporters in response to mycorrhizal symbiosis [103,104]. Symbiotic Pi uptake by plants is mediated by plasma-membrane-localized Pi transporters. Plant-encoded H+/Pi symporters located in the periarbuscular membrane (PAM), are expressed only in arbuscule-containing cortex cells to absorb Pi [64,105,106].
Specific phosphate transporters from mycorrhized plants are involved in the absorption of Pi during symbiosis [104,107–111]. The rice phosphate transporters OsPT11 and OsPT13 are expressed only in roots in symbiosis with AM fungi [106,112]. Yang et al. [106,112] reported that OsPT11 is only necessary for symbiotic Pi uptake, while OsPT13 does not contribute to symbiotic Pi transport.
Indeed, phosphate transporters has been observed in symbiotic as well as asymbiotic plants [113]. The tomato genes LePT3, LePT4 and LePT5 are strong expressed during symbiosis in low Pi conditions; while the LePT5 gene was also low expressed in distinct cells harboring AM fungal structures [114]. Furthermore, the mycorrhizal-inducible potato StPT3 gene is strongly expressed in mycorrhizal roots and moderately in non-mycorrhizal roots [109]. Similarly, OsPT11/MtPT4 orthologs in corn (ZmPT6) accumulate in both colonized roots and non-mycorrhizal roots of Pi-deficient corn [115]. These examples suggest gene duplications that may give rise to new functions with respect to Pi.
Changes in symbiotic and asymbiotic root architecture is one of the common physiological responses in nutrient signaling pathways. Lateral root density increases during Pi starvation in colonized and in non-mycorrhizal roots [116]. On the subject of phosphate transporters in different types of roots, there are still many unresolved questions since both, symbiotic and asymbiotic roots, have structural changes to absorb Pi. Furthermore, Pht1 have some expression in non-mycorrhized roots [107,109,115].
Moreover, the formation of symbiosis and the expression of mycorrhizal-induced Pht1 members in arbuscule-containing cells, is often accompanied by the down-regulation of other Pht1 transporters located in the epidermis. This down-regulation of Pht1 transporters has been observed in potatoes [107]; rice [117]; birch [68], among others. Therefore, there is a balance in the Pi uptake pathways from the root epidermis and the fungus, result of excellent communication between both.
Most studies on Pi transporters during AM symbiosis focus on Pht1 transporters, since members of the Pht2 family of Pi transporters are involved in plant-wide Pi allocation [119] and have very little or no responses at the level of Pi or AM symbiosis [102].
5.2 Arbuscular Mycorrhizal (AM) Fungal Pi Transporters
Among the first Pht1 phosphate transporters that were isolated are those from mycorrhizal fungi [120]. Mycorrhiza-specific Pht1 members are expressed in response to the AM symbiosis, [109]. AM fungi possess high affinity Pi: H + symporters involved in symbiotic transport. In 1999 Roewarne et al. [121] identified the LePT1 gene highly expressed in arbuscules-containing cortical cells in tomato mycorrhized plants; in non-mycorrhized plants they were found in the stele and bark.
The expression and regulation of the GiPT gene present in extra-radical mycelium of Medicago truncatula roots colonized by Glomus intraradices, increased in response to low Pi concentrations [122]. Liu et al. [123] also reported in Medicago truncatula genes MtPT1 and MtPT2, in mycorrhized roots by Glomus versiforme, under low Pi concentration. The expression of different coding genes for Pi transporters has been described in other crops; in barley (Hordeum vulgare) the relationship between the increase in expression of HvPT8 and the presence in the root of Glomus intraradices was found [124]; in corn ZmPht1;6 and the ortholog gen MtPT4 in Medicago truncatula were identified in the peri-arbuscular membrane [115,125,126]. The Pi transporter gene TaPTH1.2 in wheat is expressed in roots during the early stages of colonization, later, it stabilizes with the appearance of mycorrhizal structures (arbuscules) and acid phosphatases activity [127].
Javot et al. [128] reported that the three identified AM Pi transporters are all expressed in extra-radical hyphae, while only one of them (GmosPT) also shows significant expression in intra-radical hyphae. When Pi enters the fungus cytoplasm, it accumulates in the vacuole [129]. Experiments have shown that when a fungus is transferred from a medium deficient in Pi to a medium rich in Pi, the Pi accumulates and is converted in less than 3 h, in large amounts of polyphosphates [130]. Thus, polyphosphates accumulated in the vacuole can be transferred from the extra-radical hyphae to the intra-radical hyphae [131], possibly by cytoplasmic flow [13].
Ezawa et al. [132] and Rasmussen et al. [133] observed a wide distribution of long polyphosphates chains in extra-radical hyphae, and shorter polyphosphates chains in intra-radical hyphae [134]; Ohtomo et al. [131] suggesting that polyphosphates is hydrolyzed in the intra-radical hyphae to release Pi to the plant. The transfer of Pi occurs through the apoplastic space when Pi is discharged by the fungus arbuscules and it is loaded into the plant’s root symplasm, in the membrane that surrounds the arbuscule [45]. Then, Pi is absorbed through the plant periarbuscular membrane by plant phosphate transporters [135].
In tomato mycorrhized roots, three transporters of Pi were identified in arbuscules: LYCes; Pht1; 4 and LYCes; Pht1; 3 and 5 [109]. Walder et al. [136] mention that depending on the plant species, the acquisition of mycorrhizal Pi could be more regulated by the fungus or by the plant. The amount of Pi that the fungus contributes to the plant depends on each plant/fungus combination [75].
Apparently some plants and AMF can reduce the transfer of nutrients to their symbiotic partner, therefore, it has hypothesized that this may occur when they do not receive reciprocal benefit [137,138]. In the study by Walder et al. [136] sorghum was an important carbon provider for AMF and, the transfer of Pi from AMF to sorghum did not appear to be the reciprocal. However, the authors mention that the Pi could have been in the periarbuscular space, available when the plant requires it.
Although the absorption of Pi in the plant is increasingly understood, the mechanisms of Pi release from the fungus to the interfacial apoplast are still obscure. The mechanisms of Pi release from fungi to the periarbuscular space requires more research [139] and there is still much to investigate on the expression of Pi transporters of plants, and fungi at the arbuscule level.
5.3 Pi Transporters of Ectomycorrhizal Fungi (EMF)
Inorganic phosphate is absorbed by the cells of the ectomycorrhizal fungus through Pi transporters [69,140]. The specialized symbiotic interaction involves the transport of nutrients from the plasma membrane of the fungal cell to the plant [141]. The release of fungal Pi at the symbiotic interface is carried out by the release of free orthophosphate ions (Pi) through phosphate transporters (PT) [142].
Several genes have been identified in ectomycorrhizal fungi that code for Pi transporters. Three in Hebeloma cylindrosporum (HcPT1.1, HcPT1.2 and HcPT2), in Paxillus involutus (PiPT1-PiPT3), and in Tuber melanosporum (TmPT1-TmPT3); five in Laccaria bicolor (LbPT1-LbPT5) and in Amanita muscaria (AmPT1-AmPT5) [69].
All these Pi transporters are grouped into two discrete sequence groups, one characterized as Na+/Pi symporters and another group characterized as H+/Pi symporters [143]. Most of these transporters belong to the Pht1 subfamily or H+/Pi transporters, except for T. melanosporum which has Na+/Pi transporters. However, of all these transporters identified in EMF only HcPT1, HcPT2 have been characterized by heterologous expression in yeast. These two high-affinity Pi symporters are located in the plasma membrane [69,144].
Pi starvation increases the levels of HcPT1 transcripts as demonstrated in studies of cultured H. cylindrosporum hyphae [145,146], as well as in the homologs in the yeast Saccharomyces cerevisiae [147] and in Glomus versiforme [120]. Therefore, HcPT1.1 transcription is dependent on the external availability of Pi and is mainly found in mycelium mantle and external hyphae of EMF [146].
Conversely, HcPT2 is not regulated by the external availability of Pi and is located in extra-radical hyphae and in the Hartig net [148]. HcPT2 is decisive for the establishment of the ectomycorrhizal symbiosis and for the supply of Pi towards symbiotic interface [142,149], determining the entrance of Pi into the mycelium as the exit towards the roots of the plant, as demonstrated by Becquer et al. [148] in Pinus pinaster. The same authors suggest the possibility that HcPT2 could participate in the exit of Pi from the hyphae of the fungus towards the Hartig network.
These transporters have different affinities for Pi with Km values of 55 µM (HcPT1) and 4 µM (HcPT2). In summary, H. cylindrosporum uses the HcPT1 transporter to mediate Pi uptake when Pi availability in the soil is low and HcPT2 when Pi availability is high [69]. Other recently characterized genes are BePT from Boletus editus, RlPT from Rhizopogon luteolus and LbPT from Leucocortinarius bulbiger [142]. BePT belongs to the largest facilitator superfamily and has a high level of sequence identity with the Pht1 family. In a study conducted by Wang et al. [143] they confirm that members of the Pht1 family, which have been cloned from various organisms, are highly conserved through evolution and share protein sequence identities [143]. In summary, for most EMF, the import of Pi from the plasma membrane is dependent on the proton symporter [144].
Phosphate fertilizers are becoming more and more expensive and the use of phosphorus in agriculture is necessary due to the increasing demand for food. Due to the interaction of plants with mycorrhizal fungi in Pi-deficient soils, it is possible to develop crops with optimal Pi absorption efficiencies. Therefore, it is essential to understand the interaction processes of plants in symbiosis with mycorrhizal fungi and the direct absorption of Pi by the plant, to design crop management programs and achieve sustainable global food security.
Further studies are needed to better understand the interesting interrelationships between the natural populations of AM or EM fungi with various agricultural and fruit crops on different types of soils. Furthermore, little is known about the diversity of mycorrhizal fungal species in annual and semi-perennial agricultural crops, and in fruit trees, besides other benefits that these contribute to crops, such as protection against plant pathogens as reported by Newsham et al. [150].
There are still considerable gaps in our knowledge about the absorption and transport of Pi and in the release of the element in the symbiotic interface. As this knowledge advances, we will be able to develop production strategies in regions with phosphorus deficiency and other types of plant stress. The public availability of genomes of crop plants, endo and ectomycorrhizal fungal, will be a valuable tool for future research of the molecular identities of Pi transporters, cell location, mechanism, kinetic properties, and regulation.
Acknowledgement: We are indebted to Guillermo Morel de la Garza for the elaboration of the figures.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present review.
1. Bieleski, R. L. (1973). Phosphate pools, phosphate transport, and phosphate availability. Annual Review of Plant Physiology, 24(1), 225–252. DOI 10.1146/annurev.pp.24.060173.001301. [Google Scholar] [CrossRef]
2. Vance, C. P., Uhde-Stone, C., Allan, D. L. (2003). Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytologist, 157(3), 423–447. DOI 10.1046/j.1469-8137.2003.00695.x. [Google Scholar] [CrossRef]
3. Richardson, A. E., Simpson, R. J. (2011). Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiology, 156(3), 989–996. DOI 10.1104/pp.111.175448. [Google Scholar] [CrossRef]
4. Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., Nasrulhaq Boyce, A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability—A review. Molecules, 21(5), 573. DOI 10.3390/molecules21050573. [Google Scholar] [CrossRef]
5. Owen, D., Williams, A. P., Griffith, G. W., Withers, P. J. (2015). Use of commercial bio-inoculants to increase agricultural production through improved phosphrous acquisition. Applied Soil Ecology, 86, 41–54. DOI 10.1016/j.apsoil.2014.09.012. [Google Scholar] [CrossRef]
6. Raliya, R., Tarafdar, J. C., Biswas, P. (2016). Enhancing the mobilization of native phosphorus in the mung bean rhizosphere using ZnO nanoparticles synthesized by soil fungi. Journal of Agricultural and Food Chemistry, 64(16), 3111–3118. DOI 10.1021/acs.jafc.5b05224. [Google Scholar] [CrossRef]
7. Corrales-Ramírez, L. C., Arévalo, Z. Y., Moreno, V. E. (2014). Solubilización de fosfatos: Una función microbiana importante en el desarrollo vegetal. Nova, 12(21), 67–79. DOI 10.22490/24629448.997. [Google Scholar] [CrossRef]
8. Chiu, C. H., Paszkowski, U. (2019). Mechanisms and impact of symbiotic phosphate acquisition. Cold Spring Harbor Perspectives in Biology, 11(6), a034603. DOI 10.1101/cshperspect.a034603. [Google Scholar] [CrossRef]
9. Richardson, A. E., Barea, J. M., McNeill, A. M., Prigent-Combaret, C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant and Soil, 321(1), 305–339. DOI 10.1007/s11104-009-9895-2. [Google Scholar] [CrossRef]
10. Hayat, R., Ali, S., Amara, U., Khalid, R., Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: A review. Annals of Microbiology, 60(4), 579–598. DOI 10.1007/s13213-010-0117-1. [Google Scholar] [CrossRef]
11. Tsuzuki, S., Handa, Y., Takeda, N., Kawaguchi, M. (2016). Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Molecular Plant-Microbe Interactions, 29(4), 277–286. DOI 10.1094/MPMI-10-15-0234-R. [Google Scholar] [CrossRef]
12. Carotenuto, G., Chabaud, M., Miyata, K., Capozzi, M., Takeda, N. et al. (2017). The rice LysM receptor-like kinase Os CERK 1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytologist, 214(4), 1440–1446. DOI 10.1111/nph.14539. [Google Scholar] [CrossRef]
13. Smith, S. E., Read, D. J. (2008). Mycorrhizal symbiosis. Third edition. New York, USA: Elsevier. [Google Scholar]
14. Schnepf, A., Jones, D., Roose, T. (2011). Modelling nutrient uptake by individual hyphae of arbuscular mycorrhizal fungi: Temporal and spatial scales for an experimental design. Bulletin of Mathematical Biology, 73(9), 2175–2200. DOI 10.1007/s11538-010-9617-1. [Google Scholar] [CrossRef]
15. Mohammadi, K., Khalesro, S., Sohrabi, Y., Heidari, G. (2011). A review: Beneficial effects of the mycorrhizal fungi for plant growth. Journal of Applied Environmental and Biological Sciences, 1(9), 310–319. [Google Scholar]
16. Muñoz-Márquez, E., Macías-López, C., Franco-Ramírez, A., Sánchez-Chávez, E., Jiménez-Castro, J. et al. (2009). Identificación y colonización natural de hongos micorrízicos arbusculares en nogal. Terra Latinoamericana, 27(4), 355–361. [Google Scholar]
17. Al-Qarawi, A. A., Mridha, M. A. U., Dhar, P. P. (2013). Report of Funneliformis mosseae (Nicol. & Gerd.) Gerd. and Trappe from Rangeland soil of Saudi Arabia. Research Journal of Biotechnology, 8(2), 93–96. [Google Scholar]
18. Wyatt, G. A., Kiers, E. T., Gardner, A., West, S. A. (2014). A biological market analysis of the plant-mycorrhizal symbiosis. Evolution, 68(9), 2603–2618. DOI 10.1111/evo.12466. [Google Scholar] [CrossRef]
19. Daguerre, Y., Plett, J. M., Veneault-Fourrey, C. (2016). Signaling pathways driving the development of ectomycorrhizal symbiosis. Molecular mycorrhizal symbiosis. USA: John Wiley & Sons, Inc. [Google Scholar]
20. Finlay, R. D., Read, D. J. (1986). The structure and function of the vegetative mycelium of ectomycorrhizal plants I. Translocation of 14C-labelled carbon between plants interconnected by common mycelium. New Phytologist, 103(1), 143–156. DOI 10.1111/j.1469-8137.1986.tb00603.x. [Google Scholar] [CrossRef]
21. Horton, T., Bruns, T. D. (2001). The molecular revolution in ectomycorrhizal ecology: Peeking into the black-box. Molecular Ecology, 10(8), 1855–1871. DOI 10.1046/j.0962-1083.2001.01333.x. [Google Scholar] [CrossRef]
22. Martin, F., Kohler, A., Murat, C., Veneault-Fourrey, C., Hibbett, D. S. (2016). Unearthing the roots of ectomycorrhizal symbioses. Nature Reviews Microbiology, 14(12), 760–773. DOI 10.1038/nrmicro.2016.149. [Google Scholar] [CrossRef]
23. Peterson, R. L., Bonfante, P. (1994). Comparative structure of vesicular-arbuscular mycorrhizas and ectomycorrhizas. Plant and Soil, 159(1), 79–88. DOI 10.1007/BF00000097. [Google Scholar] [CrossRef]
24. Clasen, B. E., Silveira, A. O., Baldoni, D. B., Montagner, D. F., Jacques, R. J. S. et al. (2018). Characterization of ectomycorrhizal species through molecular biology tools and morphotyping. Scientia Agricola, 75(3), 246–254. DOI 10.1590/1678-992x-2016-0419. [Google Scholar] [CrossRef]
25. Kurth, F., Feldhahn, L., Bönn, M., Herrmann, S., Buscot, F. et al. (2015). Large scale transcriptome analysis reveals interplay between development of forest trees and a beneficial mycorrhiza helper bacterium. BMC Genomics, 16(1), 1–13. DOI 10.1186/s12864-015-1856-y. [Google Scholar] [CrossRef]
26. Holmer, R., Rutten, L., Kohlen, W., Velzen, R., Geurts, R. (2017). Commonalities in symbiotic plant-microbe signalling. Advances in Botanical Research, 82, 187–221. [Google Scholar]
27. Boivin, S., Fonouni-Farde, C., Frugier, F. (2016). How auxin and cytokinin phytohormones modulate root microbe interactions. Frontiers in Plant Science, 7(575), 1240. DOI 10.3389/fpls.2016.01240. [Google Scholar] [CrossRef]
28. Sukumar, P., Legué, V., Vayssiéres, A., Martin, F., Tuskan, G. A. et al. (2013). Involvement of auxin pathways in modulating root architecture during beneficial plant-microorganism interactions. Plant, Cell and Environment, 36(5), 909–919. DOI 10.1111/pce.12036. [Google Scholar] [CrossRef]
29. Vayssières, A., Pencík, A., Felten, J., Kohler, A., Ljung, K. (2015). Development of the poplar-Laccaria bicolor ectomycorrhiza modifies root auxin metabolism, signaling, and response. Plant Physiology, 169(1), 890–902. DOI 10.1104/pp.114.255620. [Google Scholar] [CrossRef]
30. Ditengou, F. A., Lapeyrie, F. (2000). Hypaphorine from the ectomycorrhizal fungus Pisolithus tinctorius counteracts activities of indole-3-acetic acid and ethylene but not synthetic auxins in eucalypt seedlings. Molecular Plant-Microbe Interactions, 13(2), 151–158. DOI 10.1094/MPMI.2000.13.2.151. [Google Scholar] [CrossRef]
31. Miura, K., Tada, Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Frontiers in Plant Science, 5, 4. DOI 10.3389/fpls.2014.00004. [Google Scholar] [CrossRef]
32. de Ollas, C.,Arbona, V., Gómez-Cadenas, A. (2015). Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant, Cell & Environment, 38(10), 2157–2170. DOI 10.1111/pce.12536. [Google Scholar] [CrossRef]
33. Calvo-Polanco, M., Armada, E., Zamarreño, A. M., García-Mina, J. M., Aroca, R. (2019). Local root ABA/cytokinin status and aquaporins regulate poplar responses to mild drought stress independently of the ectomycorrhizal fungus Laccaria bicolor. Journal of Experimental Botany, 70(21), 6437–6446. DOI 10.1093/jxb/erz389. [Google Scholar] [CrossRef]
34. Balestrini, R., Lanfranco, L. (2006). Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza, 16(8), 509–524. DOI 10.1007/s00572-006-0069-2. [Google Scholar] [CrossRef]
35. López-Arredondo, D. L., Leyva-González, M. A., González-Morales, S. I., López-Bucio, J., Herrera-Estrella, L. (2014). Phosphate nutrition: Improving low-phosphate tolerance in crops. Annual Review of Plant Biology, 65(1), 95–123. DOI 10.1146/annurev-arplant-050213-035949. [Google Scholar] [CrossRef]
36. Asaff-Torres, A., Armendáriz-Ruiz, M., Kirchmayr, M., Rodríguez-Heredia, R., Orozco, M. et al. (2017). Rhizospheric microbiome profiling of Capsicum annuum L. cultivated in amended soils by 16S and Internal Transcribed Spacer 2 rRNA amplicon metagenome sequencing. Genome Announcements, 5(30), 463. DOI 10.1128/genomeA.00626-17. [Google Scholar] [CrossRef]
37. Crovadore, J., Torres, A. A., Rodriguez-Heredia, R., Cochard, B., Chablais, R. et al. (2017). Metagenomes of soils samples from an established perennial cropping system of asparagus related with biostimulants in Southern France. Genome Announcements, 5(24), e00511–17. [Google Scholar]
38. Smith, S. E., Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annual Review of Plant Biology, 62(1), 227–250. DOI 10.1146/annurev-arplant-042110-103846. [Google Scholar] [CrossRef]
39. Guetsky, R., Kobiler, I., Wang, X., Perlman, N., Gollop, N. et al. (2005). Metabolism of the flavonoid epicatechin by laccase of Colletotrichum gloeosporioides and its effect on pathogenicity on avocado fruits. Phytopathology, 95(11), 1341–1348. DOI 10.1094/PHYTO-95-1341. [Google Scholar] [CrossRef]
40. García-Avila, C. J., Valenzuela-Tirado, G. A., Florencio-Anastasio, J. G., Ruiz-Galván, I., Moreno-Velázquez, M. et al. (2018). Organisms associated with damage to post-harvest potato tubers. Revista Mexicana de Fitopatología, 36(2), 308–320. [Google Scholar]
41. Guenoune, D., Galili, S., Phillips, D. A., Volpin, H., Chet, I. et al. (2001). The defense response elicited by the pathogen Rhizoctonia solani is suppressed by colonization of the AM-fungus Glomus intraradices. Plant Science, 160(5), 925–932. DOI 10.1016/S0168-9452(01)00329-6. [Google Scholar] [CrossRef]
42. Tian, H., Wang, R., Li, M., Dang, H., Solaiman, Z. M. (2019). Molecular signal communication during arbuscular mycorrhizal formation induces significant transcriptional reprogramming of wheat (Triticum aestivum) roots. Annals of Botany, 124(6), 1109–1119. [Google Scholar]
43. Luginbuehl, L., Oldroyd, G. E. (2016). Calcium signaling and transcriptional regulation in arbuscular mycorrhizal symbiosis. Molecular mycorrhizal symbiosis. USA: John Wiley & Sons, Inc. [Google Scholar]
44. Ortiz-Berrocal, M., Lozano, L., Sánchez-Flores, A., Nava, N., Hernández, G. et al. (2017). Expression in rice of an autoactive variant of Medicago truncatula DMI3, the Ca+2/calmodulin-dependent protein kinase from the common symbiotic pathway modifies root transcriptome and improves mycorrhizal colonization. Plant Biotechnology Reports, 11(5), 271–287. DOI 10.1007/s11816-017-0449-4. [Google Scholar] [CrossRef]
45. Berruti, A., Borriello, R., Lumini, E., Scariot, V., Bianciotto, V. et al. (2013). Application of laser microdissection to identify the mycorrhizal fungi that establish arbuscules inside root cells. Frontiers in Plant Science, 4, 135. DOI 10.3389/fpls.2013.00135. [Google Scholar] [CrossRef]
46. Siciliano, V., Genre, A., Balestrini, R., de Wit, P. J. G. M., Bonfante, P. (2007). Pre-penetration apparatus formation during AM infection is associated with a specific transcriptome response in epidermal cells. Plant Signaling & Behavior, 2(6), 533–535. DOI 10.4161/psb.2.6.4745. [Google Scholar] [CrossRef]
47. Genre, A., Chabaud, M., Faccio, A., Barker, D. G., Bonfante, P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell, 20(5), 1407–1420. DOI 10.1105/tpc.108.059014. [Google Scholar] [CrossRef]
48. Koch, A. M., Antunes, P. M., Maherali, H., Hart, M. M., Klironomos, J. N. (2017). Evolutionary asymmetry in the arbuscular mycorrhizal symbiosis: Conservatism in fungal morphology does not predict host plant growth. New Phytologist, 214(3), 1330–1337. DOI 10.1111/nph.14465. [Google Scholar] [CrossRef]
49. Pozo, M. J., Cordier, C., Dumas-Gaudot, E., Gianinazzi, S., Barea, J. M. et al. (2002). Localized versus systemic effect of arbuscular mycorrhizal fungi on defence responses to Phytophthora infection in tomato plants. Journal of Experimental Botany, 53(368), 525–534. DOI 10.1093/jexbot/53.368.525. [Google Scholar] [CrossRef]
50. Zhang, H., Franken, P. (2014). Comparison of systemic and local interactions between the arbuscular mycorrhizal fungus Funneliformis mosseae and the root pathogen Aphanomyces euteiches in Medicago truncatula. Mycorrhiza, 24(6), 419–430. DOI 10.1007/s00572-013-0553-4. [Google Scholar] [CrossRef]
51. Mohan, V., Nivea, R., Menon, S. (2015). Evaluation of ectomycorrhizal fungi as potential bio-control agents against selected plant pathogenic fungi. Journal of Artificial Intelligence Research, 3(9), 408–412. [Google Scholar]
52. Reyes-Tena, A., Rincón-Enríquez, G., López-Pérez, L., Quiñones-Aguilar, E. E. (2017). Effect of mycorrhizae and actinomycetes on growth and bioprotection of Capsicum annuum L. against Phytophthora capsici. Pakistan Journal of Agricultural Sciences, 54(3), 513–522. DOI 10.21162/PAKJAS/17.4245. [Google Scholar] [CrossRef]
53. Rubio, V., Bustos, R., Irigoyen, M. L., Cardona-López, X., Rojas-Triana, M. et al. (2009). Plant hormones and nutrient signaling. Plant Molecular Biology, 69(4), 361–373. DOI 10.1007/s11103-008-9380-y. [Google Scholar] [CrossRef]
54. Chiou, T. J., Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology, 62(1), 185–206. DOI 10.1146/annurev-arplant-042110-103849. [Google Scholar] [CrossRef]
55. Lei, M., Zhu, C., Liu, Y., Karthikeyan, A. S., Bressan, R. A. et al. (2011). Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytologist, 189(4), 1084–1095. DOI 10.1111/j.1469-8137.2010.03555.x. [Google Scholar] [CrossRef]
56. Siddiqi, K. S., Husen, A. (2017). Plant response to strigolactones: Current developments and emerging trends. Applied Soil Ecology, 120, 247–253. DOI 10.1016/j.apsoil.2017.08.020. [Google Scholar] [CrossRef]
57. Visentin, I., Vitali, M., Ferrero, M., Zhang, Y., Ruyter-Spira, C. et al. (2016). Low levels of strigolactones in roots as a component of the systemic signal of drought stress in tomato. New Phytologist, 212(4), 954–963. DOI 10.1111/nph.14190. [Google Scholar] [CrossRef]
58. Yoneyama, K., Xie, X., Kisugi, T., Nomura, T., Yoneyama, K. (2013). Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta, 238(5), 885–894. DOI 10.1007/s00425-013-1943-8. [Google Scholar] [CrossRef]
59. Yoneyama, K., Xie, X., Kim, H. I., Kisugi, T., Nomura, T. et al. (2012). How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta, 235(6), 1197–1207. DOI 10.1007/s00425-011-1568-8. [Google Scholar] [CrossRef]
60. Xie, X., Yoneyama, K., Yoneyama, K. (2010). The strigolactone story. Annual Review of Phytopathology, 48(1), 93–117. DOI 10.1146/annurev-phyto-073009-114453. [Google Scholar] [CrossRef]
61. Fusconi, A. (2014). Regulation of root morphogenesis in arbuscular mycorrhizae: What role do fungal exudates, phosphate, sugars and hormones play in lateral root formation? Annals of Botany, 113(1), 19–33. DOI 10.1093/aob/mct258. [Google Scholar] [CrossRef]
62. Ruyter-Spira, C., Kohlen, W., Charnikhova, T., van Zeijl, A.,van Bezouwen, L. et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiology, 155(2), 721–734. DOI 10.1104/pp.110.166645. [Google Scholar] [CrossRef]
63. Kretzschmar, T., Kohlen, W., Sasse, J., Borghi, L., Schlegel, M. et al. (2012). A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature, 483(7389), 341–344. DOI 10.1038/nature10873. [Google Scholar] [CrossRef]
64. Garcia, K., Doidy, J., Zimmermann, S. D., Wipf, D., Courty, P. E. (2016). Take a trip through the plant and fungal transportome of mycorrhiza. Trends in Plant Science, 21(11), 937–950. DOI 10.1016/j.tplants.2016.07.010. [Google Scholar] [CrossRef]
65. Kohlen, W., Charnikhova, T., Liu, Q., Bours, R., Domagalska, M. A. et al. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiology, 155(2), 974–987. DOI 10.1104/pp.110.164640. [Google Scholar] [CrossRef]
66. Bhandari, P., Garg, N. (2017). Arbuscular mycorrhizal symbiosis: A promising approach for imparting abiotic stress tolerance in crop plants. Plant-microbe interactions in agro-ecological perspectives. Singapore: Springer. [Google Scholar]
67. Oldroyd, G. E. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology, 11(4), 252–263. DOI 10.1038/nrmicro2990. [Google Scholar] [CrossRef]
68. Wright, D. P., Johansson, T., Le Quéré, A., Söderström, B., Tunlid, A. et al. (2005). Spatial patterns of gene expression in the extramatrical mycelium and mycorrhizal root tips formed by the ectomycorrhizal fungus Paxillus involutus in association with birch (Betula pendula) seedlings in soil microcosms. New Phytologist, 167(2), 579–596. DOI 10.1111/j.1469-8137.2005.01441.x. [Google Scholar] [CrossRef]
69. Casieri, L., Lahmidi, N. A., Doidy, J., Veneault-Fourrey, C., Migeon, A. et al. (2013). Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza, 23(8), 597–625. DOI 10.1007/s00572-013-0496-9. [Google Scholar] [CrossRef]
70. Nygren, C. M. R., Rosling, A. (2009). Localisation of phosphomonoesterase activity in ectomycorrhizal fungi grown on different phosphorus sources. Mycorrhiza, 19(3), 197–204. DOI 10.1007/s00572-008-0223-0. [Google Scholar] [CrossRef]
71. Ruess, R. W., Swanson, M. M., Kielland, K., McFarland, J. W., Olson, K. D. (2019). Phosphorus mobilizing enzymes of Alnus-associated ectomycorrhizal fungi in an Alaskan boreal floodplain. Forests, 10(7), 554. DOI 10.3390/f10070554. [Google Scholar] [CrossRef]
72. van Aarle, I. M., Plassard, C. (2010). Spatial distribution of phosphatase activity associated with ectomycorrhizal plants is related to soil type. Soil Biology and Biochemistry, 42(2), 324–330. DOI 10.1016/j.soilbio.2009.11.011. [Google Scholar] [CrossRef]
73. Vilches, A. P., Norström, S. H., Olofsson, M. A., Fransson, P., Bylund, D. (2018). Biofuel ash addition increases ectomycorrhizal fungal exudation in pure culture. Environmental Chemistry, 15(8), 481–492. DOI 10.1071/EN18146. [Google Scholar] [CrossRef]
74. Gordon-Weeks, R., Tong, Y., Davies, T. E., Leggewie, G. (2003). Restricted spatial expression of a high-affinity phosphate transporter in potato roots. Journal of Cell Science, 116(15), 3135–3144. DOI 10.1242/jcs.00615. [Google Scholar] [CrossRef]
75. Smith, S. E., Smith, F. A., Jakobsen, I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiology, 133(1), 16–20. DOI 10.1104/pp.103.024380. [Google Scholar] [CrossRef]
76. Raghothama, K. G. (1999). Phosphate acquisition. Annual Review of Plant Biology, 50(1), 665–693. DOI 10.1146/annurev.arplant.50.1.665. [Google Scholar] [CrossRef]
77. Ticconi, C. A., Delatorre, C. A., Lahner, B., Salt, D. E., Abel, S. (2004). Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant Journal, 37(6), 801–814. DOI 10.1111/j.1365-313X.2004.02005.x. [Google Scholar] [CrossRef]
78. Zakhleniuk, O. V., Raines, C. A., Lloyd, J. C. (2001). pho3: A phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta, 212(4), 529–534. DOI 10.1007/s004250000450. [Google Scholar] [CrossRef]
79. Bayle, V., Arrighi, J. F., Creff, A., Nespoulous, C., Vialaret, J. et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell, 23(4), 1523–1535. DOI 10.1105/tpc.110.081067. [Google Scholar] [CrossRef]
80. Cox, G., Tinker, P. B. (1976). Translocation and transfer of nutrients in vesicular-arbuscular mycorrhizas. I. The arbuscule and phosphorus transfer: A quantitative ultrastructural study. New Phytologist, 77(2), 371–378. DOI 10.1111/j.1469-8137.1976.tb01526.x. [Google Scholar] [CrossRef]
81. Sharif, M., Ahmad, E., Sarir, M. S., Muhammad, D., Shafi, M. et al. (2011). Response of different crops to arbuscular mycorrhiza fungal inoculation in phosphorus-deficient soil. Communications in Soil Science and Plant Analysis, 42(19), 2299–2309. DOI 10.1080/00103624.2011.605488. [Google Scholar] [CrossRef]
82. Chiou, T. J., Liu, H., Harrison, M. J. (2001). The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant Journal, 25(3), 281–293. DOI 10.1046/j.1365-313x.2001.00963.x. [Google Scholar] [CrossRef]
83. Li, X. L., George, E., Marschner, H. (1991). Phosphorus depletion and pH decrease at the root-soil and hyphae-soil interfaces of VA mycorrhizal white clover fertilized with ammonium. New Phytologist, 119(3), 397–404. DOI 10.1111/j.1469-8137.1991.tb00039.x. [Google Scholar] [CrossRef]
84. Karandashov, V., Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends in Plant Science, 10(1), 22–29. DOI 10.1016/j.tplants.2004.12.003. [Google Scholar] [CrossRef]
85. Wu, Z., Zhao, J., Gao, R., Hu, G., Gai, J. et al. (2011). Molecular cloning, characterization and expression analysis of two members of the Pht1 family of phosphate transporters in Glycine max. PLoS One, 6(6), e19752. DOI 10.1371/journal.pone.0019752. [Google Scholar] [CrossRef]
86. Johri, A. K., Oelmüller, R., Dua, M., Yadav, V., Kumar, M. et al. (2015). Fungal association and utilization of phosphate by plants: Success, limitations, and future prospects. Frontiers in Microbiology, 6(1241), 984. DOI 10.3389/fmicb.2015.00984. [Google Scholar] [CrossRef]
87. Sánchez-Calderón, L., Chacon-López, A., Pérez-Torres, C. A., Herrera-Estrella, L. (2010). Phosphorus: Plant strategies to cope with its scarcity. Cell biology of metals and nutrients. Germany: Springer. [Google Scholar]
88. Fang, Z., Shao, C., Meng, Y., Wu, P., Chen, M. (2009). Phosphate signaling in Arabidopsis and Oryza sativa. Plant Science, 176(2), 170–180. DOI 10.1016/j.plantsci.2008.09.007. [Google Scholar] [CrossRef]
89. Preuss, C. P., Huang, C. Y., Gilliham, M., Tyerman, S. D. (2010). Channel-like characteristics of the low-affinity barley phosphate transporter PHT1; 6 when expressed in Xenopus oocytes. Plant Physiology, 152(3), 1431–1441. DOI 10.1104/pp.109.152009. [Google Scholar] [CrossRef]
90. Daram, P., Brunner, S., Amrhein, N., Bucher, M. (1998). Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta, 206(2), 225–233. DOI 10.1007/s004250050394. [Google Scholar] [CrossRef]
91. Liu, C., Muchhal, U. S., Mukatira, U., Kononowicz, A. K., Raghothama, K. G. (1998). Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiology, 116(1), 91–99. DOI 10.1104/pp.116.1.91. [Google Scholar] [CrossRef]
92. Drew, M. C., Saker, L. R. (1984). Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: Evidence of non-allosteric regulation. Planta, 160(6), 500–507. DOI 10.1007/BF00411137. [Google Scholar] [CrossRef]
93. Anghinoni, I., Barber, S. A. (1980). Phosphorus influx and growth characteristics of corn roots as influenced by phosphorus supply. Agronomy Journal, 72(4), 685–688. DOI 10.2134/agronj1980.00021962007200040028x. [Google Scholar] [CrossRef]
94. Clarkson, D. T. (1993). Roots and the delivery of solutes to the xylem. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 341(1295), 5–17. DOI 10.1098/rstb.1993.0086. [Google Scholar] [CrossRef]
95. Epstein, E. (1953). Mechanism of ion absorption by roots. Nature, 171(4341), 83–84. DOI 10.1038/171083a0. [Google Scholar] [CrossRef]
96. Epstein, E., Hagen, C. E. (1952). A kinetic study of the absorption of alkali cations by barley roots. Plant Physiology, 27(3), 457–474. DOI 10.1104/pp.27.3.457. [Google Scholar] [CrossRef]
97. Mimura, T. (1999). Regulation of phosphate transport and homeostasis in plant cells. International Review of Cytology, 191, 149–200. [Google Scholar]
98. Mimura, T., Sakano, K., Shimmen, T. (1996). Studies on the distribution, re-translocation and homeostasis of inorganic phosphate in barley leaves. Plant, Cell and Environment, 19(3), 311–320. DOI 10.1111/j.1365-3040.1996.tb00253.x. [Google Scholar] [CrossRef]
99. Gutjahr, C., Sawers, R. J. H., Marti, G., Andrés-Hernández, L., Yang, S. Y. et al. (2015). Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proceedings of National Academy of Sciences, 112(21), 6754–6759. DOI 10.1073/pnas.1504142112. [Google Scholar] [CrossRef]
100. Chen, J., Liu, Y., Ni, J., Wang, Y., Bai, Y. et al. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiology, 157(1), 269–278. DOI 10.1104/pp.111.181669. [Google Scholar] [CrossRef]
101. Mudge, S. R., Rae, A. L., Diatloff, E., Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant Journal, 31(3), 341–353. DOI 10.1046/j.1365-313X.2002.01356.x. [Google Scholar] [CrossRef]
102. Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N. et al. (1999). Pht2; 1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell, 11(11), 2153–2166. DOI 10.1105/tpc.11.11.2153. [Google Scholar] [CrossRef]
103. Hata, S., Kobae, Y., Banba, M. (2010). Interactions between plants and arbuscular mycorrhizal fungi. International Review of Cell and Molecular Biology, 281, 1–48. [Google Scholar]
104. Loth-Pereda, V., Orsini, E., Courty, P. E., Lota, F., Kohler, A. et al. (2011). Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiology, 156(4), 2141–2154. DOI 10.1104/pp.111.180646. [Google Scholar] [CrossRef]
105. Javot, H., Penmetsa, R. V., Terzaghi, N., Cook, D. R., Harrison, M. J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences of the United States of America, 104(5), 1720–1725. DOI 10.1073/pnas.0608136104. [Google Scholar] [CrossRef]
106. Yang, S. Y., Grønlund, M., Jakobsen, I., Grotemeyer, M. S., Rentsch, D. et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell, 24(10), 4236–4251. DOI 10.1105/tpc.112.104901. [Google Scholar] [CrossRef]
107. Rausch, C., Daram, P., Brunner, S., Jansa, J., Laloi, M. et al. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature, 414(6862), 462–465. DOI 10.1038/35106601. [Google Scholar] [CrossRef]
108. Harrison, M. J., Dewbre, G. R., Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell, 14(10), 2413–2429. DOI 10.1105/tpc.004861. [Google Scholar] [CrossRef]
109. Nagy, R., Karandashov, V., Chague, V., Kalinkevich, K., Tamasloukht, M. B. et al. (2005). The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant Journal, 42(2), 236–250. DOI 10.1111/j.1365-313X.2005.02364.x. [Google Scholar] [CrossRef]
110. Xie, X., Huang, W., Liu, F., Tang, N., Liu, Y. et al. (2013). Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytologist, 198(3), 836–852. DOI 10.1111/nph.12188. [Google Scholar] [CrossRef]
111. Walder, F., Brulé, D., Koegel, S., Wiemken, A., Boller, T. et al. (2015). Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytologist, 205(4), 1632–1645. DOI 10.1111/nph.13292. [Google Scholar] [CrossRef]
112. Yang, W. T., Baek, D., Yun, D. J., Lee, K. S., Hong, S. Y. et al. (2018). Rice OsMYB5P improves plant phosphate acquisition by regulation of phosphate transporter. PLoS One, 13(3), e0194628. DOI 10.1371/journal.pone.0194628. [Google Scholar] [CrossRef]
113. Xu, G. H., Chague, V., Melamed-Bessudo, C., Kapulnik, Y., Jain, A. et al. (2007). Functional characterization of LePT4: A phosphate transporter in tomato with mycorrhiza-enhanced expression. Journal of Experimental Botany, 58(10), 2491–2501. DOI 10.1093/jxb/erm096. [Google Scholar] [CrossRef]
114. Chen, A., Chen, X., Wang, H., Liao, D., Gu, M. et al. (2014). Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biology, 14(1), 61. DOI 10.1186/1471-2229-14-61. [Google Scholar] [CrossRef]
115. Nagy, R., Vasconcelos, M. J. V., Zhao, S., McElver, J., Bruce, W. et al. (2006). Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biology, 8(2), 186–197. DOI 10.1055/s-2005-873052. [Google Scholar] [CrossRef]
116. Chen, W., Li, J., Zhu, H., Xu, P., Chen, J. et al. (2017). Arbuscular mycorrhizal fungus enhances lateral root formation in Poncirus trifoliata (L.) as revealed by RNA-Seq analysis. Frontiers in Plant Science, 8, 327. DOI 10.3389/fpls.2017.02039. [Google Scholar] [CrossRef]
117. Paszkowski, U., Kroken, S., Roux, C., Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Science, 99(20), 13324–13329. DOI 10.1073/pnas.202474599. [Google Scholar] [CrossRef]
118. Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A., Kornberg, R. D. (2001). Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292(5523), 1876–1882. DOI 10.1126/science.1059495. [Google Scholar] [CrossRef]
119. Versaw, W. K., Harrison, M. J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell, 14(8), 1751–1766. DOI 10.1105/tpc.002220. [Google Scholar] [CrossRef]
120. Harrison, M. J., van Buuren, M. L. (1995). A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature, 378(6557), 626–629. DOI 10.1038/378626a0. [Google Scholar] [CrossRef]
121. Roewarne, G. M., Berker, S. J., Smith, S. E., Smith, F. A., Schachtman, D. P. (1999). A Lycopersicon esculentum phosphate transporter (LePT1) involved in phosphorus uptake from avesicular-arbuscular mycorrhizal fungus. New Phytologist, 144(3), 507–516. DOI 10.1046/j.1469-8137.1999.00536.x. [Google Scholar] [CrossRef]
122. Maldonado-Mendoza, I. E., Dewbre, G. R., Harrison, M. J. (2001). A phosphate transporter gene from the extra-radical mycelium of an arbuscular mycorrhizal fungus Glomus intraradices is regulated in response to phosphate in the environment. Molecular Plant-Microbe Interactions, 14(10), 1140–1148. DOI 10.1094/MPMI.2001.14.10.1140. [Google Scholar] [CrossRef]
123. Liu, H., Trieu, A. T., Blaylock, L. A., Harrison, M. J. (1998). Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Molecular Plant-Microbe Interactions, 11(1), 14–22. DOI 10.1094/MPMI.1998.11.1.14. [Google Scholar] [CrossRef]
124. Grace, E. J., Cotsaftis, O., Tester, M., Smith, F. A., Smith, S. E. (2009). Arbuscular mycorrhizal inhibition of growth in barley cannot be attributed to extent of colonization, fungal phosphorus uptake or effects on expression of plant phosphate transporter genes. New Phytologist, 181(4), 938–949. DOI 10.1111/j.1469-8137.2008.02720.x. [Google Scholar] [CrossRef]
125. Wang, C., White, J. P., Li, C. (2016). Colonization and community structure of arbuscular mycorrhizal fungi in maize roots at different depths in the soil profile respond differently to phosphorus inputs on a long-term experimental site. Mycorrhiza, 27(4), 369–381. DOI 10.1007/s00572-016-0757-5. [Google Scholar] [CrossRef]
126. Sawers, R. J. H., Svane, S. F., Quan, C., Gronlund, M., Wozniak, B. et al. (2017). Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytologist, 214(2), 632–643. DOI 10.1111/nph.14403. [Google Scholar] [CrossRef]
127. Deng, Y., Teng, W., Tong, Y. P., Chen, X. P., Zou, C. Q. (2018). Phosphorus efficiency mechanisms of two wheat cultivars as affected by a range of phosphorus levels in the field. Frontiers in Plant Science, 9, 1614. DOI 10.3389/fpls.2018.01614. [Google Scholar] [CrossRef]
128. Javot, H., Pumplin, N., Harrison, M. J. (2007). Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant, Cell & Environment, 30(3), 310–322. DOI 10.1111/j.1365-3040.2006.01617.x. [Google Scholar] [CrossRef]
129. Ezawa, T., Smith, S. E., Smith, F. A. (2001). Enzyme activity involved in glucose phosphorylation in two arbuscular mycorrhizal fungi: Indication that polyphosphates is not the main phosphagen. Soil Biology and Biochemistry, 33(9), 1279–1281. DOI 10.1016/S0038-0717(01)00007-4. [Google Scholar] [CrossRef]
130. Ezawa, T., Cavagnaro, T. R., Smith, S. E., Smith, F. A., Ohtomo, R. (2004). Rapid accumulation of polyphosphate in extraradical hyphae of an arbuscular mycorrhizal fungus as revealed by histochemistry and a polyphosphate kinase/luciferase system. New Phytologist, 161(2), 387–392. DOI 10.1046/j.1469-8137.2003.00966.x. [Google Scholar] [CrossRef]
131. Ohtomo, R., Saito, M. (2005). Polyphosphate dynamics in mycorrhizal roots during colonization of an arbuscular mycorrhizal fungus. New Phytologist, 167(2), 571–578. DOI 10.1111/j.1469-8137.2005.01425.x. [Google Scholar] [CrossRef]
132. Ezawa, T., Kuwahara, S. Y., Sakamoto, K., Yoshida, T., Saito, M. (1999). Specific inhibitor and substrate specificity of alkaline phosphatase expressed in the symbiotic phase of the arbuscular mycorrhizal fungus, Glomus etunicatum. Mycologia, 91(4), 636–641. DOI 10.1080/00275514.1999.12061062. [Google Scholar] [CrossRef]
133. Rasmussen, N., Lloyd, D. C., Ratcliffe, R. G., Hansen, P. E., Jakobsen, I. (2000). 31P NMR for the study of Pi metabolism and translocation in arbuscular mycorrhizal fungi. Plant and Soil, 226(2), 245–253. DOI 10.1023/A:1026411801081. [Google Scholar] [CrossRef]
134. Solaiman, M. Z., Ezawa, T., Kojima, T., Saito, M. (1999). Polyphosphates in intraradical and extraradical hyphae of an arbuscular mycorrhizal fungus, Gigaspora margarita. Applied and Environmental Microbiology, 65(12), 5604–5606. DOI 10.1128/AEM.65.12.5604-5606.1999. [Google Scholar] [CrossRef]
135. Bucher, M. (2007). Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytologist, 173(1), 11–26. DOI 10.1111/j.1469-8137.2006.01935.x. [Google Scholar] [CrossRef]
136. Walder, F., Boller, T., Wiemken, A., Courty, P. E. (2016). Regulation of plants’ phosphate uptake in common mycorrhizal networks: Role of intraradical fungal phosphate transporters. Plant Signaling & Behavior, 11(2), e1131372. DOI 10.1080/15592324.2015.1131372. [Google Scholar] [CrossRef]
137. Kiers, E. T., Duhamel, M., Beesetty, Y., Mensah, J. A., Franken, O. et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science, 333(6044), 880–882. DOI 10.1126/science.1208473. [Google Scholar] [CrossRef]
138. Fellbaum, C. R., Mensah, J. A., Cloos, A. J., Strahan, G. E., Pfeffer, P. E. et al. (2014). Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytologist, 203(2), 646–656. DOI 10.1111/nph.12827. [Google Scholar] [CrossRef]
139. Smith, S. E., Jakobsen, I., Gronlund, M., Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant phosphorus nutrition: Interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiology, 156(3), 1050–1057. DOI 10.1104/pp.111.174581. [Google Scholar] [CrossRef]
140. Plassard, C., Dell, B. (2010). Phosphorus nutrition of mycorrhizal trees. Tree Physiology, 30(9), 1129–1139. DOI 10.1093/treephys/tpq063. [Google Scholar] [CrossRef]
141. Cairney, J. W. G., Burke, R. M. (1996). Physiological heterogeneity within fungal mycelia: An important concept for a functional understanding of the ectomycorrhizal symbiosis. New Phytologist, 134(4), 685–695. DOI 10.1111/j.1469-8137.1996.tb04934.x. [Google Scholar] [CrossRef]
142. Plassard, C., Becquer, A., Garcia, K. (2019). Phosphorus transport in mycorrhiza: How far are we? Trends in Plant Science, 24(9), 794–801. DOI 10.1016/j.tplants.2019.06.004. [Google Scholar] [CrossRef]
143. Wang, J., Li, T., Wu, X., Zhao, Z. (2014). Molecular cloning and functional analysis of a H+-dependent phosphate transporter gene from the ectomycorrhizal fungus Boletus edulis in southwest China. Fungal Biology, 118(5–6), 453–461. DOI 10.1016/j.funbio.2014.03.003. [Google Scholar] [CrossRef]
144. Nehls, U., Plassard, C. (2018). Nitrogen and phosphate metabolism in ectomycorrhizas. New Phytologist, 220(4), 1047–1058. DOI 10.1111/nph.15257. [Google Scholar] [CrossRef]
145. Tatry, M. V., El Kassis, E., Lambilliotte, R., Corratgé, C., van Aarle, I. et al. (2009). Two differentially regulated phosphate transporters from the symbiotic fungus Hebeloma cylindrosporum and phosphorus acquisition by ectomycorrhizal Pinus pinaster. Plant Journal, 57(6), 1092–1102. DOI 10.1111/j.1365-313X.2008.03749.x. [Google Scholar] [CrossRef]
146. Garcia, K., Haider, M. Z., Delteil, A., Corratgé-Faillie, C., Conéjero, G. et al. (2013). Promoter-dependent expression of the fungal transporter HcPT1. 1 under Pi shortage and its spatial localization in ectomycorrhiza. Fungal Genetics and Biology, 58, 53–61. DOI 10.1016/j.fgb.2013.06.007. [Google Scholar] [CrossRef]
147. Secco, D., Wang, C., Shou, H., Whelan, J. (2012). Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Letters, 586(4), 289–295. DOI 10.1016/j.febslet.2012.01.036. [Google Scholar] [CrossRef]
148. Becquer, A., Garcia, K., Amenc, L., Rivard, C., Doré, J. et al. (2018). The Hebeloma cylindrosporum HcPT2 Pi transporter plays a key role in ectomycorrhizal symbiosis. New Phytologist, 220(4), 1185–1199. DOI 10.1111/nph.15281. [Google Scholar] [CrossRef]
149. Zheng, R., Wang, J., Liu, M., Duan, G., Gao, X. et al. (2016). Molecular cloning and functional analysis of two phosphate transporter genes from Rhizopogon luteolus and Leucocortinarius bulbiger, two ectomycorrhizal fungi of Pinus tabulaeformis. Mycorrhiza, 26(7), 633–644. DOI 10.1007/s00572-016-0702-7. [Google Scholar] [CrossRef]
150. Newsham, K. K., Fitter, A. H., Watkinson, A. R. (1995). Multi-functionality and biodiversity in arbuscular mycorrhizas. Trends in Ecology & Evolution, 10(10), 407–411. DOI 10.1016/S0169-5347(00)89157-0. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |