
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016134
ARTICLE
S-Nitrosoglutathion Reductase Activity Modulates the Thermotolerance of Seeds Germination by Controlling ABI5 Stability under High Temperature
Shanghai Key Laboratory of Bio-Energy Crops, School of Life Sciences, Shanghai University, Shanghai, 200444, China
*Corresponding Author: Ping Li. Email: liping80@shu.edu.cn
Received: 10 February 2021; Accepted: 16 March 2021
Abstract: Seed germination or dormancy status is strictly controlled by endogenous phytohormone and exogenous environment signals. Abscisic acid (ABA) is the important phytohormone to suppress seed germination. Ambient high temperature (HT) also suppressed seed germination, or called as secondary seed dormancy, through upregulating ABI5, the essential component of ABA signal pathway. Previous result shows that appropriate nitric oxide (NO) breaks seed dormancy through triggering S-nitrosoglutathion reductase (GSNOR1)-dependent S-nitrosylation modification of ABI5 protein, subsequently inducing the degradation of ABI5. Here we found that HT induced the degradation of GSNOR1 protein and reduced its activity, thus accumulated more reactive nitrogen species (RNS) to damage seeds viability. Furthermore, HT increased the S-nitrosylation modification of GSNOR1 protein, and triggered the degradation of GSNOR1, therefore stabilizing ABI5 to suppress seed germination. Consistently, the ABI5 protein abundance was lower in the transgenic line overexpressing GSNOR1, but higher in the gsnor mutant after HT stress. Genetic analysis showed that GSNOR1 affected seeds germination through ABI5 under HT. Taken together, our data reveals a new mechanism by which HT triggers the degradation of GSNOR1, and thus stabilizing ABI5 to suppress seed germination, such mechanism provides the possibility to enhance seed germination tolerance to HT through genetic modification of GNSOR1.
Keywords: Seed germination; ambient high temperature; GSNOR1; S-nitrosoglutathion; ABI5
Seed germination is an important period during the plant life cycle, it germinates at suitable environment, or enters into dormancy status when the environment becomes harsh. Once seed germination or dormancy is strictly controlled by the endogenous hormone and environment signal, the phytohormone Abscisic acid (ABA) and gibberellic acid (GA) are two important hormone molecules that antagonistically control seed germination or dormancy, GA stimulates seed germination, while ABA suppresses seed germination and arrest post-germination growth under unfavorable environment [1–5]. Genetic screening reveals that a series of mutant deficiency in GA or ABA metabolism affect seed germination or dormancy process, the components associated with GA/ABA perceiving and signal transduction also modulate seed germination [2,6]. The basic leucine zipper-type t transcriptional factor ABI5 plays a central role to control the seed germination or dormancy status in response to ABA stimulation [7,8]. The ABA receptor PYR/PYL/PCAR can sense and bind ABA to inactive protein phosphatase 2C(PP2C), and then release SNF-REALATIVE PROTEIN KINASE (SnRK2) activity which phosphorylate ABI5 and other downstream substrate such as ABA-responsive element binding factor, consequently activate the expression of ABA-responsive gene to initiate ABA response [9,10]. DELLA protein, mainly RGL2, as the negative regulator of GA signal also modulates seed germination, as the mutant of rgl2 can rescue the germination ability of GA biosynthesis mutant ga1 without additional GA [11]. RGL2 can bind the promoter of ABI5 to activate the expression of ABI5, simultaneously stabilize ABI5, to suppress seed germination, suggesting the reciprocal interaction of GA and ABA to coordinate seed germination [12–14]. Other phytohormone, such as ethylene, BR or auxin also affects seeds germination directly or indirectly through altering GA/ABA metabolism or signal transduction [15–18].
Besides phytohormone molecule, small molecular signal, such as NO and CO also affect the seed germination [19–21]. We previous reported that CO and H2S affected the seed germination after high temperature stress [21,22]. NO is a key short-lived, endogenous gas signaling molecular that is associated with various developmental processes and stress response in both animals and plants [23,24]. And development in plants, the main bioactivity of NO is performed via protein S-nitrosylation modification, an oxidative process in which NO covalently reacting with reactive cysteine thiol residue to form an S-nitrosothiol (SNO). Such process has been demonstrated to alter protein conformation, DNA-protein interaction, protein stability and enzyme activities. NO and its derivations can also react with glutathione (GSH) to form S-nitrosoglutathione (GSNO), which is a major stable cellular reservoir of NO to sustain S-nitrosylated protein level [12,25]. NO signal induces the protein S-nitrosylation of ABI5 for its degradation to promote seed germination and seedling growth. In plants, GSNO is irreversibly degraded by the evolutionary conserved GSNO reductase 1 (GSNOR1), which catalyzes the NADH-dependent conversion of GSNO to GSSG and NH3. There exist a single gene (At5g43940) encoding GSNOR1 (also named HOT5) in Arabidopsis genome, and the corresponding mutant defect in GSNOR1 function displays high protein S-nitrosylation level to damage plant development, immunity and abiotic stress response et al. [26–29]. Meanwhile, NO also trigger the S-nitrosylation of GSNOR1 itself at multiple conserved Cys sites, thus attenuate GSNOR1 activity and stability. Hypoxia stress induce S-nitrosylation of GSNOR1, such modification changes the conformation of GSNOR1 to exposure the ATG8-interacxtion motif (AIM), such motif can be specifically recognized by autophagy machinery, subsequently GSNOR1 is recruited into the autophagosome for degradation in an AIM-dependent manner [30]. It is reported that ambient high temperature induces the hypocotyl elongation, also called thermomorphogenesis [31,32], but such effect was compromise in the gsnor mutant, but the detail mechanism by which GSNOR1 regulates seed germination under high temperature remains elusive.
In this study we investigate the possible function of GSNOR1 during seed germination under HT stress, and found that HT suppressed seed germination, also increased the expression of ABI5. We also found that HT induced the degradation of GSNOR1, subsequently reduced the activity of GSNOR1 to enhance the content of SNOs, further analysis revealed that overexpressing GSNOR1 increased the S-nitrosylation of ABI5 to induce ABI5 degradation under HT stress, on the control the S-nitrosylation level of ABI5 was lower in the mutant deficiency in GSNOR1. Genetic analysis also showed that ABI5 functions downstream of GSNOR1 to control seed germination under HT stress. Thus, our data reveals a new function of GSNOR1 in coordinating the thermoinhibition of seed germination, and HT trigger the degradation of GSNOR1 to stabilize ABI5 by attenuating the S-nitrosylation level of ABI5, as a result, suppress seed germination.
2.1 Arabidopsis Materials and Growth
Arabidopsis thaliana ecotype Columbia (Col-0) plants were used as the wild type. The hot5-1 (CS66011) and gsnor1-3 (CS66012) mutants were obtained from the Arabidopsis Biological Resource Center. The gsnor1-1 and abi5 mutant was described previously [33,34]. Seeds of each genotype were surface sterilized and sown on 1/2 Murashige and Skoog medium consisting of 0.7% agar, stratified at 4°C in the dark for 3 days, and transferred to a growth chamber at 22°C under constant light. After 10 days, the seedlings were transferred to soil (3:1 [v/v] potting soil:vermiculite) and grown in a greenhouse under a 16-h white light (100–120 μmol m–2 s–1)/8-h dark cycle. Seeds used for dormancy and germination rate analysis were simultaneously harvested from plants grown in the same batches at 22°C under long-day (16-h light/8-h dark) conditions. Dry seeds were stored at room temperature for 2–5 months before germination analysis.
2.2 Seed Dormancy and Germination Assays
For the seed dormancy experiments, approximately 100–200 freshly harvested or after-ripened seeds from an individual plant were sown directly on a 0.7% (w/v) agar plate or germination paper moistened with deionized water. The plates were incubated in a growth cabinet (22°C, 70% relative humidity, and a 16-h light (white light at 80 μmol m–2 s–1]/8-h dark cycle) for 5 days and the germination frequency determined. Germination was scored as “radicle emergence” and was observed under a dissecting stereomicroscope. At least three different batches from different plant pools (biological replicates) were used for each genotype in each experiment.
2.3 Generation of Transgenic Plants
To generate transgenic plants overexpressing GSNOR1, the full-length coding sequence of this gene was amplified using the primers listed in Supplemental Table 1. The amplified fragments were cloned into the NdeI/EcoRI (New England Biolabs) site of the pRI101-6Flag vector [6 × Flag tag inserted in the EcoRI/SacI site of the pRI101-AN vector (3262; Takara)] to generate the pRI101-GSNOR1-6FLAG construct, using an In-fusion HD Cloning Kit (638911, Clontech). These constructs were transformed into Arabidopsis plants using the Agrobacterium tumefaciens-mediated floral dip method.
Total RNA was extracted from the seed samples using a Plant Total RNA extraction kit (NEB) and reverse-transcribed to cDNA using the reverse transcriptase kit (Takara Bio) according to the manufacturer’s instructions [35]. RT-qPCR was performed using SYBR Premix Ex Taq (Perfect Real Time; Takara Bio) on a LightCycler 96 (Roche). The gene expression results were normalized to the expression of PP2A as the internal control. All experiments were performed three biological replicates and technical repeats. Primers are listed in Supplemental Table 1.
2.5 Western Blotting and Cell-Free Analysis
For immunoblot analysis, freshly harvested or after-ripened seeds were rapidly frozen in liquid nitrogen and ground in extraction buffer (50 mM sodium phosphate, pH 7.0, 100 mM NaCl, 5 mM EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] sodium deoxycholate, and protease inhibitor tablet; Roche). Following centrifugation at 14,000 g for 5 min at 4°C, the supernatant was collected. Approximately 10 mg of total protein was separated on a 12% (w/v) SDS–PAGE gel and transferred to a nitrocellulose membrane, which was then probed with the appropriate primary antibody (anti-FLAG, 1:3000, Sigma-Aldrich; anti-Tubulin, 1:3000, Abmart) and horseradish-conjugated goat anti-mouse secondary antibody (1:3000, Promega). Signals were detected using an ECL Kit (Invitrogen). For cell-free analysis, the total protein extracts prepared from Col, GSNOR1-6FLAG or gsnor1 mutant were adjusted to equal concentrations in the degradation buffer for each assay. Purified GST-ABI5 from E. coli was incubated at 22°C with the Arabidopsis extracts in degradation buffer. Samples were taken at the indicated intervals for determination of ABI5 protein abundance by immunoblots.
2.6 GSNOR Enzyme Activity and SNO Content Analysis
Total SNO content was determined using a previously reported method with minor modifications [36]. Briefly, SNO measurement was based on the reductive decomposition of nitroso species by an iodine/triiodide mixture, and the released NO was measured using a gas-phase chemiluminescence method and a NO analyzer (Model 410, 2B Technologies, Boulder, CO, USA). SNOs are sensitive to mercury-induced decomposition, in contrast to nitroso species such as nitrosamine and nitrosyl hemes. Samples (10 ml) after exposure to different treatments were centrifuged at 3000 × g for 10 min at 25°C, and the pellet was homogenized in 1 ml of extraction buffer (50 mM Hepes–KOH, pH 7.5, 1 mM DTT, 1 mM EDTA, 7 mM cysteine, 100 μM diethylenetriaminepentaacetic acid, DTPA) (1:5; w/v), and centrifuged at 3000 g for 10 min at 25°C. The supernatants were then incubated with an equal volume of 10 mM N-ethylmaleimide (NEM) for 15 min at 4°C. For each sample, two aliquots were prepared; aliquot (1) was treated with 10 mM sulphanilamide for 15 min at 4°C to eliminate nitrite and aliquot (2) was treated with 10 mM sulphanilamide and 7.3 mM HgCl2 for 15 min at 4°C to eliminate nitrite and SNOs, respectively. These samples were analyzed using the NO analyzer. The data from aliquots (1) and (2) represent the total SNO concentration. The entire procedure was performed under a red safety light to protect the SNOs from light-dependent decomposition.
GSNOR1 activity was determined spectrophotometrically at 25°C by monitoring the oxidation of NADH at 340 nm [36]. The treated samples (5 ml) were centrifuged at 3000 g for 15 min at 4°C, the pellet was then quickly homogenized in liquid nitrogen and extracted with assay mixture (20 mM Tris–HCl (pH 8.0), 0.2 mM NADH, and 0.5 mM EDTA) at 4°C and then centrifuged at 3000 g for 10 min at 25°C. The supernatants were subjected to further assays, and the reaction was initiated by adding GSNO (Calbiochem, San Diego, CA, USA) to the supernatants at a final concentration of 400 mM. The activity was expressed as nanomoles NADH consumed per minute per milligram of protein (e340 6.22 mM−1 cm−1).
2.7 Determination of S-Nitrosylated Proteins
The SNO content was measured using the Saville–Griess assay as previously described [36]. The identification of the S-nitrosylated proteins was performed as previously described with some modifications. The seeds were ground in liquid nitrogen and homogenized in HEN buffer (25 mM HEPES-NaOH, pH 7.7, 50 mM NaCl, 0.1 mM EDTA, and 0.1 mM neocuproine). The supernatant was then blocked with 20 mM methyl methanethiosulphonate and 2.5% SDS in HEN buffer. After acetone precipitation, the pellet was re-suspended in HEN buffer containing 1% SDS. The proteins were labelled with 2 mM biotin-HPDP, and the reaction was initiated by 200 mM sodium ascorbate. Proteins untreated with biotin-HPDP and sodium ascorbate were used as a control. Finally, the pellet was analysed using either immunoblotting with anti-biotin antibody (Cayman).
The GUS staining was manipulated as previous method. The germinated seeds or seedling of pGSNOR1:GUS was incubated in 0.1 m sodium phosphate buffer containing 50 mm K3Fe(CN)6, 50 mm K4Fe(CN)6, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C for 12 h. GUS staining was examined with a stereomicroscope, and images were captured with a digital camera (Zeiss).
3.1 GSNOR1 Modulates Seed Germination after Ambient High Temperature Stress
Previous study showed that GSNOR1 controls the hypocotyl elongation in response to ambient HT [29]. Ambient HT also suppresses seed germination, or induces seed secondary dormancy [33]. Here we checked the possible function of GSNOR1 in seed germination under HT. At first, we generated the transgenic line expressing GUS marker gene under the control of GSNOR1 promoter (pGSNOR1: GUS). As shown in Fig. 1A.
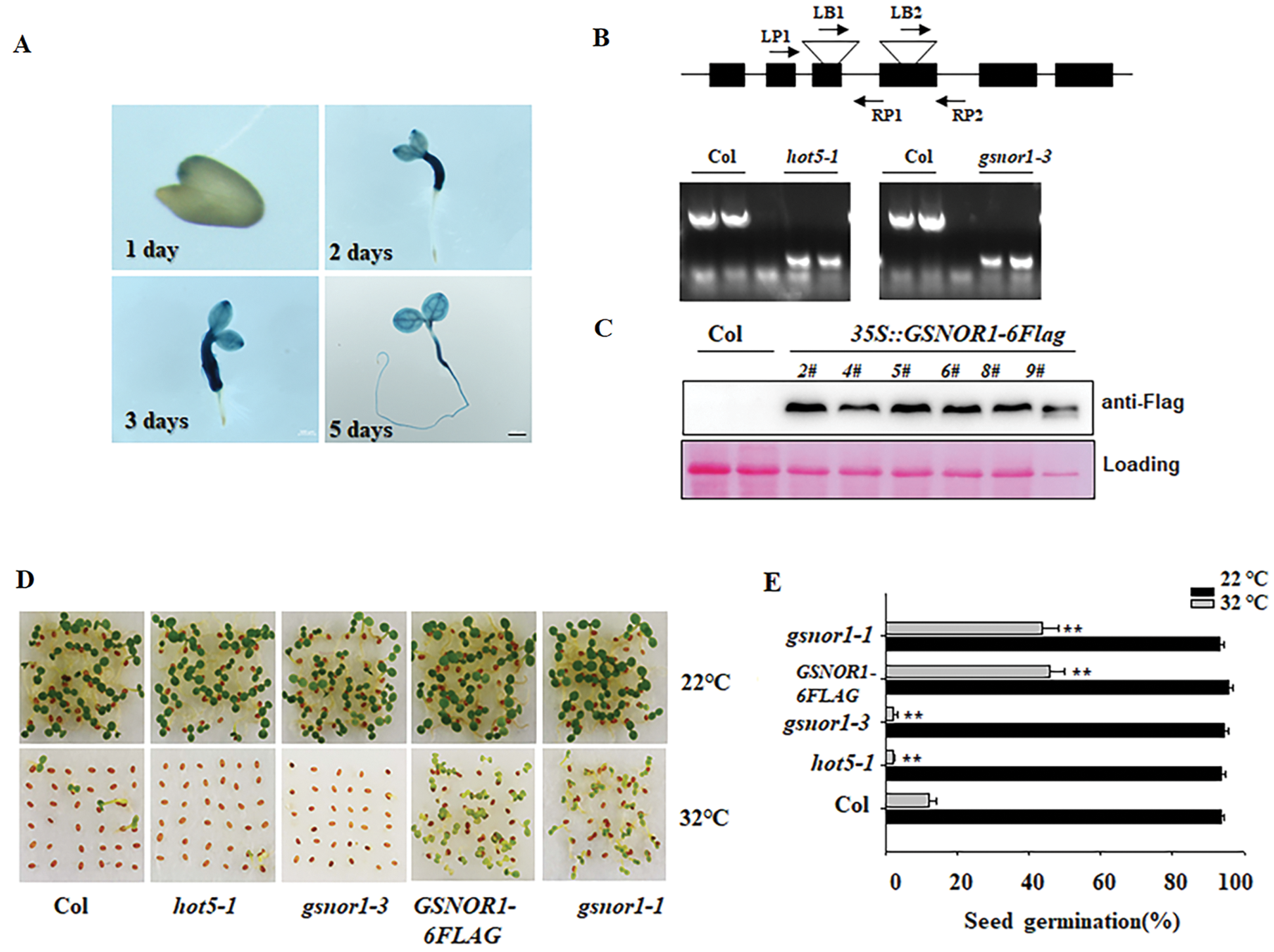
Figure 1: GSNOR1 mediates the seeds germination under HT. (A) GUS staining profile of transgenic pGSNOR1: GUS seeds. The transgenic pGSNOR1: GUS seeds were cold stratificated for 3 days at 4°C and then placed at 22°C for 1 day, 2 days, 3 days and 5 days for GUS staining, Bar = 200 μm. (B) Validating the T-DNA insertion position by corresponding primers and PCR analysis. T-DNA is inserted into the third and fourth exon of GSNOR1 genomic in hot5-1 and gsnor1-3 mutant. The top diagram indicates the genomic structure of GSNOR1 and the T-DNA insertion position. (C) Western blotting analysis of the GSNOR1-6FLAG accumulation in individual transgenic GSNOR1-6FLAG line. The total protein was extracted from individual transgenic line for western blotting analysis, the immunoblotting signal was detected by anti-Flag antibody, and the Ponceau S staining was used as the loading control. (D & E) Different germination percentage of wild-type Col, hot5-1, gsnor1-1, gsnor1-3 and GSNOR1-6FLAG seed were recorded after HT stress. All of these was sowed on the wet filter paper and placed under 32°C for HT stress, or placed at 22°C as the control. After 5 day of treatment, the photo was taken. Three biological repeated experiments were done and obtained the similar result. The picture from one group of experiment was presented. The seed germination percentage was calculated. The Values are shown as means SD of triplicate experiments. Asterisks indicate significant difference by Student’s t test (**p < 0.01).
GUS staining could be detected in the 1-day germination seeds, and became stronger in 2-day and 3-day germination seeds. GSNOR1 expression could also be detected in the silique by GUS staining (Data not shown). These data suggest the potential role of GSNOR1 in controlling seed germination. To further reveal the role of GSNOR1, we then used gsnor1-3 mutant and hot5-1 (cs66012) mutant, their genotypes were also validated by corresponding primers (Fig. 1B), both of them was reported to loss the GSNOR activity and showed insensitive to HT-induced hypocotyl elongation. We also generated the transgenic line overexpression GSNOR1-6FLAG fusion under the control of 35S promoter (35S:GSNOR1-6FLAG, abbreviated as GSNOR1-6FLAG). Western blotting also showed the strong immunoblotting blot in several individual transgenic GSNOR1-6FLAG lines (Fig. 1C). Under normal condition, all of these seeds grown well, but ambient HT obviously suppressed the seed germination in wild-type Col, as well as gsnor1-3 and hot5-1 seeds. On the contrary, the GSNOR1-6FLAG seed still showed relative higher seed germination under HT stress (Fig. 1D). The dominant mutant gsnor1-1 showed more expression of GSNOR1 [34], our data also showed that the gsnor1-1 mutant showed higher seed germination under HT stress. These data suggest that GSNOR1 acts as the positive regulator to control seed germination.
3.2 HTs Decreased the Expression of GSNOR1 and Its Activity
To determine the underlying mechanism by which GSNOR1 modulate seed germination under HT, we then monitored the protein abundance of GSNOR1 in response to HT. To this end, we first obtained the antibody against endogenous GSNOR1 protein, western blotting also reveals this antibody can specially recognize the endogenous GSNOR1, as not any band could be detected in the total protein extracted from the gnsor1-3 or hot5-1 mutant seeds, but high accumulation in gsnor1-1 mutant (Fig. 2A). Using this antibody, we observed that ambient HT gradually reduced the protein accumulation of GSNOR1 (Fig. 2B). In agreement with it, HT also gradually decreased the enzyme activity of GSNOR1 (Fig. 2C). It is known that GSNOR1 can efficiently degrade the endogenous RNS content that is normally induced by harsh environmental stress. Thus, we also checked the level of SNO, the main RNS component in planta, in loss-of-function mutants of GSNOR1 or its overexpression lines. In agreement with the above result, we observed SNO content was obviously higher in the hot5-1 or gsnor1-3 mutant, but lower in the GSNOR1-6FLAG seeds, compared with those of wild-type Col seeds (Fig. 2D).
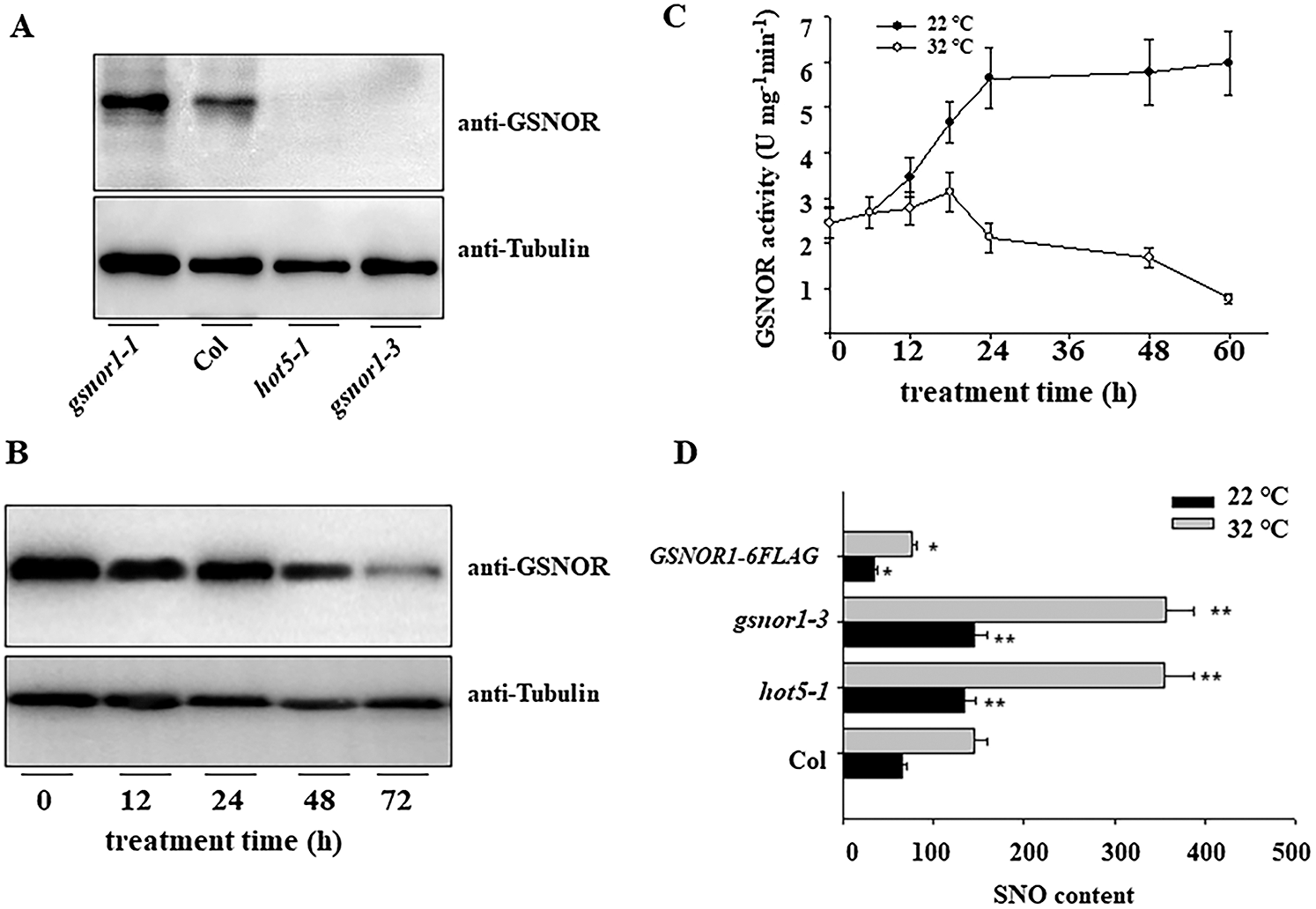
Figure 2: HT regulates the expression of GSNOR and its activity. (A) Specially recognize endogenous GSNOR1 by anti-GSNOR1 antibody. The total protein was extracted from wild-type Col, the gsnor1-1, hot5-1 and gsnor1-3 mutant seedling for western blotting analysis and anti-GSNOR1 antibody. The immunoblotting blot could be specially detected in the total protein from wild-type Col seed but not from gsnor mutant seedling. (B) HT reduced the protein accumulation of GSNOR protein in the imbibition seeds of wild type Col. The imbibition seeds were treated for HT at 32°C for indicated time and the total protein was extracted for western blotting analysis by anti-GSNOR antibody, the anti-Actin was used as the loading control. (C) HT reduced GSNOR activity in the imbibition seeds of wild-type Col. The imbibition seeds were treated for HT at 32°C for indicated time and GSNOR activity was measured at indicated time. The values are shown as means ± SD of triplicate experiments. (D) HT induced the overaccumulation of SNO in the imbibition seed of Col, hot5-1, gsnor1-3 and GSNOR1-6FLAG line. The imbibition seeds were treated for HT at 32°C for indicated time and the content of SNO was measured. The values are shown as means ± SD of triplicate experiments. Asterisks indicate significant difference by Student’s t test (**p < 0.01).
3.3 GSNOR1 Controls Seed Germination through ABI5 in Response to High Temperature Stress
ABI5 act as the negative regulator for seed germination in response to ABA stress [7]. It is reported that low concentration of NO donor, such as GSNO or SNAP, induces the seed germination by trigger the S-nitrosylation modification of ABI5, thereby inducing the degradation of ABI5 [37]. Here also checked the ABI5 expression in the imbibition seed after HT stress. As shown in Fig. 3A, HT increased the protein expression of ABI5, such increasement was more obvious in hot5-1 or gsnor1-3 mutant but lower in the transgenic GSNOR1-6FLAG seeds overexpressing GSNOR1, or the dominant mutant of gsnor1-1 (Fig. 3B), hinting that GSNOR1 regulates seed germination under HT through ABI5. Our previous study showed that the seed germination of abi5 showed more tolerance to HT stress. To test such possibility that GNSOR modulates the degradation of ABI5 for seed germination under HT, we crossed gsnor1-3 or GSNOR1-6FLAG with abi5 to obtain the crossed gsnro1-3/abi5 and GSNOR1-6FLAG/abi5 line. We could detect the strong expression of GSNOR1-6FLAG protein in GSNOR1-6FLAG/abi5 line, and could not detect the transcript of ABI5 in the gsnro1-3/abi5 and GSNOR1-6FLAG/abi5 line, but more transcripts in GSNOR1-6FLAG/abi5 (Fig. 3C), these data validated the successfully crossing in gsnro1-3/abi5 and GSNOR1-6FLAG/abi5. We then treated these parents or crossed lines, and found that both of gsnro1-3/abi5 and the GSNOR1-6FLAG/abi5 showed high germination percentage under HT, similar to abi5 (Fig. 3D). These data suggest that ABI5 is required for GSNOR1-dependent regulation of seed germination under HT treatment.
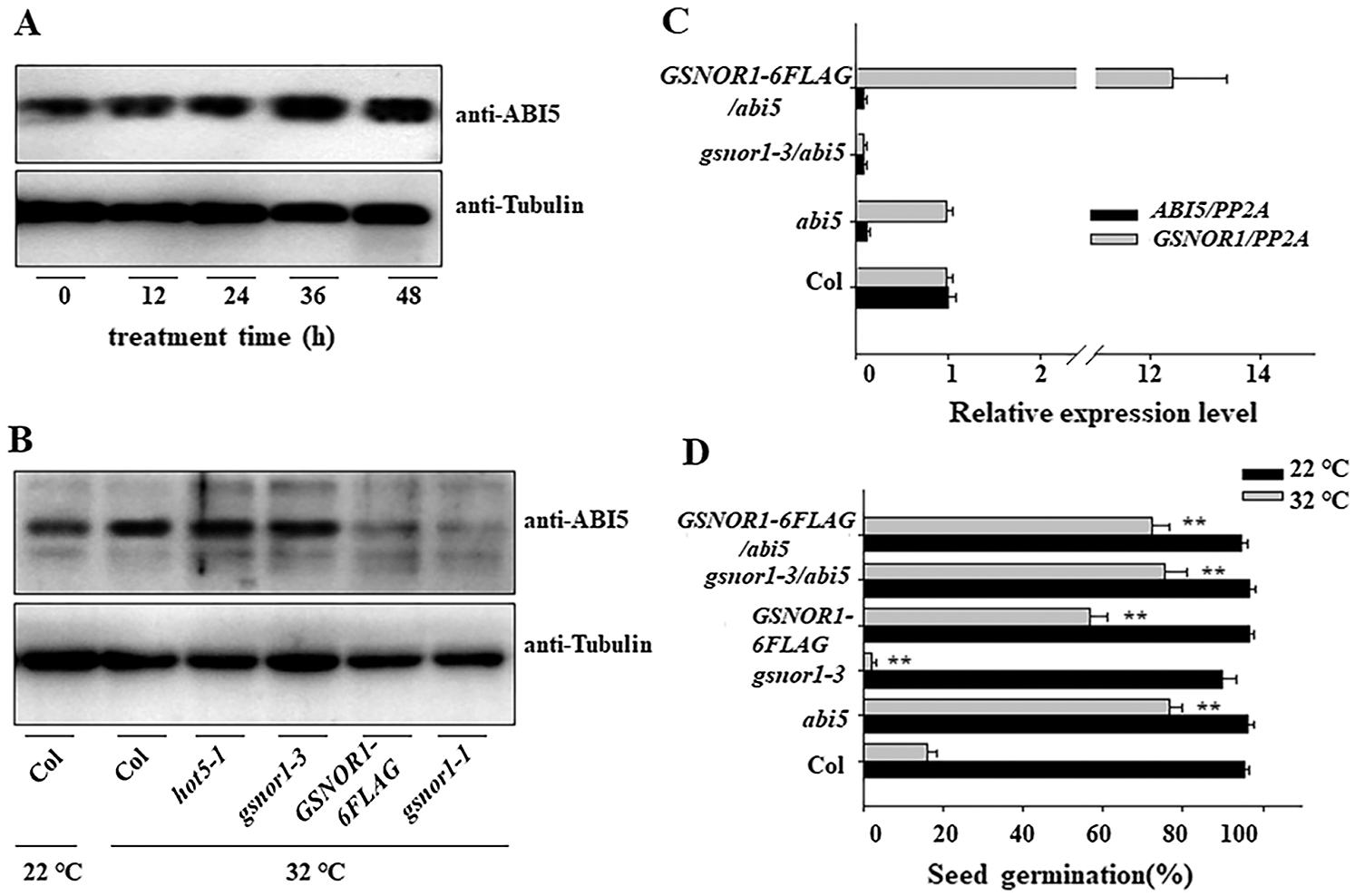
Figure 3: HT increased the protein abundance of ABI5 and reduced its S-nitrosylation modification level. (A) HT increased the protein abundance of ABI5. The imbibition seeds of Col were treated with HT at 32°C for indicated time, and the protein abundance of ABI5 was measured by western blotting and anti-ABI5 antibody. The anti-Tubulin was used as the loading control. (B) Different protein accumulation of ABI5 in different mutant after HT stress. The imbibition seeds of Col, hot5-1, gsnor1-3, GSNOR1-6FLAG and gsnor1-1 lines were treated with HT at 32°C for 24 h, and the protein abundance of ABI5 was measured by western blotting and anti-ABI5 antibody. The anti-Tubulin was used as the loading control. (C) Checking the transcript of GSNOR1 and ABI5 in the crossed lines by RT-qPCR analysis. The total RNA was extracted from Col, gsnor1-3, abi5, GSNOR1-6FLAG, gsnor1-3/abi5 and GSNOR1-6FLAG/abi5, and the transcriptional level of GSNOR1 and ABI5 was measured by RT-qPCR analysis. PP2A was used for normalization. Values are shown as means ± SD from three biological replicates. (D) Different seed germination in the imbibition seeds of Col, hot5-1, gsnor1-3, GSNOR1-6FLAG and gsnor1-1 lines after HT treatment. These imbibition seeds were treated with HT at 22°C or 32°C for 5 days, and the seed germination was calculated. The Values are shown as means ± SD of triplicate experiments. Asterisks indicate significant difference by Student’s t test (**p < 0.01).
3.4 Overexpressing GSNOR Triggers the Degradation of ABI5 under HT
It is reported that hypoxia stress induced the S-nitrosylation of GSNOR1 for its autophagic degradation [30], and our above results showed that HT induced the accumulation of ABI5 [33], but the degradation of GSNOR1, hinting the possibility that HT increased the S-nitrosylation modification of GSNOR1. To test such possibility, we measured the S-nitrosylation level of ABI5 before or after HTs stress. As shown in Fig. 4A, biotin-switch analysis HT obviously increased the S-nitrosylation modification level of GSNOR1 in the wild-type Col imbibed seeds, suggesting the correlation between S-nitrosylation modification of GSNOR1 and increased ABI5 abundance. It is possible HT induced the S-nitrosylation modification of GSNOR1, leading to the degradation of GSNOR1 for ABI5 increase. In other word, it is possible that overexpressing GSNRO should induce the degradation of ABI5, because the transgenic GSNOR1-6FLAG seeds showed more tolerance to HT with lower ABI5 protein abundance. Thus, we performed cell-free analysis to check the GST-ABI5 degradation speed by incubating the recombinant GST-ABI5 purified from bacteria with the total extracted protein from the transgenic germinated seeds of GSNOR1-6FLAG. As shown in Fig. 4B, GST-ABI5 was rapidly degraded, suggesting GSNOR1 mediates the degradation of GST-ABI5 in planta. However, the degraded speed of GST-ABI5 was obviously retarded once incubating the extract protein for the transgenics GSNOR1-6FLAG after 24 h of HT treatment, or incubating GST-ABI5 with the protein extract from the hot5-1 or gsnor1-3 seeds (Fig. 4C). Thus, these data confirm that HT accelerates the S-nitrosylation of GSNOR1 and its degradation, thereby increases RNS level to stabilize ABI5 and seed thermoinhibition.
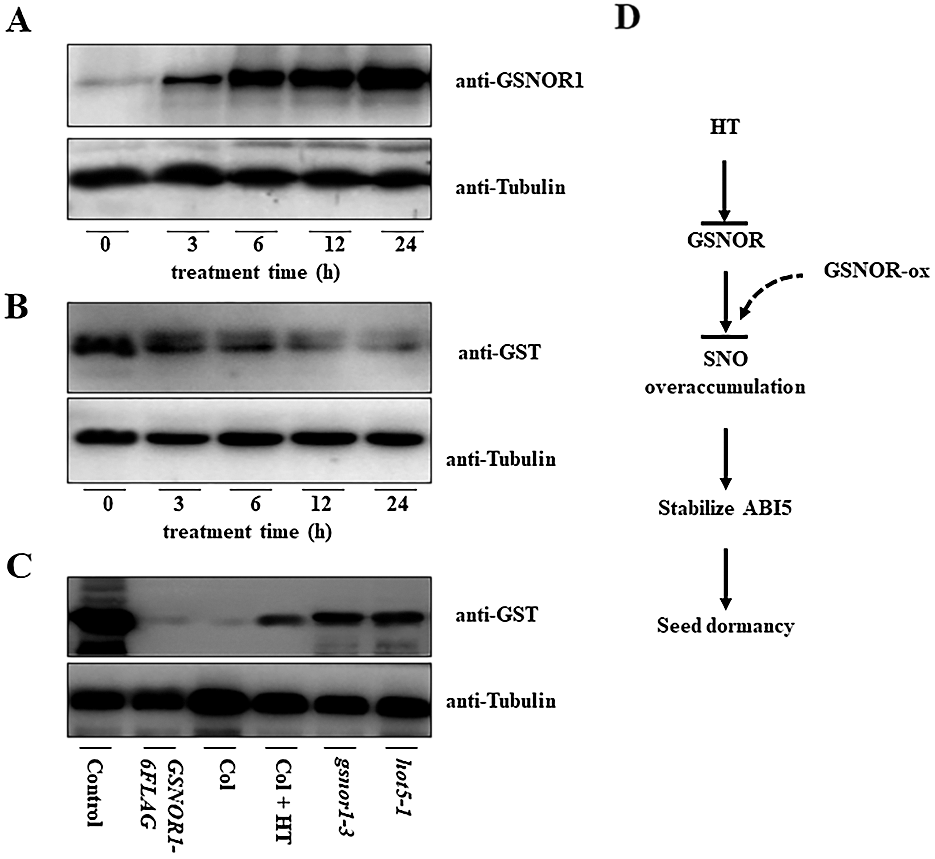
Figure 4: HT induced the S-nitrosylation of GSNOR1 for its degradation, thereby increasing ABI5 protein abundance. (A) HT increased the S-nitrosylation level of GSNOR1. The wild-type Col seed was treated with HT at 32°C for indicated time, and the S-nitrosylation level of GSNOR1 was measured by biotin-switch analysis. The anti-biotin antibody was used to immunoprecipitate the S-nitrosylation protein, and S-nitrosylated GSNOR1 protein was measured by anti-GSNOR1 antibody. The anti-Tubulin was used as the loading control. (B) Cell-free analysis of the degradation of GST-ABI5. The total protein was extracted from the imbibition seeds of wild-type Col line, and the purified GST-ABI5 was incubated for indicated time. The degradation speed of GST-ABI5 was measured by anti-GST antibody. The anti-tubulin was used as the loading control. (C) HT and GSNOR1 protein regulated the degradation of GST-ABI5. The total protein was extracted from different imbibition seeds with or without HT treatment, and the purified GST-ABI5 protein was incubated for 6 h, and the degradation of GST-ABI5 was measured by anti-GST antibody. The anti-tubulin was used as the loading control. Control: the purified GST-ABI5; Col: the total protein extracted from the wild-type Col seed under 22°C; Col + HT: the total protein extracted from the wild-type Col seed under 32°C for 48 h; gsnor1-3: the total protein extracted from the gsnor1-3 mutant seed under 22°C; hot5-1: the total protein extracted from the hot5-1 mutant seed under 22°C. (D) The propose model to illustrate the possible function of GSNOR1 in regulating ABI5 protein stability and seed germination under HT stress. HT induced the S-nitrosylation modification of GSNOR1 for its degradation, leading to the overaccumulation of SNO to stabilize ABI5, ultimately suppress seeds germination. As for the transgenic GSNOR1-6FLAG seeds, high level of GSNOR scavenges HT-induced SNO level, thus the appropriate SNO level causes the degradation of ABI5 to trigger the seed germination under HT stress.
Plant has evolved specific strategy to adapt environment stress. Environment high temperature induces the longer petioles, up-ward leave and hypocotyl elongation for young seedling, such phenotype is called as thermomorphogenesis. Previous study demonstrates that the key regulatory enzyme in ABA biosynthesis, 9-cisEPOXYCAROTENOID DIOXYGENASE4 (NCED4), is required for thermoinhibition of lettuce seeds [38]. Besides ABA biosynthesis, ABI5 as the core component of ABA signal transduction is also important to mediate seed germination [7]. Our previous study showed that ABI5 mediated seed germination thermoinhibition, the negative factor AFP2 enhances seed germination under HT through interacting with ABI5 to antagonize ABI5 activity [33]. Our now experiment also confirmed that HT induced the accumulation of ABI5 protein to suppress seed germination, as the abi5 mutant seeds presented high germination under HT. Post-translational modification, such as protein phosphorylation, can affect the protein stability of ABI5, for example the proteasome-mediated protein degradation, on the contrary, more sumolyation modification level can stabilize ABI5 [39]. Like protein phosphorylation, protein S-nitrosylation modification also affects its target protein stability. During seeds germination, NO signal triggers the S-nitrosylation medication at the Cys153 site of ABI5, thus induce proteasome-mediated degradation of ABI5 [37]. Here our cell-free analysis showed that HT treatment obviously suppressed the S-nitrosylation level of ABI5, which can explain the higher accumulation of ABI5 to suppress seed germination in response to HT. Consistently, we did not observe the obviously change of S-nitrosylation level in the transgenic line overexpression ABI5C153S after HT treatment, such transgenic seeds also showed more sensitive to HT treatment. Therefore, these data suggest that HT reduces the S-nitrosylation modification level of ABI5 to avoid the degradation of ABI5. As a result, more ABI5 accumulation induces the seed thermoinhibition.
During seed germination, the endogenous NO as the signal triggers the S-nitrosylation of ABI5 to induce ABI5 degradation, during this process, CUL3-based E3 ligase mediates the degradation of ABI5. Under environment stress, such as HT, induces the overaccumulation of NO, which forms the inappropriately higher level of RNS, to damage plant viability [37]. However, GSNOR can efficiently scavenge overaccumulation of RNS to prevent plant from environment stress. Consequently, the gsnor mutant show more sensitive to HT stress, for example, the gsnor mutant presents shorter hypocotyl than wild type Col seedling after HT stress [29]. In this study we observed HT suppressed the seed germination of wild-type Col or gsnor mutant seeds, however, overexpressing GSNOR1 significantly enhances the seed germination thermotolerance. HT seriously reduced the activity of GSNOR1, which caused more accumulation of RNS. Consistently, the transgenic seed overexpressing GSNOR1 showed lower RNS accumulation, while gsnor mutant showed overaccumulation of RNS. What is more, additional GSNO treatment induced high RNS level in the seeds, thus reversed the high germination percentage of transgenic GSNOR1-6FLAG seeds. These data suggest that GSNOR1 acts as the critical regulator for seed germination under HT stress. Though GSNOR1 regulates the protein s-nitrosylation level of downstream several target proteins, such as ABI5, NR or OTS1, etc. GSNOR1 itself experiences S-nitrosylation modification at Cys10 site and causes the local conformation change, and exposed its ATG8-interacting motif (AIM) for autophagy-mediated protein degradation under hypoxia stress [30]. Here we found HT treatment induced the quick degradation of GSNOR1, Biotin-switch analysis also revealed that HT triggered the protein S-nitrosylation modification of GSNOR1, and more S-nitrosylated GSNOR1 could be detected after HT stress, which can explain why HT induced the rapid degradation of GSNOR. Previous study identified a non-canonical catalase ROG1 that mediates the S-nitrosylation modification process, and targets GSNOR1 for its S-nitrosylation modification at Cys-10 site, ultimately altering GSNOR1 activity [28]. It is possible that such catalase is involved in HT-induced GSNOR1 degradation through transnitrosylation process, though more investigation work needs to be done in future. Our cell-free analysis showed that overexpressing GSNOR1 triggers the degradation of ABI5, and HT treatment delayed GSNOR1-mediated ABI5 degradation. These data support our opinion that HT induces more accumulation of RNS, which trigger the S-nitrosylation of GSNOR1 degradation, thus stabilizing ABI5 to suppress seed germination.
In a summary, our research investigated the underlying mechanism by which HT suppress seed germination through modulating GSNOR1 and its target ABI5 protein. Based on our data we propose a new model to illustrate the critical function of GSNOR1 in controlling seed germination thermoinhibition (Fig. 4D). As shown in Fig. 4C, under normal condition, GSNOR1 efficiently scavenges the overaccumulation of RNS and coordinates the appropriate endogenous NO level to degrade ABI5 through protein S-nitrosylation, leading seeds germination. However, HT induced the accumulation of RNS, which triggers S-nitrosylation of GSNOR1 for its degradation; as a result, lacking GSNOR1 induces overaccumulation of RNS, which stabilize ABI5 to suppress seeds germination. In agreement with it, overexpressing GSNOR1 in the transgenic GSNOR1-6FLAG seeds efficiently diminishes the endogenous RNS level under HT, subsequently suppressing ABI5 accumulation to enhance seed germination tolerance to HT stress. Therefore, our data reveal the novel function of GSNOR1 in enhancing seeds germination under HT via scavenging HT-induced RNS overaccumulation and triggering ABI5 degradation (Fig. 4D), this finding suggest that GSNOR1 is a new target protein that could be genetically engineered to cultivate HT-tolerance crops.
Funding Statement: This work was funded by the National Natural Science Foundation of China (Grants No. 31970289).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Graeber, K., Nakabayashi, K., Miatton, E., Leubner-Metzger, G., Soppe, W. J. (2012). Molecular mechanisms of seed dormancy. Plant, Cell & Environment, 35(10), 1769–1786. DOI 10.1111/j.1365-3040.2012.02542.x. [Google Scholar] [CrossRef]
2. Shu, K., Liu, X. D., Xie, Q., He, Z. H. (2016). Two faces of one seed: Hormonal regulation of dormancy and germination. Molecular Plant, 9(1), 34–45. DOI 10.1016/j.molp.2015.08.010. [Google Scholar] [CrossRef]
3. Yang, L., Liu, S., Lin, R. (2020). The role of light in regulating seed dormancy and germination. Journal of Integrative Plant Biology, 62(9), 1310–1326. DOI 10.1111/jipb.13001. [Google Scholar] [CrossRef]
4. Holdsworth, M. J., Bentsink, L., Soppe, W. J. J. (2008). Molecular networks regulating arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist, 179(1), 33–54. DOI 10.1111/j.1469-8137.2008.02437.x. [Google Scholar] [CrossRef]
5. Li, P., Ni, H. H., Ying, S. B., Wei, J. L., Hu, X. Y. (2020). Teaching an old Dog a New trick: Multifaceted strategies to control primary seed germination by DELAY of GERMINATION 1 (DOG1). Phyton-International Journal of Experimental Botany, 89, 1–12. DOI 10.32604/phyton.2020.09817. [Google Scholar] [CrossRef]
6. Finch-Savage, W. E., Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytologist, 171(3), 501–523. DOI 10.1111/j.1469-8137.2006.01787.x. [Google Scholar] [CrossRef]
7. Finkelstein, R. R., Lynch, T. J. (2000). The arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell, 12(4), 599–609. DOI 10.1105/tpc.12.4.599. [Google Scholar] [CrossRef]
8. Yadukrishnan, P., Datta, S. (2020). Light and abscisic acid interplay in early seedling development. New Phytologist, 229(2). DOI 10.1111/nph.16963. [Google Scholar] [CrossRef]
9. Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: Emergence of a core signaling network. Annual Review of Plant Biology, 61, 651–679. DOI 10.1146/annurev-arplant-042809-112122. [Google Scholar] [CrossRef]
10. Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes & Development, 24(16), 1695–1708. DOI 10.1101/gad.1953910. [Google Scholar] [CrossRef]
11. Peng, J., Harberd, N. P. (2002). The role of GA-mediated signalling in the control of seed germination. Current Opinion in Plant Biology, 5(5), 376–381. DOI 10.1016/S1369-5266(02)00279-0. [Google Scholar] [CrossRef]
12. Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y. et al. (2008). The gibberellic acid signaling repressor RGL2 inhibits arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell, 20(10), 2729–2745. DOI 10.1105/tpc.108.061515. [Google Scholar] [CrossRef]
13. Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y. et al. (2016). The NF-yC-rGL2 module integrates GA and ABA signalling to regulate seed germination in arabidopsis. Nature Communication, 7, 12768. DOI 10.1038/ncomms12768. [Google Scholar] [CrossRef]
14. He, D., Deng, G., Ying, S., Yang, W., Wei, J. et al. (2020). Carbon monoxide signal breaks primary seed dormancy by transcriptional silence of DOG1 in arabidopsis thaliana. Phyton-International Journal of Experimental Botany, 89(3), 633–643. DOI 10.32604/phyton.2020.010498. [Google Scholar] [CrossRef]
15. Li, X., Chen, T., Li, Y., Wang, Z., Cao, H. et al. (2019). ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-tPL complex on DELAY of GERMINATION1 expression. Plant Cell, 31(4), 832–847. DOI 10.1105/tpc.18.00449. [Google Scholar] [CrossRef]
16. Liu, X., Zhang, H., Zhao, Y., Feng, Z., Li, Q. et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 110(38), 15485–15490. DOI 10.1073/pnas.1304651110. [Google Scholar] [CrossRef]
17. Wang, Z., Chen, F., Li, X., Cao, H., Ding, M. et al. (2016). Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nature Communication, 7, 13412. DOI 10.1038/ncomms13412. [Google Scholar] [CrossRef]
18. Hu, Y., Yu, D. (2014). BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in arabidopsis. Plant Cell, 26(11), 4394–4408. DOI 10.1105/tpc.114.130849. [Google Scholar] [CrossRef]
19. Arc, E., Sechet, J., Corbineau, F., Rajjou, L., Marion-Poll, A. (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Frontiers in Plant Science, 4, 63. DOI 10.3389/fpls.2013.00063. [Google Scholar] [CrossRef]
20. Xu, H., Lantzouni, O., Bruggink, T., Benjamins, R., Lanfermeijer, F. et al. (2020). A molecular signal integration network underpinning arabidopsis seed germination. Current Biology, 30(19), 3703–3712, DOI 10.1016/j.cub.2020.07.012. [Google Scholar] [CrossRef]
21. Jia, Y., Li, R., Yang, W., Chen, Z., Hu, X. (2018). Carbon monoxide signal regulates light-initiated seed germination by suppressing SOM expression. Plant Science, 272, 88–98. DOI 10.1016/j.plantsci.2018.04.009. [Google Scholar] [CrossRef]
22. Chen, Z., Huang, Y., Yang, W., Chang, G., Li, P. et al. (2019). The hydrogen sulfide signal enhances seed germination tolerance to high temperatures by retaining nuclear COP1 for HY5 degradation. Plant Science, 285, 34–43. DOI 10.1016/j.plantsci.2019.04.024. [Google Scholar] [CrossRef]
23. Wilson, I. D., Neill, S. J., Hancock, J. T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell & Environment, 31(5), 622–631. DOI 10.1111/j.1365-3040.2007.01761.x. [Google Scholar] [CrossRef]
24. Hancock, J. T., Neill, S. J. (2019). Nitric oxide: Its generation and interactions with other reactive signaling compounds. Plants (Basel8(2), 41. DOI 10.3390/plants8020041. [Google Scholar] [CrossRef]
25. Neill, S., Bright, J., Desikan, R., Hancock, J., Harrison, J. et al. (2008). Nitric oxide evolution and perception. Journal of Experimental Botany, 59(1), 25–35. DOI 10.1093/jxb/erm218. [Google Scholar] [CrossRef]
26. Chen, R., Sun, S., Wang, C., Li, Y., Liang, Y. et al. (2009). The arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Research, 19(12), 1377–1387. DOI 10.1038/cr.2009.117. [Google Scholar] [CrossRef]
27. Lubega, J., Umbreen, S., Loake, G. J. (2020). Recent advances in the regulation of plant immunity by S-nitrosylation. Journal of Experimental Botany. DOI 10.1093/jxb/eraa454. [Google Scholar] [CrossRef]
28. Shi, Y. F., Wang, D. L., Wang, C., Culler, A. H., Kreiser, M. A. et al. (2015). Loss of GSNOR1 function leads to compromised auxin signaling and polar auxin transport. Molecular Plant, 8(9), 1350–1365. DOI 10.1016/j.molp.2015.04.008. [Google Scholar] [CrossRef]
29. Lee, U., Wie, C., Fernandez, B. O., Feelisch, M., Vierling, E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in arabidopsis. Plant Cell, 20(3), 786–802. DOI 10.1105/tpc.107.052647. [Google Scholar] [CrossRef]
30. Zhan, N., Wang, C., Chen, L., Yang, H., Feng, J. et al. (2018). S-Nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Molecular Cell, 71(1), 142–154. DOI 10.1016/j.molcel.2018.05.024. [Google Scholar] [CrossRef]
31. Hayes, S., Schachtschabel, J., Mishkind, M., Munnik, T., Arisz, S. A. (2020). Hot topic: Thermosensing in plants. Plant Cell Environment. DOI 10.1111/pce.13979. [Google Scholar] [CrossRef]
32. Quint, M., Delker, C., Franklin, K. A., Wigge, P. A., Halliday, K. J. et al. (2016). Molecular and genetic control of plant thermomorphogenesis. Nature Plants, 2, 15190. DOI 10.1038/nplants.2015.190. [Google Scholar] [CrossRef]
33. Chang, G., Wang, C., Kong, X., Chen, Q., Yang, Y. et al. (2018). AFP2 as the novel regulator breaks high-temperature-induced seeds secondary dormancy through ABI5 and SOM in arabidopsis thaliana. Biochemical and Biophysical Research Communications, 501(1), 232–238. DOI 10.1016/j.bbrc.2018.04.222. [Google Scholar] [CrossRef]
34. Feechan, A., Kwon, E., Yun, B. W., Wang, Y., Pallas, J. A. et al. (2005). A central role for S-nitrosothiols in plant disease resistance. Proceedings of the National Academy of Sciences of the United States of America, 102(22), 8054–8059. DOI 10.1073/pnas.0501456102. [Google Scholar] [CrossRef]
35. Li, P., Zhang, Q., He, D., Zhou, Y., Ni, H. et al. (2020). AGAMOUS-Like67 cooperates with the histone mark reader EBS to modulate seed germination under high temperature. Plant Physiology, 184(1), 529–545. DOI 10.1104/pp.20.00056. [Google Scholar] [CrossRef]
36. Cheng, T. L., Chen, J. H., Abd Allah, E. F., Wang, P. K., Wang, G. P. et al. (2016). Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta, 243(4), 1081. DOI 10.1007/s00425-016-2494-6. [Google Scholar] [CrossRef]
37. Albertos, P., Romero-Puertas, M. C., Tatematsu, K., Mateos, I., Sanchez-Vicente, I. et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nature Communication, 6, 8669. DOI 10.1038/ncomms9669. [Google Scholar] [CrossRef]
38. Huo, H., Dahal, P., Kunusoth, K., McCallum, C. M., Bradford, K. J. (2013). Expression of 9-cis-ePOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell, 25(3), 884–900. DOI 10.1105/tpc.112.108902. [Google Scholar] [CrossRef]
39. Yu, F., Wu, Y., Xie, Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends in Plant Science, 20(9), 569–575. DOI 10.1016/j.tplants.2015.05.004. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |