
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.015508
REVIEW
Overview of Strain Characterization in Relation to Serological and Molecular Detection of Citrus tristeza Closterovirus
1Department of Plant Pathology, College of Agriculture, University of Sargodha, Sargodha, 40100, Pakistan
2Institute of Horticultural Sciences, University of Agriculture Faisalabad, Faisalabad, 38000, Pakistan
3State Key Laboratory of Agricultural Microbiology and Provincial Key Laboratory of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, 430070, China
4Department of Soil Science, Faculty of Agricultural Sciences and Technology, Bahauddin Zakariya University, Multan, 60800, Pakistan
*Corresponding Authors: Yasir Iftikhar. Email: yasir.iftikhar@uos.edu.pk; Muhammad Zafar-ul-Hye. Email: zafarulhyegondal@yahoo.com
Received: 23 December 2020; Accepted: 26 January 2021
Abstract: Tristeza is a devastating viral disease in all the citrus growing countries throughout the world and has killed millions of citrus trees in severely affected orchards. The citrus species grafted on sour orange rootstock are affected by this disease. Predominantly, the sweet orange, grapefruit and lime trees grafted on sour orange exhibit severe symptoms like quick decline, vein clearing, pin holing, bark scaling and degeneration leading to variable symptoms. Symptomatic expression of Citrus tristeza virus (CTV) in different hosts has been attributed to virus isolates which are from severe to mild. Different serological and molecular assays have been deployed to differentiate the strains of CTV. Citrus tristeza virus is diversified towards its strains on the basis of biological, serological and molecular characterization. Phenotypic expression is due to genetic alteration and different molecular basis have now been adopted for strain differentiation. This review will give a brief idea about the different CTV isolates, their characterization based on nucleic acid and serological assays. Different methods along with salient features for strain characterization has also been reviewed. This review will also open the new aspects towards formulation of management strategies through different detection techniques.
Keywords: Citrus tristeza virus; symptomology; detection; serology; isolates and host range
Virus and virus-like pathogens are major contributor towards the citrus decline. The most obvious impact of these diseases is their lethal effects on specific hosts. Among virus and virus-like diseases, Citrus tristeza Closterovirus (CTV) is one of the most destructive and widely distributed citrus virus diseases in citrus growing areas of the world. CTV has been declared as a potential threat for the citrus [1]. This virus probably originated in Asia and has been spread to all citrus growing areas by infected germplasm movement [2–4]. Economic losses are hard to be estimated but millions of trees have been died due to CTV in different part of the world. CTV is widely distributed to all citrus growing areas of the world. In South Africa epidemics of CTV were first reported in the early part of the 20th century due to imports of CTV infected material aphid vector [5]. Brown citrus aphid (Toxoptera citricida Kirkaldy) is the most efficient vector of CTV and has caused the death of millions of trees in Brazil, Spain, and Argentina. It was mainly sweet orange and mandarins grafted on sour orange were victimized by CTV [6]. The death of millions of citrus trees, over two decades, i.e., 1930’s & 1940’s, was the result of CTV in South American countries [3,4,7]. In southern California alone, approximately three million citrus trees on sour orange rootstock were killed [8,9]. The Tristeza was first reported in Florida in 1959 and by 1980’s it had become a serious threat, causing tree decline on sour orange and citrus macrophylla rootstock [10].
In New Zealand and Australia quick decline was first time identified in 1940 and then similar symptoms were found in South Africa in 1900, Java in 1928, Argentina in 1930, Brazil in 1937 and California in 1939. It was then considered to be incompatibility problem due to complete failure of sour orange rootstock. Finally, in 1946, quick decline problem was found due to virus [11,12]. Tristeza disease become notorious because of emergence of quick decline strain. In 1981, a total world loss of 50 million trees was estimated due to CTV and by 1991; CTV quick decline had destroyed 100 million trees in Argentina, Brazil, Spain, California, Venezuela and other areas [13,14]. Today, CTV is widespread in Israel, Morocco, China, Japan, Southern California, Florida, Argentina, Brazil, South Africa, Australia and Southern Spain and now moving to Northern Spain. Italy is supposed to be free from CTV infection despite the main rootstock used in Italy is sour orange [3]. Although this disease has been reviewed by many researchers in the world, but this review has been written with the objective; how strain differentiation not only helps in disease diagnoses but also virus can be characterized on the basis of genotype and serotype. Moreover, this review will highlight the geographic distribution of different strains of CTV.
The CTV genome, the largest plant virus reported as a monopartite, single stranded positive sense RNA molecule around 19.3 kb in length, with 12 open reading frames (ORFs) that encode at least 19 proteins. the genome is encapsulated in a flexuous rod 2000 nm long particles composed of coat protein subunits of 25 KDA [15,16]. It is phloem limited and belongs to genus Closterovirus, family Closteroviridae. Genetic studies of different strains of CTV revealed the existence of seven distinct genetic lineages worldwide, identified as VT, T3, T30, T36, T68 and the resistance breaking genotype, described for the first time in New Zealand.
There is an extensive genetic variation among various isolates of CTV and many genetic groups of viruses have been recognized. Variation in strains also exists within each group [17]. The CTV isolates have been grouped on the basis of serology, sequence homology, phenotypic expression and genetic alteration. The interlink of phenotypic expression and genotype is not clear therefore strain identification on the basis of symptoms are not advised (Fig. 1). The major groups of strains are mild strains that cause barely detectable clearing of leaf veins in Mexican lime; decline-inducing strains cause death of trees when propagated on sour orange rootstock. Stem pitting strains cause mild to severe pitting of stems and branches of grapefruit and orange resulting in low yield [10,17,18]. Mild and decline strains of CTV were detected in mainland Portugal and severe stem pitting strains on Madeira Island also detected. In Florida the presence of infections reacted with probes is against from stem pitting strains. Mild strains of CTV which because symptoms show clearing or flecking of the leaf veins of Mexican Lime. Some strains cause yellowing of seedlings, but their significance in the field is uncertain There is also biological and biochemical evidence that individual citrus trees may be infected with several different strains of CTV [18]. Different CTV strains, generally referred as seedling yellows (CTV-SY), tristeza (CTV-T), stem-pitting (CTV-SP), and a mild type, have been widespread for many years [19].
Symptom expression of CTV in citrus hosts is highly variable and depends upon host species (rootstock and scion combination), virulence of CTV isolates and soil or environmental conditions. Characteristics symptoms of CTV are vein clearing, decline, stem pitting, stunting and leaf corking on different citrus hosts like sweet orange, grapefruit, grafted on sour range root stock [1,20,21]. Severity of infection and symptoms expression on cultivars vary from mild to severe isolates [21]. Behavior of different citrus species is diversified against commonly occurring stains of CTV [18]. Different isolates of CTV like decline, stem pitting and seedling yellows induce different symptoms on citrus cultivars. Major group of symptoms by different isolates such as mild, seedling yellows, quick decline, stem pitting of grape fruit and sweet orange on sour orange is attributed to CTV strains which have synergistic effect of more than one strain attacking the host [22,23].
Diversity of symptoms associated with CTV infection is attributed towards CTV strains ranging from mild to severe isolates. Mild isolates may not cause noticeable symptoms but severe strains may cause seedling yellows, decline on sour orange rootstock, stem pitting of grapefruit and sweet orange. Specific strains as CTV-D causes death of phloem at bud union which leads to over growth of the scion, girding effect, stunting, yellowing of leaves, reduced fruit size, poor growth, die back and wilting etc. In case of other infective strains, stem pitting occurs and the twigs may brittle. Sometime stem pitting strains of grape fruit causes rope-like external symptoms [24]. CTV isolates classified in 3 categories on the basis of phylogenetic analysis and expression of symptoms i) mild, inducing only in lime, ii) severe, cause stem pitting in sweet orange and grape fruit and iii) an atypical, induce variable symptoms when grafted on sour orange [25].
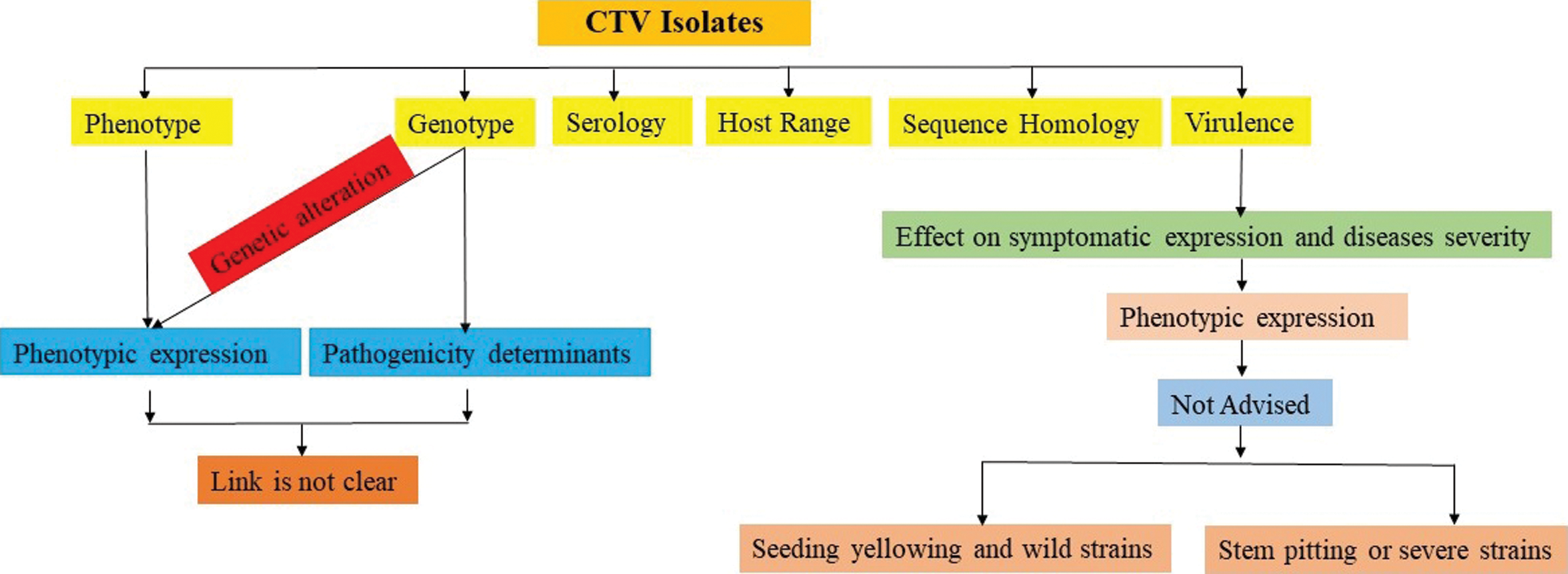
Figure 1: Relationship of differential basis for grouping of CTV isolates
Citrus tristeza virus is prevailing throughout all the citrus growing areas of the world. This virus has a great biodiversity on the basis of its strains. Strain differentiation shows an image in its mutation ability. Therefore, strain differentiation is the prerequisite for the correct diagnosis of CTV. Serological, biological and molecular techniques for the indexing are major contributors for the strain identification and characterization of this virus. Keeping in view the different aspects of regarding incidence (detection), transmissibility through vector multiplication in the ecosystem, Formulation of a comprehensive strategy for the management of CTV is the need of time. Different indexing assays for the virus detection and strain characterizations are the pre-requisite for the integrated disease management. Besides the detection, biological, molecular and serological indexing procedures have different sensitivity and limitations (Fig. 2).
Different methods for the assessment of CTV have been described. Sampling groups of citrus trees were made and recorded the groups as either CTV positive (one or more infected trees) or CTV negative (no infected trees). Symptoms produced by different CTV strains such as mild, decline and stem pitting can be characterized in the field through indicator plants and by molecular assays in the laboratory. Mexican lime (Citrus aurantifolia) is used as a good indicator plant for many CTV strains [26]. Indexing includes biological, serological and molecular methods, which are the common procedures according to the reliability, sensitivity and duration to detect the CTV. Disease incidence was then calculated at the scale of the individual tree by means of a formula-involving incidence at the group scale and the number of trees/groups.
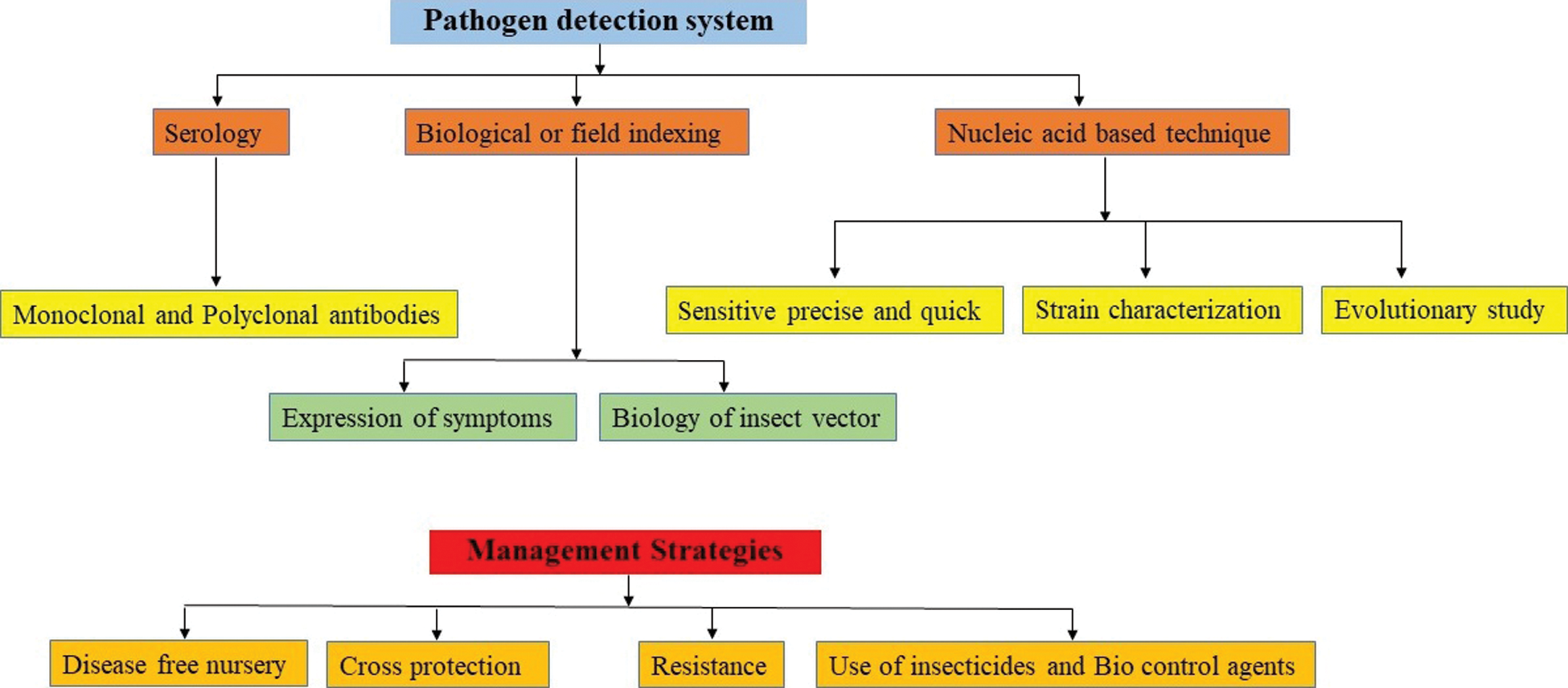
Figure 2: Pathogen detection system leading the formulation of management strategies
5 Serological Characterization of CTV
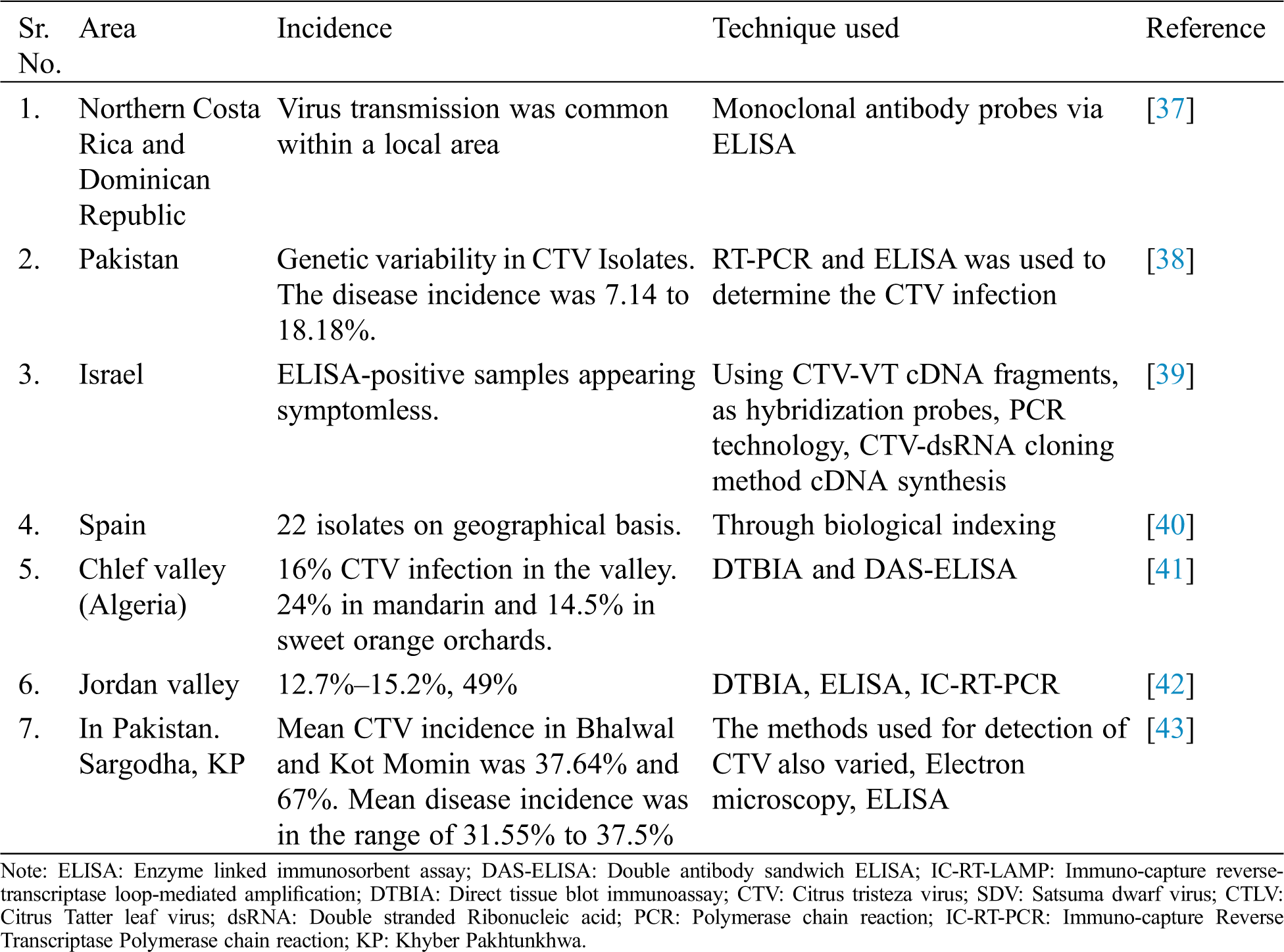
The serological indexing is usually based on ELISA and its variants. ELISA is used commonly to detect CTV [27] and is being used at large scale in routine and preliminary detection. Some of the serological indexing to detect the CTV has been summarized in Tab. 1. In Israel, great variation was found with the help of repeated Indirect ELISA procedure in the distribution of CTV throughout the canopies even of the declining trees CTV is wide spread in Iran but there was no information about the distribution and genetic diversity of CTV isolates. Therefore, a study was conducted on distribution of CTV and ELISA showed 50% positive samples and CP and K17 region was amplified through PCR. On the basis of CP and K17 amplification, results revealed the higher homology of Iranian isolates with California stem-pitting (SY568) and Japanese seedling yellows (NUagA) isolates [28]. Serological assay DAS-ELISA was used to detect the CTV incidence in Spain. The incidence was varied in orange grafted on citrange and on mandarin and grape fruit grafted on citrange [29]. Development and improvement of new serological methods has made a remarkable progress for the detection of CTV. Polyclonal and monoclonal antibodies have been developed to enhance the serological studies in relation to advantages, disadvantages, relativity sensitivity and applications, etc. [30]. A wide range of CTV strains have been characterized on the basis of symptoms, serology and molecular assays. The symptoms were quick decline, vein clearing stem pitting and seedling yellows. Mexican lime was a good indicator plant for symptoms and asymptomatic strains. MCA-13 was used to differentiate the isolates [31]. Studies on the recombinant single-chain variable fragment antibodies (SCFV) showed that bind specifically to tristeza virus (CTV), which represents the most detrimental viral disease in the citrus industry. The engineered antibodies were successfully used for CTV diagnosis in plants by tissue print and DAS-ELISA. It was highly specific and sensitive as much as conventional ELISAs performed with the parental monoclonal antibodies and showed the usefulness of recombinant antibodies for routine detection of virus in woody plants for the first time [32].
The incidence of CTV through DTBIA and ELISA was found 12.7%–15.2% and 49% respectively in the Jordan valley. Comparison was studied between the direct tissue blot immunoassay (DTBIA) with DAS-ELISA for the detection of 10 different Turkish isolates of CTV by employing monoclonal and polyclonal antibodies. Two different monoclonal antibodies 3E10 and MCA-13 were used. Two hundred and fifty-eight satsuma trees were tested using ELISA and DTBIA. All polyclonal (1212-1,981-1,908-7) and monoclonal (ECTV-175, ECTV-176, 11B1-3 & 3E10-6) antibodies reacted to CTV isolates, except MCA-13, which only reacted to Idgir & Cyprus CTV isolates. DAS-ELISA was also positive for all CTV isolates that reacted to DTBIA. Assays of 258 satsuma trees by ELISA and DTBIA indicated similar rates of CTV infection. Seven trees were found infected by both assays. DTBIA was proved a rapid, sensible and reliable procedure for the detection of CTV. It requires little sample preparation and tissue blots can be stored at 25°C for at least 4 weeks prior to assay and can easily be sent safely to another place for testing [33].
Orange and grapefruit were indexed through DASI-ELISA against CTV in the selected areas of Cuba. Aphid population was also studied. Coat protein of 12 local CTV isolates of the Cyprus were analyzed through PCR amplification and compared with the known CTV isolates of the world. Some of them caused the stem pitting on the branches of sweet orange and grapefruit and that isolate was not observed often in the Mediterranean basin [34]. ELISA and Western blots detected the Chinese CTV isolate (HB1) when polyclonal antiserum (PAb-L5). However, it did not react with PAb-I raised to the Italian CTV isolate and ten Mabs to the Chinese isolate S4 (MAbs-S4).
Trials were conducted for the detection of CTV by different antisera, two of which were monoclonal and two polyclonal. They were compared in enzyme-linked immunosorbent assay (ELISA) against different CTV isolates from four Mediterranean countries: Italy, Turkey, Moracco & Cyprus. All the four antisera were effective in CTV diagnosis except one monoclonal antiserum, which failed to recognize a few viruses isolates from Turkey and Cyprus. Therefore, ELISA procedure can be employed for routine testing in certification programmers [35]. Uniform positive and negative controls have been developed for the use in ELISA assays. The availability of such controls can enable more direct comparisons of data among the laboratories and quality control of ELISA results for tristeza virus. These controls vary greatly from laboratory to laboratory. Even in the same laboratory, there can be a variation in results. With the help of these studies 80 standardized positive and negative controls were developed which could be stable over a long period of time [36]. The section give a brief overview of different serological assays used for the detection of CTV with ease.
Table 1: Serological indexing of CTV in different citrus growing areas of the world
6 Molecular Characterization of CTV Isolates
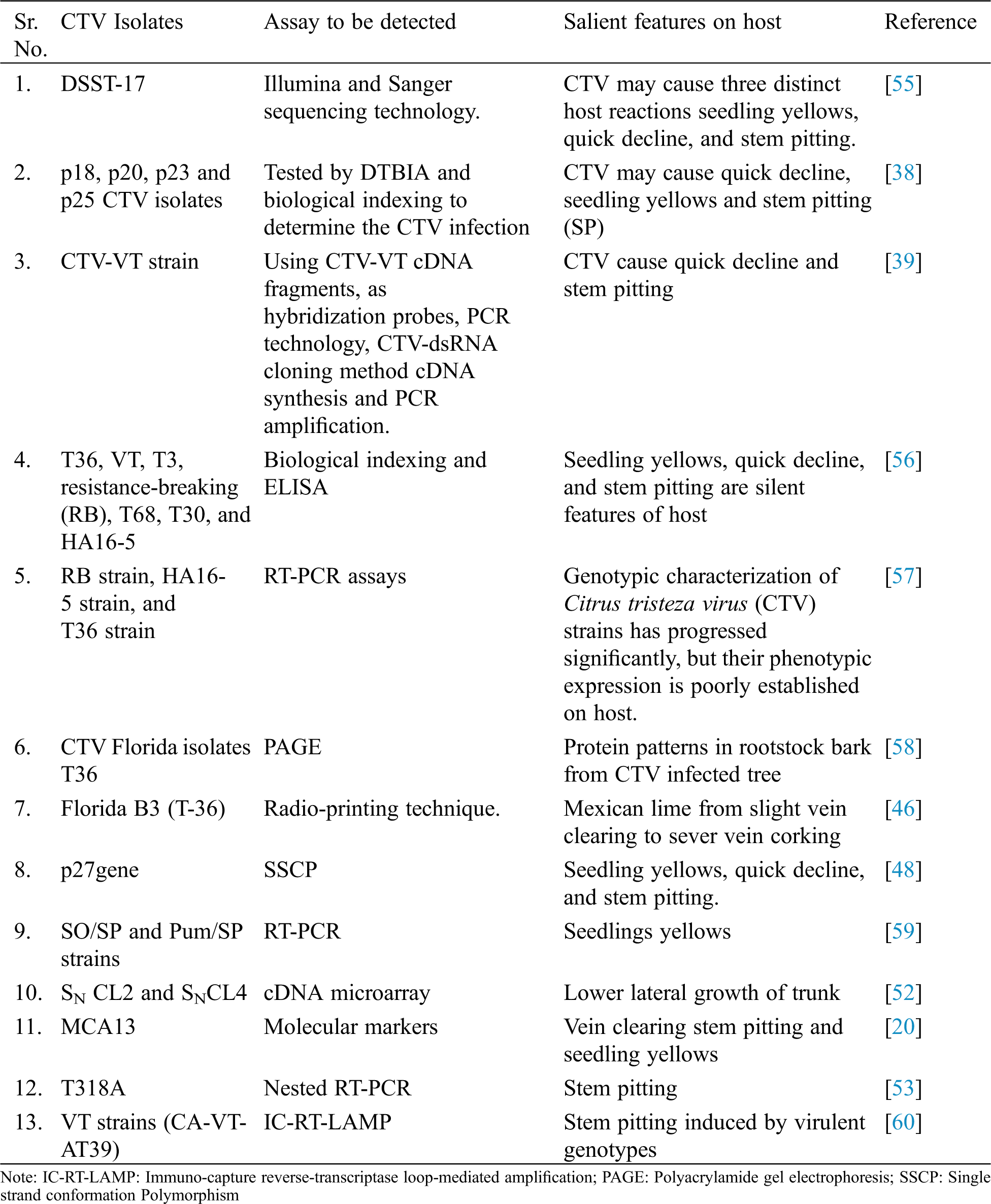
Different nucleic acid base indexing methods have been developed for the quick detection of CTV (Tab. 2). The adaptability of these methods depends upon the reliability, time duration and sensitivity. Alzhanova et al. [44] were analyzed Protein patterns in rootstock bark from CTV infected tree were studied through PAGE. There was clear modification in protein pattern but not in CTV free trees. Similarly, Northern blot technique was used to compare dsDNAs extracted from CTV infected and CTV free plants and hybridized. Seven isolates were detected using hybridization; cDNA probe was generated from dsDNA and then labeled with radioactive nucleotide using reverse transcriptase. Nucleic acid of CTV was detected in the clarified sap by hybridization with end labeled dsRNA probes were labeled with 5 ends radioactive nucleotides and using polynucleotide kinase [45]. The complete sequence of CTV Florida isolates T36 was analyzed and found that genome encodes 12 open reading frames (ORFs) to code at least 17 protein products.
In Italy diversified CTV isolates were characterized using molecular hybridization cDNA probes. In this hybridization signals were obtained from crude sap and viral dsRNA as well using radio-printing technique. Citrus tristeza virus was detected in the three aphid species through RT-PCR. Seven CTV isolates were chosen for acquisition feeding of aphids from worldwide isolate collection. The amplified products from the aphid fed on citrus infected with isolate B3 were confirmed as the CTV coat protein gene by digesting with various restriction enzymes. This method was considered to be useful in investigations of CTV-vector-plant interaction and CTV epidemiology [46].
Twenty-one dominant PCR-based DNA markers were identified as linked to CTV (a dominant gene that control CTV resistance in Poncirus trifoliate) by bulked segregate analysis of the 11 closet markers to CTV, only 2 segregated in all populations. Gene bank database revealed high sequence similarities between 2 markers and known plant disease resistance genes indicating that a resistance gene cluster exists in the CTV region in P. trifoliata [8]. Complete gene sequence of the Spanish mild isolate T385 of citrus tristeza virus was studied. This isolate was compared with genomes of severe isolates T36 (Florida), VT (Israel) and SY568 (California). The genome of T385 varies in length with other genome but their organization was identical [47].
Sequence variability in p27gene of CTV was studied through single strand conformation Polymorphism (SSCP) [48]. DIG-labelled cDNA probes (Non isotopic hybridization) and different types of target RNA were used to differentiate the isolates of CTV from Spain [49]. In Taiwan RT-PCR is a rapid and sensitive assay than other serological methods but one step RT-PCR is more sensitive and detects the CTV when virus concentration is very low. Three strains seedlings yellows, SO/SP and Pum/SP strains were detected [50].
Two strains viz., SN CL2 and SNCL4 of CTV were characterized on the Lisbon lemon in Cameron. Lower lateral growth of trunk was induced in response of virulent strain (SN CL2) on the combination of Mexican lime/citrange Troyer after three years of inoculation of these strains [51]. cDNA microarray was used to analyse the changes in gene expression of Mexican lime when infected with severe isolate (T305) or mild strain (T385) of CTV. There was significant change in the 334 genes of T305 whereas T385 was without significant changes [52].
Dot blot hybridization using specific DNA probes differentiated 24% samples for non-decline inducing CTV isolates and 16% for the decline inducing strains out of 58 samples. Some modifications were introduced in PCR-ELISA to increase its sensitivity and reduced the costs of detection. PCR-ELISA is the immune detection of PCR products and effective for detection and differentiation of plant viral nucleic acids. PCR-ELISA being a laborious and expensive method was modified and simplified by using asymmetric PCR.
In Sudan CTV was detected by tissue printing in arid and semi-arid zones of Sudan. CTV was in thirteen samples of orange trees. In addition to this, a mandarin and lime tree also reacted positively. In a nested RT-PCR approach from ten samples a specific PCR product was amplified showing the presence of CTV in four trees (three oranges, one lime tree), which were presumably tested, positive by tissue print of a specific PCR product proved the presence of CTV. This indicated the presence of at least two different CTV-strains in citrus orchards in Sudan [1]. During the comparison of isolates from different origins with CTV-stem pitting isolate from Spain through complete nucleotide sequence (T318A) showed strong sequence similarities with the severe isolates Sy568 from California and Nuaga from Japan and distant relationships with wild non-stem pitting T385 from Spain and T30 from Florida. In contrast to other severe CTV isolates, T318A had a predominant sequence variant highly variable 5-terminal untranslated region; in w/c a unique sequence variant associated with severe stem pitting isolates was detected. The high homogeneity of the T318A population suggests that the sequence obtained is probably responsible for symptoms induction and is a useful tool to delimit pathogenicity determinants [53].
Three microscopy procedures for detecting CTV were compared which provided additional alternatives for very rapid CTV indexing, such as the use of EM, SSEM and light microscopy. In light microscopy inclusion were found in young phloem tissues of all CTV-infected hosts examined. Inclusions appeared similar for all strains of CTV. In the case of EM of negatively stained extracts, particles of CTV were detected from plants infected with more than 20 isolates producing mild to very severe symptoms. Some isolates were checked only in a limited number of hosts. Similarly, in SSEM, virus particles were found on grids prepared with antiserum and extracts from infected tissue [54].
Table 2: Molecular detection of CTV isolates in different areas of the world
7 Conclusions and Future Thrust
Tristeza disease is widely distributed in citrus growing areas of the world. CTV leads to quick decline of the tree and is transmitted through insect and vegetative propagation. Infected plants exhibit variable symptoms because of mild to severe strains of this virus. Different serological, biological and molecular assays for the detection of this virus are being used based on their sensitivity and reliability. These assays have their advantages and disadvantages. Most of the nucleic acid based molecular assays are more sensitive than serological and biological, whereas serological and biological assays are helpful in routine detection of virus in bulks of samples. Although different detection methods of CTV strains are being employed but based on literature reviewed, the implication of these assays in relation to certification programs should be studied and incorporated. Moreover, symptomatic expressions in relation to detection of specific strain should be coupled with molecular assays. Detections methods should be friendly user for different management practices.
Compliance with Ethical Standards: The authors declare that the review is in compliance with ethical.
Research Involving Human Participants and/or Animals: The authors declare that the manuscript does not contain research involving Human Participants and/or Animals.
Authors Contribution: All the authors have equally contributed in gathering literature and manuscript write up and formatting, standards of the journal.
Funding Statement: The authors declare no competing financial interests.
Conflicts of Interest: The authors declare no conflict of interests and give their consent for publishing the material.
1. Fuchs, M., Bar-Joseph, M., Candresse, T., Maree, H. J., Martelli, G. P. et al. (2020). ICTV virus taxonomy profile: Closteroviridae. Journal of General Virology, 101(4), 364. [Google Scholar]
2. Dawson, W. O., Garnsey, S. M., Tatineni, S., Folimonova, S. Y., Harper, S. J. et al. (2013). Citrus tristeza virus-host interactions. Frontiers in Microbiology, 4, 88. DOI 10.3389/fmicb.2013.00088. [Google Scholar] [CrossRef]
3. Yokomi, R., Selvaraj, V., Maheshwari, Y., Chiumenti, M., Saponari, M. et al. (2018). Molecular and biological characterization of a novel mild strain of citrus tristeza virus in California. Archives of Virology, 163(7), 1795–1804. DOI 10.1007/s00705-018-3799-5. [Google Scholar] [CrossRef]
4. Harper, S. J., Yokomi, R., Dawson, W. O. (2016). Citrus tristeza virus-aphid interactions. In: Brown, J. K. (eds.Vector-mediated transmission of plant pathogens, pp. 121–130. St. Paul, MN: APS Press. [Google Scholar]
5. Mann, K. S., Sanfaçon, H. (2019). Expanding repertoire of plant positive-strand RNA virus proteases. Viruses, 11(1), 66. DOI 10.3390/v11010066. [Google Scholar] [CrossRef]
6. Qiao, W., Medina, V., Kuo, Y. W., Falk, B. W. (2018). A distinct, non-virion plant virus movement protein encoded by a crinivirus essential for systemic infection. mBio, 9(6), 154. DOI 10.1128/mBio.02230-18. [Google Scholar] [CrossRef]
7. Harper, S. J., Cowell, S. J., Dawson, W. O. (2015). With a little help from my friends: Complementation as a survival strategy for viruses in a long-lived host system. Virology, 478(15), 123–128. DOI 10.1016/j.virol.2014.12.041. [Google Scholar] [CrossRef]
8. Dawson, W. O., Bar-Joseph, M., Garnsey, S. M., Moreno, P. (2015). Citrus tristeza virus: Making an ally from an enemy. Annual Review of Phytopathology, 53(1), 137–155. DOI 10.1146/annurev-phyto-080614-120012. [Google Scholar] [CrossRef]
9. Harper, S. J., Cowell, S. J., Dawson, W. O. (2017). Isolate fitness and tissue-tropism determine superinfection success. Virology, 511(1), 222–228. DOI 10.1016/j.virol.2017.08.033. [Google Scholar] [CrossRef]
10. Futch, S. H., Brlansky, R. H. (2005). Field diagnosis of citrus tristeza virus. HS996, one of a series of the Horticultural services department, Florida cooperative extension service. USA: IFAS, University of Florida. [Google Scholar]
11. Folimonova, S. Y. (2020). Citrus tristeza virus: A large RNA virus with complex biology turned into a valuable tool for crop protection. PLoS Pathogens, 16(4), e1008416. DOI 10.1371/journal.ppat.1008416. [Google Scholar] [CrossRef]
12. Fu, S., Shao, J., Paul, C., Zhou, C., Hartung, J. S. (2017). Transcriptional analysis of sweet orange trees co-infected with ‘Candidatus Liberibacter asiaticus’ and mild or severe strains of Citrus tristeza virus. BMC Genomics, 18(1), 183. DOI 10.1186/s12864-017-4174-8. [Google Scholar] [CrossRef]
13. Yokomi, R. (2019). CTV vectors and interactions with the virus and host plants. In: Citrus tristeza virus, pp. 29–53. New York, USA: Humana. [Google Scholar]
14. Folimonova, S. Y. (2013). Developing an understanding of cross-protection by Citrus tristeza virus. Frontiers in Microbiology, 4, 76. DOI 10.3389/fmicb.2013.00076. [Google Scholar] [CrossRef]
15. Niblett, C. L., Genc, H., Cevik, B., Halbert, S., Brown, L. et al. (2000). Progress on strain differentiation of Citrus tristeza virus and its application to the epidemiology of citrus tristeza disease. Virus Research, 71(1–2), 97–106. DOI 10.1016/S0168-1702(00)00191-X. [Google Scholar] [CrossRef]
16. Suastika, G., Natsuaki, T., Terui, H., Kano, T., Leki, H. et al. (2001). Nucleotide sequence of citrus tristeza virus seedling yellows isolates. Journal of General Plant Pathology, 67(1), 73–77. DOI 10.1007/PL00012992. [Google Scholar] [CrossRef]
17. Chung, K. R., Brlansky, R. H. (2006). Citrus diseases exotic to Florida: Citrus Tristeza Virus-Stem Pitting (CTV-SP). Plant pathology department, florida cooperative extension service, institute of food and agricultural sciences. Florida, USA: University of Florida. [Google Scholar]
18. Niblett, C. L., Genc, H., Cevik, B., Halbert, S., Brown, L. et al. (2000). Progress on strain differentiation of Citrus tristeza virus and its application to the epidemiology of citrus tristeza disease. Virus Research, 71(1–2), 97–106. DOI 10.1016/S0168-1702(00)00191-X. [Google Scholar] [CrossRef]
19. Atta, S., Zhou, C. Y., Yan, Z., Cao, M. J., Wang, X. F. (2012). Distribution and research advances of Citrus tristeza virus. Journal of Integrative Agriculture, 11(3), 346–358. DOI 10.1016/S2095-3119(12)60019-7. [Google Scholar] [CrossRef]
20. Brlansky, R. H., Damsteegt, V. D., Howd, D. S., Roy, A. (2003). Molecular analysis of Citrus tristeza virus subisolates separated by aphid transmission. Plant Disease, 87(4), 397–401. DOI 10.1094/PDIS.2003.87.4.397. [Google Scholar] [CrossRef]
21. Harper, S. J. (2013). Citrus tristeza virus: Evolution of complex and varied genotypic groups. Frontiers in Microbiology, 4, 93. DOI 10.3389/fmicb.2013.00093. [Google Scholar] [CrossRef]
22. Cambra, M., Serra, J., Bonet, J. C., Moreno, P. (1988). Present status of the citrus tristeza virus in the valencian community. In: Timmer, L. W., Garnsey, S. M., Navarro, L. (eds.Proceeding of 10th International Organization of Citrus Virologists. California Riverside, USA. [Google Scholar]
23. Hilf, M. E. (2004). Citrus tristiza virus: Evolution in a host-limited pathosystem. Phytopathology, 94, 131. [Google Scholar]
24. Roberts, P. D., Mcgovern, R. J., Lee, R. F., Niblett, C. L. (2001). Tristeza. CH089. Series of Plant Pathology Department Florida Cooperative Extension Service, Institution of Food and Agriculture Sciences. USA: University of Florida. [Google Scholar]
25. Sambade, A., Lopez, C., Rubio, L., Flores, R., Guerri, J. et al. (2003). Polymorphism of a specific region in gene p23of citrus tristeza virus allows discrimination between mild and severe isolates. Archives of Virology, 148(12), 2325–2340. DOI 10.1007/s00705-003-0191-9. [Google Scholar] [CrossRef]
26. Flores, R., Di Serio, F., Navarro, B., Duran-Vila, N. U. R. I. A., Owens, R. A. (2011). Viroids and viroid diseases of plants. Studies in Viral Ecology: Microbial and Botanical Host Systems, 1, 311–346. [Google Scholar]
27. Iftikhar, Y., Khan, M. A., Rashid, A., Mughal, S. M., Iqbal, Z. et al. (2009). Occurrence and distribution of Citrus Tristeza Closterovirus in the Punjab and NWFP. Pakistan Pakistan Journal of Botany, 41(1), 373–380. [Google Scholar]
28. Ahmadi, S., Afsharifar, A., Niazi, A., Izadpanah, K. (2007). Distribution and genetic diversity of citrus tristeza virus (CTV) isolates in kerman province. Iranian Journal of Plant Pathology, 43, 131–134. [Google Scholar]
29. Rocha-Pena, M. A., Lee, R. F. (1991). Serological techniques for detection of citrus tristeza virus. Journal of Virological Methods, 34(3), 311–331. DOI 10.1016/0166-0934(91)90109-D. [Google Scholar] [CrossRef]
30. Guzman, M., Verniere, C., Niblett, C., Bove, J. M. (2001). Biological, serological and molecular characterization of Isolates of the citrus tristeza virus (CTV) in ornamental Kumquat and calamondino plants on the Island of Corcega. Fitopatologia Colombiana, 25, 23–27. [Google Scholar]
31. Terrada, E., Kerschbaumer, R. J., Giunta, G., Galeffi, P., Himmler, G. et al. (2000). Fully recombinant enzyme-linked immunosorbent assays using genetically engineered single-chain antibody fusion proteins for detection of Citrus tristeza virus. Phytopathology, 90(12), 1337–1344. DOI 10.1094/PHYTO.2000.90.12.1337. [Google Scholar] [CrossRef]
32. Korkmaz, S. (2001). Application of Direct Tissue Blot Immunoassay in comparison with DAS-ELISA for detection of Turkish isolates of citrus tristeza closterovirus (CTV). Turkish Journal of Agriculture and Forestry, 26, 203–209. [Google Scholar]
33. Papayiannis, L. C., Santos, C., Kyriakou, A., Kapari, T., Nolasco, G. (2007). Molecular characterization of citrus tristeza virus isolates from cyprus on the basis of the coat protein gene. Journal of Plant Pathology, 89, 291–295. [Google Scholar]
34. Boubker, J., Kyriakou, A., Onghia, A. D., Baloglu, S., Ylmaz, M. A. (1998). Comparative trials for the detection of CTV by antisera from different sources. Options Mediterraneennes, 21, 113–117. [Google Scholar]
35. Lee, R. F., Dekkers, M. G. H., Bar-Joseph, M. (2004). Development of stable, uniform controls for use in ELISA assays for tristeza virus. Abstract in program, 16th Conference of International Organization of Citrus Virologist, Monterrey, Mexico. [Google Scholar]
36. Moreno, P., Guerri, J., Ortiz, J. (1989). Alteration of bark proteins associated with citrus tristeza virus (CTV) infection on susceptible citrus species and scion-rootstock combinations. Journal of Phytopathology, 125(1), 55–66. DOI 10.1111/j.1439-0434.1989.tb01056.x. [Google Scholar] [CrossRef]
37. Gottwald, T. R., Garnsey, S. M., Borbón, J. (1998). Increase and patterns of spread of citrus tristeza virus infections in costa rica and the dominican republic in the presence of the brown citrus aphid, toxoptera citricida. Phytopathology, 88(7), 621–636. DOI 10.1094/PHYTO.1998.88.7.621. [Google Scholar] [CrossRef]
38. Atta, S., Cao, M., Umar, U., Zhou, Y., Yang, F. et al. (2017). Biological indexing and genetic analysis of Citrus tristeza virus in Pakistan. Journal of General Plant Pathology, 83(6), 382–389. DOI 10.1007/s10327-017-0737-4. [Google Scholar] [CrossRef]
39. Bar-Joseph, M. (2019). A Short Note on Reflections and Publications on Citrus tristeza virus (CTV) Methodologies. In: Citrus Tristeza Virus, pp. 1–6. New York, USA: Humana. [Google Scholar]
40. Ballester-Olmos, J. F., Pina, J., Carbonell, E., Moreno, P., Mendoza, A. et al. (1993). Biological diversity of citrus tristeza virus (CTV) isolates in Spain. Plant Pathology, 42, 219–229. [Google Scholar]
41. Arous, S. A., Guenaqui, Y., Djelouah, K. (2017). Current status of citrus tristeza virus (CTV) and its potential Aphid’s vectors in Chlef Valley (Algeria). In: VIII International Scientific Agriculture Symposium, Agrosym, Jahorina, Faculty of Agriculture. University of East Sarajevo, Bosnia and Herzegovina, Book of Proceedings, 1064–1070. [Google Scholar]
42. Anfoka, G., Abhary, M., Fattash, I., Nakhla, M. (2005). Occurrence and distribution of Citrus tristeza virus (CTV) in the Jordan Valley. Phytopathologia Mediterranea, 44(1), 17–23. [Google Scholar]
43. Naseem, S., Mahmood, S., Ali, Z. (2016). Occurrence of citrus tristeza virus in Pakistan: A GIS based approach combining host distribution and disease reports. Pakistan Journal of Agriculture Science, 1, 53. [Google Scholar]
44. Alzhanova, D. V., Prokhnevsky, A. I., Peremyslov, V. V., Dolja, V. V. (2007). Virion tails of Beet yellows virus: Coordinated assembly by three structural proteins. Virology, 359(1), 220–226. DOI 10.1016/j.virol.2006.09.007. [Google Scholar] [CrossRef]
45. Mehrotra, R. S. (2013). Fundamentals of plant pathology. USA: Tata McGraw-Hill Education. [Google Scholar]
46. Vives, M. C., Rubio, L., Lopez, C., Navas-Castillo, J., Albiach Marti, M. R. et al. (1999). The complete genome sequence of the major component of a mild citrus tristeza virus isolate. Journal of General Virology, 80(3), 811–816. DOI 10.1099/0022-1317-80-3-811. [Google Scholar] [CrossRef]
47. Gago-Zachert, R., Costa, N., Semorile, L., Grau, O. (1999). Sequence variability in p27 gene of citrus tristiza virus (CTV) revealed by SSCP. Electron Journal of Biotechnology, 2, 16–25. [Google Scholar]
48. Narvaez, G., Skander, B. S., Ayllon, M. A., Rubio, L., Guerri, J. et al. (2000). A new procedure to differentiate citrus tristeza virus isolates by hybridization with digoxigenin-labelled cDNA probes. Journal of Virological Methods, 85(1–2), 83–92. DOI 10.1016/S0166-0934(99)00158-5. [Google Scholar] [CrossRef]
49. Selvaraj, V., Maheshwari, Y., Hajeri, S., Yokomi, R. (2019). A rapid detection tool for VT isolates of Citrus tristeza virus by immunocapture-reverse transcriptase loop-mediated isothermal amplification assay. PLoS One, 14(9), e0222170. DOI 10.1371/journal.pone.0222170. [Google Scholar] [CrossRef]
50. Killiny, N., Harper, S. J., Alfaress, S., El Mohtar, C., Dawson, W. O. (2016). Minor coat and heat shock proteins are involved in the binding of citrus tristeza virus to the foregut of its aphid vector, Toxoptera citricida. Applied and Environmental Microbiology, 82(21), 6294–6302. DOI 10.1128/AEM.01914-16. [Google Scholar] [CrossRef]
51. Gandia, M., Conesa, A., Ancillo, G., Gadea, J., Forment, J. et al. (2007). Transcriptional response of Citrus aurantifolia to infection by Citrus tristeza virus. Virology, 367(2), 298–306. DOI 10.1016/j.virol.2007.05.025. [Google Scholar] [CrossRef]
52. Ruiz-Ruiz, S., Moreno, P., Guerri, J., Ambros, S. (2006). The complete nucleotide sequence of a severe stem pitting isolate of CTV from spain: Comparison with isolates from different origins. Archives of Virology, 151(2), 387–398. DOI 10.1007/s00705-005-0618-6. [Google Scholar] [CrossRef]
53. Flores, R., Ruiz, S., Soler, N., Sánchez-Navarro, J., Fagoaga, C. et al. (2013). Citrus tristeza virus p23: A unique protein mediating key virus-host interaction. Frontiers in Microbiology, 4(98), 1–9. [Google Scholar]
54. Maheshwari, Y., Selvaraj, V., Hajeri, S., Ramadugu, C., Keremane, M. L. et al. (2017). On-site detection of Citrus tristeza virus (CTV) by lateral flow immunoassay using polyclonal antisera derived from virions produced by a recombinant CTV. Phytoparasitica, 45(3), 333–340. DOI 10.1007/s12600-017-0591-0. [Google Scholar] [CrossRef]
55. Benítez-Galeano Jose, M., Vallet, T., Carrau, L., Hernández-Rodríguez, L., Bertalmío, A. et al. (2018). Complete genome sequence of a novel recombinant citrus tristeza virus, a resistance-breaking isolate from Uruguay. Gen Genome Announcements, 22, e00442–18. [Google Scholar]
56. Yokomi, R. K., Selvaraj, V., Maheshwari, Y., Saponari, M., Giampetruzzi, A. et al. (2017). Identification and characterization of Citrus tristeza virus isolates breaking resistance in trifoliate orange in California. Phytopathology, 107(7), 901–908. DOI 10.1094/PHYTO-01-17-0007-R. [Google Scholar] [CrossRef]
57. Cook, G., van Vuuren, S. P., Breytenbach, J. H., Burger, J. T., Maree, H. J. (2016). Expanded strain-specific RT-PCR assay for differential detection of currently known citrus tristeza virus strains: A useful screening tool. Journal of Phytopathology, 164(10), 847–851. DOI 10.1111/jph.12454. [Google Scholar] [CrossRef]
58. Ruiz-Ruiz, S., Navarro, B., Peña, L., Navarro, L., Moreno, P. et al. (2019). Citrus tristeza virus: Host RNA silencing and virus counteraction. Methods in Molecular Biology, 2015(4–5), 195–207. DOI 10.1007/978-1-4939-9558-5_14. [Google Scholar] [CrossRef]
59. Almasi, M. A. (2017). Development of a colorimetric reverse transcription loop-mediated isothermal amplification assay for the detection of Mirafiori lettuce big-vein virus. Archives of Virology, 162(9), 2775–2780. DOI 10.1007/s00705-017-3404-3. [Google Scholar] [CrossRef]
60. Selvaraj, V., Maheshwari, Y., Hajeri, S., Yokomi, R. (2019). A rapid detection tool for VT isolates of Citrus tristeza virus by immunocapture-reverse transcriptase loop-mediated isothermal amplification assay. PLOS ONE, 14(9), e0222170. DOI 10.1371/journal.pone.0222170. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |